
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 02 November 2017
Sec. Extreme Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02140
This article is part of the Research TopicThermophilic and Halophilic Extremophiles in Eurasian EnvironmentsView all 12 articles
 Alexander G. Elcheninov1*†
Alexander G. Elcheninov1*† Peter Menzel2,3†
Peter Menzel2,3† Soley R. Gudbergsdottir2
Soley R. Gudbergsdottir2 Alexei I. Slesarev4
Alexei I. Slesarev4 Vitaly V. Kadnikov5
Vitaly V. Kadnikov5 Anders Krogh2
Anders Krogh2 Elizaveta A. Bonch-Osmolovskaya1
Elizaveta A. Bonch-Osmolovskaya1 Xu Peng2
Xu Peng2 Ilya V. Kublanov1,6*
Ilya V. Kublanov1,6*Xanthan gum, a complex polysaccharide comprising glucose, mannose and glucuronic acid residues, is involved in numerous biotechnological applications in cosmetics, agriculture, pharmaceuticals, food and petroleum industries. Additionally, its oligosaccharides were shown to possess antimicrobial, antioxidant, and few other properties. Yet, despite its extensive usage, little is known about xanthan gum degradation pathways and mechanisms. Thermogutta terrifontis, isolated from a sample of microbial mat developed in a terrestrial hot spring of Kunashir island (Far-East of Russia), was described as the first thermophilic representative of the Planctomycetes phylum. It grows well on xanthan gum either at aerobic or anaerobic conditions. Genomic analysis unraveled the pathways of oligo- and polysaccharides utilization, as well as the mechanisms of aerobic and anaerobic respiration. The combination of genomic and transcriptomic approaches suggested a novel xanthan gum degradation pathway which involves novel glycosidase(s) of DUF1080 family, hydrolyzing xanthan gum backbone beta-glucosidic linkages and beta-mannosidases instead of xanthan lyases, catalyzing cleavage of terminal beta-mannosidic linkages. Surprisingly, the genes coding DUF1080 proteins were abundant in T. terrifontis and in many other Planctomycetes genomes, which, together with our observation that xanthan gum being a selective substrate for many planctomycetes, suggest crucial role of DUF1080 in xanthan gum degradation. Our findings shed light on the metabolism of the first thermophilic planctomycete, capable to degrade a number of polysaccharides, either aerobically or anaerobically, including the biotechnologically important bacterial polysaccharide xanthan gum.
Planctomycetes is a bacterial phylum, comprising only a few cultivated species, while a large number of ribosomal RNA sequences from various uncultured planctomycetes have been observed in the SILVA Ref database (release 128, Quast et al., 2013). Altogether, around a hundred sequenced planctomycetes genomes are available today, including 38 genome sequences of cultivated and ca. twice that amount of uncultivated ones (IMG database, March 2017, Markowitz et al., 2012). Cultivated planctomycetes with validly published names comprise 2 classes, 3 orders, 5 families, 25 genera, and 29 species. Until recently, all of these were characterized as strictly aerobic, heterotrophic and peptidoglycan-less microorganisms, which reproduce by budding and grow at mesophilic and slightly psychrophilic conditions (Liesack et al., 1986; Fuerst and Sagulenko, 2011). However, all of these features were reconsidered during the past few years. Their cell walls have been shown to contain a uniquely thin peptidoglycan layer (Jeske et al., 2015), representatives of the novel class Phycisphaerae divide by binary fission (Fukunaga et al., 2009; Kovaleva et al., 2015) instead of budding, and, finally, a few thermophilic and facultative anaerobic representatives were recently isolated (Kovaleva et al., 2015; Slobodkina et al., 2015). Even though no autotrophic planctomycetes were isolated and cultivated so far, members of the third class-level lineage, represented by uncultivated anammox planctomycetes (van de Graaf et al., 1995), are thought to fix CO2 via the acetyl-CoA pathway (Strous et al., 2006).
Thermogutta terrifontis was characterized as the first thermophilic representative of the phylum Planctomycetes (Slobodkina et al., 2015). Among two species of the genus one, T. hypogea, was isolated from a subsurface environment of a Beatrix gold mine, SAR, while the second, T. terrifontis, was isolated from a microbial mat, developed in a terrestrial hot spring of Kunashir island (Far-East of Russia). As other cultivated planctomycetes, T. terrifontis grew well on various carbohydrates including oligo- and polysaccharides. At the same time its capability of anaerobic growth by either fermentation or anaerobic respiration was a novel finding among the representatives of this phylum. Yet, nothing is known on the mechanisms underlying these novel capabilities. T. terrifontis strain R1 has been shown to grow on xanthan gum (Slobodkina et al., 2015) – a complex polysaccharide synthesized by Xanthomonas campestris, comprising a beta-1,4-glucan backbone and mannosyl–glucuronyl–mannose side chains. Xanthan gum itself has numerous biotechnological applications in cosmetics, agriculture pharmaceuticals, food and petroleum industries (García-Ochoa et al., 2000). Moreover, its oligosaccharides were described as elicitors, stimulating plants in their defense response against pathogens (Liu et al., 2005); as antimicrobial compounds (Qian et al., 2006) and as antioxidants (Xiong et al., 2013). At the moment, not much is known about its decomposition pathways, especially on mechanisms and involved proteins. A few key enzymes needed to break xanthan gum side chains (xanthan lyases, beta-glucuronyl hydrolases, and alpha-mannosidases) are known, yet still no glycosidases acting on the glucan backbone of xanthan gum have been characterized. Here, we reconstructed the central carbohydrate metabolism of T. terrifontis strain R1 using genomic and transcriptomic sequencing with a special emphasis on xanthan gum degradation.
Thermogutta terrifontis R1 cells were grown under microaerophilic conditions for 2–4 days in glass bottles, sealed with butyl rubber plug with aluminum cap, on modified Pfenning medium supplemented with xanthan gum or trehalose as a growth substrate. Mineral growth medium (Podosokorskaya et al., 2011) was prepared aerobically; 50 mg l-1 of yeast extract (Sigma) was added as a source of indefinite growth factors. Atmospheric air was in the gas phase and no reducing agents or resazurin were added. pH was adjusted to 6.1 with 5 M NaOH. Xanthan gum (1 g/l) [KELTROL®T (food grade Xanthan Gum, Lot#2F5898K) CP Kelco] or trehalose (2 g/l) (Sigma, T9531) were added from sterile stock solutions before inoculation.
For genome sequencing 2000 ml of T. terrifontis grown culture was centrifuged at 17664 G, and cells pellet was harvested. Total DNA was extracted from the cell pellet by freezing and thawing in TNE buffer (Tris 20 mM, NaCl 15 mM, EDTA 20 mM). After treating with lysozyme, RNase A, SDS, and proteinase K the DNA was extracted with phenol/chloroform and precipitated with EtOH and dissolved in 2 mM TE buffer (Gavrilov et al., 2016).
For the transcriptomic experiment, cultures on both xanthan gum and trehalose were grown for 4 days. Then, three samples of each culture (cultivated on xanthan gum and trehalose) were used for RNA extraction. RNA was extracted using TRI-reagent (Sigma), following standard protocol with the addition of two freeze and thaw cycles of the cells in TRI-reagent, as well as addition of chloroform and washing twice with 75% EtOH. RNA pellet was dissolved in RNAase-free water.
