
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 October 2017
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.02094
The plasmid-borne colistin-resistant gene mcr-1 has rapidly become a worldwide public health concern. This study aims to determine the host bacterial strains, plasmids, and genetic contexts of mcr-1 in hospital sewage. A 1-ml hospital sewage sample was cultured. Colistin-resistant bacterial colonies were selected on agar plates and were subjected to whole genome sequencing and subsequent analysis. The transfer of mcr-1 between bacterial strains was tested using conjugation. New variants of mcr-1 were cloned to test the impact of variations on the function of mcr-1. Plasmids carrying mcr-1 were retrieved from GenBank for comparison based on concatenated backbone genes. In the sewage sample, we observed that mcr-1 was located in various genetic contexts on the chromosome, or plasmids of four different replicon types (IncHI2, IncI2, IncP, and IncX4), in Klebsiella pneumoniae, Kluyvera spp. and seven Escherichia coli strains of six different sequence types (ST10, ST34, ST48, ST1196, ST7086, and ST7087). We also identified two new variants of mcr-1, mcr-1.4 and mcr-1.7, both of which encode an amino acid variation from mcr-1. mcr-1-carrying IncX4 plasmids, which have a global distribution across the Enterobacteriaceae, are the result of global dissemination of a single common plasmid, while IncI2 mcr-1 plasmids appear to acquire mcr-1 in multiple events. In conclusion, the unprecedented remarkable diversity of species, strains, plasmids, and genetic contexts carrying mcr-1 present in a single sewage sample from a single healthcare site highlights the continued evolution and dynamic transmission of mcr-1 in healthcare-associated environments.
Colistin (also known as polymycin E) is an antibiotic and has long been one of the last resort treatments for infections caused by multi-drug resistant Gram-negative bacteria. However, bacterial strains that acquired resistance to colistin resistance have emerged worldwide (Olaitan et al., 2014). The mechanisms mediating resistance to colistin are mainly due to mutations or insertions in the chromosomal genes such as the phoP-Q and pmrA-B and ccrA-B two-component systems and the regulator gene mgrB (Olaitan et al., 2014). A plasmid-borne colistin resistance gene, mcr-1, has recently been found in Escherichia coli and in a lesser extent Klebsiella pneumonia (Liu et al., 2016). mcr-1 encodes a phosphoethanolamine (PEA) transferase enzyme that is capable of adding PEA to the lipid A moiety of lipopolysaccharides (LPSs), the initial target of colistin (Liu et al., 2016). Besides E. coli and K. pneumonia, mcr-1 has been detected in various species of the Enterobacteriaceae in many countries (Schwarz and Johnson, 2016), imposing an emerging threat for human and animal health. During a screening study for colistin-resistant bacterial isolates in hospital sewage, we found that mcr-1 genes including two new variants were carried by plasmids of various replicon types in multiple E. coli strains.
Sewage (1 ml) was collected from the influx of the wastewater treatment plant at West China Hospital in November 2015. West China Hospital is a 5,000-bed tertiary teaching hospital and serves as one of the major referral medical centers in western China. All sewages produced in the hospital were processed in the plant. The sewage sample was mixed with 100 ml brain heart infusion broth (Oxoid, Basingstoke, United Kingdom) in a 500-ml flask. After overnight incubation at 37°C, the culture suspension was diluted to 0.5 McFarland standard and an 100 μl aliquot was plated onto a CHROMAgar Orientation agar plate (CHROMAgar, Paris, France) containing 4 μg/ml colistin and 64 μg/ml linezolid. The plate was then incubated at 37°C overnight. Pink colonies that represent E. coli were screened for mcr-1 as described previously (Liu et al., 2016). Species identification of the colonies was established by partially sequencing the gyrB gene as described previously (Yamamoto and Harayama, 1995). MICs of amikacin, cefotaxime, ciprofloxacin, colistin, imipenem, polymycin B, and tigecycline were determined using the microdilution broth method following recommendations of the Clinical Laboratory Standards Institute (CLSI) (CLSI, 2013).
The complete coding sequence of mcr-1.1, mcr-1.4, and mcr-1.7 was amplified with primers mcr1-up1 (TGCCGTAATTATCCCACCGT) and mcr1-dw1 (ACCAATCAGCGACCATCAGA) using PrimeSTAR Max DNA Polymerase (Takara, Dalian, China). Amplicons were ligated to the pMD20-T vector using the Mighty TA-cloning kit (Takara). The ligated fragments were transformed into E. coli DH5α. pMD20-T::mcr-1.1-, pMD20-T::mcr-1.4-, or pMD20-T::mcr-1.7-containing transformants were selected on LB agar plates containing 2 μg/mL colistin. The presence of mcr-1.1, mcr-1.4, or mcr-1.7 in transformants was confirmed by PCR and sequencing. MICs of colistin were determined for transformants carrying pMD20-T::mcr-1.1, pMD20-T::mcr-1.4, or pMD20-T::mcr-1.7 using the broth microdilution method (CLSI, 2013).
Pulsed-field gel electrophoresis (PFGE) was performed using the protocol for non-O157 E. coli of PulseNet International1. E. coli strains were assigned to the phylogenetic groups A, B1, B2, and D using PCR as described previously (Clermont et al., 2000).
Conjugation experiments were carried out in BHI broth and on filter. The azide-resistant E. coli strain J53 was used as the recipient and transconjugants were selected on LB agar plates containing 2 μg/ml colistin plus 150 μg/ml sodium azide. The presence of mcr-1.1, mcr-1.4, or mcr-1.7 in transconjugants was confirmed using PCR and sequencing.
Genomic DNA was prepared using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Purified DNA was 150-bp paired-end whole genome sequenced to around 200× coverage using the HiSeq X10 Sequencer (Illumina, San Diego, CA, United States). Reads were de novo assembled into contigs using SPAdes (Bankevich et al., 2012). In addition, strain WCHEC1613 was also sequenced using the long-read PacBio RSII Sequencer (Pacific Biosciences, Menlo Park, CA, United States). The assembly was initially built from the PacBio reads alone using program Canu (Koren et al., 2017) with default settings. To obtain high-quality reads for correction, the Illumina reads were trimmed using Trimmomatic (Bolger et al., 2014) with 3, 25, and 50 as the size of sliding window, threshold of mean quality, and minimum length of reads, respectively. The filtered reads were then mapped against the initial assembly to obtain a coordinate sorted BAM file and subsequently a filtered VCF file (minDP10 and minQ30) using Smalt2 (version 0.7.4), SAMtools (version 1.3.1) (Li et al., 2009), and VCFtools (version 0.1.14) (Danecek et al., 2011). The final assembly of WCHEC1613 was created by correcting SNPs and indels from the BAM file using PacBio-utilities3.
