- 1Retroviral Immunology, The Francis Crick Institute, London, United Kingdom
- 2Retrovirus-Host Interactions, The Francis Crick Institute, London, United Kingdom
- 3Department of Medicine, Faculty of Medicine, Imperial College London, London, United Kingdom
In addition to evolutionarily-accrued sequence mutation or deletion, endogenous retroelements (EREs) in eukaryotic genomes are subject to epigenetic silencing, preventing or reducing their transcription, particularly in the germplasm. Nevertheless, transcriptional activation of EREs, including endogenous retroviruses (ERVs) and long interspersed nuclear elements (LINEs), is observed in somatic cells, variably upon cellular differentiation and frequently upon cellular transformation. ERE transcription is modulated during physiological and pathological immune cell activation, as well as in immune cell cancers. However, our understanding of the potential consequences of such modulation remains incomplete, partly due to the relative scarcity of information regarding genome-wide ERE transcriptional patterns in immune cells. Here, we describe a methodology that allows probing RNA-sequencing (RNA-seq) data for genome-wide expression of EREs in murine and human cells. Our analysis of B cells reveals that their transcriptional response during immune activation is dominated by induction of gene transcription, and that EREs respond to a much lesser extent. The transcriptional activity of the majority of EREs is either unaffected or reduced by B cell activation both in mice and humans, albeit LINEs appear considerably more responsive in the latter host. Nevertheless, a small number of highly distinct ERVs are strongly and consistently induced during B cell activation. Importantly, this pattern contrasts starkly with B cell transformation, which exhibits widespread induction of EREs, including ERVs that minimally overlap with those responsive to immune stimulation. The distinctive patterns of ERE induction suggest different underlying mechanisms and will help separate physiological from pathological expression.
Introduction
Vertebrate genomes contain a considerable number of endogenous retroelements (EREs) with various degrees of open reading frame (ORF) integrity and replication autonomy. Occupying approximately a fifth of the mouse and human genomes, long interspersed nuclear elements (LINEs) are the largest group of EREs (Lander et al., 2001; Mouse Genome Sequencing Consortium et al., 2002). LINEs are still capable of autonomous retrotransposition in both host species. They also provide the reverse-transcriptase (RT) activity and retrotransposition machinery for mobilization of other EREs that lack long terminal repeats (LTRs), collectively known as non-LTR elements, and occasionally also of processed RNAs from cellular genes (Burns and Boeke, 2012). Distinguished by the presence of LTRs flanking the proviral genome, endogenous retroviruses (ERVs) and mammalian apparent LTR-retrotransposons (MaLRs), together comprise approximately 9.8 and 8.5% of mouse and human genomes, respectively (Lander et al., 2001; Mouse Genome Sequencing Consortium et al., 2002). ERVs may still possess and express ORFs encoding functional RT, which is necessary for the replication of non-autonomous LTR elements, such as MaLRs (Mccarthy and Mcdonald, 2004; Mager and Stoye, 2015). However, only few distinct ERVs are still replication-competent in mice (Mccarthy and Mcdonald, 2004; Mager and Stoye, 2015), and ERV replication has not been demonstrated to date in humans (Kassiotis and Stoye, 2017).
In addition to loss of replication competence as a result of sequence mutation or deletion sustained over long evolutionary periods, EREs are subject to epigenetic silencing preventing or reducing their transcription, which may otherwise produce nucleic acid and protein products with significant effects on host physiology and pathology (Kassiotis and Stoye, 2016). Epigenetic silencing of EREs is particularly potent in the germplasm, but is thought to be less effective when somatic cells alter their gene expression patterns, as part of the physiological process of their differentiation or response to stimuli or as part of the pathological process of cellular transformation (Slotkin and Martienssen, 2007). Increased ERE expression has frequently been reported as a hallmark of murine and human cancer (Kassiotis and Stoye, 2017). However, ERE induction is also characteristic of the physiological lymphocyte response to stimulation. For example, the transcriptional induction of certain groups of endogenous murine leukaemia viruses (MLVs) upon lipopolysaccharide (LPS) stimulation of murine B cells has been well documented over 3 decades ago and has been linked to B cell differentiation (Stoye and Moroni, 1983). Moreover, transcriptional induction of EREs was also described in B cells from Multiple Sclerosis (MS) patients, which were found to express elevated surface levels of ERV envelope glycoproteins (Brudek et al., 2009).
Thus, this transcriptional regulation of EREs in B cells or other hematopoietic cells may influence immune function, and both beneficial and detrimental effects have been proposed (Kassiotis and Stoye, 2016). However, what remains an open question is the degree of specificity of ERE induction during physiological or pathological conditions. Understanding the degree of overlap between those EREs that are induced as part of the normal processes of cellular activation and differentiation and those that signify cellular transformation or other pathological conditions requires detailed knowledge of ERE transcriptional patterns on a genome-wide scale, which is currently lacking.
Previous analyses of ERE expression have frequently employed PCR-based assays or microarrays, which rarely afforded element-specific or genome-wide resolution. For instance, although there are 100s or 1000s of EREs represented on commercial microarray probesets, these amount to only 0.25 and 0.04% of all genomic LTR elements and non-LTR elements, respectively (Young et al., 2014). The recent advent of RNA-sequencing (RNA-seq) techniques and the increasing availability of public RNA-seq datasets now provides the opportunity to study genome-wide ERE transcriptional regulation under a range of physiological or pathological conditions (Haase et al., 2015; Sokol et al., 2016). Here, we have analyzed ERE modulation in RNA-seq data from murine and human B cells, covering physiological B cell responses to in vitro and in vivo stimulation, as well as chronic diseases, including B cell lymphoma. Our results reveal distinct patterns of limited ERE induction during B cell cellular activation, contrasting with wide-spread ERE upregulation during B cell transformation, which indicates different underlying mechanisms.
Materials and Methods
Repeat Region Annotation
The precise annotation of repetitive regions is central to the accurate assessment of their activities. Until recently, this has relied upon the use of manually curated consensus sequences (Bao et al., 2015) with BLASTn-based search methods to define regions of interest. In place of these flattened representations, hidden Markov models (HMMs) can now also be used to represent repeat families, better representing the full range and variability of their sequence space (Hubley et al., 2016). This profile-based masking improves both accuracy and sensitivity, and annotates an additional 5.5 and 5.1% of the mouse and human genomes, respectively (Hubley et al., 2016). Using this method, the mouse and human genomes (GRCm38.78 and GRCh38.78, respectively) were masked using RepeatMasker1 configured with nhmmer (Wheeler and Eddy, 2013) in sensitive mode using the Dfam 2.0 library (v150923). RepeatMasker annotates LTR and internal regions separately, complicating the summation of reads spanning these divides. Tabular outputs were, therefore, parsed to merge adjacent annotations for the same element and to produce gene transfer format (GTF) files compatible with popular read-counting programs. GTF files for both genomes are freely available upon request.
Read Mapping and Counting
The expression data used in this study have been previously described and are publicly available. Ethical review, experimental and methodological details relating to study design and data acquisition can be found in the original reports. The following accessions were used: E-MTAB-2499; GSE61608; GSE60927; GSE68769; GSE65422; GSE60424; GSE72420 and GSE62241, which are a mixture of single-end and paired-end Illumina RNA-seq reads. Adapter contamination, assessed with FastQC2, was removed using Trimmomatic (Bolger et al., 2014), with additional quality trimming (Q20) and subsequent length filtering (both reads of a pair ≥ 35 nts). The resulting read pairs were aligned with HISAT2 (Kim et al., 2015) and primary mappings counted with featureCounts (Subread, Liao et al., 2014) using standard GTFs for annotated genes and the curated RepeatMasker GTFs for repeat regions. For accuracy and to prevent ambiguity, only reads that could be uniquely assigned to a single feature were counted. This may underestimate total expression in certain situations, but ensures confident count allocation to individual features. Features with no assigned reads across all samples within an experiment were discarded. Those remaining were normalized to account for variable sequencing depth between samples using DESeq2 (Love et al., 2014). In comparison to the use of normalization to transcripts-per million (TPM), for example, normalized read counts do not facilitate comparison of individual feature expression levels between experiments, but are nevertheless preferable for the assessment of repetitive element expression. Methods normalizing expression to TPM or reads per kilobase million, RPKM, require the accurate knowledge of transcript lengths, which cannot simply be determined for repetitive elements and are, in fact, often variable between treatments and systems. Normalized read counts were subsequently imported into Qlucore Omics Explorer (Qlucore, Lund, Sweden) for all downstream analysis and visualization. This included all statistical comparisons, calculation of fold-changes in transcript abundance, computation of Z-Scores (the number of standard deviations from the mean of each variable for each data point), and plotting either Z-Scores or log2 fold-changes in heat-map form.
