
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 July 2017
Sec. Food Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01357
Objectives: Citrobacter spp. especially Citrobacter freundii, is frequently causing nosocomial infections, and increasingly becoming multi-drug resistant (MDR). In this study, we aimed to determine the genetic diversity and relationships of Citrobacter spp. from diarrheal patients and food sources, their antimicrobial resistance profiles and in vitro virulence properties.
Methods: Sixty two Citrobacter isolates, including 13 C. freundii, 41 C. youngae and eight C. braakii isolates, were obtained from human diarrheal patients and food sources. Multilocus Sequence Typing (MLST) of seven housekeeping genes and antimicrobial susceptibility testing using the broth microdilution method according to CLSI recommendations were carried out. Adhesion and cytotoxicity to HEp-2 cells were performed. PCR and sequencing were used to identify blaCTX−M, blaSHV, blaTEM and qnr genes.
Results: The 62 isolates were divided into 53 sequence types (STs) with all STs being novel, displaying high genetic diversity. ST39 was a predominant ST shared by 5 C. youngae strains isolated from four foods and a diarrheal patient. All isolates were resistant to cefoxitin, and sensitive to imipenem, meropenem and amikacin. The majority of Citrobacter isolates (61.3%) were MDR of three or more antibiotics out of the 22 antibiotics tested. Two C. freundii isolates each carried the blaTEM−1 gene and a variant of qnrB77. Three Citrobacter isolates each carried qnrS1 and aac(6')-Ib-cr genes. Seven isolates that showed strong cytotoxicity to HEp-2 cells were MDR.
Conclusions: Citrobacter spp. from human and food sources are diverse with variation in virulence properties and antibiotic resistance profiles. Food may be an important source of Citrobacter species in transmission to humans. C. freundii and C. youngae are potential foodborne pathogens.
Citrobacter spp. are commensal inhabitants of the intestinal tract of humans and other animals. They have also been recovered from water, sewage, and soil (Nada et al., 2004; Bae et al., 2010). Citrobacter spp. are opportunistic pathogens of humans and have been associated with a range of infections including urinary tract infections (UTIs), gastroenteritis, wound infections, pneumonia, brain abscesses, septicaemia, meningitis, and endocarditis, in particular in neonates and immunocompromised hosts (Doran, 1999). Citrobacter freundii is the most common Citrobacter species causing infections (Mohanty et al., 2007; Samonis et al., 2009; Bai et al., 2012), C. youngae and C. braakii are rarely a cause of infections. Some C. freundii isolates have acquired virulence traits and caused food poisoning or diarrhea in humans (Bai et al., 2012). The main virulence factors found in diarrhea-associated C. freundii are toxins, including Shiga-like toxins, heat stable toxins and a cholera toxin B subunit homolog (Bae et al., 2010). In our previous study, we identified one cytotoxic and aggregative C. freundii strain and found strains causing diarrheal infections in humans belonged to four sequence types (STs) (Bai et al., 2012). C. braakii has been associated with infections, such as hospital-acquired bacteremia and UTIs, making it an opportunistic pathogen (Arens and Verbist, 1997). It was reported that C. braakii caused an acute peritonitis in peritoneal dialysis patients (Bai et al., 2012). Moreover, C. braakii has been isolated from raw ground beef samples and pork products (Basra et al., 2015; Kwak et al., 2015).
Citrobacter spp. as a bacterial contaminant, has been partly responsible for the cause of food-borne diseases, and often transmitted through food and water (Ifeadike et al., 2012). Accordingly, food-handlers with poor personal hygiene could be potential sources of infections by these microorganisms (Ifeadike et al., 2012; Settanni et al., 2013). Citrobacter has been isolated from a range of foods (Tassew et al., 2010; Saba and Gonzalez-Zorn, 2012; Kouame et al., 2013) and food poisoning and diarrhea caused by foods contaminated by Citrobacter had been reported (Warner et al., 1991; Tschape et al., 1995; Doulgeraki et al., 2011; Giammanco et al., 2011).
Extended spectrum β-lactamases (ESBLs) producing Citrobacter strains have been reported. The prevalence of ESBLs varied among countries and Citrobacter spp. with reports of 4.9–20.6%, 0.2–4.6%, and 0.9% of C. freundii isolates from Korea, Japan and USA, respectively; and 3.5 and 60.0% of C. koseri isolates from USA and Japan, respectively (Park et al., 2005; Moland et al., 2006; Choi et al., 2007). Among Citrobacter spp. various CTX−M types, SHV and TEM have been reported worldwide (Kanamori et al., 2011).
Plasmid-mediated quinolone resistance genes including qnr and aac(6′)-Ibcr have been reported in Citrobacter spp. (Park et al., 2007; Zhang et al., 2012). The qnr and aac(6′)-Ibcr genes were present in 72.8 and 11.6% of clinical C. freundii isolates from China, respectively (Zhang et al., 2012). The prevalence of qnr genes was found in 38.4% of C. freundii isolates in Korea (Park et al., 2007). Numerous qnrB alleles have been detected, which seem to be more common than other qnr genes (Jacoby et al., 2014). About 40 qnrB variants are located on the chromosome of Citrobacter spp. especially C. freundii (Liao et al., 2015). Of the clinical C. freundii isolates with the qnr gene, 63.1% carried qnrB (Bae et al., 2010).
In this study, we analyzed the genetic diversity by Multilocus Sequence Typing (MLST) and antimicrobial resistance profiles of Citrobacter isolates from diarrheal patients, food and food-handlers in Maanshan Anhui Province, China, investigated the prevalence of blaCTX−M, blaSHV, blaTEM and qnr genes and determined the adhesion and cytotoxicity to HEp-2 cells of the isolates.