Sequence type (ST) was assigned using the genomic sequence to query the Enterobase database4. Antimicrobial resistance genes were predicted using ResFinder5. Plasmid sequences carrying mcr-1 were completely circularized by PCR and Sanger sequencing. For mcr-1 that was not carried by plasmid, its chromosomal location was confirmed by PCR to link the contig containing mcr-1 and those containing housekeeping genes belonging to the chromosome.
The sequence of all available IncX4, IncI2, and IncHI2 plasmids regardless of the carriage of mcr-1 were retrieved from the GenBank (Supplementary Tables 1–3). Genes present on all analyzed IncX4, IncI2, or IncHI2 plasmids were considered as backbone genes, which were identified using OrthoFinder (Emms and Kelly, 2015). Sequences of backbone genes were concatenated and were then aligned to construct a phylogenetic tree for IncX4, IncI2, or IncHI2 plasmids, respectively, using RAxML (Stamatakis, 2014) with a 1,000-bootstrap test.
Reverse PCR was performed to amplify mcr-1 and its surroundings in strain WCHKP_1511 (Zhao et al., 2017), which contains an intact ISApl1 upstream of mcr-1 and an interrupted ISApl1 downstream as described previously (Li et al., 2017) and the amplicon was sequenced.
A total of nine pink colonies (indicative of E. coli) were recovered on CHROMAgar agar plates containing 4 μg/ml colistin and 64 μg/ml linezolid from sewage. The nine isolates were designated WCHEC1604, WCHEC1606, WCHEC1609, WCHEC1612, WCHEC1613, WCHEC1614, WCHEC1615, WCHEC1618, and WCHEC1622 here. All of the isolates were identified as E. coli, were resistant to colistin (MICs, 4 or 8 μg/ml) and polymyxin B (MIC, 4 μg/ml), and were found to carry mcr-1 by PCR.
The nine isolates displayed seven different PFGE patterns (data not shown) with two pairs of isolates (WCHEC1612/WCHEC1613, WCHEC1614/WCHEC1615) having identical PFGE patterns, suggesting that the nine isolates belonged to seven strains. Therefore, seven isolates were included for further studies with WCHEC1612 and WCHEC1615 being excluded. All seven strains were susceptible to amikacin, ceftazidime, ciprofloxacin, imipenem, and tigecycline except one strain (WCHEC1604) that was resistant to ciprofloxacin (MIC, 8 μg/ml) and one (WCHEC1609) that was intermediate to ceftazidime (MIC, 8 μg/ml). Nonetheless, all seven strains carried multiple antimicrobial resistant genes (Table 1).
Draft genome sequences of the seven selected isolates were generated by Illumina whole genome sequencing, which were assembled into 141–242 contigs (83–134 contigs ≥ 1,000 bp in length; N50, 97,014–253,501 bp) with a 50.27–50.80% GC content, respectively (Table 2). Strain WCHEC1613 was also sequenced using PacBio, which generated 48,400 reads and 451,334,091 bases. A hybrid assembly of the PacBio data with the Illumina reads formed four circular contigs representing one chromosome and three plasmids for strain WCHEC1613.
The seven strains were belonged to six STs, ST10, ST34, ST48, ST1196, ST7086, and ST7087 with the latter two being new types, which have not been identified before. ST7086 has a single allele (fumC) different from ST155, while ST7087 differs from ST165 by one allele (mdh). Of note, strains WCHEC1614 and WCHEC1618 belonged to the same ST (ST34) but there were 554 single nucleotide polymorphisms (SNPs) between their genomes, suggesting that the two strains are likely divergent over a manner of years rather than days or weeks (Stoesser et al., 2016). Therefore, in a single sewage sample, we identified seven E. coli strains belonging to six different STs. Previous reports of mcr-1 gene carriage in E. coli have identified a similarly diverse range of STs carrying the resistance gene (Kong et al., 2016; El Garch et al., 2017; Quan et al., 2017; Wang et al., 2017), though none have reported such diversity in a single confined sample type. This indicates that the dissemination of mcr-1 is not due to expansions of high-risk clones, but rather that mcr-1 is frequently being acquired across the E. coli population in multiple independent events. In the same hospital sewage sample, there were also two blue colonies that were found to carry mcr-1. The two blue colonies were identified as Kluyvera spp. and K. pneumoniae and have been reported elsewhere (Zhao and Zong, 2016; Zhao et al., 2017). Nonetheless, hospital sewage accumulates high density of bacteria, while antibiotics, disinfectants, and their metabolic products are disposed of into hospital sewage and impose selection pressure in favor of the existence of antimicrobial resistant bacteria (Kummerer, 2004), which might explain the diversity of mcr-1-carrying isolates seen here.
Sequencing the whole coding sequence of mcr-1 revealed the original mcr-1, designated mcr-1.1 here, in eight isolates, one of which (strain WCHEC1604) contained two mcr-1 variants including mcr-1.1 and a new variant, designated mcr-1.7 here. The remaining strain, WCHEC1606, had another new variant of mcr-1.1, designated mcr-1.4 here. Both mcr-1.4 and mcr-1.7 have a single nucleotide substitution (G1318T and G643A, respectively) compared to mcr-1.1, resulting in an amino acid substitution (G1318T and A215T, respectively). In addition to mcr-1.4 and mcr-1.7, there are seven variants of mcr-1 in GenBank. Four variants have a single amino acid variation from mcr-1.1, while the remaining three have two or three amino acid substitutions compared with mcr-1.1 (Table 3).
The mcr-1 contains a transmembrane domain and a PEA transferase domain with 8α, 12β, and 12η units (Gao et al., 2016). The variations of mcr-1.4 and mcr-1.7 occurred in the region prior to the β1 unit and in the α6 unit of the PEA transferase domain, respectively, both of which have been found not to influence the function of mcr-1 (Gao et al., 2016). MICs of colistin against transformants containing pMD20-T::mcr-1.4 and pMD20-T::mcr-1.7 were both 4 μg/ml. Among other mcr-1.1 variants, only mcr-1.2 has been characterized at present. MIC of colistin against an E. coli transconjugant containing mcr-1.2 was 8 μg/ml (Di Pilato et al., 2016). Therefore, mcr-1.2, mcr-1.4, and mcr-1.7 have unaltered activity against colistin compared to mcr-1.1. The impact of amino acid substitutions seen in other mcr-1 variants on the function of mcr-1 remains unclear and warrants further investigations.
