
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 November 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02255
This article is part of the Research Topic Rhizosphere Bacterial Community and Bacteria-Plant Interactions View all 14 articles
Rhizobia induce root nodules and fix atmospheric N2 for most legume species in exchange for carbon. However, the diverse endophytic non-rhizobial bacteria in legume nodules that co-exist with rhizobia are often ignored because they are difficult to cultivate using routine cultivation approaches. To enhance our understanding of the incidence and diversity of legume–bacteria associations, a high-throughput sequencing analysis of bacterial 16S rRNA genes was used to examine the bacterial community in the rhizospheres and root nodules of Dalbergia odorifera seedlings that were uninoculated or inoculated with Bradyrhizobium elkanii H255, Rhizobium multihospitium–like HT221, or Burkholderia pyrrocinia–like H022238, in two growth media (nitrogen [N]-supplied soil or N-omitted potting mix). Seedlings inoculated with Bradyrhizobium had significantly more nodules than seedlings in the other inoculation conditions, regardless of growth media. Using the 15N natural abundance method, it was shown that the inoculated plants had significantly higher N2 fixation efficiency (48–57%) and specific nodule activity [269–313 μg N mg−1 of dry weight (dwt) nodule] compared to the uninoculated plants (203 μg N mg−1 dwt nodule). The 16S rRNA gene analysis showed that there was generally a higher bacterial diversity in the rhizosphere than in the nodules in the corresponding condition. Both rhizobial inoculation and media status significantly altered the bacterial communities in the rhizospheres and nodules (P < 0.05), with the exception of the inoculated soil rhizospheres. Regarding non-rhizobial bacteria, three genera, i.e., Lactococcus, Bacillus, and Pseudomonas, were consistently enriched in the rhizosphere and Bradyrhizobium, Chloroplast norank (which belongs to Cyanobacteria), and Lactococcus were commonly found in the nodules. In contrast, common rhizobial genera (including Rhizobium, Mesorhizobium, and Burkholderia) were only present in the nodules at low relative abundances (0.01–3.41%). Regarding non-rhizobial bacteria, 32 genera were found in the nodules, with non-rhizobial bacteria being predominant in the N omitted potting mix (with a relative abundance of 56–87%). This study suggests that legume nodules are inhabited by a high diversity of non-rhizobial bacteria, which may play a vital role in nodulation and N2 fixation in the host plants.
Plants grow in intimate association with heterotrophic microorganisms, and they provide carbohydrates for the growth of these microorganisms. Reciprocally, some types of microbes [including free-living nitrogen (N2) fixers, intercellular endophytic bacteria, and symbiotic bacteria] are able to fix atmospheric N2 in exchange for carbon from their plant hosts (Mus et al., 2016; Sprent et al., 2017). To adapt to this habitat, a limited range of microbes, e.g., endophytes and symbionts, have evolved the ability to colonize external or internal plant tissues with little or no host damage (Bulgarelli et al., 2013).
The N2-fixing symbiosis between most legumes and bacteria is a well-studied example of the formation of root nodules, and occasionally stem nodules, which are induced and subsequently invaded by specific rhizobia. Phylogenetic studies have shown that, based on a diverse range of legume species, over 150 rhizobial species belonging to 12 genera of Alphaproteobacteria and two genera of Betaprotebacteria occupied the root nodules tested (Peix et al., 2015; Zahran, 2017). In various legumes, in addition to rhizobia that are responsible for nodulation and N2 fixation, other endophytic bacteria, called non-rhizobial bacteria, are also found in the nodules (Sachs and Simms, 2008; Wu et al., 2011; Busby et al., 2016). For example, 71 genera, including Dyella, Enterobacter, Pseudomonas, and Steroidobacter, were observed in Lespedeza nodules (Busby et al., 2016); eight genera comprising Paenibacillus, Bacillus, Klebsiella, Ensifer, Agrobacterium, Blastobacter, Dyadobacter, and Chitinophaga were isolated from the nodules of field-grown Vigna radiate (Pandya et al., 2013); and a wide variety of non-rhizobial bacteria (e.g., Agrobacterium, Enterobacter, Paenibacillus, and Phyllobacterium) can colonize Glycyrrhiza nodules (Li et al., 2012). Some of these non-rhizobial bacteria have proven beneficial to their legume hosts, as they enhance plant growth by producing plant hormones, fixing atmospheric N2, and solubilizing phosphate (Peix et al., 2015). Nevertheless, these non-rhizobial bacteria have been disregarded in the past during the isolation of rhizobia from legumes [using the traditional cultivation method involving the use of yeast-mannitol-based agar (YMA)]. This led to a lack of information on the true diversity of non-rhizobial bacteria coexisting with rhizobia in the nodules, and on their possible essential roles in nodulation and N2 fixation. Current metagenomic approaches can be used to shed light on the situation by allowing the detection of both rhizobia and non-rhizobial bacteria in nodules. This approach may help to accurately determine the roles of bacterial species in the nodulation and N2 fixation processes.
Dalbergia odorifera T. Chen is one of the extremely precious rosewoods that are native to Southern China and that have both medicinal and commercial value (Ma et al., 2013; Lee et al., 2014). When it is effectively fixing N2, D. odorifera has been shown to be a suitable host for the hemiparasite Santalum album (Indian sandalwood) (Lu et al., 2013, 2014). Unlike other species of Dalbergia, D. odorifera is distinguished by its capacity to form N2-fixing nodules after the invasion of rhizobia (Gao et al., 1994; Rasolomampianina et al., 2005; Lu et al., 2011, 2012). We recently isolated a number of bacterial strains (belonging to Agrobacterium, Bradyrhizobium, Ensifer, Rhizobium, and Burkholderia) from D. odorifera nodules obtained from various locations in Southern China (Lu et al., 2011, 2012). Identification of bacteria in nodules has traditionally relied on their cultivability when streaked on YMA plates. Thus, in the past, complete information on the diversity of uncultured rhizobia and non-rhizobial bacteria was not obtained.
This study used three strains of rhizobia (originally isolated from D. odorifera) to inoculate D. odorifera seedlings, and the effect of the inoculation on host performance and N2 fixation was assessed. We hypothesized that (i) inoculation influences host N2 fixation; (ii) inoculation has a significant impact on bacterial communities in both the rhizosphere and nodules; and (iii) non-rhizobial bacteria are relatively abundant compared to rhizobia. The outcomes of this study could improve our understanding of legume–bacteria associations.
A randomized split-plot design was used, which involved four inoculation conditions (non-inoculation and inoculation with three rhizobial strains) as the first factor and two growth media [N-supplied soil (N+) and N-omitted potting mix (N−)] as the second factor. For each treatment combination, six replicates (plants) were assessed in a greenhouse study.
