- Department of Biochemical Engineering and Biotechnology, Indian Institute of Technology, New Delhi, India
Stationary phase is the stage when growth ceases but cells remain metabolically active. Several physical and molecular changes take place during this stage that makes them interesting to explore. The characteristic proteins synthesized in the stationary phase are indispensable as they confer viability to the bacteria. Detailed knowledge of these proteins and the genes synthesizing them is required to understand the survival in such nutrient deprived conditions. The promoters, which drive the expression of these genes, are called stationary phase promoters. These promoters exhibit increased activity in the stationary phase and less or no activity in the exponential phase. The vectors constructed based on these promoters are ideal for large-scale protein production due to the absence of any external inducers. A number of recombinant protein production systems have been developed using these promoters. This review describes the stationary phase survival of bacteria, the promoters involved, their importance, regulation, and applications.
Introduction
The majority of the microorganisms around us in air, sea water, and soil are predominantly present in stationary phase (Gefen et al., 2014). The natural habitat of microorganisms often contains limited nutrients due to which rapid growth is usually hampered. Apart from nutrient deprivation, there are other conditions, including physical and chemical stresses, which result in unbalanced growth. All these events result in many changes at the molecular level. These molecular changes are comparable to those observed during the stationary phase of bacteria as witnessed in laboratory studies. The entry of bacteria to the stationary phase can be caused by different factors, including limitation of a specific essential nutrient, accumulation of toxic by-products, presence of stress factors such as changes in pH, temperature, osmolarity, etc. As the cell enters this phase, there is a reduction in cell size and the DNA/protein ratio is said to increase during transition to stationary phase (Nystrom, 2004). The stationary phase has received much attention due to the pattern of protein synthesis in this phase and also because of survival strategies adopted by bacteria. Numerous physiological, morphological, and gene expression changes are observed when a growing cell enters the stationary phase. These are discussed in the following sections.
Physiology of the Stationary Phase
In the stationary phase, the cells become spherical and smaller with a rigid cell envelope, the cell wall is highly cross-linked, membrane fluidity reduces, and cells activate the stringent response mechanism in order to survive the calamity. The activation of this mechanism allows the bacteria to reprogram the gene expression pattern to adapt to different stresses. Two key components of the bacterial stringent response are ppGpp and pppGpp (which are explained in a later section). As a consequence, the cells divert their resources away from growth toward synthesizing amino acids so as to promote survival till nutrient conditions improve.
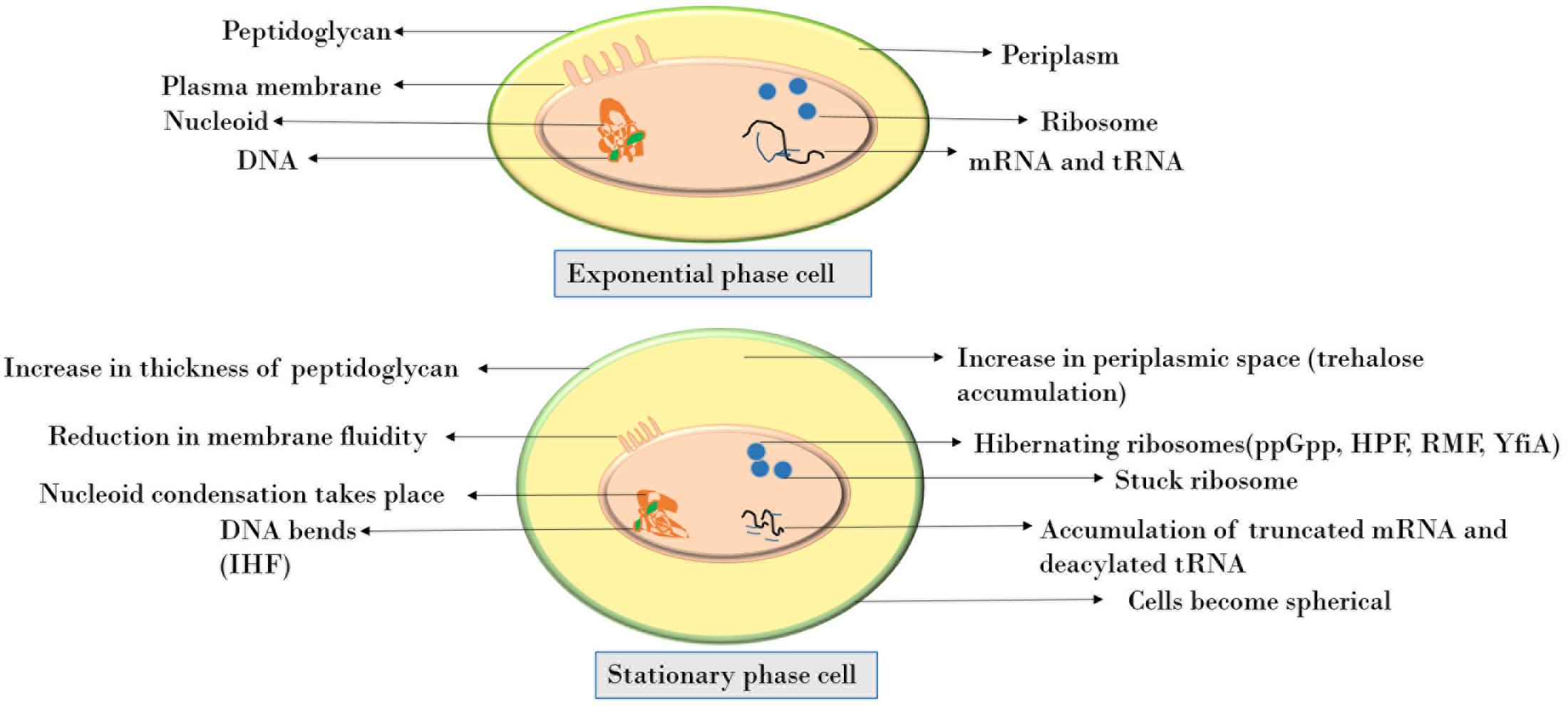
Figure 1 depicts the various changes observed in a cell when it enters the stationary phase. The peptidoglycan layer, being the stress-bearing component of cell, increases in thickness. It accounts for 0.7–0.8% of cell’s dry weight in exponential phase cells whereas in stationary phase it increases up to 1.4–1.9% (Mengin-Lecreulx and van Heijenoort, 1985). At the subcellular level, nucleoid condensation occurs for DNA protection, the cytoplasm gets condensed with an overall decrease in protein synthesis as a consequence of stress or stationary phase (Navarro Llorens et al., 2010). At the translational level, the 70S ribosomes are converted into inactive 100S ribosome dimers by associating with ribosome modulation factor (Wada, 1998). This process, termed as ribosome hibernation, is thought to be a mechanism to fine-tune the translation process according to environmental conditions (McKay and Portnoy, 2015). Recently, 16S rRNA fragmentation at the tip of helix 6 has been shown to attenuate the activity of 30S ribosomal subunit and thereby protein synthesis (Luidalepp et al., 2016). Also, during limited nutrient availability, accumulation of truncated mRNA and deacylated tRNA occurs. The ribosomes become stuck on these mRNAs and owing to the absence of a stop codon, the ribosome is unable to get released (Pletnev et al., 2015). These mechanisms are understood to be the defense response upon starvation. As a result of the various morphological, metabolic, transcriptional, or translational alterations, the stationary phase cells become resistant to high temperature, high concentrations of H2O2, and very high medium osmolarity.
Cells in exponential, stationary, and long-term stationary phases have different fates (Figure 2). As a consequence of starvation, many bacteria including the genera Bacillus and Clostridium form resistant spores helping them withstand the harsh surrounding environment. Non-optimal growth conditions also lead to the formation of biofilm in many bacterial species. Physiologically, biofilm bacteria are similar to stationary phase bacteria. One key transition is the formation of persisters induced during stationary phase, in biofilms, and also as a consequence of a general stress response. These cells could also arise in exponential growth by the activation of ppGpp as a consequence of sub-lethal antibiotic concentration. The formation of these bacterial persisters is understood to be the reason behind relapsing infections and is a major cause of drug resistance (Harms et al., 2016).
