- 1Quorum Sensing Laboratory, Centre for Research on Infectious Diseases, School of Chemical and Biotechnology, SASTRA University, Thanjavur, India
- 2Department of Chemistry, School of Chemical and Biotechnology, SASTRA University, Thanjavur, India
Staphylococcus aureus is a widely acknowledged Gram-positive pathogen for forming biofilm and virulence gene expressions by quorum sensing (QS), a cell to cell communication process. The quorum regulator SarA of S. aureus up-regulates the expression of many virulence factors including biofilm formation to mediate pathogenesis and evasion of the host immune system in the late phases of growth. Thus, inhibiting the production or blocking SarA protein might influence the down-regulation of biofilm and virulence factors. In this context, here we have synthesized 2-[(Methylamino)methyl]phenol, which was specifically targeted toward the quorum regulator SarA through in silico approach in our previous study. The molecule has been evaluated in vitro to validate its antibiofilm activity against clinical S. aureus strains. In addition, antivirulence properties of the inhibitor were confirmed with the observation of a significant reduction in the expression of representative virulence genes like fnbA, hla and hld that are governed under S. aureus QS. Interestingly, the SarA targeted inhibitor showed negligible antimicrobial activity and markedly reduced the minimum inhibitory concentration of conventional antibiotics when used in combination making it a more attractive lead for further clinical tests.
Introduction
Staphylococcus aureus is an opportunistic pathogen widely acknowledged for its various biofilm-related infections (Lister and Horswill, 2014). The prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infections is increasing evidently and continues to be a menace in healthcare (Lowy, 2003; Chambers and Deleo, 2009; Liu, 2009). The constraint in perfusion of antimicrobial agents into biofilm pose threat to treat biofilm-related infections (Singh et al., 2016). Moreover, cell to cell communication in bacteria which is referred as quorum sensing (QS) enables them to reach a threshold in their population and express various virulence factors not limited only to biofilm formation. Thus, the attenuation of QS by modulating transcriptional regulators to prevent biofilm formation and inhibit virulence factors offers an attractive strategy to treat such infections (Hentzer and Givskov, 2003). More importantly, little or no selective pressure in QS inhibitory approach which does not confer resistance is an added benefit when compared to the conventional antimicrobials (Defoirdt et al., 2010). However, in spite of these claims the future of QS targeted medical therapy as a sole alternative to conventional antibiotics is uncertain at the present scenario (Sully et al., 2014). Hence, considering the combined use of antiquorum compounds and conventional antibiotics would be more rationale to address bacterial infections in a superior way.
Out of the identified and established QS mechanisms in S. aureus, Sar locus governs the inception of agr locus and in addition independently triggers some of the virulence genes expression (Chien and Cheung, 1998; Chien et al., 1999). Particularly, SarA is an important transcriptional regulator that activates agr operon and also modulates the expression of many virulence genes (Dunman et al., 2001). For instance, cytotoxic virulence factors like α and δ hemolysins are coded by hla and hld genes, respectively, and play a vital role in severe Staphylococcal infections. Also, SarA encompasses an open reading frame of 375 bp that codes for DNA-binding protein which is essential for the optimal transcription of RNA III (Cheung and Manna, 2005). Thus, interference with SarA activity signifies an alternative therapeutic approach to Staphylococcal infections. In this context, we proposed to screen molecules against the quorum regulator, SarA in our earlier in silico structure-assisted screening approach (Arya and Princy, 2013) and had identified several lead molecules. One amongst them, 2-[(Methylamino)methyl]phenol termed as SarABI-12 with a molecular weight of 137.18, ALogP of 1.125, 2 rotatable bonds, 2 hydrogen acceptors and 2 hydrogen donors exhibited LibDock binding score of 53.8786 to quorum regulator of S. aureus, SarA, at residues Glu-89 and Arg-90 (Arya and Princy, 2013). The present study involves the synthesis of 2-[(Methylamino)methyl]phenol and further validation of its antibiofilm activity and down-regulation of virulence gene expressions. As we mentioned earlier, combined use of antiquorum compounds and conventional antibiotics were carried out which showed good results. To our knowledge, this is the first time 2-[(Methylamino)methyl]phenol has been evaluated and reported for its antibiofilm and antivirulence activity.
Materials and Methods
Reagents and Chemicals
Microbiological media and antibiotic disks were bought from HiMedia, Mumbai. Antibiotic powders were purchased from SRL fine chemicals, India. Chemical reagents for compound synthesis were purchased from Sigma–Aldrich, United States. LIVE/DEAD BacLight Bacterial Viability Kit (L-7012) was purchased from Thermo Fisher Scientific, United States. Qiagen RNeasy Mini kit was purchased from Qiagen, Germany and iScript cDNA synthesis kit was purchased from Bio-Rad, Hercules, CA, United States. SYBR Green PCR Master Mix was purchased from Thermo Fisher Scientific, United States. Primers used in this study were from Integrated DNA Technologies, United States.
Chemical Synthesis of 2-[(Methylamino)methyl]phenol
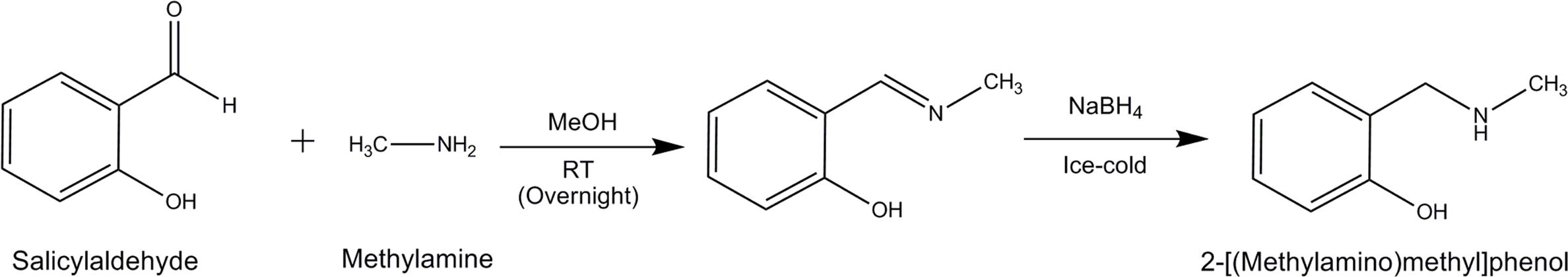
The scheme of chemical synthesis is provided in Figure 1. Briefly, Salicylaldehyde (5 mmol) and methylamine (7 mmol) were stirred in methanol (5 ml) at room temperature overnight. The intermediate imine formed was reduced to amine by the addition of sodium borohydride (5 mmol) at ice-cold condition. The reaction mixture was stirred continuously at room temperature for an overnight period. Further, the reaction mixture was diluted with water and extracted thrice with dichloromethane. The organic phases were pooled together, dried over anhydrous sodium sulfate and evaporated to dryness under reduced pressure in a rotary evaporator. The crude product was purified on a silica column with a methanol: DCM system. The purity and identity of the recovered product were supported by 1H-NMR, 13C-NMR and FTIR spectroscopy.
