
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 February 2018
Sec. Infectious Agents and Disease
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00099
 Qi Zhai1,2
Qi Zhai1,2 Zhiqiang Fu2
Zhiqiang Fu2 Yang Hong2*
Yang Hong2* Xingang Yu1,2
Xingang Yu1,2 Qian Han2
Qian Han2 Ke Lu2
Ke Lu2 Hao Li2
Hao Li2 Xuefeng Dou2
Xuefeng Dou2 Chuangang Zhu2
Chuangang Zhu2 Jinming Liu2
Jinming Liu2 Jiaojiao Lin2*
Jiaojiao Lin2* Guoqing Li1*
Guoqing Li1*Schistosomiasis japonicum is one of the most severe zoonotic diseases in China. Water buffalo and yellow cattle are important reservoir hosts and the main transmission sources of Schistosoma japonicum in endemic areas. The susceptibility of these two hosts to schistosome infection is different, as water buffaloes are less susceptible to S. japonicum than yellow cattle. In this study, iTRAQ-coupled LC-MS/MS was applied to compare the protein expression profiles of adult schistosomes recovered from water buffalo with those of yellow cattle. A total of 131 differentially expressed proteins (DEPs) were identified, including 46 upregulated proteins and 85 downregulated proteins. The iTRAQ results were confirmed by Western blotting and quantitative real-time PCR. Further analysis indicated that these DEPs were primarily involved in protein synthesis, transcriptional regulation, protein proteolysis, cytoskeletal structure and oxidative stress response processes. The results revealed that some of the differential expression molecules may affect the development and survival of schistosomes in these two natural hosts. Of note, this study provides useful information for understanding the interplay between schistosomes and their final hosts.
Schistosomiasis is caused by infection with trematodes of the genus Schistosoma and affects approximately 260 million people worldwide (WHO, 2016). Among the six species of Schistosoma, S. japonicum is popular mainly in China, Philippines and small pockets of Indonesia. Schistosomiasis control in China has been remarkably successful, and this disease has been efficiently controlled and even eliminated in many areas (Li et al., 2016). However, there were still 77,194 cases of schistosomiasis in China by the end of 2015 (Zhang L. et al., 2016). Endemic areas of this disease are distributed in the lake, marshland, and mountainous region of China (Liu et al., 2016; Zhang S. Q. et al., 2016). The results of an epidemiological survey indicate that yellow cattle and water buffalo are the most important reservoir hosts and the main source of transmission for schistosomiasis in China (Wang et al., 2005; Liu et al., 2012), because these animal species graze freely in areas where it is endemic and may be frequently in contact with schistosome-infested water, the infected animals spread more eggs into the environment than human and other animal hosts (Van Dorssen et al., 2017). Also water buffalo and yellow cattle may pose great threat to human infection because of the constant and close contact with humans in agricultural areas. Therefore, the effective intervention of bovine schistosomiasis will greatly further the process of controlling the disease in China.
The susceptibility of different hosts to infection with schistosomes varies. In the high Schistosoma mansoni transmission areas, some residents are chronical infected, while some individuals are considered “Putative Resistants” who are constantly exposed to S. mansoni infection but remained egg-negative (Viana et al., 1995; Gaze et al., 2014). Of the rodents, the hamster is permissive to S. mansoni and Schistosoma haematobium infection (Draz et al., 2008; Tallima et al., 2015), whereas the rat is semi-permissive host to S. mansoni, in which the worms do not cause typical granuloma in liver and cannot develop well (Hsu et al., 1973; Khalife et al., 2000; Amaral et al., 2016), and the mice show lower permissiveness to S. haematobium infection along with schistosomula’s delay in migrating through the lungs and adult worms’ low recovery rate (Dean et al., 1996; Rheinberg et al., 1998). In contrast to the other Schistosoma species, S. japonicum is a true zoonotic parasite. There are 46 species of mammals which have been reported to be naturally infected with S. japonicum, such as rabbits, cats, horses, goats, dogs, cattle, rats, and pigs. The susceptibility of different hosts to S. japonicum was also varied. Water buffaloes, rats, horses, and pigs are less susceptible to infection than yellow cattle, goats, and rabbits (He et al., 2001). Representative observations have shown differences in worm recovery rate, egg production, pathogenicity and immunological responses to parasite infection between these hosts (Jiang et al., 2010; Peng et al., 2011; Yang et al., 2012b). Moreover, the parasite self-cure phenomenon has been observed in water buffaloes and pigs, where parasites establish but are then eliminated in a self-cure response (Li et al., 2014).
Previous studies from our research group using microarray and comparative proteomic analyses revealed that schistosomula from susceptible BALB/c mice, less susceptible Wistar rats, and resistant Microtus fortis voles had different expression profiles (Hong et al., 2011; Peng et al., 2011). Another study compared the gene expression profiles of S. japonicum from water buffaloes, mice, and rabbits, and identified a panel of differentially expressed genes in worms from different hosts (Liu et al., 2015). This study suggested that the data exhibiting in worms from laboratory animals were not completely available to the worms from natural host animals. Since water buffaloes and yellow cattle are the most important sources of transmission of schistosomiasis in China and have distinct susceptibilities to S. japonicum infection, the use of these natural hosts as experimental animal models in the investigation of the schistosome–host interaction may contribute to the identification of key molecules involved in worm development, and screening of vaccine candidates or new drug targets, and consequently lead to the effective control of schistosomiasis in China.
For better understanding the development mechanism of schistosome in their natural hosts, water buffaloes and yellow cattle, we have compared the gene expression profiles of schistosomes from these two hosts in our lab, several differential expression genes which may be important for the development and survival of schistosome were identified (Yang et al., 2012a). However, the difference in protein expression levels in schistosomes from these natural hosts remains unknown to date.
Proteomics is a powerful tool for the high-throughput characterization of proteins combined with mass spectrometry and bioinformatics. As a quantitative method, the isobaric tag for relative and absolute quantitation (iTRAQ), is more suitable and sensitive for the comparison and identification of differentially expressed proteins (DEPs) compared to 2-D gel electrophoresis (Wu et al., 2006). In this study, water buffalo and yellow cattle were infected with S. japonicum and the iTRAQ technique was used to detect and quantify DEPs in adult worms from these two natural hosts. Our results may provide valuable information for screening potential vaccine candidates or new drug targets and consequently for the control of schistosomiasis in natural hosts in endemic areas.
All animal-handling procedures complied with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The animal use protocol was approved by the Institutional Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, People’s Republic of China.
Male yellow cattle and male water buffaloes aged 15–18 months, free of parasitic helminth infections and other infectious diseases, were obtained from schistosome non-endemic areas and used for experimental infection. All animals were housed in covered pens and received adequate care. S. japonicum (Chinese mainland strain) was routinely maintained in Oncomelamia hupensis at the Shanghai Veterinary Research Institute, CAAS. Yellow cattle and water buffaloes were challenged percutaneously with S. japonicum cercariae, respectively, on the upper back using the cover glass method as described previously (Da’Dara et al., 2008). Adult worms were perfused from the hepatic portal vein of the animals at 8 weeks post-challenge and the worm recovery rate was calculated. The worms were manually washed thoroughly in PBS to remove residual host debris. The equal amounts of worms obtained from water buffalo and yellow cattle were pooled and designated as B and C group, respectively. Then the worms were stored in liquid nitrogen until use.
STD buffer (pH 8.0) containing 4%SDS, 150 mM Tris-HCl, 1 mM DTT and 0.1% v/v protease inhibitor mixture (Merck) were added to the adult worm samples and homogenized using a Dounce homogenizer. The mixture was heated at 100°C for 5 min and then sonicated on ice. The crude extract was heated again at 100°C for 5 min. After centrifugation, the supernatants were collected and protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, United States). The protein samples were analyzed by SDS-PAGE and CBB staining, then samples suitable for subsequent analysis was determined.
Proteins were digested according to the filter-aided sample preparation procedure, as previously described (Wiśniewski et al., 2009). Briefly, 200 μg of protein for each sample was diluted in 200 μL of uric acid (UA) buffer (8 M Urea, 150 mM Tris-HCl pH 8.0) and transferred onto a 10-kDa ultrafiltration filter. DTT and other low molecular-weight components were removed by centrifugation at 14,000 × g for 15 min and an additional washing step with 200 μL of UA buffer. Then, 100 μL of 50 mM iodoacetamide in UA buffer was added and the reaction tubes were vortexed for 1 min at 600 rpm. The samples were incubated for 30 min in the dark and centrifuged at 14,000 × g for 10 min. The filters were washed three times with 100 μL UA buffer and then twice with 100 μL dissolution buffer (50 mM triethylammonium bicarbonate at pH 8.5). The protein suspensions were digested with 40 μL trypsin buffer (2 μg trypsin in 40 μL dissolution buffer) and vortexed for 1 min at 600 rpm. The samples were incubated at 37°C for 16–18 h and centrifuged at 14,000 × g for 10 min. The resulting peptides were collected and the filters were rinsed with 40 μL dissolution buffer.
Peptide concentration was determined at an OD of 280 nm. iTRAQ labeling was performed using the iTRAQ Reagent-8 plex Multiplex Kit according to the manufacturer’s instructions (Applied Biosystems). The proteins of worms from water buffalo were labeled with reagents 113, 114, 115, and 116 whereas the proteins of worms from yellow cattle were labeled with reagents 117, 118, 119, and 121. Four independent repetitions were made for each group sample.