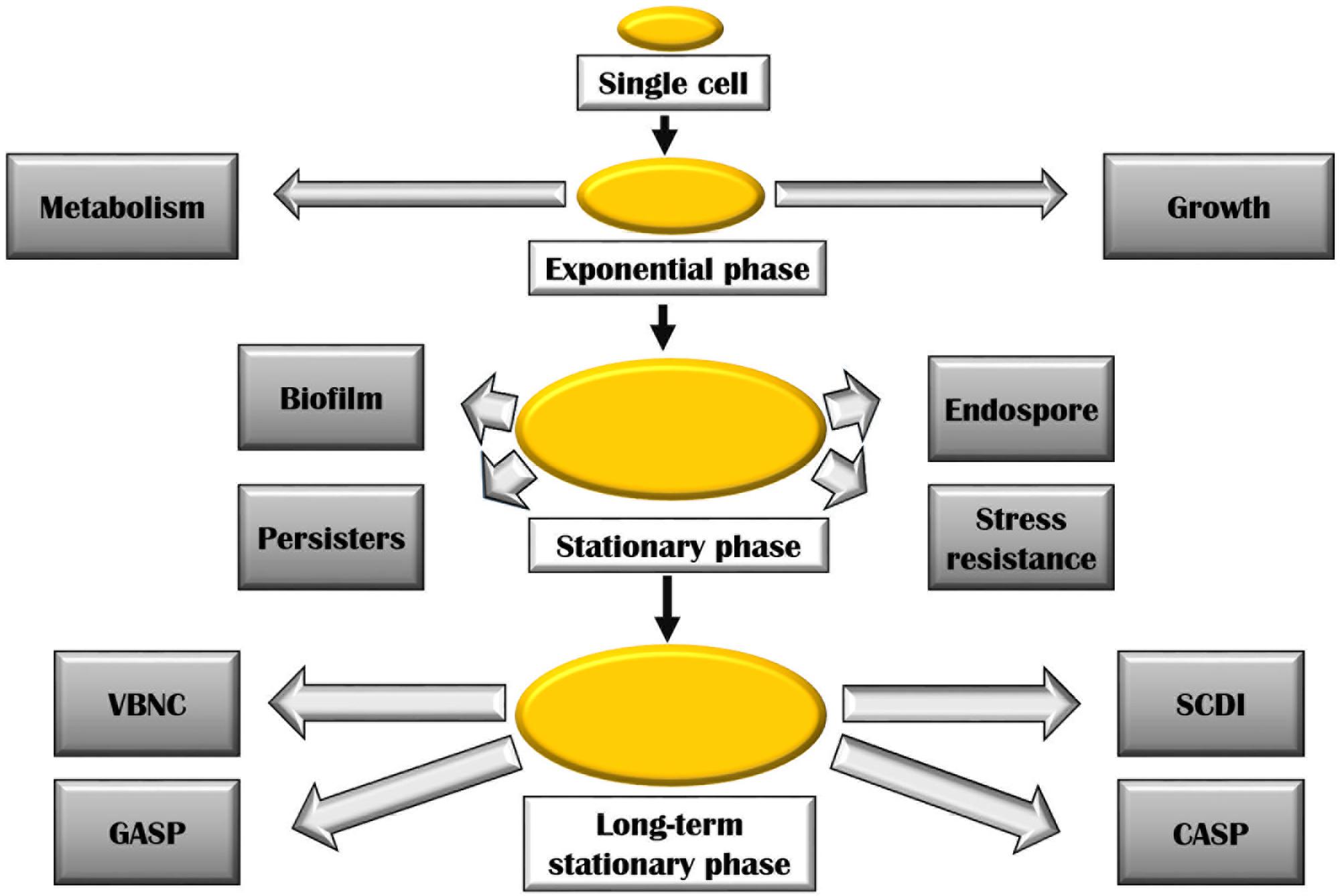
FIGURE 2. Various bacterial adaptations at stationary and long-term stationary phase. Abbreviations are described in the text.
During the late stationary phase sometimes referred to as long-term stationary phase, several remarkable adaptations take place. On continued starvation, one of the survival strategies includes bacteria entering a viable but non-culturable state (VBNC). In this state, bacteria remain metabolically active but fail to form colonies on bacteriological media. Several bacteria including Rhodococcus biphenylivorans (Su et al., 2015), Escherichia coli, Agrobacterium tumefaciens, Helicobacter pylori, Lactococcus lactis, many Vibrio species, and Pseudomonas species have been shown to enter the VBNC state (Oliver, 2005). The VBNC state poses a serious health risk as the dormant bacterial species could remain undetected in culturable conditions, though having the ability to cause infections (Navarro Llorens et al., 2010). A variety of stresses is said to lead to the manifestation of VBNC state (Pletnev et al., 2015). Prolonged starvation also results in Growth Advantage in Stationary Phase (GASP) phenotype. The GASP phenomenon is a result of mutations in the rpoS allele (described later) which confers a gainful ability to continue growing during starvation conditions, thus replacing the parental population (Navarro Llorens et al., 2010). These mutations allow the mutants to effectively scavenge the nutrients released by dead cells (Zambrano and Kolter, 1996). A number of Gram-positive bacteria such as Listeria monocytogenes (Bruno and Freitag, 2011), Staphylococcus aureus, Enterococcus faecalis, and Bacillus globigii (Finkel et al., 1997) and Gram-negative bacteria including Campylobacter, Geobacter, Vibrio, E. coli, Pseudomonas, etc., have been found to enter the GASP state (Chen and Chen, 2014). Gefen et al. (2014) coined the term ‘constant activity stationary phase’ (CASP) to describe the phenomenon of constant rate of protein synthesis observed in non-growing bacteria that have undergone over more than 60 h of starvation. On studying the protein production at this stage, they have found that both the protein synthesis machinery including ribosomes, RNA polymerases, etc., and resources such as amino acids, nucleotides, etc., remain constant at CASP. Finally, constant promoter activity was observed in this experiment for up to 10 h of starvation. Another interesting phenomenon experienced by bacterial population in stationary phase is the ‘stationary phase contact-dependent inhibition’ (SCDI). It requires physical contact between the evolved and original bacteria (Lemonnier et al., 2008). In this process, it was observed that the evolved strains either killed or inhibited the growth of bacteria that they were derived from. The inhibiting ability of these strains is attributed to mutations within a single gene involved in glycogen synthesis pathway: glgC (encoding ADP-glucose pyrophosphorylase). Astonishingly, all evolved strains overproduced glycogen which seemed to be necessary for SCDI to occur (Navarro Llorens et al., 2010).
Alternative Sigma Factors Active At Stationary Phase
A key regulator of stationary phase gene expression in E. coli is the transcription factor σS [a product of rpoS (katF) gene]. The E. coli genome was found to contain two genes katE and katG encoding for HPII and HP1w1-4x catalases. The expression of HPII was highest in stationary phase and has been shown to be completely dependent on katF gene product. The latter serves as sigma factor for RNA polymerase and therefore named as rpoS or σS or σ38 or stationary phase sigma factor or starvation sigma factor (Tanaka et al., 1997).
The amount of σS remains relatively low in the growing phase of cells but increases markedly when the cell encounters stress, starvation or enters stationary phase. The role of this protein is to aid in survival and improved resistance to stressful conditions. Induction of σS is observed under conditions of low pH, heat or cold shock, UV-induced DNA damage, nutrient starvation, high cell density, high osmolarity, etc. (Hengge, 2011). The σS-dependent genes have been attributed to morphological changes (Hengge, 2011), induction of starvation proteins (Alexander and St. John, 1994), iron uptake, carbohydrate metabolism, amino acid transport, and so on, at the onset of stationary phase (Lacour and Landini, 2004).
