- 1Department of Entomology, Institute of Plant Protection, Agricultural Research Organization, Rishon LeZion, Israel
- 2Faculty of Agricultural, Food and the Environmental Quality Sciences, The Hebrew University of Jerusalem, Rehovot, Israel
- 3Department of Entomology – Center for Pollinator Research – Huck Institutes of the Life Sciences, Pennsylvania State University, University Park, PA, United States
- 4Department of Plant Pathology and Weed Research, Institute of Plant Protection, Agricultural Research Organization, Rishon LeZion, Israel
- 5Institute of Bee Health, Vetsuisse Faculty, University of Bern, Bern, Switzerland
The viral ecology of bee communities is complex, where viruses are readily shared among co-foraging bee species. Additionally, in honey bees (Apis mellifera), many viruses are transmitted – and their impacts exacerbated – by the parasitic Varroa destructor mite. Thus far, the viruses found to be shared across bee species and transmitted by V. destructor mites are positive-sense single-stranded RNA viruses. Recently, a negative-sense RNA enveloped virus, Apis rhabdovirus-1 (ARV-1), was found in A. mellifera honey bees in Africa, Europe, and islands in the Pacific. Here, we describe the identification – using a metagenomics approach – of ARV-1 in two bee species (A. mellifera and Bombus impatiens) and in V. destructor mites from populations collected in the United States and Israel. We confirmed the presence of ARV-1 in pools of A. mellifera, B. impatiens, and V. destructor from Israeli and U.S. populations by RT-PCR and found that it can reach high titers in individual honey bees and mites (107–108 viral genomic copies per individual). To estimate the prevalence of ARV-1 in honey bee populations, we screened 104 honey bee colonies across Israel, with 21 testing ARV-1-positive. Tagged-primer-mediated RT-PCR analysis detected the presence of the positive-sense ARV-1 RNA in A. mellifera and V. destructor, indicating that ARV-1 replicates in both hosts. This is the first report of the presence of ARV-1 in B. impatiens and of the replication of a rhabdovirus in A. mellifera and V. destructor. Our data suggest that Varroa mites could act as an ARV-1 vector; however, the presence of ARV-1 in B. impatiens (which are not parasitized by Varroa) suggests that it may not require the mite for transmission and ARV-1 may be shared among co-foraging bee species. Given that ARV-1 is found in non-Apis bee species, and because “ARV” is used for the Adelaide River virus, we propose that this virus should be called bee rhabdovirus 1 and abbreviated BRV-1. These results greatly expand our understanding of the diversity of viruses that can infect bee communities, though further analysis is required to determine how infection with this virus impacts these different hosts.
Introduction
Global populations of pollinator species have been experiencing serious declines (Biesmeijer, 2006; Potts et al., 2010; Goulson et al., 2015). In managed honey bee populations in the United States and Europe, beekeepers typically experience heavy colony losses every year, averaging 40% in the United States (Steinhauer et al., 2017). Pollinators are vital to production of many key agricultural crops, with about 75% of the major crops that are used worldwide for human consumption benefiting from insect – primarily bee – pollination (Klein et al., 2007); therefore, bee decline can have a long-term impact on food production (Klein et al., 2007; Winfree, 2008; Potts et al., 2010; Singh et al., 2010). It was estimated that the contribution of insects to agriculture was €153 billion in 2009 (Gallai et al., 2009), and these values are likely significantly higher if economic impact on downstream industrial sectors is taken into consideration (Chopra et al., 2015).
Viral infections have been implicated as a major factor underpinning colony decline and loss in honey bees (Rosenkranz et al., 2010; Cornman et al., 2012; Dainat et al., 2012a,b; Martin et al., 2012; Nazzi et al., 2012; Gisder and Genersch, 2015; McMenamin and Genersch, 2015). Honey bees are hosts to more than 20 viruses, most of which are positive-sense single-stranded RNA viruses (McMenamin and Genersch, 2015). Moreover, these positive-sense RNA viruses – such as acute bee paralysis virus (ABPV), deformed wing virus (DWV), Israeli acute paralysis virus (IAPV), Kashmir bee virus (KBV), Sacbrood virus (SBV), and Lake Sinai virus (LSV) (Bowen-Walker et al., 1999; Chen et al., 2004; Shen et al., 2005; Chen and Siede, 2007; de Miranda and Genersch, 2010; Di Prisco et al., 2011; Ravoet et al., 2015) – can be vectored by an ectoparasitic mite (Varroa destructor), a key parasite of honey bees. V. destructor parasitization exacerbates the negative effects of virus infections on a honey bee colony (Nazzi and Le Conte, 2016), it has been shown to enhance ABPV’s prevalence in Apis mellifera colonies (reviewed in Genersch and Aubert, 2010), and is associated with significant increases in DWV titers as well as in promoting lower diversity of viral genotypes (Yang and Cox-Foster, 2007; Martin et al., 2012; Nazzi et al., 2012; Mondet et al., 2014; Ryabov et al., 2014; Wilfert et al., 2016). Furthermore, DWV infection leads to increased V. destructor reproduction on infected bees (Di Prisco et al., 2016). V. destructor’s synergistic interactions with DWV has been strongly correlated with A. mellifera colony losses (Dainat et al., 2012a; Nazzi et al., 2012). Additionally, it has been suggested that increasing infections with RNA viruses have led to substantial decrease in mite infestation thresholds leading to honey bee colony loss (Sumpter and Martin, 2004; Le Conte et al., 2010).
Importantly, many viruses first detected in honey bees have been shown to be able to infect other pollinator species (reviewed in Tehel et al., 2016). Again, primarily positive-sense RNA viruses have been shown to infect non-honey bee species, including the same viruses that are transmitted by V. destructor to honey bees, namely ABPV, DWV, IAPV, KBV, SBV, and LSV (Tehel et al., 2016). Viral transmission appears to occur via co-foraging on infected flowers (Singh et al., 2010; Fürst et al., 2014). As generalist foragers with large foraging ranges and high population numbers, honey bees could be sharing pathogens with multiple pollinator species in any given area (Geslin et al., 2017). The degree to which these viruses negatively impact other pollinators remains to be determined (Genersch et al., 2006; Dolezal et al., 2016; Tehel et al., 2016; Melathopoulos et al., 2017).
Little information is available on negative-sense RNA viruses in pollinator communities. Recently, Remnant et al. (2017) describe the identification of two negative-sense strand viruses, Apis rhabdovirus-1 (ARV-1) and Apis rhabdovirus-2 (ARV-2) in A. mellifera colonies from three locations (Netherlands, South Africa, and South Pacific) as well in Varroa mites associated with the colonies. Rhabdoviruses are negative-sense RNA enveloped viruses, whose particles are 100–430 nm long and 45–100 nm in diameter. The viral genome encodes five structural proteins: RNA-dependent RNA-polymerase (L), a phosphorylated phosphoprotein (P), a nucleoprotein (N), the matrix protein (M), and the envelope glycoprotein (G) (Kuzmin et al., 2009). It was reported that rhabdovirus genomes bear additional putative proteins from alternative or overlapping open-reading frames (ORFs) within the major structural protein genes or from independent ORFs between the structural protein genes (reviewed in Walker et al., 2015). Analysis using the L protein sequences of ARV-1 and ARV-2, that displayed 30% and 23% of identity to that of the Farmington virus of birds (FARV-1; Palacios et al., 2013), showed that they form a monophyletic group with FARV-1 (Remnant et al., 2017).
To gain insight about the presence and distribution of negative-sense RNA viruses in honey bees and other bees, we used high-throughput sequencing to examine the transcriptomes and viromes of honey bee (Apis mellifera) populations in the United States and Israel, a bumble bee (Bombus impatiens) population in the United States, as well as populations of the ectoparasitic mite of honey bees V. destructor in Israel. We identified ARV-1 in these samples, and characterized its distribution, prevalence, and infectivity. We confirmed the presence of viral copies in honey bees (from both continents), B. impatiens and V. destructor, and estimated the viral prevalence among 104 honey bee colonies as well as its titers in individual A. mellifera bees and V. destructor mites. Importantly, we examined whether ARV-1 is actively replicating in A. mellifera and V. destructor mites. Replication of single-stranded sense RNA viruses requires the synthesis of the complementary positive-sense RNA, which can be detected by using strand-specific RT-PCR (Horsington and Zhang, 2007). Using this approach we found evidence that ARV-1 was able to replicate in A. mellifera and V. destructor mites.