We sought to determine whether there are signatures of plasmid dissemination through the E. coli population present in our sewage sample. In two strains (WCHEC1609 and WCHEC1614), mcr-1 was located on the chromosome, while in the remaining five strains, mcr-1 was carried by a plasmid belonging to four different replicon types including IncHI2, IncI2, IncP, and IncX4 (Table 1). Of note, mcr-1.1 was located on an IncX4 plasmid and mcr-1.7 was on an IncI2 plasmid in strain WCHEC1604. It is remarkable that mcr-1-carrying plasmids of four replicon types were found in a single 1 ml sewage sample at one site. All IncI2, IncP, and IncX4 mcr-1-carrying plasmids in this study could be transferred by conjugation at a frequency of 10-5 to 10-7 cells per recipient cell by mating, while the mcr-1-carrying IncHI2 plasmid was not.
The mcr-1.1 gene was carried by IncX4 plasmids in two strains, WCHEC1604 and WCHEC1618, designated pMCR_1604-IncX4 and pMCR_WCHEC1618, respectively. mcr-1.4 in strain WCHEC1606 was also located on an IncX4 plasmid (designated pMCR_WCHEC1606). pMCR_1604-IncX4 is 31,229 bp in length and is 2,080 bp less than the 33,309-bp pMCR_WCHEC1618 and pMCR_WCHEC1606, which was likely due to homologous recombination between the two copies of the dnaJ-containing region. pMCR_WCHEC1606 differed from pMCR_WCHEC1618 by only a single nucleotide substitution, which was the one defining mcr-1.4, suggesting that mcr-1.4 evolved from mcr-1.1 by a point mutation on the IncX4 plasmid. IncX4 plasmids carrying mcr-1 have been found in E. coli or K. pneumoniae strains in Africa (South Africa), Asia (China), Europe (Estonia, Italy, Netherlands, and Switzerland), and North (United States) and South America (Brazil), suggesting a global distribution. Phylogenetic analysis based on all 17 backbone genes of IncX4 plasmids (Supplementary Table 1) revealed that all mcr-1-carrying IncX4 plasmids formed a clade with several non-mcr-1-carrying IncX4 plasmids (Figure 1), suggesting that the mcr-1-carrying IncX4 plasmids were likely from a common ancestor and the acquisition of mcr-1 onto the IncX4 backbone occurred recently. By contrast, mcr-2-carrying IncX4 plasmid pKP37-BE was distinct from mcr-1-carrying IncX4 plasmids (Figure 1).
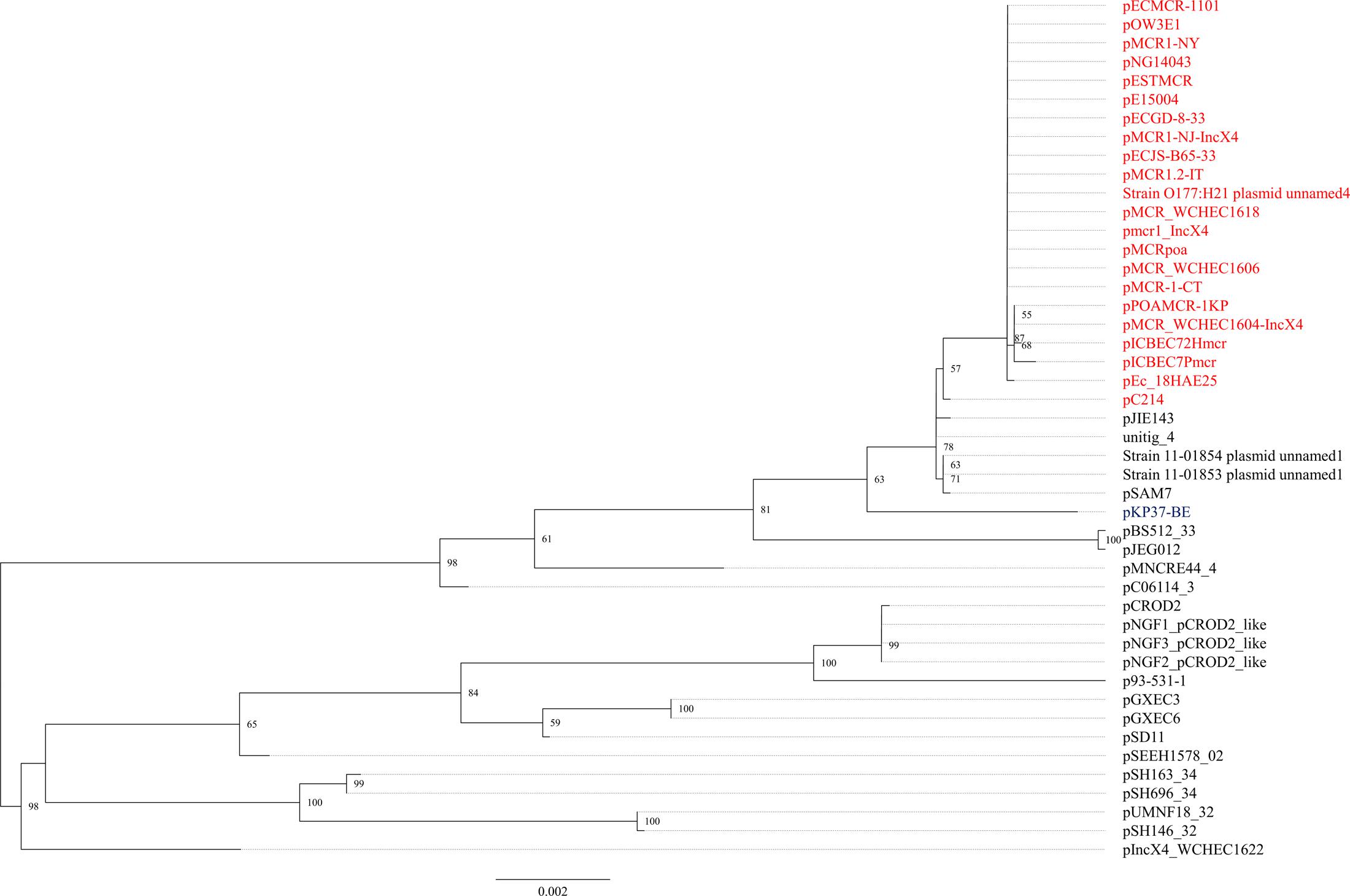
FIGURE 1. Phylogenetic analysis of IncX4 plasmids. The accession numbers of the plasmids are shown in Supplementary Table 1. Those carrying mcr-1 are indicated in red, while pKP37-BE carrying mcr-2 is shown in blue. The tree was constructed using concatenated sequences of 17 genes belonging to the IncX4 backbone.