This study was reviewed and approved by the ethics committee of National Institute for Communicable Disease Control and Prevention, China CDC. Human fecal pecimens were acquired with the written informed consent of the diarrheal patients and food-handlers with the approval of the ethics committee of National Institute for Communicable Disease Control and Prevention, according to the medical research regulations of Ministry of Health (permit number 2007-17-3).
Sixty two Citrobacter isolates, including 13 C. freundii, eight C. braakii and 41 C. youngae isolates were obtained from patients and food samples from 2007 to 2011 in Maanshan Anhui Province, China. Among these 62 isolates, 18 C. youngae and two C. freundii isolates were obtained from diarrheal patients. The diarrheal patients harbored no other known enteric bacterial pathogens. Viral causes were not investigated. 42 isolates, including 23 C. youngae, 11 C. freundii and eight C. braakii were isolated from foods (including chicken, pork, fish and vegetables) and food-handlers (Table 1). The identity of each isolate was determined using API 20E test strips (bioMérieux, La Balme les Grottes, France) at the time of isolation, and they were stored as glycerol stocks at −80°C. Bacteria were grown in Luria-Bertani (LB) broth or on LB and Mueller–Hinton agar plates (pH 7.4) at 37°C.
The Citrobacter MLST scheme (http://pubmlst.org/cfreundii/) was used. The seven housekeeping genes for MLST were aspC, clpX, fadD, mdh, arcA, dnaG and lysP, and the MLST primers were as previously described (Bai et al., 2012) and synthesized by Shanghai Sangon Biological Engineering Technology and Services (Shanghai, China). PCR products were verified on 1% agarose gels and purified. DNA sequence was determined using Sanger sequencing in both directions (Shanghai Sangon Biological Engineering Technology and Services, China). Sequences were analyzed using SeqMan 7.0 software.
Antimicrobial susceptibility testing was carried out using the broth microdilution method according to CLSI recommendations. Minimum inhibitory concentration (MIC) results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. The antibiotics were serially diluted 2-fold in 50 μL of cation-adjusted Mueller-Hinton broth. The bacterial suspension was prepared from actively growing bacteria in 5 mL of cation-adjusted Mueller-Hinton broth, and diluted to a bacterial cell density of 106 colony forming units (CFU)/mL. Five microliter of bacterial suspension was then added to wells containing 100 μL of serially diluted antimicrobial agents to yield a final inoculum of approximately 5 × 104 CFU/mL. The MICs were read after overnight incubation (18–24 h) at 35°C. Quality control for MICs was performed using the reference E. coli ATCC 25922.
All the isolates were screened for the following genes, qnrA, qnrB, qnrS, qnrC, qnrD, aac(6′)-Ib-cr, qepA, blaCTX−M, blaSHV, and blaTEM by PCR using primers listed in Table S1. Primers of qnrA, qnrB, qnrS, qnrC, qnrD, aac(6′)-Ib-cr, and qepA were from Shao et al (Shao et al., 2011), primers for screening blaCTX−M, blaSHV and blaTEM genes were from Zhang et al. (2011). All primers were synthesized by Shanghai Sangon Biological Engineering Technology and Services (Shanghai, China). Positive PCR products were confirmed by sequencing.
In vitro adhesion to host cells was performed using the human epidermoid carcinoma cell line HEp-2 (CCC0068; Beijing Union Medical College cell resource center), as previously described (Bai et al., 2012). An adhesion index (<1; >1 and <50; >50) describing the mean number of bacteria per HEp-2 after examination of 10 visual fields was determined (Bai et al., 2012). Infections were repeated three times in duplicate.
The lactate dehydrogenase (LDH) released by the HEp-2 cells was determined using the Cytotox96 kit (Promega) according to the manufacturer's instructions. The relative amount of cytotoxicity was expressed as follows: (experimental release–spontaneous release)/(maximum release–spontaneous release)X100, where the spontaneous release was the amount of LDH activity in the supernatant of uninfected cells and the maximum release was that when cells were lysed with the lysis buffer provided by the manufacturer. All experiments were performed two times in duplicate (Bai et al., 2012).
The 62 Citrobacter isolates including 13 C. freundii, 41 C. youngae and eight C. Braakii isolates were divided into 53 STs by MLST (Table 1). The 41 C. youngae isolates were divided into 32 STs, 13 C. freundii isolates into 12 STs and eight C. Braakii isolates into 8 STs. Four STs (ST25, ST28, ST39, and ST59), all belonging to C. youngae, contained multiple isolates from two to five isolates. ST25 and ST28 each contained two isolates from diarrheal patients. ST39 contained five isolates with one from a diarrheal patient and four from foods. All three ST59 isolates were from foods.
A phylogenetic tree for the 62 isolates and representative isolates for ST1 to ST6 reported previously (Bai et al., 2012) was constructed using the neighbor-joining algorithm based on the concatenated sequences of the seven housekeeping genes (Figure 1). Salmonella LT2 was used as an outgroup. The tree could be divided into four clusters with robust bootstrap support of the major divisions. Cluster 1 is comprised of all C. freundii isolates; cluster 2 is comprised of all C. braakii isolates; and Cluster 3 and cluster 4 are comprised of all C. youngae isolates. It is interesting to note that clusters 3 and 4 are not grouped together. Rather, cluster 3 is grouped with clusters 1 and 2 with 90% bootstrap support, suggesting that cluster 3 should be a separate species from cluster 4. However, more isolates are needed to get a better understanding of the diversity of these 3 species and their relationships.
