- 1Center for Reproductive Medicine and Andrology, Martin Luther University of Halle-Wittenberg, Halle, Germany
- 2Department of Pediatrics I, Martin Luther University of Halle-Wittenberg, Halle, Germany
- 3Department of General, Visceral and Thoracic Surgery, Städtische Klinikum Dessau, Dessau-Roßlau, Germany
- 4Division Molecular Urology, Department of Urology and Pediatric Urology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany
A wide variety of endogenous retroviral sequences has been demonstrated in the human genome so far, divided into several different families according to the sequence homology to viral strains. While increased expression of human endogenous retrovirus (HERV) elements has already been linked to unfavorable prognosis in hepatocellular carcinoma, breast cancer, and ovarian carcinoma yet less is known about the impact of the expression of different HERV elements on sarcomagenesis in general as well as the outcome of soft tissue sarcoma (STS) patients. Therefore, in this study the association between expression of HERV-K and HERV-F and the clinicopathological characteristics in a cohort of STSs as well as the patients’ prognosis was evaluated. HERV-K and HERV-F expression was assessed by quantitative real-time PCR in 120 patient specimens. HERV-K and HERV-F expression was significantly correlated (rS = 0.5; p = 6.4 × 10-9; Spearman’s rank bivariate correlation). Also, tumor diameter exhibited a significant negative association to HERV-K and HERV-F expression. Levels of several hypoxia-related RNAs like HIF-1α and miR-210 showed a significant positive correlation with both HERV-K and HERV-F expression. Although in survival analyses no impact of HERV expression on disease-specific survival could be detected, patients with elevated HERV-K expression had a significantly shorter relapse-free survival (p = 0.014, log-rank analysis). In conclusion, we provide evidence for the first time that the increased expression of HERV-K in tumors is associated with STS patients’ prognosis.
Introduction
Soft tissue sarcomas (STSs) are a heterogeneous group of tumors classified by the somatic tissue they resemble, with over 50 subtypes that can be distinguished (Ducimetière et al., 2011). The incidence of STS is relatively low – with estimated 4–5 cases per 100,000 per year (Stiller et al., 2013) – but the 5-year survival is only around 50% (Ferrari et al., 2011). Although treatment of STS usually consists of a wide resection of the tumor, followed by radio- or chemotherapy in selected cases (Casali and Blay, 2010), relapses and metastases are still an urgent clinical issue (Steinestel and Wardelmann, 2015). Due to the heterogeneity in genetics and phenotypes of the different STS entities, many of the proposed laboratory biomarkers and clinical factors still are insufficient for a satisfying prognostic evaluation of the individual patient’s outcome.
Human endogenous retroviruses (HERVs) are a class of retroviral sequences acquired during evolution by integration of viral genes in the host genome. They became non-infectious by mutation or loss of relevant genes for replication or virus release (Vargiu et al., 2016). They comprise an estimated 8% of the human genome (Lander et al., 2001), and at least 22 independent families based on their homology to known mammalian retroviruses exist (Griffiths, 2001).
Among the different families, retroviral sequences of the HERV-K family (HML-2) were the latest acquired, therefore they are the most complete and biologically active family (Bannert and Kurth, 2004; Hughes and Coffin, 2004, 2005). HERV-K expression was detected in several tumor entities, among them chronic myeloid leukemia (Brodsky et al., 1993), renal cell carcinoma (Florl et al., 1999; Kreimer et al., 2013), breast carcinoma (Wang-Johanning et al., 2003), prostate carcinoma (Wallace et al., 2014), pancreatic cancer (Li et al., 2017), and melanoma (Serafino et al., 2009). In melanoma, HERV-K expression is suggested to be an early event in tumorigenesis, seemingly enhancing the pathological process of tumor formation (Serafino et al., 2009; Cegolon et al., 2013). Most recently, it was demonstrated that shRNA-mediated downregulation of HERV-K in pancreatic cancer cell lines suppressed growth rates and metastases as well as the expression of several proliferation-related genes (Li et al., 2017). However, it is worth mentioning that the above described associations between HERV expression and tumor tissues are correlative. It has also been demonstrated, that healthy tissues of different origins express HERV sequences, especially during early embryo development and placentation or in the innate immune response (reviewed in Meyer et al., 2017).
HERV-F is another family of HERVs originally identified by cloning of human genomic DNA sequences and homology analyses (Kjellman et al., 1999a,b). HERV-F expression was described in leukemia cell lines (Patzke et al., 2002) and in a wide range of other tumor cell lines (Yi and Kim, 2004). On the contrary, in adult somatic tissues HERV-F is only expressed in placenta (Kjellman et al., 1999a; Yi and Kim, 2004). Recently, constitutive HERV-F expression was reported in a cohort of breast cancer patients, with an increased expression of HERV-F members in comparison to normal breast tissue (Frank et al., 2008).
The knowledge on the expression and clinical impact of HERV-K and HERV-F family in STS is scarce. Schiavetti et al. (2002) reported a robust expression of HERV-K-MEL in human sarcoma specimens, which was higher than in other tumor entities and comparable to the expression in bladder and breast carcinoma, but lower than in melanoma samples. We hypothesized that a detectable HERV mRNA expression could be a common feature in STS and might be related to the patients’ clinical outcome. Therefore, the aim of this study was the quantification of HERV-K and HERV-F family mRNA expression in a cohort of 120 STS samples and the correlation to clinicopathological and prognostic data of the patients. Furthermore, as a secondary end point we analyzed the correlation of the mRNA expression of HERV-K and HERV-F with the RNA expression of known apoptosis-related [B-cell cll/lymphoma 2 (BCL2)] or hypoxia-related (miR-210, miR-199a, Hypoxia inducible factor 1a) genes as well as known epigenetically regulated genes (miR-203, H2A.Bbd).