The labeled peptides were pooled and fractionated on an AKTA Purifier 100 system (GE Healthcare) using a SCX column (PolyLCInc, Columbia, MD, United States). Briefly, the peptides were dried in a vacuum concentrator, dissolved in 2 mL of buffer A (10 mM KH2PO4 in 25% of ACN, pH 3.0) and loaded onto a 4.6 mm × 100 mm Polysulfoethyl column (5 μm, 200 A, PolyLC Inc.) at a flow rate of 1 mL/min. The peptides were eluted at a flow rate of 1 mL/min with a gradient of 0–10% buffer B (500 mM KCl, 10 mM KH2PO4 in 25% of ACN, pH 3.0) for 7 min, 10–20% buffer B for 10 min, 20–45% buffer B for 5 min, and 45–100% buffer B for 5 min. The eluted peptides were collected and desalted using an offline fraction collector. The collected fractions were combined into 15 pools, vacuum freeze-dried, and desalted on C18 cartridges (Sigma). All collected fractions were stored at –80°C until further analysis.
The samples were separated using an Easy-nLC in the nano HPLC system. The column was equilibrated with 95% buffer A (0.1% formic acid). Each dried peptide was resuspended in 10 μL buffer A and loaded onto a Thermo Scientific EASY column (2 cm × 100 μm, 5 μm C18) using an autosampler. The peptides were eluted onto an analytical Thermo Scientific EASY column (75 μm × 100 mm, 3 μm C18) at a flow rate of 300 nL/min and separated using a 160-min gradient elution. The gradient was 50 min in 0–35% buffer B (0.1% formic acid, 84% acetonitrile), 50 min in 35–100% buffer B, followed by maintenance in 100% buffer B for 60 min. The HPLC eluates were directly electrosprayed into the mass spectrometer.
MS data acquisition was performed by using Q-Exactive (Thermo Finnigan, United States). Positive ion detection mode was used. Full-scan MS spectra (m/z of 300–1800) were obtained at a resolution of 70,000, m/z of 200, maximum ion accumulation time of 10 ms, and AGC target value of 3e6. The number of scan ranges was 1 with a 40-s dynamic exclusion. MS2 scans were performed using high-energy collisional dissociation (HCD) activation type at a resolution of 17,500, m/z of 200, and isolation window of 2 m/z. The microscans number was 1; the normalized collision energy was 30 eV; the maximum allowed ion accumulation times were 60 ms; and the under-fill ratio was defined as 0.1%. Twenty fraction profiles were collected after a full scan.
All LC-MS/MS data were combined and converted using Proteome Discoverer software version 1.4 (Thermo Fisher Scientific Inc., United States). The fragmentation spectra were searched using the Mascot software version 2.2 (Matrix Science, United States). The S. japonicum protein sequence data used in the search were retrieved from UniprotKB (16511 sequences, release time 20161203). The following search parameters were used: MS/MS ion search; trypsin was specified as the digestion enzyme with two max missed cleavage; monoisotopic mass values; fixed modifications of carbamidomethyl (C), iTRAQ8plex (N-term) and iTRAQ8plex (K); variable modification of oxidation (M); peptide mass tolerance at ±20 ppm; MS/MS tolerance at 0.1 Da; unrestricted protein mass. All reported data were based on false discovery rate (FDR) of less than 1% confidence for protein identification.
Differentially expressed proteins were analyzed for significant up- or downregulation using the software Proteome Discoverer version 1.4. The parameters were set as follows: use only unique peptide and normalize on protein median. Ratios were used to assess the fold-change of the abundant proteins of worms collected from water buffalo and yellow cattle and Student’s t-tests were used to evaluate the significant (P-value < 0.05). A fold-change (water buffalo/yellow cattle) higher than 1.5 (P < 0.05) or lower than 0.67 (P < 0.05) considering the average of four replicates was considered significantly upregulated or downregulated. To assess the reproducibility of the MS data, hierarchical cluster analysis on quantification of identified proteins from replicates was conducted using R with average linkage method and Euclidean distance.
Protein sequences were clustered using the CD-HIT program with default parameters (sequence identity ≥ 95%) (Li et al., 2001; Huang et al., 2010). Isoelectric point (pI) and molecular weight were calculated using the Pepstats program in EMBOSS. Functional analysis of DEPs was performed using gene ontology (GO) analysis, and the DEPs were categorized according to the biological process, molecular function, and cellular component (Conesa et al., 2005). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to classify and group the DEPs (Kanehisa and Goto, 2000). Protein–protein interaction (PPI) networks were built using the database STRING version 10 (Szklarczyk et al., 2015).
Two DEPs, SjGST and SjPDI, which were identified as upregulated in worms from water buffalo, were used to confirm the proteomic results by Western blotting. A pre-processed equivalent amount of protein was obtained from each sample, separated by 12% SDS-PAGE, and transferred to 0.45-μm pore size nitrocellulose membranes (Merck Millipore). The membranes were blocked overnight at 4°C with 5% non-fat milk in 0.05% Tween 20-PBS and incubated with monoclonal antibodies specific to mouse β-actin (Beyotime, China, 1:500 dilution), and mouse sera specific to SjPDI or SjGST (1:100), respectively. The membranes were washed three times and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Beyotime, China, 1:2000 dilution). Moreover, the samples were detected using an automatic chemiluminescence imaging analysis system (Tanon, China).
The genes encoding eight DEPs were investigated at the transcriptional level by qRT-PCR. The total RNA was isolated from mixed parasite samples from water buffalo or yellow cattle using TRIZOL Reagent (Invitrogen, Carlsbad, CA, United States) and transcribed into cDNA using the PrimeScript RT reagent Kit (TaKaRa, Osaka, Japan) according to the manufacturer’s instructions. Nicotinamide adenine dinucleotide (NADH) was used as a reference gene (Gobert et al., 2009) and the primers used in this study are presented in Table 1. PCR amplification was conducted using the SYBR Premix Ex TaqTM kit (TaKaRa) in an ABI 7500 Real-time System (Life Technologies). The relative expression level of the genes was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Student’s t-tests were used to evaluate deviations of protein quantification and worm recovery rate between two groups of worm samples by using SPSS for Windows version 22 (SPSS, Chicago, IL, United States), and P-values of less than 0.05 were considered statistically significant. In addition, the cluster analysis of protein quantification was performed in the R environment using gplots package. Fisher’s exact test was used to assess the KEGG enrichment analysis and P-values of less than 0.05 were considered significant.
The average worm recovery rate from water buffalo and yellow cattle was 10.8 and 68.8%, respectively (Supplementary Table 1). Each sample containing equivalent proteins was separated on 12% SDS-PAGE (Figure 1) and clearly distinct band patterns were visualized. The protein isolation was suitable for trypsin digestion and LC-MS/MS analysis.
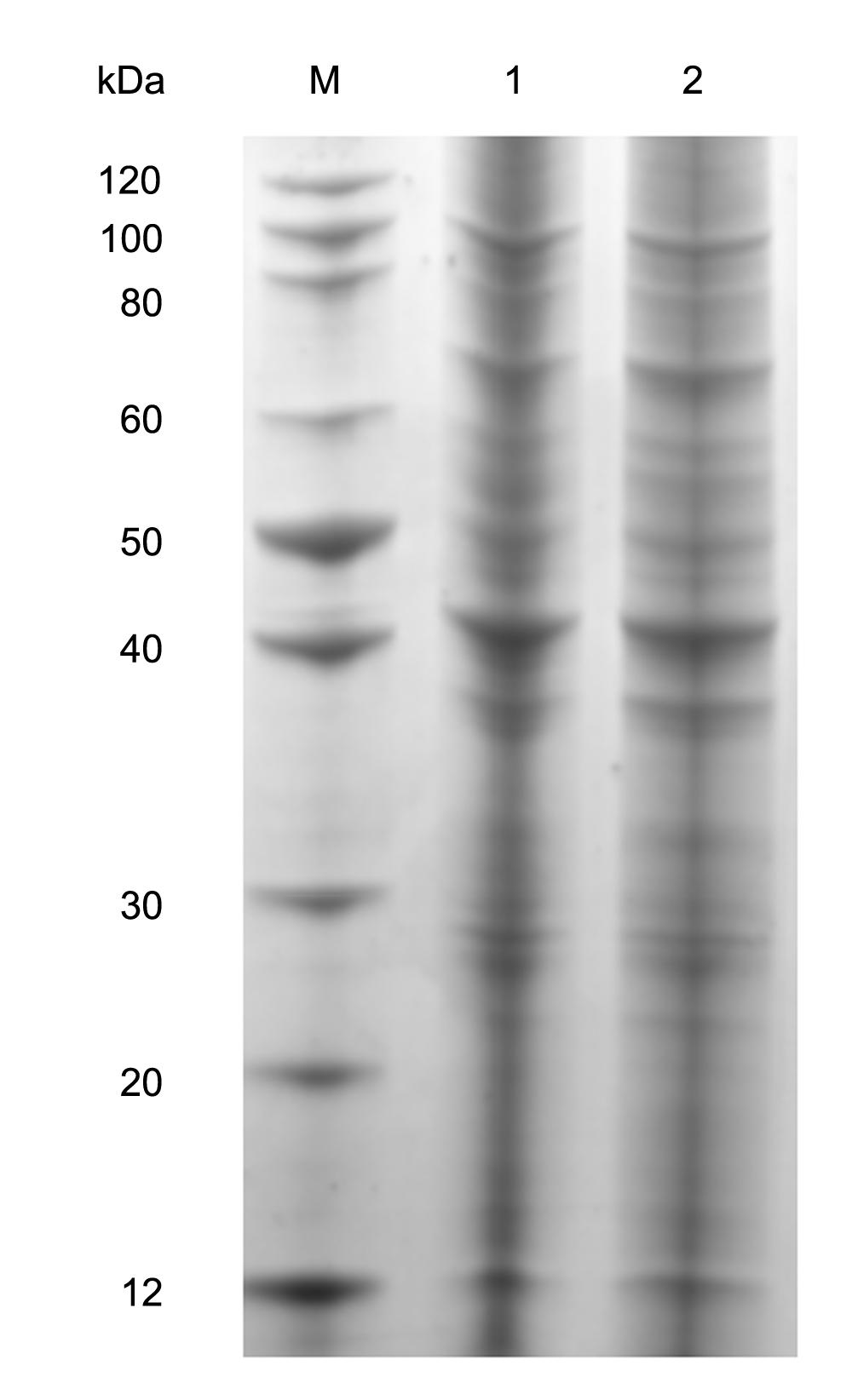
FIGURE 1. Separation of complete proteins of adult schistosomes from water buffalo and yellow cattle. 1: from yellow cattle; 2: from water buffalo.
