- Department of Plant Pathology, College of Plant Protection, Nanjing Agricultural University, and Key Laboratory of Integrated Management of Crop Diseases and Pests, Ministry of Education, Nanjing, China
Guanylate kinases (GKs), which convert guanosine monophosphate into guanosine diphosphate (GDP), are important for growth and mannose outer chain elongation of cell wall N-linked glycoproteins in yeast. Here, we identified the ortholog of Saccharomyces cerevisiae GK Guk1, named MoGuk1 and a novel family of fungal GKs MoGuk2 in the rice blast fungus Magnaporthe oryzae. MoGuk1 contains 242 aa with an C-terminal GuKc domain that very similar to yeast Guk1. MoGuk2 contains 810 amino acids with a C-terminal GuKc domain and an additional N-terminal efThoc1 domain. Expression of either MoGuk1 or MoGuk2 in heterozygote yeast guk1 mutant could increase its GDP level. To investigate the biological role of MoGuk1 and MoGuk2 in M. oryzae, the gene replacement vectors were constructed. We obtained the ΔMoguk2 but not ΔMoguk1 mutant by screening over 1,000 transformants, indicating MoGuk1 might be essential for M. oryzae. The ΔMoguk2 mutant showed weak reductions in vegetative growth, conidial germination, appressorial formation, and appressorial turgor, and showed significant reductions in sporulation and pathogenicity. Moreover, the ΔMoguk2 mutant failed to produce perithecia and was sensitive to neomycin and a mixture of neomycin-tunicamycin. Exogenous GDP and ATP partially rescued the defects in conidial germination, appressorial formation, and infectious growth of the mutant. Further analysis revealed that intracellular GDP and GTP level was decreased, and GMP level was increased in the mutant, suggesting that MoGuk2 exhibits enzymatic activity. Structural analysis proved that the efThoc1, GuKc, and P-loop domains are essential for the full function of MoGuk2. Taken together, our data suggest that the guanylate kinase MoGuk2 is involved in the de novo GTP biosynthesis pathway and is important for infection-related morphogenesis in the rice blast fungus.
Introduction
Rice blast, caused by Magnaporthe oryzae, is one of the most devastating rice diseases worldwide (Yan and Talbot, 2016; Zhang et al., 2016). Outbreaks of rice blast are a constant threat to global rice production and could result in an annual loss of rice capable of feeding more than half of the global population (Zeigler, 1998; Talbot, 2003; Zhang et al., 2016). The control of rice blast is problematic because of the volatility of rice blast physiological races (Zeigler et al., 1994). An improved understanding of the developmental and pathogenic molecular mechanisms of the fungus will facilitate research into targeted fungicides and disease management (Wilson and Talbot, 2009). A key step in the disease cycle of M. oryzae is the formation of a highly specialized infection structure known as an appressorium, which differentiates from the asexual spores (Saunders et al., 2010; Oses-Ruiz et al., 2017). Following maturation, enormous turgor pressure is generated in the appressorium, which forces the penetration peg to physically puncture the plant cuticle (Howard et al., 1991; Dejong et al., 1997). After invading host cells, infectious hyphae (IH) differentiate and rapidly extend into plant cells, with a new disease cycle reinitiating in 5–7 days (Foster et al., 2017). To complete the infection cycle, M. oryzae evolved genetic regulatory systems that allow it to respond to available nutrients in the plant host and also encounter nitrogen-starved environments at the start of the infection cycle (Snoeijers et al., 2000; Wilson et al., 2007; Fernandez et al., 2014). The regulatory systems include nitrogen metabolite repression (NMR) and carbon catabolite repression (CCR) to ensure the use of preferred sources of nitrogen (ammonium and L-glutamine) and carbon (glucose), respectively (Wilson et al., 2007, 2012; Wilson and Talbot, 2009; Fernandez et al., 2012). Many regulators (such as Nut1, Npr1/2, Nir1, Rbp35, Snf1, Tps1, Nmr1-3, and Mdt1) involved in these systems were characterized to be important for appressorial formation and penetration, as well as infection in M. oryzae (Froeliger and Carpenter, 1996; Lau and Hamer, 1996; Wilson et al., 2007, 2010, 2012; Yi et al., 2008; Franceschetti et al., 2011; Fernandez et al., 2012).
The purine metabolic pathway provides the cell with adenosine triphosphate (ATP) and guanosine triphosphat (GTP), which are critical to cellular processes (Elion, 1989). Purine nucleotides are synthesized by both de novo and salvage pathways in organisms (Nyhan, 2005). The biochemistry of de novo purine nucleotide biosynthesis and salvage is a broadly conserved and well-understood component of cellular metabolism (Beck et al., 2003). The de novo purine nucleotide biosynthesis pathway is the pre-dominant method for purine nucleotide generation, whereby phosphoribosyl pyrophosphate (PRPP) is the initial substrate and, via a series of enzymatic reactions, and formed the purine nucleotide inosine monophosphate (IMP) (Liechti and Goldberg, 2012). IMP is the central product of both de novo and salvage pathways and central to the interconversion to adenine and guanine nucleotides (Hedstrom, 2009; Sunohara et al., 2013). IMP could be utilized to generate xanthomine monophosphate (XMP) and then to guanine monophosphate (GMP) by the dual reactions of IMP dehydrogenase and GMP synthase (Sunohara et al., 2013), or to produce adenosine monophosphate (AMP) catalyzed by the enzymes of adenylosuccinate synthase and adenylosuccinate lyase (Liechti and Goldberg, 2012; Morrow et al., 2012; Guibinga et al., 2013). The other pathway for purine nucleotide biosynthesis is the purine salvage pathway (Figure 1). The process of purine salvage can generate GMP from guanine and AMP from adenine directly, or generate IMP or XMP from hypoxanthine and xanthine indirectly. These nucleotides can then be fed into the nucleotide biosynthesis pathway, where they can then be used to generate GMP or AMP. In addition, based on the established pathway, enzymes such as adenine deaminase, and guanine deaminase allow the cycling of nucleotides bypasses the de novo and salvage pathways, thereby allowing for various purine bases to be used for generating both GMP and AMP (Pettersson et al., 2007).
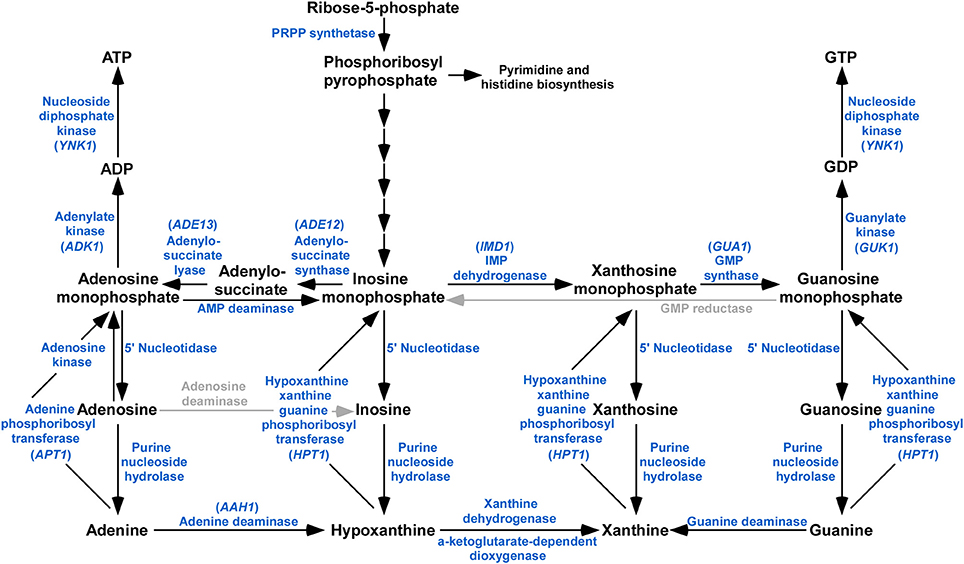
Figure 1. Schematic representation of purine metabolism pathway identified in M. oryzae. BLASTp analysis using S. cerevisiae and C. neoformans orthologs reveals that majority of the components of the purine biosynthetic pathway are present in the genome of M. oryzae. Enzymes or activities missing are grayed.
Guanylate kinase (GK) is an essential enzyme that catalyzes the transfer of a phosphate from ATP to GMP (Sekulic et al., 2002). The enzyme is indispensable for converting GMP to GDP and therefore synthesis of GTP (Li et al., 1996). Plants have two distinct types of GKs, which are essential for growth and development (Nomura et al., 2014). Both yeast and humans have a single essential GK involved in GDP biosynthesis (Konrad, 1992; Jain et al., 2016). Here, we identified and characterized the GK MoGuk1 and MoGuk2 in the phytopathogen M. oryzae. We proved that MoGuk1 likely was essential for M. oryzae, and MoGuk2 is involved in the de novo GTP biosynthesis pathway and is important for development and virulence in the rice blast fungus.
