- Ministry of Health Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
There are four human coronaviruses (HCoVs), distributed worldwide, that are associated with a range of respiratory symptoms. The discovery of severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV shows that HCoVs pose a significant threat to human health. Our work aims to develop a sensitive method (mCoV-MS) which can not only identify known HCoVs accurately, but also have the ability to provide clues for the emerging HCoVs. The method was performed using a MassARRAY matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) system. We developed a 17-plex analysis to detect six HCoVs in Panel A and another 17-plex analysis to detect Alphacoronavirus and Betacoronavirus in Panel B. All tested primers and probes for the mCoV-MS method were effective, with no cross-reactivity observed with other common respiratory viruses. To confirm the usefulness of the mCoV-MS method we screened 384 pharyngeal and/or anal swab samples collected from bats/rodents, and 131 nasal and throat swabs from human patients. The results showed good concordance with the results of metagenomic analysis or PCR-sequencing. The validation test showed mCoV-MS method can detect potentially pathogenic CoVs in Alphacoronavirus and Betacoronavirus and provide convincingly phylogenetic evidences about unknown CoVs. The mCoV-MS method is a sensitive assay that is relatively simple to carry out. We propose that this method be used to complement next generation sequencing technology for large-scale screening studies.
Introduction
Coronaviruses (CoVs) are large, enveloped, positive-sense RNA viruses that can cause respiratory, enteric, hepatic, and neurological diseases in a range of animals, including humans (Lai et al., 2007). According to the latest criteria of International Committee on Taxonomy of Viruses, CoVs are classified into four genera, including Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus. There are six human coronaviruses (HCoVs), HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) which belong to Alphacoronavirus and Betacoronavirus. HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 are usually associated with relatively mild respiratory symptoms and the detection rate ranges from 0.3 to 4.5% in respiratory specimens according to different studies (Gaunt et al., 2010; Gadsby et al., 2016; Trombetta et al., 2016). In 2003, SARS emerged from China as a rapidly spreading respiratory illness with a fatality rate close to 10%. The SARS-CoV was found to be a lineage B Betacoronavirus and was confirmed as the etiological agent of SARS (Drosten et al., 2003; Ksiazek et al., 2003; Lee et al., 2003; Peiris et al., 2003). In 2012, MERS-CoV was detected in humans, which is a lineage C Betacoronavirus that differs from other HCoVs. This virus has caused severe acute respiratory infection (SARI) in patients with a fatality rate close to 40% and led to a new awareness of the medical importance of HCoVs (van Boheemen et al., 2012; Zaki et al., 2012; de Groot et al., 2013). As of 15 June 2017, there were 2015 laboratory confirmed cases of MERS-CoV infection and 703 deaths in 27 countries (http://www.who.int/emergencies/mers-cov/en/). The emergence of SARS-CoV and MERS-CoV suggests that highly pathogenic HCoVs remain a key threat to human health. Because of the severity and communicability of SARS and SARI, rapid and sensitive diagnostic methods are essential for the timely implementation of effective infection control measures to prevent further transmission. Millions of pilgrims from across the globe perform the Hajj annually, increasing the risk of global transmission of MERS-CoV (Sharif-Yakan and Kanj, 2014). The large number of tourists traveling to the Middle East also increases the risk of transmitting MERS-CoV to other areas. Thus, it is important to develop a high-throughput and sensitive method that can detect HCoVs, especially MERS-CoV.
Isolation of virus from clinical specimens by cell culture is regarded as the gold standard for laboratory diagnosis of respiratory virus infection. However, for HCoVs, attempting to isolate virus has long turnaround time and some HCoVs are difficult to culture, which makes the detection results lack timeliness. Compared with cell culture, molecular tests, such as reverse transcription polymerase chain reaction (RT-PCR), nested RT-PCR, and quantitative real-time RT-PCR assays (qPCR) are more rapid, and have superior sensitivity (van der Hoek et al., 2004; Bellau-Pujol et al., 2005; Vijgen et al., 2005; Vabret et al., 2006; Gaunt et al., 2010). To date, molecular tests are accepted worldwide as an important method for diagnosing HCoV infection. However, most of the current molecular methods target a single HCoV gene, which limits their application because of the mutations that can occur in HCoV genomes. Thus, false negative results can be returned because they are only targeting one gene.
In view of the lessons we learned from MERS and SARS outbreak, CoVs have the ability of interspecies transmission and may emerge as novel pathogens (Chan et al., 2013; Ge et al., 2013). Therefore, an ideal diagnosis method should not only identify known HCoVs accurately, but also have the ability to provide clues for the emerging HCoVs. However, most of the current molecular assays only target some of HCoVs, which may cause a false negative result when facing novel HCoV infections. In this study, we designed and evaluated a multiplexed CoVs test, mCoV-MS, which can detect 6 known HCoVs and have the potential to discover new HCoVs that haven't been clearly described yet. Our new method consists of two panels. Panel A is designed to cover six known HCoVs. Panel B is designed to cover all the known CoVs in Alphacoronavirus and Betacoronavirus which is developed to expand our detection spectrum. Our method is based on the MassARRAY matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) system (Agena Bioscience, Inc., San Diego, CA, USA), which has been used for pathogen detection successfully, including common respiratory viruses, human papillomavirus, human polyomavirus, human enterovirus and Staphylococcus aureus (Yang et al., 2005; Soderlund-Strand et al., 2008; Basu et al., 2011; Syrmis et al., 2011; Li et al., 2013, 2015; Peng et al., 2013a,b, 2014, 2016b,a; Zhang et al., 2015). Likewise, the mCoV-MS method uses multiplex PCR conjugated MALDI-TOF MS technology to achieve high-throughput CoVs detection. The method was divided into three stages: multiplex PCR, primer extension and MALDI-TOF MS identification. Briefly, all the targets are amplified via multiplexed PCR followed by incubation with shrimp alkaline phosphatase (SAP) to dephosphorylate any unincorporated dNTPs. In primer extension stage, site-specific short oligonucleotide primers will bind the respective amplicons and are extended a single base, as the substrate used in this reaction is ddNTPs. Finally, MALDI-TOF MS is used to measure the masses of extended primers accurately, thus receiving a positive or negative result for every target (Oeth et al., 2009).
