
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 July 2017
Sec. Microbial Symbioses
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01287
Probiotics have been advocated as a novel therapeutic approach to respiratory disease, but knowledge of how oral administration of probiotics influences the respiratory microbiota is needed. Using 16S rRNA amplicon sequencing of bacterial DNA our objective was to determine whether oral probiotics changed the composition of the upper and lower airway, rectal, and blood microbiota. We hypothesized that oral probiotics would modulate the respiratory microbiota in healthy cats, demonstrated by the detection and/or increased relative abundance of the probiotic bacterial species and altered composition of the microbial population in the respiratory tract. Six healthy young research cats had oropharyngeal (OP), bronchoalveolar lavage fluid (BALF), rectal, and blood samples collected at baseline and 4 weeks after receiving oral probiotics. 16S rRNA gene amplicon libraries were sequenced, and coverage, richness, and relative abundance of representative operational taxonomic units (OTUs) were determined. Hierarchical and principal component analyses (PCA) demonstrated relatedness of samples. Mean microbial richness significantly increased only in the upper and lower airways. The number of probiotic OTUs (out of 5 total) that significantly increased in relative abundance vs. baseline was 5 in OP, 3 in BAL and 2 in feces. Using hierarchical clustering, BALF and blood samples grouped together after probiotic administration, and PERMANOVA supported that these two sites underwent significant changes in microbial composition. PERMANOVA revealed that OP and rectal samples had microbial population compositions that did not significantly change. These findings were visualized via PCA, which revealed distinct microbiomes in each site; samples clustered more tightly at baseline and had more variation after probiotic administration. This is the first study describing the effect of oral probiotics on the respiratory microbiota via detection of probiotic species in the airways. Finding bacterial species present in the oral probiotics in the upper and lower airways provides pilot data suggesting that oral probiotics could serve as a tool to target dysbiosis occurring in inflammatory airway diseases such as feline asthma, a disease in which cats serve as an important comparative and translational model for humans.
Prior to the advent of culture-independent microbiological techniques, the lower airways were considered sterile, and the primary role of bacteria in respiratory disorders was assumed to be as pathogens leading to development, persistence, exacerbation, and/or progression of respiratory diseases (Beigelman et al., 2014). However, recent studies using sequencing of microbial 16S rRNA amplicons (Sidiq et al., 2016) have demonstrated that healthy humans (Charlson et al., 2012), dogs (Ericsson et al., 2016), and cats (Vientos-Plotts et al., 2017) harbor rich, complex commensal microbial populations in the respiratory tract. There is increasing evidence that commensal microbes play a critical role in the maturation, education, and function of the immune system and, in particular, regulation of inflammatory responses. Additionally, dysbiosis, or deviations from the healthy microbiota, is associated with overgrowth of pathogens, decreased biodiversity, and stimulation of the host immune system that contributes to disease development (Gollwitzer and Marsland, 2014; Fujimura and Lynch, 2015). Dysbiosis has been documented in a variety of human respiratory diseases such as asthma (Huang, 2015; Ling et al., 2016), chronic obstructive pulmonary disease (COPD) (Gomez and Chanez, 2016; Huang et al., 2017), cystic fibrosis (Hogan et al., 2016; Madan, 2016), and respiratory tract infections (Pirrone et al., 2016; Yin et al., 2017).
Dietary supplementation in the form of probiotics, live microorganisms that provide a health benefit to the host when administered in adequate amounts (Guarner et al., 2012), beneficially alter the gut-lung axis (Forsythe, 2011; Kalyuzhin et al., 2016). Lactobacillus, Streptococcus, and Bifidobacterium species are some of the most common organisms considered probiotics. The specific mechanisms of action for health benefits of different probiotic strains are poorly understood. Probiotic strains may exert their effects on dysbiosis directly by changing the composition of the host microbiome or indirectly by interacting with the host through the common mucosal immune system (CMIS) (Neish, 2014; Thaiss et al., 2016). The CMIS is a unique branch of the immune system shared by mucosal sites; importantly, it enables cross-talk between the gastrointestinal (GI) and respiratory tracts (Budden et al., 2016; Ghaisas et al., 2016). In humans, orally administered probiotics modulate immune responses in the lung, promoting a tolerogenic environment of benefit in allergic disease (Rautava and Isolauri, 2002).
To our knowledge, no studies have evaluated the effect of oral probiotics on the airway microbiota of healthy cats, a species of importance for translational models of respiratory disease including allergic asthma (Norris Reinero et al., 2004; Reinero et al., 2009). The objective of the current study was to determine the effect of oral probiotics on the composition of the upper and lower airway, rectal, and blood microbiota. We hypothesized that probiotics would modulate the respiratory microbiota in healthy cats, demonstrated by the detection and/or increased relative abundance of the probiotic bacterial species and altered composition of the commensal microbial population in the respiratory tract after treatment. Furthermore, we postulated that alterations in rectal and blood microbiota might provide clues for mucosal or hematogenous transport of probiotic species.
All studies were performed in accordance with the Guide for the Use and Care of Laboratory Animals, and were approved by the University of Missouri Institutional Animal Care and Use Committee (MU IACUC protocol #7891).
Cats were bred from a colony (Comparative Internal Medicine Laboratory, University of Missouri, Columbia, MO), were all sexually intact and were aged <1 year (31–41 weeks by end of study). Cats were housed by sex with 2 males and 4 females housed in separate, large runs with elevated platforms for climbing and enrichment toys. Cats were transitioned to a commercial dry diet formulated for growth (Purina growth formula, St. Louis, MO) at 4 weeks of age, and remained on this diet for the duration of the study. Access to food and clean drinking water were provided ad libitum. In order to reduce risk of aspiration, cats were fasted for at least 12 h prior to anesthesia for sample collection.
Cats were determined to be healthy by absence of respiratory clinical signs, a normal physical examination by a board-certified veterinary internal medicine specialist and lack of cytologic evidence of infection or inflammation from bronchoalveolar lavage fluid (BALF) samples. Euthanasia was not an endpoint of the study; all cats were subsequently adopted into private homes.
Cats were administered one capsule of VSL#3 probiotic mixed in their food twice daily (225 × 109 CFU/day), for 4 weeks. Cats were individually fed a portion of their meal with the probiotic to ensure complete consumption before receiving the remainder of the meal.
Rectal swabs, oropharyngeal (OP) swabs, BALF and blood were collected at the beginning of the study (baseline), and immediately after 4 weeks of receiving probiotics. Samples were collected as previously described (Vientos-Plotts et al., 2017). Briefly, cats were anesthetized and a sterile cotton-tip swab was inserted rectally, while avoiding contact with the perianal area. After induction, but prior to intubation, a second moistened sterile swab was used to rub the caudodorsal aspect of the oropharynx, while avoiding contact with the rest of the oral cavity. The OP and rectal swabs were each added to 5 mL of sterile saline. Cats were carefully intubated using a sterile 3.5–4 French endotracheal tube. To collect BALF, a 20 mL aliquot of sterile saline was instilled via a sterile 8 French red rubber catheter that was threaded through the endotracheal tube. Four milliliters of whole blood were obtained by jugular venipuncture (site shaved of fur and prepared with ethanol) into sterile tubes with the anticoagulant EDTA. Immediately after collection, all samples were placed on ice and transported to the laboratory.