Bacterial Strains and Culture Conditions
Two clinical isolates of S. aureus (P1966 and AB459) were received from a collaborator at JSS Medical College, Mysore, India. Received strains were cultured in mannitol salt agar to ascertain the viability and identity of S. aureus. Pure colonies obtained were maintained as glycerol stocks in -80°C. SarA deletion mutant, i.e., S. aureus ALC637 strain (ΔsarA::Tn917LTV1) was a kind gift from Professor Christiane Wolz. Strains were grown in tryptic soy broth (TSB) supplemented with 0.5% glucose and incubated at 37°C under the stationary condition for subsequent assays.
Antibiotic Susceptibility Testing for Drug Resistance
Staphylococcus aureus isolates were cultured to early exponential phase (1.4 × 107 CFU/ml). Antibiotic susceptibility assay was performed to determine the drug resistance of S. aureus to various antibiotics that are commonly administered for S. aureus infections (Supplementary Table S1). A bacterial suspension of each strain was swabbed onto Mueller-Hinton agar plates and allowed to dry for 10 min. Antibiotic disks were placed on the plates and incubated for 24 h at 37°C. After the incubation period, the zone of inhibition diameter was measured and interpreted using the standard chart.
Minimum Inhibitory Concentration (MIC) and Minimum Biofilm Inhibitory Concentration (MBIC) Determination
The overnight bacterial culture was inoculated in 10 ml of sterile TSB media and incubated at 37°C for 6 h to reach log phase culture. Ten microlitre of the diluted inoculum (1:200 dilutions, approximately containing 2.1 × 106 CFU/ml) was inoculated into 96 well polystyrene microtitre plates containing different concentrations of the compound (0.075–1500 μM in twofold dilutions) in TSB supplemented with 0.5% (w/v) glucose. The plates were incubated at 37°C for 24 h and growth was measured at OD595. The lowest concentration of the compound that prevents a visible growth was considered as minimum inhibitory concentration (MIC).
Similarly, biofilm inhibition was determined with another set of 96 well microtitre plate treated as above. Briefly, the planktonic cells were removed after the incubation period, and the wells were washed with 200 μl PBS twice to remove unbound or loosely adhered cells. The cells were then stained with 100 μl of 0.2% crystal violet for 20 min. After removing the excess stain by washing, the plates were air dried. The bound crystal violet was eluted with 33% acetic acid and the optical reading was taken at OD595 in ELISA plate reader (BioRad i-Mark, Japan). Minimum biofilm-inhibiting concentration (MBIC) is the lowest concentration of the compound in which the formation of biofilm is limited to a level ≥90% (MBIC90) or ≥50% (MBIC50) in comparison with the untreated control cultures. All assays were carried out in triplicates.
Visualization of Biofilm Inhibition by Confocal Microscopy Imaging
Biofilm inhibition was visually confirmed by confocal microscopy imaging. Clean sterile glass slides were placed in 15 ml of TSB medium in the absence (positive control) and the presence of the test compound (0.6, 1.25, and 2.5 μM) in a 50 ml centrifuge tube. One percent (100 μl) of the diluted bacterial culture (2.1 × 106 CFU/ml) was added and incubated for 6 h at 37°C. After 6 h, the slides were rinsed with sterile PBS to remove non-adherent planktonic cells and stained with BacLight Bacterial Viability Kit (L7012) as per the kit protocol. Two and three-dimensional images were captured using a confocal laser scanning microscope with a 40× objective lens to show biofilm distribution (Olympus FLUOVIEW, FV1000).
Quantification of Gene Expression Using qRT-PCR
Staphylococcus aureus isolates (P1966 and AB459) were grown in TSB in the presence (1.25 μM) and absence of test compound for 6 and 24 h at 37°C. After the incubation period, the walls of the test tubes were scrapped with a sterile cell scraper to bring the adhered cells into suspension. The total cell population (planktonic and biofilm) were harvested by centrifugation from grown cultures (0.5 ml) at log phase (6 h) and stationary phase (24 h) and immediately stored at -80°C. RNA was isolated using a Qiagen RNeasy Mini kit in accordance with the manufacturer’s instructions. RNA concentrations were determined by optical density measurements in a spectrophotometer at OD260 (Thermo Scientific NanoDrop, United States). cDNA synthesis was carried out using the iScript cDNA synthesis kit according to the manufacturer’s instructions. Briefly, the reaction mixture was incubated for annealing at 25°C for 5 min, extension at 42°C for 30 min and inactivation of samples at 85°C for 5 min.
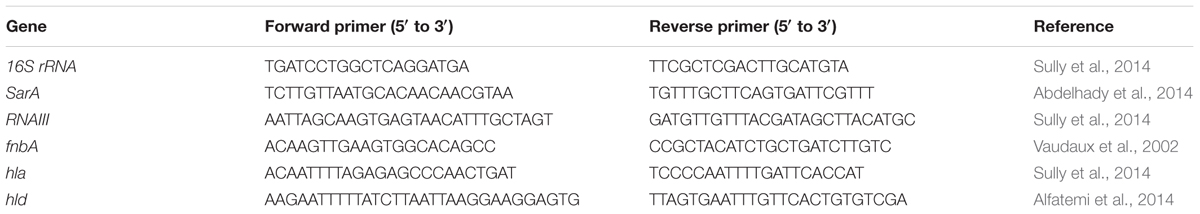
qRT-PCR was used to assess the transcriptional expression levels of SarA in the bacterial strains taken and down-regulation of representative virulence genes (RNAIII, fnbA, hla and hld) when treated with the compound. Sequences of primers used in this study are furnished in Table 1. The reaction mixture in a total volume of 20 μl, consisted 10 μl 2× SYBR Green PCR Master Mix, forward and reverse primers (1 μl each), 4 μl of nuclease free water and 4 μl of 20× diluted cDNA. PCR conditions included an initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation (95°C for 15 s), annealing (55.8°C for 15 s), and extension (72°C for 20 s). To ensure the samples were free from contamination, negative controls containing nuclease-free water instead of cDNA were run in parallel. The relative gene expression was analyzed using the 2-ΔΔCT method with 16S rRNA as a reference gene.