Results
ERE Modulation during Murine B Cell Activation
Induction of endogenous MLVs in LPS-stimulated B cells provided one of the earliest examples of ERE modulation upon immune cell activation (Stoye and Moroni, 1983). We therefore focused on murine B cells to examine the transcriptional response of LTR and non-LTR elements to B cell stimulation. To this end, we analyzed RNA-seq data (E-MTAB-2499) from mature B cells, isolated from the spleens of C57BL/6 (B6) mice and stimulated in vitro with LPS, a-IgM antibodies, or a combination of CD40 ligand (CD40L) and IL-4 (Hartweger et al., 2014).
As expected, analysis of this dataset highlighted a strong modulation of a great number of non-viral gene transcripts, with just over half of responding genes (53.6%) upregulated upon stimulation, relative to unstimulated cells (Figure 1A). In contrast, under the same conditions of stimulation, the majority of LTR element and LINE transcripts (85.8 and 89.3%, respectively) were proportionally downregulated (Figure 1A). This apparent reduction in ERE transcription is likely the effect of the increase in overall gene transcription in response to stimulation. Closer inspection of the top 31 LTR elements that were induced in these B cell upon stimulation, revealed several different groups of LTR elements (Figure 1B). However, notable was the over-representation of xenotropic endogenous MLVs (Supplementary Table 1). These included two closely integrated proviruses on chromosome 1, Xmv41 (LTR/ERV1|MuLV-int∼RLTR4_Mm|1|171481146|171489815) and Xmv43 (LTR/ERV1|MuLV-int∼RLTR4_Mm|1|170941521|170950177), which have been previously shown to be LPS-responsive (Young et al., 2014), as well as a previously unlocalized provirus, Xmv45 on chromosome 5 (LTR/ERV1|MuLV-int∼RLTR4_Mm|5|23700579|23709245) (Supplementary Table 1). Four additional xenotropic proviruses, previously uncharacterized due to their location of the Y chromosome (Frankel et al., 1989), were also significantly upregulated (Figure 1B and Supplementary Table 1). However, these were highly homologous with Xmv41, Xmv43 and Xmv45 (Supplementary Figure 1), making it difficult to discern whether the Y-linked proviruses are genuinely expressed or whether they report expression of Xmv41, Xmv43 or Xmv45.
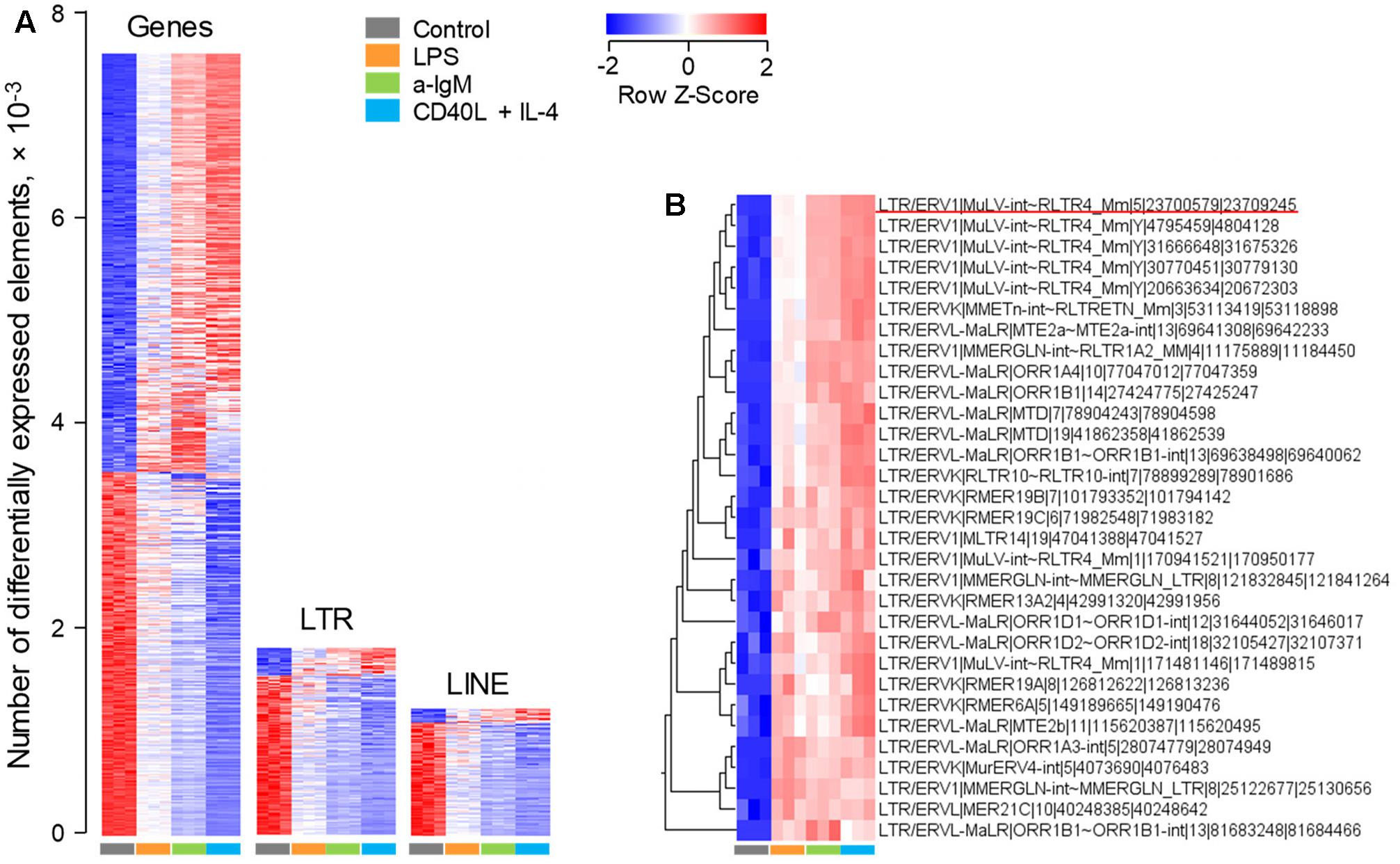
FIGURE 1. Modulation of LTR element and LINE expression upon murine B cell stimulation in vitro. Transcriptional analysis of purified splenic follicular B cells before and after 6-h in vitro stimulation with a-IgM (10 μg/ml), LPS (10 μg/ml) or a combination of CD40L (1 μg/ml) and IL-4 (0.1 μg/ml) (E-MTAB-2499). (A) Number of gene, LTR and LINE transcripts that are differentially expressed (≥2-fold change; p < 0.05) between in vitro activated and directly ex vivo isolated B cells. (B) The top 31 LTR EREs induced by 6-h B cell activation. In (A,B) each column is an independent sample. The underlined element in (B) is Xmv45.