Materials and Methods
Sample Collection
Israeli Samples
The honey bee colonies (A.mellifera ligustica) from the Agricultural Research Organization (ARO) used for transcriptome and virome analysis in Israel were maintained without treatment against Varroa for 30 months before sampling and received seasonal sugar feeding and Fumagilin treatment against Nosema. For validation, prevalence, and replication studies, nurse honey bees were collected from apiaries located at the North (Galilee, Kibbutz Dan Apiary), the Center (Kfar Ruth), and the South (Kibbutz Yad Mordechai) of the country.
U.S. Samples
Honey bees (A. mellifera) and bumble bees (B. impatiens) were collected in State College, PA, United States (40.821419, -78.14635), while co-foraging on flowering plants. Individuals from the different bee species were collected in separate tubes to ensure that the species were not mixed. At least 20 individuals were collected onto ice-cold 95% ethanol, and ultimately stored at -20°C until the samples were processed. Additionally, honey bees that exhibited symptoms of DWV infection were collected from a single colony from a Pennsylvania State University apiary in State College using the same protocol.
Preparation of U.S. and IL Samples
Israeli Samples
RNA was extracted from honey bees and V. destructor mites samples from four ARO colonies and subsequently pooled (30 bees and 310 mites for A. mellifera and V. destructor cDNA libraries, respectively). RNA was extracted using TRI reagent® (Sigma–Aldrich) according to the manufacturer’s instructions in a Geno/grinder homogenizer (Metuchen, NJ, United States) following further purification by precipitation with 2.5 M lithium chloride as described previously (Levin et al., 2016). The quality and quantity of the extracted RNA was evaluated using an Agilent bio-Bioanalyzer (Agilent Technologies).
U.S. Samples
Ten individuals from each sample group (field-collected B. impatiens, field-collected A. mellifera, and colony-collected A. mellifera with deformed wings) were placed individually in 2.0 ml nuclease-free microcentrifuge tubes with molecular grade H2O with three to five sterile glass beads and homogenized for 45 s in a FastPrep FP120 Cell Disruptor (Thermo-Fisher Scientific, Waltham, MA, United States). Samples were stored on ice for 10 min, and the previous step was repeated to ensure adequate homogenization. Hundred microliters of the homogenate from each of the 10 samples was pooled in a common 1.5 ml microcentrifuge tube. The resulting homogenate was passed through a 0.2-μm cell filter (Corning, Tewksbury, MA, United States) to purify the virus extracts as in Hunter et al. (2010) and Liu et al. (2010). One hundred and twenty-five microliters of each sample was treated with a nuclease cocktail (including 14 U Turbo DNase I, 25 U Benzonase, 20 U RNase I, and 10× DNase buffer) and incubated at 37°C for 1.5 h to remove all nucleic acid that is not protected by a viral capsid as in He et al. (2013). Nucleic acids (both RNA and DNA) from the purified encapsulated viruses were extracted using a MagMAX Viral Isolation Kit (Thermo-Fisher Scientific, Waltham, MA, United States), according to the manufacturer’s protocol.
Transcriptome and Virome Analysis
Israeli Samples
High-throughput sequencing of the RNA from the honey bees and V. destructor mites samples from Israeli colonies was described previously (Levin et al., 2016). Complementary Sanger sequencing was performed at the Weizmann Institute of Science (Rehovot, Israel) with primers indicated in Supplementary Table S1. Note that Levin et al. (2016) focused on the identification of viruses that were unique to V. destructor mites, and thus information related to ARV-1 (which was found in both samples) is newly described in the current manuscript.
U.S. Samples
A previously established protocol was used to obtain unbiased random amplification of the extracted nucleic acids (Ng et al., 2012). Briefly, the first strand cDNA was synthesized from 30 μl of viral nucleic acid using a random primer design (consisting of a 20 base oligonucleotide sequence followed by a randomized octamer sequence: GACCATCTAGCGACCTCCACNNNNNNNN) in a reverse transcriptase (RT) reaction using a High-Capacity cDNA Reverse Transcription Kit (Thermo-Fisher Scientific, Waltham, MA, United States). To synthesize the second strand, the 19 μl of the initial cDNA was denatured at 95°C for 2 min and cooled to 4°C prior to the addition of Klenow Fragment DNA Polymerase (New England Biolabs, Ipswich, MA, United States) for fragment extension at 37°C for 60 min. Finally, PCR amplification was performed for 40 cycles using the above primer without the randomized octamer sequence (GACCATCTAGCGACCTCCAC) and 2 μl of dsDNA template. The PCR reactions were then purified using a MSB Spin PCRapace Purification Kit (Invitek, Berlin, Germany) using the manufacturer’s standard protocol. Quality and concentration of the samples were assessed using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, United States). The samples were then submitted to the Genome Core Facility at Pennsylvania State University for sequencing on the Illumina MiSeq, resulting in ∼1 × 106 reads of 150 nucleotides each per sample.
Bioinformatic Identification of ARV-1 Contigs
Israeli Samples
The transcriptome and virome data were analyzed as described before (Levin et al., 2016). The transcriptome and virome files of V. destructor and A. mellifera were uploaded to SRA database under accession numbers PRJNA329427 and PRJNA329428, respectively. The sequences were cleaned from remains of adaptor sequences and low quality reads using Trimmomatic Software (Bolger et al., 2014) and assembled de novo using Trinity (Haas et al., 2013). The assembled contigs were subsequently translated and aligned to the GenBank nr database (all nr database without filtering) by BLASTx (Altschul et al., 1997) (with a cut-off value of <1e-5) (Corpet, 1988).
U.S. Samples
The quality of the sequencing run was assessed using FastQC (Andrews, 2010) to ensure only high-quality sequences were used for further analyses. Adaptor sequences and reads with low-quality scores were removed using Trimmomatic (Bolger et al., 2014). Processed reads were assembled into longer contigs using three de novo assembly programs; SPAdes (Bankevich et al., 2012), Velvet/Oases (Schulz et al., 2012), and Trinity (Grabherr et al., 2011; Haas et al., 2013). To improve the overall assembly, the results from these three assemblies were consolidated into a single assembly using Mix (Soueidan et al., 2013). To identify homologous sequences, the assembled contigs were compared to the viral sequences in the NCBI nr database (Pruitt et al., 2007) using BLASTx (Altschup et al., 1990) using an E-value cutoff of 10-10. ORF-finder (Rombel et al., 2002) was used to identify potential ORF within the contigs with significant BLAST hits to known viruses to further characterize the viral contigs from each sample. Contig sequences with significant BLAST hits for ARV-1 and Farmington virus are reported here, the remaining sequences will be reported in a future study (Galbraith et al., in preparation).
Molecular Analysis
The complete nucleotides and predicted ORF of ARV-1 from the four sample groups (U.S. A. mellifera, Israeli A. mellifera, U.S. B. impatiens, and Israeli V. destructor) were used for the analysis (Figure 1 and Supplementary Figure S1). Protein Alignment was done using the web interface of MultAlin1 (Corpet, 1988). The identified rhabdovirus sequences were deposited in NCBI GenBank (accession numbers MF114348–MF114351).
