- State Key Laboratory of Applied Microbiology Southern China, Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangdong Institute of Microbiology, Guangzhou, China
The aim of this study was to characterize the subtypes and virulence profiles of 69 Staphylococcus aureus isolates obtained from retail ready-to-eat food in China. The isolates were analyzed using multilocus sequence typing (MLST) and polymerase chain reaction (PCR) analysis of important virulence factor genes, including the staphylococcal enterotoxin (SE) genes (sea, seb, sec, sed, see, seg, seh, sei, sej), the exfoliative toxin genes (eta and etb), the toxic shock syndrome toxin-1 gene (tst), and the Panton-Valentine leucocidin-encoding gene (pvl). The isolates encompassed 26 different sequence types (STs), including four new STs (ST3482, ST3484, ST3485, ST3504), clustered in three clonal complexes and 17 singletons. The most prevalent STs were ST1, ST6, and ST15, constituting 34.8% of all isolates. Most STs (15/26, 57.7%) detected have previously been associated with human infections. All 13 toxin genes examined were detected in the S. aureus isolates, with 84.1% of isolates containing toxin genes. The three most prevalent toxin genes were seb (36.2%), sea (33.3%), and seg (33.3%). The classical SE genes (sea–see), which contribute significantly to staphylococcal food poisoning (SFP), were detected in 72.5% of the S. aureus isolates. In addition, pvl, eta, etb, and tst were found in 11.6, 10.1, 10.1, and 7.2% of the S. aureus isolates, respectively. Strains ST6 carrying sea and ST1 harboring sec-seh enterotoxin profile, which are the two most common clones associated with SFP, were also frequently detected in the food samples in this study. This study indicates that these S. aureus isolates present in Chinese ready-to-eat food represents a potential public health risk. These data are valuable for epidemiological studies, risk management, and public health strategies.
Introduction
The facultative anaerobe Staphylococcus aureus is the leading cause of both nosocomial infections and community-acquired infections worldwide, causing many serious illnesses (Rodríguez-Lázaro et al., 2015; Yang et al., 2016). In addition, high rates of human nasal carriage, a high incidence of airborne spread, and the ability to survive for long periods on fomites contribute to making S. aureus an effective foodborne pathogen. Of particular concern is the fact that S. aureus is increasingly showing resistance to multiple antimicrobial agents. Methicillin-resistant S. aureus (MRSA) strains, including healthcare-acquired MRSA (HA-MRSA), community-acquired MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA) strains, exhibit resistance to all β-lactam antibiotics through acquisition of the mobile staphylococcal cassette chromosome mec (SCCmec) and constitute a major health concern.
Staphylococcus aureus possesses various virulence factors that are implicated in pathogenesis, such as the staphylococcal enterotoxins (SEs), toxic shock syndrome toxin-1 (TSST-1), the exfoliative toxins A and B (ETA and ETB), and Panton-Valentine leukocidin (PVL) (Wang et al., 2012, 2014; Song et al., 2015). Among these, SEs and TSST-1 belong to a family of staphylococcal superantigens that cause the abnormal activation of T-cells (Yamamoto et al., 2013). The heat-stable SEs are associated with staphylococcal food poisoning (SFP) and other severe diseases (Yan et al., 2012). To date, at least 24 SEs (SEA–E, SEG–V, and SEX) have been identified (Yamamoto et al., 2013). Of these, types SEA–SEE are referred to as the classical SE types, and are reported to cause about 95% of SFP cases (Wang et al., 2012). The TSST-1 causes toxic shock syndrome (TSS) and neonatal TSS-like exanthematous disease (He et al., 2013; Yamamoto et al., 2013). Furthermore, ETA and ETB are epidermolytic proteases that digest the desmoglein 1 component of desmosomes, which disrupt stratum granulosum and subsequently cause blister formation and exfoliation of the epidermis (Yamamoto et al., 2013), a condition known as staphylococcal scalded skin syndrome (Song et al., 2015). Finally, PVL causes necrotizing pneumonia, sepsis, and severe tissue damage (Lina et al., 1999; Yamamoto et al., 2013). The presence of these virulence factors in S. aureus isolates from food may therefore pose a threat to public health.
Multilocus sequence typing (MLST) has been widely used to investigate the epidemiology of microbial populations (Xu et al., 2015; Wu et al., 2016). The method is based on the sequencing of a number of different housekeeping genes, with the resulting data made publicly available online. This allows the comparison of different isolates and their sequence types (STs) between laboratories, thus producing a powerful resource for global epidemiology (Maiden et al., 1998). Owing to these advantages, MLST has become an important and reliable technique for epidemiological analysis of S. aureus. For HA-MRSA, CA-MRSA, and LA-MRSA strains, they produce different STs, which help to identify them. The New York/Japan (ST5/SCCmec II) and Brazilian/Hungarian (ST239/SCCmec III) clones are pandemic HA-MRSA lineages (Yamamoto et al., 2013), while the Taiwanese (ST59/SCCmec IV or V), USA300 (ST8/SCCmec IV), European (ST80/SCCmec IV), and USA400 (ST1) clones are always associated with community-acquired infections (Yamamoto et al., 2013).