The rpoS sigma factor is selectively utilized in stationary phase. The major sigma factor rpoD (σ70) is inhibited by a regulator of sigma D (Rsd). The rationale for σS selectivity in vivo is not completely understood, but it is known that many promoters can exhibit both σS and σ70 mediated expression in vitro. It is well known that σ70 is affected by changes in spacer region and consensus –10 and –35 positions, but the alternative σS is shown to be less affected by changes in these regions, thus making it more selective in vivo (Hengge, 2011). Another observation by Tanaka et al., 1995 indicates that the –35 region is not always required for stationary-phase expression (Tanaka et al., 1995). In this study, the fic promoter was shown to function with promoter sequences downstream from –17. Also, the promoters recognized by RpoS are found to contain curved DNA region. Hence, the absence of consensus –35 and the presence of curved DNA region imparted σS dependence to galP1 and galP2 promoters, whereas the presence of –35 sequence in the same promoter changed the specificity toward σ70 (Kolb et al., 1995). Thus, the general belief is that the σS promoters lack a –35 consensus sequence. However, some authors have suggested CTGCAA (Bohannon et al., 1991) or CCGACA (Wise et al., 1996) as the –35 consensus sequence. Similarly for –10, Hengge-Aronis (1993) has suggested a consensus sequence of TATACT, which was later changed to CTATACT (Espinosa-Urgel et al., 1996). More recently, a long consensus sequence KCTAYRCTTAA for –10 region has been proposed, where K could be T or G, Y could be T or C, and R could be A or G (Weber et al., 2005). Not all the stationary-phase induced genes depend on σS, and out of the many genes that show higher level of expression in the stationary phase, only 10% is known to be dependent on σS (Rava et al., 1999). Out of the genes induced in stationary phase, those that show σS independent behavior are dnaK, groEL, htpG which depend on σ32 (Kolter et al., 1993).
Several other alternative sigma factors have been reported. In Salmonella typhimurium, σE has been suggested to serve a complementary role in stationary phase survival. Mutants deficient in rpoH gene coding for σE have been shown to be susceptible to oxidative stress (Testerman et al., 2002).
The number of sigma factors varies from 1 in Mycoplasma genitalium (Dorman, 2011), 6 in Gordonia sp. IITR100 (Jaishankar et al., 2017), 7 in E. coli (Ishihama, 1997), 18 in B. subtilis (Gruber and Gross, 2003), 24 in Pseudomonas aeruginosa (Potvin et al., 2008), and 65 in Streptomyces coelicolor (Kim et al., 2008). Table 1 gives a list of various sigma factors in well-known bacterial species and the types of sigma factors upregulated at stationary phase.
Regulation of RpoS
The RpoS is regulated at post-transcriptional level by rpoS mRNA secondary structure, small RNAs, Hfq, and HU proteins, ClpXP protease and RssB (phosphorylation-modulated RpoS recognition factor) (Hengge-Aronis, 2002). The rpoS mRNA is stimulated by regulatory factors such as Hfq (HF-1) protein and DsrA (small regulatory RNA) and repressed by H-NS (histone-like protein) and oxyS RNA. The 5′ UTR of rpoS mRNA forms a loop which represses its translation. This loop can be disrupted by non-coding RNAs such as dsrA, rprA, and arcA (Gaida et al., 2013). Another sRNA which positively regulates rpoS mRNA is gcvB (Jin et al., 2009).
The turnover of RpoS protein in exponential phase is very high with a half-life of 1.4 min (Lange and Hengge-Aronis, 1994). The RpoS protein is stable in stationary phase.
The levels of RpoS are also controlled by a number of other factors. These include both positive regulators such as ppGpp and polyphosphate (polyp) and negative regulators such as cAMP and UDP glucose.
The availability of ppGpp is dependent on RelA, a ppGpp synthase that is associated with ribosomes. In stationary phase, when the uncharged tRNAs accumulate due to decreased availability of amino acids, relA is turned on and synthesizes ppGpp. This turns on the promoters involved in amino acid biosynthesis and uptake (Barker et al., 2001). It has been shown that 6S RNA regulates relA gene expression, which leads to alteration in ppGpp levels in stationary phase (Cavanagh et al., 2010). The rRNA genes are turned off by ppGpp. Many stationary phase promoters (SPPs) are also regulated by 6S RNA, even in the absence of ppGpp.
In B. subtilis, it has been demonstrated that cells entering in stationary phase have small GTP and GDP pools. This is possibly due to conversion of GTP to (p)ppGpp or due to the lack of sufficient precursors available for nucleotide synthesis. Lopez and coworkers demonstrated that treatment of cells with decoyinine, an inhibitor of GMP synthase, can result in induction of stationary phase genes (Ratnayake-Lecamwasam et al., 2001).
The intracellular levels of certain compounds such as trehalose, glycine betaine, glycogen, and polyphosphate are high under stress conditions. Some of these compounds modulate function of the RpoS holoenzyme. For example, glutamate and trehalose modulate the holoenzyme binding to promoters. Similarly, altered promoter selectivity has been observed in E. coli when RpoS associates with inorganic polyphosphate. The inhibition due to PolyP is relieved by high concentrations of potassium glutamate (Shimada et al., 2004). Bacterial pheromone, Homoserine lactone (HSL), a small molecule responsible for communication between bacteria, also affects the concentration of σS in the cell. Mutants in the biosynthetic pathway for synthesis of HSL loose the ability to induce σS (Zambrano and Kolter, 1996).
Expression of Genes in Stationary Phase
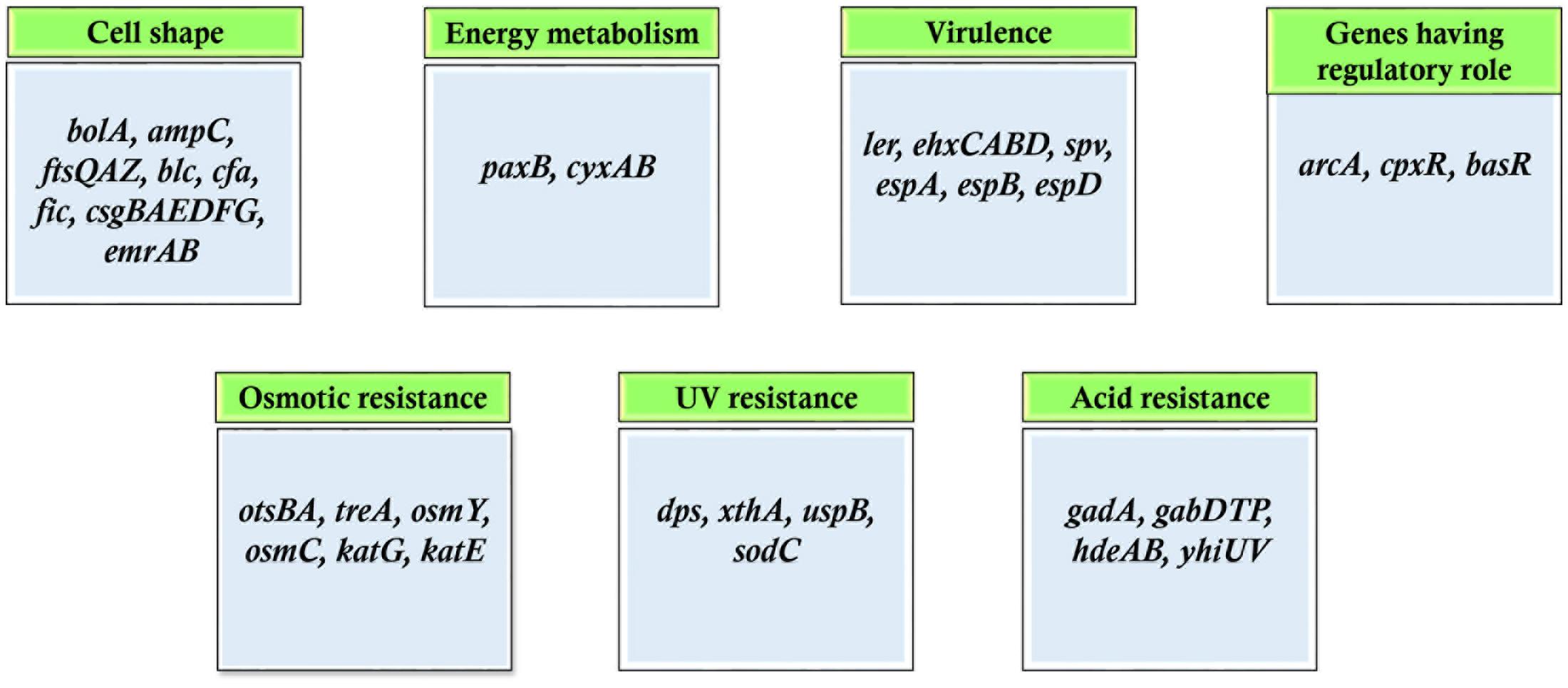
When the cells are growing, the metabolism-linked genes are highly expressed, and get turned off when the cells enter stationary phase. The stationary phase is a period of no growth, however, genes essential for survival of organisms are expressed at this stage. Around 20% of the genes of E. coli are found to express at higher level in the stationary phase (Rava et al., 1999). These genes are directly linked to many key events including DNA repair, glycogen production, thermotolerance, osmotolerance, etc. (Bohannon et al., 1991; Ishihama, 1997). Transcriptome profiling/expression analysis of E. coli in stationary phase revealed upregulation of genes which are involved in survival during osmotic stress (ots, tre, osm), long-term survival (e.g., bolA, dps, cbpA, and glgS), periplasmic shock (rpoE and rseA), cold shock (csp genes), etc. Other genes include carbon storage regulator (csrA), trp repressor binding protein (wrbA) and universal stress protein (uspA) (Chang et al., 2002). Moreover, several antibiotics including lactocin B of lactic acid bacteria, alfatoxin of Aspergillus species are produced mainly in stationary phase (Matin, 1992).