The mcr-1.7 in strain WCHEC1604 was carried by a 62,098-bp IncI2 plasmid, designated pMCR_1604-IncI2 here. Phylogenetic analysis based on all 27 backbone genes of IncI2 plasmids (Supplementary Table 2) revealed that mcr-1-carrying plasmids belonged to a number of clades and mixed with plasmids without mcr-1, suggesting that IncI2 plasmids were likely to have acquired mcr-1 in multiple events rather than a single plasmid expansion into different strains (Figure 2). pMCR_1604-IncI2 was most closely related (99% identity and 93% coverage) to pECJS-61-63 (GenBank accession no. KX254342) in an E. coli isolated from a pig in China and it is likely that mcr-1.7 evolved from mcr-1.1 on an IncI2 plasmid.
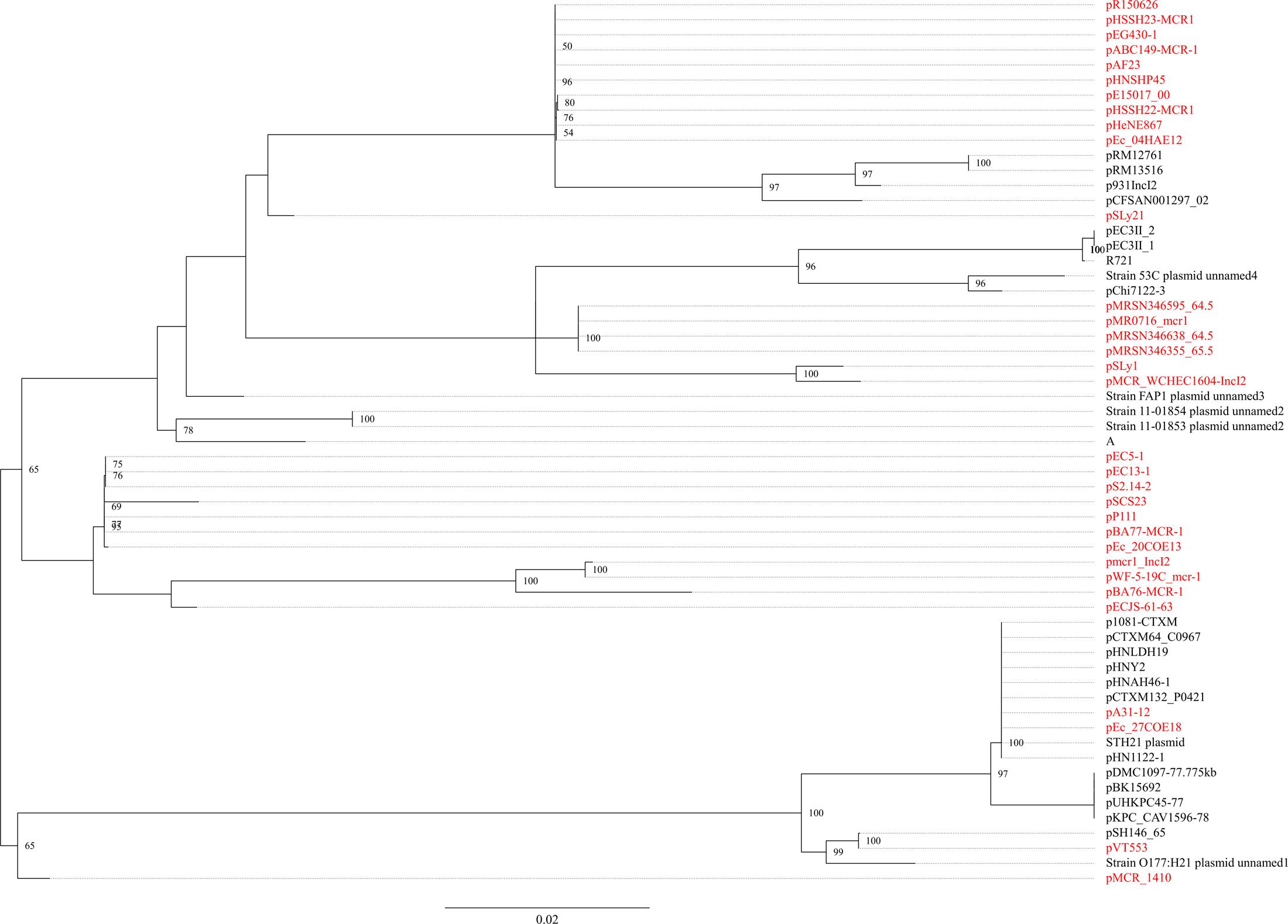
FIGURE 2. Phylogenetic analysis of IncI2 plasmids. The accession numbers of the plasmids are shown in Supplementary Table 2. Those carrying mcr-1 are indicated in red. The tree was constructed using concatenated sequences of 27 genes belonging to the IncI2 backbone.
In strain WCHEC1613, mcr-1.1 was carried by a 300-kb large plasmid (designated pMCR_WCHEC1613) containing both IncHI2 and IncN replicons. pMCR_WCHEC1613 was most closely related (83% coverage and 99% identity) to the IncHI2 plasmid pHNSHP45-2 (GenBank accession no. KU341381) carrying mcr-1.1 from E. coli strain SHP45 in China (Liu et al., 2016). In addition, a large part (94,689 bp; positions 73,442–170,084) of pMCR_WCHEC1613 was nearly identical to the counterpart of a plasmid containing both IncFIB and IncN replicons, pMR0516mcr (GenBank accession no. KX276657), carrying mcr-1.1 from E. coli in United States (McGann et al., 2016). It is likely that pMCR_WCHEC1613 was formed by the fusion of two plasmids, which contain IncHI2 and IncN replicon, respectively. Phylogenetic analysis based on all 33 backbones genes of IncHI2 plasmids (Supplementary Table 3) revealed that all mcr-1-carrying IncHI2 plasmids were clustered together with a few non-mcr-1-carrying IncHI2 plasmids (Figure 3), suggesting that the mcr-1-carrying IncHI2 plasmids arose from a common ancestor and the acquisition of mcr-1 onto the IncHI2 backbone occurred recently.
