- 1Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA, United States
- 2HGF MPG Joint Research Group for Deep Sea Ecology and Technology, Max Planck Institute for Marine Microbiology, Bremen, Germany
- 3Department for Microbiology, Max Planck Institute for Marine Microbiology, Bremen, Germany
- 4Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, United States
- 5Genome Center, Department of Evolution and Ecology, Department of Medical Microbiology and Immunology, University of California, Davis, Davis, CA, United States
- 6School of Integrative Plant Science, Cornell University, Ithaca, NY, United States
- 7Department of Microbiology, Cornell University, Ithaca, NY, United States
Microbial interactions driving key biogeochemical fluxes often occur within multispecies consortia that form spatially heterogeneous microenvironments. Here, we describe the “green berry” consortia of the Sippewissett salt marsh (Falmouth, MA, United States): millimeter-sized aggregates dominated by an uncultured, diazotrophic unicellular cyanobacterium of the order Chroococcales (termed GB-CYN1). We show that GB-CYN1 is closely related to Crocosphaera watsonii (UCYN-B) and “Candidatus Atelocyanobacterium thalassa” (UCYN-A), two groups of unicellular diazotrophic cyanobacteria that play an important role in marine primary production. Other green berry consortium members include pennate diatoms and putative heterotrophic bacteria from the Alphaproteobacteria and Bacteroidetes. Tight coupling was observed between photosynthetic oxygen production and heterotrophic respiration. When illuminated, the green berries became supersaturated with oxygen. From the metagenome, we observed that GB-CYN1 encodes photosystem II genes and thus has the metabolic potential for oxygen production unlike UCYN-A. In darkness, respiratory activity rapidly depleted oxygen creating anoxia within the aggregates. Metagenomic data revealed a suite of nitrogen fixation genes encoded by GB-CYN1, and nitrogenase activity was confirmed at the whole-aggregate level by acetylene reduction assays. Metagenome reads homologous to marker genes for denitrification were observed and suggest that heterotrophic denitrifiers might co-occur in the green berries, although the physiology and activity of facultative anaerobes in these aggregates remains uncharacterized. Nitrogen fixation in the surface ocean was long thought to be driven by filamentous cyanobacterial aggregates, though recent work has demonstrated the importance of unicellular diazotrophic cyanobacteria (UCYN) from the order Chroococcales. The green berries serve as a useful contrast to studies of open ocean UCYN and may provide a tractable model system to investigate microbial dynamics within phytoplankton aggregates, a phenomenon of global importance to the flux of particulate organic carbon and nitrogen in surface waters.
Introduction
Fixed nitrogen is often a limiting nutrient for primary productivity in the surface ocean, and consequently influences the dynamics of oceanic carbon sequestration (Karl et al., 2002). Nitrogen (N2) fixation by marine cyanobacteria is an important source of oceanic fixed nitrogen, adding an estimated 100–200 Tg-N annually to open ocean ecosystems (Karl et al., 2002; Galloway et al., 2004). This nitrogen fixation is often associated with cyanobacterial trichomes or aggregates colonized by heterotrophic bacteria, picoeukaryotes and metazoans (Paerl et al., 1989; Hewson et al., 2009; Ploug et al., 2010). Respiratory activity within these so-called ‘pseudo-benthic’ environments can create ephemeral suboxic to anoxic zones, establishing a niche for facultative anaerobes within otherwise oxygenated surface waters (Paerl and Prufert, 1987; Ploug et al., 2011; Klawonn et al., 2015). Emerging evidence suggests that denitrification occurs within these anoxic habitats, coupling processes of nitrogen-fixation and loss at the microscale (Ploug et al., 2011; Wyman et al., 2013; Klawonn et al., 2015).
While initial studies of marine biological nitrogen fixation focused on colonial filamentous Trichodesmium species (Capone et al., 1997) and symbiotic, heterocystous Richelia species (Foster and Zehr, 2006), more recent work has demonstrated the importance of unicellular diazotrophic cyanobacteria (UCYN) from the order Chroococcales (Montoya et al., 2004; Zehr et al., 2007). Diazotrophic UCYN have been studied extensively in the global oceans by surveys of the nitrogenase gene nifH diversity, which revealed three phylogenetically distinct clades (A-C) (Zehr et al., 2001; Langlois et al., 2005; Foster et al., 2007). UCYN-A are small (circa 1 μm), metabolically streamlined, uncultured cyanobacteria that lack the oxygen-producing photosystem II and live as endosymbionts within haptophytes, a lineage of eukaryotic algae (Zehr et al., 2008; Tripp et al., 2010; Thompson et al., 2012; Hagino et al., 2013). UCYN clades B and C are larger (>2 μm), free-living cyanobacteria and include cultured representatives, such as Crocosphaera watsonii and Cyanothece sp. ATCC51142.
Studies of aggregate-associated nitrogen fixation have focused predominantly on Trichodesmium sp. colonies and rafts (Paerl et al., 1989), or filamentous heterocystous cyanobacterial colonies (Ploug, 2008; Ploug et al., 2010; Klawonn et al., 2015). However, some Crocosphaera watsonii strains have been observed to produce copious quantities of exopolysaccharides and have been linked to the formation of transparent exopolymer particles (TEP) (Webb et al., 2009; Sohm et al., 2011). These gel-like particles provide microhabitats for other microorganisms, and thus have the potential to play an important role in marine biogeochemical cycling (Passow, 2002).
Here, we report a new species of uncultured, unicellular cyanobacteria from the order Chroococcales which forms millimeter-sized aggregates together with diatoms and other putatively heterotrophic bacteria. These macroscopic aggregates, which we call “green berries,” are found in the muddy, intertidal pools of Little and Great Sippewissett salt marshes (Falmouth, MA, United States). They are found interspersed with previously described, sulfur-cycling “pink berry” consortia (Seitz et al., 1993; Wilbanks et al., 2014). Using a combination of metagenomic sequencing and ecophysiological measurements, we demonstrate that the green berries are characterized by diazotrophy and rapid rates of photosynthesis and respiration that produce steep oxygen gradients. Heterotrophic bacteria within the green berries are closely related to other marine epiphytic marine strains and encode key genes in the denitrification pathway.
Results and Discussion
Morphological Description of the Aggregates
The green berries are found in the same organic-rich, intertidal pools of Little Sippewissett salt marsh on Cape Cod (MA, United States) where both multicellular magnetotactic bacteria and pink berries have been previously studied (Seitz et al., 1993; Shapiro et al., 2011; Wilbanks et al., 2014). Though less abundant than the pink berries found in these pools (Figure 1A), the green berries form similar irregular ellipsoid aggregates measuring 1–8 mm in diameter, with an average equivalent spherical diameter of 1.7 mm ± 0.1 mm (standard deviation, Figures 1A,D). Green berries were dense and compact aggregates that were typically observed at the sediment-water interface, but were occasionally found to float at the water surface when suspended by bubbles. Microscopic observation of the green berries revealed abundant coccoid unicellular cyanobacteria 5–7 μm in diameter (which we call GB-CYN1, “green berry cyanobacteria 1”), interspersed with pennate diatoms (Figures 1B,C). Filamentous cyanobacteria were observed occasionally, but were rare compared to the unicellular GB-CYN1 morphotype. A clear, extracellular matrix (putatively exopolysaccharides) coated these aggregates of phototrophic cells, and was colonized by a variety of smaller bacteria (Figures 1B,C). GB-CYN1 exhibited absorption maxima at 620, 660, and 680 nm corresponding to the presence of phycocyanin, allophycocyanin and chlorophyll a, respectively.