Persister cell formation has also been attributed to genes differentially expressed in stationary phase. These cells are recalcitrant to antibiotic treatments and often are the major cause of drug resistance. Several polyamines including putrescine, spermidine, and cadaverine direct persister formation through upregulation of genes such as rpoS, rmf, yqjD (Tkachenko et al., 2017). This observation suggests that polyamine metabolism participates in the regulation of persister cells formation. To determine the genes upregulated at stationary phase microarray was done in Mycobacterium smegmatis grown under conditions of glycerol and glucose depletion. Different subset of genes were identified that were preferentially upregulated at stationary phase. The categories of genes included those involved in metabolism of sulfur, sigma factors including sigB, sigE, and sigH, fatty acid degradation, anaerobic respiration, etc. (Hampshire et al., 2004). Also, of key interest in this study is the presence of stationary phase operons involving many gene clusters that were significantly upregulated in stationary phase. On investigating further, the presence of other such operons were also found. The pdh operon of Streptococcus mutans is expressed only in the stationary phase. This operon was observed to be transcribed only by a subpopulation of bacteria in stationary phase and was vital for survival during long periods of sugar starvation. The pdh operon consists of four genes that are transcribed as an operon: pdhD, pdhA, pdhB, pdhC, which encode the components of PDH (pyruvate dehydrogenase) complex, i.e., pyruvate dehydrogenase (two subunits encoded by pdhA and pdhB), dihydrolipoyl transacetylase (pdhC), and dihydrolipoyl dehydrogenase (pdhD). The inactivation of the first gene: pdhD resulted in impaired survival in both batch cultures and biofilms (Busuioc et al., 2010). Similarly, phage shock protein operon (pspABCE) of E. coli was reported to be critical for survival under prolonged stationary phase at alkaline conditions. This operon was expressed strongly under extreme stressful conditions and remained significant for survival under nutrient-limited conditions (Weiner and Model, 1994). Categories of genes that are preferentially upregulated in stationary phase is shown in Figure 3. Studies have demonstrated that starved cells exhibit more protective resistance to different stresses as compared to resistance induced during growing stage by non-lethal exposure of stresses (Kolter et al., 1993).
Stationary Phase Promoters
The genes expressed in stationary phase are controlled by promoters, which result in induction of stationary phase. The promoters, which are turned on, are called SPPs. They are recognized by RNA polymerase holoenzyme containing σS and therefore called RpoS.
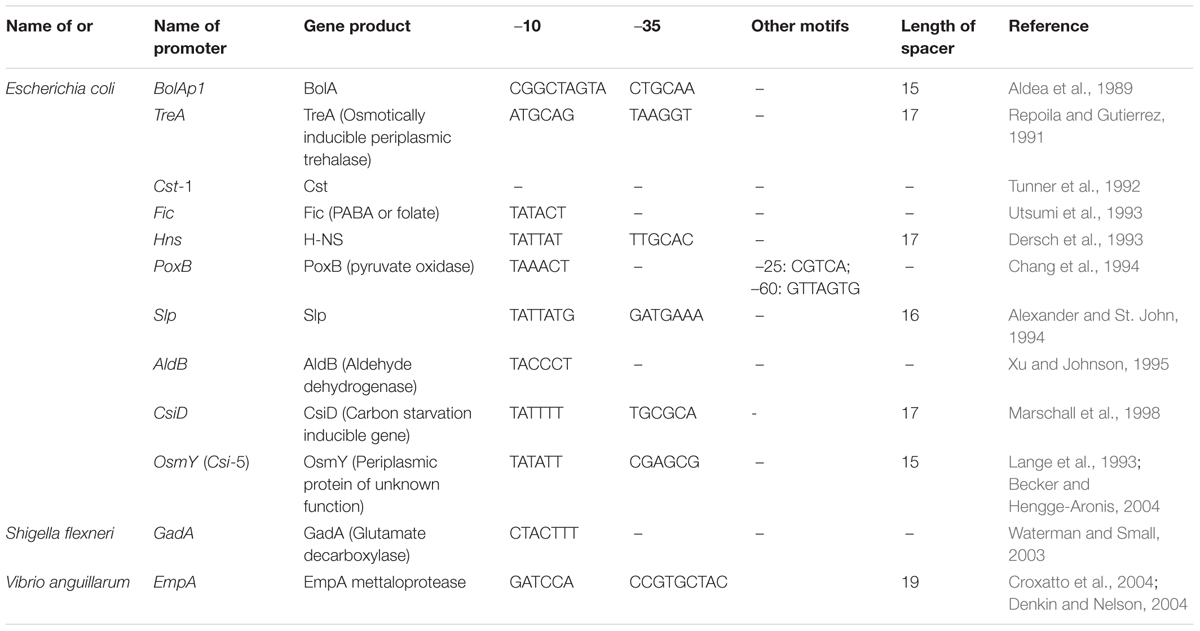
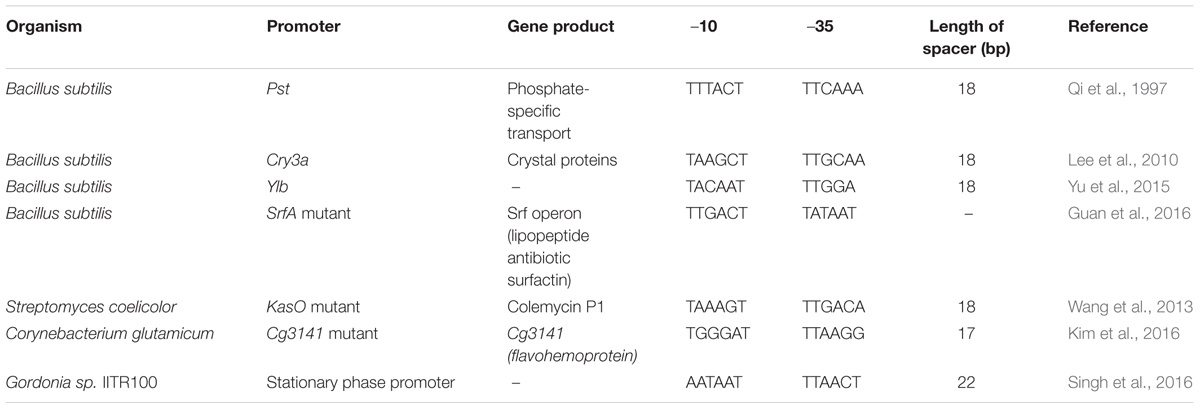
The vast importance of SPPs had been realized way back in 1980s with the study of mcbA promoter and bolA P1 promoter of E. coli (Connell et al., 1987; Aldea et al., 1989). The mcbA promoter causes increased level of transcription initiation for Microcin B17, a DNA replication inhibitor. Promoter mcbA-LacZ fusion showed the induction of transcription in nitrogen, phosphate, and carbon starvation conditions. Similarly, bolA-lacZ fusion demonstrated an increase in expression of approximately 10- to 20-fold during transition to stationary phase. Since then many SPPs have been isolated and characterized in both Gram-positive and Gram-negative bacteria (Tables 2, 3). Particularly regarding E. coli and B. subtilis, the stationary-phase-specific gene regulation has been intensively studied (Hengge, 2011).