Materials and Methods
Patients
One hundred and twenty STS patients agreed to participate in this study. An overview of the patient cohort is given in Table 1. Patients underwent tumor surgical resection between 1998 and 2001 at the Department of Surgery, University of Leipzig (Leipzig, Germany) without prior adjuvant treatment. Thirty-nine patients exhibited metastases (32.5%). Fresh tumor tissue was snap-frozen immediately after excision and stored at -80°C until RNA isolation. The study was approved by the local ethics committee of the Medical Faculty of the Martin Luther University of Halle-Wittenberg and the Medical Faculty of the University of Leipzig. According to the Helsinki Declaration, all patients gave written informed consent. Patient cohort composition as well as tissue cryopreservation was as described previously (Kappler et al., 2001; Würl et al., 2002).
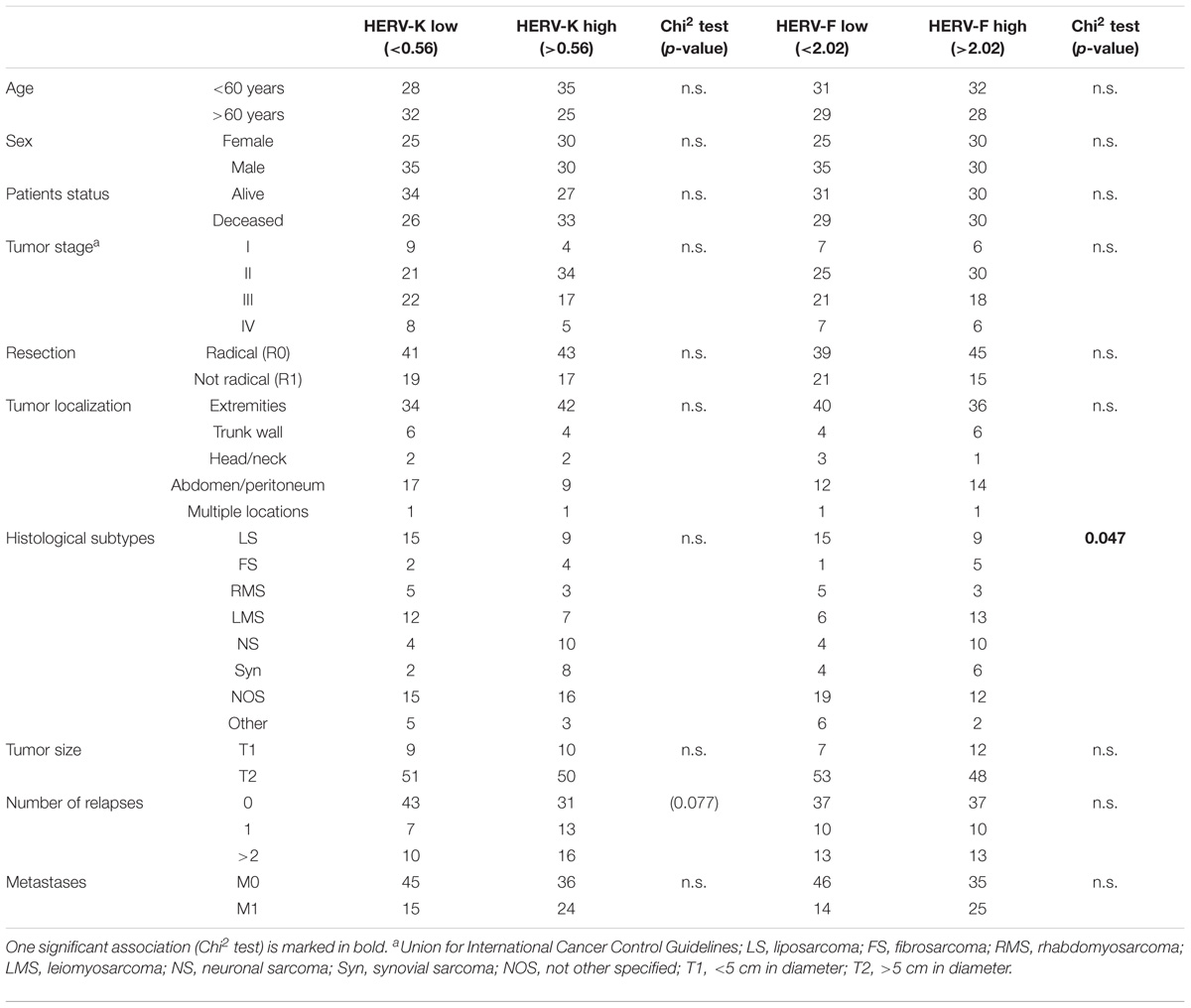
TABLE 1. Clinical and histopathological characteristics in relation to HERV-K and HERV-F mRNA expression.
RNA Isolation
Tissue specimens were partly processed on a cryotome in 5 μm tissue slices, and RNA was isolated from 20 tissue slices. The slices were incubated in Trizol (Thermo Fisher Scientific, Waltham, MA, United States) for 5 min at room temperature and subsequently mixed with chloroform (AppliChem, Darmstadt, Germany). After centrifugation, aqueous phase was collected and treated with DNase (Qiagen, Hilden, Germany). Total RNA was precipitated with isopropanol (AppliChem, Darmstadt, Germany) for 12 h at 4°C, washed with different ice-cooled ethanol solutions (96 and 70%) and finally dissolved in RNase-free water (Qiagen, Hilden, Germany). RNA concentrations were assessed spectrometrically.