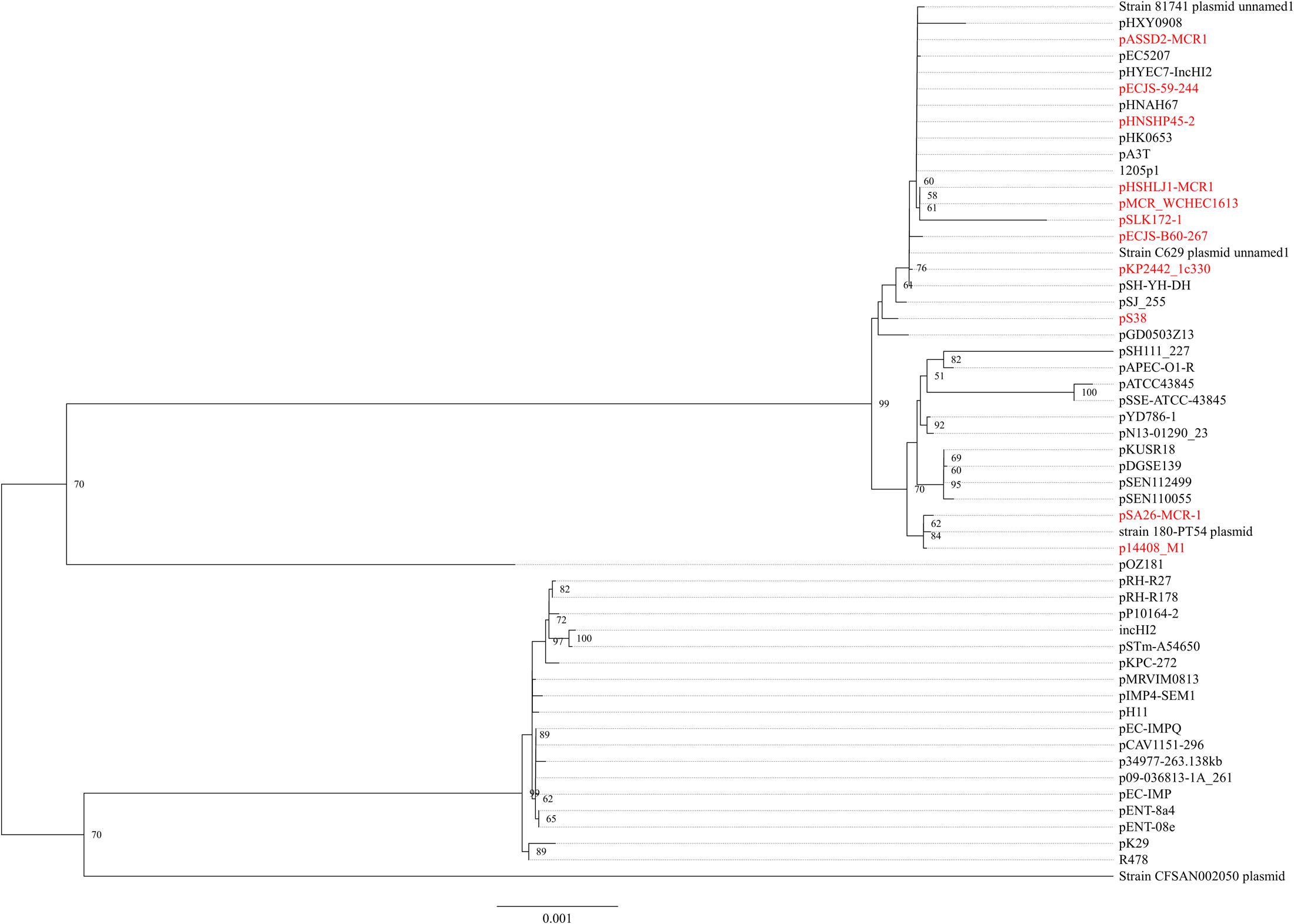
FIGURE 3. Phylogenetic analysis of IncHI2 plasmids. The accession numbers of the plasmids are shown in Supplementary Table 3. Those carrying mcr-1 are indicated in red. The tree was constructed using concatenated sequences of 27 genes belonging to the IncI2 backbone.
In WCHEC1622, mcr-1.1 was carried by a 49,897-bp IncP plasmid, designated pMCR_WCHEC1622, which was almost identical (100% coverage and 99% identity) to pMCR_1511 (Figure 4), an IncP plasmid recovered from K. pneumoniae in the same sewage sample (Zhao et al., 2017). Like pMCR_1511, pMCR_WCHEC1622 belongs to a new clade of IncP (Zhao et al., 2017). As IncP plasmids are broad-host-range, the carriage of mcr-1 on IncP plasmids has the potential to mediate the dissemination of mcr-1 from the Enterobacteriaceae to other bacterial species.