Ready-to-eat (RTE) food, such as cooked meat and poultry, cold vegetable dishes, cold noodles, and fried rice, are becoming more popular in China and their consumption has increased significantly. In our previous work, 69 S. aureus isolates were identified from Chinese RTE food samples, and the clonal lineages of the MRSA isolates were determined, including CA-MRSA [ST59/SCCmec IVa (n = 2), ST338/SCCmec V, ST1/SCCmec V], and LA-MRSA (ST9) (Yang et al., 2016). To assess the potential virulence and risk posed by these 69 S. aureus isolates from RTE food, we provide a phylogenetic framework for these isolates in the current study, based on MLST analysis and screened for the presence of virulence-associated genes.
Materials and Methods
Staphylococcus aureus Strains
We analyzed a total of 69 S. aureus isolates from 550 RTE food samples (cooked pork, cooked chicken, cooked duck, cold vegetable dishes in sauce, cold noodles, and fried rice/sushi) collected from markets in 24 Chinese cities from December 2011 to May 2014 (Yang et al., 2016). Of the 69 S. aureus isolates, seven were identified as methicillin-resistant, of which six were shown to be mecA-positive (Yang et al., 2016). All isolates were stored at -40°C until further analysis.
Multilocus Sequence Typing
Of the 69 S. aureus isolates, the STs of the six mecA-positive MRSA isolates were determined in our previous work (Yang et al., 2016). In this study, the remaining 63 S. aureus isolates [62 methicillin-susceptible S. aureus (MSSA) and 1 mecA-negative MRSA] were characterized by MLST analysis. Chromosomal DNA was prepared using a Genomic DNA Extraction kit (Dongsheng Biotech, Guangzhou, China) following the manufacturer’s protocol, and MLST was carried out using previously reported primers that are specific for seven housekeeping genes: arcC, aroE, glpF, gmk, pta, tpi, and yqiL. The ST of each isolate was assigned according to the MLST database1. The associated clonal complex (CC) was calculated using the eBURST algorithm, which is used to cluster related STs2. A phylogenetic tree was generated using the unweighted pair-group method with arithmetic mean from the sequence data using MEGA 6.06.
Detection of Virulence-Associated Genes
Thirteen toxin genes with reported contributions to virulence were selected, including nine major SE genes (sea, seb, sec, sed, see, seg, seh, sei, sej), the TSST-1 toxin gene (tst), the exfoliative toxin genes (eta and etb), and the PVL-encoding gene (pvl). All examined 69 S. aureus isolates were screened for the presence of 13 toxin genes using previously described primers (Lina et al., 1999; Noguchi et al., 2006; Peles et al., 2007; Wang et al., 2012; Supplementary Table S1). The 25 μL reaction mixtures contained 12.5 μL of 2× GoldStar Taq MasterMix (Cwbio, Beijing, China), 0.5 μL of each primer (10 μM), 4 μL of extracted genomic DNA, and sterile deionized water. Polymerase chain reaction (PCR) was performed using a T-Professional thermocycler (Biometra, Gottingen, Germany) under the following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 60 s, and a final extension step at 72°C for 10 min. A 6 μL aliquot of each amplicon was resolved on a 1.5% agarose gel (Invitrogen, Paisley, United Kingdom) with GoldView Nucleic Acid Stain (SBS Genetech Co., Ltd., Beijing, China). Gels were examined using the ImageQuant 350 system (GE Healthcare, Chalfont St. Giles, United Kingdom).
Results
Multilocus Sequence Typing of Staphylococcus aureus Isolates
In addition to the previously determined STs of the six mecA-positive MRSA isolates, a total of 26 distinct STs were identified, including four novel STs (ST3482, ST3484, ST3485, and ST3504) (Table 1). The number of alleles for the seven housekeeping genes ranged from 1 to 330. The most commonly detected allelic profiles were ST1, ST6, and ST15 (8/69, 11.6% for each). With the exception of ST9, ST72, ST121, ST338, ST630, ST1094, ST2196, ST3239, and the four novel STs, the remainder of the STs included more than one isolate. Of these, ST1, ST59, and ST25 included both MRSA and MSSA isolates. Based on eBURST analysis, three CCs were identified, including CC1 (ST1, ST2592), CC2483 (ST2483, ST2196, ST3482, ST3484, ST3504), and CC59 (ST59, ST338). The two predominant CCs (CC1 and CC2483) included 14.5% (9/62) and 11.3% (7/62) of the MSSA isolates, respectively, while CC59 accounted for the majority of MRSA isolates (3/7, 42.9%). A phylogenetic tree based on the seven concatenated sequences generated during MLST showed clustering among the 69 RTE food isolates and indicated the relatedness between the STs (Figure 1).