cDNA Synthesis and qPCR
cDNA synthesis was carried out with RevertAid First strand synthesis kit (Thermo Fisher Scientific, Waltham, MA, United States) according to manufacturer’s protocol. One μg total RNA was applied for cDNA synthesis per tissue specimen. The complete elimination of genomic DNA was controlled by mock-RT PCR reactions (see Supplementary Figures S1, S2). cDNA was quantified with Maxima SyBR Green Kit (Thermo Fisher Scientific, Waltham, MA, United States) in a quantitative real-time-PCR reaction. The applied primer sequences for the PCR reaction were: HERV-K forward: 5′-GGC CAT CAG AGT CTA AAC CAC G-3′; HERV-K reverse: 5′-CTG ACT TTC TGG GGG TGG CCG-3′; HERV-F forward: 5′-CCT CCA GTC ACA ACA ACT C-3′; HERV-F reverse: 5′-TAT TGA AGA AGG CGG CTG G-3′ (Seifarth et al., 2005); H2A.Bbd forward: 5′-TCG TTT TCA GTA GCC AGG T-3′; H2A.Bbd reverse: 5′-CAG AAT TAA TGA AGG CCC AAG-3′; HPRT forward: 5′-TTG CTG ACC TGC TGG ATT AC-3′; HPRT reverse: 5′-CTT GCG ACC TTG ACC ATC TT-3′. Samples were run on a MyIQ cycler (BioRad, Hercules, CA, United States) and HERV-K or HERV-F expression calculated according to the 2-ΔCT method (Schmittgen and Livak, 2008) with HPRT as reference gene. Linearity of the qPCR reaction for both HERV-K and HERV-F was analyzed by dilution series of the gel-extracted amplicon (see Supplementary Figure S3). All amplicons were analyzed by qPCR melt analyses on the occurrence of a single, distinct peak (see Supplementary Figures S4, S5). Representative PCR products were purified by agarose gel electrophoresis and subsequently sequenced (see Table 2). Analysis of the sequenced PCR products with RepeatMasker1 demonstrated that the used primers amplified sequences from HERV-K and HERV-Fb (HERVFH21). Expression analyses for BCL2 mRNA, miR-203 and miR-210 (Greither et al., 2012), HIF-1α mRNA (Kessler et al., 2010) and miR-199a (Keßler et al., 2016) were carried out as previously described.
Statistical Analyses
Statistical analyses were performed with SPSS 20.0 (IBM Statistics, Ehingen, Germany). HERV expression data were analyzed with bivariate correlation analyses (Spearman rank correlation) and Chi2 tests. Survival analyses were performed with Kaplan–Meier analyses and multivariate Cox’s Regressions analyses adjusted for resection status, localization of the tumor, tumor entity, and tumor stage (inclusion).
Results
HERV-K and -F Expression in Soft Tissue Sarcoma Samples
