- 1Department of Immunology and Pathogen Biology, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 2State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Sciences, Fudan University, Shanghai, China
- 3Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 4Shanghai Centre for Clinical Laboratory, Shanghai, China
Objective: The aim of the present study was to explore the potential biological role of Rv2629 in Mycobacterium smegmatis and Mycobacterium tuberculosis.
Methods: Recombinant wild type and mutant Rv2629 strains were constructed. Rv2629 expression was evaluated by real-time PCR and western blot. Microarray and interaction network analyses were used to identify the gene interactions associated with wild type and mutant Rv2629. Bacterial growth was assessed in Balb/c mice infected with wild type and mutant Rv2629 strains using CFU assay and histological analysis of the organs.
Results: Overexpression of Rv2629 could delay the entry of the Mycobacterium tuberculosis cells into the log-phase, while Rv2629 decreased the number of ribosomes and the expression of uridylate kinase in Mycobacterium smegmatis. The Gene Ontology (GO) and pathway analysis indicated that 122 genes correlated with wild type Rv2629, whereas the Rv2629 mutation led to decrease in the ribosome production, oxidative phosphorylation, and virulence in Mycobacterium tuberculosis. Overexpression of Rv2629 slightly enhanced the drug resistance of Mycobacterium smegmatis to antibiotics, and increased its survival and pathogenicity in Balb/c mice.
Conclusion: It is suggested that Rv2629 is involved in the survival of the clinical drug-resistant strain via bacterial growth repression and bacterial persistence induction.
Introduction
Mycobacterium tuberculosis (M. tuberculosis) infection is the major cause of TB. In 2016, 9287 new TB cases were reported in the United States and the successful elimination of TB in the United States is not expected to be achieved during this century (Doosti-Irani et al., 2016). The successful survival and evolution of M. tuberculosis is attributed to a large extent to its ability to persist for long periods within the human body in a latent and/or dormant state (Peddireddy et al., 2017). During latent infection, M. tuberculosis bacilli are retained within granulomas, which offers a niche with increased concentration of nitric oxide, low oxygen, and absence of nutrients (Bartek et al., 2009; Trauner et al., 2012). The non-replicating persistent form of M. tuberculosis aids the escape of the pathogen from the host defense mechanisms and provides additional advantage to the bacilli as regards the effect of standard anti-TB drugs. This process results in the long term survival of the bacteria for several decades (Voskuil et al., 2003; Keshari et al., 2017).
Genes such as dosR, icl, acr (hspx), and lsr2, which were demonstrated to be stimulated by hypoxia (Boon and Dick, 2002; Florczyk et al., 2003; Park et al., 2003; Starck et al., 2004; Fu and Fu-Liu, 2007), are expressed at low levels and/or are absent in the log-phase of bacterial growth. These genes were shown to be associated with dormancy of Mycobacterium (Boon and Dick, 2002; Park et al., 2003). Among the aforementioned genes, the dormancy survival regulon, regulated by the response regulator DosR, appears to be essential for hypoxic survival in a vast number of mycobacterial species, including M. tuberculosis (Park et al., 2003; Domenech et al., 2017), Mycobacterium bovis BCG (Alonso et al., 2011), and Mycobacterium smegmatis (M. smegmatis) (Homolka et al., 2009). DosR was shown to initiate transcription of an array of genes known as the dosR regulon, which comprises 48 genes containing the DosR-binding site and an ACP family sequence motif (Florczyk et al., 2003). The expression of these genes allows long-term survival of M. tuberculosis under anaerobic conditions in the host (Errey and Blanchard, 2005; Homolka et al., 2009; Alonso et al., 2011).
Rv2629 is a member of the dosR dormancy regulon (Park et al., 2003) due to the presence of an ACP motif predicted by the Gibbs Recursive Sampler and SCAN (Florczyk et al., 2003). The Rv2629 gene is conserved with regard to its expression in several environmental mycobacteria and related strains of M. tuberculosis (Wang et al., 2007). Rv2629 has been reported to be induced by hypoxia and nitric oxide, while it is upregulated under dormancy conditions (Boon and Dick, 2002; Park et al., 2003; Voskuil et al., 2003; Starck et al., 2004), suggesting an association with the dormant state of M. tuberculosis (Starck et al., 2004). Rv2629 was further identified as a drug-resistant protein by 2D-gel electrophoresis and MS analysis (Wang et al., 2007). A previous study determined the Rv2629 sequence variations in 58 MDR strains and in 55 strains from a reference collection that represented the major M. tuberculosis complex pathogens namely, M. tuberculosis Beijing, M. africanum West African 1, M. tuberculosis (H37Rv; ATCC 27294), M. bovis (ATCC 19210), and M. africanum (ATCC 25420) (Homolka et al., 2009). Among these 58 MDR strains, 36 (62%) had the 191A/C mutation (codon 64, Asp to Ala) in Rv2629 that supported an association with rifampicin resistance (Chakravorty et al., 2008; Homolka et al., 2009). The 191C allele was present in the Beijing strains when the MDR strains were stratified according to different phylogenetic lineages that indicated a classical Beijing IS6110 restriction fragment length polymorphism pattern (Chakravorty et al., 2008; Homolka et al., 2009). The IS6110-restriction fragment length polymorphism (IS6110-RFLP) has been applied for the identification of the Beijing strains based on their hybridization patterns (Alonso et al., 2011). The Beijing strains evade the innate immune response and exhibit high capacity to cause and spread TB in humans compared with other M. tuberculosis lineages. Nevertheless, the exact mechanism of this action is yet to be determined (Alonso et al., 2011). Although the biological function of Rv2629 remains unclear, it is speculated that its association with drug resistance involves maintenance of the persistent state of M. tuberculosis.
The present study aimed to investigate the function of the Rv2629 protein, notably the relationship between Rv2629 over-expression and bacterial physiological changes.
Materials and Methods
Strains and Growth Media
Mycobacterium smegmatis (MC2 155) and M. tuberculosis H37Ra were grown in Middlebrook 7H9 broth or Middlebrook 7H10 agar (DIFCO, United States) supplemented with 10% ADC (BD, United States) and kept in a biosafety P1 laboratory. The media for recombinant M. smegmatis and M. tuberculosis H37Ra that over-expressed Rv2629 were supplemented with 50 μg/mL kanamycin, while for recombinant M. smegmatis that expressed low levels of MSMEG_1130, the medium was supplemented with 50 μg/mL kanamycin and 0.1% acetamide (w/v). Transformed Escherichia coli DH5α was cultured in Luria-Bertani broth and/or agar with the appropriate antibiotic (50 μg/mL kanamycin).
Strains isolated from clinical drug-resistant patients (strains 1570, 934, 945, 1573, 1176, 522, 1219, and 1221, all of the Beijing type) were obtained and cultured in a P2 laboratory at the Shanghai Pulmonary Hospital. All clinical strains included were rifampicin-resistant. Strain 934 was rifampicin (RIF) resistant with a minimum inhibitory concentrations (MIC) of 32 μg/mL. Strains 1570, 945, 1573, 1176, 1219, 522, and 1221 were MDR. All strains were identified and drug-susceptibility was evaluated as previously described (Ramaswamy et al., 2003; Wang et al., 2007). Clinical isolates and M. tuberculosis H37Rv were cultured under the same conditions as M. tuberculosis H37Ra (Kwon et al., 2017).
Cell Culture
THP-1 cells were grown in house and were cultured in RPMI-1640 (Gibco, Foster City, CA, United States) supplemented with 10% (w/v) fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin (Gibco). THP-1 cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Cloning and Transformation
Recombinant plasmids encoding wild type Rv2629 (Rv2629W) and the 191C mutant (Rv2629M) were constructed. The full length Rv2629 genes were amplified from genomic DNA extracted from H37Rv (for Rv2629W) and from clinical strains (for Rv2629Mgenotype). Recombinant plasmids encoding the antisense strand of MSMEG_1130, an ortholog of Rv2629 in M. smegmatis, was constructed. The amplification was conducted by PCR using the appropriate primers (listed in Supplementary Table S1). The PCR products were digested with the corresponding enzymes (NEB, United States) (Supplementary Table S1) for 3 h at 37°C. Plasmid pMV261 (for over-expression) and/or pACT (an acetamide-inducible expression vector, for reduced expression) (Invitrogen, Waltham, MA, United States) were digested with the same restriction enzymes. The two fragments were ligated, transformed into E. coli DH5α and plated on LB agar containing kanamycin (50 μg/mL). Following overnight incubation at 37°C, single colonies were randomly selected and grown in LB broth. The plasmids were isolated from E. coli DH5α culture using an Axygen Miniprep Kit (Axygen, Union City, CA, United States) and confirmed by DNA sequencing.
The recombinant pMV261 and/or pACT plasmids were transformed into the M. smegmatis strain MC2 155 (MS) and/or M. tuberculosis H37Ra, respectively by electroporation (Reed et al., 2007). The study of M. tuberculosis genes in recombinant M. smegmatis is a recognized approach (Falcone et al., 1995; Daugelat et al., 2003; Andreu et al., 2004; Zimhony et al., 2004; Altaf et al., 2010; Sweeney et al., 2011; Bae et al., 2017; Sun et al., 2017; Yu et al., 2017; Wang et al., 2017). Recombinant M. smegmatis over-expressing Rv2629 was identified as MSW (Rv2629 wild type) and MSM (Rv2629 mutant). Recombinant M. tuberculosis H37Ra over-expressing Rv2629 was identified as RaW (Rv2629 wild type) and RaM (Rv2629 mutant). Recombinant M. smegmatis expressing low levels of MSMEG_1130 was identified as MSL. Recombinant strains with the plasmid pMV261and/or pACT were constructed (MSP, RaP, or MS pACT) and used as controls. The types of strains with the corresponding genes and plasmids are presented in Supplementary Table S2.
RNA Extraction and Real Time PCR (RT-qPCR)
Total RNA was isolated from M. smegmatis and M. tuberculosis using an RNA extraction kit (Omega, United States), according to the manufacturer’s instructions. Contaminating DNA was digested with RNase-free DNase I (Omega, United States) for 15 min at 37°C. Prior to further analysis the quality check of the RNA was determined by the estimation of the A260/A280 ratio. For RT-qPCR, cDNA was synthesized according to the manufacturer’s instructions (ReverTraAce qPCR RT kit, Toyobo, Japan).
qPCR analysis was conducted using Rv2629-specific primers (see Supplementary Table S1) with SYBR Green Real-Time PCR Master Mix (QPK-201) (Toyobo, Japan). 16S rRNA and Rv2703 (SigA) were used as reference sequences. The reaction conditions were: 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, 56 to 65°C for 45 s, and 72°C for 30 s. A standard curve of copy number (range: 101–108) was constructed. Data were collected using a BIO-RAD iCycler (BIO-RAD, United States). RT-negative (without reverse transcriptase) reactions were used to account for residual DNA, and the copy numbers were normalized to those of 16S rRNA. The normalized values were used to determine the relative fold change in expression. The estimation of the relative expression of each gene was carried out using the ddCT method. The experiments were carried out in triplicate, and the results are expressed as mean ± SD.
Western Blot Analysis
Whole cells were harvested from 20 mL cultures of M. smegmatis strains and M. tuberculosis isolates. The bacterial pellets were washed twice with 0.02 M PBS, resuspended, and sonicated in 200 μL of 0.02M PBS (supplemented with 1 mM PMSF and 10 mM EDTA) and centrifuged. The supernatant was treated with lysis buffer (8 M urea, 2 M thiourea, 140 mM DTT, 0.5% Biolyte pH 4–7, and 4% CHAPs). The protein concentration was estimated using the Bradford assay. Equal amounts of protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12% gel), transferred to a PVDF membrane and detected with anti-Rv2629 rabbit antiserum (in house preparation) and/or the FtsZ monoclonal antibody (4°C, overnight) followed by a goat anti-rabbit IgG antibody conjugated with alkaline phosphatase (Sigma, United States) (37°C, 1 h). The blots were visualized with BCIP/NBT solution (room temperature, 20 min). The reaction was terminated by addition of deionized water. FtsZ was used as a loading control.
Minimum Inhibitory Concentration Assay
Minimum inhibitory concentrations assay of recombinant M. smegmatis was carried out to detect the relationship between over-expressing Rv2629 proteins and the susceptibility to anti-TB drugs (streptomycin, isoniazid, rifampicin, ethambutol, ofloxacin, levofloxacin, moxifloxacin, amikacin, kanamycin, and capreomycin). Antibiotic dilutions and 96-well plate preparations (Falcon 3072; Becton Dickinson, Lincoln Park, NJ, United States) were carried out as described by Caviedes et al. (2002). Briefly, Rv2629 overexpressing strains were grown in 7H9/ADC at 37°C until an OD600 value of 0.5 was obtained. The bacterial suspensions were diluted 1000 times in 7H9 broth and the final diluted broth was dispensed into each well of a 96-well cell culture plate (100 μL/well). The drug storage solutions were diluted to prepare two-fold serial diluted solutions in 7H9 broth and subsequently added to the wells containing the bacterial suspensions. The plate was incubated for 48 h at 37°C. The wells containing bacterial pellets were denoted as the positive wells, whereas the wells containing clear broth were denoted as the negative wells. The minimum drug concentrations of the negative wells were the MICs of the drugs. All the experiments were carried out in triplicate.
