
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 21 December 2017
Sec. Infectious Agents and Disease
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02602
This article is part of the Research Topic Bacterial Colonization: Bystander Microbiota or the Initial Step of Chronic Disease? View all 15 articles
 Nikolai Siemens1,2*
Nikolai Siemens1,2* Sonja Oehmcke-Hecht3
Sonja Oehmcke-Hecht3 Thomas C. Mettenleiter4
Thomas C. Mettenleiter4 Bernd Kreikemeyer3
Bernd Kreikemeyer3 Peter Valentin-Weigand5
Peter Valentin-Weigand5 Sven Hammerschmidt1
Sven Hammerschmidt1Bacterial and viral co-infections of the respiratory tract are life-threatening and present a global burden to the global community. Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes are frequent colonizers of the upper respiratory tract. Imbalances through acquisition of seasonal viruses, e.g., Influenza A virus, can lead to bacterial dissemination to the lower respiratory tract, which in turn can result in severe pneumonia. In this review, we summarize the current knowledge about bacterial and viral co-infections of the respiratory tract and focus on potential experimental models suitable for mimicking this disease. Transmission of IAV and pneumonia is mainly modeled by mouse infection. Few studies utilizing ferrets, rats, guinea pigs, rabbits, and non-human primates are also available. The knowledge gained from these studies led to important discoveries and advances in understanding these infectious diseases. Nevertheless, mouse and other infection models have limitations, especially in translation of the discoveries to humans. Here, we suggest the use of human engineered lung tissue, human ex vivo lung tissue, and porcine models to study respiratory co-infections, which might contribute to a greater translation of the results to humans and improve both, animal and human health.
In recent years the human microbiota is more and more recognized to play a crucial role in pathogenesis of many diseases (Weinstock, 2012). The upper respiratory tract is a natural niche for potentially pathogenic bacteria embedded in commensal communities forming the nasopharyngeal microbiome. In particular, the microbial communities of the nasopharynx (Hilty et al., 2012) are associated with respiratory diseases, i.e., severe pneumonia, which are responsible for substantial mortality and morbidity in humans worldwide (Prina et al., 2016). The composition of the nasopharyngeal microbiome is highly dynamic (Biesbroek et al., 2014a,b,c) and many factors, including environmental and host factors, can affect microbial colonization (Koppen et al., 2015). Recent studies on neonates have shown that the respiratory microbiota develops from initially maternally transmitted mixed flora with predominance of Streptococcus viridans species to niche-specific bacterial profiles containing mostly Staphylococcus aureus at around 1 week of age (Bosch et al., 2016a). Between 2 weeks and 6 months after birth, the staphylococcal predominance declines and colonization with Streptococcus pneumoniae (pneumococci) as a predominant pathobiont emerges (Miller et al., 2011; Bosch et al., 2016a,b). The dynamic microbiome composition is guaranteed through the interplay between bacterial species, other microbes, and changing environmental conditions, as well as host–bacteria interactions (Blaser and Falkow, 2009). Most of the time, the microbiome and its interplay with the human host are believed to be beneficial for both (Pettigrew et al., 2008; Murphy et al., 2009). However, imbalances in microbial composition can lead to acquisition of new viral or bacterial species and invasion of potential pathogens, which in turn can become detrimental, especially in elderly people and children with an exhausted or immature immune system (Pettigrew et al., 2008; Blaser and Falkow, 2009; Murphy et al., 2009).
One particular example showing imbalances introduced by single dosage of antibiotics was demonstrated by Ichinohe and colleagues (Ichinohe et al., 2011). While commensal respiratory microbiota facilitated immune-support against Influenza A virus infection (IAV), oral treatment with antibiotics resulted not only in a shift of bacterial composition, but also in impaired CD4 T-, CD8 T-, and B-cell immunity following infection with IAV in mice (Ichinohe et al., 2011). Analyses of human oropharyngeal microbiomes during the 2009 H1N1 IAV pandemic revealed that at the phylum level, the abundance of Fermicutes and Proteobacteria was augmented in pneumonia patients as compared to healthy controls (Leung et al., 2013). However, another study published in the same year contradicted these results (Chaban et al., 2013). Chaban and colleagues analyzed microbiomes of 65 patients from H1N1 IAV outbreak in 2009. Although the phylogenetic composition of pneumonia patients was dominated by Fermicutes, Proteobacteria, and Actinobacteria, no significant differences between the patients and healthy controls or any other variables tested, including age and gender, were observed (Chaban et al., 2013).
In this review we discuss secondary bacterial infections of the respiratory tract after primary infection by IAV with a focus on mechanisms by which these interactions are potentially mediated, and we will provide insight into the host contribution and immunological consequences. We further focus on potential animal models suitable for mimicking asymptomatic bacterial colonization and disease progression and thus, enabling to study adaptation strategies, viral-bacterial interactions, and immune responses in these highly lethal co-infections.
Influenza A viruses belong to the family of Orthomyxoviridae and based on the antigenicity of their haemagglutinin (HA) and neuraminidase (NA) they are classified into 16 classical HA and 9 classical NA subtypes (Neumann et al., 2009). The 8-segmented genomes of influenza A viruses are characterized by a significant plasticity. Due to point mutations and re-assortment events new variants or strains with epidemic or pandemic potential emerge (Neumann et al., 2009). In addition, influenza can be transmitted between animals, including swine, birds, horses, and humans, making it a zoonotic disease (van der Meer et al., 2010). Seasonal influenza usually resolves without consequences in healthy individuals. However, it is estimated that seasonal influenza effects 5–10% of the world’s population resulting in about 250,000 to 500,000 deaths annually (Tjon-Kon-Fat et al., 2016). At greater risk to develop secondary bacterial pneumonia are individuals with comorbidities, elderly people (age > 65), pregnant women, and children under the age of one (Rothberg et al., 2008).
For a long time it was considered that the H1N1 strain, an avian-like H1N1 virus, directly caused most of the fatalities during the 1918–1919 pandemic (Spanish Flu), often from a hemorrhagic pneumonitis rapidly progressing to acute respiratory distress syndrome and death (Osterholm, 2005; Gerberding, 2006; Oxford et al., 2006). The pandemic killed around 50 million people worldwide and remains unique in its severity compared to other big outbreaks. However, many of the findings have been reinterpreted in recent years (Brundage and Shanks, 2007; Chien et al., 2009). It is estimated that around 95% of all severe cases and deaths were attributed to secondary infections with bacterial pathogens, most predominantly by Streptococcus pneumoniae (Morens et al., 2008). Individual studies limited to certain regions identified also other pathogens commonly colonizing the respiratory tract, including Staphylococcus aureus, group A streptococcus (GAS) and Haemophilus influenzae (Brundage and Shanks, 2008). During the next two pandemics (H2N2 Asian Flu 1957-1958 and H3N2 Hong Kong Flu 1968-1969) bacterial co-infections were less likely the cause of death compared to the Spanish Flu (Giles and Shuttleworth, 1957; Trotter et al., 1959). Still, pneumonia accounted for about 44% of deaths during the Asian Flu (Giles and Shuttleworth, 1957). Most fatalities resulting from pneumonia occurred in individuals with chronic conditions, i.e., chronic lung diseases, rheumatic carditis, and hypertension (Giles and Shuttleworth, 1957). In 1957–1958, S. aureus was predominantly isolated from fatal pneumonia cases (Hers et al., 1957, 1958; Robertson et al., 1958; Martin et al., 1959), whereas S. pneumoniae returned as predominant cause of severe pneumonia during the Hong Kong Flu (Sharrar, 1969; Bisno et al., 1971; Burk et al., 1971; Schwarzmann et al., 1971). Forty years later in 2009, a novel H1N1 virus of swine origin emerged and caused again a pandemic (Dawood et al., 2009, 2012). In contrast to Asian and Hong Kong Flu, mortality rates were rather low, but most deaths occurred in healthy young individuals with no underlying conditions (Reichert et al., 2010; Monsalvo et al., 2011; Dawood et al., 2012). About 25–50% of severe or fatal cases were linked to complications due to bacterial pneumonia (Dominguez-Cherit et al., 2009; Estenssoro et al., 2010; Mauad et al., 2010; Shieh et al., 2010). Although regional variations occurred, pneumococci and S. aureus were the most frequently isolated bacterial species (Mauad et al., 2010; Shieh et al., 2010; Rice et al., 2012). Group A streptococcus was absent in many local pneumonia outbreaks associated with viruses, but was predominant in others (Brundage and Shanks, 2008; Ampofo et al., 2010). When it does appear, it is typically third in incidence (Chaussee et al., 2011). Overall, data on pandemic outbreaks suggest that disease severity and mortality can be linked to secondary bacterial pathogens with variations depending on regions and state of immunity of the population (Brundage and Shanks, 2008; Shanks et al., 2010, 2011; McCullers, 2013).
There is increasing evidence that the nasopharyngeal microbiota plays an important role in the pathogenesis of acute viral respiratory infections (Teo et al., 2015; de Steenhuijsen Piters et al., 2016; Rosas-Salazar et al., 2016a,b). Respiratory viruses, including IAV, have been shown to alter bacterial adherence and colonization leading to an increased risk of secondary bacterial infections (Tregoning and Schwarze, 2010). Pneumococci, S. aureus, and GAS are important human Gram-positive pathogens. All of them are frequent colonizers of the human nasopharynx and they share many features including pathogenic mechanisms and clinical aspects (Figure 1). However, they also have unique properties.
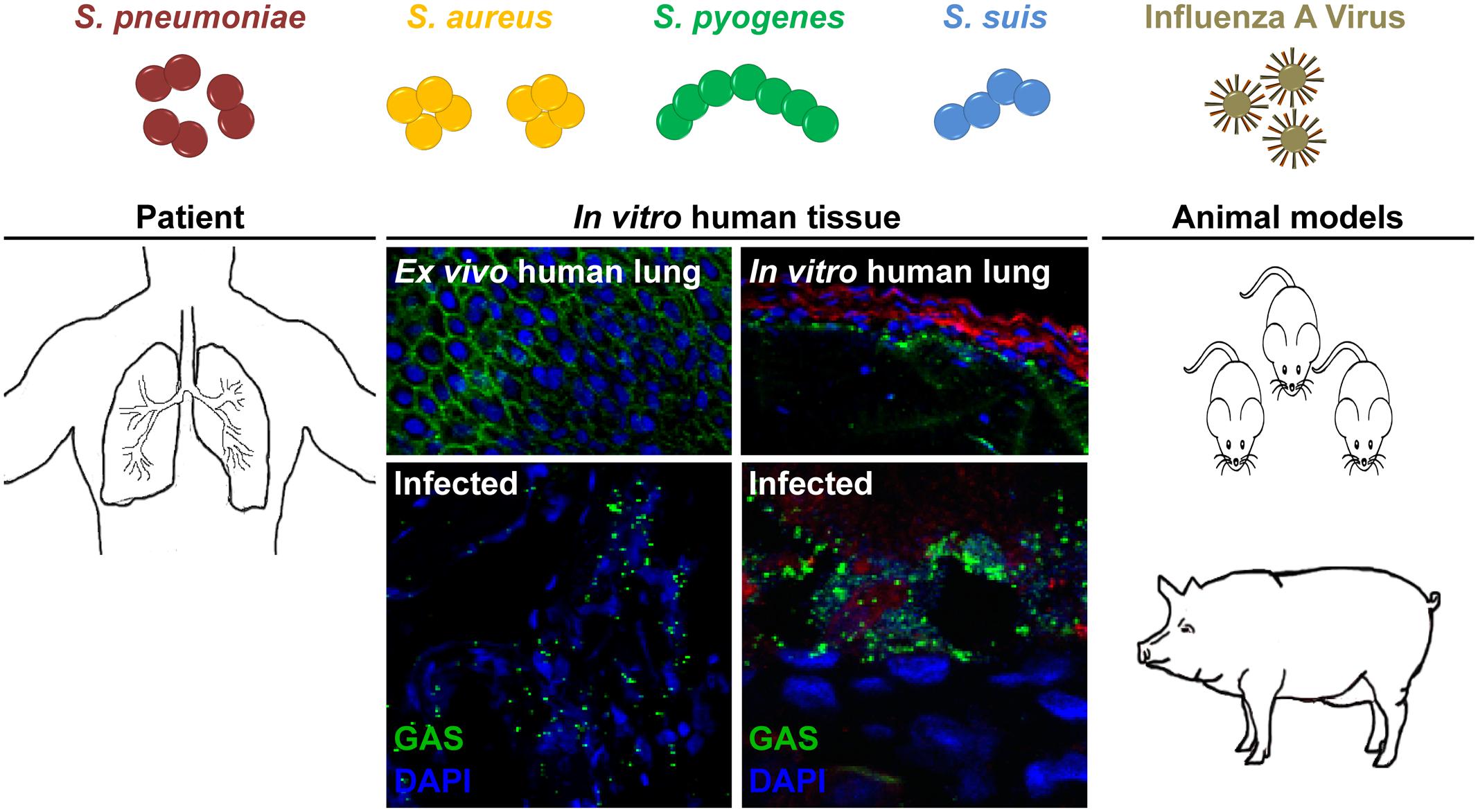
FIGURE 1. Potential models to study bacterial and viral co-infections of the respiratory tract. S. pneumoniae, S. aureus, S. pyogenes, and S. suis are frequent colonizers of the upper respiratory tract. Seasonal IAV infection can lead to an increased risk of secondary bacterial infections, i.e., pneumonia. Several experimental models can be used for studying these severe infections. Patient samples, including ex vivo lung tissue are materials of choice, but they are rare due to ethical considerations. Tissue engineering approaches closely resemble the 3D architecture, cellular composition, and matrix complexity of the respective organ and were proven as useful tool to study infectious diseases. In vivo bacterial and viral co-infections are mainly performed in mice, which does not necessarily resemble the human physiology and immune system. Thus, we suggest using the porcine model, which nearly resembles over 80% of the human immune system.
Staphylococcus aureus colonizes persistently about 30% of the human population and typical niches include nares, axillae, and skin (Peacock et al., 2001; von Eiff et al., 2001; van Belkum et al., 2009). They cause a variety of clinical manifestations ranging from mild skin infections to fatal necrotizing pneumonia. In the last decades, the pathogen became resistant to an increasing number of antibiotics and methicillin-resistant S. aureus (MRSA) is now a major cause of hospital acquired infections (Hartman and Tomasz, 1984; Ubukata et al., 1989; Zetola et al., 2005). Also the rise of community-acquired S. aureus strains is of special concern, because certain clones are associated with very severe infections (Rasigade et al., 2010). Recent prospective studies demonstrated an increase in proportion of community-acquired methicillin-sensitive S. aureus in severe pneumonia cases (McCaskill et al., 2007; Sicot et al., 2013).