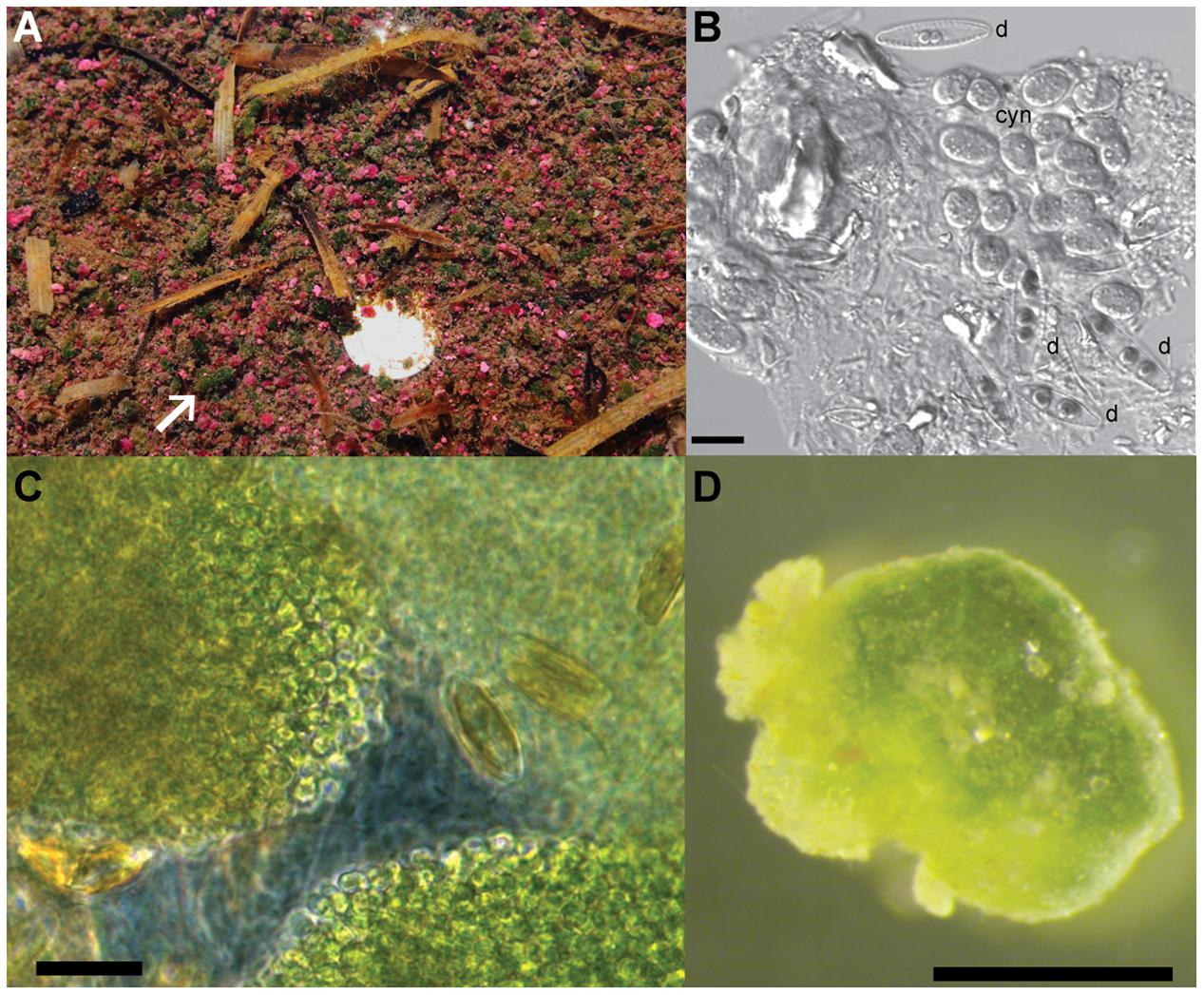
FIGURE 1. Morphology of the green berries. (A) Pink and green berries at the sediment water interface of intertidal pools in the Little Sippewissett salt marsh. White arrow indicates a large green berry and a dime (bright spot) is provided for scale. (B) Differential interference contrast (DIC) micrograph of cells from a homogenized green berry showing two different pennate diatom morphotypes (d) and clumps of coccoid unicellular cyanobacteria 5–7 μm in diameter (cyn). Scale bar = 10 μm. (C) Phase contrast image of an intact green berry aggregate compressed under a coverslip. Note the dense clumps of coccoid cyanobacterial cells and pennate diatoms interspersed in a clear exopolymer matrix. Scale bar = 10 μm. (D) Dissecting microscope image of a single green berry aggregates. Scale bar = 1 mm.
Microbial Diversity of the Green Berry Aggregates
Sequencing of 18S rRNA genes from the green berries indicated that the eukaryotic community was predominantly made up of two different pennate diatom species related to Navicula cari strain AT-82.04c (96% sequence identity) and Amphora pediculus strain AT-117.11 (95% sequence identity; Supplementary Figure S1). These same diatom species were also the dominant eukaryotic 18S rRNA gene sequences recovered from pink berry aggregates, though diatoms were more abundant in green berries than in pink berries, as observed by microscopy and the relative abundance of 16S rRNA chloroplast sequences (Wilbanks et al., 2014).
Bacterial 16S rRNA gene sequences amplified from the green berries were dominated by sequences related to either diatom chloroplasts (phylum Bacillariophyta, 18/92 clones, representing 4 OTUs at 97% similarity threshold) or Chroococcales unicellular cyanobacteria (18/92 clones, representing 2 OTUs; Figure 2). Unassembled metagenomic sequence reads assigned to rRNA sequences and protein-coding regions support the observed abundance of Chroococcales (accounting for up to 37% relative sequence abundance), but did not recover comparable proportions of diatom chloroplasts (<5% relative sequence abundance; Figure 2).
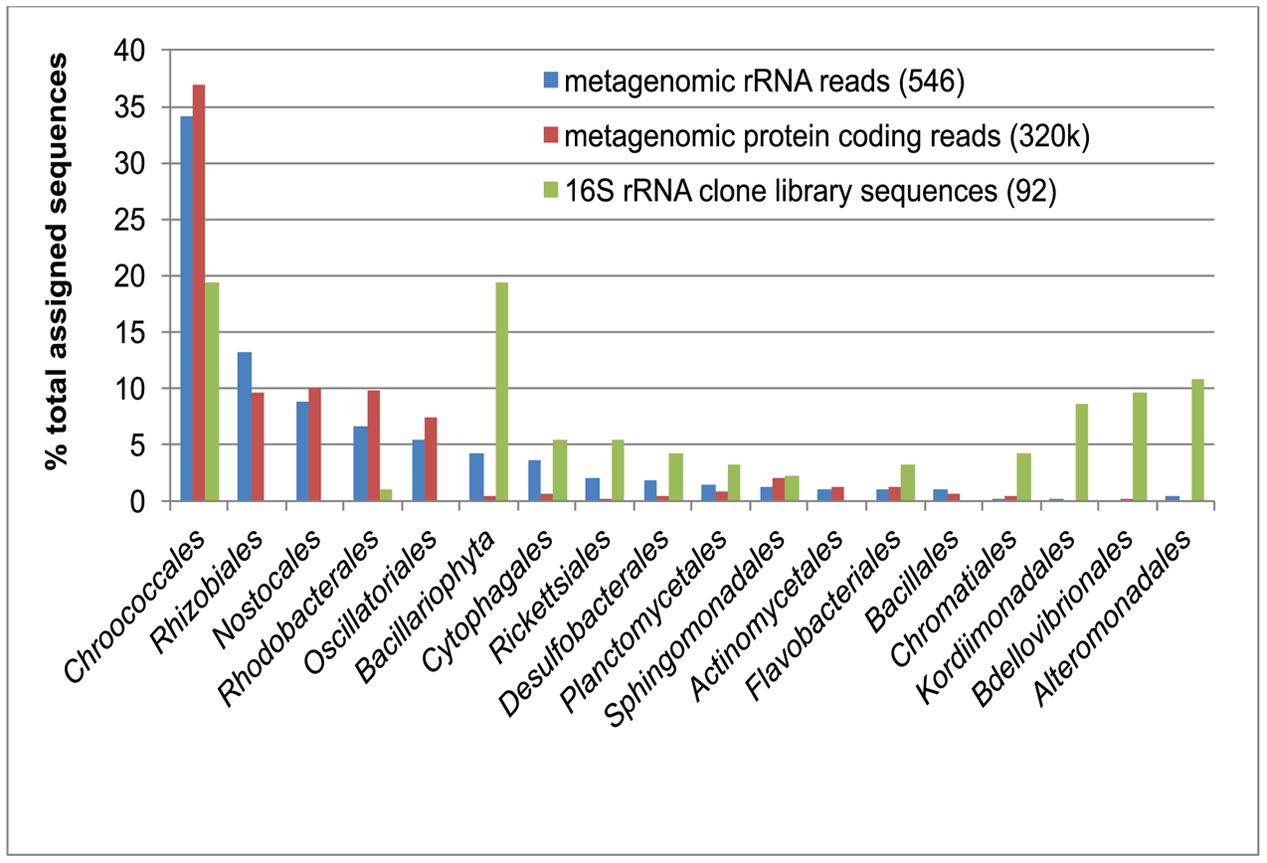
FIGURE 2. Comparison of green berry bacterial diversity estimates from16S rDNA PCR amplified clones library (green bars; 92 clone sequences) with unassembled Roche 454 metagenomic sequence reads. Taxonomic assignment of metagenomic reads matching ribosomal RNA reads (16S/18S/23S) was conducted using the M5RNA database in MG-RAST (blue bars; 546 assigned reads). A similar taxonomic assignment was conducted with metagenomic reads matching protein coding sequences in the M5NR database (red bars; 319,692 assigned reads). Note that the 16S rRNA clone library abundance data for the Bacillariophyta (green bar, asterisk) was obtained from diatom chloroplasts sequences, which are likely present in multiple copies in the cell and thus not directly comparable to metagenomic 18S rRNA sequences for this group (blue bar).
The overall bacterial community structure of the green berries was significantly different from coexisting pink berry consortia (unweighted UniFrac analysis, Bonferroni corrected p-value < 0.01). Some abundant taxa from the pink berries co-occurred in the green berries as rare OTUs, such as the purple sulfur bacterial species Thiohalocapsa sp. PB-PSB1 (1/92 clones), and a Winogradskyella species (Flavobacteriales, 2/92 clones) (Wilbanks et al., 2014). The persistence of these distinct, co-occurring pink and green berry consortia suggests that the process of macroscopic aggregation enables niche partitioning between oxygenic (green berries) and anoxygenic (pink berries) phototrophs in these marsh pools.