Microarray Analysis
Total RNA from each sample was quantified using the NanoDrop 1000 (Thermo Fisher, United States). RNA integrity was assessed using standard denaturing agarose gel electrophoresis. For microarray analysis, the Agilent Array platform was used according to the manufacturer’s standard protocols for sample preparation and microarray hybridization. Briefly, total RNA (1 μg) was amplified and transcribed into fluorescent cRNA, following the manufacturer’s Quick Amp Labeling protocol (Version 5.7, Agilent Technologies). The labeled samples were hybridized toward the M. tuberculosis H37Ra custom Oligo Array (8x15K, Agilent array) and were scanned using the Agilent Scanner G2505B (Agilent Technologies, United States). The Agilent Feature Extraction Software (version 10.5.1.1) (Agilent Technologies, United States) was used to analyze acquired array images. The median normalization and subsequent data processing was carried out using the GeneSpring GX v11.0 software package (Agilent Technologies, United States). Following Median normalization of the raw data, genes of at least one out of nine samples that exhibited flags in Present and/or Marginal (“All Targets Value”) were selected for further data analysis. Differentially expressed genes were identified using Volcano Plot filtering. Pathway and GO analyses were conducted in order to reveal the biological functions of this subset of differentially expressed genes. Finally, hierarchical clustering was carried out in order to show distinguishable gene expression profiling among samples.
Transmission Electron Microscopy
For ultrathin cryo sections, log-phase M. Smegmatis cultures were harvested (5 mL). The cells were washed twice with 0.1 M phosphate buffer (PB) and fixed with 2.5% glutaraldehyde in 0.1 M PB (2 h). The log phase was estimated as determined previously (Belanger and Hatfull, 1999). Notably, a culture of M. smegmatis mc2155 was grown at 37°C, until an OD600 reading of approximately 0.8 was obtained. The cells were rinsed three times (15 min per wash) with 0.1 M PBS. After fixation in 1% osmium tetroxide (3 h), the cells were rinsed as previously described prior to dehydration with solutions containing increasing concentrations of ethanol and/or acetone at 4°C. The samples were subjected to gradual infiltration with Epon resin (3 days). Subsequently, the samples were solidified by incubation at increasing temperature for a total period of 2 days. Thin sections (70–80 nm) were prepared and stained with 3% uranyl acetate and lead citrate prior to examination under a Philips CM-120 transmission electron microscope (TEM) (Philips, United States).
Growth Curve
To determine whether Rv2629 expression levels among strains would lead to observable phenotypic growth and survival differences, recombinant M. tuberculosis H37Ra and M. smegmatis strains were subcultured thrice in Middlebrook 7H9 broth supplemented with 10% ADC and 0.05% Tween-80 (OD600 = 0.4). The M. smegmatis cultures were diluted when an OD600 value of 0.05 was obtained. Aliquots of 50 mL were dispensed in conical flasks and grown under continuous shaking at 220 rpm and aerobic conditions. The growth and survival were assayed by optical density readings (OD600) every 3 h following inoculation. The experiments were performed in triplicate.
The growth curve of recombinant M. tuberculosis H37Ra was detected using the BACTEC MGIT 960 Mycobacterial Detection System (Becton, Dickinson and Company, United States), as determined by the manufacturer’s instructions.
Infection of THP-1 Cells with M. tuberculosis Strains and M. smegmatis Strains
THP-1 cells (5 × 105) were stimulated with 5 ng/mL PMA (Sigma Chemical Co, St. Louis, MO, United States) for 24 h prior to infection. The infection was conducted using H37Rv, clinical M. tuberculosis strain 934, and recombinant M. smegmatis in the log-phase at a multiplicity of infection (MOI) value of 1:10 (37°C, 5% CO2, 4 h). Extracellular bacilli were removed by washing cells three times with PBS and the cells were cultured for 1 h in complete medium in the presence of 100 μg/mL gentamycin. The medium was replaced every 12 h during incubation, and the cells were lysed using 1% Triton X-100 solution at specific time points (0, 24, 48, 72, and 96 h). The precipitates were collected by centrifugation (2000 g, 20 min) for total RNA extraction, as described above. The number of CFU of intracellular mycobacteria and the viable bacteria present in the supernatant were determined on Middlebrook 7H10 agar with 10 % ADC.
Mouse Infection
The culture of MSP and MSW was conducted in 5 mL of Middlebrook7H9 medium supplemented with 10% ADC and 500 mg sterile 2-mm glass beads at 37°C under continuous shaking. The cultures isolated during the log phase were centrifuged at 500 g in order to remove clumps, and the cells were washed twice with sterile saline. The cell suspension was adjusted to an OD600 of 0.8, corresponding to a concentration of 5 × 107 CFU/mL. Female Balb/c mice (6–8-week-old) were infected intravenously with 0.1 mL of the bacterial suspension. The size of inoculum was confirmed by CFU counts.
The ability of survival and dissemination of each strain in vivo was assessed in six mice per group that were sacrificed by cervical dislocation on 0, 3, 6, 9, 12, 15, 20, and 35 days after infection. Lung, spleen, and kidney tissue specimens were weighed and homogenized in saline. The specimens were cultured in Middlebrook 7H10 medium (Becton Dickinson, United States) supplemented with 10% OADC following appropriate dilutions. CFUs were enumerated following 3–4 days of incubation at 37°C. All of the animal procedures were approved by the animal care committee of the Fudan University.
2D Gel Electrophoresis and MS Analysis
Whole cell extracts were prepared from 100 mL of cultures of each M. smegmatis strain in the log-phase. Bacterial pellets were washed twice and centrifuged (15°C, 8000 rpm, 15 min) in 0.02 M PBS (pH 7.4). The cells were sonicated in 500 μL of 0.02 PBS (0.02 M supplemented with 1 mM PMSF and 10 mM EDTA) and centrifuged at 12,000 rpm for 20 min at 4°C. The cells were treated with lysis buffer (8 M urea, 2 M thiourea, 140 mM DTT, 0.5% Biolyte pH 4-7, and 4% CHAPs) for 1 h and centrifuged (4°C, 12,000 rpm, 15 min). The supernatant was harvested and the protein concentration was estimated using the Bradford assay. Total protein (80 μg) were loaded on linear pH 4-7 IPG strips (Bio-Rad, United States) that were allowed to rehydrate at 50 V for 13 h. Isoelectric focusing was carried out using a Protean IEF (Bio-Rad, United States). The proteins were separated using 2-dimensional SDS-PAGE (12% gels) and stained with silver nitrate, as reported previously (Scheler et al., 1998; Gharahdaghi et al., 1999). The gels were scanned using Molecular Image FX (Bio-Rad, United States) and analyzed with the PD Quest 6.0 software (Bio-Rad, United States). The proteins of interest were excised from the silver-stained 2D gels and identified by MALDI-TOF-MS (Fu and Fu-Liu, 2007).
Statistical Analysis
Statistical analysis was conducted using SPSS 11.5 (SPSS Inc., Chicago, IL, United States). Statistical differences of the continuous variables were analyzed by one-way analysis of variance (ANOVA) and the Student–Newman–Keuls test and/or Student t-test. The statistical level of significance was set at P < 0.05.
Results
Expression of Rv2629 in Different Clinical Strains
Previous studies have demonstrated an increase in the expression of Rv2629 at the transcriptional and/or translational levels induced by hypoxia and nitric oxide in clinical isolates of M. tuberculosis (Voskuil et al., 2003; Kassa et al., 2012; Zhang et al., 2014). Thus, we investigated the expression of the Rv2629 protein and mRNA levels among clinical isolates of M. tuberculosis. Quantitative RT-PCR was conducted using cDNA synthesized from total RNA isolated from bacilli from seven clinical strains (1573, 1570, 945, 1176, 1219, 522, and 1221) and the M. tuberculosis H37Rv control strain in the log and/or stationary-phases respectively (Figure 1A). The stationary phase was observed notably following 10 days of growth and the bacterial cultures were harvested at 3 h intervals for further analysis. It was observed that the average transcriptional level of Rv2629 in clinical isolates during the logarithmic phase was approximately two-fold lower than that noted in M. tuberculosis H37Rv, while during the stationary phase, this level was higher in the clinical isolates than in the control strain. The expression of Rv2629 at the protein level was further measured by Western blot analysis (Figures 1B,C). The cytoplasmic fraction of the cell extracts obtained from the clinical isolates indicated a two-fold increase in the stationary phase compared to that noted in M. tuberculosis H37Rv. The over-expression of Rv2629 at both the protein and the mRNA levels in clinical M. tuberculosis isolates was related to the stationary phases of the strains. Rv2629 exhibited reduced expression during the log-phase, when bacilli were actively duplicating.
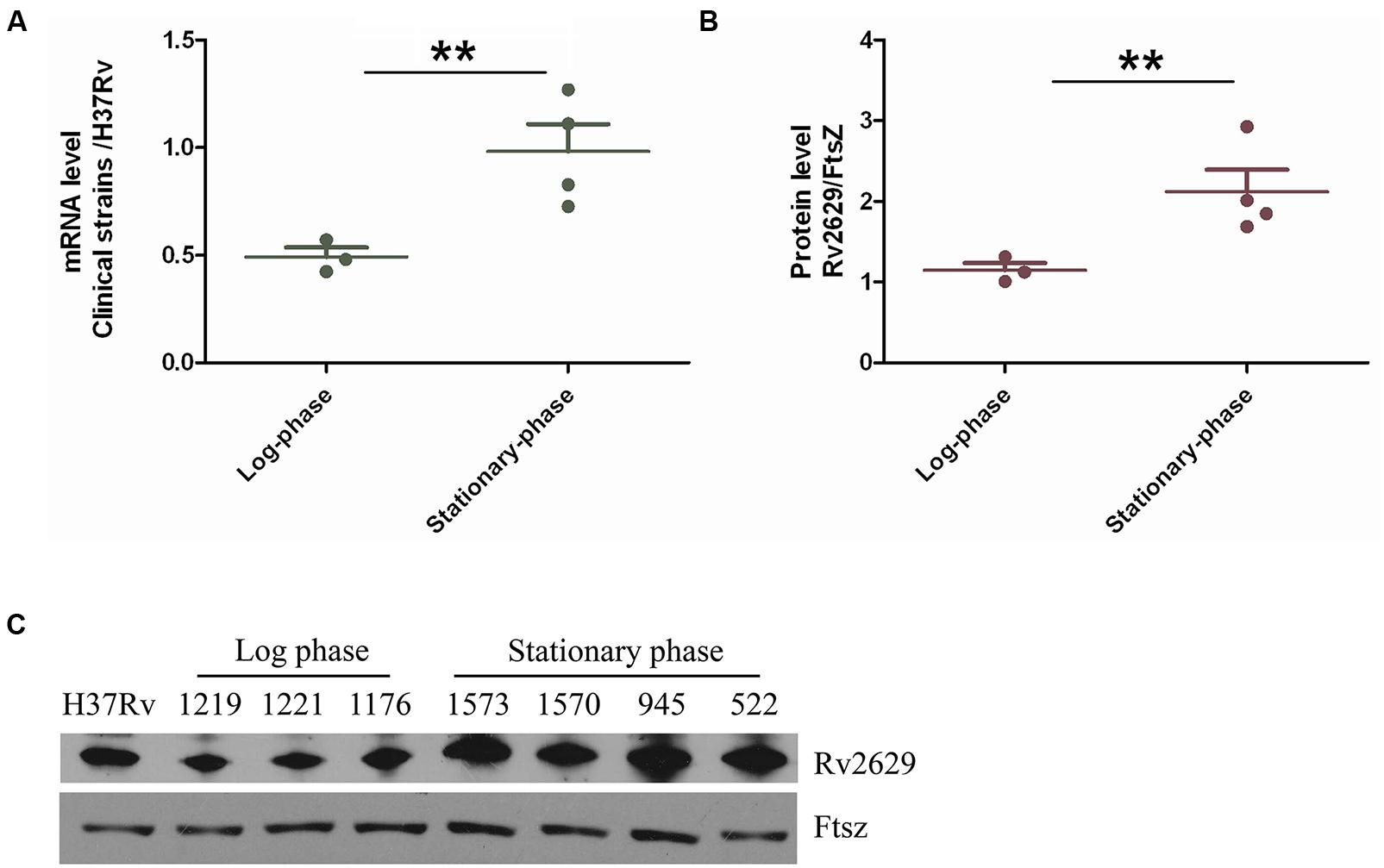
FIGURE 1. The expression of Rv2629 was upregulated and down-regulated in clinical multidrug resistant (MDR) isolates in the stationary and log phases, respectively. (A) Total RNA was extracted from different drug-resistant clinical M. tuberculosis isolates in the log and stationary phases and was used RT-qPCR analysis. 16s RNA was used as the reference transcript. (B) Western blot analysis of Rv2629 in the clinical MDR isolates at the log and stationary phases. (C) FtsZ was used as the loading control. The results are presented as the ratio of mRNA and/or protein level of Rv2629 in clinical isolates compared with the corresponding ratio in H37Rv. The data are indicated as mean ± SEM. ∗∗P < 0.01.
Expression of Rv2629 in Clinical M. tuberculosis Strain 934 in THP-1 Cells
In order to investigate the association of Rv2629 with dormancy, the THP-1 human leukemic monocyte cell line was infected with H37Rv and/or a clinical RIF-resistant isolate 934 in the log-phase at MOI = 10, following stimulation with 5 ng/mL PMA for 24 h. The mRNA levels of Rv2629 expressed in bacteria were measured at 0, 3, 24, and 72 h. Strain 934 was cultured in the absence of THP-1 cells and served as control. Rv2629 gene intracellular expression was further determined during several time periods (Figure 2). The results indicated that the ratios of the Rv2629 mRNA levels of strain 934 to H37Rv at 3 and 24 h were higher in the extracellular compared with the intracellular compartment (P < 0.01), whereas the opposite was noted for the 72 h period (P < 0.001) (Figure 2). Moreover, a high upregulation (45-fold) of intracellular Rv2629 expression in the clinical strain was observed at 72 h post-infection compared with the extracellular control (Figure 2), supporting the hypothesis that intracellular bacilli can remain in a dormant phenotype up to day 3 by over-expressing Rv2629.