HERV-K mRNA expression was detected in 120 patients samples with a mean expression of 4.1 (range: 0.06–95.6; 2-ΔCT value). HERV-F mRNA expression was also detected in 120 patients sarcoma specimen with a mean expression of 6.0 (range: 1 × 10-5–99.3; 2-ΔCT value, see Figures 1A,B). Additionally, we measured the HERV-K and HERV-F mRNA expression in normal skeletal muscle tissue. HERV-K mRNA expression was determined at 0.093 (2-ΔCT value) and HERV-F mRNA expression at 0.296 (2-ΔCT value). Skeletal muscle tissue therefore exhibited lower HERV mRNA values than 76.6 and 78.3% of STS samples, respectively. For survival analyses, HERV-K or HERV-F mRNA expression were classified according to the median as cut-off value (HERV-K: 0.56; HERV-F: 2.02). In Chi2 tests, low or elevated HERV-K expressions exhibited no correlation to demographic (age, sex) or clinical parameters (tumor entity or localization, resection type, tumor size, number of relapses, and patients status). In contrast, a significant correlation of HERV-F expression with the histological subtype of the STS was observed (p = 0.047).
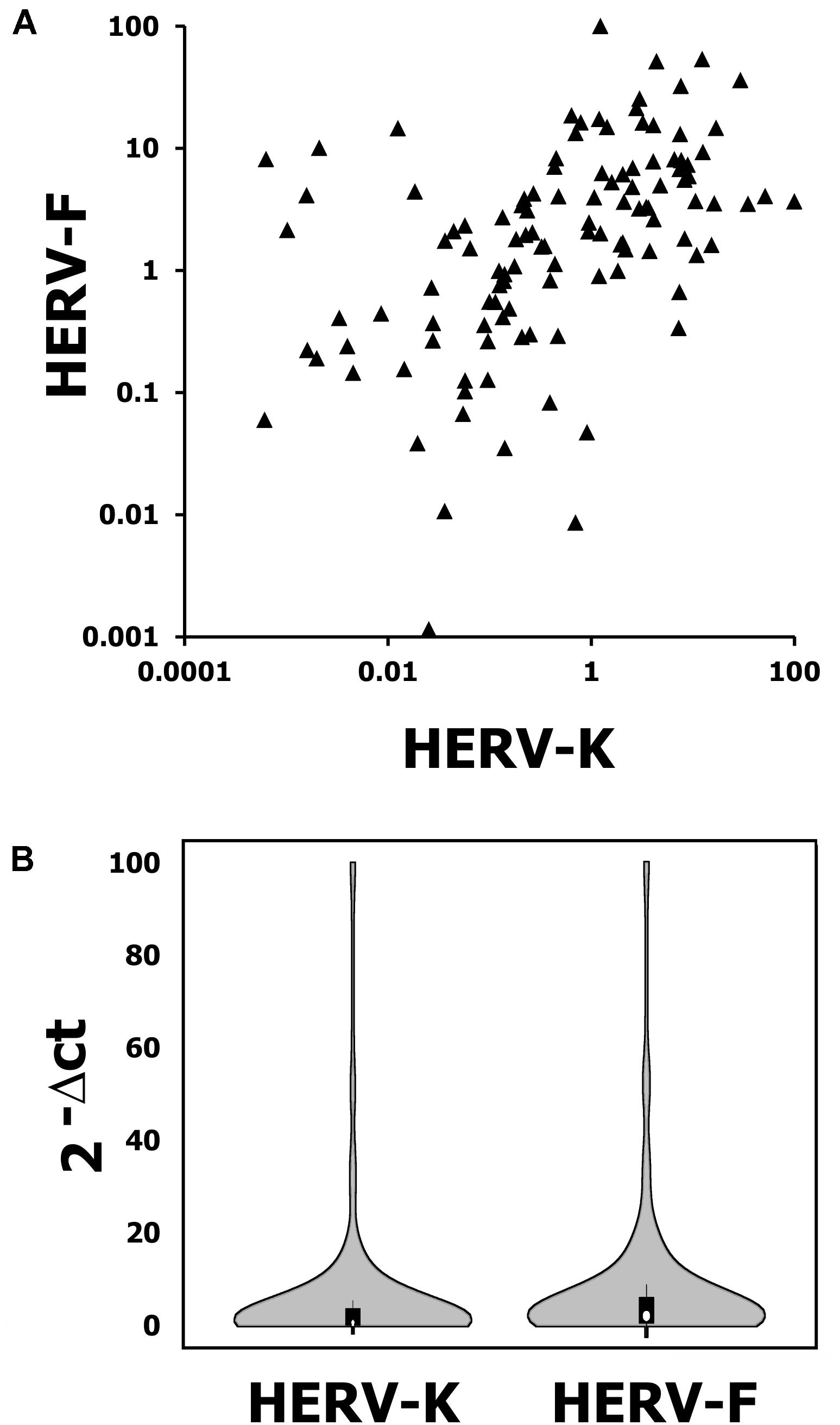
FIGURE 1. K vs. F blot (A) and violin blot (B) exhibiting the distribution of HERV-K and HERV-F mRNA expression in 120 analyzed soft tissue sarcoma (STS) specimens. Violin plot was generated with BoxPlotR (http://shiny. chemgrid.org/boxplotr/).
Association of HERV Expression with Clinicopathological Parameters
In bivariate regression analyses, the association between HERV-K or HERV-F expression and several clinicopathological parameters were tested (see Table 3). Interestingly, HERV-F and HERV-K expression was significantly associated (rS = 0.499; p = 6.4 × 10-9). Both HERV-F and HERV-K expression exhibited a significant inverse association with the actual tumor diameter at surgery (rS = -0.309; p = 0.001 and rS = -0.467; p = 7.4 × 10-8; respectively). Moreover, HERV-K mRNA expression was significantly inversely associated with BCL2 mRNA expression (rS = -0.408; p = 0.0002) and miR-199a (rS = 0.361; p = 0.0004) expression, while solely HERV-F expression was significantly associated with miR-203 expression (rS = 0.333; p = 0.005). Intriguingly, both HERV-K and HERV-F expression were significantly associated with levels of hypoxia-related genes like HIF-1α mRNA expression (rS = 0.444; p = 3.0 × 10-6 and 0.359; p = 0.0002; respectively) or miR-210 expression (rS = 0.399; p = 0.001 and rS = 0.366; p = 0.002; respectively). Additionally, both HERV-K and HERV-F expression were significantly associated to H2A.Bbd mRNA expression (rS = 0.456; p = 0.0009 and rS = 0.302; p = 0.012, respectively).
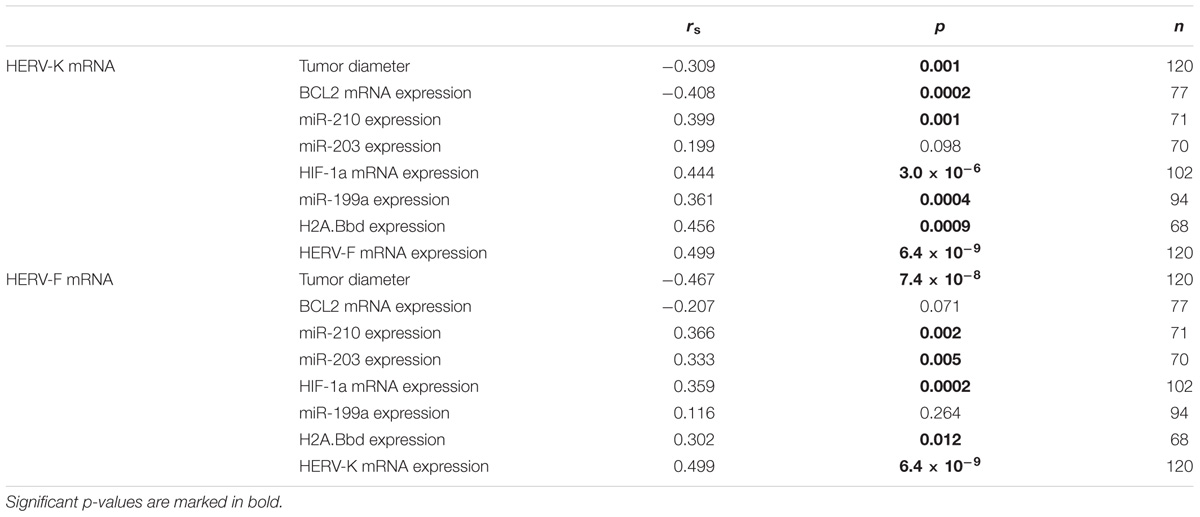
TABLE 3. Bivariate correlations (Spearman’s rank test; rs) of HERV-K or HERV-F mRNA expression with several clinicopathological and molecular parameters.