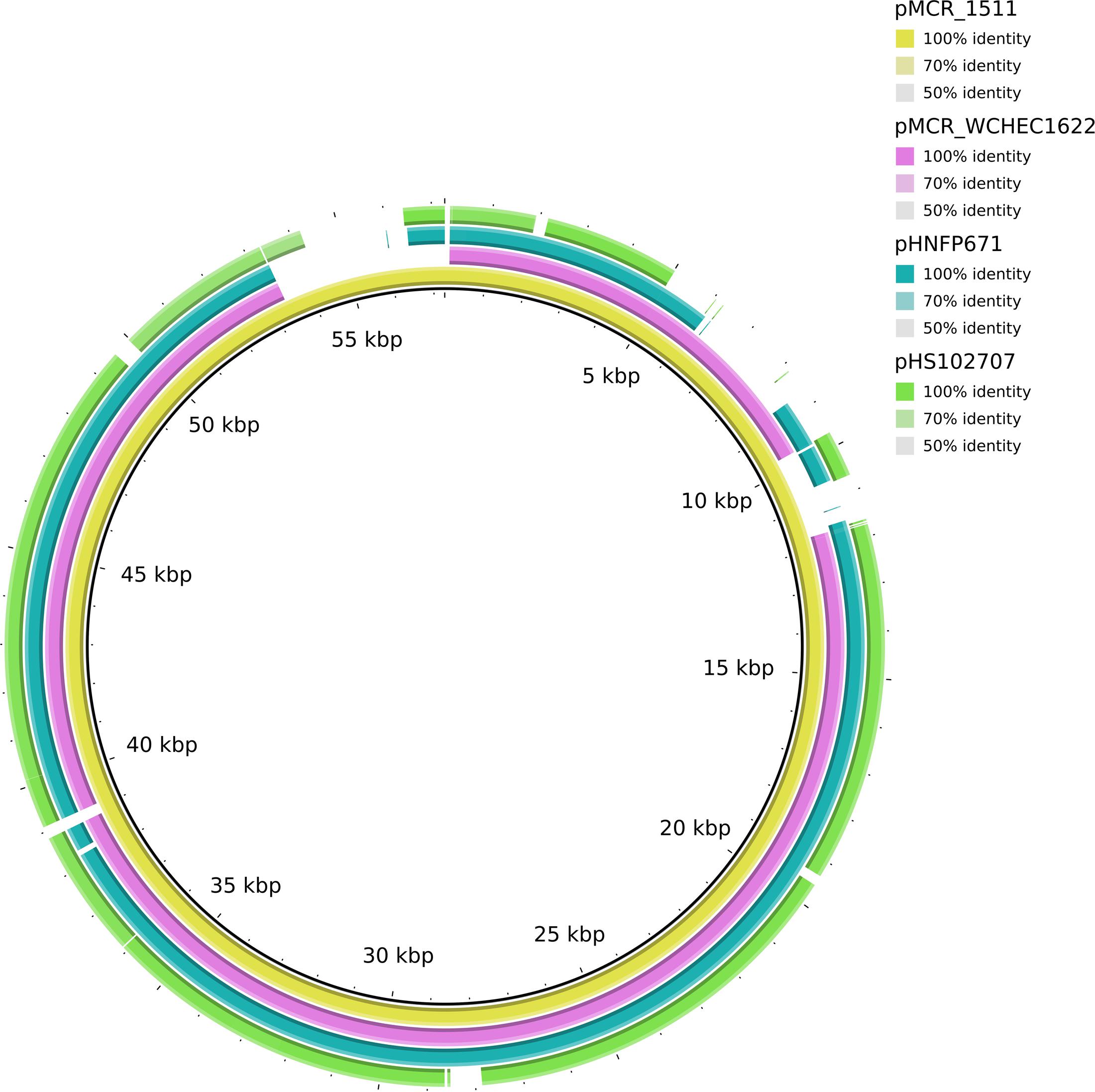
FIGURE 4. Alignment of pMCR_WCHEC1622 and closely related IncP plasmids. pMCR_1511 (Zhao et al., 2017) was used as a reference. The alignment is a pairwise BLASTn alignment performed using BRIG. pMCR_1511, pMCR_WCHEC1622, pHNFP671 (GenBank accession no. KP324830), pHS102707 (GenBank accession no. KF701335), pMCR16_P053 (GenBank accession no. KY352406), and pJJ1886_4 (GenBank accession no. CP006788) belong to a new unnamed clade of IncP plasmids (Zhao et al., 2017).
The mcr-1 has been typically seen in three types of genetic contexts, i.e., mcr-1-pho, ISApl1-mcr-1-pho, and ISApl1-mcr-1-pho-ISApl1 (Gao et al., 2016; Snesrud et al., 2016; Li et al., 2017) with pho referring to a gene (also called pap in some publications) encoding a putative phosphoesterase and ISApl1 being an insertion sequence of the IS30 family. All three types of the mcr-1 genetic context were seen in the seven E. coli strains here (Figure 5). Two copies of ISApl1 bracketing mcr-1 and pho (ISApl1-mcr-1-pho-ISApl1) could form a composite transposon termed Tn6330, which is able to generate a circular intermediate (ISApl1-mcr-1-pho) by excision from a plasmid and the intermediate could then insert into another plasmid (Li et al., 2017) and possibly could also insert into chromosome. A previous study (Li et al., 2017) found that the circular intermediate is formed by the ISApl1 downstream of mcr-1. However, in this study, the ISApl1-mcr-1-pho circular intermediate was obtained from pMCR_1511, in which the ISApl1 downstream of mcr-1 was interrupted (Figure 6). This suggests that the ISApl1 upstream of mcr-1 participated in the formation of the ISApl1-mcr-1-pho circular intermediate by its own or via recombination with the ISApl1 downstream of mcr-1.