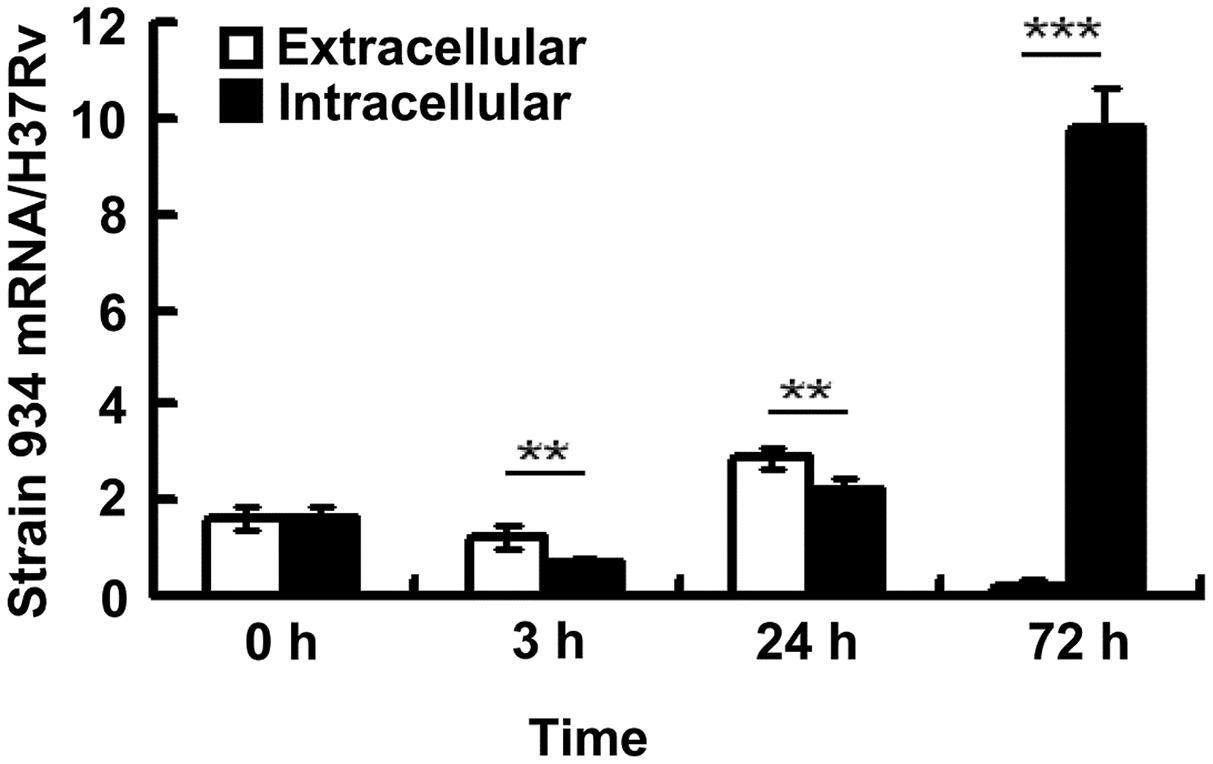
FIGURE 2. Rv2629 mRNA was upregulated in RIF-resistant clinical M. tuberculosis strains following infection in THP-1 cells. A total of 5 × 105 THP-1 cells were infected with H37Rv or, a clinical rifampicin-resistant isolate (strain 934) in the log-phase at multiplicity of infection (MOI) = 10, following stimulation with 5 ng/mL of PMA for 24 h. The mRNA levels of Rv2629 were measured at 0, 3, 24, and 72 h (intracellular group). Strain 934 was cultured in the absence of THP-1 cells and served as control (extracellular group). The results are presented as the ratio of mRNA levels of Rv2629 in strains 934 and H37Rv. The expression of intracellular Rv2629 mRNA was 45-fold higher than that noted in the extracellular 72 h post-infection. The data are presented as mean ± SEM. ∗∗P < 0.01, ∗∗∗P < 0.001. The experiments were performed in triplicate.
Confirmation of Overexpression and Reduced-Expression of Rv2629 in Mycobacteria
M. smegmatis over-expressing Rv2629W (MSW) and Rv2629M (MSM) were produced in order to simulate Rv2629 expression levels in clinical M. tuberculosis strains and investigate the function of the Rv2629 protein. Western blot and RT-qPCR analysis of four M. smegmatis strains during the log-phase confirmed the increased expression of Rv2629 in the MSW and MSM compared with the control MS and MSP strains (Figures 3A,B).
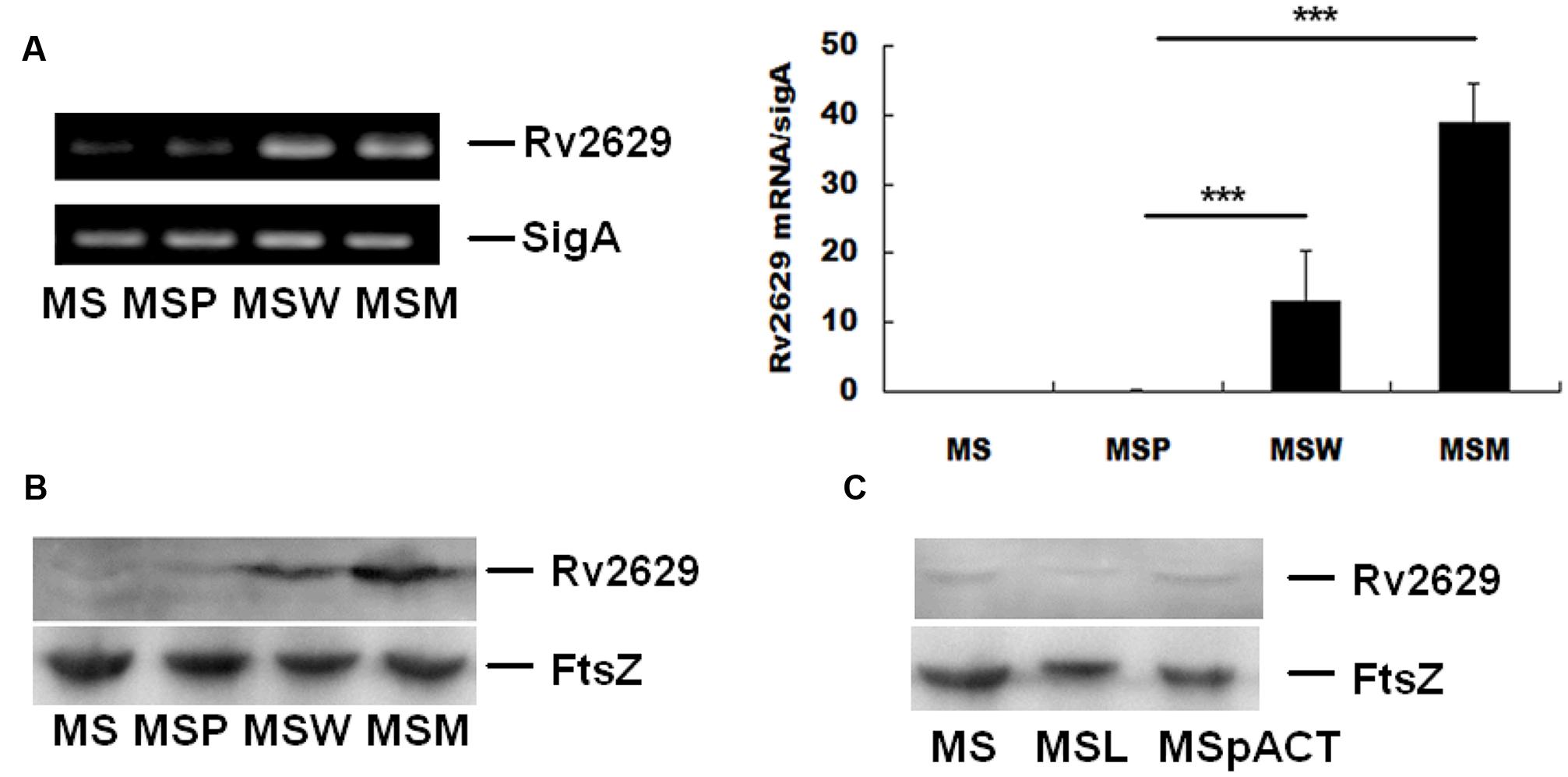
FIGURE 3. Overexpression and reduced expression of wild type Rv2629 and/or mutant Rv2629 in M. smegmatis. (A) qRT-PCR using total RNA extracted from four M. smegmatis strains, namely, MSW (wild type Rv2629), MSM (191A/C mutant), MS, and MSP (control strains). (B) Western blot analysis of over-expression of Rv2629 in the four M. smegmatis strains. (C) Western blot analysis of reduced expression of Rv2629 in MSL and MSpACT. The data are presented as mean ± SEM. ∗∗∗P < 0.001. The experiments were performed in triplicate.
The ortholog of Rv2629 in H37Ra and M. smegmatis strains are MRA_2657 and MSMEG_1130, respectively. The DNA and amino acid sequences of MRA_2657 are completely identical with Rv2629, while MSMEG_1130 and Rv2629 are highly homologous, in which the overlap value between MSMEG_1130 and Rv2629 was 366 based on the data of KEGG database. M. smegmatis MSL corresponded to a bacterial strain expressing low levels of MSMEG_1130. The strain was constructed to investigate the function of Rv2629, whereas the plasmid pACT, an acetamide-inducible expression vector, served as control. For generating the MSL strain, the plasmid pACT was subjected to recombination with the DNA fragment, whose mRNA production could complement with the MSMEG_1130 mRNA sequence reversely and thus could inhibit the expression of MSMEG_1130 gene. Western blot analysis in the two M. smegmatis strains during log-phase confirmed low expression of MSMEG_1130 in MSL compared with control MSpACT (Figure 3C). The use of M. smegmatis as a parental strain for the overexpression of Rv2629 has been reported in a previous study (Wang et al., 2007).
Minimum Inhibitory Concentration Assay of Recombinant M. smegmatis
In order to investigate the association of Rv2629 with drug-resistance, 10 anti-TB drugs were selected including four first-line drugs (streptomycin, isoniazid, rifampicin, and ethambutol) and six second-line drugs (ofloxacin, levofloxacin, moxifloxacin, amikacin, kanamycin, and capreomycin). The MIC of recombinant M. Smegmatis was determined. The results are shown in Table 1. The Rv2629 over-expressing strains enhanced drug-resistance of recombinant M. smegmatis to ethambutol, moxifloxacin, and levofloxacin at least two-fold compared with the MSP strain.
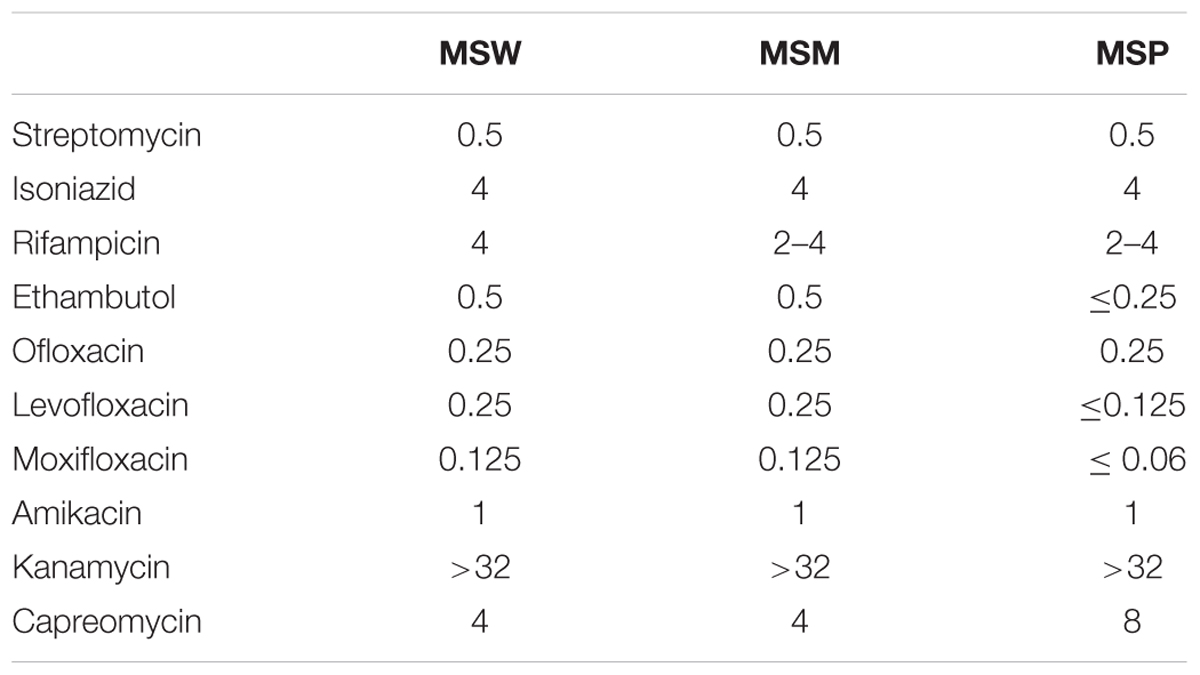
TABLE 1. Minimal inhibition concentrations (MICs) of recombinant M. smegmatis against first- and second-line drug treatment.