Cytotoxicity Assay
The MTT assay was carried out to determine any cytotoxicity effect of 2-[(Methylamino)methyl]phenol. Eagle’s Minimum Essential Medium supplemented with non-essential amino acids, 10% fetal bovine serum and penicillin-streptomycin (100 U/ml–100 μg/ml)) was used as culture medium. In brief, 200 μl of viable Human epithelial type 2 (HEp-2) cells in a concentration of 1.5 × 104 cells/ml were seeded in 96 well tissue culture plates. Varying concentrations (1.25, 12.5, and 125 μM) of the synthesized compound were added to each well when cells reached 80% confluence. The plate was incubated at 37°C for 24 h with 5% CO2. Further 20 μl of MTT solution (5 mg/ml) were added to each well and incubated for 3 h. Following the incubation period, 200 μl of DMSO was added to the wells and OD was taken at OD570. The percentage cell viability was calculated with reference to the untreated control cells using the below formula.
Percentage cell viability
Combinatorial Studies
Combinatorial studies were carried out between selected antibiotics that are commonly used for Staphylococcal infections and the test compound 2-[(Methylamino)methyl]phenol at the MBIC50 concentration. The S. aureus strains (P1966 and AB459) were cultured in TSB along with test compound at the fixed MBIC concentration, 1.25 μM and varying concentration of antibiotics in twofold dilutions at 37°C for 24 h at the stationary condition. At the end of incubation, absorbance was measured at OD595 and MIC was noted.
Statistical Analysis
Statistical analysis was carried out using GraphPad Prism software version 6.05 (GraphPad Software Inc., San Diego, CA, United States). One-way ANOVA and multiple comparisons were carried out to test the significance. The minimum level of significance was set at p ≤ 0.05. All the assays were carried out in triplicates and the results were expressed as mean ± SD.
Results
Synthesis of 2-[(Methylamino)methyl]phenol
2-[(Methylamino)methyl]phenol was synthesized as shown in the reaction scheme (Figure 1). The obtained compound had a yield of 82%, golden yellow in appearance. 1H-NMR (300 MHz, CDCl3) δ2.48 (s, 3H), 3.97(s, 2H), 6.75–6.80 (m, 1H), 6.83 (d, J = 8.1 Hz, 1H), 6.99 (d, J = 7.2 Hz, 1H), 7.14–7.29 (m, 1H). 13C-NMR (75 MHz, CDCl3) 35.1, 54.5, 122.2, 128.5, 116.3, 118.9, 128.8, 158.3. IR (Neat) 3430.7, 3001.7, 1646.0, 1434.8, 1411.6, 1261.2, and 1023.1 cm-1. Spectra of 1H-NMR, 13C-NMR and FTIR are provided in Supplementary Figures S1–S3, respectively.
Antibiotic Susceptibility Testing for Drug Resistance
The resistance pattern to various conventionally used antibiotics against S. aureus infections was shown in Supplementary Table S1. The clinical isolates exhibited multidrug resistance to most of the tested antibiotics that include gentamicin, azithromycin, erythromycin, co-trimoxazole, ciprofloxacin, doxycycline, methicillin, oxacillin, cloxacillin, cefuroxime, cephalexin, vancomycin and trimethoprim.
Antibacterial and Antibiofilm Activity
The expression level of SarA in the clinical isolates taken in this study was evaluated by qRT-PCR which confirms that the strains express SarA in the log and late phases of growth and is valid for further gene expression studies with the drug at corresponding phases (Figure 2A). The SarA deletion mutant ALC637 did not show any significant difference in biofilm when treated with different concentrations of the SarA targeted compound, 2-[(Methylamino)methyl]phenol (Figure 2B). Antibiofilm activity was evaluated for the SarA target-specific compound 2-[(Methylamino)methyl]phenol against the SarA positive clinical S. aureus strains and the results were presented in Figures 2C,D. Significant biofilm inhibition (>70%) was observed at a concentration of 1.25 μM. Moreover, 2-[(Methylamino)methyl]phenol did not show any antibacterial activity in the entire test range of concentrations up to 1500 μM. The compound did not exhibit disruption of preformed biofilm of both the strains (data not shown).
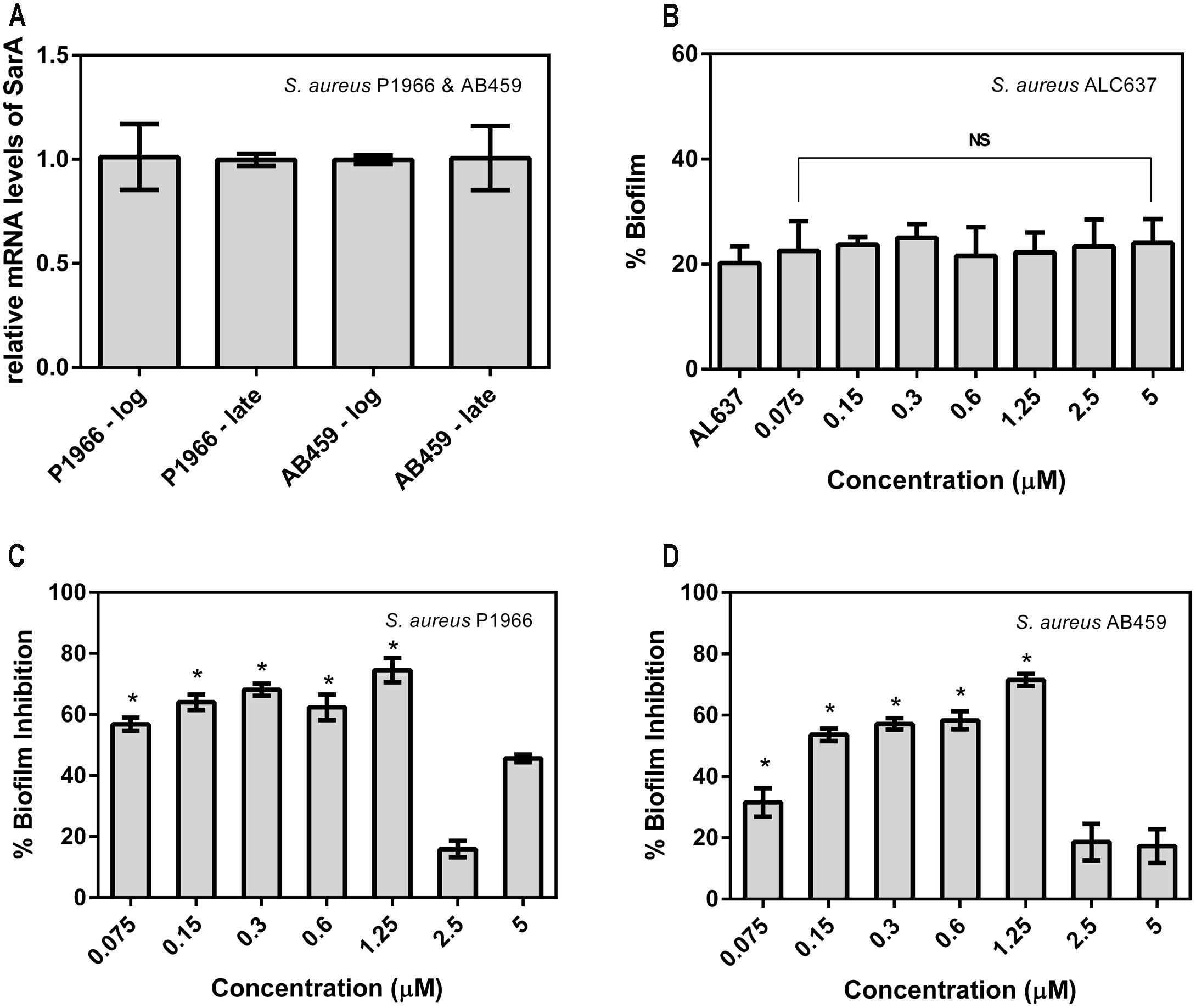
FIGURE 2. Percentage biofilm formed and inhibited in Staphylococcus aureus. (A) Expression levels of SarA in the bacterial strains taken in this study at the log and late phases of growth. (B) Percentage biofilm formation of SarA mutant ALC637 (ΔsarA::795 Tn917LTV1) on treatment and no treatment with 2-[(Methylamino)methyl]phenol by crystal violet method. (C,D) Percentage biofilm inhibition in clinical S. aureus isolates P1966 and AB459 by 2-[(Methylamino)methyl]phenol. All the assays were done in triplicates and the values were expressed as mean ± SD. ∗Indicates significantly different (p ≤ 0.05) when compared to untreated (control) with 2-[(Methylamino)methyl]phenol. NS denotes not significant (P > 0.05).