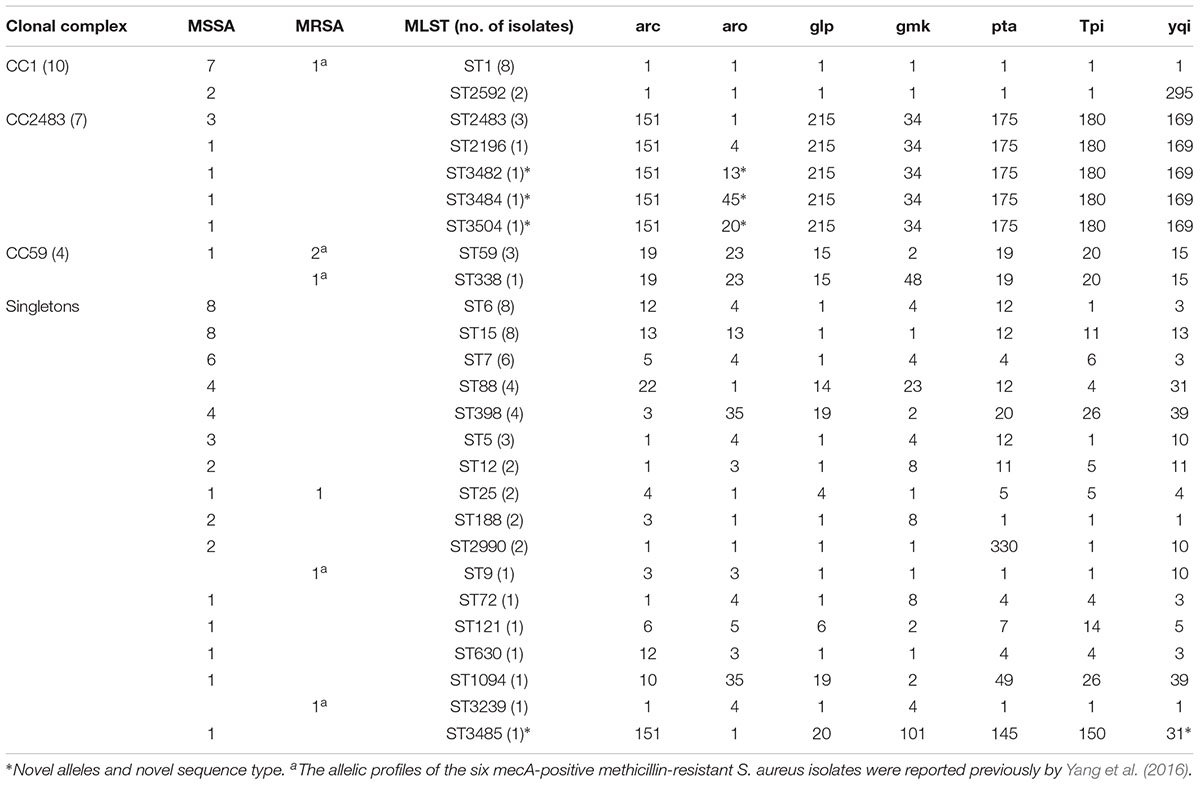
TABLE 1. Allelic profiles of Staphylococcus aureus isolates from ready-to-eat food as determined by multilocus sequence typing.
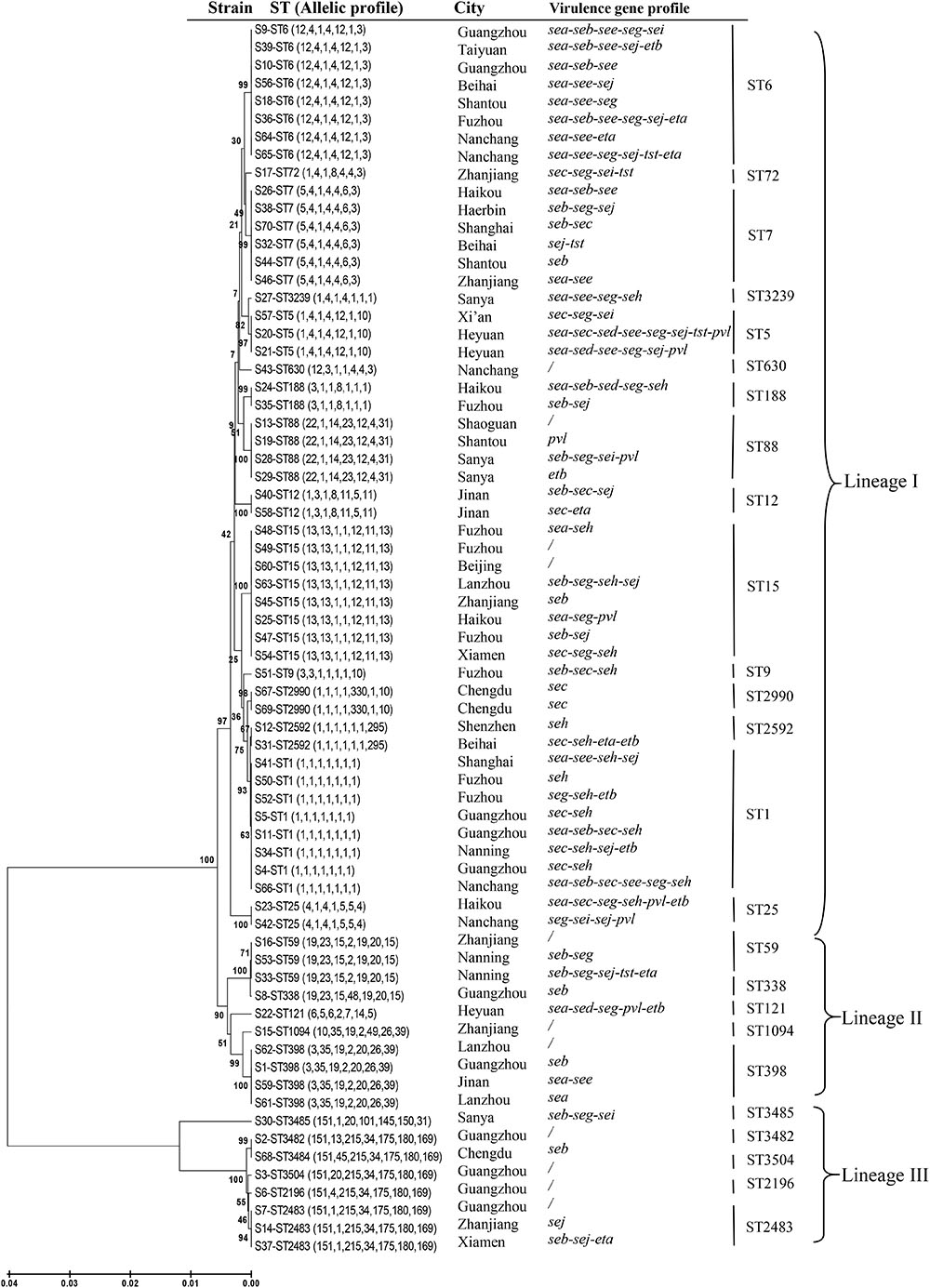
FIGURE 1. Unweighted pair-group method with arithmetic mean tree of the seven gene fragments examined during multi-locus sequence typing of Staphylococcus aureus isolates from ready-to-eat food in China.
Distribution of Toxin Genes
All 13 toxin genes examined were detected in the 69 S. aureus isolates. In total, toxin genes were identified in 84.1% (58/69) of S. aureus isolates, and 49.3% (34/69) of isolates contained more than three toxin genes. As shown in Table 2, 81.2% of S. aureus isolates harbored SE genes and the three most prevalent were seb (36.2%, 25/69), sea (33.3%, 23/69), and seg (33.3%, 23/69). In addition, pvl, eta, etb, and tst were found in 11.6, 10.1, 10.1, and 7.2% of the S. aureus isolates, respectively.
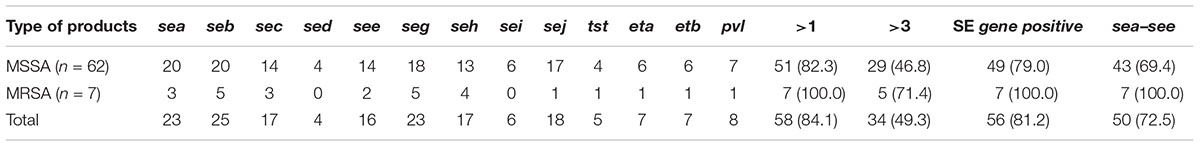
TABLE 2. Distribution of 13 toxin genes in Staphylococcus aureus isolates From retail ready-to-eat food in China.