The genome was sequenced using a combination of Illumina GA II, and Roche/454 sequencing platforms. All general aspects of 500 bp paired-end (PE) library construction and sequencing, as well as 454 single-end sequencing can be found at the corresponding company website. A hybrid assembly of Illumina and 454 datasets was done using the Newbler assembler (Margulies et al., 2005). The initial Newbler assembly consisting of seven unique contigs was used to identify repeat regions that were subsequently screened out. At this point a 2 kb mate-pair Illumina library was constructed and sequenced and obtained paired end information was used to arrange multiple screened contigs into a single scaffold using the Phred/Phrap/Consed software package (Gordon et al., 1998). This package was also used for further sequence assembly and quality assessment in the subsequent finishing process. Sequence gaps between contigs that represented repeats were filled with Dupfinisher (Han and Chain, 2006), and a single scaffold was manually created and verified using available paired-end information. Illumina reads were used to correct potential base errors and increase consensus quality. Together, the combination of the Illumina and 454 sequencing platforms provided 320× coverage of the genome.
The assembled chromosome was uploaded to the RAST server (Aziz et al., 2008) for de novo gene prediction using Glimmer-3 (Delcher et al., 2007) and initial detection of homologs. Furthermore, the predicted genes were searched against the protein databases Pfam 27.0 (Finn et al., 2014), COG 2003–2014 (Galperin et al., 2015), MEROPS 9.12 (Rawlings et al., 2016), CAZy/dbCAN (Yin et al., 2012; Lombard et al., 2014), and TCDB (Saier et al., 2014) in order to expand the initial RAST annotation. The positive hits from these searches were manually curated using Uniprot/NCBI BLASTp. Signal peptides were predicted with the SignalP 4.1 web server (Petersen et al., 2011), and transmembrane helices were predicted with the TMHMM 2.0 web server (Krogh et al., 2001). Infernal 1.1.1 (Nawrocki and Eddy, 2013) was used in conjunction with the covariance models from Rfam 12.0 (Nawrocki et al., 2015) to search for non-coding RNA genes.
Extracted total RNA was converted to cDNA by reverse transcriptase. Total cDNA was sequenced from the three samples of each culture by strand-specific paired-end Illumina sequencing using an insert size of 270 bp and read length of 90 bp. RNA-seq reads were mapped to the genome using BWA ver 0.7.8 (Li and Durbin, 2009) requiring properly mapped pairs. Read assignment to genes was done using the featureCounts program from the Subread package (Liao et al., 2013). Only uniquely assigned read pairs were counted. Differential gene expression between the two groups was measured using edgeRun (Dimont et al., 2015) and genes were called differentially expressed using a BH-corrected p-value of 0.05.
Phylogenetic analyses were performed according to Sorokin et al. (2016) using the maximum likelihood method in MEGA6 (Tamura et al., 2013). Initial multiple amino acid sequence alignments were done in Mafft 7 (Katoh and Standley, 2013).
The genome of T. terrifontis strain R1 was sequenced and assembled into a single circular chromosome with a length of 4,810,751 bp and GC content of 57.34%. Genome annotation was performed using the RAST server and Infernal. In total, 4,504 protein coding genes were found in the genome, of which 2,412 could not be annotated by our database search and are therefore designated as “hypothetical protein.”
Both RAST and Infernal identified the same set of the 3 rRNAs and 46 tRNAs. Additionally, the ribonuclease P (RNase P), SRP, and tmRNA genes were identified by Infernal. No homolog was found for the non-coding 6S RNA gene. A recent computational screen for 6S RNA across all bacterial phyla (Wehner et al., 2014) reported the absence of 6S RNA in Pirellula staleyi and Rhodopirellula baltica, which are the closest related species to T. terrifontis in the 16S rRNA phylogeny (Slobodkina et al., 2015). This suggests that this gene is also likely to be absent in T. terrifontis. The Infernal search also revealed three riboswitches: cyclic di-GMP-I (RF01051), cobalamin (RF00174), and fluoride (RF01734).
The genome was submitted to GenBank with the accession number CP018477.
Thermogutta terrifontis R1 cells were cultured in growth media containing trehalose or xanthan gum, each in triplicates (see section “Materials and Methods”). Transcriptome sequencing using Illumina paired-end sequencing resulted in between 11.5 and 12.1 m read pairs for the 2 × 3 replicates. Across these, between 91.3 and 98.5% of the read pairs could be mapped uniquely to the genome. Differential expression analysis reported that 665 genes are up- and 617 genes are down-regulated on xanthan gum compared to trehalose-grown culture (Figure 1 and Supplementary Table S1).
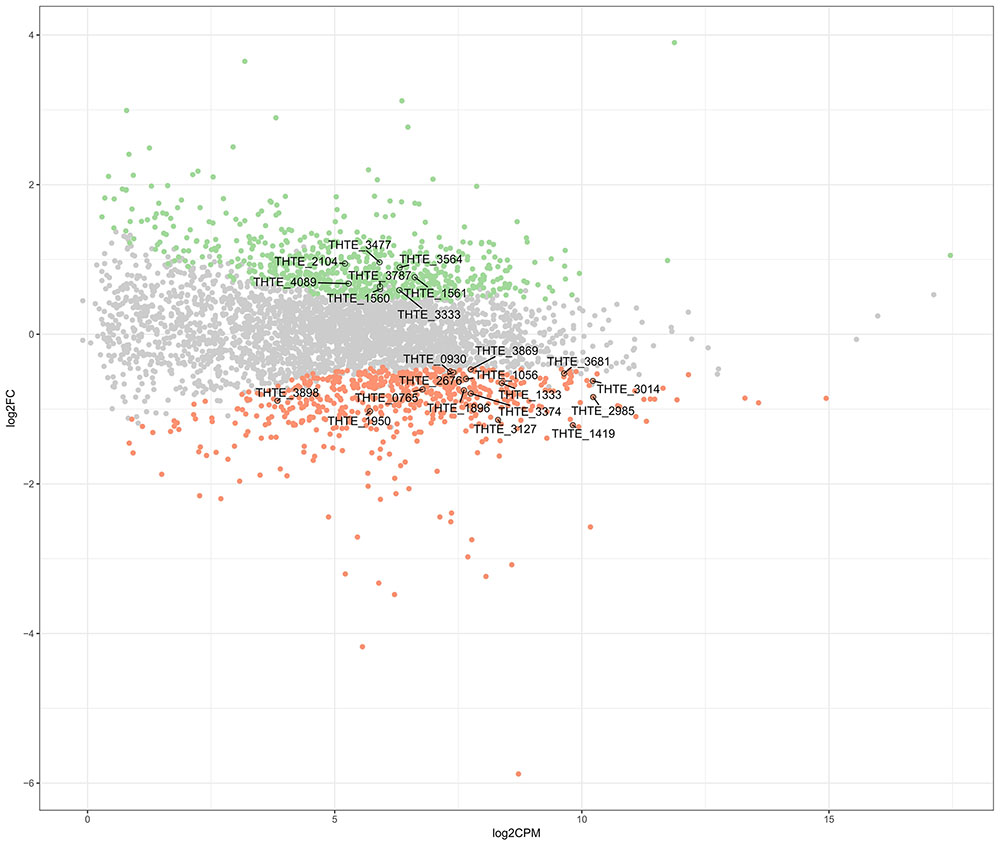
FIGURE 1. Differently expressed genes between xanthan gum-grown and trehalose-grown Thermogutta terrifontis R1 cells. Green and red dots indicate significantly up-regulated and down-regulated genes in xanthan gum culture compared with trehalose culture (P-value < 0.05). Annotated data points show locus tag identifiers of differentially expressed genes encoding proteins that are involved either in xantham gum / trehalose degradation pathways or in the central carbon metabolism. Y axis: log2 Fold Change between trehalose and xanthan gum-grown cultures. X axis: Read count of each gene per million mapped reads, averaged across all samples.