Materials and Methods
Strains and Culture Conditions
The wild type Guy11 (ATCC201236) and mutant strains of M. oryzae were cultured on complete medium (CM) agar plates at 28°C. Fungal mycelia were cultured and harvested from 2-day-old liquid CM medium by filtration through one layer of Miracloth, and used for genomic DNA and total RNA extraction. Protoplasts preparation and M. oryzae transformation was performed as described previously (Sweigard et al., 1992). For vegetative growth, mycelial plugs (3 × 3 mm) were placed onto CM, minimal medium (MM), oatmeal medium (OM), and straw decoction and corn (SDC) medium and cultured at 28°C for 7 days (Talbot et al., 1993; Song et al., 2010; Zhang et al., 2010, 2014). Each experiment was performed in triplicate and repeated three times.
Construction of MoGUK1 and MoGUK2 Null Mutant, Domain Deletion, and Complementation Vectors
The gene deletion construct was generated according to the homologous recombination principle as described (Tang et al., 2015). In brief, a flanking sequence fragment of MoGUK1 and MoGUK2 (1-kb upstream and 1-kb downstream) was amplified from M. oryzae genomic DNA and cloned into the pCX62 vector to flank the hygromycin phosphotransferase cassette (HPH). The resulting pCX62::MoGUK1::HPH and pCX62::MoGUK2::HPH vectors were confirmed by sequence analysis and used for protoplast transformation. For domain deletion and full length complemented constructs generation, the entire MoGUK2 gene or domain deletion fragments and its native promoter region (1,500 bp upstream from the starting codon) was amplified by PCR and cloned into the pYF11(bleomycin resistance) vector using a yeast gap repair approach, respectively (Bruno et al., 2004). The final constructs were sequenced and transformed into the ΔMoguk2 mutant to obtain the full length complemented transformant and domain deletion transformants. The primers used in this section are listed in Table S1.
Conidiation, Sexual Reproduction, Appressorial Formation, and Turgor Assays
For conidial development on glass slides assay, 5 × 5 mm mycelial plugs were cut from the indicated strains cultured on SDC plates for 7 days, and placed on glass slides to induce conidiation under black light for 20 h, and observed under a light microscope. For conidial quantification assay, the indicated strains were inoculated on SDC plates to promote conidiation as described (Zhang et al., 2009). Conidia were collected in distilled water and filtered through with one layer Miracloth (Millipore, USA). Conidial number was counted with a hemocytometer under a microscope. For sexual reproduction, plugs of the ΔMoguk2 mutant and the wild type Guy11 (MAT1-2) and the mating partner strain TH3 (MAT1-1) were point-inoculated 3 cm apart on oatmeal agar medium and cultured at 20°C with continuous light for 3–4 weeks as described (Zhang et al., 2011). Mature perithecia were crushed to examine the asci and ascospores and photographed. Appressorial formation was performed by placing conidial suspensions (1 × 104 spores/ml) on cover glass (Fisher-brand, UK) (hydrophobic surface) and cultured at 28°C. Conidial germination and appressorial formation was examined under a microscope by time course. Appressorium turgor was measured by incipient cytorrhysis (cell collapse) assay using a 1–4 molar concentration of glycerol solution as described previously (Howard et al., 1991). All experiments were performed in triplicate and repeated three times.
Plant Infection and Infectious Hyphal Growth Assays
Plant infection was conducted on 2-week old rice seedlings and 1-week old detached barley leaves as described previously (Zhang et al., 2011; Li et al., 2016). Diseased leaves were observed daily and photographed at 5 (barley) or 7 (rice) days post-inoculation (dpi). For infectious hyphal growth assay, the rice leaf sheaths were inoculated with 100 μl conidial suspensions (1 × 105 spores/ml), and incubated in a humid chamber at 28°C. Infectious hyphal growth was observed at 30 or 40 h post-inoculation (hpi) under a microscope.
GDP, GTP, and GMP Extraction and HPLC Analysis
All of the strains were cultured on CM medium at 28°C, cut into 1 × 1 mm square and cultured in liquid CM for 2 days. Filtering to collect mycelium and quickly ground into powder. Every 1 mg mycelium mixed with 20 μl 6% TCA solution, turbulenced. Samples were centrifuged (4,000 rpm, 15 min), the top layer were collected and washed with five times the volume of anhydrous ether twice. The pellet was collect for HPLC (High Performance Liquid Chromatography). HPLC analysis was done with a programmable Aglient Technology zorbax 1200 series liquid chromatograph. The solvent system consisted of methanol (90%): water (10%), at a flow rate of 1 ml/min. Solution (GDP, GTP, and GMP) was eluted through the column (SB-C18, 5 μl, 4.6 × 250 mm) and detected at 259 nm UV. Each sample through the column in turn and detected the peak value with the same time as the standard.
Yeast Complementation Assay
The full length cDNA of MoGUK1 and MoGUK2 was amplified using primers (Table S1), and cloned into the yeast expression vector pYES2 plasmid. The resulting plasmids were verified by sequencing and transformed into yeast mutant strain (BY4743; MAT a/alpha; hisΔ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met 15Δ0; uraΔ0/uraΔ0; YDR454c::kanMX4/YDR454c) which bought from EUROSCARF. Yeast protein extraction was conducted as follows: Cells were cultured in liquid SDgal media to mid log phase and centrifuged at 5,000 rpm, 4°C, 5 min; Washed once in 1 ml cold 0.85% NaCl; Re-suspended the cells in 200 μl lysis buffer, added 100 μl of glass beads, and vortex 5 min at 4°C; then centrifuged at 4°C, 13,000 rpm for 10 min and transferred supernatant to new tubes. The samples were used immediately for HPLC analysis as performed above.
Nucleic Acid Manipulation, Quantitative RT-PCR, and Southern Blot Assays
The MoGUK2 gene probe and HPH probe were amplified with the primer pairs, respectively. Probe labeling, hybridization, and detection were performed with a DIG High Prime DNA Labeling and Detection Starter Kit (Roche). Total RNA was isolated from frozen fungal mycelia using an RNA extraction kit (TaKaRa, China). Total RNA was pretreated with DNase I and was then reverse transcribed (TaKaRa, China). Quantitative RT-PCR was performed on an ABI 7500 real-time PCR system (Applied Biosystems, USA) according to the manufacturer's instructions. The stable expression ACTIN gene (MGG_03982) was used as internal control. All primers used in this section are listed in Table S1.
Results
Identification and Domain Prediction of MoGUK Proteins in M. oryzae
S. cerevisiae ScGuk1 protein sequence was used to search the M. oryze genome database using blastp (1e−40 as an e-value cut-off) in FungiDB (http://fungidb.org), and yield the ortholog MoGuk1 (MGG_06764) and a protein with similarity to ScGuk1, named MoGuk2 (MGG_06394). Phylogenetic analysis for the homologs of MoGuk1 and MoGuk2 in some fungal species indicated that MoGuk1, as well as ScGuk1, formed a conserved group that contained one ortholog in each analyzed species; while MoGuk2 orthologs were only found in a few species, but also formed a conserved group that different from Guk1 group, indicating MoGuk2 is a novel family of GKs in M. oryzae (Figure 2A).
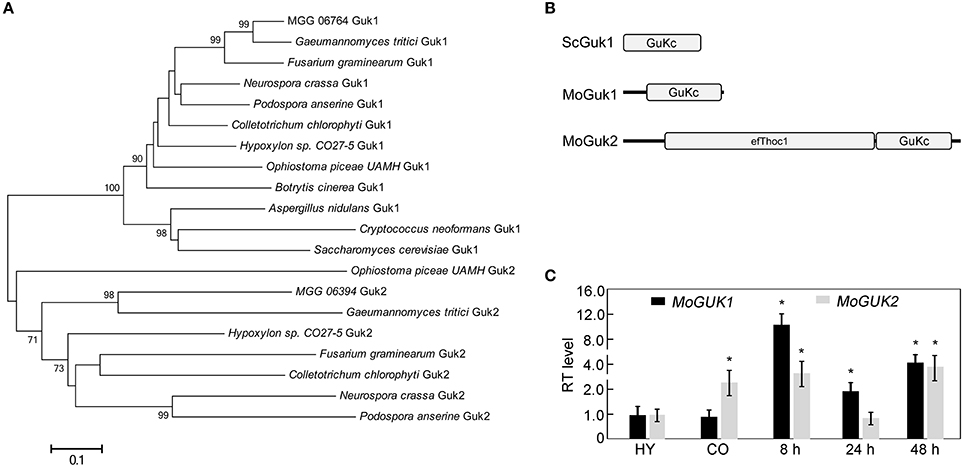
Figure 2. Phylogenetic analysis of Guk proteins from different organisms. (A) The homologs of ScGuk1 in M. oryzae were identified using blastp program and 1e-40 as an e-value cut-off. To construct the tree, homolog sequences retrieved from GuKc domains of MoGuk1 and MoGuk2 families in other indicated species were identified using blastp program default parameters. The tree was constructed using MEGA 5, with the Neighbor-Joining method and default parameters, bootstrap values >65 were displayed beside the braches. (B) The GuKc domains were predicted using SMART server (http://smart.embl-heidelberg.de). (C) Quantitative RT-PCR analysis the expression of MoGUK1 and MoGUK2. Total RNA was extracted from mycelia (MY), conidia (CO), and infectious hyphae of the wild type Guy11. Error bars represent standard deviation and asterisks represent significant differences at P < 0.01.