Microarray Analysis of M. tuberculosis H37Ra Overexpressing Rv2629
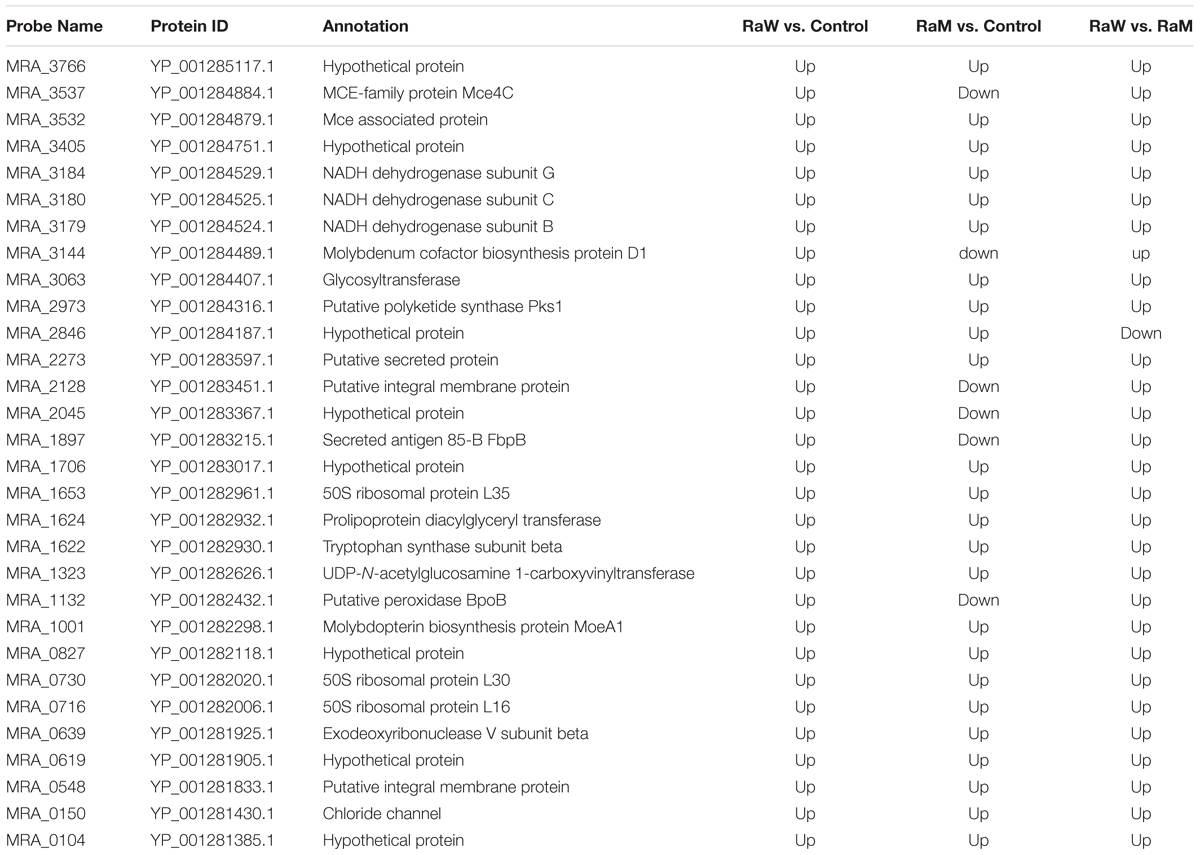
The present study aimed to investigate the differences in gene expression caused by Rv2629 overexpression in M. tuberculosis strains and the potential interplay in the biological function of the Rv2629 gene. Microarray analysis was conducted on recombinant M. tuberculosis H37Ra overexpressing Rv2629 in the log phase. The results indicated that recombinant M. tuberculosis H37Ra overexpressing Rv2629 (RaW) and H37Ra overexpressing 191 A/C mutated Rv2629 (RaM) remained in the log phase of growth, respectively. In addition, 122 genes correlated with wild type Rv2629 (Supplementary Table S3), whereas 92 genes correlated with 191A/C mutated Rv2629. The genes from the two strains were shown in different sizes (Figure 4). As shown in Figure 4, two genes are connected if the correlation between them is greater than a selected threshold (0.9). The correlation is based on a correlation coefficient between each particular gene and the two modules. A total of 30 genes correlated with wild type Rv2629 and to a lesser extent with 191 A/C mutated Rv2629 (Table 2). The correlation coefficient was lower than the selected threshold in RaM. The remaining 92 genes correlated with Rv2629 in the two bacterial strains.
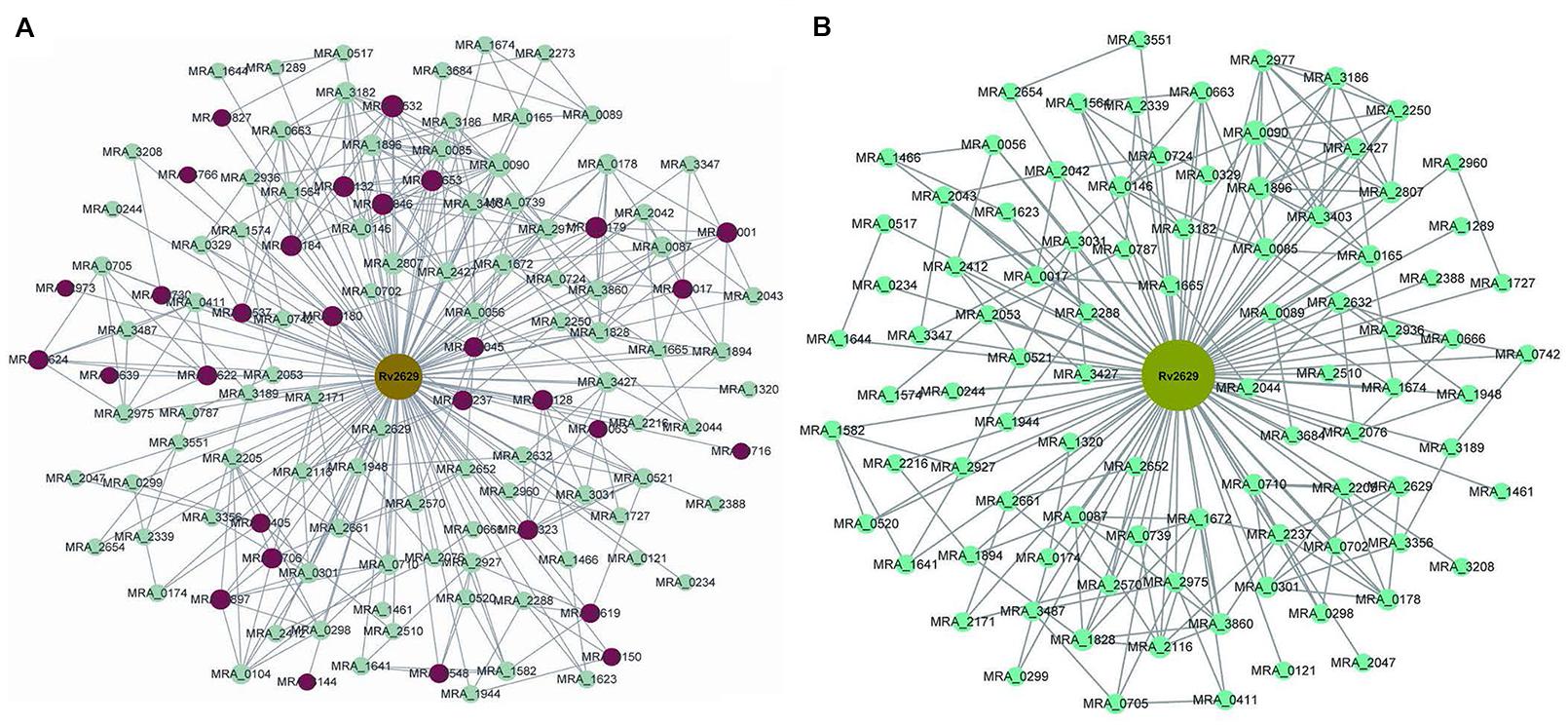
FIGURE 4. Interaction network of genes that correlated with Rv2629 and/or 191A/C mutated Rv2629 in RaW and/or RaM strains. (A) Module A indicates the interaction of the genes associated with the wild type Rv2629 in RaW. (B) Module B indicates the interaction of genes associated with the mutated Rv2629 (191A/C mutation) RaM strain. Taking into account the correlation coefficient between each particular gene and these two modules, two genes are connected if the correlation is greater than a selected threshold (0.9). Genes are shown in different sizes. A larger size (122 genes) indicates more connections with other modules; a smaller size (92 genes) indicates fewer connections. Different types of genes are in diverse colors. Rv2629 is the target gene of the present study. Green nodes are included in two conditions. Red nodes represent genes only in RaW and not in RaM strains.
The GO and pathway analysis indicated that 122 genes correlated with wild type Rv2629; they were involved in ribosome construction, toluene degradation, arginine and proline metabolism, oxidative phosphorylation, and homologous recombination. The genes were notably related to the ribosome construction and the Fisher P-value was 4.28 × 10-10 (Supplementary Table S4). The genes that solely correlated with wild type Rv2629 were notably related to ribosome production, oxidative phosphorylation, and virulence (Table 2). The results indicated that the 191A/C mutation in Rv2629 could influence the function of wild type Rv2629 partially. The mutation may lead to the decreased effect of Rv2629 on ribosome production, oxidative phosphorylation, and virulence.
Reduced Ribosome Content in M. smegmatis Overexpressing Rv2629
The effects of the Rv2629 gene on the morphological features of MSW and MSM at the log-phase were investigated by TEM, as demonstrated by previous studies (Tumminia et al., 1991). No difference was noted in cell wall thickness and morphology, although recombinant M. smegmatis MSW revealed apparent differences in their cytoplasmic component compared to the MSM, MSP, and MS strains (Figure 5). The cytoplasm of MSW cells exhibited lower electron density and the cytoplasmic ribosomal content was reduced significantly compared to the MS and MSP strains, with the exception of the MSM strain (Figure 5).
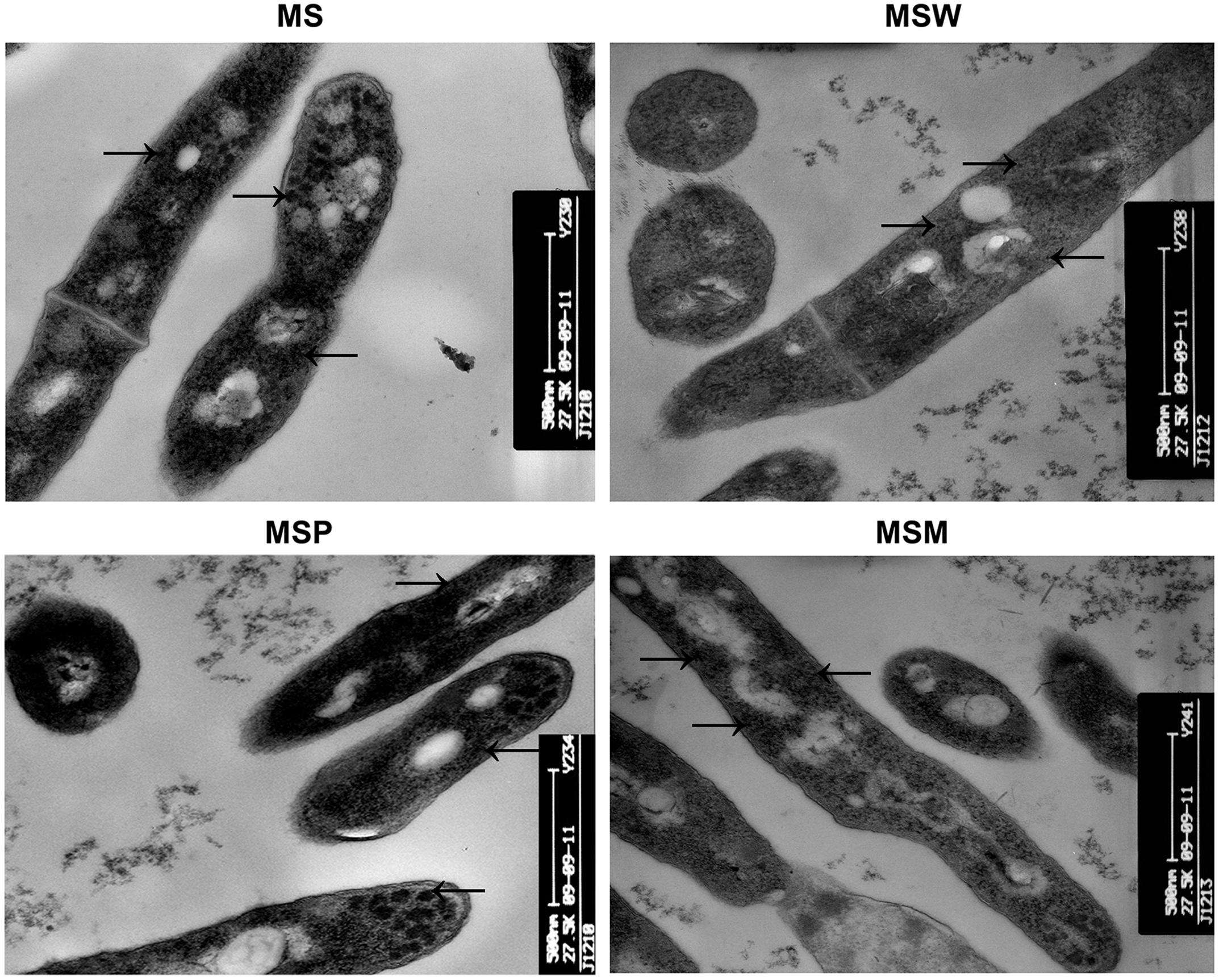
FIGURE 5. Transmission electron microscope analysis of ribosomal content. The ribosomal content (arrows) of MS, MSW, MSP, and MSM strains. The data are representative of two independent experiments with three samples in each experiment. Five random fields were observed.