Thermogutta terrifontis R1 was shown to be able to grow using the following oligo- and polysaccharides as substrates: sucrose, trehalose, cellobiose, starch, xylan, pectin, or xanthan gum (Slobodkina et al., 2015). No growth was detected when maltose, lactose, agarose, alginate, cellulose, chitin, or inulin were added to the medium as sole carbon sources (Slobodkina et al., 2015).
Our analysis of the T. terrifontis R1 genome revealed 101 genes encoding glycosidases (GHs), 14 genes encoding polysaccaride lyases (PLs) and 3 genes encoding carbohydrate esterases (CEs) (Supplementary Table S2). Among these, 54 genes encode proteins that were predicted to be secreted outside the cells (whether anchored on the cells surface or being released into the culture broth). No dominant CAZy (GH, PL, or CE) families (Lombard et al., 2014) were observed among T. terrifontis R1 CAZymes, yet the most numerous were GH5 (10 proteins) and putative glycosidases (9 proteins), including DUF1080 domain. Detailed analysis of the CAZymes specificities revealed following activities: trehalose can be degraded by trehalose synthase (Qu et al., 2004) acting in opposite direction (THTE_2039). Sucrose hydrolysis may occur by the action of intracellular fructosidase (THTE_0696). Alpha-1,4-bonds and alpha-1,6-bonds in starch can be hydrolyzed by a number of GH13 and GH77 glycosidases (THTE_1477, THTE_2143, THTE_3153, and THTE_3783), producing maltooligosaccharides and finally D-glucose. Cellobiose can be hydrolyzed by the putative beta-glucosidase (THTE_0963), or one of the GH2 and GH5 glycosidases with currently uncertain function. Xylan can be decomposed to xylooligosaccharides by means of endoxylanases (THTE_2600 and THTE_3961) and to xylose by beta-xylosidases (THTE_0688, THTE_1819, THTE_1884, and THTE_2108). Pectin degradation occurs, most probably, by the action of a pectate lyase (THTE_1993) and several polygalacturonases (THTE_0436, THTE_1516, and THTE_2121), releasing D-galacturonic residues, that are further metabolized to D-glyceraldehyde 3-phosphate and pyruvate (see below). A large number of glycosidases was predicted to be involved in xanthan gum hydrolysis (see section “Xanthan Gum and Trehalose Utilization Pathways, Revealed by Comparative Genomic and Transcriptomic Analyses”).
According to the results of genome analysis, the final products of oligo- and polysaccharides decomposition were predicted to comprise glucose, fructose, mannose, xylose, galacturonate, and glucuronate. Some of these (glucose, mannose, xylose) as well as galactose were also shown to be used as growth substrates by T. terrifontis R1 according to Slobodkina et al., 2015. D-Glucose and D-fructose oxidation seems to occur via the Embden-Meyerhof (EM) pathway (Figures 2, 3 and Supplementary Table S3). Interestingly, the genome contains four genes encoding phosphofructokinases: one ATP-dependent (THTE_2190) and three pyrophosphate-dependent (THTE_0093, THTE_1056, THTE_2629). Since no genes for fructose-1,6-bisphosphatase were found, and PPi-dependent phosphofructokinases are thought to be reversible, at least one of them should be a part of gluconeogenesis. Additionally, analysis of the nearest characterized homologs supports two of them to be involved in xylose utilization (see below). The Entner-Doudoroff pathway seems to be inoperative due to the absence of the gene encoding 6-phosphogluconate dehydratase, a key enzyme of the pathway. Glucose-1-dehydrogenase, gluconokinase, and gluconate dehydratase genes are also absent in the genome.
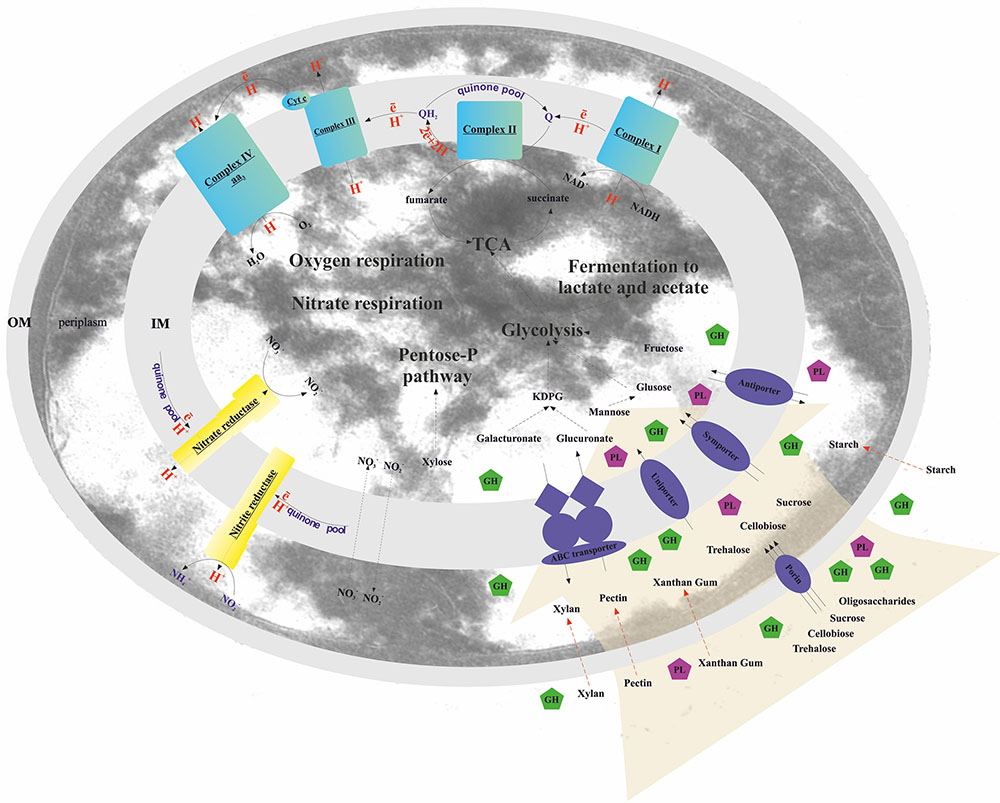
FIGURE 2. Schematic reconstruction of the T. terrifontis R1 catabolism. OM, outer membrane; IM, inner membrane; KDPG, 2-keto-3-deoxyphosphogluconate; TCA, tricarboxylic acid cycle; Cyt c, cytochrome c; GH, glycoside hydrolase; PL, polysaccharide lyase. Red dotted arrows indicate unique planctomycetal direct uptake of polysaccharides into periplasm (Boedeker et al., 2017), possibly occurred in T. terrifontis R1.
