
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 July 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01327
This article is part of the Research Topic Rhizosphere Bacterial Community and Bacteria-Plant Interactions View all 14 articles
Azorhizobium caulinodans ORS571 is a motile soil bacterium that interacts symbiotically with legume host Sesbania rostrata, forming nitrogen-fixing root and stem nodules. Bacterial chemotaxis plays an important role in establishing this symbiotic relationship. To determine the contribution of chemotaxis to symbiosis in A. caulinodans ORS571-S. rostrata, we characterized the function of TlpA1 (transducer-like protein in A. caulinodans), a chemoreceptor predicted by SMART (Simple Modular Architecture Research Tool), containing two N-terminal transmembrane regions. The tlpA1 gene is located immediately upstream of the unique che gene cluster and is transcriptionally co-oriented. We found that a ΔtlpA1 mutant is severely impaired for chemotaxis to various organic acids, glycerol and proline. Furthermore, biofilm forming ability of the strain carrying the mutation is reduced under certain growth conditions. Interestingly, competitive colonization ability on S. rostrata root surfaces is impaired in the ΔtlpA1 mutant, suggesting that chemotaxis of the A. caulinodans ORS571 contributes to root colonization. We also found that TlpA1 promotes competitive nodulation not only on roots but also on stems of S. rostrata. Taken together, our data strongly suggest that TlpA1 is a transmembrane chemoreceptor involved in A. caulinodans-S. rostrata symbiosis.
Bacterial motility and chemotaxis are thought to play an important role in adaptation to soil and rhizosphere. Chemotaxis is an important factor in establishing the initial chemical and physical contact between the bacteria and host roots (Caetano-Anollés et al., 1988b; Miller et al., 2007; Scharf et al., 2016). Chemotaxis towards root exudates has been suggested to be the first step in bacterial colonization of plant roots (Vande Broek and Vanderleyden, 1995; Gupta, 2003; Greer-Phillips et al., 2004). Bacterial biofilm formation also contributes to effective colonization of plant roots (Timmusk et al., 2005; Li et al., 2013). In the rhizobia-legume association, chemotaxis towards chemoattractants released from roots can promote infection and nodulation by guiding rhizobia to proper site for nodulation (Ames and Bergman, 1981; Bauer and Caetano-Anollés, 1990; Greer-Phillips et al., 2004). Chemoreceptors allow bacteria to detect attractants and repellents, thereby directing bacteria to find favorable conditions (Broek and Vanderleyden, 1995). Previous research has suggested that various species of rhizobia show chemotaxis toward plant root or seed exudates (Aguilar et al., 1988; Caetano-Anollés et al., 1988a; Barbour et al., 1991; Webb et al., 2014, 2017) and that certain mutants impaired for chemotaxis are also less competitive than wild-type in root colonization and nodulation (Dharmatilake and Bauer, 1992; Yost et al., 1998; Miller et al., 2007; Jiang et al., 2016a).
Motile bacteria such as Escherichia coli regulate chemotactic behavior based on environmental signals by a two-component signal transduction network. The molecular mechanism of chemotaxis has been best studied in the model organism, E. coli, which encodes a single chemotaxis system comprised of a histidine kinase CheA and its response regulator CheY (Falke et al., 1997). CheA is coupled to the transmembrane chemoreceptor by CheW scaffold protein. Transmembrane chemoreceptors provide input for initiation of the chemotaxis signal transduction cascade (Wuichet et al., 2007) and regulate cell motility by modulating histidine kinase activity of CheA. Phosphorylated CheA transfers its phosphoryl group to CheY, forming phospho-CheY, which influences the rotational direction of the flagella (Hazelbauer et al., 2008). However, chemosensory signaling systems in many other bacteria, including plant-associated bacteria, are more complex. Many motile bacteria possess two or more chemotaxis systems and several chemoreceptors (Mukherjee et al., 2016). Chemoreceptors typically contain two distinct structural and functional domains: a variable N-terminal periplasmic ligand-binding domain (LBD), which is responsible for sensing environmental stimuli, and a conserved C-terminal domain that transmits signals to the downstream chemotactic machinery. The C-terminal domain of membrane-bound chemoreceptor can be further differentiated into a HAMP (Histidine kinases, Adenylyl cyclases, Methyl-accepting proteins and Phosphatases) domain and a MA (methyl-accepting chemotaxis-like) domain. The HAMP domain transmits transmembrane ligand binding signals to the MA domain (Collins et al., 2014). The MA domain interacts with CheW and CheA (Zhulin, 2001; Collins et al., 2014).
Azorhizobium caulinodans ORS571 is a motile alpha-proteobacterium, capable of fixing nitrogen. It can also form a nitrogen-fixing symbiotic relationship with the host plant Sesbania rostrata (Trinick, 1979; Dreyfus et al., 1983, 1988; Tsien et al., 1983), which is a small semi-aquatic leguminous tree (Cook et al., 2005; Capoen et al., 2010). This symbiotic relationship leads to the formation of nitrogen fixing nodules in the S. rostrata (Capoen et al., 2010). While nodules normally occur on roots of leguminous plants, stem nodules can also be formed at the site of adventitious root primordia following crack-entry invasion by A. caulinodans (Tsien et al., 1983). Chemoreceptors of nitrogen-fixing rhizobia play a crucial role in establishing a symbiotic relationship with the host plant (Yost et al., 1998).
We used the MiST2 (Microbial Signal Transduction) and SMART (Simple Modular Architecture Research Tool) databases to identify 43 chemoreceptors in A. caulinodans ORS571 (Jiang et al., 2016b) based on its sequenced genome (Lee et al., 2008). Of these, 30 were localized to the cytoplasmic membrane fraction as they were predicted to contain two transmembrane spanning regions. Four of the chemoreceptors contained only one transmembrane domain, while nine internal chemotaxis proteins lacked a transmembrane domain. Jiang et al. (2016a) have identified a PAS-containing soluble internal chemotaxis protein IcpB in A. caulinodans that is important for nodulation and nitrogen fixation upon symbiosis with S. rostrata (Jiang et al., 2016a). In this work, we characterize a transmembrane chemoreceptor TlpA1 in A. caulinodans and describe its role in chemotaxis and competitive nodule formation on roots and stems of host plants.
All bacterial strains and plasmids used in this study are listed Supplementary Table S1. Wild-type A. caulinodans ORS571 and its mutant derivatives were grown in L3 minimal medium (Jiang et al., 2016a), TY medium or on agar plates at 37°C containing 25 μg ml-1 nalidixic acid and 100 μg ml-1 ampicillin. E. coli was grown in LB broth or on LB agar plates at 37°C with the following concentrations of antibiotics as required: 50 μg ml-1 kanamycin, 100 μg ml-1 ampicillin, 50 μg ml-1 gentamycin, 10 μg ml-1 tetracycline.
A tlpA1-deficient strain was constructed as follows: A 693-bp fragment spanning a region immediately upstream of tlpA1 and the 5′ end of the gene (Supplementary Figure S1A) was amplified by PCR from the genomic DNA of A. caulinodans ORS571 using primer pair TlpA1UF and TlpA1UR (Supplementary Table S2). A 653-bp fragment encompassing 501-bp at 3′ end of tlpA1, the stop codon and a 152-bp region immediately downstream of the gene (Supplementary Figure S1A) was amplified from the genomic DNA of the wild-type strain using the primer pair TlpA1DF and TlpA1DR (Supplementary Table S2). The 693-bp fragment was digested with KpnI and NdeI and ligated into KpnI and NdeI sites of the vector pCM351 (Marx and Lidstrom, 2002). One of the resulting recombinant plasmid was designated as pCM351::UF. The 653-bp fragment was digested with ApaI-SacI enzymes and cloned into ApaI-SacI sites of the recombinant plasmid pCM351::UF. The ligation mixtures were transformed into E. coli DH5α to recover individual clones. One of the resulting recombinant plasmids, designated as pCM351::UF::DF, was verified by sequencing. The recombinant plasmid pCM351::UF::DF was transferred into wild-type A. caulinodans ORS571 by tri-parental conjugation with a helper plasmid pRK2013 (Figurski and Helinski, 1979). Colonies were screened for loss of the recombinant plasmid and for double homologous recombination. Potential mutants were obtained from on TY plates by selecting for gentamicin-resistance (from pCM351 vector) and tetracycline-sensitivity (Marx and Lidstrom, 2002). The gentamicin gene was then deleted by introduction of plasmid pCM157 (Marx and Lidstrom, 2002). Mutations in the strains were verified by PCR with primer pair TlpA1F and TlpA1R. A confirmed mutation was named ΔtlpA1.