Promptly after collection, rectal, OP, BALF, and blood samples were centrifuged to pellet bacterial cells. Red blood cells (RBCs) were lysed prior to centrifugation of the blood samples. To lyse RBCs, 3 mL of blood was mixed with 27 mL sterile water and incubated at room temperature for 1 min before adding 3.3 mL of 10 × PBS and centrifuging for 20 min at 2,000 × g. In all cases, supernatant was discarded and pellets were resuspended in 800 μL lysis buffer adapted from Yu et al. (4% sodium dodecyl sulfate, 50 mM EDTA, 500 mM NaCl, and 50 mM Tris-HCl pH 8.0) (Yu and Morrison, 2004). All samples were banked at −80°C until the end of the study, and DNA was extracted as a single batch.
DNA from feces, OP, and BALF, and blood was extracted using the column method as previously described (Ericsson et al., 2016; Vientos-Plotts et al., 2017). Library construction and sequencing was completed at the University of Missouri DNA Core facility as previously described (Ericsson et al., 2016). Briefly, libraries were generated using single-indexed universal primers (U515F/806R) flanked by Illumina adapter sequences and PCR was performed using the following parameters: 98°C(3:00) + [98°C(0:15) + 50°C(0:30) + 72°C(0:30)] × 25 cycles + 72°C(7:00). Amplicons were pooled for sequencing on the Illumina MiSeq platform using the V2 chemistry and 2 × 250 paired-end reads. No samples returning less than 200 reads for BALF or blood, less than 1,350 for OP swabs, or less than 10,000 reads for rectal swabs were included. Assembly, filtering, binning, and annotation of DNA sequences was performed at the MU Informatics Research Core Facility as previously reported (Vientos-Plotts et al., 2017). Briefly, DNA contigs were assembled using FLASH software (Magoc and Salzberg, 2011) and culled if found to be short after trimming for base quality below 31. Qiime v1.8 (Kuczynski et al., 2011) was used to perform de novo and reference-based chimera detection and removal, and remaining contigs were assigned to operational taxonomic units (OTUs) via de novo OTU clustering using BLAST (Altschul et al., 1997) against the Greengenes database (DeSantis et al., 2006) of 16S rRNA sequences and taxonomy. Principal coordinate analysis of Bray-Curtis distances (1/4 root-transformed data) was performed using Past 3.13 (http://folk.uio.no/ohammer/past/).
Statistical analysis was performed using Sigma Plot 12.3 (Systat Software Inc., Carlsbad, CA). Normality was first tested using the Shapiro-Wilk method and equal variance was tested using the Brown-Forsyth method. Depending on normality, t-test or non-parametric Mann-Whitney rank sum test was used to test for differences between baseline and post-administration in coverage, richness (number of distinct sequences and Chao1 index), α-diversity (Shannon and Simpson indices), and relative abundance of all operational taxonomic units (OTUs) detected at greater than 0.5% of samples from any one sample site. Prior to testing of all metrics except coverage, data were rarefied via a reiterative random subsampling (10×) of data to the lowest read count obtained for any sample from that site (both time-points included). Mean values from subsampling iterations were then used for testing. Fisher's exact test was used to compare the number of cats in which the probiotic strains were present at week 4 compared to baseline. Testing via PERMANOVA, using Past 3.13, was performed to identify differences in β-diversity between time points and samples sites. Results were considered statistically significant for p-values ≤ 0.05.
Coverage (total number of sequences detected) varied by site. Pre-treatment, rectal and OP samples yielded the highest coverage (mean ± SEM of 73,260 ± 8,105 and 36,213 ± 80,395 sequences/sample, respectively), with no significant difference in coverage after probiotic administration (p = 0.59; p = 0.54, respectively). BALF and blood had lower coverage compared to other sites, presumably due to lower starting microbial biomass. However, the primary comparisons of interest were between baseline and post-VSL3 administration values, rather than between sample sites. Thus, to avoid rarefaction of data from samples such as rectal swabs to the lower read counts obtained from samples such as blood while still accounting for differences in coverage within sample sites, sequence data were rarefied independently for each sample site to the lowest read count obtained for any sample from that sample site. Overall, there was a pattern of decreasing richness (represented by mean number of number of distinct sequences and Chao1 index) progressing from rectal swabs to OP swabs to BALF to blood (Figure 1). Testing for differences in richness within each site revealed significant increases following probiotic administration in samples from OP and BALF, but not blood or rectal swabs (Figure 1). With regard to α-diversity (a metric incorporating both richness and evenness of distribution), time-dependent differences were detected only in BALF samples, with VSL3 administration correlating with increased diversity. Collectively, these data suggest that oral probiotic administration is associated with increased richness and diversity of the airway microbiota, with no concurrent changes detected in the fecal or blood microbiota.
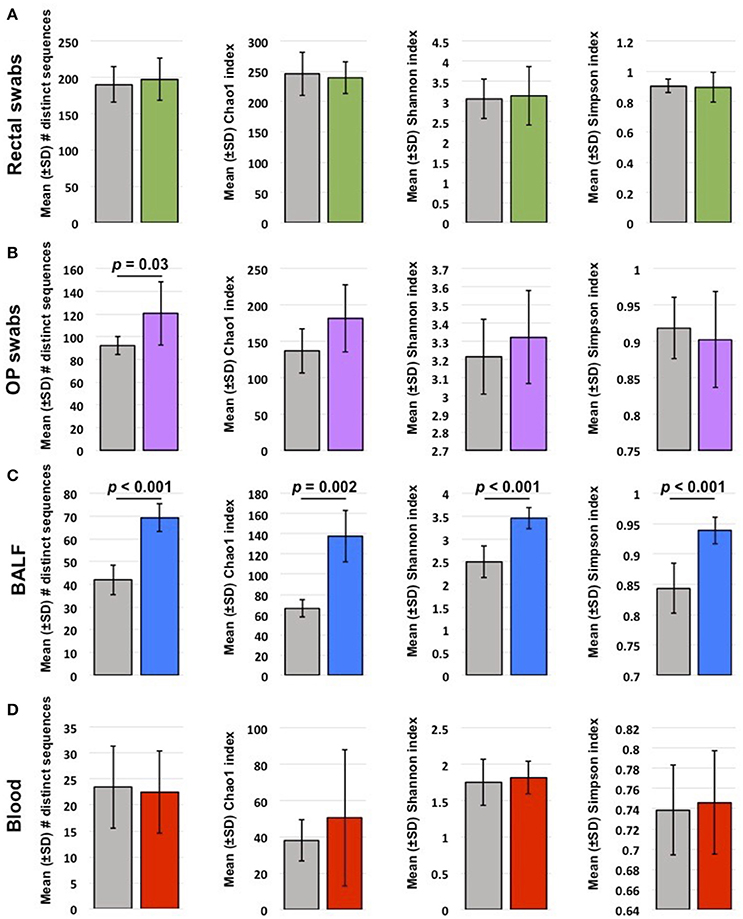
Figure 1. Mean ± standard deviation (SD) richness and diversity of rectal swabs (A), oropharyngeal (OP) swabs (B), bronchoalveolar lavage fluid (BALF) (C), and blood samples (D) collected from 6 healthy young cats at baseline (gray bars) and after probiotic administration (colored bars). Richness is represented by the number of distinct sequences detected and Chao1 index; α-diversity is represented by Shannon and Simpson indices. Significant (p < 0.05) differences noted above bars (t-test or Mann-Whitney rank sum test based on normality).
