- 1School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Department of Medical Microbiology and Immunology, University of Wisconsin-Madison, Madison, WI, United States
- 3Department of Plant Pathology, University of Wisconsin-Madison, Madison, WI, United States
- 4Department of Bacteriology, University of Wisconsin-Madison, Madison, WI, United States
Aspergillus flavus is a saprophytic soil fungus that poses a serious threat worldwide as it contaminates many food and feed crops with the carcinogenic mycotoxin called aflatoxin. This pathogen persists as sclerotia in the soil which enables fungal survival in harsh environmental conditions. Sclerotia formation by A. flavus depends on successful cell communication and hyphal fusion events. Loss of LaeA, a conserved developmental regulator in fungi, abolishes sclerotia formation in this species whereas overexpression (OE) of laeA results in enhanced sclerotia production. Here we demonstrate that sclerotia loss and inability to form heterokaryons in A. flavusΔlaeA is mediated by homologs of the Neurospora crassa ham (hyphal anastomosis) genes termed hamE-I in A. flavus. LaeA positively regulates ham gene expression and deletion of hamF, G, H, or I phenocopies ΔlaeA as demonstrated by heterokaryon and sclerotia loss and reduced aflatoxin synthesis and virulence of these mutants. Deletion of hamE showed a less severe phenotype. hamE-I homologs are positively regulated by the clock controlled transcription factor ADV-1 in N. crassa. Similarly, the ADV-1 homolog NosA regulates hamE-I expression in A. flavus, is required for sclerotial development and is itself positively regulated by LaeA. We speculate that a putative LaeA>NosA>fusion cascade underlies the previously described circadian clock regulation of sclerotia production in A. flavus.
Introduction
Most plant pathogenic fungi have developed both asexual and sexual modes of reproduction with specific roles in life and disease cycles. A critical developmental stage of several plant pathogenic fungi is formation of the survival structure, the sclerotium. Sclerotia are large, multicellular structures visible by the eye that are formed by branching and fusion of interwoven hyphae (Erental et al., 2008). There are three overlapping stages in the development of sclerotia: initiation, when hyphae begin to fusion together to form small, discrete initials; development, the sclerotial size enlarges with white coloration; and maturation, the surface becomes pigmented and harder (Willetts and Bullock, 1992). Under appropriate environmental conditions and presence of opposite mating types, sclerotia of most fungal species will develop sexual spores (Terhem and van Kan, 2014; Horn et al., 2016) which can also serve as inocula for some sclerotial producing fungi (Jurick and Rollins, 2007). Sclerotia can also germinate directly to form infective hyphae. The longevity of sclerotia in agricultural soils is a concern in crop protection and contributes to outbreaks of disease by sclerotial fungi (Coley-Smith and Cooke, 1971).
Aspergillus flavus is a well known sclerotia forming fungus, infamous for production of the carcinogenic mycotoxin, aflatoxin (reviewed in Amare and Keller, 2014). Field studies of the related species, A. parasiticus, demonstrated sclerotia inoculum was more effective than conidial inoculum in invasion of peanut pods (Horn et al., 1994). Both conidia and sclerotia contain aflatoxin (Wicklow and Shotwell, 1983) and studies have positively correlated sclerotial development with aflatoxin production (Brown et al., 2009) although one process is not necessarily dependent on the other (Chang et al., 2016, 2017). Despite the importance of sclerotia in A. flavus survival and pathogenesis and its correlation with toxin synthesis, little is known about the genetic program leading to sclerotia formation.
One clear example of genetic linkage of sclerotia and aflatoxin formation is demonstrated by the transcriptional regulator LaeA, a member of the conserved transcriptional regulatory Velvet Complex (Bayram et al., 2008). Deletion and overexpression of laeA results in loss and over production of both aflatoxin and sclerotia, respectively (Kale et al., 2008; Amaike and Keller, 2009). LaeA (Lae1) is a conserved protein in ascomycete fungi that coordinately connects secondary metabolism with sexual development (Wu et al., 2012). Similar to laeA loss in A. flavus, deletion of the Botrytis cinerea ortholog, BcLae1, results in loss of sclerotial production in this species (Schumacher et al., 2015). Sclerotia are the progenitor tissues of sexual stage development in B. cinerea (Terhem and van Kan, 2014). However, how LaeA/Lae1 regulates sclerotial formation is unknown. Other studies have implicated hyphal fusion - also known as anastomosis – as important in sclerotial formation with microscopic evidence of anastomosis events in initial developmental stages (Kohn et al., 1990; Erental et al., 2008). Loss of BcNoxD, BcNoxB, or BcNoxA, members of the NADPH oxidase complex, impair both conidial anastomosis tube (CAT) fusion and sclerotial formation in B. cinerea (Segmüller et al., 2008; Siegmund et al., 2015) and hyphal fusion is required for heterokaryotic sclerotia formation in A. oryzae (Wada et al., 2014), a non-aflatoxigenic clade of A. flavus (Varga et al., 2011).
Hyphal fusion is best characterized in the non-sclerotial model fungus Neurospora crassa (Fu et al., 2011; Herzog et al., 2015; Fleißner and Herzog, 2016; Daskalov et al., 2017), with many of the genes required for fusion events called ham (hyphal anastomosis) genes (Xiang et al., 2002). In N. crassa, MAP kinases, the striatin-interacting protein phosphatase and kinase (STRIPAK) complex and the NADPH oxidase complex, together with fungal specific proteins are wired into an intricate signaling network to mediate hyphal fusion events (Herzog et al., 2015). The MAK-2 protein and the fungal specific protein SO/HAM-1 participate in cellular crosstalk known as “ping-pong signaling” where both proteins are recruited to the plasma membrane of the growing tips in an oscillatory manner, with one phase lasting between 6 and 12 min (Fleissner et al., 2009; Serrano et al., 2017). HAM-5 functions as a scaffold-like protein proposed to link the activation of the MAK-2 cascade to upstream factors and proteins involved in hyphal fusion (Jonkers et al., 2014). HAM-2 (Xiang et al., 2002), HAM-3 and HAM-4 (Simonin et al., 2010) are members of the STRIPAK complex that governs multiple aspects of fungal development including fusion and sexual development (Dettmann et al., 2013). HAM-6, a homolog of PRO41 in Sordaria macrospora (Nowrousian et al., 2007), BcNoxD in B. cinerea (Siegmund et al., 2015) and PaNoxD in Podospora anserina (Lacaze et al., 2015), is an endoplasmic reticulum membrane protein and member of the NADPH oxidase complex essential for hyphal fusion in filamentous fungi. HAM-7 acts as a membrane receptor for the MAK-1 pathway during cell to cell signaling and hyphal fusion in N. crassa (Maddi et al., 2012). There is limited knowledge about HAM-8 and HAM-9 although phosphorylation studies showed that HAM-8 together with HAM-6 and HAM-7 regulate the MAK-1 pathway, and HAM-9 regulates crosstalk between MAK-1 and MAK-2 pathway during vegetative growth in N. crassa (Fu et al., 2014).
We present in this communication our finding that LaeA regulation of sclerotial formation in A. flavus is mediated by through transcriptional regulation of homologs of N. crassa fusion genes. LaeA positively regulates expression of hamE, F, G, H, and I (homologs of ham-5, 6, 7, 8, and 9 in N. crassa, respectively) with deletions of the latter four resulting in inability of A. flavus to form heterokaryons or sclerotia, the same phenotype as ΔlaeA. Moreover, we find that LaeA regulation of the ham cascade is likely signaled through the transcription factor NosA, a homolog of the circadian regulated ADV-1 also required for cell fusion in N. crassa (Fu et al., 2011). NosA is required for hamE-I expression, cell fusion and sclerotial formation and is itself positively regulated by LaeA. Furthermore, ΔhamF-I and ΔnosA result in significantly decreased aflatoxin synthesis and decreased virulence on corn seed.
Materials and Methods
Strains and Culture Conditions
Strains used in this research are listed in Table 1 and stored as glycerol stocks at -80°C. All strains were grown on glucose minimal medium (GMM) (Shimizu and Keller, 2001) for spore production at 29°C. In some cases, 0.56 g/L uracil and 1.26 g/L uridine, 1 g/L arginine or all three supplements were added and denoted as “+UU,” “+A,” or “+AUU,” respectively. YEP (6% peptone, 2% yeast extract, pH 5.8) medium, YES (6% sucrose, 2% yeast extract, pH 5.8) medium and GMM were used for Northern blot analysis. GMM + 0.1 M sorbitol was used for sclerotial formation. Heterokaryon analysis was assessed on GMM+0.25% Triton X-100 medium (Tsukasaki et al., 2014). For genomic DNA extraction, strains were grown on liquid GMM plus 0.5% yeast extract with appropriate supplements. Sorbitol minimal medium (GMM + 1.2 M sorbitol) (Lim et al., 2012) with appropriate supplements were used for transformant selection.
Strain Construction
All primers used for strain construction are listed in Supplementary Table S1. All PCR generated flanks ranged in size from 1 to 1.3 kb. All DNA transformation constructs were made by double joint PCR (Lim et al., 2012) or single joint PCR and transformed individually into the appropriate parental strain. Transformation of fungal strains was carried out according to the protocol of (Szewczyk et al., 2007) and (Yang et al., 2016) with the following modifications: 108 spores were inoculated into 100 ml of rich growth media (2 g glucose, 0.5 g yeast extract, 100 μL trace element, 0.56 g/L uracil and 1.26 g/L uridine, 1g/L arginine) for 11 h at 37°C and 150 rpm, the mycelia were then collected through a sterile miracloth (Calbiochem), transferred to a 250 ml flask, and resuspended in 20 mL protoplast solution composed of 20 mM NaH2PO4 pH5.8, 20 mM CaCl2, 200 μL β-glucuronidase (85000 U/mL, Sigma), 200 mg lysing enzymes from Trichoderma harzianum (Sigma), 50 mg Driselase from Basidiomycetes sp. (Sigma) in 1.2 M NaCl. Protoplasting was performed at 100 rpm, 30°C for 5–6 h. After transformation, the protoplasts were plated on sorbitol minimal medium (above) plus appropriate supplements. At least 10 independent isolates were screened by Southern analysis and the 5′ and 3′ fragments used in the double joint or single joint PCR reaction to amplify specific gene fragments were used as probes labeled with dCTP αP32. Details for each primer and method to create each mutant are described below:
Parental Strain TXZ21.3
TXZ21.3 was created by deleting A. fumigatus pyrG which had been placed in the argB locus in strain TJES20.1 (Δku70,ΔargB::A.fumigatus pyrG, pyrG-). Primers 1 and 2 were used to amplify the argB 5′ flank, primers 3 and 4 were used to amplify the argB 3′ flank. Single joint PCR were used to amplify the fusion construct by primers 1 and 4, and the construct was transferred into TJES20.1 to get TXZ21.3. The transformants were selected on SMM+UUA with 1.5 mg/ml 5- Fluoroorotic Acid (FOA, Thermo Fisher Scientific). One of resulting double auxotrophic mutants, Δku70 ΔargB pyrG-, called TXZ21.3, was used as the parental control for the following deletion strains. A schematic of how TXZ21.3 was derived is shown in Supplementary Figure S1. The southern of the TXZ21.3 can be found in Supplementary Figure S2.