The pneumococcus is a typical colonizer of the human nasopharynx. About 20–50% of healthy children and 8–30% of healthy adults are asymptomatically colonized (McCullers, 2006). Pneumococci cause diseases ranging from mild, i.e., sinusitis, conjunctivitis, and otitis media, to more severe and potentially life-threatening infections, including community-acquired pneumonia, bacteraemia, and meningitis (Bogaert et al., 2004; Valles et al., 2016). This bacterium is associated with high morbidity and mortality rates in risk groups such as immunocompromised individuals, children, and elderly (Black et al., 2010; Valles et al., 2016).
Group A streptococci colonize the mouth and upper respiratory tract in about 2–5% of world’s population (Okumura and Nizet, 2014). The most common, non-invasive and mild infections caused by GAS are tonsillitis and pharyngitis with estimated 600 million cases per year (Carapetis et al., 2005). Listed as number nine in the list of global killers with around 500,000 deaths annually (Carapetis et al., 2005), it is obvious that this pathogen can cause severe invasive infections, including pneumonia, sepsis, streptococcal toxic shock syndrome, and necrotizing skin infections (Cunningham, 2000; Carapetis et al., 2005).
Although all three pathogens are able to cause highly lethal diseases, the most fatal remains the pneumococcus, estimated to cause ca. 10% of all deaths in children below 5 years of age (O’Brien et al., 2009), in the elderly (Marrie et al., 2017), and in immuno-compromised individuals (Baxter et al., 2016).
Influenza A virus binds via HA to either α2,3- or α2,6-linked sialic acid at the surface of epithelial cells of the upper and lower respiratory tract (Webster et al., 1992). Seasonal strains show usually affinity to α2,6-linked sialic acids that are expressed in the human trachea, whereas avian-like viruses preferentially bind to α2,3-linked sialic acids of alveolar type II cells (Shinya et al., 2006; van Riel et al., 2007, 2010). The release of viral genomic RNA into the cytosol activates different immune response pathways. Binding of viral RNA to retinoic acid inducible gene 1 induces the expression of type I and III interferons and activates transcription factor NF-κB, which in turn activates the release of pro-inflammatory cytokines (Durbin et al., 2013; Iwasaki and Pillai, 2014). In addition, inflammasome activation leads to the release of IL-1β and IL-18 (Pothlichet et al., 2013; Iwasaki and Pillai, 2014). All these responses are supposed to promote viral clearance. However, the presence of viral proteins during infection induces also direct activation of the intrinsic or indirectly the activation of the extrinsic apoptotic pathway via production of inflammatory cytokines, resulting in apoptosis or even necrosis of the epithelium (Korteweg and Gu, 2008). Furthermore, aberrant coagulation induced by virus infection causes a hyper-inflammatory response (Yang and Tang, 2016). All these events contribute to lung tissue injury (Imai et al., 2008; Davidson et al., 2014). The epithelial damage due to viral replication provides a beneficial environment for initial bacterial attachment (Plotkowski et al., 1993). On the other hand, already colonized bacteria might enhance influenza virus virulence either by directly secreting proteases that cleave and activate HA (Figure 2) (Bottcher-Friebertshauser et al., 2013) or, indirectly, by activating host proteases such as plasminogen, which increases replication rates and infectivity of the virus (Scheiblauer et al., 1992; Tse and Whittaker, 2015).
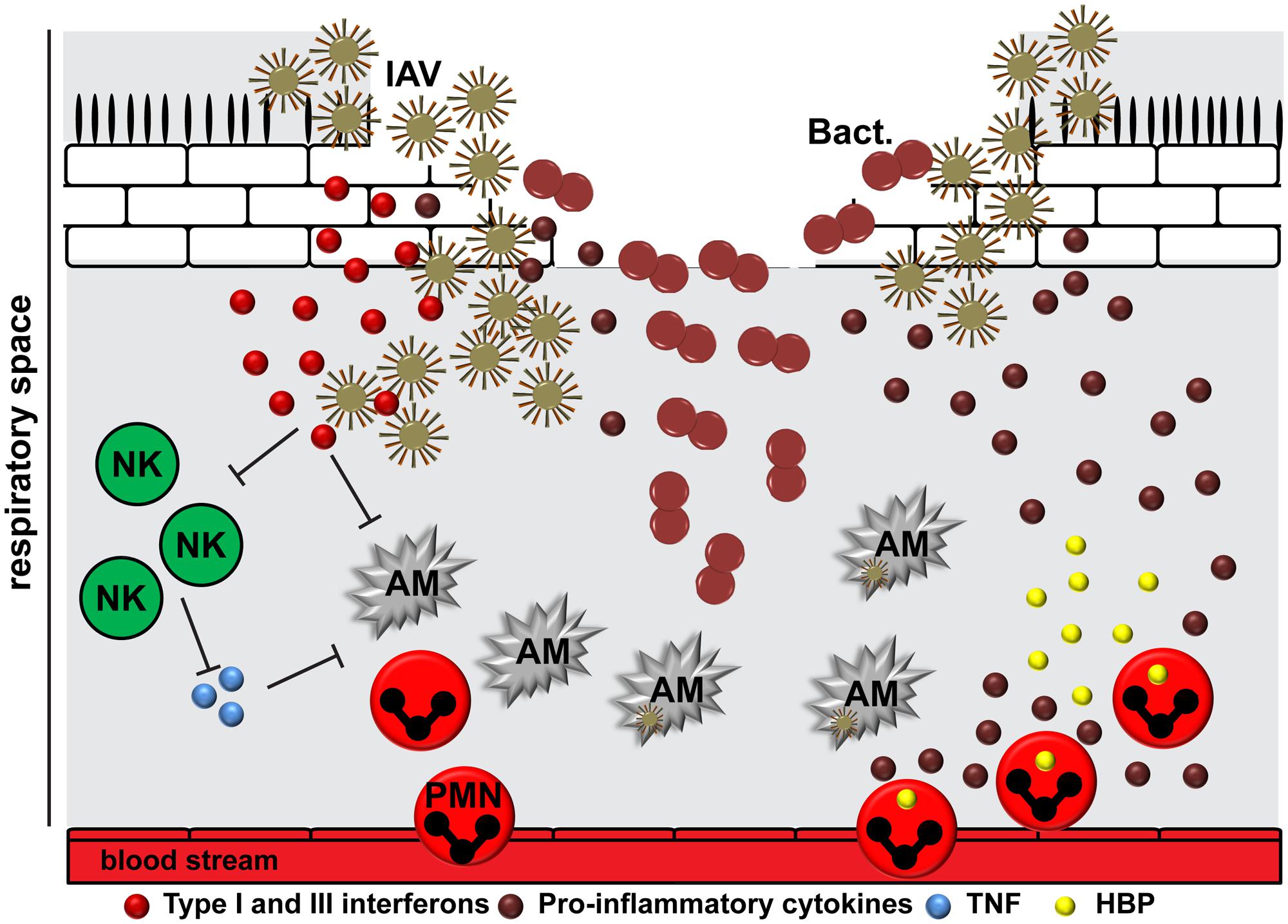
FIGURE 2. The interplay between IAV, bacteria, and the human host. The epithelial damage due to viral replication provides a beneficial environment for bacterial (Bact.) attachment. IAV is able to induce suppression and killing of resident alveolar macrophages (AM), which in turn delays viral clearance. The release of viral RNA activates different immune response pathways resulting in cytokine storm. Type I and III interferons compromise the immune recognition of Gram-positive bacteria by neutrophils and macrophages. In addition, they might suppress natural killer cell function (NK), including release of TNF, which activates alveolar macrophages. After initial inflammation, the situation might worsen due to cellular infiltration of the lungs by neutrophils (PMN), leading to an increased degranulation and tissue damage by effector molecules, including heparin-binding protein (HBP).
Potentially pathogenic bacteria, including the three species mentioned above, express an arsenal of virulence factors responsible for attachment to human host structures. Microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), such as PspC, PspA, and PsaA in pneumococci (Hammerschmidt, 2006), SPA, FnbA, ClfA, and ClfB in S. aureus (Bartlett and Hulten, 2010; Otto, 2010), and M-protein, PrtF1, and PrtF2 in GAS (Cunningham, 2000), respectively, and so-called moon-lightning proteins expressed by all three species, e.g., GAPDH, enolase or PGK (Fulde et al., 2013), enable the bacteria to attach to damaged cells or molecules of the extracellular matrix, including fibronectin, fibrin, fibrinogen, and collagens, or fibrinolytic proteins like plasminogen (McCullers and Rehg, 2002; Bergmann and Hammerschmidt, 2007; Linke et al., 2012; Siemens et al., 2012; Voss et al., 2012). Once the initial attachment occurs, bacterial cytotoxins including pneumolysin of pneumococci (Garcia-Suarez Mdel et al., 2007; Zahlten et al., 2015), α-hemolysin and leukocidins of S. aureus (Mairpady Shambat et al., 2015), and Streptolysins S and O and Streptococcal pyrogenic exotoxin B of S. pyogenes (Tsai et al., 1998; Gurel et al., 2013; Siemens et al., 2015, 2016), can synergize with viral counterparts to further increase lung tissue pathology. Additional potential mechanisms by which the initial colonization of the lower respiratory tract and lung tissue damage might occur include potentiation of the development of pneumonia by IAV neuraminidase through enzymatic removal of sialic acid from the lung, thus exposing host receptors for pneumococcal adherence (McCullers and Bartmess, 2003). The host inflammatory state in response to viral infection can alter presentation of receptors on the surface, thus allowing bacterial invasion (Cundell and Tuomanen, 1994). As the patient begins to recover from viral infection, secondary bacterial infections might occur (Louria et al., 1959) due to the incomplete wound healing and exposure of host membrane components, including laminin, collagens type I and IV to classical bacterial MSCRAMMs (Louria et al., 1959; Puchelle et al., 2006).
Epithelial cells are the first responders to infections in the lung, followed by the tissue resident alveolar macrophages. They promote viral clearance via phagocytosis, efferocytosis, and release of cytokines and chemokines to promote immune responses (Hashimoto et al., 2007; Kumagai et al., 2007; Wang et al., 2012; Hillaire et al., 2013). Respiratory viruses like IAV are able to induce suppression and killing of the resident alveolar macrophages (Figure 2) (Ghoneim et al., 2013). These cells are usually replaced by differentiation of recruited blood derived monocytes into macrophages of different polarization patterns. This in turn creates a delay in pathogen clearance and opens a window for host susceptibility to secondary bacterial infections, colloquially named superinfections (Ghoneim et al., 2013). In addition, induction of interferons as a response to viral infection compromises the immune sensing of Gram-positive bacteria by neutrophils and macrophages, which would normally clear the bacteria from the lungs (Figure 2) (Sun and Metzger, 2008; Tian et al., 2012). The exact mechanism underlying this phenomenon is still not understood. Several studies suggested that viral RNA activates Toll-like receptors (TLR) 2 and TLR4 and, consequently, the production of type I interferons to promote an antiviral state (Shahangian et al., 2009). The subsequent infection with Gram-positive bacteria, e.g., pneumococci, enhances the type I interferon expression, which in turn suppresses production of the CCL2 chemokine and recruitment of macrophages (Nakamura et al., 2011). Another study by Shahangian et al. (2009) revealed that the antiviral state leads to impaired production of neutrophil chemoattractants CXCL1 and CXCL2, which in turn promotes less effective immune responses due to attenuated neutrophil functions during the early phase of pneumococcal invasion. Other studies found that IAV exposed lungs had impaired natural killer (NK) cell responses in the airway to subsequent S. aureus infection (Small et al., 2010). Reduced TNFα production by NK cells was identified as a crucial upstream mechanism of depressed antimicrobial activities by alveolar macrophages (Figure 2) (Small et al., 2010). It seems likely that IAV NA is also able to activate host cell receptors in a TGF-β dependent manner, which in turn promotes GAS invasion and subsequent lung pathology (Li et al., 2015). In vitro studies on the interplay between IAV-pneumococci and human dendritic cells revealed TLR3 as a crucial sensor of viral and bacterial RNA leading to enhanced IL-12p70 production, which in turn might promote an anti-viral state by upregulation of interferons (Yamamoto et al., 2004; Spelmink et al., 2016). However, it should be noted that depending on the bacterial species the disease manifestation and underlying innate immune responses might vary (Sharma-Chawla et al., 2016).
A lot of the experimental studies on disease mechanisms and immune responses are based on a subsequent bacterial infection within hours or a few days post IAV infection. However, bacterial infiltrations of the lungs might occur much later, i.e., during the onset of wound healing after partial clearance of IAV, which has been reported in most studies performed in recent years (Snelgrove et al., 2008; Hussell and Cavanagh, 2009). These processes are characterized by a general anti-inflammatory state and suppression of mechanisms involved in pathogen clearance due to increased interleukin-10 production (van der Sluijs et al., 2004; Metzger and Sun, 2013). The anti-inflammatory state suppresses the expression of pattern recognition receptors (PRR) on professional phagocytes leading to impaired phagocytosis and killing of microbes. These events might allow bacterial overgrowth in the lungs and tissue pathology (Sun and Metzger, 2008; Goulding et al., 2011).
Like other severe infectious diseases caused by single agents, pneumonia is characterized by hyper-inflammatory conditions of the lungs at the onset of infection followed by a hypo-inflammatory state with immune paralysis (Morton et al., 2014). In co-infections, after initial inflammation in response to viral infection the situation might worsen due to bacterial invasion and enhanced cellular infiltration of the lungs by neutrophils, leading to an increased tissue damage and cytokine storm (Figure 2) (Conenello et al., 2007; McAuley et al., 2007, 2010; Porto and Stein, 2016). Furthermore, the coagulation system becomes activated and contributes to the pathophysiological response to infection (van der Poll and Herwald, 2014). Bacteria like pneumococci, S. aureus, and GAS can activate and modulate the coagulation system, leading to extensive expression of tissue factor and increasing the risk of severe coagulopathy (Nguyen et al., 2012; Shannon et al., 2013; Walters et al., 2016).
Bacterial pathogens also express a variety of cytolytic toxins that can contribute to inflammation and tissue pathology. Pneumolysin, a pneumococcal pore-forming toxin with low affinity to lung epithelial cells, can damage neutrophils by utilizing P2X7 receptor (Domon et al., 2016). Staphylococcal cytotoxins (α-toxin and leukocidins, including Panton-Valentine leucocidin, PVL) are associated with severe tissue pathology, strong upregulation of chemokines, and increased neutrophil influx of the lungs (Mairpady Shambat et al., 2015). GAS toxins, including SLO and SpeB, are capable of directly causing tissue damage and promoting pro-inflammatory states through neutrophil lysis (Snall et al., 2016; Uhlmann et al., 2016). The cytolytic effects caused by bacterial toxins might synergize with the outcome of IAV cytotoxic accessory protein, PB1-F2, mediated tissue pathology leading to enhanced cytokine production (Ramos and Fernandez-Sesma, 2012). Taken together, most likely synergistic effects of the pathways that are involved in bacterial and viral inflammation lead to enhanced immune activation and higher morbidity and mortality (Joyce et al., 2009; Koppe et al., 2012; Ramos and Fernandez-Sesma, 2012; Bucasas et al., 2013; Kuri et al., 2013). Figure 2 summarizes the interplay between virus, bacteria, and host.