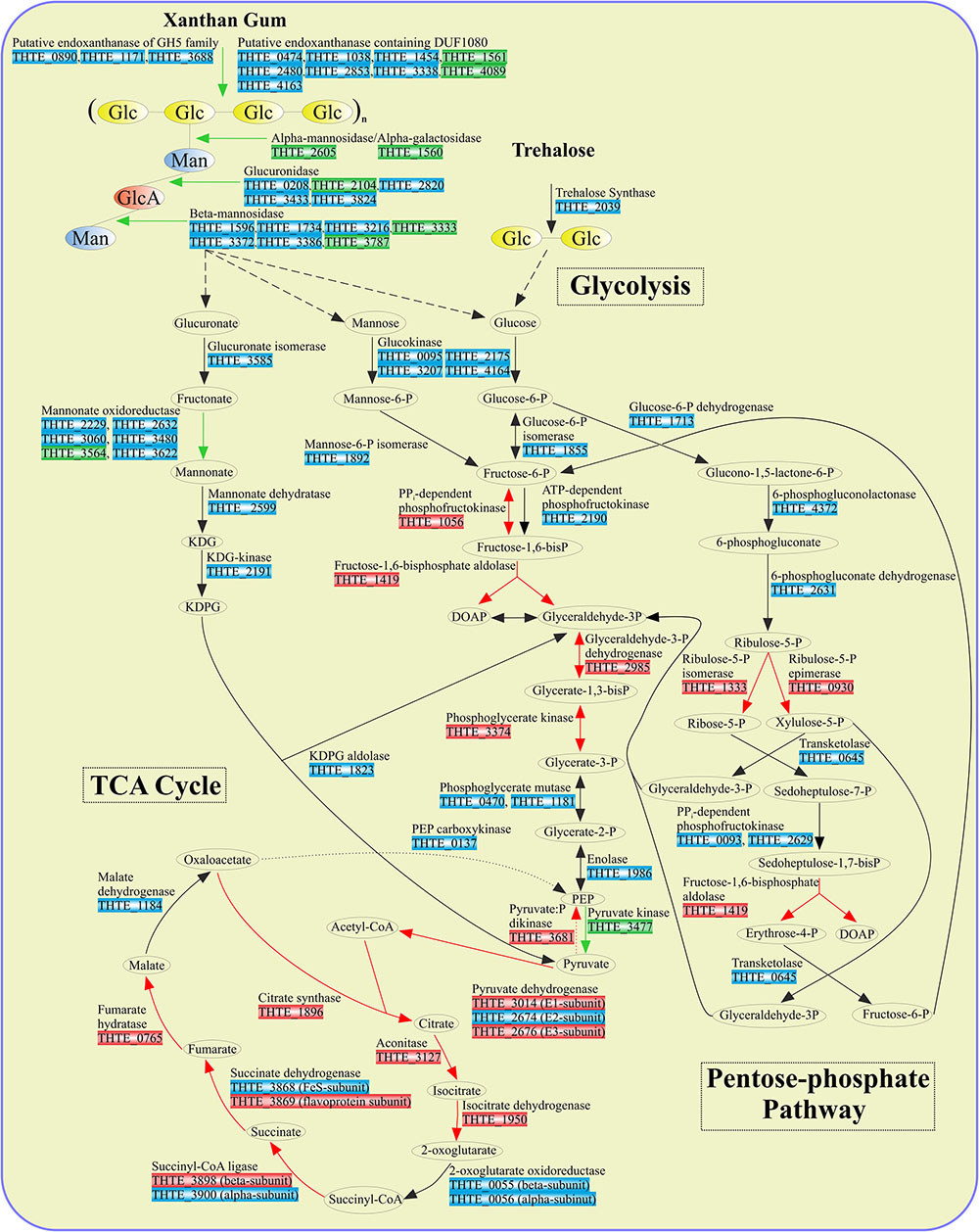
FIGURE 3. Enzymes involved in sugar turnover. Color boxes indicate the differential expression of the genes, indicated by the transcriptomic analysis. Green: up-regulation with xanthan gum; Red: down-regulation with xanthan gum; Blue: genes are not significantly differentially expressed between xanthan gum and trehalose.
The first step of galactose utilization – phosphorylation to galactose-1-phosphate – is catalyzed by galactokinase (THTE_0177). Next, the putative galactose-1-P-uridyltransferase (THTE_3784) transfers the UDP-group from UDP-glucose to galactose-1-P, producing UDP-galactose and glucose-1-P. This protein belongs to the type 1 galactose-1-P-uridyltransferases family, however, its closest characterized homolog was ADP-glucose:phosphate adenylyltransferase (UniProt ID Q9FK51). While phylogenetic analysis (Supplementary Figure S1) supports this finding, the sequence identity of these two proteins is rather low (Identity 35%, Coverage 97%), leaving the function of this enzyme unclear. However, since no other putative galactose-1-P-uridyltransferase genes were found, the assignment of the function to THTE_3784 remains plausible. Finally UDP-galactose is converted to UDP-glucose by the UDP-glucose 4-epimerase (THTE_2863), whereas the glucose-1-phosphate is converted to glucose-6-phosphate by the phosphoglucomutase (THTE_3829).
Xylose utilization was predicted to occur as follows: xylose is isomerized to xylulose by xylose isomerase (THTE_2111); xylulokinase (THTE_0598) phosphorylates xylulose to xylulose-5-phosphate, which finally enters the pentose-phosphate pathway. All genes encoding proteins of both oxidative and synthetic parts of this pathway were found in the genome with transaldolase gene as an exception (Figure 3 and Supplementary Table S4). Yet, sedoheptulose-7-phosphate (S-7-P), formed under action of transketolase, could be phosphorylated by PPi-dependent phosphofructokinases THTE_0093 and THTE_2629, of which the nearest characterized homolog from Methylococcus capsulatus (UniProt Q609I3, Reshetnikov et al., 2008) was shown to reversibly phosphorylate S-7-P with higher activity and affinity than fructose-6-phosphate (F-6-P). The resulting sedoheptulose-1,7-bisphosphate could be eliminated to erythrose-4-phosphate and dihydroxyacetone-phosphate by fructose-1,6-bisphosphate aldolase (THTE_1419) as it was proposed by Susskind et al. (1982) and Schellenberg et al. (2014).
D-Galacturonate, released in the course of pectin degradation, is presumably oxidized to glyceraldehyde-3-phosphate and pyruvate through a number of reactions (Supplementary Figure S2) catalyzed by uronate isomerase (THTE_3585), putative altronate oxidoreductase (see below), altronate dehydratases (THTE_0455 and THTE_0456), KDG kinase (THTE_2191), and KDPG aldolase (THTE_1823). No genes encoding altronate oxidoreductase belonging to the polyol-specific long-chain dehydrogenase/reductase family (Klimacek et al., 2003) were found. However, the genome contains several genes (THTE_0865, THTE_1784, THTE_2229, THTE_2632, THTE_3060, THTE_3480, and THTE_3564), probably encoding proteins of the short-chain dehydrogenase/reductase family (Jörnvall et al., 1995). One of its biochemically characterized representatives, an oxidoreductase UxaD from the hyperthermophilic anaerobic bacterium Thermotoga maritima, was shown to possess mannonate oxidoreductase activity (Rodionova et al., 2012).
The metabolism of xanthan gum degradation products – glucose, glucuronate, and mannose – is described in Section “Xanthan Gum and Trehalose Utilization Pathways, Revealed by Comparative Genomic and Transcriptomic Analyses.”
Pyruvate, generated in the course of degradation of sugars and sugar acids, is further oxidized to acetyl-CoA in the reactions catalyzed by the pyruvate dehydrogenase complex (Figure 3): pyruvate dehydrogenase (E1) (THTE_3014), dihydrolipoamide acetyltransferase (E2) (THTE_2674), and lipoamide dehydrogenase (E3) (THTE_2676).