Resolved to the taxonomic level of phylum, the probiotic consisted of 59.27% Actinobacteria and 40.67% Firmicutes. The phylum Actinobacteria was predominantly represented by genus Bifidobacterium (35.93%) and Bifidobacterium animalis (23.29%) in the Bifidobacteriaceae family. The phylum Firmicutes was represented by genus Streptococcus (25.53%) in the Streptococcaceae family, and Lactobacillus zeae (11.64%) and genus Lactobacillus (3.21%) in the Lactobacillaceae family.
In rectal samples, B. animalis and L. zeae increased in relative abundance after probiotic administration (p = 0.015 and p = 0.004, respectively) but other probiotic OTUs did not change significantly. In OP samples, all probiotic OTUs significantly increased in relative abundance compared to baseline (Table 1). In BALF samples, relative abundance of genus Streptococcus (p = 0.015), L. zeae, (p = 0.015), and B. animalis (p = 0.002) significantly increased. There was no significant difference in relative abundance of the predominant probiotic OTUs in blood samples. The number of cats in which the predominant probiotic OTUs were found in each site at baseline and after probiotic administration is listed in Table 2.
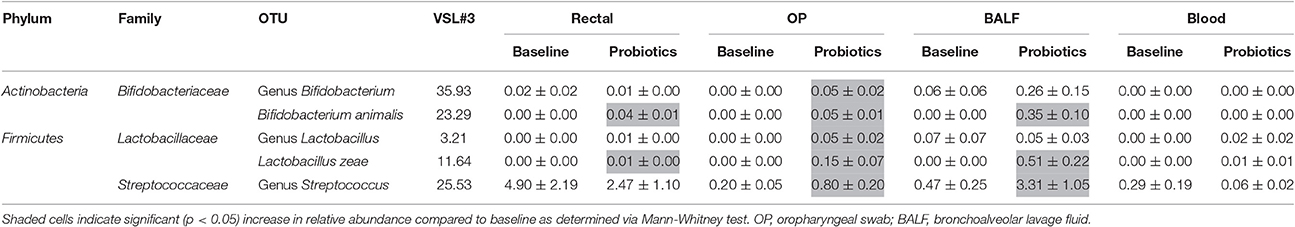
Table 1. Relative abundance (mean ± SEM) of the predominant probiotic operational taxonomic units (OTUs) in each site.
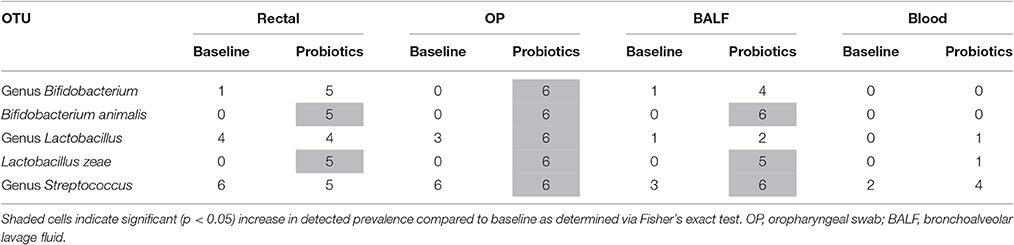
Table 2. Number of cats (out of 6) in which the predominant probiotic operational taxonomic units (OTUs) were sequenced at baseline and after probiotic administration.
In addition to introducing some of the predominant probiotic OTUs, probiotic administration was associated with a change in the overall composition of the microbial communities at each site. All taxa that significantly changed after probiotic administration and were detected at greater than 0.50% mean relative abundance in at least one sample site are listed in Supplementary Table 1. The most significant changes in rectal samples included a significant decrease in Bacteroides ovatus (mean ± SEM relative abundance of 8.96± 3.50% to 0%; p = 0.004), and concurrent increase in Bacteroides fragilis (from 2.37 ± 0.86% to 14.31 ± 7.82%; p = 0.009), among others. In the upper airways, the following OTUs in the phylum Proteobacteria were no longer detected after probiotic administration: Lautropia sp. (from 0.95 ± 0.19 to 0%; p = 0.004), Acinetobacter johnsonii (from 0.02 ± 0.00%; p = 0.004) and unclassified (UC) Moraxellaceae family (from 1.59 ± 0.31 to 0%; p = 0.004). Conversely, there was a significant increase in relative abundance in Porphyromonas endodontalis (from 2.47 ± 0.39 to 4.67 ± 0.47%; p = 0.004).
Of all samples, the lower airway microbiota underwent the most drastic changes after administration of probiotics. There was a significant decrease in UC Sphingobacteriaceae family (from 47.15 ± 5.48 to 13.09 ± 5.69%; p = 0.026) and A. johnsonii (from 0.85 ± 0.24 to 0 ± 0%; p = 0.002). The relative abundance of UC Sphingobacteriaceae also significantly decreased in blood samples (from 64.25 ± 2.38 to 33.02 ± 5.20%; p = 0.002).
The relative abundance of Acinetobacter sp. significantly increased in OP (from 0 ± 0 to 0.17 ± 0.08%; p = 0.004), BALF (from 0.07 ± 0.07 to 0.66 ± 0.22%; p = 0.015), and blood (from 0 ± 0 to 17.67 ± 8.92%; p = 0.015), but not in rectal swabs. This was the only OTU that changed significantly in more than two sites. Larger percentage changes in relative abundance observed in blood samples, and to a lesser extent BALF, are likely a reflection of the lower biomasses in these sites.
Neighbor joining, hierarchical clustering dendograms were used to visualize the compositional dissimilarity between populations. Regardless of the individual, after treatment, BALF and blood samples were more similar to each other than they were at their corresponding baseline, and were therefore grouped together in the dendogram. In contrast, rectal and OP samples had varied composition among samples independent of treatment (Figure 2). PERMANOVA was used to test for differences in community structure at baseline and after probiotic administration. There were significant differences in microbial community composition in lower airway (p = 0.0028; F = 4.655) and blood samples (p = 0.0022; F = 8.233). Interestingly, there was no significant difference in community composition in rectal (p = 0.4431; F = 0.9843) or OP swabs (p = 0.0765; F = 1.25) after probiotic administration.
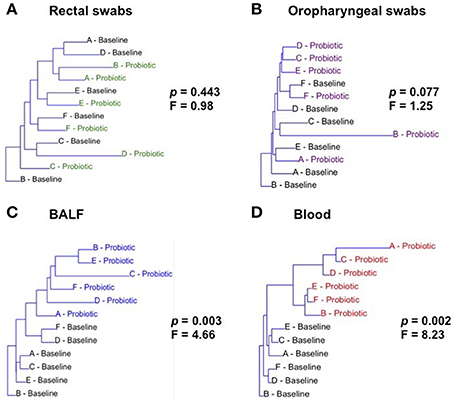
Figure 2. Hierarchical clustering of samples at baseline and after treatment (Probiotic) for all sites: rectal swabs (A), oropharyngeal swabs (OP) (B), bronchoalveolar lavage fluid (BALF) (C), and blood (D). Letters A through F represent each of the cats. p and F values determined via one way PERMANOVA of Bray-Curtis distances.