For complementation, the tlpA1 ORF with 737-bp upstream non-coding sequence was amplified by PCR using primer pair TlpA1comF and TlpA1comR (Supplementary Table S2). The amplified 2250-bp fragment was inserted into HindIII and BamHI sites of the broad-host-range cloning vector pBBR1MCS-2 (Kovach et al., 1995). The ligation mixture was transformed into E. coli DH5α. DNA sequences from individual clones were sequenced and one verified clone was designated as pBBR1MCS-2-tlpA1-com. This recombinant plasmid was then introduced into the tlpA1 mutant via tri-parental mating and the transformants were recovered by selection for kanamycin resistance. One of the resulting strains was designated as tlpA1-com.
To construct a cheA deletion mutant of A. caulinodans ORS571, a 696-bp upstream fragment (UF) was amplified by PCR using primer pair CheAUF and CheAUR (Supplementary Table S2), and a 675-bp downstream fragment (DF) was amplified by PCR using primer pair CheADF and CheADR (Supplementary Table S2). The upstream PCR product was digested with NsiI-NdeI and cloned into NsiI-NdeI sites of vector pCM351. The resulting plasmid was designated as pCM351::UF. The downstream PCR product was digested with AgeI-SacI and cloned into AgeI-SacI sites of pCM351::UF. The final plasmid pCM351::UF::DF was recovered from the ligation mixture by transforming into E. coli DH5α and verified by sequencing. The plasmid was transferred into A. caulinodans ORS571 by a tri-parental mating with the aid of a helper plasmid pRK2013. The gentamicin gene replaces the cheA gene by double homologous recombination in the potential mutant, which was then removed by introducing the vector pCM157 (Marx and Lidstrom, 2002). The cheA-deletion mutant was thus generated by deletion of an internal 2340-bp fragment (Supplementary Figure S1B) and was verified by PCR with primer pair CheAF and CheAR. The confirmed mutant was named ΔcheA.
Strains were grown overnight in TY medium containing 25 μg ml-1 nalidixic acid and 100 μg ml-1 ampicillin. Cultures were diluted either into L3 medium containing 10 mM succinate and 10 mM NH4Cl or into TY medium to an initial OD600 of 0.02 and grown at 200 rpm and 37°C. All data were depicted as means and standard deviations from three individual repetitions.
For the soft agar assay, overnight cultures were washed and resuspended in chemotaxis buffer (Jiang et al., 2016a). The cultures were then adjusted to an optical density at 600 nm (OD600) of 1.0. To compare chemotaxis response to different carbon sources, 5 μl aliquots of cell suspensions of the wild-type, mutant and complemented strains were inoculated on L3 soft agar plates (0.3% agar) containing 10 mM various carbon sources (succinate, citrate, proline, tartrate, malate, and glycerol) with or without 10 mM NH4Cl. The inoculated plates were incubated for three days at 37°C.
A quantitative capillary chemotaxis assay was carried out as described in Reyes-Darias et al. (2016) with a few modifications. The wild-type and ΔtlpA1 strains were grown overnight in TY medium at 200 rpm and 37°C. The cells were then washed with chemotaxis buffer and the cell pellets were resuspended to a final density of OD600 = 0.05. 200 μl of bacterial suspension mixtures (containing 100 μl wild-type and 100 μl mutant) were added into each well of a 96-well plate. Open ends of the capillaries containing chemotaxis buffer or 10 mM succinate as carbon source were then inserted into the wells. After incubation for an hour, the portions of the capillaries that were in contact with bacteria were rinsed with sterile water. The sealed ends were broken and the contents were transferred into microfuge tubes containing 1 ml sterile water. From each of these tubes, five individual 20 μl aliquots were placed onto TY plates and incubated at 37°C for 48 h prior to cell counting. Cells on TY plates were tested by PCR using primer pair TlpA1F and TlpA1R.
Total RNA was isolated from cultured free-living cells. cDNA was generated using GoScriptTM Reverse Transcription Kit (Promega) and diluted 10-fold (analysis of 16S rRNA expression) and 500-fold (analysis of the cheA gene expression) for subsequent PCR amplification. Quantitative PCR was performed using GoTaq® qPCR Master Mix kit (Promega) with gene-specific primer pairs (TlpA1QF and TlpA1QR for tlpA1, CheAQF and CheAQR for cheA, 16SQF and 16SQR for 16S rRNA). Genomic DNA of ORS571 strain was used as template to construct a standard curve for determination of gene copy numbers. Transcript levels of the cheA gene were normalized against 16S rRNA levels. Real-time PCR was carried out on a Bio-Rad CFX96TM real-time PCR system. The program for amplification included an initial denaturation step at 95°C for 2 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Specificity of the reaction was monitored by melting curve analysis.
Single colonies were inoculated into L3 liquid medium and incubated at 37°C and 200 rpm. The cell culture (OD600 around 2.5) was washed once with L3 medium and resuspended in an equal volume of L3 medium. For biofilm formation assay, 1.5 ml L3 medium was added to each glass tube, into which 150 μl cell cultures was added and incubated at 37°C for 3–5 days. For Crystal Violet (CV) staining, cultures were removed carefully with a pipette, and the tubes were washed 3–5 times with ddH2O and 1.6 ml of 0.5% CV was added into each tube. After incubation for 20 min, CV was removed and the tubes were washed five times with ddH2O. Finally, 1.5 ml of 30% acetic acid was added to each tube to wash off the “CV ring”. The OD value of each tube was determined at 570 nm after CV staining.
Sesbania rostrata seeds were treated as described previously (Suzuki et al., 2007) with a few modifications. Seeds were immersed in concentrated sulfuric acid for 30 min to induce uniform germination, rinsed five times with sterile water, and soaked in sterile water for two days at 37°C in dark. Cells were grown in TY medium and harvested by centrifugation at mid-log phase (OD600 of 0.5–0.6). The cells were washed thrice and resuspended in sterile chemotaxis buffer (Greer-Phillips et al., 2004) to an OD600 of 0.5. The concentration of the cell suspensions (OD600 = 0.5) was further verified by serial dilution and plating onto TY plates. 50 ml cultures of each strain were used to soak (50 rpm, 28°C) two day-old seedlings of S. rostrata for 4 and 24 h. The seedlings were then washed four times with sterile water at 100 rpm for 1 min each in a rotary bath. Equal-sized roots from five seedlings were then dispersed in sterile water by using a homogenizer. Serial dilutions from this suspension were spread onto TY agar plates and incubated for two days at 37°C. The adsorption pattern of root surface was examined by analysis of the wild-type and ΔtlpA1 colonies on TY plates. In competition adsorption experiments, surface-sterilized seedlings were co-inoculated with wild-type ORS571 and ΔtlpA1 strains at approximately 1:1 and 1:10 ratios for 4 and 24 h. Adsorbed bacteria were detected by PCR using the primer pair TlpA1F and TlpA1R. A non-inoculated flask was used as a control for all experiments.
Nodulation and competitive nodulation assays of S. rostrata were performed as previously described in Yost et al. (1998) with a few modifications. For nodulation assays on roots, surface-sterilized seedlings were co-inoculated with wild-type ORS571 or ΔtlpA1 strains at an OD600 of approximately 0.5. For nodulation assays on stem, S. rostrata seeds were germinated and the seedlings were transferred to vermiculite in moistened pots containing a low-N nutrient (Callow and Vincent, 1971). After 3–4 weeks of plant growth, S. rostrata stems were inoculated with either the wild-type or ΔtlpA1 strains at an OD600 of approximately 0.5. For nodulation competition assays on roots and stems, surface-sterilized seedlings and stems were co-inoculated with the wild-type ORS571 and ΔtlpA1 mixtures at approximately 1:1 and 1:10 ratios. All S. rostrata plants were grown in the greenhouse at 27°C. Nodules were harvested 3 weeks post inoculation and bacteria were re-isolated from root and stem nodules. Competitive abilities of the wild-type and ΔtlpA1 strains were analyzed by PCR using the primer pair TlpA1F and TlpA1R. Non-inoculated plants served as controls.
To measure symbiotic ARA, 50 root or stem nodules were weighed and immediately placed into individual 3 ml sealed tubes. 300 μl acetylene (C2H2) gas was added to each tube up to 10% (vol/vol) and incubated at 37°C for 4 h. 100 μl gas was taken out from each tube and analyzed using a gas chromatograph (Agilent Technologies 7890A). Nitrogen-fixing ability was expressed as μmol C2H4 produced h-1 g -1 of fresh nodules. Each measurement was repeated at least three times.