On analysis of the different SPPs, our observation is that there is not much variation between this class of promoters and σ70 promoters. It is the sequence outside the –10 and –35 regions that distinguish between σ70- and σS-dependent promoters. Figures 4A,B shows the –10, –35 and spacer region of few SPPs from Gram-positive and Gram-negative bacteria and the consensus sequence at the –10 region is shown as a logo designed using WebLogo software available online (Crooks et al., 2004).

FIGURE 4. Sequence alignment of (A) Gram-positive and (B) Gram-negative stationary phase promoters. –10 and –35 are underlined and shown in red and green, respectively. The conserved bases are shown below.
Among the SPPs exist a special class of promoters known as the gearbox promoters which include mcbAp, bolAp1, ftsQp to name a few. This class of promoters has been studied in several Gram-negative bacteria including E. coli. Two different highly conserved consensus –10 and –35 sequence have been proposed by Aldea et al. (1993) for this class of promoters: CTGCAA or GTTAAGC at –35 position and CGGCAAGTA or CGTCC at –10 position. Gearbox promoter-induced gene expression seems to correlate inversely with growth rate and these promoters may or may not depend on σS.
Energy Reserves Consumed During Stationary Phase and Source of Nutrients for Protein Production
During unfavorable conditions of growth, reprogramming the cellular machinery for sustaining viability is a natural process of adaptation. Reserve polymers like glycogen and poly-β-hydroxybutyric acid that are accumulated by bacteria during growth are rapidly consumed during conditions of carbon starvation to ensure survival. In case of bacteria that do not accumulate these polymers, cellular RNA is rapidly degraded for energy generation (Matin, 1992). Among RNA, rRNA is preferentially degraded (Deutscher, 2003). Besides, 50% of ribosomes synthesized during exponential growth are degraded during entry to stationary phase (Piir et al., 2011). What is surprising is that, when in stationary phase, these ribosomes are fairly stable and so degradation occurs only in between the stages.
The yield of protein production from stationary phase systems is as high as 121% as compared to their log phase counterparts (Ou et al., 2004). This raises a very important question: What makes protein synthesis possible at stationary phase?
Balaban and coworkers devised a microfluidic device and followed the production of fluorescent proteins at stationary phase. They found that cells after entering stationary phase continue to produce proteins for several days (Gefen et al., 2014). It has been suggested that cells continue to produce proteins at stationary phase by reusing amino acids from degraded proteins. Moreover, the biosynthetic pathway of a few amino acids including serine, aspartate/asparagine, glutamine/glutamate, and alanine were shown to be active during stationary phase (Shaikh et al., 2010). In addition, it is shown that each condition resulting in starvation results in induction of specific set of proteins (Kolter et al., 1993).
Development of Gene Expression Systems Using Stationary Phase Promoters
A strong promoter is the key for developing efficient gene expression systems. For recombinant protein production, several bacterial hosts have been used as cell factories, with features such as easy purification, improved protein folding and secretion, high production of membrane proteins, etc. (Ferrer-Miralles and Villaverde, 2013). To develop more such expression systems in bacteria, it is necessary to ensure proper selection of a promoter that would drive the expression of genes at the right time and with maximum amount.
Promoters could be classified as constitutive or inducible, growth-stage limited, tissue specific, etc. Inducible promoters can further be classified into inducer-specific and auto-inducible promoters. Constitutive promoters are not useful for toxic proteins. Inducer-specific promoters involve the cost of inducer. Also, some chemical inducers such as Isopropyl-β-D-1-thiogalactopyranoside (IPTG) are expensive and toxic (Cao and Xian, 2011). Further, the addition of external inducers often requires growth monitoring which is vital for productivity and hence lead to difficulty in fermentation.
Auto-inducible promoters are ideal for large-scale protein production as they are induced at late log phase or stationary phase. Such promoters induce expression of the recombinant gene without any inducer and thus are economical. However, most of them have low activity (Yu et al., 2015). In B. subtilis, Fan and coworkers successfully identified a strong SPP Pylb by microarray approach (Yu et al., 2015). The β-galactosidase activities were observed to be up to 5000 miller units. The authors have proposed that such a promoter will be useful for protein production. A SPP-based auto-inducible gene expression system has been constructed using cry3Aa promoter. The Pcry drives the expression of crystal proteins in B. thuringiensis. The promoter cry3Aa was tested in B. subtilis and the wild type have the LacZ levels up to 1000 miller units and on mutagenesis resulted in levels up to 5200 miller units (Lee et al., 2010). Similarly, in another Gram-positive bacteria, Gordonia sp. IITR100, a SPP was identified and the β-galactosidase activities were up to 600 miller units (Singh et al., 2016). However, the β-galactosidase activities vary with respect to strain, copy number of plasmid, growth medium, temperature, etc., so it is difficult to assess the strength of promoter based on Miller units alone. In future, a study of such promoters based on the number of transcripts would be useful to compare the strength.
In Corynebacterium glutamicum, promoter of cg3141 gene coding for flavohemoprotein was found to show higher inducibility in the stationary phase. Then, a synthetic promoter library was prepared to change the spacer and flanking regions in the promoter, to obtain a range of promoter strengths (Kim et al., 2016). At the end, one of the synthetic promoters that showed up to 20-fold higher strength compared to the original cg3141 promoter was obtained and demonstrated for fed-batch cultivation of glutathione S-transferase in a 5L reactor. Table 4 depicts the list of SPP-based expression vectors constructed till date. Studies like these indicate that the potential of SPPs is phenomenal. In Streptomyces, a high-level recombinant protein expression system has been patented (US Patent No. 7,316,914).