In addition, principal coordinate analysis (PCA) was used to assess the β-diversity of the microbial communities found at all sample sites collectively and at each site independently. When samples from all sites at baseline and after probiotic administration were included in the analysis, principal coordinates 1 and 2 (PC1 and PC2), accounting for 34.38 and 19.52% of the variation respectively, showed complete separation of the rectal, OP, and blood bacterial populations while BALF communities overlapped all of the other sites (Supplementary Figure 1). Of note, there was one rectal swab sample and one OP swab sample, both collected post probiotic administration that demonstrated a markedly different composition than the others in the group. These samples were not from the same cat and the reason for this divergence is unknown. Testing for main effects via two-way PERMANOVA found significant main effects of both sample site (p = 0.0001; F = 22.05) and time-point (p = 0.0041; F = 4.31), as well as a significant interaction (p = 0.01; F = 2.25). Recognizing that the inherent and well-known compositional differences between microbial communities present in the sites tested could obscure time-dependent differences within each sample site, we also performed PCA of samples from each site independently (Figure 3). Confirming what was seen in the hierarchical clustering analysis, there was complete separation of baseline and post-probiotic BALF and blood samples along PC1. The discordant post-administration samples within the rectal swab and OP swab sites skewed the PCA plots and any apparent separation of pre- and post-probiotic samples within those sample sites occurred along PC2 (accounting for a modest 10.6 and 11.1% variation respectively) and did not achieve significance. Collectively, these analyses suggest that while there were negligible, if any, probiotic-dependent effects on the composition of the rectal or OP microbial communities, oral VSL3 administration was associated with significant compositional changes in the microbial DNA detected in the BALF and blood. Taking these data in the context of the richness and α-diversity data reported above, we believe these findings to support the hypothesis that oral administration of certain probiotic species can significantly alter the resident airway microbiota, both in overall composition and richness and diversity.
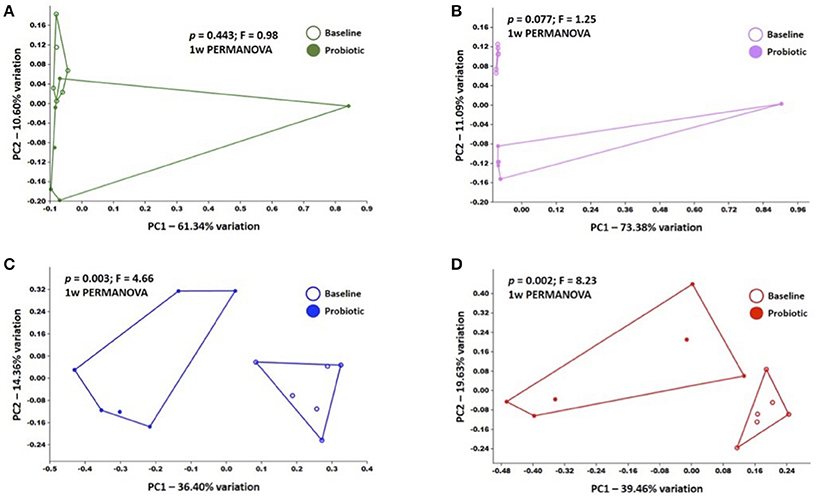
Figure 3. Principal coordinate analyses of baseline and post-administration (Probiotic) samples within each sample site, including rectal swabs (A), OP swabs (B), bronchoalveolar lavage fluid (BALF) (C), and blood (D); legends at right of each plot. p and F values determined via one way PERMANOVA of Bray-Curtis distances.
This is the first study to document that in the healthy cat, orally administered probiotics have the potential to change microbial communities at sites distant from the gastrointestinal (GI) tract. Traditionally, probiotics have been employed as a way to modulate the GI microbiota to promote local health (Lievin-Le Moal and Servin, 2014). In conjunction with the resident microbiota, probiotics can help inhibit colonization and/or proliferation of pathogens by interfering with adhesion to the intestinal mucosa and outcompeting pathogens for shared nutrients (Guarner et al., 2012). There is growing evidence that oral probiotics not only influence the GI tract directly, but also have immunomodulatory effects at distant sites, including the urogenital and respiratory tracts, oral cavity/dentition, mammary gland and skin. Although oral probiotics can prevent and/or decrease frequency or severity of diseases outside of the gastrointestinal tract (i.e., an indirect effect) (Alberda et al., 2007; Lappin et al., 2009; Di Nardo et al., 2014; Esposito et al., 2014; Lee et al., 2015; Zuccotti et al., 2015; Fuchs-Tarlovsky et al., 2016; Gruner et al., 2016; Vieira et al., 2016; Zamani et al., 2016), few studies have directly evaluated the appearance of orally administered probiotic species or changes in host microbial communities at distant sites (De Alberti et al., 2015; Mastromarino et al., 2015; Treven et al., 2015). A recent study in pre-menopausal women demonstrated an increase in orally administered probiotic species in vaginal swabs using qPCR analysis, setting the stage for use of oral probiotics to prevent or treat urogenital infections (Mezzasalma et al., 2017). A combination of bovine lactoferrin and two probiotics administered to healthy women led to detection of the probiotic species in vaginal swabs, important pilot data for future studies of bacterial vaginosis (De Alberti et al., 2015). Oral probiotics given to mice during pregnancy and lactation modulated the composition of bacterial communities in the mesenteric lymph nodes, mammary gland and milk with implications for neonatal health (Treven et al., 2015). Results were similar in women administered oral probiotics from late pregnancy through lactation showing significant increases in probiotic species in colostrum and milk (Mastromarino et al., 2015). A study relevant to prevention of dental caries employed 16S rRNA pyrosequencing to demonstrate changes in the composition of the microbiome in the oral cavity, specifically tooth biofilms, after 12 weeks of supplementation (Romani Vestman et al., 2015).
Probiotics have appeal, as there is growing evidence that they can improve health in patients with dysbiosis. The existence of a gut-lung axis allowing cross-talk through the CMIS has been proposed as a potential explanation for how microbes in the GI tract might influence immune functions in the lungs relevant in health and disease (Marsland et al., 2015; He et al., 2017). One potential mechanism is through interactions between host cell pattern recognition receptors (PRRs) and gut-derived pathogen-associated molecular patterns (PAMPs) leading to innate immune responses as well as systemic and CMIS immunoregulation (Neish, 2014; Davies and Abreu, 2015). Another mechanism is through production of bacterial metabolites such as short-chain fatty acids, which provide broad local and systemic immune effects (Meijer et al., 2010; Marsland and Salami, 2015; Budden et al., 2016). Additionally, dendritic cells that reside in the GI lamina propria or gut associated lymphoid tissue can sample members of the gut microbiota; interactions allow for maturation and migration of DCs to draining lymph nodes key for homeostatic adaptive immune responses (Samuelson et al., 2015). DCs activate various T cells subsets, which in turn are imprinted with chemokine receptors allowing homing to distal mucosal sites such as the respiratory tract (Samuelson et al., 2015). In addition to the gut contributing to immune homeostasis and immune responses in the lung, there is bi-directional exposure of microbiota at each site: inhaled microbes (as demonstrated by nasal cavity deposition in mice) consistently appear in the GI tract, and microaspiration of GI microbes is an important route by which the respiratory microbiota is established (Southam et al., 2002; Beigelman et al., 2014; Dickson and Huffnagle, 2015; Adar et al., 2016).