Growth Characteristic of Recombinant M. smegmatis
The microarray results indicated that Rv2629 might be related to ribosome production. Consequently, the growth curves of the MSW and MSM strains were investigated by harvesting cultures at 3-h intervals and by measuring the OD600. MS and MSP served as controls. The results of the two independent experiments demonstrated comparatively slow growth of MSW (Figure 6A) and indicated that Rv2629 overexpression inhibits bacterial growth and delays entry in the log phase. The growth of MSM was more rapid compared with MSW and slower than MSP and MS, which suggested that the 191A/C mutation of the Rv2629 gene weakens the inhibitory function of Rv2629. A similar effect was noted in the RaW and RaM strains (Supplementary Figure S1).
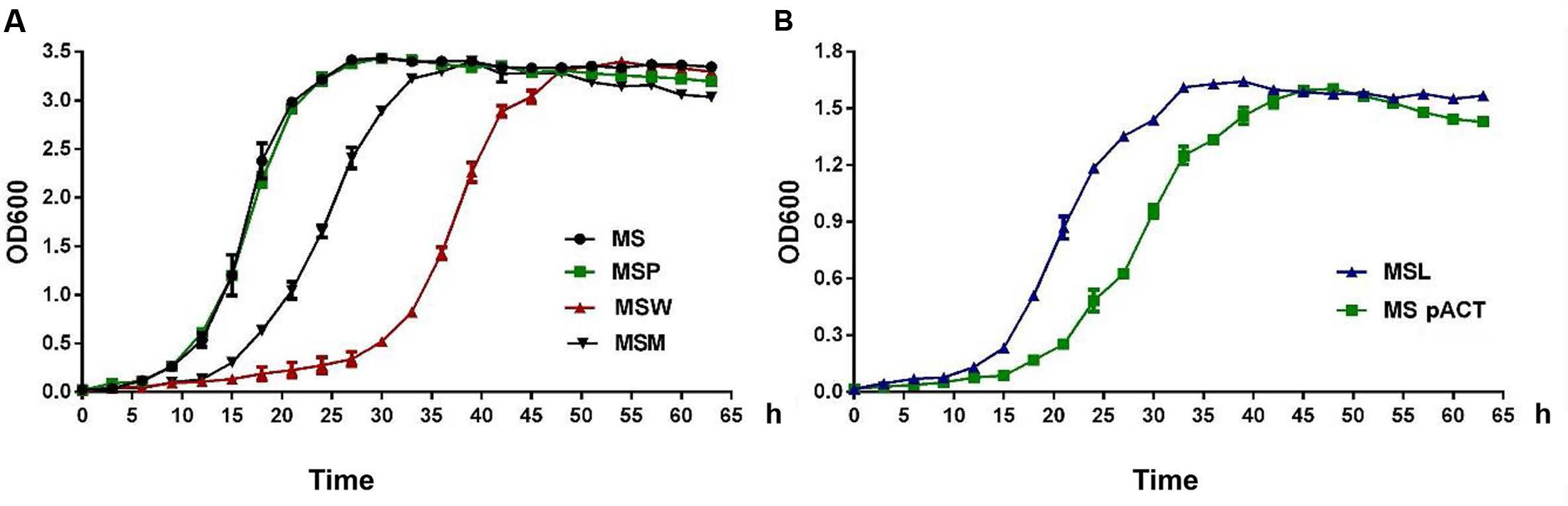
FIGURE 6. The expression of Rv2629 modulates the growth of M. smegmatis under aerobic conditions. The growth curve of recombinant M. smegmatis strains that exhibited reduced expression of Rv2629 and overexpression of the Rv2629W and/or the Rv2629M genes, respectively. The growth of the strains was measured by harvesting cultures at 3-h intervals and subsequent measurement of the absorbance values at 600 nm (OD600). The abbreviations were: MS (wild type), MSP (control strain transformed with the pMV261 plasmid), and MSpACT (control strain transformed with the pACT plasmid). The results indicated (A) comparatively slow growth of MSW. (B) Comparatively rapid growth of MSL. The data are representative of two independent experiments with two samples in each experiment which shows similar results.
MSL strains that expressed low levels of MSMEG_1130 were investigated, as described above. MSpACT was used as the control. The results of two independent experiments indicated comparatively rapid growth of MSL (Figure 6B), while the low expression of MSMEG_1130 promoted bacterial growth and entry to the log phase.
The enhanced and/or attenuated in vitro growth phenotypes of recombinant M. smegmatis prompted the examination of the correlation between Rv2629 expression and dormant phenotype. THP-1 cells were infected with the bacteria. No major differences in the intracellular number of viable bacteria were observed at 24 h, suggesting that recombinant M. smegmatis strains that expressed high and low levels of Rv2629 exhibited similar rates of growth to that of control strains within the first day after the infection (Figure 7A). Nevertheless, the bacterial growth of the over-expressing Rv2629 strains was impaired at day 2 post-infection, whereas the viable bacilli number at the turn point of growth from lag to the log-phase was decreased. This result is consistent to that found in the in vitro experiments. Furthermore, over-expression of Rv2629 decreased the growth rate of the bacilli and further prevented them from achieving higher cell density in the stationary phase. This conclusion was further confirmed by comparison of the growth capability observed in THP1 cells of low-expressing the MSMEG_1130 M. smegmatis strains. The bacilli showed increased growth rate compared with the control samples from 48 to 96 h post-infection (Figure 7B). The growth of MSL in THP-1 cells was evaluated at MOI values of 1:5 and 1:20. The results were consistent with the results observed at MOI of 1:10 (Supplementary Figure S2).
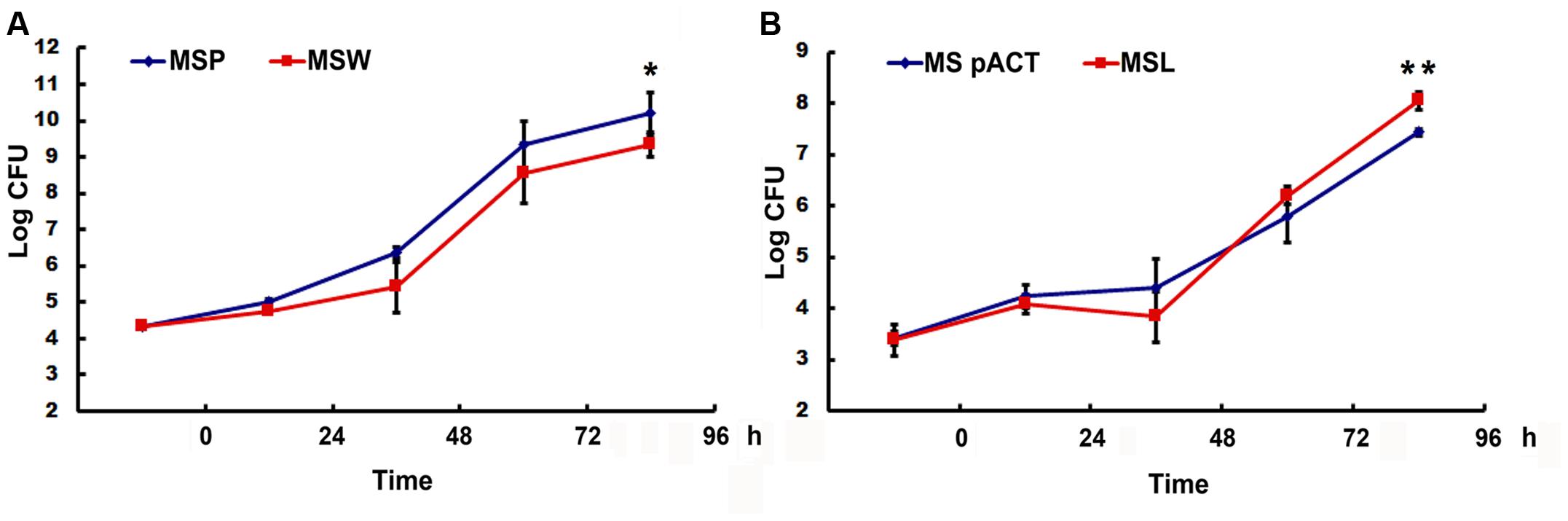
FIGURE 7. The expression of Rv2629 influences the growth of M. smegmatis in human THP-1 cells. (A) M. smegmatis over-expression of Rv2629 (MSW) resulted in the slow growth of THP-1 following infection (P < 0.01, 96 h). (B) M. smegmatis low-expressing MSMEG_1130 (MSL) strain exhibited a rapid growth in THP-1 cells at 96 h post-infection, P < 0.01 (96 h). The data of each time point was analyzed by the Student’s t-test after homogeneity test of variance, n = 4. The experiments were performed in triplicate.
Growth and Pathogenicity of Rv2629 Overexpressing M. smegmatis Strains in Mice
The attenuated reproductive capacity of MSW in macrophage infection prompted us to investigate whether MSW could further decrease the virulence and pathogenicity of host tissues. Balb/c mice were infected with MSP, MSW, and MSM in order to identify the different pathogenicity and survival ability among these strains in vivo. The CFU values of bacteria in the lung, spleen, and kidney tissues were counted. The results indicated that the MSW strain exhibited a longer survival time, notably in the lung tissues compared with MSP, while with regard to the MSW strain the detection of the bacterial growth was evident in the lungs of the mice following 20 days of infection (Figure 8A). This observation occurred despite the absence of CFU counts between MSP and MSW at each time point. The acid-fast staining of lung tissue indicated that overexpression of wild type Rv2629 could promote the survival of M. smegmatis in granulomas of the lung. No significant differences among the three strains were noted at the 15th day after infection. The number of MSW strains that overexpressed the wild type Rv2629 increased significantly compared to the other two strains in the granuloma of the lung at the 20th day after infection. No visible MSP and/or MSM could be found in the diseased location at the 35th day post-infection, although a high number of MSW strains formed granulomas in the lung (Figure 8B). Additional pathological tests indicated that the pathogenicity of MSW increased and additional damage was caused in the lung tissues compared to the MSP and/or MSM strains. The pulmonary alveolus of mice infected with MSP was damaged slightly and it was the sole cause of bleeding in the bronchial lumen in the absence of granuloma formation during the 15th day following infection. Sporadic granuloma could be found in the small part of the lungs at the 20th day after infection, whereas the inflammatory exudate was absorbed and the main part of the lung recovered to the normal structure 35 days post-infection (Figure 9A). Apparent changes could be observed in the lung of MSW-infected mice. The normal structure of the major part of the pulmonary alveolus disappeared, which was permeated with inflammatory exudate and associated with the infiltration of a great number of inflammatory cells. The alveolar wall indicated congestion and incrassation, whereas apparent granuloma could be found in the lungs 15 days post-infection. The area and degree of damage increased further 20 days post-infection, and the normal structure of the complete part of the pulmonary alveolus disappeared. The structure of the majority of the pulmonary alveolus recovered, although a certain part of granuloma could be found in the lung (Figure 9B). The pathogenicity that was caused by the Rv2629 expressing strains decreased when a missense mutation (Asp64 to Ala64, A/C) was present at position 191 of the genetic sequence of Rv2629. The pulmonary alveolus of mice infected with MSM was damaged partly and sporadic granuloma could be found in certain parts of the lungs 15 days post-infection. The damaged structure of pulmonary alveolus recovered gradually and the majority of the lung tissue recovered to the normal structure at the 20th and the 35th day week post-infection, respectively, as noted for the MSP strain (Figure 9C). Consequently, the overexpression of the wild type Rv2629 could increase the survival and pathogenicity of M. smegmatis, whereas the 191 A/C mutation in Rv2629 could weaken the pathogenicity of the Rv2629-expressing strains.
