
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Microbiol. , 12 December 2017
Sec. Microbial Immunology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02477
This article is part of the Research Topic Immunity to Human Fungal Pathogens: Mechanisms of Host Recognition, Protection, Pathology, and Fungal Interference View all 37 articles
The respiratory tract is a complex system that is inhabited by niche-specific communities of microbes including bacteria, fungi, and viruses. These complex microbial assemblages are in constant contact with the mucosal immune system and play a critical role in airway health and immune homeostasis. Changes in the composition and diversity of airway microbiota are frequently observed in patients with chronic inflammatory diseases including chronic rhinosinusitis (CRS), cystic fibrosis, allergy, and asthma. While the bacterial microbiome of the upper and lower airways has been the focus of many recent studies, the contribution of fungal microbiota to inflammation is an emerging research interest. Within the context of allergic airway disease, fungal products are important allergens and fungi are potent inducers of inflammation. In addition, murine models have provided experimental evidence that fungal microbiota in peripheral organs, notably the gastrointestinal (GI) tract, influence pulmonary health. In this review, we explore the role of the respiratory and GI microbial communities in chronic airway inflammatory disease development with a specific focus on fungal microbiome interactions with the airway immune system and fungal-bacterial interactions that likely contribute to inflammatory disease. These findings are discussed in the context of clinical and immunological features of fungal-mediated disease in CRS, allergy, and asthmatic patients. While this field is still nascent, emerging evidence suggests that dysbiotic fungal and bacterial microbiota interact to drive or exacerbate chronic airway inflammatory disease.
Microbial communities associated with host mucosal surfaces play essential and diverse roles in biological processes from metabolism to immune regulation and homeostasis. In return, host inflammatory responses can shape the microbial community by supporting the growth of some microbes while inhibiting others (Kumamoto, 2016). Most studies of host-associated microbiota in health and disease have focused on the bacterial microbiota, despite fungal diseases incurring a substantial infectious disease burden (Huffnagle and Noverr, 2013). Because of this, relatively little is known about the importance and function of the fungal microbiota (mycobiota). Recent advances in culture-independent sequencing approaches have helped elucidate the fungal diversity, burden, and functions associated with mucosal surfaces such as the human intestinal tract, oral cavity, skin, and respiratory tract.
In virtually every body habitat studied to date, niche-specific microbial communities have been identified, including the gastrointestinal and respiratory tracts (Human Microbiome Project Consortium, 2012). Fungi live in host niches as commensals and interact with bacteria and the host as members of the healthy microbiome. While fungal microbiota are numerically small, they are diverse, have large genomes, and potentially act as important keystone species in the microbiome (Huffnagle and Noverr, 2013). Studies have shown that disrupting commensal fungi can affect both local and peripheral immune responses and enhance disease states. In addition, members of the mycobiome can switch from commensalism to pathogenicity, often dependent upon co-colonizing microbial taxa (Cohen et al., 1969; Peleg et al., 2010). Recent studies suggest a role for airway microbial communities in maintaining airway health (Abreu et al., 2012). In this perspective, we focus on the potential role of fungal interactions with the host and bacterial co-colonizers chronic rhinosinusitis (CRS) and lower airway comorbidities.
Like the GI tract (Jeraldo et al., 2012), the respiratory tract is composed of distinct environments that vary according to mucosal architecture and immune responses. Reflective of this, the microbial communities that inhabit the respiratory tract exhibit niche specificity; compositionally distinct communities exist between the nares, sinuses, oral cavity, oropharynx and lower airway in healthy individuals (Lemon et al., 2010; Bassis et al., 2015). The sinonasal cavity, nasopharynx, and trachea are lined with a pseudostratified columnar ciliated epithelium and mucus-producing goblet cells. Lung bronchi and bronchioles are covered by stratified columnar ciliated epithelia and few goblet cells. Mucus secretion and immune effector molecules protect the host from insults by trapping and removing inhaled pathogens and irritants, a process orchestrated by ciliary beating (Cohen, 2006). Airway epithelial cells express pattern recognition receptors (PRRs), including toll-like receptors (TLR) and non-TLR PRRs such as Dectin-1, that can recognize and respond to components of microorganisms (Ryu et al., 2013; Liu et al., 2015).
Chronic airway inflammatory diseases including asthma, rhinosinusitis, and rhinitis are substantial health problems resulting in significant healthcare expenditures. Chronic rhinosinusitis (CRS) alone affects approximately 15% of the adult population annually; recent estimates suggest that costs associated with CRS exceed $65 billion each year accounting for 5% of the US healthcare budget (Caulley et al., 2015). Several epidemiological studies suggest that concomitant upper and lower airway disease result from shared pathophysiologic mechanisms (Passalacqua et al., 2001; Ciprandi et al., 2012; Licari et al., 2014).