A total of 18,288 unique peptides corresponding to 3,605 proteins were identified by iTRAQ and LC-MC/MC analysis. Using CD-HIT filtration with sequence identity ≥95%, 3,407 proteins were identified, of which 3,398 were quantified (Table 2). The reproducibility of the MS data were compared, which indicated that the two group protein samples separated into two main clusters (Supplementary Figure 1). Proteins with a statistically significant (p < 0.05) fold-change of 1.5 or greater were considered differentially expressed. Compared with schistosomes from yellow cattle, 131 proteins were found to be differentially expressed, of which 46 were upregulated and 85 were downregulated (Figure 2A and Supplementary Table 2). The molecular mass of DEPs ranged from 6.7 kDa to 113.8 kDa and the pI of DEPs ranged from 4.25 to 11.78 (Figure 2B and Supplementary Tables 3, 4). Ribosomes, transcription regulation-associated proteins, and cytoskeletal structure-associated proteins were the most abundant among the downregulated DEPs. Lysosomal proteins and oxidative stress response proteins were the most abundant among the upregulated DEPs. Some of the DEPs were listed in Table 3.
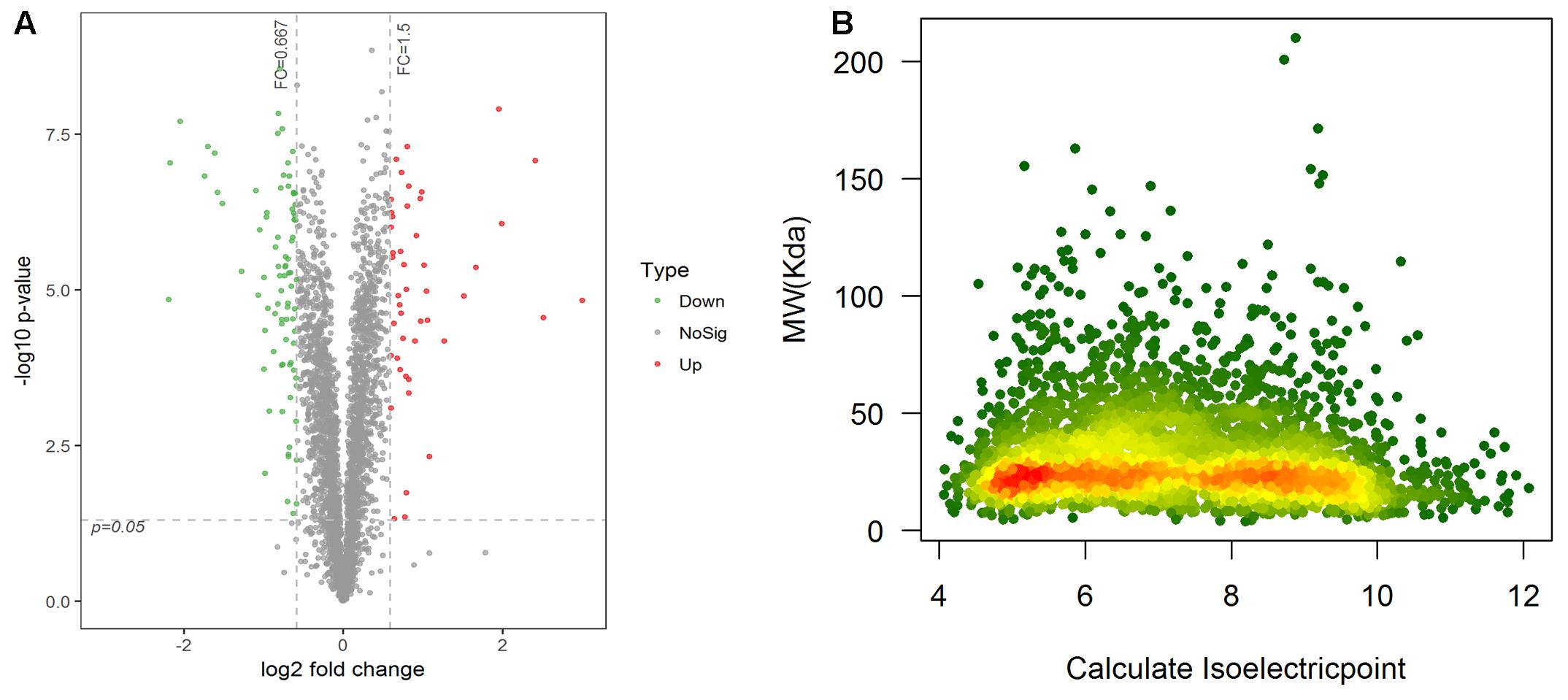
FIGURE 2. (A) Ratio distribution of all quantitative proteins. Differentially expressed protein marked in red or green (red indicates upregulation and green indicates downregulation). (B) Theoretical 2-D (pI, MW) distribution of identified DEPs.
Gene ontology analysis was used to identify significantly enriched functional terms of DEPs (Figure 3 and Supplementary Tables 3, 4). The results showed that DEPs were primarily involved in cellular, metabolic, and single-organism processes; biological regulation; localization; cellular component organization or biogenesis, and other biological processes. With regard to the cellular components, most DEPs were associated with cell composition, macromolecular complexes, organelles, membrane structure, and other components. Under the category of molecular function, most DEPs were correlated with binding, catalysis, transport, and structure, among others.
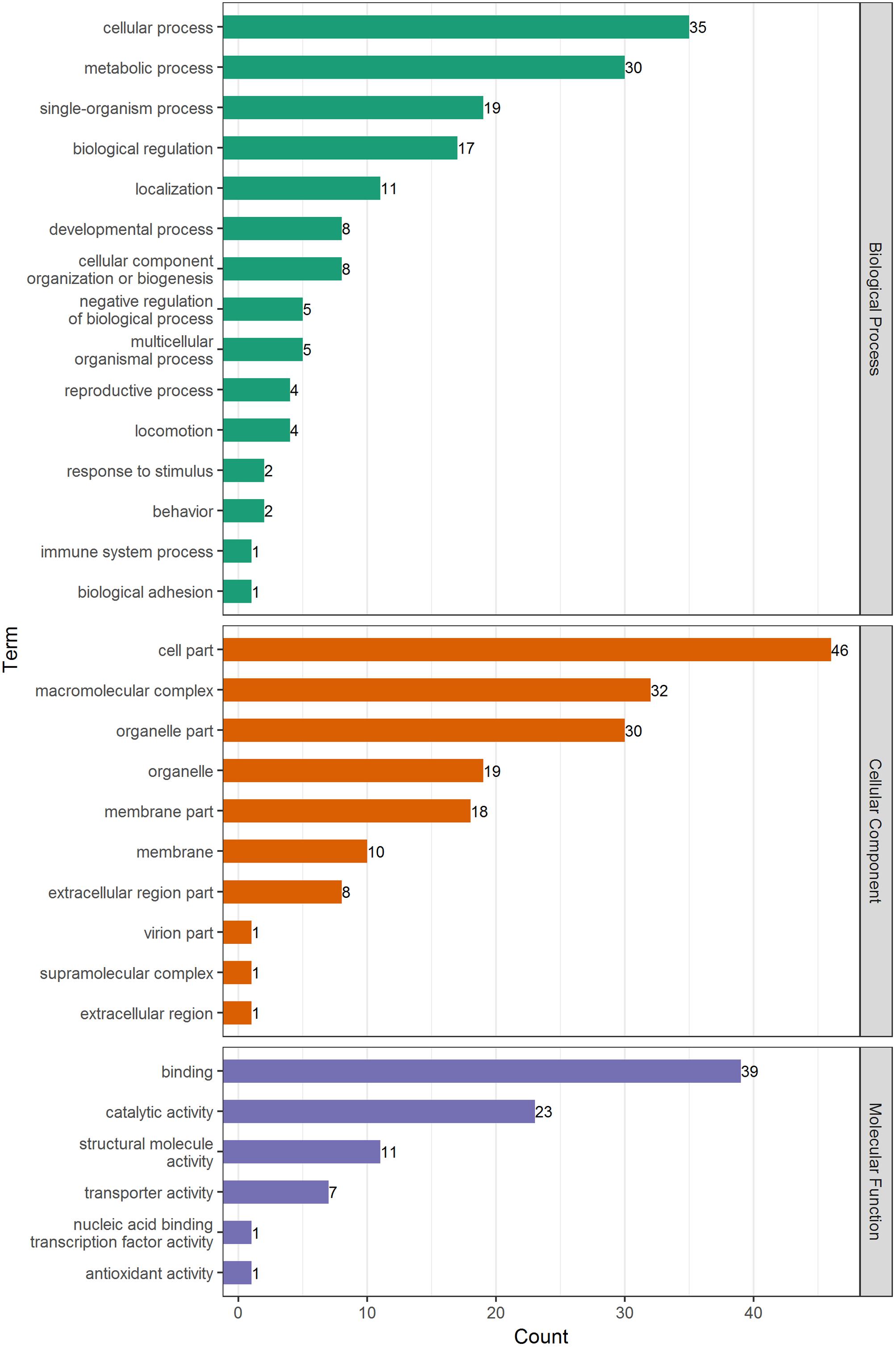
FIGURE 3. GO analysis of DEPs in adult schistosome from water buffalo and yellow cattle. Proteins were classified into three main categories: biological process, cellular component, and molecular function.