Most of the non-cyanobacterial sequences in the green berry consortia are related to aerobic and facultatively anaerobic marine heterotrophs from the Bacteroidetes, Alphaproteobacteria, and Gammaproteobacteria (Figure 2). Many of these sequences (e.g., taxa from the Rhodobacterales, Kordiimonadales, Sphingomonadales, and Flavobacterales) were most closely related to environmental 16S rRNA sequences associated with aggregates of oxygenic phototrophs. Examples of such habitats included phytodetrital aggregates (marine snow) collected from euphotic and hadal environments (DeLong et al., 1993; Eloe et al., 2011), and epiphytes of marine macroalgae (Burke et al., 2011; Fernandes et al., 2012). The occurrence of related phylotypes in such environments suggests that taxa may be well adapted to an attached lifestyle, degradation of photosynthate, and the fluctuating oxygen conditions in an aggregate environment.
Metagenomic data indicate that the orders Rhizobiales and Rhodobacterales of the Alphaproteobacteria are abundant in the green berry consortia. While these groups were rarely detected in the PCR-based 16S rRNA survey, we have previously observed this same PCR bias from the 8F primer during studies of the pink berry consortia (Wilbanks et al., 2014). We find the abundance of these clades in the green berries particularly interesting as they include lineages of marine denitrifying bacteria. For example, pelagic Rhizobiales (e.g., Labrenzia and Roseibium species) have been linked to denitrification when found in association with macroscopic Trichodesmium sp. aggregates in oxic waters bordering oxygen minimum zones (Wyman et al., 2013).
Phylogenomic Analysis of the Green Berry Cyanobacteria, GB-CYN1
The cyanobacterial 16S rRNA gene sequences from the green berries grouped into two closely related OTUs (97% similarity threshold), GB-CYN1a and GB-CYN1b, that can be confidently placed in the order Chroococcales (Figure 3). The GB-CYN1 monophyletic cluster formed a clade basal to the UCYN-A clade (96% sequence identity to CP001842, “Candidatus Atelocyanobacterium thalassa” isolate ALOHA). Using 29 concatenated single-copy phylogenetic marker genes (Wu et al., 2013) assembled from the metagenome (Supplementary Table S1), we reconstructed a phylogenetic tree that placed the GB-CYN1 within a clade including Crocosphaera watsonii and Cyanothece sp. ATCC 51142 as a sister taxa to “Candidatus Atelocyanobacterium thalassa” isolate ALOHA (Supplementary Figure S2).
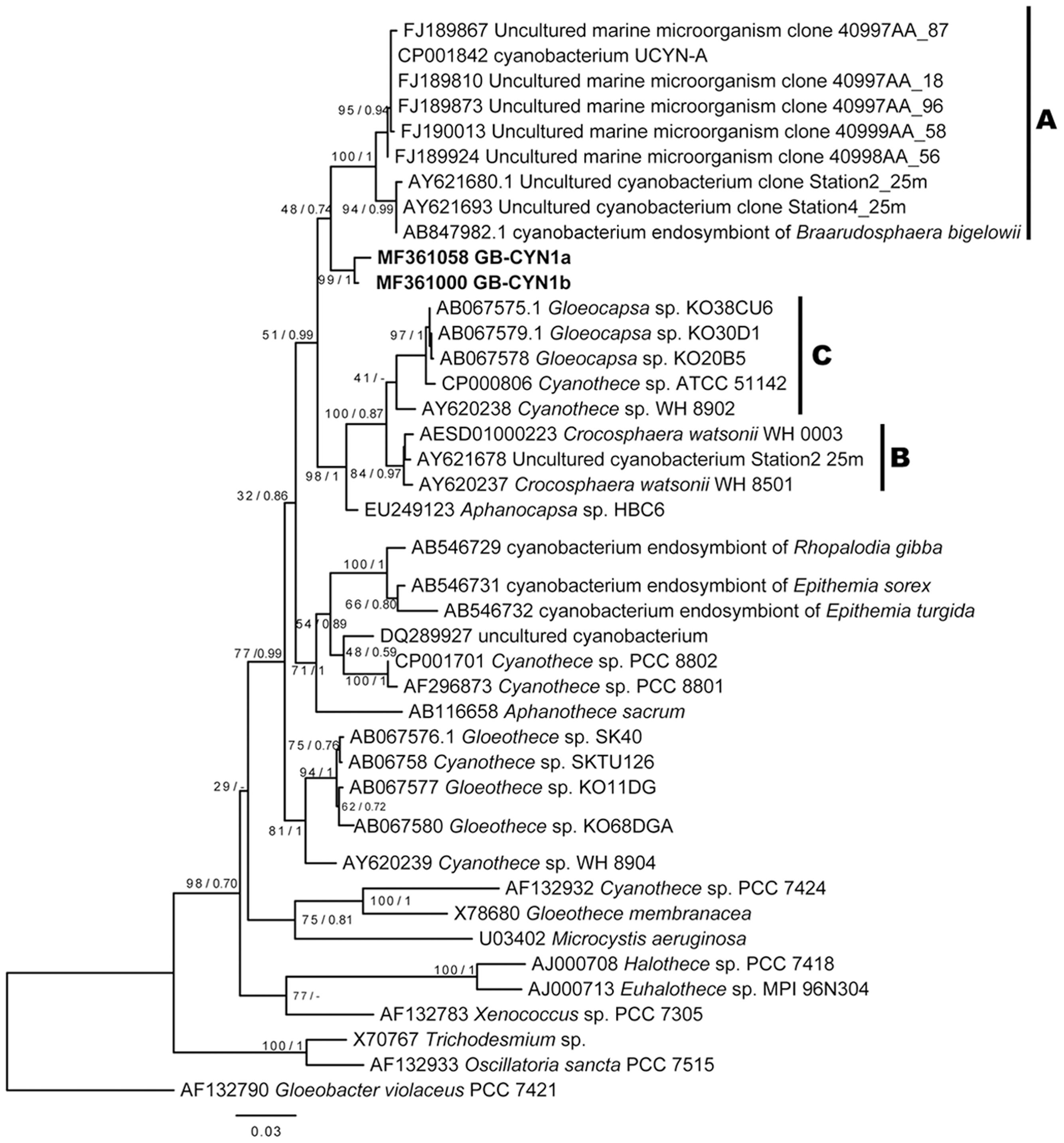
FIGURE 3. Maximum likelihood and Bayesian 16S rRNA gene phylogenies of GB-CYN1 species and related cyanobacteria. Reference sequences were full length and GB-CYN1 OTUs (bold) were partial (∼700–800 bp). Constructed from (A) 16S rRNA sequences from (partial sequences, ∼700–800 bp, bold) and Numbers at the nodes indicate bootstrap values from maximum likelihood analysis (1000 replicates) and the posterior probability of the Bayesian tree inference. The Bayesian consensus tree was concordant with maximum likelihood tree topology except where indicated by a dash. Scale bar represents the mean number of nucleotide substitutions per site. The major clades of unicellular cyanobacteria (UCYN-A-C) are labeled according to (Zehr et al., 2001; Langlois et al., 2005; Foster et al., 2007).
A phylogenetic tree inferred from nifH gene sequences reveals that the near full-length nifH gene recovered from the GB-CYN1 metagenomic data affiliated with the UCYN-B clade, and was most closely related to Cyanothece sp. 8801/8802 and Crocosphaera watsonii (Figure 4). We conclude that the observed discordance between 16S rRNA, concatenated, and nifH gene phylogenies involving species such as Cyanothece sp. 8801, Gloeothece sp. KO68DGA, and the cyanobacterial endosymbiont of Rhopalodia gibba is most likely due to lateral gene transfer of the nifH gene. Lateral transfer of nifH has been observed in many other species, including mat-forming filamentous cyanobacteria (Cantera et al., 2004; Bolhuis et al., 2010; Parker, 2012; Gaby and Buckley, 2014; Meyer and Huber, 2014).
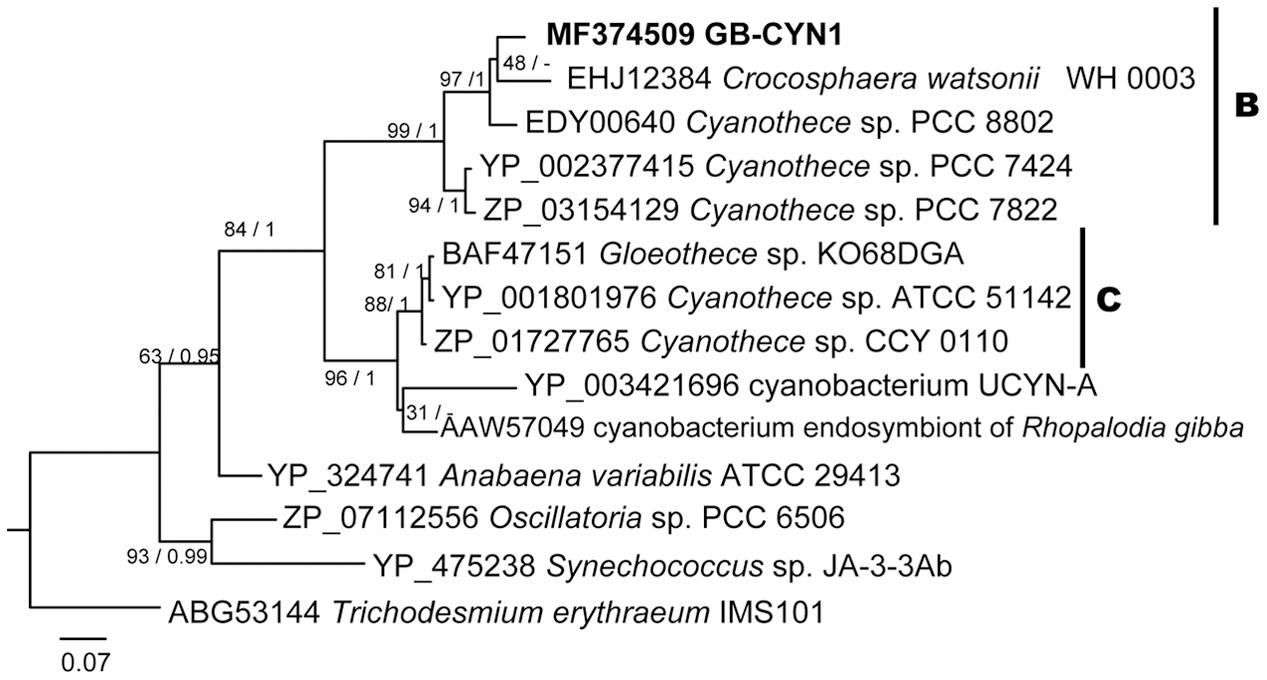
FIGURE 4. Maximum likelihood and Bayesian NifH phylogenies of GB-CYN1 species and related cyanobacteria. Near-full length NifH amino acid sequence from GB-CYN1 (267aa, from assembled metagenomic contig) was aligned with full length reference sequences. Numbers at the nodes indicate bootstrap values from maximum likelihood analysis (1000 replicates) and the posterior probability of the Bayesian tree inference. The Bayesian consensus tree was concordant with maximum likelihood tree topology except where indicated by a dash. Scale bar represents the mean number of amino acid substitutions per site. The major clades of unicellular cyanobacteria (UCYN-A-C) are labeled according to Zehr et al. (2001), Langlois et al. (2005), and Foster et al. (2007).