Overall, the 69 S. aureus isolates displayed 59 different toxin gene profiles, with one to eight toxin genes found in each isolate in different combinations. The most prevalent toxin gene profile was the single enterotoxin gene seb (7.2%, 5/69). The sea-see combination, with or without additional toxin genes, occurred in 16 isolates, and it was the most common combination observed. As for the distribution of toxin genes among the S. aureus isolates based on the food categories, the SE genes, except sed and sei, were detected in isolates from all six food categories. All S. aureus isolates contained the classic SE genes in samples collected from cold vegetable dishes in sauce and fried rice/sushi, and these two kinds of food, along with cooked chicken, were also the major sources of isolates containing multiple toxin genes (Table 3).
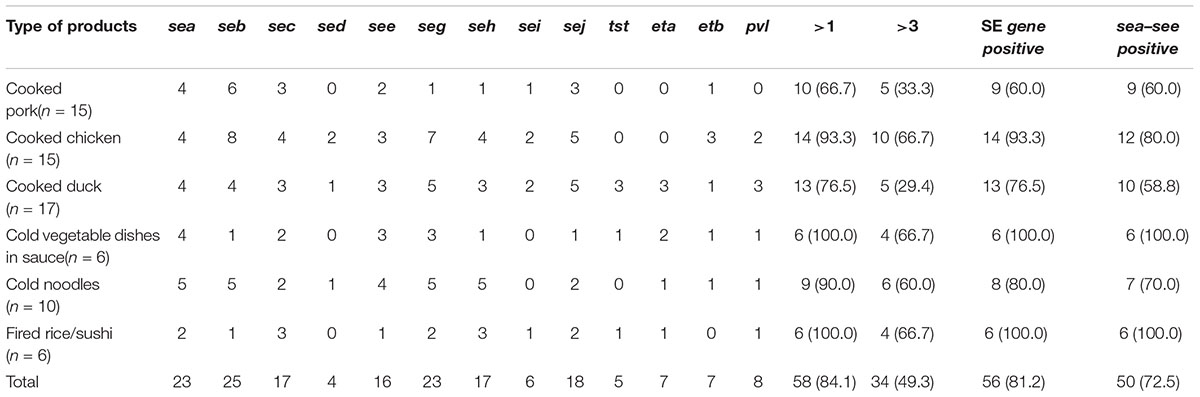
TABLE 3. Distribution of 13 toxin genes in Staphylococcus aureus isolates arranged by different food categories.