Deletion of ham E-I, nosA, and laeA
Genes were deleted by replacing the target gene with A. flavus argB in the parental strain TXZ21.3 (Δku70,ΔargB, pyrG-) to create pyrG- single auxotrophs or TJES19.1 (Δku70, pyrG-) or TJES20.1 (Δku70,ΔargB::A. fumigatus pyrG, pyrG-) to create prototrophs. Primers 7 and 8 were used to amplify the argB gene from A. flavus wild type NRRL3357. To delete hamE (AFLA_095770) in TXZ21.3, primers 17 and 18 were used to amplify the hamE 5′ flank, primers 19 and 20 were used to amplify the hamE 3′ flank, then primers 15 and 16 were used to amplify the fusion construct. To delete hamF (AFLA_033600), primers 23 and 24 were used to amplify the hamF 5′ flank, primers 25 and 26 were used to amplify the hamF 3′ flank, then primers 21 and 22 were used to amplify the fusion construct. To delete hamG (AFLA_099760), primers 29 and 30 were used to amplify the hamG 5′ flank, primers 31 and 32 were used to amplify the hamG 3′ flank, then primers 27 and 28 were used to amplify the fusion construct. To delete hamH (AFLA_131310), primers 35 and 36 were used to amplify the hamH 5′ flank, primers 37 and 38 were used to amplify the hamH 3′ flank, then primers 33 and 34 were used to amplify the fusion construct. To delete hamI (AFLA_021920), primers 41 and 42 were used to amplify the hamI 5′ flank, primers 43 and 44 were used to amplify the hamI 3′ flank, then primers 39 and 40 were used to amplify the fusion construct. To delete laeA (AFLA_033290), primers 63 and 64 were used to amplify the laeA 5′ flank, primers 65 and 66 were used to amplify the laeA 3′ flank, then primers 61 and 62 were used to amplify the fusion construct. To delete nosA (AFLA_025720), primers 73 and 74 were used to amplify the nosA 5′ flank, primers 75 and 76 were used to amplify the nosA 3′ flank, then primers 71 and 72 were used to amplify the fusion construct. All constructs were individually transformed into TXZ21.3 to get deletion mutants as a pyrG auxotroph and the nosA construct was also transformed into TJES20.1 to obtain the prototrophic deletion mutant, and southern analyses are showed in Supplementary Figure S2A.
Complement Strains
TXZ21.3 was complemented to generate a prototrophic isogenic “wild type” strain TXZ21.3.7, for comparison with other mutants. To achieve this, primers 7 and 8 were used to amplify the argB gene from A. flavus NRRL 3357 genomic DNA, primers 9 and 10 were used to amplify the A. fumigatus pyrG gene from A. fumigatus Af293 genomic DNA, then primers 7 and 10 were used to fuse A. flavus argB and A. fumigatus pyrG together by single joint PCR. Primers 5 and 6 were used to amplify the KU70 5′ flank, primers 11 and 12 were used to amplify the KU70 3′ flank, then primers 13 and 14 were used to amplify the fusion construct by double joint PCR. The construct was transformed into TXZ21.3 to create TXZ21.3.7, southern analyses are showed in Supplementary Figure S2B.
Deletion strains of hamE-H were complemented with A. fumigatus pyrG into the KU70 gene locus to created prototrophs. To achieve this, primers 45 and 46 were used to amplify the KU70 5′ flank, primers 47 and 48 were used to amplify the KU70 3′ flank, and the A. fumigatus pyrG gene was amplified from A. fumigatus Af293 genomic DNA using primers No. 61 and 62 from (Affeldt et al., 2014). Then primers 13 and 14 were used to amplify the fusion construct, and the construct have been transformed into ΔhamE -TXZ5.2, ΔhamF -TXZ6.2, ΔhamG -TXZ7.2, ΔhamH -TXZ8.1 mutants to obtain prototrophs, southern analyses are showed in Supplementary Figure S2B. hamI and nosA were deleted in TJES19.1 and TJES20.1 to obtain prototrophs, respectively. For hamI deletion in TJES19.1, primers T1 and T2 were used to amplify the hamI 5′ flank, primers T3 and T4 were used to amplify the hamI 3′ flank, and the pyrG marker was the same as used above for the complemented strains. Primers 39 and 40 were used to get the construct by fusion these three fragments together through double joint PCR. For the nosA deletion in TJES20.1, the same construct was used for as for nosA deletion in TXZ21.3. Southern analyses are showed in Supplementary Figure S2A.
Overexpression Mutants
Because an earlier study showed that adding an extra copy of laeA to the A. flavus genome resulted in overexpression of laeA transcripts (Amaike and Keller, 2009), we replicated that method here. To create an overexpression strain of laeA, primers 67 and 68 were used to amplify A. flavus laeA (including promoter, coding region and terminator) from A. flavus NRRL 3357 genomic DNA, then fused with the argB marker (the same marker gene used for gene deletion) to get the argB-laeA fragment by single joint PCR with primers 7 and 68. Primers 13 and 14 were used to fuse this fragment (argB-laeA) with KU70 5′ flank (primers 45 and 46) and 3′ flank (primers 47 and 48) together as a construct, and then transformed into the KU70 gene locus in TXZ21.3 to obtain a pyrG- OE::laeA strain (TXZ16.1). For overexpression of the nosA gene, primers 77 and 78 were used to amplify the A. nidulans gpdA promoter from plasmid pJMP9.1 (Soukup et al., 2012), which was then fused with the A. fumigatus pyrG gene to get the pyrG-gpdA promoter fragment. Primers 73 and 79 were used to amplify the 5′flank for OE::nosA, primers 80 and 81 were used to amplify the 3′flank for OE::nosA. Then these two flanks were fused with the pyrG-gpdA promoter fragment by using primers 71 and 82, the construct was transformed into TJES19.1 to get OE::nosA (TXZ22.5).
RNA Extraction and Northern Analysis
Gene expression was examined in all strains (NRRL 3357), ΔlaeA (TJW71.1), OE::laeA (TJW79.13), TXZ21.3.7 (as wild type control), ΔnosA (TXZ19.1) and OE::nosA (TXZ22.5) following the same culture condition as in (Georgianna et al., 2010): 106 conidia/mL were inoculated into 50 ml liquid YEP medium (aflatoxin-repressing), mycelia were collected after shaking with 250 rpm at 29°C for 24 h, washed with ddH2O and then transferred to 50 ml YES medium (aflatoxin-promoting), shaking with 220 rpm at 29°C for 6 and 24 h, respectively. Each strain and treatment had three replicates. Mycelia were collected, washed and lyophilized, and total RNA was extracted using QIAzol Lysis Reagent (Qiagen). 15–20 μg of RNA was loaded for northern analysis. The primers in Supplementary Table S1 were used to amplify the probes for hamE (primers 49 and 50), hamF (primers 51 and 52), hamG (primers 53 and 54), hamH (primers 55 and 56), hamI (primers 57 and 58), nosA (primers 69 and 70), and actin (primers 59 and 60).
Sclerotial Formation, Aflatoxin Production, and Spore Enumeration
For sclerotial formation, 106 conidia/plate were mixed with 3 ml GMM+0.1 M sorbitol medium (0.5% top agar), and poured onto 10 ml GMM+0.1 M sorbitol medium (1.5% agar), cultures were grown for 5 days at 29°C under dark, each strain had six replicates. Three plates were sprayed with 70% ethyl alcohol (EtOH) to wash away the spores, and then the plates were scanned for analysis sclerotial formation.
Aflatoxin was extracted from the other three replicates. A 14-mm-diameter core was punched from the center of each plate and homogenized in 3 mL 0.01% Tween 20. Three mL of ethyl acetate was added to each tube and the tubes were shaken vigorously and spun at 3,000 rpm for 15 min. The organic layer was collected, dried down, and resuspended in 500 μL 20% acetonitrile with 1% Formic acid (FA). Samples were filtered through an Acrodisc syringe filter with a nylon membrane (0.45 μm; Pall Corporation), and the samples were separated on a ZORBAX Eclipse XDB-C18 column (Agilent, 4.6 mm by 150 mm with a 5 μm particle size) by using a binary gradient of 1% (v/v) FA as solvent A and 1% FA in Acetonitrile as solvent B using a Flexar Binary Liquid Chromatography (LC) Pump (PerkinElmer) coupled to a Flexar LC Autosampler (PerkinElmer) and a Flexar Fluorescence Light (FL) Detector (PerkinElmer) with the excitation wavelength at 365 nm and the emission wavelength at 455 nm. The binary gradient started with an isocratic step at 80% A for 1 min followed by a linear gradient to 35% A in 10 min and an additional linear gradient to 100% B in 0.5 min. After each run the column was washed for 3 min using 100% B and was equilibrated for 3 min using 80% A. The flow rate was set to 1.5 mL/min. Identification and quantification of aflatoxin B1 was performed using Chromera Manager (PerkinElmer) by comparison to a standard curve derived from an aflatoxin B1 standard (Sigma–Aldrich, United States). Aflatoxin B1 was detected by a fluorescent detector with an emission wavelength of 455 nm and excitation wavelength of 365 nm.