HERV-K or -F Expression and Patients’ Survival
In Kaplan–Meier analyses, while HERV-F mRNA expression showed no significant correlation to patients’ disease-specific survival, a lower HERV-K mRNA expression was in trend associated with a worsened survival (p = 0.08; log rank test). Further, in a multivariate Cox’s regression analysis adjusted to the confounders resection type, tumor localization, tumor histotype and staging, there was no significant correlation between HERV-K or HERV-F mRNA expression and patient survival (see Supplementary Figure S6). Interestingly, when analyzing the relapse-free survival in Kaplan–Meier analyses, patients with a lower HERV-K mRNA expression exhibited a significantly longer relapse-free survival (p = 0.014; log-rank test, see Figure 2A). A comparable effect was observed in patients with lower HERV-F mRNA expression; however, there it was not significant (p = 0.22, see Figure 2B). Additionally, when analyzing the effect of the HERV-K mRNA expression on the relapse-free survival in a multivariate Cox’s regression analysis, an elevated HERV-K mRNA expression was in trend associated with a 1.78-fold increased risk for a relapse (p = 0.08). Furthermore, when comparing only patients exhibiting low HERV-F and HERV-K (n = 43) with patients exhibiting both elevated HERV-F and HERV-K expression (n = 43) in a multivariate Cox’s regression analysis, an elevated expression of HERVs was in trend significantly associated with a 2.08-fold increased relative risk for a relapse (p = 0.066).
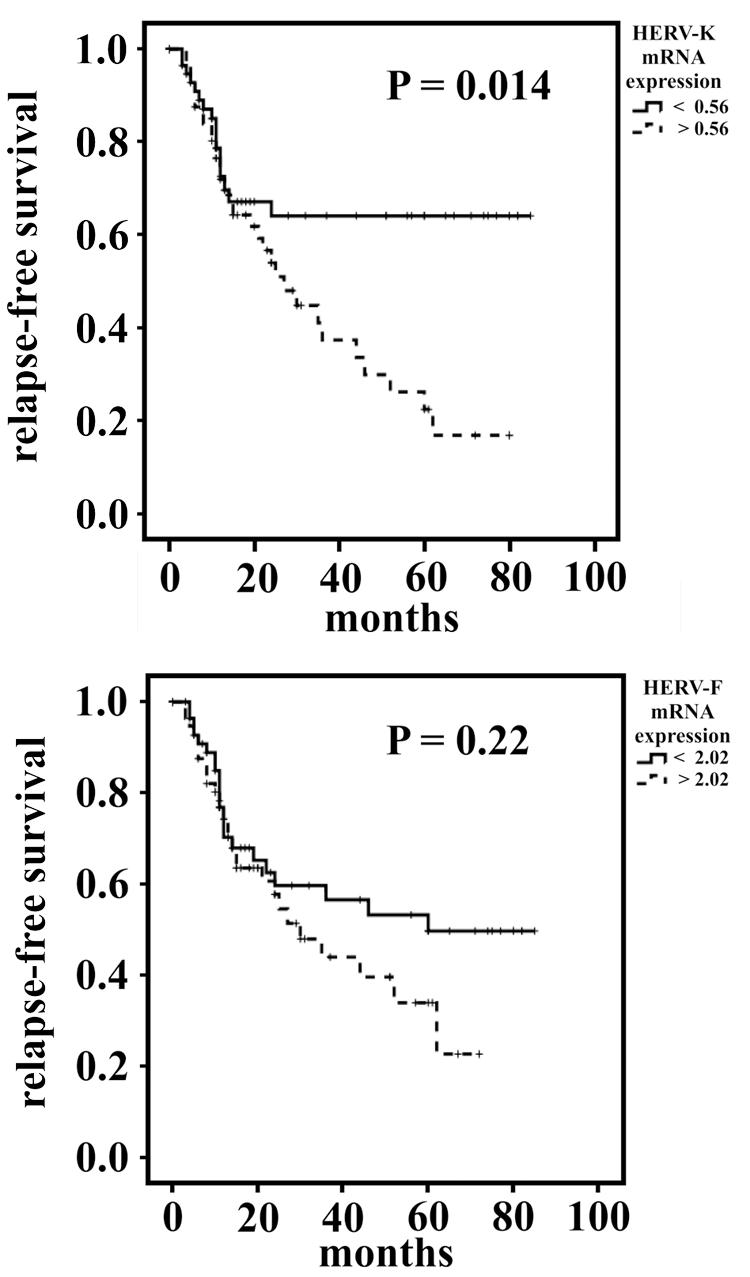
FIGURE 2. Survival analyses of STS patients with low or elevated intratumoral HERV-K (A) or HERV-F (B) expression. Kaplan–Meier survival blots for the relapse-free survival.
Discussion
In this study, we demonstrated that a robust mRNA expression of HERV-K and HERV-F in a cohort of 120 STS samples is detectable, and that the expression of HERV-K and HERV-F is correlated with clinicopathological features and hypoxia-related gene expression. Furthermore, an elevated HERV-K mRNA expression was significantly associated with a shorter relapse-free survival.
There are only few data on the expression of HERVs in STS. Schiavetti and colleagues studied the expression of HERV-K-MEL in sarcoma in comparison to the expression in a patient’s sample of melanoma cells. This transcript was detectable in 9/23 (39.1%) of sarcoma samples (Schiavetti et al., 2002). There is no data on the expression of HERV-F in sarcoma, however, one report shows a wide expression of HERV-F in tumor cell lines originating from mamma carcinoma, ovarian carcinoma, pancreatic adenocarcinoma, prostate carcinoma, glioblastoma, and others (Yi and Kim, 2004). Interestingly, HERV-F expression was not detected in any somatic tissue tested, with the exception of placenta (Yi and Kim, 2004). These reports are consistent with the assumption, that HERV sequences are normally methylated and therefore transcriptionally inactive, but are hypomethylated and activated during carcinogenesis (Kreimer et al., 2013; Hurst and Magiorkinis, 2017). Concordantly, the treatment with 5′-azacytdidine, a known DNA methyltransferase inhibitor, activates HERV sequences in diverse tumor cell lines (Strissel et al., 2012; Laska et al., 2013; Chiappinelli et al., 2015). Therefore, we propose that re-induction of HERVs may also occur during the multi-step process of sarcomagenesis.