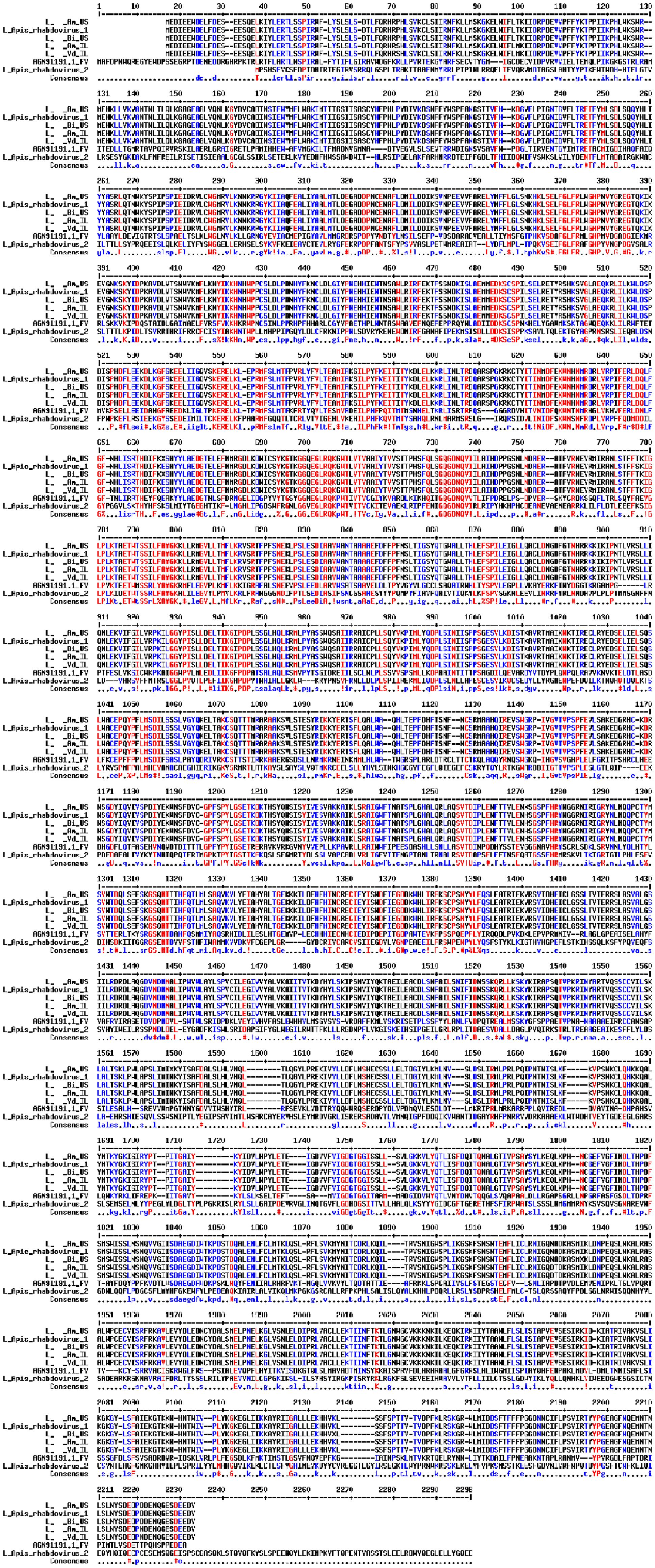
FIGURE 1. Alignment of the L proteins of the rhabdovirus from A. mellifera (Am_US and Am_ IL), B. impatiens (Bm_US), and V. destructor (Vd_IL) to ARV-1, ARV-2, and Farmington virus of birds (GenBank KY354232.1, KY354234.1, and YP_009091822.1, respectively). Amino acid residues in red, blue, and black are fully, partially, and not identical among all the aligned sequences.
RT-PCR Samples
Israeli Samples
Nurse bees and V. destructor mites were collected and RNA was extracted from individuals using TRI reagent as described above. cDNA was prepared using Maxima-RT (Fermentas-Thermo, Fisher Scientific, Burlington, Canada) using oligo-dT and random primers according to the manufacturer’s instructions. Hundred nanograms of template RNA was used for screening and a second re-screening was performed for honey bee samples with 200 ng of the same RNA. RT-conditions: incubation of RNA and primers at 65°C 5 min, followed by addition of buffer containing 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 2 mM MgCl2, 5 mM DTT, 4 units of RNase inhibitor Ribolock® (Thermo Scientific), the RT enzyme (200 units) in a 20-μl volume, and further incubation at 55°C for 30 min. The reaction was terminated by heating at 85°C for 5 min.
PCRs were performed using LongAmp Taq Polymerase (New England Biolabs, Ipswich, MA, United States) and specific primers (see Supplementary Table S1) in a BioER GenePro TC-E-96G apparatus (Hangzhou Bori Technology Co., Ltd., P.R. China). For long templates amplification 1 μl cDNA was used with 2.5 units of Taq DNA polymerase with enzyme buffer, 10 mM dNTP, 2 mM MgCl2, 2% DMSO, and 0.2 μM of each forward and reverse primer in a final reaction volume of 25 μl. The protocol used was 95°C for 4 min, then 35 cycles of 94°C for 20 s, 55°C for 1 min, 65°C for 8 min, and a final extension step of 65°C for 10 min. PCR validations were performed with GoTaq® (Promega Corporation, United States) with 1.5 mM MgCl2, 1 μl cDNA template, and 0.2 μM of each forward and reverse primer in a 20 μl reaction with the following conditions: 95°C for 4 min, 30 cycles at 94°C for 30 s, then 58°C for 50 s, 72°C for 2 min, and a final extension step of 72°C for 10 min. Amplification and validation primers are provided in Supplementary Table S1, and in the corresponding legends of the figures. All the PCR reactions included non-template controls (NTC). PCR products were evaluated by conventional agarose electrophoresis.
Additionally, Sanger sequencing (performed at the Biological Services Unit of the Weizmann Institute of Science, Israel) was used to confirm that the amplified products indeed corresponded to the expected ARV-1 sequences. Also, these sequences allowed us to validate gaps in sequence data obtained from small contigs and to compare it to the nucleotide sequence obtained from the largest contig of 14606 (the primer sequences used to analyze the termini and subsequent gaps are provided in Supplementary Table S1). The sequencing perfectly matched the large nucleotide sequence obtained in the above large contig.
U.S. Samples
The RT-PCR was performed on the same individuals originally homogenized for the virome analysis (described above). For each individual bee used in the virome analysis, 50 μl of the homogenate was used for RNA extraction using an RNAeasy Kit (Qiagen, Valencia, CA, United States). cDNA was prepared using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, United States) using the standard protocol provided by the manufacturer. Specifically, 200 ng of template RNA was mixed with 2 μl 10× RT Buffer, 3.2 mM dNTP Mix, 2 μl 10× RT Random Primers, 50 units of MultiScribe Reverse TranscriptaseTM, 20 units of RNase inhibitor, and additional H2O to bring the reaction volume to 20 μl. The reaction was performed according to the manufacturer’s protocol: the solution was heated to 25°C for 10 min, 37°C for 120 min, 85°C for 5 min, and then cooled to 4°C.
The cDNA product was then amplified with PCR using primers described in Supplementary Table S1. According to the manufacturer’s protocol, 2 μl of cDNA template was mixed with 0.2 μM of each Forward and Reverse primer, 2.5 μl 10× Taq buffer, 0.2 mM dNTPs, 0.75 units of Taq DNA polymerase (New England Biolabs, Ipswich, MA, United States), and 18.875 μl of H2O, in a final reaction volume of 25 μl. The solution was heated to 95°C for 4 min, followed by 30 cycles at 94°C for 30 s, 58°C for 50 s, and 72°C for 2 min, with a final extension step of 72°C for 10 min, and finally cooled to 4°C. All the PCR reactions included NTCs. PCR products were evaluated by conventional agarose electrophoresis to identifying PCR products at the expected sizes.
qRT-PCR Assay
cDNA was prepared from individual nurse bees and V. destructor mites as described in the “RT-PCR – Israeli samples” section above. Quantitative PCR amplifications were performed on a PikoReal96 machine (Thermo-Fisher Scientific, Waltham, MA, United States) using a standard protocol (95°C 2 min; 40 cycles: 95°C 10 s, 60°C 20 s, 72°C 20 s). Each quantitative PCR analysis was performed in triplicate, in a 96-well PCR plate sealed with an optical adhesive cover (Thermo-Fisher Scientific, Waltham, MA, United States). Non-template controls (water) were included in triplicates in each assay. The KAPA SYBR FAST qPCR Master Mix (2×) Universal (Kapa Biosystems) was used in 10 μl final volume. For each analysis, 2 μl of the diluted cDNA was used (dilution factor of 4) and specific primers BRV-qRT-F1 and BRV-qRT-R1 concentration were 0.25 μM (Supplementary Table S1). The efficiency of the PCR reaction was as follows: E = 97%, R2 = 0.9979, and the slope = -3.387. The specificity of the amplicons synthesized during the PCR run was ascertained by performing a dissociation curve protocol from 60 to 95°C.
Amplicons of 182 bp containing the ARV-1 target sequence were obtained by performing PCR with the primers BRV-qRT-F1 and BRV-qRT-R1. A 10-point standard curve was prepared [fourfold serial dilutions of the obtained amplicon with known concentrations from 4 pg (Cq = 8.5) and up to 1.5 × 10-5 pg (Cq = 27.5)]. To establish a calibration curve for the quantification of the V. destructor mRNA for cytoplasmic actin reference, a 336 bp V. destructor DNA fragment was amplified with the primers VcytoactinF and VcytoactinR (Supplementary Table S1). After 34 amplification cycles, the DNA fragments were gel purified and aliquots were used for measuring their DNA concentration using a NanoDrop (Thermo-Fisher Scientific, Waltham, MA, United States). For each quantitative PCR assay, n2 fold serial dilutions in water were made and each dilution was processed in triplicates on the same 96-well PCR plate where the samples were deposited. For each sample, both ARV-1 and V. destructor mRNA for cytoplasmic actin quantitative analysis were performed in separate wells. To set up a calibration curve of A. mellifera, the housekeeping primers were RPL8 F and RPL8 R (Evans et al., 2006) as described before (Zioni et al., 2011) (primers in Supplementary Table S1).