Experimental animal models are a useful tool to study in vivo effects of different infectious agents and they represent approximately 3% of all pneumonia research published in peer-review journals (Hraiech et al., 2015). However, the constant increase of animal studies in the last decades is in contrast to their reproducibility in humans (Hackam and Redelmeier, 2006). Hackam and colleagues identified 2,000 articles published between 1980 and 2006 in seven leading scientific journals that regularly publish animal studies (Hackam and Redelmeier, 2006). Seventy-six out of 2,000 were highly cited with a median citation count of 889. Out of these 76 studies 28 were replicated in human randomized trials, 14 were contradicted, and 34 remained untested (Hackam and Redelmeier, 2006). Only 1.4% of the animal studies published in high-impact journals were translated in human randomized trials (Hackam and Redelmeier, 2006), whereas about 44% replication rate was reported for highly cited human studies (Ioannidis, 2005). In pneumonia models, mammalians are mostly used because of their anatomical and physiological proximity to humans (Hraiech et al., 2015). To monitor extensive physiological studies, larger mammalian species, including ferrets, dogs, rabbits, pigs, and baboons are the models of choice (Mizgerd and Skerrett, 2008). However, rodents and in particular mice are used more frequently as a pneumonia model organisms. Rapid reproductive rate, small size, less complicated handling, the ability to reproduce and compare results with already published bacterial and viral mono-infections, detailed knowledge of genetics and immune responses, and a plethora of available reagents to study infections in mice are reasons for the use of these animals. To avoid variations in responses due to genetic diversity inbred mice strains are useful tools for studies aiming to elucidate molecular mechanisms of diseases. In addition, genetic engineering allowed to generate a wide variety of mouse variants with gain-of-function, loss-of-function or reporter genes (Mizgerd and Skerrett, 2008).
As outlined above, many in vivo mice studies on bacterial and viral co-infections provided useful insights into severe pneumonia, including (i) the fact that viral infection primes the host for bacterial susceptibility leading to severe secondary infection (Hashimoto et al., 2007; Shahangian et al., 2009; Chaussee et al., 2011; Nakamura et al., 2011), (ii) pathogen synergism (Tsai et al., 1998; McCullers and Rehg, 2002; Garcia-Suarez Mdel et al., 2007; Gurel et al., 2013; Mairpady Shambat et al., 2015; Zahlten et al., 2015), (iii) enhanced inflammatory response at the onset of infection (Korteweg and Gu, 2008; Durbin et al., 2013; Pothlichet et al., 2013; Iwasaki and Pillai, 2014) leading to increased alveolar damage followed by immune paralysis with defective clearance of microorganisms (Shinya et al., 2006; van Riel et al., 2007, 2010), and (iv) host receptor availability for sustained bacterial infection (Louria et al., 1959; Plotkowski et al., 1993; Cundell and Tuomanen, 1994; Puchelle et al., 2006; Korteweg and Gu, 2008). However, mouse models for bacterial and/or viral infections have several limitations. Most of the bacterial and viral species under study are human pathogens. In recent years it was also shown that host genetic variations and sex differences have an impact on predisposition, severity, and outcome of infection (Chella Krishnan et al., 2015, 2016) While C57BL/6 and BALB/c mice are characterized by a higher resistance, DBA/2 strains are more susceptible and permissive to bacterial and viral strains (Alymova et al., 2011; Chella Krishnan et al., 2015, 2016). In addition, transmission of IAV and bacteria is inefficient in adult mice, thus requiring alternative animal models, including neonatal mice or ferrets (Diavatopoulos et al., 2010; McCullers et al., 2010). IAV was shown to be essential for pneumococcal transmission from colonized mice to their naive littermates and the transmission occurred only when all mice were infected with IAV (Diavatopoulos et al., 2010).
The facilitated transmission of pneumococci after IAV infection was confirmed by Mc Cullers et al. in ferrets (Mustela putorius furo) (McCullers et al., 2010). The pneumococcal disease manifestation and transmission between animals was enhanced if animals had previously been infected with IAV (McCullers et al., 2010). Ferrets are naturally susceptible to IAV isolated from different species, including humans, birds, and swine (Thangavel and Bouvier, 2014). The infection of ferrets with human seasonal IAV isolates results in an upper respiratory tract infection similar to human influenza infection (Tripp and Tompkins, 2009). In contrast to mice, non-adapted human IAV can be used for the infection. Unfortunately, there are only few reports on bacterial and IAV co-infections in this model organism. A report by Sanford and Ramsay showed enhanced staphylococcal colonization of the upper respiratory tract in IAV infected animals as compared to non-infected, while no difference between both groups was observed in group B streptococcal infection (Sanford and Ramsay, 1987). In contrast, Smith and Mc Cullers reported lack of establishment of staphylococcal infection even when ferrets were pre-infected with IAV (Smith and McCullers, 2014). The biggest advantages of using ferrets as a model include (i) their susceptibility to non-adapted human pathogens, (ii) efficiency in transmitting IAV and bacteria from one individual to another, and (iii) presentation of the clinical signs of disease manifestation akin to human influenza infection. Unfortunately, their limited availability, complex husbandry, and limited accessibility to ferret-specific reagents makes this research difficult to perform (Bouvier and Lowen, 2010).
In recent years, the guinea pig (Cavia porcellus) was also used in pneumonia research. The physiology and anatomy of the guinea pig lung resembles to a certain extent the human lung and this model organism is often used in non-infectious lung diseases, including asthma and chronic obstructive pulmonary disease (Canning and Chou, 2008). In addition, its commercial availability, ease of husbandry, the ability to work with non-adapted pathogens and the efficiency of transmission are reasons for using this in vivo model (Bouvier and Lowen, 2010). Guinea pigs are susceptible to human, avian, and swine influenza viruses. Although viral replication can be readily detected upon intranasal inoculation in the upper respiratory tract and the lungs, guinea pigs exhibit only minor clinical symptoms (Lowen et al., 2006; Gabbard et al., 2014). However, the lung pathology of human IAV infected guinea pigs correlates with the clinical severity of human infection (Gabbard et al., 2014). Transmission of pneumococci in guinea pigs is promoted by co-infection with Sendai virus (Saito et al., 1988). Guinea pigs infected with pneumococci alone and cage-mated with non-treated contact animals transmitted the bacteria only in 7% of cases, while Sendai-virus infected, co-housed guinea pigs acquired pneumococcal infection in 83% of contacts (Saito et al., 1988). Another study evaluated antibiotic efficacy in invasive pulmonary infection caused by penicillin resistant pneumococcus (Ponte et al., 1996). Intra-tracheal instillation of 3 × 109 CFU of S. pneumoniae induced a fatal pneumonia and bacteremia in 85% of untreated animals within 46 h (Ponte et al., 1996). As with ferrets, there is a paucity of data describing immune responses to pulmonary infectious agents. This is in parts due to the lack of species specific reagents, which is a disadvantage in using this model organism.
Recently, the cotton rat (Sigmodon hispidus) was reported to be susceptible to IAV. Nasal and pulmonary infection in adult inbred cotton rats did not require viral adaptation (Ottolini et al., 2005). The infection led to increased breathing rates accompanied by weight loss and decreased body temperature. Replication of IAV was more extensive in nasal tissues than the lung, and persisted for six consecutive days. Tissue pathology included damage of bronchiolar epithelium and the animals developed pneumonia which persisted for nearly 3 weeks (Ottolini et al., 2005). In bacteriological studies rats are more frequently used. There are numerous rat models investigating the impact of diabetes (Oliveira et al., 2016), metabolic syndromes (Feng et al., 2015), cirrhosis (Preheim et al., 1991), pharmaco-kinetics and dynamics (Antonopoulou et al., 2015; Hoover et al., 2015), intoxication (Davis et al., 1991), immunization (Iinuma and Okinaga, 1989), and general bacterial virulence factors (Shanley et al., 1996) on development of pneumococcal, streptococcal, and staphylococcal pneumonia and lung pathology. Unfortunately, there are only few studies on bacterial and viral co-infections in rats. The first was performed by Harford et al., 1946 (Harford et al., 1946). The authors concluded that the secondary bacterial pneumonia does not convert the sub-lethal viral infection to a lethal outcome (Harford et al., 1946). Another study on human respiratory syncytial virus and S. pneumoniae revealed that rats were easily colonized with pneumococci, but viral replication after subsequent infection was strain dependent. In addition, neither pneumococci nor the virus spread from the upper to the lower respiratory tract, and neither pathogen was transmitted to naive cage mates (Nguyen et al., 2015). Although rats share a lot of immune features with humans, including nitric oxide production by macrophages (Carsillo et al., 2009), the biggest disadvantages are low animal availability, aggressiveness of the species, and the lack of specific reagents.
Rabbits (Oryctolagus cuniculus) are well known for their use in studying cardiovascular diseases, antibody production, and eye research. Rabbits were also employed to study pneumonia, although only a few models are available. Typical read-out parameters include survival, leukocyte infiltration of the lungs, lung pathology, and assessment of drug concentration in serum. One of the first studies on pneumococcal pneumonia in rabbits was performed in Kline and Winternitz (1913). This study revealed that rabbits possess an active immunity if they have recovered from one attack of experimental pneumonia and they may subsequently resist repeated intra-tracheal dosages of pneumococci (Kline and Winternitz, 1913). In 1926 an infection by inhalation of Type I pneumococci was established in rabbits (Stillman and Branch, 1926). The bacteria infiltrated easily the lower respiratory tract and pneumococci which reached the lungs usually disappeared within hours and fatal septicemia appeared in some of the animals (Stillman and Branch, 1926). Most recent rabbit models of pneumococcal and staphylococcal pneumonia are based on intra-bronchial or intra-pulmonary infections which make them useful for pathogenesis (Diep et al., 2010, 2017), as well as drug efficiency and efficacy studies (Cabellos et al., 1992; Croisier-Bertin et al., 2011). However, this infection route requires surgery and species-specific reagents are scarce. In IAV research rabbits are frequently used for antibody production and for studies on antibody kinetics following single or multiple IAV administrations (Loza-Tulimowska et al., 1977). Also, rabbits are used for safety investigations of vaccines (e.g., CoVaccine HT or Aflunov) (Heldens et al., 2010; Gasparini et al., 2012). In recent years the shedding of avian IAV by cottontails (Sylvilagus spp.) was investigated revealing that nasally and orally inoculated cottontails shed relatively large quantities of viral RNA (Root et al., 2014). Notably, low viral titers were found to be sufficient to initiate viral replication in cottontails (Root et al., 2017). However, despite their susceptibility to IAV infection, rabbits are only rarely used as model for IAV pathogenesis since they offer no improvement over other established infection models.
Macaques represent the major non-human primate for studying infectious diseases. They are omnivorous and adaptable. The species most commonly used are rhesus macaques (Macaca mulatta) and cynomolgus macaques (Macaca fasciluraris). Although it was shown early that macaques were susceptible to IAV (Saslaw et al., 1946), the animal models of choice remained ferrets and mice. Recently, macaques have been used to compare the pathogenesis of highly virulent 1918 pandemic IAV and the pathogenic bird flu strain (H5N1) with a conventional H1N1 strain (Rimmelzwaan et al., 2001). Cynomolgus macaques infected with highly pathogenic H5N1 developed acute respiratory distress syndrome, fever, and necrotizing pneumonia (Rimmelzwaan et al., 2001). The 1918 IAV strain induced dysregulation of the antiviral response leading to insufficient protection of the host, which in turn resulted in acute respiratory distress and a fatal outcome (Kobasa et al., 2007). The 2009 pandemic H1N1 US isolate caused severe pathological lesions in the lungs of the macaques (Itoh et al., 2009). The three studies mentioned above used combined intra-tracheal delivery of high doses of virus. A recent study by Marriott et al. analyzed the outcome of challenge routes, including inhaled aerosol and intra-nasal instillation with low to moderate doses of H1N1 in cynomolgus macaques (Marriott et al., 2016). Virus replication was detected in all challenge groups, although the disease remained sub-clinical.
In bacteriological studies non-human primates are rarely used. For group A streptococcal infection longitudinal transcriptome analyses were performed in experimental pharyngitis (Virtaneva et al., 2005) and lower respiratory tract infection in cynomolgus macaques (Olsen et al., 2010a). The lower respiratory tract disease observed in macaques after GAS infection mimicked the clinical and pathological features of severe bronchopneumonia in humans (Olsen et al., 2010a). Another study by Olsen and colleagues analyzed the contribution of PVL of a highly virulent USA300 S. aureus strain in respiratory infection (Olsen et al., 2010b). Although the lower respiratory tract disease observed in monkey mimicked the clinical and pathological features of early mild to moderate pneumonia in humans, no involvement of PVL in lung pathology or immune cell influx of the lungs could be detected (Olsen et al., 2010b). The same research group has developed a non-lethal IAV (H3N2)-S. aureus co-infection model in cynomolgus macaques (Kobayashi et al., 2013). Pneumonia progression was monitored by clinical parameters assessment, blood chemistry, nasal swabs, and pathology of the lungs. Seasonal IAV infection in healthy cynomolgus macaques caused mild pneumonia, but did not predispose the animals to subsequent severe infection with the USA300 clone (Kobayashi et al., 2013).
Although macaques are frequently used for evaluation of pneumococcal vaccine efficacy, including testing the impact of 13-valent pneumococcal conjugate vaccine and 23-valent pneumococcal polysaccharide vaccine on antigen-specific memory B cell repertoires (Jia et al., 2017), only two studies on pneumococcal carriage and pneumonia were conducted in the last decade. In 2013, Philipp and colleagues analyzed the carriage rate of pneumococcus in 158 colony animals. None of the surveyed rhesus macaques carried S. pneumoniae in the nasopharynx (Philipp et al., 2012). The authors concluded that rhesus macaque is probably not a natural host of pneumococci. But, when infants were colonized with 19F strain via nasopharyngeal instillation, the colonization was induced in eight of eight infants, lasted for 2 weeks in all animals and for 7 weeks in more than 60% (Philipp et al., 2012). The same group tested detoxified pneumolysin (dPly) and pneumococcal histidine triad protein D (PhtD) as potential vaccine candidates to prevent pneumonia (Denoel et al., 2011). After immunization the rhesus macaques were challenged with a 19F pneumococcal strain. AS02-adjuvanted PhtD-dPly vaccine protected the animals against S. pneumoniae-induced pneumonia, which was linked to the capacity (i) to greatly reduce bacterial load within the first week post-challenge and (ii) the levels of PhtD- and Ply-specific antibodies (Denoel et al., 2011). Although only a few macaque studies on pneumonia exist, due to the close proximity to humans in terms of physiology and immunity, these animals can be a good model in the context of translational studies evaluating therapeutics and prophylaxis.