We used MiST2 and SMART databases and identified a chemotaxis gene cluster in the genome of A. caulinodans ORS571. This cluster encodes homologs of known chemotaxis proteins, which we termed as che gene cluster (Jiang et al., 2016b). This unique che gene cluster contains cheA, cheW, cheY, cheB, and cheR genes with a chemotaxis receptor, which we named tlpA1 (transducer-like protein in A. caulinodans), located upstream of cheA and predicted to be transcriptionally co-oriented (Figure 1A). TlpA1 is predicted to be a ∼60 kDa protein. N-terminal domain of TlpA1 consists of two transmembrane helices (TM) and a periplasmic ligand-binding domain (LBD), while the C-terminal region consists of a HAMP and a highly conserved MA domain (Figure 1B).
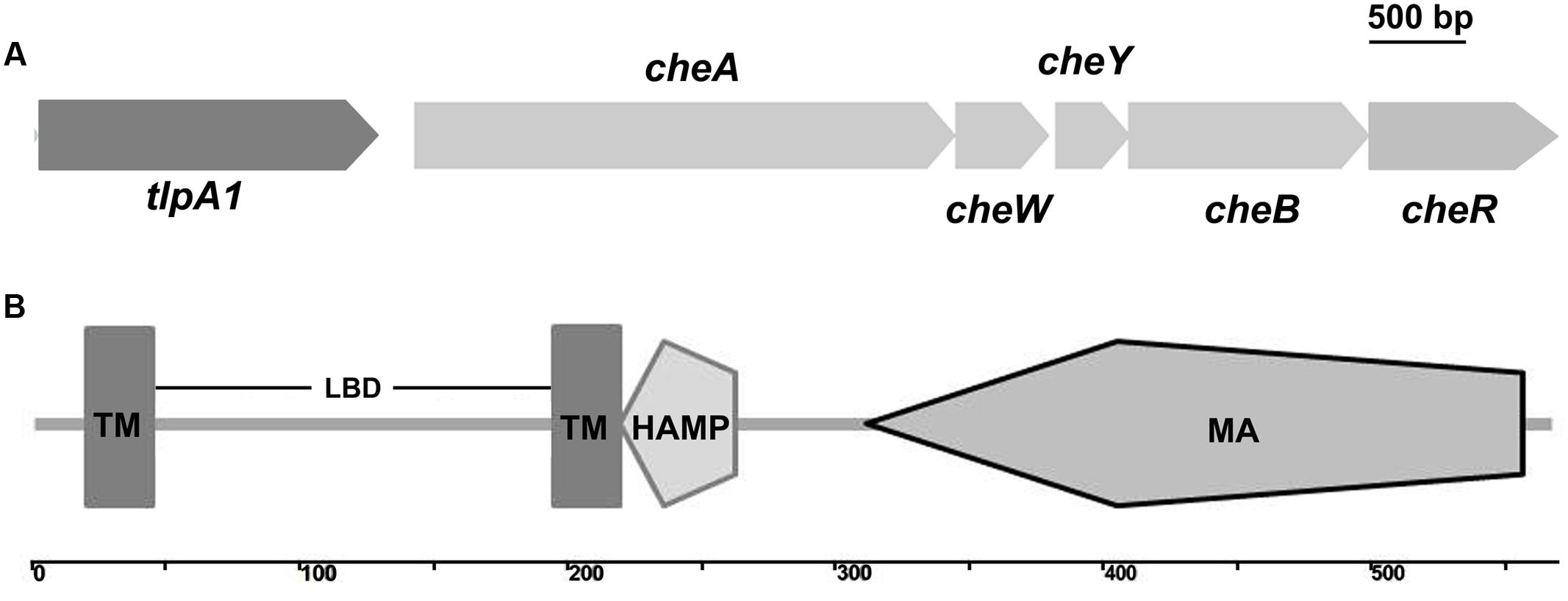
FIGURE 1. Schematic diagram showing the genome neighborhood of the tlpA1 gene and domain architecture of the TlpA1. (A) Genome neighborhood of the tlpA1 (AZC_0660) gene. Arrows indicate the direction of transcription. cheA (AZC_0661), cheW (AZC_0662), cheY (AZC_0663), cheB (AZC_0664), cheR (AZC_0665). (B) Domain architecture of the predicted TlpA1 protein, as defined by the SMART database: TM, transmembrane regions; LBD, periplasmic ligand binding domain; HAMP domain (histidine kinases, adenylyl cyclases, methyl binding proteins, and phosphatases); MA, methyl-accepting chemotaxis-like domain.
Similarity searches using BLASTP revealed that TlpA1 is homologous to chemoreceptors from closely related alpha-proteobacteria such as Azorhizobium doebereinerae, Afifella, Ancylobacter, and Chelatococcu (Supplementary Figure S2A). Sequence similarity searches with the N-terminal periplasmic ligand-binding region of TlpA1 followed by multiple sequences alignment of related proteins showed that this region is found exclusively in the N-terminal variable extracellular regions of chemoreceptors from various distantly related bacterial species (Supplementary Figure S2B). The presence of a conserved LBD domain in various classes of chemoreceptors suggests a possible fitness advantage to bacteria in different environments. Interestingly, 30 other chemoreceptors of A. caulinodans ORS571 also contain two transmembrane regions in the N-terminal region, and HAMP and chemoreceptor signaling domains in their C-terminal regions as predicted by MiST2 and SMART. However, sequence similarity searches using BLASTP indicated that neither the entire TlpA1 protein sequence nor the N-terminal periplasmic region is homologous to the other 30 transmembrane chemoreceptors (Supplementary Figure S2).
To determine the role of the chemotaxis receptor TlpA1 in A. caulinodans ORS571, we generated strain containing a tlpA1 deletion mutant (named ΔtlpA1). Soft agar plates were used to assess chemotaxis towards carbon sources such as succinate, citrate, tartrate, malate, proline, and glycerol, which are strong attractants of A. caulinodans (Jiang et al., 2016a). The ΔtlpA1 mutant was significantly impaired (∼25 to 40% of wild-type) in chemotaxis towards all carbon sources tested, regardless of presence or absence of combined nitrogen (Figures 2A,B). We complemented the ΔtlpA1 strain with a plasmid carrying the wild-type tlpA1 gene. The chemotaxis defect of the ΔtlpA1 mutant was partially rescued by expression of the wild-type tlpA1 gene driven by its native promoter (Figures 2A,B).
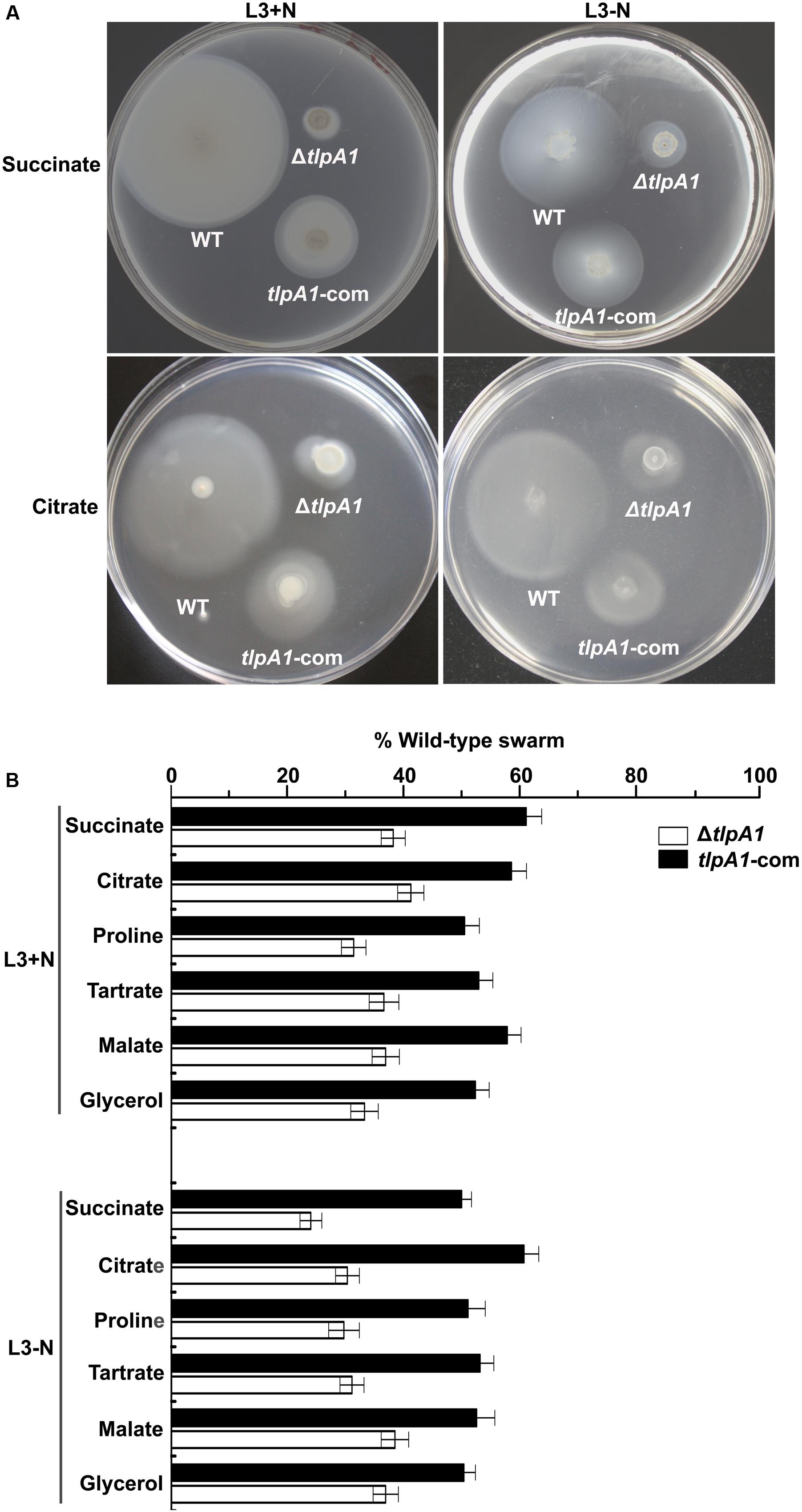
FIGURE 2. The ΔtlpA1 is deficient for chemotaxis. (A) Chemotactic behaviors of the wild-type (WT) A. caulinodans ORS571 (WT), ΔtlpA1 strain and complemented strains (tlpA1-com) were compared by soft agar plate assay. Pictures shown are representative plates for these strains on L3+N/-N with succinate and citrate as sole carbon sources. (B) Average diameters on L3+N/-N plates were expressed as percentages relative to that of the wild-type strain (defined as 100%). Error bars represent standard deviations (SD) from the mean calculated from at least three repetitions.