Individual ARV-1 loads of 100 ng total RNA extracted from the individual sample were calculated by plotting Ct values against the logarithm of the RNA copy number using the PikoRealTM Software 2.2 (Thermo-Fisher Scientific, Waltham, MA, United States). These values were used to calculate the ARV-1 copy number in the total RNA extracted from the individual sample.
Replication Assay
Using the Israeli samples from individual nurse honey bees and V. destructor mites collected for the RT-PCR analysis, a replication analysis (presence of the positive strand-sense RNA) was performed. To avoid artifactual positive results caused by false priming (Vashist et al., 2012), we synthesized tagged primers and used them in the detection of the positive strand by RT-PCR as described before for analysis of replication of the RNA virus DWV of bees (Yue and Genersch, 2005). The negative-strand cDNA from the RNA samples was produced using the tagged-primer BRV-8711F-TAG and subsequently performing PCR with primers BRV-10356-R and TAG-D_F (Supplementary Table S1). cDNA produced without any primer was used as control followed by PCR with the same primers from above. PCR was performed at 95°C for 4 min, 30 cycles at 94°C for 30 s, then 59°C for 50 s, 72°C for 1.5 min, and a final extension step of 72°C for 10 min. A second PCR reaction was performed using only the primer BRV-10356-R to control for the absence of unspecific priming by presence of residual BRV-8711F-TAG primer from the RT reaction (Supplementary Figure S4). The identity of the amplified fragment was confirmed by Sanger sequencing (performed at the Biological Services Unit of the Weizmann Institute of Science, Israel).
Results
Identification of Apis Rhabdovirus-1 (ARV-1) in Honey Bees, Bumble Bees, and Varroa Mites
BLASTx analysis revealed the presence of a large contig homologous to a rhabdovirus in the transcriptomes of A. mellifera bees sampled from Israeli honey bee colonies. A similar parallel analysis evaluating the viral transcriptomes of honey bees (A. mellifera) and bumble bees (B. impatiens) sampled from populations in the United States also detected this rhabdovirus. Furthermore, exploring the virome of V. destructor mites sampled in Israel, we were able to identify the virus in the mites. The identified sequences were most homologous to the nucleotide sequence of ARV-1 identified in A. mellifera [(GenBank KY354232.1, Remnant et al., 2017); see below].
Assembly of the sequences in the individual data sets generated viral contigs from the A. mellifera samples of 14,606 nucleotides (in the Israeli samples, the U.S. samples had a slightly smaller contig size of 13,842 nucleotides), a viral contig from the B. impatiens sample of 14,590 nucleotides, and a viral contig from the V. destructor sample of 14,589 nucleotides.
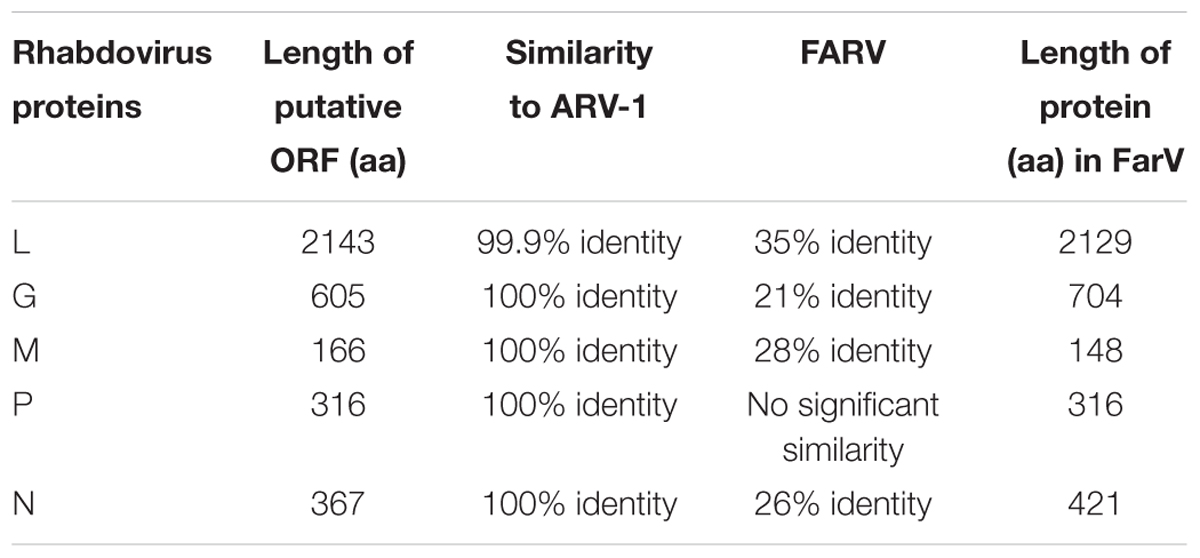
Alignment among the putative proteins generated from the viral contigs of Israeli honey bees, U.S. honey bees, U.S. bumble bees, and Israeli V. destructor mites showed 100% identity for the L, M, and P proteins and only one amino acid change for each of the N proteins (amino acid 255, K in the U.S. samples, and R in the Israeli samples) and G protein (amino acid 141, N in the U.S. samples, and S in the Israeli samples) (Figure 1 and not shown). The honey bee, bumble bee, and Varroa rhabdoviruses L protein exhibited 99.9% identity to the L-protein of ARV-1 (only two changes amino acids 777 T to S and 981 V to I compared to ARV-1, respectively), and 35% identity for the L-protein of FARV (Figure 1). The proteins of the rhabdovirus we found were identical to that of ARV-1 (summarized in Table 1). BLASTn showed that the rhabdovirus genomes that we found had 99% similarity to that of ARV-1 and very low similarity (2% of the virus sequence with e-value of 0.16) to that of ARV-2 (GenBank KY354234.1, Remnant et al., 2017; and not shown). Thus, we concluded that the rhabdovirus we identified in honey bees, bumble bees, and V. destructor mites was ARV-1.
Confirmation of ARV-1 Presence Using RT-PCR
We collected two additional pools of nurse honey bees and their associated V. destructor mites from two Israeli honey bee colonies and screened these for the presence of ARV-1 using RT-PCR. We utilized two different pairs of primers that anneal at different regions of the viral genome (nucleotides 2683–4195 and 10245–11804, Figure 2A, lanes BP1, VP1 and BP2, and VP2, respectively). This analysis clearly identified the presence of ARV-1 sequences in these pools (Figure 2A, lanes BP and VP, respectively).