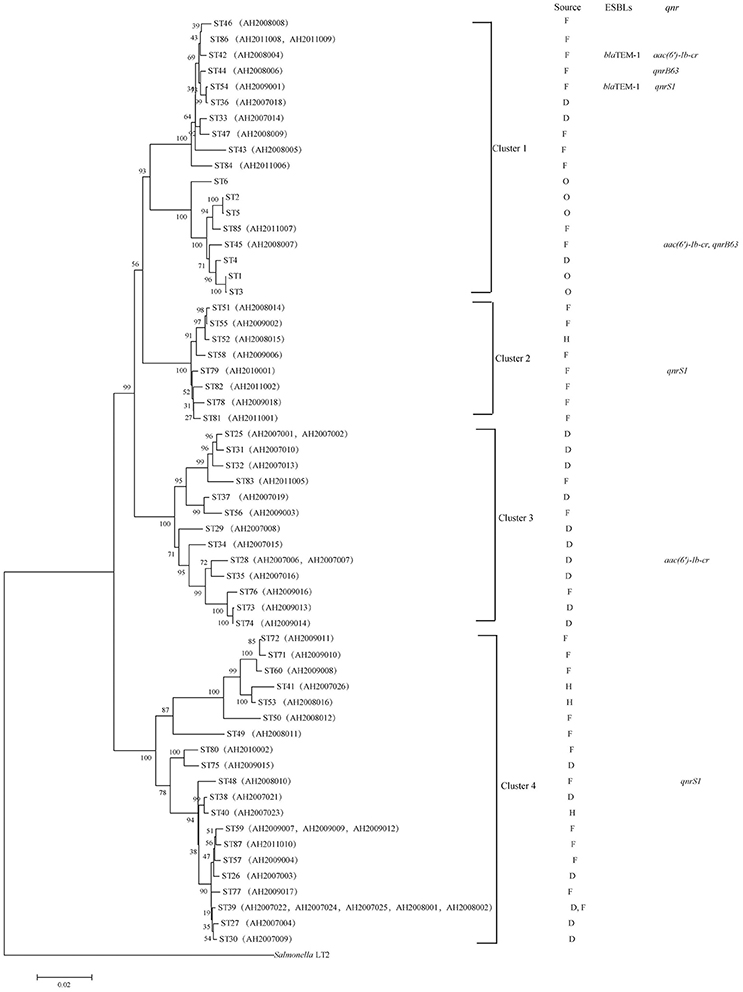
Figure 1. Phylogenetic relationships as determined by MLST data. The presence of ESBLs and qnr genes among the Citrobacter isolates is shown on the right. The tree was constructed using neighbor joining algorithm. For each ST, F, D, H, and O indicate isolates from foods, diarrheal patients, food-handlers and animals respectively. Cluster divisions are marked. Numbers on near the nodes are bootstrap values from 1,000 replicates.
The 62 Citrobacter isolates were tested for susceptibility to 22 antibiotics using the broth microdilution method according to CLSI recommendations (Table 2). All were resistant to cefoxitin (CFX), and sensitive to imipenem (IMI), meropenem (MEM) and amikacin (AMI). Non-susceptibility to β-lactams ranged from 0% to 100%; non-susceptibility to the three quinolones tested ranged from 12.9% to 27.4%; and non-susceptibility to other antibiotics included aminoglycosides (0–12.9%), phenicols (12.9%), sulfonamides (12.9–25.8%), tetracyclines (25.8%), and macrolide (3.2%) (Table 2).
Among the 62 Citrobacter isolates tested for MIC to 22 antibiotics, six C. youngae, seven C. freundii, and four C. braakii isolates were highly resistant to NAL, with MICs > 128 μg/mL and were multidrug resistant, with resistance to ≥3 antibiotics. Among 17 NAL resistant isolates, 14 isolates were from food and three were from diarrheal patients. These isolates belonged to different phylogenetic clusters, seven in cluster 1, four in cluster 2, one in cluster 3 and five in cluster 4. Three C. youngae isolates (one in cluster 3 and two in cluster 4) had a CTX MIC of > 16 μg/mL, and were multidrug resistant, with resistance to ≥8 antibiotics and were not closely related by MLST (Figure 1 and Table 3). There are six isolates (three C. youngae, two C. freundii, and one C. braakii) that had high MIC to CHL (>32 μg/mL), STR (>32 μg/mL), Sul (>512 μg/mL), TET (>32 μg/mL) and SXT (>8/152 μg/mL) (Table 3). The three highly multidrug resistant C. youngae isolates were separated on the tree but all in cluster 4 (Figure 1 and Table 3).
Two C. freundii isolates (AH2008004 and AH2009001) were found to harbor a blaTEM−1 gene by PCR and sequencing, both of which were resistant to AMP, CLP, LEV, SXT, CTX, NAL, CHL, STR, SUL, TET, CFX, and DOX. However, the two blaTEM−1 positive isolates belonged to two different STs with AH2008004 belonging to ST42 and AH2009001 belonging to ST54 (Figure 1 and Table 3).
Three isolates were positive for qnrS1 including one C. youngae (AH2008010), one C. freundii (AH2009001) and one C. braakii isolate (AH2010001). One C. youngae (AH2007006) and two C. freundii isolates (AH2008004 and AH2008007) were found to harbor an aac(6′)-Ib-cr gene. These two C. freundii isolates belonged to two different STs (Figure 1 and Table 1).