The KEGG pathway and enrichment analysis indicated that the DEPs were highly enriched in transcription, translation, and transport and catabolic pathways, including spliceosome, ribosome, lysosome, mRNA surveillance, and peroxisome (Figure 4 and Supplementary Tables 3, 4).
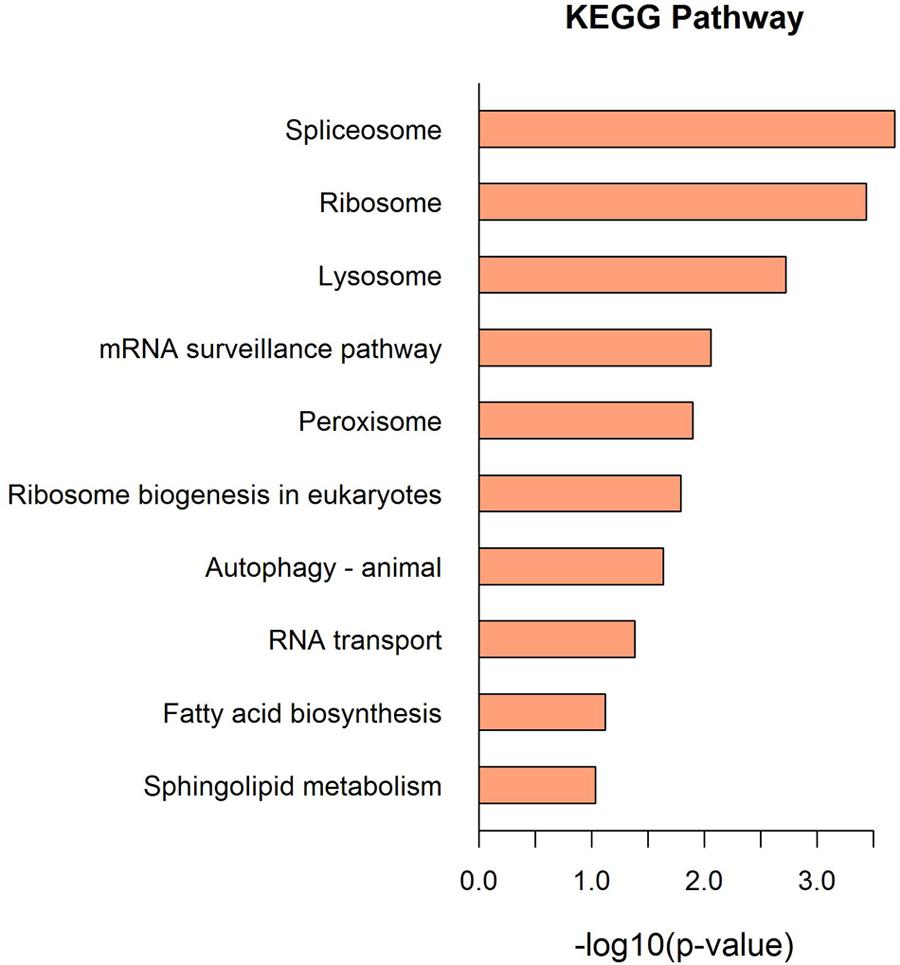
FIGURE 4. The KEGG pathway enrichment analysis of the DEPs. Top 10 enrichment in KEGG pathway maps of the DEPs. P-value was calculated using Fisher’s exact test.
The PPI networks of DEPs were built and analyzed using STRING version 10. There were 18 nodes and 20 edges in the PPI network (Figure 5). Three distinct clusters with a high combined score were found among the downregulated proteins, and these clusters were associated with or involved in protein synthesis (I), transcription regulation (II), and cytoskeleton-associated proteins (III). No significant protein–protein interactions were observed among the upregulated proteins.
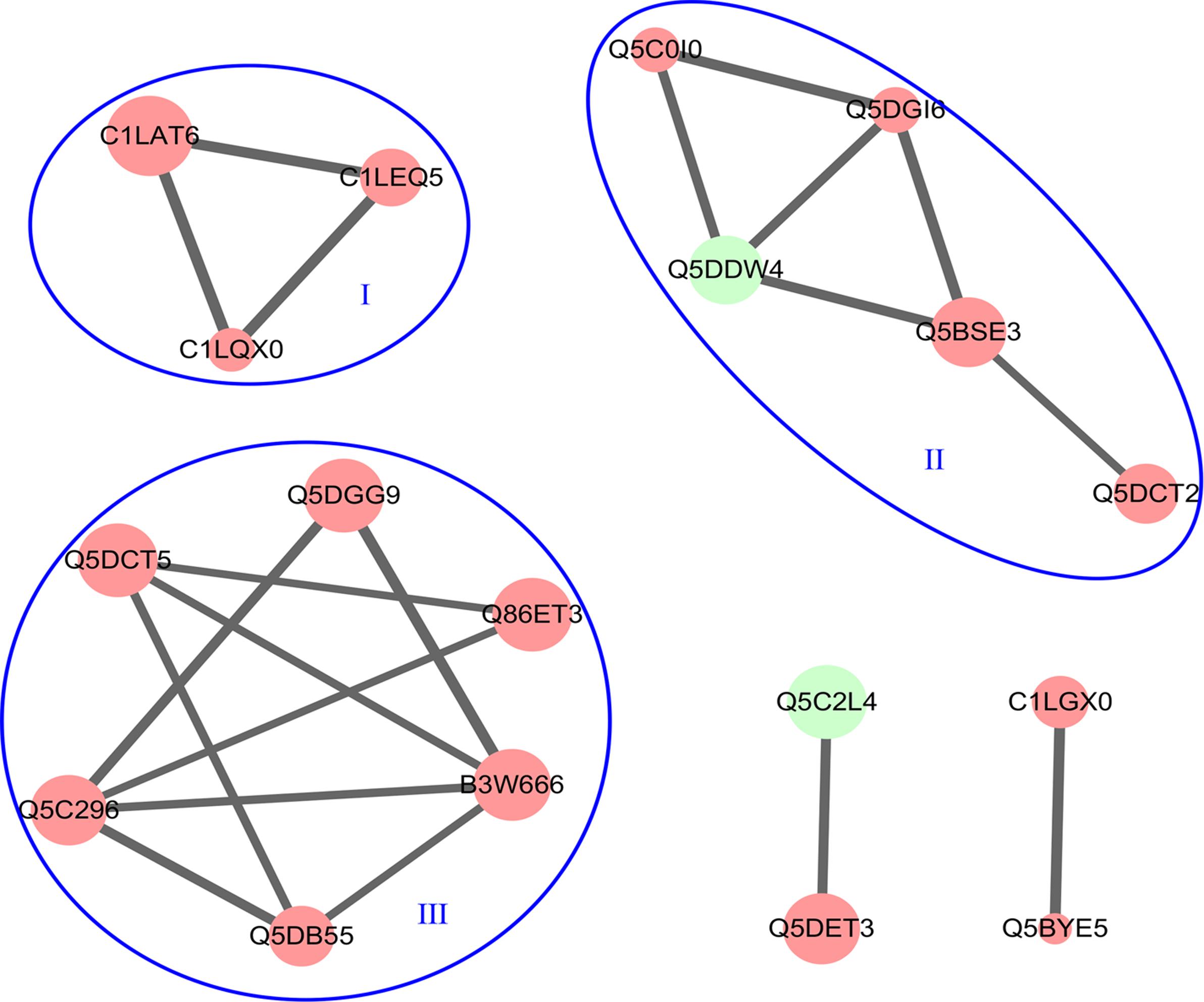
FIGURE 5. The PPI networks of all DEPs. The PPI networks were built using STRING version 10.0 with a high combined score >700. The node color indicates downregulated proteins (red) and upregulated proteins (green).