CRS is clinically defined by duration of symptoms and endoscopic or radiographic evidence of sinus inflammation. Multiple subtypes of CRS have been described based on clinical (Akdis et al., 2013; Orlandi et al., 2016), immunological (Tomassen et al., 2016) and microbiological (Cope et al., 2017) heterogeneity. The role of fungi in the pathophysiology of CRS has long been controversial. While some authors argued that fungi drove inflammation in nearly all CRS patients (Ponikau et al., 2000), this finding has been recently disputed (Orlandi and Marple, 2010; Fokkens et al., 2012). Fungi clearly play a central role in several discrete subtypes of CRS, such as allergic fungal sinusitis and development of fungus balls (deShazo et al., 1997). Fungi found in culture-based studies of CRS patients include members of Aspergillus, Alternaria, Candida, Cladosporium, and Penicillium (Ponikau et al., 1999). Molecular studies have expanded this list to include fastidious fungi Malassezia, Curvularia, Schizophyllum, and Neocosmospora (Cleland et al., 2014; Gelber et al., 2016; Zhao et al., 2017). While there are few studies on host-fungal interactions in the upper airways, recent research suggests that defects in epithelial genes might compromise immune barrier function and lead to dysfunctional host immune responses to bacterial or fungal colonization (Tieu et al., 2009). Additionally, fungal glycans (e.g., β-glucan) interact with airway epithelial cells via surface receptors and can act as determinants of allergenicity or induce inflammation (Roy and Klein, 2013). In allergic bronchopulmonary aspergillosis (ABPA), structural abnormalities in the airway epithelium allow fungal spores to breach immune defenses and germinate into hyphae (Chaudhary and Marr, 2011). Upon germination, fungal spores secrete proteases which can disrupt epithelial barrier integrity (Chen et al., 2011). Epithelial barrier dysfunction is frequently observed in CRS and asthma (Tieu et al., 2009; Lambrecht and Hammad, 2012). The presence of bacterial and fungal biofilms increased mucosal IgE in CRS patients, suggesting that these biofilms interact with the host immune system and perpetuate chronic inflammation (Foreman et al., 2012).
CRS is phenotypically classified into two groups based on the presence or absence of nasal polyps. This clinical classification of CRS patients was initially reflected immunologically with a predominance of TH2 cells and eosinophils in CRS patients with polyps (CRSwNP) and a predominance of TH1 cells and neutrophils in CRS patients without polyps (CRSsNP). However, recent studies using molecular methods have demonstrated up to 10 distinct endotypes of patients using markers of TH1, TH2, TH17, eosinophil, and neutrophil activation (Tomassen et al., 2016). A separate study found four distinct groups of CRS patients characterized by distinct bacterial communities, each conferring a unique immune response (Cope et al., 2017). Corynebacteriaceae-dominated communities conferred increased IL-5 gene expression and a higher risk for nasal polyps. While these findings are helpful, it is also important to understand how the sinus microbiota drive or exacerbate observed inflammatory endotypes so new therapeutics can target the initiation of disease.
CRS often exists in the setting of concomitant lower airway disease, including asthma, and is frequently preceded by rhinitis. Upper respiratory infections can exacerbate asthma symptoms; bronchial hyperresponsiveness often occurs along with rhinitis (Leynaert et al., 2000; Passalacqua et al., 2001). Between 20 and 60% of CRS patients with nasal polyps have asthma (Larsen, 1996; Klossek et al., 2005). These observations have led to the “unified airway” hypothesis, which treats the respiratory tract as one organ rather than distinct organs affected by specific diseases. Supporting this hypothesis, medical and surgical treatment in the upper airways for CRS frequently results in reduced asthma symptoms. Therefore, respiratory inflammatory disease can be considered a disorder of the entire respiratory tract. Dysbiotic (altered) microbial communities have been described in the upper airways of CRS patients and in the lower airways of asthmatic patients, however, the microbiota of these two sites have not yet been examined in parallel. Below, we will discuss recent findings in CRS and asthmatic microbiota and potential interactions between these important microbial populations.
The upper and lower airways harbor niche-specific microbial communities that relate to health status (Lemon et al., 2010; Huang and Boushey, 2014). Recent studies of the sinonasal microbiome generally demonstrate a loss of bacterial diversity and concomitant enrichment of pathobionts, resident microbes with pathogenic potential, in the sinuses of patients with CRS (Biswas et al., 2015; Cope et al., 2017; Lal et al., 2017; Wagner Mackenzie et al., 2017). Abreu and colleagues found that the sinus microbiome was characterized by reduced microbial richness and depletion of taxa associated with healthy individuals, such as lactic acid bacteria (Abreu et al., 2012). This depleted CRS microbiome was enriched with the pathobiont Corynebacterium tuberculostearicum. In a murine model, C. tuberculostearicum increased host mucin secretion, potentially increasing factors such as nutrient availability and adherence. Larger studies of upper airway microbiota in CRS patients demonstrate a heterogeneous, compositionally distinct microbiota which relate to distinct host immune responses (Cope et al., 2017; Lal et al., 2017). Studies comparing the lung microbiome of asthmatics and healthy controls found increased bacterial burden and diversity among asthmatics and a compositional shift in the bacterial microbiome composition characterized by enriched Proteobacteria (Huang et al., 2011; Han et al., 2012; Durack et al., 2017). Polymicrobial biofilms consisting of bacteria and fungi have been observed on sinonasal mucosa using fluorescence in situ hybridization, demonstrating that these taxa exist in mixed-species mucosal communities (Sanderson et al., 2006; Doble et al., 2007). More research is required on the upper and lower airway microbiota, and, in particular, on the relationship between sinonasal and pulmonary microbiota and mycobiota. The function and composition of these communities may shed light on the heterogeneity observed in CRS.