Association Between STs and Toxin Genes
Some associations were noted between certain STs and toxin genes. For example, each isolate belonging to CC1 contained seh, while the sec-seh combination, with or without additional toxin genes, occurred in 50% of the CC1 isolates. The combination of sea-see with additional toxin genes was found in all eight isolates assigned to ST6. Furthermore, half of the ST6 isolates also harbored eta or etb. Eight isolates contained pvl and mainly belonged to three STs (ST5, ST25, and ST88). In addition, all of the isolates belonging to ST5, ST6, or ST25 carried more than three toxin genes.
Discussion
Human infections caused by foodborne S. aureus strains have been previously reported (Jones et al., 2002). Therefore, the potential role of food in the dissemination of successful S. aureus lineages cannot be ignored. Our previous study reported the existence of two lineages of MRSA in these RTE food: CA-MRSA [ST59/SCCmec IVa (n = 2), ST338/SCCmec V, ST1/SCCmec V], and LA-MRSA (ST9) (Yang et al., 2016). Both ST59 and ST338 belong to CC59, which is the prevalent CA-MRSA clone in China and other Asian countries (Li et al., 2013; Chen and Huang, 2014). Along with three CC59 MRSA isolates, we also identified a single ST59 MSSA isolate among the RTE samples. In China, ST59 and ST338 are the first and second most dominant STs in cases of pediatric community-acquired pneumonia (Geng et al., 2010).
The strain ST398 was the first detected and most widespread LA-MRSA clone in Europe and North America (Petinaki and Spiliopoulou, 2012), in contrast, ST9 is the most prevalent LA-MRSA clone in most Asian countries (Cui et al., 2009; Wagenaar et al., 2009; Petinaki and Spiliopoulou, 2012) and ST398 MSSA is predominant in Asian countries, including China (Asai et al., 2012; Li et al., 2015). This was confirmed in our study. In addition to the previously identified ST9 LA-MRSA isolate, we identified four ST398 MSSA isolates, but no ST398 MRSA isolates were detected. The identification of ST398 MSSA isolates is of particular concern because human infections caused by MSSA ST398 strains have recently been described (David et al., 2013; Li et al., 2015).
The strain ST1 was the most common S. aureus clone causing SFP in Korea (Cha et al., 2006). However, in China, the ST6 strain carrying sea was the most dominant clone among isolates associated with SFP outbreaks and the ST1 strain with sec-seh enterotoxin profile was also frequently detected (Yan et al., 2012). In the present study, ST6 was found to be one of the most common STs. Furthermore, each isolate belonging to ST6 contained sea, and half of them also harbored eta and etb. In addition to the ST1 MRSA isolate, seven ST1 MSSA isolates were also identified in this study. These eight ST1 isolates, together with the two ST2592 MSSA isolates, constituted the predominant CC1. Moreover, half of the CC1 isolates exhibited the sec-seh enterotoxin profile with or without additional toxin genes. These results suggest that food is a potential source of S. aureus strains, which is significantly relevant on a clinical basis. With the exception of CC2483, ST9, ST12, ST1094, ST2592, ST2990, ST3239, and ST3485, all of the STs or CCs identified in this study have also been linked to bacteremia in China (He et al., 2013). Thus, these types of S. aureus isolates found in Chinese RTE food in the current study may represent a risk to human health.
Toxins are generally regarded as one of the main factors in the virulence of S. aureus worldwide, and for this reason, it is important to determine their prevalence in isolates from food with respect to assessing public health risks. In the current study, the prevalence of S. aureus isolates containing SE genes was higher than in several previous studies from food products in both China and other countries (Hammad et al., 2012; Vázquez-Sánchez et al., 2012; Wang et al., 2012, 2014). The classical SE genes (sea–see), which contribute significantly to SFP, were detected in 72.5% of the S. aureus isolates.
As for the single SE gene, seb, sea, and seg were the most frequently detected, followed by sej, seh, and sec in this study. Notably, SEB has been listed as a biological warfare agent because it can be toxic at very low concentrations (Varshney et al., 2009; Yamamoto et al., 2013). Recent studies indicated that SEB showed lethality in a rabbit model of pneumonia, and may suppress the motility of polymorphonuclear neutrophils, allowing MRSA to invade and damage tissues (Yamamoto et al., 2013). Infection by S. aureus strains that produce SEB may lead to serious diseases (Varshney et al., 2009). It is a concern that seb was prevalent in both the MSSA and MRSA isolates in the present study; likewise, seb was predominantly distributed in S. aureus isolates in the previous studies from food products in China and Vietnam (Huong et al., 2010; Wang et al., 2014).