As an independent confirmation of the observed pattern of LTR element transcriptional activation, we analyzed a second set of RNA-seq data (GSE61608) from mature B cells, isolated from spleens of B6 mice and stimulated in vitro with LPS or a-IgM antibodies (Fowler et al., 2015). Again, 12 out of the top 31 LTR elements identified in the previous set, were also significantly induced at the earlier time-point of 2 h in this set and, notably, these included Xmv45 (Figure 2A). Furthermore, Xmv45 was also significantly induced in a third set of RNA-seq data (GSE60927) by longer in vitro stimulation of B6-derived B cells with LPS or a combination of CD40L, IL-4 and IL-5 (Shi et al., 2015) (Figure 2B). More importantly, Xmv45 seems also to be transcriptionally induced in vivo, as splenic plasma cells assessed directly ex vivo, which represent a state of recent B cell activation, showed elevated Xmv45 transcription and clustered closely with in vitro LPS-stimulated B cells (Figure 2B). Lastly, further support for transcriptional induction of Xmv45 in vivo was provided by analysis of RNA-seq data (GSE68769) from B cells isolated from the lymph nodes of mice responding to Influenza A virus vaccination. Indeed, Xmv45 was one of the few LTR elements that were significantly induced over the course of vaccination, despite the fact that Influenza A-specific B cells should constitute only a small fraction of total lymph node B cells that were analyzed (Figure 2C).
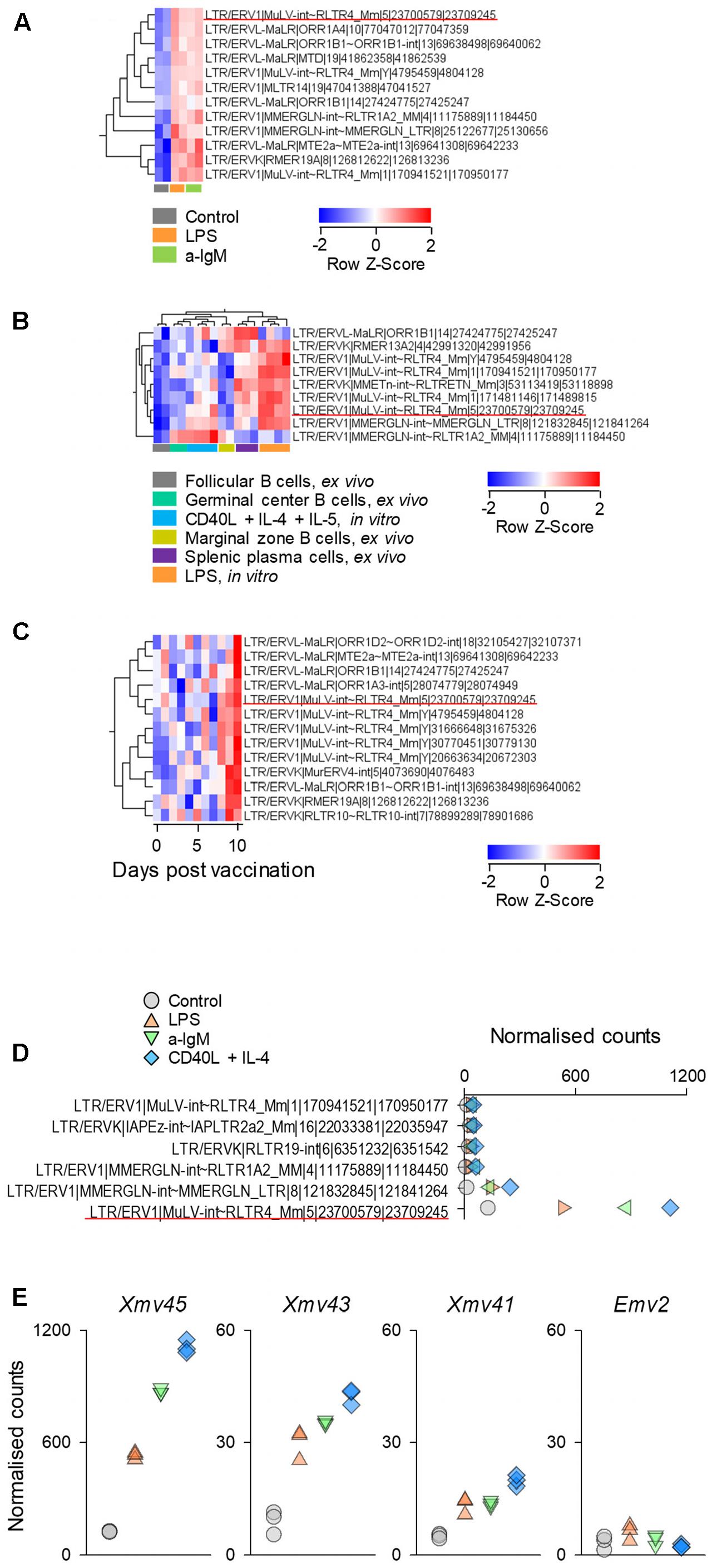
FIGURE 2. Xmv45 induction during in vitro and in vivo murine B cell stimulation. Expression pattern of the 31 includible LTR elements identified in Figure 1B in three independent datasets. Significantly induced LTR elements were identified in each study separately (≥2-fold change; p < 0.05, and the elements shared with in vitro stimulated cells (Figure 1B) are shown. (A) Transcriptional analysis of purified splenic follicular B cells before and after 2-h in vitro stimulation with a-IgM (10 μg/ml) or LPS (25 μg/ml) (GSE61608), depicting the significantly induced LTR elements. (B) Transcriptional analysis of purified splenic follicular B cells before and after in vitro stimulation with LPS for 3 days or a combination of CD40L, IL-4 and IL-5 for 4 days (GSE60927). Also included in the comparison are ex vivo analyzed splenic germinal center B cells, marginal zone B cells and plasma cells. The heat map depicts the significantly induced LTR elements and unsupervised hierarchical clustering of samples according to their expression. (C) Mice were primed by intramuscular injection of inactivated influenza A/New Caledonia/20/99 virus and were boosted with intramuscular injection of seasonal (2006–2007) trivalent inactivated influenza vaccine 30 days later (GSE68769). The figure shows the transcriptional analysis of purified lymph node B cells, pooled from 3 mice for each of the indicated time-points after boost, depicting the significantly induced LTR elements. In (A–C) each column is an independent pool and the underlined element is Xmv45. (D) Normalized counts for the 6 LTR elements with the highest expression in dataset described in Figure 1A. Symbols represent the mean values of triplicate samples. The underlined element is Xmv45. (E) Normalized counts of the indicated proviruses in the same dataset described in Figure 1A. Each symbol is an independent sample.
We next explored whether the consistency with which Xmv45 transcriptional induction was detected in multiple datasets was explained by the degree of this induction. Indeed, transcription of Xmv45 in stimulated B cells was much higher than any other induced LTR element (Figure 2D) and dwarfed transcription of other MLV proviruses that are either weakly (Emv2) or strongly (Xmv41 and Xmv43) inducible by LPS stimulation (Figure 2E; please note that Xmv45 expression is plotted on a scale that is 20-times higher than the rest). Together, these data suggest that transcription of a small selection of LTR elements, exemplified by Xmv45, is consistently induced in murine B cells by a multitude of in vitro and in vivo stimuli and validate the capacity of our analysis to detect this induction in multiple datasets.
ERE Modulation in Murine B Cell Lymphoma
We next explored whether ERE transcriptional modulation as observed during physiological B cell activation overlapped with modulation that may occur following B cell transformation. For this purpose, we compared RNA-seq data (GSE65422) from non-transformed B cells (resting splenic B cells and germinal center B cells analyzed directly ex vivo; and B cells activated in vitro with a-CD40 and a-IgM antibodies) with B cells resembling diffuse large B cell lymphoma (DLBCL) (Zhang et al., 2015). The latter were obtained from mice that develop spontaneous B cell lymphomas as a result of deregulated expression of BCL6 under the immunoglobulin (Ig) heavy chain Iμ promoter and of deregulated activation of the alternative NF-κB pathway by expression of the NF-κB inducing kinase (NIK) under the ROSA26 promoter (Zhang et al., 2015). Both these genetic alterations were restricted to the germinal center lineage by conditional mutagenesis, using the Cγ1-cre transgene (Zhang et al., 2015).