Despite the wide use of different animal models, the optimal in vivo model for human pneumonia remains to be identified. Small mammals including rodents are well known from a biological, genetic, and immunological point of view and are easy to maintain. The choice of these particular animals for infectious disease studies is often a result of a compromise between technical and financial options. However, they are also far from humans’ anatomy, physiology, immunology, and susceptibility to exclusively human pathogens. The experimental animal model should be chosen based on responses comparable to humans. Primates are usually legally reserved to specific topics. In this case, pigs could be an appropriate model system for studying infectious diseases including pneumonia (Figure 1). The composition and size of the porcine genome is comparable to that of humans (Hart et al., 2007). In addition, human and porcine organs have many common features and functions (Swindle et al., 2012). The upper respiratory tract of humans and pigs, including the lymphoid tissue in the nasopharynx, is anatomically similar. Furthermore, like humans, pigs possess tonsils, which are absent in mice (Horter et al., 2003). A major advantage of studying infectious diseases by utilizing pigs as a host organism is that pigs have a full set of innate and adaptive immune effectors. According to whole genome sequencing results the porcine immune system resembles over 80% of the human immune system, whereas mice share less than 10% with humans (Dawson et al., 2016). Most of the immune cell compartments identified in humans are also present in pigs (Piriou-Guzylack and Salmon, 2008; Fairbairn et al., 2011). In contrast to mice and similar to humans, pigs have 50–70% of circulating polymorph nuclear cells (Fairbairn et al., 2011). In addition, all functional cytokines or orthologs involved in Th1, Th2, Th17, and Treg paradigm and corresponding immune cells have been described in pigs (Murtaugh et al., 2009; Kaser et al., 2011; Kiros et al., 2011). Especially the very prominent human pro-inflammatory chemo-attractant, CXCL8, is present as an ortholog in pigs, whereas there is no homologue in mice (Fairbairn et al., 2011). In contrast to human monocytes, which can be divided in three subclasses (classical CD14+CD16-, non-classical CD14+CD16+, and intermediate CD14++CD16+), porcine monocytes consist of four subclasses (Chamorro et al., 2005; Fairbairn et al., 2013). Like human monocytes they express adhesion molecules, such as VLA-4 and LFA-1 and co-stimulatory molecules, including CD80 and CD86 (Chamorro et al., 2005).
The pig has previously been used to mimic a number of human infectious diseases. Examples for S. aureus infections with this model organism are wound infections (Sanden et al., 1989; Svedman et al., 1989), osteomyelitis (Jensen et al., 2010), and sepsis (Nielsen et al., 2009). Intravenous inoculation of piglets with pneumococci led to bacteremia during a 5 days period and was associated with fever and septic arthritis. Intranasal inoculation of piglets led to colonization for at least six consecutive days without causing clinical signs (De Greeff et al., 2016). In addition, research on respiratory infections of pigs by human pathogens including S. aureus (Luna et al., 2009), Mycobacterium tuberculosis (Gil et al., 2010), Bordetella pertussis (Elahi et al., 2007), Pseudomonas aeruginosa (Luna et al., 2009), and IAV (Khatri et al., 2010), was performed in recent years. The fact that pigs and humans are infected with identical subtypes of IAV (H1N1, H3N2), and show similar clinical presentation and pathogenesis, makes pigs an ideal model organism for studies on respiratory co-infections (Van Reeth et al., 1998). Especially IAV infections are already well established in swine (Van Reeth et al., 1998, 2002a,b; Jung et al., 2007; Khatri et al., 2010; Barbe et al., 2011).
In addition to the limited number of publications on pigs and human pathogens, a lot can be translated and learned from studies on the porcine zoonotic pathogen Streptococcus suis. S. suis usually inhabits mucosal surfaces of tonsils, nares, genital and alimentary tract of piglets. Once the microbial balance is disturbed, the bacteria can cause meningitis, septicemia, arthritis, and pneumonia in pigs (Staats et al., 1997). Some S. suis strains are considered to be hyper-virulent and others hypo- or avirulent. In general, serotype 2 is most frequently isolated from diseased pigs (Staats et al., 1997). S. suis can also cause severe diseases in humans including septicemia, meningitis, arthritis, and streptococcal toxic shock syndrome (Tang et al., 2006; Yu et al., 2006; Gottschalk et al., 2007). Although many in vivo studies on S. suis have been performed by utilizing mice as a model organism (Seitz et al., 2012; Auger et al., 2016), several other studies have shown the advantage of using swine as a natural host for S. suis (Bi et al., 2014; Ferrando et al., 2015). A recent publication by Lin and colleagues on H1N1 and S. suis co-infected piglets demonstrated the synergistic effects of both pathogens (Lin et al., 2015). Co-infected piglets had more severe clinical presentation and pathological changes in the lung, as compared to animals infected with single pathogens (Lin et al., 2015). In addition, genes associated with immune responses, inflammatory cytokine production, and apoptotic pathways were highly overexpressed in the co-infected group (Lin et al., 2015). Although the porcine model seems to be ideal to mimic human infectious diseases, there are also disadvantages, including, e.g., requirement for specialized experimental animal facilities, time consuming management, high maintenance costs, and limited availability of transgenic animals.
Although the use of animals contributes greatly to our understanding of infectious diseases, human 3D-organotypic tissue models and ex vivo organ tissues should be considered, as they are most valuable tools to study host–pathogen interactions in a more complex setting (Figure 1). Tissue engineering approaches were originally focused on regenerative medicine (Langer and Vacanti, 1993). In contrast to standard monolayer cell cultures, tissue models much more closely resemble the 3D architecture, cellular composition, and matrix complexity of the respective organ. In recent years tissue engineering was also successfully employed in a number of studies in infectious diseases, including Zika virus infections of cerebral organoids (Lancaster et al., 2013; Dang et al., 2016), Helicobacter pylori infections of gastric epithelial organoids (McCracken et al., 2014; Schlaermann et al., 2016), Escherichia coli and Rotavirus infections of gastrointestinal and small intestinal enteroids (Saxena et al., 2015; VanDussen et al., 2015), Entamoeba histolytica or Hepatitis B virus infections of hepatic sinusoid tissue (Petropolis et al., 2014, 2016), group A and G streptococcal or staphylococcal infections of skin tissue models (Siemens et al., 2015, 2016; Mairpady Shambat et al., 2016), and staphylococcal and Andes hantavirus infections of human lung tissue (Mairpady Shambat et al., 2015; Sundstrom et al., 2016). The adaptability of these tissue-engineered models to multiple pathogens suggests a great potential for studies of infectious diseases. For instance, the lung tissue model relevant for pneumonia consists of lung fibroblasts embedded in a collagen matrix with a stratified epithelial layer on top (Nguyen Hoang et al., 2012). The engineered tissue is suitable for implanting and studying immune cells, including dendritic cells, monocytes, macrophages, and even peripheral blood mononuclear cells (Nguyen Hoang et al., 2012; Mairpady Shambat et al., 2015). A recent publication demonstrated a two-hit-event of lung pathology in staphylococcal necrotizing pneumonia (Mairpady Shambat et al., 2015). While the α-toxin had direct damaging effect on the lung epithelium, PVL induced lung pathology indirectly through the lysis of neutrophils (Mairpady Shambat et al., 2015). All the studies mentioned above highlight a significant progress in the field of infectious diseases not only from a scientific point of view but also by contributing to the three R principle of animal experimentation (Russell, 1995).
On these terms, the use of cultured ex vivo human organ biopsies, which are rare due to ethical considerations, is an additional option to study host–pathogen interactions. This ex vivo system may overcome even the limitations of the engineered tissue. In recent years human ex vivo lung tissue infections with various microorganisms, including pneumococci (Szymanski et al., 2012; Fatykhova et al., 2015), Bacillus anthracis (Chakrabarty et al., 2007), Haemophilus influenzae (Zhang et al., 2016), and IAV (Nicholls et al., 2007; Chan et al., 2009), were performed. In the human setting, most of the work focused on tropism, severity of infections, release of inflammatory mediators, and replication rates of the microorganisms. In addition, recently also experiments on swine influenza virus (SIV) and S. suis co-infections of the porcine ex vivo lung slices were reported. Meng and colleagues showed that SIV promotes subsequent bacterial infections in a two-step process of which the first initial step was dependent on capsule expression, whereas the second step of bacterial invasion into deeper layers was capsule-independent and required virus-mediated damage (Meng et al., 2015). However, this is just a beginning and more investigations are needed to unravel the complexity underlying these highly invasive infections.
In summary, bacterial and viral co-infections of the respiratory tract are highly lethal and present a dramatic burden for the global health system. The synergy between bacterial and viral infectious agents is related to a variety of factors, including epithelial barrier damage, exaggerated innate immune response, and cytokine storm. Despite many advances in recent years, more knowledge on mechanisms and immunology of disease progression is needed. The synergistic mechanisms between viruses and bacteria leading to enhanced morbidity and mortality are poorly understood. In vivo characterizations of these severe infections are mainly performed in mice which poorly resemble the human physiology and immune system. Several efforts have been made to establish other models, including ferrets, guinea pigs, rabbits, rats, and non-human primates. However, all have limitations. Here, we suggest using the porcine model, which provides obvious advantages in studies of human infectious diseases and should be considered much more frequent for future studies on severe infectious diseases, including pneumonia.
NS conceived the concept for this review article. NS and SO-H wrote the manuscript. NS, SO-H, TCM, BK, PV-W, and SH read, edited, and reviewed the manuscript.
This research is supported by Federal Excellence Initiative of Mecklenburg Western Pomerania and European Social Fund (ESF) Grant KoInfekt (ESF_14-BM-A55-00xx_16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alymova, I. V., Green, A. M., Van De Velde, N., Mcauley, J. L., Boyd, K. L., Ghoneim, H. E., et al. (2011). Immunopathogenic and antibacterial effects of H3N2 influenza A virus PB1-F2 map to amino acid residues 62, 75, 79, and 82. J. Virol. 85, 12324–12333. doi: 10.1128/JVI.05872-11
Ampofo, K., Herbener, A., Blaschke, A. J., Heyrend, C., Poritz, M., Korgenski, K., et al. (2010). Association of 2009 pandemic influenza A (H1N1) infection and increased hospitalization with parapneumonic empyema in children in Utah. Pediatr. Infect. Dis. J. 29, 905–909. doi: 10.1097/INF.0b013e3181df2c70
Antonopoulou, A., Tsaganos, T., Tzepi, I. M., Giamarellou, H., and Giamarellos-Bourboulis, E. J. (2015). Comparative efficacy of tigecycline VERSUS vancomycin in an experimental model of soft tissue infection by methicillin-resistant Staphylococcus aureus producing Panton-Valentine leukocidin. J. Chemother. 27, 80–86. doi: 10.1179/1973947814Y.0000000171
Auger, J. P., Fittipaldi, N., Benoit-Biancamano, M. O., Segura, M., and Gottschalk, M. (2016). Virulence studies of different sequence types and geographical origins of Streptococcus suis serotype 2 in a mouse model of infection. Pathogens 5:E48. doi: 10.3390/pathogens5030048
Barbe, F., Atanasova, K., and Van Reeth, K. (2011). Cytokines and acute phase proteins associated with acute swine influenza infection in pigs. Vet. J. 187, 48–53. doi: 10.1016/j.tvjl.2009.12.012
Bartlett, A. H., and Hulten, K. G. (2010). Staphylococcus aureus pathogenesis: secretion systems, adhesins, and invasins. Pediatr. Infect. Dis. J. 29, 860–861. doi: 10.1097/INF.0b013e3181ef2477
Baxter, R., Yee, A., Aukes, L., Snow, V., Fireman, B., Atkinson, B., et al. (2016). Risk of underlying chronic medical conditions for invasive pneumococcal disease in adults. Vaccine 34, 4293–4297. doi: 10.1016/j.vaccine.2016.07.003
Bergmann, S., and Hammerschmidt, S. (2007). Fibrinolysis and host response in bacterial infections. Thromb. Haemost. 98, 512–520. doi: 10.1160/TH07-02-0117
Bi, Y., Li, J., Yang, L., Zhang, S., Li, Y., Jia, X., et al. (2014). Assessment of the pathogenesis of Streptococcus suis type 2 infection in piglets for understanding streptococcal toxic shock-like syndrome, meningitis, and sequelae. Vet. Microbiol. 173, 299–309. doi: 10.1016/j.vetmic.2014.08.010
Biesbroek, G., Bosch, A. A., Wang, X., Keijser, B. J., Veenhoven, R. H., Sanders, E. A., et al. (2014a). The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 190, 298–308. doi: 10.1164/rccm.201401-0073OC
Biesbroek, G., Tsivtsivadze, E., Sanders, E. A., Montijn, R., Veenhoven, R. H., Keijser, B. J., et al. (2014b). Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 190, 1283–1292. doi: 10.1164/rccm.201407-1240OC
Biesbroek, G., Wang, X., Keijser, B. J., Eijkemans, R. M., Trzcinski, K., Rots, N. Y., et al. (2014c). Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. Emerg. Infect. Dis. 20, 201–210. doi: 10.3201/eid2002.131220
Bisno, A. L., Griffin, J. P., Van Epps, K. A., Niell, H. B., and Rytel, M. W. (1971). Pneumonia and Hong Kong influenza: a prospective study of the 1968-1969 epidemic. Am. J. Med. Sci. 261, 251–263. doi: 10.1097/00000441-197105000-00004