Furthermore, SEA has been considered the most common cause of SFP worldwide (Pinchuk et al., 2010; Vázquez-Sánchez et al., 2012). The dominance of sea observed in the present study is consistent with previous findings (Vázquez-Sánchez et al., 2012; Zhang et al., 2013) and provides further evidence that this gene is widely distributed among S. aureus strains from food. In China, sea was reported to be the most commonly detected toxin gene in clinical S. aureus isolates associated with food poisoning outbreaks (Yan et al., 2012). In addition, the high prevalence of seg observed in this study was similar to the previous reports in China (Wang et al., 2012, 2014). However, seh and sej, which were also commonly identified in the present study, were not detected in the studies of Wang et al. (2012, 2014).
In this study, 11.6% of S. aureus isolates carried pvl. Among them, only one MRSA isolate, which was mecA-negative, contained pvl. As for pvl carriage by MRSA isolates, the previously reported rate varied greatly. While an absence of pvl in MRSA isolates has been reported (Fessler et al., 2011; Vestergaard et al., 2012), other studies show a high pvl carriage rate among MRSA isolates from food in China (Wang et al., 2014) and other countries (Pu et al., 2009; Hanson et al., 2011). Because pvl-positive S. aureus strains can cause necrotizing pneumonia, bloodstream infection, and soft-tissue pyogenic infection (Hammad et al., 2012), their potential to cause infections in humans through the food chain needs attention.
A novel finding in this study is the observed high rate of S. aureus strains carrying eta, etb, and tst. Previous studies in China have never reported the identification of etb and tst in S. aureus isolates from food, and the detected eta level was very low (Wang et al., 2012, 2014; Song et al., 2015). It has been reported that S. aureus clones containing eta, etb, and tst are increasingly responsible for severe infections (Xie et al., 2011; He et al., 2013; Yamamoto et al., 2013; Song et al., 2015). Thus, the presence of these isolates in RTE food indicates a potential risk to public health.
Nowadays, it is well established that the production of SEs in food is affected by several factors and the formation of SEs in food environments is significantly different from that in cultures of pure S. aureus (Schelin et al., 2011). In our study, we only used PCR analysis to determine the presence of toxin genes in pure S. aureus strains. These data are valuable and can represent virulence potential of S. aureus isolates to some extent.
Additionally, it was reported that food poisoning by S. aureus toxins does not occur until the level of the pathogen reaches 100,000 to 1,000,000/g (Food and Drug Administration [FDA], 2001; Hammad et al., 2012). The S. aureus contamination levels of the positive food samples had been determined in our previous work, these were mostly in the range of 0.3–10 most probable number (MPN)/g, with five samples exceeding 10 MPN/g (Yang et al., 2016). However, the RTE foods of this study are consumed without further treatment, the microbial load will increase over time, which represents a potential health hazard to humans.
Conclusion
In summary, to the best of our knowledge, this is the first comprehensive study of the subtypes and presence of virulence-associated genes in Staphylococcus aureus isolates from retail ready-to-eat (RTE) food in most regions in China. Our findings revealed a high prevalence of S. aureus isolates containing toxin genes in RTE food in China, and most of the sequence types detected in this study have previously been associated with human infections. The three most common toxin genes were sea, seb, and seg, and other important toxin genes, including pvl, eta, etb, and tst, were also detected in these S. aureus isolates. Overall, our study indicated that these S. aureus isolates present in Chinese RTE food represents a potential public health risk.
Author Contributions
Conceived and designed the experiments: XY, QW, and JZ. Performed the experiments: XY and SY. Analyzed the data: XY, SY, SW, and DR.
Funding
This work was supported by GDAS’ Special Project of Science and Technology Development (2017GDASCX-0817; 2017GDASCX-0201), China’s Post-doctoral Science Fund (2017M612623), and Science and Technology Projects of Guangdong (2017A070702018).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00197/full#supplementary-material
Footnotes
References
Asai, T., Hiki, M., Baba, K., Usui, M., Ishihara, K., and Tamura, Y. (2012). Presence of Staphylococcus aureus ST398 and ST9 in Swine in Japan. Jpn. J. Infect. Dis. 65, 551–552. doi: 10.7883/yoken.65.551
Cha, J. O., Lee, J. K., Jung, Y. H., Yoo, J. I., Park, Y. K., Kim, B. S., et al. (2006). Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J. Appl. Microbiol. 101, 864–871. doi: 10.1111/j.1365-2672.2006.02957.x
Chen, C. J., and Huang, Y. C. (2014). New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 20, 605–623. doi: 10.1111/1469-0691.12705
Cui, S., Li, J., Hu, C., Jin, S., Li, F., Guo, Y., et al. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
David, M. Z., Siegel, J., Lowy, F. D., Zychowski, D., Taylor, A., Lee, C. J., et al. (2013). Asymptomatic carriage of sequence type 398, spa type t571 methicillin susceptible Staphylococcus aureus in an urban jail: a newly emerging, transmissible pathogenic strain. J. Clin. Microbiol. 51, 2443–2447. doi: 10.1128/JCM.01057-13
Fessler, A. T., Kadlec, K., Hassel, M., Hauschild, T., Eidam, C., Ehricht, R., et al. (2011). Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 77, 7151–7157. doi: 10.1128/AEM.00561-11
Food and Drug Administration [FDA] (2001). Chapter 12: Pathogen Growth and Toxin Formation (other than Clostridium botulinum) as a Result of Time/Temperature Abuse (A Biological Hazard). Fish and Fishery Products Hazards and Controls Guide, 3rd Edn. Available at: http://www.fda.gov/Food/GuidanceComplianceRegulatoryInformation/GuidanceDocuments/Seafood/FisandFisheriesProductsHazardsandControlsGuide/ucm092162.htm [accessed April 15, 2011].