Two C. freundii isolates (AH2008006 and AH2008007) were found to harbor a qnrB gene. This qnrB allele has two in-phase ATG start codons. Wang et al. reported that two in-phase ATG start codons are present in many qnrB alleles (qnrB1, qnrB3, and qnrB5). However, in qnrB2 and qnrB4, the first ATG is out of phase with the remainder of the reading frame, the translation may be initiated at the second ATG codon (Wang et al., 2009). If sequence analysis from the ATG at position 37 (the second ATG codon), our qnrB allele has an identical qnrB sequence as qnrB77. But qnrB77 (GenBank accession no. KM985470.1) did not contain this 36 bp region. The 36 bp in our qnrB contained a LexA binding site (Wang et al., 2009). Therefore, we suggest that our qnrB allele is a variant of qnrB77.
These two qnrB positive isolates AH2008006 and AH2008007 belonged to two different STs, ST44 and ST45, respectively, suggesting that these isolates were epidemiologically unrelated (Figure 1).
Adhesion is an essential virulence property of bacterial pathogens. In vitro assays have been widely used to assess this property (Mange et al., 2006). We tested the 62 isolates for adhesion to HEp-2 cells and categorized the extent of adhesion using the adhesive index (Mange et al., 2006) (Table 1). Four isolates (including three C. youngae and one C. freundii) showed the strongest adhesion, with adhesion indexes >50. Twenty-five isolates showed intermediate adhesion, with an adhesion index between 1 and 50. Nineteen isolates showed little adhesion, with an adhesion index of <1. The remaining isolates showed ambivalent adhesion or no adhesion.
The adhesion rate was lower for C. braakii (25%) than C. youngae (88%) and C. freundii (77%). No difference was evident (P > 0.05) when adhesion behavior was compared in view of the source (human and food) of the Citrobacter isolates.
The 62 Citrobacter isolates were tested for Cytotoxicity to cultured HEp-2 cells by measuring the amount of lactate dehydrogenase (LDH) released by HEp-2 cells. We tested all isolates at 8 h. The released LDH levels ranged from 0.1–60.0% (Table 3). C. freundii strain CF74 were used as a positive control of cytotoxicity and C. freundii strain CF72 was used as a negative control (Bai et al., 2012). The levels of LDH released by CF74 and CF72 were 25.7 and 12.8% respectively. Seven isolates (including five C. youngae and two C. freundii isolates) released LDH more than 24%, showing strong cytotoxicity (Table 1). Among these seven isolates, three isolates showed strongest adherence; four isolates showed intermediate adhesion (Figure 2). Another seven isolates (including six C. youngae and one C. freundii isolates) released LDH from 18.7 to 22.4% and are considered intermediate cytotoxic. The remaining 48 isolates showed LDH release <16.7% and are likely to be non-cytotoxic.
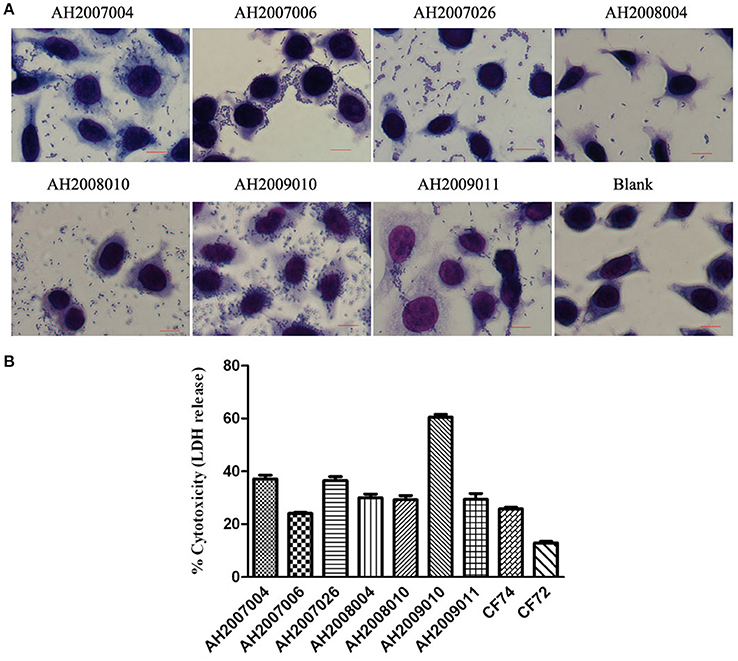
Figure 2. HEp-2 cell adhesion and cytotoxicity of Citrobacter isolates. (A) Light micrographs show the adherence patterns displayed by seven cytotoxic Citrobacter isolates. Blank as negative control. Bar: 10 μm. (B) Cytotoxicity was based the LDH released from HEp-2 cells after exposure to cytotoxic Citrobacter isolatesat 8 h. CF72 and CF74 were control strains.
Seven strongly cytotoxic isolates were multidrug resistant, with resistance to ≥3 antibiotics (Tables 1, 3). Four isolates (AH2008004, AH2008010, AH2009010, and AH2009011) showed multi-drug resistant (MDR) to nine antibiotics (CFX, NAL, CLP, LEV, CHL, STR, TET, SXT, and DOX). Moreover, AH2007006 harbored an aac(6′)-Ib-cr gene, AH2008004 harbored a blaTEM−1 gene and an aac(6′)-Ib-cr gene, and AH2008010 harbored a qnrS1 gene.
Four intermediate cytotoxic isolates (including AH2007001, AH2007008, AH2008002 and AH2009007) were resistant to AMP, CAZ and CFX (Tables 1, 3).