The mechanisms of action by which oral probiotics modify differing populations of microbial communities at various body sites and/or influence the host immune system are multifactorial and not fully understood. Our study supports the appearance of probiotic bacterial species in the upper and lower airways that were absent or in decreased abundance prior to the probiotic trial. We speculate that clinically silent microaspiration may have contributed to seeding of probiotic species in the respiratory tract. Compositional similarities between the blood and lower airway microbiome bring into question the possibility of GI tract bacterial translocation from the gut through the bloodstream as a route of entry; however, the bacterial communities in the GI tract were significantly different from those in blood and BAL. Perhaps as has been suggested in other studies, a systemic effect of oral probiotics (e.g., alteration of cytokine concentrations) may have modulated the respiratory microbiota (Vitali et al., 2012; Dai et al., 2013; Hepburn et al., 2013; Mastromarino et al., 2015).
In line with multiple other studies (Madan et al., 2012; Morris et al., 2013; Budden et al., 2016; Dickson et al., 2016) and as mentioned above, our results support microaspiration as a primary route of microbial colonization of the lower airways. In the current study, all of the predominant probiotic OTUs significantly increased in relative abundance in the upper airways, genus Streptococcus increased in the lower airways and L. zeae, and B. animalis increased in the lower airways and GI tract post-treatment. In healthy individuals, the respiratory microbiota is postulated to be established by a balance between entry (inhalation, microaspiration) and removal (e.g., via the mucociliary apparatus) of microbes and unlike the gut, there is little evidence for long-term colonization from local microbial reproduction (Dickson et al., 2016). This, along with the lower biomass, may explain why the probiotic OTUs more significantly altered the lower airways compared with the GI tract.
Several studies have reported benefits of probiotics to lung health (Forsythe, 2011, 2014; Yoda et al., 2012). Understanding the influence of oral probiotics on distant respiratory tract microbiota is crucial for future studies evaluating therapeutic applications in respiratory diseases like allergic asthma that affect humans and cats. Spontaneous and experimentally induced feline asthma shares the hallmark features of human allergic asthma, such as airway eosinophilia, airway hyper-responsiveness, and airway remodeling, making cats a valuable translational model for this disease (Reinero et al., 2009; Williams and Roman, 2016). Allergic asthma results from stimulation and amplification of a local inflammatory response mediated by allergen-specific T helper 2 (TH2) cells that occurs in genetically susceptible individuals exposed to environmental triggers. Recent studies in humans have provided evidence that dysbiosis occurs in asthma (Hilty et al., 2010; Goleva et al., 2013; Huang, 2013; Gollwitzer and Marsland, 2014).
The original hygiene hypothesis stated that improved hygiene and reduced exposure of the immune system to microbial stimulus during infancy and early childhood predisposes to impaired immunoregulation and an increase in prevalence of allergic diseases (Beigelman et al., 2014). Allergic diseases are driven by TH2 cells and many childhood infections trigger a TH1 immune response, therefore by having fewer childhood infections the immune system is thought to be polarized toward TH2 immune responses, promoting allergies. However, allergic diseases are not the only type of inflammatory diseases that have been on the rise; other TH1-driven chronic inflammatory responses, such as inflammatory bowel disease, have been increasing in developed countries (Haahtela et al., 2013; McCoy and Koller, 2015). This has led to the evolution of the hygiene hypothesis to the biodiversity or the microbiota hypothesis. The microbiota hypothesis proposes that dysbiosis or disruption of the host commensal microbiota disrupts the relationship between the microbes and the immune system, leading to immune dysfunction (Haahtela et al., 2013).
Dysbiosis leads to multiple important consequences: overgrowth of pathogens that compete with commensal microbes for host binding sites, loss of commensal microbial diversity, and a host inflammatory response that contributes to disease development (Fujimura et al., 2010). Chronic inflammation further disrupts the microbiota leading to a self-perpetuating inflammatory state. Return of the microbiota to a normal/healthy state has appeal in treatment of various disorders and studies have shown, for instance, that therapeutic alteration of the GI microbiota can promote remission of disease. Fecal microbiota transplantation is a classic example in which microbiota from healthy individuals are transplanted into patients with Clostridium difficile overgrowth, thereby inducing disease remission (Borody et al., 2004). Several studies in murine models of allergic airway disease indicate that probiotics can promote a tolerogenic environment in the lungs via induction of CD4+CD25+FoxP3+ T regulatory cells (Feleszko et al., 2007; Karimi et al., 2009), altering TH2 cytokine profiles (Forsythe et al., 2007; Fujimura et al., 2014) and modulating allergen-specific antibody production (Alvarez et al., 2001). In addition, multiple studies in experimental asthma models using oral probiotics have reported reduction of the asthmatic phenotype including airway inflammation and airflow limitation (Forsythe et al., 2007; Inoue et al., 2009; Karimi et al., 2009; Hougee et al., 2010; Li et al., 2010).
This is the first study to evaluate the effect of orally administered probiotics on the microbial communities in the airways and blood by determining changes in relative abundance of predominant probiotic OTUs. These data support microaspiration being one of the routes for lower airway microbial colonization and seeding of probiotic species. After oral administration, not only were the predominant probiotic OTUs present in distant sites, but they were also associated with changes in richness and the overall composition of colonizing microbial populations of the respiratory tract and blood.
We acknowledge there was a significant difference in coverage and richness at baseline compared to after treatment. Aside from this being secondary to probiotic administration, it could be argued that maturation of the cats may play a role in both richness and coverage, and this differential coverage could lead to changes in richness. However, we would argue that the changes in coverage and richness do not influence whether or not a particular OTU would be detected in a sample, and would speculate that this change is secondary to probiotic administration. These data support the idea that orally administered probiotics could be suited for modulation of upper and lower airway microbiota diseases relevant to both human and veterinary medicine.
All sequence data have been deposited in the NCBI Short Read Archive (SRA) under the accession number PRJNA376735.
AV participated in the conception and design of the study, sample collection, DNA extraction, data interpretation and drafted manuscript. AE participated in the conception and design of the study, interpreted sequence data and helped to draft the manuscript. CR participated in sample collection, the conception and design of the study and data interpretation, and helped to draft the manuscript. HR assisted with DNA extraction, sample collection, and study coordination. All authors read and approved the final manuscript.
AVP, resident in the MU College of Veterinary Medicine (CVM) Department of Veterinary Medicine and Surgery, is a veterinarian currently completing specialty training in small animal internal medicine. AE, Research Assistant Professor in the MU CVM Department of Veterinary Pathobiology, is a veterinarian with specialty training in laboratory animal and comparative medicine, and Director of the University of Missouri Metagenomics Center. CR, Associate Professor in the MU CVM Department of Veterinary Medicine and Surgery, is a veterinarian with specialty training in small animal internal medicine, and Director of the MU Comparative Internal Medicine Laboratory. HR is a Senior Research Associate in the Comparative Internal Medicine Laboratory.