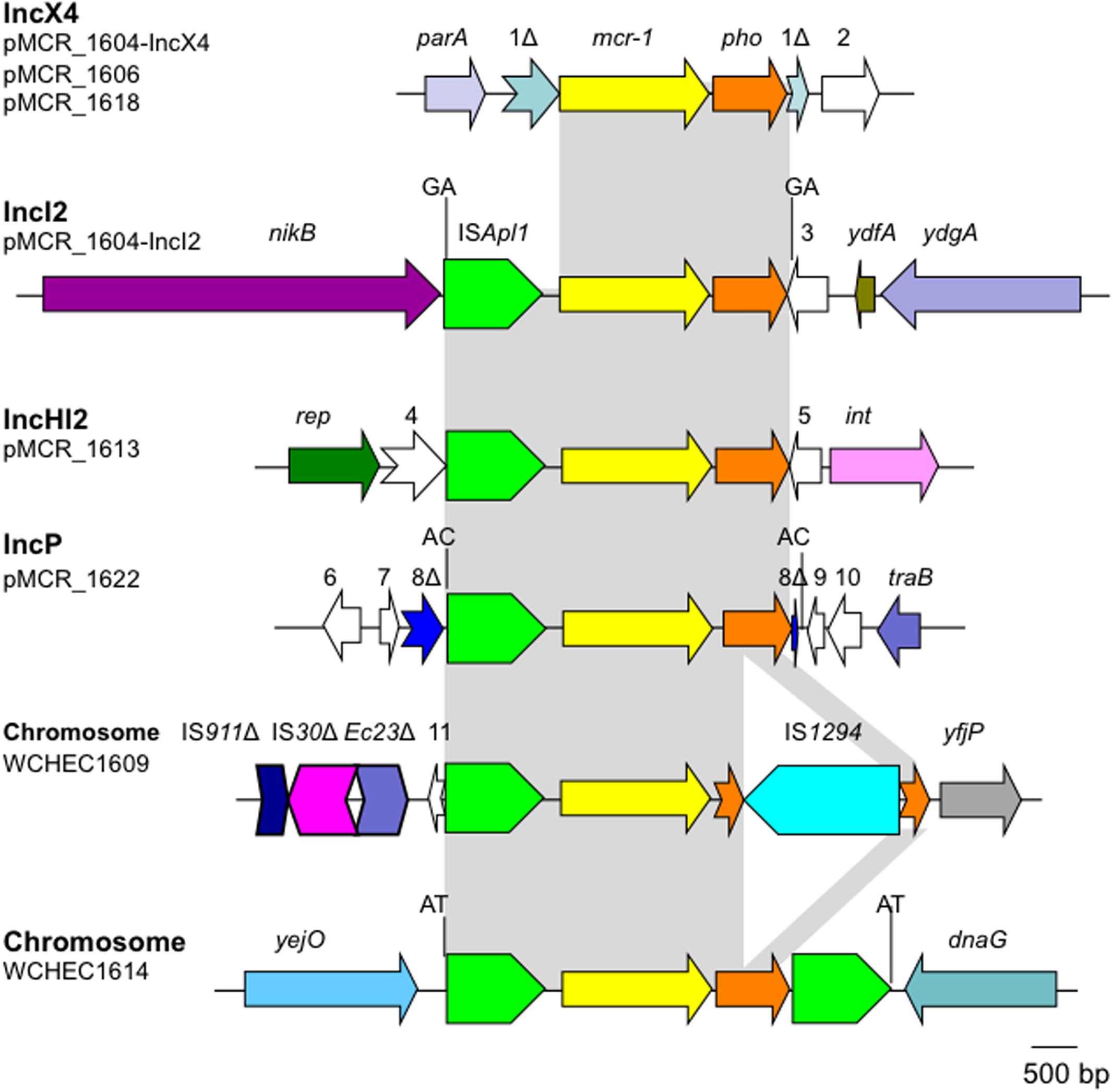
FIGURE 5. Genetic contexts of mcr-1 in the seven E. coli strains. orfs that encode hypothetical proteins with unknown function are indicated in white except that those interrupted by the element carrying mcr-1. Orfs are indicated with numbers, 1–11. Δ represents interrupted or truncated. Other genes are dnaG (encoding a DNA primase), int (encoding a recombinase), parA (encoding partition), nikB (encoding a relaxase of the plasmid), rep (encoding the replication protein), traB (encoding a conjugative protein), ydfA (a transcriptional regulator), ydgA (encoding a DNA topoisomerase III), yejO (an outer membrane β-barrel domain-containing protein), and yfjP (encoding a 50S ribosome-binding GTPase). The 2-bp direct repeat (DR) is shown if it is present.
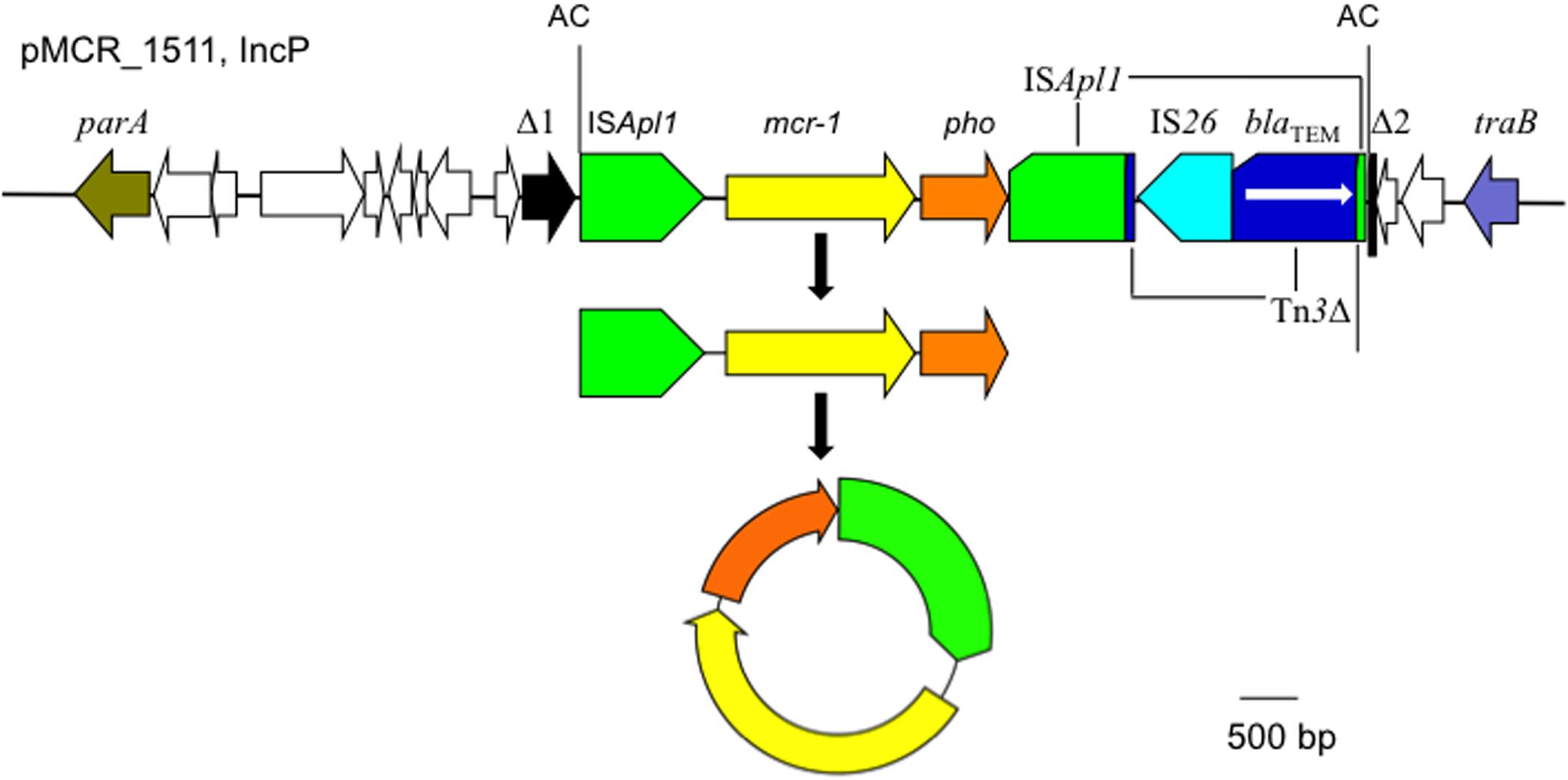
FIGURE 6. The circular intermediate formed by ISApl1. Genetic context of mcr-1 on the IncP plasmid pMCR_1511 is shown. Orfs that encode hypothetical proteins with unknown function are indicated in white except that the one disrupted by the ISApl1-formed composite transposon on pMCR_1511 is shown in black (Δ1 and Δ2). The 2-bp direct repeat (AC) abutting the ISApl1-formed composite transposon on pMCR_1511 is shown. On pMCR_1511, the ISApl1 downstream of mcr-1 was interrupted by Tn3, which was interrupted by IS26.