Materials and Methods
Design of the mCoV-MS Method
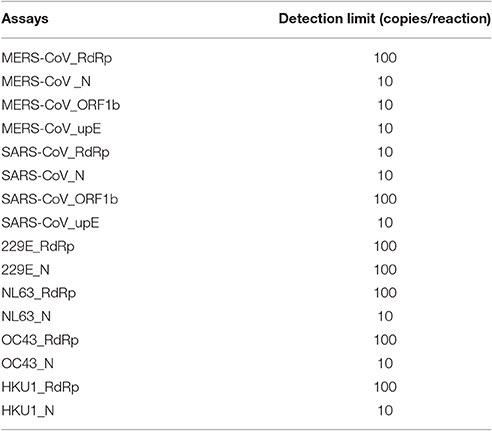
Representative HCoV strains used in the design of the mCoV-MS method are shown in Supplementary Table 1. The CoV genome sequences used for primer and extension probe design were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Specific primers and extension probes (Supplementary Table 2) used in this study were designed using Assay Design v4.0 (Agena Bioscience, Inc.). In this study, we developed a 17-plex analysis to detect six HCoVs in Panel A and another 17-plex analysis to detect Alphacoronavirus and Betacoronavirus in Panel B. Panel A: including specific primers and extension probes targeting the RNA-dependent RNA polymerase (RdRp) and nucleocapsid (N) genes of six HCoVs, and ribonuclease P (as an internal control). In addition, for SARS-CoV and MERS-CoV, we also designed primers and extension probes targeting the ORF1b gene and regions upstream of the E gene (upE). Panel B: including specific primers and extension probes targeting RdRp gene of CoVs in Alphacoronavirus and Betacoronavirus.
Experiment Procedures of mCoV-MS Method
The primary PCR and iPLEX reaction of the mCoV-MS method was performed in a 384-plate by using a ProFlex PCR system (Applied Biosystems, USA) according to our modified protocol as previously described (Peng et al., 2013a). The PCR mixes (5 μl) contained 2 μl template and final concentrations of 500 μM nucleotide mix (dATP, dGTP, dCTP, and dUTP), 100 nM primer mix, 1 U DNA polymerase enzyme, 2 mM MgCl2, and 0.2 U uracil-DNA glycosylase. The reaction conditions of primary PCR were: 45°C for 2 min; a denaturing step of 2 min at 95°C followed by 45 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 1 min; and then a final extension step at 72°C for 5 min. Sterile water was used as negative control. Then SAP was used to dephosphorylate the primary PCR mixes as follows: 37°C for 40 min and then 85°C for 5 min. After SAP treatment, the iPLEX reaction (single-base extension reaction) mix which included 0.2 μl terminator mix, 0.94 μl extension probe mix, 0.2 μl iPLEX Pro buffer, and 0.041 μl ThermoSequenase enzyme was added. The iPLEX reaction, desalting and sample dispensing were performed according to standard procedures (Agena Bioscience, Inc.). All data were acquired and analyzed using Typer software, v4.0.3 (Agena Bioscience, Inc.).
Test of the mCoV-MS Method
We used the plasmids containing nearly the full-length sequence of target genes of HCoVs to determine the analytical sensitivity of the mCoV-MS method. The plasmids were diluted to a series of concentrations of 1, 10, 100 1,000, and 10,000 copies per reaction. The test of each plasmid concentration was performed in triplicate.
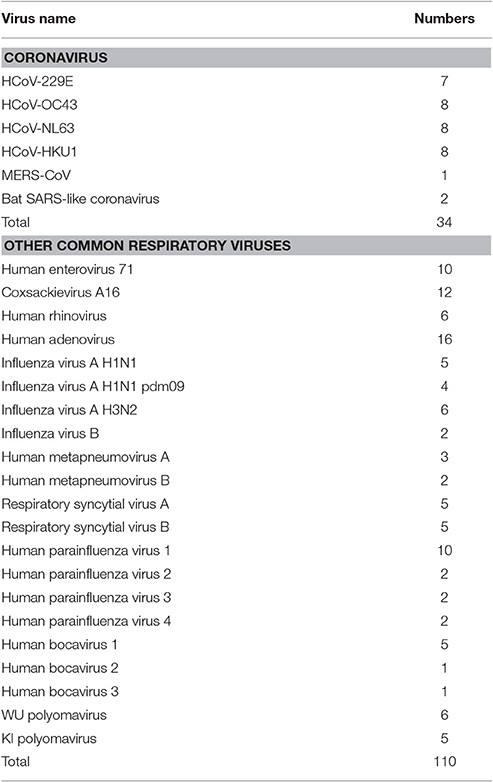
All samples used in this study were obtained from collections at our institute. We isolated total RNA and DNA from all samples using the Qiagen DNA/RNA isolation kits (Qiagen, Germany) according to the manufacturer's instructions. To evaluate the specificity of the mCoV-MS method, we used 32 samples that had previously been confirmed positive for HCoV (Ren et al., 2009; Zhang et al., 2014), and two bat SARS-like coronavirus isolated from bats (Yang et al., 2013). A panel of common respiratory viruses (Ren et al., 2009; n = 110) was also used to determine the specificity of the mCoV-MS method (Table 1).