Citrobacter spp. especially C. freundii, is recognized as an emerging opportunistic pathogen and is known to cause a variety of infections (UTIs, wound infections, gastrointestinal infections, septicemia, meningitis), especially in immunocompromised patients and in hospital settings (Joaquin et al., 1991; Brenner et al., 1993; Gupta et al., 2003; Samonis et al., 2009; Ranjan and Ranjan, 2013; Leski et al., 2016a). This emergence has coincided with the finding that C. freundii is often resistant to multiple classes of antibiotics, suggesting that both clinical and environmental strains may be a reservoir of antimicrobial resistance determinants (Pepperell et al., 2002; Nada et al., 2004; Yim et al., 2013; Feng et al., 2015; Leski et al., 2016a; Sheppard et al., 2016). A recent survey of outpatients in Bo, Sierra Leone, revealed that a surprisingly high number of C. freundii isolates from UTIs were highly MDR (Leski et al., 2016b). In this study, we surveyed Citrobacter species from diarrheal patients and foods to provide a better understanding of their genetic diversity, antibiotic resistance profile, virulence properties and their potential as foodborne pathogens.
The worldwide prevalence of ESBLs in Citrobacter spp. was reported to be 0.5–36% (Ali et al., 2004; Fernandes et al., 2014; Praharaj et al., 2016). In India, 80.9% of Citrobacter isolates from hospitalized patients were ESBL producers (Praharaj et al., 2016). In this study, we did not test for ESBL phenotype but screened by PCR for BlaCTX−M, blaTEM and blaSHV genes. We found that a very low percentage of our isolates were blaTEM−1 positive (3.2%) and none carried BlaCTX−M and blaSHV. In contrast to a study in India, Shahid (Shahid, 2010) found that BlaCTX−M, blaTEM and blaSHV were found in 67.5%, 40%, and 25% of Citrobacter isolates from human clinical infections, respectively. However, most of our isolates were from food sources.
The prevalence of qnr and aac(6′)-Ib-cr genes varied. A Korean study showed that 38.4% of C. freundii isolates harbored qnr determinants (Park et al., 2007). A study from China showed prevalence of qnr and aac(6′)-Ib-cr genes at 63.3% and 26.7% in C. freundii isolates, respectively (Yang et al., 2008), while another Chinese study showed the prevalence of qnr and aac(6′)-Ib-cr in C. freundii at 72.8% and 68.9%, respectively (Zhang et al., 2012). The latter study also reported the prevalence of qnr and aac(6′)-Ib-cr in C. braakii at 42.9% and 42.9%, respectively (Zhang et al., 2012). We found much lower prevalence of qnr and aac(6′)-Ib-cr genes at 23.1% and 15.4% in C. freundii isolates; 2.4% and 2.4% in C. youngae isolates, and 12.5% and 0% in C. braakii isolates, respectively.
QnrB is the most common of the five qnr families and has the greatest number of allelic variants (Jacoby et al., 2011). We found a variant of qnrB77 in two C. freundii isolates. The variant contained a 36 bp sequence upstream of the qnrB77 start codon with an in-phase ATG codon at the beginning and a LexA binding site within the sequence, similar to several other qnrB alleles. The study by Wang et al. showed that the LexA binding site renders the qnrB under SOS control leading to its higher expression in response to ciprofloxacin or mitomycin C treatment (Wang et al., 2009). However, it should be noted that the qnrB77 first reported has no upstream sequence available in the GenBank entry and therefore it cannot be ascertained whether the sequence was absent or not reported.
QnrB-carrying C. freundii isolates do not always show high level of quinolone resistance (Zhang et al., 2012). However, our two qnrB-carrying C. freundii had a high MIC for NAL (>128 μg/mL). C. freundii carrying qnrS and aac(6′)-Ib-cr have been shown to have a higher MIC for quinolones (Zhang et al., 2012). Our results are consistent with this observation. One aac(6′)-Ib-cr-carrying C. freundii and three qnrS1-carrying Citrobacter isolates had high MIC of three quinolones (NAL, >128 μg/mL; CLP, >32 μg/mL; LEV, >16 μg/mL).
High prevalence of multidrug resistant Citrobacter has been reported (Moges et al., 2014; Leski et al., 2016b). Moges et al found that 13 MDR Citrobacter spp. were isolated from waste water in hospital and non-hospital environments (Moges et al., 2014). Twenty-two MDR C. freundii isolates from outpatient urine samples were resistant to >7 antibiotics out of the 11 tested, and 81.8% of the C. freundii isolates produced ESBLs (Leski et al., 2016b). In this study, 61.3% Citrobacter isolates were resistant to ≥3 antibiotics out of the 22 tested, and seven MDR isolates were strongly cytotoxic and four were intermediately cytotoxic. Moreover, two of the seven strongly cytotoxic and MDR isolates (from C. youngae) were obtained from diarrheal patients. The cytotoxic property of these isolates implies that they may cause more severe disease while the MDR properties limit clinical therapeutic options.
Citrobacter youngae is rarely a cause of infections. It has been reported to cause peritonitis (Chen et al., 2013). However, C. younage has not been recognized as a diarrheal pathogen. We found that 50% of the isolates showed moderate to strong adhesion and 15% of the isolates also showed strong cytoxicity. Nearly half of the C. younage isolates were from diarrheal patients. However, not all human isolates were adhesive or cytotoxic. Three of the six adhesive and cytotoxic isolates were obtained from diarrheal patients, suggesting that such strains are likely to cause diarrheal disease. STs from both human and food isolates were diverse with most STs being only isolated once. However, three STs were isolated more than once. Interestingly one ST was isolated from food as well as from a diarrheal patient. These findings suggest that C. youngae is a potential foodborne diarrheal pathogen.