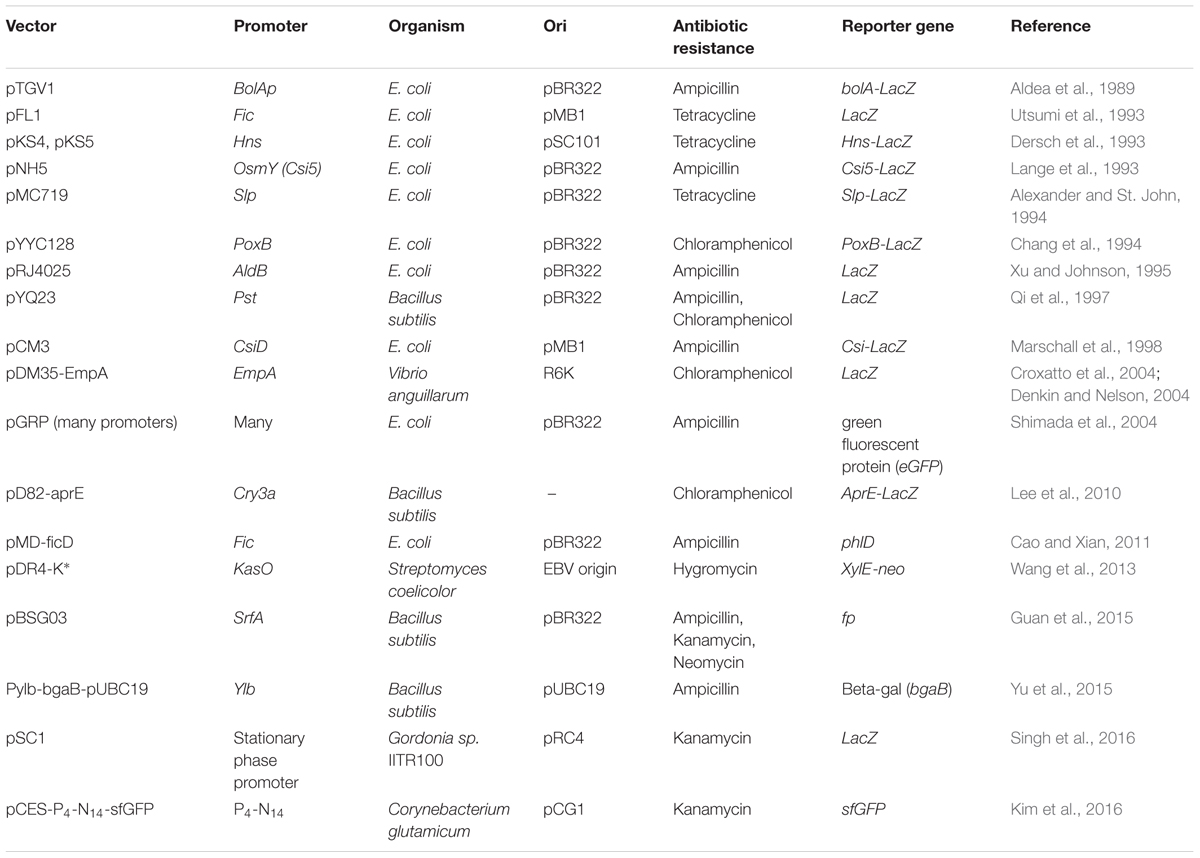
TABLE 4. Stationary phase promoter–based gene expression systems reported from Gram-negative and Gram-positive bacteria.
Applications
SPPs have immense potential for use in many industries (Figure 5).
Recombinant production of toxins whose overproduction is detrimental to the growth of cells needs controlled conditions for expression. In such cases, the use of SPP is advantageous as the overproduction will not affect the growth of the host cells. Many bacteria have been used to demonstrate the utility of cell-density-dependent expression systems for heterologous protein production. Metabolic engineering of bacteria for enhanced production of industrially important chemicals has been carried out since a long time. The fic promoter of E. coli was used to express phlD gene at a higher titer in stationary phase, without the addition of any inducer, for the production of phloroglucinol, which has utility in pharmaceutical industry and plant tissue culture. After 20 h of cultivation in a flask with shaking, 9% of glucose supplied had converted to phloroglucinol with a productivity of 0.014g/l h (Cao and Xian, 2011). B. subtilis has been engineered for overproduction of aminopeptidase using a mutated PsrfA system and has resulted in 87.89 U/ml of enzyme activity (Guan et al., 2015). Using B. subtilis, a cry-promoter-based system was developed wherein cellulose and alkaline protease were produced with a higher yield as compared to the wild-type cry3A promoter (Lee et al., 2010).
It is a well-known fact that the non-growing phase of lactic acid bacteria accounts for a major proportion of flavor production in lactic acid bacteria (van de Bunt et al., 2014). Therefore, engineering bacterial cells in such a way that they are expressed at high levels, during the ripening process, by using SPPs would enhance their applicability in food industry.
In the bioremediation industry, microorganisms have routinely been employed for removing pollutants. Due to low nutrient availability in polluted sites, genetic engineering of cells resulting in higher enzymatic activities at lower growth rates have been shown to be highly efficient for bioremediation process. On studying the phenol degradation capability of two non-growing recombinant E. coli strains, it was found that the groEL-promoter-driven gene expression system caused 75% phenol degradation while the tac-promoter-driven expression could cause only 15% degradation of phenol (Matin, 1992). As suggested by Tunner et al. (1992), it is possible to use starvation-induced promoters for chemical waste biodegradation wherein enzymes can be induced naturally by bacteria due to the occurrence of nutrient-limited conditions in the environment. This could save the cost of induction thereby increasing the efficiency of the process.
In a very interesting experiment, Rhodospirillum rubrum cells grown photoheterotrophically, evolved hydrogen for about 70 h after growth ceased (Melnicki et al., 2008). Similarly, a purple non-sulfur photosynthetic bacterium, Rhodopseudomonas palustris under nitrogen starvation conditions, produced hydrogen gas for over 4000 h thus paving way for creation of ‘artificial leaves’ (Gosse et al., 2010).
Conclusion and Future Prospects
Stationary phase survival is a means of bacterial adaptation by which bacteria survive under conditions of stress or starvation. The ugly aspect of this is that such a mechanism results in the persistence of pathogenic bacteria which can cause relapsing of infections. However, the good side is represented by the various biotechnological applications that have come up recently based on the promoters of the genes which are upregulated at stationary phase. In the present review, we have discussed not only the changes at the cellular and molecular levels at stationary phase, but also the various promoters characterized, their regulation and the gene expression systems developed. There are still many unknowns. For example, very little is known about the proteins which are involved in chromosome organization and their interaction with DNA at stationary phase. Such proteins could be important players in regulating gene expression at stationary phase. Further very few SPPs have been experimentally characterized till date. Such promoters should be highly useful for protein production as the growth and protein production phase can be uncoupled. This will pave way toward constructing improved gene expression systems for recombinant protein production.
Author Contributions
JJ and PS wrote and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Department of Biotechnology, Government of India for the financial support.
References
Aldea, M., Garrido, T., Hernandez-Chico, C., Vicente, M., and Kushner, S. R. (1989). Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 8, 3923–3931.
Aldea, M., Garrido, T., and Tormo, A. (1993). Gearbox gene expression and growth rate. World J. Microbiol. Biotechnol. 9, 414–420. doi: 10.1007/BF00328029
Alexander, D. M., and St. John, A. C. (1994). Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol. Microbiol. 11, 1059–1071. doi: 10.1111/j.1365-2958.1994.tb00383.x
Barker, M. M., Gaal, T., Josaitis, C. A., and Gourse, R. L. (2001). Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305, 673–688. doi: 10.1006/jmbi.2000.4327
Becker, G., and Hengge-Aronis, R. (2004). What makes an Escherichia coli promoter σS dependent? Role of the -13/-14 nucleotide promoter positions and region 2.5 of σS. Mol. Microbiol. 39, 1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x
Bohannon, D. E., Connell, N., Keener, J., Tormo, A., Espinosa-Urgel, M., Zambrano, M. M., et al. (1991). Stationary-phase-inducible ”gearbox” promoters: differential effects of katF mutations and role of sigma 70. J. Bacteriol. 173, 4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991
Bruno, J. C. Jr., and Freitag, N. E. (2011). Listeria monocytogenes adapts to long-term stationary phase survival without compromising bacterial virulence. FEMS Microbiol. Lett. 323, 171–179. doi: 10.1111/j.1574-6968.2011.02373.x
Busuioc, M., Buttaro, B. A., and Piggot, P. J. (2010). The pdh operon is expressed in a subpopulation of stationary-phase bacteria and is important for survival of sugar-starved Streptococcus mutans. J. Bacteriol. 192, 4395–4402. doi: 10.1128/JB.00574-10
Cao, Y., and Xian, M. (2011). Production of phloroglucinol by Escherichia coli using a stationary-phase promoter. Biotechnol. Lett. 33, 1853–1858. doi: 10.1007/s10529-011-0638-0
Cavanagh, A. T., Chandrangsu, P., and Wassarman, K. M. (2010). 6S RNA regulation of relA alters ppGpp levels in early stationary phase. Microbiology 156(Pt 12), 3791–3800. doi: 10.1099/mic.0.043992-43990
Chang, D. E., Smalley, D. J., and Conway, T. (2002). Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45, 289–306. doi: 10.1046/j.1365-2958.2002.03001.x
Chang, Y. Y., Wang, A. Y., and Cronan, J. E. Jr. (1994). Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol. Microbiol. 11, 1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x
Chen, H., and Chen, C. Y. (2014). Starvation induces phenotypic diversification and convergent evolution in Vibrio vulnificus. PLOS ONE 9:e88658. doi: 10.1371/journal.pone.0088658
Connell, N., Han, Z., Moreno, F., and Kolter, R. (1987). An E. coli promoter induced by the cessation of growth. Mol. Microbiol. 1, 195–201. doi: 10.1111/j.1365-2958.1987.tb00512.x
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Croxatto, A., Pride, J., Hardman, A., Williams, P., Camara, M., and Milton, D. L. (2004). A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol. Microbiol. 52, 1677–1689. doi: 10.1111/j.1365-2958.2004.04083.x
Denkin, S. M., and Nelson, D. R. (2004). Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 70, 4193–4204. doi: 10.1128/AEM.70.7.4193-4204.2004
Dersch, P., Schmidt, K., and Bremer, E. (1993). Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8, 875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x
Deutscher, M. P. (2003). Degradation of stable RNA in bacteria. J. Biol. Chem. 278, 45041–45044. doi: 10.1074/jbc.R300031200
Dorman, C. J. (2011). Regulation of transcription by DNA supercoiling in Mycoplasma genitalium: global control in the smallest known self-replicating genome. Mol. Microbiol. 81, 302–304. doi: 10.1111/j.1365-2958.2011.07718.x
Espinosa-Urgel, M., Chamizo, C., and Tormo, A. (1996). A consensus structure for sigma S-dependent promoters. Mol. Microbiol. 21, 657–659. doi: 10.1111/j.1365-2958.1996.tb02573.x
Ferrer-Miralles, N., and Villaverde, A. (2013). Bacterial cell factories for recombinant protein production; expanding the catalogue. Microb. Cell Fact. 12:113. doi: 10.1186/1475-2859-12-113
Finkel, S. E., Zinser, E., Gupta, S., and Kolter, R. (1997). “Life and Death in stationary phase,” in Molecular Microbiology. NATO ASI Series (Series H: Cell Biology), Vol. 103, eds S. J. W. Busby, C. M. Thomas, and N. L. Brown (Berlin: Springer).