While the bacterial communities in airway inflammatory disease have been more extensively studied, fungal microbiota are still poorly characterized. This is due, in part, to the challenges faced by researchers who study the airway mycobiome. Low fungal biomass in the airways and databases that are curated from GI or environmental sources affect sample collection, DNA extraction, library preparation, and sequence analysis. In other body sites such as the GI tract, fungal dysbiosis has been implicated in diseases including inflammatory bowel disease. Although outside the scope of this perspective, gut mycobiome dysbiosis and interaction with the host immune response has been extensively reviewed (Underhill and Iliev, 2014; Mukherjee et al., 2015). In the airways, similarities between allergic fungal rhinosinusitis and ABPA illustrate the ability of fungi to act as antigens and invade the respiratory tract.
Studies on the fungal microbiome in CRS and asthma suggest that fungal communities in the airways likely influence host health. Malassezia, a known pathobiont on skin (Findley et al., 2013), is the predominant fungal genus in the sinonasal cavity in healthy individuals, CRS patients, and patients with allergic rhinitis (Cleland et al., 2014; Jung et al., 2015; Gelber et al., 2016). Lower abundance fungi in the sinuses of CRS patients include Aspergillus, Alternaria, Fusarium, and Saccharomyces (Cleland et al., 2014). A recent study using ITS gene sequencing found fungal presence in the sinuses of 63% of CRS patients and the predominant fungal genus in this study was Aspergillus (Zhao et al., 2017). PCR evaluations of Aspergillus, however, have demonstrated the presence of this fungus only in the sinuses of CRS patients with a known fungal subtype of CRS (Gelber et al., 2016). These discrepancies may be due to challenges related to fungal databases for human-associated ITS sequences. Analysis of the nasal vestibule of patients with AR and healthy individuals demonstrated increased diversity of fungal communities in AR characterized by preponderance of Malassezia (91–99% of 18S rRNA gene sequences) with lower abundance Aspergillus and Alternaria (Jung et al., 2015). In asthma, the lung mycobiome composition is altered between patients with severe asthma, ABPA, asthma with fungal sensitization (SAFS), and mild asthmatics. In ABPA, Pseudomonas abundance and fungal diversity both increased. Severe asthmatics were characterized by enrichment of Aspergillus; the relative abundance of Aspergillus increased approximately 15-fold compared to mild asthmatics (Chishimba et al., 2015). These results suggest that bacterial and fungal communities change in parallel in subsets of human disease. Future studies examining paired upper and lower airway microbiota (bacterial and fungal) in individuals with CRS and asthma are warranted.
Aspergillus fumigatus is often involved in the development of allergic fungal rhinosinusitis (AFRS) specific subtype CRS that accounts for an estimated 6–9% of all CRS patients undergoing surgery (Schubert, 2004). Patients with AFRS present with hyperplastic sinus disease, characterized by chronic eosinophilic-lymphocytic inflammation and nasal polyps. In AFRS, fungal hyphae are found within allergic mucin. Sinus cultures obtained in surgical patients reveal the presence of fungal taxa such as Bipolaris spicifera, Alternaria, and Aspergillus. AFRS bears many similarities to ABPA in its immunopathology, treatment, and outcomes. ABPA and AFS patients have elevated levels of serum IgE, and ABPA patients have elevated A. fumigatus-specific IgE and IgG. Allergic mucin in AFRS patients is histologically identical to the bronchial mucus plugs in ABPA patients (Schubert, 2004). In ABPA, immunological hypersensitivity is higher relative to AFS, possibly due to differences in the diseased organ or etiological fungus.
Concurrent upper and lower airway disease may be a singular inflammatory process caused or mediated by microbial communities. Microbes or their metabolites might translocate between the upper and lower airways. Interactions between microbes, the environment, and the host inflammatory response could alter the mucus-associated microbial communities, causing dysbiosis. Fungi involved in these allergic fungal disorders, along with associated bacteria such as S. aureus, viruses, or other microbes, may act as the source of microbial T-cell superantigens. Fungal contributions to chronic inflammatory airway diseases are an emerging research interest, one which may advance the understanding of such unified airway disorders and treatment for patients with allergic airway disease.
Bacteria and fungi co-inhabit the human body and mounting evidence suggests that microbial interactions in a given niche can influence human health and disease. Bacterial co-colonizers can directly affect fungal morphology (Peters et al., 2012), survival (Romano and Kolter, 2005; Harriott and Noverr, 2009), growth (Kerr et al., 1999), virulence (Schlecht et al., 2015), and attachment to host epithelia or other microbes (El-Azizi et al., 2004; Morales and Hogan, 2010). Current investigations have focused on bacterial interactions with the opportunistic pathogen Candida albicans. In the oral cavity, C. albicans selectively attaches to the bacterium Streptococcus gordonii to increase attachment and growth of the fungi (Holmes et al., 1996). In vitro experimental studies have demonstrated that S. aureus and P. aeruginosa selectively attach to C. albicans hyphae but not yeast (Peters et al., 2012; Schlecht et al., 2015), and these interactions have distinct outcomes. S. aureus attachment to Candida hyphae results in increased invasion of tissue (Schlecht et al., 2015), while P. aeruginosa forms biofilms on the hyphal cells, killing the fungus (Hogan and Kolter, 2002). Physical sensing and Quorum sensing (QS) through small molecules that allow microbes to respond to environmental stimuli play important roles in mediating these interactions. For example, the C. albicans QS molecule farnesol inhibits Pseudomonas quinolone signal (PQS) and virulence factor production (Cugini et al., 2010). C. albicans can respond to P. aeruginosa QS molecules, specifically 3-3-oxo-C12 homoserine lactone, which prevents hyphal formation in the fungus (Hogan et al., 2004). These species-specific interactions through physical contact and QS molecules likely affect microbial virulence or biofilm formation in host upper or lower airways. Ongoing research in our lab seeks to explore these interactions.