Black, R. E., Cousens, S., Johnson, H. L., Lawn, J. E., Rudan, I., Bassani, D. G., et al. (2010). Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 375, 1969–1987. doi: 10.1016/S0140-6736(10)60549-1
Blaser, M. J., and Falkow, S. (2009). What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7, 887–894. doi: 10.1038/nrmicro2245
Bogaert, D., De Groot, R., and Hermans, P. W. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154. doi: 10.1016/S1473-3099(04)00938-7
Bosch, A. A. T. M., Levin, E., Van Houten, M. A., Hasrat, R., Kalkman, G., Biesbroek, G., et al. (2016a). Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. Ebiomedicine 9, 336–345. doi: 10.1016/j.ebiom.2016.05.031
Bosch, A. A. T. M., Van Houten, M. A., Bruin, J. P., Wijmenga-Monsuur, A. J., Trzcinski, K., Bogaert, D., et al. (2016b). Nasopharyngeal carriage of Streptococcus pneumoniae and other bacteria in the 7th year after implementation of the pneumococcal conjugate vaccine in the Netherlands. Vaccine 34, 531–539. doi: 10.1016/j.vaccine.2015.11.060
Bottcher-Friebertshauser, E., Klenk, H. D., and Garten, W. (2013). Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 69, 87–100. doi: 10.1111/2049-632X.12053
Bouvier, N. M., and Lowen, A. C. (2010). Animal models for influenza virus pathogenesis and transmission. Viruses 2, 1530–1563. doi: 10.3390/v20801530
Brundage, J. F., and Shanks, G. D. (2007). What really happened during the 1918 influenza pandemic? The importance of bacterial secondary infections. J. Infect. Dis. 196, 1717–1718; author reply 1718–1719. doi: 10.1086/522355
Brundage, J. F., and Shanks, G. D. (2008). Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg. Infect. Dis 14, 1193–1199. doi: 10.3201/eid1408.071313
Bucasas, K. L., Mian, A. I., Demmler-Harrison, G. J., Caviness, A. C., Piedra, P. A., Franco, L. M., et al. (2013). Global gene expression profiling in infants with acute respiratory syncytial virus broncholitis demonstrates systemic activation of interferon signaling networks. Pediatr. Infect. Dis. J. 32, e68–e76. doi: 10.1097/INF.0b013e318278b4b3
Burk, R. F., Schaffner, W., and Koenig, M. G. (1971). Severe influenza virus pneumonia in the pandemic of 1968-1969. Arch. Intern. Med. 127, 1122–1128. doi: 10.1001/archinte.1971.00310180138019
Cabellos, C., Macintyre, D. E., Forrest, M., Burroughs, M., Prasad, S., and Tuomanen, E. (1992). Differing roles for platelet-activating factor during inflammation of the lung and subarachnoid space. The special case of Streptococcus pneumoniae. J. Clin. Invest. 90, 612–618. doi: 10.1172/JCI115900
Canning, B. J., and Chou, Y. (2008). Using guinea pigs in studies relevant to asthma and COPD. Pulm. Pharmacol. Ther. 21, 702–720. doi: 10.1016/j.pupt.2008.01.004
Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694. doi: 10.1016/S1473-3099(05)70267-X
Carsillo, M., Kutala, V. K., Puschel, K., Blanco, J., Kuppusamy, P., and Niewiesk, S. (2009). Nitric oxide production and nitric oxide synthase type 2 expression by cotton rat (Sigmodon hispidus) macrophages reflect the same pattern as human macrophages. Dev. Comp. Immunol. 33, 718–724. doi: 10.1016/j.dci.2008.12.004
Chaban, B., Albert, A., Links, M. G., Gardy, J., Tang, P., and Hill, J. E. (2013). Characterization of the upper respiratory tract microbiomes of patients with pandemic H1N1 influenza. PLOS ONE 8:e69559. doi: 10.1371/journal.pone.0069559
Chakrabarty, K., Wu, W., Booth, J. L., Duggan, E. S., Nagle, N. N., Coggeshall, K. M., et al. (2007). Human lung innate immune response to Bacillus anthracis spore infection. Infect. Immun. 75, 3729–3738. doi: 10.1128/IAI.00046-07
Chamorro, S., Revilla, C., Alvarez, B., Alonso, F., Ezquerra, A., and Dominguez, J. (2005). Phenotypic and functional heterogeneity of porcine blood monocytes and its relation with maturation. Immunology 114, 63–71. doi: 10.1111/j.1365-2567.2004.01994.x
Chan, R. W., Chan, M. C., Wong, A. C., Karamanska, R., Dell, A., Haslam, S. M., et al. (2009). DAS181 inhibits H5N1 influenza virus infection of human lung tissues. Antimicrob. Agents Chemother. 53, 3935–3941. doi: 10.1128/AAC.00389-09
Chaussee, M. S., Sandbulte, H. R., Schuneman, M. J., Depaula, F. P., Addengast, L. A., Schlenker, E. H., et al. (2011). Inactivated and live, attenuated influenza vaccines protect mice against influenza: Streptococcus pyogenes super-infections. Vaccine 29, 3773–3781. doi: 10.1016/j.vaccine.2011.03.031
Chella Krishnan, K., Mukundan, S., Alagarsamy, J., Hur, J., Nookala, S., Siemens, N., et al. (2016). Genetic architecture of group A streptococcal necrotizing soft tissue infections in the mouse. PLOS Pathog. 12:e1005732. doi: 10.1371/journal.ppat.1005732
Chella Krishnan, K., Mukundan, S., Alagarsamy, J., Laturnus, D., and Kotb, M. (2015). Host genetic variations and sex differences potentiate predisposition, severity, and outcomes of group A Streptococcus-mediated necrotizing soft tissue infections. Infect. Immun. 84, 416–424. doi: 10.1128/IAI.01191-15
Chien, Y. W., Klugman, K. P., and Morens, D. M. (2009). Bacterial pathogens and death during the 1918 influenza pandemic. N. Engl. J. Med. 361, 2582–2583. doi: 10.1056/NEJMc0908216
Conenello, G. M., Zamarin, D., Perrone, L. A., Tumpey, T., and Palese, P. (2007). A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLOS Pathog. 3:1414–1421. doi: 10.1371/journal.ppat.0030141
Croisier-Bertin, D., Piroth, L., Charles, P. E., Larribeau, A., Biek, D., Ge, Y., et al. (2011). Ceftaroline versus ceftriaxone in a highly penicillin-resistant pneumococcal pneumonia rabbit model using simulated human dosing. Antimicrob. Agents Chemother. 55, 3557–3563. doi: 10.1128/AAC.01773-09
Cundell, D. R., and Tuomanen, E. I. (1994). Receptor specificity of adherence of Streptococcus pneumoniae to human type-II pneumocytes and vascular endothelial cells in vitro. Microb. Pathog. 17, 361–374. doi: 10.1006/mpat.1994.1082
Cunningham, M. W. (2000). Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511. doi: 10.1128/CMR.13.3.470-511.2000
Dang, J., Tiwari, S. K., Lichinchi, G., Qin, Y., Patil, V. S., Eroshkin, A. M., et al. (2016). Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 19, 258–265. doi: 10.1016/j.stem.2016.04.014
Davidson, S., Crotta, S., Mccabe, T. M., and Wack, A. (2014). Pathogenic potential of interferon alphabeta in acute influenza infection. Nat. Commun. 5:3864. doi: 10.1038/ncomms4864
Davis, C. C., Mellencamp, M. A., and Preheim, L. C. (1991). A model of pneumococcal pneumonia in chronically intoxicated rats. J. Infect. Dis. 163, 799–805. doi: 10.1093/infdis/163.4.799
Dawood, F. S., Dalton, C. B., Durrheim, D. N., and Hope, K. G. (2009). Rates of hospitalisation for acute respiratory illness and the emergence of pandemic (H1N1) 2009 virus in the Hunter New England area health service. Med. J. Aust. 191, 573–574.
Dawood, F. S., Iuliano, A. D., Reed, C., Meltzer, M. I., Shay, D. K., Cheng, P. Y., et al. (2012). Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 12, 687–695. doi: 10.1016/S1473-3099(12)70121-4
Dawson, H. D., Smith, A. D., Chen, C., and Urban, J. F. Jr. (2016). An in-depth comparison of the porcine, murine and human inflammasomes; lessons from the porcine genome and transcriptome. Vet. Microbiol. 202, 2–15. doi: 10.1016/j.vetmic.2016.05.013
De Greeff, A., Van Selm, S., Buys, H., Harders-Westerveen, J. F., Tunjungputri, R. N., De Mast, Q., et al. (2016). Pneumococcal colonization and invasive disease studied in a porcine model. BMC Microbiol. 16:102. doi: 10.1186/s12866-016-0718-3
de Steenhuijsen Piters, W. A., Heinonen, S., Hasrat, R., Bunsow, E., Smith, B., Suarez-Arrabal, M. C., et al. (2016). Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am. J. Respir. Crit. Care Med. 194, 1104–1115. doi: 10.1164/rccm.201602-0220OC
Denoel, P., Philipp, M. T., Doyle, L., Martin, D., Carletti, G., and Poolman, J. T. (2011). A protein-based pneumococcal vaccine protects rhesus macaques from pneumonia after experimental infection with Streptococcus pneumoniae. Vaccine 29, 5495–5501. doi: 10.1016/j.vaccine.2011.05.051
Diavatopoulos, D. A., Short, K. R., Price, J. T., Wilksch, J. J., Brown, L. E., Briles, D. E., et al. (2010). Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 24, 1789–1798. doi: 10.1096/fj.09-146779
Diep, B. A., Chan, L., Tattevin, P., Kajikawa, O., Martin, T. R., Basuino, L., et al. (2010). Polymorphonuclear leukocytes mediate Staphylococcus aureus panton-valentine leukocidin-induced lung inflammation and injury. Proc. Natl. Acad. Sci. U.S.A. 107, 5587–5592. doi: 10.1073/pnas.0912403107
Diep, B. A., Hilliard, J. J., Le, V. T., Tkaczyk, C., Le, H. N., Tran, V. G., et al. (2017). Targeting alpha toxin to mitigate its lethal toxicity in ferret and rabbit models of Staphylococcus aureus necrotizing pneumonia. Antimicrob. Agents Chemother. 61, e02456-16. doi: 10.1128/AAC.02456-16
Dominguez-Cherit, G., Lapinsky, S. E., Macias, A. E., Pinto, R., Espinosa-Perez, L., De La Torre, A., et al. (2009). Critically Ill patients with 2009 influenza A(H1N1) in Mexico. JAMA 302, 1880–1887. doi: 10.1001/jama.2009.1536
Domon, H., Oda, M., Maekawa, T., Nagai, K., Takeda, W., and Terao, Y. (2016). Streptococcus pneumoniae disrupts pulmonary immune defence via elastase release following pneumolysin-dependent neutrophil lysis. Sci. Rep. 6:38013. doi: 10.1038/srep38013
Durbin, R. K., Kotenko, S. V., and Durbin, J. E. (2013). Interferon induction and function at the mucosal surface. Immunol. Rev. 255, 25–39. doi: 10.1111/imr.12101
Elahi, S., Holmstrom, J., and Gerdts, V. (2007). The benefits of using diverse animal models for studying pertussis. Trends Microbiol. 15, 462–468. doi: 10.1016/j.tim.2007.09.003
Estenssoro, E., Rios, F. G., Apezteguia, C., Reina, R., Neira, J., Ceraso, D. H., et al. (2010). Pandemic 2009 influenza a in Argentina: a study of 337 patients on mechanical ventilation. Am. J. Respir. Crit. Care Med. 182, 41–48. doi: 10.1164/201001-0037OC
Fairbairn, L., Kapetanovic, R., Beraldi, D., Sester, D. P., Tuggle, C. K., Archibald, A. L., et al. (2013). Comparative analysis of monocyte subsets in the pig. J. Immunol. 190, 6389–6396. doi: 10.4049/jimmunol.1300365
Fairbairn, L., Kapetanovic, R., Sester, D. P., and Hume, D. A. (2011). The mononuclear phagocyte system of the pig as a model for understanding human innate immunity and disease. J. Leukoc. Biol. 89, 855–871. doi: 10.1189/jlb.1110607
Fatykhova, D., Rabes, A., Machnik, C., Guruprasad, K., Pache, F., Berg, J., et al. (2015). Serotype 1 and 8 pneumococci evade sensing by inflammasomes in human lung tissue. PLOS ONE 10:e0137108. doi: 10.1371/journal.pone.0137108
Feng, X., Maze, M., Koch, L. G., Britton, S. L., and Hellman, J. (2015). Exaggerated acute lung injury and impaired antibacterial defenses during Staphylococcus aureus infection in rats with the metabolic syndrome. PLOS ONE 10:e0126906. doi: 10.1371/journal.pone.0126906
Ferrando, M. L., De Greeff, A., Van Rooijen, W. J., Stockhofe-Zurwieden, N., Nielsen, J., Wichgers Schreur, P. J., et al. (2015). Host-pathogen interaction at the intestinal mucosa correlates with zoonotic potential of Streptococcus suis. J. Infect. Dis. 212, 95–105. doi: 10.1093/infdis/jiu813
Fulde, M., Steinert, M., and Bergmann, S. (2013). Interaction of streptococcal plasminogen binding proteins with the host fibrinolytic system. Front. Cell Infect. Microbiol. 3:85. doi: 10.3389/fcimb.2013.00085
Gabbard, J. D., Dlugolenski, D., Van Riel, D., Marshall, N., Galloway, S. E., Howerth, E. W., et al. (2014). Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J. Virol. 88, 1502–1512. doi: 10.1128/JVI.02959-13
Garcia-Suarez Mdel, M., Florez, N., Astudillo, A., Vazquez, F., Villaverde, R., Fabrizio, K., et al. (2007). The role of pneumolysin in mediating lung damage in a lethal pneumococcal pneumonia murine model. Respir. Res. 8:3.
Gasparini, R., Amicizia, D., Lai, P. L., and Panatto, D. (2012). Aflunov((R)): a prepandemic influenza vaccine. Exp. Rev. Vaccines 11, 145–157. doi: 10.1586/erv.11.170
Gerberding, J. L. (2006). Pandemic preparedness: pigs, poultry, and people versus plans, products, and practice. J. Infect. Dis. 194(Suppl. 2), S77–S81. doi: 10.1086/507563
Ghoneim, H. E., Thomas, P. G., and Mccullers, J. A. (2013). Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J. Immunol. 191, 1250–1259. doi: 10.4049/jimmunol.1300014
Gil, O., Diaz, I., Vilaplana, C., Tapia, G., Diaz, J., Fort, M., et al. (2010). Granuloma encapsulation is a key factor for containing tuberculosis infection in minipigs. PLOS ONE 5:e10030. doi: 10.1371/journal.pone.0010030
Giles, C., and Shuttleworth, E. M. (1957). Postmortem findings in 46 influenza deaths. Lancet 273, 1224–1225. doi: 10.1016/S0140-6736(57)90180-0
Gottschalk, M., Segura, M., and Xu, J. (2007). Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim. Health Res. Rev. 8, 29–45. doi: 10.1017/S1466252307001247
Goulding, J., Godlee, A., Vekaria, S., Hilty, M., Snelgrove, R., and Hussell, T. (2011). Lowering the threshold of lung innate immune cell activation alters susceptibility to secondary bacterial superinfection. J. Infect. Dis. 204, 1086–1094. doi: 10.1093/infdis/jir467
Gurel, V., Lambert, K., Page, A. E., Loynachan, A. T., Huges, K., Timoney, J. F., et al. (2013). Streptolysin-O/antibiotics adjunct therapy modulates site-specific expression of extracellular matrix and inflammatory genes in lungs of Rhodococcus equi infected foals. Vet. Res. Commun. 37, 145–154. doi: 10.1007/s11259-013-9557-y
Hackam, D. G., and Redelmeier, D. A. (2006). Translation of research evidence from animals to humans. JAMA 296, 1731–1732. doi: 10.1001/jama.296.14.1731
Hammerschmidt, S. (2006). Adherence molecules of pathogenic pneumococci. Curr. Opin. Microbiol. 9, 12–20. doi: 10.1016/j.mib.2005.11.001
Harford, C. G., Smith, M. R., and Wood, W. B. (1946). Sulfonamide chemotherapy of combined infection with influenza virus and bacteria. J. Exp. Med. 83, 505–518. doi: 10.1084/jem.83.6.505
Hart, E. A., Caccamo, M., Harrow, J. L., Humphray, S. J., Gilbert, J. G., Trevanion, S., et al. (2007). Lessons learned from the initial sequencing of the pig genome: comparative analysis of an 8 Mb region of pig chromosome 17. Genome Biol. 8:R168. doi: 10.1186/gb-2007-8-8-r168
Hartman, B. J., and Tomasz, A. (1984). Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158, 513–516.