Sequence analysis revealed that MoGUK1 comprises 845 base pairs (bp) separated by one intron, encoding a 242-amino-acid (aa) protein with a Guanylate_kin domain (PFAM PF00625) that catalyzes ATP-dependent phosphorylation of GMP to GDP, which is similar to ScGuk1. MoGUK2 comprises 2,646 bp separated by three introns, encoding an 810 aa protein with a C-terminal Guanylate_kin domain, and an additional N-terminal efThoc1 domain (PFAM PF11957), which is required for transcription elongation through genes containing tandemly repeated DNA sequences and is also involved in mRNA export from nucleus (Figure 2B). Protein sequence alignment exhibited that GuKc domain showed high identities among MoGuk1, MoGuk2, and their orthologs, therefore driven a question that whether the Guk1 and Guk2 groups proteins had similar functions (Figure S1). To test the hypothesis, we constructed the yeast expression vectors pYES2-MoGUK1 and pYES2-MoGUK2 and introduced into the diploid yeast strain since GUK1 is an essential gene in yeast. The result showed that intracellular GDP level of the yeast strain expressed MoGUK1 (3.54 ± 0.12) or MoGUK2 (6.72 ± 0.06) was increased compared to the strain expressed empty vector (2.92 ± 0.14), indicating both MoGuk1 and MoGuk2 have GK activity like yeast Guk1. Expression profile analysis revealed that the transcription level of both MoGUK1 and MoGUK2 was increased at the early infection stage in comparison to the mycelial stage (Figure 2C). MoGUK2 also showed high transcription level in conidial stage (Figure 2C). These results implicated that MoGUK1 and MoGUK2 plays potential roles in the development and infection of M. oryzae.
MoGuk2 Is Involved in Vegetative Growth
To investigate the role of MoGuk1 and MoGuk2 in M. oryzae, gene knock-out constructs pCX62::MoGUK1 U&D::HPH and pCX62::MoGUK2 U&D::HPH was generated by replacing MoGUK1 or MoGUK2 with a hygromycin phosphotransferase cassette (HPH), respectively (Figure S2A). The final constructs were transformed into protoplasts of the wild type strain, Guy11. The resulting hygromycin-resistant transformants were first screened by PCR. However, we failed to get a MoGUK1 null mutant by screening over 1,000 transformants, indicating MoGUK1 might be an essential gene in M. oryzae. Fortunately, four ΔMoguk2 mutants were obtained from 210 transformants, which further confirmed by Southern blot analysis (Figure S2B). The complemented transformant ΔMoguk2/MoGUK2 was generated by re-introducing the wild type MoGUK2 gene into the ΔMoguk2 mutant.
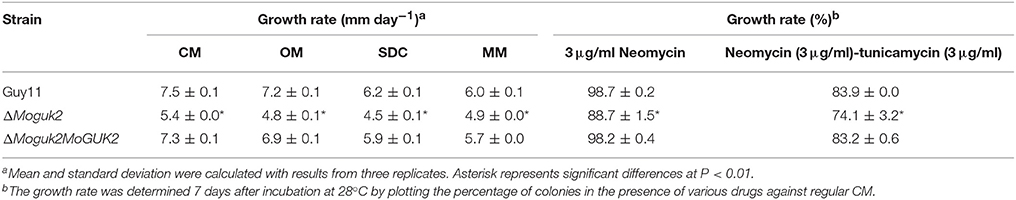
We first analyzed the role of MoGuk2 in vegetative growth. Wild type Guy11, ΔMoguk2 mutant and ΔMoguk2/MoGUK2 were inoculated onto complete medium (CM), oatmeal medium (OM), straw decoction and corn medium (SDC), and minimal medium (MM) agar plates for 7 days. The mutant displayed no difference in colony morphology in comparison with the wild type or complemented transformant (Figure 3A); however, the growth rate of the ΔMoguk2 mutant was decreased on these four media. The percentage of reduction in growth was 28.0, 33.3, 27.4, and 18.3% in CM, OM, SDC, and MM, respectively (Table 1). Therefore, MoGuk2 plays a role in vegetative growth of the rice blast fungus.
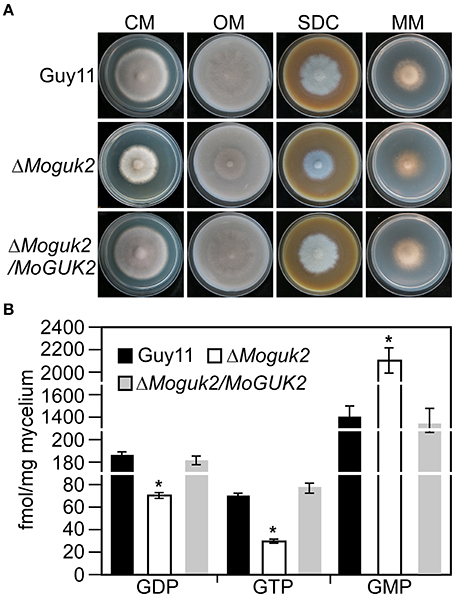
Figure 3. The ΔMoguk2 mutant decreased vegetative growth and intracellular GDP, GTP level, and increased GMP level. (A) Colony morphology was examined on CM, SDC, OM, and MM ager plates, and photographed after culture at 28°C in darkness for 7 days. (B) Intracellular GDP, GTP, and GMP measurement. Error bars are standard deviations and asterisks represent significant differences at P < 0.01.
Intracellular GDP and GTP Level was Decreased, and GMP was Increased in the ΔMoguk2 Mutant
Guk1 is an enzyme that indispensable for converting GMP to GDP and therefore synthesis of GTP (Li et al., 1996). To investigate whether MoGuk2 have the similar role and indeed involving in GTP biosynthesis pathway, we measured the intracellular GDP, GTP, and GMP levels in the ΔMoguk2 mutant. The results showed that GDP and GTP level was significantly decreased in the ΔMoguk2 mutant compared to the wild type Guy11, and decreased 60.1 and 63.7%, respectively. In contrast, intracellular GMP was remarkably increased in the mutant, and increased 34.0% compared to the wild type (Figure 3B). These results indicated that MoGuk2 plays a role in converting GMP to GDP and therefore synthesis of GTP in M. oryzae.
MoGuk2 Is Important for Asexual and Sexual Development
Conidiation is one of the key steps in the disease cycle of M. oryzae (Lee et al., 2006). Therefore, the role of MoGuk2 in conidial production was examined. The three strains were inoculated onto SDC plates to promote conidiation. The ΔMoguk2 mutant formed normal conidiophores but with fewer conidia compared with the wild type and complemented transformant. The conidial number was decreased 87.4% in the mutant (Figure 4A). We next quantified the numbers of conidia produced by the indicated strains cultured for 10 days on SDC plates. Consistent with the above mentioned result, conidial production by the ΔMoguk2 mutant was significantly decreased to 12.7% of that by the wild type. In addition, the conidial morphology of the mutant was altered; 53.0% of conidia showed an abnormal morphology with one or two septa, compared with 98.0% normal conidia with two septa in the wild type strain (Figure 4B). Regarding sexual development, the ability of the ΔMoguk2 mutant to produce perithecia was completely abolished. No perithecia were observed in the mutant, while numerus mature perithecia and asci were observed in the wild type and complemented transformant (Figure 4C). The results above suggest that MoGuk2 plays an important role in conidiogenesis and is essential for sexual reproduction.