Citrobacter freundii is the most common cause of Citrobacter infections (Mohanty et al., 2007; Samonis et al., 2009) and has been implicated in gastroenteritis associated outbreaks (Guerrant et al., 1976; Warner et al., 1991; Tschape et al., 1995; Doulgeraki et al., 2011; Giammanco et al., 2011) and foodborne outbreaks (Ifeadike et al., 2012; Settanni et al., 2013). We only obtained two isolates from diarrheal patients. Neither isolate was adhesive and one of them was intermediately cytotoxic, questioning its role in diarrhea in these cases. However, five isolates from foods were adhesive or strongly cytotoxic, suggesting that food isolates serve as a potential foodborne pathogen. The STs from this study were compared with six STs (ST1-ST6) from our previous study and 28 STs from the Citrobacter MLST database of global isolates, all STs found in this study were novel STs, showing high diversity of C. freundii from different regions and countries.
Citrobacter braakii is commonly found in water, soil, food, and the intestinal tracts of animals and humans (Basra et al., 2015). C. braakii is an opportunistic pathogen and has been isolated from hospital infections and UTIs (Arens and Verbist, 1997). C. braakii can cause acute peritonitis in peritoneal dialysis patients (Chao et al., 2013). All eight C. Braakii isolates from this study were isolated from foods. It requires further study to determine whether C. braakii contributes to diarrheal disease.
We analyzed 13 C. freundii, 41 C. youngae, and eight C. braakii isolates from Maanshan Anhui Province, China, isolated from human diarrheal patients and foods for their genetic diversity, antibiotic sensitivity and in vitro virulence phenotype. The 62 isolates were divided into 53 STs with all STs being novel, displaying high genetic diversity. Half of the isolates were MDR of three or more antibiotics. The blaTEM−1 gene was detected in two C. freundii isolates, while qnrS1 and aac(6′)-Ib-cr genes were detected in three Citrobacter isolates, respectively. We found seven isolates that showed strong cytotoxicity to HEp-2 cells, all of which were MDR. We also found a variant of qnrB77 that contained a LexA site in two C. freundii isolates. Our data suggest that food is an important source of Citrobacter species in transmission to humans and C. freundii and C. youngae are potential foodborne pathogens. Further studies are required to determine their public health significance.
LyL and JX designed the project; YlW carried out the sampling work; YZ carried out the experiments; YtW, LqL, and RL analyzed data; LyL and RL drafted the manuscript. All authors have read and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grants from National Natural Science Foundation of China (No. 81301401).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01357/full#supplementary-material
Ali, A. M., Rafi, S., and Qureshi, A. H. (2004). Frequency of extended spectrum beta lactamase producing gram negative bacilli among clinical isolates at clinical laboratories of Army Medical College, Rawalpindi. J. Ayub Med. Coll. Abbottabad 16, 35–37.
Arens, S., and Verbist, L. (1997). Differentiation and susceptibility of Citrobacter isolates from patients in a university hospital. Clin. Microbiol. Infect. 3, 53–57. doi: 10.1111/j.1469-0691.1997.tb00251.x
Bae, I. K., Park, I., Lee, J. J., Sun, H. I., Park, K. S., Lee, J. E., et al. (2010). Novel variants of the qnrB gene, qnrB22 and qnrB23, in Citrobacter werkmanii and Citrobacter freundii. Antimicrob. Agents Chemother. 54, 3068–3069. doi: 10.1128/AAC.01339-09
Bai, L., Xia, S., Lan, R., Liu, L., Ye, C., Wang, Y., et al. (2012). Isolation and characterization of cytotoxic, aggregative Citrobacter freundii. PLoS ONE 7:e33054. doi: 10.1371/journal.pone.0033054
Basra, P., Koziol, A., Wong, A., and Carrillo, C. D. (2015). Complete genome sequences of citrobacter braakii strains GTA-CB01 and GTA-CB04, isolated from ground beef. Genome Announc 3:e01307–14. doi: 10.1128/genomeA.01307-14
Brenner, D. J., Grimont, P. A., Steigerwalt, A. G., Fanning, G. R., Ageron, E., and Riddle, C. F. (1993). Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three unnamed Citrobacter genomospecies. Int. J. Syst. Bacteriol. 43, 645–658. doi: 10.1099/00207713-43-4-645
Chao, C. T., Lee, S. Y., Yang, W. S., Chen, H. W., Fang, C. C., Yen, C. J., et al. (2013). Citrobacter peritoneal dialysis peritonitis: rare occurrence with poor outcomes. Int. J. Med. Sci. 10, 1092–1098. doi: 10.7150/ijms.6251
Chen, K. J., Chen, T. H., and Sue, Y. M. (2013). Citrobacter youngae and Pantoea agglomerans peritonitis in a peritoneal dialysis patient. Perit. Dial. Int. 33, 336–337. doi: 10.3747/pdi.2012.00151
Choi, S. H., Lee, J. E., Park, S. J., Kim, M. N., Choo, E. J., Kwak, Y. G., et al. (2007). Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp. Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 26, 557–561. doi: 10.1007/s10096-007-0308-2