Gaida, S. M., Al-Hinai, M. A., Indurthi, D. C., Nicolaou, S. A., and Papoutsakis, E. T. (2013). Synthetic tolerance: three noncoding small RNAs, DsrA, ArcZ and RprA, acting supra-additively against acid stress. Nucleic Acids Res. 41, 8726–8737. doi: 10.1093/nar/gkt651
Gefen, O., Fridman, O., Ronin, I., and Balaban, N. Q. (2014). Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc. Natl. Acad. Sci. U.S.A. 111, 556–561. doi: 10.1073/pnas.1314114111
Gosse, J. L., Engel, B. J., Hui, J. C. H., Harwood, C. S., and Flickinger, M. C. (2010). Progress toward a biomimetic leaf: 4,000 h of hydrogen production by coating-stabilized nongrowing photosynthetic Rhodopseudomonas palustris. Biotechnol. Prog. 26, 907–918. doi: 10.1002/btpr.406
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Guan, C., Cui, W., Cheng, J., Zhou, L., Guo, J., Hu, X., et al. (2015). Construction and development of an auto-regulatory gene expression system in Bacillus subtilis. Microb. Cell Fact. 14, 150. doi: 10.1186/s12934-015-0341-2
Guan, C., Cui, W., Cheng, J., Zhou, L., Liu, Z., and Zhou, Z. (2016). Development of an efficient autoinducible expression system by promoter engineering in Bacillus subtilis. Microb. Cell Fact. 15, 66. doi: 10.1186/s12934-016-0464-0
Hampshire, T., Soneji, S., Bacon, J., James, B. W., Hinds, J., Laing, K., et al. (2004). Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis 84, 228–238. doi: 10.1016/j.tube.2003.12.010
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aa4268. doi: 10.1126/science.aaf4268
Hengge, R. (2011). Stationary-phase gene regulation in Escherichia coli. EcoSal Plus 4. doi: 10.1128/ecosalplus.5.6.3
Hengge-Aronis, R. (1993). Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72, 165–168. doi: 10.1016/0092-8674(93)90655-A
Hengge-Aronis, R. (2002). Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395. doi: 10.1128/MMBR.66.3.373-395.2002
Ishihama, A. (1997). Adaptation of gene expression in stationary phase bacteria. Curr. Opin. Genet. Dev. 7, 582–588. doi: 10.1016/S0959-437X(97)80003-2
Jaishankar, J., Singh, P., and Srivastava, P. (2017). Draft genome sequence of a biodesulfurizing bacterium, Gordonia sp. strain IITR100. Genome Announc. 5:e00230-17. doi: 10.1128/genomeA.00230-17
Jin, Y., Watt, R. M., Danchin, A., and Huang, J. D. (2009). Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genomics 10:165. doi: 10.1186/1471-2164-10-165
Kim, E. S., Song, J. Y., Kim, D. W., Chater, K. F., and Lee, K. J. (2008). A possible extended family of regulators of sigma factor activity in Streptomyces coelicolor. J. Bacteriol. 190, 7559–7566. doi: 10.1128/JB.00470-08
Kim, M. J., Yim, S. S., Choi, J. W., and Jeong, K. J. (2016). Development of a potential stationary-phase specific gene expression system by engineering of SigB-dependent cg3141 promoter in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 100, 4473–4483. doi: 10.1007/s00253-016-7297-y
Kolb, A., Kotlarz, D., Kusano, S., and Ishihama, A. (1995). Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23, 819–826. doi: 10.1093/nar/23.5.819
Kolter, R., Siegele, D. A., and Tormo, A. (1993). The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47, 855–874. doi: 10.1146/annurev.mi.47.100193.004231
Lacour, S., and Landini, P. (2004). SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004
Lange, R., Barth, M., and Hengge-Aronis, R. (1993). Complex transcriptional control of the sigma s-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J. Bacteriol. 175, 7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993
Lange, R., and Hengge-Aronis, R. (1994). The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13, 733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x
Lee, S. J., Pan, J. G., Park, S. H., and Choi, S. K. (2010). Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J. Biotechnol. 149, 16–20. doi: 10.1016/j.jbiotec.2010.06.021
Lemonnier, M., Levin, B. R., Romeo, T., Garner, K., Baquero, M. R., Mercante, J., et al. (2008). The evolution of contact-dependent inhibition in non-growing populations of Escherichia coli. Proc. Biol. Sci. 275, 3–10. doi: 10.1098/rspb.2007.1234
Luidalepp, H., Berger, S., Joss, O., Tenson, T., and Polacek, N. (2016). Ribosome shut-down by 16S rRNA fragmentation in stationary-phase Escherichia coli. J. Mol. Biol. 428(10 Pt B), 2237–2247. doi: 10.1016/j.jmb.2016.01.033
Marschall, C., Labrousse, V., Kreimer, M., Weichart, D., Kolb, A., and Hengge-Aronis, R. (1998). Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on sigma s and requires activation by cAMP-CRP. J. Mol. Biol. 276, 339–353. doi: 10.1006/jmbi.1997.1533
Matin, A. (1992). Physiology, molecular biology and applications of the bacterial starvation response. Soc. Appl. Bacteriol. Symp. Ser. 21, 49S–57S. doi: 10.1111/j.1365-2672.1992.tb03624.x
McKay, S. L., and Portnoy, D. A. (2015). Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 59, 6992–6999. doi: 10.1128/AAC.01532-15
Melnicki, M., Bianchi, L., Dephilippis, R., and Melis, A. (2008). Hydrogen production during stationary phase in purple photosynthetic bacteria. Int. J. Hydrogen Energy 33, 6525–6534. doi: 10.1016/j.ijhydene.2008.08.041
Mengin-Lecreulx, D., and van Heijenoort, J. (1985). Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J. Bacteriol. 163, 208–212.