We are interested in interactions between sinonasal-associated fungi and bacterial co-colonizers, including Malassezia sp. (Mowat et al., 2010; Jung et al., 2015; Gelber et al., 2016). Malassezia, a dimorphic yeast implicated in a variety of conditions including dandruff and atopic dermatitis, is present in the upper airways (Cleland et al., 2014; Gelber et al., 2016). Malassezia species are known to be immunomodulatory and encode at least 13 different allergens; these fungi can exist as commensals and can be immunosuppressive (Ashbee and Evans, 2002) or as pathobionts and can become immunostimulatory (Gaitanis et al., 2012). Similar to CRS, Malassezia dermatis often co-occurs with S. aureus in atopic dermatitis, perhaps interacting with bacteria in driving skin inflammation. Further research is required to identify these Malassezia at the species or strain level and to compare populations between healthy subjects and those with allergic airway disease. Mechanistic studies are needed to reveal the interactions between Malassezia, bacterial community members, and the host immune system.
Airway epithelial cells and phagocytes express pattern recognition receptors, including C-type lectins, that sense and respond to components of the fungal cell wall. Dectin-1, a pattern recognition receptor expressed by dendritic cells, neutrophils, and macrophages, recognizes the fungal polysaccharide β-1,3 glucan motif found on fungal cell walls. Dectin-1 may mediate host immune responses to these fungi. In mice, loss of dectin-1 resulted in more severe colitis compared to wild-type mice. DSS-induced colitis was associated with increased Candida and Trichosporon and decreased Saccharomyces. Fungi were also found to invade inflamed tissues in dectin-1 KO mice, but not in wild-type mice. When given the antifungal fluconazole, dectin-1 KO mice had milder symptoms (Iliev et al., 2012). These results indicate that fungi contribute to the aggravation of inflammatory responses in colitis, and dectin-1 is an immune mechanism that regulates fungal community composition. Another C-type lectin that specifically recognizes Malassezia has recently been described (Yamasaki et al., 2009). When expressed on macrophages, Mincle (also called Clec4e and Clecsf9) sensing of Malassezia induces pro-inflammatory responses (Yamasaki et al., 2009), although this receptor has also been implicated in TH2-polarization upon activation (Geijtenbeek and Gringhuis, 2016). Mincle-deficient mice showed a 2-4 fold reduction of IL-10, TNF-alpha, and MIP1 when challenged with Malassezia but not A. fumigatus (Yamasaki et al., 2009) Host-associated commensal fungi, therefore, specifically interact with the mucosal immune system, maintaining host and microbial homeostasis.
Fungal microbial communities in the GI tract can also affect the development of respiratory disease. Mice treated with antifungals exhibited increased development of allergic airway disease along with increased disease severity in models of colitis (Wheeler et al., 2016). The microbiomes of these mice revealed a restructuring of fungal and bacterial communities characterized by decreased Candida (2.25-fold) and increased Aspergillus (2.5-fold), Wallemia (6-fold), and Epicoccum (4-fold). Oral supplements of A. amstelodami, W. sebi, and E. nigrum recapitulated the effects of antifungal drugs to exacerbate allergic airway disease as measured by increased pulmonary eosinophils and lymphocytes and elevated serum IgE (Wheeler et al., 2016). Colonization of the gut by Candida albicans has also previously been shown to influence allergic airway disease and asthma in a mouse model, perhaps through secretion of prostaglandin-like immunomodulatory molecules (Noverr et al., 2005; Huffnagle, 2010; Marsland and Salami, 2015). Thus, fungal dysbiosis in the gut can affect disease development and exacerbation in the respiratory tract, indicating that a healthy mycobiome mediates immune responses throughout the body. Whether these effects reflect a direct action of fungi on the host or occur indirectly by altering the bacterial microbiome composition is unknown. Further studies are needed to elucidate the mechanisms for these responses and to determine whether fungal communities in other organs can influence allergic airway disease.
The evidence for the influence of dysbiotic fungal and bacterial microbiota interactions to drive or exacerbate chronic airway inflammatory disease is compelling. These findings open up the potential for targeted manipulation of the airway or gastrointestinal tract microbiota to improve airway health and manage airway disease in patients with CRS, asthma, and cystic fibrosis, among others. Current mycobiome studies focus on comparisons in healthy vs. diseased models, but more research needs to occur to understand the interactions between fungal and bacterial communities with the host immune system in host body sites. Fungi produce diverse secondary metabolites that can affect bacteria, while bacteria can keep the mycobiome in check by producing substances that inhibit the yeast to hyphae transition of fungal pathobionts such as in C. albicans.