Three replicates of Western blot experiments were performed to confirm the iTRAQ results. The Western blot analysis indicated that two proteins, SjGST and SjPDI, were highly expressed in schistosomes infecting water buffaloes (Figure 6 and Supplementary Figure 2), which was consistent with the results of iTRAQ, although there were slight differences in the mean values between the iTRAQ and Western blotting. These results further confirmed the changes in DEPs detected by iTRAQ.
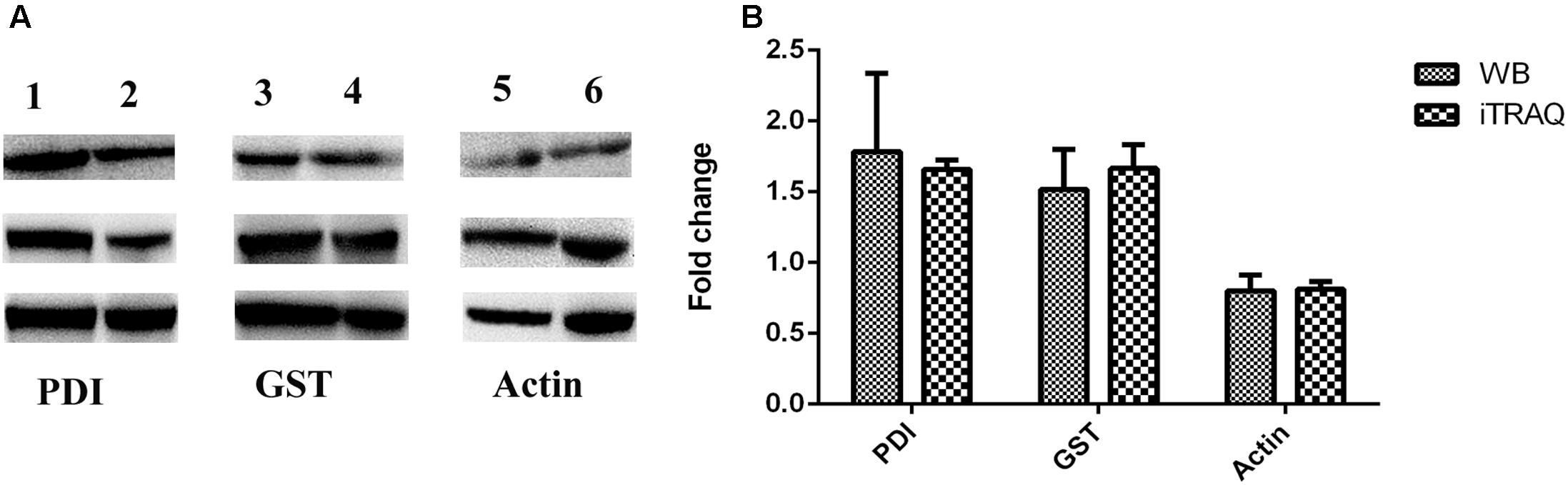
FIGURE 6. Confirmation of differentially expressed proteins by Western blotting. Each experiment was repeated three times. (A) Immunoblot analysis of proteins (SjPDI, SjGST, and actin) in adult schistosome from water buffalo and yellow cattle. Lane 1, SjPDI of water buffalo group. Lane 2, SjPDI of yellow cattle group. Lane 3, SjGST of water buffalo group. Lane 4, SjGST of yellow cattle group. Lane 5, actin of water buffalo group. Lane 6, actin of yellow cattle group. (B) WB ratios (water buffalo group/yellow cattle group) were consistent with those obtained by iTRAQ.
Since the specific antibodies available for DEPs in our laboratory is limited, qRT-PCR was performed to further verify the reliability of the iTRAQ results at the transcriptional level. The mRNA transcription levels of eight DEPs were measured and six of them showed expression patterns similar to those of iTRAQ whereas the transcriptional pattern of the other two DEPs differed from the protein expression pattern (Table 1).
It has been shown that S. japonicum can infect more than 40 species of mammals, including mice, rabbit, goat, and yellow cattle, which are susceptible to infection, whereas other species such as rat, pig, and water buffalo are less susceptible, and the decreased susceptibility is indicated by the smaller worm size, fewer eggs laid, and low developmental rate (He et al., 2001; Yang et al., 2012b). A previous study from our lab has shown that the development of schistosomes (considering worm length, worm recovery rate, and ultrastructural structure) was significantly better than that in water buffaloes, and the liver pathological damage caused by schistosome infections in yellow cattle was more severe than that of water buffaloes (Yang et al., 2012b). In the present study, the average worm recovery rate from water buffalo was also significantly lower than that from yellow cattle. These results suggest that water buffaloes are less susceptible to S. japonicum infection than yellow cattle. Our group also compared the transcriptional profiles of adult schistosomes infecting yellow cattle and water buffalo and the results revealed that several genes involved in transcription, transport, lipid metabolism, energy metabolism, nucleotide, and energy and signaling pathways were differentially expressed in worms from these two hosts (Yang et al., 2012a). In this study, we compared the protein expression profiles of adult worms recovered from yellow cattle and water buffalo using iTRAQ. The iTRAQ technique in combination with LC-MS/MS is an effective method for investigating changes in protein levels and provides a more uniform coverage of proteins with unique physical properties (high/low hydrophobicity, pI, and Mw) than traditional 2-DE. The aim of this study was to elucidate the differences between adult S. japonicum infecting yellow cattle and water buffalo at the proteome level. The findings may provide valuable information for better understanding the interplay between S. japonicum and its natural hosts. A total of 131 DEPs were identified, of which 46 proteins were upregulated and 85 proteins were downregulated. The results were further confirmed by Western blot analysis and qRT-PCR. The bioinformatics analysis revealed that the identified DEPs were mainly involved in the protein synthesis, transcription regulation, protein proteolysis, cytoskeletal structure and oxidative stress response processes.
Ribosomes synthesize proteins that sustain the growth of biological organisms and are assembled from both ribosomal proteins and rRNA (Meyuhas, 2000). Approximately 80 different ribosomal proteins have been described in eukaryotic cells. Any loss of core ribosomal proteins or any defect in ribosomal components can change gene expression patterns (Thomas, 2000; Kondrashov et al., 2011). Our results indicated that the ribosomal pathway was significantly enriched among the DEPs and a large number of ribosome-associated proteins, including 40S ribosomal protein S24, ribosomal protein L29, ribosomal protein L35, and U3 small nucleolar RNA-associated protein 14, were downregulated in adult schistosomes recovered from water buffalo. This result may explain why schistosomes grow and develop better in yellow cattle than in water buffalo, as ribosomes are responsible for protein synthesis, and the expression of ribosomal proteins is higher in rapidly dividing cells (Pearson and Haber, 1980; Ju and Warner, 1994). A previous study revealed that the pairing of female and male schistosomula triggered a particular ribosomal protein expression profile, and more ribosomal genes or proteins were upregulated in schistosomula from double-sex infections than those from single-sex infections (Sun et al., 2015). We have reported that schistosomes from water buffalo present more pairing retardation than worms from yellow cattle (Yang et al., 2012b). This result suggests that the difference in the expression of ribosomal proteins may affect the development and even the paring of schistosomes in natural final hosts.
Spliceosomes are responsible for splicing the pre-mRNA, which is transcribed from DNA and has exons and introns. After that, the introns are spliced and the remaining exons are joined together to form a mature mRNA, which instructs to build a protein (de Longevialle et al., 2010). RNA splicing is an important posttranscriptional event and is crucial for the regulation of gene expression. The spliceosome consists of five small nuclear ribonucleoprotein particles (U1, U2, U4, U5, and U6 snRNPs) and non-snRNP proteins (Will and Luhrmann, 2011). The association of U1 snRNP with the 5′ splice site is an early event in the spliceosomal pathway and the base-pairing of U1 snRNA to the 5′ splice site occurs by the interaction between pre-mRNA and U1 small nuclear ribonucleoprotein C (U1-C) (Du and Rosbash, 2002; Wahl et al., 2009). The small nuclear ribonucleoprotein F belongs to the pre-mRNA-binding protein family and is involved in the alternative splicing of various genes (Han et al., 2010; Decorsiere et al., 2011; Talukdar et al., 2011). In this study, U1 small nuclear ribonucleoprotein C, small nuclear ribonucleoprotein F, and other spliceosome-associated proteins were downregulated in adult worms from water buffalo, suggesting that the lower expression of these spliceosome-related molecules might affect transcriptional processes in schistosomes infecting water buffaloes and lead to the poor development of the worms.
Cyclophilins have been found in cells from all studied organisms and have peptidyl-prolyl cis–trans isomerase activity (Wang and Heitman, 2005). Parasite-encoded cyclophilin proteins act as molecular chaperones by affecting protein folding and assembly and also contribute to RNA splicing (Bell et al., 2006). Previous studies showed that SjCyclophilin A was an important regulator of schistosome growth in the host environment and played a role in the host-parasite interplay (Han et al., 2012; Li et al., 2013). Histone proteins and DNA are packaged into nucleosomes and form a macromolecular complex known as chromatin. Moreover, the biogenesis of histones is tightly coupled to DNA replication (Anderson et al., 2012). In schistosomes, histone modifications, including acetylation, methylation, phosphorylation, ubiquitinylation, and sumoylation (Liu, 2016), have been reported to play critical roles in DNA repair, chromatin remodeling, and the transcriptional regulation of gene expression (Hong et al., 2016). The histone H4 is a core histone. The downregulation of cyclophilin A and histone H4 observed in the present study may affect transcription and translation in schistosomes infecting water buffaloes.