Three rhizobial strains, Bradyrhizobium elkanii (H255), Rhizobium multihospitium–like (HT221), or Burkholderia pyrrocinia–like (H022238), isolated from the root nodules of D. odorifera obtained from various geographic locations in China, were used in this inoculation experiment (Table 1). The GenBank accession numbers for the 16S rRNA, recA, and nodC gene sequences are listed in Table 1. Phylogenetic analysis of the 16S rRNA gene showed that strains H255, HT221, and H022238 were most closely related to Bradyrhizobium elkanii USDA76T (100%), Rhizobium miluonense CCBAU41251T (99.2%) and Burkholderia pyrrocinia JK-SH007 (98.4%), respectively (Figure S1).
Bacterial suspensions (107-109 cfu/ml in sterilized 0.75% NaCl) of each of the three strains cultivated for 3–5 days on yeast-mannitol media in 500 ml triangular flasks at 28°C (centrifuged at 4,000 rpm, 5 min) were used as the rhizobia inocula in this study.
The experiment was carried out in a temperature-controlled glasshouse at the Research Institute of Tropical Forestry, Chinese Academy of Forestry, Guangzhou, China (23°11′N, 113°23′E), with day/night temperatures of 31/23°C and a relative humidity of 68/80%. Seeds of D. odorifera were surface-sterilized with 3% sodium hypochlorite for 5 min, and then washed three times with distilled water. The seeds were then incubated on sterile moistened filter paper in Petri dishes at 28°C in the dark until germination (~3 weeks).
Germinated D. odorifera seedlings with 2-cm radicals were soaked in the rhizobia preparations for 5 min before transplanting them into pots. A further 2 ml/pot of the rhizobia inoculum was placed around the taproot area 3 days after transplantation. Regarding the uninoculated condition, the seedlings were administered the same quantity of sterilized 0.75% NaCl solution.
Two media conditions were used to study N2 fixation efficiency and to compare the bacterial communities in different growth media conditions. The seedlings were transplanted into plastic pots (maximum diameter, 14 cm, depth, 12 cm). For the N+ condition, the pots were filled with 1 kg field soil (Utisol) that was collected from a forest within the grounds of the Research Institute of Tropical Forestry (the soil contained 5.16 g·kg−1 organic matter, 0.30 g·kg−1 total N, 0.15 g·kg−1 total P, and 7.19 g·kg−1 total K, and it had a pH of 4.5 in deionized water). For the N− condition, the pots were filled with 250 g potting mix (vermiculite: perlite, 2:1, v/v, pH 6.5), which was organic-matter free and had almost no N.
The plants were grown for 8 months (December 2013 to August 2014). Each pot was watered three times per week with tap water and fertilized monthly with 50 ml N-free Jensen nutrient solution (Burdon et al., 1999). For the N+ condition, 1 ml 0.5% (NH4)2SO4 solution was supplied monthly.
The N2 fixation efficiency was calculated using Equation 1 (Knowles and Blackburn, 1993):
where B is the δ15N value of the nodulated N2-fixing plants grown under N-free conditions. Two local indigenous woody plants, the first being Bischofia polycarpa (Levl.) Airy Shaw and the second being Michelia macclurei Dandy, were chosen as the non-N2-fixing reference plants to calculate the averaged foliar δ15N values for the equation.
Eight months after transplantation, the leaves and nodules of D. odorifera, along with the rhizosphere soils, were collected. The plant leaves were oven-dried (70°C for 72 h) and ground in a ball mill (Retsch GmbH & Co. KG, Haan, Germany) to analyze the total N and δ15N using a spectrometer (Isotope Ratio Mass Spectrometer, Thermo Fisher Scientific Inc., Waltham, MA, USA).
For each treatment combination, fresh nodules (0.5 g) from three plant replicates were surface sterilized (in 95% ethanol for 45 s followed by immersion in 1% HgCl2 for 3 min), washed with sterile water, and then stored at −80°C until DNA extraction.
For each treatment combination, the total genomic DNA was extracted from 0.5 g of nodule and rhizosphere samples from three plant replicates using an E.Z.N.A. soil DNA kit (Omega Biotek, Norcross, GA, USA) following the manufacturer's protocol. The presence of DNA was evaluated by carrying out electrophoresis on 1% agarose gel (Biowest, Barcelona, Spain) with a 1-kb DNA ladder (Pomega, Madison, WI, USA). The DNA concentration was determined using a spectrophotometer (Nanodrop 2000, Thermo Fisher Scientific), and samples with values >0.1 μg/μl were used in subsequent analyses. Extracted DNA were used as templates for subsequent PCR. As there were eight treatment combinations and two replicates per treatment combination, there were 16 rhizosphere and 16 nodule samples, comprising a total of 32 samples.
The DNA was PCR-amplified using universal bacterial primers: 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). These primers were selected to amplify the V4+V5 variable region of the 16S rRNA gene in the bacteria, creating an amplicon of ~390 bp.
Each PCR mixture contained 0.8 μl of each primer (5 μM), 2 μl 2.5 mM deoxynucleotide triphosphates (dNTPs; BBI, Ontario, Canada), 0.4 μl FastPfu polymerase (TransGen Biotech, Beijing, China), 10 ng genomic DNA, and 4 μl 5 × FastPfu Buffer (TransGen Biotech, Beijing, China), with a final reaction volume of 20 μl. The amplifications were carried out using GeneAmp 9700 PCR (Applied Biosystems, Carlsbad, CA, USA) using the following program: 95°C for 3 min, then 27 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, ending with 72°C for 10 min.
The amplicons were extracted using 2% agarose gels and purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer's instructions, and they were then quantified using QuantiFluor™-ST (Promega, Madison, WI, USA). The purified amplicons were pooled in equimolar quantities and paired-end sequenced (2 × 250) using an Illumina MiSeq platform according to the standard protocols. The raw reads were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database (BioProject ID: PRJNA322091).
All the sequence reads were checked for quality. Using QIIME (version 1.17), poor-quality reads and primer dimers were discarded. Using Usearch (version 7.1), operational taxonomic unit (OTU) clustering was performed, with a 97% similarity threshold. Singletons and chimeric sequences were filtered out. The taxonomy of each 16S rRNA gene sequence was analyzed using the RDP Classifier (http://rdp.cme.msu.edu/) against the SILVA (SSU115) 16S rRNA database, based on a confidence threshold of 70% (Amato et al., 2013). To reduce the influence of sequencing depth on the estimated effect sizes, a subsample of 10,461 sequences was selected for each sample.