Conidia production was quantified on the 3rd and 5th days of culture. Ten mL of 0.5% (w/v) top agar of GMMUU medium containing 106 spores/plate were overlaid on 20 mL GMMUU agar plates, then plates were cultured at 29°C. To count conidia, 7 mm plug from each plate was homogenized in 2 mL 0.01% Tween 20 water, diluted 10-fold and counted with a hemocytometer. For each strain, conidia counts were performed using three replicates.
Heterokaryotic Colony Assay
Hyphal fusion of laeA, ham, and nosA mutants were processed following Tsukasaki et al. (2014) with the following modification: conidia of the argB auxotrophic strain (TJES20.1) and pyrG auxotrophic strains (TJES19.1, ΔlaeA -TXZ15.1, OE::laeA -TXZ16.1, ΔhamE -TXZ5.2, ΔhamF -TXZ6.2, ΔhamG -TXZ7.2, ΔhamH -TXZ8.1, ΔhamI -TXZ9.16, and ΔnosA -TXZ20.1) were collected from GMM medium supplemented with arginine or uridine/uracil as needed. Equal numbers of conidia from the argB auxotrophic strain and pyrG auxotrophic strains were mixed, and 107 conidia/mL were spotted onto the agar media containing arginine (1 g/L) and uracil (5 mM)/uridine (5 mM). After the incubation at 29°C for 5 days, the newly formed conidia were collected, and 105 conidia were spread onto the GMM+0.25% Triton X-100 medium (the 0.25% Triton X-100 restricts colony diameter to help with precise colony counts). Heterokaryotic colonies were counted after incubation for 3 days at 29°C.
This protocol required presence of conidia which could contain more than one nuclei. Previous work has shown that A. flavus conidia contain either one or two nuclei (Runa et al., 2015). We confirmed this using the nuclear stain DAPI (Supplementary Figure S3).
Pathogenicity Assays
Pathogenicity assays were conducted according to a published protocol (Christensen et al., 2012) with some modifications. Corn kernels (Blue River organic hybrid) were washed in 70% EtOH for 5 min, rinsed with sterile water, and shaken in bleach (100 rpm) for 10 min. After three rinses in sterile water to remove the bleach, the kernels were blotted dry on sterile paper towels and a sterile needle was used to puncture a small hole in the embryo of each kernel. Four kernels were placed into a sterile scintillation vial, weighed, and inoculated with 200 μL of a 106 conidia/mL suspension of spores in 0.01% Tween 20. A mock control inoculated with just 0.01% Tween 20 was also included. After inoculation, the vials were vortexed 5 s, the cap then loosened (to allow air circulation) and the vials were placed in a plastic box containing moist paper towels. The box was covered with Press’n Seal (Glad) and placed in a 29°C incubator with 12 h light/dark cycles (beginning on dark) for 5 days. Five replicates per strain were included.
After the 5-day period, 3 mL 0.01% Tween 20 were added to each vial, vortexed thoroughly for 1 min and then 100 μL were removed from the vials for spore enumeration using a hemocytometer. For aflatoxin extraction, 3 mL of ethyl acetate was added to the vials which were then shaken vigorously and spun at 3,000 rpm for 15 min. The organic layer was removed, dried down, and resuspended in 300 μL 20% acetonitrile with 1% FA. Then samples will be prepared as above (section of sclerotial formation) for aflatoxin HPLC analysis.
Statistical Analysis
The GraphPad Prism software (La Jolla, CA, United States) was used for statistical analysis. Statistically significant differences were determined by an unpaired Student’s t-test with a two-tailed distribution and P < 0.05. The error bars in all figures indicate the standard error of the mean.
Results
LaeA Regulates ham Gene Expression
LaeA, although originally identified as a global regulator of fungal secondary metabolites (Bok and Keller, 2004), has since been associated with diverse fungal developmental processes including sclerotial production in A. flavus where loss of laeA (ΔlaeA) and overexpression of laeA (OE::laeA) result in loss and overproduction of sclerotia, respectively, (Kale et al., 2008; Amaike and Keller, 2009). Sclerotia are overwintering structures formed through extensive hyphal fusion events and in A. flavus their formation is associated with aflatoxin synthesis. To try to gain insight into mechanism(s) by which LaeA regulates sclerotial synthesis, we re-examined previous microarray data of wild type, ΔlaeA and OE::laeA strains of A. flavus (Georgianna et al., 2010) and identified a set of five LaeA regulated genes that are homologs of ham genes involved in hyphal fusion in N. crassa. Supplementary Table S2 shows the results of the A. flavus microarray where five putative ham genes, hamE (AFLA_095770), hamF (AFLA_033600), hamG (AFLA_099760), hamH (AFLA_131310), hamI (AFLA_021920) (homologs gene of ham-5, 6, 7, 8, and 9 in N. crassa, respectively) were significantly down regulated in ΔlaeA and upregulated in OE::laeA. Other N. crassa ham homologs were not regulated by LaeA.
To confirm LaeA regulation of these five genes, RNA was extracted from the same strains – WT (NRRL3357), ΔlaeA (TJW79.1), OE::laeA (TJW79.13) – and grown identically as in the microarray study (Georgianna et al., 2010) for northern blot analysis. Our results (Figure 1) showed that all five genes were positively regulated by LaeA with near loss in ΔlaeA and higher expression in the OE::laeA strain.
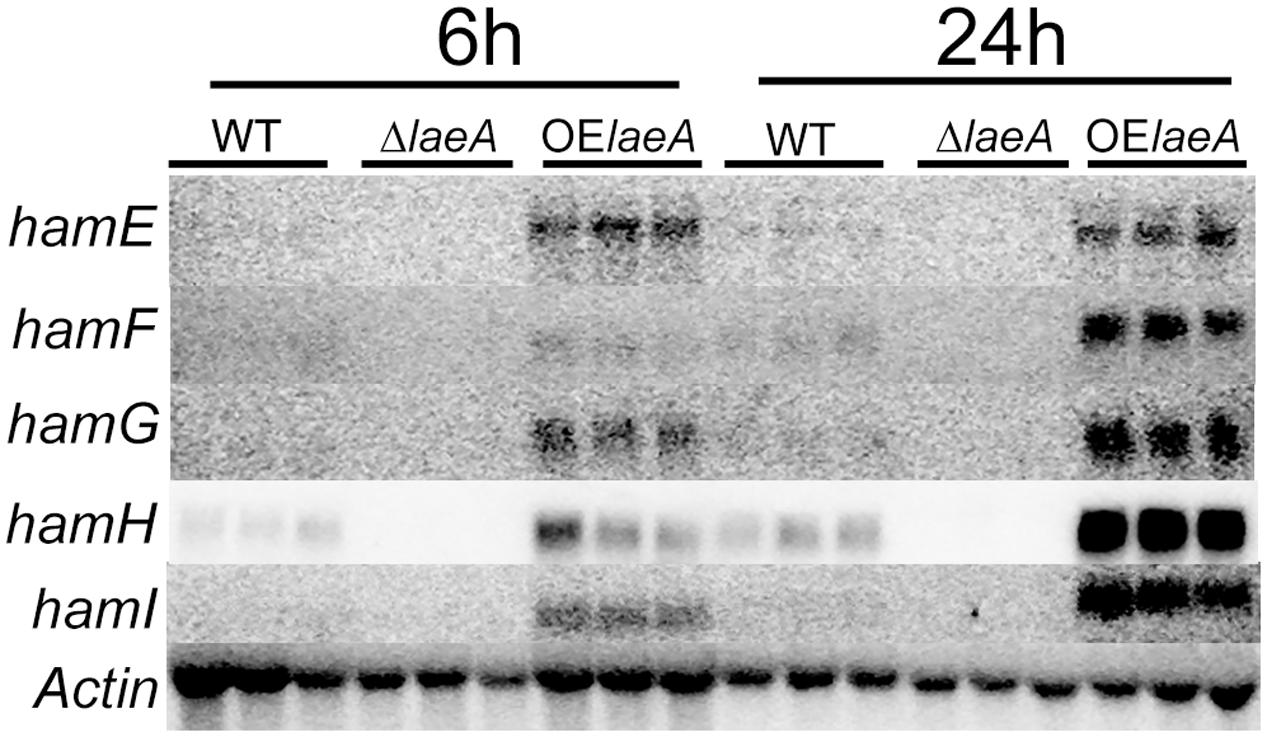
FIGURE 1. Northern result of the ham genes in WT, ΔlaeA and OE::laeA strains. 50 ml liquid YEP medium (6% peptone, 2% yeast extract) was inoculated with 106 conidia/ml of A. flavus NRRL3357, an laeA deletion strain or an laeA overexpression strain in 125 ml flasks, incubated with shaking at 250 rpm at 29°C. Treatments were triplicated. After 24 h, mycelia were collected and incubated in the aflatoxin-stimulating YES (6% sucrose, 2% yeast extract) medium for 6 h and 24 h (220 rpm, 29°C), respectively. hamE (AFLA_095770), hamF (AFLA_033600), hamG (AFLA_099760), hamH (AFLA_131310), hamI (AFLA_021920).
Deletion of ham Genes Recapitulates ΔlaeA Phenotype of Sclerotia and Aflatoxin Loss
Studies of sclerotial formation have largely focused on morphological development, with several studies illustrating initial fusion events in sclerotia development (Kohn et al., 1990; Erental et al., 2008). We thus considered it possible that deletion of ham genes could decrease or eliminate sclerotial production and possibly aflatoxin synthesis due to its association with sclerotial production (Duran et al., 2006; Kale et al., 2008). hamE, F, G, and H were individually deleted in an A. flavus double mutant strain (TXZ21.3, ΔKU70,ΔargB, pyrG-) by replacing the ham encoding sequence with argB of A. flavus. hamI was deleted in an A. flavus single mutant strain (TJES19.1, ΔKU70, pyrG-) by replacing the hamI encoding sequence with A. fumigatus pyrG. Deletion mutants were confirmed by Southern blotting (Supplementary Figure S2A), and then one confirmed deletion mutant of each gene (excepting the hamI mutant as it was already a prototroph) was complemented with the A. fumigatus pyrG gene to create a prototroph as confirmed by Southern blotting (Supplementary Figure S2B).