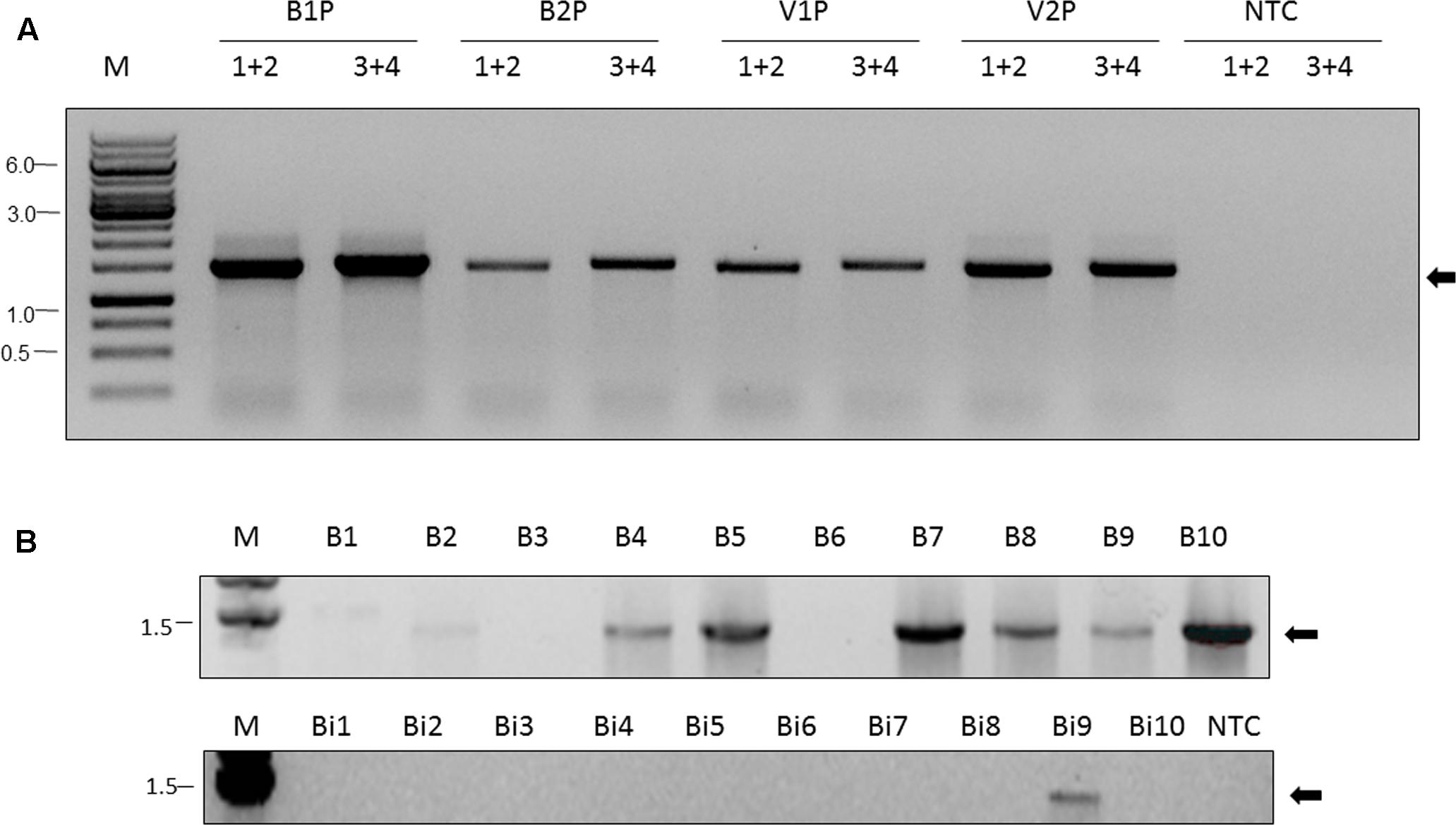
FIGURE 2. Apis rhabdovirus-1 is detected in A. mellifera, B. impatiens, and V. destructor by RT-PCR. (A) Pools of nurse bees (BP) and mites (VP), respectively, from Israeli colonies 1 and 2 (B1P, V1P and B2P, and V2P, respectively, indicated above the figure). 1+2 (primers BRV-1-2683-F and BRV-1-4195-R) and 3+4 (primers BRV-1-10245-F and BRV-1-11804-R) in conserved regions used in the PCR reaction, see the section “Materials and Methods” and Table 1. M, GeneRuler Marker 1 kb DNA Ladder (Thermo Scientific Inc.); NTC, non-template control; Arrow, ARV-1. (B) Individual honey bees (B1, … , B10) and bumble bees (Bi1, … , Bi10) from U.S. samples. NTC, non-template control. Primers used 1+2 (BRV-1-2683-F and BRV-1-4195-R). Arrow, ARV-1.
Additionally, we analyzed the collected U.S. bee samples for the presence of ARV-1 using RT-PCR with the same primers. For the virome analysis, described above we evaluated pooled samples (10 bees), and the pools of field-collected bumble bees and colony-collected honey bees were positive for ARV-1. For the RT-PCR analysis, we examined the 10 individuals from each of these pooled samples. ARV-1 amplified in 7 of the 10 honey bees and 1 of 10 bumble bees (Figure 2B).
Prevalence, Quantification, and Replication of ARV-1 in Individuals
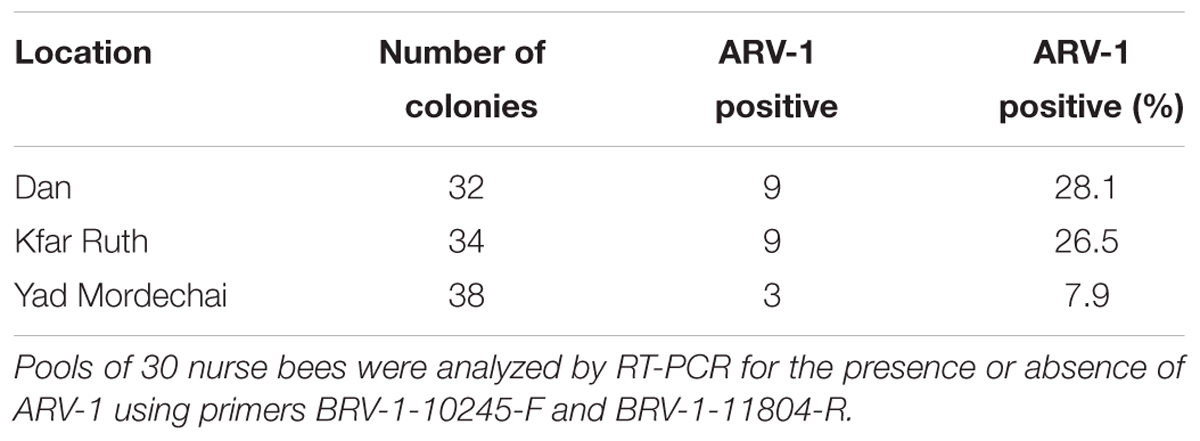
To estimate the prevalence of ARV-1 in Israeli honey bee populations, we screened for its presence in colonies from apiaries located at the North, Center, and South of Israel (Dan, Kfar Ruth, and Yad Mordechai, respectively). We found that 21 of the 104 colonies analyzed were positive for the virus (Table 2).
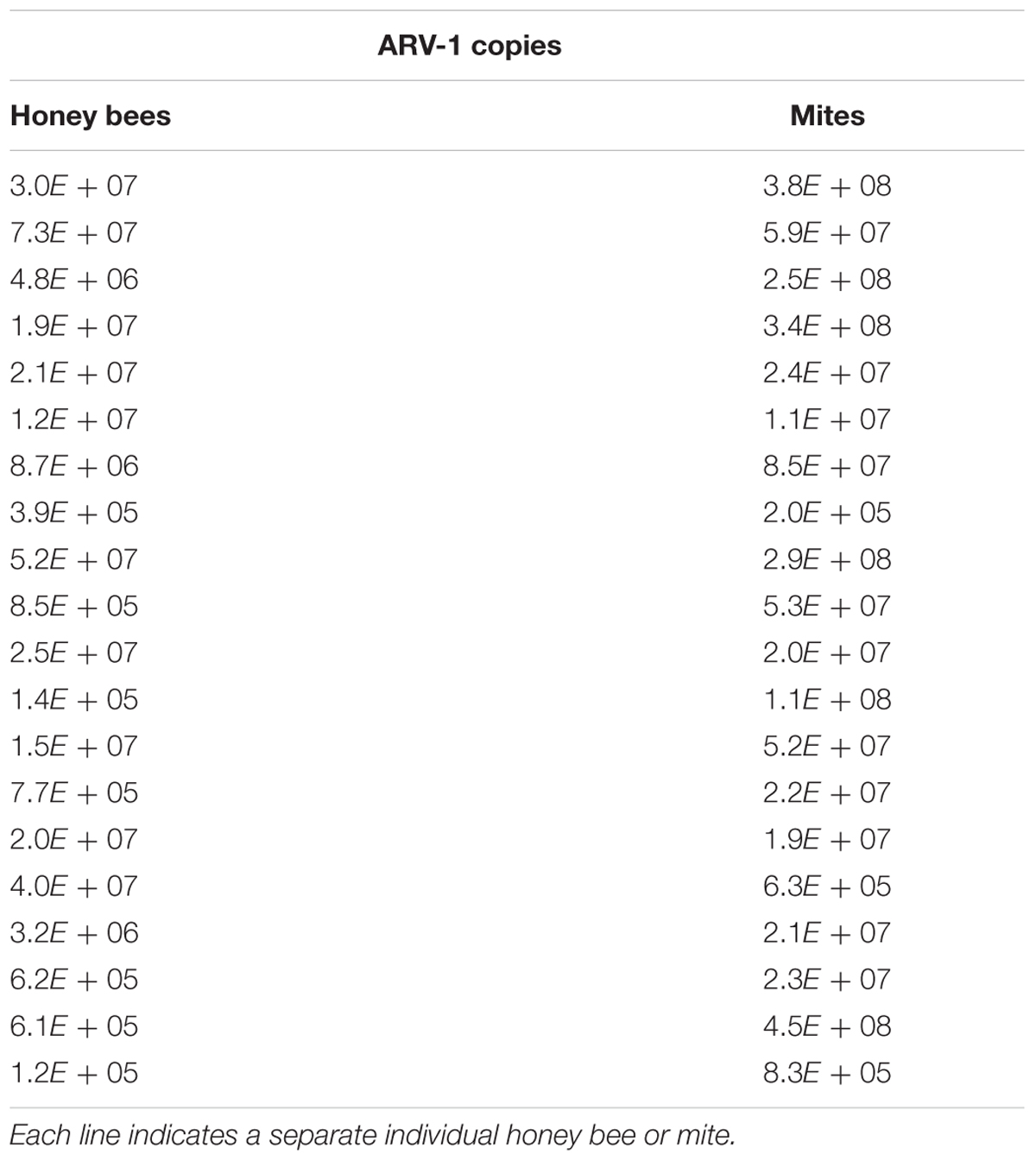
RT-PCR screening of small subsamples of individual nurse bees (n = 84) and V. destructor mites (n = 21) from these Israeli colonies demonstrated that the viruses were not present in all the individuals from the same colony. Thus, in random sampling of individual bees and mites from four ARV-1-positive colonies we found the virus in 4.8% of bees and in 76.2% of the mites evaluated. Moreover, the number of genomic copies of ARV-1 estimated by RT-qPCR was similar among bees and V. destructor mites collected from these colonies: 1.2 × 105–7.3 × 107 and 2.0 × 105–4.5 × 108, respectively (Table 3).