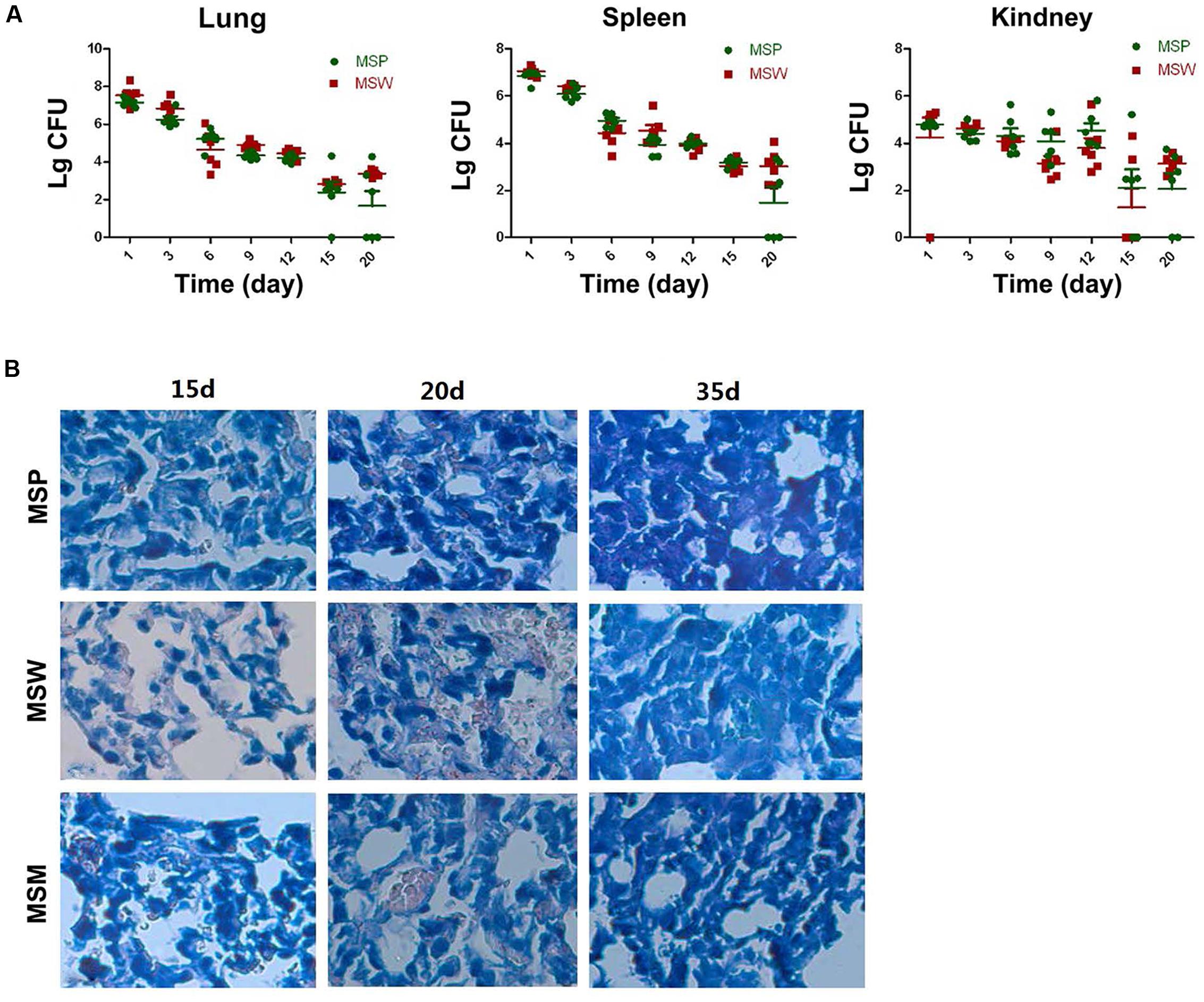
FIGURE 8. The growth of M. smegmatis strains in different organs of Balb/c mice. (A) Colony forming units (CFU) of M. smegmatis in lung, spleen, and kidney tissues of mice infected with M. smegmatis at different time points (0, 3, 6, 9, 12, 15, and 20 days post-infection). (B) Acid-fast staining of lung tissues of mice infected with different M. smegmatis strains (400×) at different time points (15, 20, and 35 days post-infection). The data are representative of two independent experiments, n = 6.
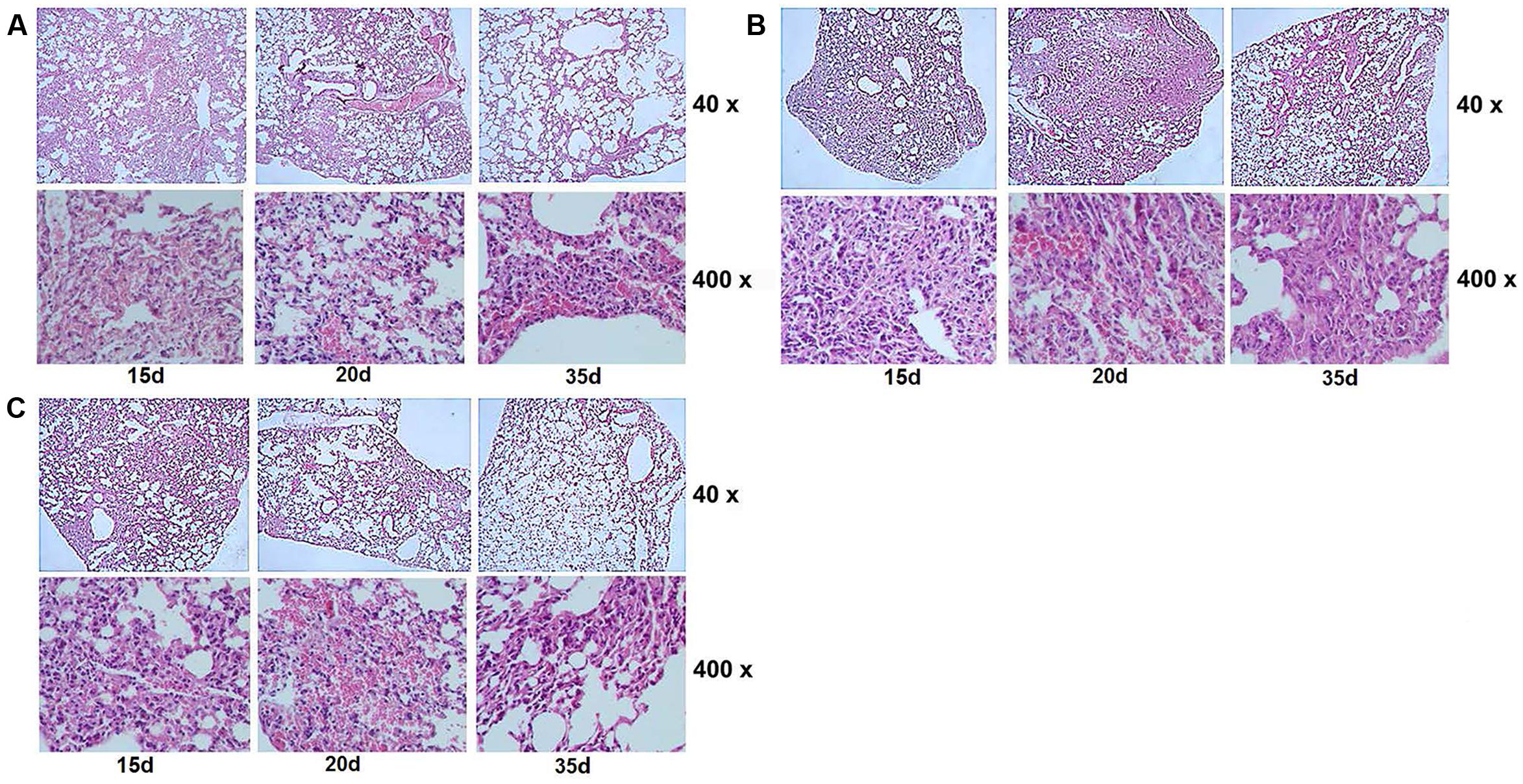
FIGURE 9. Pathological changes in the lung tissues of each group at different time points (15, 20, and 35 d) post-infection. Images (A–C) indicate the lung tissues of mice infected with MSP, MSW, and MSM, respectively. The structure of pulmonary alveolus of the mice infected by MSW was permeated with inflammatory exudates and infiltrated by inflammatory cells. The alveolar wall indicated congestion and incrassation, with apparent granuloma formation in the lungs at 15 and 20 days post-infection. The data are representative of two independent experiments, n = 6.
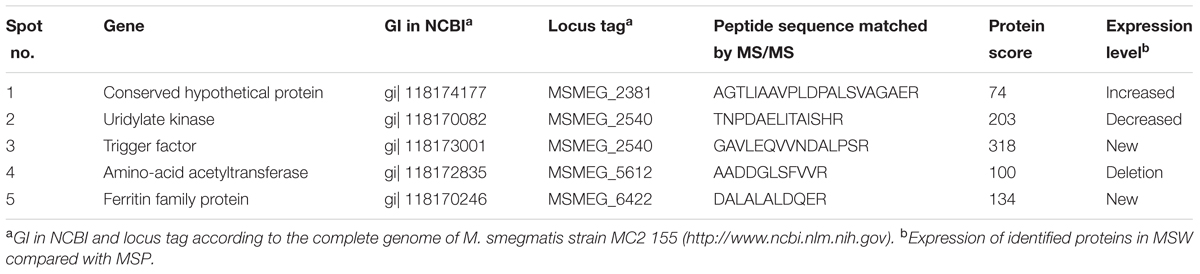
Identification of Protein Expression by MALDI- TOF-MS in M. smegmatis Overexpressing Rv2629
Protein expression maps of M. smegmatis overexpressing Rv2629 were obtained by 2-DE (Figure 10). A total of eight differential proteins were found by comparing the protein differential expressing map on 2-DE gels. A total of five proteins, namely, CHP, UK, TF, AA, and FFP, were identified by MALDI-TOF-MS (Table 3). Nevertheless, the expression pattern of these proteins differed among the strains. TF and FFP were present only on the MSW 2-DE gel, as opposed to the MSP gel. AA was not detected on the MSW 2-DE gel. The expression of UK decreased, while the expression of CHP increased in the MSW compared with the MSP strains.
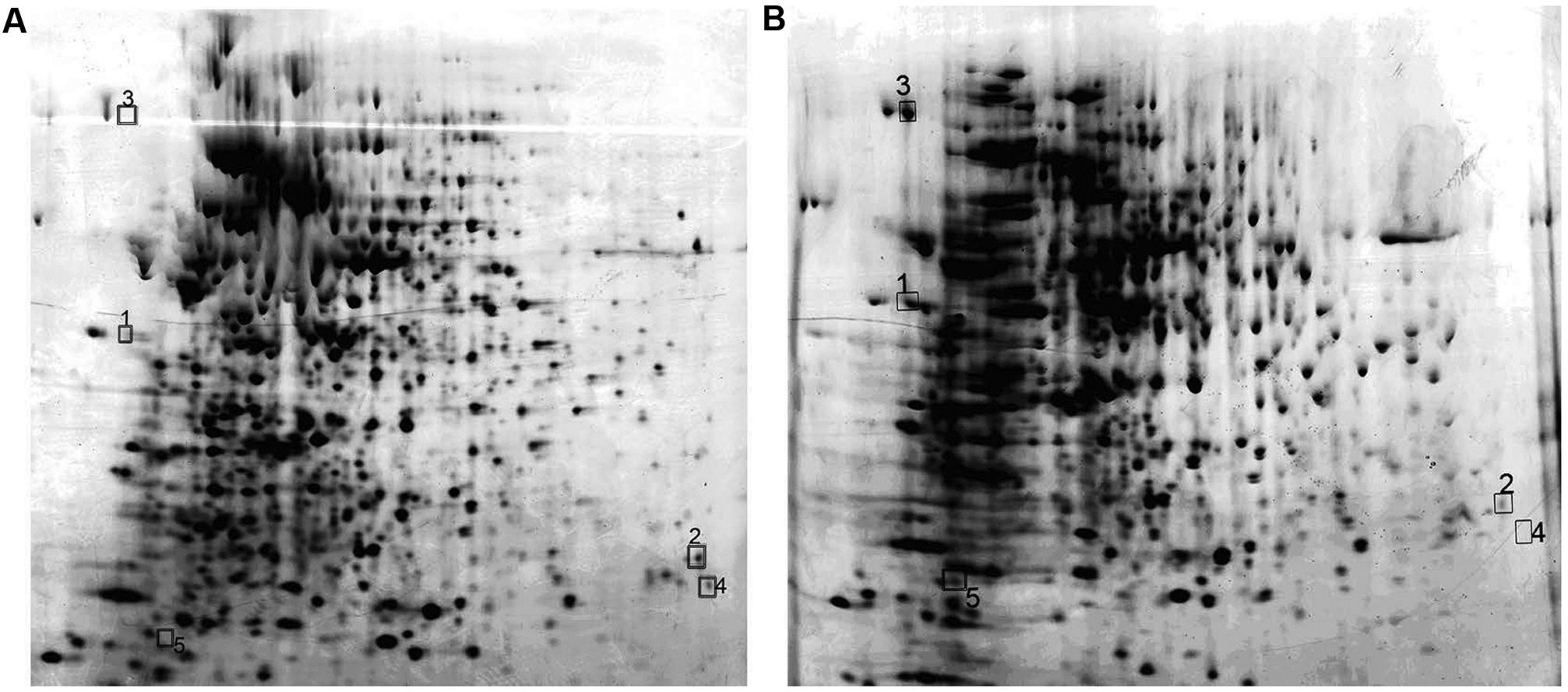
FIGURE 10. 2-DE patterns of whole cell supernatant proteins from MSP and MSW strains that were conducted in the pH range of 4–7. The proteins from MSP (A) and MSW (B) were visualized by silver staining during the late log phase of bacterial growth. The spot numbers on the reverse image correspond to proteins identified by MALDI-TOF-MS, as listed in Table 3. The data are representative of two independent experiments.
Discussion
In the present study, an expression analysis of the Rv2629 native protein was conducted in recombinant M. smegmatis and M. tuberculosis strains and in clinical isolates. In addition, the growth characteristics of recombinant M. smegmatis, expressing Rv2629 wild type and mutant genes were evaluated in vitro and in vivo. It is suggested that Rv2629 is correlated with the survival of the clinical drug-resistant M. tuberculosis strains via the repression of bacterial growth and the induction of bacterial persistence.