This research was partially funded by the Phi Zeta Veterinary Research Society – Pi Chapter.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to acknowledge Megan E. Grobman, Amber Graham, Kaitlin Bishop and Leah A. Cohn, for their assistance in data collection and Karen Clifford for assistance formatting figures.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01287/full#supplementary-material
Supplementary Figure 1. β-diversity as shown via principal coordinate analysis of Bray-Curtis distances between samples from all four sample sites (rectal swab, OP swab, BALF, and blood); legend at right.
Supplementary Table 1. Relative abundance of all taxa detected at greater than 0.50% mean relative abundance in at least one sample site (feces, oropharyngeal swabs (OP), bronchoalveolar lavage (BAL) or blood), that changed significantly after probiotic administration, annotated to the level of phylum, family, and operational taxonomic unit (OTU). Data presented as mean ± standard error of the mean (SEM). *Shaded cells indicate p = <0.05 compared to baseline.
BALF, bronchoalveolar lavage fluid; CMIS, common mucosal immune system; GI, gastrointestinal; OP, oropharyngeal; OTU, operational taxonomic unit; UC, unclassified; PCA, principal component analysis; TH, T helper.
Adar, S. D., Huffnagle, G. B., and Curtis, J. L. (2016). The respiratory microbiome. an underappreciated player in the human response to inhaled pollutants? Ann. Epidemiol., 26, 355–359. doi: 10.1016/j.annepidem.2016.03.010
Alberda, C., Gramlich, L., Meddings, J., Field, C., McCargar, L., Kutsogiannis, D., et al. (2007). Effects of probiotic therapy in critically ill patients: a randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 85, 816–823.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Lipman, D. J., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Alvarez, S., Herrero, C., Bru, E., and Perdigon, G. (2001). Effect of Lactobacillus casei and yogurt administration on prevention of Pseudomonas aeruginosa infection in young mice. J. Food Prot., 64, 1768–1774. doi: 10.4315/0362-028X-64.11.1768
Beigelman, A., Weinstock, G. M., and Bacharier, L. B. (2014). The relationships between environmental bacterial exposure, airway bacterial colonization, and asthma. Curr. Opin. Allergy Clin. Immunol. 14, 137–142. doi: 10.1097/ACI.0000000000000036
Borody, T. J., Warren, E. F., Leis, S. M., Surace, R., Ashman, O., and Siarakas, S. (2004). Bacteriotherapy using fecal flora: toying with human motions. J. Clin. Gastroenterol., 38, 475–483. doi: 10.1097/01.mcg.0000128988.13808.dc
Budden, K. F., Gellatly, S. L., Wood, D. L., Cooper, M. A., Morrison, M., Hansbro, P. M., et al. (2016). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Charlson, E. S., Bittinger, K., Chen, J., Diamond, J. M., Li, H., Collman, R. G., et al. (2012). Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS ONE 7:e42786. doi: 10.1371/journal.pone.0042786
Dai, C., Zheng, C. Q., Meng, F. J., Zhou, Z., Sang, L. X., and Jiang, M. (2013). VSL#3 probiotics exerts the anti-inflammatory activity via PI3k/Akt and NF-κB pathway in rat model of DSS-induced colitis. Mol. Cell. Biochem., 374, 1–11. doi: 10.1007/s11010-012-1488-3
Davies, J. M., and Abreu, M. T. (2015). Host-microbe interactions in the small bowel. Curr. Opin. Gastroenterol., 31, 118–123. doi: 10.1097/MOG.0000000000000143
De Alberti, D., Russo, R., Terruzzi, F., Nobile, V., and Ouwehand, A. C. (2015). Lactobacilli vaginal colonisation after oral consumption of Respecta(®) complex: a randomised controlled pilot study. Arch. Gynecol. Obstet. 292, 861–867. doi: 10.1007/s00404-015-3711-4
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Andersen, G. L., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dickson, R. P., Erb-Downward, J. R., Martinez, F. J., and Huffnagle, G. B. (2016). The microbiome and the respiratory tract. Annu. Rev. Physiol.,78, 481–504. doi: 10.1146/annurev-physiol-021115-105238
Dickson, R. P., and Huffnagle, G. B. (2015). The lung microbiome. new principles for respiratory bacteriology in health and disease. PLoS Pathog. 11:e1004923. doi: 10.1371/journal.ppat.1004923
Di Nardo, G., Oliva, S., Menichella, A., Pistelli, R., De Biase, R. V., Stronati, L., et al. (2014). Lactobacillus reuteri ATCC55730 in cystic fibrosis. J. Pediatr. Gastroenterol. Nutr. 58, 81–86. doi: 10.1097/MPG.0000000000000187
Ericsson, A. C., Personett, A. R., Grobman, M. E., Rindt, H., and Reinero, C. R. (2016). Composition and predicted metabolic capacity of upper and lower airway microbiota of healthy dogs in relation to the fecal microbiota. PLoS ONE 11:e0154646. doi: 10.1371/journal.pone.0154646
Esposito, S., Rigante, D., and Principi, N. (2014). Do children's upper respiratory tract infections benefit from probiotics? BMC Infect. Dis. 14:194. doi: 10.1186/1471-2334-14-194
Feleszko, W., Jaworska, J., Rha, R. D., Steinhausen, S., Avagyan, A., Jaudszus, A., et al. (2007). Probiotic-induced suppression of allergic sensitization and airway inflammation is associated with an increase of T regulatory-dependent mechanisms in a murine model of asthma. Clin. Exp. Aller. 37, 498–505. doi: 10.1111/j.1365-2222.2006.02629.x
Forsythe, P. (2014). Probiotics and lung immune responses. Ann. Am. Thorac. Soc. 11(Suppl. 1), S33-7. doi: 10.1513/AnnalsATS.201306-156MG
Forsythe, P., Inman, M. D., and Bienenstock, J. (2007). Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 175, 561–569. doi: 10.1164/rccm.200606-821OC
Fuchs-Tarlovsky, V., Marquez-Barba, M. F., and Sriram, K. (2016). Probiotics in dermatologic practice. Nutrition 32, 289–295. doi: 10.1016/j.nut.2015.09.001
Fujimura, K. E., Demoor, T., Rauch, M., Faruqi, A. A., Jang, S., Lynch, S. V. et al. (2014). House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. U.S.A. 111, 805–810. doi: 10.1073/pnas.1310750111
Fujimura, K. E., and Lynch, S. V. (2015). Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 17, 592–602. doi: 10.1016/j.chom.2015.04.007
Fujimura, K. E., Slusher, N. A., Cabana, M. D., and Lynch, S. V. (2010). Role of the gut microbiota in defining human health. Expert Rev. Anti Infect. Ther. 8, 435–454. doi: 10.1586/eri.10.14