The effects of the rhizobial inoculation and media conditions were assessed using two-way analysis of variance (ANOVA). Where the effects were significant, a Tukey–Kramer honestly significant difference (HSD) test was used for pairwise comparisons of the means. One-way ANOVA was used to compare the effects of rhizobial inoculation on N2 fixation efficiency and specific nodule activity. A P < 0.05 was considered significant. The data were analyzed using PASW Statistics software version 18 (SPSS Inc., Chicago, IL, USA).
Using QIIME, an Adonis analysis based on Bray–Curtis dissimilarity was used to evaluate the significance of the differences in the bacterial communities between the treatment combinations and between each pair of samples in a given treatment combination group. For each treatment combination, the mean number of OTUs of the two replicates was used for the subsequent bioinformatic analyses. Diversity indexes [abundance-based coverage estimator (ACE), Chao1, and the Shannon index] were assessed using MOTHUR. The relative bacterial abundances and a cluster tree were generated, and a principal component analysis (PCA) was performed, using the R statistical platform (version 3.2.4).
In both media conditions, reddish-brown nodules were observed on the roots of D. odorifera (Figure 1). In general, significantly more nodules were observed on N− seedlings than on the N+ seedlings (P < 0.01) (Table S1), with the exception of those in the Burkholderia inoculation condition (Table 2). Irrespective of media status, D. odorifera inoculated with Bradyrhizobium had significantly more nodules than the plants in the other inoculation conditions (P < 0.01). Plants inoculated with Burkholderia had significantly lower foliar δ15N values than those in the other inoculation conditions, regardless of media status (P < 0.01).
For each media condition, the tissue N concentrations were similar between inoculation conditions (Table 2). The N+ seedlings always had a higher tissue N concentration than the N− seedlings. Among the plants in the four inoculation conditions, N2 fixation supplied 45–57% of the total N requirement, and the specific nodule activity was 203–313 μg N mg−1 of dry weight (dwt) nodule (Table 2). Significantly higher N2 fixation efficiency and specific nodule activity were observed in the inoculated plants compared to the uninoculated plants (P < 0.05) (Table S2).
The 32 samples collected from the rhizospheres and nodules provided a total of 878,192 raw reads. After quality filtering and chimera removal, 854,805 reads remained to be analyzed, with a mean length of 248 bp. The number of sequences per sample varied from 14,871 to 39,015. Using a 97% sequence similarity threshold, 2,949 bacterial OTUs were identified after randomly resampling the 16S rRNA gene sequence reads to the same depth (10,461 sequences per sample). However, this sequencing depth was not sufficient to assess the vast diversity of the bacterial communities in this study (Figure S2). The 32 samples exhibited a clear and significant (P = 0.001, Adonis) separation. The OTU data on the nodules differed significantly (P = 0.01–0.03, Adonis) from the data on the rhizospheres, regardless of the inoculation or media conditions. Rhizobial inoculation significantly influenced the nodule and rhizosphere bacterial communities (P = 0.01–0.04, Adonis), except in the N+ rhizosphere samples (P = 0.10, Adonis).
The ACE and Chao1 richness estimators, Goods' coverage, and Shannon index were calculated based on a 3% genetic distance for each sample (Table 3). The Chao1 estimator revealed a significant difference in bacterial community richness between the rhizosphere and nodule samples. Indeed, the estimated number of OTUs was 1.90–2.97 times lower in nodules (with means of 322 and 405 OTUs for the N+ and N− nodules, respectively) than in the corresponding rhizospheres (with means of 899 and 703 OTUs for the N+ and N− rhizospheres, respectively). The rhizosphere diversity indices (ACE, Chao1, and Shannon index) were generally higher than those for the nodule samples, with one exception: the N+ nodules in the uninoculated condition had a similar Shannon index to the corresponding rhizospheres. Rhizobial inoculation showed no clear pattern regarding bacterial community richness.
The distribution of phyla and genera in the nodules and rhizospheres subjected to the eight treatment combinations are presented in Figure 2. Ten phyla, i.e., Acidobacteria, Actinobacteria, Armatimonadetes, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, Planctomycetes, Proteobacteria, and Verrucomicrobia, and 32 genera were identified. The genera were Acetobacter, Acinetobacter, Aquicella, Armatimonas, Arthrobacter, Bacillus, Bradyrhizobium, Caulobacter, Chloroplast norank, Comamonas, Devosia, Dyella, Enhydrobacter, Escherichia, Gaiellales norank, Lactococcus, Lysinibacillus, Mesorhizobium, Mucilaginibacter, Mycobacterium, Opitutus, Phenylobacterium, Piscinibacter, Planctomyces, Pseudomonas, Psychrobacter, Reyranella, Rhodocyclus, Shigella, Solibacillus, Streptococcus, and Xanthomonas.
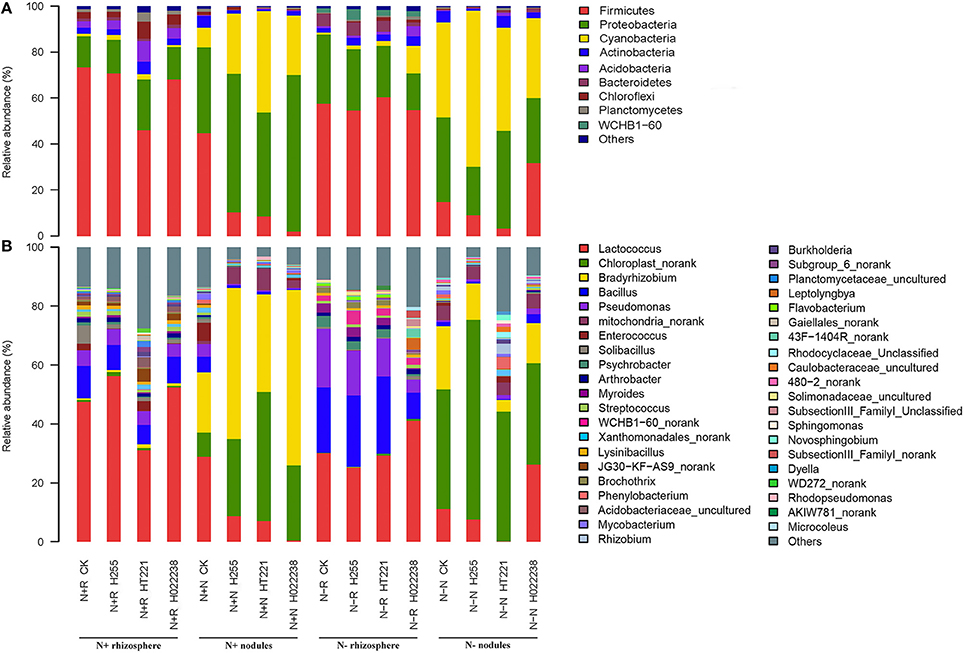
Figure 2. Relative abundance of the dominant phyla (A) and genera (B) in the rhizosphere and nodules of D. odorifera seedlings. N + R and N–R: D. odorifera rhizospheres in N-supplied soil or N-omitted potting mix, respectively; N + N and N–N: D. odorifera nodules in N-supplied soil or N-omitted potting mix, respectively; CK, no inoculation; H255, HT221, and H022238, inoculation with Bradyrhizobium elkanii H255, Rhizobium multihospitium–like HT221, and Burkholderia pyrrocinia–like H022238, respectively.