Potential biohazardous materials were handled in required levels of biosafety laboratories. All experiments (collection of samples, sample handing, operator training and protection, etc.) were carried out according to the approved biosafety standard guidelines set by The Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College.
Application of the mCoV-MS Method
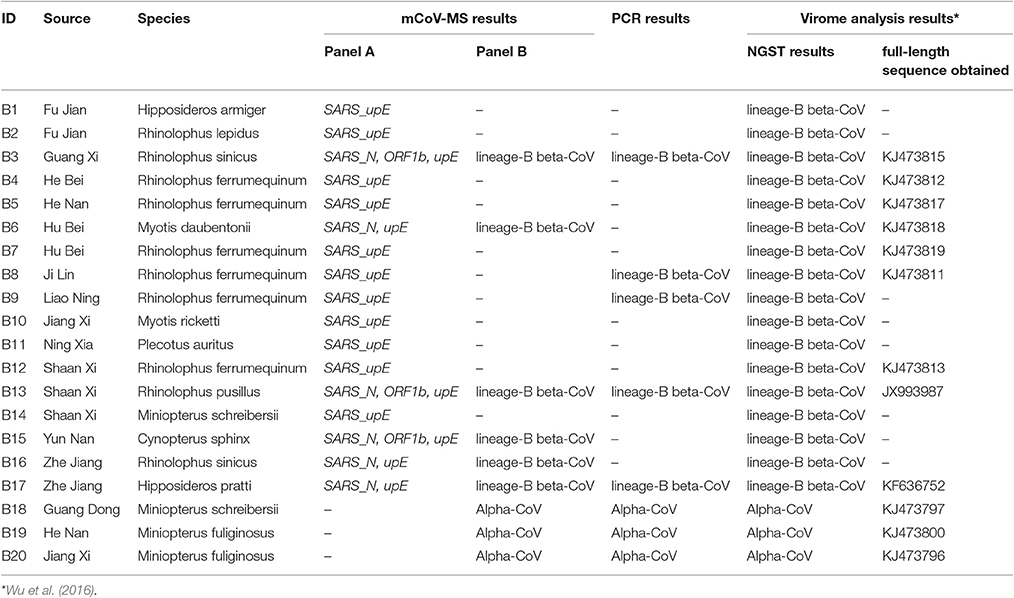
Further testing of the mCoV-MS method involved 384 pharyngeal and/or anal swab samples collected from bats (n = 352) or rodents (n = 32) which were part of our bat and rodent virome project. Alphacoronavirus and Betacoronavirus in 384 samples of bats and rodents were also screened using a RT-PCR of the RdRp gene of CoVs (Lau et al., 2012). For comparison, the next generation sequencing technology (NGST) results of 352 bat samples in another study were also included (Wu et al., 2016).
We screened 131 nasal and throat swabs obtained from patients for HCoVs using the mCoV-MS method. The results were compared with those of other methods as described follows. Four common HCoVs were also screened by using PCR or nested PCR assays, followed by sequencing analysis according to previously reported methods (van der Hoek et al., 2004; Bellau-Pujol et al., 2005; Vabret et al., 2006). To screen for SARS-CoV and SARS-like CoV, we used the method described by the World Health Organization (http://www.who.int/csr/sars/primers/en/). To screen for MERS-CoV in samples, we used a qPCR assay developed by Corman et al. (2012a,b).
Statistical Analysis
We tested the differences between the detection rates of the two different methods using the χ2 test. A P < 0.05 was considered statistically significant.
Results
Performance of the mCoV-MS Method
All primers and probes used in the mCoV-MS method were evaluated using BLASTN searches against the non-redundant nucleotide database of the National Center for Biotechnology Information. Panel A and B test can be carried out simultaneously or separately according to actual needs. Sensitivity tests showed that the panel A of mCoV-MS method can accurately detected all six targeted HCoVs. The limit of detection of the mCoV-MS method was ~10–100 copies per assay panel (Table 2). For example, for the MERS-CoV panels, we were able to detect 10 copies of N, ORF1b and upE, and 100 copies of RdRp (Figure 1). As expected, negative controls yielded no positive results for the mCoV-MS method.