Comparison of a-CD40 and a-IgM in vitro activated B cells with resting B cells in this dataset (Figure 3A), revealed a picture comparable with that from the previous dataset (Figure 1A), with transcriptional activation favoring gene induction, and to a lesser extent LTR element and LINE transcription. In contrast, LTR elements and LINEs were dominating the transcriptional differences between resting B cell and B cell lymphomas, with minimal overlap between B cell lymphomas and activated B cells (Figure 3B). Of note, whereas Xmv45 was still the most induced provirus upon in vitro B cell activation, B cell lymphomas where characterized by significantly elevated expression of Emv2 (Figure 3C). In fact, expression of Emv2 in lymphomas was ∼50-times higher than of Xmv45 (Figure 3C; please note that Emv2 expression is plotted on a scale that is 10-times higher than that of Xmv45). The elevated ecotropic MLV expression likely reflects restoration of Emv2 infectivity, which has been previously observed in cancer cell lines (Li et al., 1999) and immunodeficient animals (Young et al., 2012). In contrast to Xmv45, Emv2 transcription in immune cells is only weakly inducible by LPS, but strongly inducible by the epigenetic derepression through BrdU treatment (Young et al., 2014), suggesting that the primary cause of its upregulation in B cell lymphomas is loss of epigenetic repression, followed by restoration of infectivity.
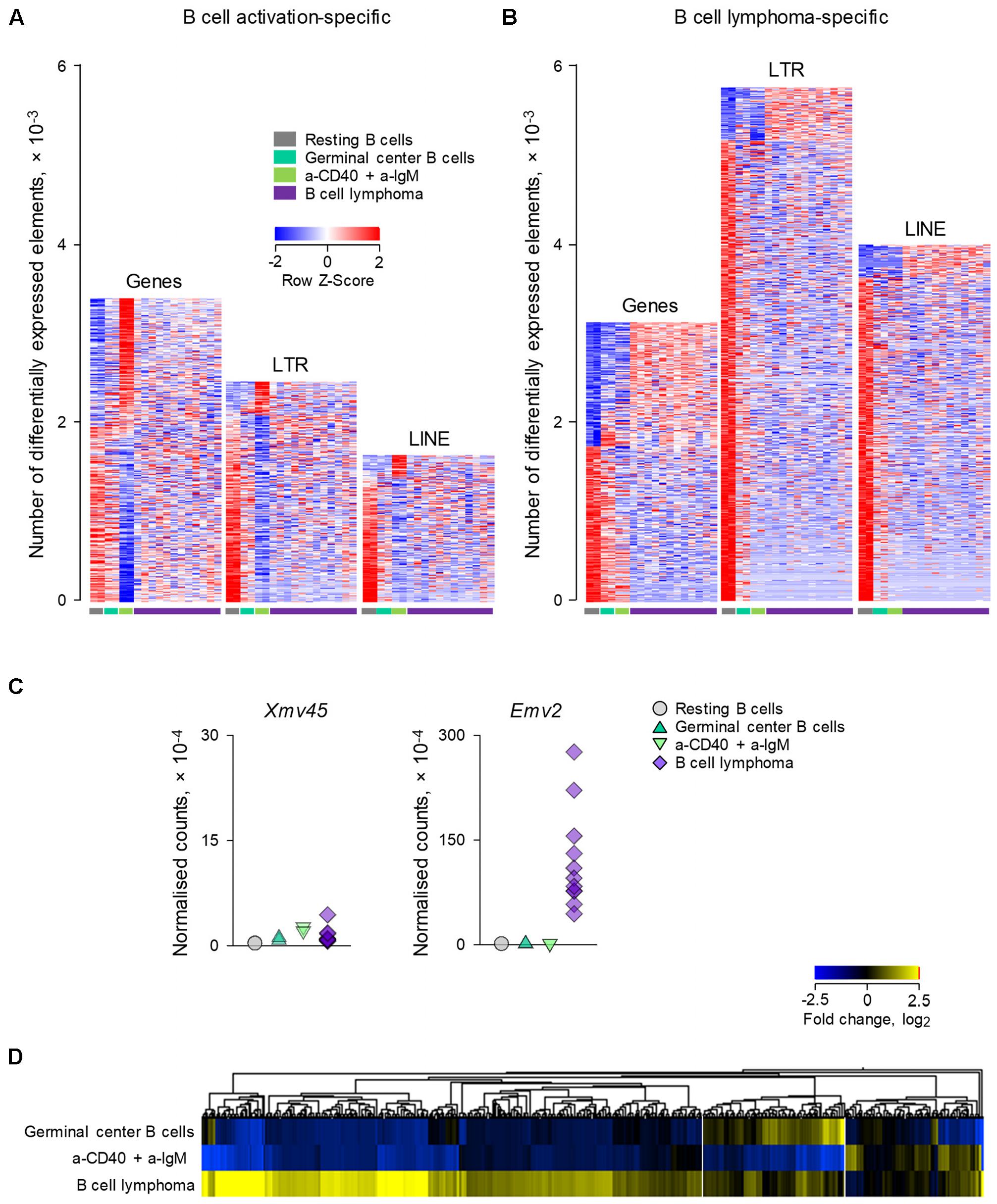
FIGURE 3. Modulation of LTR and LINE EREs following B cell transformation. (A) Number of gene, LTR and LINE transcripts that are differentially expressed (≥2-fold change; p < 0.05) between resting B cells and B cells activated in vitro with a-CD40 and a-IgM antibodies for 24 h (GSE65422). Also shown for comparison are germinal center B cells from wild-type mice and B cell lymphomas from mice with germinal center B cell-specific deregulation of BCL6 and NIK. (B) Number of gene, LTR and LINE transcripts that are differentially expressed (≥2-fold change; p < 0.05) in B cell lymphomas in the same dataset described above in (A). Each column is an independent sample. (C) Normalized counts of Xmv45 and Emv2 transcripts in the same samples described above in (A). Symbols represent individual samples. (D) Hierarchical clustering of LTR elements that are significantly induced in either in vitro activated B cells (53 transcripts) or B cell lymphomas (606 transcripts) in the same dataset as in (A), in comparison with resting B cells. Mean fold changes from resting B cells are plotted.
Consistent with different mechanistic origins of LTR element modulation during B cell activation and B cell transformation, approximately one-third of LTR elements that were transcriptionally induced in B cell lymphomas were also induced either in germinal center B cells or in in vitro activated B cells (in equal proportions between the two), whereas the majority (two-thirds) were unique to B cell lymphomas (Figure 3D).
ERE Modulation in Human B Cells under Physiological and Pathological Conditions
Given the evolutionary divergence between EREs in different host species, we next asked whether the specificity with which EREs are modulated in murine B cells in distinct conditions, also characterized ERE modulation in B cells from a different host, namely the human. We started by investigating gene and ERE transcriptional modulation in RNA-seq data (GSE60424), generated from peripheral blood B cells, isolated from healthy individuals and those with infectious, degenerative or autoimmune diseases, including Sepsis, Amyotrophic Lateral Sclerosis (ALS), Type 1 Diabetes (T1D) and MS (Linsley et al., 2014).
As might be expected by its acute and severe nature, Sepsis accounted for the majority of the 2,159 genes that were differentially regulated between the studied conditions, with a smaller, but clearly evident signature in MS patients shortly after the first treatment with IFNβ (Figure 4). A comparable number of LTR elements were also differentially expressed between the conditions (Figure 4). Interestingly, transcription of LTR elements appeared more distinct between the conditions, with the exception of T1D, than overall gene expression, with a particularly strong signature in the IFNβ-treated subset of MS patients (Figure 4). Moreover, B cells from these individuals differentially expressed more than twice the number of LINEs than of genes, with clusters of LINEs clearly distinguishing the different conditions, again with a very strong signature evident in the IFNβ-treated subset of MS patients (Figure 4).