Cytoskeletal proteins play an important role in the maintenance of cell movement and cell morphology. Myosins are a critical component of the cytoskeleton and bind actin to form microfilaments (Reymann et al., 2012). Tropomyosin is best known for its role in the regulation of the contractile system in muscle and an important cytoskeletal component of non-muscle cells (Gunning et al., 2015). The movement of schistosomes is achieved through good muscular arrangements that are mainly performed by microfilaments and microtubules (Mair et al., 2003). A previous study showed that actin was downregulated in schistosomula from less susceptible rats and resistant M. fortis voles compared with susceptible mice (Hong et al., 2011). In the present study, several myosin-related proteins and tropomyosin proteins were downregulated in adult worms from water buffalo, which is consistent with the poor surface topography and internal structures at the ultrastructural level (Yang et al., 2012a). We hypothesize that this process may limit the development of muscle systems and in turn limit the growth of worms. In contrast, the results of iTRAQ and Western blotting indicated that β-actin levels were not significantly different between the two groups, suggesting that the growth and development of schistosomes in natural host animals is different from those of rodents.
The STRING analysis reveals the relations hidden behind the changes of protein levels by means of a computer-assisted analytical approach (Xu et al., 2017). In this study, the protein–protein relationships within the DEPs were also examined using STRING. It was of note that the downregulated proteins formed three subsets of protein interaction networks: protein synthesis, transcription regulation, and cytoskeleton-associated proteins. This result suggests that the downregulation of these proteins maybe affect the growth and development of schistosomes. Furthermore, there was no significant interactions among the upregulated proteins and this result needs to be further investigated.
Lysosomes are lytic vacuoles that contain acid hydrolase enzymes involved in the cleavage of waste materials, including cellular debris and exogenous proteins (Turk et al., 2002). Proteolysis in lysosomes is an important protein degradation pathway. The tetraspanin protein 2, also known as CD63, is a lysosomal membrane protein that plays an important role in the formation of phagosomes (Pols and Klumperman, 2009; Reymann et al., 2012). Cathepsin L is a lysosomal endopeptidase involved in terminal protein degradation, and the upregulation of cathepsin L has been reported in a wide range of malignancies (Dou and Carruthers, 2011; Sudhan and Siemann, 2015). The lysosomal protein saposin B is critical for lipid degradation metabolism (Huta et al., 2016), and several proteins containing the saposin B domain were reported to act as lysosome carrier proteins that bind sphingolipids to facilitate their degradation in Schistosome mansoni (Hall et al., 2011). Autophagy is the primary pathway for the degradation of dysfunctional macromolecules and lysosomal processes (Hong et al., 2016). The water buffalo can develop the phenomenon of self-cure of S. japonicum infection and the worms usually do not grow in the animals for 1–2 years post infection (Angeles et al., 2015). The upregulation of three lysosome-associated proteins—tetraspanin protein 2, cathepsin L, and saposin B domain-containing protein—in schistosomes from water buffalo might be partially correlated with this phenomenon.
Reactive oxygen species (ROS) are found in normal living organisms where they are constantly being produced under the oxidative stress caused by toxic heavy metals, heat shock, inflammation, ionizing irradiation, immune responses and environmental stimuli (Yu, 1994; Pushpamali et al., 2008). In schistosomes, ROS has been reported to be highly toxic, and parasites have developed antioxidant defense systems to protect themselves against ROS (Kazura et al., 1981; Hong et al., 2013). Thioredoxin peroxidase-1 (TPx-1) belongs to the family of thioredoxin peroxidases and is a major contributor to the detoxification of hydrogen peroxide in helminth parasites (Kumagai et al., 2006). Protein disulfide isomerase (PDI) belongs to the thioredoxin (Trx) superfamily and contains several Trx domains with redox active thiol/disulfide motifs (Gilbert, 1998). SjPDI may protect schistosomes from the oxidative damage induced by ROS, which is produced by host immune cells during oxygen metabolism (Cao et al., 2014). Glutathione S-transferases (GSTs) are a family of isoenzymes involved in the detoxification of potentially harmful electrophilic compounds by binding a range of hydrophobic ligands or by conjugation to glutathione (GSH) (Luo et al., 2009). There are 26 kDa and 28 kDa two isoenzyme GST molecules exist in schistosome. The 26-kDa GST of S. japonicum has been well characterized and several studies have demonstrated that rSj26GST could induce a strong protection against S. japonicum infection in mice, pig, sheep, water buffalo, and other animals (Wu et al., 2005). In the present study, SjTPx-1, SjPDI, and Sj26GST were significantly upregulated in schistosomes from water buffaloes, which were less susceptible hosts. We hypothesize that the overexpression of the three proteins may be necessary for the parasite to adapt to the host environment and enable survival and development in the less susceptible host.
We observed that the function of some identified DEPs was unknown or unannotated. Although the genome of S. japonicum was sequenced and assembled in 2009 (Zhou et al., 2009), the annotation of coding genes and proteins is still being developed. Therefore, the mechanisms involved in the susceptibility of natural hosts to schistosomes at the protein level need to be better explained with the gradual improvement of genome annotation.
Western blotting is a technique suitable to confirm the iTRAQ results but is limited by the availability of only a few specific antibodies. For this reason, qRT-PCR was carried out to verify the reliability of the iTRAQ data at the transcriptional level. The results revealed that the protein expression pattern of two selected DEPs did not coincide with the transcriptional expression pattern. This discrepancy may be due to the higher sensitivity of iTRAQ compared to qRT-PCR or posttranscriptional regulation (Vogel and Marcotte, 2012; Lv et al., 2017). Consistent with the results in this study, spliceosome and a mRNA surveillance pathway were significantly enriched among the DEPs. In addition, miRNAs play important roles in gene regulation and usually bind to the 3′UTR of their target mRNAs and cause mRNA degradation or translational inhibition (Liu, 2016). We previously found that miRNAs were differentially expressed between schistosomes from susceptible and less susceptible rodent hosts (Han et al., 2015a,b). This mechanism may also occur in worms from different natural hosts.
The DEPs information from the current study is fundamental to understand the different susceptibility to S. japonicum infection in water buffalo and yellow cattle. In the next work, we will silence some genes of S. japonicum in the in vitro cultural environment by RNAi according to the data of this study. We expect to further understand the host–parasite relationships in water buffalo and yellow cattle and gain useful information for screening bovine schistosome vaccine candidates. If the veterinary vaccine can be successfully developed, the contributions to the elimination of S. japonicum in China might be amazing.
In the present study, we used iTRAQ to identify DEPs in adult schistosomes from a S. japonicum-susceptible natural host (yellow cattle) and a less-susceptible natural host (water buffalo). The bioinformatics analysis indicated that some DEPs might have affected the survival and development of schistosomes. The identification of these proteins provides a new basis to understand the developmental mechanism of schistosomes and the interplay between schistosomes and their natural hosts.
