- 1State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3Department of Plant Pathology and Microbiology, The Robert H. Smith Faculty of Agriculture, Food and Environment, The Hebrew University of Jerusalem, Rehovot, Israel
Fungi transcriptionally upregulate expression of azole efflux pumps and ergosterol biosynthesis pathway genes when exposed to antifungal agents that target ergosterol biosynthesis. To date, these transcriptional responses have been shown to be dependent on the presence of the azoles and/or depletion of ergosterol. Using an inducible promoter to regulate Neurospora crassa erg11, which encodes the major azole target, sterol 14α-demethylase, we were able to demonstrate that the CDR4 azole efflux pump can be transcriptionally activated by ergosterol biosynthesis inhibition even in the absence of azoles. By analyzing ergosterol deficient mutants, we demonstrate that the transcriptional responses by cdr4 and, unexpectedly, genes encoding ergosterol biosynthesis enzymes (erg genes) that are responsive to azoles, are not dependent on ergosterol depletion. Nonetheless, deletion of erg2, which encodes C-8 sterol isomerase, also induced expression of cdr4. Deletion of erg2 also induced the expression of erg24, the gene encoding C-14 sterol reductase, but not other tested erg genes which were responsive to erg11 inactivation. This indicates that inhibition of specific steps of ergosterol biosynthesis can result in different transcriptional responses, which is further supported by our results obtained using different ergosterol biosynthesis inhibitors. Together with the sterol profiles, these results suggest that the transcriptional responses by cdr4 and erg genes are associated with accumulation of specific sterol intermediate(s). This was further supported by the fact that when the erg2 mutant was treated with ketoconazole, upstream inhibition overrode the effects by downstream inhibition on ergosterol biosynthesis pathway. Even though cdr4 expression is associated with the accumulation of sterol intermediates, intra- and extracellular sterol analysis by HPLC-MS indicated that the transcriptional induction of cdr4 did not result in efflux of the accumulated intermediate(s). This study demonstrates, by detailed genetic and chemical analysis, that transcriptional responses by a major efflux pump and genes of the ergosterol biosynthesis pathway to ergosterol biosynthesis inhibitors can be independent of the presence of the drugs and are linked with the accumulation of ergosterol intermediate(s).
Introduction
Fungal diseases in crops significantly contribute to yield loss and mycotoxin contaminations (Dean et al., 2012), while invasive fungal infections in immunodeficient patients are often the cause for mortality (Brown et al., 2012). For decades, antifungal azoles have been prominently used in the control of detrimental fungi in the clinic and in agriculture due to their broad antifungal spectra, low toxicity and low cost. However, azole resistance, which accompanies the long-term drug use of these compounds, has made fungal pathogen control more challenging in recent years in both clinic and agriculture (Cowen et al., 2014; Price et al., 2015).
Antifungal azoles block ergosterol biosynthesis by inhibiting sterol 14α-demethylase. Inhibition by azoles commonly leads to depletion of ergosterol and accumulation of other sterols, such as lanosterol, eburicol and the toxic 14α-methyl-3,6-diol, within fungal cell membranes (Watson et al., 1989; Shapiro et al., 2011; Chen et al., 2016). Two important systems shown to be critical to azole resistance are the efflux pump and ergosterol homeostasis systems. The efflux pump system, which is comprised of pumps located in the cell membrane and their regulators, is present in almost all species and plays an important roles in drug resistance in bacteria, fungi and human cancer cells by efflux of the drugs (Golin et al., 2007; Blair et al., 2014; Cowen et al., 2014; Paul and Moye-Rowley, 2014; Tanwar et al., 2014; Kathawala et al., 2015). Ergosterol homeostasis, which is tightly maintained by several important regulators, is imperative for ergosterol biosynthesis and cell membrane functions (Bien and Espenshade, 2010; Maguire et al., 2014). Upon azole stress, fungi can respond with a change in the transcription of a variety of genes. The most commonly observed azole-responsive genes include the gene encoding the azole target sterol 14α-demethylase and those encoding efflux pumps such as the Saccharomyces cerevisiae Pdr5p, the Candida albicans Cdr1/2p and Mdr1p and the N. crassa CDR4 (NCU05591), as well as other genes in ergosterol biosynthesis, including ERG2 (encoding C-8 sterol isomerase), ERG5 (encoding C-22 sterol desaturase), ERG6 (encoding C-24 sterol methyl transferase) and ERG24 (encoding C-14 sterol reductase) in S. cerevisiae and their homologs in other fungi (Agarwal et al., 2003; Liu et al., 2005, 2010; Ferreira et al., 2006; Yu et al., 2007; Hoehamer et al., 2010; Florio et al., 2011; Sun et al., 2014), indicating the two systems are transcriptionally activated by azoles. In clinical and agricultural azole-resistant isolates, overexpression of sterol 14α-demethylase or azole efflux pumps are among the major causes for azole resistance (White, 1997; Perea et al., 2001; Perea et al., 2002; Redding et al., 2003; Cools et al., 2013; Price et al., 2015). The responsive C-22 sterol desaturase coding gene was also demonstrated to be important for the basal resistance to azoles in N. crassa and Fusarium verticillioides (Sun et al., 2013). In addition, transcription factors, such as Tac1p that regulates the azole efflux pumps and Upc2p that regulates ergosterol biosynthesis genes in C. albicans, govern the expression of many azole-responsive genes and confer azole resistance (Coste et al., 2004; Coste et al., 2006; Dunkel et al., 2008; Heilmann et al., 2010; Flowers et al., 2012; Vasicek et al., 2014). The phenomena described above indicate that activation of the transcriptional responses to antifungal azoles is an important strategy for azole resistance.
For transcriptional responses of azole efflux pumps, the azole drug itself was shown to activate gene expression of azole efflux pumps. In S. cerevisiae, the transcription factors Pdr1p and Pdr3p, which regulate expression of efflux pumps, can be activated by directly interacting with xenobiotics, including ketoconazole, and thus might act as sensors of xenobiotics (Thakur et al., 2008). However, it is unknown whether the sterol content changes caused by azole inhibition can result in the activation of azole efflux pumps.
For azole-activated transcriptional responses by genes for ergosterol biosynthesis, several lines of evidence suggest that ergosterol depletion might be a cause. In Schizosaccharomyces pombe, it was reported that ergosterol depletion activated the expression of ergosterol biosynthesis genes and their regulator Sre1p (Hughes et al., 2005; Porter et al., 2010). In S. cerevisiae, transcription factor Upc2p regulates expression of ergosterol biosynthesis genes. Upc2p directly interacts with ergosterol and its activity was regulated by ergosterol levels (Yang et al., 2015). Thus, azole-caused ergosterol depletion might activate transcriptional response via Upc2p. Other conditions causing ergosterol depletion, by ergosterol biosynthesis inhibitors other than azoles, or mutation of some genes in ergosterol biosynthesis pathway, also activate the expression of genes for ergosterol biosynthesis in S. cerevisiae and Candida species (Bammert and Fostel, 2000; Henry et al., 2000). All above lines of evidence suggest that ergosterol depletion resulted from azole stress might be a cause for the transcriptional responses by ergosterol biosynthesis genes, but some direct experimental data in other fungi are still required for making a definite conclusion. In addition, the accumulated toxic 14α-methyl-3,6-diol was also thought to cause membrane stresses and activate stress responses with unclear mechanisms (Watson et al., 1989; Shapiro et al., 2011). Thus, the accumulation of sterol intermediates is also possible to induce the transcriptional responses.
In this study, based on genetic and chemical analysis in N. crassa, we demonstrate that abnormal ergosterol biosynthesis, causing accumulation of sterol intermediates but not ergosterol depletion, can result in transcriptional responses by efflux pump CDR4, as well as ergosterol biosynthesis genes, similar to those under azole stress.
Materials and Methods
Strains and Culture Conditions
Neurospora crassa strains used in this study and their sources are listed in Table 1. Designations of ergosterol biosynthesis genes followed a previous study in N. crassa, in which all of the genes were named with those of their orthologs in yeast (Sun et al., 2013). A suggested partial ergosterol biosynthesis pathway in N. crassa is shown in Figure 1. However, as different N. crassa designations have been used by various authors in the past, we have compiled a list of comparative erg gene designations for clarification (Supplementary Table 1). Strains were cultured on Vogel’s medium with 1.5% agar [Vogel’s minimum medium, supplemented with 2% (w/vol) sucrose for slants or glucose for plates] (Vogel, 1956). Liquid Vogel’s minimum medium supplemented with 2% glucose was used for preparing samples for RNA, protein and sterol extraction. Sorbose plates (Vogel’s minimum medium, supplemented with 20 g/L sorbose, 0.5 g/L fructose and 0.5 g/L glucose) were used for electroporation transformation (Davis and de Serres, 1970). All cultures were grown at 28°C.
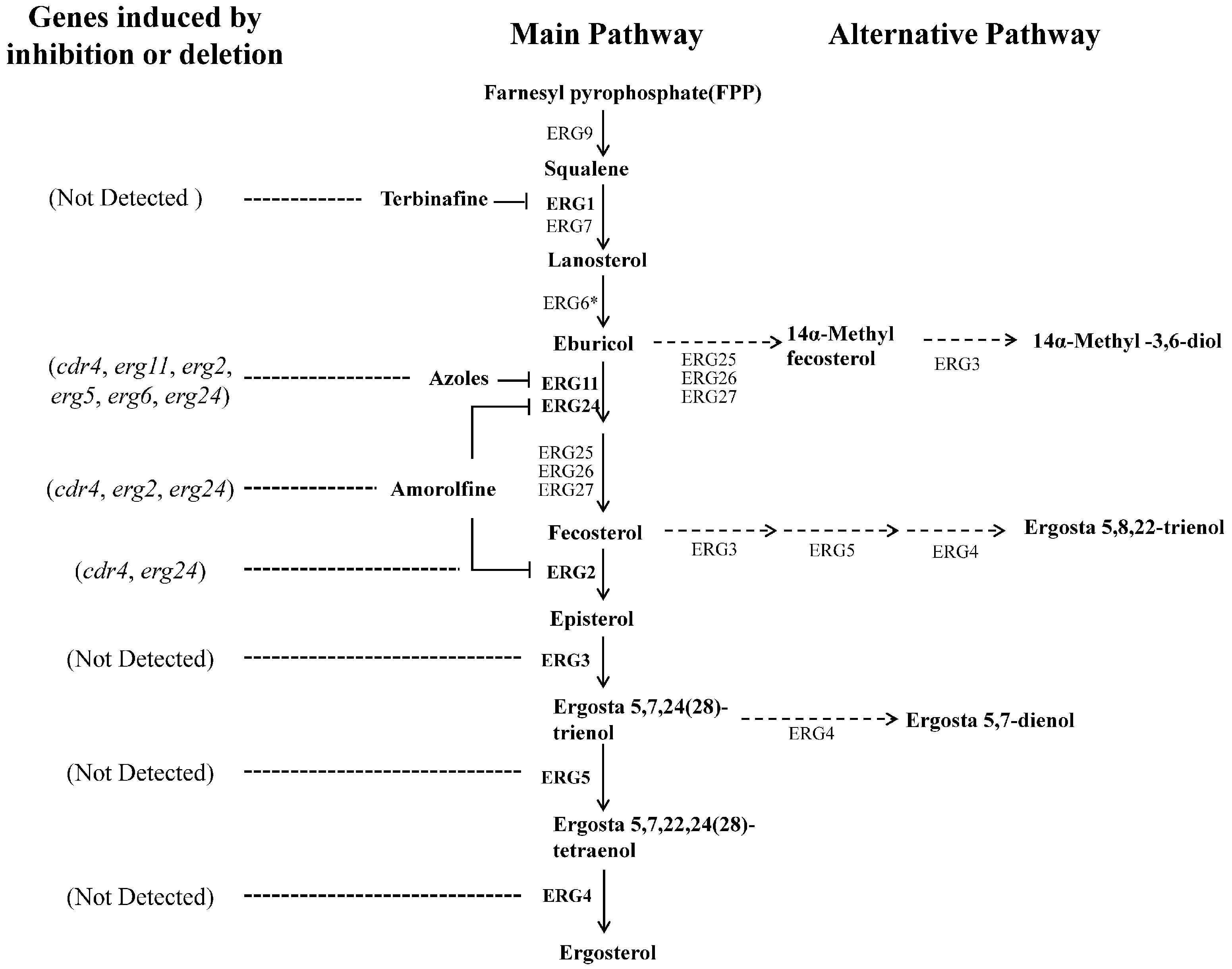
FIGURE 1. Suggested ergosterol biosynthesis pathway in N. crassa and transcriptional responses to disruption at certain steps. Each enzyme relates to one step of the ergosterol biosynthesis pathway in N. crassa. The most marked difference between the N. crassa and S. cerevisiae is the step catalyzed by ERG6 (marked by an asterisk). This step follows the reactions catalyzed by ERG25, ERG26 and ERG27 in yeast. Corresponding genes encoding enzymes in bold were analyzed in this study. Dotted arrows show the alternative biosynthesis pathways when the main pathway is impaired. Dotted lines showed the genes transcriptional induced by ergosterol biosynthesis inhibitors or erg gene deletion.