It has long been speculated that a latent and/or dormant state may play a role in drug tolerance of M. tuberculosis (Florczyk et al., 2003; Starck et al., 2004; Errey and Blanchard, 2005; Bartek et al., 2009; Peddireddy et al., 2017). The first-line antibiotics used to treat TB are all active against replicating bacteria (Hoffmann et al., 2010) but not against anaerobic dormant bacilli (Ramaswamy et al., 2003; Fu and Fu-Liu, 2007). The fact that the DosR regulon is required for long-term survival under anaerobic conditions has led to the notion that DosR-regulated genes may contribute to the higher level of M. tuberculosis drug tolerance during infection (Boon and Dick, 2002; Florczyk et al., 2003; Ramaswamy et al., 2003; Starck et al., 2004; Errey and Blanchard, 2005; Bartek et al., 2009; Alonso et al., 2011; Peddireddy et al., 2017). The present study indicated that Rv2629 expression was decreased in clinical drug-resistant isolates during the log-phase compared with H37Rv, although the expression of the protein was increased during the stationary-phase. It can be speculated that upregulation of Rv2629 in the stationary-phase is beneficial to the survival of drug-resistant strains in vivo, and therefore the protein may be indirectly associated with drug resistance. The drug tolerance observed in the present study may be due to alternative mechanisms, such as the survival of the bacilli in a non-replicating and/or low metabolic state environment.
The present study indicated that overexpression of Rv2629 may inhibit the biosynthesis of M. smegmatis in the lag-phase, during which the synthesis of the enzymes and proteins required for metabolism and rapid reproduction occurs. TEM indicated that the ribosome content was decreased in MSW in the log phase compared with the MSP strain. Furthermore, 2D-gel electrophoresis indicated a decrease of UK and a deletion of AA proteins in MSW compared with the MSP strain. The enzymes are known to be involved in pyridine synthesis and arginine metabolism in M. tuberculosis (Errey and Blanchard, 2005; Rostirolla et al., 2011). UKs, also known as UMP kinases, are key enzymes in the synthesis of nucleoside triphosphates. The members of the UMP kinase family catalyze the conversion of UMP to UDP, an essential step in the pyrimidine metabolic pathway in a variety of bacteria (Yoshida et al., 2012). In many bacterial genomes, the gene tends to be located immediately downstream of elongation factor T and upstream of ribosome recycling factor. This enzyme has been shown to be essential in bacteria. The product of pyrH (Rv2883) gene, a member of the UMP kinase family, has been shown to be essential for M. tuberculosis growth by the rapid screening method (Lee et al., 2007; Rostirolla et al., 2011). Genetic studies have provided evidence that UMP kinases are essential for the growth of both Gram-negative (e.g., E. coli) and Gram-positive bacteria (e.g., Streptococcus pneumoniae) (Yamanaka et al., 1992; Fassy et al., 2004). Therefore, it can be speculated that Rv2629 delays entry in the log-phase by inhibiting protein biosynthesis. Delayed growth under conditions of dormancy is likely to be beneficial for the survival of bacteria in the host, and therefore may contribute to the increased pathogenicity of the strain. Although Rv2629 has been characterized as a dormancy protein that is induced under anaerobic conditions of M. tuberculosis, the main focus of research has been toward the ability of this protein to confer resistance to bacterial strains (Florczyk et al., 2003; Wang et al., 2007). The current study investigates the novel hypothesis that the reduced growth and dormancy-type characteristics of M. smegmatis and M. tuberculosis are not entirely dependent on antibiotic resistance. The drug tolerance observed in the present study may be due to alternative mechanisms, such as the survival of the bacilli in a non-replicating and/or low metabolic state. Similarly, it has been reported that the phenotypic tolerance acquired in M. tuberculosis to isoniazid treatment is via the activation of mycobacterial DNA activating protein (MDP1) and the regulation of kat G transcription (Niki et al., 2012).
The aforementioned results supported the functional overlap of Rv2629 between M. smegmatis and M. tuberculosis. The overlap between the two strains has been proposed by a previous study (Homolka et al., 2009). The dosR gene exhibited a severe survival defect in M. smegmatis under hypoxic conditions and this phenotype could be complemented by the dosR gene from M. tuberculosis, which supported the functional overlap between DosR from M. smegmatis and M. tuberculosis (Boon and Dick, 2002; Park et al., 2003; Homolka et al., 2009).
The pathogenicity of the Rv2629 overexpressing M. smegmatis strain in the tissues of BalB/c mice was investigated and it was found that the overexpression of Rv2629 increased the bacterial survival in the lung and spleen organs. The exact cause of this effect remains unknown. Microarray analysis indicated that the mRNA levels of several proteins, namely, MCE-family protein Mce4C (MRA_3537), secreted antigen 85-B FbpB (MRA_1897), and Mce associated protein (MRA_3532), were upregulated in the recombinant strain wild type Rv2629 compared with the Rv2629 mutant strain, which might be associated with the increased pathogenicity and dormancy resistance. Mce4 encodes a cholesterol transporter that is associated with cholesterol metabolism and is essential for mycobacterial persistence. The disruption of mce4 results in failure of M. tuberculosis to maintain chronic infection in mice, while retaining full virulence during the acute phase (Mohn et al., 2008; Pandey and Sassetti, 2008). In contrast to these findings, the MCE family protein and the Mce-associated protein are generally considered as virulence factors, and are associated with lipid transport and host cell invasion (Rodriguez et al., 2015; Perkowski et al., 2016). Ag85B may also contribute to the adherence, invasion, and dissemination of organisms in the host tissue, which can bind to Fn and play a critical role in mycobacterial adherence to the host cells (Kassa et al., 2012). Furthermore, Ag85B serves as an important colonization factor potentially contributing to mycobacterial virulence (Kuo et al., 2012). To date, no proteomic studies have been conducted with regard to the Rv2629 mutant strain of M. smegmatis and/or M. tuberculosis. The 2D-gel electrophoresis indicated that the proteins TF and FFP were expressed in MSW, but not in MSP strains. TF is a FK506 binding protein (FKBP)-type peptidyl-prolyl cis-trans isomerase (PPIase) that is considered a highly conserved ribosome-associated chaperone found in a vast number of bacteria (Hoffmann et al., 2010). This protein has been reported to be essential for protein synthesis and survival under conditions of stress (Wu et al., 2011). The present study indicated that overexpression of Rv2629 induced the expression of TF and FFP and might improve bacterial survival in the persistence state.
The success of the pathogenicity of M. tuberculosis is largely attributed to its ability to manipulate the host immune responses. The genome of M. tuberculosis encodes multiple immune-modulatory proteins that have been implicated in the pathogenicity of this strain. In agreement with the main concept of the current study, it has been shown that the immune-modulatory protein PE_PGRS41 boosted the survival of M. smegmatis within macrophages and was accompanied by enhanced cytotoxic cell death via inhibition of apoptosis and autophagy. This indicated that specific bacterial proteins can act as virulence factors that enhance the pathogenic properties of M. smegmatis (Deng et al., 2017). To the best of our knowledge the implication of the antigenic protein Rv2629 in the virulence of M. smegmatis has not been reported to date. The number of studies that have examined the contribution of this antigen in the immune response elicited against M. tuberculosis is very limited (eight in total). The majority of the studies on Rv2629 have focused on the contribution of this protein to the immune response elicited against M. tuberculosis as well as in the resistance developed against agents such as Rifampicin (Chakravorty et al., 2008; Louw et al., 2009; Niki et al., 2012). In addition, polymorphisms of Rv2629 have been used as potential markers for the identification process of certain M. tuberculosis genotypes (Alonso et al., 2011; Zhang et al., 2014). Consequently, the current study offers novel evidence on the potential interplay of the Rv2629 protein regarding the survival of M. tuberculosis.
Based on KEGG database, the overlap value between MSMEG_1130 and Rv2629 was 366. Moreover, Blastn data showed that there are 54% identical nucleotide sequences and Blastp data showed that there are 59% similar or identical amino acid sequences between them. The nucleotide and amino acid sequences of two genes are similar, there was certain homology of nucleotide and amino acid sequences between the two genes. Therefore, MSMEG_1130 was the most homologous protein with Rv2629 in M. smegmatis strains, with more than 50% amino acids identical or similar with Rv2629. To further clarify the problem, other low-expression models such as the MRA_2657 low-expression H37Ra strain should be further investigated. Furthermore, there was no experimental data supporting the different function of MSMEG 1130 from Rv2629. On the contrary, MSMEG_1130 might have similar function with Rv2629 related with the growth of M. smegmatis, for our data show MSMEG_1130 low-expression strain exhibited a rapid growth compared with the control. MSMEG_1130 was also considered as a protein related with the growth of M. smegmatis in other study (Lauten et al., 2010).
Conclusion
The present study strongly suggests that the overexpression of the wild type Rv2629 protein could delay the growth of the M. smegmatis by modifying the expression of specific proteins involved in bacterial metabolism. The overexpression of both WT and mutant Rv2629 did not affect the susceptibility of M. smegmatis to antibiotics, although it increased its survival and pathogenicity in vivo. The findings propose a potential explanation regarding the association of the protein Rv2629 with the survival of the clinical drug resistant M. tuberculosis strain via the repression of bacterial growth and the induction of bacterial persistence in vivo. Nevertheless, the biological function and signaling pathway of Rv2629 warrants further investigation.
Author Contributions
QW and LZ conceived and supervised the study; DL, KH, WW, CP, and YD performed experiments. RJ and WX provided new tools and reagents; HyW and HhW analyzed data and helped to design experiments; DL, QW and LZ wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant No. 81401300), the China’s 13th Five Year Programs for the prevention and cure of great infectious diseases (2017ZX10201301-005 and 2017ZX10301301-001-005), the Shanghai Municipal Natural Science Foundation (16ZR1402800), the International cooperation and exchange program of Shanghai Municipal (16430724000), and the Shanghai Municipal Natural Science Foundation (13ZR1441800).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02231/full#supplementary-material
FIGURE S1 | Expression levels of Rv2629 modulate the growth of H37Ra strains under aerobic conditions. The growth curve of recombinant H37Ra overexpressing the Rv2629W (RaW) and/or Rv2629M (RaM) genes were measured using the BD BACTEC MGIT 960 Mycobacterium growth and detection system. The results are shown as the fluorescence intensity acquired automatically. RaP (transformed with the pMV261 plasmid) was used as a control. The results indicated comparatively slow growth of RaW. Data are representative of two experiments.
FIGURE S2 | The growth of MSL in THP-1 cells infected at different multiplicity of infections (MOI). (A) The M. smegmatis low-expressing MSMEG_1130 (MSL) strain exhibited a rapid growth in THP-1 cells at 72 and 96 h post-infection, when infected at a MOI value of 1:5, P < 0.001(72 and 96 h). (B) M. smegmatis low-expressing MSMEG_1130 (MSL) strain exhibited a rapid growth in THP-1 cells at 72 and 96 h post-infection, when infected at a MOI value of 1:20, P < 0.05 (72 and 96 h). The data of each time point was analyzed by the Student’s t-test after homogeneity test of variance, n = 4.
Abbreviations
AA, amino-acid acetytransferase; ACP, acr-coregulated promoter; ADC, albumin-dextrose-catalase; BCIP/NBT, 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium; CHP, conserved hypothetical protein; FFP, ferritin family protein; FtsZ, filamentous temperature-sensitive protein Z; GO, Gene Ontology; MDR, multidrug resistance; MS, mass spectrometry; SD, standard deviation; TB, tuberculosis; TF, trigger factor; UK, uridylate kinase.
References
Alonso, M., Navarro, Y., Barletta, F., Martinez Lirola, M., Gotuzzo, E., Bouza, E., et al. (2011). A novel method for the rapid and prospective identification of Beijing Mycobacterium tuberculosis strains by high-resolution melting analysis. Clin. Microbiol. Infect. 17, 349–357. doi: 10.1111/j.1469-0691.2010.03234.x
Altaf, M., Miller, C. H., Bellows, D. S., and O’Toole, R. (2010). Evaluation of the Mycobacterium smegmatis and BCG models for the discovery of Mycobacterium tuberculosis inhibitors. Tuberculosis 90, 333–337. doi: 10.1016/j.tube.2010.09.002
Andreu, N., Soto, C. Y., Roca, I., Martin, C., and Gibert, I. (2004). Mycobacterium smegmatis displays the Mycobacterium tuberculosis virulence-related neutral red character when expressing the Rv0577 gene. FEMS Microbiol. Lett. 231, 283–289. doi: 10.1016/S0378-1097(04)00008-4
Bae, H. J., Lee, H. N., Baek, M. N., Park, E. J., Eom, C. Y., Ko, I. J., et al. (2017). Inhibition of the DevSR two-component system by overexpression of Mycobacterium tuberculosis PknB in Mycobacterium smegmatis. Mol. Cells 40, 632–642. doi: 10.14348/molcells.2017.0076
Bartek, I. L., Rutherford, R., Gruppo, V., Morton, R. A., Morris, R. P., Klein, M. R., et al. (2009). The DosR regulon of M. tuberculosis and antibacterial tolerance. Tuberculosis 89, 310–316. doi: 10.1016/j.tube.2009.06.001
Belanger, A. E., and Hatfull, G. F. (1999). Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J. Bacteriol. 181, 6670–6678.