We compared the ability of the five prototrophic Δham strains to produce sclerotia in comparison to ΔlaeA, OE::laeA and wild type (TXZ21.3.7) on sclerotial inducing medium (Kale et al., 2008). As seen in Figure 2A, similar to ΔlaeA, deletions of ham F, G, H, and I mutants yielded strains unable to produce sclerotia. However, the ΔhamE mutant still produced sclerotia in this culture condition. As reported previously, OE::laeA produced more sclerotia than WT (Kale et al., 2008). Aflatoxin synthesis followed that of sclerotia production on this same media with no detectable aflatoxin in ΔlaeA and Δham F-I strains but a similar amount in ΔhamE as wild type and increased synthesis in OE::laeA (Figure 2B). In contrast to the effects ham gene loss had on sclerotial and aflatoxin production, we found no difference in asexual spore production on growth medium (Supplementary Figure S5).
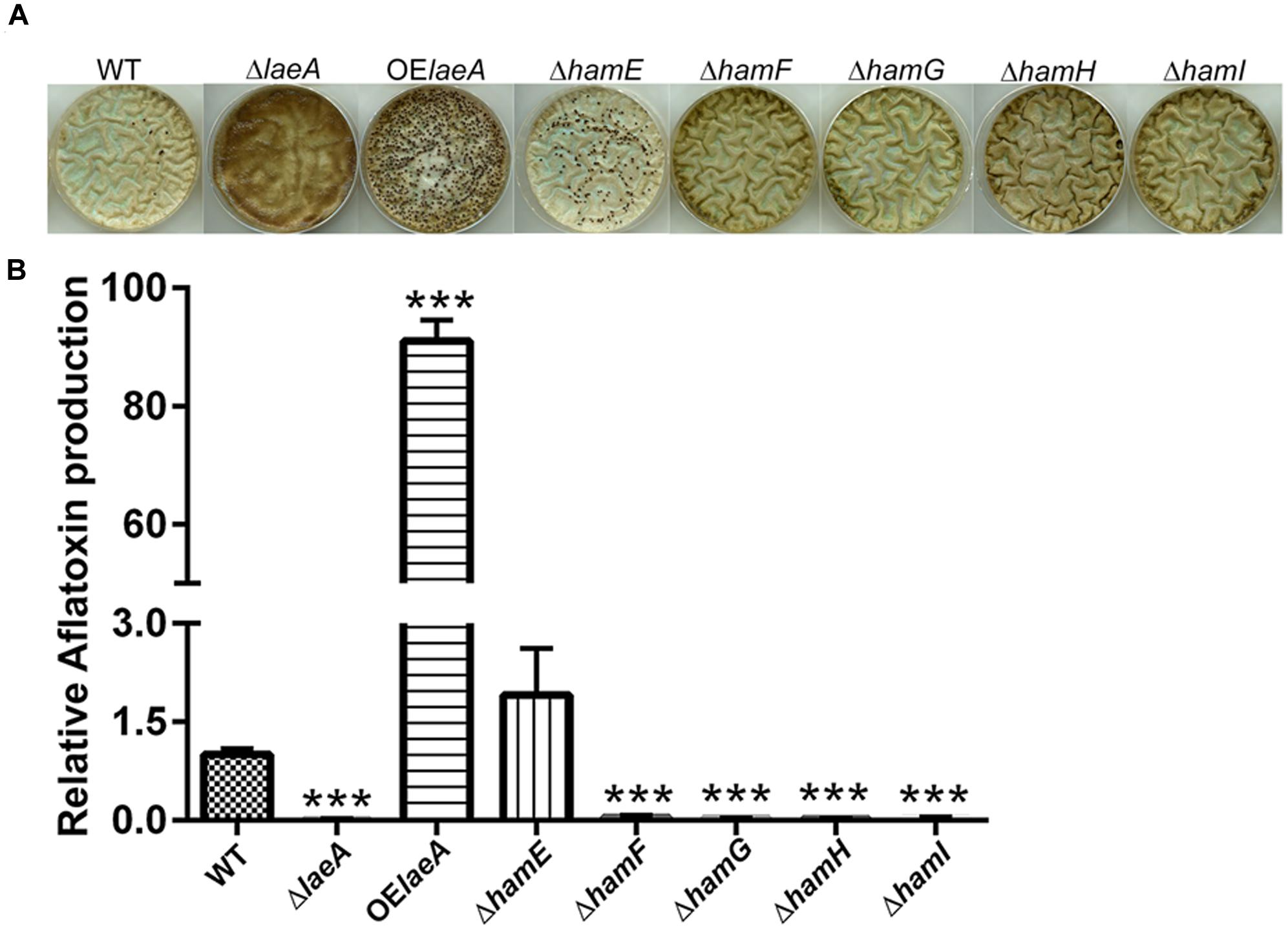
FIGURE 2. Sclerotia formation (Panel A) and aflatoxin production (Panel B) of ham mutants in GMM+2% sorbitol medium. (A) Sclerotia. (B) Aflatoxin. ∗∗∗P < 0.001. 106 spores/plate were cultured in GMM+2% sorbitol under dark for 5 days at 29°C, spores were washed away by 70% EtOH and pictures taken to observe sclerotial formation. 1.4 cm cores were excised from each plate and extracted for analysis of aflatoxin levels via HPLC. Average of three plates for each strain.
LaeA and HamF, G, H, and I Are Required for Heterokaryon Formation
Heterokaryon formation also requires hyphal fusion (Glass et al., 2004) and can be the prelude to sclerotial formation between strains of the opposite mating type (Wada et al., 2014). To test for heterokaryon formation, we followed a scheme developed by Tsukasaki et al. (2014) for heterokaryon formation in the fungus A. oryzae with some small modifications (Figure 3A). Equal numbers of conidia from pyrG auxotrophic and argB auxotrophic strains were mixed together and grown on GMM medium supplemented with uracil/uridine and arginine. Conidia were collected after 5 days and spread onto GMM medium lacking supplementation where only conidia generated from heterokaryons could grow. Figure 3B,C illustrate successful heterokaryon formation in the ham and laeA wild type controls (TJES19.1, a pyrG auxotroph, crossed to TJES20.1, an argB auxotroph). Crosses with OE::laeA and ΔhamE mutants also led to heterokaryon formation although heterokaryon colonies were decreased in ΔhamE compared to both wild type and OE::laeA. In contrast, no heterokaryons formed in the ΔlaeA and ΔhamF, G, H, and I mutants which correlated with their inability to produce sclerotia (Figure 2).
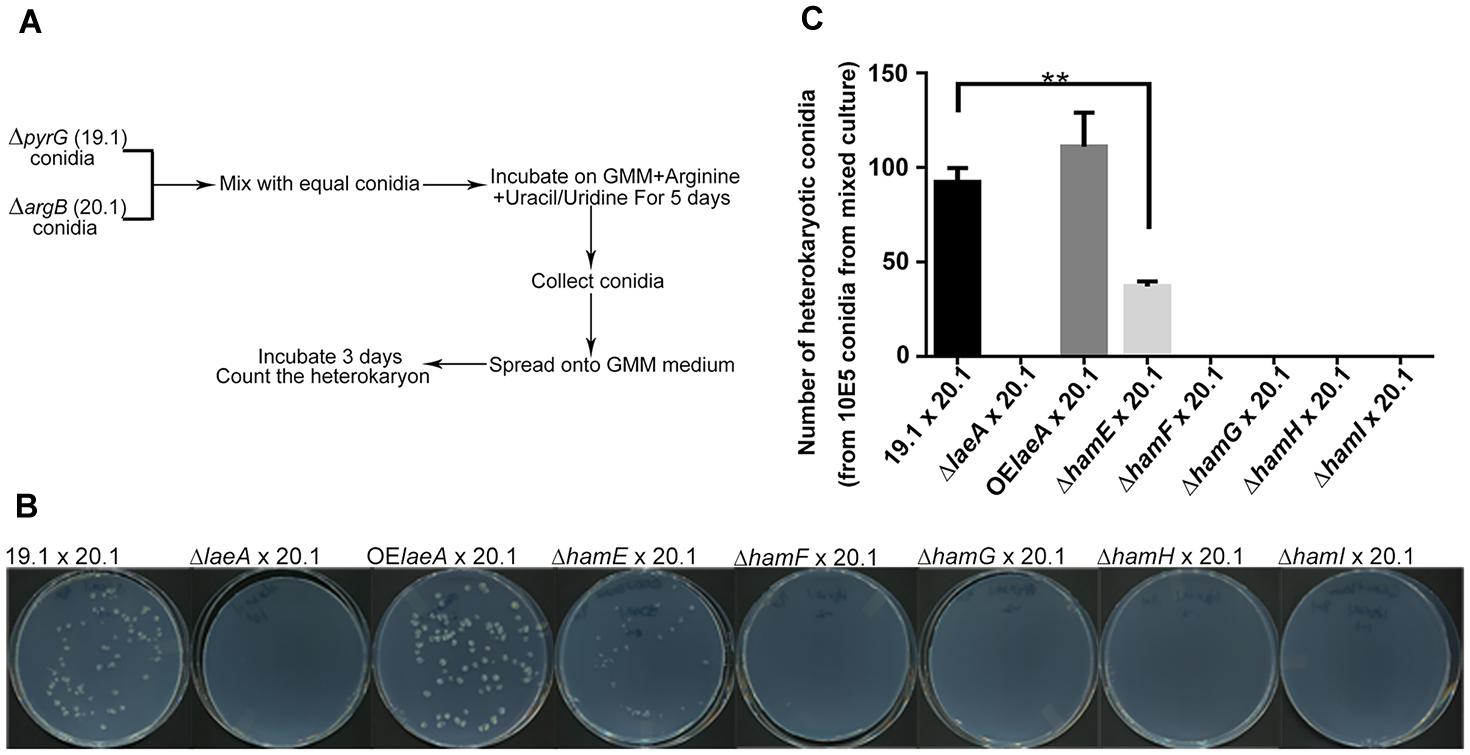
FIGURE 3. Hyphal fusion of ham mutants. (A). Outline of the method to measure hyphal fusion in Aspergillus oryzae (Tsukasaki et al., 2014) which was modified for this work. (B) Photographs of heterokaryon formation or lack of formation in crosses. Plates of mixed cultures were spread and incubated at 29°C for 3 days. (C) The number of heterokaryotic colonies formed for each cross. ∗∗P < 0.01. ΔlaeA (TXZ15.1), OElaeA (TXZ16.1), ΔhamE (TXZ5.2), ΔhamF (TXZ6.2), ΔhamG (TXZ7.2), ΔhamH (TXZ8.1) and ΔhamI (TXZ9.16) were uracil/uridine auxotrophic. 19.1 (TJES19.1), uracil/uridine auxotrophic. 20.1 (TJES20.1), arginine auxotrophic.