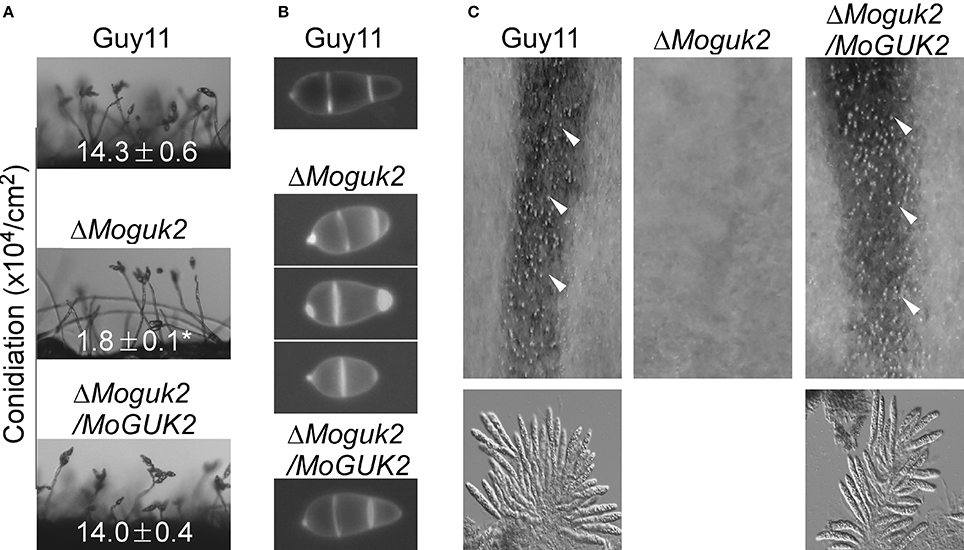
Figure 4. The ΔMoguk2 mutant showed defects in asexual and sexual development. (A) Conidial development was examined under a light microscope 20 h after induction on glass slides. ±SD was calculated from three repeated and asterisks represent significant differences at P < 0.01. (B) Conidial morphology of the ΔMoguk2 mutant. (C) Perithecia and asci formed by the indicated strains were photographed 3 weeks after inoculation. White arrows indicate peritheria.
MoGuk2 Is Indispensable for Full Virulence of M. oryzae
To evaluate the role of MoGuk2 in pathogenicity, conidial suspensions prepared from Guy11, the ΔMoguk2 mutant and the complemented transformant were sprayed onto 2-week-old rice seedlings. At 7 days after inoculation, the mutant caused few small lesions on rice leaves in comparison with the numerous typical lesions caused by the wild type and complemented transformant (Figure 5A). The lesion number was decreased 73.9% compared to the wild type (Figure 5B). Disease severity was also quantified by a lesion-type scoring assay (Valent et al., 1991). The number of lesions of types 1–3 produced by the ΔMoguk2 mutant was markedly decreased compared with the wild type; moreover, the mutant produced no type 4 or 5 lesions. The average lesion size of the mutant was decreased 45.5% compared to the wild type Guy11 (Figure 5B). Further detached barley infection assays showed similar results; i.e., the mutant was reduced in virulence, and the average lesion size was reduced 26.1% compared to the wild type in detached barley leaf assay (Figure 5C).
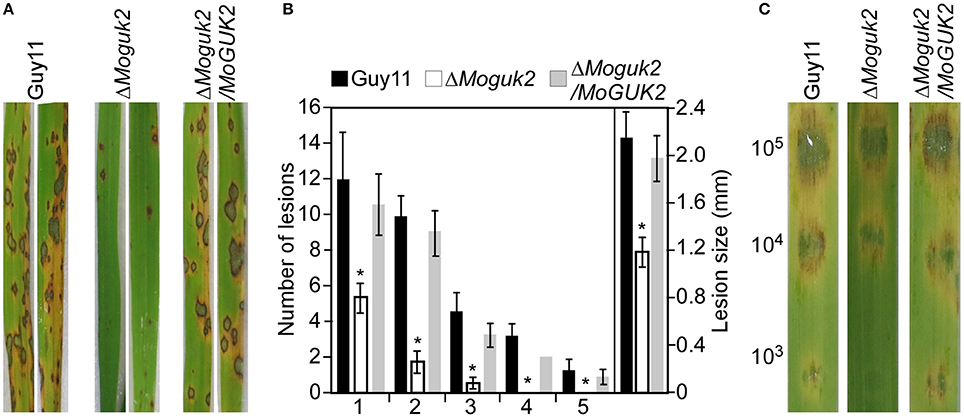
Figure 5. The ΔMoguk2 mutant attenuates virulence. (A) Conidial suspensions (5 × 104 spores/ml) from the wild type, the ΔMoguk2 mutant and the complemented transformant were sprayed onto 2-week-old rice seedlings, and photographed at 7 days post-inoculation (dpi). (B) Statistical analysis of the number of lesions in each type, and the average lesion size of the indicated strains. Lesions on 10 diseased rice leaves were counted per replicate, and the experiment was repeated three times. Error bars are standard deviations and asterisks represent significant differences at P < 0.01. (C) Conidial suspensions (1 × 105, 1 × 104, and 1 × 103 spores/ml) from the indicated strains were dropped onto 1-week-old detached barley leaves, and photographed at 5 dpi.
MoGuk2 Plays Roles in Conidial Germination, Appressorial Formation, and Turgor Generation
Appressoria are specialized structures that facilitate penetration of the rice blast fungus (Saunders et al., 2010). Because the ΔMoguk2 mutant caused only a few lesions on rice leaves, we speculated that the mutant might have defects in appressorial formation or penetration. To assess this possibility, we first observed conidial germination and appressorial formation on inductive surfaces. The results showed that both of these two processes were delayed by 6 h in the mutant compared to the wild type (Figure 6A, Table 2), indicating MoGuk2 plays a role in conidial germination and appresssorial formation in M. oryzae. We also measured the appressorial turgor, the mutant showed decreased turgor pressure compared with the wild type, suggestive of a defect in appressorial function (Figure 6B).
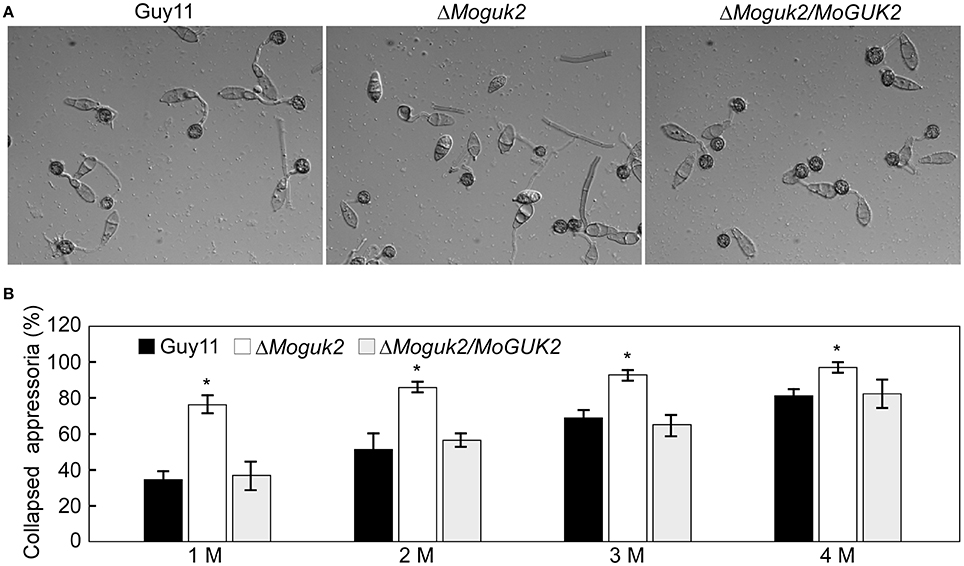
Figure 6. The ΔMoguk2 mutant showed defects in appressorial formation and turgor. (A) Appressorial formation was observed on hydrophobic surfaces under a microscope at 24 hpi. (B) Appressorial turgor measurement. Error bars are standard deviations and asterisks represent significant differences at P < 0.01. All graphs represent three biological and three technical replicate observations.
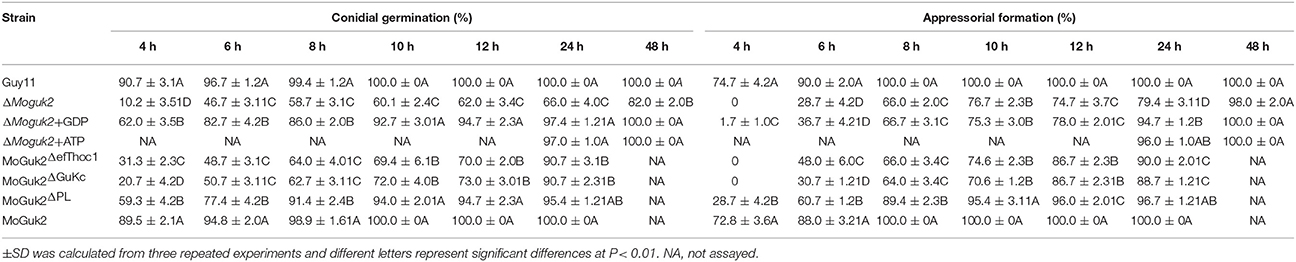
Table 2. Conidial germination and appressorial formation of the indicated strains with or without exogenous GDP or ATP.