Additional research is needed to define the functional effects of the mycobiome (Figure 1). Rather than focusing on taxonomic diversity, studies comparing microbial community function are potentially more useful. Further understanding of the contribution of the mycobiome and bacterial-fungal interactions should move beyond the GI tract into respiratory tract, where fungal dysbiosis has been shown to influence disease development and exacerbate symptoms. A variety of methods, combining metagenomics and sequencing approaches, experimental models, and functional studies should be used to clarify mechanisms by which the mycobiome impacts health and disease. In particular, focusing on microbial interactions with mucosal surfaces and their influence on local immune response likely will provide additional insights into the role of microbes in inflammatory disease of the upper and lower airways. Because fungal dysbiosis can have as great an effect on the host as bacterial dysbiosis, we cannot overlook the contributions of these largely unexplored fungal communities in our search to understand the microbial mechanisms behind health and disease.
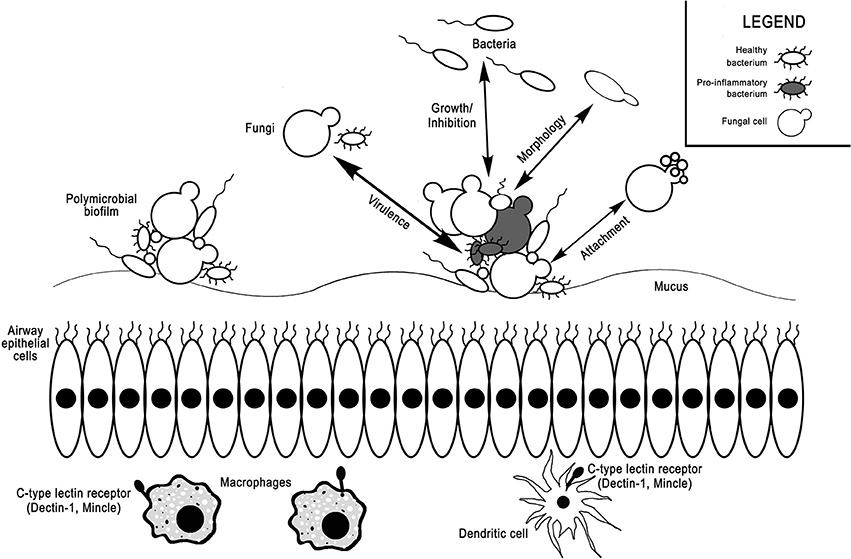
Figure 1. Interactions between the bacterial microbiome, mycobiome, and host immune system in the airways. Bacteria and fungi coexist in mucosa-attached polymicrobial biofilms in the airways. Fungi can selectively induce or inhibit growth of various bacterial taxa, increase bacterial virulence factors, alter bacterial morphology, or act as attachment sites for bacteria. Vice versa, bacteria can also alter fungal growth, virulence, morphology, and attachment. C-type lectin receptors on macrophages and dendritic cells, such as dectin-1 and Mincle, can sense fungi and mediate host inflammatory responses.
IZ drafted and revised the manuscript. EC conceived, wrote, and revised the manuscript. BB conceived, wrote and revised the manuscript. AG conceived and revised the manuscript. SP conceived, wrote, and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abreu, N. A., Nagalingam, N. A., Song, Y., Roediger, F. C., Pletcher, S. D., Goldberg, A. N., et al. (2012). Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci. Transl. Med. 4:151ra124. doi: 10.1126/scitranslmed.3003783
Akdis, C. A., Bachert, C., Cingi, C., Dykewicz, M. S., Hellings, P. W., Naclerio, R. M., et al. (2013). Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the european academy of allergy and clinical immunology and the american academy of allergy, asthma & immunology. J. Allergy Clin. Immunol. 131, 1479–1490. doi: 10.1016/j.jaci.2013.02.036
Ashbee, H. R., and Evans, E. G. V. (2002). Immunology of diseases associated with Malassezia species. Clin. Microbiol. Rev. 15, 21–57. doi: 10.1128/CMR.15.1.21-57.2002
Bassis, C. M., Erb-Downward, J. R., Dickson, R. P., Freeman, C. M., Schmidt, T. M., Young, V. B., et al. (2015). Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6:e00037-15. doi: 10.1128/mBio.00037-15
Biswas, K., Hoggard, M., Jain, R., Taylor, M. W., and Douglas, R. G. (2015). The nasal microbiota in health and disease: variation within and between subjects. Front. Microbiol. 9:134. doi: 10.3389/fmicb.2015.00134
Caulley, L., Thavorn, K., Rudmik, L., Cameron, C., and Kilty, S. J. (2015). Direct costs of adult chronic rhinosinusitis by using 4 methods of estimation: results of the US medical expenditure panel survey. J. Allergy Clin. Immunol. 136, 1517–1522. doi: 10.1016/j.jaci.2015.08.037
Chaudhary, N., and Marr, K. A. (2011). Impact of Aspergillus fumigatus in allergic airway diseases. Clin. Transl. Allergy 1:4. doi: 10.1186/2045-7022-1-4
Chen, J.-C., Chuang, J.-G., Su, Y.-Y., Chiang, B.-L., Lin, Y.-S., and Chow, L.-P. (2011). The protease allergen Pen c 13 induces allergic airway inflammation and changes in epithelial barrier integrity and function in a murine model. J. Biol. Chem. 286, 26667–26679. doi: 10.1074/jbc.M110.193987