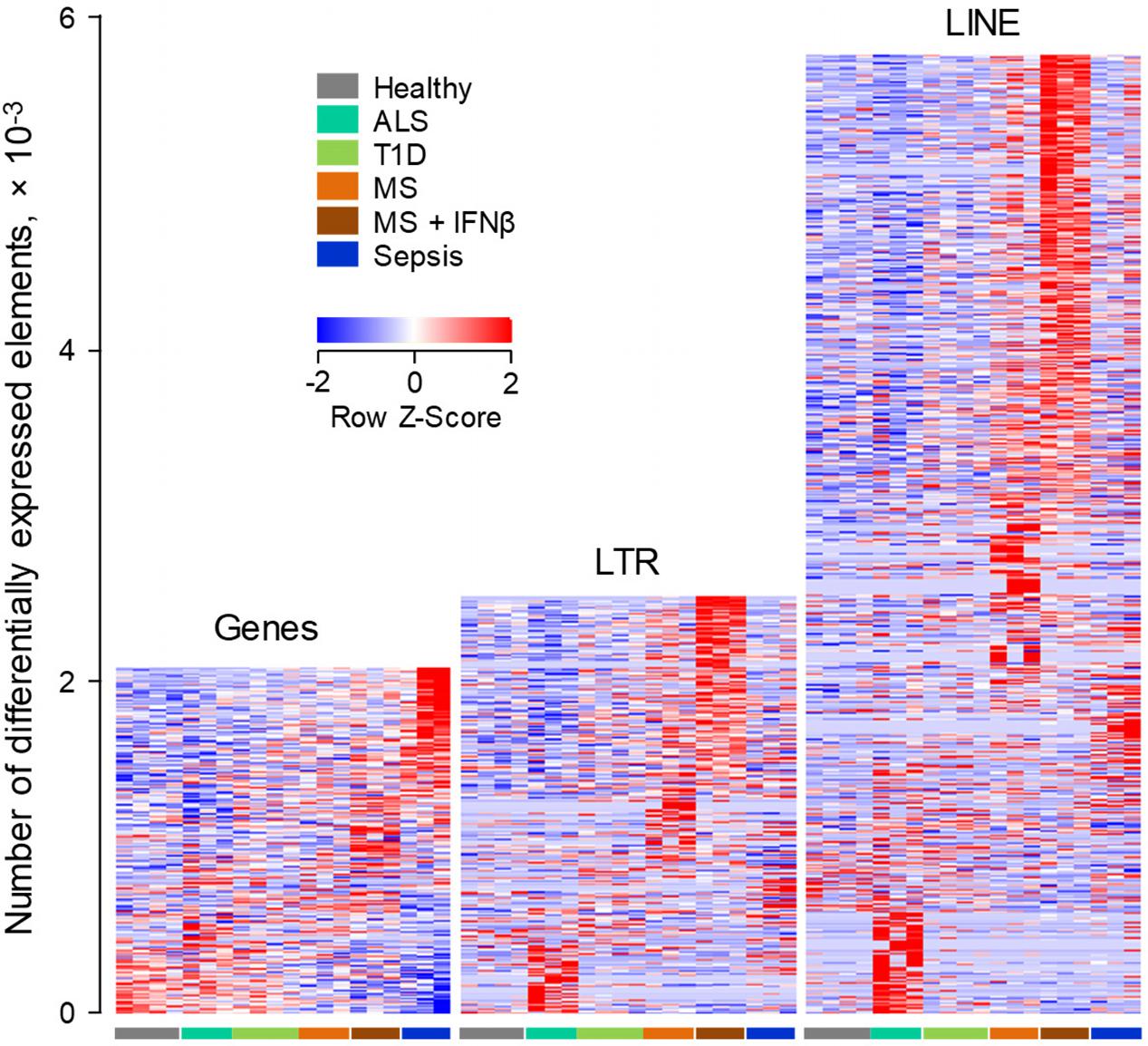
FIGURE 4. ERE modulation in human B cells from infectious, degenerative or autoimmune disease. Transcriptional analysis by RNA-seq of peripheral blood B cells isolated from healthy individuals, from patients with Sepsis, ALS or T1D and from MS patients before and 24 h after the first treatment with IFNβ (GSE60424). Heat-maps show the number of gene, LTR and LINE transcripts that are differentially expressed (p < 0.05) between the groups. Each column is an independent sample.
To probe further the IFNβ-responsiveness of EREs, we first examined whether the pattern observed in purified B cells was also present when the entire complement of blood cell types was analyzed. We focused on LTR elements as they include phylogenetically more diverse groups than LINEs. Indeed, a sizeable set of the IFNβ-inducible LTR elements upregulated in B cells from IFNβ-treated MS patients was also detectable and highly induced in the same patient group, when RNA-seq data from whole-blood was analyzed (131 of 779 elements, Figure 5A).
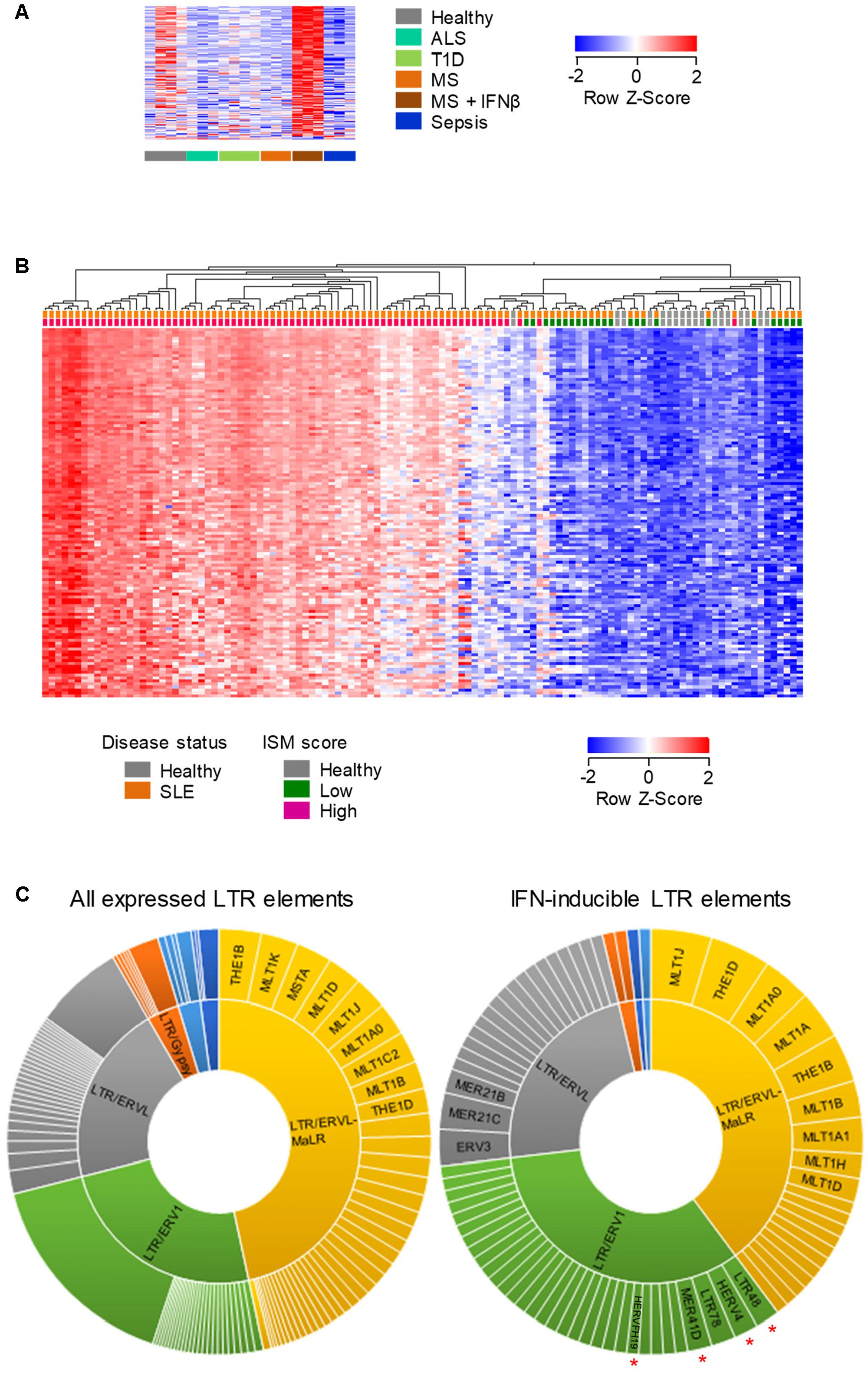
FIGURE 5. Human LTR elements induced by IFN I in whole-blood. (A) Expression profile of 131 LTR elements that are transcriptionally induced specifically by IFNβ treatment of MS patients, in whole-blood RNA-seq data (GSE60424). (B) Hierarchical clustering of healthy individuals and SLE patients according to the expression of 219 LTR elements that were induced in B cells by IFNβ treatment of MS patients and were also upregulated in RNA-seq data (GSE72420) from whole-blood samples from SLE patients as a group, compared with those from healthy individuals (≥2-fold change; p < 0.05). In (A,B) each column is an independent sample. (C) Diversity of the LTR elements that are expressed in peripheral blood cells (left) and of the 108 IFN I-inducible LTR elements that were common between purified B cells and whole-blood samples from IFNβ-treated MS patients and SLE patients. Slice widths are proportional to the frequency of each member. Significantly enriched groups (p < 0.05, χ2 with multiple comparison correction) are indicated by red asterisks.