Diazotrophy in the Green Berries
A full suite of nitrogenase genes were found in the green berry metagenome and were consistently assigned to GB-CYN1, indicating the metabolic potential for nitrogen fixation characteristic of other members of the UCYN A-C clades (Supplementary Table S1). Diazotrophy in the green berries was detected in whole aggregates by acetylene reduction. In two separate experiments (containing 5 berries each), we measured rates of 11 and 20 nanomoles acetylene reduced per hour per milligram of aggregate dry weight (nmol mg-1 hr-1). These rates are comparable, though faster than the rates of 3–6 nmol mg-1 hr-1 reported in macroscopic aggregates of filamentous cyanobacteria from Bogue Sound, North Carolina (Paerl and Prufert, 1987). Rates measured from actively growing Cyanothece cultures (Reddy et al., 1993) were two orders of magnitude larger (∼1000 nmol mg-1 hr-1) than the rates in the green berries.
The total aggregate elemental composition was analyzed using elemental analyzer isotope ratio mass spectrometry (EA-IRMS). The mean observed C:N ratio in the green berries, 7.1 ± 0.6 (n = 6), falls within range of the Redfield molar ratio (Redfield et al., 1963). This observed C:N ratio is higher than that the ratio of 5.4 ± 0.4 (n = 50) observed in similarly large, anoxic, diazotrophic Nodularia spumigena aggregates from the Baltic Sea (Klawonn et al., 2015). Cultures of Crocosphaera watsonii exhibit wide diel fluctuations in C:N ratios (∼5 at dawn to ∼9 at dusk) as a result of temporal partitioning of carbon (day) and nitrogen (night) fixation activities (Dron et al., 2012, 2013). Our samples, collected in the late afternoon on a 14 h light/10 h dark photoperiod, are comparable to reports of C:N = 7 from C. wastsonii at similar late afternoon times in a 16 h light/8 h dark photoperiod (Dron et al., 2013). Future studies investigating the temporal partitioning for such activities in the green berry aggregate would be informative to elucidate the dynamics of carbon and nitrogen flow in the consortia.
Respiration and Photosynthesis
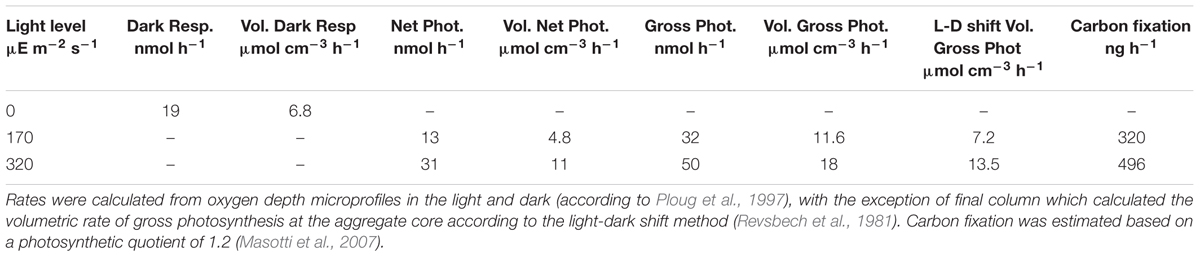
Oxygen microsensors were used to characterize the balance between respiration and photosynthesis in the green berry aggregates. Aggregates examined were relatively symmetric ellipsoids of similar size with an equivalent spherical diameter (ESD) of 1.7 ± 0.1 mm (n = 5, average ± standard deviation). Photosynthesis produced supersaturated oxygen concentrations within the aggregates: 380 μM O2 with illumination at 170 μE m-2 s-1 (one lamp) and 520 μM O2 at 320 μE m-2 s-1 (two lamps; Figure 5). During these experiments, bubbles were not observed on aggregate surfaces and the aggregates never floated. Oxygen production rates per aggregate (net photosynthesis) were calculated from these profiles as 13 and 31 nmol O2 per hour at 170 μE m-2 s-1 and 320 μE m-2 s-1, respectively (Table 1). We did not rigorously determine the saturating light intensity; however, we observed that illumination with a third lamp failed to stimulate increased oxygen production beyond that with two lamps (320 μE m-2 s-1), suggesting that the saturating light intensity lies in the range of 170 – 320 μE m-2 s-1 (data not shown).
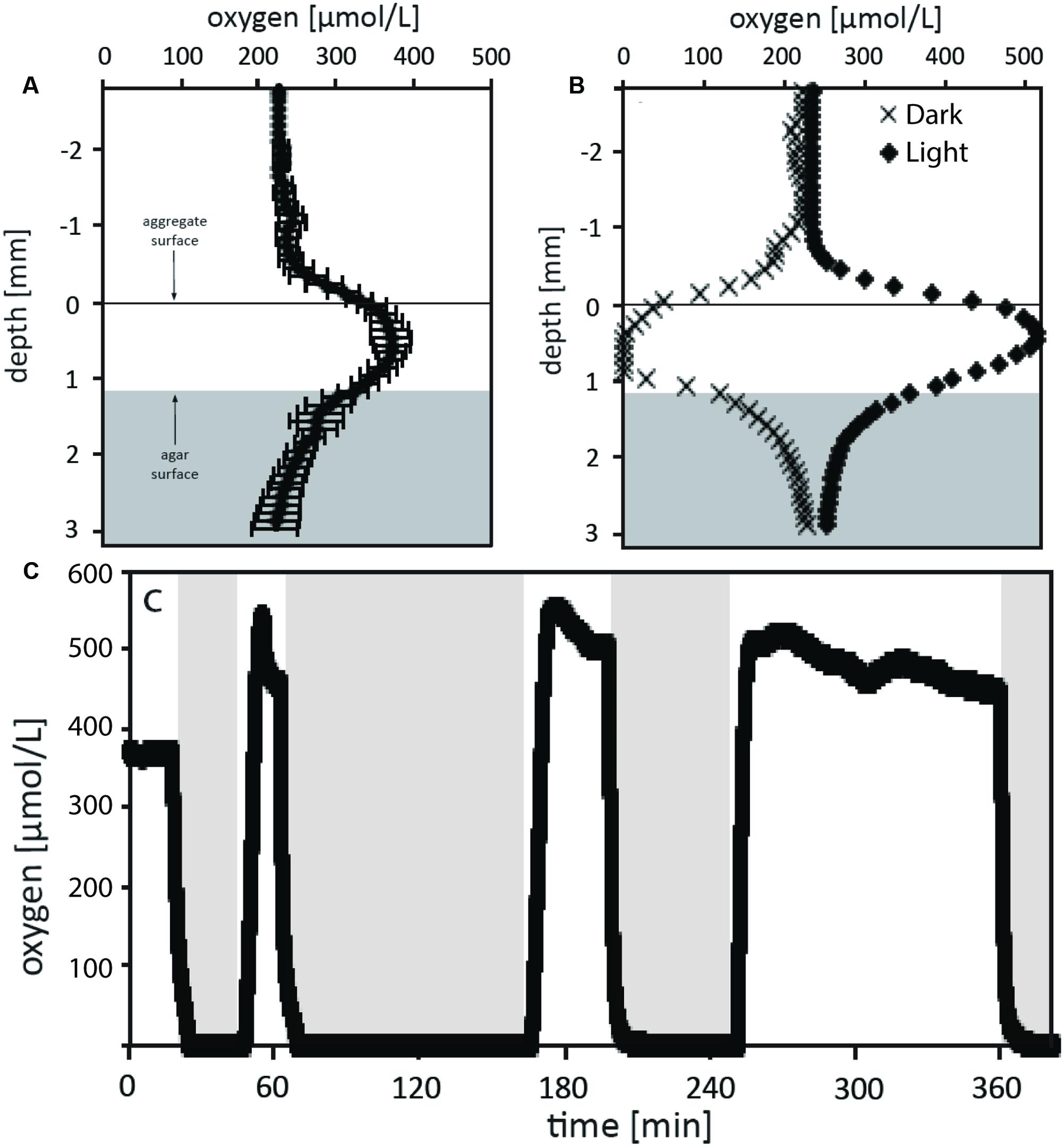
FIGURE 5. Oxygen measurements of the green berry aggregates in light and dark conditions. (A) Depth microprofiles with oxygen microsensors were conducted for 3 similar aggregates under steady state illumination with 170 μE m-2 s-1. The average O2 concentration versus depth relative to aggregate surface is plotted for these aggregates. (B) O2 profiles were measured for an aggregate under conditions of steady state illumination at 320 μE m-2 s-1 (black diamonds) and steady state darkness (black Xs). (C) The O2 microsensor was positioned at the center of an aggregate which was then exposed to series of different illumination conditions over a time course while concentration was monitored. Initial conditions recorded were at 170 μE m-2 s-1 followed by shift to darkness, and three subsequence rounds of illumination at 320 μE m-2 s-1 followed by dark shifts. Dark periods are indicated by the position of the light gray boxes.