Visualization of Biofilm Inhibition by Confocal Microscopy Imaging
Visual confirmation of the biofilm inhibition by the compound was checked by Live/Dead staining in a confocal laser scanning microscope (Figure 3). The maximum biofilm inhibited concentration of the compound in microtitre plate assays (1.25 μM), also showed more inhibition in the captured images compared to that of the lower (0.6 μM) and higher (2.5 μM) concentrations. The results were in concordance with our crystal violet biofilm inhibition studies. The absence of dead cells (presented by red fluorescence) in Live/Dead stained images indicate the exclusion of any antibacterial activity possessed by the compound.
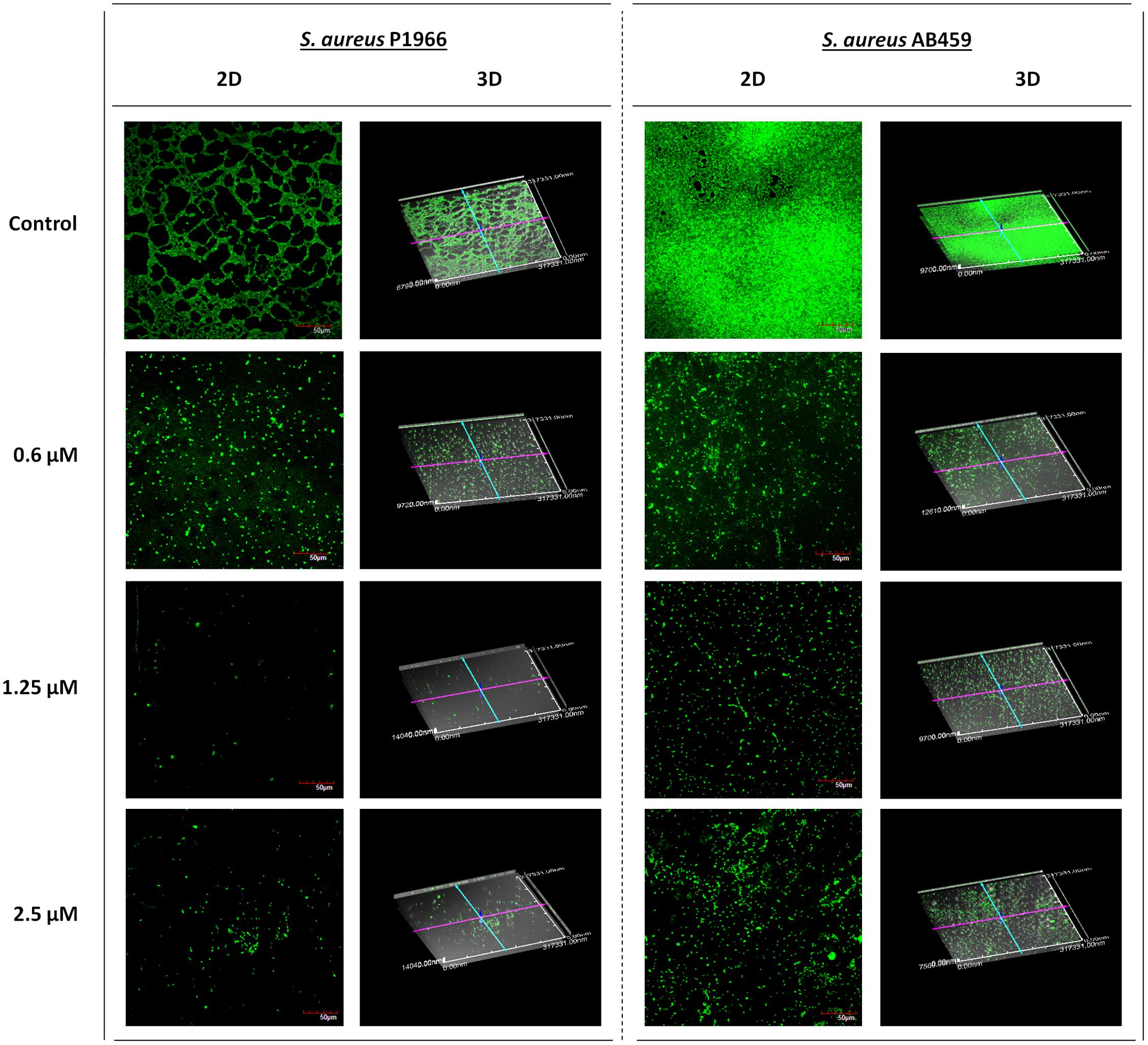
FIGURE 3. Confocal laser scanning microscopy imaging. Representative images showing the biofilm inhibition effects of 2-[(Methylamino)methyl]phenol at varying concentrations on Staphylococcus aureus P1966 and AB459 during log phase of growth (6 h). The results were in concordance with microtitre plate quantitative assays where biofilm inhibition was more at 1.25 μM. Scale bar in images represents 50 μm.