To confirm replication of ARV-1 in our samples, we screened for the presence of the positive-sense RNA strand of the virus using RNA-strand sense-specific primer-tagged-RT-PCR (see the section “Materials and Methods”). A predicted size fragment of about 1650 nucleotides corresponding to the ARV-1 positive-sense-strand RNA size comprised between nucleotides 8711 and 10356 was found in all the tested samples from individual Varroa mites or nurse bees that showed ARV-1 copies in the above RT-qPCR assay (Figure 3, lanes V1, V2, …, etc. and B1, B2, …, etc.). No amplification was observed in control samples when PCR was performed with cDNA of the same individuals prepared from RNA without any oligonucleotide primer in the RT reaction (Figure 3, lanes V1C, V2C, …, etc. and B1C, B2C, …, etc.; and Replication assay in the section “Materials and Methods”).
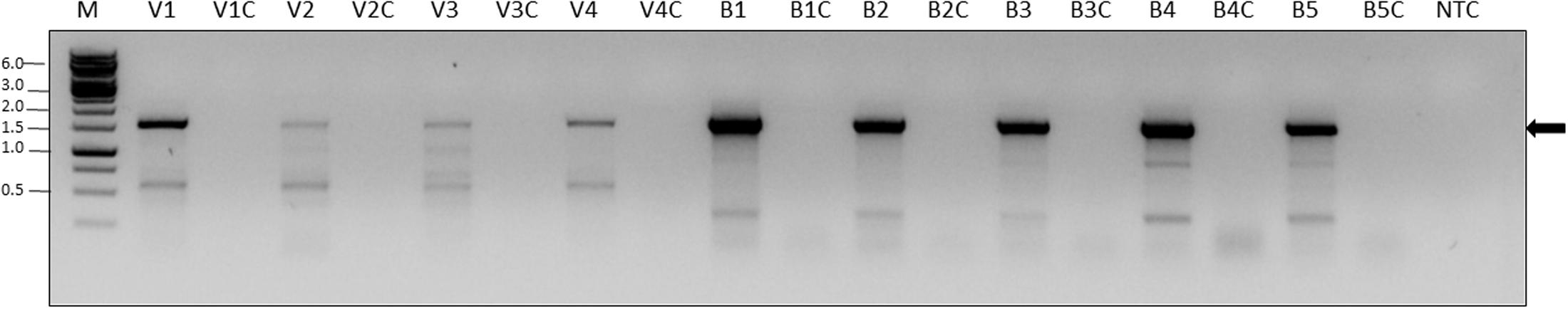
FIGURE 3. Detection of the ARV-1 positive-sense RNA strand in V. destructor (V) and A. mellifera (B). The numbers indicate the individual tested. Suffix C indicates PCR control reaction from the same individual RNA performed on cDNA produced without any primer (see the section “Materials and Methods” and Table 1). PCR primers: BRV-1F-10356-R and TAG-D_F. M, GeneRuler Marker 1 kb DNA Ladder (Thermo Scientific Inc.); NTC, non-template control; Arrow, ARV-1 amplicon that was confirmed by sequencing (see the section “Materials and Methods”).
Individuals nurse honey bees and V. destructor mites that were negative in the above RT-qPCR did not show amplification (not shown). Moreover, using Sanger DNA sequencing of the above specific-primer-tagged amplicons, we confirmed that it was identical to the ARV-1 sequence comprised between the nucleotides 8711 and 10356 of the viral genome.
Discussion
Currently, there have been more than 20 viruses identified from A. mellifera honey bees around the world (McMenamin and Genersch, 2015). At least 11 of these viruses have been observed in other bee species (Tehel et al., 2016), and many of these viruses are also found in – and vectored by – V. destructor mites, a key parasite of A. mellifera honey bees. The vast majority of viruses identified in A. mellifera and found, thus far, to be circulating in other bee species and transmitted by V. destructor mites are positive-sense single-strand RNA viruses. Here, we found that ARV-1, a negative-sense single-strand RNA virus, that was recently found in honey bees and Varroa mites (Remnant et al., 2017), was present in two communities of bee pollinators.
Members of the Rhabdoviridae family of viruses typically infect animals and plants, but are mostly transmitted by arthropods (Kuzmin et al., 2009). Because viruses can be transmitted through pollen (Singh et al., 2010) and bees actively collect pollen, there is concern that viruses identified in bee samples may simply represent contamination from pollen. Indeed, in metagenomics studies of bee viruses, many viruses known to infect plants are often found (Schoonvaere et al., 2016). In our study, we demonstrated that ARV-1 is actively replicating in its A. mellifera honey bee and V. destructor mite hosts, by identifying the positive-sense RNA strand of the virus using RNA strand sense-specific-primer-tagged-RT-PCR, and confirming the product via Sanger sequencing. Furthermore, the fact that ARV-1 has been identified in A. mellifera populations from five distinct regions (Europe, North America, Middle East, Africa, and South Pacific) suggests that it is indeed infecting honey bee populations and is not simply a plant contaminant.
We identified ARV-1 in A. mellifera honey bee populations in 21 out of 104 colonies screened from three distinct locations in Israel. Although ARV-1 was found in ∼20% of the colonies screened, it was only present in 4.8% of the sampled bees and 76.2% of the mites collected from ARV-1-positive colonies (Table 2). These data suggest that this virus is not highly infectious. However, many factors – including age, physiological state, nutritional status, co-infection with other parasites and pathogens, and pesticide exposure – can influence the ability of a honey bee to tolerate or clear a viral infection (McMenamin et al., 2016). Thus, more detailed monitoring and experimental studies are needed to understand the infection dynamics of ARV-1.
While ARV-1 was detected in honey bees with deformed wings in the U.S. samples, there was no indication of any symptoms in the Israeli samples. It remains to be determined if ARV-1 causes symptoms in honey bees. It is known that bees can carry viruses asymptomatically and virulent infections can be induced by various stresses (Genersch and Aubert, 2010; Nazzi and Pennacchio, 2014). Other bee viruses such as LSV are not associated with any known symptom in individual bees (Daughenbaugh et al., 2015). Additional controlled studies are necessary to determine if ARV-1 infection causes any negative symptoms in its bee or mite hosts, under a wide range of ecologically relevant conditions. Moreover, some viruses are in mutualistic relationships with their hosts, and can improve their hosts’ resilience and fitness under certain conditions (Roossinck, 2011).
Remnant et al. (2017; Roossinck, 2011) named the rhabdoviruses they found ARV-1 and ARV-2, however, they were likely unaware that the ARV abbreviation they applied was previously attributed to another rhabdovirus Adelaide River virus (Wang et al., 1995), a fact that may create confusion while analyzing data. Furthermore, our results indicate that this virus is found in bumble bee species as well. Thus, we propose that these viruses should be called bee rhabdoviruses and abbreviated BRV-1 and BRV-2, instead of ARV-1 and -2, respectively.
In addition to BRV-1 and BRV-2, recent studies have identified several negative-sense RNA viruses: two putative Bunya-like virus of A. mellifera, A. mellifera bunyavirus-1 and -2 (ABV-1 and ABV-2, respectively; Remnant et al., 2017), Scaldis River bee virus (SRBV), and Ganda bee virus (GABV) in Osmia cornuta bees (Schoonvaere et al., 2016). The latter appear to belong the proposed new family Chuviridae of the order Mononegavirales and the new family Phasmaviridae of the order Bunyavirales.