Antifungal compounds were added as needed. The antifungal agents include ketoconazole (KTC, Sigma), amorolfine (AMOR, Tokyo Chemical Industry) and terbinafine (TERB, J&K), dissolved in DMSO, Methanol and Methanol, respectively. Media were amended with the drugs to a final concentration of 2 mg/L, 0.375 mg/L and 3 mg/L, respectively, which resulted in N. crassa growth inhibition of at least 50%. Hygromycin B (Amresco) at a final concentration of 150 mg/L was used for screening fungal transformants and added as required.
Mutant Construction
In order to generate a strain in which transcription of erg11 (NCU02624, encoding sterol 14α-demethylase, Figure 1) could be controlled, the native promoter of erg11 was replaced with the copper responsive promoter Ptcu-1, based on a previously described strategy (Lamb et al., 2013). The construct diagram was shown in Figure 2A. First, a dominant selectable marker cassette was fused to the 5′ terminus of the tcu-1 promoter. The appropriate fragments of the tcu-1 promoter (1716 bp) and the Hygromycin B resistance gene hph, flanked by the trpC promoter and terminator (2169 bp), were amplified using primers Ptcu-1F/R and HphR-F/R, respectively (Supplementary Table 2). A homologous recombination technology-based kit (ClonExpress MultiS One Step Cloning Kit; Vazyme, Nanjing, China) was used to insert the two fragments into pBluescript II SK, resulting in the formation of plasmid pSKII-hph-Ptcu-1. Second, a split marker method was used to construct the mutant and two rounds of PCR were performed to obtain the recombination fragments. In the first round, four fragments, comprised of the 565 bp upstream homologous region of erg11, 834 bp downstream homologous region of erg11 (erg11N), upstream half of hph-Ptcu-1 and downstream half of hph-Ptcu-1, were amplified using primer pairs exPerg-up-F/R, exPerg-down-F/R, hph-Ptcu-up-F/R and hph-Ptcu-down-F/R, respectively (Supplementary Table 2). In addition, the downstream homologous region of erg11 was amplified from a construct containing a 5 × myc-6 × his-tagged version of the gene (Hu et al., unpublished). In the second round of amplification, the two upstream fragments and the two downstream fragments were joined, by fusion PCR, using primer pairs exPerg-up-F, hph-Ptcu-up-R and hph-Ptcu-down-F, exPerg-down-R, respectively (Supplementary Table 2) and resulted in 5′ recombination fragment and 3′ recombination fragment (Figure 2A). The obtained two recombination fragments were purified and then directly co-transformed into an available strain that was Ku70 deficient in a Ras-1bd background by electroporation (He et al., 2006). Transformants were screened on plates containing 50 mg/L bathocuproinedisulfonic acid (BCS, Sigma) and 150 mg/L Hygromycin B and verified by PCR using primer pair exPerg-vF/R (Supplementary Table 2). After grown for several generations on slants containing 150 mg/L Hygromycin B, homozygous strains were screened and verified by PCR and used for the following experiments.
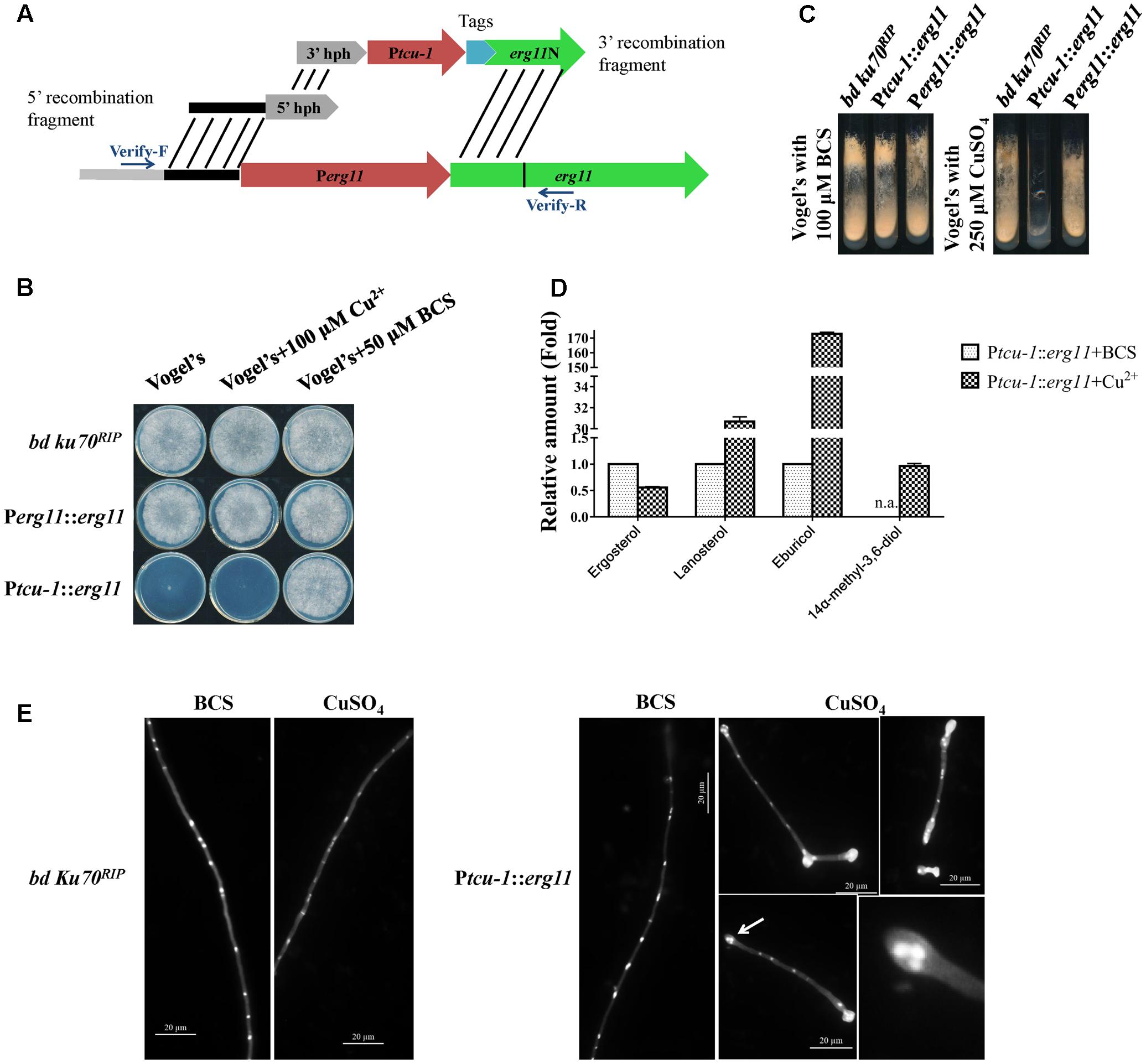
FIGURE 2. erg11 is an essential gene in N. crassa. (A) Schematic diagram of the process of Ptcu-1::erg11 (NCU02624, encoding sterol 14α-demethylase) strain construction. (B,C) erg11 is required for fungal growth. Conidia of N. crassa bd Ku70RIP strain (the parental strain), Ptcu-1::erg11 mutant and the complemented Perg11::erg11 mutant were inoculated on Vogel’s plates (B) and Vogel’s slants (C) supplemented with or without CuSO4 or Cu2+ chelator BCS and grown at 28°C for 7 days (B) or 30 h (C), respectively. (D) erg11 inactivation resulted in sterol content changes in N. crassa. After grown for 13.5 h in liquid Vogel’s medium amended with BCS, the mycelium was then transferred to liquid Vogel’s medium amended with BCS and CuSO4 and treated for 22 h. The sterols were then extracted and subjected to HPLC-MS analysis. The abundance of sterols was calculated on the basis of the chromatogram peak area and normalized using an internal control and sample weight. The results presented here are means of two biological replicates. (E) Conidia of bd Ku70RIP and Ptcu-1::erg11 were germinated in Vogel’s minimal medium supplemented with BCS or CuSO4 for 10 h. Hyphae were imaged after fixing and staining with calcofluor white and DAPI. Clustered nuclei are indicated by arrows and enlarged (below panel on the right).
The erg11 inactivation strain was complemented by replacing the tcu-1 promoter with the native promoter of erg11. Briefly, three fragments, comprised of the terminator region of trpC, native promoter of erg11 and 5 × myc-6 × his tagged coding region of erg11, were amplified using primers TtrpC-F/R, Perg11F/R, and exPerg-down-F’/R’, respectively (Supplementary Table 2). The three fragments were then joined, by fusion PCR, using primer pairs TtrpC-F and exPerg-down-R’ (Supplementary Table 2). The obtained fragment was purified and then directly transformed into the erg11 inactivation strain Ptcu-1::erg11. Since the Ptcu-1::erg11 strain cannot grow on media containing Cu2++, Cu2+ were used to screen the transformants. The genetic alterations introduced to the transformants were then verified by PCR.
The erg2 gene (NCU04156, encoding C-8 sterol isomerase, Figure 1) was deleted as described (Colot et al., 2006). Briefly, a 1153 bp promoter region and 1304 bp terminator region of erg2, as well as a 1416 bp fragment containing the Hygromycin B resistant gene, were amplified using primer pairs Perg2-up-F/R, Perg2-down-F/R and hph-F/R, respectively (Supplementary Table 2). The three fragments were subjected to fusion PCR using primers Perg2-up-F and Perg2-down-R (Supplementary Table 2). The obtained fragment was purified and then transformed into the wild-type strain FGSC #4200 by electroporation transformation as previously reported (Wang et al., 2015). Transformants were screened on plates containing 150 mg/L Hygromycin B and the gene replacement was verified by PCR.