Since the tlpA1 gene is located directly upstream of cheA and transcribed in same direction (Figure 1A), we determined whether deletion of tlpA1 affected the cheA promoter region and thus cheA gene expression. We thus compared cheA gene expression in the wild-type and ΔtlpA1 strains and found that cheA was expressed in both strains (Figures 3A,B). Furthermore, there was no difference in growth rates of the mutant and wild-type cells in TY medium (Supplementary Figure S3A) or in L3 minimal medium (Supplementary Figure S3B). These data suggested that the diminished diameters observed on the soft agar plate are related to defects in chemotaxis.
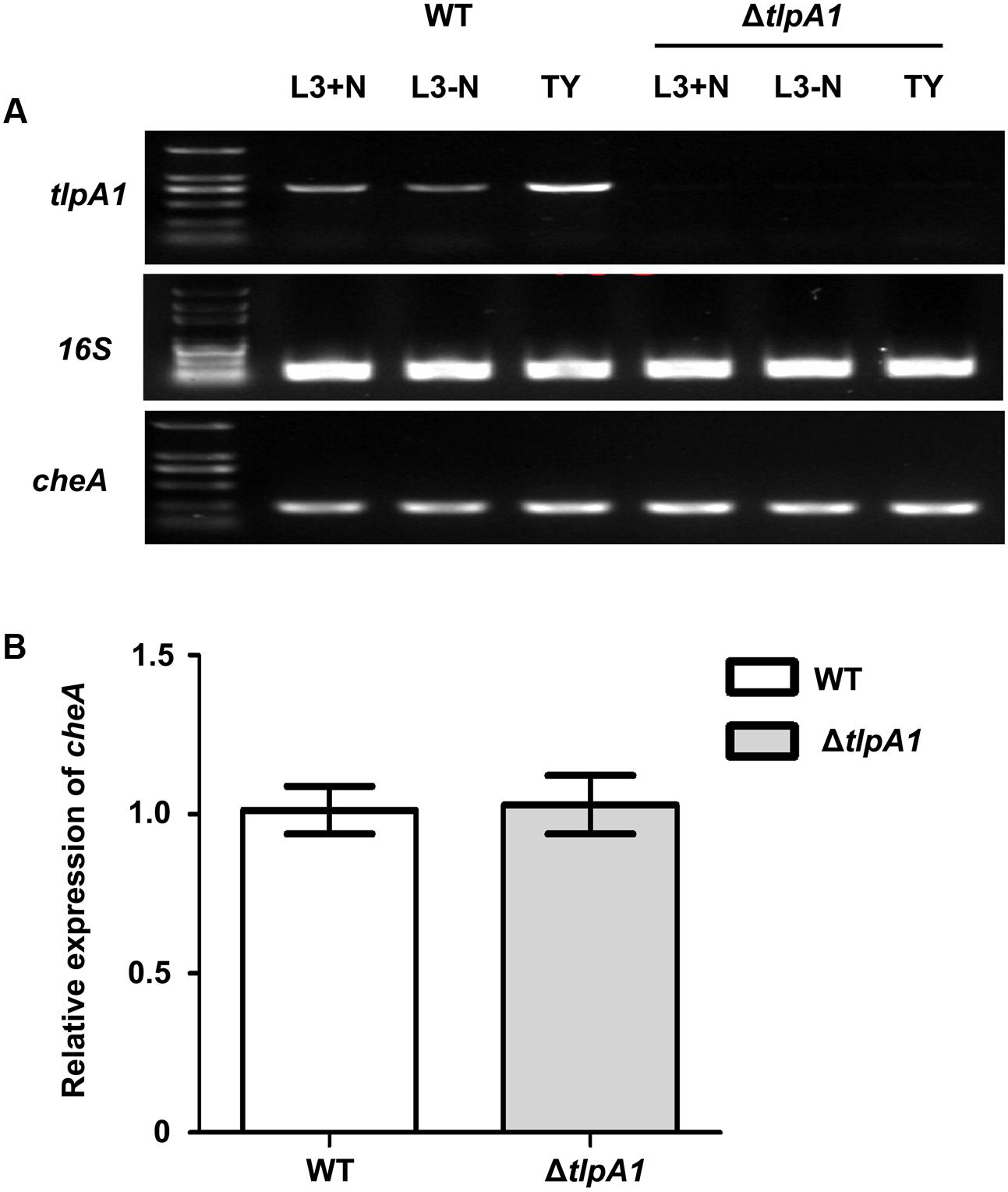
FIGURE 3. Expression of the cheA gene in the wild-type and ΔtlpA1 strains. (A) The cheA and tlpA1 genes were detected by PCR using the WT and ΔtlpA1 cDNA as templates; 16S rRNA gene was amplified as a positive control. (B) Expression of the cheA gene in the WT and ΔtlpA1 strains was detected by qRT-PCR, with 16S rRNA gene as internal standard. All data were derived from three independent experiments and are expressed as mean and SD. The WT and ΔtlpA1 strains were grown in TY medium (OD600 = 0.8).
To further assess chemotaxis and motility of the ΔtlpA1 mutant, competitive quantitative capillary assays of the wild-type and ΔtlpA1 strains were performed. In this assay, capillaries filled with chemoeffector are immersed into a bacterial suspension, which causes cells to preferentially swim into the capillary filled with chemoattractant. Buffer without chemoattractant was used as a control (Figure 4A). After this assay, the number of colony forming units in the capillaries was determined. For competitive quantitative capillary chemotaxis assay, a bacterial suspension containing a mixture of the wild-type and ΔtlpA1 strains at an approximate 1:1 ratio was used. Figure 4A shows competitive quantitative capillary chemotaxis assay with buffer and succinate as the attractants for the wild-type and ΔtlpA1 mutant. The cell count of the ΔtlpA1 from capillaries filled with buffer was similar to that of the wild-type (about 1:1) (Figure 4B). This observation indicates that swim motility behavior of the free-swimming cells is not affected by the ΔtlpA1 mutant. However, cell count of the ΔcheA mutant from capillaries filled with buffer was significantly less than that of the wild-type cells (Supplementary Figure S4). With succinate as the attractant, the wild-type cell count was almost twice that of the ΔtlpA1 mutant (Figure 4B). This data shows that the ΔtlpA1 mutant is impaired in chemotaxis and that TlpA1 is essential for chemotaxis but not for cell motility in A. caulinodans ORS571.