Intriguingly, we identified a significant association of the HERV-K and -F expression with those of the hypoxia-related genes HIF-1α and miR-210. HIF-1α is a key regulator of the hypoxic response, and miR-210 is the most prominent microRNA upregulated by hypoxia (Kulshreshtha et al., 2007). There is little knowledge about interactions between HERV expression and hypoxic response. It has been described, that the hypoxia-mimetic CoCl2 increases the expression of ERV3 in Hodgkin’s lymphoma cell lines, and that this increase in ERV3 expression might be associated with a pro-apoptotic reaction (Kewitz and Staege, 2013). Other groups demonstrated upregulation of the HERV-W expression in neuroblastoma cell lines due to hypoxic conditions (Hu et al., 2016) or upregulation of the HERV-E expression in renal cell carcinomas due to inactivation of the von Hippel-Lindau factor and subsequent stabilization of the oxygen sensor protein HIF-1α (Cherkasova et al., 2011). In our ex vivo samples, we detected an inverse association between HERV-K and BCL2 mRNA expression, implying that HERV-K overexpression could be associated with apoptosis-induction. However, other reports performed on in vitro cell cultures demonstrate that the HERV-K family exerts an anti-apoptotic role (Broecker et al., 2016). Further research on this contradiction is warranted.
Furthermore, HERV-K and HERV-F expression were both significantly correlated to the mRNA expression of H2A.Bbd, a histone A2 variant encoded on the X chromosome, which is found to be associated with the nucleosomes of transcriptionally active genomic regions (Chadwick and Willard, 2001). H2A.Bbd was further shown to induce a more relaxed structure of the DNA by destabilizing the nucleosome (Bao et al., 2004; Doyen et al., 2006), which resembles the genomic reorganization induced by histone acetylation in a modification-independent manner (Eirín-López et al., 2008). A recent report showed H2A.Bbd to localize temporarily on replication-active DNA regions. By this mechanism, H2A.Bbd is holding the DNA in a more decondensed state, thereby increasing S-phase progression (Sansoni et al., 2014). Thus, it can be speculated that an H2A.Bbd overexpression in patient samples may be associated with a more transcriptionally active genome resulting in an increased chance of reactivation and expression HERV species.
In our patient cohort, we detected a significant association between a lower HERV-K expression and a longer relapse-free survival. This is concordant with previous reports describing a better overall prognosis for patients with a lower HERV-K expression in breast cancer (Golan et al., 2008; Zhao et al., 2011) or hepatocellular carcinoma (Ma et al., 2016). Additionally, hypomethylation and subsequent HERV induction was also demonstrated in ovarian carcinoma, and specifically the extent HERV-K hypomethylation was associated with a poor prognosis and therapy resistance in ovarian carcinoma patients (Menendez et al., 2004; Iramaneerat et al., 2011).
Conclusion
To the best of our knowledge we present the first report suggesting an involvement of the HERV-K expression in the clinical course of STS. From our ex vivo data, we also suggest that HERV-K and -F expression may be regulated directly or indirectly by tumor hypoxia. Furthermore, HERV-K was associated to apoptosis in our samples, therefore may be an interesting therapeutic target.
Author Contributions
MG performed the data analysis and revised the manuscript; SB, LO, and MK carried out the clinical sample processing and the qPCR measurements; PW recruited the patients and collected the tissue specimen; TG, HT, and MS conceived the study design, prepared and revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to thank Ines Volkmer for excellent technical assistance. They also acknowledge the financial support of the Open Access Publication Fund of the Martin Luther University of Halle-Wittenberg.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00211/full#supplementary-material
Footnotes
References
Bannert, N., and Kurth, R. (2004). Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U.S.A. 101(Suppl. 2), 14572–14579. doi: 10.1073/pnas.0404838101
Bao, Y., Konesky, K., Park, Y.-J., Rosu, S., Dyer, P. N., Rangasamy, D., et al. (2004). Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 23, 3314–3324. doi: 10.1038/sj.emboj.7600316
Brodsky, I., Foley, B., and Gillespie, D. (1993). Expression of human endogenous retrovirus (HERV-K) in chronic myeloid leukemia. Leuk. Lymphoma 11(Suppl. 1), 119–123. doi: 10.3109/10428199309047874
Broecker, F., Horton, R., Heinrich, J., Franz, A., Schweiger, M.-R., Lehrach, H., et al. (2016). The intron-enriched HERV-K(HML-10) family suppresses apoptosis, an indicator of malignant transformation. Mob. DNA 7:25. doi: 10.1186/s13100-016-0081-9
Casali, P. G., and Blay, J.-Y. (2010). Soft tissue sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 21(Suppl. 5), v198–v203. doi: 10.1093/annonc/mdq209