The relative abundances of three abundant bacterial phyla (Firmicutes, Proteobacteria, and Cyanobacteria) differed significantly between the rhizosphere and nodule samples. Firmicutes were significantly enriched in the rhizospheres (relative abundance, 46–73%) compared to the corresponding nodules (2–45%). In contrast, Proteobacteria and Cyanobacteria were significantly enriched in the nodules (21–68% and 9–68%, respectively) compared to the corresponding rhizospheres (14–30% and 1–12%, respectively).
Overall, the most abundant genus in the rhizosphere was Lactococcus (31–56% of total sequences for N+ and 25–41% for N−). The Bradyrhizobium genus was dominant in the N+ nodules, corresponding to 51% and 60% of total sequences in the N+ Bradyrhizobium and N+ Burkholderia nodules, respectively. The Chloroplast norank group was dominant in N+ Rhizobium nodules (44%), and it was also the most prevalent group in the N− nodules, corresponding to 68, 44, 34, and 41% in the Bradyrhizobium, Rhizobium, Burkholderia, and uninoculated nodules, respectively. In addition, the Lactococcus genus was always enriched in the N+ uninoculated nodules.
Interestingly, three α-rhizobia (Bradyrhizobium, Mesorhizobium, and Rhizobium) and two β-rhizobia (Burkholderia and Cupriavidus) were detected in most of the rhizosphere and nodule samples. The exceptions involved Cupriavidus, which was absent from the nodules in the N+ media inoculated with Bradyrhizobium and Rhizobium conditions, and the N− media inoculated with Burkholderia condition; and Rhizobium, which was absent from the rhizospheres in the N+ media inoculated with Bradyrhizobium condition.
At the species level, the non-rhizobial Lactococcus piscium was the dominant species in the rhizosphere, regardless of media status. Overall, in the 32 samples, the most abundant rhizobia species was Bradyrhizobium elkanii, with a relative abundance of 0.5–1.3% and 20–60% in the N+ rhizosphere and nodules, respectively, and 0.02–0.12% and 12–40% in the N− rhizosphere and nodules, respectively (Table 4). Unclassified Rhizobium and Mesorhizobium plurifarium were also found in each rhizosphere sample (0.02–0.11%) and nodule sample (0.03–3.41%), with the exception that no Unclassified Rhizobium was detected in the N+ Bradyrhizobium rhizospheres (Table 4). However, Unclassified Bradyrhizobium was only found in the nodules (0.01–0.03%). Rhizobium huautlense was detected in both N− Bradyrhizobium nodules and rhizospheres (≤ 0.03% for both). Paraburkholderia mimosarum was found in all N+ rhizospheres (0.01–0.04%), and in the N+ nodules inoculated with Rhizobium and Burkholderia (0.01 and 0.04%, respectively) (Table 4). Nevertheless, these rhizobia (including Unclassified Bradyrhizobium and Rhizobium, Mesorhizobium plurifarium and P. mimosarum) were not dominant species in all nodules. A species of Cyanobacteria, which was previously isolated from Phaseolus acutifolius (the Tepary bean), was the dominant species in all the N− nodules (34–68%).
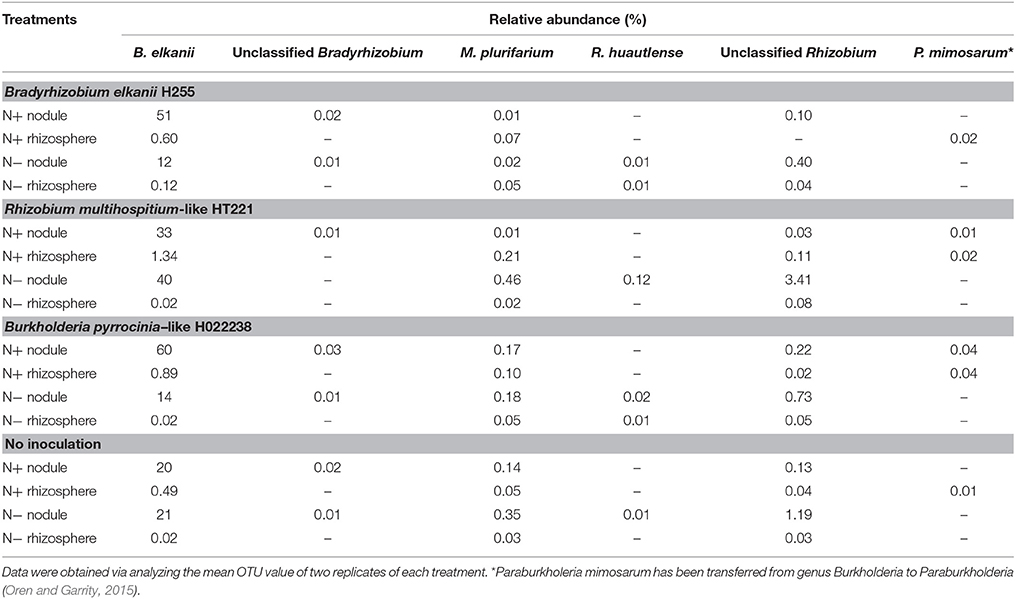
Table 4. Relative abundance of common rhizobia detected in the rhizosphere and root nodule samples of D. odorifera seedlings.
Across all 32 samples, 28 OTUs and 125,564 reads were shared, at a 3% genetic distance (Table 5). The N+ rhizosphere samples shared the greatest number of OTUs (356), most of which belonged to the genus Lactococcus (Table 5). Among the N− rhizosphere samples, 221 OTUs were shared. Among the N+ and N− nodule samples, 96 and 103 OTUs were shared, respectively (Table 5).