Geng, W., Yang, Y., Wu, D., Huang, G., Wang, C., Deng, L., et al. (2010). Molecular characteristics of community-acquired, methicillin-resistant Staphylococcus aureus isolated from Chinese children. FEMS Immunol. Med. Microbiol. 58, 356–362. doi: 10.1111/j.1574-695X.2010.00648.x
Hammad, A. M., Watanabe, W., Fujii, T., and Shimamoto, T. (2012). Occurrence and characteristics of methicillin-resistant and -susceptible Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci from Japanese retail ready-to-eat raw fish. Int. J. Food Microbiol. 156, 286–289. doi: 10.1016/j.ijfoodmicro.2012.03.022
Hanson, B. M., Dressler, A. E., Harper, A. L., Scheibel, R. P., Wardyn, S. E., Roberts, L. K., et al. (2011). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) on retail meat in Iowa. J. Infect. Public Health 4, 169–174. doi: 10.1016/j.jiph.2011.06.001
He, W., Chen, H., Zhao, C., Zhang, F., Li, H., Wang, Q., et al. (2013). Population structure and characterisation of Staphylococcus aureus from bacteraemia at multiple hospitals in China: association between antimicrobial resistance, toxin genes and genotypes. Int. J. Antimicrob. Agents 42, 211–219. doi: 10.1016/j.ijantimicag.2013.04.031
Huong, B. T. M., Mahmud, Z. H., Neogi, S. B., Kassu, A., Nhien, N. V., Mohammad, A., et al. (2010). Toxigenicity and genetic diversity of Staphylococcus aureus isolated from Vietnamese ready-to-eat foods. Food Control 21, 166–171. doi: 10.1016/j.foodcont.2009.05.001
Jones, T. F., Kellum, M. E., Porter, S. S., Bell, M., and Schaffner, W. (2002). An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 8, 82–84. doi: 10.3201/eid0801.010174
Li, G., Wu, C., Wang, X., and Meng, J. (2015). Prevalence and characterization of methicillin susceptible Staphylococcus aureus ST398 isolates from retail foods. Int. J. Food Microbiol. 196, 94–97. doi: 10.1016/j.ijfoodmicro.2014.12.002
Li, J., Wang, L., Ip, M., Sun, M., Sun, J., Huang, G., et al. (2013). Molecular and clinical characteristics of clonal complex 59 methicillin-resistant Staphylococcus aureus infections in Mainland China. PLoS One 8:e70602. doi: 10.1371/journal.pone.0070602
Lina, G., Piémont, Y., Godail-Gamot, F., Bes, M., Peter, M. O., Gauduchon, V., et al. (1999). Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132. doi: 10.1086/313461
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Noguchi, N., Nakaminami, H., Nishijima, S., Kurokawa, I., So, H., and Sasatsu, M. (2006). Antimicrobial agent of susceptibilities and antiseptic resistance gene distribution among methicillin-resistant Staphylococcus aureus isolates from patients with impetigo and staphylococcal scalded skin syndrome. J. Clin. Microbiol. 44, 2119–2125. doi: 10.1128/JCM.02690-05
Peles, F., Wagner, M., Varga, L., Hein, I., Rieck, P., Gutser, K., et al. (2007). Characterization of Staphylococcus aureus strains isolated from bovine milk in Hungary. Int. J. Food Microbiol. 118, 186–193. doi: 10.1016/j.ijfoodmicro.2007.07.010
Petinaki, E., and Spiliopoulou, I. (2012). Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin. Microbiol. Infect. 18, 626–634. doi: 10.1111/j.1469-0691.2012.03881.x
Pinchuk, I. V., Beswick, E. J., and Reyes, V. E. (2010). Staphylococcal enterotoxins. Toxins 2, 2177–2197. doi: 10.3390/toxins2082177
Pu, S., Han, F., and Ge, B. (2009). Isolation and characterization of methicillin resistant Staphylococcus aureus strains from Louisiana retail meats. Appl. Environ. Microbiol. 75, 265–267. doi: 10.1128/AEM.01110-08
Rodríguez-Lázaro, D., Ariza-Miguel, J., Diez-Valcarce, M., Fernández-Natal, I., Hernández, M., and Rovira, J. (2015). Foods confiscated from non-EU flights as a neglected route of potential methicillin-resistant Staphylococcus aureus transmission. Int. J. Food Microbiol. 209, 29–33. doi: 10.1016/j.ijfoodmicro.2014.08.016
Schelin, J., Wallin-Carlquist, N., Cohn, M. T., Lindqvist, R., Barker, G. C., and Rådström, P. (2011). The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2, 580–592. doi: 10.4161/viru.2.6.18122