Cegolon, L., Salata, C., Weiderpass, E., Vineis, P., Palù, G., and Mastrangelo, G. (2013). Human endogenous retroviruses and cancer prevention: evidence and prospects. BMC Cancer 13:4. doi: 10.1186/1471-2407-13-4
Chadwick, B. P., and Willard, H. F. (2001). A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol. 152, 375–384. doi: 10.1083/jcb.152.2.375
Cherkasova, E., Malinzak, E., Rao, S., Takahashi, Y., Senchenko, V. N., Kudryavtseva, A. V., et al. (2011). Inactivation of the von Hippel-Lindau tumor suppressor leads to selective expression of a human endogenous retrovirus in kidney cancer. Oncogene 30, 4697–4706. doi: 10.1038/onc.2011.179
Chiappinelli, K. B., Strissel, P. L., Desrichard, A., Li, H., Henke, C., Akman, B., et al. (2015). Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986. doi: 10.1016/j.cell.2015.07.011
Doyen, C.-M., Montel, F., Gautier, T., Menoni, H., Claudet, C., Delacour-Larose, M., et al. (2006). Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 25, 4234–4244. doi: 10.1038/sj.emboj.7601310
Ducimetière, F., Lurkin, A., Ranchère-Vince, D., Decouvelaere, A.-V., Péoc’h, M., Istier, L., et al. (2011). Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLOS ONE 6:e20294. doi: 10.1371/journal.pone.0020294
Eirín-López, J. M., Ishibashi, T., and Ausió, J. (2008). H2A.Bbd: a quickly evolving hypervariable mammalian histone that destabilizes nucleosomes in an acetylation-independent way. FASEB J. 22, 316–326. doi: 10.1096/fj.07-9255com
Ferrari, A., Sultan, I., Huang, T. T., Rodriguez-Galindo, C., Shehadeh, A., Meazza, C., et al. (2011). Soft tissue sarcoma across the age spectrum: a population-based study from the surveillance epidemiology and end results database. Pediatr. Blood Cancer 57, 943–949. doi: 10.1002/pbc.23252
Florl, A. R., Löwer, R., Schmitz-Dräger, B. J., and Schulz, W. A. (1999). DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br. J. Cancer 80, 1312–1321. doi: 10.1038/sj.bjc.6690524
Frank, O., Verbeke, C., Schwarz, N., Mayer, J., Fabarius, A., Hehlmann, R., et al. (2008). Variable transcriptional activity of endogenous retroviruses in human breast cancer. J. Virol. 82, 1808–1818. doi: 10.1128/JVI.02115-07
Golan, M., Hizi, A., Resau, J. H., Yaal-Hahoshen, N., Reichman, H., Keydar, I., et al. (2008). Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia 10, 521–533. doi: 10.1593/neo.07986
Greither, T., Würl, P., Grochola, L., Bond, G., Bache, M., Kappler, M., et al. (2012). Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int. J. Cancer 130, 1230–1235. doi: 10.1002/ijc.26109
Griffiths, D. J. (2001). Endogenous retroviruses in the human genome sequence. Genome Biol. 2, reviews1017.1–reviews1017.5. doi: 10.1186/gb-2001-2-6-reviews1017
Hu, L., Uzhameckis, D., Hedborg, F., and Blomberg, J. (2016). Dynamic and selective HERV RNA expression in neuroblastoma cells subjected to variation in oxygen tension and demethylation. APMIS 124, 140–149. doi: 10.1111/apm.12494
Hughes, J. F., and Coffin, J. M. (2004). Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. U.S.A. 101, 1668–1672. doi: 10.1073/pnas.0307885100
Hughes, J. F., and Coffin, J. M. (2005). Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171, 1183–1194. doi: 10.1534/genetics.105.043976
Hurst, T. P., and Magiorkinis, G. (2017). Epigenetic control of human endogenous retrovirus expression: focus on regulation of long-terminal repeats (LTRs). Viruses 9:E130. doi: 10.3390/v9060130
Iramaneerat, K., Rattanatunyong, P., Khemapech, N., Triratanachat, S., and Mutirangura, A. (2011). HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int. J. Gynecol. Cancer. 21, 51–57. doi: 10.1097/IGC.0b013e3182021c1a
Kappler, M., Köhler, T., Kampf, C., Diestelkötter, P., Würl, P., Schmitz, M., et al. (2001). Increased survivin transcript levels: an independent negative predictor of survival in soft tissue sarcoma patients. Int. J. Cancer 95, 360–363.
Kessler, J., Hahnel, A., Wichmann, H., Rot, S., Kappler, M., Bache, M., et al. (2010). HIF-1α inhibition by siRNA or chetomin in human malignant glioma cells: Effects on hypoxic radioresistance and monitoring via CA9 expression. BMC Cancer 10:605. doi: 10.1186/1471-2407-10-605
Kewitz, S., and Staege, M. S. (2013). Expression and regulation of the endogenous retrovirus 3 in Hodgkin’s lymphoma cells. Front. Oncol. 3:179. doi: 10.3389/fonc.2013.00179
Keßler, J., Rot, S., Bache, M., Kappler, M., Würl, P., Vordermark, D., et al. (2016). miR-199a-5p regulates HIF-1α and OSGIN2 and its expression is correlated to soft-tissue sarcoma patients’ outcome. Oncol. Lett. 12, 5281–5288. doi: 10.3892/ol.2016.5320
Kjellman, C., Sjögren, H. O., Salford, L. G., and Widegren, B. (1999a). HERV-F (XA34) is a full-length human endogenous retrovirus expressed in placental and fetal tissues. Gene 239, 99–107.
Kjellman, C., Sjögren, H. O., and Widegren, B. (1999b). HERV-F, a new group of human endogenous retrovirus sequences. J. Gen. Virol. 80(Pt 9), 2383–2392.