Gene Expression Profiling Using qRT-PCR
Representative genes took in the present study (RNAIII, fnbA, hla and hld) under the control of quorum regulator SarA were down-regulated significantly except hld at both the log and late phases of growth in the case of S. aureus P1966. Whereas, in the case of S. aureus AB459, downregulation was more in the log growth phase compared to late growth phase (Figure 4).
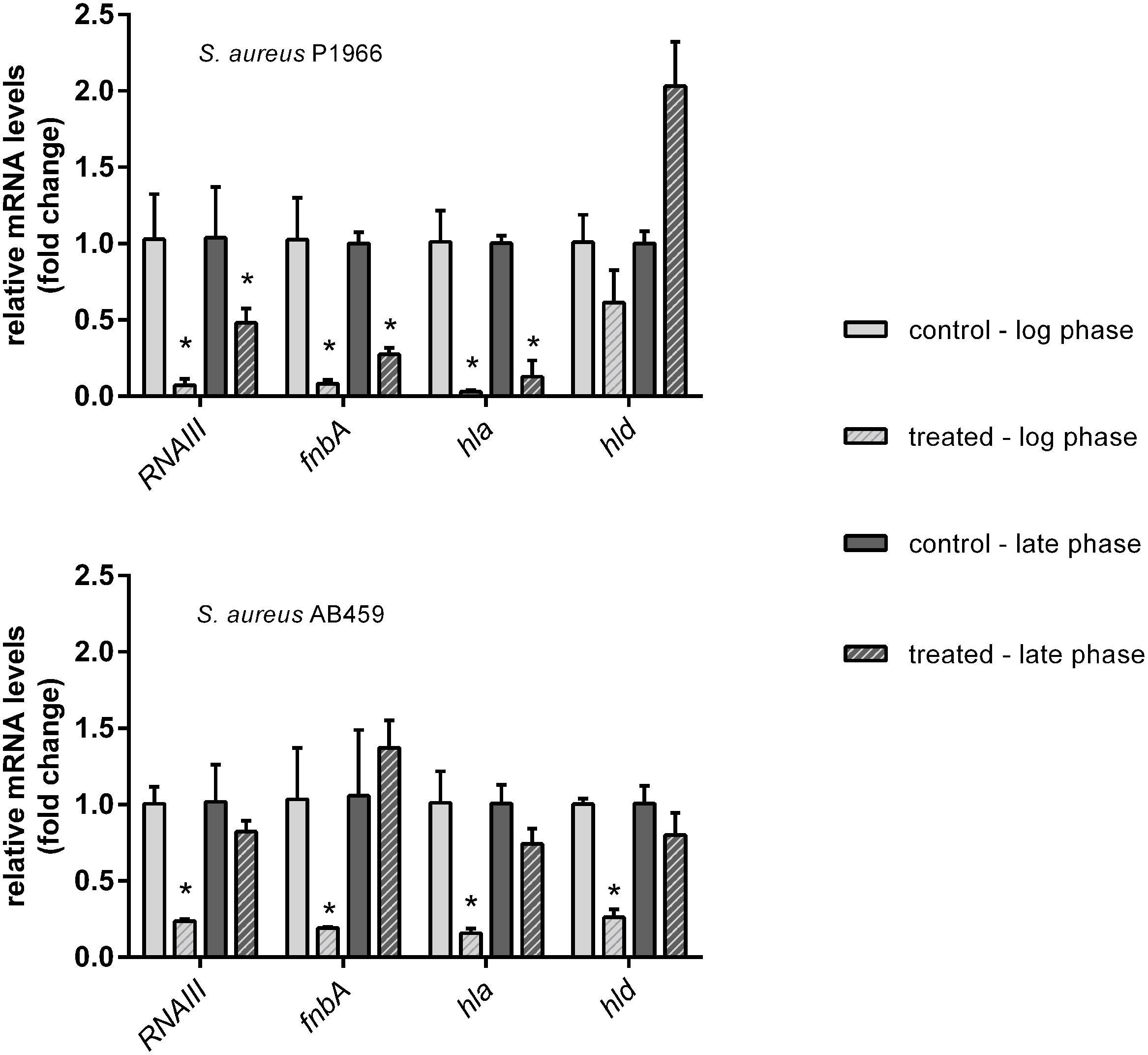
FIGURE 4. qRT-PCR analyses of representative virulence genes under the control of SarA. Downregulation of virulence genes (RNAIII, fnbA, hla and hld) in S. aureus P1966 and AB459 treated with 1.25 μM of 2-[(Methylamino)methyl]phenol at the log and late phases of growth are expressed a fold change by the 2-ΔΔCT method. 16S rRNA was used as reference gene for data normalization. ∗Indicates significantly different (p ≤ 0.05) when compared to untreated (control) with 2-[(Methylamino)methyl]phenol.
Cytotoxicity Assay
MTT assay data showed that the drug neither inhibits the proliferation of cells nor gives toxic effect to the HEp-2 cells even after incubating the cells with the compound at its MBIC concentration. Percentage of cell viability was equal to that of control (untreated cells) even after cells were exposed to 10× and 100× MBIC concentrations of the compound (Figure 5).