Phenotypic Analysis of Mutants
For macroscopic and microscopic analysis of mutants, Vogel’s minimal medium dishes were inoculated with 2 μL aliquot of conidial suspension (2 × 106 /mL) and the strains were cultured at 28°C. Growth media either contained or lacked CuSO4 or was amended with BCS supplements. The final concentrations of CuSO4 and BCS were 100 and 50 μM, respectively. Growth was documented after growing for at least 28 h and colony edges were subjected to microscopic examination. Spore suspensions were also inoculated onto Vogel’s slants with or without CuSO4 or BCS supplements and incubated at 28°C. After 5–7 days, aerial hyphal formation and conidiation were analyzed.
Calcofluor white (0.5 g/L, Fluka, Sigma) and DAPI (Sango Biotech) were used to visualize cell wall and nucleus, respectively. Briefly, conidia of each strain were inoculated into liquid media to a final concentration of 2 × 106 conidia per mL and allowed to germinate and grow for 10 h. The cultures were centrifuged and the pellets were fixed in 10% formalin solution for 30 min. The fixed mycelia were then washed with phosphate-buffered saline (PBS) for 2–3 times. After that, 5 μL of concentrated culture was mixed with calcofluor white and DAPI to a final concentration of 500 μg/L and 3 μg/mL, respectively, and stained for 10 min. Fluorescent microscopic observation was performed with a Zeiss ImagerA2 microscope equipped with a digital camera and DAPI Zeiss filter set was used for visualizing.
RNA Extraction and Gene Expression Analysis Using qRT-PCR
Samples for RNA extraction were prepared as previously described (Zhang et al., 2012) with some modifications. Briefly, conidia were first inoculated on plates containing Vogel’s liquid medium and grew for at least 24 h. The mycelial mat was then cut into fragments of about 2–4 mm2. About 10–12 such fragments were used to inoculate 150 ml flasks containing 100 ml liquid Vogel’s medium. The cultures were incubated at 28°C at 200 rpm. After growing for 13.5 h, antifungal agents were added and the cultures were further grown for 22 h. Since erg11 is essential, the inactivation strain and its parental strain were grown in liquid Vogel’s medium containing 50 μM BCS during the first 13.5 h and then transferred to two new 150 ml flasks which were either unamended (control) or supplemented 200 μM CuSO4, respectively. The control flasks, but not the 200 μM CuSO4 flask also contained 50 μM BCS to support growth.
After 22 h of drug treatment, mycelial samples were harvested, flash frozen in liquid nitrogen, and ground into fine powder. Total RNA extraction was performed according to the standard TRIzol protocol (Invitrogen, Carlsbad, CA, United States). The cDNAs were synthesized from 3 μg total RNA using a cDNA synthesis kit (FastQuant RT Kit (with gDNase); TIANGEN, China) according to the manufacturer’s protocol.
Gene-specific primers were designed using the online tools PrimerQuest or Primer 6 and are listed in Supplementary Table 3. qRT-PCR was performed on a CFX96 multicolor real-time PCR detection system (Bio-Rad, Hercules, CA, United States) with SYBR green detection (KAPA SYBR® FAST qPCR Kits; KAPA BIOSYSTEMS, Boston, MA, United States) according to the manufacturer’s instructions. Three independent experiments were carried out and each cDNA sample was analyzed at least in duplicate. The average threshold cycle (CT) values were used to calculate relative expression levels normalized to β-tubulin using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The significance of the differences between each two samples were evaluated by an unpaired two-tailed Student’s t-test using GraphPad Prism 7.0 and a p-value of less than 0.05 was considered significant.
ERG11 Detection by Western Blotting
For protein extraction, the collected mycelia were immediately frozen and then ground into fine powder for extraction. Proteins were extracted as previously reported (He et al., 2005) and quantified with the 2-D Quant Kit (GE, United States). Forty microgram protein of each sample was loaded for electrophoresis with SDS-PAGE and then transferred to a PVDF membrane (Merck Millipore, Germany). The tagged ERG11 protein level was detected using anti-myc antibodies (Abbkine, Redlands, CA, United States) using a standard protocol (Garceau et al., 1997).
HPLC-MS Analysis of Sterol Contents and Ketoconazole
For analyzing intracellular sterols, mycelial samples were heat-dried at 80°C and then ground into fine powder for extraction. Extraction and analysis were performed as previously described (Sun et al., 2013) using ergosterol (J&K Scientific Ltd.) and KTC standards.
For analyzing extracellular sterols, the cultured medium was extracted by chloroform after lyophilization. The extractions were then analyzed by HPLC-MS as described (Sun et al., 2013).
The significance of the differences between each two samples were evaluated by an unpaired two-tailed Student’s t-test using GraphPad Prism 7.0 and a p-value of less than 0.05 was considered as significant.
Results
Generation and Characterization of a Strain with Inducible erg11 Expression
In order to test whether transcriptional responses to azoles can be induced by disruption of ergosterol biosynthesis, we first used a genetic approach to impair the function of an essential enzyme in the pathway –sterol 14α-demethylase. To do so, we generated a strain, Ptcu-1::erg11, in which the native N. crassa erg11 promoter was replaced by the promoter of a copper responsive gene tcu-1 (NCU00830, encoding the Ctr-domain containing copper transporter). When controlled by this promoter, expression of the target gene is repressed by high copper concentration while addition of the copper chelator BCS can release the suppression by copper (Ouyang et al., 2015).
The Ptcu-1::erg11 phenotype was characterized and compared to its parental bd ku70RIP strain. As shown in Figure 2B, on Vogel’s plates containing 50 μM BCS or 100 μM CuSO4, the growth of the parental strain was not affected, similar to that on normal Vogel’s plates. The growth of Ptcu-1::erg11 was severely inhibited on normal Vogel’s plate which containing 1 μM Cu2+, and only a small visible colony was formed (Figure 2B). The formed colonies of Ptcu-1::erg11 on the Vogel’s plates were also examined microscopically and clustered hyphae were observed (Supplementary Figure 1A). When 100 μM CuSO4 were added to Vogel’s plates, no visible growth of Ptcu-1::erg11 was detected and only short germlings were observed under microscope (Figure 2B and Supplementary Figure 1A). When copper chelator BCS were added to Vogel’s plates, the growth of Ptcu-1::erg11 was indistinguishable from that of the parental strain. Similar results were observed on Vogel’s slants amended with 100 μM BCS or 250 μM CuSO4 (Figure 2B).
When cultured in liquid medium containing 100 μM CuSO4, conidia of Ptcu-1::erg11 geminated normally (Figure 2E). However, after about 2 rounds of nuclear division, hyphal tips started to swell and grew no further. Nonetheless, nuclear division was not arrested (Figure 2E). As a result, multiple nuclei clustered together at the swollen hyphal tips. In the medium amended with the Cu2+ chelator BCS, hyphal growth of Ptcu-1::erg11 was not arrested and nuclei were distributed evenly along the germ tubes (Figure 2E), as observed in the parental strain (Figure 2E). This result indicates that erg11 is required for hyphal elongation but not germination.
Since only the hyphal elongation was found to be affected in Ptcu-1::erg11 treated with Cu2+, the growth of the strains was examined by shifting the mycelial plugs from BCS to CuSO4 plates. As shown in Supplementary Figure 1B, after shifting to Vogel’s plates containing 100 μM CuSO4, the mycelium of Ptcu-1::erg11 strain grew for several hours and then ceased further colony growth, while the growth of parental strain was not affected. This result further verified that erg11 is required for hyphal elongation, although the mycelium could grow for a while, possibly due to the presence of sufficient quantities of erg11 mRNA or sterols within the mycelium to support growth.
To verify that sterol 14α-demethylase ERG11 is affected in the Ptcu-1::erg11 strain treated with CuSO4, the sterol profile of this strain treated with CuSO4 was compared to that of the strain treated with BCS. As shown in Figure 2D, CuSO4 treatment resulted in the reduction of ergosterol to a level of approximately 56% of that treated with BCS along with the accumulation of lanosterol, eburicol and 14α-methyl-3,6-diol, similar to that found in N. crassa treated with azoles (Chen et al., 2016). The transcription and protein levels of erg11 and ERG11 in Ptcu-1::erg11 in the presence of BCS and CuSO4, were also determined. In the presence of BCS, erg11 expression was as high as 50 folds as that of the parental strain and the protein was easily detected (Figure 3). Mycelium pre-grown in the medium with BCS was transferred to the medium with CuSO4. The growth of the Ptcu-1::erg11 strain became slower than the parental strain. After 22 h at the time for RNA analysis, its growth was less than 50% that of the parental strain. Its erg11 expression was twofold lower than the parental strain and the protein was undetectable (Figure 3).
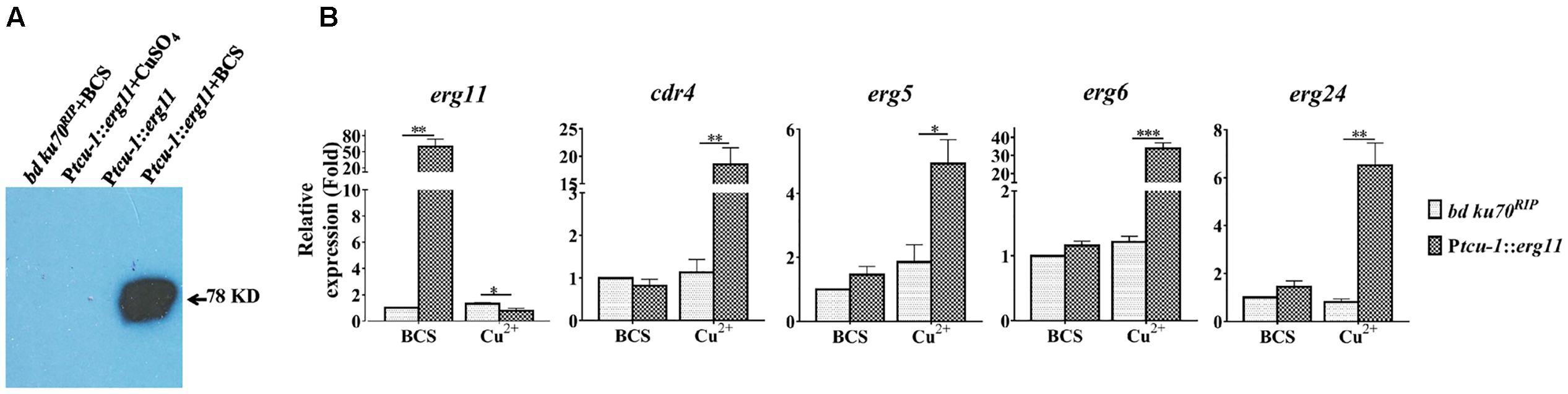
FIGURE 3. Transcriptional induction of cdr4 and erg genes following erg11 inactivation. (A) Detection of ERG11 (sterol 14α-demethylase) by Western Blotting in the Ptcu-1::erg11 strain grown in Vogel’s medium supplemented with Cu2+ or BCS using anti-myc antibodies. The parental strain (bd Ku70RIP) grown in Vogel’s medium supplemented with BCS and the Ptcu-1::erg11 strain grown in normal Vogel’s medium was used as controls. (B) After grown in medium containing BCS for 13.5 h, mycelium were then shifted to indicated conditions to grow for additional 22 h. The transcript levels of selected genes were examined in the N. crassa bd Ku70RIP strain and Ptcu-1::erg11 strain treated with 50 μM BCS and 100 μM CuSO4 for 22 h. Transcript levels of cdr4 (NCU05591, encoding azole efflux pump CDR4), erg5 (NCU05278, encoding C-22 sterol desaturase), erg6 (NCU03006, encoding sterol C-24 methyl transferase), erg11 (NCU02624, encoding sterol 14α-demethylase) and erg24 (NCU08762, encoding C-14 sterol reductase) were measured by quantitative real-time polymerase chain reaction (qRT-PCR), and the expression was calculated by 2-ΔΔCt method and normalized to β-tubulin. The results presented here are means of three biological replicates, and the significant levels between the two strains in each treatment were calculated by t-test and marked as ∗(p < 0.05), ∗∗(p < 0.01) or ∗∗∗(p < 0.001).