MoGuk2 Plays a Role in IH Growth
As described above, the ΔMoguk2 mutant was defective in appressorial turgor and caused small lesions on rice seedlings, suggesting defects in penetration and IH growth. Therefore, penetration and IH growth of the mutant were examined in rice sheath cells. After incubation for 24 h, 33% of the appressoria of the mutant were unable to penetrate rice epidermal cells (type 1), 67% of IH were restricted to one cell with no (type 2) or two branches (type 3), and almost no IH extended to neighboring cells with three or more branches (type 4) In contrast, almost all appressoria penetrate into the cells and showed severe IH growth (Figure 7). When observed at 48 hpi, IH growth became severe in the mutant, percentage IH of type 3 and type 4 increased to 76%. (Figure 7), indicating that IH growth was delayed by 24 h in the ΔMoguk2 mutant compared to the wild type.
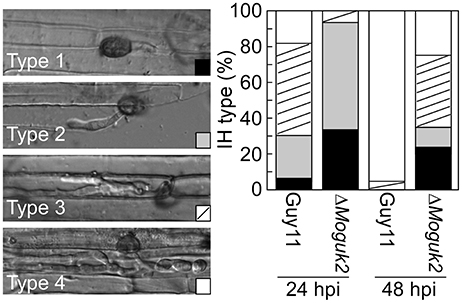
Figure 7. The ΔMoguk2 mutant was defective in infectious hyphal growth. Statistical analysis of the percentage of infectious hyphae (IH) types in rice sheath cells at 24 and 48 hpi (type 1: no infectious hyphae; type 2: one infectious hyphae; type 3: two or three branches restricted in one cell; type 4: more than three branches extended to a neighboring cell; N = 100).
Exogenous GDP and ATP Partially Restored the Defects of the ΔMoguk2 Mutant
To determine whether MoGuk2 plays a role in de novo GTP biosynthesis, the ΔMoguk2 mutant was treated with GDP, the immediate product of GK. Addition of 10 mM exogenous GDP restored conidial germination in the ΔMoguk2 mutant (Table 2). Appressorial formation was also restored, albeit not to the wild type level, at 24 hpi (Figure 8A, Table 2). Observation of IH growth revealed that ΔMoguk2+GDP displayed longer IH with more branches (40% type 3 and 12% type 4), in comparison with 33% type 1 (no penetration) and 55% type 2 (with no branches) in the ΔMoguk2 mutant (Figure 8B). At 40 hpi, the ΔMoguk2 mutant showed short IH without branches, and ΔMoguk2+GDP showed longer IH, with one or two branches restricted to one plant cell. In contrast, the wild type Guy11 exhibited IH with many branches extending to neighboring cells (Figure 8C). We also treated the ΔMoguk2 mutant with exogenous ATP, addition of 2.5 mM ATP restored the conidial germination, appressorial formation as well as infectious growth defects of the mutant (Figures 8A–C; Table 2). However, both GDP and ATP failed to restore vegetative growth, conidiation and conidial morphology of the mutant (data not shown). We also added exogenous GTP in the ΔMoguk2 conidial suspensions to test the virulence on barley leaves, and found 2.5 mM GTP could restore the virulence defect of the mutant (Figure S3). These results indicate that MoGuk2 plays a role in the de novo GTP biosynthesis pathway in M. oryzae.
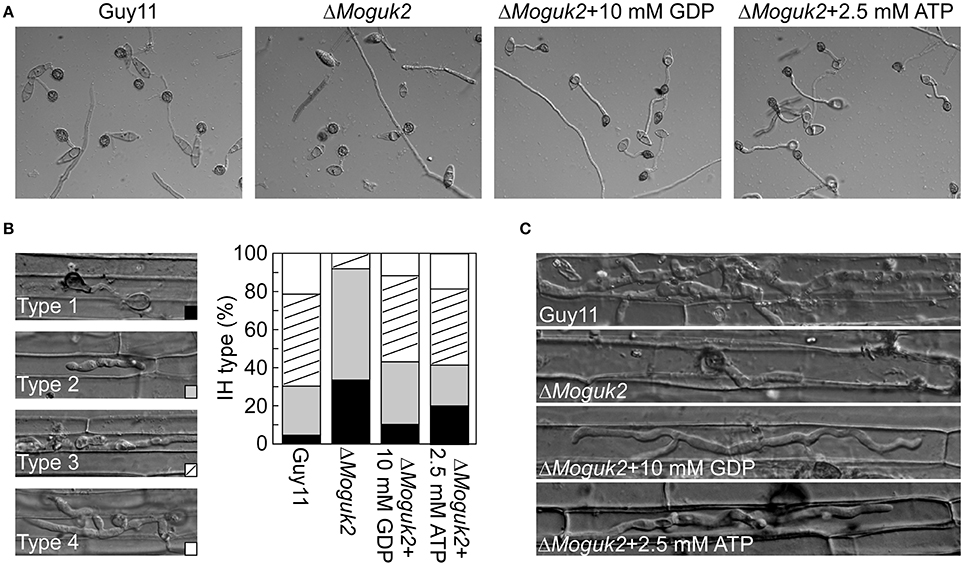
Figure 8. Exogenous GDP and ATP partially restored the defects of the ΔMoguk2 mutant. (A) Appressorial formation of the ΔMoguk2 mutant with or without exogenous GDP or ATP, and photographed at 24 hpi. (B) Statistical analysis the infectious hyphae (IH) type of the ΔMoguk2 mutant in rice leaf sheath with or without exogenous GDP or ATP, and photographed at 30 hpi. (C) IH observation of the ΔMoguk2 mutant with or without exogenous GDP or ATP at 40 hpi.
The ΔMoguk2 Mutant Is Sensitive to Neomycin and Neomycin-Tunicamycin
In yeast, guanylate kinase Guk1 mutants displayed a strong defect in protein N-glycosylation and cell wall organization that leads to a hypersensivity to neomycin, and a mixture of neomycin-tunicamycin (Shimma et al., 1997). To investigate whether MoGuk2 has a similar role in M. oryzae, the wild type Guy11, ΔMoguk2 mutant and complemented transformant were inoculated onto CM agar plates with neomycin and neomycin-tunicamycin (an inhibitor of N-glycosylation) mixture. After 7 days incubation, the results showed that the mutant was more sensitive to the drugs compared to the wild type and complemented transformant. The growth rate was decreased 11.3 and 25.9% on neomycin, neomycin-tunicamycin, compared to 1.3 and 16.1% in the wild type, respectively (Table 1).
The Domains of MoGuk2 Are Essential for Its Full Function
To investigate the contributions of its various domains to the function of MoGuk2, efThoc1, GuKc, and PL domain-deleted constructs (MoGuk2ΔefThoc1, MoGuk2ΔGuKc, and MoGuk2ΔPL, respectively, Figure 9A) were generated and transformed into the ΔMoguk2 mutant. The resulting transformants were screened by quantitative RT-PCR (Figure S4). All three transformants showed similar phenotypes to that of the ΔMoguk2 mutant, and only a few defects were partially restored. The vegetative growth of all three transformants was indistinguishable from that of the ΔMoguk2 mutant (Figure 9B). Conidial production was partially rescued in the transformants, particularly in MoGuk2ΔPL (Figure 9B). Conidial morphology and virulence were not rescued in MoGuk2ΔefThoc1 but were partially rescued in MoGuk2ΔGuKc and MoGuk2ΔPL. Moreover, MoGuk2ΔPL caused more severe lesions on rice leaves in comparison with MoGuk2ΔGuKc (Figures 9C,D). Disease severity was also quantified by the lesion-type scoring assay. MoGuk2ΔefThoc1 showed similar lesion number and types to the ΔMoguk2 mutant. While MoGuk2ΔGuKc and MoGuk2ΔPL displayed more type 2 and type 3 lesions compared to the mutant (Figure 9E). Conidial germination and appressorial formation were also partially rescued in the three transformants, to the greatest degree in MoGuk2ΔPL (Table 2). These results indicate that the domains are critical for MoGuk2 functions in conidiogenesis, appressorial formation, and virulence. The efThoc1 and GuKc domains are more important than the PL domain of MoGuk2.