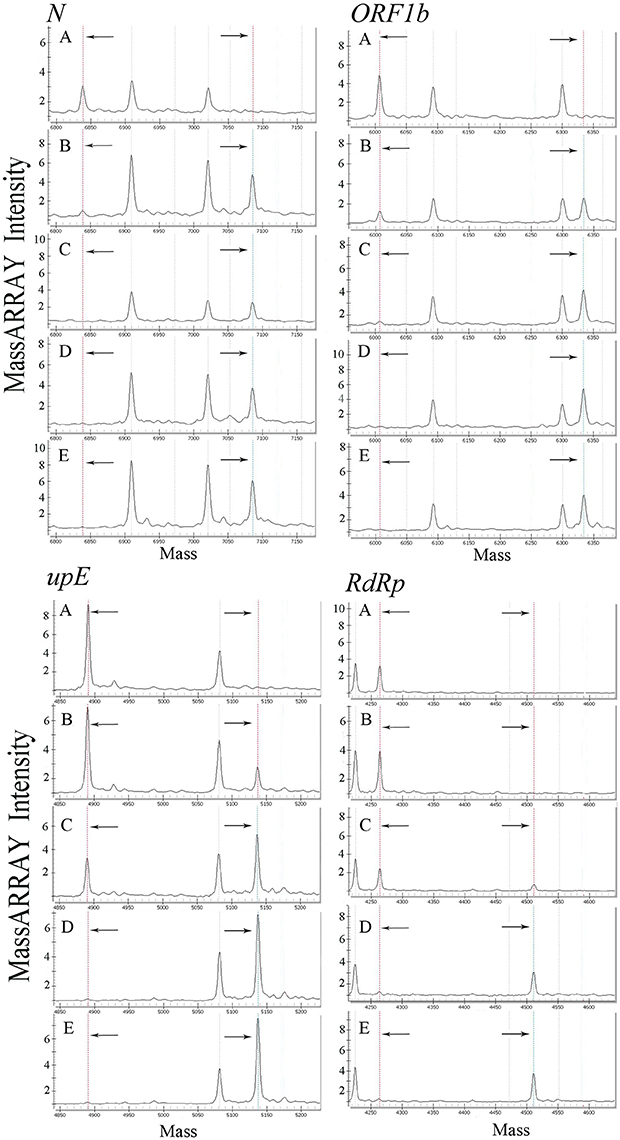
Figure 1. Analysis of a dilution series of MERS-CoV plasmids using mCoV-MS method. (A) 1 copy/reaction, (B) 10 copies/reaction, (C) 100 copies/reaction, (D) 1,000 copies/reaction, (E) 10,000 copies/reaction. In the mass spectrometry mass spectra, the dotted lines in the left and the dotted lines in the right represent the unextended primers and the extended primers of the assay, respectively.
We used 32 HCoV-positive samples to evaluate the common HCoV assays and MERS-CoV assays, and two SARS-like CoV isolates to evaluate the SARS-CoV assays (Table 1). Our results showed that all combinations of primers and probes were effective (Supplementary Figures S1–S5), with no observed cross-reactivity with other HCoVs.
A panel of common respiratory viruses (influenza A virus, influenza B virus, adenovirus, respiratory syncytial virus A, respiratory syncytial virus B, parainfluenza virus types 1–4, human metapneumovirus A, human metapneumovirus B, rhinovirus, human enterovirus 71, coxsackievirus A16, human bocavirus 1–3, WU polyomavirus, and KI polyomavirus) were also used in this study (Table 1). Our results clearly showed no cross-reactivity with other common respiratory viruses.
To Screen Bat and Rodent Samples Using the mCoV-MS Method
We used 352 pools of bat pharyngeal and anal swab samples to validate the performance of mCoV-MS. These samples were collected in 22 provinces which is a part of our bat virome project (Wu et al., 2016). Among the 352 pools, 20 (5.68%) were tested positive by our method. Seventeen pools (4.83%) were identified by panel A assays as SARS-CoV, which showed good concordance with the results of SARS-like CoVs by means of metagenomic analysis (Wu et al., 2016; Table 3). Six of the 17 (35.3%) pools were also confirmed as lineage-B beta-CoV by panel B assays. We found that these samples obtained two or more positive results by panel A assays. It is obvious that multiple target using in panel A avoid the false negative results. Five of the 17 (29.4%) pools were confirmed as lineage-B beta-CoV by PCR-sequencing method. It seemed that the mCoV-MS method were more sensitive than PCR method (P = 0.01 by χ2 test). Interestingly, all the 17 pools obtained SARS-upE-positive results, while only 8 (47.1%) of them were detected by the panel B assays or PCR method targeting CoV's RdRp gene. Our results are consistent with the earlier research demonstrating that upE assay is the most sensitive test for coronavirus detection (Corman et al., 2012a; Chan et al., 2015). The remaining three pools (0.9%) were identified as Alphacoronavirus by panel B assays and were ascertained by PCR-sequencing method.
Another 32 rodent pharyngeal and anal swab pools collected in eight provinces across China were also screened by our novel method. A total of 4 (12.5%) pools were classified as lineage-A Betacoronavirus by panel B assays and PCR-sequencing verified two of them (Table 4).
To Screen Clinical Samples Using the mCoV-MS Method
Using the mCoV-MS method, we analyzed 131 nasal and throat swabs. The mCoV-MS method detected HCoVs in 22.14% (29/131) of samples. This result was identical to that when we used a combination of PCR and sequencing techniques. We detected HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1 in 9.9% (13/131), 6.9% (9/131), 3.1% (4/131) and 2.3% (3/131) of specimens, respectively. SARS-CoV and MERS-CoV were not detected in any of these samples.