Decreased Virulence of ham Mutants on Host Seed
As LaeA is required for A. flavus virulence on host seed, we were curious to see if loss of any ham gene also impacted host colonization. All five Δham mutants were compared to wild type, ΔlaeA and OE::laeA in ability to colonize (as assessed by spore production) and produce aflatoxin on corn seed 5 days post inoculation. The ΔhamE strain presented a similar infection pattern and aflatoxin production as both wild type and OE::laeA (Figure 4). However, all the other four ham mutants, ΔhamF- I, showed reduced sporulation compared to wild type (Figure 4B). Aflatoxin production was reduced in ΔhamF, G, H, and I infected corn samples compared to wild type although these mutants produced slightly more aflatoxin than the ΔlaeA mutant (Figure 4C).
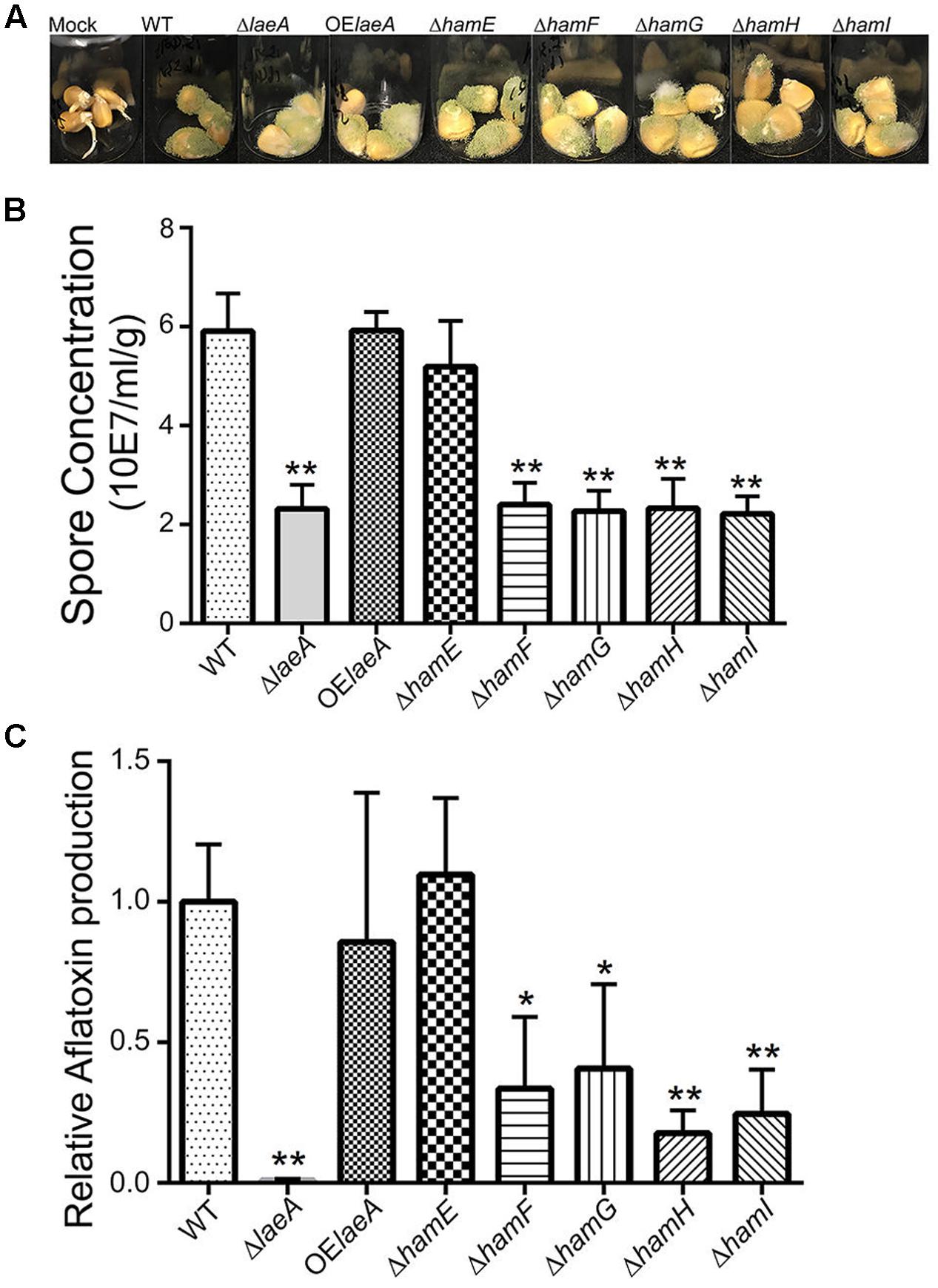
FIGURE 4. Corn infection with ham mutants. (A) 200 μl of 106 spore/ml suspension of spores in 0.01% Tween 20 were inoculated on corn and compared to a mock control of 200 μl 0.01% Tween 20. The vials were kept in a moist chamber and placed in 29°C incubator with 12 h light/dark cycling and incubated for 5 days (Christensen et al., 2012). (B) The spores were washed off the seed with 3 ml 0.01% Tween 20 and counted. Each treatment was replicated three times. (C) Relative aflatoxin production of these mutants compared to WT (TXZ21.3.7). P-value, ∗p < 0.05, ∗∗p < 0.01.
NosA Connects LaeA to the Ham Cascade
Recent studies have found that ham-5, 6, 7, 8, and 9, the same subset of ham genes regulated by LaeA in A. flavus, were coordinately regulated by the transcription factor in N. crassa ADV-1, (Dekhang et al., 2017) and its homolog Pro1 in S. macrospora (Steffens et al., 2016). The orthologous transcription factor in Aspergillus is NosA, which is involved in sexual development in A. nidulans (Vienken and Fischer, 2006) and regulated by LaeA in A. fumigatus (Soukup et al., 2012). An examination of the A. flavus microarray data (Georgianna et al., 2010) showed LaeA regulated nosA in a similar fashion as hamE-I (Supplementary Table S2) which we confirmed by northern analysis (Figure 5A).
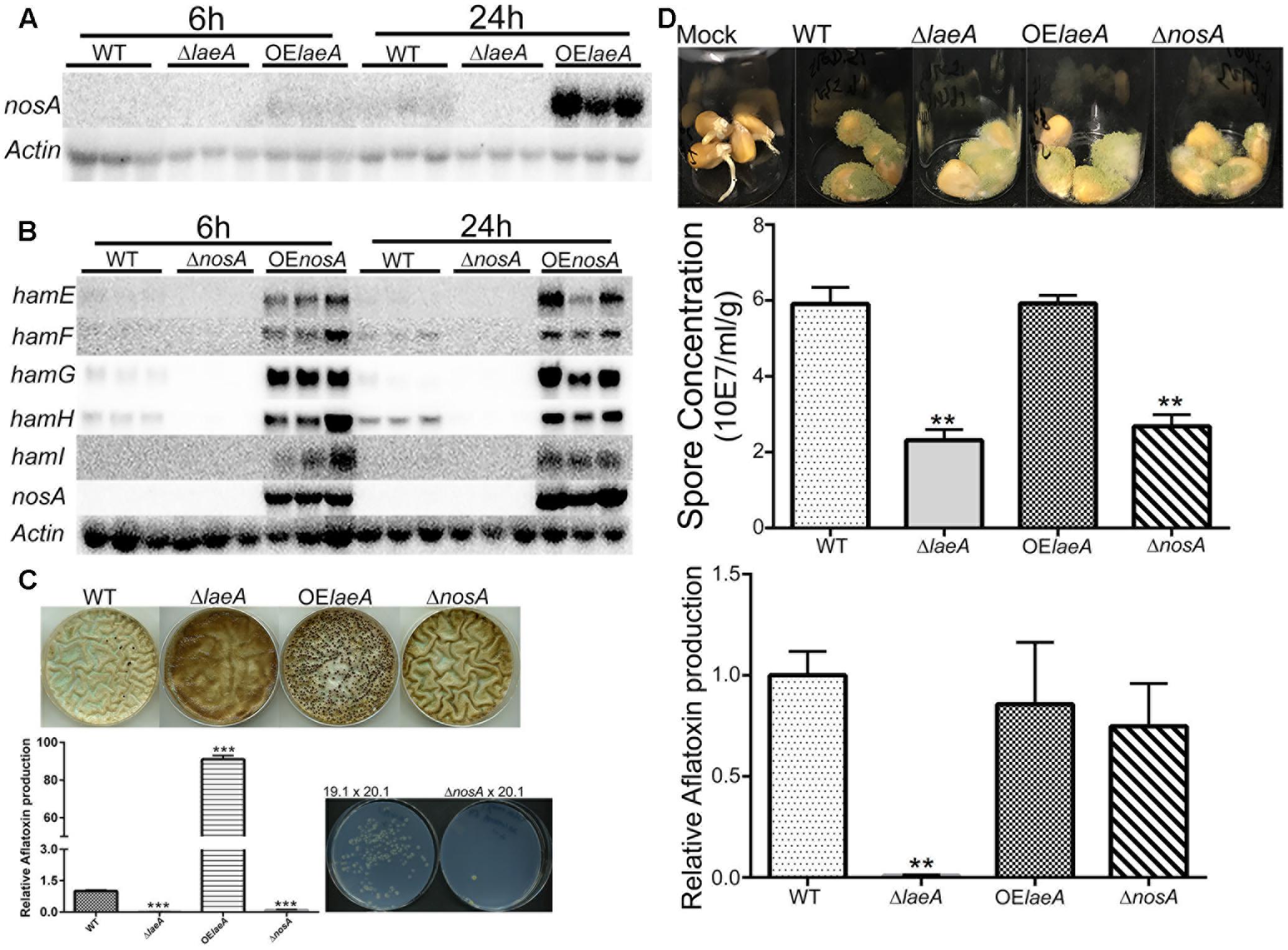
FIGURE 5. NosA regulation of ham genes and heterokaryon, virulence and aflatoxin production for ΔnosA mutants (A). LaeA positively regulates nosA expression. The same northern blot as Figure 1 was used for nosA expression. nosA (AFLA_025720). (B) NosA positively regulates hamE-I (using the same culture conditions as Figure 1). A. flavus WT (TXZ21.3.7), ΔnosA (TXZ19.1), and OE::nosA (TXZ22.5). (C) Deletion of nosA (TXZ19.1) results in total or near loss of sclerotia formation, aflatoxin production and heterokaryon formation. NosA deletion was assessed with the strains in Figure 2 and the pictures of WT, ΔlaeA and OE::laeA are from Figure 2. (D) Deletion of nosA (TXZ19.1) produces less spores but is not affected in aflatoxin biosynthesis on corn. NosA deletion was assessed with the strains in Figure 4 and the pictures of WT, ΔlaeA and OE::laeA are from Figure 4. Each treatment was replicated three times. P-value, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next deleted and overexpressed nosA (Supplementary Figure S2A). As shown in Figure 5B and Supplementary Figure S4, ham gene expression was down regulated in the ΔnosA mutant and upregulated in OE::nosA mutant, respectively, in two different media. An examination of the ΔnosA strain showed that, similar to ΔlaeA and ΔhamF-I, the mutant was unable to form heterokaryons or sclerotia and was impaired in aflatoxin synthesis on media (Figure 5C). The ΔnosA strain showed a decreased ability to infect corn (as measured by sporulation) although it showed no difference in ability to produce aflatoxin in seed (Figure 5D).