We next examined the overlap between LTR elements that are induced by IFNβ treatment of MS patients and those that might be naturally induced in a setting of elevated levels of endogenously produced type I IFN (IFN I). To this end, we analyzed RNA-seq data (GSE72420) from whole-blood samples obtained from Systemic Lupus Erythematosus (SLE) patients (Hung et al., 2015), an autoimmune disease with a strong IFN I signature (Obermoser and Pascual, 2010). The intersection of LTR elements that were upregulated in purified B cells in response to IFNβ treatment of MS patients and those induced in SLE patients as a group, identified 219 common LTR elements (Figure 5B). Importantly, significantly elevated expression of these LTR elements was present in the vast majority (88%; 66 of 75) of SLE patients with a high Interferon Signature Metric (ISM) score, but not in any of the SLE patients with a low ISM score (0/24) or any healthy individuals (0/18) (Figure 5B). These results indicated that an overlapping set of LTR elements (Supplementary Table 2) were responsive to IFN I both in IFNβ-treated MS patients and in SLE patients with a high ISM score.
To explore whether IFN I was preferentially inducing certain LTR groups, we compared the composition of all LTR elements expressed in MS or SLE patients with that of the IFN I-inducible LTR elements shared between the two conditions (Supplementary Table 2). This comparison uncovered significant enrichment for the ERV1 groups as a whole, with members of the LTR48, HERV4, MER41D and HERVFH19 subgroups being frequently responsive to IFN I (Figure 5C). Thus, transcriptional induction of LTR elements by exogenously administered or endogenously produced IFN I displays a certain degree of specificity.
Lastly, we investigated how human B cell transformation might influence ERE transcriptional behavior. To achieve this, we compared ERE transcription in RNA-seq data (GSE62241) from follicular B cell lymphoma and from non-transformed B cells (Koues et al., 2015). The groups included B cells purified from follicular lymphoma biopsies; centrocytes, the non-cycling fraction of germinal center B cells, isolated ex vivo from tonsillar tissues; and activated B cells, isolated from peripheral blood samples and stimulated in vitro using a combination of IL-4, a-CD40, a-IgM, and a-IgD (Koues et al., 2015). Consistent with their original description (Koues et al., 2015), follicular lymphoma B cells differentially expressed a substantial number of genes, in comparison with non-transformed activated B cells or centrocytes (Figure 6A). Notably, a much larger number of EREs were dysregulated in follicular lymphoma B cells, with twice as many LTR elements and four-times as many LINEs upregulated in follicular lymphoma B cells as genes (Figure 6A). More importantly, investigation of the overlap between LTR elements upregulated in follicular lymphoma B cells with those induced in B cells from diseases other than cancer uncovered distinguishable, non-overlapping patterns, with the majority of induced LTR elements specific to B cell lymphoma (Figure 6B). Together, these results suggested that distinct LTR elements are transcriptionally activated in cancer and in other diseases.
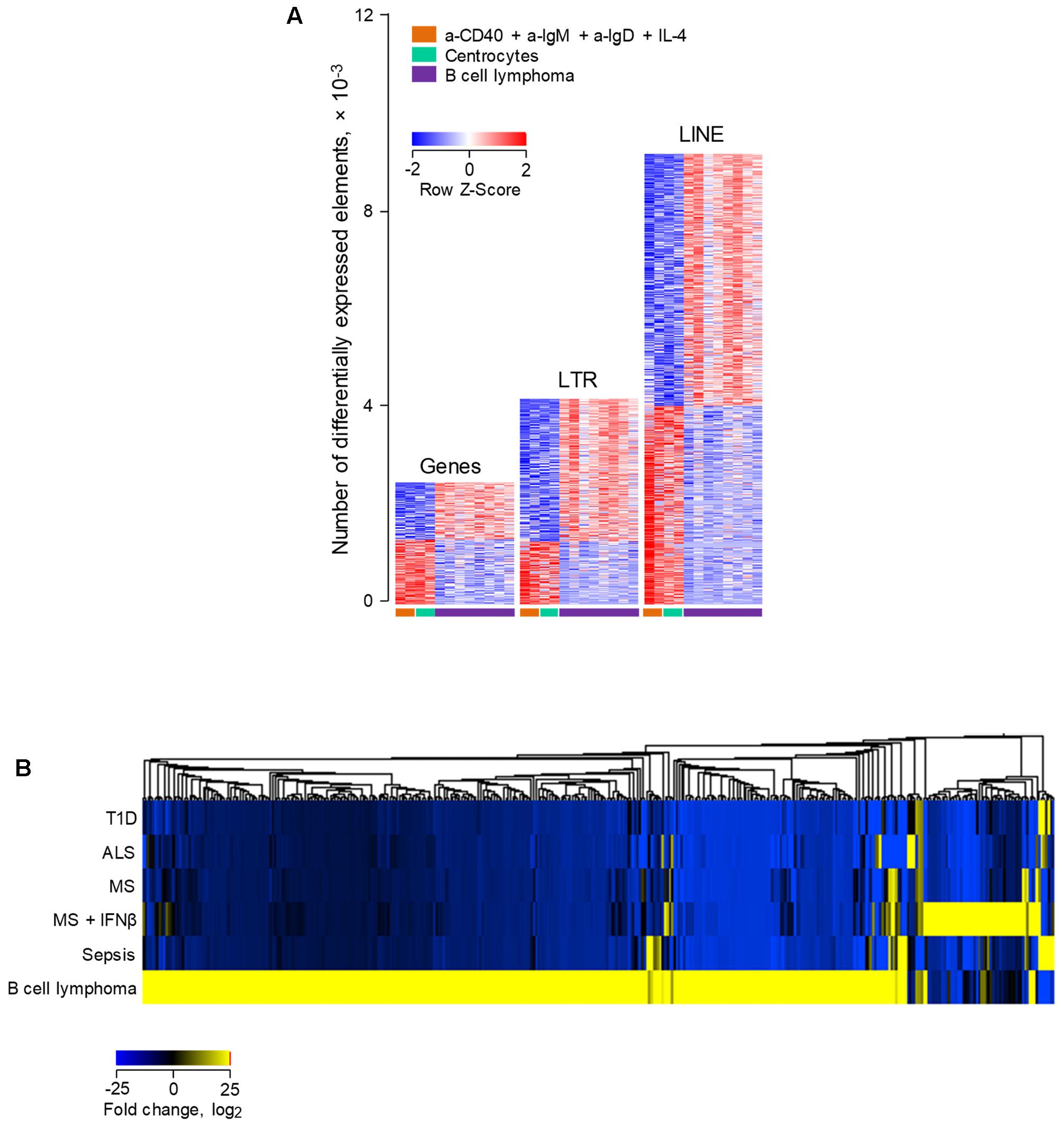
FIGURE 6. Differential modulation of LTR and LINE EREs following human B cell activation or transformation. (A) Number of gene, LTR and LINE transcripts that are differentially expressed (≥2-fold change; p < 0.01) between ex vivo isolated follicular lymphoma B cells and either peripheral blood B cells activated in vitro with IL-4, a-CD40, a-IgM, and a-IgD antibodies or ex vivo isolated tonsillar centrocytes (GSE62241). Each column is an independent sample. (B) Hierarchical clustering of a total of 395 LTR elements that are significantly induced in either ex vivo isolated follicular lymphoma B cells or peripheral blood B cells from infectious, degenerative or autoimmune diseases described in Figure 4. Mean fold changes from the respective B cell control are plotted.