The green berries were anoxic in darkness with a dark respiration rate of 19 nmol per aggregate per hour (Figure 5B). This rate, equivalent to a volumetric rate of 6.8 μmol cm-3 hr-1, is well above the theoretical threshold for respiration rates capable of causing oxygen diffusional limitation from surrounding seawater (5.0 μmol cm-3 hr-1 for ESD = 1.7 mm; calculated after Ploug et al., 1997). In light-dark shifts, we observed a rapid response where the aggregate core transitioned from steady state supersaturation to full anoxia in 5–8 min (Figure 5C).
Volumetric gross photosynthetic rates were calculated by two methods: (1) from depth microprofiles via the sum of net photosynthesis and dark respiration and (2) via the light-dark shift technique (Revsbech et al., 1981) performed at a single point in the aggregate center (Table 1). At both light intensities examined, rates calculated via the light-dark shift method were found to be 4.5 μmol cm-3 hr-1, lower than those from depth microprofiles. While this difference could arise from biological variability between aggregates, we suspect that the light-dark shift rates measured at the aggregate core were lower than those we might have measured closer to the aggregate surface. Future depth integrated studies of photosynthetic rates will help to clarify this difference and allow better characterization of respiratory activity in the light.
Comparing the green berries’ dark respiration and gross photosynthesis to other photosynthetic mats and aggregates, we find them similar to the high rates measured for large (>1 mm), filamentous aggregates of the heterocystous cyanobacterium, Nodularia spumigena from the Baltic Sea (Ploug et al., 2011). Indeed, our estimates of carbon fixation (Table 1) are close to prediction of 349 ng C per aggregate per hour calculated using Ploug et al.’s (2011) regression of volume to gross photosynthesis from a 2009 Nodularia bloom. The green berry dark respiration rate (19 nmol agg-1 h-1) was similar, though slightly higher than that observed for Nodularia aggregates of similar diameter; however, the ratio of dark respiration to gross photosynthesis of 0.38 (for an aggregate volume of 2.8 mm3) was identical to that observed for 7 mm3 aggregates found late in the bloom (Ploug et al., 2011). In large Nodularia aggregates, dark anoxia is associated with active dissimilatory nitrogen cycling, including denitrification, dissimilatory reduction of nitrate to ammonia (DNRA), and significant rates of cryptic nitrification (Klawonn et al., 2015).
Ecophysiological Implications from Metagenome Data
Our metagenomic data analysis predicts genes in GB-CYN1 involved in the Calvin cycle, TCA cycle, and photosystems I and II (Supplementary Table S1). Sequence identities for these genes in GB-CYN1 and published Chroococcales genomes were high, ranging from 75 to 99% (average of 85% for assembled contigs, Supplementary Table S1). The observation of photosystem II genes in GB-CYN1 indicates the metabolic potential for oxygenic photosynthesis, unlike UCYN-A which lacks photosystem II, RuBisCo and the TCA cycle. These findings confirm that GB-CYN1 resembles free-living Chroococcales of the clades B and C (Welsh et al., 2008; Bandyopadhyay et al., 2011; Bench et al., 2011, 2013) rather than the metabolically streamlined, endosymbiotic UCYN-A clade (Zehr et al., 2008; Tripp et al., 2010; Thompson et al., 2012; Hagino et al., 2013).
Related Chroococcales species are known to produce copious quantities of exopolysaccharides (EPS) (Pereira et al., 2009; Sohm et al., 2011; Mota et al., 2013), and EPS produced by Crocosphaera watsonii has been linked to the formation of transparent exopolymeric particles in pelagic environments (Passow, 2002; Webb et al., 2009; Sohm et al., 2011). A cassette of genes required for EPS production has been identified by comparative genomic analysis of related Crocosphaera watsonii strains (Webb et al., 2009; Bench et al., 2011, 2013); however, we did not recover homologs of these genes in our metagenomic sequence (either assembled contigs or sequence reads). We conclude that the absence of these sequences is most likely due to our incomplete sampling of the GB-CYN1 genome, though it could also indicate either an unknown pathway for EPS synthesis in GB-CYN1, or an alternate source of the green berry exopolymeric matrix (e.g., diatoms).
Consistent with the dynamic oxygen conditions within the green berries, we found sequences in the metagenomic data that suggest the metabolic potential for aerobic and anaerobic respiration, and anaerobic fermentative pathways assigned to several different phyla of bacteria (data not shown). Abundant fixed nitrogen from diazotrophy and transient anoxia presents an ecological opportunity for dissimilatory nitrogen metabolism, a process that could cause concomitant nitrogen fixation and loss over rapid spatiotemporal scales. Indeed, previous studies of diazotrophic Nodularia spumigena aggregates measured significant rates of both nitrification and denitrification (Klawonn et al., 2015).
We specifically investigated potential marker genes for dissimilatory nitrogen metabolism within the green berries. Metagenomic sequences homologous to the nitrite reductase gene nirK were found in six unassembled reads and on a corresponding 753 bp long contig (Supplementary Table S2). A single read homologous to nitrous oxide reductase nosZ was also found suggesting the presence of a denitrification pathway (Supplementary Table S2). The top database matches to these nirK and nosZ sequences belonged to marine phytoplankton epiphytes from the Alphaproteobacteria (nirK) and Flavobacterales (nosZ). In the case of the nirK metagenomic sequences, the best match was to Roseibium sp. TrichSKD4, an alphaproteobacterial species isolated from a nitrogen-fixing Trichodesmium aggregate in the Atlantic Ocean (Mann and Barbeau, 2014). Homologs to the napA periplasmic nitrate reductase and the nirB nitrite reductase were identified from six and five unassembled reads, respectively, and were most similar to database sequences from marine heterotrophs in the Alphaproteobacteria (Rhodobacterales, Rhizobiales) and Gammaproteobacteria (Oceanospirillales; Supplementary Table S2). We did not detect sequences supporting the presence of nitrifying bacteria or archaea, though given our limited sequence depth, this could be a function of missing data.
Conclusion
Cyanobacterial nitrogen fixation in the global oceans is frequently aggregate-associated, as with Trichodesmium sp. colonies and rafts (Paerl et al., 1989), filamentous heterocystous cyanobacterial colonies (Ploug, 2008; Ploug et al., 2010), or Crocosphaera watsonii associated with TEP (Webb et al., 2009; Sohm et al., 2011). The green berries of the Sippewissett Salt Marsh are nitrogen-fixing macroscopic consortia of unicellular cyanobacteria (GB-CYN1), diatoms and heterotrophic bacteria. While nitrogen and carbon fixation mediated by the green berries is unlikely to play a major role in the overall marsh ecosystem due to their low abundance and patchy distribution (Carpenter et al., 1978; Howarth et al., 1988), these consortia provide an interesting comparative system to investigate the dynamics nitrogen flux within diazotrophic cyanobacterial aggregates. More broadly, studies of coastal marine estuarine sediments have indicated complex dynamics and close spatial coupling of co-occurring nitrogen fixation and denitrification processes (Fulweiler et al., 2013).
In other oceanic diazotrophic cyanobacterial aggregates, similarly rapid respiratory rates create transient anoxic zones within the aggregate core (Paerl and Bebout, 1988; Ploug et al., 2011; Klawonn et al., 2015), and create a heterogeneous microenvironment where both aerobic and anaerobic metabolisms co-exist. This parallel is not merely conceptual: the green berry heterotrophic bacteria were similar, both phylogenetically and in their metabolic marker genes, to those observed to colonize other marine phytoplankton aggregates. The recovery of denitrification marker gene sequences in metagenome suggests that there might be heterotrophic denitrifiers in the green berries with the potential to exploit this suboxic niche. However, further studies including rate measurements and better genome reconstructions are needed to clarify the importance of denitrification in the berries.