Ghaisas, S., Maher, J., and Kanthasamy, A. (2016). Gut microbiome in health and disease. Linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 158, 52–62. doi: 10.1016/j.pharmthera.2015.11.012
Goleva, E., Jackson, L. P., Harris, J. K., Robertson, C. E., Sutherland, E. R., Hall, C. F., et al. (2013). The effects of airway microbiome on corticosteroid responsiveness in asthma. Am. J. Respir. Crit. Care Med. 188, 1193–1201. doi: 10.1164/rccm.201304-0775OC
Gollwitzer, E. S., and Marsland, B. J. (2014). Microbiota abnormalities in inflammatory airway diseases - potential for therapy. Pharmacol. Ther. 141, 32–39. doi: 10.1016/j.pharmthera.2013.08.002
Gomez, C., and Chanez, P. (2016). The lung microbiome. the perfect culprit for COPD exacerbations? Eur. Respir. J. 47, 1034–1036. doi: 10.1183/13993003.00270-2016
Gruner, D., Paris, S., and Schwendicke, F. (2016). Probiotics for managing caries and periodontitis. Systematic review and meta-analysis. J. Dent. 48, 16–25. doi: 10.1016/j.jdent.2016.03.002
Guarner, F., Khan, A. G., Garisch, J., Eliakim, R., Gangl, A., Thomson, A., et al. (2012). World gastroenterology organisation global guidelines. probiotics and prebiotics. J. Clin. Gastroenterol. 46, 468–481. doi: 10.1097/mcg.0b013e3182549092
Haahtela, T., Holgate, S., Pawankar, R., Akdis, C. A., and Benjaponpitak, S., Caraballo, L., et al. (2013). The biodiversity hypothesis and allergic disease: world allergy organization position statement. World Aller. Organ. J. 6:3. doi: 10.1186/1939-4551-6-3
He, Y., Wen, Q., Yao, F., Xu, D., Huang, Y., and Wang, J. (2017). Gut-lung axis: the microbial contributions and clinical implications. Crit. Rev. Microbiol. 43, 81–95. doi: 10.1080/1040841X.2016.1176988
Hepburn, N. J., Garaiova, I., Williams, E. A., Michael, D. R., and Plummer, S. (2013). Probiotic supplement consumption alters cytokine production from peripheral blood mononuclear cells: a preliminary study using healthy individuals. Benef. Microb. 4, 313–317. doi: 10.3920/BM2013.0012
Hilty, M., Burke, C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., et al. (2010). Disordered microbial communities in asthmatic airways. PLoS ONE 5:0008578. doi: 10.1371/journal.pone.0008578
Hogan, D. A., Willger, S. D., Dolben, E. L., Hampton, T. H., Stanton, B. A., Ashare, A., et al. (2016). Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS ONE 11:e0149998. doi: 10.1371/journal.pone.0149998
Hougee, S., Vriesema, A. J., Wijering, S. C., Knippels, L. M., Folkerts, G., Nijkamp, F. P., et al. (2010). Oral treatment with probiotics reduces allergic symptoms in ovalbumin-sensitized mice: a bacterial strain comparative study. Int. Arch. Aller. Immunol. 151, 107–117. doi: 10.1159/000236000
Huang, Y. J. (2013). Asthma microbiome studies and the potential for new therapeutic strategies. Curr. Aller. Asthma Rep. 13, 453–461. doi: 10.1007/s11882-013-0355-y
Huang, Y. J. (2015). The respiratory microbiome and innate immunity in asthma. Curr. Opin. Pulm. Med. 21, 27–32. doi: 10.1097/MCP.0000000000000124
Huang, Y. J., Erb-Downward, J. R., Dickson, R. P., Curtis, J. L., Huffnagle, G. B., and Han, M. K. (2017). Understanding the role of the microbiome in chronic obstructive pulmonary disease. principles challenges and future directions. Transl. Res. 179, 71–83. doi: 10.1016/j.trsl.2016.06.007
Inoue, Y., Iwabuchi, N., Xiao, J. Z., Yaeshima, T., and Iwatsuki, K. (2009). Suppressive effects of bifidobacterium breve strain M-16V on T-helper type 2 immune responses in a murine model. Biol. Pharm. Bull., 32, 760–763. doi: 10.1248/bpb.32.760
Kalyuzhin, O. V., Afanasyev, S. S., and Bykov, A. S. (2016). [Probiotics as stimulators of immune response against pathogens in the respiratory tract]. Ter. Arkh. 88, 118–124. doi: 10.17116/terarkh2016885118-124
Karimi, K., Inman, M. D., Bienenstock, J., and Forsythe, P. (2009). Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med., 179, 186–193. doi: 10.1164/rccm.200806-951OC
Kuczynski, J., Stombaugh, J., Walters, W. A., Gonzalez, A., Caporaso, J. G., and Knight, R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformat. Chapter 10:Unit 10 7. doi: 10.1002/0471250953.bi1007s36
Lappin, M. R., Veir, J. K., Satyaraj, E., and Czarnecki-Maulden, G. (2009). Pilot study to evaluate the effect of oral supplementation of Enterococcus faecium SF68 on cats with latent feline herpesvirus 1. J. Feline Med. Surg. 11, 650–654. doi: 10.1016/j.jfms.2008.12.006
Lee, D. E., Huh, C. S., Ra, J., Choi, I. D., Jeong, J. W., Kim, S. H., et al. (2015). Ahn. clinical evidence of effects of Lactobacillus plantarum HY7714 on skin aging. A randomized, double blind, placebo-controlled study. J. Microbiol. Biotechnol. 25, 2160–2168. doi: 10.4014/jmb.1509.09021
Li, C. Y., Lin, H. C., Hsueh, K. C., Wu, S. F., and Fang, S. H. (2010). Oral administration of Lactobacillus salivarius inhibits the allergic airway response in mice. Can. J. Microbiol. 56, 373–379. doi: 10.1139/W10-024
Lievin-Le Moal, V., and Servin, A. L. (2014). Anti-infective activities of lactobacillus strains in the human intestinal microbiota. from probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin. Microbiol. Rev. 27, 167–199. doi: 10.1128/CMR.00080-13
Ling, Z., Liu, X., Cheng, Y., and Li, L. (2016). Potential roles of disordered airway microbiota in patients with severe asthma. J. Allergy Clin. Immunol. 137:648. doi: 10.1016/j.jaci.2015.09.043
Madan, J. C. (2016). Neonatal gastrointestinal and respiratory microbiome in cystic fibrosis. potential interactions and implications for systemic Health Clin Ther. 38, 740–746. doi: 10.1016/j.clinthera.2016.02.008
Madan, J. C., Koestler, D. C., Stanton, B. A., Davidson, L., Moulton, L. A., Housman, M. L., et al. (2012). Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio 3: pii: e00251-12. doi: 10.1128/mBio.00251-12
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marsland, B. J., and Salami, O. (2015). Microbiome influences on allergy in mice and humans. Curr. Opin. Immunol. 36, 94–100. doi: 10.1016/j.coi.2015.07.005
Marsland, B. J., Trompette, A., and Gollwitzer, E. S. (2015). The gut-lung axis in respiratory disease. Ann. Am. Thorac. Soc. 12(Suppl. 2), S150–S156. doi: 10.1513/AnnalsATS.201503-133AW