Hashimoto, Y., Moki, T., Takizawa, T., Shiratsuchi, A., and Nakanishi, Y. (2007). Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J. Immunol. 178, 2448–2457. doi: 10.4049/jimmunol.178.4.2448
Heldens, J. G., Glansbeek, H. L., Hilgers, L. A., Haenen, B., Stittelaar, K. J., Osterhaus, A. D., et al. (2010). Feasibility of single-shot H5N1 influenza vaccine in ferrets, macaques and rabbits. Vaccine 28, 8125–8131. doi: 10.1016/j.vaccine.2010.09.097
Hers, J. F., Goslings, W. R., Masurel, N., and Mulder, J. (1957). Death from asiatic influenza in the Netherlands. Lancet 273, 1164–1165. doi: 10.1016/S0140-6736(57)92066-4
Hers, J. F., Masurel, N., and Mulder, J. (1958). Bacteriology and histopathology of the respiratory tract and lungs in fatal Asian influenza. Lancet 2, 1141–1143. doi: 10.1016/S0140-6736(58)92404-8
Hillaire, M. L., Haagsman, H. P., Osterhaus, A. D., Rimmelzwaan, G. F., and Van Eijk, M. (2013). Pulmonary surfactant protein D in first-line innate defence against influenza a virus infections. J. Innate Immun. 5, 197–208. doi: 10.1159/000346374
Hilty, M., Qi, W., Brugger, S. D., Frei, L., Agyeman, P., Frey, P. M., et al. (2012). Nasopharyngeal microbiota in infants with acute otitis media. J. Infect. Dis. 205, 1048–1055. doi: 10.1093/infdis/jis024
Hoover, J., Lewandowski, T., Straub, R. J., Novick, S. J., Demarsh, P., Aubart, K., et al. (2015). Pharmacokinetics/Pharmacodynamics of peptide deformylase inhibitor GSK1322322 against Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in rodent models of infection. Antimicrob. Agents Chemother. 60, 180–189. doi: 10.1128/AAC.01842-15
Horter, D. C., Yoon, K. J., and Zimmerman, J. J. (2003). A review of porcine tonsils in immunity and disease. Anim. Health Res. Rev. 4, 143–155. doi: 10.1079/AHRR200358
Hraiech, S., Papazian, L., Rolain, J. M., and Bregeon, F. (2015). Animal models of polymicrobial pneumonia. Drug Des. Devel. Ther. 9, 3279–3292. doi: 10.2147/DDDT.S70993
Hussell, T., and Cavanagh, M. M. (2009). The innate immune rheostat: influence on lung inflammatory disease and secondary bacterial pneumonia. Biochem. Soc. Trans. 37, 811–813. doi: 10.1042/BST0370811
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108
Iinuma, H., and Okinaga, K. (1989). Prevention of pneumococcal bacteremia by immunization with type 6 pneumococcal capsular polysaccharide vaccine in splenectomized rats. J. Infect. Dis. 160, 66–75. doi: 10.1093/infdis/160.1.66
Imai, Y., Kuba, K., Neely, G. G., Yaghubian-Malhami, R., Perkmann, T., Van Loo, G., et al. (2008). Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133, 235–249. doi: 10.1016/j.cell.2008.02.043
Ioannidis, J. P. (2005). Contradicted and initially stronger effects in highly cited clinical research. JAMA 294, 218–228. doi: 10.1001/jama.294.2.218
Itoh, Y., Shinya, K., Kiso, M., Watanabe, T., Sakoda, Y., Hatta, M., et al. (2009). In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460, 1021–1025. doi: 10.1038/nature08260
Iwasaki, A., and Pillai, P. S. (2014). Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328. doi: 10.1038/nri3665
Jensen, H. E., Nielsen, O. L., Agerholm, J. S., Iburg, T., Johansen, L. K., Johannesson, E., et al. (2010). A non-traumatic Staphylococcus aureus osteomyelitis model in pigs. In Vivo 24, 257–264.
Jia, B., Mcneil, L. K., Dupont, C. D., Tsioris, K., Barry, R. M., Scully, I. L., et al. (2017). Longitudinal multiparameter single-cell analysis of macaques immunized with pneumococcal protein-conjugated or unconjugated polysaccharide vaccines reveals distinct antigen specific memory B cell repertoires. PLOS ONE 12:e0183738. doi: 10.1371/journal.pone.0183738
Joyce, E. A., Popper, S. J., and Falkow, S. (2009). Streptococcus pneumoniae nasopharyngeal colonization induces type I interferons and interferon-induced gene expression. BMC Genomics 10:404. doi: 10.1186/1471-2164-10-404
Jung, K., Alekseev, K. P., Zhang, X., Cheon, D. S., Vlasova, A. N., and Saif, L. J. (2007). Altered pathogenesis of porcine respiratory coronavirus in pigs due to immunosuppressive effects of dexamethasone: implications for corticosteroid use in treatment of severe acute respiratory syndrome coronavirus. J. Virol. 81, 13681–13693. doi: 10.1128/JVI.01702-07
Kaser, T., Gerner, W., and Saalmuller, A. (2011). Porcine regulatory T cells: mechanisms and T-cell targets of suppression. Dev. Comp. Immunol. 35, 1166–1172. doi: 10.1016/j.dci.2011.04.006
Khatri, M., Dwivedi, V., Krakowka, S., Manickam, C., Ali, A., Wang, L., et al. (2010). Swine influenza H1N1 virus induces acute inflammatory immune responses in pig lungs: a potential animal model for human H1N1 influenza virus. J. Virol. 84, 11210–11218. doi: 10.1128/JVI.01211-10
Kiros, T. G., Van Kessel, J., Babiuk, L. A., and Gerdts, V. (2011). Induction, regulation and physiological role of IL-17 secreting helper T-cells isolated from PBMC, thymus, and lung lymphocytes of young pigs. Vet. Immunol. Immunopathol. 144, 448–454. doi: 10.1016/j.vetimm.2011.08.021
Kline, B. S., and Winternitz, M. C. (1913). Studies upon experimental pneumonia in rabbits : Vi. Studies in immunity. J. Exp. Med. 18, 61–74. doi: 10.1084/jem.18.1.61
Kobasa, D., Jones, S. M., Shinya, K., Kash, J. C., Copps, J., Ebihara, H., et al. (2007). Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323. doi: 10.1038/nature05495
Kobayashi, S. D., Olsen, R. J., Lacasse, R. A., Safronetz, D., Ashraf, M., Porter, A. R., et al. (2013). Seasonal H3N2 influenza A virus fails to enhance Staphylococcus aureus co-infection in a non-human primate respiratory tract infection model. Virulence 4, 707–715. doi: 10.4161/viru.26572
Koppe, U., Suttorp, N., and Opitz, B. (2012). Recognition of Streptococcus pneumoniae by the innate immune system. Cell Microbiol. 14, 460–466. doi: 10.1111/j.1462-5822.2011.01746.x
Koppen, I. J. N., Bosch, A. A. T. M., Sanders, E. A. M., Van Houten, M. A., and Bogaert, D. (2015). The respiratory microbiota during health and disease: a paediatric perspective. Pneumonia 6, 90–100. doi: 10.15172/pneu.2015.6/656
Korteweg, C., and Gu, J. (2008). Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 172, 1155–1170. doi: 10.2353/ajpath.2008.070791
Kumagai, Y., Takeuchi, O., Kato, H., Kumar, H., Matsui, K., Morii, E., et al. (2007). Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27, 240–252. doi: 10.1016/j.immuni.2007.07.013
Kuri, T., Sorensen, A. S., Thomas, S., Karlsson Hedestam, G. B., Normark, S., Henriques-Normark, B., et al. (2013). Influenza A virus-mediated priming enhances cytokine secretion by human dendritic cells infected with Streptococcus pneumoniae. Cell Microbiol. 15, 1385–1400. doi: 10.1111/cmi.12122
Lancaster, M. A., Renner, M., Martin, C. A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Langer, R., and Vacanti, J. P. (1993). Tissue engineering. Science 260, 920–926. doi: 10.1126/science.8493529
Leung, R. K., Zhou, J. W., Guan, W., Li, S. K., Yang, Z. F., and Tsui, S. K. (2013). Modulation of potential respiratory pathogens by pH1N1 viral infection. Clin. Microbiol. Infect 19, 930–935. doi: 10.1111/1469-0691.12054
Li, N., Ren, A., Wang, X., Fan, X., Zhao, Y., Gao, G. F., et al. (2015). Influenza viral neuraminidase primes bacterial coinfection through TGF-beta-mediated expression of host cell receptors. Proc. Natl. Acad. Sci. U.S.A. 112, 238–243. doi: 10.1073/pnas.1414422112
Lin, X., Huang, C., Shi, J., Wang, R., Sun, X., Liu, X., et al. (2015). Investigation of pathogenesis of H1N1 influenza virus and swine Streptococcus suis serotype 2 co-infection in pigs by microarray analysis. PLOS ONE 10:e0124086. doi: 10.1371/journal.pone.0124086
Linke, C., Siemens, N., Oehmcke, S., Radjainia, M., Law, R. H., Whisstock, J. C., et al. (2012). The extracellular protein factor Epf from Streptococcus pyogenes is a cell surface adhesin that binds to cells through an N-terminal domain containing a carbohydrate-binding module. J. Biol. Chem. 287, 38178–38189. doi: 10.1074/jbc.M112.376434
Louria, D. B., Blumenfeld, H. L., Ellis, J. T., Kilbourne, E. D., and Rogers, D. E. (1959). Studies on influenza in the pandemic of 1957-1958. II. Pulmonary complications of influenza. J. Clin. Invest. 38, 213–265. doi: 10.1172/JCI103791
Lowen, A. C., Mubareka, S., Tumpey, T. M., Garcia-Sastre, A., and Palese, P. (2006). The guinea pig as a transmission model for human influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 103, 9988–9992. doi: 10.1073/pnas.0604157103
Loza-Tulimowska, M., Michalak, T., Semkow, R., and Nowoslawski, A. (1977). Autoantibodies in sera of rabbits immunized with influenza virus. Acta Virol. 21, 495–498.
Luna, C. M., Sibila, O., Agusti, C., and Torres, A. (2009). Animal models of ventilator-associated pneumonia. Eur. Respir. J. 33, 182–188. doi: 10.1183/09031936.00046308
Mairpady Shambat, S., Chen, P., Nguyen Hoang, A. T., Bergsten, H., Vandenesch, F., Siemens, N., et al. (2015). Modelling staphylococcal pneumonia in a human 3D lung tissue model system delineates toxin-mediated pathology. Dis. Model. Mech. 8, 1413–1425. doi: 10.1242/dmm.021923
Mairpady Shambat, S., Siemens, N., Monk, I. R., Mohan, D. B., Mukundan, S., Krishnan, K. C., et al. (2016). A point mutation in AgrC determines cytotoxic or colonizing properties associated with phenotypic variants of ST22 MRSA strains. Sci. Rep. 6:31360. doi: 10.1038/srep31360
Marrie, T. J., Tyrrell, G. J., Majumdar, S. R., and Eurich, D. T. (2017). Effect of age on the manifestations and outcomes of invasive pneumococcal disease in adults. Am. J. Med. 131, 100.e1–100.e7. doi: 10.1016/j.amjmed.2017.06.039
Marriott, A. C., Dennis, M., Kane, J. A., Gooch, K. E., Hatch, G., Sharpe, S., et al. (2016). Influenza A virus challenge models in cynomolgus macaques using the authentic inhaled aerosol and intra-nasal routes of infection. PLOS ONE 11:e0157887. doi: 10.1371/journal.pone.0157887
Martin, C. M., Kunin, C. M., Gottlieb, L. S., and Finland, M. (1959). Asian influenza A in Boston, 1957-1958. II. Severe staphylococcal pneumonia complicating influenza. AMA Arch. Intern. Med. 103, 532–542. doi: 10.1001/archinte.1959.00270040018002
Mauad, T., Hajjar, L. A., Callegari, G. D., Da Silva, L. F., Schout, D., Galas, F. R., et al. (2010). Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Respir. Crit. Care Med. 181, 72–79. doi: 10.1164/rccm.200909-1420OC
McAuley, J. L., Chipuk, J. E., Boyd, K. L., Van De Velde, N., Green, D. R., and Mccullers, J. A. (2010). PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLOS Pathog. 6:e1001014. doi: 10.1371/journal.ppat.1001014
McAuley, J. L., Hornung, F., Boyd, K. L., Smith, A. M., Mckeon, R., Bennink, J., et al. (2007). Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2, 240–249. doi: 10.1016/j.chom.2007.09.001
McCaskill, M. L., Mason, EO Jr, Kaplan, S. L., Hammerman, W., Lamberth, L. B., and Hultén, K. G. (2007). Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections. Pediatr. Infect. Dis. J. 26, 1122–1127. doi: 10.1097/INF.0b013e31814536e0
McCracken, K. W., Cata, E. M., Crawford, C. M., Sinagoga, K. L., Schumacher, M., Rockich, B. E., et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516, 400–404. doi: 10.1038/nature13863
McCullers, J. A. (2006). Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 19, 571–582. doi: 10.1128/CMR.00058-05
McCullers, J. A. (2013). Do specific virus-bacteria pairings drive clinical outcomes of pneumonia? Clin. Microbiol. Infect. 19, 113–118. doi: 10.1111/1469-0691.12093
McCullers, J. A., and Bartmess, K. C. (2003). Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187, 1000–1009. doi: 10.1086/368163
McCullers, J. A., Mcauley, J. L., Browall, S., Iverson, A. R., Boyd, K. L., and Henriques Normark, B. (2010). Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J. Infect. Dis. 202, 1287–1295. doi: 10.1086/656333
McCullers, J. A., and Rehg, J. E. (2002). Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 186, 341–350. doi: 10.1086/341462
Meng, F., Wu, N. H., Nerlich, A., Herrler, G., Valentin-Weigand, P., and Seitz, M. (2015). Dynamic virus-bacterium interactions in a porcine precision-cut lung slice coinfection model: swine influenza virus paves the way for Streptococcus suis infection in a two-step process. Infect. Immun. 83, 2806–2815. doi: 10.1128/IAI.00171-15
Metzger, D. W., and Sun, K. (2013). Immune dysfunction and bacterial coinfections following influenza. J. Immunol. 191, 2047–2052. doi: 10.4049/jimmunol.1301152
Miller, E., Andrews, N. J., Waight, P. A., Slack, M. P., and George, R. C. (2011). Herd immunity and serotype replacement 4 years after seven-valent pneumococcal conjugate vaccination in England and Wales: an observational cohort study. Lancet Infect. Dis. 11, 760–768. doi: 10.1016/S1473-3099(11)70090-1
Mizgerd, J. P., and Skerrett, S. J. (2008). Animal models of human pneumonia. Am. J. Physiol. Lung. Cell Mol. Physiol. 294, L387–L398.