Chishimba, L., Niven, R., Fraczek, M., Bowyer, P., Smyth, L., Simpson, A., et al. (2015). Lung microbiome is associated with asthma severity in fungal associated asthma. Eur. Respir. J. 46:OA1462. doi: 10.1183/13993003.congress-2015.OA1462
Ciprandi, G., Caimmi, D., Miraglia Del Giudice, M., La Rosa, M., Salpietro, C., and Marseglia, G. L. (2012). Recent developments in United airways disease. Allergy Asthma Immunol. Res. 4, 171–177. doi: 10.4168/aair.2012.4.4.171
Cleland, E. J., Bassiouni, A., Bassioni, A., Boase, S., Dowd, S., Vreugde, S., et al. (2014). The fungal microbiome in chronic rhinosinusitis: richness, diversity, postoperative changes and patient outcomes. Int. Forum Allergy Rhinol. 4, 259–265. doi: 10.1002/alr.21297
Cohen, N. A. (2006). Sinonasal mucociliary clearance in health and disease. Ann. Otol. Rhinol. Laryngol. Suppl. 196, 20–26. doi: 10.1177/00034894061150S904
Cohen, R., Roth, F. J., Delgado, E., Ahearn, D. G., and Kalser, M. H. (1969). Fungal flora of the normal human small and large intestine. N. Engl. J. Med. 280, 638–641. doi: 10.1056/NEJM196903202801204
Cope, E. K., Goldberg, A. N., Pletcher, S. D., and Lynch, S. V. (2017). Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome 5:53. doi: 10.1186/s40168-017-0266-6
Cugini, C., Morales, D. K., and Hogan, D. A. (2010). Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156, 3096–3107. doi: 10.1099/mic.0.037911-0
Doble, P., Kern, R. C., Healy, D. Y., Leid, J., Sanderson, A. R., and Hunsaker, D. H. (2007). 11:06: biofilms with fungi in chronic rhinosinusitis. Otolaryngol. Head Neck Surg. 137, P40–P40. doi: 10.1016/j.otohns.2007.06.033
Durack, J., Lynch, S. V., Nariya, S., Bhakta, N. R., Beigelman, A., Castro, M., Dyer, A.-M., Israel, E., Kraft, M., Martin, R. J., et al. (2017). Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 140, 63–75. doi: 10.1016/j.jaci.2016.08.055
El-Azizi, M. A., Starks, S. E., and Khardori, N. (2004). Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J. Appl. Microbiol. 96, 1067–1073. doi: 10.1111/j.1365-2672.2004.02213.x
Findley, K., Oh, J., Yang, J., Conlan, S., Deming, C., Meyer, J. A., Schoenfeld, D., Nomicos, E., and Park, M. (2013). NIH intramural sequencing center comparative sequencing program, et al. topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370. doi: 10.1038/nature12171
Fokkens, W. J., van Drunen, C., Georgalas, C., and Ebbens, F. (2012). Role of fungi in pathogenesis of chronic rhinosinusitis: the hypothesis rejected. Curr. Opin. Otolaryngol. Head Neck Surg. 20, 19–23. doi: 10.1097/MOO.0b013e32834e9084
Foreman, A., Boase, S., Psaltis, A., and Wormald, P.-J. (2012). Role of bacterial and fungal biofilms in chronic rhinosinusitis. Curr. Allergy Asthma Rep. 12, 127–135. doi: 10.1007/s11882-012-0246-7
Gaitanis, G., Magiatis, P., Hantschke, M., Bassukas, I. D., and Velegraki, A. (2012). The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 25, 106–141. doi: 10.1128/CMR.00021-11
Geijtenbeek, T. B. H., and Gringhuis, S. I. (2016). C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 16, 433–448. doi: 10.1038/nri.2016.55
Gelber, J. T., Cope, E. K., Goldberg, A. N., and Pletcher, S. D. (2016). Evaluation of Malassezia and common fungal pathogens in subtypes of chronic rhinosinusitis. Int. Forum Allergy Rhinol. 6, 950–955. doi: 10.1002/alr.21777
Han, M. K., Huang, Y. J., Lipuma, J. J., Boushey, H. A., Boucher, R. C., Cookson, W. O., et al. (2012). Significance of the microbiome in obstructive lung disease. Thorax 67, 456–463. doi: 10.1136/thoraxjnl-2011-201183
Harriott, M. M., and Noverr, M. C. (2009). Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob. Agents Chemother. 53, 3914–3922. doi: 10.1128/AAC.00657-09
Hogan, D. A., and Kolter, R. (2002). Pseudomonas-candida interactions: an ecological role for virulence factors. Science 296, 2229–2232. doi: 10.1126/science.1070784
Hogan, D. A., Vik, Å., and Kolter, R. (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54, 1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x
Holmes, A. R., McNab, R., and Jenkinson, H. F. (1996). Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64, 4680–4685.