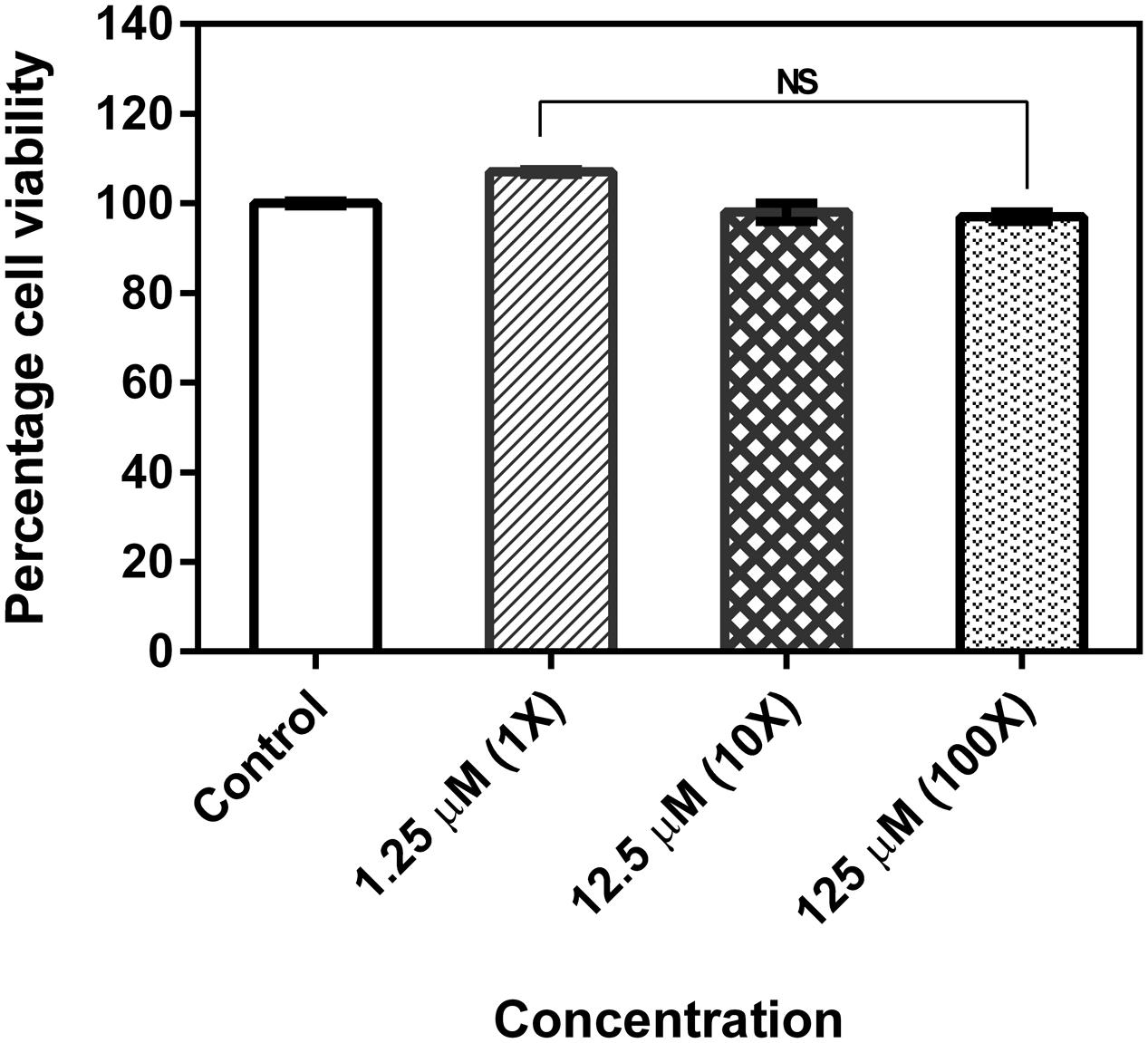
FIGURE 5. Cell viability of human epithelial type 2 (HEp-2) cells treated with 2-[(Methylamino)methyl]phenol. No significant difference between untreated and treated with varying concentrations of the test compound by MTT assay. The assay was done in triplicates and the values were expressed as mean ± SD. NS denotes not significant (P > 0.05).
Combinatorial Studies
Effect of the test compound in reducing the MIC values of conventionally used antibiotics against S. aureus was tested by combinatorial usage of both. In the case of S. aureus P1966, except for Azithromycin and Cephalexin, all other antibiotics taken showed a marked reduction in MIC values when used in combination with 1.25 μM of the test compound. Whereas in the case of S. aureus AB459 only a few antibiotics (Cloxacillin, Cephalexin and Vancomycin) showed marked MIC reduction and other antibiotics MICs were as same as that of when used alone (Table 2).
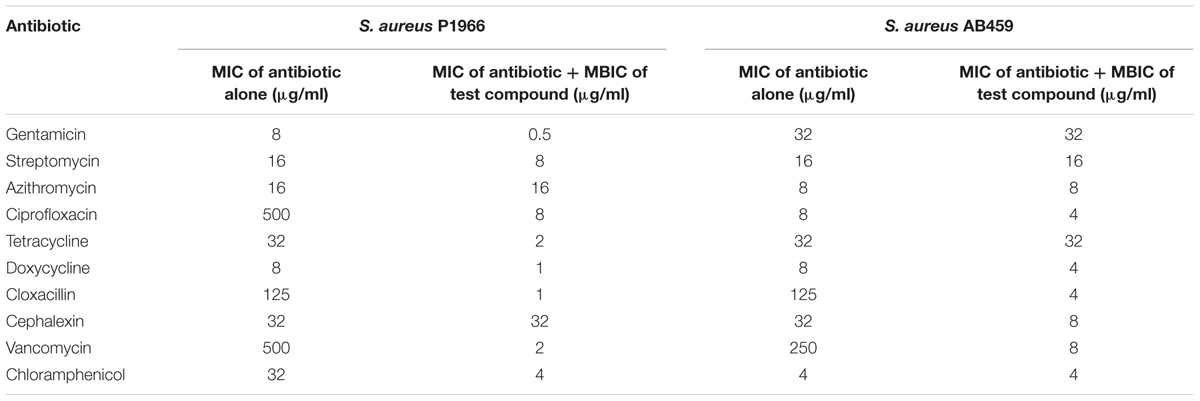
TABLE 2. Minimum inhibitory concentration of antibiotics alone and in combination with the test compound.
Discussion
Ineffectual usage of antibiotics has raised multidrug resistance to most of the commonly administered drugs against bacterial infections. In Staphylococcus aureus, methicillin resistance and vancomycin resistance are a major advanced and persistent concern (Loomba et al., 2010). In the present study resistance to most of the current generation antibiotics were observed, especially to the aforesaid antibiotics, underscoring the importance of S. aureus as a potential clinical threat. Alternatives to antibiotic therapies are the need of the hour to overcome pathologies stemming from multidrug-resistant microbes.
One of the alternative approaches to overcome multidrug resistance is tantamount to pursue drugs that do not impose survival stress on the pathogenic organism. Exploration of antibiofilm and antivirulence compounds for alternative therapy is of recent focus. The importance of the quorum regulator SarA as an alternative drug target has been detailed in the introduction and to date, not much of small molecule inhibitors are reported for SarA inhibition. However, our own research group has reported previously a range of SarA inhibitors through in silico approach and evaluated a top-scored SarA inhibitor, 4-[(2,4-diflurobenzyl)amino] cyclohexanol (Arya and Princy, 2013; Arya et al., 2015). In this context, one of the SarA targeted inhibitor 2-[(Methylamino)methyl]phenol that was identified in our previous in silico study (Arya and Princy, 2013) was evaluated for biofilm inhibition and downregulation of virulence genes.