Discussion
Endogenous retroelements constitute a sizeable fraction of the genome and their transcription has considerable potential to affect cellular physiology or contribute to pathology. However, their precise contribution can only be accurately assessed with detailed knowledge of their transcriptional behavior at the genome-wide level with the resolution of individual ERE integrations (Kassiotis and Stoye, 2016). The technological advances of transcriptional profiling by RNA-seq now afford a means of addressing genome-wide ERE modulation in health and disease. Using such methodology, we uncovered unique patterns of ERE modulation characteristic of physiological activation or pathological transformation of murine and human B cells.
Study of murine B cell responses to physiological innate immune, cytokine, or BCR stimuli highlighted the responsiveness predominantly of gene transcription and the lack of widespread induction of LTR elements or LINEs. In fact, most B cell-expressed EREs appeared downregulated in murine B cells upon activation, likely due to the overshadowing induction of strong gene transcription under these conditions. Prior analysis of immune cell stimulation by microarray methods had also indicated that transcription of different ERE groups could either increase or, indeed, decrease upon activation (Young et al., 2014), but lacked the resolution of RNA-seq data analysis. Our current analysis identified a small number of distinct LTR elements that are consistently activated in stimulated murine B cells, with a single endogenous MLV provirus, Xmv45, expressed at higher levels than all other LTR elements together. It would be interesting to explore the reasons for this unique inducibility of Xmv45, as well as its potential consequences for B cell function.
Contrasting the strong induction of a very limited number of ERE proviruses following B cell stimulation, expression of a much larger number of EREs was found altered following B cell transformation. Gene expression profiling of DLBCL has revealed patterns associated either with an activated B cell phenotype or with a germinal center phenotype (Lenz and Staudt, 2010). Each subtype is characterized by a different frequency of mutations in pathways affecting cellular activation, such as mutations promoting constitutive NF-κB activation, or differentiation, such as BCL6 or its antagonist Blimp-1 (Lenz and Staudt, 2010). Consistent with these observations, we found that one-third of LTR elements that were induced in B cell lymphoma cells were shared with either activated B cells or germinal center B cells. Induction of these shared LTR elements is likely to be induced by the same transcriptional regulators induced in activated B cells (e.g., NF-κB) or germinal center B cells (e.g., BCL6), which are also overexpressed in B cell lymphomas.
NF-κB, together with IRF1, have been incriminated for the transcriptional activation of HERV-K(HML2) proviruses in ALS brain tissue and human astrocytes and neurons treated with inflammatory cytokines (Manghera et al., 2016). These two immune activation-induced transcription factors are part of a longer list of nearly 40 host transcription factors that are suspected to directly drive transcription of HERVK LTRs (Manghera and Douville, 2013). Indeed, this relatively high affinity of ERV LTRs for host transcription factors seems to be an intrinsic, evolutionarily shared property (Dunn et al., 2005), and underlies their ability to establish and rewire host gene regulatory networks (Rebollo et al., 2012). Whether BCL6 or Blimp-1 directly affect transcription of ERVs is not currently known, but Blimp-1 has been reported to repress expression of HIV-1 proviruses in T cells (Kaczmarek Michaels et al., 2015). Therefore, BCL6 may induce expression of ERVs indirectly, through its established role in repressing the repressor Blimp-1 (Crotty et al., 2010).
Given the common pathways that drive B cell activation, germinal center response and B cell transformation, it was surprising to observe that the majority of LTR elements that were significantly upregulated in B cell lymphoma were unique to this condition and were not shared with physiologically activated B cells. This was exemplified by the observed expression of Emv2, which surpassed the highly inducible Xmv45 by two orders of magnitude, to become the single most expressed MLV in B cell lymphoma. Whereas Xmv45 is highly inducible by LPS, cytokine or antigenic stimulation of B cells, Emv2 is primarily responsive to epigenetic modifiers (Young et al., 2014), implicating epigenetic changes in the altered MLV expression profile in B cell lymphoma cells. It should be noted that although expression of ecotropic MLV found in RNA-seq data is attributed here to the germline copy of Emv2, based on sequence identity, it may also arise from new somatically acquired integrations of an Emv2-derived infectious retrovirus. Indeed, the dysregulation of Emv2 alongside the expression of complementary viruses may support the production of infectious recombinant retroviruses, further increasing observed expression, particularly in B cell lymphomas (Young et al., 2012). The same is also true for other mobile EREs, such as intracisternal A particle (IAP) elements in mice and LINEs in both humans and mice, where the reported expression is the combination of transcription of germline copies and any somatically acquired additional copies.
Akin to murine B cells, distinct patterns of LTR element and LINE expression characterized B cells isolated from patients suffering from different autoimmune, infectious, degenerative or neoplastic diseases. However, interesting differences between the two host species were also observed. Whereas the transcriptional response of murine B cells was overshadowed by gene expression changes, this was not the case in human B cells where transcription of EREs was far more responsive to the influence of the diseases studied here. This was particularly visible for LINEs, which indeed were nearly three-times more numerous than non-viral genes in the transcriptional difference between B cells from the different diseases. These findings may indicate higher overall transcriptional activity of LINEs in humans (Hancks and Kazazian, 2012).
Regardless of its origin, the enhanced transcriptional responsiveness of human EREs provides a considerably more detailed map of transcriptional activity across the genome than annotated genes alone. For example, the transcriptional response to IFN I treatment of MS patients was more evident in LTR element or LINE transcription than in gene transcription overall. The sheer number of transcribed EREs allows for increased statistical power, revealing differences that may be too subtle to detect otherwise.
Also similar to the specificity of LTR element expression in stimulated murine B cells, human B cells upregulated a select list of LTR elements in conditions of IFN I stimulation. IFN I-inducible LTR elements, shared between purified B cells and whole-blood samples and between IFNβ-treated MS patients an SLE patients, were enriched for ERV1 class elements and included LTR48, HERV4 and MER41D members. Several IFN-induced transcription factors, including NF-κB, IRFs and STATs are predicted to bind HERV-K LTRs (Manghera and Douville, 2013), indicating direct responsiveness of at least certain ERV groups to IFN stimulation. Interestingly, members of the MER41 group were recently shown to confer IFNγ-responsiveness to the AIM2 gene and to contribute to the activation of other immune-related genes, by providing binding sites for the transcription factors IRF1 and STAT1 (Chuong et al., 2016). It is, therefore, conceivable that the IFN I responsiveness of LTR48, HERV4 and MER41D elements is mediated by IFN I-induced transcription factors.
In comparison with resting or activated B cells, the most pronounced induction of ERE transcription was witnessed in human B cell lymphoma cells, affecting thousands of LTR elements and LINEs. More importantly, as was the case with murine B cell lymphoma cells, the LTR elements that were activated in human B cell lymphoma cells exhibited minimal overlap with those expressed in non-transformed B cells from any of the conditions studied. Together, these results suggest that cellular transformation, at least in the B cell lineage, is associated with dysregulation of a non-random set of EREs that are not typically found dysregulated in other conditions. The distinctive patterns of ERE induction will help separate physiological from pathological expression, as well as provide targets for possible intervention.
Author Contributions
GY developed the bioinformatics pipeline. JA, GY, and GK analyzed the data. JA, GY, JS, and GK wrote the manuscript. JS and GK supervised the study.
Funding
This work was supported by the Francis Crick Institute (FC001099 and FC001162), which receives its core funding from Cancer Research UK, the UK Medical Research Council and the Wellcome Trust; and by the Wellcome Trust (102898/B/13/Z).
Conflict of Interest Statement
GK is a scientific co-founder of and consulting for ERVAXX and a member of its scientific advisory board. GK, GY, and JA may receive royalties through their institution from ERVAXX.