Discussion
The persistence of sclerotia in adverse environmental conditions represents a clear fitness advantage for fungi. Not only can sclerotia withstand environmental extremes but they contain numerous bioactive secondary metabolites proposed to act as insect feeding deterrents (Laakso et al., 1994; Whyte et al., 1996). Despite their importance in plant pathology and potentially drug discovery (Lee et al., 2017), very little is known about the genetic pathways regulating their formation. Following leads from studies showing a requirement of LaeA (Lae1) for sclerotia production (Kale et al., 2008; Amaike and Keller, 2009; Schumacher et al., 2015), we present our findings supporting a model where LaeA regulation of a cell fusion cascade underscores a critical cellular mechanism required for sclerotial formation in fungi.
Hyphal fusion plays a key role in establishment of the mycelial colony, tissue development, heterokaryon formation and nutrition communication in filamentous fungi including N. crassa, S. macrospora, Fusarium oxysporum, and B. cinerea (Lichius and Lord, 2014; Fleißner and Serrano, 2016; Shahi et al., 2016). The majority of our genetic understanding of hyphal fusion is obtained from a series of N. crassa studies where genes involved in fusion events have been termed hyphal anastomosis genes, ham (Herzog et al., 2015; Fleißner and Herzog, 2016; Daskalov et al., 2017). Here we present data showing that five A. flavus homologs (HamE-I) of N. crassa Ham proteins (HAM-5-9) share the same regulatory cascade as in N. crassa with four of them (HamF-I) to exhibit clear roles in fusion events in A. flavus. Cellular fusion in A. flavus is most clearly observed in inability to form heterokaryons or sclerotia. The consequence of heterokaryon formation and sclerotial loss is also observed in decreased ability to synthesize aflatoxin in these mutants.
The HamE (HAM-5) mutant showed the least difference to wild type A. flavus. HAM-5 was originally isolated in a UV screen to isolate N. crassa mutants in hyphal fusion, as measured by loss of conidial anastomosis tube formation, or CAT formation (Aldabbous et al., 2010). More recently HAM-5 has been characterized as a putative scaffold protein for the MAK-2 MAP kinase complex, physically interacting with NRC-1, MEK-2, and MAK-2 (Jonkers et al., 2014). The protein is an important player in oscillation kinetics during CAT formation in N. crassa (Jonkers et al., 2014). The homolog of HAM-5 in P. anserina, IDC1, is a member of the PaMpk1 signaling pathway and required for the movement of a MAP kinase into the nucleus (Jamet-Vierny et al., 2007). The A. flavus hamE mutant was deficient in heterokaryon formation (Figure 2, Figure 3) although fusion was not completely eliminated in contrast to the other four ham mutants. Also, this mutant was still able to produce sclerotia and aflatoxin and demonstrated equal virulence as wild type. If HamE is a member of homologous MAK-2 Map kinase pathway in A. flavus, these results may suggest this pathway is not heavily involved in fusion dynamics.
In contrast to HamE loss, deletion of HamF-I (HAM-6-9) showed near identical phenotypes to ΔlaeA. HAM-6, HAM-7 and HAM-8 were all identified in a CAT fusion screen separate from the HAM-5 screen (Fu et al., 2011). Their sequence suggested they might form a cell membrane complex with experimental evidence to link such a complex to the MAK-1 cell wall integrity MAPK pathway due to a dramatic decrease of MAK-1 phosphorylation in Δham-6,Δham-7, and Δham-8 N. crassa mutants (Fu et al., 2014). However, a clear picture of such a complex still needs to be resolved. BcNoxD/Pro41/PaNoxD are the homologs of HAM-6 in B. cinerea (Siegmund et al., 2015), S. macrospora (Nowrousian et al., 2007), and P. anserina (Lacaze et al., 2015), respectively. These proteins are thought to be homologs of the p22phox NADPH subunit in fungi and found located at the ER (Lacaze et al., 2015; Scott, 2015). The p22phox subunit interacts with the gp91phox protein to form the NADPH oxidase complex in fungi and mutants of gp91phox display similar phenotypes as p22phox mutants. In particular, the deletion of the gp91phox equivalent in A. nidulans, NoxA, results in loss of sexual fruit body (cleistothecium) formation in that fungus (Lara-Ortíz et al., 2003). Cleistothecia are the developmental equivalent to sclerotia in A. flavus, thus fitting in with our observation of sclerotial loss in ΔhamF mutants of A. flavus. We would predict that deletion of the noxA homolog in A. flavus or deletion of hamF in A. nidulans would result in loss of sclerotia and cleistothecia in each species, respectively.
There is limited knowledge about HAM-7, HAM-8, and HAM-9. HAM-7, a GPI-anchored cell wall protein, is required for activating the MAK-1 MAP kinase cascade and regulates cell wall integrity and hyphal anastomosis (Maddi et al., 2012; Fu et al., 2014). As mentioned earlier, HAM-8 is proposed to function in this same signaling network (Fu et al., 2014). This same study suggested that HAM-9 might play slightly different roles in the network as its loss did not generate all of the same physiological patterns as HAM-6, HAM-7, and HAM-8 mutants. The authors suggested HAM-9 might mediate the communication of the two MAPK pathways during hyphal fusion (Fu et al., 2014). The phenotypes of deletions of all four ham equivalents (e.g., hamF-I) in A. flavus were identical in all parameters tested in this study with identity to ΔlaeA in lack of heterokaryon fusion (Figure 3) and sclerotial development (Figure 2A) although the ΔlaeA mutant showed a more extreme aflatoxin loss during colonization of corn (Figure 4C).
Relatively few studies have examined the impact of loss/impaired fusion on virulence of pathogenic fungi but, of those published, anastomosis appears to be important for virulence. Deletion of the SO homolog (Aso1) in Alternaria brassicicola yielded a strain lacking the ability to fuse and greatly impaired in ability to form lesions on cabbage leaves (Craven et al., 2008). The F. verticillioides SO mutant (FvSO) was essential for infection of corn ears, talks and seedling (Guo et al., 2015). A striatin mutant in the corn pathogen Colletotrichum graminicola was defective in stalk rot and leaf blight infections, showed reduced hyphal fusion and was defective in sexual development (Wang et al., 2016). The orthologous mutant in F. verticillioides likewise was significantly impaired in virulence (Zhang et al., 2017). The B. cinerea noxD (ham-6/hamF) mutant was less virulence when it was inoculated on French bean plants (Phaseolus vulgaris) (Siegmund et al., 2015). Our results support a requirement for hyphal fusion for wild type virulence of A. flavus on corn seed and, additionally, production of aflatoxin (Figure 4C).
In addition to LaeA, other parameters have been shown to regulate sclerotia production in A. flavus including a circadian oscillator where sclerotia production peaks at early evening (Greene et al., 2003). The Aspergillus clock components remain unresolved as this genus does not contain an ortholog of frequency, the central clock protein in N. crassa (Montenegro-Montero et al., 2015). Recently, the N. crassa clock controlled transcription factor ADV-1 (Dekhang et al., 2017), and the homolog protein Pro1 in S. macrospora (Steffens et al., 2016), both were shown to regulate the same ham genes (ham-5, -6, -7, -8, and -9) as regulated by LaeA in A. flavus. The work in N. crassa further demonstrated that ADV-1 transduces clock driven rhythmic expression of these ham genes and thus hyphal fusion events. The homolog of ADV-1/Pro1 in Aspergillus, NosA, controls fruiting body formation in A. nidulans (Vienken and Fischer, 2006). Intriguingly, we find A. flavus NosA to regulate the same five ham genes as ADV-1/Pro1 and LaeA and loss of ΔnosA to exhibit the majority of the ΔlaeA phenotypes similarly to deletion of hamF-I. Furthermore, nosA itself is under the transcriptional control of LaeA (Figure 5). Thus we speculate that the circadian output of sclerotial formation in A. flavus, reported in Greene et al. (2003), may in part be regulated through a LaeA>NosA>fusion cascade, a topic of further investigation in our laboratory.