All the phenotypic features described above indicate that erg11 is an essential gene in N. crassa. To further verify that the observed effects were all due to the introduction of the inducible erg11 cassette, Ptcu-1::erg11 was complemented by replacing the Ptcu-1 promoter with the native erg11 promoter. Since Ptcu-1::erg11 could be inhibited by excess Cu2+, the transformants were screened on the medium containing 250 μM CuSO4 and verified by PCR. The complemented strains restored wild-type characteristics to the Ptcu-1::erg11 strain (Figures 2B,C and Supplementary Figure 1), verifying the essential roles of erg11 for fungal growth.
Inactivation of erg11 Causes Transcriptional Responses Similar to Those Following Ketoconazole Treatment
To determine the relationship between the transcriptional response by the efflux-pump CDR4 encoding gene and ergosterol biosynthesis inhibition, we examined changes in cdr4 expression when erg11 was inactivated. In addition, we monitored the transcriptional changes of several N. crassa genes for ergosterol biosynthesis whose responses to azoles have been previously described (Sun et al., 2013). We compared the transcriptional responses between the Ptcu-1::erg11 strain and its parental counterpart when cultured in Vogel’s medium with BCS or CuSO4. When compared to the parental strain, BCS amendment did not affect most of the tested genes except for erg11 in the Ptcu-1::erg11 strain. The presence of CuSO4 did not alter the abundance of any of the tested gene in the parental strain. However, down-regulation was observed in erg11 expression measured in the Ptcu-1::erg11 strain cultured in the presence of the CuSO4 amendment, indicating that suppression of transcription of the gene regulated by the tcu-1 promoter was functional (Figure 3B, left panel). However, a marked difference in transcript abundance of other tested genes was observed between the two strains grown in the presence of CuSO4. Transcript levels of cdr4 as well as those of ergosterol biosynthesis genes were all significantly higher in the Ptcu-1::erg11 strain, in spite of the lack of KTC in the medium. While the growth of Ptcu-1::erg11 was inhibited after CuSO4 was added to the medium, the mRNA levels of cdr4, erg5 (NCU05278, encoding sterol C-22 desaturase), erg24 (NCU08762, encoding sterol C-14 reductase) and erg6 (NCU03006, encoding sterol C-24 methyltransferase) were dramatically induced, by at least 28, 5, 6.5, and 33-fold, respectively, in a manner similar to the levels of induction imposed by the KTC treatment (Figure 3B). Hence, the presence of the drug was not a prerequisite for the transcriptional response observed.
Ergosterol Depletion Is Not the Cause for Activating Stress Response
In order to determine whether the transcriptional responses are caused by ergosterol depletion in N. crassa, similar to that of other fungi, we took advantage of the available Δerg3 (NCU06207, encoding sterol C-5 desaturase), Δerg4 [NCU01333, encoding sterol C-24(28) reductase] and Δerg5 strains and produced another Δerg2 strain (Table 1). These genes encode enzymes involved in last few steps of ergosterol biosynthesis, and deletion or mutation in any of these genes results in ergosterol depletion (Grindle and Farrow, 1978; Sun et al., 2013; Weichert et al., 2016). All the mentioned gene deletion mutants are viable (Figure 4A), similarly to that described in S. cerevisiae (Lees et al., 1995). We first determined the extent of ergosterol deficiency in these mutants by sterol profiling. Similar as wild type, ergosterol was easily detected in Δerg3 strain, possibly due to the present of a second copy of erg3 (Weichert et al., 2016). While in the Δerg2, Δerg4 and Δerg5 strains, no ergosterol were detected (Figure 4B). Rather, ergosta-5,7,22,24(28)-tetraenol was accumulated in the Δerg4 strain and ergosta-5,7,24(28)-trienol and ergosta-5,7-dienol were accumulated in the Δerg5 strain when compared to wild type (Figure 4B and Supplementary Table 4). In the Δerg2 strain, ergosta-5,8,22-trienol and fecosterol were accumulated (Supplementary Table 4).
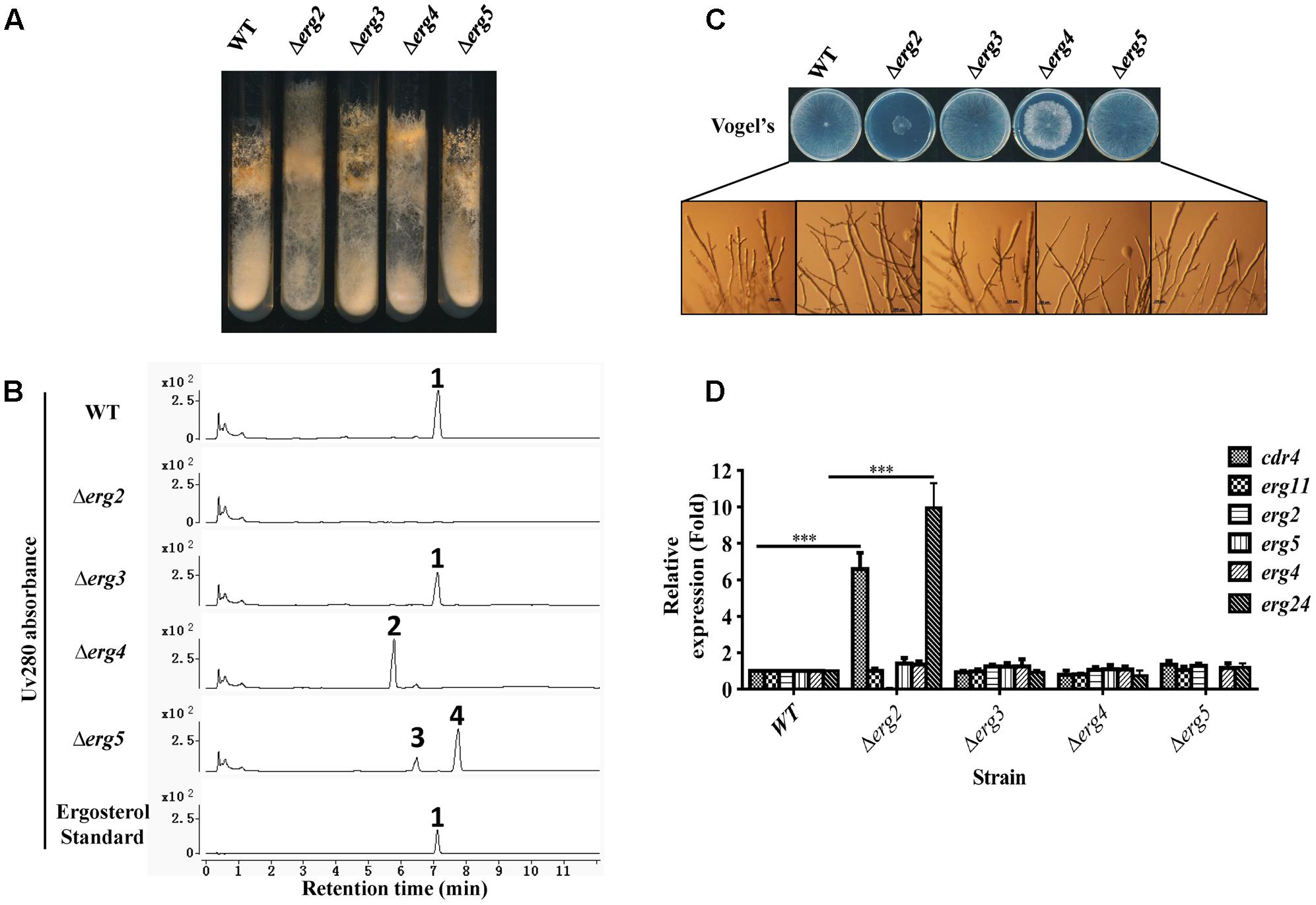
FIGURE 4. Phenotypic analysis and gene expression analysis of ergosterol deficient mutants. Conidia of N. crassa wild type, Δerg2 (NCU04156, encoding C-8 sterol isomerase), Δerg3 (NCU06207, encoding C-5 sterol desaturase), Δerg4 (NCU01333, encoding C-24(28) sterol reductase), and Δerg5 (NCU05278, encoding C-22 sterol desaturase) mutants were inoculated on Vogel’s slants (A) and Vogel’s plates (C) and grown at 28°C for 7 days (A) or 30 h (C), respectively. (B) Major sterols in ergosterol deficient mutants. Mycelium grown in liquid Vogel’s medium for 36 h was used for sterol extraction. The extracted sterols were then subjected to HPLC-MS analysis. Ergosterol was identified by UV absorbance at 280 nm and referred to an ergosterol standard. Other sterols accumulated were further checked by MS and listed in Supplementary Table 4. The sterols marked with numbers are ergosterol for 1, ergosta-5,7,22,24(28)-tetraenol for 2, ergosta-5,7,24(28)-trienol for 3, ergosta-5,7-dienol for 4. (D) Transcript levels of cdr4 (NCU05591, encoding azole efflux pump CDR4), erg2, erg5, erg6 (NCU03006, encoding sterol C-24 methyl transferase), erg11 (NCU02624, encoding sterol 14α-demethylase) and erg24 (NCU08762, encoding C-14 sterol reductase) were measured by quantitative real-time polymerase chain reaction (qRT-PCR) in the selected mutants. The expression was calculated by 2-ΔΔCt method and normalized to β-tubulin. The results presented here are means of three biological replicates, and the significant levels of cdr4 and erg24 between WT and Δerg2 were calculated by t-test and marked as ∗(p < 0.05), ∗∗(p < 0.01) or ∗∗∗(p < 0.001).
When cultured in slants, a marked reduction in conidial abundance was observed in the Δerg2 strain. Furthermore, aerial hyphae grew higher along the slant sides and in a more compact manner (Figure 4A). The Δerg2 strain grew extremely slow on Vogel’s plates, exhibiting a growth of about 25% of wild type (Figure 4C). A few more branches at the hyphal tips of this mutant were also observed (Figure 4C). Deletion of erg4 also resulted in slow hyphal growth, albeit in a less severe manner than Δerg2, as this strain grew approximately 80% of wild type, yet its colony morphology was more compact and fluffy in appearance (Figure 4C). Thus, deletions in two of the analyzed genes encoding enzymes catalyzing last steps in ergosterol biosynthesis resulted in growth defects, while deletion of the others (erg3 and erg5) did not.
The expression of azole responsive genes including cdr4, erg11, erg2, erg5, erg6 and erg24 were then examined in these mutants by qRT-PCR. Interestingly, in a Δerg2 background, expression of cdr4 and erg24 were increased by 7 and 10-folds, respectively, while other genes tested were not affected (Figure 4D). This provides additional evidence that cdr4 responds to ergosterol biosynthesis inhibition. However, no change in gene expression was evident in the mutants of erg3, erg4 and erg5, which are downstream of erg2 (Figure 4D). Thus, the stress responses to azoles by cdr4 and ergosterol biosynthesis genes are not activated by ergosterol depletion.