Huang, Y. J., and Boushey, H. A. (2014). The microbiome and asthma. Ann. Am. Thorac. Soc. 11, S48–S51. doi: 10.1513/AnnalsATS.201306-187MG
Huang, Y. J., Nelson, C. E., Brodie, E. L., Desantis, T. Z., Baek, M. S., Liu, J., et al. (2011). Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 127, 372–381.e1–3. doi: 10.1016/j.jaci.2010.10.048
Huffnagle, G. B. (2010). The microbiota and allergies/asthma. PLoS Pathog. 6:e1000549. doi: 10.1371/journal.ppat.1000549
Huffnagle, G. B., and Noverr, M. C. (2013). The emerging world of the fungal microbiome. Trends Microbiol. 21, 334–341. doi: 10.1016/j.tim.2013.04.002
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Iliev, I. D., Funari, V. A., Taylor, K. D., Nguyen, Q., Reyes, C. N., Strom, S. P., et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. doi: 10.1126/science.1221789
Jeraldo, P., Sipos, M., Chia, N., Brulc, J. M., Dhillon, A. S., Konkel, M. E., et al. (2012). Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc. Natl. Acad. Sci. U.S.A. 109, 9692–9698. doi: 10.1073/pnas.1206721109
Jung, W. H., Croll, D., Cho, J. H., Kim, Y. R., and Lee, Y. W. (2015). Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses 58, 167–172. doi: 10.1111/myc.12296
Kerr, J. R., Taylor, G. W., Rutman, A., Høiby, N., Cole, P. J., and Wilson, R. (1999). Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52, 385–387.
Klossek, J. M., Neukirch, F., Pribil, C., Jankowski, R., Serrano, E., Chanal, I., et al. (2005). Prevalence of nasal polyposis in France: a cross-sectional, case-control study. Allergy 60, 233–237. doi: 10.1111/j.1398-9995.2005.00688.x
Kumamoto, C. A. (2016). The fungal mycobiota: small numbers, large impacts. Cell Host Microbe 19, 750–751. doi: 10.1016/j.chom.2016.05.018
Lal, D., Keim, P., Delisle, J., Barker, B., Rank, M. A., Chia, N., et al. (2017). Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects. Int. Forum Allergy Rhinol. 7, 561–569. doi: 10.1002/alr.21934
Lambrecht, B. N., and Hammad, H. (2012). The airway epithelium in asthma. Nat. Med. 18, 684–692. doi: 10.1038/nm.2737
Larsen, K. (1996). The clinical relationship of nasal polyps to asthma. Allergy Asthma Proc. 17, 243–249. doi: 10.2500/108854196778662255
Lemon, K. P., Klepac-Ceraj, V., Schiffer, H. K., Brodie, E. L., Lynch, S. V., and Kolter, R. (2010). Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 1. doi: 10.1128/mBio.00129-10
Leynaert, B., Neukirch, F., Demoly, P., and Bousquet, J. (2000). Epidemiologic evidence for asthma and rhinitis comorbidity. J. Allergy Clin. Immunol. 106, S201–S205. doi: 10.1067/mai.2000.110151
Licari, A., Caimmi, S., Bosa, L., Marseglia, A., Marseglia, G. L., and Caimmi, D. (2014). Rhinosinusitis and asthma: a very long engagement. Int. J. Immunopathol. Pharmacol. 27, 499–508. doi: 10.1177/039463201402700405
Liu, Z.-C., Wang, M., Sun, W.-K., Xia, D., Tan, M.-M., Ding, Y., et al. (2015). Up-regulation of Dectin-1 in airway epithelial cells promotes mice defense against invasive pulmonary aspergillosis. Int. J. Clin. Exp. Med. 8, 17489–17497.
Marsland, B. J., and Salami, O. (2015). Microbiome influences on allergy in mice and humans. Curr. Opin. Immunol. 36, 94–100. doi: 10.1016/j.coi.2015.07.005
Morales, D. K., and Hogan, D. A. (2010). Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 6:e1000886. doi: 10.1371/journal.ppat.1000886
Mowat, E., Rajendran, R., Williams, C., McCulloch, E., Jones, B., Lang, S., et al. (2010). Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 313, 96–102. doi: 10.1111/j.1574-6968.2010.02130.x
Mukherjee, P. K., Sendid, B., Hoarau, G., Colombel, J.-F., Poulain, D., and Ghannoum, M. A. (2015). Mycobiota in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 12, 77–87. doi: 10.1038/nrgastro.2014.188
Noverr, M. C., Falkowski, N. R., McDonald, R. A., McKenzie, A. N., and Huffnagle, G. B. (2005). Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect. Immun. 73, 30–38. doi: 10.1128/IAI.73.1.30-38.2005
Orlandi, R. R., Kingdom, T. T., and Hwang, P. H. (2016). International consensus statement on allergy and rhinology: rhinosinusitis executive summary. Int. Forum Allergy Rhinol. 6(Suppl. 1), S3–S21. doi: 10.1002/alr.21694
Orlandi, R. R., and Marple, B. F. (2010). Fungus and chronic rhinosinusitis: weighing the evidence. Otolaryngol. Head Neck Surg. 143, 611–613. doi: 10.1016/j.otohns.2010.07.002
Passalacqua, G., Ciprandi, G., and Canonica, G. W. (2001). The nose-lung interaction in allergic rhinitis and asthma: united airways disease. Curr. Opin. Allergy Clin. Immunol. 1, 7–13.