The other author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors are grateful for assistance from the Scientific Computing Facility at the Francis Crick institute.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02489/full#supplementary-material
Footnotes
References
Bao, W., Kojima, K. K., and Kohany, O. (2015). Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6:11. doi: 10.1186/s13100-015-0041-9
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brudek, T., Christensen, T., Aagaard, L., Petersen, T., Hansen, H. J., and Moller-Larsen, A. (2009). B cells and monocytes from patients with active multiple sclerosis exhibit increased surface expression of both HERV-H Env and HERV-W Env, accompanied by increased seroreactivity. Retrovirology 6:104. doi: 10.1186/1742-4690-6-104
Burns, K. H., and Boeke, J. D. (2012). Human transposon tectonics. Cell 149, 740–752. doi: 10.1016/j.cell.2012.04.019
Chuong, E. B., Elde, N. C., and Feschotte, C. (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. doi: 10.1126/science.aad5497
Crotty, S., Johnston, R. J., and Schoenberger, S. P. (2010). Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11, 114–120. doi: 10.1038/ni.1837
Dunn, C. A., Van De Lagemaat, L. N., Baillie, G. J., and Mager, D. L. (2005). Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene 364, 2–12. doi: 10.1016/j.gene.2005.05.045
Fowler, T., Garruss, A. S., Ghosh, A., De, S., Becker, K. G., Wood, W. H., et al. (2015). Divergence of transcriptional landscape occurs early in B cell activation. Epigenetics Chromatin 8:20. doi: 10.1186/s13072-015-0012-x
Frankel, W. N., Stoye, J. P., Taylor, B. A., and Coffin, J. M. (1989). Genetic analysis of endogenous Xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J. Virol. 63, 1763–1774.
Haase, K., Mosch, A., and Frishman, D. (2015). Differential expression analysis of human endogenous retroviruses based on ENCODE RNA-seq data. BMC Med. Genomics 8:71. doi: 10.1186/s12920-015-0146-5
Hancks, D. C., and Kazazian, H. H. (2012). Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 22, 191–203. doi: 10.1016/j.gde.2012.02.006
Hartweger, H., Schweighoffer, E., Davidson, S., Peirce, M. J., Wack, A., and Tybulewicz, V. L. (2014). Themis2 is not required for B cell development, activation, and antibody responses. J. Immunol. 193, 700–707. doi: 10.4049/jimmunol.1400943
Hubley, R., Finn, R. D., Clements, J., Eddy, S. R., Jones, T. A., Bao, W., et al. (2016). The Dfam database of repetitive DNA families. Nucleic Acids Res. 44, D81–D89. doi: 10.1093/nar/gkv1272
Hung, T., Pratt, G. A., Sundararaman, B., Townsend, M. J., Chaivorapol, C., Bhangale, T., et al. (2015). The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 350, 455–459. doi: 10.1126/science.aac7442
Kaczmarek Michaels, K., Natarajan, M., Euler, Z., Alter, G., Viglianti, G., and Henderson, A. J. (2015). Blimp-1, an intrinsic factor that represses HIV-1 proviral transcription in memory CD4+ T cells. J. Immunol. 194, 3267–3274. doi: 10.4049/jimmunol.1402581
Kassiotis, G., and Stoye, J. P. (2016). Immune responses to endogenous retroelements: taking the bad with the good. Nat. Rev. Immunol. 16, 207–219. doi: 10.1038/nri.2016.27
Kassiotis, G., and Stoye, J. P. (2017). Making a virtue of necessity: the pleiotropic role of human endogenous retroviruses in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160277 doi: 10.1098/rstb.2016.0277
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Koues, O. I., Kowalewski, R. A., Chang, L. W., Pyfrom, S. C., Schmidt, J. A., Luo, H., et al. (2015). Enhancer sequence variants and transcription-factor deregulation synergize to construct pathogenic regulatory circuits in B-cell lymphoma. Immunity 42, 186–198. doi: 10.1016/j.immuni.2014.12.021
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Lenz, G., and Staudt, L. M. (2010). Aggressive lymphomas. N. Engl. J. Med. 362, 1417–1429. doi: 10.1056/NEJMra0807082
Li, M., Huang, X., Zhu, Z., and Gorelik, E. (1999). Sequence and insertion sites of murine melanoma-associated retrovirus. J. Virol. 73, 9178–9186.
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Linsley, P. S., Speake, C., Whalen, E., and Chaussabel, D. (2014). Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLOS ONE 9:e109760. doi: 10.1371/journal.pone.0109760
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mager, D. L., and Stoye, J. P. (2015). Mammalian endogenous retroviruses. Microbiol. Spectr. 3:MDNA3-0009-2014.
Manghera, M., and Douville, R. N. (2013). Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10:16. doi: 10.1186/1742-4690-10-16
Manghera, M., Ferguson-Parry, J., Lin, R., and Douville, R. N. (2016). NF-κB and IRF1 induce endogenous retrovirus K expression via interferon-stimulated response elements in its 5’ long terminal repeat. J. Virol. 90, 9338–9349. doi: 10.1128/JVI.01503-16
Mccarthy, E. M., and Mcdonald, J. F. (2004). Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 5:R14. doi: 10.1186/gb-2004-5-3-r14
Mouse Genome Sequencing Consortium, Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. doi: 10.1038/nature01262
Obermoser, G., and Pascual, V. (2010). The interferon-alpha signature of systemic lupus erythematosus. Lupus 19, 1012–1019. doi: 10.1177/0961203310371161
Rebollo, R., Romanish, M. T., and Mager, D. L. (2012). Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 46, 21–42. doi: 10.1146/annurev-genet-110711-155621
Shi, W., Liao, Y., Willis, S. N., Taubenheim, N., Inouye, M., Tarlinton, D. M., et al. (2015). Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16, 663–673. doi: 10.1038/ni.3154
Slotkin, R. K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Sokol, M., Jessen, K. M., and Pedersen, F. S. (2016). Utility of next-generation RNA-sequencing in identifying chimeric transcription involving human endogenous retroviruses. APMIS 124, 127–139. doi: 10.1111/apm.12477
Stoye, J. P., and Moroni, C. (1983). Endogenous retrovirus expression in stimulated murine lymphocytes. Identification of a new locus controlling mitogen induction of a defective virus. J. Exp. Med. 157, 1660–1674. doi: 10.1084/jem.157.5.1660
Wheeler, T. J., and Eddy, S. R. (2013). nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489. doi: 10.1093/bioinformatics/btt403
Young, G. R., Eksmond, U., Salcedo, R., Alexopoulou, L., Stoye, J. P., and Kassiotis, G. (2012). Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491, 774–778. doi: 10.1038/nature11599
Young, G. R., Mavrommatis, B., and Kassiotis, G. (2014). Microarray analysis reveals global modulation of endogenous retroelement transcription by microbes. Retrovirology 11:59. doi: 10.1186/1742-4690-11-59
Keywords: endogenous retroviruses, endogenous retroelements, transcription, genetic, B lymphocyte activation, B cell lymphoma, autoimmunity, cancer
Citation: Attig J, Young GR, Stoye JP and Kassiotis G (2017) Physiological and Pathological Transcriptional Activation of Endogenous Retroelements Assessed by RNA-Sequencing of B Lymphocytes. Front. Microbiol. 8:2489. doi: 10.3389/fmicb.2017.02489
Received: 29 September 2017; Accepted: 29 November 2017;
Published: 12 December 2017.
Edited by:
Martin Sebastian Staege, Martin Luther University of Halle-Wittenberg, GermanyReviewed by:
Tara Patricia Hurst, Abcam, United KingdomYukihito Ishizaka, National Center for Global Health and Medicine, Japan
Copyright © 2017 Attig, Young, Stoye and Kassiotis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: George Kassiotis, Z2VvcmdlLmthc3Npb3Rpc0Bjcmljay5hYy51aw==
†These authors have contributed equally to this work.
 Jan Attig
Jan Attig George R. Young
George R. Young Jonathan P. Stoye
Jonathan P. Stoye George Kassiotis
George Kassiotis