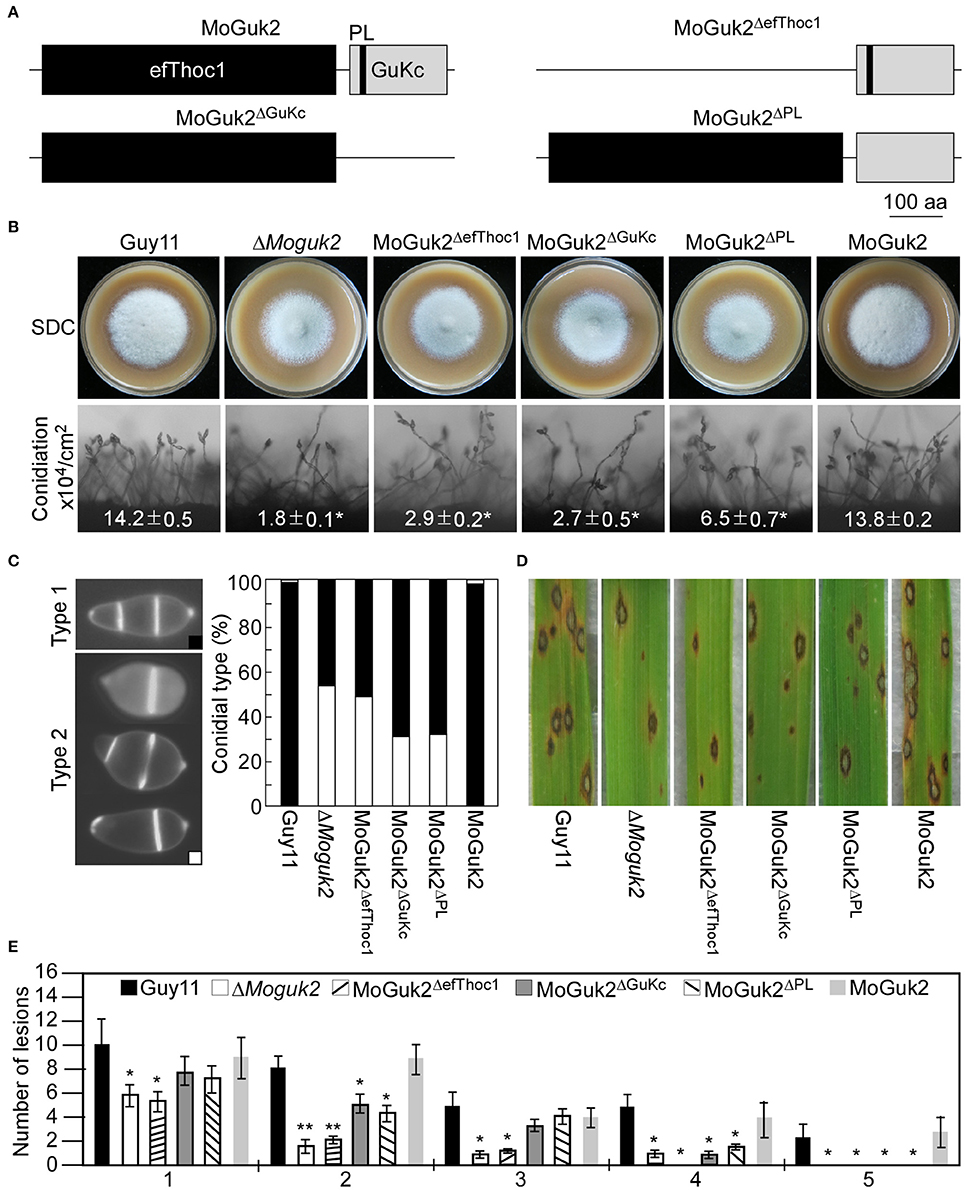
Figure 9. The efThoc1, GuKc, and P-loop domains are required for the full function of MoGuk2. (A) Domain deletion strategy and diagram of MoGuk2. (B) Vegetative growth and conidiation of the indicated strains. (C) Statistical analyzes the abnormal conidia of the indicated strains. (D) Spraying assay. Conidial suspensions prepared from the indicated strains were sprayed onto 2-week-old rice seedlings, and photographed at 7 dpi. (E) Statistical analysis of the number of lesions in each type. Error bars are standard deviations and asterisks represent significant differences at P < 0.01.
Discussion
The GTP de novo biosynthesis and salvage pathways play critical roles in the growth, development and virulence of diverse organisms (Kirsch and Whitney, 1991; Rodriguez-Suarez et al., 2007; Jiang et al., 2010; Morrow et al., 2012). The key enzymes of these pathways are potential targets of anticryptococcal drugs (Morrow et al., 2012). In this study, we identified and characterized the yeast Guk1 ortholog MoGuk1 and a novel family of guanylate kinase MoGuk2 in M. oryzae that involved in GTP de novo biosynthesis pathway. Phylogenetic analysis revealed that both MoGuk1 and MoGuk2 and its orthologs were well conserved in different species, but Guk1 and Guk2 proteins clustered in different group, indicating MoGuk2 belongs to a novel family of GK proteins. MoGuk1 showed very similar structure to yeast Guk1 which is an essential gene in yeast, indicating MoGuk1 might also be essential for M. oryzae, since we failed to obtain a null mutant by screening over 1,000 transformants. However, further experiments such as performing transformation in the presence of GTP or to perform promoter replacement need to be conducted to confirm this hypothesis.
Deletion of MoGuk2 resulted in a reduced growth rate on different media, including nutrient-rich (CM) and minimal media, this is different from the case in C. neoformans, in which purine bases are required for growth of mutants in the purine biosynthesis pathway (Morrow et al., 2012). One explanation might be the intracellular GDP and GTP levels of the mutant could still maintain its vegetative growth, though GDP and GTP levels were decreased in the mutant. However, exogenous GDP and ATP partially restored the conidial germination, appressorial formation, and IH growth defects of the mutant, but not conidial production and morphology, suggesting that MoGuk2 plays a role in the de novo GTP biosynthesis pathway which important for conidial germination, appressorial formation, and IH growth. Nevertheless, conidial production and morphology alteration was not observed in the related mutants that similar to the ΔMoguk2 mutant in these organisms. The possible explanation is that there is another GK (MoGuk1) that is functional in the ΔMoguk2 mutant. Like in yeast, the ΔMoguk2 mutant was hypersensitivity to the drugs neomycin and neomycin-tunicamycin, indicating MoGuk2 probably plays a role in protein N-glycosylation and cell wall organization.
Appressoria and their normal function are critical for M. oryzae penetration of host cells (Howard et al., 1991; Saunders et al., 2010). More than 20% of the mutant conidia were unable to germinate on an inductive surface even at 48 hpi, possibly due to a defect in sensing surface signals or cell viability. However, infectious growth in host cells was restricted. Although the mutant had defect in appressorial turgor, the mutant could not restore the virulence on wounded barley leaves (Figure S5A), indicating that the attenuation of the ΔMoguk2 mutant mainly due to IH growth defect. In plants, the generation of reactive oxygen species (ROS) is regarded as one of the first responses to fungal invasion (Mellersh et al., 2002). For plant pathogens, ROS also play an important role and loss of the ability to eliminate intracellular ROS can result in functional defects (Egan et al., 2007). Our result showed that the ΔMoguk2 mutant did not induce plant defense response and ROS accumulation (Figure S5B), indicating the IH growth defects was not due to plant defense. In M. oryzae, virulence defects of the mutants with deletions in amino acid or uridine biosynthesis genes—such as STR3/MET6/MET13, ILV1/2/6, LYS2/20, CPA2/ARG, ADE1, and PYR5, could be restored by exogenous amino acids or uridine (Wilson et al., 2012; Du et al., 2013; Fernandez et al., 2013; Yan et al., 2013; Chen et al., 2014; Zhang et al., 2014, 2015; Saint-Macary et al., 2015; Liu et al., 2016; Qi et al., 2016). Similar to these mutants, IH growth defects of the ΔMoguk2 mutant could be restored by exogenous GDP and ATP, suggesting GTP biosynthesis pathway plays a critical role in IH growth in M. oryzae.
Structural analysis revealed that MoGuk2 contains an efThoc1, a GuKc, and a PL domain. Deletion of each domain proved that all three domains are essential for the full function of MoGuk2. The efThoc1 domain seemed to be more important than the GuKc and PL domains in terms of conidial morphology and virulence. Although the PL motif contributes to the function of Guk2, other functional motifs are also present in the GuKc domain; PL was found to play a less important role in conidiation, appressorial formation, and infection than was GuKc. These three domains are likely involved in the de novo GTP biosynthesis pathway, and they likely affect the development and virulence of M. oryzae. Domain deletion might affect protein conformation and decrease or abolish its activity, thereby affecting the function of MoGuk2. MoGuk2 is an atypical GK and play important roles in many developmental processes, such as vegetative growth, asexual/sexual development, conidial morphology and germination, appressorial formation, and IH growth which closely related to GDP or ATP. These results are consistent to the findings in other fungi that mutants defective in the GTP biosynthesis are autotrophic and could be rescued by guanine (Konrad, 1992; Rodriguez-Suarez et al., 2007; Morrow et al., 2012). MoGuk1 and MoGuk2 likely functional redundancy in some respects, such as GDP biosynthesis, but MoGuk1 seems is a more important protein, as it was likely essential for M. oryzae.