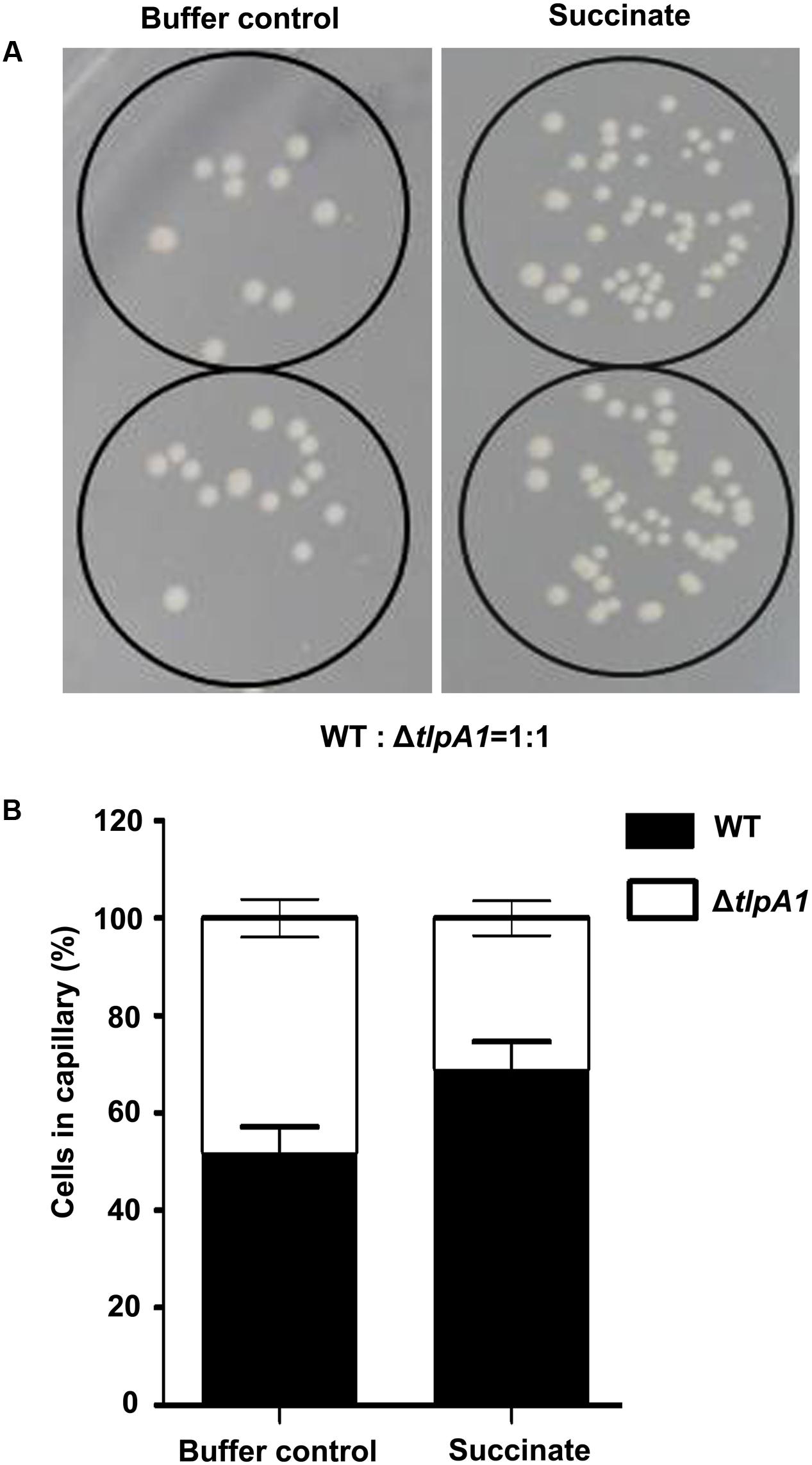
FIGURE 4. Competitive quantitative capillary chemotaxis assays of the WT and ΔtlpA1 mutant. (A) TY agar plates used for cell counting in the quantitative capillary chemotaxis assay. The capillaries filled with buffer control (left) or 10 mM succinate as the carbon source (right) were inserted into wells containing the WT and ΔtlpA1 (1:1 ratio) bacterial suspension mixture. (B) Statistical analysis of the WT and ΔtlpA1 mutant cell ratios in the capillary. The error bars represent the SE of data from three independent experiments.
Chemosensory systems have been previously implicated in regulation of biofilm formation (Hickman et al., 2005). To test whether deletion of tlpA1 affected biofilm formation, we compared biofilm formation ability of the ΔtlpA1 with the wild-type. Quantitative data reveal that biofilm formation in the ΔtlpA1 mutant is less than that in the wild-type (Figures 5A,B). These data indicate that TlpA1 may directly modulate biofilm formation under certain growth conditions.
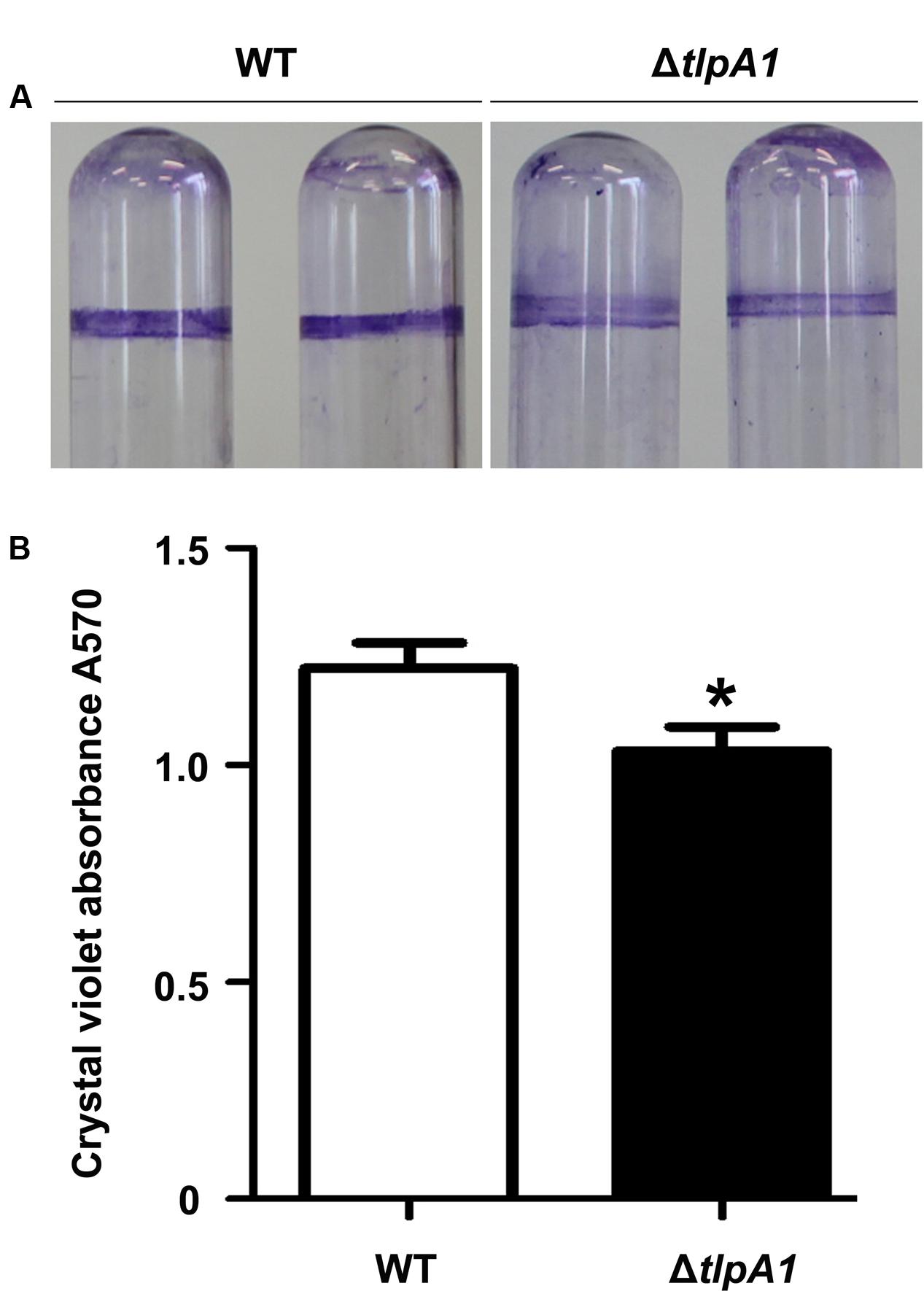
FIGURE 5. The ΔtlpA1 mutant is impaired for biofilm formation. (A) Biofilm morphology upon crystal violet staining of the WT and ΔtlpA1 mutant. (B) Biofilm formation was quantified by crystal violet staining method. The error bars represent SE of means from five independent experiments. Asterisks (∗) indicate statistically significant differences (P < 0.05) between the wild-type and ΔtlpA1 mutant.