Biofilm formation is a significant event in a bacterial life cycle that offers many benefits like environmental protection from pH fluctuations, desiccation, and physical insults, while also acting as an antibiotic diffusion barrier (Paraje, 2011). Eventually, this protection mechanism of bacteria from antimicrobials becomes problematic in clinical settings as the pathogen will endure treatments and emerge as a multidrug resistant strain. Hence, inhibiting bacterial biofilm is a vital concern and here we have observed significant biofilm inhibition in a dose-dependent manner up to 1.25 μM and after that a decrease in inhibition (Figures 2C,D). Similar results were observed qualitatively in microscopy imaging results also (Figure 3). A possible explanation for this could be because of the formation of insoluble macroaggregates of the compound at higher concentrations, which had been visibly observed on the treatment wells. These insoluble macro aggregates cannot pass through the cell membrane of bacteria to interact with the SarA protein and the very few left molecules would have entered the cell thus producing a reduced/no significant effect further in higher concentrations. In addition, the absence of antibacterial effect till the tested concentration of 1500 μM suggest that the compound could be a perfect choice for combating multidrug resistance in S. aureus, as it will not pose any survival stress or selection pressure on the bacteria forcing it to develop resistance. No difference in biofilm when the SarA deletion mutant S. aureus ALC637 treated with 2-[(Methylamino)methyl]phenol reveals the possible action of the compound on the specific target SarA as we could observe significant biofilm inhibition in SarA expressing clinical strains.
Our gene expression studies on representative virulence genes state that 2-[(Methylamino)methyl]phenol has antivirulence effect which was evidenced from the reduced transcriptional expressions. Fibronectin binding protein (FnBP) is one of the essential virulence factors that favor S. aureus for its anchorage with integrin α5β1 in a variety of non-professional phagocytic cells for successful invasion (Hudson et al., 1995; Yao et al., 1995; Almeida et al., 1996). The roles of extracellular adherence protein (Eap), plasmin-sensitive protein (Pls) and wall teichoic acid (WTA) in S. aureus cell invasion cannot be ruled out, but the importance of these proteins pale in comparison to FnBP-mediated adhesion (Weidenmaier et al., 2004; Hussain et al., 2008, 2009). It has been established that FnBP makes a molecular bridge between the bacterial microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) and the host cell β1 integrins, leading to bacterial uptake (Fowler et al., 2000). Twofold higher levels of adhesion and 20-fold higher levels of invasion have been observed in microbial cells at exponential phase in comparison with those cells at stationary phase, during a study of bovine mammary gland invasion by S. aureus (Lammers et al., 1999). Our results are in concordance with this study where fnbA transcription down-regulation is highly significant at log phase of growth as compared to the stationary phase of growth, for combating bacterial adhesion. In our present study, both the strains of S. aureus showed better down-regulation of α-hemolysin compared to δ-hemolysin. Staphylococcal α-hemolysin toxins are secreted as an outcome of virulence mechanisms that are under the QS control in S. aureus and α–toxin is believed to be involved in the phagolysosomal escape as it is a pore-forming toxin (Jarry et al., 2008). Many earlier studies have reported the role of SarA and its effect when it is mutated making it non-functional. A mutational study on SarA has shown reduced accumulation of extracellular toxins like α–toxin and phenol soluble modulins (Mrak et al., 2012). Similarly, another study showed reduced ability to induce septic arthritis and osteomyelitis in murine models of musculoskeletal infection when SarA is mutated (Abdelnour et al., 1993; Nilsson et al., 1996; Blevins et al., 2003). All these signifies the importance of Sar A role in virulence factor expressions and scores as a potential therapeutic target.
Physical barrier to antibiotics in biofilm related infections can be tackled by the use of antibiofilm agents in combination where these would resuscitate the potency of the antibiotics. The antibiofilm agent will inhibit biofilm formation and thus exposing the planktonic cells to sub-inhibitory concentration of antibiotic that can eliminate the bacterial population by easy penetration. Combined use of the test compound and conventionally used antibiotics against S. aureus was tested and the results showed a significant reduction in MIC values when antibiotics were used alone. This could be attributed to the fact that, reduction in biofilm by 2-[(Methylamino)methyl]phenol would have enhanced the antibiotic action by complete diffusion into the biofilms. Such a significant reduction in MIC levels possibly have a way of reducing the over usage of antibiotic concentrations in therapeutic treatments.
Conclusion
Our experimental results prove that 2-[(Methylamino)methyl]phenol has antibiofilm and antivirulence properties. Moreover, the SarA targeted inhibitor is specific in targeting QS of S. aureus and does not have cytotoxicity, making it an alternative therapeutic choice over conventional antibiotics. Further, in vivo studies are needed to evaluate the clinical effectiveness as a drug molecule. Thus we report, to the best of our knowledge, the first observation of antibiofilm and antivirulence properties of 2-[(Methylamino)methyl]phenol against S. aureus.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SAP acknowledges the support of the Indian Council of Medical Research, New Delhi for funding this research work (58/1/2013-BMS). The authors sincerely thank SASTRA University and its management for providing the required infrastructure to complete the research work. We also acknowledge the DST-FIST program (SR/FST/LSI-058/2010) for confocal laser scanning microscope imaging facility. We are also thankful to Professor Christiane Wolz (Department of Medical Microbiology and Hygiene, University of Tübingen, Germany) for gifting S. aureus ALC637 strain and Professor Sumana (Department of Microbiology, JSS Medical College and JSS University, India) for the clinical S. aureus strains.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01290/full#supplementary-material
FIGURE S1 | 1H-NMR spectrum of 2-[(Methylamino)methyl]phenol.
FIGURE S2 | 13C-NMR spectrum of 2-[(Methylamino)methyl]phenol.
FIGURE S3 | FTIR spectrum of 2-[(Methylamino)methyl]phenol.
References
Abdelhady, W., Bayer, A. S., Seidl, K., Moormeier, D. E., Bayles, K. W., Cheung, A., et al. (2014). Impact of vancomycin on sarA-mediated biofilm formation: role in persistent endovascular infections due to methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 209, 1231–1240. doi: 10.1093/infdis/jiu007
Abdelnour, A., Arvidson, S., Bremell, T., Ryden, C., and Tarkowski, A. (1993). The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61, 3879–3885.