Discussion
The emergence of SARS-CoV and MERS-CoV has tested our ability to design new diagnostic methods for their rapid detection. Lesson from SARS showed that we were not adequately prepared for the first pandemic of the twenty-first century (de Wit et al., 2016). It took several months to confirm the causative agent as SARS-CoV. Fortunately, advances in molecular technology, especially NGST, allows us to timely and accurately identify MERS-CoV (Zaki et al., 2012). It is very important that these viruses are rapidly and accurately identified in order to assist in commencing an appropriate treatment regimen for patients. The emergence of MERS-CoV has once again highlighted the impact of HCoVs on human health (Chan et al., 2015). Several studies have shown that HCoVs likely originate from wild animal hosts, such as bats (Calisher et al., 2006; Wang et al., 2006; Woo et al., 2012). Shi and colleagues found a SARS-like CoV in Chinese horseshoe bats, suggesting that bats are the natural reservoir of SARS-CoV, and palm civets are an incidental host (Li et al., 2005; Ge et al., 2013). Huynh et al. found that HCoV-NL63 might share common ancestry with Alphacoronaviruses of the North American tricolored bat (Huynh et al., 2012). Several groups have shown that MERS-CoV also likely originates in bats (Annan et al., 2013; Ithete et al., 2013; Memish et al., 2013). Based on neutralizing serum antibody tests, Reusken et al. showed that MERS-CoV, or a related virus, has infected camels (Reusken et al., 2013). A multidisciplinary team of virologists isolated MERS-CoV from dromedary camels (Haagmans et al., 2014). Alagaili et al. also showed that MERS-CoV has been circulating in camels for nearly 20 years; however there is no evidence of MERS-CoV infection in domestic goats or sheep (Alagaili et al., 2014). The identification of SARS-CoV and MERS-CoV indicates that viral cross-species transmission is a real threat to human health (Chan et al., 2013; de Wit et al., 2016). Recently, Menachery et al. suggested that a cluster of SARS-like CoV circulating in bat populations shows potential for human emergence (Menachery et al., 2015). It is possible there are additional CoVs circulating in the wild that could infect humans. To confirm this, a large-scale study screening samples from various wild animals all over the world needs to be conducted. However, it is possible that false negative results would be seen using conventional detection methods that target one gene, because of the genetic diversity of HCoVs.
In clinical laboratories, it has been shown that qPCR assays are sensitive detection methods, which are also quantitative (Vijgen et al., 2005; Gaunt et al., 2010). Most qPCR-based methods target a single gene of a virus. Gaunt et al. performed a large-scale screening of four common HCoVs using a 4-plex qPCR assay, thereby significantly improving HCoV diagnosis (Gaunt et al., 2010). It has been shown that qPCR assays have played an important role in the detection and identification of MERS-CoV. Multiple methods targeting different genes of MERS-CoV have been developed and used to detect MERS-CoV. Corman et al. successfully developed several qPCR assays targeting upE, ORF1b and ORF1a (Corman et al., 2012a,b). In another study, Lu et al. developed a multiplex qPCR assay targeting the N gene and the upE region (Lu et al., 2014). However, the availability of fluorescent dyes limits the multiplexing capacity of their qPCR assay. The mCoV-MS method we developed targets two or four genes, making it a powerful method for screening samples from humans and other animals. Our results also showed that multiple target using in panel A avoid the false negative results. The validation test showed our method can target potentially pathogenic CoVs in Alphacoronavirus and Betacoronavirus and provide convincingly phylogenetic evidences about unknown CoVs. Thus, our method is an effective way to expand CoVs detection spectrum.
NGST is able to generate a large amount of data by random sequencing, and has been widely used in pathogen detection (Haagmans et al., 2009), including deciphering the bat virome by our group (Wu et al., 2016). For CoVs, the large RNA genome has a high frequency of mutation and recombination. Conventional molecular diagnosis requires prior knowledge of the target whereas NGST provides an unbiased result. From this point, NGST is more sensitive than PCR, especially for screening RNA virus with a high frequency of variation. However, the high cost, long turn-around time, and sophisticated bioinformatic analysis required, limits the number of samples that can be sequenced. Our mCoV-MS method can be used to complement NGST methods in large-scale screening studies. On one hand, we can screen a large number samples using mCoV-MS method. Then, the selected positive samples or suspicious positive samples were analyzed using the NGST method. On the other hand, new assays can be incorporated into the mCoV-MS method when we obtain new information regarding CoVs, such as the information we get from NGST. The mCoV-MS method is flexible and can be modified as necessary. As an example, we can use the SARS-CoV or MERS-CoV panels on their own.
There are 10 countries in or near the Arabian Peninsula with laboratory-confirmed MERS cases, and 17 countries with travel-associated MERS cases (http://www.cdc.gov/coronavirus/mers/index.html). There is no border for virus. Therefore, nasopharyngeal samples collected from pilgrims have been screened for MERS-CoV (Gautret et al., 2013). Memish et al. stated that their results only represented a relatively small proportion of the total number of pilgrims (Memish et al., 2014). The largest known outbreak of MERS outside the Arabian Peninsula occurred in the Republic of Korea in 2015. The outbreak was associated with a single person returning from the Middle East which involved 16 hospitals and 186 patients (Korea Centers for Disease Control and Prevention, 2015). The WHO encourages all member states to continue their surveillance for acute respiratory infections, in particular SARI (http://www.who.int/csr/don/03-february-2015-mers/en/). The United Kingdom has established a surveillance system to screen people from the Middle East for MERS-CoV (Thomas et al., 2014). Our mCoV-MS method is a suitable choice for this type of screening, as it is possible for one technician to analyze two 384-well plates within 8 h.