To investigate the role of TlpA1 in symbiosis, the colonization ability on surface of roots was assessed by a quantitative assay. When inoculated individually, cell number of the ΔtlpA1 strain re-isolated from S. rostrata root surfaces was not significantly different from that of the wild-type (Supplementary Figure S5). This result suggested that the ΔtlpA1 is able to colonize on root surface. Surface-sterilized seedlings were co-inoculated with the wild-type ORS571 and ΔtlpA1 at approximately 1:1 and 1:10 ratios and cell numbers of the wild-type and ΔtlpA1 strains on the root surface of S. rostrata seedlings were determined after inoculation. The results indicated that colonization efficiency of the ΔtlpA1 mutant was significantly reduced when compared to the wild-type strain (Figure 6). These data therefore suggest that the ΔtlpA1 mutant affects competitive colonization of host root surfaces.
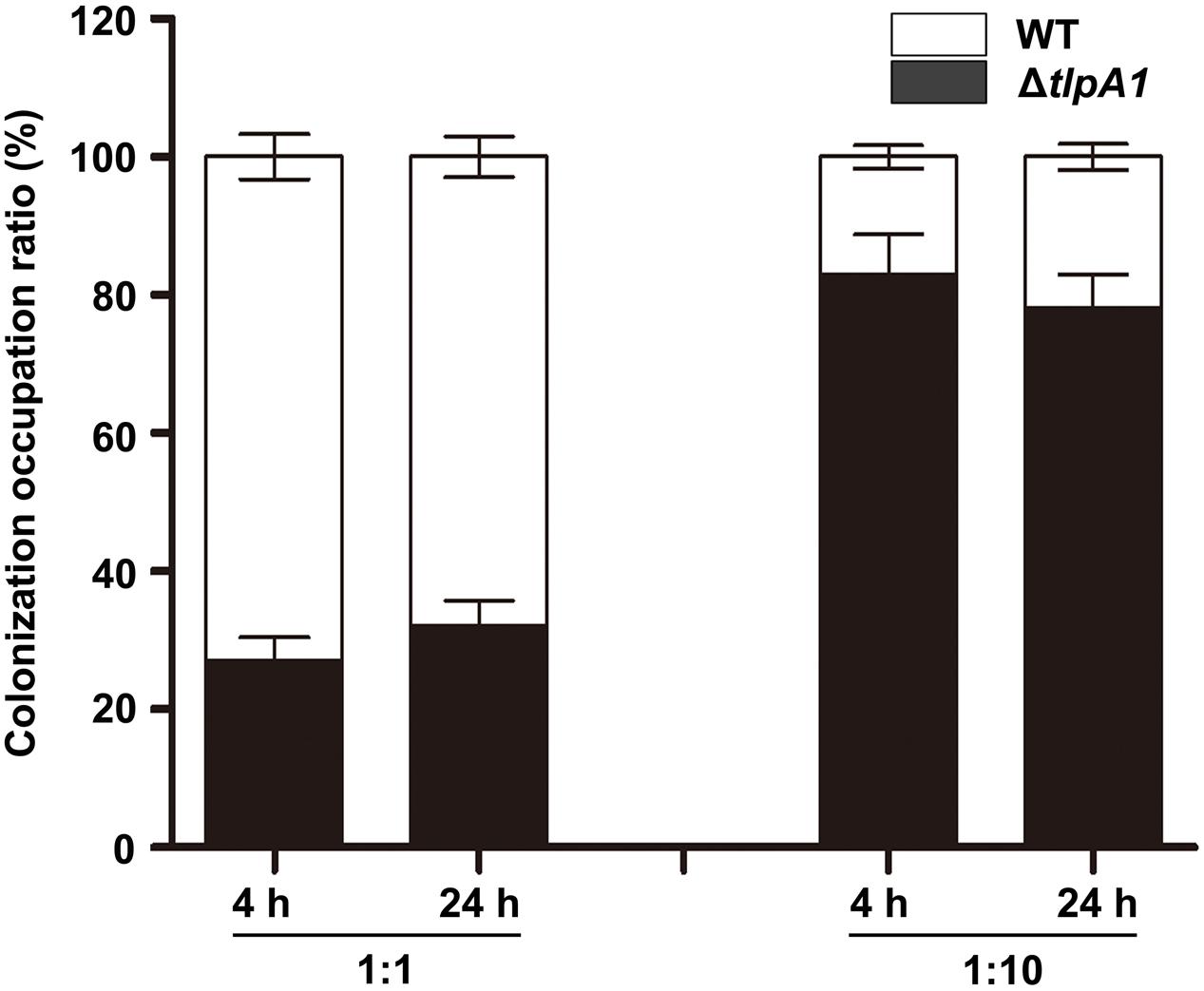
FIGURE 6. The ΔtlpA1 is impaired for competitive colonization of S. rostrata root surfaces. Colonization ratios of the WT and ΔtlpA1 strains were analyzed. The pattern of competitive colonization of the WT and ΔtlpA1 strains for adsorption to the surface of roots of S. rostrata seedlings is shown at 4 h and 24 h after inoculation. Mixed inocula at 1:1 and 1:10 ratios were used and analyzed by PCR with the primer pair TlpA1F and TlpA1R. Similar results were obtained in three independent experiments. The error bars represent standard errors of mean calculated for three independent experiments.
Root colonization is an important step in the establishment of symbiosis between rhizobia and host plants (Benizri et al., 2001). To further test whether the ΔtlpA1 mutant is defective for symbiosis and nodulation inducing ability in host, the wild-type and ΔtlpA1 strains were inoculated individually or in combination onto roots and stems of S. rostrata. The nodule occupancy of each strain was then determined. As shown in Supplementary Figures S6A–C, the ΔtlpA1 mutant induced functional nodules on roots and stems when inoculated alone, with no differences in nodule number and morphology as compared to the wild-type. Leghemoglobin of the stem nodules induced by the ΔtlpA1 mutant was orange–brown 20 days post inoculation (Supplementary Figure S6B). This result indicates that TlpA1 does not affect leghemoglobin production of the host plant in mature nodules. Likewise, nitrogenase activities of the root and stem nodules formed by the ΔtlpA1 strain were similar to the wild-type (Supplementary Figure S7). When we tested the ability of the mutant to induce nodule formation on roots in competition with the wild-type, we found that it was impaired in its ability to form nodules (Figure 7). The ΔtlpA1 mutant could not successfully compete for nodule formation with the wild-type even at 10-fold excess. We also tested the ability of the mutant to form stem nodules in competition with the wild-type. The results were similar to those obtained for root nodules (Figure 7). Taken together, these results indicate that the TlpA1 chemoreceptor is implicated in the processes related to competitive nodulation.