JjL and GL conceived and designed the study. QZ, ZF, YH, XY, QH, KL, HL, XD, CZ, and JmL performed the experiments. QZ, ZF, and YH analyzed the data. QZ, JjL, and GL wrote the paper. QZ and YH revised the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Key Research and Development Program of China (2017YFD0501306) and the National Natural Science Foundation of China (Grant Nos. 31472188, 31402192, 31672541).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00099/full#supplementary-material
Amaral, K. B., Silva, T. P., Malta, K. K., Carmo, L. A., Dias, F. F., Almeida, M. R., et al. (2016). Natural Schistosoma mansoni infection in the wild reservoir Nectomys squamipes leads to excessive lipid droplet accumulation in hepatocytes in the absence of liver functional impairment. PLOS ONE 11:e0166979. doi: 10.1371/journal.pone.0166979
Anderson, L., Pierce, R. J., and Verjovski-Almeida, S. (2012). Schistosoma mansoni histones: from transcription to chromatin regulation: an in silico analysis. Mol. Biochem. Parasitol. 183, 105–114. doi: 10.1016/j.molbiopara.2012.03.001
Angeles, J. M. M., Leonardo, L. R., Goto, Y., Kirinoki, M., Villacorte, E. A., Hakimi, H., et al. (2015). Water buffalo as sentinel animals for Schistosomiasis surveillance. Bull World Health Organ. 93, 511–512. doi: 10.2471/BLT.14.143065
Bell, A., Monaghan, P., and Page, A. P. (2006). Peptidyl-prolyl cis–trans isomerases (immunophilins) and their roles in parasite biochemistry, host–parasite interaction and antiparasitic drug action. Int. J. Parasitol. 36, 261–276. doi: 10.1016/j.ijpara.2005.11.003
Cao, X., Hong, Y., Zhang, M., Han, Y., Wu, M., Wang, X., et al. (2014). Cloning, expression and characterization of protein disulfide isomerase of Schistosoma japonicum. Exp. Parasitol. 146, 43–51. doi: 10.1016/j.exppara.2014.09.004
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Da’Dara, A. A., Li, Y. S., Xiong, T., Zhou, J., Williams, G. M., McManus, D. P., et al. (2008). DNA-based vaccines protect against zoonotic Schistosomiasis in water buffalo. Vaccine 26, 3617–3625. doi: 10.1016/j.vaccine.2008.04.080
de Longevialle, A. F., Small, I. D., and Lurin, C. (2010). Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant 3, 691–705. doi: 10.1093/mp/ssq025
Dean, D. A., Mangold, B. L., Harrison, R. A., and Ricciardone, M. D. (1996). Homologous and heterologous protective immunity to Egyptian strains of Schistosoma mansoni and S. haematobium induced by ultraviolet-irradiated cercariae. Parasite Immunol. 18, 403–410. doi: 10.1046/j.1365-3024.1996.d01-129.x
Decorsiere, A., Cayrel, A., Vagner, S., and Millevoi, S. (2011). Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3’-end processing and function during DNA damage. Genes Dev. 25, 220–225. doi: 10.1101/gad.607011
Dou, Z., and Carruthers, V. B. (2011). Cathepsin proteases in Toxoplasma gondii. Adv. Exp. Med. Biol. 712, 49–61. doi: 10.1007/978-1-4419-8414-2_4
Draz, H. M., Ashour, E., Shaker, Y. M., Khattab, H. M., Wu, C. H., and Wu, G. Y. (2008). Host susceptibility to schistosomes: effect of host sera on cell proliferation of Schistosoma mansoni schistosomula. J. Parasitol. 94, 1249–1252. doi: 10.1645/GE-1607.1
Du, H., and Rosbash, M. (2002). The U1 snRNP protein U1C recognizes the 5’-splice site in the absence of base pairing. Nature 419, 86–90. doi: 10.1038/nature00947
Gaze, S., Driguez, P., Pearson, M. S., Mendes, T., Doolan, D. L., Trieu, A., et al. (2014). An immunomics approach to schistosome antigen discovery: antibody signatures of naturally resistant and chronically infected individuals from endemic areas. PLOS Pathog. 10:e1004033. doi: 10.1371/journal.ppat.1004033
Gilbert, H. F. (1998). Protein disulfide isomerase. Methods Enzymol. 290, 26–50. doi: 10.1016/S0076-6879(98)90005-2
Gobert, G. N., Moertel, L., Brindley, P. J., and Mcmanus, D. P. (2009). Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics 10:128. doi: 10.1186/1471-2164-10-128
Gunning, P. W., Hardeman, E. C., Lappalainen, P., and Mulvihill, D. P. (2015). Tropomyosin - master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 128, 2965–2974. doi: 10.1242/jcs.172502
Hall, S. L., Braschi, S., Truscott, M., Mathieson, W., Cesari, I. M., and Wilson, R. A. (2011). Insights into blood feeding by schistosomes from a proteomic analysis of worm vomitus. Mol. Biochem. Parasitol. 179, 18–29. doi: 10.1016/j.molbiopara.2011.05.002
Han, H., Peng, J., Hong, Y., Fu, Z., Lu, K., Li, H., et al. (2015a). Comparative analysis of microRNA in schistosomula isolated from non-permissive host and susceptible host. Mol. Biochem. Parasitol. 204, 81–88. doi: 10.1016/j.molbiopara.2015.11.005
Han, H., Peng, J., Hong, Y., Fu, Z., Lu, K., Li, H., et al. (2015b). Comparative characterization of microRNAs in Schistosoma japonicum schistosomula from Wistar rats and BALB/c mice. Parasitol. Res. 114, 2639–2647. doi: 10.1007/s00436-015-4468-1
Han, H., Peng, J., Hong, Y., Fu, Z., Xu, J., Lin, J., et al. (2012). Molecular cloning and characterization of a cyclophilin A homologue from Schistosoma japonicum. Parasitol. Res. 111, 807–817. doi: 10.1007/s00436-012-2903-0
Han, S. P., Tang, Y. H., and Smith, R. (2010). Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430, 379–392. doi: 10.1042/BJ20100396
He, Y. X., Salafsky, B., and Ramaswamy, K. (2001). Host–parasite relationships of Schistosoma japonicum in mammalian hosts. Trends Parasitol. 17, 320–324. doi: 10.1016/S1471-4922(01)01904-3
Hong, Y., Cao, X., Han, Q., Yuan, C., Zhang, M., Han, Y., et al. (2016). Proteome-wide analysis of lysine acetylation in adult Schistosoma japonicum worm. J. Proteomics 148, 202–212. doi: 10.1016/j.jprot.2016.08.008
Hong, Y., Han, Y., Fu, Z., Han, H., Qiu, C., Zhang, M., et al. (2013). Characterization and expression of the Schistosoma japonicum thioredoxin peroxidase-2 gene. J. Parasitol. 99, 68–76. doi: 10.1645/GE-3096.1
Hong, Y., Peng, J., Jiang, W., Fu, Z., Liu, J., Shi, Y., et al. (2011). Proteomic analysis of Schistosoma japonicum schistosomulum proteins that are differentially expressed among hosts differing in their susceptibility to the Infection. Mol. Cell. Proteomics 10:M110.006098. doi: 10.1074/mcp.M110.006098
Hsu, S. Y., Hsu, H. F., Lust, G. L., Davis, J. R., and Eveland, L. K. (1973). Comparative studies on the lesions caused by eggs of Schistosoma japonicum and Schistosoma mansoni in the liver of hamsters, guinea pigs, and albino rats. Ann. Trop. Med. Parasitol. 67, 349–356. doi: 10.1080/00034983.1973.11686897
Huang, Y., Niu, B., Gao, Y., Fu, L., and Li, W. (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. doi: 10.1093/bioinformatics/btq003
Huta, B. P., Mehlenbacher, M. R., Nie, Y., Lai, X., Zubieta, C., Bou-Abdallah, F., et al. (2016). The Lysosomal protein saposin B binds chloroquine. ChemMedChem 11, 277–282. doi: 10.1002/cmdc.201500494
Jiang, W., Hong, Y., Peng, J., Fu, Z., Feng, X., Liu, J., et al. (2010). Study on differences in the pathology, T cell subsets and gene expression in susceptible and non-susceptible hosts infected with Schistosoma japonicum. PLOS ONE 5:e13494. doi: 10.1371/journal.pone.0013494
Ju, Q., and Warner, J. R. (1994). Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10, 151–157. doi: 10.1002/yea.320100203
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kazura, J. W., Fanning, M. M., Blumer, J. L., and Mahmoud, A. A. F. (1981). Role of cell-generated hydrogen peroxide in granulocyte-mediated killing of schistosomula of Schistosoma mansoni in vitro. J. Clin. Invest. 67, 93–102. doi: 10.1172/JCI110037
Khalife, J., Cetre, C., Pierrot, C., and Capron, M. (2000). Mechanisms of resistance to S. mansoni infection: the rat model. Parasitol. Int. 49, 339–345. doi: 10.1016/S1383-5769(00)00059-3
Kondrashov, N., Pusic, A., Stumpf, C. R., Shimizu, K., Hsieh, A. C., Xue, S., et al. (2011). Ribosome-mediated specificity in hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. doi: 10.1016/j.cell.2011.03.028
Kumagai, T., Osada, Y., and Kanazawa, T. (2006). 2-Cys peroxiredoxins from Schistosoma japonicum: the expression profile and localization in the life cycle. Mol. Biochem. Parasitol. 149, 135–143. doi: 10.1016/j.molbiopara.2006.05.004
Li, J., Zhuang, W., Cong, L., Shi, W., Cai, X., Huang, F., et al. (2013). Cyclophilin A from Schistosoma japonicum promotes a Th2 response in mice. Parasit. Vectors 6:330. doi: 10.1186/1756-3305-6-330
Li, S. Z., Utzinger, J., Bergquist, R., and Zhou, X. N. (2016). Preface: elimination of Schistosomiasis japonica in the People’s Republic of China: the last leg. Adv. Parasitol. 92, xix–xxii. doi: 10.1016/S0065-308X(16)30052-5
Li, W., Jaroszewski, L., and Godzik, A. (2001). Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283. doi: 10.1093/bioinformatics/17.3.282
Li, Y., McManus, D. P., Lin, D., Williams, G. M., Harn, D. A., Ross, A. G., et al. (2014). The Schistosoma japonicum self-cure phenomenon in water buffaloes: potential impact on the control and elimination of Schistosomiasis in China. Int. J. Parasitol. 44, 167–171. doi: 10.1016/j.ijpara.2013.10.007
Liu, J., Zhu, C., Shi, Y., Li, H., Wang, L., Qin, S., et al. (2012). Surveillance of Schistosoma japonicum infection in domestic ruminants in the Dongting Lake region, Hunan province, China. PLOS ONE 7:e31876. doi: 10.1371/journal.pone.0031876
Liu, S., Zhou, X., Piao, X., Wu, C., Hou, N., and Chen, Q. (2015). Comparative analysis of transcriptional profiles of adult Schistosoma japonicum from different laboratory animals and the natural host, water buffalo. PLOS Neglect. Trop. D 9:e0003993. doi: 10.1371/journal.pntd.0003993
Liu, W. (2016). Epigenetics in Schistosomes: what we know and what we need know. Front. Cell. Infect. Microbiol. 6:149. doi: 10.3389/fcimb.2016.00149
Liu, Y., Zhou, Y. B., Li, R. Z., Wan, J. J., Yang, Y., Qiu, D. C., et al. (2016). Epidemiological features and effectiveness of Schistosomiasis control programme in mountainous and hilly region of the people’s republic of China. Adv. Parasitol. 92, 73–95. doi: 10.1016/bs.apar.2016.02.019
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, Q., Qiao, Z., Zhou, Y., Li, X., Zhong, Z., Yu, Y., et al. (2009). Application of signaling protein 14-3-3 and 26kDa glutathione-S-transferase to serological diagnosis of Schistosomiasis japonica. Acta Trop. 112, 91–96. doi: 10.1016/j.actatropica.2009.06.011
Lv, L., Wang, Y., Feng, W., Hernandez, J. A., Huang, W., Zheng, Y., et al. (2017). iTRAQ-based differential proteomic analysis in Mongolian gerbil brains chronically infected with Toxoplasma gondii. J. Proteomics 160, 74–83. doi: 10.1016/j.jprot.2017.03.012
Mair, G. R., Maule, A. G., Fried, B., Day, T. A., and Halton, D. W. (2003). Organization of the musculature of schistosome cercariae. J. Parasitol. 89, 623–625. doi: 10.1645/0022-3395(2003)089[0623:OOTMOS]2.0.CO;2
Meyuhas, O. (2000). Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267, 6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x
Pearson, N. J., and Haber, J. E. (1980). Changes in regulation of ribosomal protein synthesis during vegetative growth and sporulation of Saccharomyces cerevisiae. J. Bacteriol. 143, 1411–1419.