Monsalvo, A. C., Batalle, J. P., Lopez, M. F., Krause, J. C., Klemenc, J., Hernandez, J. Z., et al. (2011). Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat. Med. 17, 195–199. doi: 10.1038/nm.2262
Morens, D. M., Taubenberger, J. K., and Fauci, A. S. (2008). Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970. doi: 10.1086/591708
Morton, B., Pennington, S. H., and Gordon, S. B. (2014). Immunomodulatory adjuvant therapy in severe community-acquired pneumonia. Exp. Rev. Respir. Med. 8, 587–596. doi: 10.1586/17476348.2014.927736
Murphy, T. F., Bakaletz, L. O., and Smeesters, P. R. (2009). Microbial interactions in the respiratory tract. Pediatr. Infect. Dis. J. 28, S121–S126. doi: 10.1097/INF.0b013e3181b6d7ec
Murtaugh, M. P., Johnson, C. R., Xiao, Z., Scamurra, R. W., and Zhou, Y. (2009). Species specialization in cytokine biology: is interleukin-4 central to the T(H)1-T(H)2 paradigm in swine? Dev. Comp. Immunol. 33, 344–352. doi: 10.1016/j.dci.2008.06.014
Nakamura, S., Davis, K. M., and Weiser, J. N. (2011). Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 121, 3657–3665. doi: 10.1172/JCI57762
Neumann, G., Noda, T., and Kawaoka, Y. (2009). Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459, 931–939. doi: 10.1038/nature08157
Nguyen, D. T., Louwen, R., Elberse, K., Van Amerongen, G., Yuksel, S., Luijendijk, A., et al. (2015). Streptococcus pneumoniae enhances human respiratory syncytial virus infection in vitro and in vivo. PLOS ONE 10:e0127098. doi: 10.1371/journal.pone.0127098
Nguyen, T., Kyle, U. G., Jaimon, N., Tcharmtchi, M. H., Coss-Bu, J. A., Lam, F., et al. (2012). Coinfection with Staphylococcus aureus increases risk of severe coagulopathy in critically ill children with influenza A (H1N1) virus infection. Crit. Care Med. 40, 3246–3250. doi: 10.1097/CCM.0b013e318260c7f8
Nguyen Hoang, A. T., Chen, P., Juarez, J., Sachamitr, P., Billing, B., Bosnjak, L., et al. (2012). Dendritic cell functional properties in a three-dimensional tissue model of human lung mucosa. Am. J. Physiol. Lung. Cell Mol. Physiol. 302, L226–L237. doi: 10.1152/ajplung.00059.2011
Nicholls, J. M., Bourne, A. J., Chen, H., Guan, Y., and Peiris, J. S. (2007). Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir. Res. 8:73. doi: 10.1186/1465-9921-8-73
Nielsen, O. L., Iburg, T., Aalbaek, B., Leifsson, P. S., Agerholm, J. S., Heegaard, P., et al. (2009). A pig model of acute Staphylococcus aureus induced pyemia. Acta Vet. Scand. 51:14. doi: 10.1186/1751-0147-51-14
O’Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., Deloria-Knoll, M., Mccall, N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. doi: 10.1016/S0140-6736(09)61204-6
Okumura, C. Y., and Nizet, V. (2014). Subterfuge and sabotage: evasion of host innate defenses by invasive gram-positive bacterial pathogens. Annu. Rev. Microbiol. 68, 439–458. doi: 10.1146/annurev-micro-092412-155711
Oliveira, T. L., Candeia-Medeiros, N., Cavalcante-Araujo, P. M., Melo, I. S., Favaro-Pipi, E., Fatima, L. A., et al. (2016). SGLT1 activity in lung alveolar cells of diabetic rats modulates airway surface liquid glucose concentration and bacterial proliferation. Sci. Rep. 6:21752. doi: 10.1038/srep21752
Olsen, R. J., Ashraf, M., Gonulal, V. E., Ayeras, A. A., Cantu, C., Shea, P. R., et al. (2010a). Lower respiratory tract infection in cynomolgus macaques (Macaca fascicularis) infected with group A Streptococcus. Microb. Pathog. 49, 336–347. doi: 10.1016/j.micpath.2010.06.012
Olsen, R. J., Kobayashi, S. D., Ayeras, A. A., Ashraf, M., Graves, S. F., Ragasa, W., et al. (2010b). Lack of a major role of Staphylococcus aureus Panton-Valentine leukocidin in lower respiratory tract infection in nonhuman primates. Am. J. Pathol. 176, 1346–1354. doi: 10.2353/ajpath.2010.090960
Osterholm, M. T. (2005). Preparing for the next pandemic. N. Engl. J. Med. 352, 1839–1842. doi: 10.1056/NEJMp058068
Otto, M. (2010). Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64, 143–162. doi: 10.1146/annurev.micro.112408.134309
Ottolini, M. G., Blanco, J. C., Eichelberger, M. C., Porter, D. D., Pletneva, L., Richardson, J. Y., et al. (2005). The cotton rat provides a useful small-animal model for the study of influenza virus pathogenesis. J. Gen. Virol. 86, 2823–2830. doi: 10.1099/vir.0.81145-0
Oxford, J. S., Lambkin, R., Elliot, A., Daniels, R., Sefton, A., and Gill, D. (2006). Scientific lessons from the first influenza pandemic of the 20th century. Vaccine 24, 6742–6746. doi: 10.1016/j.vaccine.2006.05.101
Peacock, S. J., De Silva, I., and Lowy, F. D. (2001). What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9, 605–610.
Petropolis, D. B., Faust, D. M., Deep Jhingan, G., and Guillen, N. (2014). A new human 3D-liver model unravels the role of galectins in liver infection by the parasite Entamoeba histolytica. PLOS Pathog. 10:e1004381. doi: 10.1371/journal.ppat.1004381
Petropolis, D. B., Faust, D. M., Tolle, M., Riviere, L., Valentin, T., Neuveut, C., et al. (2016). Human liver infection in a dish: easy-to-build 3D liver models for studying microbial infection. PLOS ONE 11:e0148667. doi: 10.1371/journal.pone.0148667
Pettigrew, M. M., Gent, J. F., Revai, K., Patel, J. A., and Chonmaitree, T. (2008). Microbial interactions during upper respiratory tract infections. Emerg. Infect. Dis. 14, 1584–1591. doi: 10.3201/eid1410.080119
Philipp, M. T., Doyle, L. A., Martin, D. S., Plauche, G. B., Phillippi-Falkenstein, K. M., and Bohm, R. P. Jr. (2012). A rhesus macaque model of Streptococcus pneumoniae carriage. J. Med. Primatol. 41, 60–66. doi: 10.1111/j.1600-0684.2011.00512.x
Piriou-Guzylack, L., and Salmon, H. (2008). Membrane markers of the immune cells in swine: an update. Vet. Res. 39:54. doi: 10.1051/vetres:2008030
Plotkowski, M. C., Bajolet-Laudinat, O., and Puchelle, E. (1993). Cellular and molecular mechanisms of bacterial adhesion to respiratory mucosa. Eur. Respir. J. 6, 903–916.
Ponte, C., Parra, A., Nieto, E., and Soriano, F. (1996). Development of experimental pneumonia by infection with penicillin-insensitive Streptococcus pneumoniae in guinea pigs and their treatment with amoxicillin, cefotaxime, and meropenem. Antimicrob. Agents Chemother. 40, 2698–2702.
Porto, B. N., and Stein, R. T. (2016). Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front. Immunol. 7:311. doi: 10.3389/fimmu.2016.00311
Pothlichet, J., Meunier, I., Davis, B. K., Ting, J. P., Skamene, E., Von Messling, V., et al. (2013). Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLOS Pathog. 9:e1003256. doi: 10.1371/journal.ppat.1003256
Preheim, L. C., Gentry, M. J., and Snitily, M. U. (1991). Pulmonary recruitment, adherence, and chemotaxis of neutrophils in a rat model of cirrhosis and pneumococcal pneumonia. J. Infect. Dis. 164, 1203–1206. doi: 10.1093/infdis/164.6.1203
Prina, E., Ceccato, A., and Torres, A. (2016). New aspects in the management of pneumonia. Crit. Care 20:267. doi: 10.1186/s13054-016-1442-y
Puchelle, E., Zahm, J. M., Tournier, J. M., and Coraux, C. (2006). Airway epithelial repair, regeneration, and remodeling after injury in chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 3, 726–733. doi: 10.1513/pats.200605-126SF
Ramos, I., and Fernandez-Sesma, A. (2012). Cell receptors for influenza a viruses and the innate immune response. Front. Microbiol. 3:117. doi: 10.3389/fmicb.2012.00117
Rasigade, J. P., Laurent, F., Lina, G., Meugnier, H., Bes, M., Vandenesch, F., et al. (2010). Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981-2007. J. Infect. Dis. 201, 1589–1597. doi: 10.1086/652008
Reichert, T., Chowell, G., Nishiura, H., Christensen, R. A., and Mccullers, J. A. (2010). Does glycosylation as a modifier of original antigenic sin explain the case age distribution and unusual toxicity in pandemic novel H1N1 influenza? BMC Infect. Dis. 10:5. doi: 10.1186/1471-2334-10-5
Rice, T. W., Rubinson, L., Uyeki, T. M., Vaughn, F. L., John, B. B., Miller, R. R., et al. (2012). Critical illness from 2009 pandemic influenza A virus and bacterial coinfection in the United States. Crit. Care Med. 40, 1487–1498. doi: 10.1097/CCM.0b013e3182416f23
Rimmelzwaan, G. F., Kuiken, T., Van Amerongen, G., Bestebroer, T. M., Fouchier, R. A., and Osterhaus, A. D. (2001). Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 75, 6687–6691. doi: 10.1128/JVI.75.14.6687-6691.2001
Robertson, L., Caley, J. P., and Moore, J. (1958). Importance of Staphylococcus aureus in pneumonia in the 1957 epidemic of influenza A. Lancet 2, 233–236. doi: 10.1016/S0140-6736(58)90060-6
Root, J. J., Shriner, S. A., Bentler, K. T., Gidlewski, T., Mooers, N. L., Spraker, T. R., et al. (2014). Shedding of a low pathogenic avian influenza virus in a common synanthropic mammal–the cottontail rabbit. PLOS ONE 9:e102513. doi: 10.1371/journal.pone.0102513
Root, J. J., Shriner, S. A., Ellis, J. W., Vandalen, K. K., and Sullivan, H. J. (2017). Low viral doses are sufficient to infect cottontail rabbits with avian influenza A virus. Arch. Virol. 162, 3381–3388. doi: 10.1007/s00705-017-3493-z
Rosas-Salazar, C., Shilts, M. H., Tovchigrechko, A., Chappell, J. D., Larkin, E. K., Nelson, K. E., et al. (2016a). Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood asthma risk. Am. J. Respir. Crit. Care Med. 193, 1180–1183. doi: 10.1164/rccm.201512-2350LE
Rosas-Salazar, C., Shilts, M. H., Tovchigrechko, A., Schobel, S., Chappell, J. D., Larkin, E. K., et al. (2016b). Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J. Infect. Dis. 214, 1924–1928.
Rothberg, M. B., Haessler, S. D., and Brown, R. B. (2008). Complications of viral influenza. Am. J. Med. 121, 258–264. doi: 10.1016/j.amjmed.2007.10.040
Saito, M., Nakayama, K., Matsubara, J., Matsuno, K., and Nakagawa, M. (1988). Enhanced multiplication and transmission of Streptococcus pneumoniae in guinea pigs by Sendai virus infection. Jikken Dobutsu 37, 59–65. doi: 10.1538/expanim1978.37.1_59
Sanden, G., Ljungh, A., Wadstrom, T., Banck, G., Miedzobrodzki, J., and Svedman, P. (1989). Staphylococcal wound infection in the pig: Part II. Inoculation, quantification of bacteria, and reproducibility. Ann. Plast. Surg. 23, 219–223. doi: 10.1097/00000637-198909000-00005
Sanford, B. A., and Ramsay, M. A. (1987). Bacterial adherence to the upper respiratory tract of ferrets infected with influenza A virus. Proc. Soc. Exp. Biol. Med. 185, 120–128. doi: 10.3181/00379727-185-42525
Saslaw, S., Wilson, H. E., Doan, C. A., Woolpert, O. C., and Schwab, J. L. (1946). Reactions of monkeys to experimentally induced influenza virus a infection : an analysis of the relative roles of humoral and cellular immunity under conditions of optimal or deficient nutrition. J. Exp. Med. 84, 113–125. doi: 10.1084/jem.84.2.113
Saxena, K., Blutt, S. E., Ettayebi, K., Zeng, X. L., Broughman, J. R., Crawford, S. E., et al. (2015). Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol. 90, 43–56. doi: 10.1128/JVI.01930-15
Scheiblauer, H., Reinacher, M., Tashiro, M., and Rott, R. (1992). Interactions between bacteria and influenza A virus in the development of influenza pneumonia. J. Infect. Dis. 166, 783–791. doi: 10.1093/infdis/166.4.783
Schlaermann, P., Toelle, B., Berger, H., Schmidt, S. C., Glanemann, M., Ordemann, J., et al. (2016). A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 65, 202–213. doi: 10.1136/gutjnl-2014-307949
Schwarzmann, S. W., Adler, J. L., Sullivan, R. J. Jr., and Marine, W. M. (1971). Bacterial pneumonia during the Hong Kong influenza epidemic of 1968-1969. Arch. Intern. Med. 127, 1037–1041. doi: 10.1001/archinte.1971.00310180053006
Seitz, M., Beineke, A., Seele, J., Fulde, M., Valentin-Weigand, P., and Baums, C. G. (2012). A novel intranasal mouse model for mucosal colonization by Streptococcus suis serotype 2. J. Med. Microbiol. 61, 1311–1318. doi: 10.1099/jmm.0.043885-0
Shahangian, A., Chow, E. K., Tian, X., Kang, J. R., Ghaffari, A., Liu, S. Y., et al. (2009). Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119, 1910–1920. doi: 10.1172/JCI35412
Shanks, G. D., Mackenzie, A., Mclaughlin, R., Waller, M., Dennis, P., Lee, S. E., et al. (2010). Mortality risk factors during the 1918-1919 influenza pandemic in the Australian army. J. Infect. Dis. 201, 1880–1889. doi: 10.1086/652868
Shanks, G. D., Mackenzie, A., Waller, M., and Brundage, J. F. (2011). Low but highly variable mortality among nurses and physicians during the influenza pandemic of 1918-1919. Influenza Other Respir. Viruses 5, 213–219. doi: 10.1111/j.1750-2659.2010.00195.x
Shanley, T. P., Schrier, D., Kapur, V., Kehoe, M., Musser, J. M., and Ward, P. A. (1996). Streptococcal cysteine protease augments lung injury induced by products of group A streptococci. Infect. Immun. 64, 870–877.