Navarro Llorens, J. M., Tormo, A., and Martinez-Garcia, E. (2010). Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34, 476–495. doi: 10.1111/j.1574-6976.2010.00213.x
Nystrom, T. (2004). Stationary-phase physiology. Annu. Rev. Microbiol. 58, 161–181. doi: 10.1146/annurev.micro.58.030603.123818
Ou, J., Wang, L., Ding, X., Du, J., Zhang, Y., Chen, H., et al. (2004). Stationary phase protein overproduction is a fundamental capability of Escherichia coli. Biochem. Biophys. Res. Commun. 314, 174–180. doi: 10.1016/j.bbrc.2003.12.077
Pátek, M., and Nešvera, J. (2013). “Promoters and plasmid vectors of Corynebacterium glutamicum,” in Corynebacterium glutamicum. Microbiology Monographs, Vol. 23, eds H. Yukawa and M. Inui (Berlin: Springer), 51–88.
Piir, K., Paier, A., Liiv, A., Tenson, T., and Maivali, U. (2011). Ribosome degradation in growing bacteria. EMBO Rep. 12, 458–462. doi: 10.1038/embor.2011.47
Pletnev, P., Osterman, I., Sergiev, P., Bogdanov, A., and Dontsova, O. (2015). Survival guide: Escherichia coli in the stationary phase. Acta Nat. 7, 22–33.
Potvin, E., Sanschagrin, F., and Levesque, R. C. (2008). Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32, 38–55. doi: 10.1111/j.1574-6976.2007.00092.x
Qi, Y., Kobayashi, Y., and Hulett, F. M. (1997). The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 179, 2534–2539. doi: 10.1128/jb.179.8.2534-2539.1997
Ratnayake-Lecamwasam, M., Serror, P., Wong, K. W., and Sonenshein, A. L. (2001). Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15, 1093–1103. doi: 10.1101/gad.874201
Rava, P. S., Somma, L., and Steinman, H. M. (1999). Identification of a regulator that controls stationary-phase expression of catalase-peroxidase in Caulobacter crescentus. J. Bacteriol. 181, 6152–6159.
Repoila, F., and Gutierrez, C. (1991). Osmotic induction of the periplasmic trehalase in Escherichia coli K12: characterization of the treA gene promoter. Mol. Microbiol. 5, 747–755. doi: 10.1111/j.1365-2958.1991.tb00745.x
Shaikh, A. S., Tang, Y. J., Mukhopadhyay, A., Martin, H. G., Gin, J., Benke, P. I., et al. (2010). Study of stationary phase metabolism via isotopomer analysis of amino acids from an isolated protein. Biotechnol. Prog. 26, 52–56. doi: 10.1002/btpr.325
Shimada, T., Makinoshima, H., Ogawa, Y., Miki, T., Maeda, M., and Ishihama, A. (2004). Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 186, 7112–7122. doi: 10.1128/JB.186.21.7112-7122.2004
Singh, P., Chachan, S., Singhi, D., and Srivastava, P. (2016). Isolation and molecular characterization of a stationary phase promoter useful for gene expression in Gordonia. Gene 591, 153–160. doi: 10.1016/j.gene.2016.07.018
Su, X., Sun, F., Wang, Y., Hashmi, M. Z., Guo, L., Ding, L., et al. (2015). Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci. Rep. 5:18590. doi: 10.1038/srep18590
Tanaka, K., Handel, K., Loewen, P. C., and Takahashi, H. (1997). Identification and analysis of the rpoS-dependent promoter of katE, encoding catalase HPII in Escherichia coli. Biochim. Biophys. Acta 1352, 161–166. doi: 10.1016/S0167-4781(97)00044-4
Tanaka, K., Kusano, S., Fujita, N., Ishihama, A., and Takahashi, H. (1995). Promoter determinants for Escherichia coli RNA polymerase holoenzyme containing sigma 38 (the rpoS gene product). Nucleic Acids Res. 23, 827–834. doi: 10.1093/nar/23.5.827
Testerman, T. L., Vazquez-Torres, A., Xu, Y., Jones-Carson, J., Libby, S. J., and Fang, F. C. (2002). The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol. Microbiol. 43, 771–782. doi: 10.1046/j.1365-2958.2002.02787.x
Tkachenko, A. G., Kashevarova, N. M., Tyuleneva, E. A., and Shumkov, M. S. (2017). Stationary-phase genes upregulated by polyamines are responsible for the formation of Escherichia coli persister cells tolerant to netilmicin. FEMS Microbiol. Lett. 364:fnx084. doi: 10.1093/femsle/fnx084
Tripathi, L., Zhang, Y., and Lin, Z. (2014). Bacterial sigma factors as targets for engineered or synthetic transcriptional control. Front. Bioeng. Biotechnol. 2:33. doi: 10.3389/fbioe.2014.00033
Tunner, J. R., Robertson, C. R., Schippa, S., and Matin, A. (1992). Use of glucose starvation to limit growth and induce protein production in Escherichia coli. Biotechnol. Bioeng. 40, 271–279. doi: 10.1002/bit.260400211
Utsumi, R., Kusafuka, S., Nakayama, T., Tanaka, K., Takayanagi, Y., Takahashi, H., et al. (1993). Stationary phase-specific expression of the fic gene in Escherichia coli K-12 is controlled by the rpoS gene product (sigma 38). FEMS Microbiol. Lett. 113, 273–278. doi: 10.1016/0378-1097(93)90217-P
van de Bunt, B., Bron, P. A., Sijtsma, L., de Vos, W. M., and Hugenholtz, J. (2014). Use of non-growing Lactococcus lactis cell suspensions for production of volatile metabolites with direct relevance for flavour formation during dairy fermentations. Microb. Cell Fact. 13:176. doi: 10.1186/s12934-014-0176-2
Wada, A. (1998). Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3, 203–208. doi: 10.1046/j.1365-2443.1998.00187.x
Wang, W., Li, X., Wang, J., Xiang, S., Feng, X., and Yang, K. (2013). An engineered strong promoter for streptomycetes. Appl. Environ. Microbiol. 79, 4484–4492. doi: 10.1128/AEM.00985-13
Waterman, S. R., and Small, P. L. (2003). Identification of the promoter regions and sigma(s)-dependent regulation of the gadA and gadBC genes associated with glutamate-dependent acid resistance in Shigella flexneri. FEMS Microbiol. Lett. 225, 155–160. doi: 10.1016/S0378-1097(03)00508-1
Weber, H., Polen, T., Heuveling, J., Wendisch, V. F., and Hengge, R. (2005). Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005
Weiner, L., and Model, P. (1994). Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. U.S.A. 91, 2191–2195. doi: 10.1073/pnas.91.6.2191
Wise, A., Brems, R., Ramakrishnan, V., and Villarejo, M. (1996). Sequences in the -35 region of Escherichia coli rpoS-dependent genes promote transcription by E sigma S. J. Bacteriol. 178, 2785–2793. doi: 10.1128/jb.178.10.2785-2793.1996
Xu, J., and Johnson, R. C. (1995). aldB, an RpoS-dependent gene in Escherichia coli encoding an aldehyde dehydrogenase that is repressed by Fis and activated by Crp. J. Bacteriol. 177, 3166–3175. doi: 10.1128/jb.177.11.3166-3175.1995
Yu, X., Xu, J., Liu, X., Chu, X., Wang, P., Tian, J., et al. (2015). Identification of a highly efficient stationary phase promoter in Bacillus subtilis. Sci. Rep. 5:18405. doi: 10.1038/srep18405
Keywords: stationary phase promoters, stationary phase gene expression, plasmid vectors, sigma factor, stationary phase
Citation: Jaishankar J and Srivastava P (2017) Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 8:2000. doi: 10.3389/fmicb.2017.02000
Received: 31 July 2017; Accepted: 28 September 2017;
Published: 16 October 2017.
Edited by:
Tatiana Venkova, University of Texas Medical Branch, United StatesReviewed by:
Grzegorz Wegrzyn, University of Gdansk, PolandSusana Brom, National Autonomous University of Mexico, Mexico
Jan Nesvera, Institute of Microbiology of the Czech Academy of Sciences, Czechia
Copyright © 2017 Jaishankar and Srivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Preeti Srivastava, preeti@dbeb.iitd.ac.in; preetisrivastava@hotmail.com
 Jananee Jaishankar
Jananee Jaishankar Preeti Srivastava
Preeti Srivastava