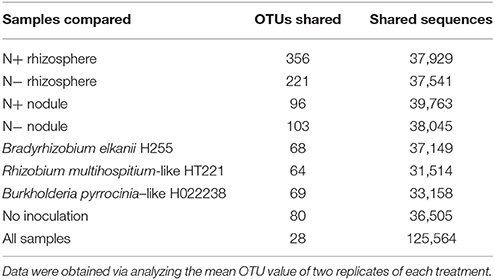
Table 5. The number of shared OTUs and sequences across different samples at a 3% genetic distance generated using a shared OTU table.
Regarding inoculation, the uninoculated group shared 80 OTUs, which was higher than the three inoculated groups (Table 5). The Bradyrhizobium, Rhizobium, and Burkholderia groups shared 68, 64, and 69 OTUs, with 37,149, 31,514, and 33,158 sequences, respectively (Table 5).
The clustering of the 32 samples was evaluated using unweighted Unifrac (Figure 3). The rhizosphere and nodule samples could be separated clearly. Regardless of rhizobial inoculation or media status, the different treatment combination groups of rhizospheres and nodules were each clustered independently.
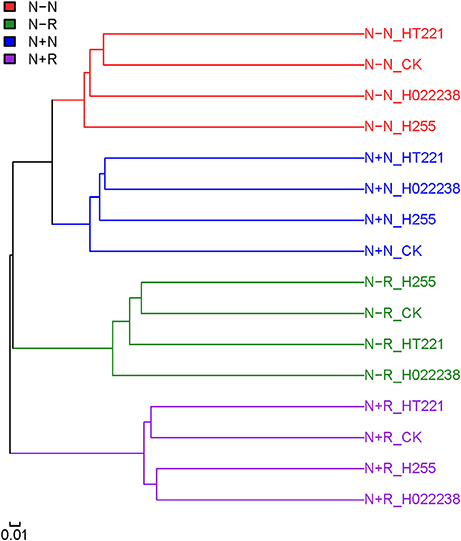
Figure 3. Clustering of rhizosphere and nodule samples of D. odorifera seedlings. Based on the abundance of operational taxonomic units (OTUs), an unweighted Unifrac test was performed using QIIME software to verify the sample structure via clustering. Refer to Figure 2 for treatment combination details.
The PCA of the 16 treatment combinations showed that the first principal component explained 71.5% of the variance in the OTU data (Figure 4). The PCA demonstrated that the treatment combination-specific bacterial samples could be separated using the OTU data, indicating that the 16 treatment combinations produced significantly different bacterial compositions. Specifically, according to the PCA, OTU4279 (Lactococcus) and OTU1501 (Bacillus) were enriched in the rhizospheres, while OTU1702 (Chloroplast norank) and OTU898 (Bradyrhizobium) were more abundant in the nodules (Figure 4).
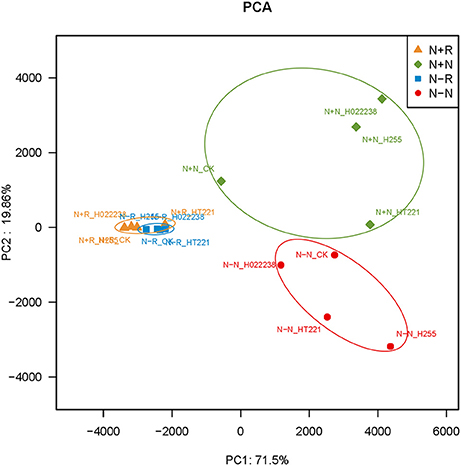
Figure 4. Principal components analysis of operational taxonomic unit (OUT) abundance data for the rhizosphere and nodules of D. odorifera seedlings. Refer to Figure 2 for treatment combination details.
Our study used a high-throughput sequencing approach to determine the bacterial community compositions in the rhizosphere and root nodules of D. odorifera seedlings under four inoculation conditions with two growth media. The experiment permitted us to examine the influence of rhizobial inoculation on bacterial communities in rhizospheres and root nodules under greenhouse conditions. Due to the small nodule number, the nodule samples were not sufficient for the DNA extraction-related analyses. We only had two replicates from 6 plants in each treatment condition for the high-throughput sequencing analysis, and so our data must be considered with caution. However, this process allowed us to assess rhizobial biodiversity associated with legume–rhizobium mutualism.
To assess the influence of rhizobial inoculation on nodulation and N2 fixation in D. odorifera seedlings, this study used three rhizobial strains as inocula, which led to enhanced N2 fixation efficiency compared with the uninoculated condition (Table 2). The seedlings inoculated with Bradyrhizobium had significantly more nodules than those inoculated with the other two inocula (Table 2). Bradyrhizobium has been reported to be an efficient microsymbiont with respect to most legumes, and it is widely acknowledged to be an ecologically and economically important organism (Parker, 2003; Rasolomampianina et al., 2005; Yao et al., 2014; VanInsberghe et al., 2015; Sprent et al., 2017).
The N− potting mix condition induced more nodules than the N+ soil condition (Table 2). Soil N concentration together with other soil properties, including pH and texture, played a vital role in influencing nodulation in this study. Previous research has revealed that high rates of rhizobial nodulation and N2 fixation occur in plants in low-N environments (Menge et al., 2015), and the N concentration is known to have a major impact on the symbiosis between rhizobia and host plants. In addition, soil pH could be an important factor influencing legume nodulation and N2 fixation (Zahran, 1999). Most legumes require neutral or slightly acidic soil for growth. This is because soil acidity limits rhizobial sustainability, and ultimately their survival, in soils, and thus it reduces nodulation, constrains symbiotic N2 fixation, and even shifts gene expression related to various cellular functions in the plant host (Hellweg et al., 2009; Ferguson et al., 2013; Abd-Alla et al., 2014). In the present study, we assume that the media pH could be the determinant factor that influenced nodulation in this pot study, although different media and N levels may also have had some effects on nodulation. Nevertheless, the influences of N status are also worthy of further study.
The 15N natural abundance method is an effective approach for estimating N2 fixation by potted plants and in agroforestry ecosystems (Unkovich et al., 2008; Denton et al., 2013; Peoples et al., 2015). In our study, 45–57% of the tissue N in the 8-month-old nodulated D. odorifera seedlings was derived from N2 fixation (Table 2), with the rest being derived from the seeds and growth media, as previously described by Unkovich et al. (2008). These N2 fixation rates are comparable to those calculated in our previous study, in which 41–44% of tissue N in 6-month-old D. odorifera seedlings was derived from fixation (Lu et al., 2013).