With the advent of high-throughout sequencing, improved bioinformatics approaches, and expanded genomic databases, it is now possible to readily identify the microbial communities from field-collected populations of animals and plants. Bee viruses are particularly fascinating, as they can infect diverse species, are transmitted via multiple routes (contaminated flowers, social interactions, and vectors), and their pathogenicity and virulence are influenced by a number of biotic and abiotic factors. While previous studies have identified and focused on positive-sense single-strand RNA viruses in honey bees and the broader bee community, our results, together with those of Schoonvaere et al. (2016), Remnant et al. (2017), and those of Li et al. (2015) – who found numerous negative-sense RNA viruses by metagenomics analysis of 70 species of insects, spiders, centipedes, etc. in China – suggest that negative-sense strand RNA viruses may also be circulating and broadly distributed among honey bee populations and bee communities. It remains to be determined if these negative-sense RNA viruses are functionally distinct from positive-sense RNA viruses in terms of their pathogenicity and virulence.
Author Contributions
NC, SL, NS, TE, DG, and CG conceived and designed the experiments. SL, TE, and DG performed the experiments. NS, SL, and DG bioinformatic analysis. NC, SL, DG, and CG data analysis. NC, NS, SL, TE, DG, and CG wrote the paper and prepared the figures and tables. NC, DG, and CG edited the manuscript. NC, NS, SL, TE, DC, and CG revised and approved the manuscript.
Funding
NC was supported by a USAID Grant number TA-MOU-11-M32-035 and a Grant of the Chief Scientist of the Ministry of Agriculture of Israel number 131-1857. DG and CG (Penn State) were supported by funding from U.S. Department of Agriculture’s Animal and Plant Inspection Service (USDA-APHIS, Agreement No. 15-8130-0501-CA).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer PW and handling Editor declared their shared affiliation.
Acknowledgments
The authors thank Dr. Victoria Soroker, Mr. Yossef Kamer, and Mrs. Nurit Eliash for their assistance with the honey bee colonies from the Zrifin Apiary. They also like to thank Mario Padilla (Penn State) for collection of State College samples and Joyce Sakamoto (Penn State) for guidance on the viral purification of the State College samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02482/full#supplementary-material
Footnotes
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Altschup, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Biesmeijer, J. C. (2006). Parallel declines in pollinators and insect-pollinated plants in Britain and Netherlands. Science 313, 351–354. doi: 10.1126/science.1127863
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowen-Walker, P., Martin, S., and Gunn, A. (1999). The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 73, 101–106. doi: 10.1006/jipa.1998.4807
Chen, Y., Pettis, J. S., Evans, J. D., Kramer, M., and Feldlaufer, M. F. (2004). Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie 35, 441–448. doi: 10.1051/apido:2004031
Chen, Y. P., and Siede, R. (2007). Honey bee viruses. Adv. Virus Res. 70, 33–80. doi: 10.1016/S0065-3527(07)70002-7
Chopra, S. S., Bakshi, B. R., and Khanna, V. (2015). Economic dependence of U.S. industrial sectors on animal-mediated pollination service. Environ. Sci. Technol. 49, 14441–14451. doi: 10.1021/acs.est.5b03788
Cornman, R. S., Tarpy, D. R., Chen, Y., Jeffreys, L., Lopez, D., Pettis, J. S., et al. (2012). Pathogen webs in collapsing honey bee colonies. PLOS ONE 7:e43562. doi: 10.1371/journal.pone.0043562
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. doi: 10.1093/nar/16.22.10881
Dainat, B., Evans, J. D., Chen, Y. P., Gauthier, L., and Neumann, P. (2012a). Dead or alive: deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 78, 981–987. doi: 10.1128/AEM.06537-11
Dainat, B., Evans, J. D., Chen, Y. P., Gauthier, L., and Neumann, P. (2012b). Predictive markers of honey bee colony collapse. PLOS ONE 7:e32151. doi: 10.1371/journal.pone.0032151
Daughenbaugh, K. F., Martin, M., Brutscher, L. M., Cavigli, I., Garcia, E., Lavin, M., et al. (2015). Honey bee infecting Lake Sinai viruses. Viruses 7, 3285–3309. doi: 10.3390/v7062772
de Miranda, J. R., and Genersch, E. (2010). Deformed wing virus. J. Invertebr. Pathol. 103(Suppl.), S48–S61. doi: 10.1016/j.jip.2009.06.012
Di Prisco, G., Annoscia, D., Margiotta, M., Ferrara, R., Varricchio, P., Zanni, V., et al. (2016). A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proc. Natl. Acad. Sci. U.S.A. 113, 201523515. doi: 10.1073/pnas.1523515113
Di Prisco, G., Pennacchio, F., Caprio, E., Boncristiani, H. F., Evans, J. D., and Chen, Y. (2011). Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 92, 151–155. doi: 10.1099/vir.0.023853-0
Dolezal, A. G., Hendrix, S. D., Scavo, N. A., Carrillo-Tripp, J., Harris, M. A., Wheelock, M. J., et al. (2016). Honey bee viruses in wild bees: viral prevalence, loads, and experimental inoculation. PLOS ONE 11:e0166190. doi: 10.1371/journal.pone.0166190
Evans, J. D., Aronstein, K., Chen, Y. P., Hetru, C., Imler, J.-L., Jiang, H., et al. (2006). Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656. doi: 10.1111/j.1365-2583.2006.00682.x
Fürst, M. A., McMahon, D. P., Osborne, J. L., Paxton, R. J., and Brown, M. J. F. (2014). Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506, 364–366. doi: 10.1038/nature12977
Gallai, N., Salles, J. M., Settele, J., and Vaissière, B. E. (2009). Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 68, 810–821. doi: 10.1016/j.ecolecon.2008.06.014
Genersch, E., and Aubert, M. (2010). Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Vet. Res. 41:54. doi: 10.1051/vetres/2010027
Genersch, E., Yue, C., Fries, I., and de Miranda, J. R. (2006). Detection of Deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J. Invertebr. Pathol. 91, 61–63. doi: 10.1016/j.jip.2005.10.002
Geslin, B., Gauzens, B., Baude, M., Dajoz, I., Fontaine, C., Henry, M., et al. (2017). “Chapter Four - Massively introduced managed species and their consequences for plant-pollinator interactions,” in Advances in Ecological Research, ed. H. Caswell (Cambridge: Academic Press), 147–199. doi: 10.1016/bs.aecr.2016.10.007
Gisder, S., and Genersch, E. (2015). Special issue: honey bee viruses. Viruses 7, 5603–5608. doi: 10.3390/v7102885
Goulson, D., Nicholls, E., Botías, C., and Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1255957. doi: 10.1126/science.1255957
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
He, B., Li, Z., Yang, F., Zheng, J., Feng, Y., Guo, H., et al. (2013). Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel mammalian viruses. PLOS ONE 8:e61950. doi: 10.1371/journal.pone.0061950
Horsington, J., and Zhang, Z. (2007). Analysis of foot-and-mouth disease virus replication using strand-specific quantitative RT-PCR. J. Virol. Methods 144, 149–155. doi: 10.1016/j.jviromet.2007.05.002
Hunter, W., Ellis, J., Vanengelsdorp, D., Hayes, J., Westervelt, D., Glick, E., et al. (2010). Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLOS Pathog. 6:e1001160. doi: 10.1371/journal.ppat.1001160
Klein, A.-M., Vaissière, B. E., Cane, J. H., Steffan-Dewenter, I., Cunningham, S. A., Kremen, C., et al. (2007). Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 274, 303–313. doi: 10.1098/rspb.2006.3721
Kuzmin, I. V., Novella, I. S., Dietzgen, R. G., Padhi, A., and Rupprecht, C. E. (2009). The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect. Genet. Evol. 9, 541–553. doi: 10.1016/j.meegid.2009.02.005
Le Conte, Y., Ellis, M., and Ritter, W. (2010). Varroa mites and honey bee health: can Varroa explain part of the colony losses? Apidologie 41, 353–363. doi: 10.1051/apido/2010017
Levin, S., Sela, N., and Chejanovsky, N. (2016). Two novel viruses associated with the Apis mellifera pathogenic mite Varroa destructor. Sci. Rep. 6:37710. doi: 10.1038/srep37710