It has been shown by Slobodkina et al. (2015), that the products of T. terrifontis R1 glucose fermentation were hydrogen, lactate and acetate. Lactate could be produced from pyruvate by lactate dehydrogenase (THTE_3348), while the mechanism of acetate formation remains unclear. Although two acetate kinases were found (THTE_1319 and THTE_2274), no genes coding for phosphate-acetyl transferase were detected in the genome. It is therefore possible that acetate could be formed due to the action of CoA-acylating aldehyde dehydrogenase (THTE_1321), catalyzing the NADH-dependent reduction of acetyl-CoA to acetaldehyde (Toth et al., 1999), and aldehyde dehydrogenase (THTE_2212) catalyzing the oxidation of acetaldehyde to acetate along with formation of NADH (Ho and Weiner, 2005). Finally, acetate could be formed under the action of putative ADP-forming acetyl-CoA synthetase (THTE_2996), as it was shown for few hyperthermophilic archaea (Musfeldt et al., 1999; Musfeldt and Schönheit, 2002). Surprisingly, the genome encoded an ATP-dependent acetyl-CoA synthase (THTE_1589), which catalyzes the irreversible activation of acetate, whereas acetate was not listed among the substrates, supporting the growth of T. terrifontis R1 in Slobodkina et al. (2015).
Hydrogen formed by T. terrifontis R1 in the course of fermentation apparently results from the operation of group 3c [NiFe]-hydrogenase (Vignais and Billoud, 2007) THTE_4311-4313 and/or [FeFe]-hydrogenases (Vignais and Billoud, 2007) THTE_2884, THTE_2882, THTE_2881, THTE_3842-THTE_3844. On the other hand, according to our analysis, all the genomes of planctomycetes, available in the IMG database (34 genomes of planctomycetes with assigned genus and species names. The analysis was performed 03.07.17), lack genes of [FeFe]-hydrogenase. Since, T. terrifontis is the first planctomycete known to synthesize hydrogen in the course of fermentation, and it is currently the only one in which genes for [FeFe]-hydrogenases have been found, at least some of its [FeFe]-hydrogenases could be involved in hydrogen production.
All genes, coding the citrate cycle (TCA cycle) enzymes were found in the T. terrifontis R1 genome (Figures 2, 3 and Supplementary Table S5).
Genes for all three subunits of the respiratory cytoplasmic nitrate reductase Nar (Simon and Klotz, 2013) were found in the genome. The alpha subunit (NarG) THTE_1509 belongs to the deep lineage within the Nar-DMSO cluster (Supplementary Figure S3) of the molybdopterine superfamily (Duval et al., 2008). THTE_1508 and THTE_1507 encode the other two subunits NarH and NarI, respectively, while THTE_1506 encodes a chaperon subunit (TorD). The NarGHI complex might form a supercomplex with an electrogenic membrane-bound NADH dehydrogenase (Simon and Klotz, 2013, Figure 2 and Supplementary Table S6). No diheme subunit NarC was found, yet the genes of cytochrome b/c1 complex (complex III, THTE_1510-THTE_1512) are located in close vicinity to the NarGHI genes (THTE_1509-THTE_1507), what might reflect the involvement of the complex III in the electron and proton transfer during T. terrifontis anaerobic growth with nitrate. Nitrite is reduced to ammonium by means of non-electrogenic periplasmic membrane-bound nitrite reductase Nrf, the catalytic subunit NrfA and the membrane-bound subunit NrfH (Simon and Klotz, 2013) of which are encoded by THTE_1450 and THTE_1449, respectively.
The complete aerobic respiratory electron transfer chain (ETC), including H+-translocating NADH-dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome b/c1-complex (complex III), and terminal cytochrome c oxidase aa3-type (complex IV) was found (Figure 2 and Supplementary Table S6). We did not find the typical cytochrome c gene or the plastocyanin gene, involved in transferring electrons from complex III to complex IV, yet THTE_3354 encodes a putative large (258 amino acids) cytochrome c containing two monoheme domains. No genes encoding terminal quinol oxidases (bd-type, bo3-type, or ba3-type) were found in the genome.
In order to decipher the mechanisms of xanthan gum degradation, T. terrifontis R1 was grown on xanthan gum and trehalose (as the control), and the transcriptomes were sequenced and analyzed for genes that are up-regulated in the cultures with xantham gum as the substrate. Trehalose is a disaccharide consisting of 1-1-alpha-linked glucose molecules, and it was chosen as one of the simple sugars, supporting growth of the strain. Interestingly, T. terrifontis R1 genomic analysis revealed no genes coding for known trehalose-hydrolyzing enzymes of GH15, GH37, and GH65 families. Furthermore, two GH13 proteins (THTE_1477 and THTE_3153) have no trehalose-converting enzymes among their nearest characterized relatives. Therefore, the only remaining reasonable candidate involved in decomposition of trehalose is a trehalose synthase of GT4 family (THTE_2039), acting in reverse direction, leading to a release of D-glucose and NDP-D-glucose molecules. The level of its expression in cells, grown on trehalose and xanthan-gum was similar (Figure 3), what could be explained by reversibility of its action (Qu et al., 2004; Ryu et al., 2005) at various growth conditions: trehalose degradation when trehalose is being sole substrate and trehalose synthesis when other substrates are used.
Despite its ubiquitous usage in pharmaceutical and food industries, not much is known about xanthan gum (beta-1,4-glucan with mannosyl–glucuronyl–mannose side chains) degradation mechanisms. For the complete hydrolysis of the molecule the following linkages should be broken: β-mannose-1–4-α-glucuronate, β-glucuronate-1–2-α-mannose, α-mannose-1–3-β-glucose linkages in side chains, and β-1,4-glucosidic linkages in polyglucose backbone (Figure 4).
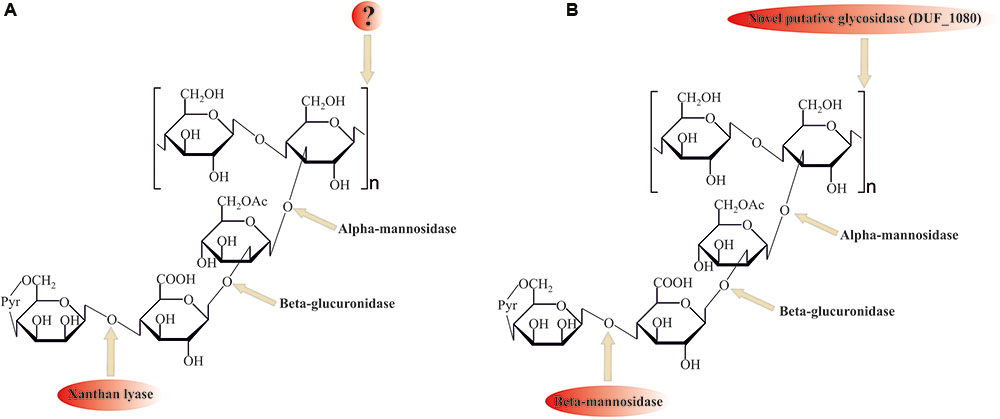
FIGURE 4. Comparison of (A) known (with xanthan lyase) and (B) proposed novel (without xanthan lyase) xanthan gum degradation pathways. ? question mark indicates no sequence data available for these enzymes. Please, see the text for details.