In general, microbial inoculation can change, at least temporarily, the native bacterial community in the soil (Schlaeppi and Bulgarelli, 2015). Bacterial communities in the soil consist of a complex mix of endemic and newly introduced taxa, with a degree of genetic exchange between rhizobia and other soil bacteria (Nandasena et al., 2007). Our study showed that rhizobial inoculation altered the bacterial communities in the rhizosphere and nodules regardless of growth media status, with a particularly high Shannon index for nodules in the N+ inoculated conditions (Table S3). Differences in the physiology of bacteria (including their ability to establish diverse bacteria–bacteria interactions) influence the composition of microbial communities in the soil, rhizosphere, and legume nodules (Masson-Boivin et al., 2009; Peiffer et al., 2013; Peix et al., 2015; Shi et al., 2015). Denton et al. (2013) demonstrated that inoculation of faba beans with sufficient rhizobia can increase N2 fixation in the faba beans in the presence of massive quantities of soil rhizobia, suggesting that inoculant rhizobia and soil bacteria might form an association for effective nodulation and N2 fixation. However, little is known about the buffering capacity of the soil and plant microbiota. Hence, understanding the persistence of inoculated bacteria in the field and their effects on the native microbial communities has theoretical and practical significance with respect to the use of microbial inoculants in agroforestry.
Microbes in the rhizosphere play key roles in the growth and ecological fitness of host plants. Common physiological processes exhibited by these microbes include phytopathogenic activity, phytohormone production, atmospheric N2 fixation, geochemical mineral cycling, and plant colonization (Buée et al., 2009; Peiffer et al., 2013). Members of the phyla Firmicutes and Proteobacteria are well-known rhizosphere colonizers and have generally been characterized as fast-growing r-strategists that respond positively to plant root exudates (e.g., low-molecular-weight substrates) (Shi et al., 2015). This is consistent with our results, which showed that a few taxa were consistently enriched in the rhizosphere of D. odorifera regardless of media status, such as the phyla Firmicutes and Proteobacteria, and the genera Lactococcus, Bacillus, and Pseudomonas. The genus Lactococcus has been shown to be enriched in the rhizosphere, seed and rice silage (Ennahar et al., 2003; Fhoula et al., 2013; Adam et al., 2016), and bacteria in this genus are known to produce biological control agents (see Adam et al., 2016). Recent studies of lettuce and sweet potato have also noted enrichment of Bacillus and Pseudomonas in the rhizosphere (Marques et al., 2014; Schreiter et al., 2014), both of which play an important role in plant health by suppressing plant pathogens (Santoyo et al., 2012).
Using a traditional cultivation method, our recent research verified that D. odorifera can establish N2-fixing symbiosis with Bradyrhizobium, Ensifer, Rhizobium, and Burkholderia (Lu et al., 2011, 2012), suggesting that D. odorifera can efficiently establish symbioses with a wide range of rhizobial species. A study by Rasolomampianina et al. (2005) also showed that several Dalbergia species endemic in Madagascar were able to form symbioses with bacteria in the genera Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Phyllobacterium (which belong to the Alphaproteobacteria class), as well as with bacteria in the genera Burkholderia and Ralstonia (which belong to the Betaproteobacteria class) (Rasolomampianina et al., 2005). In the present study, the genera enriched in the nodules included Bradyrhizobium, Chloroplast norank (belonging to Cyanobacteria), Lactococcus, Mycobacterium, and Bacillus, whereas common rhizobia genera, such as Rhizobium, Mesorhizobium, and Burkholderia were found at low relative abundances in the nodules (Figure 2). This study, which relied on a metagenomic approach, provides the first report on the full diversity of bacteria in D. odorifera nodules, especially regarding large relative abundance of Cyanobacteria.
Legume nodules represent a distinctive ecological niche, with an adapted program for the accommodation of compatible soil microbes. Apart from rhizobia, legume nodules are often occupied by a phylogenetically diverse bacterial microbiome. These co-evolved bacteria may impact plant growth and health or even nodule formation and N2 fixation, but their ecological roles remain unknown (Bai et al., 2003; Sachs and Simms, 2008; Buée et al., 2009; Peix et al., 2015). Although the bacterial diversity in root nodules was significantly lower than that in the corresponding rhizospheres, the species diversity and relative abundance of non-rhizobial bacteria in the nodules were greater than expected. The most abundant rhizobia were distributed among five genera, with Bradyrhizobium being the predominant genus. In addition to the abovementioned rhizobia, 32 genera of non-rhizobial bacteria were found in all the D. odorifera nodule samples. These results indicate the coexistence of diverse non-rhizobial bacteria with rhizobia in the D. odorifera nodules in this study.
Non-rhizobial bacteria are considered to be competent to infect and multiply within legume nodules. However, it has been assumed that rhizobia symbionts dominate the interior of the nodules, with an adapted ability to outcompete other bacteria and to efficiently communicate with the host during infection (Zgadzaj et al., 2015). In contrast, Sachs and Simms (2008) found that non-rhizobial bacteria may dominate, encompassing up to 99% of the total bacterial population in legume nodules. In our study, all the nodules of the N- samples contained non-rhizobial bacteria as the predominant population (56–87% of the total bacterial population), while nearly half of the bacterial population in the nodules of the N+ samples were non-rhizobial bacteria. The ubiquity of non-rhizobia taxa raises the question of whether they exploit the opportunity to invade nodules as an adaptive strategy. Some researchers believe that the non-rhizobial bacteria might invade nodules that are formed by nodulating rhizobia (Wu et al., 2011; Mus et al., 2016), which have improved fitness compared to other soil bacteria or a better ability to communicate with the legume host (Zgadzaj et al., 2015). However, recent studies have shown that non-rhizobial bacteria in nodules are not passive players (Zgadzaj et al., 2015; Gano-Cohen et al., 2016). They may even control the host plant to some degree, using exopolysaccharides to subtly influence the host during their chronic infection of the nodules (Zgadzaj et al., 2015). In addition, some non-rhizobial bacteria help rhizobium to extend their host range (Liu et al., 2010) and improve the nodulation and N2 fixation of legume–rhizobia symbionts (Peix et al., 2015). In contrast to these positive effects, some non-rhizobial bacteria may be able to reduce the fitness of nodulating rhizobia via competitive exclusion in the rhizosphere (Gano-Cohen et al., 2016).
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors acknowledge the support provided by the Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (CAFYBB2017ZB001), the National Natural Science Foundation of China (31170582, 31722012, and 31471946), and the Chinese Academy of Sciences (“100 Talent” Program, A315021449).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02255/full#supplementary-material
Abd-Alla, M. H., Issa, A. A., and Ohyama, T. (2014). “Impact of harsh environmental conditions on nodule formation and dinitrogen fixation of legumes,” in Advances in Biology and Ecology of Nitrogen Fixation, ed T. Ohyama (Rijeka: InTech.), 978–953.