Kreimer, U., Schulz, W. A., Koch, A., Niegisch, G., and Goering, W. (2013). HERV-K and LINE-1 DNA methylation and reexpression in Urothelial Carcinoma. Front. Oncol. 3:255. doi: 10.3389/fonc.2013.00255
Kulshreshtha, R., Ferracin, M., Wojcik, S. E., Garzon, R., Alder, H., Agosto-Perez, F. J., et al. (2007). A microRNA signature of hypoxia. Mol. Cell. Biol. 27, 1859–1867. doi: 10.1128/MCB.01395-06
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Laska, M. J., Nissen, K. K., and Nexø, B. A. (2013). (Some) cellular mechanisms influencing the transcription of human endogenous retrovirus, HERV-Fc1. PLOS ONE 8:e53895. doi: 10.1371/journal.pone.0053895
Li, M., Radvanyi, L., Yin, B., Li, J., Chivukula, R., Lin, K., et al. (2017). Downregulation of human endogenous retrovirus type K (HERV-K) viral env RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin. Cancer Res. 23, 5892–5911. doi: 10.1158/1078-0432.CCR-17-0001
Ma, W., Hong, Z., Liu, H., Chen, X., Ding, L., Liu, Z., et al. (2016). Human endogenous retroviruses-K (HML-2) expression is correlated with prognosis and progress of hepatocellular Carcinoma. BioMed Res. Int. 2016:8201642. doi: 10.1155/2016/8201642
Menendez, L., Benigno, B. B., and McDonald, J. F. (2004). L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol. Cancer 3:12.
Meyer, T. J., Rosenkrantz, J. L., Carbone, L., and Chavez, S. L. (2017). Endogenous retroviruses: with us and against us. Front. Chem. 5:23. doi: 10.3389/fchem.2017.00023
Patzke, S., Lindeskog, M., Munthe, E., and Aasheim, H. C. (2002). Characterization of a novel human endogenous retrovirus, HERV-H/F, expressed in human leukemia cell lines. Virology 303, 164–173. doi: 10.1006/viro.2002.1615
Sansoni, V., Casas-Delucchi, C. S., Rajan, M., Schmidt, A., Bönisch, C., Thomae, A. W., et al. (2014). The histone variant H2A.Bbd is enriched at sites of DNA synthesis. Nucleic Acids Res. 42, 6405–6420. doi: 10.1093/nar/gku303
Schiavetti, F., Thonnard, J., Colau, D., Boon, T., and Coulie, P. G. (2002). A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 62, 5510–5516.
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seifarth, W., Frank, O., Zeilfelder, U., Spiess, B., Greenwood, A. D., Hehlmann, R., et al. (2005). Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 79, 341–352. doi: 10.1128/JVI.79.1.341-352.2005
Serafino, A., Balestrieri, E., Pierimarchi, P., Matteucci, C., Moroni, G., Oricchio, E., et al. (2009). The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 315, 849–862. doi: 10.1016/j.yexcr.2008.12.023
Steinestel, K., and Wardelmann, E. (2015). Metastasierung und Progressionsmechanismen von Weichteiltumoren. Pathologe 36(Suppl. 2), 167–170. doi: 10.1007/s00292-015-0072-5
Stiller, C. A., Trama, A., Serraino, D., Rossi, S., Navarro, C., Chirlaque, M. D., et al. (2013). Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur. J. Cancer 49, 684–695. doi: 10.1016/j.ejca.2012.09.011
Strissel, P. L., Ruebner, M., Thiel, F., Wachter, D., Ekici, A. B., Wolf, F., et al. (2012). Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: emergence of new molecular targets. Oncotarget 3, 1204–1219. doi: 10.18632/oncotarget.679
Vargiu, L., Rodriguez-Tomé, P., Sperber, G. O., Cadeddu, M., Grandi, N., Blikstad, V., et al. (2016). Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13:7. doi: 10.1186/s12977-015-0232-y
Wallace, T. A., Downey, R. F., Seufert, C. J., Schetter, A., Dorsey, T. H., Johnson, C. A., et al. (2014). Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis 35, 2074–2083. doi: 10.1093/carcin/bgu114
Wang-Johanning, F., Frost, A. R., Jian, B., Epp, L., Lu, D. W., and Johanning, G. L. (2003). Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 22, 1528–1535. doi: 10.1038/sj.onc.1206241
Würl, P., Kappler, M., Meye, A., Bartel, F., Köhler, T., Lautenschläger, C., et al. (2002). Co-expression of survivin and TERT and risk of tumour-related death in patients with soft-tissue sarcoma. Lancet 359, 943–945. doi: 10.1016/S0140-6736(02)07990-4
Yi, J.-M., and Kim, H.-S. (2004). Expression analysis of endogenous retroviral elements belonging to the HERV-F family from human tissues and cancer cells. Cancer Lett. 211, 89–96. doi: 10.1016/j.canlet.2004.01.026
Keywords: soft tissue sarcoma, HERV-K, HERV-Fb, prognosis, relapse
Citation: Giebler M, Staege MS, Blauschmidt S, Ohm LI, Kraus M, Würl P, Taubert H and Greither T (2018) Elevated HERV-K Expression in Soft Tissue Sarcoma Is Associated with Worsened Relapse-Free Survival. Front. Microbiol. 9:211. doi: 10.3389/fmicb.2018.00211
Received: 26 October 2017; Accepted: 30 January 2018;
Published: 13 February 2018.
Edited by:
Gkikas Magiorkinis, National and Kapodistrian University of Athens, GreeceReviewed by:
George Robert Young, Francis Crick Institute, United KingdomCésar López-Camarillo, Universidad Autónoma de la Ciudad de México, Mexico
Tara Patricia Hurst, Abcam, United Kingdom
Copyright © 2018 Giebler, Staege, Blauschmidt, Ohm, Kraus, Würl, Taubert and Greither. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Greither, dGhvbWFzLmdyZWl0aGVyQG1lZGl6aW4udW5pLWhhbGxlLmRl
†These authors have contributed equally to this work.
 Maria Giebler
Maria Giebler Martin S. Staege
Martin S. Staege Sindy Blauschmidt1
Sindy Blauschmidt1 Helge Taubert
Helge Taubert Thomas Greither
Thomas Greither