The presence of such a pathway for nitrogen loss in the green berries is speculative, given the fragmented metagenome and absence of activity measurements, but remains an interesting direction for future work. The existence of closely coupled nitrogen cycling within diazotrophic cyanobacterial aggregates has been explored previously, though initial studies demonstrating the association of heterocystous cyanobacterial aggregates with key bacterial species and marker genes for nitrification and denitrification measured only low to negligible rates (Hietanen et al., 2002; Tuomainen et al., 2003, 2006). However, more recent investigations of in vitro rates and in situ marker gene transcription indicates that denitrification within anoxic cyanobacterial aggregates could contribute significantly to nitrogen loss in hypoxic waters, where bulk oxygen concentrations would normally inhibit such activity (Wyman et al., 2013; Klawonn et al., 2015). In the light of modeling studies showing tight spatiotemporal coupling of nitrogen fixation and denitrification processes in the global ocean (Deutsch et al., 2007), we postulate that this coupling may be occurring at the microscale within ephemeral, aggregate-associated niches. The potential for such interactions emphasizes the need to examine biogeochemical cycles from the microbial perspective (nanometers to millimeters) in the spatially heterogeneous marine environment.
Experimental Procedures
Sampling and Microscopy
Green berries were sampled in June and July of 2010 (for all studies except O2 microsensor work) or July and August 2014 (for oxygen microsensors) from a single intertidal pool formed in the Little Sippewissett Salt Marsh, Falmouth, MA United States (41°34′33.01″N, 70°38′21.24″W). These aggregates, however, have also been observed at other locations throughout Little and Great Sippewissett marshes. Berries were collected from the sediment-water interface by sieving (1 mm mesh size) and were washed three times in 0.2 μm filter sterilized marsh water. Imaging of the berries was conducted using a Zeiss Axio IMAGER MZ epifluorescence microscope equipped with a color camera (AxioCam HRc, Zeiss) and a Zeiss LSM 710 spectral confocal scanning laser microscope (Carl Zeiss, Oberkochen, Germany).
DNA Sequencing and Assembly
DNA extraction, 16S/18S rRNA PCR, and clone libraries were performed as described previously (Wilbanks et al., 2014). Roche GC Titanium 454 metagenomic sequencing was performed as described previously (Wilbanks et al., 2014), generating 100 megabases (Mbp) of sequence data after quality filtering (275,263 reads of average length = 365 bp). Metagenomic data was assembled using Newbler 2.3 (Margulies et al., 2005), generating a 4.8 Mbp assembly in 6,113 contigs of N50 equal to 941 bp and a maximum contig size of 7,845 bp. Data are publicly available at MG-RAST (reads: mgm4454183.3; assembly: mgm4454167.3). 16S/18S rRNA gene sequence from clone libraries available in NCBI GenBank (MF360994-MF361083; MF372423-MF372506). Metagenome data is associated with NCBI BioProject number PRJNA390846 and raw sequence read data is available in the NCBI SRA (SRR5710041). For the assembled metagenome, a Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession NJIW00000000. The version described in this paper is version NJIW01000000.
Sequence Analysis and Phylogenetic Reconstruction
The MG-RAST 3.3 pipeline was used to classify unassembled metagenomic sequence reads as ribosomal RNA and protein coding sequences using the M5RNA and M5NR databases to provide a diversity description shown in Figure 2 (Meyer et al., 2008). Metagenomic sequence data (both assembled sequences and unassembled reads) was mined using MG-RAST. Functions of interest were mined using the hierarchical function assignment and were assigned to GB-CYN1 when the best hits (amino acid similarity search) were to sequenced Chroococcales genomes. Marker genes were further investigated via profile HMM (Eddy, 2011) and BLASTX searches (Altschul et al., 1990).
PCR amplified SSU rRNA gene sequences were aligned to the SILVA 115 database (Quast et al., 2013) using SINA (Pruesse et al., 2012) and curated using ARB (Ludwig et al., 2004). 16S rRNA phylogenies were inferred using the GTRGAMMA rate approximation. For functional genes of interest that were found to have frameshift sequencing errors (nifH, nosZ), the sequences were corrected using Framebot (Wang et al., 2013). The metagenomic nifH amino acid sequence was aligned to references sequences from the nifH database available from the Zehr research group (Zehr and Jenkins, 2003) using the ARB software package (Ludwig et al., 2004). ProtTest3 was used to select the fixed WAG model of amino acid evolution with an inverse gamma rate approximation (Darriba et al., 2011) for the nifH phylogeny, though similar topologies were recovered using related models.
Phylogenetic reconstruction for both 16S rRNA and nifH genes were conducted as follows: maximum likelihood phylogeny was constructed with RAxML 7.2.8 (Stamatakis, 2006) with 1000 rapid bootstrap inferences and Bayesian phylogeny with MrBayes 3.3 (Ronquist et al., 2012). For Bayesian tree inferences, MCMC was run with default parameters and convergence was assessed when the reported average standard deviation of split frequencies fell below 0.01.
Hidden Markov model (HMM) profiles of 40 phylogenetic marker genes (phyeco markers) for Bacteria and Archaea (Wu et al., 2013) were used to search the green berry metagenomic peptide sequence database (from six frame translated assembled nucleotide sequence data) using the trusted cutoffs. For each of the 34 markers that have green berry hits, green berry peptide sequences were aligned with all the bacterial and archaeal references sequences by hmmalign in HMMER3 (Eddy, 2011). A maximum likelihood tree was built by Fastree2 (Price et al., 2010) for each alignments, and the alignments and trees were examined. As a result, 33 green berry peptide sequences were selected for further analysis because they branched with Cyanobacteria with good alignments.
These sequences were further filtered to include only those 29 phyeco gene families with a single copy, cyanobacterial green berry hit. Single-copied reference sequences from 126 Cyanobacteria genomes of the 29 markers and the green berry cyanobacterial metagenomic sequences were retrieved from the alignments built in the previous step and were concatenated into a large alignment. A maximum likelihood tree was built using PHYML3.0 with the LG substitution model (Guindon et al., 2010). Tree topology and branch lengths were optimized by the program and SH statistics was used for branch support estimation.
Acetylene Reduction Assay
Whole-aggregate in vivo acetylene reduction assays were conducted on two 30 mL serum bottles of five washed green berries each. Acetylene, generated by the hydration of calcium carbide in an evacuated 150 mL serum-vial, was added to 10% of the headspace in a 30 mL serum-vial containing the berries in 5 mL of anoxic 0.2 μm filtered in situ marsh water under an 90% N2-CO2 atmosphere (90:10). Experiments were incubated on a 14 h light, 10 h dark cycle with full spectrum illumination at 30°C for 2 days. Acetylene and ethylene were quantified using a Varian 2400 series gas chromatograph (Varian Instrument Group, Walnut Creek, CA, United States) with an H2 flame ionization detector, as described previously (Lobo and Zinder, 1988). Rates were calculated over the 2 days incubation period with the assumption that nitrogen fixation was restricted to the dark hours of incubation, as previously described for unicellular cyanobacteria (Dron et al., 2013).
Analysis of C:N Content
Six green berries sampled from the acetylene reduction experiment (control conditions without acetylene added) in late afternoon were dried at 55°C and crimped in aluminum foil. Samples were analyzed using a Europa 20-20 elemental analyzer interfaced with a Europa ANCA-SL mass analyzer (Sercon Limited, Cheshire, United Kingdom) under contract with the Stable Isotope Lab at the Marine Biological Laboratory’s Ecosystem Center (Woods Hole, MA, United States).
Microelectrode Measurements
Individual aggregates were placed on an agar plate and covered with filter-sterilized seawater collected from the marsh pool. An airstream was directed at the water so that slight ripples were visible on the surface. The water had a salinity of 3.5 psu and the temperature during measurements was either 24 or 27°C, which was taken into account for calibration. We used Clark-type oxygen microsensors (tip diameter 50 μm, 90% response time < 1 s; Unisense A/S, Aarhus, Denmark), and calibrated before and after measurements in air-saturated (by bubbling) and anaerobic (0.1 M acetate, 0.1 M NaOH) 3.5% NaCl. The micro-profiling apparatus and software Sensor Trace Suite was also provided from Unisense. Starting at the water surface, microprofiles through individual aggregates were measured in the light, and light-dark shifts. Time-lapse light-dark-shift recordings included sensor tips placed directly above the aggregate (in the water phase); and inserting the tip into the central core of the aggregate. Recordings of the oxygen signal were taken every second. Light sources were 65 W halogen lamps and experiments were conducted with either 170 μmol photons m-2 s-1 (one lamp), or 320 μmol photons m-2 s-1 (two lamps). Dark conditions were realized by switching off the lamps, removing them from the table, and carefully placing a carton box over the entire profiling setup to avoid residual light from the room. Background light intensities under the box were < 1 μmol m-2 s-1.
Theoretical limits of oxygen and DIC flux and whole aggregate O2 flux calculations were calculated from depth concentration profiles according to Ploug et al. (1997), with a diffusion coefficient for O2 in 3.5% saline water of 2.175 × 10-5 cm2 s-1 at 24°C and 2.3535 × 10-5 cm2 s-1 at 27°C. Inside the aggregate, the apparent diffusivity of O2 was assumed to be 0.95 (Ploug and Passow, 2007). Carbon fixation was estimated based on a photosynthetic quotient of 1.2 (Masotti et al., 2007). The diffusion of oxygen in agar (0.2–2%) was not found to be different than in water over a wide range of salinities (Revsbech, 1989).
Author Contributions
Experiments were conducted by EW, VS-C, UJ, and PH. EW analyzed data and prepared the manuscript with VS-C. JE, DB, and SZ advised and assisted with experiments and edited and revised the manuscript.