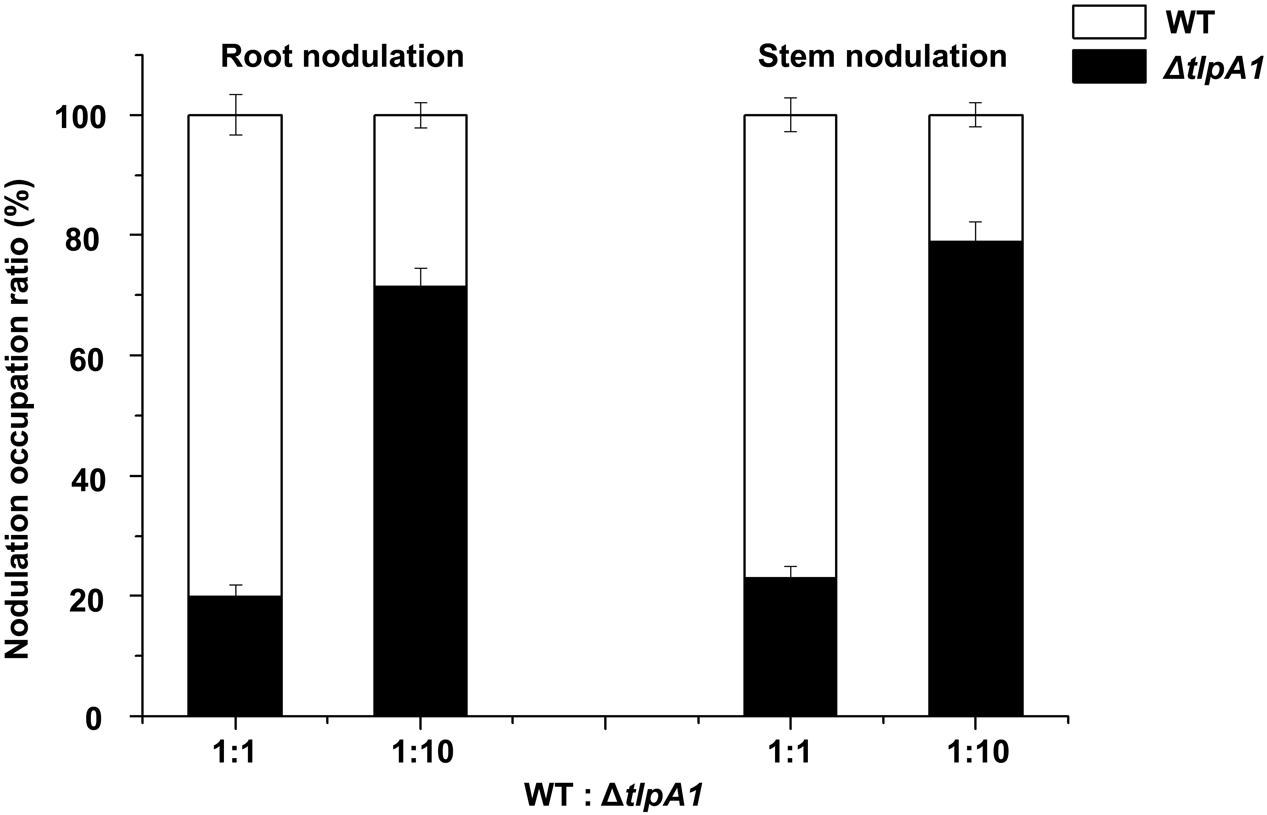
FIGURE 7. Competitive nodulation tests between the WT and ΔtlpA1 mutant. Cell numbers of the ΔtlpA1 mutant and WT were determined on root (left) and stem (right) nodules. The x-axis indicates ratios between the WT and ΔtlpA1 mutant. Error bars represent standard errors of the mean calculated from three independent experiments.
Bacterial chemotaxis is initiated by transmembrane receptor proteins that undergo conformational changes upon ligand binding (Falke and Hazelbauer, 2001). Chemoreceptors link environmental stimuli to bacterial motility via regulation of a CheA/CheY two-component system. Our results demonstrate that deficiency of the transmembrane chemoreceptor TlpA1 results in obvious defects in chemotaxis towards multiple carbon sources, suggesting that it plays a major role in chemotaxis of A. caulinodans ORS571. The tlpA1-deficient strain has a phenotype similar to that of a mcpB mutant in Rhizobium leguminosarum that also affects chemotaxis towards all carbon sources tested (Yost et al., 1998). The tlpA1 and mcpB mutant strains appear to have a generalized defect in chemotaxis, although the reasons remain unclear. Interestingly, this chemotaxis phenotype does not completely conform to the E. coli model where a given chemoreceptor is responsive to a specific set of ligands, indicating that the role of certain chemoreceptors in alpha-proteobacteria may substantially differ from those of E. coli. In addition, compared to the loss of a soluble cytoplasmic chemotaxis receptor IcpB in A. caulinodans ORS571 (Jiang et al., 2016a), loss of TlpA1 more detrimental to chemotaxis. Furthermore, the swim motility behavior of free-swimming cells is not affected in the ΔtlpA1 mutant (Figure 4) and is different from that of the cheA1-deficient strain (Supplementary Figure S4).
In the tlpA1-deficient strain, cells are impaired for chemotaxis in various carbon source chemical gradients, including creation of gradients on the swim plates or on the host root. Root adsorption and nodulation tests suggested that the tlpA1-deficient strain could colonize roots and induce nodules when inoculated alone. Furthermore, the ΔtlpA1 mutant can induce functional nodules on roots and stems without any significant difference in nodule number, morphology and nitrogenase activities as compared to the wild-type. However, the competitive colonization and nodulation efficiency of ΔtlpA1 was significantly lower than that of the wild-type (Figure 7). These findings point to an important role of chemotaxis in host root colonization and nodulation. Similarly, an Azospirillum brasilense mutant lacking an energy-sensing chemoreceptor is severely compromised for wheat root surface colonization (Greer-Phillips et al., 2004). In addition, nodulation competition experiments with R. leguminosarum suggested that two chemoreceptors MCPB and MCPC are important for pea nodulation (Yost et al., 1998). It has been speculated that chemotaxis may play a role in competition between rhizobia strains in legume rhizosphere. Some studies have shown that non-motile or non-chemotactic mutant strains cannot compete effectively with the wild-type strain (Ames and Bergman, 1981; Siuti et al., 2011). We found that cell motility of the ΔtlpA1 mutant is similar to that of the wild-type. Therefore, colonization and competitive nodulation of host plants appear to be affected by chemotaxis.
Root colonization ability is mainly determined by chemotaxis and biofilm formation (Hölscher et al., 2015). Although there is evidence suggesting that chemosensory pathways are involved in biofilm formation process, it remains unclear as to how chemoreceptors and signals modulate this process. In this study, we showed that biofilm formation ability of the ΔtlpA1 mutant was reduced when compared to that of the wild-type. In Pseudomonas putida KT2440, four chemoreceptor mutants had significantly altered biofilm phenotypes. Similarly, a strain carrying the mutation PP1488 showed a slight reduction in bacterial spread radii on soft agar as well as had a reduced capacity to form biofilms (Corral-Lugo et al., 2016). Further work will be done to understand how biofilm formation affected by chemoreceptors.
In summary, our data demonstrate that the ΔtlpA1 mutant is impaired for chemotaxis, biofilm formation, and competitive colonization and nodulation when tested on roots and stems. Our results suggest that impaired competitiveness to host roots apparently results in reduced nodule occupancy. These findings indicate that environmental signals or other stimuli trigger chemotaxis by the transmembrane receptor TlpA1 of A. caulinodans cells towards roots and stems of S. rostrata.
WL and ZX conceived and designed the experiments. WL, JY, YS, XL, performed the experiments. YL and ZZ provided technical support. WL and ZX were responsible for the drafting the manuscript. All authors contributed to manuscript revisions.
This work is financed by the National Natural Science Foundation of China (31370108, and 31570063), the Key Research Program of the Chinese Academy of Sciences (Grant NO. KZZD-EW-14), One Hundred-Talent Plan of Chinese Academy of Sciences (CAS), the Shandong Independent Innovation and Achievement Transformation Pro-gram (2014ZZCX07303), Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (+CXGC2016B10), Shandong Province Science Foundation for Youths (ZR2016CB44).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Toshihiro Aono, Shunpeng Li, and Zhentao Zhong for kindly providing A. caulinodans ORS571, S. rostrata seeds. We thank Mary E. Lidstrom for kindly providing pCM351 and pCM157 plasmids. We thank Gladys Alexandre and Claudine Elmerich for their suggestions on the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01327/full#supplementary-material
Aguilar, J. M. M., Ashby, A. M., Richards, A. J. M., Loake, G. J., Watson, M. D., and Shaw, C. H. (1988). Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. Microbiology 134, 2741–2746. doi: 10.1099/00221287-134-10-2741
Ames, P., and Bergman, K. (1981). Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J. Bacteriol. 148, 728–908.
Barbour, W. M., Hattermann, D. R., and Stacey, G. (1991). Chemotaxis of Bradyrhizobium japonicum to soybean exudates. Appl. Environ. Microbiol. 57, 2635–2639.
Bauer, W. D., and Caetano-Anollés, G. (1990). Chemotaxis, induced gene expression and competitiveness in the rhizosphere. Plant Soil. 129, 45–52. doi: 10.1007/BF00011690
Benizri, E., Baudoin, E., and Guckert, A. (2001). Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol. Sci. Technol. 11, 557–574. doi: 10.1080/09583150120076120
Broek, A. V., and Vanderleyden, J. (1995). Review: genetics of the azospirillum-plant root association. Crit. Rev. Plant Sci. 14, 445–466. doi: 10.1080/07352689509701932
Caetano-Anollés, G., Crist-Estes, D. K., and Bauer, W. D. (1988a). Chemotaxis of Rhizobium meliloti to the plant flavone luteolin requires functional nodulation genes. J. Bacteriol. 170, 3164–3169.
Caetano-Anollés, G., Wall, L. G., De Micheli, A. T., Macchi, E. M., Bauer, W. D., and Favelukes, G. (1988b). Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol. 86, 1228–1235.