Among the enzymes currently known to be involved in xanthan gum decomposition, there are few removing terminal mannose residues xanthan lyases (Hashimoto et al., 1998; Ruijssenaars et al., 1999), belonging to the PL8 family, and an endoxanthanase (xanthan-specific endoglucanase, Li et al., 2009), hydrolyzing the glucan backbone (Figure 4A). The latter has been biochemically characterized, yet its sequence is still unknown, preventing structure analysis and evolutionary reconstructions using sequence comparison.
No homologs of PL8 family lyases, to which all known xanthan lyases belong, were found in the in silico translated T. terrifontis R1 proteome. They were probably replaced by several putative endomannanases/beta-mannosidases of GH5 family (THTE_1596, THTE_1734, THTE_3216, THTE_3333, THTE_3372, THTE_3386 and THTE_3787, Supplementary Figure S4), cleaving Man(1β–4α)GlcA linkages. Four of them (THTE_1596, THTE_3216, THTE_3333, and THTE_3386) were predicted to be extracellular. Transcriptomic analysis showed two genes THTE_3333 and THTE_3787 to be up-regulated in the xanthan gum cultures, assuming their involvement in its decomposition.
For cleavage of the GlcA(β1–2α)Man linkage the action of beta-glucuronidase is needed. All known beta-glucuronidases belong to the GH1, GH2, GH30, and GH79 families. While no genes encoding GH1, GH30, and GH79 proteins were found, five genes encoding GH2 family glycosidases (THTE_0208, THTE_2104, THTE_2820, THTE_3433, and THTE_3824) were revealed in the T. terrifontis R1 genome. The family GH2 contains a number of enzymes with various specificities. We compared T. terrifontis GH2s with the previously characterized members of the GH2 family, to predict whether the putative five T. terrifontis GH2 proteins act as beta-glucuronidases. All five genes formed a monophyletic group, adjoined to the cluster with characterized beta-galactosidases and beta-glucuronidases (Supplementary Figure S5). Given that one of these (THTE_2104) was significantly up-regulated in the cells growing on xanthan gum, the beta-glucuronidase activity seems to be characteristic of at least this one, yet possibly all five GH2 from this monophyletic group possess this activity. It should be noted that THTE_2104 and also THTE_0208 were predicted to be secreted.
The Man(1α–β3)Glc linkage could be hydrolyzed by an alpha-mannosidase of GH38 family (THTE_2605). Another option is an extracellular putative alpha-galactosidase of GH36 family (THTE_1560), whose characterized homologs are known to hydrolyze a number of oligosaccharides of various structures (Merceron et al., 2012). Both THTE_2605 and THTE_1560 genes were up-regulated during the growth on xanthan gum.
Finally, the hydrolysis of a Glc(β1–4)Glc linkage in xanthan gum backbone could be catalyzed by the GH5 enzymes THTE_0890, THTE_1171 and THTE_3688, however, only one of them was predicted to be extracellular (THTE_1171) and none of them were up-regulated on xanthan gum.
Search of the other putative glycosidases of T. terrifontis R1 revealed nine proteins (THTE_0474, THTE_1038, THTE_1454, THTE_1561, THTE_2480, THTE_2853, THTE_3338, THTE_4089, and THTE_4163) containing a domain of unknown function (DUF1080). All of these proteins, except THTE_1454 and THTE_3338, were predicted to be extracellular, and two of them (THTE_1561 and THTE_4089) were up-regulated on xanthan gum. These proteins may be representatives of a novel family of glycosidases according to TOPSAN annotation1. Such a high number of genes encoding putative glycosidases with unknown function in a xanthan gum degrading microorganism might be an indication on their involvement in the process, most probably for hydrolysis of the backbone linkage (Figure 4B). Interestingly, proteins containing DUF1080 are highly overrepresented among all Planctomycetes (Supplementary Table S7) in comparison with other organisms. This, together with our observation that xanthan gum being a selective substrate for many planctomycetes, suggest an important role of DUF1080 proteins in xanthan gum degradation. Finally, five of nine T. terrifontis R1 DUF1080 proteins, including THTE_1561, have a CBM66 domain, which was found mainly among Firmicutes representatives and helps binding the terminal fructoside residue in fructans (Cuskin et al., 2012). Yet, the DUF1080 and CBM66 domains overlap each other, indicating two different designations of the same domain occurred.
The predicted products of xanthan gum degradation are mannose, glucuronic acid, and glucose. Although no mannokinase genes were found in the T. terrifontis R1 genome, mannose could be phosphorylated to mannose-6-P by the variety of its putative glucokinases from the ROK family (THTE_0095, THTE_2175, THTE_3207, THTE_4164), representatives of which are known to be capable of acting on various hexoses (Conejo et al., 2010; Nakamura et al., 2012). Mannose-6-phosphate upon conversion to fructose-6-phosphate by mannose-6-phopshate isomerase (THTE_1892) enters the EM pathway (Figures 2, 3). However, none of the genes coding these proteins were up-regulated on xanthan gum in our experiment, possibly due to their wide specificity and hence constitutive expression.
An oxidation of glucuronate, released during xanthan gum hydrolysis, might occur through formation of fructonate, followed by reduction to mannonate, dehydration to 2-keto-3-desoxy-6-phosphogluconate (KDPG) and its elimination to pyruvate and glyceraldehyde-3-phosphate (Figure 3). All genes encoding the respective proteins, except mannonate oxidoreductase, were found in the genome. As it was hypothesized for altronate oxidoreductases (see above), we suggest that some of short-chain reductases/dehydrogenases (THTE_2229, THTE_2632, THTE_3060, THTE_3480, THTE_3564 and THTE_3622) with unknown specificity, especially the up-regulated THTE_3564, may act as a mannonate oxidoreductase.
In most cases, the majority of enzymes involved in both the trehalose and xanthan gum-dependent pathways of the central carbohydrate metabolism were expressed on the same level or were down-regulated in the cells grown on xanthan gum. This could be due to the lower structural complexity of trehalose in comparison with xanthan gum, which requires fewer degradation steps and determines easier import into the cell: one transporter and one enzymatic step are enough to transport and decompose trehalose to the basic metabolites (D-glucose and NTP-α-D-glucose) compared with most certainly multiple transporters and four steps of xanthan gum decomposition, coupled with two- and five-step mannose and glucuronate, respectively, conversions to EM pathway metabolites (Figure 3). Finally, the majority of flagellar, as well as pili IV and secretion system proteins, were up-regulated (Supplementary Table S1) in the cells grown on xanthan gum. This might be a reflection of viscosity of the substrate, which force cells to be more agile and capable of binding to the substrate.
The central carbohydrate metabolism of T. terrifontis R1 was deciphered using genomic and transcriptomic approaches, and the novel xanthan gum degradation pathway was proposed. The pathway involves endomannanases/beta-mannosidases instead of xanthan lyases as well DUF1080 proteins for the hydrolysis of xanthan gum backbone. Surprisingly, the genes coding DUF1080 proteins were highly abundant in T. terrifontis R1, as well as in many other Planctomycetes genomes. These results are relevant due to lack of the information on microorganisms degrading xanthan gum and its degradation pathways, which has so far been limited to few representatives of Actinobacteria and Firmicutes and their enzymes. Yet, further studies including proteomics of xanthan gum-growing cultures, as well as, purification and characterization of the respective enzymes are needed to verify the predicted pathway.