Funding
This work was supported by NSF grant DEB-1310168, and award to EW from the NSF Graduate Research Fellowship, UC Davis Dissertation Year Fellowship, P.E.O. Scholar Award, and the NAI/APS Lewis and Clark Fund in Astrobiology. This research was performed by participants in the MBL Microbial Diversity course, which was supported in part by the Howard Hughes Medical Foundation, the Gordon and Betty Moore Foundation (#2493), the NSF (DEB-0917499), the US DOE (DE-FG02-10ER13361), and the NASA Astrobiology Institute.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the excellent work of many students, teaching fellows, and instructors who have investigated these aggregates in the Microbial Diversity course at the Marine Biological Laboratory. In particular, we thank Margret Gentile for her pioneering studies, James R. Henriksen for microscopy, and Nanelle R. Barash and Douglas C. Nelson for insightful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01623/full#supplementary-material
TABLE S1 | Summary of metagenomic data for the unicellular cyanobacteria GB-CYN1. Analysis includes assignment of phylogenetic marker genes to GB-CYN1 and analysis of genes from different metabolic pathways of interest.
TABLE S2 | Summary of metagenomic analysis of marker genes for nitrate and nitrite reduction.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bandyopadhyay, A., Elvitigala, T., and Welsh, E. (2011). Novel metabolic attributes of the genus Cyanothece, comprising a group of unicellular nitrogen-fixing cyanobacteria. mBio 2:e00214-11. doi: 10.1128/mBio.00214-11
Bench, S. R., Heller, P., Frank, I., Arciniega, M., Shilova, I. N., and Zehr, J. P. (2013). Whole genome comparison of six Crocosphaera watsonii strains with differing phenotypes. J. Phycol. 49, 786–801. doi: 10.1111/jpy.12090
Bench, S. R., Ilikchyan, I. N., Tripp, H. J., and Zehr, J. P. (2011). Two strains of Crocosphaera watsonii with highly conserved genomes are distinguished by strain-specific features. Front. Microbiol. 2:261. doi: 10.3389/fmicb.2011.00261
Bolhuis, H., Severin, I., Confurius-Guns, V., Wollenzien, U. I. A., and Stal, L. J. (2010). Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISME J. 4, 121–130. doi: 10.1038/ismej.2009.99
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Cantera, J. J. L., Kawasaki, H., and Seki, T. (2004). The nitrogen-fixing gene (nifH) of Rhodopseudomonas palustris: a case of lateral gene transfer? Microbiology 150, 2237–2246.
Capone, D. G., Zehr, J. P., Paerl, H. W., Bergman, B., and Carpenter, E. J. (1997). Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229. doi: 10.1126/science.276.5316.1221
Carpenter, J., Raalte, D. V., and Valiela, I. (1978). Nitrogen fixation by algae in a Massachusetts salt marsh. Limnol. Oceanogr. 23, 318–327. doi: 10.4319/lo.1978.23.2.0318
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
DeLong, E. F., Franks, D. G., and Alldredge, A. L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934. doi: 10.4319/lo.1993.38.5.0924
Deutsch, C., Sarmiento, J. L., Signman, D. M., Gruber, N., and Dunne, J. P. (2007). Spatial coupling of nitrogen inputs and losses in the ocean. Nature 445, 163–167. doi: 10.1038/nature05392
Dron, A., Rabouille, S., Claquin, P., Le Roy, B., Talec, A., and Sciandra, A. (2012). Light-dark (12:12) cycle of carbon and nitrogen metabolism in Crocosphaera watsonii WH8501: relation to the cell cycle. Environ. Microbiol. 14, 967–981. doi: 10.1111/j.1462-2920.2011.02675.x
Dron, A., Rabouille, S., Claquin, P., Talec, A., Raimbault, V., and Sciandra, A. (2013). Photoperiod length paces the temporal orchestration of cell cycle and carbon-nitrogen metabolism in Crocosphaera watsonii. Environ. Microbiol. 15, 3292–3304. doi: 10.1111/1462-2920.12163
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Eloe, E. A., Shulse, C. N., Fadrosh, D. W., Williamson, S. J., Allen, E. E., and Bartlett, D. H. (2011). Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ. Microbiol. Rep. 3, 449–458. doi: 10.1111/j.1758-2229.2010.00223.x
Fernandes, N., Steinberg, P., Rusch, D., Kjelleberg, S., and Thomas, T. (2012). Community structure and functional gene profile of bacteria on healthy and diseased thalli of the red seaweed Delisea pulchra. PLoS ONE 7:e50854. doi: 10.1371/journal.pone.0050854
Foster, R. A., Subramaniam, A., Mahaffey, C., Carpenter, E. J., Capone, D. G., and Zehr, J. P. (2007). Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical north Atlantic Ocean. Limnol. Oceanogr. 52, 517–532. doi: 10.4319/lo.2007.52.2.0517
Foster, R. A., and Zehr, J. P. (2006). Characterization of diatom-cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences. Environ. Microbiol. 8, 1913–1925. doi: 10.1111/j.1462-2920.2006.01068.x
Fulweiler, R. W., Brown, S. M., Nixon, S. W., and Jenkins, B. D. (2013). Evidence and a conceptual model for the co-occurrence of nitrogen fixation and denitrification in heterotrophic marine sediments. Mar. Ecol. Prog. Ser. 482, 57–68. doi: 10.3354/meps10240
Gaby, J. C., and Buckley, D. H. (2014). A comprehensive aligned nifH gene database: a multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014:bau001. doi: 10.1093/database/bau001
Galloway, J. N., Dentener, F. J., Capone, D. G., Boyer, E. W., Howarth, R. W., Seitzinger, S. P., et al. (2004). Nitrogen cycles: past, present, and future. Biogeochemistry 70, 153–226. doi: 10.1007/s10533-004-0370-0
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hagino, K., Onuma, R., Kawachi, M., and Horiguchi, T. (2013). Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae). PLoS ONE 8:e81749. doi: 10.1371/journal.pone.0081749
Hewson, I., Poretsky, R. S., Dyhrman, S. T., Zielinski, B., White, A. E., Tripp, H. J., et al. (2009). Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 3, 1286–1300. doi: 10.1038/ismej.2009.75
Hietanen, S., Moisander, P. H., Kuparinen, J., and Tuominen, L. (2002). No sign of denitrification in a Baltic Sea cyanobacterial bloom. Mar. Ecol. Prog. Ser. 242, 73–82. doi: 10.3354/meps242073
Howarth, R. W., Marina, R., Lane, J., and Cole, J. J. (1988). Nitrogen fixation in freshwater, estuarine, and marine ecosystems. 1. Rates and importance. Limnol. Oceanogr. 33, 669–687. doi: 10.4319/lo.1988.33.4_part_2.0669
Karl, D. M., Michaels, A., Bergman, B., Capone, D. G., Carpenter, E. J., Letelier, R., et al. (2002). Dinitrogen fixation in the world’s oceans. Biogeochemistry 5, 47–98. doi: 10.1023/A:1015798105851
Klawonn, I., Bonaglia, S., Brüchert, V., and Ploug, H. (2015). Aerobic and anaerobic nitrogen transformation processes in N2-fixing cyanobacterial aggregates. ISME J. 9, 1456–1466. doi: 10.1038/ismej.2014.232
Langlois, R. J., Laroche, J., and Raab, P. A. (2005). Diazotrophic diversity and distribution in the tropical and subtropical Atlantic Ocean. Appl. Environ. Microbiol. 71, 7910–7919. doi: 10.1128/AEM.71.12.7910-7919.2005
Lobo, A. L., and Zinder, S. H. (1988). Diazotrophy and nitrogenase activity in the archaebacterium Methanosarcina barkeri 227. Appl. Environ. Microbiol. 54, 1656–1661.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Mann, E. L., and Barbeau, K. A. (2014). Data from: Roseibium sp. TrichSKD4: GOLD Card Gi03577. Genomes Online Database. (2014). Available at: http://genomesonline.org/cgi-bin/GOLD/bin/GOLDCards.cgi?goldstamp=Gi03577
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. doi: 10.1038/nature03959
Masotti, I., Ruiz Pino, D., and Le Bouteiller, A. (2007). Photosynthetic characteristics of Trichodesmium in the southwest Pacific Ocean: importance and significance. Mar. Ecol. Prog. Ser. 338, 47–59. doi: 10.3354/meps338047
Meyer, F., Paarmann, D., D’souza, M., Olson, R., Glass, E. M., Kubal, M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386
Meyer, J. L., and Huber, J. A. (2014). Strain-level genomic variation in natural populations of Lebetimonas from an erupting deep-sea volcano. ISME J. 8, 867–880. doi: 10.1038/ismej.2013.206
Montoya, J. P., Holl, C. M., Zehr, J. P., Hansen, A., Villareal, T. A., and Capone, D. G. (2004). High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature 430, 1027–1031. doi: 10.1038/nature02824
Mota, R., Guimarães, R., Büttel, Z., Rossi, F., Colica, G., Silva, C. J., et al. (2013). Production and characterization of extracellular carbohydrate polymer from Cyanothece sp. CCY 0110. Carbohydr. Polym. 92, 1408–1415. doi: 10.1016/j.carbpol.2012.10.070
Paerl, H. W., and Bebout, B. M. (1988). Direct measurement of O2-depleted microzones in marine Oscillatoria: relation to N2 fixation. Science 241, 442–445. doi: 10.1126/science.241.4864.442
Paerl, H. W., Bebout, B. M., and Prufert, L. E. (1989). Bacterial associations with marine Oscillatoria sp. (Trichodesmium sp.) populations: ecophysiological implications. J. Phycol. 25, 773–784. doi: 10.1111/j.0022-3646.1989.00773.x
Paerl, H. W., and Prufert, L. E. (1987). Oxygen-poor microzones as potential sites of microbial N2 fixation in nitrogen-depleted aerobic marine waters. Appl. Environ. Microbiol. 53, 1078–1087.