Differential Responses to Ergosterol Biosynthesis Inhibition in N. crassa
Above results indicate that inhibition at different steps in ergosterol biosynthesis, such as deletion of erg2 or inactivation of erg11, could cause transcriptional responses by different genes. To further see transcriptional responses upon inhibition at more steps in ergosterol biosynthesis, we examined the transcriptional response of several genes to three different sterol biosynthesis inhibitors, KTC, AMOR and TERB. Different from azoles, TERB targets squalene epoxidase ERG1 (NCU08280), an enzyme upstream of sterol 14α-demethylase ERG11, while AMOR targets two enzymes downstream of ERG11 in the ergosterol biosynthetic pathway, sterol C8-C7 isomerase ERG2 and C-14 sterol reductase ERG24 (Lupetti et al., 2002) (Figure 1).
As cdr4 expression can be induced by inactivation of erg11 or deletion of erg2, it was not surprising that AMOR or KTC treatments led to an almost 5- and 13-fold increase in cdr4 expression, respectively (Figure 5), verifying that sterol inhibition does, in fact, induce cdr4 transcription. TERB had no effect on cdr4 expression (Supplementary Figure 2), suggesting that this pump may not be involved in expulsion of the compound from the cell, or, the inhibition of ergosterol biosynthesis by TERB cannot induce the expression of this pump.
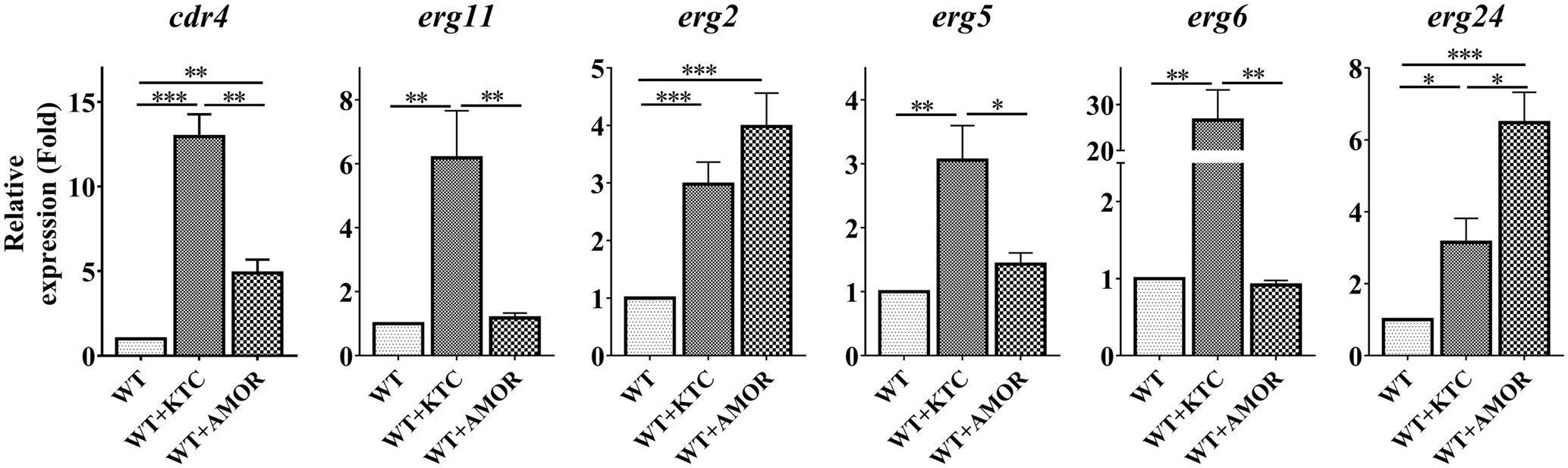
FIGURE 5. Differential transcriptional responses to different ergosterol biosynthesis inhibitors. After grown in liquid media for 13.5 h, the N. crassa wild-type strain were treated with 2 mg/L ketoconazole and 0.375 mg/L Amorolfine for 22 h, same amount dissolvent treatment was used as a control. Transcript levels of cdr4 (NCU05591, encoding azole efflux pump CDR4), erg2 (NCU04156, encoding C-8 sterol isomerase), erg5 (NCU05278, encoding C-22 sterol desaturase), erg6 (NCU03006, encoding sterol C-24 methyl transferase), erg11 (NCU02624, encoding sterol 14α-demethylase) and erg24 (NCU08762, encoding C-14 sterol reductase) were measured by quantitative real-time polymerase chain reaction (qRT-PCR), and the expression was calculated by the 2-ΔΔCt method and normalized to β-tubulin. The results presented here are means of three biological replicates. The significant levels were calculated by t-test and marked as ∗(p < 0.05), ∗∗(p < 0.01) or ∗∗∗(p < 0.001).
We also analyzed the expression of several genes in ergosterol biosynthesis pathway, as affected by these antifungal drugs. As demonstrated above, transcription of these genes was induced by KTC (Figure 5). Similar to that observed in the Δerg2 strain, AMOR treatment only induced erg24 by 6 folds but not other erg genes (Figure 5). AMOR treatment also resulted in 4-fold induction of erg2, the gene encoding one of the AMOR targets. Similar to that observed in the case of cdr4, no significant transcriptional responses of these tested erg genes to TERB were detected (Supplementary Figure 2). These results together indicate the responsive genes are different upon inhibition at different steps of the ergosterol biosynthesis pathway.
We then examined the sterol content changes in N. crassa treated with different ergosterol biosynthesis inhibitors. In the presence of all the inhibitors tested, ergosterol levels were reduced by at least two folds, excluding TERB which resulted in ergosterol depletion to only 80% of wild-type levels (Supplementary Figure 3 and Supplementary Table 4). Along with the depletion of ergosterol, KTC treatment resulted in accumulation of lanosterol, eburicol and 14α-methyl-3,6-diol, AMOR treatment resulted in accumulation of ignosterol and ergosta-5,8,22-trienol, while TERB treatment resulted in accumulation of squalene and an unknown compound (Supplementary Table 4). Thus, inhibition at different steps in ergosterol biosynthesis pathway resulted in accumulation of different sterol intermediates in addition to depletion of ergosterol.
Stress Response Is Associated with Accumulation of Sterol Intermediates in N. crassa
Based on above results, it is possible that the responses by different genes could be associated with accumulation of different sterol intermediates. If this is the case, when the accumulated intermediates are reduced or other intermediates are additionally accumulated, the transcriptional responses caused may be reduced or changed.
To examine the possibility, we treated the Δerg2 strain with KTC (Figure 1). The sterol profiles were firstly compared between the two strains with or without KTC. When treated with KTC, the accumulation of lanosterol, eburicol and 14α-methyl-3,6-diol along with the depletion of ergosterol was observed in wild type (Supplementary Figure 4 and Supplementary Table 4). Deletion of erg2 resulted in the accumulation of ergosta-5,8,22-trienol as its major sterol, while KTC treatment of this strain also lead to accumulation of lanosterol, eburicol and 14α-methyl-3,6-diol along with the depletion of ergosta-5,8, 22-trienol (Supplementary Figure 4 and Supplementary Table 4). Overall, KTC treatment in the Δerg2 strain could result in the depletion of sterol intermediates accumulated normally in the Δerg2 strain and the accumulation of sterol intermediates resulted from azole inhibition.
We then measured the expression of cdr4 and erg24 which respond to erg2 deletion, erg11 inactivation and corresponding antifungals. The expression of three additional genes, erg11, erg5 and erg6 which respond to only erg11 inactivation or KTC inhibition, was also analyzed. As shown in Figure 6, transcription of cdr4 was induced in a Δerg2 background. Yet when KTC was applied, the quantitative difference between the transcriptional responses of wild type and the mutant was minimal. Thus, it appears that the response of wild type to the drug is not dependent on a functional erg2. The expression of erg24 in the Δerg2 mutant treated with KTC was reduced when compared to that in the Δerg2 mutant (nine folds) and similar to that observed in wild type treated with the drug. The expression of erg5, erg6 and erg11 in the Δerg2 mutant treated with KTC also showed a similar response to KTC induction on its own, which is very different from that in the non-treated Δerg2 strain. All the results indicate that the transcriptional responses by cdr4 and genes in ergosterol biosynthesis pathway are associated with accumulation of sterol intermediates.
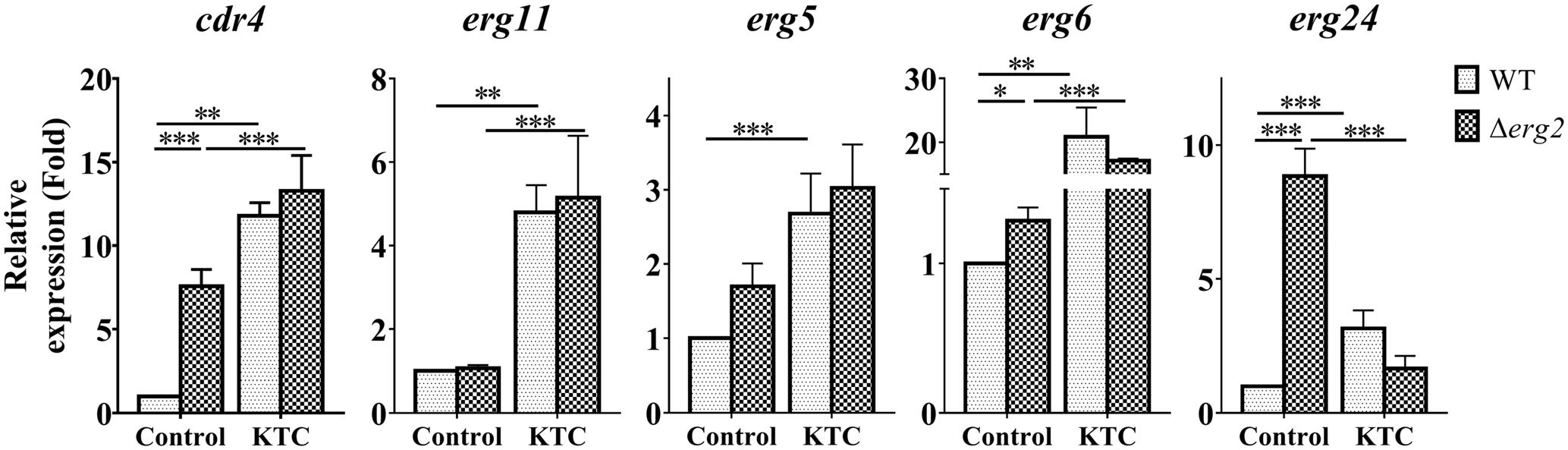
FIGURE 6. ERG11 inhibition overrides the effects of erg2 deletion. After grown in liquid media for 13.5 h, the N. crassa wild-type and Δerg2 strains were treated with 2 mg/L ketoconazole for 22 h and same amount DMSO was used as a control. Transcript levels of cdr4 (NCU05591, encoding azole efflux pump CDR4), erg5 (NCU05278, encoding C-22 sterol desaturase), erg6 (NCU03006, encoding sterol C-24 methyl transferase), erg11 (NCU02624, encoding sterol 14α-demethylase) and erg24 (NCU08762, encoding C-14 sterol reductase) were measured by quantitative real-time polymerase chain reaction (qRT-PCR), and the expression was calculated by 2-ΔΔCt method and normalized to β-tubulin. The results presented here are means of three biological replicates. The significant levels were calculated by t-test and marked as ∗(p < 0.05), ∗∗(p < 0.01), or ∗∗∗(p < 0.001).