Callow, J. A., and Vincent, J. M. (1971). A manual for the practical study of root nodule bacteria. J. Appl. Ecol. 8:977. doi: 10.2307/2402718
Capoen, W., Oldroyd, G., Goormachtig, S., and Holsters, M. (2010). Sesbania rostrata: a case study of natural variation in legume nodulation. New Phytol. 186, 340–345. doi: 10.1111/j.1469-8137.2009.03124.x
Collins, K. D., Lacal, J., and Ottemann, K. M. (2014). Internal sense of direction: sensing and signaling from cytoplasmic chemoreceptors. Microbiol. Mol. Biol. Rev. 78, 672–684. doi: 10.1128/MMBR.00033-14
Cook, B. G., Pengelly, B. C., Brown, S. D., Donnelly, J. L., Eagles, D. A., Franco, M. A., et al. (2005). Tropical Forages: An Interactive Selection Tool. Brisbane, QD: CSIRO
Corral-Lugo, A., Jesús, D. L. T., Matilla, M. A., Fernández, M., Morel, B., Espinosa-Urgel, M., et al. (2016). Assessment of the contribution of chemoreceptor-based signalling to biofilm formation. Environ. Microbiol. 18, 3355–3372. doi: 10.1111/1462-2920.13170
Dharmatilake, A. J., and Bauer, W. D. (1992). Chemotaxis of Rhizobium meliloti towards nodulation gene-inducing compounds from Alfalfa roots. Appl. Environ. Microbiol. 58, 1153–1158.
Dreyfus, B., Garcia, J. L., and Gillis, M. (1988). Characterization of Azorhizobium caulinodans gen. nov., sp.nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Bacteriol. 38, 633–639. doi: 10.1099/00207713-38-1-89
Dreyfus, B. L., Elmerich, C., and Dommergues, Y. R. (1983). Free-living Rhizobium strains able to grow on N2 as sole nitrogen source. Appl. Environ. Microbiol. 45, 711–713.
Falke, J. J., Bass, R. B., Butler, S. L., Chervitz, S. A., and Danielson, M. A. (1997). The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13, 457–512. doi: 10.1146/annurev.cellbio.13.1.457
Falke, J. J., and Hazelbauer, G. L. (2001). Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26, 257–265. doi: 10.1016/S0968-0004(00)01770-9
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Greer-Phillips, S. E., Stephens, B. B., and Alexandre, G. (2004). An energy taxis transducer promote root colonization by Azospirillum brasilense. J. Bacteriol. 186, 6595–6604. doi: 10.1128/JB.186.19.6595-6604.2004
Gupta, S. S. (2003). Chemotactic response of plant-growthpromoting bacteria towards roots of vesicular-arbuscular mycorrhizal tomato plants. FEMS Microbiol. Ecol. 45, 219–227. doi: 10.1016/S0168-6496(03)00155-7
Hazelbauer, G. L., Falke, J. J., and Parkinson, J. S. (2008). Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19. doi: 10.1016/j.tibs.2007.09.014
Hickman, J. W., Tifrea, D. F., and Harwood, C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427. doi: 10.1073/pnas.0507170102
Hölscher, T., Bartels, B., Lin, Y. C., Gallegos-monterrosa, R., Price-whelan, A., Kolter, R., et al. (2015). Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J. Mol. Biol. 427, 3695–3708. doi: 10.1016/j.jmb.2015.06.014
Jiang, N., Liu, W., Li, Y., Wu, H. L., Zhang, Z. H., Alexandre, G., et al. (2016a). A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-sesbania rostrata symbiosis. Appl. Environ. Microbiol. 82, 3174–3184. doi: 10.1128/AEM.00230-16
Jiang, N., Liu, W., Li, Y., and Xie, Z. H. (2016b). Comparative genomic and protein sequence analyses of the chemotaxis system of Azorhizobium caulinodans. Acta Microbiol. Sin. 56, 1256–1265.
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, RM 2nd, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lee, K. B., De Backer, P., Aono, T., Liu, C. T., Suzuki, S., Suzuki, T., et al. (2008). The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271
Li, S., Zhang, N., Zhang, Z., Luo, J., Shen, B., Zhang, R., et al. (2013). Antagonist Bacillus subtilis HJ5 controls Verticillium wilt of cotton by root colonization and biofilm formation. Biol. Fertil. Soils 49, 295–303. doi: 10.1007/s00374-012-0718-x
Marx, C. J., and Lidstrom, M. E. (2002). Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacterium. Biotechniques 33, 1062–1067.
Miller, L. D., Yost, C. K., Hynes, M. F., and Alexandre, G. (2007). The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 63, 348–362. doi: 10.1111/j.1365-2958.2006.05515.x
Mukherjee, T., Kumar, D., Burriss, N., Xie, Z. H., and Alexandre, G. (2016). Azospirillum brasilense chemotaxis depends on two signaling pathways regulating distinct motility parameters. J. Bacteriol. 198, 1764–1772. doi: 10.1128/JB.00020-16
Reyes-Darias, J. A., García, V., Rico-Jiménez, M., Corral-Lugo, A., and Krell, T. (2016). Identification and characterization of bacterial chemoreceptors using quantitative capillary and gradient plate chemotaxis assays. Bio Protoc. 6:e1789. doi: 10.21769/BioProtoc.1789
Scharf, B. E., Hynes, M. F., and Alexandre, G. M. (2016). Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Boil. 90, 549–559. doi: 10.1007/s11103-016-0432-4
Siuti, P., Green, C., Edwards, A. N., Doktycz, M. J., and Alexandre, G. (2011). The chemotaxis-like Che1 pathway has an indirect role in adhesive cell properties of Azospirillum brasilense. FEMS Microbiol. Lett. 323, 105–112. doi: 10.1111/j.1574-6968.2011.02366.x
Suzuki, S., Aono, T., Lee, K. B., Suzuki, T., Chi-Te Liu, C. T., Miwa, H., et al. (2007). Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis. Appl. Environ. Microbiol. 73, 6650–6659. doi: 10.1128/AEM.01514-07
Timmusk, S., Grantcharova, N., and Wagner, E. G. (2005). Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 71, 7292–7300. doi: 10.1093/jxb/54.381.325
Trinick, M. J. (1979). Structure of nitrogen-fixing nodules formed by Rhizobium on roots of Parasponia andersonii Planch. Can. J. Microbiol. 25, 565–578. doi: 10.1139/m79-082
Tsien, H. C., Dreyfus, B. L., and Schmidt, E. L. (1983). Initial stages in the morphogenesis of nitrogen-fixing stem nodules of Sesbania rostrata. J. Bacteriol. 156, 888–897.
Vande Broek, A., and Vanderleyden, J. (1995). The role of bacterial motility, chemotaxis, and attachment in bacteria-plant interactions. Mol. Plant Microbe. Interact. 8, 800–810. doi: 10.1094/MPMI-8-0800
Webb, B. A., Compton, K. K., Castañeda Saldaña, R., Arapov, T. D., Ray, W. K., Helm, R. F., et al. (2017). Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor ChemoreceptorX. Mol. Microbiol. 103, 333–346. doi: 10.1111/mmi.13561
Webb, B. A., Hildreth, S., Helm, R. F., and Scharf, B. E. (2014). Sinorhizobium meliloti chemoreceptor ChemoreceptorU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl. Environ. Microbiol. 80, 3404–3415. doi: 10.1128/AEM.00115-14
Wuichet, K., Alexander, R. P., and Zhulin, I. B. (2007). Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 422, 3–31. doi: 10.1016/S0076-6879(06)22001-9
Yost, C. K., Rochepeau, P., and Hynes, M. F. (1998). Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology 144, 1945–1956. doi: 10.1099/00221287-144-7-1945
Keywords: chemotaxis, Azorhizobium caulinodans, chemoreceptor, symbiosis, nodulation
Citation: Liu W, Yang J, Sun Y, Liu X, Li Y, Zhang Z and Xie Z (2017) Azorhizobium caulinodans Transmembrane Chemoreceptor TlpA1 Involved in Host Colonization and Nodulation on Roots and Stems. Front. Microbiol. 8:1327. doi: 10.3389/fmicb.2017.01327
Received: 13 January 2017; Accepted: 30 June 2017;
Published: 13 July 2017.
Edited by:
Jan Dirk Van Elsas, University of Groningen, NetherlandsReviewed by:
Christian Staehelin, Sun Yat-sen University, ChinaCopyright © 2017 Liu, Yang, Sun, Liu, Li, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Xie, emh4aWVAeWljLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.