Shannon, O., Herwald, H., and Oehmcke, S. (2013). Modulation of the coagulation system during severe streptococcal disease. Curr. Top. Microbiol. Immunol. 368, 189–205. doi: 10.1007/82_2012_283
Sharma-Chawla, N., Sender, V., Kershaw, O., Gruber, A. D., Volckmar, J., Henriques-Normark, B., et al. (2016). Influenza A virus infection predisposes hosts to secondary infection with different Streptococcus pneumoniae serotypes with similar outcome but serotype-specific manifestation. Infect. Immun. 84, 3445–3457. doi: 10.1128/IAI.00422-16
Sharrar, R. G. (1969). National influenza experience in the USA, 1968-69. Bull. World Health Organ. 41, 361–366.
Shieh, W. J., Blau, D. M., Denison, A. M., Deleon-Carnes, M., Adem, P., Bhatnagar, J., et al. (2010). 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 177, 166–175. doi: 10.2353/ajpath.2010.100115
Shinya, K., Ebina, M., Yamada, S., Ono, M., Kasai, N., and Kawaoka, Y. (2006). Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436. doi: 10.1038/440435a
Sicot, N., Khanafer, N., Meyssonnier, V., Dumitrescu, O., Tristan, A., Bes, M., et al. (2013). Methicillin resistance is not a predictor of severity in community-acquired Staphylococcus aureus necrotizing pneumonia–results of a prospective observational study. Clin. Microbiol. Infect. 19, E142–E148. doi: 10.1111/1469-0691.12022
Siemens, N., Chakrakodi, B., Shambat, S. M., Morgan, M., Bergsten, H., Hyldegaard, O., et al. (2016). Biofilm in group A streptococcal necrotizing soft tissue infections. JCI Insight 1:e87882. doi: 10.1172/jci.insight.87882
Siemens, N., Fiedler, T., Normann, J., Klein, J., Munch, R., Patenge, N., et al. (2012). Effects of the ERES pathogenicity region regulator Ralp3 on Streptococcus pyogenes serotype M49 virulence factor expression. J. Bacteriol. 194, 3618–3626. doi: 10.1128/JB.00227-12
Siemens, N., Kittang, B. R., Chakrakodi, B., Oppegaard, O., Johansson, L., Bruun, T., et al. (2015). Increased cytotoxicity and streptolysin O activity in group G streptococcal strains causing invasive tissue infections. Sci. Rep. 5:16945. doi: 10.1038/srep16945
Small, C. L., Shaler, C. R., Mccormick, S., Jeyanathan, M., Damjanovic, D., Brown, E. G., et al. (2010). Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J. Immunol. 184, 2048–2056. doi: 10.4049/jimmunol.0902772
Smith, A. M., and McCullers, J. A. (2014). Secondary bacterial infections in influenza virus infection pathogenesis. Curr. Top. Microbiol. Immunol. 385, 327–356. doi: 10.1007/82_2014_394
Snall, J., Linner, A., Uhlmann, J., Siemens, N., Ibold, H., Janos, M., et al. (2016). Differential neutrophil responses to bacterial stimuli: Streptococcal strains are potent inducers of heparin-binding protein and resistin-release. Sci. Rep. 6:21288. doi: 10.1038/srep21288
Snelgrove, R. J., Goulding, J., Didierlaurent, A. M., Lyonga, D., Vekaria, S., Edwards, L., et al. (2008). A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 9, 1074–1083. doi: 10.1038/ni.1637
Spelmink, L., Sender, V., Hentrich, K., Kuri, T., Plant, L., and Henriques-Normark, B. (2016). Toll-like receptor 3/TRIF-dependent IL-12p70 secretion mediated by Streptococcus pneumoniae RNA and its priming by influenza A virus coinfection in human dendritic cells. mBio 7:e00168-16. doi: 10.1128/mBio.00168-16
Staats, J. J., Feder, I., Okwumabua, O., and Chengappa, M. M. (1997). Streptococcus suis: past and present. Vet. Res. Commun. 21, 381–407. doi: 10.1023/A:1005870317757
Stillman, E. G., and Branch, A. (1926). Susceptibility of rabbits to infection by the inhalation of virulent pneumococci. J. Exp. Med. 44, 581–587. doi: 10.1084/jem.44.5.581
Sun, K., and Metzger, D. W. (2008). Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 14, 558–564. doi: 10.1038/nm1765
Sundstrom, K. B., Nguyen Hoang, A. T., Gupta, S., Ahlm, C., Svensson, M., and Klingstrom, J. (2016). Andes hantavirus-infection of a 3D human lung tissue model reveals a late peak in progeny virus production followed by increased levels of proinflammatory cytokines and VEGF-A. PLOS ONE 11:e0149354. doi: 10.1371/journal.pone.0149354
Svedman, P., Ljungh, A., Rausing, A., Banck, G., Sanden, G., Miedzobrodzki, J., et al. (1989). Staphylococcal wound infection in the pig: Part I. course. Ann. Plast. Surg. 23, 212–218. doi: 10.1097/00000637-198909000-00004
Swindle, M. M., Makin, A., Herron, A. J., Clubb, F. J., and Frazier, K. S. (2012). Swine as models in biomedical research and toxicology testing (vol 49, pg 344, 2012). Vet. Pathol. 49, 738–738. doi: 10.1177/0300985811402846
Szymanski, K. V., Toennies, M., Becher, A., Fatykhova, D., N’guessan, P. D., Gutbier, B., et al. (2012). Streptococcus pneumoniae-induced regulation of cyclooxygenase-2 in human lung tissue. Eur. Respir. J. 40, 1458–1467. doi: 10.1183/09031936.00186911
Tang, J., Wang, C., Feng, Y., Yang, W., Song, H., Chen, Z., et al. (2006). Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLOS Med. 3:e151.
Teo, S. M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., et al. (2015). The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715. doi: 10.1016/j.chom.2015.03.008
Thangavel, R. R., and Bouvier, N. M. (2014). Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods 410, 60–79. doi: 10.1016/j.jim.2014.03.023
Tian, X., Xu, F., Lung, W. Y., Meyerson, C., Ghaffari, A. A., Cheng, G., et al. (2012). Poly I:C enhances susceptibility to secondary pulmonary infections by gram-positive bacteria. PLOS ONE 7:e41879. doi: 10.1371/journal.pone.0041879
Tjon-Kon-Fat, R., Meerhoff, T., Nikisins, S., Pires, J., Pereyaslov, D., Gross, D., et al. (2016). The potential risks and impact of the start of the 2015-2016 influenza season in the WHO European Region: a rapid risk assessment. Influenza Other Respir. Viruses 10, 236–246. doi: 10.1111/irv.12381
Tregoning, J. S., and Schwarze, J. (2010). Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin. Microbiol. Rev. 23, 74–98. doi: 10.1128/CMR.00032-09
Tripp, R. A., and Tompkins, S. M. (2009). Animal models for evaluation of influenza vaccines. Curr. Top. Microbiol. Immunol. 333, 397–412. doi: 10.1007/978-3-540-92165-3_19
Trotter, Y. Jr., Dunn, F. L., Drachman, R. H., Henderson, D. A., Pizzi, M., and Langmuir, A. D. (1959). Asian influenza in the United States, 1957-1958. Am. J. Hyg. 70, 34–50.
Tsai, P. J., Kuo, C. F., Lin, K. Y., Lin, Y. S., Lei, H. Y., Chen, F. F., et al. (1998). Effect of group A streptococcal cysteine protease on invasion of epithelial cells. Infect. Immun. 66, 1460–1466.
Tse, L. V., and Whittaker, G. R. (2015). Modification of the hemagglutinin cleavage site allows indirect activation of avian influenza virus H9N2 by bacterial staphylokinase. Virology 482, 1–8. doi: 10.1016/j.virol.2015.03.023
Ubukata, K., Nonoguchi, R., Matsuhashi, M., and Konno, M. (1989). Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J. Bacteriol. 171, 2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989
Uhlmann, J., Siemens, N., Kai-Larsen, Y., Fiedler, T., Bergman, P., Johansson, L., et al. (2016). Phosphoglycerate kinase-A novel streptococcal factor involved in neutrophil activation and degranulation. J. Infect. Dis. 214, 1876–1883. doi: 10.1093/infdis/jiw450
Valles, J., Diaz, E., Martin-Loeches, I., Bacelar, N., Saludes, P., Lema, J., et al. (2016). Evolution over a 15-year period of the clinical characteristics and outcomes of critically ill patients with severe community-acquired pneumonia. Med. Intensiva 40, 238–245. doi: 10.1016/j.medin.2015.07.005
van Belkum, A., Melles, D. C., Nouwen, J., Van Leeuwen, W. B., Van Wamel, W., Vos, M. C., et al. (2009). Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 9, 32–47. doi: 10.1016/j.meegid.2008.09.012
van der Meer, F. J., Orsel, K., and Barkema, H. W. (2010). The new influenza A H1N1 virus: balancing on the interface of humans and animals. Can. Vet. J. 51, 56–62.
van der Poll, T., and Herwald, H. (2014). The coagulation system and its function in early immune defense. Thromb. Haemost. 112, 640–648. doi: 10.1160/TH14-01-0053
van der Sluijs, K. F., Van Elden, L. J., Nijhuis, M., Schuurman, R., Pater, J. M., Florquin, S., et al. (2004). IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J. Immunol. 172, 7603–7609. doi: 10.4049/jimmunol.172.12.7603
Van Reeth, K., Nauwynck, H., and Pensaert, M. (1998). Bronchoalveolar interferon-alpha, tumor necrosis factor-alpha, interleukin-1, and inflammation during acute influenza in pigs: a possible model for humans? J. Infect. Dis. 177, 1076–1079. doi: 10.1086/517398
Van Reeth, K., Van Gucht, S., and Pensaert, M. (2002a). Correlations between lung proinflammatory cytokine levels, virus replication, and disease after swine influenza virus challenge of vaccination-immune pigs. Viral Immunol. 15, 583–594.
Van Reeth, K., Van Gucht, S., and Pensaert, M. (2002b). In vivo studies on cytokine involvement during acute viral respiratory disease of swine: troublesome but rewarding. Vet. Immunol. Immunopathol. 87, 161–168.
van Riel, D., Den Bakker, M. A., Leijten, L. M., Chutinimitkul, S., Munster, V. J., De Wit, E., et al. (2010). Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am. J. Pathol. 176, 1614–1618. doi: 10.2353/ajpath.2010.090949
van Riel, D., Munster, V. J., De Wit, E., Rimmelzwaan, G. F., Fouchier, R. A., Osterhaus, A. D., et al. (2007). Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171, 1215–1223. doi: 10.2353/ajpath.2007.070248
VanDussen, K. L., Marinshaw, J. M., Shaikh, N., Miyoshi, H., Moon, C., Tarr, P. I., et al. (2015). Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920. doi: 10.1136/gutjnl-2013-306651
Virtaneva, K., Porcella, S. F., Graham, M. R., Ireland, R. M., Johnson, C. A., Ricklefs, S. M., et al. (2005). Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. U.S.A. 102, 9014–9019. doi: 10.1073/pnas.0503671102
von Eiff, C., Becker, K., Machka, K., Stammer, H., and Peters, G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N. Engl. J. Med. 344, 11–16. doi: 10.1056/NEJM200101043440102
Voss, S., Gamez, G., and Hammerschmidt, S. (2012). Impact of pneumococcal microbial surface components recognizing adhesive matrix molecules on colonization. Mol. Oral. Microbiol. 27, 246–256. doi: 10.1111/j.2041-1014.2012.00654.x
Walters, K. A., D’agnillo, F., Sheng, Z. M., Kindrachuk, J., Schwartzman, L. M., Kuestner, R. E., et al. (2016). 1918 pandemic influenza virus and Streptococcus pneumoniae co-infection results in activation of coagulation and widespread pulmonary thrombosis in mice and humans. J. Pathol. 238, 85–97. doi: 10.1002/path.4638
Wang, J., Nikrad, M. P., Travanty, E. A., Zhou, B., Phang, T., Gao, B., et al. (2012). Innate immune response of human alveolar macrophages during influenza A infection. PLOS ONE 7:e29879. doi: 10.1371/journal.pone.0029879
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M., and Kawaoka, Y. (1992). Evolution and ecology of influenza A viruses. Microbiol. Rev. 56, 152–179.
Weinstock, G. M. (2012). Genomic approaches to studying the human microbiota. Nature 489, 250–256. doi: 10.1038/nature11553
Yamamoto, N., Kawakami, K., Kinjo, Y., Miyagi, K., Kinjo, T., Uezu, K., et al. (2004). Essential role for the p40 subunit of interleukin-12 in neutrophil-mediated early host defense against pulmonary infection with Streptococcus pneumoniae: involvement of interferon-gamma. Microbes Infect. 6, 1241–1249. doi: 10.1016/j.micinf.2004.08.007
Yang, Y., and Tang, H. (2016). Aberrant coagulation causes a hyper-inflammatory response in severe influenza pneumonia. Cell Mol. Immunol. 13, 432–442. doi: 10.1038/cmi.2016.1
Yu, H., Jing, H., Chen, Z., Zheng, H., Zhu, X., Wang, H., et al. (2006). Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12, 914–920. doi: 10.3201/eid1206.051194
Zahlten, J., Kim, Y. J., Doehn, J. M., Pribyl, T., Hocke, A. C., Garcia, P., et al. (2015). Streptococcus pneumoniae-induced oxidative stress in lung epithelial cells depends on Pneumococcal autolysis and is reversible by resveratrol. J. Infect. Dis. 211, 1822–1830. doi: 10.1093/infdis/jiu806
Zetola, N., Francis, J. S., Nuermberger, E. L., and Bishai, W. R. (2005). Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5, 275–286. doi: 10.1016/S1473-3099(05)70112-2
Keywords: pneumonia, co-infections, Influenza A virus, Gram-positive bacteria, Streptococcus pneumoniae, Staphylococcus aureus, Streptococcus pyogenes, animal models
Citation: Siemens N, Oehmcke-Hecht S, Mettenleiter TC, Kreikemeyer B, Valentin-Weigand P and Hammerschmidt S (2017) Port d’Entrée for Respiratory Infections – Does the Influenza A Virus Pave the Way for Bacteria? Front. Microbiol. 8:2602. doi: 10.3389/fmicb.2017.02602
Received: 20 September 2017; Accepted: 13 December 2017;
Published: 21 December 2017.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Michael S. Chaussee, University of South Dakota, United StatesCopyright © 2017 Siemens, Oehmcke-Hecht, Mettenleiter, Kreikemeyer, Valentin-Weigand and Hammerschmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nikolai Siemens, bmlrb2xhaS5zaWVtZW5zQHVuaS1ncmVpZnN3YWxkLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.