Author Contributions
XZ carried out the experimentation of this work and with NK conceived experiments and wrote the manuscript. JS, JB, and TV helped create strains for this research. Z-MH helped revise this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (grant no. 31470198) to Z-MH and the China Scholarship Council to XZ and in part by support by the National Institute of Food and Agriculture, United States Department of Agriculture, Hatch project 232876 and 1006780 to NK and Food Research Institute support to NK as well as an NSF Graduate Research Fellowship under grant no. DGE-1256259 to JS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01925/full#supplementary-material
FIGURE S1 | History of Aspergillus flavus double mutant TXZ21.3. A. flavus NRRL3357.5 (pyrG-) from He et al. (2007). KU70 (AFLA_061470) ORF was replaced by A. parasiticus pyrG to get TJW149.27, then the ku70 gene locus pyrG was knockout to get TJES19.1. A. fumigatus pyrG was used to replace the argB ORF in TJES19.1 to get TJES20.1, and then the fumigatus pyrG in the argB locus was deleted again to get the TXZ21.3.
FIGURE S2 | Southern blot analysis of the mutants in this research. (A) Southern blot of the deletion and overexpression strains. (B) Southern blot of the auxotroph mutants complement to prototroph.
FIGURE S3 | DAPI stain of the ΔnosA and Δham mutants. Fresh spores were harvested from the strains with 0.05% Triton X-100 and centrifuged at 3000 rpm for 5 min. The supernatant was removed and conidia were washed twice with 1X Phosphate Buffered Saline (PBS). The pellet was re-suspended with 200 μl DAPI stain solution. DAPI stain solution was prepared by adding 5 μl of 10-mg/ml stocks of DAPI (40, 60-diamino-2-phenylinodole, Sigma–Aldrich, United States) plus 100- μl antifade PPD (P-phenylenediamine, Sigma–Aldrich, United States, stock solution 1 ng/lL) plus 900 μl of 70% glycerol. Conidia were incubated in the solution at room temperature in the dark for 20–30 min. Stained conidia were washed twice with 1X PBS buffer and then twice with water. The conidia were observed under a fluorescence microscope (Zeiss AxioMagerA10) using a DAPI filter set. (A) ΔnosA (TXZ20.1). (B) ΔhamE (TXZ5.2). (C) ΔhamF (TXZ6.2). (D) ΔhamG (TXZ7.2). (E) ΔhamH (TXZ8.1). (F) ΔhamI (TXZ9.16).
FIGURE S4 | NosA positively regulates hamE-I in GMM medium. 107 spores were inoculated into 50 ml GMM liquid medium for 48 h at 220 rpm at 29°C. RNA was extracted for Northern blot analysis using the probes described in section “Materials and Methods.”
FIGURE S5 | Sporulation of ΔhamE-I and ΔnosA mutants in GMMUU medium. 10 ml top agar GMMUU medium [0.5% (w/v) agar] containing 106 spores were overlaid on 20 ml GMMUU agar plates, then plates were inoculated at 29°C. The spores were counted from cores taken on 3 and 5 days separately.
References
Affeldt, K. J., Carrig, J., Amare, M., and Keller, N. P. (2014). Global survey of canonical Aspergillus flavus G protein-coupled receptors. mBio 5:e01501-14. doi: 10.1128/mBio.01501-14
Aldabbous, M. S., Roca, M. G., Stout, A., Huang, I. C., Read, N. D., and Free, S. J. (2010). The ham-5, rcm-1 and rco-1 genes regulate hyphal fusion in Neurospora crassa. Microbiology 156, 2621–2629. doi: 10.1099/mic.0.040147-0
Amaike, S., and Keller, N. P. (2009). Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 8, 1051–1060. doi: 10.1128/EC.00088-09
Amare, M. G., and Keller, N. P. (2014). Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 66, 11–18. doi: 10.1016/j.fgb.2014.02.008
Bayram, O., Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3, 527–535. doi: 10.1128/EC.3.2.527-535.2004
Brown, S. H., Scott, J. B., Bhaheetharan, J., Sharpee, W. C., Milde, L., Wilson, R. A., et al. (2009). Oxygenase coordination is required for morphological transition and the host–fungus interaction of Aspergillus flavus. Mol. Plant Microbe Interact. 22, 882–894. doi: 10.1094/MPMI-22-7-0882
Chang, P.-K., Scharfenstein, L. L., Ehrlich, K. C., and Diana Di Mavungu, J. (2016). The Aspergillus flavus fluP-associated metabolite promotes sclerotial production. Fungal Biol. 120, 1258–1268. doi: 10.1016/j.funbio.2016.07.010
Chang, P.-K., Scharfenstein, L. L., Li, R. W., Arroyo-Manzanares, N., De Saeger, S., and Diana Di Mavungu, J. (2017). Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet. Biol. 104, 29–37. doi: 10.1016/j.fgb.2017.04.006
Christensen, S., Borrego, E., Shim, W.-B., Isakeit, T., and Kolomiets, M. (2012). Quantification of fungal colonization, sporogenesis, and production of mycotoxins using kernel bioassays. J. Vis. Exp. 62:3727. doi: 10.3791/3727
Coley-Smith, J. R., and Cooke, R. C. (1971). Survival and germination of fungal sclerotia. Annu. Rev. Phytopathol. 9, 65–92. doi: 10.1146/annurev.py.09.090171.000433
Craven, K. D., Vélëz, H., Cho, Y., Lawrence, C. B., and Mitchell, T. K. (2008). Anastomosis is required for virulence of the fungal necrotroph Alternaria brassicicola. Eukaryot. Cell 7, 675–683. doi: 10.1128/EC.00423-07
Daskalov, A., Heller, J., Herzog, S., Fleißner, A., and Glass, N. L. (2017). Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol. Spectrum. 5:FUNK-0015-2016. doi: 10.1128/microbiolspec.FUNK-0015-2016
Dekhang, R., Wu, C., Smith, K. M., Lamb, T. M., Peterson, M., Bredeweg, E. L., et al. (2017). The Neurospora transcription factor ADV-1 transduces light signals and temporal information to control rhythmic expression of genes involved in cell fusion. G3 7, 129–142. doi: 10.1534/g3.116.034298
Dettmann, A., Heilig, Y., Ludwig, S., Schmitt, K., Illgen, J., Fleißner, A., et al. (2013). HAM-2 and HAM-3 are central for the assembly of the Neurospora STRIPAK complex at the nuclear envelope and regulate nuclear accumulation of the MAP kinase MAK-1 in a MAK-2-dependent manner. Mol. Microbiol. 90, 796–812. doi: 10.1111/mmi.12399
Duran, R. M., Cary, J. W., and Calvo, A. M. (2006). Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. doi: 10.1007/s00253-006-0581-5
Erental, A., Dickman, M. B., and Yarden, O. (2008). Sclerotial development in Sclerotinia sclerotiorum: awakening molecular analysis of a “Dormant” structure. Fungal Biol. Rev. 22, 6–16. doi: 10.1016/j.fbr.2007.10.001
Fleissner, A., Leeder, A. C., Roca, M. G., Read, N. D., and Glass, N. L. (2009). Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. U.S.A. 106, 19387–19392. doi: 10.1073/pnas.0907039106
Fleißner, A., and Herzog, S. (2016). Signal exchange and integration during self-fusion in filamentous fungi. Semin. Cell Dev. Biol. 57, 76–83. doi: 10.1016/j.semcdb.2016.03.016
Fleißner, A., and Serrano, A. (2016). “7 The art of networking: vegetative hyphal fusion in filamentous ascomycete fungi,” in Growth, Differentiation and Sexuality, eds U. Kües and R. Fischer (Cham: Springer International Publishing), 133–153. doi: 10.1007/978-3-319-25844-7_7
Fu, C., Ao, J., Dettmann, A., Seiler, S., and Free, S. J. (2014). Characterization of the Neurospora crassa cell fusion proteins, HAM-6, HAM-7, HAM-8, HAM-9, HAM-10, AMPH-1 and WHI-2. PLOS ONE 9:e107773. doi: 10.1371/journal.pone.0107773
Fu, C., Iyer, P., Herkal, A., Abdullah, J., Stout, A., and Free, S. J. (2011). Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10, 1100–1109. doi: 10.1128/EC.05003-11
Georgianna, D. R., Fedorova, N. D., Burroughs, J. L., Dolezal, A. L., Bok, J. W., Horowitz-Brown, S., et al. (2010). Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11, 213–226. doi: 10.1111/j.1364-3703.2009.00594.x
Glass, N. L., Rasmussen, C., Roca, M. G., and Read, N. D. (2004). Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12, 135–141. doi: 10.1016/j.tim.2004.01.007
Greene, A. V., Keller, N., Haas, H., and Bell-pedersen, D. (2003). A circadian oscillator in Aspergillus spp. regulates daily development and gene expression. Eukaryot. Cell 2, 231–237. doi: 10.1128/EC.2.2.231
Guo, L., Wenner, N., and Kuldau, G. A. (2015). FvSO regulates vegetative hyphal fusion, asexual growth, fumonisin B1 production, and virulence in Fusarium verticillioides. Fungal Biol. 119, 1158–1169. doi: 10.1016/j.funbio.2015.08.013
He, Z.-M., Price, M. S., Obrian, G. R., Georgianna, D. R., and Payne, G. A. (2007). Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 7:104. doi: 10.1186/1471-2180-7-104
Herzog, S., Schumann, M. R., and Fleißner, A. (2015). Cell fusion in Neurospora crassa. Curr. Opin. Microbiol. 28, 53–59. doi: 10.1016/j.mib.2015.08.002
Horn, B. W., Dorner, J. W., Greene, R. L., Blankenship, P. D., and Cole, R. J. (1994). Effect of Aspergillus parasiticus soil inoculum on invasion of peanut seeds. Mycopathologia 125, 179–191. doi: 10.1007/BF01146524