Alfatemi, S. M. H., Motamedifar, M., Hadi, N., and Saraie, H. S. E. (2014). Analysis of virulence genes among methicillin resistant Staphylococcus aureus (MRSA) strains. Jundishapur J. Microbiol. 7:e10741. doi: 10.5812/jjm.10741
Almeida, R. A., Matthews, K. R., Cifrian, E., Guidry, A. J., and Oliver, S. P. (1996). Staphylococcus aureus invasion of bovine mammary epithelial cells. J. Dairy Sci. 79, 1021–1026. doi: 10.3168/jds.S0022-0302(96)76454-8
Arya, R., and Princy, S. A. (2013). Computational approach to design small molecule inhibitors and identify SarA as a potential therapeutic candidate. Med. Chem. Res. 22, 1856–1865. doi: 10.1007/s00044-012-0185-9
Arya, R., Ravikumar, R., Santhosh, R. S., and Princy, S. A. (2015). SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front. Microbiol. 6:416. doi: 10.3389/fmicb.2015.00416
Blevins, J. S., Elasri, M. O., Allmendinger, S. D., Beenken, K. E., Skinner, R. A., Thomas, J. R., et al. (2003). Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71, 516–523. doi: 10.1128/IAI.71.1.516-523.2003
Chambers, H. F., and Deleo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. doi: 10.1038/nrmicro2200
Cheung, A. L., and Manna, A. C. (2005). Role of the distal sarA promoters in SarA expression in Staphylococcus aureus. Infect. Immun. 73, 4391–4394. doi: 10.1128/IAI.73.7.4391-4394.2005
Chien, Y. T., and Cheung, A. L. (1998). Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273, 2645–2652. doi: 10.1074/jbc.273.5.2645
Chien, Y. T., Manna, A. C., Projan, S. J., and Cheung, A. L. (1999). SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274, 37169–37176. doi: 10.1074/jbc.274.52.37169
Defoirdt, T., Boon, N., and Bossier, P. (2010). Can bacteria evolve resistance to quorum sensing disruption? PLOS Pathog. 6:e1000989. doi: 10.1371/journal.ppat.1000989
Dunman, P. Á., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S., et al. (2001). Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agrand/or sarA loci. J. Bacteriol. 183, 7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001
Fowler, T., Wann, E. R., Joh, D., Johansson, S., Foster, T. J., and Hook, M. (2000). Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79, 672–679. doi: 10.1078/0171-9335-00104
Hentzer, M., and Givskov, M. (2003). Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Invest. 112, 1300–1307. doi: 10.1172/JCI20074
Hudson, M. C., Ramp, W. K., Nicholson, N. C., Williams, A. S., and Nousiainen, M. T. (1995). Internalization of Staphylococcus aureus by cultured osteoblasts. Microb. Pathog. 19, 409–419. doi: 10.1006/mpat.1995.0075
Hussain, M., Haggar, A., Peters, G., Chhatwal, G. S., Herrmann, M., Flock, J. I., et al. (2008). More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host cell invasion but not for leukocyte activation. Infect. Immun. 76, 5615–5623. doi: 10.1128/IAI.00480-08
Hussain, M., Schäfer, D., Juuti, K. M., Peters, G., Haslinger-Löffler, B., Kuusela, P. I., et al. (2009). Expression of Pls (plasmin sensitive) in Staphylococcus aureus negative for pls reduces adherence and cellular invasion and acts by steric hindrance. J. Infect. Dis. 200, 107–117. doi: 10.1086/599359
Jarry, T. M., Memmi, G., and Cheung, A. L. (2008). The expression of alpha-haemolysin is required for Staphylococcus aureus phagosomal escape after internalization in CFT-1 cells. Cell. Microbiol. 10, 1801–1814. doi: 10.1111/j.1462-5822.2008.01166.x
Lammers, A., Nuijten, P. J., and Smith, H. E. (1999). The fibronectin binding proteins of Staphylococcus aureus are required for adhesion to and invasion of bovine mammary gland cells. FEMS Microbiol. Lett. 180, 103–109. doi: 10.1111/j.1574-6968.1999.tb08783.x
Lister, J. L., and Horswill, A. R. (2014). Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 4:178. doi: 10.3389/fcimb.2014.00178
Liu, G. Y. (2009). Molecular pathogenesis of Staphylococcus aureus infection. Pediatr. Res. 65, 71R–77R. doi: 10.1203/PDR.0b013e31819dc44d
Loomba, P. S., Taneja, J., and Mishra, B. (2010). Methicillin and vancomycin resistant S. aureus in hospitalized patients. J. Glob. Infect. Dis. 2, 275–283.
Lowy, F. D. (2003). Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 111, 1265–1273. doi: 10.1172/JCI18535
Mrak, L. N., Zielinska, A. K., Beenken, K. E., Mrak, I. N., Atwood, D. N., Griffin, L. M., et al. (2012). saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS ONE 7:e38453. doi: 10.1371/journal.pone.0038453
Nilsson, I. M., Bremell, T., Ryden, C., Cheung, A. L., and Tarkowski, A. (1996). Role of the staphylococcal accessory gene regulator (sar) in septic arthritis. Infect. Immun. 64, 4438–4443.
Paraje, M. G. (2011). “Antimicrobial resistance in biofilms,” in Science against Microbial Pathogens: Communicating Current Research and Technological Advances, Vol. 2, ed. A. Méndez-Vilas (Badajoz: Formatex Research Center), 736–744.
Singh, R., Sahore, S., Kaur, P., Rani, A., and Ray, P. (2016). Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain-and antibiotic-specific differences. Pathog. Dis. 74:ftw056. doi: 10.1093/femspd/ftw056
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Brian, M. G., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Vaudaux, P., Francois, P., Bisognano, C., Kelley, W. L., Lew, D. P., Schrenzel, J., et al. (2002). Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70, 5428–5437. doi: 10.1128/IAI.70.10.5428-5437.2002
Weidenmaier, C., Kokai-Kun, J. F., Kristian, S. A., Chanturiya, T., Kalbacher, H., and Gross, M. (2004). Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10, 243–245. doi: 10.1038/nm991
Keywords: Staphylococcus aureus, SarA, antibiotics, quorum sensing, RT-PCR, multidrug resistance, gene expression
Citation: Balamurugan P, Praveen Krishna V, Bharath D, Lavanya R, Vairaprakash P and Adline Princy S (2017) Staphylococcus aureus Quorum Regulator SarA Targeted Compound, 2-[(Methylamino)methyl]phenol Inhibits Biofilm and Down-Regulates Virulence Genes. Front. Microbiol. 8:1290. doi: 10.3389/fmicb.2017.01290
Received: 13 April 2017; Accepted: 27 June 2017;
Published: 11 July 2017.
Edited by:
Sara María Soto, Barcelona Institute for Global Health, SpainReviewed by:
Devendra Hiraman Dusane, The Ohio State University Columbus, United StatesGiovanna Batoni, University of Pisa, Italy
Copyright © 2017 Balamurugan, Praveen Krishna, Bharath, Lavanya, Vairaprakash and Adline Princy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Adline Princy, YWRsaW5lcHJpbnp5QGJpb3RlY2guc2FzdHJhLmVkdQ==
 P. Balamurugan
P. Balamurugan V. Praveen Krishna
V. Praveen Krishna D. Bharath
D. Bharath Raajaraam Lavanya
Raajaraam Lavanya Pothiappan Vairaprakash
Pothiappan Vairaprakash S. Adline Princy
S. Adline Princy