We found some limitations of mCoV-MS method just like we mentioned it in previous report (Peng et al., 2013a). When the HCoV load is very low in a sample, the mCoV-MS method may fail to detect it because of the volume of sample used was 2 μl. So we should use a larger primary PCR volume or several primary PCR reactions for those samples. It should be pointed out that our method is based on the known CoVs sequence, as far as possible covers the known CoVs. It is difficult to detect a new HCoV only use mCoV-MS method. However, it can provide some clues for new HCoV detection compared to current methods.
Ethics Statement
This study was carried out in accordance with the recommendations of national ethics regulations and approved by the ethics committee of Institute of Pathogen Biology with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the ethics committee of Institute of Pathogen Biology.
Author Contributions
JP conceived the experiments, analyzed the results and wrote the manuscript. LX and CZ conducted the experiments and analyzed the results. ZW collected specimens. All authors reviewed the manuscript.
Conflict of Interest Statement
Junping Peng, Junhua Guo, and Qi Jin have a patent (ZL201310309972.4, People's Republic of China) related to this report.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JP was funded by grants from the Ministry of Science and Technology of the People's Republic of China (2014ZX10004001, 2016YFC1202704), National Health and Family Planning Commission of the People's Republic of China (201402001), the CAMS Innovation Fund for Medical Sciences (2016-I2M-3-021), the Program for Changjiang Scholars and Innovative Research Team in University (IRT13007), PUMC Youth Fund, and the Fundamental Research Funds for the Central Universities (3332013118). We give special thanks to Junhua Guo, Lili Ren, Jiang Du, and Guangyu Zhao for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01510/full#supplementary-material
References
Alagaili, A. N., Briese, T., Mishra, N., Kapoor, V., Sameroff, S. C., de Wit, E., et al. (2014). Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio 5:e01002-14. doi: 10.1128/mBio.00884-14
Annan, A., Baldwin, H. J., Corman, V. M., Klose, S. M., Owusu, M., Nkrumah, E. E., et al. (2013). Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerging Infect. Dis. 19, 456–459. doi: 10.3201/eid1903.121503
Basu, P., Chandna, P., Bamezai, R. N., Siddiqi, M., Saranath, D., Lear, A., et al. (2011). MassARRAY spectrometry is more sensitive than PreTect HPV-Proofer and consensus PCR for type-specific detection of high-risk oncogenic human papillomavirus genotypes in cervical cancer. J. Clin. Microbiol. 49, 3537–3544. doi: 10.1128/JCM.00354-11
Bellau-Pujol, S., Vabret, A., Legrand, L., Dina, J., Gouarin, S., Petitjean-Lecherbonnier, J., et al. (2005). Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods 126, 53–63. doi: 10.1016/j.jviromet.2005.01.020
Calisher, C. H., Childs, J. E., Field, H. E., Holmes, K. V., and Schountz, T. (2006). Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19, 531–545. doi: 10.1128/CMR.00017-06
Chan, J. F., Lau, S. K., To, K. K., Cheng, V. C., Woo, P. C., and Yuen, K. Y. (2015). Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 28, 465–522. doi: 10.1128/CMR.00102-14
Chan, J. F., To, K. K., Tse, H., Jin, D. Y., and Yuen, K. Y. (2013). Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 21, 544–555. doi: 10.1016/j.tim.2013.05.005
Corman, V. M., Eckerle, I., Bleicker, T., Zaki, A., Landt, O., Eschbach-Bludau, M., et al. (2012a). Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 17:20285.
Corman, V. M., Muller, M. A., Costabel, U., Timm, J., Binger, T., Meyer, B., et al. (2012b). Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 17:20334.
de Groot, R. J., Baker, S. C., Baric, R. S., Brown, C. S., Drosten, C., Enjuanes, L., et al. (2013). Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 87, 7790–7792. doi: 10.1128/JVI.01244-13
de Wit, E., van Doremalen, N., Falzarano, D., and Munster, V. J. (2016). SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534. doi: 10.1038/nrmicro.2016.81
Drosten, C., Gunther, S., Preiser, W., van der Werf, S., Brodt, H. R., Becker, S., et al. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976. doi: 10.1056/NEJMoa030747
Gadsby, N. J., Russell, C. D., McHugh, M. P., Mark, H., Conway Morris, A., Laurenson, I. F., et al. (2016). Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin. Infect. Dis. 62, 817–823. doi: 10.1093/cid/civ1214
Gaunt, E. R., Hardie, A., Claas, E. C., Simmonds, P., and Templeton, K. E. (2010). Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 48, 2940–2947. doi: 10.1128/JCM.00636-10
Gautret, P., Charrel, R., Belhouchat, K., Drali, T., Benkouiten, S., Nougairede, A., et al. (2013). Lack of nasal carriage of novel corona virus (HCoV-EMC) in French Hajj pilgrims returning from the Hajj 2012, despite a high rate of respiratory symptoms. Clin. Microbiol. Infect. 19:E315–E317. doi: 10.1111/1469-0691.12174
Ge, X. Y., Li, J. L., Yang, X. L., Chmura, A. A., Zhu, G., Epstein, J. H., et al. (2013). Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538. doi: 10.1038/nature12711
Haagmans, B. L., Al Dhahiry, S. H., Reusken, C. B., Raj, V. S., Galiano, M., Myers, R., et al. (2014). Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 14, 140–145. doi: 10.1016/S1473-3099(13)70690-X
Haagmans, B. L., Andeweg, A. C., and Osterhaus, A. D. (2009). The application of genomics to emerging zoonotic viral diseases. PLoS Pathog. 5, e1000557. doi: 10.1371/journal.ppat.1000557
Huynh, J., Li, S., Yount, B., Smith, A., Sturges, L., Olsen, J. C., et al. (2012). Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 86, 12816–12825. doi: 10.1128/JVI.00906-12
Ithete, N. L., Stoffberg, S., Corman, V. M., Cottontail, V. M., Richards, L. R., Schoeman, M. C., et al. (2013). Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerging Infect. Dis. 19, 1697–1699. doi: 10.3201/eid1910.130946
Korea Centers for Disease Control and Prevention (2015). Middle East respiratory syndrome coronavirus outbreak in the Republic of Korea, 2015. Osong Public Health Res. Perspect. 6, 269–278. doi: 10.1016/j.phrp.2015.08.006
Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., et al. (2003). A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966. doi: 10.1056/NEJMoa030781
Lai, M. M. C., Perlman, S., and Anderson, L. J. (2007). “Coronaviridae,” in Fields Virology, 5th Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Lippincott Williams & Wilkins), 1306–1335.