The T. terrifontis R1 genome is available via GenBank accession number CP018477.
IK, XP, and EB-O conceived the study. SG and PM contributed to cultivation and transcriptome sequencing. AS, VK, PM, and AK contributed to genome sequencing and assembly. AE, PM, and IK contributed to genome and transcriptome analysis. AE, PM, XP, and IK wrote the manuscript.
The work was supported by the European Union 7th Framework Programme FP7/2007-2013 under grant agreement no. 265933 (Hotzyme). The work of AE, EB-O and IK was supported by RSF grant 17-74-30025. The work of VK was supported by the Federal Agency for Scientific Organisations of Russia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Drs. S. N. Gavrilov, O. L. Kovaleva and A. V. Lebedinsky for their valuable comments and advices. This work was performed using the scientific equipment of Core Research Facility “Bioengineering”.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02140/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Boedeker, C., Schüler, M., Reintjes, G., Jeske, O., van Teeseling, M. C. F., Jogler, M., et al. (2017). Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 8:14853. doi: 10.1038/ncomms14853
Conejo, M. S., Thompson, S. M., and Miller, B. G. (2010). Evolutionary bases of carbohydrate recognition and substrate discrimination in the ROK protein family. J. Mol. Evol. 70, 545–556. doi: 10.1007/s00239-010-9351-1
Cuskin, F., Flint, J. E., Gloster, T. M., Morland, C., Baslé, A., Henrissat, B., et al. (2012). How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 20889–20894. doi: 10.1073/pnas.1212034109
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Dimont, E., Shi, J., Kirchner, R., and Hide, W. (2015). edgeRun: an R package for sensitive, functionally relevant differential expression discovery using an unconditional exact test. Bioinformatics 31, 2589–2590. doi: 10.1093/bioinformatics/btv209
Duval, S., Ducluzeau, A.-L., Nitschke, W., and Schoepp-Cothenet, B. (2008). Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol. Biol. 8:206. doi: 10.1186/1471-2148-8-206
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, 222–230. doi: 10.1093/nar/gkt1223
Fuerst, J. A., and Sagulenko, E. (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413. doi: 10.1038/nrmicro2578
Fukunaga, Y., Kurahashi, M., Sakiyama, Y., Ohuchi, M., Yokota, A., and Harayama, S. (2009). Phycisphaera mikurensis gen. nov., sp nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov and Phycisphaerae classis nov in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275. doi: 10.2323/jgam.55.267
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded Microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
García-Ochoa, F., Santos, V. E., Casas, J. A., and Gómez, E. (2000). Xanthan gum: production, recovery, and properties. Biotechnol. Adv. 18, 549–579. doi: 10.1016/S0734-9750(00)00050-1
Gavrilov, S. N., Stracke, C., Jensen, K., Menzel, P., Kallnik, V., Slesarev, A., et al. (2016). Isolation and characterization of the first xylanolytic hyperthermophilic euryarchaeon Thermococcus sp. strain 2319x1 and its unusual multidomain glycosidase. Front. Microbiol. 7:552. doi: 10.3389/fmicb.2016.00552
Gordon, D., Abajian, C., and Green, P. (1998). Consed: a graphical tool for sequence finishing. Genome Res. 8, 195–202. doi: 10.1101/gr.8.3.195
Han, C., and Chain, P. (2006). “Finishing repetitive regions automatically with dupfinisher,” in Proceeding of the 2006 International Conference on Bioinformatics & Computational Biology, eds H. R. Arabnia and H. Valafar (Las Vegas, NV: REA Press), 141–146.
Hashimoto, W., Miki, H., Tsuchiya, N., Nankai, H., and Murata, K. (1998). Xanthan lyase of Bacillus sp. strain GL1 liberates pyruvylated mannose from xanthan side chains. Appl. Environ. Microbiol. 64, 3765–3768.
Ho, K. K., and Weiner, H. (2005). Isolation and characterization of an aldehyde dehydrogenase encoded by the aldB gene of Escherichia coli isolation and characterization of an aldehyde dehydrogenase encoded by the aldB gene of Escherichia coli. J. Bacteriol. 187, 1067–1073. doi: 10.1128/JB.187.3.1067
Jeske, O., Schüler, M., Schumann, P., Schneider, A., Boedeker, C., Jogler, M., et al. (2015). Planctomycetes do possess a peptidoglycan cell wall. Nat. Commun. 6:7116. doi: 10.1038/ncomms8116
Jörnvall, H., Persson, B., Krook, M., Atrian, S., Gonzàlez-Duarte, R., Jeffery, J., et al. (1995). Short-chain dehydrogenases/reductases(SDR). Biochemistry 34, 6003–6013. doi: 10.1021/bi00018a001
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klimacek, M., Kavanagh, K. L., Wilson, D. K., and Nidetzky, B. (2003). Pseudomonas fluorescens mannitol 2-dehydrogenase and the family of polyol-specific long-chain dehydrogenases/reductases: sequence-based classification and analysis of structure-function relationships. Chem. Biol. Interact. 14, 559–582. doi: 10.1016/S0009-2797(02)00219-3
Kovaleva, O. L., Merkel, A. Y., Novikov, A. A., Baslerov, R. V., Toshchakov, S. V., and Bonch-Osmolovskaya, E. A. (2015). Tepidisphaera mucosa gen. nov., sp. nov., a moderately thermophilic member of the class Phycisphaerae in the phylum Planctomycetes, and proposal of a new family, Tepidisphaeraceae fam. nov., and a new order, Tepidisphaerales ord. nov. Int. J. Syst. Evol. Microbiol. 65, 549–555. doi: 10.1099/ijs.0.070151-0
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Li, B., Guo, J., Chen, W., Chen, X., Chen, L., Liu, Z., et al. (2009). Endoxanthanase, a novel β-D-Glucanase hydrolyzing backbone linkage of intact xanthan from newly isolated Microbacterium sp. XT11. Appl. Biochem. Biotechnol. 159, 24–32. doi: 10.1007/s12010-008-8439-1
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liao, Y., Smyth, G. K., and Shi, W. (2013). The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. doi: 10.1093/nar/gkt214
Liesack, W., König, H., Schlesner, H., and Hirsch, P. (1986). Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch. Microbiol. 145, 361–366. doi: 10.1007/BF00470872
Liu, H., Huang, C., Dong, W., Du, Y., Bai, X., and Li, X. (2005). Biodegradation of xanthan by newly isolated Cellulomonas sp. LX, releasing elicitor-active xantho-oligosaccharides-induced phytoalexin synthesis in soybean cotyledons. Process Biochem. 40, 3701–3706. doi: 10.1016/j.procbio.2005.05.006
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome sequencing in open microfabricated high density picoliter reactors. Nat. Biotechnol. 437, 376–380. doi: 10.1038/nature03959
Markowitz, V. M., Chen, I. M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, 115–122. doi: 10.1093/nar/gkr1044
Merceron, R., Foucault, M., Haser, R., Mattes, R., Watzlawick, H., and Gouet, P. (2012). The molecular mechanism of thermostable α-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies. J. Biol. Chem. 287, 39642–39652. doi: 10.1074/jbc.M112.394114
Musfeldt, M., and Schönheit, P. (2002). Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic Archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J. Bacteriol. 184, 636–644. doi: 10.1128/JB.184.3.636
Musfeldt, M., Selig, M., and Schönheit, P. (1999). Acetyl coenzyme A synthetase (ADP forming) from the hyperthermophilic archaeon Pyrococcus furiosus: identification, cloning, separate expression of the encoding genes, acdAI and acdBI, in Escherichia coli, and in vitro reconstitution of the active heterotetrameric enzyme from its recombinant subunits. J. Bacteriol. 181, 5885–5888.