Peng, J., Han, H., Gobert, G. N., Hong, Y., Jiang, W., Wang, X., et al. (2011). Differential gene expression in Schistosoma japonicum schistosomula from Wistar rats and BALB/c mice. Parasit. Vectors 4:155. doi: 10.1186/1756-3305-4-155
Pols, M. S., and Klumperman, J. (2009). Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315, 1584–1592. doi: 10.1016/j.yexcr.2008.09.020
Pushpamali, W. A., De Zoysa, M., Kang, H., Oh, C. H., Whang, I., Kim, S. J., et al. (2008). Comparative study of two thioredoxin peroxidases from disk abalone (Haliotis discus discus): cloning, recombinant protein purification, characterization of antioxidant activities and expression analysis. Fish Shellfish Immunol. 24, 294–307. doi: 10.1016/j.fsi.2007.11.016
Reymann, A. C., Boujemaa-Paterski, R., Martiel, J. L., Guerin, C., Cao, W., Chin, H. F., et al. (2012). Actin network architecture can determine myosin motor activity. Science 336, 1310–1314. doi: 10.1126/science.1221708
Rheinberg, C. E., Mone, H., Caffrey, C. R., Imbert-Establet, D., Jourdane, J., and Ruppel, A. (1998). Schistosoma haematobium, S. intercalatum, S. japonicum, S. mansoni, and S. rodhaini in mice: relationship between patterns of lung migration by schistosomula and perfusion recovery of adult worms. Parasitol. Res. 84, 338–342. doi: 10.1007/s004360050407
Sudhan, D. R., and Siemann, D. W. (2015). Cathepsin L targeting in cancer treatment. Pharmacol. Ther. 155, 105–116. doi: 10.1016/j.pharmthera.2015.08.007
Sun, J., Li, C., and Wang, S. (2015). The Up-regulation of ribosomal proteins further regulates protein expression profile in female Schistosoma japonicum after pairing. PLOS ONE 10:e0129626. doi: 10.1371/journal.pone.0129626
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Tallima, H., Dalton, J. P., and El, R. R. (2015). Induction of protective immune responses against Schistosomiasis haematobium in hamsters and mice using cysteine peptidase-based vaccine. Front. Immunol. 6:130. doi: 10.3389/fimmu.2015.00130
Talukdar, I., Sen, S., Urbano, R., Thompson, J., Yates, J. R., and Webster, N. J. (2011). hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PLOS ONE 6:e27869. doi: 10.1371/journal.pone.0027869
Thomas, G. (2000). An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2, E71–E72. doi: 10.1038/35010581
Turk, V., Turk, B., Guncar, G., Turk, D., and Kos, J. (2002). Lysosomal cathepsins: structure, role in antigen processing and presentation, and cancer. Adv. Enzyme Regul. 42, 285–303. doi: 10.1016/S0065-2571(01)00034-6
Van Dorssen, C. F., Gordon, C. A., Li, Y., Williams, G. M., Wang, Y., Luo, Z., et al. (2017). Rodents, goats and dogs - their potential roles in the transmission of Schistosomiasis in China. Parasitology 144, 1633–1642. doi: 10.1017/S0031182017000907
Viana, I. R., Correa-Oliveira, R., Carvalho, O. S., Massara, C. L., Colosimo, E., Colley, D. G., et al. (1995). Comparison of antibody isotype responses to Schistosoma mansoni antigens by infected and putative resistant individuals living in an endemic area. Parasite Immunol. 17, 297–304. doi: 10.1111/j.1365-3024.1995.tb00895.x
Vogel, C., and Marcotte, E. M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. doi: 10.1038/nrg3185
Wahl, M. C., Will, C. L., and Lührmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. doi: 10.1016/j.cell.2009.02.009
Wang, P., and Heitman, J. (2005). The cyclophilins. Genome Biol. 6:226. doi: 10.1186/gb-2005-6-7-226
Wang, T., Maria, V. J., Zhang, S., Wang, F., Wu, W., Zhang, G., et al. (2005). Transmission of Schistosoma japonicum by humans and domestic animals in the Yangtze River valley, Anhui province, China. Acta Trop. 96, 198–204. doi: 10.1016/j.actatropica.2005.07.017
WHO (2016). Schistosomiasis: number of people treated worldwide in 2014. Wkly. Epidemiol. Rec. 91, 53–60.
Will, C. L., and Luhrmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707. doi: 10.1101/cshperspect.a003707
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Wu, W. W., Wang, G., Baek, S. J., and Shen, R. (2006). Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J. Proteome Res. 5, 651–658. doi: 10.1021/pr050405o
Wu, Z., Lü, Z., and Yu, X. (2005). Development of a vaccine against Schistosoma japonicum in China: a review. Acta Trop. 96, 106–116. doi: 10.1016/j.actatropica.2005.08.005
Xu, J., Zhang, L., Hou, J., Wang, X., Liu, H., Zheng, D., et al. (2017). iTRAQ-based quantitative proteomic analysis of the global response to 17beta-estradiol in estrogen-degradation strain Pseudomonas putida SJTE-1. Sci. Rep. 7:41682. doi: 10.1038/srep41682
Yang, J., Feng, X., Fu, Z., Yuan, C., Hong, Y., Shi, Y., et al. (2012a). Ultrastructural observation and gene expression profiling of Schistosoma japonicum derived from two natural reservoir hosts, water buffalo and yellow cattle. PLOS ONE 7:e47660. doi: 10.1371/journal.pone.0047660
Yang, J., Fu, Z., Feng, X., Shi, Y., Yuan, C., Liu, J., et al. (2012b). Comparison of worm development and host immune responses in natural hosts of Schistosoma japonicum, yellow cattle and water buffalo. BMC Vet. Res. 8:25. doi: 10.1186/1746-6148-8-25
Yu, B. P. (1994). Cellular defenses against damage from reactive oxygen species. Physiol. Rev. 74, 139–162. doi: 10.1152/physrev.1994.74.1.139
Zhang, L., Xu, Z., Qian, Y., Dang, H., Lv, S., Xu, J., et al. (2016). Endemic status of Schistosomiasis in People’s Republic of China in 2015. Chin. J. Schisto. Control 28, 611–617.
Zhang, S. Q., Sun, C. S., Wang, M., Lin, D. D., Zhou, X. N., and Wang, T. P. (2016). Epidemiological features and effectiveness of Schistosomiasis control programme in lake and marshland region in the People’s Republic of China. Adv. Parasitol. 92, 39–71. doi: 10.1016/bs.apar.2016.02.018
Keywords: Schistosoma japonicum, adult worm, water buffalo, yellow cattle, iTRAQ
Citation: Zhai Q, Fu Z, Hong Y, Yu X, Han Q, Lu K, Li H, Dou X, Zhu C, Liu J, Lin J and Li G (2018) iTRAQ-Based Comparative Proteomic Analysis of Adult Schistosoma japonicum from Water Buffalo and Yellow Cattle. Front. Microbiol. 9:99. doi: 10.3389/fmicb.2018.00099
Received: 11 October 2017; Accepted: 16 January 2018;
Published: 06 February 2018.
Edited by:
Miguel Cacho Teixeira, Universidade de Lisboa, PortugalReviewed by:
Rashika El Ridi, Cairo University, EgyptCopyright © 2018 Zhai, Fu, Hong, Yu, Han, Lu, Li, Dou, Zhu, Liu, Lin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Hong, aG9uZ3lhbmdAc2h2cmkuYWMuY24= Jiaojiao Lin, ampsaW5Ac2h2cmkuYWMuY24= Guoqing Li, Z3FsaUBzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.