Deletion of the N. crassa CDR4 Efflux Pump Leads to Intracellular, But Not Extracellular, Sterol Intermediate Accumulation Following KTC Treatment
During the course of this study we have found that even though cdr4 expression can be induced by KTC and AMOR, when erg11 expression is repressed or erg2 is deleted, cdr4 expression is significantly increased, even in the absence of the sterol biosynthesis inhibitor. Since the CDR4 homologs in S. cerevisiae and C. albicans have been shown capable of transporting steroids (Kolaczkowski et al., 1996; Krishnamurthy et al., 1998), it is possible that N. crassa CDR4 may also function as a transporter of sterol intermediates. As the first step toward analyzing the possibility, we examined whether the expression of cdr4 is related to the accumulation of sterols. As inhibition or inactivation of erg11 causes accumulation of lanosterol, eburicol and the toxic sterol 14α-methyl-3,6-diol (Watson et al., 1989; Chen et al., 2016) (Supplementary Figure 4 and Supplementary Table 4), we monitored erg11 and cdr4 expression along a 24-h period after amending the growth medium with KTC. As can be seen from the results of the time course study, the expression of cdr4 was induced 3 h after KTC treatment while erg11 expression was induced within 1 h in the presence of the drug (Figure 7). This suggests that the accumulation of the excess sterols occurs before the onset of pump activity.
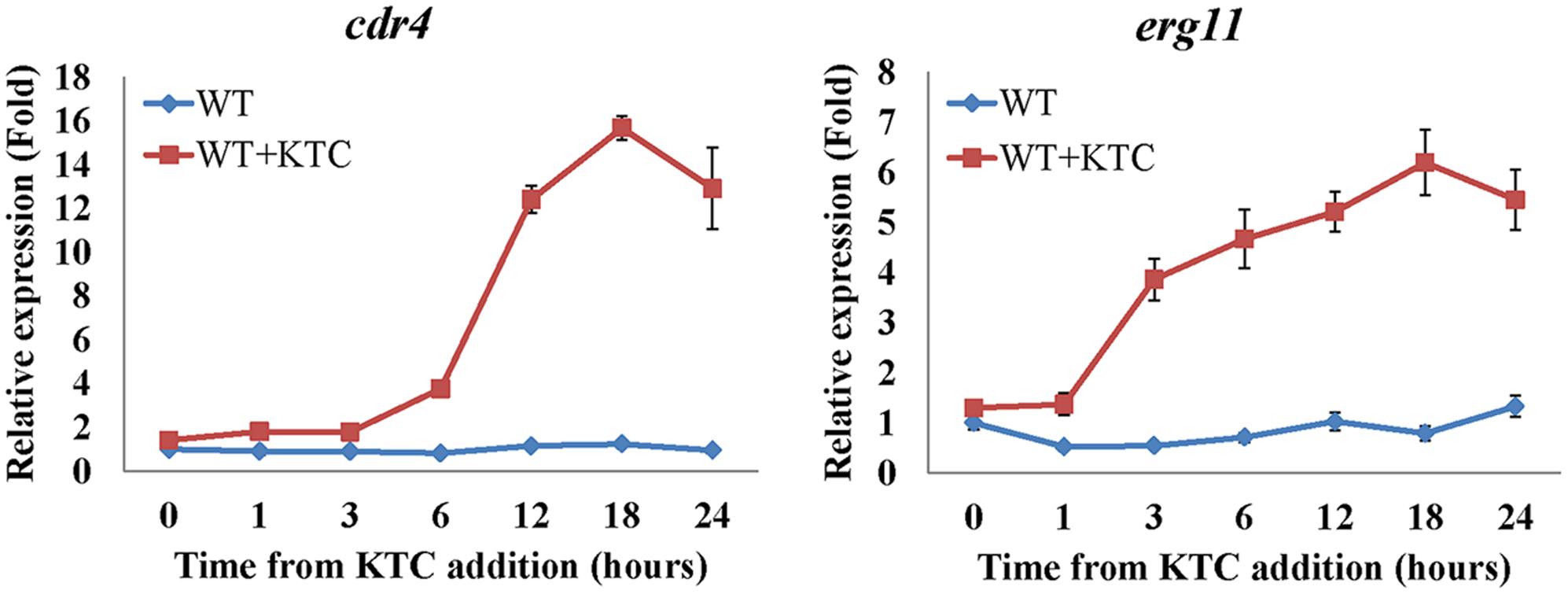
FIGURE 7. Transcriptional response of cdr4 and erg11 to 2 mg/L ketoconazole (KTC). After grown in liquid media for 13.5 h, KTC was added and grew for another 24 h. The gene expression of cdr4 (NCU05591, encoding azole efflux pump CDR4) and erg11 (NCU02624, encoding sterol 14α-demethylase) were monitored at various time points following KTC treatment. Transcript levels were measured by quantitative real-time polymerase chain reaction (qRT-PCR), and the expression was calculated with 2-ΔΔCt method and normalized to β-tubulin.
Do sterol intermediates accumulate in the Δcdr4 strain? To determine whether this is the case, we used HPLC-MS to analyze the sterol content in the cdr4 deletion mutant with or without KTC treatment. Deletion of cdr4 in N. crassa resulted in hypersensitive to azoles (Zhang et al., 2012). We firstly determined whether deletion of cdr4 would result in the accumulation of KTC. The accumulation of KTC in the Δcdr4 strain was 2.4-fold of that in wild type (Figure 8A), confirming CDR4 is the transporter of azoles, functionally the same to its ortholog Pdr5p in S. cerevisiae. As shown in Figure 8B, under normal condition, deletion of cdr4 had no obvious effect on the amount of intracellular sterols. In wild type, KTC treatment resulted in a reduction of total ergosterol content and the abnormal accumulation of sterols along the biosynthetic pathway (Figure 8B). In the Δcdr4 strain, a much more pronounced reduction (about threefolds) in total ergosterol content was evident, following KTC treatment (Figure 8B). This was accompanied by the increased accumulation of lanosterol, eburicol and the toxic sterol 14α-methyl-3,6-diol, which were 1.6-, 1.7- and 2.8-folds of that in wild type, respectively (Figure 8B).
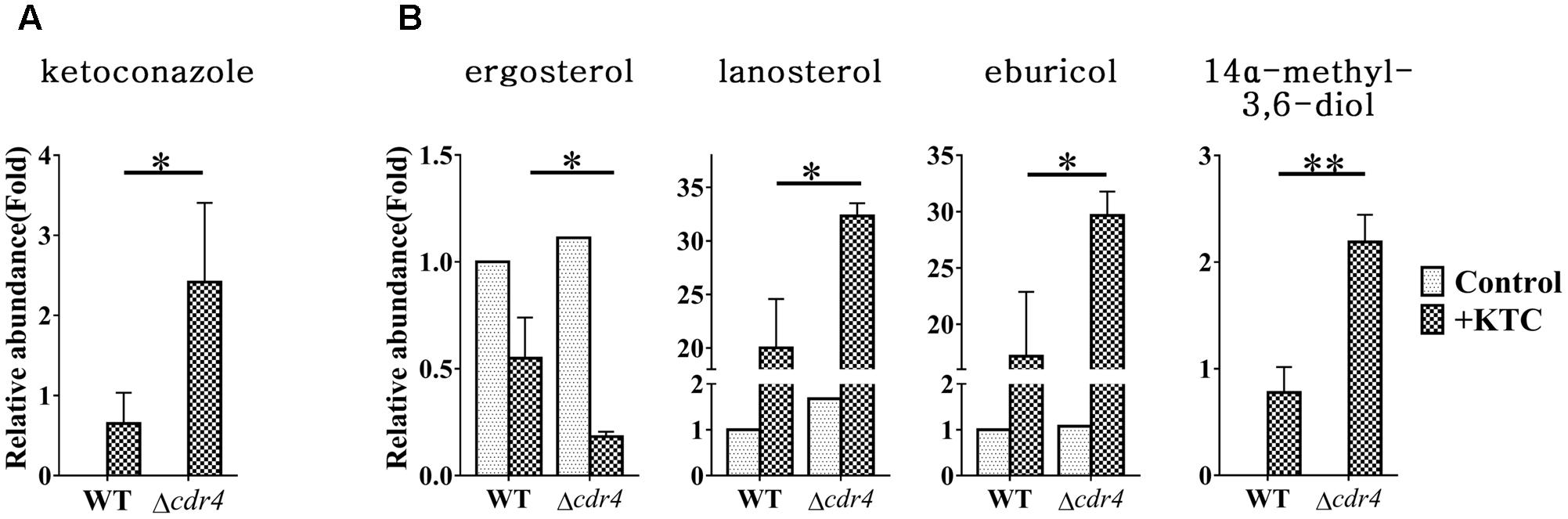
FIGURE 8. Sterol content between the N. crassa wild-type and Δcdr4 strain. After grown in liquid media for 13.5 h, the N. crassa wild-type and Δcdr4 strains were treated with 2 mg/L ketoconazole for 22 h and same amount DMSO was used as a control. KTC (A) and Sterols (B) were then extracted and measured by HPLC-MS. The abundance of sterols was calculated on the basis of the chromatogram peak area and normalized using an internal control and sample weight. The results presented here are means of three biological replicates. The significant levels were calculated by t-test and marked as ∗(p < 0.05), ∗∗(p < 0.01), or ∗∗∗(p < 0.001).
To examine whether sterol intermediates can be exported via CDR4, the sterols in the cultured medium were also analyzed and compared between wild type and the Δcdr4 strain. Neither free ergosterol, nor lanosterol, eburicol or 14α-methyl-3,6-diol was detected in the media of any of the treatments. Moreover, other sterols, such as sterol acetate, were also not detected. Thus, the intracellular accumulation of sterols in Δcdr4 mutant is not due to reduced efflux of these sterols.
Discussion
Transcriptional responses by the systems of efflux pumps and ergosterol homeostasis confer fungi the basal resistance to antifungal azoles (Cools et al., 2013; Paul and Moye-Rowley, 2014). Many studies suggest there is a linkage between the two systems in antifungal stress responses, but detailed evidence and investigation are required. Our results in this study showed that genetic disruption of erg2 and erg11, two genes encoding the targets of ergosterol biosynthesis inhibitor amorolfine and azoles, respectively, activated the expression of azole efflux pump coding gene cdr4, indicating cdr4 can make response to defects in ergosterol biosynthesis in absence of any ergosterol biosynthesis inhibitor. Azole-caused disruption in ergosterol biosynthesis should be an inductive factor for transcriptional response by cdr4 to azole stress. Thus, this study reveals the linkage between the two systems in stress responses to azoles. It was reported that the expression of yeast azole efflux pumps could be activated directly by various drugs including azoles (Thakur et al., 2008). This study adds another mechanism to transcriptional activation of azole efflux pumps: indirect activation by azole-caused defects in ergosterol biosynthesis. Our results also showed that disruption at different steps in ergosterol biosynthesis had different effects and resulted in difference in sterol profiles. Interestingly, the transcriptional responses to disruption at different steps in ergosterol biosynthesis were also different, suggesting a correlation between the sterol profiles and transcriptional responses. Moreover, we found that abnormal ergosterol biosynthesis, causing accumulation of sterol intermediates but not ergosterol depletion, induced transcriptional responses by azole efflux pump coding gene cdr4 and genes for ergosterol biosynthesis, providing a novel understanding to the mechanisms of stress responses to ergosterol biosynthesis inhibitors. Further studies by genome-wide transcriptional profiling might provide a comprehensive view on the responses and give more insights in how fungi cope with ergosterol biosynthesis inhibitors.