Nakamura, T., Kashima, Y., Mine, S., Oku, T., and Uegaki, K. (2012). Characterization and crystal structure of the thermophilic ROK hexokinase from Thermus thermophilus. J. Biosci. Bioeng. 114, 150–154. doi: 10.1016/j.jbiosc.2012.03.018
Nawrocki, E. P., Burge, S. W., Bateman, A., Daub, J., Eberhardt, R. Y., Eddy, S. R., et al. (2015). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137. doi: 10.1093/nar/gku1063
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. doi: 10.1093/bioinformatics/btt509
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Podosokorskaya, O. A., Merkel, Y. A., Kolganova, T. V., Chernyh, N. A., Miroshnichenko, M. L., Bonch-Osmolovskaya, E. A., et al. (2011). Fervidobacterium riparium sp. nov., a thermophilic anaerobic cellulolytic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 61, 2697–2701. doi: 10.1099/ijs.0.026070-0
Qian, F., An, L., He, X., Han, Q., and Li, X. (2006). Antibacterial activity of xantho-oligosaccharide cleaved from xanthan against phytopathogenic Xanthomonas campestris pv. campestris. Process Biochem. 41, 1582–1588. doi: 10.1016/j.procbio.2006.03.003
Qu, Q., Lee, S., and Boos, W. (2004). TreT, a novel trehalose glycosyltransferring synthase of the hyperthermophilic archaeon Thermococcus litoralis. J. Biol. Chem. 279, 47890–47897. doi: 10.1074/jbc.M404955200
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rawlings, N. D., Barrett, A. J., and Finn, R. (2016). Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44, D343–D350. doi: 10.1093/nar/gkv1118
Reshetnikov, A. S., Rozova, O. N., Khmelenina, V. N., Mustakhimov, I. I., Beschastny, A. P., Murrell, J. C., et al. (2008). Characterization of the pyrophosphate-dependent 6-phosphofructokinase from Methylococcus capsulatus bath. FEMS Microbiol. Lett. 288, 202–210. doi: 10.1111/j.1574-6968.2008.01366.x
Rodionova, I. A., Scott, D. A., Grishin, N. V., Osterman, A. L., and Rodionov, D. A. (2012). Tagaturonate-fructuronate epimerase UxaE, a novel enzyme in the hexuronate catabolic network in Thermotoga maritima. Environ. Microbiol. 14, 2920–2934. doi: 10.1111/j.1462-2920.2012.02856.x
Ruijssenaars, H. J., De Bont, J. A. M., and Hartmans, S. (1999). A pyruvated mannose-specific xanthan lyase involved in xanthan degradation by Paenibacillus alginolyticus XL-1. Appl. Environ. Microbiol. 65, 2446–2452.
Ryu, S., Park, C., Cha, J., Woo, E., and Lee, S. (2005). A novel trehalose-synthesizing glycosyltransferase from Pyrococcus horikoshii: molecular cloning and characterization. Biochem. Biophys. Res. Commun. 329, 429–436. doi: 10.1016/j.bbrc.2005.01.149
Saier, M. H., Reddy, V. S., Tamang, D. G., and Västermark, Å. (2014). The transporter classification database. Nucleic Acids Res. 42, 251–258. doi: 10.1093/nar/gkt1097
Schellenberg, J. J., Verbeke, T. J., McQueen, P., Krokhin, O. V., Zhang, X., Alvare, G., et al. (2014). Enhanced whole genome sequence and annotation of Clostridium stercorarium DSM8532T using RNA-seq transcriptomics and high-throughput proteomics. BMC Genomics 15:567. doi: 10.1186/1471-2164-15-567
Simon, J., and Klotz, M. G. (2013). Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim. Biophys. Acta 1827, 114–135. doi: 10.1016/j.bbabio.2012.07.005
Slobodkina, G. B., Kovaleva, O. L., Miroshnichenko, M. L., Slobodkin, A. I., Kolganova, T. V., Novikov, A. A., et al. (2015). Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., thermophilic anaerobic representatives of the phylum Planctomycetes. Int. J. Syst. Evol. Microbiol. 65, 760–765. doi: 10.1099/ijs.0.000009
Sorokin, D. Y., Kublanov, I. V., Gavrilov, S. N., Rojo, D., Roman, P., Golyshin, P. N., et al. (2016). Elemental sulfur and acetate can support life of a novel strictly anaerobic haloarchaeon. ISME J. 10, 240–252. doi: 10.1038/ismej.2015.79
Strous, M., Pelletier, E., Mangenot, S., Rattei, T., Lehner, A., Taylor, M. W., et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794. doi: 10.1038/nature04647
Susskind, B. M., Warren, L. G., and Reeves, R. E. (1982). A pathway for the interconversion of hexose and pentose in the parasitic amoeba Entamoeba histolytica. Biochem. J. 204, 191–196.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Toth, J., Ismaiel, A. A., and Chen, J. S. (1999). The ald gene, encoding a coenzyme A-acylating aldehyde dehydrogenase, distinguishes Clostridium beijerinckii and two other solvent-producing clostridia from Clostridium acetobutylicum. Appl. Environ. Microbiol. 65, 4973–4980.
van de Graaf, A., Mulder, A., Bruijn, P. D. E., Jetten, M. S. M., Robertson, L. A., and Kuenen, J. G. (1995). Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61, 1246–1251.
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272.
Wehner, S., Damm, K., Hartmann, R. K., and Marz, M. (2014). Dissemination of 6S RNA among bacteria. RNA Biol. 11, 1467–1478. doi: 10.4161/rna.29894
Xiong, X., Li, M., Xie, J., Jin, Q., Xue, B., and Sun, T. (2013). Antioxidant activity of xanthan oligosaccharides prepared by different degradation methods. Carbohydr. Polym. 92, 1166–1171. doi: 10.1016/j.carbpol.2012.10.069
Keywords: planctomycetes, thermophiles, CAZymes, xanthan gum, comparative genomics, transcriptomics, metabolism reconstruction
Citation: Elcheninov AG, Menzel P, Gudbergsdottir SR, Slesarev AI, Kadnikov VV, Krogh A, Bonch-Osmolovskaya EA, Peng X and Kublanov IV (2017) Sugar Metabolism of the First Thermophilic Planctomycete Thermogutta terrifontis: Comparative Genomic and Transcriptomic Approaches. Front. Microbiol. 8:2140. doi: 10.3389/fmicb.2017.02140
Received: 08 September 2017; Accepted: 19 October 2017;
Published: 02 November 2017.
Edited by:
Nils-Kaare Birkeland, University of Bergen, NorwayReviewed by:
Magnus Øverlie Arntzen, Norwegian University of Life Sciences, NorwayCopyright © 2017 Elcheninov, Menzel, Gudbergsdottir, Slesarev, Kadnikov, Krogh, Bonch-Osmolovskaya, Peng and Kublanov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilya V. Kublanov, a3VibGFub3YuaWx5YUBnbWFpbC5jb20= Alexander G. Elcheninov, ZWxjaGVuaW5vdi5hZ0BnbWFpbC5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.