Song, M., Bai, Y., Xu, J., Carter, M. Q., Shi, C., and Shi, X. (2015). Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 195, 1–8. doi: 10.1016/j.ijfoodmicro.2014.11.020
Varshney, A. K., Mediavilla, J. R., Robiou, N., Guh, A., Wang, X., Gialanella, P., et al. (2009). Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Environ. Microbiol. 75, 6839–6849. doi: 10.1128/AEM.00272-09
Vázquez-Sánchez, D., López-Cabo, M., Saá-Ibusquiza, P., and Rodríguez-Herrera, J. J. (2012). Incidence and characterization of Staphylococcus aureus in fishery products marketed in Galicia (Northwest Spain). Int. J. Food Microbiol. 157, 286–296. doi: 10.1016/j.ijfoodmicro.2012.05.021
Vestergaard, M., Cavaco, L. M., Sirichote, P., Unahalekhaka, A., Dangsakul, W., Svendsen, C. A., et al. (2012). SCCmec type IX element in methicillin resistant Staphylococcus aureus spatype t337 (CC9) isolated from pigs and pork in Thailand. Front. Microbiol. 3:103. doi: 10.3389/fmicb.2012.00103
Wagenaar, J. A., Yue, H., Pritchard, J., Broekhuizen-Stins, M., Huijsdens, X., Mevius, D. J., et al. (2009). Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet. Microbiol. 139, 405–409. doi: 10.1016/j.vetmic.2009.06.014
Wang, X., Li, G., Xia, X., Yang, B., Xi, M., and Meng, J. (2014). Antimicrobial susceptibility and molecular typing of methicillin-resistant staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog. Dis. 11, 281–286. doi: 10.1089/fpd.2013.1643
Wang, X., Meng, J., Zhang, J., Zhou, T., Zhang, Y., Yang, B., et al. (2012). Characterization of Staphylococcus aureus isolated from powdered infant formula milk and infant rice cereal in China. Int. J. Food Microbiol. 153, 142–147. doi: 10.1016/j.ijfoodmicro.2011.10.030
Wu, S., Wu, Q., Zhang, J., Chen, M., and Guo, W. (2016). Analysis of multilocus sequence typing and virulence characterization of Listeria monocytogenes isolates from Chinese retail ready-to-eat food. Front. Microbiol. 7:168. doi: 10.3389/fmicb.2016.00168
Xie, Y., He, Y., Gehring, A., Hu, Y., Li, Q., Tu, S. I., et al. (2011). Genotypes and toxin gene profiles of Staphylococcus aureus clinical isolates from China. PLoS One 6:e28276. doi: 10.1371/journal.pone.0028276
Xu, X., Li, C., Wu, Q., Zhang, J., Huang, J., and Yang, G. (2015). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int. J. Food Microbiol. 204, 17–23. doi: 10.1016/j.ijfoodmicro.2015.03.003
Yamamoto, T., Hung, W. C., Takano, T., and Nishiyama, A. (2013). Genetic nature and virulence of community associated methicillin-resistant Staphylococcus aureus. BioMedicine 3, 2–18. doi: 10.1016/j.biomed.2012.12.001
Yan, X., Wang, B., Tao, X., Hu, Q., Cui, Z., Zhang, J., et al. (2012). Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 78, 6637–6642. doi: 10.1128/AEM.01165-12
Yang, X., Zhang, J., Yu, S., Wu, Q., Guo, W., Huang, J., et al. (2016). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready-to-eat foods in China. Front. Microbiol. 7:816. doi: 10.3389/fmicb.2016.00816
Keywords: Staphylococcus aureus, ready-to-eat food, MLST, toxin genes, SEs
Citation: Yang X, Yu S, Wu Q, Zhang J, Wu S and Rong D (2018) Multilocus Sequence Typing and Virulence-Associated Gene Profile Analysis of Staphylococcus aureus Isolates From Retail Ready-to-Eat Food in China. Front. Microbiol. 9:197. doi: 10.3389/fmicb.2018.00197
Received: 09 November 2017; Accepted: 29 January 2018;
Published: 13 March 2018.
Edited by:
Julio Parra-Flores, University of the Bío Bío, ChileReviewed by:
Beatrix Stessl, Veterinärmedizinische Universität Wien, AustriaGiuseppe Blaiotta, Dipartimento di Agraria, Italy
Ariadnna Cruz-Córdova, Hospital Infantil de México Federico Gómez, Mexico
Copyright © 2018 Yang, Yu, Wu, Zhang, Wu and Rong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t
†These authors have contributed equally to this work.
 Xiaojuan Yang†
Xiaojuan Yang† Qingping Wu
Qingping Wu