Our results clearly revealed that cdr4 transcriptionally responds to defects in ergosterol biosynthesis. In Kluyveromyces lactis, a fungus more closely related to S. cerevisiae than to N. crassa, deletion of ERG6 (encoding C-24 sterol methyl transferase) can also activate the expression of the CDR4 homolog (Konecna et al., 2016), providing another supporting evidence to the transcriptional activation induced by defects in ergosterol biosynthesis. Thus, the transcriptional induction of cdr4 in N. crassa by compounds, such as xanthone derivatives, might be caused by their inhibition on ergosterol biosynthesis (Pinto et al., 2011; Pedro Gonçalves et al., 2015). Interestingly, DTT and avicel, which can induce ER stress, also activate the expression of cdr4 in N. crassa (Coradetti et al., 2013; Fan et al., 2015). Since transcriptional induction of cdr4 by DTT or avicel was shown to be independent on the canonical unfolded protein response (UPR) pathway that copes with ER stress (Fan et al., 2015), the transcriptional induction of cdr4 might also be caused by inhibition on ergosterol biosynthesis. How efflux pumps and ergosterol biosynthesis link each other remains elusive. One possibility is that inhibition of ergosterol biosynthesis affects the function of the efflux pumps. In C. albicans, Cdr1p, the homolog of CDR4, prefers to be localized in lipid rafts which enrich in sterols and phospholipids (Pasrija et al., 2008). Thus, inhibition of ergosterol biosynthesis may disrupt the localization of efflux pumps and affect their function. In yeast, functional impairment by deletion of multiple efflux pumps, including CDR4 homolog Pdr5p, resulted in activation of the regulatory network of efflux pumps (Khakhina et al., 2015). Therefore, the activation of efflux pumps by ergosterol biosynthesis inhibitors might be a compensation effect of its functional loss caused by alteration in the sterol composition. Our results showed that accumulation of specific sterols but not ergosterol depletion was associated with the transcriptional response by cdr4 (Figure 1). Although different sterols accumulated, those associated with the transcriptional induction of cdr4 were sterols impaired in Δ8 to Δ7 isomerization (Figure 1). The role of this structure in the formation of lipid rafts is unknown, but defects in enzymes catalyzing this steps cause severe cell membrane associated impairment (Hannich et al., 2011). How this kind of sterols is associated with transcriptional responses by cdr4 to ergosterol biosynthesis inhibitors needs further elucidation.
Genes in ergosterol biosynthesis are well known to respond to impairment in ergosterol biosynthesis. Genetic disruption of ERG2, ERG5, and ERG6 in yeast can induce transcriptional responses by genes for ergosterol biosynthesis (Bammert and Fostel, 2000; Konecna et al., 2016). Ergosterol biosynthesis inhibitors that target Erg11, Erg24, Erg2 and Erg1 can also induce the responses (Bammert and Fostel, 2000; Henry et al., 2000). Responses to disruption at other steps in ergosterol biosynthesis were previously unknown. Surprisingly, we found that genetic disruption of erg4 and erg5 in N. crassa did not cause transcriptional response by any of the tested genes in ergosterol biosynthesis, although ergosterol formation was completely abolished. In addition, genes, responding to impairments in several specific steps in ergosterol biosynthesis, were different. The difference in responsive genes to different inhibitions on ergosterol biosynthesis was also found in Trichophyton rubrum, in which ketoconazole (KTC) upregulated most of the erg genes, such as genes encoding C-24 sterol methyl transferase Erg6, sterol 14α-demethylase Erg11 and C-14 sterol reductase Erg24, while TERB downregulated several erg genes and did not affect the expression of gene encoding sterol 14α-demethylase (Yu et al., 2007; Zhang et al., 2009). These results indicate the diversity in the responses to ergosterol biosynthesis inhibition among different fungi. A possible explanation for this is that, in yeast and C. albicans, ergosterol depletion alone can induce the responses. In N. crassa, depletion alone might be not sufficient to activate the responses. Induction of the responses might require more severe defects in the sterol profile, for example, accumulation of certain sterol intermediates. In humans, lanosterol accumulation promotes the degradation of HMG CoA reductase, an enzyme upstream of cholesterol biosynthesis (Song et al., 2005). Several studies have also proposed that abnormal sterol intermediates are responsible for the fungitoxicity of sterol biosynthesis inhibitors (Debieu et al., 1998). In N. crassa, accumulation of specific intermediates resulted in specific phenotypic outcomes (Weichert et al., 2016). However, it is not known which sterol intermediates may involve in the transcriptional responses. Based on our results, we proposed that 14-methyl sterols are associated with transcriptional responses to azoles (Figure 1). It is unclear whether different sterol intermediates can also result in different transcriptional responses in yeast, but exogenous addition of sterols with different structural features into the medium exerted different supporting effects on the growth of anaerobically cultured S. cerevisiae (Nes et al., 1978). Moreover, distinct defects are found in different erg mutants in yeast (Hannich et al., 2011). Thus, lack or aberrant accumulation of different ergosterol intermediates should cause the different functional defects in fungi and induce different transcriptional responses. However, how different intermediates activate transcriptional responses need further elucidation.
In summary, firstly, our data provide a novel explanation for how ergosterol biosynthesis inhibitors activate expression of efflux pumps: the responses of efflux pumps under drug stress are associated with the accumulation of sterol intermediate(s) resulting from defects in sterol 14α-demethylase ERG11 and sterol C-8 isomerase ERG2. However, why and how the efflux pumps are activated by sterol intermediates requires further elucidation. Secondly, our data also demonstrate that stress induction of ergosterol biosynthesis genes in N. crassa is also linked with sterol intermediates. This is markedly different from those observed in several other fungi, where genes for ergosterol biosynthesis are induced by ergosterol depletion, indicating different regulatory mechanisms among fungi. Such differences among fungi, especially between yeast and filamentous fungi, have been previously observed in several hierarchical and feedback systems, such as cell polarity, carbon catabolite repression and nitrogen regulation (Gorovits and Yarden, 2003; Harris, 2006; Wong et al., 2008; Ziv et al., 2008). Thus, in spite of similarities in the machinery involved, we demonstrate that the regulation and physiological outcomes in N. crassa are different from those observed in yeast and several other fungi and thus provide a novel explanation for the described aspect of antifungal stress response.
Author Contributions
SL, CH, and OY designed the study and wrote the manuscript. CH and MZ performed the main experiments. WW performed the analysis of sterols. CH, XS, OY, and SL contributed to the data analysis and the data interpretation.
Funding
This research was supported by a grant jointly supported by National Natural Science Foundation of China (with grant no. 31461143002 to SL) and the Israel Science Foundation (with grant no. 499/14 to OY).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00009/full#supplementary-material
References
Agarwal, A. K., Rogers, P. D., Baerson, S. R., Jacob, M. R., Barker, K. S., Cleary, J. D., et al. (2003). Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278, 34998–35015. doi: 10.1074/jbc.M306291200
Bammert, G. F., and Fostel, J. M. (2000). Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44, 1255–1265. doi: 10.1128/AAC.44.5.1255-1265.2000
Bien, C. M., and Espenshade, P. J. (2010). Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell 9, 352–359. doi: 10.1128/ec.00358-09
Blair, J. M. A., Richmond, G. E., and Piddock, L. J. V. (2014). Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 9, 1165–1177. doi: 10.2217/fmb.14.66
Brown, G. D., Denning, D. W., Gow, N. A. R., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv113. doi: 10.1126/scitranslmed.3004404
Chen, X., Xue, W., Zhou, J., Zhang, Z., Wei, S., Liu, X., et al. (2016). De-repression of CSP-1 activates adaptive responses to antifungal azoles. Sci. Rep. 6:19447. doi: 10.1038/srep19447
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., et al. (2006). A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357. doi: 10.1073/pnas.0601456103
Cools, H. J., Hawkins, N. J., and Fraaije, B. A. (2013). Constraints on the evolution of azole resistance in plant pathogenic fungi. Plant Pathol. 62, 36–42. doi: 10.1111/ppa.12128
Coradetti, S. T., Xiong, Y., and Glass, N. L. (2013). Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2, 595–609. doi: 10.1002/mbo3.94
Coste, A., Turner, V., Ischer, F., Morschhäuser, J., Forche, A., Selmecki, A., et al. (2006). A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172, 2139–2156. doi: 10.1534/genetics.105.054767
Coste, A. T., Karababa, M., Ischer, F., Bille, J., and Sanglard, D. (2004). Tac1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3, 1639–1652. doi: 10.1128/ec.3.6.1639-1652.2004
Cowen, L. E., Sanglard, D., Howard, S. J., Rogers, P. D., and Perlin, D. S. (2014). Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 5:a019752. doi: 10.1101/cshperspect.a019752
Davis, R. H., and de Serres, F. J. (1970). Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17, 79–143. doi: 10.1016/0076-6879(71)17168-6
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Debieu, D., Bach, J., Lasseron, A., Malosse, C., and Leroux, P. (1998). Effects of sterol biosynthesis inhibitor fungicides in the phytopathogenic fungus, Nectria haematococca: ergosterol depletion versus precursor or abnormal sterol accumulation as the mechanism of fungitoxicity. Pestic. Sci. 54, 157–167. doi: 10.1002/(SICI)1096-9063(1998100)54:2<157::AID-PS799>3.0.CO;2-Y
Dunkel, N., Liu, T. T., Barker, K. S., Homayouni, R., Morschhäuser, J., and Rogers, P. D. (2008). A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7, 1180–1190. doi: 10.1128/ec.00103-08
Fan, F., Ma, G., Li, J., Liu, Q., Benz, J. P., Tian, C., et al. (2015). Genome-wide analysis of the endoplasmic reticulum stress response during lignocellulase production in Neurospora crassa. Biotechnol. Biofuels 8, 1–17. doi: 10.1186/s13068-015-0248-5
Ferreira, M., Malavazi, I., Savoldi, M., Brakhage, A., Goldman, M., Kim, H. S., et al. (2006). Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 50, 32–44. doi: 10.1007/s00294-006-0073-2
Florio, A., Ferrari, S., De Carolis, E., Torelli, R., Fadda, G., Sanguinetti, M., et al. (2011). Genome-wide expression profiling of the response to short-term exposure to fluconazole in Cryptococcus neoformans serotype A. BMC Microbiol. 11:97. doi: 10.1186/1471-2180-11-97
Flowers, S. A., Barker, K. S., Berkow, E. L., Toner, G., Chadwick, S. G., Gygax, S. E., et al. (2012). Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 11, 1289–1299. doi: 10.1128/ec.00215-12