Lau, S. K., Woo, P. C., Yip, C. C., Fan, R. Y., Huang, Y., Wang, M., et al. (2012). Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J. Virol. 86, 5481–5496. doi: 10.1128/JVI.06927-11
Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., et al. (2003). A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348, 1986–1994. doi: 10.1056/NEJMoa030685
Li, K., Guo, J., Zhao, R., Xue, Y., Chen, L., Yang, J., et al. (2013). The prevalence of 10 human polyomaviruses in fecal samples from children with acute gastroenteritis: a case-control study. J. Clin. Microbiol. 51, 3107–3109. doi: 10.1128/JCM.01324-13
Li, K., Zhang, C., Zhao, R., Xue, Y., Yang, J., Peng, J., et al. (2015). The prevalence of STL polyomavirus in stool samples from Chinese children. J. Clin. Virol. 66, 19–23. doi: 10.1016/j.jcv.2015.02.017
Li, W., Shi, Z., Yu, M., Ren, W., Smith, C., Epstein, J. H., et al. (2005). Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679. doi: 10.1126/science.1118391
Lu, X., Whitaker, B., Sakthivel, S. K., Kamili, S., Rose, L. E., Lowe, L., et al. (2014). Real-time reverse transcription-PCR assay panel for middle east respiratory syndrome coronavirus. J. Clin. Microbiol. 52, 67–75. doi: 10.1128/JCM.02533-13
Memish, Z. A., Assiri, A., Almasri, M., Alhakeem, R. F., Turkestani, A., Al Rabeeah, A. A., et al. (2014). Prevalence of MERS-CoV nasal carriage and compliance with the Saudi health recommendations among pilgrims attending the 2013 Hajj. J. Infect. Dis. 210, 1067–1072. doi: 10.1093/infdis/jiu150
Memish, Z. A., Mishra, N., Olival, K. J., Fagbo, S. F., Kapoor, V., Epstein, J. H., et al. (2013). Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerging Infect. Dis. 19, 1819–1823. doi: 10.3201/eid1911.131172
Menachery, V. D., Yount, B. L. Jr., Debbink, K., Agnihothram, S., Gralinski, L. E., Plante, J. A., et al. (2015). A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513. doi: 10.1038/nm.3985
Oeth, P., Del Mistro, G., Marnellos, G., Shi, T., and Van Den Boom, D. (2009). Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY). Methods Mol. Biol. 578, 307–343. doi: 10.1007/978-1-60327-411-1_20
Peiris, J. S., Lai, S. T., Poon, L. L., Guan, Y., Yam, L. Y., Lim, W., et al. (2003). Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361, 1319–1325. doi: 10.1016/S0140-6736(03)13077-2
Peng, J., Gao, L., Guo, J., Wang, T., Wang, L., Yao, Q., et al. (2013a). Type-specific detection of 30 oncogenic human papillomaviruses by genotyping both E6 and L1 genes. J. Clin. Microbiol. 51, 402–408. doi: 10.1128/JCM.01170-12
Peng, J., Li, K., Zhang, C., and Jin, Q. (2016b). MW polyomavirus and STL polyomavirus present in tonsillar tissues from children with chronic tonsillar disease. Clin. Microbiol. Infect. 22, 97.e91–93. doi: 10.1016/j.cmi.2015.08.028
Peng, J., Li, K., Zhang, C., Gao, L., and Jin, Q. (2016a). Human papillomavirus and polyomavirus coinfections among Chinese men who have sex with men. J. Infect. 72, 118–120. doi: 10.1016/j.jinf.2015.09.012
Peng, J., Wang, T., Zhu, H., Guo, J., Li, K., Yao, Q., et al. (2014). Multiplex PCR/mass spectrometry screening of biological carcinogenic agents in human mammary tumors. J. Clin. Virol. 61, 255–259. doi: 10.1016/j.jcv.2014.07.010
Peng, J., Yang, F., Xiong, Z., Guo, J., Du, J., Hu, Y., et al. (2013b). Sensitive and rapid detection of viruses associated with hand foot and mouth disease using multiplexed MALDI-TOF analysis. J. Clin. Virol. 56, 170–174. doi: 10.1016/j.jcv.2012.10.020
Ren, L., Gonzalez, R., Wang, Z., Xiang, Z., Wang, Y., Zhou, H., et al. (2009). Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005-2007. Clin. Microbiol. Infect. 15, 1146–1153. doi: 10.1111/j.1469-0691.2009.02746.x
Reusken, C. B., Haagmans, B. L., Muller, M. A., Gutierrez, C., Godeke, G. J., Meyer, B., et al. (2013). Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 13, 859–866. doi: 10.1016/S1473-3099(13)70164-6