Mastromarino, P., Capobianco, D., Miccheli, A., Pratic,ò, G., Campagna, G., Laforgia, N., et al. (2015). Administration of a multistrain probiotic product (VSL#3) to women in the perinatal period differentially affects breast milk beneficial microbiota in relation to mode of delivery. Pharmacol. Res. 95–96, 63–70. doi: 10.1016/j.phrs.2015.03.013
McCoy, K. D., and Koller, Y. (2015). New developments providing mechanistic insight into the impact of the microbiota on allergic disease. Clin. Immunol. 159, 170–176. doi: 10.1016/j.clim.2015.05.007
Meijer, K., de Vos, P., and Priebe, M. G. (2010). Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 13, 715–721. doi: 10.1097/MCO.0b013e32833eebe5
Mezzasalma, V., Manfrini, E., Ferri, E., Boccarusso, M., Di Gennaro, P., Schiano, I., et al. (2017). Orally administered multispecies probiotic formulations to prevent uro-genital infections. a randomized placebo-controlled pilot study. Arch. Gynecol. Obstet. 295, 163–172. doi: 10.1007/s00404-016-4235-2
Morris, A., Beck, J. M., Schloss, P. D., Campbell, T. B., Crothers, K., Curtis, J. L., et al. (2013). Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 187, 1067–1075. doi: 10.1164/rccm.201210-1913OC
Neish, S. (2014). Mucosal immunity and the microbiome. Ann. Am. Thorac. Soc. 11(Suppl. 1), S28–S32. doi: 10.1513/AnnalsATS.201306-161MG
Norris Reinero, C. R., Decile, K. C., Berghaus, R. D., Williams, K. J., Leutenegger, C. M., Gershwin, L. J., et al. (2004). An experimental model of allergic asthma in cats sensitized to house dust mite or bermuda grass allergen. Int. Arch. Allergy Immunol. 135, 117–131. doi: 10.1159/000080654
Pirrone, M., Pinciroli, R., and Berra, L. (2016). Microbiome, biofilms, and pneumonia in the ICU. Curr. Opin. Infect. Dis. 29, 160–166. doi: 10.1097/QCO.0000000000000255
Rautava, S., and Isolauri, E. (2002). The development of gut immune responses and gut microbiota. Effects of probiotics in prevention and treatment of allergic disease. Curr. Issues Intest. Microbiol. 3, 15–22. doi: 10.1016/j.gtc.2012.08.007
Reinero, C. R., DeClue, A. E., and Rabinowitz, P. (2009). Asthma in humans and cats: is there a common sensitivity to aeroallegens in shared environments? Environ. Res. 109, 634–640. doi: 10.1016/j.envres.2009.02.001
Romani Vestman, N., Chen, T., Lif Holgerson, P., Öhman, C., and Johansson, I. (2015). Oral Microbiota Shift after 12-Week Supplementation with Lactobacillus reuteri DSM 17938 and PTA 5289; A Randomized Control Trial. PLoS ONE 10:e0125812. doi: 10.1371/journal.pone.0125812
Samuelson, D. R., Welsh, D. A., and Shellito, J. E. (2015). Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 6:1085. doi: 10.3389/fmicb.2015.01085
Sidiq, F., Hoostal, M., and Rogers, S. O. (2016). Rapid identification of fungi in culture-negative clinical blood and respiratory samples by DNA sequence analyses. BMC Res. Notes 9:293. doi: 10.1186/s13104-016-2097-0
Southam, D. S., Dolovich, M., O'Byrne, P. M., and Inman, M. D. (2002). Distribution of intranasal instillations in mice: effects of volume, time, body position, and anesthesia. Am. J. Physiol. Lung Cell. Mol. Physiol. 282, L833–L839. doi: 10.1152/ajplung.00173.2001
Thaiss, C. A., Zmora, N., Levy, M., and Elinav, E. (2016). The microbiome and innate immunity. Nature 535, 65–74. doi: 10.1038/nature18847
Treven, P., Mrak, V., Bogovič Matijašić, B., Horvat, S., and Rogelj, I. (2015). Administration of probiotics Lactobacillus rhamnosus GG and Lactobacillus gasseri K7 during pregnancy and lactation changes mouse mesenteric lymph nodes and mammary gland microbiota. J. Dairy Sci. 98, 2114–2128. doi: 10.3168/jds.2014-8519
Vieira, A. T., Rocha, V. M., Tavares, L., Garcia, C. C., Teixeira, M. M., Nicoli, J. R., et al. (2016). Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 5(1A). Microb. Infect. 18, 180–189. doi: 10.1016/j.micinf.2015.10.008
Vientos-Plotts, A. I., Ericsson, A. C., Rindt, H., Grobman, M. E., Graham, A., Bishop, K., et al. (2017). Dynamic changes of the respiratory microbiota and its relationship to fecal and blood microbiota in healthy young cats. PLoS ONE 12:e0173818. doi: 10.1371/journal.pone.0173818
Vitali, B., Cruciani, F., Baldassarre, M. E., Capursi, T., Spisni, E., Valerii, M. C., et al. (2012). Dietary supplementation with probiotics during late pregnancy: outcome on vaginal microbiota and cytokine secretion. BMC Microbiol. 12:236. doi: 10.1186/1471-2180-12-236
Williams, K., and Roman, J. (2016). Studying human respiratory disease in animals–role of induced and naturally occurring models. J. Pathol. 238, 220–232. doi: 10.1002/path.4658
Yin, Y., Hountras, P., and Wunderink, R. G. (2017). The microbiome in mechanically ventilated patients. Curr. Opin. Infect. Dis. 30, 208–213. doi: 10.1097/QCO.0000000000000352
Yoda, K., He, F., Miyazawa, K., Kawase, M., Kubota, A., and Hiramatsu, M. (2012). Orally administered heat-killed Lactobacillus gasseri TMC0356 alters respiratory immune responses and intestinal microbiota of diet-induced obese mice. J. Appl. Microbiol., 113, 155–162. doi: 10.1111/j.1365-2672.2012.05316.x
Yu, Z., and Morrison, M. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36, 808–812. doi: 10.2144/3605A0808
Zamani, B., Golkar, H. R., Farshbaf, S., Emadi-Baygi, M., Tajabadi-Ebrahimi, M., Jafari, P., et al. (2016). Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis. a randomized, double-blind, placebo-controlled trial. Int. J. Rheum. Dis. 19, 869–879. doi: 10.1111/1756-185X.12888
Keywords: respiratory, operational taxonomic unit (OTU), 16S rRNA gene, probiotics, richness, abundance, diversity, translational research
Citation: Vientós-Plotts AI, Ericsson AC, Rindt H and Reinero CR (2017) Oral Probiotics Alter Healthy Feline Respiratory Microbiota. Front. Microbiol. 8:1287. doi: 10.3389/fmicb.2017.01287
Received: 03 May 2017; Accepted: 27 June 2017;
Published: 11 July 2017.
Edited by:
Francesca Turroni, University of Parma, ItalyReviewed by:
Susan Thibeault, University of Wisconsin–Madison, United StatesCopyright © 2017 Vientós-Plotts, Ericsson, Rindt and Reinero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aaron C. Ericsson, ZXJpY3Nzb25hQG1pc3NvdXJpLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.