Conclusion
This study identified and characterized an atypical GK MoGuk2, which plays a role in the de novo GTP biosynthesis pathway and is important for infection-related morphogenesis in the rice blast fungus. Our findings will improve our understanding of the pathogenic mechanisms of phytopathogens.
Author Contributions
XC, XZha, HZ, and ZZ: conceived and designed the experiments; XC, XZha, ML, XLi, XLiu, and XW: performed the experiments; XC, XZha, ML, XLiu, and HZ: analyzed the experiment data; XC, XZha, ML, XW, HZ, and XZhe: contributed reagents, materials, analysis tools; XC, XZha, HZ, and ZZ: wrote the paper; All authors have read and approve the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the National Key Research and Development Program of China (Grant No: 2016YFD0300700/2016YFD0300706), the Fundamental Research Funds for the Central Universities (Grant No: KYTZ201604), the Outstanding Youth Foundation of Jiangsu Province (Grant No: BK20160074 to HZ), and a grant from Innovation Team Program for Jiangsu Universities (2017). We also want to thank Dr. Wenwu Ye of Nanjing Agricultural University for the assistance to build the phylogenetic tree.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02467/full#supplementary-material
References
Beck, B. J., Huelsmeyer, M., Paul, S., and Downs, D. M. (2003). A mutation in the essential gene gmk (encoding guanlyate kinase) generates a requirement for adenine at low temperature in Salmonella enterica. J. Bacteriol. 185, 6732–6735. doi: 10.1128/JB.185.22.6732-6735.2003
Bruno, K. S., Tenjo, F., Li, L., Hamer, J. E., and Xu, J. R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3, 1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004
Chen, Y., Zuo, R. F., Zhu, Q., Sun, Y., Li, M. Y., Dong, Y. H., et al. (2014). MoLys2 is necessary for growth, conidiogenesis, lysine biosynthesis, and pathogenicity in Magnaporthe oryzae. Fungal Genet. Biol. 67, 51–57. doi: 10.1016/j.fgb.2014.04.001
Dejong, J. C., McCormack, B. J., Smirnoff, N., and Talbot, N. J. (1997). Glycerol generates turgor in rice blast. Nature 389, 244–245. doi: 10.1038/38418
Du, Y., Zhang, H. F., Hong, L., Wang, J. M., Zheng, X. B., and Zhang, Z. G. (2013). Acetolactate synthases MoIlv2 and MoIlv6 are required for infection-related morphogenesis in Magnaporthe oryzae. Mol. Plant Pathol. 14, 870–884. doi: 10.1111/mpp.12053
Egan, M. J., Wang, Z. Y., Jones, M. A., Smirnoff, N., and Talbot, N. J. (2007). Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 104, 11772–11777. doi: 10.1073/pnas.0700574104
Elion, G. B. (1989). The purine path to chemotherapy. Science 244, 41–47. doi: 10.1126/science.2649979
Fernandez, J., Marroquin-Guzman, M., and Wilson, R. A. (2014). Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu. Rev. Phytopathol. 52, 155–174. doi: 10.1146/annurev-phyto-102313-050135
Fernandez, J., Wright, J. D., Hartline, D., Quispe, C. F., Madayiputhiya, N., and Wilson, R. A. (2012). Principles of carbon catabolite repression in the rice blast fungus: Tps1, Nmr1-3, and a MATE-family pump regulate glucose metabolism during infection. PLoS Genet. 8:e1002673. doi: 10.1371/journal.pgen.1002673
Fernandez, J., Yang, K. T., Cornwell, K. M., Wright, J. D., and Wilson, R. A. (2013). Growth in rice cells requires de novo purine biosynthesis by the blast fungus Magnaporthe oryzae. Sci. Rep. 3:2398. doi: 10.1038/srep02398
Foster, A. J., Ryder, L. S., Kershaw, M. J., and Talbot, N. J. (2017). The role of glycerol in the pathogenic lifestyle of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 19, 1008–1016. doi: 10.1111/1462-2920.13688
Franceschetti, M., Bueno, E., Wilson, R. A., Tucker, S. L., Gomez-Mena, C., Calder, G., et al. (2011). Fungal virulence and development is regulated by alternative pre-mRNA 3'end processing in Magnaporthe oryzae. PLoS Pathog. 7:e1002441. doi: 10.1371/journal.ppat.1002441
Froeliger, E. H., and Carpenter, B. E. (1996). NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol. Gen. Genet. 251, 647–656.
Guibinga, G. H., Murray, F., Barron, N., Pandori, W., and Hrustanovic, G. (2013). Deficiency of the purine metabolic gene HPRT dysregulates microRNA-17 family cluster and guanine-based cellular functions: a role for EPAC in Lesch-Nyhan syndrome. Hum. Mol. Genet. 22, 4502–4515. doi: 10.1093/hmg/ddt298
Hedstrom, L. (2009). IMP Dehydrogenase: structure, mechanism, and inhibition. Chem. Rev. 109, 2903–2928. doi: 10.1021/cr900021w
Howard, R. J., Ferrari, M. A., Roach, D. H., and Money, N. P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. U.S.A. 88, 11281–11284. doi: 10.1073/pnas.88.24.11281
Jain, R., Khan, N., Menzel, A., Rajkovic, I., Konrad, M., and Techert, S. (2016). Insights into open/closed conformations of the catalytically active human guanylate kinase as investigated by small-angle X-ray scattering. Eur. Biophys. J. Biophys. Lett. 45, 81–89. doi: 10.1007/s00249-015-1079-9
Jiang, L., Zhao, J., Guo, R., Li, J., Yu, L., and Xu, D. (2010). Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. FEMS Yeast Res. 10, 199–208. doi: 10.1111/j.1567-1364.2009.00600.x
Kirsch, D. R., and Whitney, R. R. (1991). Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect. Immun. 59, 3297–3300.
Konrad, M. (1992). Cloning and expression of the essential gene for guanylate kinase from yeast. J. Biol. Chem. 267, 25652–25655.