Doran, T. I. (1999). The role of Citrobacter in clinical disease of children: review. Clin. Infect. Dis. 28, 384–394. doi: 10.1086/515106
Doulgeraki, A. I., Paramithiotis, S., and Nychas, G. J. (2011). Characterization of the Enterobacteriaceae community that developed during storage of minced beef under aerobic or modified atmosphere packaging conditions. Int. J. Food Microbiol. 145, 77–83. doi: 10.1016/j.ijfoodmicro.2010.11.030
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J. Antimicrob. Chemother. 70, 2987–2991. doi: 10.1093/jac/dkv232
Fernandes, R., Amador, P., Oliveira, C., and Prudencio, C. (2014). Molecular characterization of ESBL-producing Enterobacteriaceae in northern Portugal. Sci. World J. 2014:782897. doi: 10.1155/2014/782897
Giammanco, G. M., Aleo, A., Guida, I., and Mammina, C. (2011). Molecular epidemiological survey of Citrobacter freundii misidentified as Cronobacter spp. (Enterobacter sakazakii) and Enterobacter hormaechei isolated from powdered infant milk formula. Foodborne Pathog. Dis. 8, 517–525. doi: 10.1089/fpd.2010.0719
Guerrant, R. L., Dickens, M. D., Wenzel, R. P., and Kapikian, A. Z. (1976). Toxigenic bacterial diarrhea: nursery outbreak involving multiple bacterial strains. J. Pediatr. 89, 885–891. doi: 10.1016/S0022-3476(76)80591-4
Gupta, N., Yadav, A., Choudhary, U., and Arora, D. R. (2003). Citrobacter bacteremia in a tertiary care hospital. Scand. J. Infect. Dis. 35, 765–768. doi: 10.1080/00365540310016376
Ifeadike, C. O., Ironkwe, O. C., Adogu, P. O., Nnebue, C. C., Emelumadu, O. F., Nwabueze, S. A., et al. (2012). Prevalence and pattern of bacteria and intestinal parasites among food handlers in the Federal Capital Territory of Nigeria. Niger. Med. J. 53, 166–171. doi: 10.4103/0300-1652.104389
Jacoby, G. A., Griffin, C. M., and Hooper, D. C. (2011). Citrobacter spp. as a source of qnrB Alleles. Antimicrob. Agents Chemother. 55, 4979–4984. doi: 10.1128/AAC.05187-11
Jacoby, G. A., Strahilevitz, J., and Hooper, D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2:PLAS-0006-2013. doi: 10.1128/microbiolspec.PLAS-0006-2013
Joaquin, A., Khan, S., Russel, N., and al Fayez, N. (1991). Neonatal meningitis and bilateral cerebellar abscesses due to Citrobacter freundii. Pediatr. Neurosurg. 17, 23–24. doi: 10.1159/000120561
Kanamori, H., Yano, H., Hirakata, Y., Endo, S., Arai, K., Ogawa, M., et al. (2011). High prevalence of extended-spectrum beta-lactamases and qnr determinants in Citrobacter species from Japan: dissemination of CTX-M-2. J. Antimicrob. Chemother. 66, 2255–2262. doi: 10.1093/jac/dkr283
Kouame, A. K., Djeni, T. N., N'Guessan, F. K., and Dje, M. K. (2013). Postprocessing microflora of commercial attieke (a fermented cassava product) produced in the south of Cote d'Ivoire. Lett. Appl. Microbiol. 56, 44–50. doi: 10.1111/lam.12014
Kwak, H. L., Han, S. K., Park, S., Park, S. H., Shim, J. Y., Oh, M., et al. (2015). Development of a rapid and accurate identification method for Citrobacter species isolated from pork products using a Matrix-Assisted Laser-Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). J. Microbiol. Biotechnol. 25, 1537–1541. doi: 10.4014/jmb.1503.03071
Leski, T. A., Taitt, C. R., Bangura, U., Ansumana, R., Stenger, D. A., Wang, Z., et al. (2016a). Finished genome sequence of the highly multidrug-resistant human urine isolate Citrobacter freundii strain SL151. Genome Announc 4:e01225–16. doi: 10.1128/genomea.01225-16
Leski, T. A., Taitt, C. R., Bangura, U., Stockelman, M. G., Ansumana, R., Cooper, W. H. III., et al. (2016b). High prevalence of multidrug resistant Enterobacteriaceae isolated from outpatient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infect. Dis. 16:167. doi: 10.1186/s12879-016-1495-1
Liao, X., Fang, L., Li, L., Sun, J., Li, X., Chen, M., et al. (2015). Characterization of chromosomal qnrB and ampC alleles in Citrobacter freundii isolates from different origins. Infect. Genet. Evol. 35, 214–220. doi: 10.1016/j.meegid.2015.07.011
Mange, J. P., Stephan, R., Borel, N., Wild, P., Kim, K. S., Pospischil, A., et al. (2006). Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 6:58. doi: 10.1186/1471-2180-6-58
Moges, F., Endris, M., Belyhun, Y., and Worku, W. (2014). Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Res. Notes 7:215. doi: 10.1186/1756-0500-7-215
Mohanty, S., Singhal, R., Sood, S., Dhawan, B., Kapil, A., and Das, B. K. (2007). Citrobacter infections in a tertiary care hospital in Northern India. J. Infect. 54, 58–64. doi: 10.1016/j.jinf.2006.01.015
Moland, E. S., Hanson, N. D., Black, J. A., Hossain, A., Song, W., and Thomson, K. S. (2006). Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 44, 3318–3324. doi: 10.1128/JCM.00756-06
Nada, T., Baba, H., Kawamura, K., Ohkura, T., Torii, K., and Ohta, M. (2004). A small outbreak of third generation cephem-resistant Citrobacter freundii infection on a surgical ward. Jpn. J. Infect. Dis. 57, 181–182.