Adam, E., Bernhart, M., Müller, H., Winkler, J., and Berg, G. (2016). The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil doi: 10.1007/s11104-016-3113-9. [Epub ahead of print].
Amato, K. R., Yeoman, C. J., Kent, A., Righini, N., Carbonero, F., Estrada, A., et al. (2013). Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353. doi: 10.1038/ismej.2013.16
Bai, Y., Zhou, X., and Smith, D. L. (2003). Enhanced soybean plant growth resulting from coinoculation of strains with Bradyrhizobium japonicum. Crop Sci. 43, 1774–1781. doi: 10.2135/cropsci2003.1774
Buée, M., De Boer, W., Martin, F., Van Overbeek, L., and Jurkevitch, E. (2009). The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321, 189–212. doi: 10.1007/s11104-009-9991-3
Bulgarelli, D., Schlaeppi, K., Spaepen, S., van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Burdon, J. J., Gibson, A. H., Searle, S. D., Woods, M. J., and Brockwell, J. (1999). Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: within-species interactions. J. Appl. Ecol. 36, 398–408. doi: 10.1046/j.1365-2664.1999.00409.x
Busby, R. R., Rodriguez, G., Gebhart, D. L., and Yannarell, A. C. (2016). Native Lespedeza species harbor greater non-rhizobial bacterial diversity in root nodules compared to the coexisting invader, L. Cuneata. Plant Soil 401, 427–436. doi: 10.1007/s11104-015-2763-3
DeLong, E. F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U.S.A. 89, 5685–5689. doi: 10.1073/pnas.89.12.5685
Denton, M. D., Pearce, D. J., and Peoples, M. B. (2013). Nitrogen contributions from faba bean (Vicia faba L.) reliant on soil rhizobia or inoculation. Plant Soil 365, 363–374. doi: 10.1007/s11104-012-1393-2
Ennahar, S., Cai, Y., and Fujita, Y. (2003). Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 69, 444–451. doi: 10.1128/AEM.69.1.444-451.2003
Ferguson, B., Lin, M. H., and Gresshoff, P. M. (2013). Regulation of legume nodulation by acidic growth conditions. Plant Signal. Behav. 8:e23426. doi: 10.4161/psb.23426
Fhoula, I., Najjari, A., Turki, Y., Jaballah, S., Boudabous, A., and Ouzari, H. (2013). Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of Tunisia. BioMed Res. Int. 2013:405708. doi: 10.1155/2013/405708
Gano-Cohen, K. A., Stokes, P. J., Blanton, M. A., Wendlandt, C. E., Hollowell, A. C., Regus, J. U., et al. (2016). Nonnodulating Bradyrhizobium spp. modulate the benefits of legume-Rhizobium mutualism. Appl. Environ. Microb. 82, 5259–5268. doi: 10.1128/AEM.01116-16
Gao, J. L., Sun, J. G., Li, Y., Wang, E. T., and Chen, W. X. (1994). Numerical taxonomy and DNA relatedness of tropical rhizobia isolated from Hainan Province, China. Int. J. Syst. Evol. Microbio. 44, 151–158. doi: 10.1099/00207713-44-1-151
Hellweg, C., Pühler, A., and Weidner, S. (2009). The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbio. 9:37. doi: 10.1186/1471-2180-9-37
Knowles, R., and Blackburn, T. H. (1993). Nitrogen Isotope Techniques. San Diego, CA: Academic Press.
Laguerre, G., Nour, S. M., Macheret, V., Sanjuan, J., Drouin, P., and Amarger, N. (2001). Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147, 981–993. doi: 10.1099/00221287-147-4-981
Lee, D. S., Kim, K. S., Ko, W., Li, B., Keo, S., Jeong, G. S., et al. (2014). The neoflavonoid latifolin isolated from MeOH extract of Dalbergia odorifera attenuates inflammatory responses by inhibiting NF-κB activation via Nrf2-Mediated heme oxygenase-1 expression. Phytother. Res. 28, 1216–1223. doi: 10.1002/ptr.5119
Li, L., Sinkko, H., Montonen, L., Wei, G., Lindström, K., and Räsänen, L. A. (2012). Biogeography of symbiotic and other endophytic bacteria isolated from medicinal Glycyrrhiza species in China. FEMS Microbiol. Ecol. 79, 46–68. doi: 10.1111/j.1574-6941.2011.01198.x
Liu, J., Wang, E. T., and Chen, W. X. (2010). Mixture of endophytic Agrobacterium and Sinorhizobium meliloti strains could induce nonspecific nodulation on some woody legumes. Arch. Microbiol. 192, 229–234. doi: 10.1007/s00203-010-0543-2
Lu, J. K., He, X. H., Huang, L. B., Kang, L. H., and Xu, D. P. (2012). Two Burkholderia strains from nodules of Dalbergia odorifera T. Chen in Hainan Island, southern China. New For. 43, 397–409. doi: 10.1007/s11056-011-9290-8
Lu, J. K., Kang, L. H., He, X. H., and Xu, D. P. (2011). Multilocus sequence analysis of the Rhizobia from five woody legumes in southern China. Afr. J. Microbiol. Res. 5, 5343–5353. doi: 10.5897/AJMR11.826
Lu, J. K., Kang, L. H., Sprent, J. I., Xu, D. P., and He, X. H. (2013). Two-way transfer of nitrogen between Dalbergia odorifera and its hemiparasite Santalum album is enhanced when the host is effectively nodulated and fixing nitrogen. Tree Physiol. 33, 464–474. doi: 10.1093/treephys/tpt024
Lu, J. K., Xu, D. P., Kang, L. H., and He, X. H. (2014). Host-species-dependent physiological characteristics of hemiparasite Santalum album in association with N2-fixing and non-N2-fixing hosts native to southern China. Tree Physiol. 34, 1006–1017. doi: 10.1093/treephys/tpu073
Ma, F. Y., Gu, C. B., Li, C. Y., Luo, M., Wang, W., Zu, Y. G., et al. (2013). Microwave-assisted aqueous two-phase extraction of isoflavonoids from Dalbergia odorifera T. Chen leaves. Sep. Purif. Technol. 115, 136–144. doi: 10.1016/j.seppur.2013.05.003
Marques, J. M., da Silva, T. F., Vollu, R. E., Blank, A. F., Ding, G. C., Seldin, L., et al. (2014). Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 88, 424–435. doi: 10.1111/1574-6941.12313
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbial. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Menge, D. N., Wolf, A. A., and Funk, J. L. (2015). Diversity of nitrogen fixation strategies in Mediterranean legumes. Nat. Plants 1:15064. doi: 10.1038/nplants.2015.64