Peleg, A. Y., Hogan, D. A., and Mylonakis, E. (2010). Medically important bacterial–fungal interactions. Nat. Rev. Microbiol. 8, 340–349. doi: 10.1038/nrmicro2313
Peters, B. M., Ovchinnikova, E. S., Krom, B. P., Schlecht, L. M., Zhou, H., Hoyer, L. L., et al. (2012). Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158, 2975–2986. doi: 10.1099/mic.0.062109-0
Ponikau, J. U., Sherris, D. A., and Kern, E. B. (2000). Role of fungi in allergic fungal sinusitis and chronic Rhinosinusitis: in response. Mayo Clin. Proc. 75, 540–541. doi: 10.4065/75.5.540-a
Ponikau, J. U., Sherris, D. A., Kern, E. B., Homburger, H. A., Frigas, E., Gaffey, T. A., et al. (1999). The diagnosis and incidence of allergic fungal sinusitis. Mayo Clin. Proc. 74, 877–884. doi: 10.4065/74.9.877
deShazo, R. D., Chapin, K., and Swain, R. E. (1997). Fungal sinusitis. N. Engl. J. Med. 337, 254–259. doi: 10.1056/NEJM199707243370407
Romano, J. D., and Kolter, R. (2005). Pseudomonas-Saccharomyces interactions: influence of fungal metabolism on bacterial physiology and survival. J. Bacteriol. 187, 940–948. doi: 10.1128/JB.187.3.940-948.2005
Roy, R. M., and Klein, B. S. (2013). Fungal glycan interactions with epithelial cells in allergic airway disease. Curr. Opin. Microbiol. 16, 404–408. doi: 10.1016/j.mib.2013.03.004
Ryu, J.-H., Yoo, J.-Y., Kim, M.-J., Hwang, S.-G., Ahn, K. C., Ryu, J.-C., et al. (2013). Distinct TLR-mediated pathways regulate house dust mite-induced allergic disease in the upper and lower airways. J. Allergy Clin. Immunol. 131, 549–561. doi: 10.1016/j.jaci.2012.07.050
Sanderson, A. R., Leid, J. G., and Hunsaker, D. (2006). Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope 116, 1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54
Schlecht, L. M., Peters, B. M., Krom, B. P., Freiberg, J. A., Hänsch, G. M., Filler, S. G., et al. (2015). Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology 161, 168–181. doi: 10.1099/mic.0.083485-0
Schubert, M. S. (2004). Allergic fungal sinusitis: pathogenesis and management strategies. Drugs 64, 363–374. doi: 10.2165/00003495-200464040-00002
Tieu, D. D., Kern, R. C., and Schleimer, R. P. (2009). Alterations in epithelial barrier function and host defense responses in chronic rhinosinusitis. J. Allergy Clin. Immunol. 124, 37–42. doi: 10.1016/j.jaci.2009.04.045
Tomassen, P., Vandeplas, G., Van Zele, T., Cardell, L.-O., Arebro, J., Olze, H., et al. (2016). Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol 137, 1449–1456.e4. doi: 10.1016/j.jaci.2015.12.1324
Underhill, D. M., and Iliev, I. D. (2014). The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14, 405–416. doi: 10.1038/nri3684
Wagner Mackenzie, B., Waite, D. W., Hoggard, M., Douglas, R. G., Taylor, M. W., and Biswas, K. (2017). Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis. Environ. Microbiol. 19, 381–392. doi: 10.1111/1462-2920.13632
Wheeler, M. L., Limon, J. J., Bar, A. S., Leal, C. A., Gargus, M., Tang, J., et al. (2016). Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19, 865–873. doi: 10.1016/j.chom.2016.05.003
Yamasaki, S., Matsumoto, M., Takeuchi, O., Matsuzawa, T., Ishikawa, E., Sakuma, M., et al. (2009). C-type lectin mincle is an activating receptor for pathogenic fungus, Malassezia. Proc. Natl. Acad. Sci. U.S.A. 106, 1897–1902. doi: 10.1073/pnas.0805177106
Keywords: airway microbiome, airway fungal microbiome, mycobiome, host-microbiome interactions, bacterial-fungal interactions, biofilm
Citation: Zhang I, Pletcher SD, Goldberg AN, Barker BM and Cope EK (2017) Fungal Microbiota in Chronic Airway Inflammatory Disease and Emerging Relationships with the Host Immune Response. Front. Microbiol. 8:2477. doi: 10.3389/fmicb.2017.02477
Received: 01 October 2017; Accepted: 29 November 2017;
Published: 12 December 2017.
Edited by:
Steven Templeton, Indiana University School of Medicine - Terre Haute, United StatesReviewed by:
Jeniel E. Nett, University of Wisconsin-Madison, United StatesCopyright © 2017 Zhang, Pletcher, Goldberg, Barker and Cope. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily K. Cope, ZW1pbHkuY29wZUBuYXUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.