Park, Y. J., Park, S. Y., Oh, E. J., Park, J. J., Lee, K. Y., Woo, G. J., et al. (2005). Occurrence of extended-spectrum beta-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn. Microbiol. Infect. Dis. 51, 265–269. doi: 10.1016/j.diagmicrobio.2004.11.009
Park, Y. J., Yu, J. K., Lee, S., Oh, E. J., and Woo, G. J. (2007). Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J. Antimicrob. Chemother. 60, 868–871. doi: 10.1093/jac/dkm266
Pepperell, C., Kus, J. V., Gardam, M. A., Humar, A., and Burrows, L. L. (2002). Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 46, 3555–3560. doi: 10.1128/AAC.46.11.3555-3560.2002
Praharaj, A. K., Khajuria, A., Kumar, M., and Grover, N. (2016). Phenotypic detection and molecular characterization of beta-lactamase genes among Citrobacter species in a tertiary care hospital. Avicenna J. Med. 6, 17–27. doi: 10.4103/2231-0770.173578
Ranjan, K. P., and Ranjan, N. (2013). Citrobacter: an emerging health care associated urinary pathogen. Urol. Ann. 5, 313–314.
Saba, C. K., and Gonzalez-Zorn, B. (2012). Microbial food safety in Ghana: a meta-analysis. J. Infect. Dev. Ctries. 6, 828–835. doi: 10.3855/jidc.1886
Samonis, G., Karageorgopoulos, D. E., Kofteridis, D. P., Matthaiou, D. K., Sidiropoulou, V., Maraki, S., et al. (2009). Citrobacter infections in a general hospital: characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 28, 61–68. doi: 10.1007/s10096-008-0598-z
Settanni, L., Miceli, A., Francesca, N., Cruciata, M., and Moschetti, G. (2013). Microbiological investigation of Raphanus sativus L. grown hydroponically in nutrient solutions contaminated with spoilage and pathogenic bacteria. Int. J. Food Microbiol. 160, 344–352. doi: 10.1016/j.ijfoodmicro.2012.11.011
Shahid, M. (2010). Citrobacter spp. simultaneously harboring blaCTX-M, blaTEM, blaSHV, blaampC, and insertion sequences IS26 and orf513: an evolutionary phenomenon of recent concern for antibiotic resistance. J. Clin. Microbiol. 48, 1833–1838. doi: 10.1128/JCM.01467-09
Shao, Y., Xiong, Z., Li, X., Hu, L., Shen, J., Li, T., et al. (2011). Prevalence of plasmid-mediated quinolone resistance determinants in Citrobacter freundii isolates from Anhui province, PR China. J. Med. Microbiol. 60(Pt 12), 1801–1805. doi: 10.1099/jmm.0.034082-0
Sheppard, A. E., Stoesser, N., Wilson, D. J., Sebra, R., Kasarskis, A., Anson, L. W., et al. (2016). Nested Russian Doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 60, 3767–3778. doi: 10.1128/AAC.00464-16
Tassew, H., Abdissa, A., Beyene, G., and Gebre-Selassie, S. (2010). Microbial flora and food borne pathogens on minced meat and their susceptibility to antimicrobial agents. Ethiop. J. Health Sci. 20, 137–143.
Tschape, H., Prager, R., Streckel, W., Fruth, A., Tietze, E., and Bohme, G. (1995). Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infection source. Epidemiol. Infect. 114, 441–450. doi: 10.1017/S0950268800052158
Wang, M., Jacoby, G. A., Mills, D. M., and Hooper, D. C. (2009). SOS regulation of qnrB expression. Antimicrob. Agents Chemother. 53, 821–823. doi: 10.1128/AAC.00132-08
Warner, R. D., Carr, R. W., McCleskey, F. K., Johnson, P. C., Elmer, L. M., and Davison, V. E. (1991). A large nontypical outbreak of Norwalk virus. Gastroenteritis associated with exposing celery to nonpotable water and with Citrobacter freundii. Arch. Intern. Med. 151, 2419–2424. doi: 10.1001/archinte.1991.00400120061010
Yang, H., Chen, H., Yang, Q., Chen, M., and Wang, H. (2008). High prevalence of plasmid-mediated quinolone resistance genes qnr and aac(6′)-Ib-cr in clinical isolates of Enterobacteriaceae from nine teaching hospitals in China. Antimicrob. Agents Chemother. 52, 4268–4273. doi: 10.1128/AAC.00830-08
Yim, G., Kwong, W., Davies, J., and Miao, V. (2013). Complex integrons containing qnrB4-ampC (blaDHA−1) in plasmids of multidrug-resistant Citrobacter freundii from wastewater. Can. J. Microbiol. 59, 110–116. doi: 10.1139/cjm-2012-0576
Zhang, R., Ichijo, T., Huang, Y. L., Cai, J. C., Zhou, H. W., Yamaguchi, N., et al. (2012). High prevalence of qnr and aac(6′)-Ib-cr genes in both water-borne environmental bacteria and clinical isolates of Citrobacter freundii in China. Microbes Environ. 27, 158–163. doi: 10.1264/jsme2.ME11308
Keywords: Citrobacter, Multilocus sequence typing, Multidrug resistance, adhesion, cytotoxicity
Citation: Liu L, Lan R, Liu L, Wang Y, Zhang Y, Wang Y and Xu J (2017) Antimicrobial Resistance and Cytotoxicity of Citrobacter spp. in Maanshan Anhui Province, China. Front. Microbiol. 8:1357. doi: 10.3389/fmicb.2017.01357
Received: 11 May 2017; Accepted: 04 July 2017;
Published: 20 July 2017.
Edited by:
David Rodriguez-Lazaro, University of Burgos, SpainReviewed by:
Alberto Quesada, University of Extremadura, SpainCopyright © 2017 Liu, Lan, Liu, Wang, Zhang, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianguo Xu, eHVqaWFuZ3VvQGljZGMuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.