Therefore, in a single (1 mL) hospital sewage sample, we observed multiple Enterobacteriaceae species, multiple strains of E. coli, multiple plasmids, and multiple genetic contexts carrying multiple variants of mcr-1. This suggests that mcr-1 is undergoing rapid evolution within healthcare environments and is being rapidly disseminated across plasmids, strains, and species.
Nucleotide sequence accession numbers: Reads and the Whole Genome Shotgun Sequencing project of E. coli strain WCHEC1604, WCHEC1606, WCHEC1609, WCHEC1613, WCHEC1614, WCHEC1618, and WCHEC1622 have been deposited into DDBJ/EMBL/GenBank under accession MUWZ00000000, MSRB00000000, MSQX00000000, CP019213, MSQY00000000, MSQZ00000000, and MSRA00000000, respectively. The sequence of pMCR_1604-IncX4, pMCR_1604-IncI2, pMCR_WCHEC1606, pMCR_WCHEC1613, pMCR_WCHEC1618, and pMCR_WCHEC1622 has been deposited into DDBJ/EMBL/GenBank under accession numbers KY582848, KY829117, KY463451, CP019214, KY463454, and KY463452, respectively.
ZZ: Designed the experiments, analyzed the data, and wrote the manuscript. FZ and YF: Performed the experiments and analyzed the data. XL and AM: Contributed to analyzing the data and co-wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The work was supported by a grant from the National Natural Science Foundation of China (project no. 81572030; to ZZ) and a joint grant from the National Natural Science Foundation of China (project no. 8151101182 to ZZ) and the Newton Advanced Fellowship, Royal Society (NA015363), United Kingdom (to AM and ZZ). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02094/full#supplementary-material
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
CLSI (2013). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., Depristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Di Pilato, V., Arena, F., Tascini, C., Cannatelli, A., Henrici De Angelis, L., Fortunato, S., et al. (2016). mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512. Antimicrob. Agents Chemother. 60, 5612–5615. doi: 10.1128/AAC.01075-16
El Garch, F., Sauget, M., Hocquet, D., Lechaudee, D., Woehrle, F., and Bertrand, X. (2017). mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin. Microbiol. Infect. 23, 51.e1–51.e4. doi: 10.1016/j.cmi.2016.08.033
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157. doi: 10.1186/s13059-015-0721-2
Gao, R., Hu, Y., Li, Z., Sun, J., Wang, Q., Lin, J., et al. (2016). Dissemination and Mechanism for the MCR-1 colistin resistance. PLOS Pathog. 12:e1005957. doi: 10.1371/journal.ppat.1005957
Kong, L. H., Lei, C. W., Ma, S. Z., Jiang, W., Liu, B. H., Wang, Y. X., et al. (2016). Various sequence types of Escherichia coli co-harboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob. Agents Chemother. 61:e02167-16. doi: 10.1128/AAC.02167-16
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.215116
Kummerer, K. (2004). Resistance in the environment. J. Antimicrob. Chemother. 54, 311–320. doi: 10.1093/jac/dkh325
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Quan, J., Li, X., Chen, Y., Jiang, Y., Zhou, Z., Zhang, H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/S1473-3099(16)30528-X
Schwarz, S., and Johnson, A. P. (2016). Transferable resistance to colistin: a new but old threat. J. Antimicrob. Chemother. 71, 2066–2070. doi: 10.1093/jac/dkw274
Snesrud, E., He, S., Chandler, M., Dekker, J. P., Hickman, A. B., Mcgann, P., et al. (2016). A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60, 6973–6976. doi: 10.1128/AAC.01457-16
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stoesser, N., Sheppard, A. E., Pankhurst, L., De Maio, N., Moore, C. E., Sebra, R., et al. (2016). Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15
Unger, F., Eisenberg, T., Prenger-Berninghoff, E., Leidner, U., Ludwig, M. L., Rothe, M., et al. (2017). Imported reptiles as a risk factor for the global distribution of Escherichia coli harbouring the colistin resistance gene mcr-1. Int. J. Antimicrob. Agents 49, 122–123. doi: 10.1016/j.ijantimicag.2016.10.007
Wang, Y., Tian, G.-B., Zhang, R., Shen, Y., Tyrrell, J. M., Huang, X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/S1473-3099(16)30527-8
Yamamoto, S., and Harayama, S. (1995). PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61, 1104–1109.
Zhao, F., Feng, Y., Lu, X., Mcnally, A., and Zong, Z. (2017). IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob. Agents Chemother. 61:e02229-16. doi: 10.1128/AAC.02229-16
Keywords: colistin resistance, mcr-1, sewage, Escherichia coli, plasmid
Citation: Zhao F, Feng Y, Lü X, McNally A and Zong Z (2017) Remarkable Diversity of Escherichia coli Carrying mcr-1 from Hospital Sewage with the Identification of Two New mcr-1 Variants. Front. Microbiol. 8:2094. doi: 10.3389/fmicb.2017.02094
Received: 31 August 2017; Accepted: 12 October 2017;
Published: 25 October 2017.
Edited by:
Xian-Zhi Li, Health Canada, CanadaReviewed by:
Séamus Fanning, University College Dublin, IrelandCopyright © 2017 Zhao, Feng, Lü, McNally and Zong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Zong, em9uZ3poaXlAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.