Lau, G., and Hamer, J. E. (1996). Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell 8, 771–781 doi: 10.1105/tpc.8.5.771
Lee, K., Singh, P., Chung, W. C., Ash, J., Kim, T. S., Hang, L., et al. (2006). Light regulation of asexual development in the rice blast fungus, Magnaporthe oryzae. Fungal Genet. Biol. 43, 694–706. doi: 10.1016/j.fgb.2006.04.005
Li, M., Liu, X., Liu, Z., Sun, Y., Liu, M., Wang, X., et al. (2016). Glycoside hydrolase MoGls2 controls asexual/sexual development, cell wall integrity and infectious growth in the rice blast fungus. PLoS ONE 11:e0162243. doi: 10.1371/journal.pone.0162243
Li, Y., Zhang, Y. L., and Yan, H. G. (1996). Kinetic and thermodynamic characterizations of yeast guanylate kinase. J. Biol. Chem. 271, 28038–28044. doi: 10.1074/jbc.271.45.28038
Liechti, G., and Goldberg, J. B. (2012). Helicobacter pylori relies primarily on the purine salvage pathway for purine nucleotide biosynthesis. J. Bacteriol. 194, 839–854. doi: 10.1128/JB.05757-11
Liu, X. Y., Cai, Y. C., Zhang, X., Zhang, H. F., Zheng, X. B., and Zhang, Z. G. (2016). Carbamoyl phosphate synthetase subunit MoCpa2 affects development and pathogenicity by modulating arginine biosynthesis in Magnaporthe oryzae. Front. Microbiol. 7:2023. doi: 10.3389/fmicb.2016.02023
Mellersh, D. G., Foulds, I. V., Higgins, V. J., and Heath, M. C. (2002). H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. 29, 257–268. doi: 10.1046/j.0960-7412.2001.01215.x
Morrow, C. A., Valkov, E., Stamp, A., Chow, E. W. L., Lee, I. R., Wronski, A., et al. (2012). De novo GTP biosynthesis is critical for virulence of the fungal pathogen Cryptococcus neoformans. PLoS Pathog. 8:e1002957. doi: 10.1371/journal.ppat.1002957
Nomura, Y., Izumi, A., Fukunaga, Y., Kusumi, K., Iba, K., Watanabe, S., et al. (2014). Diversity in guanosine 3′,5′-bisdiphosphate (ppGpp) sensitivity among guanylate kinases of bacteria and plants. J. Biol. Chem. 289, 15631–15641. doi: 10.1074/jbc.M113.534768
Nyhan, W. L. (2005). Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab. 86, 25–33. doi: 10.1016/j.ymgme.2005.07.027
Oses-Ruiz, M., Sakulkoo, W., Littlejohn, G. R., Martin-Urdiroz, M., and Talbot, N. J. (2017). Two independent S-phase checkpoints regulate appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Proc. Natl. Acad. Sci. U.S.A. 114, E237–E244. doi: 10.1073/pnas.1611307114
Pettersson, J., Schrumpf, M. E., Raffel, S. J., Porcella, S. F., Guyard, C., Lawrence, K., et al. (2007). Purine salvage pathways among Borrelia species. Infect. Immun. 75, 3877–3884. doi: 10.1128/IAI.00199-07
Qi, Z. Q., Liu, M. X., Dong, Y. H., Yang, J., Zhang, H. F., Zheng, X. B., et al. (2016). Orotate phosphoribosyl transferase MoPyr5 is involved in uridine 5 '-phosphate synthesis and pathogenesis of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 100, 3655–3666. doi: 10.1007/s00253-016-7323-0
Rodriguez-Suarez, R., Xu, D., Veillette, K., Davison, J., Sillaots, S., Kauffman, S., et al. (2007). Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem. Biol. 14, 1163–1175. doi: 10.1016/j.chembiol.2007.09.009
Saint-Macary, M. E., Barbisan, C., Gagey, M. J., Frelin, O., Beffa, R., Lebrun, M. H., et al. (2015). Methionine biosynthesis is essential for infection in the rice blast fungus Magnaporthe oryzae. PLoS ONE 10:e011108. doi: 10.1371/journal.pone.0111108
Saunders, D. G. O., Aves, S. J., and Talbot, N. J. (2010). Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22, 497–507. doi: 10.1105/tpc.109.072447
Sekulic, N., Shuvalova, L., Spangenberg, O., Konrad, M., and Lavie, A. (2002). Structural characterization of the closed conformation of mouse guanylate kinase. J. Biol. Chem. 277, 30236–30243. doi: 10.1074/jbc.M204668200
Shimma, Y., Nishikawa, A., bin Kassim, B., Eto, A., and Jigami, Y. (1997). A defect in GTP synthesis affects mannose outer chain elongation in Saccharomyces cerevisiae. Mol. Gen. Genet. 256, 469–480. doi: 10.1007/s004380050591
Snoeijers, S., Pérez-García, A., Joosten, M. A. J., and De Wit, P. G. M. (2000). The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 106, 493–506. doi: 10.1023/A:1008720704105
Song, W. W., Dou, X. Y., Qi, Z. Q., Wang, Q., Zhang, X., Zhang, H. F., et al. (2010). R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS ONE 5:e13193. doi: 10.1371/journal.pone.0013193
Sunohara, K., Mitsuhashi, S., Shigetomi, K., and Ubukata, M. (2013). Discovery of N-(2,3,5-triazoyl)mycophenolic amide and mycophenolic epoxyketone as novel inhibitors of human IMPDH. Bioorg. Med. Chem. Lett. 23, 5140–5144. doi: 10.1016/j.bmcl.2013.07.016
Sweigard, J. A., Chumley, F. G., and Valent, B. (1992). Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 232, 183–190.
Talbot, N. J. (2003). On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. doi: 10.1146/annurev.micro.57.030502.090957
Talbot, N. J., Ebbole, D. J., and Hamer, J. E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. doi: 10.1105/tpc.5.11.1575
Tang, W., Ru, Y., Hong, L., Zhu, Q., Zuo, R., Guo, X., et al. (2015). System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of Magnaporthe oryzae. Environ. Microbiol. 17, 1377–1396. doi: 10.1111/1462-2920.12618
Valent, B., Farrall, L., and Chumley, F. G. (1991). Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127, 87–101.
Wilson, R. A., Fernandez, J., Quispe, C. F., Gradnigo, J., Seng, A., Moriyama, E., et al. (2012). Towards defining nutrient conditions encountered by the rice blast fungus during host infection. PLoS ONE 7:e47392. doi: 10.1371/journal.pone.0047392
Wilson, R. A., Gibson, R. P., Quispe, C. F., Littlechild, J. A., and Talbot, N. J. (2010). An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. U.S.A. 107, 21902–21907. doi: 10.1073/pnas.1006839107
Wilson, R. A., Jenkinson, J. M., Gibson, R. P., Littlechild, J. A., Wang, Z. Y., and Talbot, N. J. (2007). Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 26, 3673–3685. doi: 10.1038/sj.emboj.7601795
Wilson, R. A., and Talbot, N. J. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195. doi: 10.1038/nrmicro2032
Yan, X., Que, Y. W., Wang, H., Wang, C. C., Li, Y., Yue, X. F., et al. (2013). The MET13 methylenetetrahydrofolate reductase gene is essential for infection-related morphogenesis in the rice blast fungus Magnaporthe oryzae. PLoS ONE 8:e0076914. doi: 10.1371/journal.pone.0076914
Yan, X., and Talbot, N. J. (2016). Investigating the cell biology of plant infection by the rice blast fungus Magnaporthe oryzae. Curr. Opin. Microbiol. 34, 147–153. doi: 10.1016/j.mib.2016.10.001
Yi, M., Park, J. H., Ahn, J. H., and Lee, Y. H. (2008). MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 45, 1172–1181. doi: 10.1016/j.fgb.2008.05.003
Zeigler, R. S. (1998). Recombination in Magnaporthe grisea. Annu. Rev. Phytopathol. 36, 249–275. doi: 10.1146/annurev.phyto.36.1.249
Zeigler, R. S., Leong, S. A., and Teng, P. S. (1994). Rice Blast Disease. Wallingford, UK: CAB International.
Zhang, H., Tang, W., Liu, K., Huang, Q., Zhang, X., Yan, X., et al. (2011). Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 7:e1002450. doi: 10.1371/journal.ppat.1002450
Zhang, H., Zhao, Q., Liu, K., Zhang, Z., Wang, Y., and Zheng, X. (2009). MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol. Lett. 293, 160–169. doi: 10.1111/j.1574-6968.2009.01524.x
Zhang, H. F., Liu, K. Y., Zhang, X., Song, W. W., Zhao, Q., Dong, Y. H., et al. (2010). A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr. Genet. 56, 517–528. doi: 10.1007/s00294-010-0319-x
Zhang, H. F., Ma, H. Y., Xie, X., Ji, J., Dong, Y. H., Du, Y., et al. (2014). Comparative proteomic analyses reveal that the regulators of G-protein signaling proteins regulate amino acid metabolism of the rice blast fungus Magnaporthe oryzae. Proteomics 14, 2508–2522. doi: 10.1002/pmic.201400173
Zhang, H. F., Zheng, X. B., and Zhang, Z. G. (2016). The Magnaporthe grisea species complex and plant pathogenesis. Mol. Plant Pathol. 17, 796–804. doi: 10.1111/mpp.12342
Zhang, Y., Shi, H. B., Liang, S., Ning, G. A., Xu, N. C., Lu, J. P., et al. (2015). MoARG1, MoARG5,6 and MoARG7 involved in arginine biosynthesis are essential for growth, conidiogenesis, sexual reproduction, and pathogenicity in Magnaporthe oryzae. Microbiol. Res. 180, 11–22. doi: 10.1016/j.micres.2015.07.002
Keywords: guanylate kinase, GTP biosynthesis, conidiogenesis, appressorial formation, infectious growth
Citation: Cai X, Zhang X, Li X, Liu M, Liu X, Wang X, Zhang H, Zheng X and Zhang Z (2017) The Atypical Guanylate Kinase MoGuk2 Plays Important Roles in Asexual/Sexual Development, Conidial Septation, and Pathogenicity in the Rice Blast Fungus. Front. Microbiol. 8:2467. doi: 10.3389/fmicb.2017.02467
Received: 28 February 2017; Accepted: 28 November 2017;
Published: 11 December 2017.
Edited by:
Kostya Kanyuka, Rothamsted Research (BBSRC), United KingdomReviewed by:
Marc-Henri Lebrun, Institut National de la Recherche Agronomique (INRA), FranceZonghua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2017 Cai, Zhang, Li, Liu, Liu, Wang, Zhang, Zheng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haifeng Zhang, aGZ6aGFuZ0BuamF1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Xingjia Cai†
Xingjia Cai† Xinyu Liu
Xinyu Liu Haifeng Zhang
Haifeng Zhang Zhengguang Zhang
Zhengguang Zhang