Li, C. X., Shi, M., Tian, J. H., Lin, X. D., Kang, Y. J., Chen, L. J., et al. (2015). Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. doi: 10.7554/eLife.05378
Liu, X., Zhang, Y., Yan, X., and Han, R. (2010). Prevention of Chinese sacbrood virus infection in Apis cerana using RNA interference. Curr. Microbiol. 61, 422–428. doi: 10.1007/s00284-010-9633-2
Martin, S. J., Highfield, A. C., Brettell, L., Villalobos, E. M., Budge, G. E., Powell, M., et al. (2012). Global honey bee viral landscape altered by a parasitic mite. Science 336, 1304–1306. doi: 10.1126/science.1220941
McMenamin, A. J., Brutscher, L. M., Glenny, W., and Flenniken, M. L. (2016). Abiotic and biotic factors affecting the replication and pathogenicity of bee viruses. Curr. Opin. Insect Sci. 16, 14–21. doi: 10.1016/j.cois.2016.04.009
McMenamin, A. J., and Genersch, E. (2015). Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 8, 121–129. doi: 10.1016/j.cois.2015.01.015
Melathopoulos, A., Ovinge, L., Veiga, P. W., Castillo, C., Ostermann, D., and Hoover, S. (2017). Viruses of managed alfalfa leafcutting bees (Megachille rotundata Fabricus) and honey bees (Apis mellifera L.) in Western Canada: incidence, impacts, and prospects of cross-species viral transmission. J. Invertebr. Pathol. 146, 24–30. doi: 10.1016/j.jip.2017.04.003
Mondet, F., de Miranda, J. R., Kretzschmar, A., Le Conte, Y., and Mercer, A. R. (2014). On the front line: quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLOS Pathog. 10:e1004323. doi: 10.1371/journal.ppat.1004323
Nazzi, F., Brown, S. P., Annoscia, D., Del Piccolo, F., Di Prisco, G., Varricchio, P., et al. (2012). Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLOS Pathog. 8:e1002735. doi: 10.1371/journal.ppat.1002735
Nazzi, F., and Le Conte, Y. (2016). Ecology of Varroa destructor, the major ectoparasite of the western honey bee, Apis mellifera. Annu. Rev. Entomol. 61, 417–432. doi: 10.1146/annurev-ento-010715-023731
Nazzi, F., and Pennacchio, F. (2014). Disentangling multiple interactions in the hive ecosystem. Trends Parasitol. 30, 556–561. doi: 10.1016/j.pt.2014.09.006
Ng, T. F. F., Marine, R., Wang, C., Simmonds, P., Kapusinszky, B., Bodhidatta, L., et al. (2012). High Variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 86, 12161–12175. doi: 10.1128/JVI.00869-12
Palacios, G., Forrester, N. L., Savji, N., Travassos da Rosa, A. P. A., Guzman, H., Detoy, K., et al. (2013). Characterization of Farmington virus, a novel virus from birds that is distantly related to members of the family Rhabdoviridae. Virol. J. 10, 219. doi: 10.1186/1743-422X-10-219
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., and Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. doi: 10.1016/j.tree.2010.01.007
Pruitt, K. D., Tatusova, T., and Maglott, D. R. (2007). NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, 501–504. doi: 10.1093/nar/gkl842
Ravoet, J., De Smet, L., Wenseleers, T., and de Graaf, D. C. (2015). Genome sequence heterogeneity of Lake Sinai Virus found in honey bees and Orf1/RdRP-based polymorphisms in a single host. Virus Res. 201, 67–72. doi: 10.1016/j.virusres.2015.02.019
Remnant, E. J., Shi, M., Buchmann, G., Blacquière, T., Holmes, E. C., Beekman, M., et al. (2017). A diverse range of novel RNA viruses in geographically distinct honey bee populations. J. Virol. 91:e00158-17. doi: 10.1128/JVI.00158-17
Rombel, I. T., Sykes, K. F., Rayner, S., and Johnston, S. A. (2002). ORF-FINDER: a vector for high-throughput gene identification. Gene 282, 33–41. doi: 10.1016/S0378-1119(01)00819-8
Roossinck, M. J. (2011). The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 9, 99–108. doi: 10.1038/nrmicro2491
Rosenkranz, P., Aumeier, P., and Ziegelmann, B. (2010). Biology and control of Varroa destructor. J. Invertebr. Pathol. 103(Suppl.), S96–S119. doi: 10.1016/j.jip.2009.07.016
Ryabov, E. V., Wood, G. R., Fannon, J. M., Moore, J. D., Bull, J. C., Chandler, D., et al. (2014). A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLOS Pathog. 10:e1004230. doi: 10.1371/journal.ppat.1004230
Schoonvaere, K., De Smet, L., Smagghe, G., Vierstraete, A., Bart, P., and De Graaf, D. C. (2016). Unbiased RNA shotgun metagenomics in social and solitary wild bees detects associations with eukaryote parasites and new viruses. PLOS ONE 11:e0168456. doi: 10.1371/journal.pone.0168456
Schulz, M. H., Zerbino, D. R., Vingron, M., and Birney, E. (2012). Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092. doi: 10.1093/bioinformatics/bts094
Shen, M., Cui, L., Ostiguy, N., and Cox-Foster, D. (2005). Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 86, 2281–2289. doi: 10.1099/vir.0.80824-0
Singh, R., Levitt, A. L., Rajotte, E. G., Holmes, E. C., Ostiguy, N., Vanengelsdorp, D., et al. (2010). RNA viruses in hymenopteran pollinators: evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLOS ONE 5:e14357. doi: 10.1371/journal.pone.0014357
Soueidan, H., Maurier, F., Groppi, A., Sirand-Pugnet, P., Tardy, F., Citti, C., et al. (2013). Finishing bacterial genome assemblies with Mix. BMC Bioinformatics 14:S16. doi: 10.1186/1471-2105-14-S15-S16
Steinhauer, N., Rennich, K., Caron, D. M., Ellis, J. D., Koenig, P., Kulhanek, K., et al. (2017). Colony Loss 2016–2017: Preliminary Results. Available at: https://beeinformed.org/results/colony-loss-2016-2017-preliminary-results
Sumpter, D. J. T., and Martin, S. J. (2004). The dynamics of virus epidemics in Varroa-infested honey bee colonies. J. Anim. Ecol. 73, 51–63. doi: 10.1111/j.1365-2656.2004.00776.x
Tehel, A., Brown, M. J., and Paxton, R. J. (2016). Impact of managed honey bee viruses on wild bees. Curr. Opin. Virol. 19, 16–22. doi: 10.1016/j.coviro.2016.06.006
Vashist, S., Urena, L., and Goodfellow, I. (2012). Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J. Virol. Methods 184, 69–76. doi: 10.1016/j.jviromet.2012.05.012
Walker, P. J., Firth, C., Widen, S. G., Blasdell, K. R., Guzman, H., Wood, T. G., et al. (2015). Evolution of genome size and complexity in the rhabdoviridae. PLOS Pathog. 11:e1004664. doi: 10.1371/journal.ppat.1004664
Wang, Y., Cowley, J. A., and Walker, P. J. (1995). Adelaide River virus nucleoprotein gene: analysis of phylogenetic relationships of ephemeroviruses and other rhabdoviruses. J. Gen. Virol. 76, 995–999. doi: 10.1099/0022-1317-76-4-995
Wilfert, L., Long, G., Leggett, H. C., Schmid-Hempel, P., Butlin, R., Martin, S. J. M., et al. (2016). Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 351, 594–597. doi: 10.1126/science.aac9976
Winfree, R. (2008). Pollinator-dependent crops: an increasingly risky business. Curr. Biol. 18, 968–969. doi: 10.1016/j.cub.2008.09.010
Yang, X., and Cox-Foster, D. (2007). Effects of parasitization by Varroa destructor on survivorship and physiological traits of Apis mellifera in correlation with viral incidence and microbial challenge. Parasitology 134, 405–412. doi: 10.1017/S0031182006000710
Yue, C., and Genersch, E. (2005). RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 86, 3419–3424. doi: 10.1099/vir.0.81401-0
Keywords: rhabdovirus, honey bees, bumble bees, Varroa destructor mites, pollinators
Citation: Levin S, Galbraith D, Sela N, Erez T, Grozinger CM and Chejanovsky N (2017) Presence of Apis Rhabdovirus-1 in Populations of Pollinators and Their Parasites from Two Continents. Front. Microbiol. 8:2482. doi: 10.3389/fmicb.2017.02482
Received: 04 October 2017; Accepted: 29 November 2017;
Published: 12 December 2017.
Edited by:
Ralf Georg Dietzgen, The University of Queensland, AustraliaReviewed by:
Emily J. Bailes, Royal Holloway, University of London, United KingdomPeter John Walker, The University of Queensland, Australia
Copyright © 2017 Levin, Galbraith, Sela, Erez, Grozinger and Chejanovsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nor Chejanovsky, bmluYXJAdm9sY2FuaS5hZ3JpLmdvdi5pbA==
 Sofia Levin
Sofia Levin David Galbraith
David Galbraith Noa Sela
Noa Sela Tal Erez
Tal Erez Christina M. Grozinger
Christina M. Grozinger Nor Chejanovsky
Nor Chejanovsky