Horn, B. W., Gell, R. M., Singh, R., Sorensen, R. B., Carbone, I., and Pan, Q. (2016). Sexual reproduction in Aspergillus flavus sclerotia: acquisition of novel alleles from soil populations and uniparental mitochondrial inheritance. PLOS ONE 11:e0146169. doi: 10.1371/journal.pone.0146169
Jamet-Vierny, C., Debuchy, R., Prigent, M., and Silar, P. (2007). IDC1, a Pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet. Biol. 44, 1219–1230. doi: 10.1016/j.fgb.2007.04.005
Jonkers, W., Leeder, A. C., Ansong, C., Wang, Y., Yang, F., Starr, T. L., et al. (2014). HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa. PLOS Genet. 10:e1004783. doi: 10.1371/journal.pgen.1004783
Jurick, W. M. II., and Rollins, J. A. (2007). Deletion of the adenylate cyclase (sac1) gene affects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum. Fungal Genet. Biol. 44, 521–530. doi: 10.1016/j.fgb.2006.11.005
Kale, S. P., Milde, L., Trapp, M. K., Frisvad, J. C., Keller, N. P., and Bok, J. W. (2008). Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45, 1422–1429. doi: 10.1016/j.fgb.2008.06.009
Kohn, L. M., Carbone, I., and Anderson, J. B. (1990). Mycelial interactions in Sclerotinia sclerotiorum. Exp. Mycol. 267, 255–267. doi: 10.1016/0147-5975(90)90023-m
Laakso, J. A., Narske, E. D., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1994). Isokotanins A-C: new bicoumarins from the sclerotia of Aspergillus alliaceus. J. Nat. Prod. 57, 128–133. doi: 10.1021/np50103a018
Lacaze, I., Lalucque, H., Siegmund, U., Silar, P., and Brun, S. (2015). Identification of NoxD/Pro41 as the homologue of the p22phox NADPH oxidase subunit in fungi. Mol. Microbiol. 95, 1006–1024. doi: 10.1111/mmi.12876
Lara-Ortíz, T., Riveros-Rosas, H., and Aguirre, J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50, 1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x
Lee, S. R., Lee, S., Moon, E., Park, H.-J., Park, H. B., and Kim, K. H. (2017). Bioactivity-guided isolation of anti-inflammatory triterpenoids from the sclerotia of Poria cocos using LPS-stimulated Raw264.7 cells. Bioorg. Chem. 70, 94–99. doi: 10.1016/j.bioorg.2016.11.012
Lichius, A., and Lord, K. M. (2014). Chemoattractive mechanisms in filamentous fungi. Open Mycol. J. 8, 28–57. doi: 10.2174/1874437001408010028
Lim, F. Y., Sanchez, J. F., Wang, C. C., and Keller, N. P. (2012). Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 517, 303–324. doi: 10.1016/B978-0-12-404634-4.00015-2
Maddi, A., Dettman, A., Fu, C., Seiler, S., and Free, S. J. (2012). WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa. PLOS ONE 7:e42374. doi: 10.1371/journal.pone.0042374
Montenegro-Montero, A., Canessa, P., and Larrondo, L. F. (2015). Around the fungal clock: recent advances in the molecular study of circadian clocks in Neurospora and other fungi. Adv. Genet. 92, 107–184. doi: 10.1016/bs.adgen.2015.09.003
Nowrousian, M., Frank, S., Koers, S., Strauch, P., Weitner, T., Ringelberg, C., et al. (2007). The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol. Microbiol. 64, 923–937. doi: 10.1111/j.1365-2958.2007.05694.x
Payne, G. A., Nystrom, G. J., Bhatnagar, D., Cleveland, T. E., and Woloshuk, C. P. (1993). Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 59, 156–162.
Pfannenstiel, B. T., Zhao, X., Wortman, J., Wiemann, P., Throckmorton, K., Spraker, J. E., et al. (2017). Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio 8:e01246-17. doi: 10.1128/mBio.01246-17
Runa, F., Carbone, I., Bhatnagar, D., and Payne, G. A. (2015). Nuclear heterogeneity in conidial populations of Aspergillus flavus. Fungal Genet. Biol. 84, 62–72. doi: 10.1016/j.fgb.2015.09.003
Schumacher, J., Simon, A., Cohrs, K. C., Traeger, S., Porquier, A., Dalmais, B., et al. (2015). The VELVET complex in the gray mold fungus Botrytis cinerea: impact of BcLAE1 on differentiation, secondary metabolism, and virulence. Mol. Plant Microbe Interact. 28, 659–674. doi: 10.1094/MPMI-12-14-0411-R
Scott, B. (2015). Conservation of fungal and animal nicotinamide adenine dinucleotide phosphate oxidase complexes. Mol. Microbiol. 95, 910–913. doi: 10.1111/mmi.12946
Segmüller, N., Kokkelink, L., Giesbert, S., Odinius, D., van Kan, J., and Tudzynski, P. (2008). NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol. Plant Microbe Interact. 21, 808–819. doi: 10.1094/MPMI-21-6-0808
Serrano, A., Hammadeh, H. H., Herzog, S., Illgen, J., Schumann, M. R., Weichert, M., et al. (2017). The dynamics of signal complex formation mediating germling fusion in Neurospora crassa. Fungal Genet. Biol. 101, 31–33. doi: 10.1016/j.fgb.2017.02.003
Shahi, S., Beerens, B., Bosch, M., Linmans, J., and Rep, M. (2016). Nuclear dynamics and genetic rearrangement in heterokaryotic colonies of Fusarium oxysporum. Fungal Genet. Biol. 91, 20–31. doi: 10.1016/j.fgb.2016.03.003
Shimizu, K., and Keller, N. P. (2001). Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157, 591–600.
Siegmund, U., Marschall, R., and Tudzynski, P. (2015). BcNoxD, a putative ER protein, is a new component of the NADPH oxidase complex in Botrytis cinerea. Mol. Microbiol. 95, 988–1005. doi: 10.1111/mmi.12869
Simonin, A. R., Rasmussen, C. G., Yang, M., and Glass, N. L. (2010). Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47, 855–868. doi: 10.1016/j.fgb.2010.06.010
Soukup, A. A., Farnoodian, M., Berthier, E., and Keller, N. P. (2012). NosA, a transcription factor important in Aspergillus fumigatus stress and developmental response, rescues the germination defect of a laeA deletion. Fungal Genet. Biol. 49, 857–865. doi: 10.1016/j.fgb.2012.09.005
Steffens, E. K., Becker, K., Krevet, S., Teichert, I., and Kuck, U. (2016). Transcription factor PRO1 targets genes encoding conserved components of fungal developmental signaling pathways. Mol. Microbiol. 102, 792–809. doi: 10.1111/mmi.13491
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2007). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Terhem, R. B., and van Kan, J. A. L. (2014). Functional analysis of hydrophobin genes in sexual development of Botrytis cinerea. Fungal Genet. Biol. 71, 42–51. doi: 10.1016/j.fgb.2014.08.002
Tsukasaki, W., Maruyama, J., and Kitamoto, K. (2014). Establishment of a new method to quantitatively evaluate hyphal fusion ability in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 78, 1254–1262. doi: 10.1080/09168451.2014.917262
Varga, J., Frisvad, J. C., and Samson, R. A. (2011). Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Stud. Mycol. 69, 57–80. doi: 10.3114/sim.2011.69.05
Vienken, K., and Fischer, R. (2006). The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 61, 544–554. doi: 10.1111/j.1365-2958.2006.05257.x
Wada, R., Jin, F. J., Koyama, Y., Maruyama, J., and Kitamoto, K. (2014). Efficient formation of heterokaryotic sclerotia in the filamentous fungus Aspergillus oryzae. Appl. Microbiol. Biotechnol. 98, 325–334. doi: 10.1007/s00253-013-5314-y
Wang, C.-L., Shim, W.-B., and Shaw, B. D. (2016). The Colletotrichum graminicola striatin orthologue Str1 is necessary for anastomosis and is a virulence factor. Mol. Plant Pathol. 17, 931–942. doi: 10.1111/mpp.12339
Whyte, A. C., Gloer, J. B., Wicklow, D. T., and Dowd, P. F. (1996). Sclerotiamide: a new member of the paraherquamide class with potent antiinsectan activity from the sclerotia of Aspergillus sclerotiorum. J. Nat. Prod. 59, 1093–1095. doi: 10.1021/np960607m
Wicklow, D. T., and Shotwell, O. L. (1983). Intrafungal distribution of aflatoxins among conidia and sclerotia of Aspergillus flavus and Aspergillus parasiticus. Can. J. Microbiol. 29, 1–5. doi: 10.1139/m83-001
Willetts, H. J., and Bullock, S. (1992). Developmental biology of sclerotia. Mycol. Res. 96, 801–816. doi: 10.1016/S0953-7562(09)81027-7
Wu, D., Oide, S., Zhang, N., Choi, M. Y., and Turgeon, B. G. (2012). ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus. PLOS Pathog. 8:e1002542. doi: 10.1371/journal.ppat.1002542
Xiang, Q., Rasmussen, C., and Glass, N. L. (2002). The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160, 169–180.
Keywords: hyphal fusion, aflatoxin, ham-6, ham-9, nosA, adv-1
Citation: Zhao X, Spraker JE, Bok JW, Velk T, He Z-M and Keller NP (2017) A Cellular Fusion Cascade Regulated by LaeA Is Required for Sclerotial Development in Aspergillus flavus. Front. Microbiol. 8:1925. doi: 10.3389/fmicb.2017.01925
Received: 01 August 2017; Accepted: 21 September 2017;
Published: 05 October 2017.
Edited by:
Sven Krappmann, University of Erlangen-Nuremberg, GermanyReviewed by:
Ozgur Bayram, Maynooth University, IrelandMalcolm Whiteway, Concordia University, Canada
Copyright © 2017 Zhao, Spraker, Bok, Velk, He and Keller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhu-Mei He, bHNzaGV6bUBtYWlsLnN5c3UuZWR1LmNu Nancy P. Keller, bnBrZWxsZXJAd2lzYy5lZHU=
 Xixi Zhao
Xixi Zhao Joseph E. Spraker3
Joseph E. Spraker3 Nancy P. Keller
Nancy P. Keller