Mus, F., Crook, M. B., Garcia, K., Costas, A. G., Geddes, B. A., Kouri, E. D., et al. (2016). Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 82, 3698–3710. doi: 10.1128/AEM.01055-16
Nandasena, K. G., O'Hara, G. W., Tiwari, R. P., Sezmiş, E., and Howieson, J. G. (2007). In situ lateral transfer of symbiosis islands results in rapid evolution of diverse competitive strains of mesorhizobia suboptimal in symbiotic nitrogen fixation on the pasture legume Biserrula pelecinus L. Environ. Microbiol. 9, 2496–2511. doi: 10.1111/j.1462-2920.2007.01368.x
Oren, A., and Garrity, G. M. (2015). List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 65, 2017–2025. doi: 10.1099/ijs.0.000317
Pandya, M., Naresh Kumar, G., and Rajkumar, S. (2013). Invasion of rhizobial infection thread by non-rhizobia for colonization of Vigna radiata root nodules. FEMS Microbiol. Lett. 348, 58–65. doi: 10.1111/1574-6968.12245
Parker, M. A. (2003). Genetic markers for analysing symbiotic relationships and lateral gene transfer in Neotropical bradyrhizobia. Mol. Ecol. 12, 2447–2455. doi: 10.1046/j.1365-294X.2003.01908.x
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Peix, A., Ramírez-Bahena, M. H., Velázquez, E., and Bedmar, E. J. (2015). Bacterial associations with legumes. Crit. Rev. Plant Sci. 34, 17–42. doi: 10.1080/07352689.2014.897899
Peoples, M. B., Chalk, P. M., Unkovich, M. J., and Boddey, R. M. (2015). Can differences in 15N natural abundance be used to quantify the transfer of nitrogen from legumes to neighbouring non-legume plant species? Soil Biol. Biochem. 87, 97–109. doi: 10.1016/j.soilbio.2015.04.010
Rasolomampianina, R., Bailly, X., Fetiarison, R., Rabevohitra, R., Béna, G., Ramaroson, L., et al. (2005). Nitrogen-fixing nodules from rose wood legume trees (Dalbergia spp.) endemic to Madagascar host seven different genera belonging to α-and β-proteobacteria. Mol. Ecol. 14, 4135–4146. doi: 10.1111/j.1365-294X.2005.02730.x
Sachs, J. L., and Simms, E. L. (2008). The origins of uncooperative rhizobia. Oikos 117, 961–966. doi: 10.1111/j.0030-1299.2008.16606.x
Santoyo, G., Orozco-Mosqueda, M. C., and Govindappa, M. (2012). Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: a review. Biocontrol Sci. Techn. 22, 855–872. doi: 10.1080/09583157.2012.694413
Schlaeppi, K., and Bulgarelli, D. (2015). The plant microbiome at work. Mol. Plant Microbe Interact. 28, 212–217. doi: 10.1094/MPMI-10-14-0334-FI
Schreiter, S., Ding, G. C., Grosch, R., Kropf, S., Antweiler, K., and Smalla, K. (2014). Soil type-dependent effects of a potential biocontrol inoculant on indigenous bacterial communities in the rhizosphere of field-grown lettuce. FEMS Microbiol. Ecol. 90, 718–730. doi: 10.1111/1574-6941.12430
Shi, S., Nuccio, E., Herman, D. J., Rijkers, R., Estera, K., Li, J., et al. (2015). Successional trajectories of rhizosphere bacterial communities over consecutive seasons. MBio 6, e00746–e00715. doi: 10.1128/mBio.00746-15
Sprent, J. I., Ardley, J., and James, E. K. (2017). Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 215, 40–56. doi: 10.1111/nph.14474
Unkovich, M., Herridge, D., Peoples, M., Cadisch, G., Boddey, B., Giller, K., et al. (2008). Measuring Plant-Associated Nitrogen Fixation in Agricultural Systems. Canberra, ACT: ACIAR Monograph, 131–162.
VanInsberghe, D., Maas, K. R., Cardenas, E., Strachan, C. R., Hallam, S. J., and Mohn, W. W. (2015). Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J. 9, 2435–2441. doi: 10.1038/ismej.2015.54
Vinuesa, P., León-Barrios, M., Silva, C., Willems, A., Jarabo-Lorenzo, A., Pérez-Galdona, R., et al. (2005). Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int. J. Syst. Evol. Microbiol. 55, 569–575. doi: 10.1099/ijs.0.63292-0
Wu, L. J., Wang, H. Q., Wang, E. T., Chen, W. X., and Tian, C. F. (2011). Genetic diversity of nodulating and non-nodulating rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in different ecoregions of China. FEMS Microbiol. Ecol. 76, 439–450. doi: 10.1111/j.1574-6941.2011.01064.x
Yao, Y., Wang, R., Lu, J. K., Sui, X. H., Wang, E. T., and Chen, W. X. (2014). Genetic diversity and evolution of Bradyrhizobium populations nodulating Erythrophleum fordii, an evergreen tree indigenous to the southern subtropical region of China. Appl. Environ. Microb. 80, 6184–6194. doi: 10.1128/AEM.01595-14
Zahran, H. H. (1999). Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. R. 63, 968–989.
Zahran, H. H. (2017). Plasmids impact on rhizobia-legumes symbiosis in diverse environments. Symbiosis 73, 75–91. doi: 10.1007/s13199-017-0476-5
Keywords: bacterial communities, Dalbergia odorifera, high-throughput sequencing, nitrogen fixation, non-rhizobial bacteria, rhizobia
Citation: Lu J, Yang F, Wang S, Ma H, Liang J and Chen Y (2017) Co-existence of Rhizobia and Diverse Non-rhizobial Bacteria in the Rhizosphere and Nodules of Dalbergia odorifera Seedlings Inoculated with Bradyrhizobium elkanii, Rhizobium multihospitium–Like and Burkholderia pyrrocinia–Like Strains. Front. Microbiol. 8:2255. doi: 10.3389/fmicb.2017.02255
Received: 11 April 2017; Accepted: 31 October 2017;
Published: 21 November 2017.
Edited by:
Jan Dirk Van Elsas, University of Groningen, NetherlandsReviewed by:
Munusamy Madhaiyan, Temasek Life Sciences Laboratory, SingaporeCopyright © 2017 Lu, Yang, Wang, Ma, Liang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junkun Lu, anVua3VubHVAY2FmLmFjLmNu
Yinglong Chen, eWluZ2xvbmcuY2hlbkB1d2EuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.