Sharif-Yakan, A., and Kanj, S. S. (2014). Emergence of MERS-CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 10:e1004457. doi: 10.1371/journal.ppat.1004457
Soderlund-Strand, A., Dillner, J., and Carlson, J. (2008). High-throughput genotyping of oncogenic human papilloma viruses with MALDI-TOF mass spectrometry. Clin. Chem. 54, 86–92. doi: 10.1373/clinchem.2007.092627
Syrmis, M. W., Moser, R. J., Whiley, D. M., Vaska, V., Coombs, G. W., Nissen, M. D., et al. (2011). Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 17, 1804–1810. doi: 10.1111/j.1469-0691.2011.03521.x
Thomas, H. L., Zhao, H., Green, H. K., Boddington, N. L., Carvalho, C. F., Osman, H. K., et al. (2014). Enhanced MERS coronavirus surveillance of travelers from the Middle East to England. Emerging Infect. Dis. 20, 1562–1564. doi: 10.3201/eid2009.140817
Trombetta, H., Faggion, H. Z., Leotte, J., Nogueira, M. B., Vidal, L. R., and Raboni, S. M. (2016). Human coronavirus and severe acute respiratory infection in Southern Brazil. Pathog. Glob. Health 110, 113–118. doi: 10.1080/20477724.2016.1181294
Vabret, A., Dina, J., Gouarin, S., Petitjean, J., Corbet, S., and Freymuth, F. (2006). Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 42, 634–639. doi: 10.1086/500136
van Boheemen, S., de Graaf, M., Lauber, C., Bestebroer, T. M., Raj, V. S., Zaki, A. M., et al. (2012). Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3:e00473-12. doi: 10.1128/mBio.00473-12
van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J., Wolthers, K. C., et al. (2004). Identification of a new human coronavirus. Nat. Med. 10, 368–373. doi: 10.1038/nm1024
Vijgen, L., Keyaerts, E., Moes, E., Maes, P., Duson, G., and Van Ranst, M. (2005). Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 43, 5452–5456. doi: 10.1128/JCM.43.11.5452-5456.2005
Wang, L. F., Shi, Z., Zhang, S., Field, H., Daszak, P., and Eaton, B. T. (2006). Review of bats and SARS. Emerging Infect. Dis. 12, 1834–1840. doi: 10.3201/eid1212.060401
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/JVI.06540-11
Wu, Z., Yang, L., Ren, X., He, G., Zhang, J., Yang, J., et al. (2016). Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 10, 609–620. doi: 10.1038/ismej.2015.138
Yang, H., Yang, K., Khafagi, A., Tang, Y., Carey, T. E., Opipari, A. W., et al. (2005). Sensitive detection of human papillomavirus in cervical, head/neck, and schistosomiasis-associated bladder malignancies. Proc. Natl. Acad. Sci. U.S.A. 102, 7683–7688. doi: 10.1073/pnas.0406904102
Yang, L., Wu, Z., Ren, X., Yang, F., He, G., Zhang, J., et al. (2013). Novel SARS-like betacoronaviruses in Bats, China, 2011. Emerging Infect. Dis. 19, 989–991. doi: 10.3201/eid1906.121648
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D., and Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. doi: 10.1056/NEJMoa1211721
Zhang, C., Xiao, Y., Du, J., Ren, L., Wang, J., Peng, J., et al. (2015). Application of multiplex PCR coupled with matrix-assisted laser desorption ionization-time of flight analysis for simultaneous detection of 21 common respiratory viruses. J. Clin. Microbiol. 53, 2549–2554. doi: 10.1128/JCM.00943-15
Keywords: coronavirus, human coronavirus, MALDI-TOF mass spectrometry, respiratory infection, detection
Citation: Xiu L, Zhang C, Wu Z and Peng J (2017) Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Front. Microbiol. 8:1510. doi: 10.3389/fmicb.2017.01510
Received: 16 June 2017; Accepted: 27 July 2017;
Published: 09 August 2017.
Edited by:
John W. A. Rossen, University Medical Center Groningen, NetherlandsReviewed by:
Yigang Tong, Beijing Institute of Microbiology and Epidemiology, ChinaGuido Alberto König, Instituto Nacional de Tecnología Agropecuaria (INTA), Argentina
Copyright © 2017 Xiu, Zhang, Wu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junping Peng, cGVuZ2pwQGhvdG1haWwuY29t
†These authors have contributed equally to this work.
 Leshan Xiu
Leshan Xiu Chi Zhang
Chi Zhang Zhiqiang Wu
Zhiqiang Wu Junping Peng
Junping Peng