Garceau, N. Y., Liu, Y., Loros, J. J., and Dunlap, J. C. (1997). Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89, 469–476. doi: 10.1016/S0092-8674(00)80227-5
Golin, J., Ambudkar, S. V., and May, L. (2007). The yeast Pdr5p multidrug transporter: How does it recognize so many substrates? Biochem. Biophys. Res. Commun. 356, 1–5. doi: 10.1016/j.bbrc.2007.02.011
Gorovits, R., and Yarden, O. (2003). Environmental suppression of Neurospora crassa cot-1 hyperbranching: a link between COT1 kinase and stress sensing. Eukaryot. Cell 2, 699–707. doi: 10.1128/ec.2.4.699-707.2003
Grindle, M., and Farrow, R. (1978). Sterol content and enzyme defects of nystatin-resistant mutants of Neurospora crassa. Mol. Gen. Genet. 165, 305–308. doi: 10.1007/bf00332531
Hannich, J. T., Umebayashi, K., and Riezman, H. (2011). Distribution and functions of sterols and sphingolipids. Cold Spring Harb. Perspect. Biol. 3:a004762. doi: 10.1101/cshperspect.a004762
Harris, S. D. (2006). Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 251, 41–77. doi: 10.1016/S0074-7696(06)51002-2
He, Q., Cha, J., He, Q., Lee, H.-C., Yang, Y., and Liu, Y. (2006). CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20, 2552–2565. doi: 10.1101/gad.1463506
He, Q., Cheng, P., He, Q., and Liu, Y. (2005). The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19, 1518–1531. doi: 10.1101/gad.1322205
Heilmann, C. J., Schneider, S., Barker, K. S., Rogers, P. D., and Morschhäuser, J. (2010). An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54, 353–359. doi: 10.1128/aac.01102-09
Henry, K., Nickels, J., and Edlind, T. (2000). Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44, 2693–2700. doi: 10.1128/AAC.44.10.2693-2700.2000
Hoehamer, C. F., Cummings, E. D., Hilliard, G. M., and Rogers, P. D. (2010). Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob. Agents Chemother. 54, 1655–1664. doi: 10.1128/aac.00756-09
Hughes, A. L., Todd, B. L., and Espenshade, P. J. (2005). SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120, 831–842. doi: 10.1016/j.cell.2005.01.012
Kathawala, R. J., Gupta, P., Ashby, C. R. Jr., and Chen, Z.-S. (2015). The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist. Updat. 18, 1–17. doi: 10.1016/j.drup.2014.11.002
Khakhina, S., Johnson, S. S., Manoharlal, R., Russo, S. B., Blugeon, C., Lemoine, S., et al. (2015). Control of plasma membrane permeability by ABC transporters. Eukaryot. Cell 14, 442–453. doi: 10.1128/ec.00021-15
Kolaczkowski, M., van der Rest, M., Cybularz-Kolaczkowska, A., Soumillion, J.-P., Konings, W. N., and André, G. (1996). Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271, 31543–31548. doi: 10.1074/jbc.271.49.31543
Konecna, A., Toth Hervay, N., Valachovic, M., and Gbelska, Y. (2016). ERG6 gene deletion modifies Kluyveromyces lactis susceptibility to various growth inhibitors. Yeast 33, 621–632. doi: 10.1002/yea.3212
Krishnamurthy, S., Gupta, V., Snehlata, P., and Prasad, R. (1998). Characterisation of human steroid hormone transport mediated by Cdr1p, a multidrug transporter of Candida albicans, belonging to the ATP binding cassette super family. FEMS Microbiol. Lett. 158, 69–74. doi: 10.1111/j.1574-6968.1998.tb12802.x
Lamb, T. M., Vickery, J., and Bell-Pedersen, D. (2013). Regulation of gene expression in Neurospora crassa with a copper responsive promoter. G3 3, 2273–2280. doi: 10.1534/g3.113.008821
Lees, N. D., Skaggs, B., Kirsch, D. R., and Bard, M. (1995). Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—A review. Lipids 30, 221–226. doi: 10.1007/bf02537824
Liu, T. T., Lee, R. E. B., Barker, K. S., Lee, R. E., Wei, L., Homayouni, R., et al. (2005). Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49, 2226–2236. doi: 10.1128/aac.49.6.2226-2236.2005
Liu, X., Jiang, J., Shao, J., Yin, Y., and Ma, Z. (2010). Gene transcription profiling of Fusarium graminearum treated with an azole fungicide tebuconazole. Appl. Microbiol. Biotechnol. 85, 1105–1114. doi: 10.1007/s00253-009-2273-4
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lupetti, A., Danesi, R., Campa, M., Del Tacca, M., and Kelly, S. (2002). Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8, 76–81. doi: 10.1016/S1471-4914(02)02280-3
Maguire, S. L., Wang, C., Holland, L. M., Brunel, F., Neuvéglise, C., Nicaud, J.-M., et al. (2014). Zinc finger transcription factors displaced SREBP proteins as the major sterol regulators during Saccharomycotina evolution. PLOS Genet. 10:e1004076. doi: 10.1371/journal.pgen.1004076
Nes, W. R., Sekula, B. C., Nes, W. D., and Adler, J. H. (1978). The functional importance of structural features of ergosterol in yeast. J. Biol. Chem. 253, 6218–6225.
Ouyang, S., Beecher, C. N., Wang, K., Larive, C. K., and Borkovich, K. A. (2015). Metabolic impacts of using nitrogen and copper-regulated promoters to regulate gene expression in Neurospora crassa. G3 5, 1899–1908. doi: 10.1534/g3.115.020073
Pasrija, R., Panwar, S. L., and Prasad, R. (2008). Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob. Agents Chemother. 52, 694–704. doi: 10.1128/aac.00861-07
Paul, S., and Moye-Rowley, S. (2014). Multidrug resistance in fungi: regulation of transporter-encoding gene expression. Front. Physiol. 5:143. doi: 10.3389/fphys.2014.00143
Pedro Gonçalves, A., Silva, N., Oliveira, C., Kowbel, D. J., Glass, N. L., Kijjoa, A., et al. (2015). Transcription profiling of the Neurospora crassa response to a group of synthetic (thio)xanthones and a natural acetophenone. Genomics Data 4, 26–32. doi: 10.1016/j.gdata.2015.02.001
Perea, S., López-Ribot, J. L., Kirkpatrick, W. R., McAtee, R. K., Santillán, R. A., Martıìnez, M., et al. (2001). Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45, 2676–2684. doi: 10.1128/aac.45.10.2676-2684.2001
Perea, S., López-Ribot, J. L., Wickes, B. L., Kirkpatrick, W. R., Dib, O. P., Bachmann, S. P., et al. (2002). Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal Candidiasis. Antimicrob. Agents Chemother. 46, 1695–1703. doi: 10.1128/aac.46.6.1695-1703.2002
Pinto, E., Afonso, C., Duarte, S., Vale-Silva, L., Costa, E., Sousa, E., et al. (2011). Antifungal activity of xanthones: evaluation of their effect on ergosterol biosynthesis by high-performance liquid chromatography. Chem. Biol. Drug Des. 77, 212–222. doi: 10.1111/j.1747-0285.2010.01072.x
Porter, J. R., Burg, J. S., Espenshade, P. J., and Iglesias, P. A. (2010). Ergosterol regulates sterol regulatory element binding protein (SREBP) cleavage in fission yeast. J. Biol. Chem. 285, 41051–41061. doi: 10.1074/jbc.M110.144337
Price, C. L., Parker, J. E., Warrilow, A. G. S., Kelly, D. E., and Kelly, S. L. (2015). Azole fungicides – understanding resistance mechanisms in agricultural fungal pathogens. Pest Manag. Sci. 71, 1054–1058. doi: 10.1002/ps.4029
Redding, S. W., Kirkpatrick, W. R., Saville, S., Coco, B. J., White, W., Fothergill, A., et al. (2003). Multiple patterns of resistance to fluconazole in Candida glabrata isolates from a patient with oropharyngeal Candidiasis receiving head and neck radiation. J. Clin. Microbiol. 41, 619–622. doi: 10.1128/jcm.41.2.619-622.2003
Shapiro, R. S., Robbins, N., and Cowen, L. E. (2011). Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75, 213–267. doi: 10.1128/mmbr.00045-10
Song, B.-L., Javitt, N. B., and DeBose-Boyd, R. A. (2005). Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189. doi: 10.1016/j.cmet.2005.01.001
Sun, X., Wang, K., Yu, X., Liu, J., Zhang, H., Zhou, F., et al. (2014). Transcription factor CCG-8 as a new regulator in the adaptation to antifungal azole stress. Antimicrob. Agents Chemother. 58, 1434–1442. doi: 10.1128/aac.02244-13
Sun, X., Wang, W., Wang, K., Yu, X., Liu, J., Zhou, F., et al. (2013). Sterol C-22 desaturase ERG5 mediates the sensitivity to antifungal azoles in Neurospora crassa and Fusarium verticillioides. Front. Microbiol. 4:127. doi: 10.3389/fmicb.2013.00127
Tanwar, J., Das, S., Fatima, Z., and Hameed, S. (2014). Multidrug resistance: an emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014:541340. doi: 10.1155/2014/541340
Thakur, J. K., Arthanari, H., Yang, F., Pan, S.-J., Fan, X., Breger, J., et al. (2008). A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609. doi: 10.1038/nature06836
Vasicek, E. M., Berkow, E. L., Flowers, S. A., Barker, K. S., and Rogers, P. D. (2014). UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot. Cell 13, 933–946. doi: 10.1128/ec.00221-13
Vogel, H. J. (1956). A convenient growth medium for Neurospora (medium N). Microb. Genet. Bull. 13, 42–43.
Wang, K. J., Zhang, Z. Y., Chen, X., Sun, X. Y., Jin, C., Liu, H. W., et al. (2015). Transcription factor ADS-4 regulates adaptive responses and resistance to antifungal azole stress. Antimicrob. Agents Chemother. 59, 5396–5404. doi: 10.1128/Aac.00542-15
Watson, P. F., Rose, M. E., Ellis, S. W., England, H., and Kelly, S. L. (1989). Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164, 1170–1175. doi: 10.1016/0006-291X(89)91792-0
Weichert, M., Lichius, A., Priegnitz, B.-E., Brandt, U., Gottschalk, J., Nawrath, T., et al. (2016). Accumulation of specific sterol precursors targets a MAP kinase cascade mediating cell–cell recognition and fusion. Proc. Natl. Acad. Sci. U.S.A. 113, 11877–11882. doi: 10.1073/pnas.1610527113
White, T. C. (1997). Increased mRNA Levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41, 1482–1487.
Wong, K. H., Hynes, M. J., and Davis, M. A. (2008). Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell 7, 917–925. doi: 10.1128/ec.00076-08
Yang, H., Tong, J., Lee, C. W., Ha, S., Eom, S. H., and Im, Y. J. (2015). Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 6:6129. doi: 10.1038/ncomms7129
Yu, L., Zhang, W., Wang, L., Yang, J., Liu, T., Peng, J., et al. (2007). Transcriptional profiles of the response to ketoconazole and amphotericin B in Trichophyton rubrum. Antimicrob. Agents Chemother. 51, 144–153. doi: 10.1128/aac.00755-06
Zhang, W., Yu, L., Yang, J., Wang, L., Peng, J., and Jin, Q. (2009). Transcriptional profiles of response to terbinafine in Trichophyton rubrum. Appl. Microbiol. Biotechnol. 82, 1123–1130. doi: 10.1007/s00253-009-1908-9
Zhang, Y., Zhang, Z., Zhang, X., Zhang, H., Sun, X., Hu, C., et al. (2012). CDR4 is the major contributor to azole resistance among four Pdr5p-like ABC transporters in Neurospora crassa. Fungal Biol. 116, 848–854. doi: 10.1016/j.funbio.2012.05.002
Keywords: azoles, stress response, sterol 14α-demethylase, C-8 sterol isomerase, efflux pump, sterol intermediate, tcu-1 promoter
Citation: Hu C, Zhou M, Wang W, Sun X, Yarden O and Li S (2018) Abnormal Ergosterol Biosynthesis Activates Transcriptional Responses to Antifungal Azoles. Front. Microbiol. 9:9. doi: 10.3389/fmicb.2018.00009
Received: 02 August 2017; Accepted: 05 January 2018;
Published: 17 January 2018.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Koichi Tanabe, Ryukoku University, JapanN. Louise Glass, University of California, Berkeley, United States
Dominique Sanglard, University of Lausanne, Switzerland
Copyright © 2018 Hu, Zhou, Wang, Sun, Yarden and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaojie Li, bGlzakBpbS5hYy5jbg==
 Chengcheng Hu
Chengcheng Hu Mi Zhou
Mi Zhou Wenzhao Wang1
Wenzhao Wang1 Xianyun Sun
Xianyun Sun Oded Yarden
Oded Yarden Shaojie Li
Shaojie Li