Boon, C., and Dick, T. (2002). Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184, 6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002
Caviedes, L., Delgado, J., and Gilman, R. H. (2002). Tetrazolium microplate assay as a rapid and inexpensive colorimetric method for determination of antibiotic susceptibility of Mycobacterium tuberculosis. J. Clin. Microbiol. 40, 1873–1874. doi: 10.1128/JCM.40.5.1873-1874.2002
Chakravorty, S., Aladegbami, B., Motiwala, A. S., Dai, Y., Safi, H., Brimacombe, M., et al. (2008). Rifampin resistance, Beijing-W clade-single nucleotide polymorphism cluster group 2 phylogeny, and the Rv2629 191-C allele in Mycobacterium tuberculosis strains. J. Clin. Microbiol. 46, 2555–2560. doi: 10.1128/JCM.00666-08
Daugelat, S., Kowall, J., Mattow, J., Bumann, D., Winter, R., Hurwitz, R., et al. (2003). The RD1 proteins of Mycobacterium tuberculosis: expression in Mycobacterium smegmatis and biochemical characterization. Microbes Infect. 5, 1082–1095. doi: 10.1016/S1286-4579(03)00205-3
Deng, W., Long, Q., Zeng, J., Li, P., Yang, W., Chen, X., et al. (2017). Mycobacterium tuberculosis PE_PGRS41 enhances the intracellular survival of M. smegmatis within macrophages via blocking innate immunity and inhibition of host defense. Sci. Rep. 7:46716. doi: 10.1038/srep46716
Domenech, P., Zou, J., Averback, A., Syed, N., Curtis, D., Donato, S., et al. (2017). Unique regulation of the DosR regulon in the Beijing lineage of Mycobacterium tuberculosis. J. Bacteriol. 199:e00696–16. doi: 10.1128/JB.00696-16
Doosti-Irani, A., Ayubi, E., and Mostafavi, E. (2016). Tuberculin and QuantiFERON-TB-Gold tests for latent tuberculosis: a meta-analysis. Occup. Med. 66, 437–445. doi: 10.1093/occmed/kqw035
Errey, J. C., and Blanchard, J. S. (2005). Functional characterization of a novel ArgA from Mycobacterium tuberculosis. J. Bacteriol. 187, 3039–3044. doi: 10.1128/JB.187.9.3039-3044.2005
Falcone, V., Bassey, E., Jacobs, W. Jr., and Collins, F. (1995). The immunogenicity of recombinant Mycobacterium smegmatis bearing BCG genes. Microbiology 141(Pt 5), 1239–1245. doi: 10.1099/13500872-141-5-1239
Fassy, F., Krebs, O., Lowinski, M., Ferrari, P., Winter, J., Collard-Dutilleul, V., et al. (2004). UMP kinase from Streptococcus pneumoniae: evidence for co-operative ATP binding and allosteric regulation. Biochem. J. 384(Pt 3), 619–627. doi: 10.1042/BJ20040440
Florczyk, M. A., McCue, L. A., Purkayastha, A., Currenti, E., Wolin, M. J., and McDonough, K. A. (2003). A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71, 5332–5343. doi: 10.1128/IAI.71.9.5332-5343.2003
Fu, L. M., and Fu-Liu, C. S. (2007). The gene expression data of Mycobacterium tuberculosis based on Affymetrix gene chips provide insight into regulatory and hypothetical genes. BMC Microbiol. 7:37. doi: 10.1186/1471-2180-7-37
Gharahdaghi, F., Weinberg, C. R., Meagher, D. A., Imai, B. S., and Mische, S. M. (1999). Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20, 601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6
Hoffmann, A., Bukau, B., and Kramer, G. (2010). Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 1803, 650–661. doi: 10.1016/j.bbamcr.2010.01.017
Homolka, S., Koser, C., Archer, J., Rusch-Gerdes, S., and Niemann, S. (2009). Single-nucleotide polymorphisms in Rv2629 are specific for Mycobacterium tuberculosis genotypes Beijing and Ghana but not associated with rifampin resistance. J. Clin. Microbiol. 47, 223–226. doi: 10.1128/JCM.01237-08
Kassa, D., Ran, L., Geberemeskel, W., Tebeje, M., Alemu, A., Selase, A., et al. (2012). Analysis of immune responses against a wide range of Mycobacterium tuberculosis antigens in patients with active pulmonary tuberculosis. Clin. Vaccine Immunol. 19, 1907–1915. doi: 10.1128/CVI.00482-12
Keshari, D., Singh, K. S., Sharma, R., Yadav, S., and Singh, S. K. (2017). MSMEG_5684 down-regulation in Mycobacterium smegmatis affects its permeability, survival under stress and persistence. Tuberculosis 103, 61–70. doi: 10.1016/j.tube.2017.01.004
Kuo, C. J., Bell, H., Hsieh, C. L., Ptak, C. P., and Chang, Y. F. (2012). Novel mycobacteria antigen 85 complex binding motif on fibronectin. J. Biol. Chem. 287, 1892–1902. doi: 10.1074/jbc.M111.298687
Kwon, K. W., Kim, W. S., Kim, H., Han, S. J., Hahn, M. Y., Lee, J. S., et al. (2017). Novel vaccine potential of Rv3131, a DosR regulon-encoded putative nitroreductase, against hyper-virulent Mycobacterium tuberculosis strain K. Sci. Rep. 7:44151. doi: 10.1038/srep44151
Lauten, E. H., Pulliam, B. L., DeRousse, J., Bhatta, D., and Edwards, D. A. (2010). Gene expression, bacteria viability and survivability following spray drying of Mycobacterium smegmatis. Materials (Basel) 3, 2684–2724. doi: 10.3390/ma3042684
Lee, S. E., Kim, S. Y., Kim, C. M., Kim, M. K., Kim, Y. R., Jeong, K., et al. (2007). The pyrH gene of Vibrio vulnificus is an essential in vivo survival factor. Infect. Immun. 75, 2795–2801. doi: 10.1128/IAI.01499-06
Louw, G. E., Warren, R. M., van Helden, P. D., and Victor, T. C. (2009). Rv2629 191A/C nucleotide change is not associated with rifampicin resistance in Mycobacterium tuberculosis. Clin. Chem. Lab. Med. 47, 500–501. doi: 10.1515/CCLM.2009.111
Mohn, W. W., van der Geize, R., Stewart, G. R., Okamoto, S., Liu, J., Dijkhuizen, L., et al. (2008). The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283, 35368–35374. doi: 10.1074/jbc.M805496200
Niki, M., Niki, M., Tateishi, Y., Ozeki, Y., Kirikae, T., Lewin, A., et al. (2012). A novel mechanism of growth phase-dependent tolerance to isoniazid in mycobacteria. J. Biol. Chem. 287, 27743–27752. doi: 10.1074/jbc.M111.333385
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Park, H. D., Guinn, K. M., Harrell, M. I., Liao, R., Voskuil, M. I., Tompa, M., et al. (2003). Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843. doi: 10.1046/j.1365-2958.2003.03474.x
Peddireddy, V., Doddam, S. N., and Ahmed, N. (2017). Mycobacterial dormancy systems and host responses in tuberculosis. Front. Immunol. 8:84. doi: 10.3389/fimmu.2017.00084
Perkowski, E. F., Miller, B. K., McCann, J. R., Sullivan, J. T., Malik, S., Allen, I. C., et al. (2016). An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol. Microbiol. 100, 90–107. doi: 10.1111/mmi.13303
Ramaswamy, S. V., Reich, R., Dou, S. J., Jasperse, L., Pan, X., Wanger, A., et al. (2003). Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003
Reed, M. B., Gagneux, S., Deriemer, K., Small, P. M., and Barry, C. E. III. (2007). The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J. Bacteriol. 189, 2583–2589. doi: 10.1128/JB.01670-06
Rodriguez, D. C., Ocampo, M., Varela, Y., Curtidor, H., Patarroyo, M. A., and Patarroyo, M. E. (2015). Mce4F Mycobacterium tuberculosis protein peptides can inhibit invasion of human cell lines. Pathog. Dis. 73:ftu020. doi: 10.1093/femspd/ftu020
Rostirolla, D. C., Breda, A., Rosado, L. A., Palma, M. S., Basso, L. A., and Santos, D. S. (2011). UMP kinase from Mycobacterium tuberculosis: mode of action and allosteric interactions, and their likely role in pyrimidine metabolism regulation. Arch. Biochem. Biophys. 505, 202–212. doi: 10.1016/j.abb.2010.10.019
Scheler, C., Lamer, S., Pan, Z., Li, X. P., Salnikow, J., and Jungblut, P. (1998). Peptide mass fingerprint sequence coverage from differently stained proteins on two-dimensional electrophoresis patterns by matrix assisted laser desorption/ionization-mass spectrometry (MALDI-MS). Electrophoresis 19, 918–927. doi: 10.1002/elps.1150190607
Starck, J., Kallenius, G., Marklund, B. I., Andersson, D. I., and Akerlund, T. (2004). Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150(Pt 11), 3821–3829. doi: 10.1099/mic.0.27284-0
Sun, C., Yang, G., Yuan, J., Peng, X., Zhang, C., Zhai, X., et al. (2017). Mycobacterium tuberculosis hypoxic response protein 1 (Hrp1) augments the pro-inflammatory response and enhances the survival of Mycobacterium smegmatis in murine macrophages. J. Med. Microbiol. 66, 1033–1044. doi: 10.1099/jmm.0.000511
Sweeney, K. A., Dao, D. N., Goldberg, M. F., Hsu, T., Venkataswamy, M. M., Henao-Tamayo, M., et al. (2011). A recombinant Mycobacterium smegmatis induces potent bactericidal immunity against Mycobacterium tuberculosis. Nat. Med. 17, 1261–1268. doi: 10.1038/nm.2420
Trauner, A., Lougheed, K. E., Bennett, M. H., Hingley-Wilson, S. M., and Williams, H. D. (2012). The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 287, 24053–24063. doi: 10.1074/jbc.M112.364851
Tumminia, S. J., Mandiyan, V., Wall, J. S., and Boublik, M. (1991). Heterogeneity of Escherichia coli ribosomes established by scanning transmission electron microscopy. Biochimie 73, 919–925. doi: 10.1016/0300-9084(91)90133-L
Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713. doi: 10.1084/jem.20030205
Wang, L., Zuo, M., Chen, H., Liu, S., Wu, X., Cui, Z., et al. (2017). Mycobacterium tuberculosis Lipoprotein MPT83 induces apoptosis of infected macrophages by activating the TLR2/p38/COX-2 signaling pathway. J. Immunol. 198, 4772–4780. doi: 10.4049/jimmunol.1700030
Wang, Q., Yue, J., Zhang, L., Xu, Y., Chen, J., Zhang, M., et al. (2007). A newly identified 191A/C mutation in the Rv2629 gene that was significantly associated with rifampin resistance in Mycobacterium tuberculosis. J. Proteome Res. 6, 4564–4571. doi: 10.1021/pr070242z
Wu, T., Zhao, Z., Zhang, L., Ma, H., Lu, K., Ren, W., et al. (2011). Trigger factor of Streptococcus suis is involved in stress tolerance and virulence. Microb. Pathog. 51, 69–76. doi: 10.1016/j.micpath.2010.10.001
Yamanaka, K., Ogura, T., Niki, H., and Hiraga, S. (1992). Identification and characterization of the smbA gene, a suppressor of the mukB null mutant of Escherichia coli. J. Bacteriol. 174, 7517–7526. doi: 10.1128/jb.174.23.7517-7526.1992
Yoshida, T., Nasu, H., Namba, E., Ubukata, O., and Yamashita, M. (2012). Discovery of a compound which acts as a bacterial UMP kinase PyrH inhibitor. FEMS Microbiol. Lett. 330, 121–126. doi: 10.1111/j.1574-6968.2012.02546.x
Yu, Z., Zhang, C., Zhou, M., Li, Q., Li, H., Duan, W., et al. (2017). Mycobacterium tuberculosis PPE44 (Rv2770c) is involved in response to multiple stresses and promotes the macrophage expression of IL-12 p40 and IL-6 via the p38, ERK, and NF-kappaB signaling axis. Int. Immunopharmacol. 50, 319–329. doi: 10.1016/j.intimp.2017.06.028
Zhang, L., Xu, W., Cui, Z., Liu, Y., Wang, W., Wang, J., et al. (2014). A novel method of identifying Mycobacterium tuberculosis Beijing strains by detecting SNPs in Rv0444c and Rv2629. Curr. Microbiol. 68, 381–386. doi: 10.1007/s00284-013-0487-2
Keywords: Rv2629, Mycobacterium smegmatis, Mycobacterium tuberculosis, bacterial physiology, dormancy
Citation: Liu D, Hao K, Wang W, Peng C, Dai Y, Jin R, Xu W, He L, Wang H, Wang H, Zhang L and Wang Q (2017) Rv2629 Overexpression Delays Mycobacterium smegmatis and Mycobacteria tuberculosis Entry into Log-Phase and Increases Pathogenicity of Mycobacterium smegmatis in Mice. Front. Microbiol. 8:2231. doi: 10.3389/fmicb.2017.02231
Received: 20 July 2017; Accepted: 31 October 2017;
Published: 15 November 2017.
Edited by:
Juarez Antonio Simões Quaresma, Universidade Federal do Pará, BrazilReviewed by:
Anete S. Grumach, Faculty of Medicine ABC, BrazilHridayesh Prakash, University of Hyderabad, India
Copyright © 2017 Liu, Hao, Wang, Peng, Dai, Jin, Xu, He, Wang, Wang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Zhang, emhhbmdsdTQwN0BmdWRhbi5lZHUuY24= Qingzhong Wang, emx3cXpAMTYzLmNvbQ==
 Dan Liu
Dan Liu Kewei Hao
Kewei Hao Wenjie Wang
Wenjie Wang Chao Peng
Chao Peng Yue Dai
Yue Dai Ruiliang Jin
Ruiliang Jin Wenxi Xu
Wenxi Xu Lei He
Lei He Hongyan Wang
Hongyan Wang Honghai Wang
Honghai Wang Lu Zhang
Lu Zhang Qingzhong Wang
Qingzhong Wang