Parker, M. A. (2012). Legumes select symbiosis island sequence variants in Bradyrhizobium. Mol. Ecol. 21, 1769–1778. doi: 10.1111/j.1365-294X.2012.05497.x
Passow, U. (2002). Transparent exopolymer particles (TEP) in aquatic environments. Prog. Oceanogr. 55, 287–333. doi: 10.1016/S0079-6611(02)00138-6
Pereira, S., Zille, A., Micheletti, E., Moradas-Ferreira, P., De Philippis, R., and Tamagnini, P. (2009). Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 33, 917–941. doi: 10.1111/j.1574-6976.2009.00183.x
Ploug, H. (2008). Cyanobacterial surface blooms formed by Aphanizomenon sp. and Nodularia spumigena in the Baltic Sea: small-scale fluxes, pH, and oxygen microenvironments. Limnol. Oceanogr. 53, 914–921. doi: 10.4319/lo.2008.53.3.0914
Ploug, H., Adam, B., Musat, N., Kalvelage, T., Lavik, G., Wolf-Gladrow, D., et al. (2011). Carbon, nitrogen and O2 fluxes associated with the cyanobacterium Nodularia spumigena in the Baltic Sea. ISME J. 5, 1549–1558. doi: 10.1038/ismej.2011.20
Ploug, H., Kuhl, M., Buchholz-Cleven, B., and Jørgensen, B. B. (1997). Anoxic aggregates - an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 13, 285–294. doi: 10.3354/ame013285
Ploug, H., Musat, N., Adam, B., Moraru, C. L., Lavik, G., Vagner, T., et al. (2010). Carbon and nitrogen fluxes associated with the cyanobacterium Aphanizomenon sp. in the Baltic Sea. ISME J. 4, 1215–1223. doi: 10.1038/ismej.2010.53
Ploug, H., and Passow, U. (2007). Direct measurement of diffusivity within diatom aggregates containing transparent exopolymer particles. Limnol. Oceanogr. 52, 1–6. doi: 10.4319/lo.2007.52.1.0001
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 - approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Pruesse, E., Peplies, J., and Glockner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Reddy, K. J., Haskell, J. B., Sherman, D. M., and Sherman, L. A. (1993). Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J. Bacteriol. 175, 1284–1292. doi: 10.1128/jb.175.5.1284-1292.1993
Redfield, A. C., Ketchum, B. H., and Richards, F. A. (1963). “The influence of organisms on the composition of sea-water,” in The Sea, ed. M. N. Hill (New York, NY: Wiley-Interscience), 26–77.
Revsbech, N. P. (1989). Diffusion characteristics of microbial communities determined by use of oxygen microsensors. J. Microbiol. Methods 9, 111–122. doi: 10.1016/0167-7012(89)90061-4
Revsbech, N. P., Jørgensen, B. B., and Brix, O. (1981). Primary production of microalgae in sediments measured by oxygen microprofile, H14CO3- fixation, and oxygen exchange methods. Limnol. Oceanogr. 26, 717–730. doi: 10.4319/lo.1981.26.4.0717
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Seitz, A. P., Nielsen, T. H., and Overmann, J. (1993). Physiology of purple sulfur bacteria forming macroscopic aggregates in Great Sippewissett Salt Marsh, Massachusetts. FEMS Microbiol. Ecol. 12, 225–235. doi: 10.1111/j.1574-6941.1993.tb00035.x
Shapiro, O. H., Hatzenpichler, R., Buckley, D. H., Zinder, S. H., and Orphan, V. J. (2011). Multicellular photo-magnetotactic bacteria. Environ. Microbiol. Rep. 3, 233–238. doi: 10.1111/j.1758-2229.2010.00215.x
Sohm, J. A., Edwards, B. R., Wilson, B. G., and Webb, E. A. (2011). Constitutive extracellular polysaccharide (EPS) production by specific isolates of Crocosphaera watsonii. Front. Microbiol. 2:229–229. doi: 10.3389/fmicb.2011.00229
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Thompson, A. W., Foster, R. A., Krupke, A., Carter, B. J., Musat, N., Vaulot, D., et al. (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550. doi: 10.1126/science.1222700
Tripp, H. J., Bench, S. R., Turk, K. A., Foster, R. A., Desany, B. A., Niazi, F., et al. (2010). Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, 90–94. doi: 10.1038/nature08786
Tuomainen, J., Hietanen, S., Kuparinen, J., Martikainen, P. J., and Servomaa, K. (2006). Community structure of the bacteria associated with Nodularia sp. (Cyanobacteria) aggregates in the Baltic Sea. Microb. Ecol. 52, 513–522. doi: 10.1007/s00248-006-9130-0
Tuomainen, J. M., Hietanen, S., Kuparinen, J., Martikainen, P. J., and Servomaa, K. (2003). Baltic Sea cyanobacterial bloom contains denitrification and nitrification genes, but has negligible denitrification activity. FEMS Microbiol. Ecol. 45, 83–96. doi: 10.1016/S0168-6496(03)00131-4
Wang, Q., Quensen Iii, J. F., Fish, J. A., Lee, T. K., Sun, Y., Tiedje, J. M., et al. (2013). Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 4:e00592-13. doi: 10.1128/mBio.00592-13
Webb, E. A., Ehrenreich, I. M., Brown, S. L., Valois, F. W., and Waterbury, J. B. (2009). Phenotypic and genotypic characterization of multiple strains of the diazotrophic cyanobacterium, Crocosphaera watsonii, isolated from the open ocean. Environ. Microbiol. 11, 338–348. doi: 10.1111/j.1462-2920.2008.01771.x
Welsh, E. A., Liberton, M., Stöckel, J., Loh, T., Elvitigala, T., Wang, C., et al. (2008). The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. U.S.A. 105, 15094–15099. doi: 10.1073/pnas.0805418105
Wilbanks, E. G., Jaekel, U., Salman, V., Humphrey, P. T., Eisen, J. A., Faccioti, M. T., et al. (2014). Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh. Environ. Microbiol. 16, 3398–3415. doi: 10.1111/1462-2920.12388
Wu, D., Jospin, G., and Eisen, J. A. (2013). Systematic identification of gene families for use as “markers” for phylogenetic and phylogeny-driven ecological studies of bacteria and archaea and their major subgroups. PLoS ONE 8:e77033. doi: 10.1371/journal.pone.0077033
Wyman, M., Hodgson, S., and Bird, C. (2013). Denitrifying alphaproteobacteria from the Arabian Sea that express nosZ, the gene encoding nitrous oxide reductase, in oxic and suboxic waters. Appl. Environ. Microbiol. 79, 2670–2681. doi: 10.1128/AEM.03705-12
Zehr, J. P., Bench, S. R., Carter, B. J., Hewson, I., Niazi, F., Shi, T., et al. (2008). Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322, 1110–1112. doi: 10.1126/science.1165340
Zehr, J. P., and Jenkins, B. D. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Zehr, J. P., Montoya, J. P., Jenkins, B. D., Hewson, I., Mondragon, E., Short, C. M., et al. (2007). Experiments linking nitrogenase gene expression to nitrogen fixation in the North Pacific subtropical gyre. Limnol. Oceanogr. 52, 169–183. doi: 10.4319/lo.2007.52.1.0169
Keywords: nitrogen fixation, cyanobacteria, UCYN-A, UCYN-B, biofilms, marine aggregate, unicellular cyanobacteria, salt marsh
Citation: Wilbanks EG, Salman-Carvalho V, Jaekel U, Humphrey PT, Eisen JA, Buckley DH and Zinder SH (2017) The Green Berry Consortia of the Sippewissett Salt Marsh: Millimeter-Sized Aggregates of Diazotrophic Unicellular Cyanobacteria. Front. Microbiol. 8:1623. doi: 10.3389/fmicb.2017.01623
Received: 28 June 2017; Accepted: 10 August 2017;
Published: 04 September 2017.
Edited by:
Sophie Rabouille, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Radha Prasanna, Indian Agricultural Research Institute (IARI), IndiaJennifer Glass, Georgia Institute of Technology, United States
Copyright © 2017 Wilbanks, Salman-Carvalho, Jaekel, Humphrey, Eisen, Buckley and Zinder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth G. Wilbanks, ZWxpemFiZXRoLndpbGJhbmtzQGxpZmVzY2kudWNzYi5lZHU=
†Present address: Ulrike Jaekel, Division for Science, Research Council of Norway, Oslo, Norway
 Elizabeth G. Wilbanks
Elizabeth G. Wilbanks Verena Salman-Carvalho
Verena Salman-Carvalho Ulrike Jaekel
Ulrike Jaekel Parris T. Humphrey
Parris T. Humphrey Jonathan A. Eisen5
Jonathan A. Eisen5 Daniel H. Buckley
Daniel H. Buckley Stephen H. Zinder
Stephen H. Zinder