
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Microbiol., 30 June 2017
Sec. Microbiotechnology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01220
This article is part of the Research TopicUsing Genomics, Metagenomics and Other "Omics" to Assess Valuable Microbial Ecosystem Services and Novel Biotechnological ApplicationsView all 53 articles
 Rachel A. Levin1,2,3*
Rachel A. Levin1,2,3* Christian R. Voolstra4
Christian R. Voolstra4 Shobhit Agrawal4
Shobhit Agrawal4 Peter D. Steinberg1,2,5
Peter D. Steinberg1,2,5 David J. Suggett3
David J. Suggett3 Madeleine J. H. van Oppen6,7
Madeleine J. H. van Oppen6,7Elevated sea surface temperatures from a severe and prolonged El Niño event (2014–2016) fueled by climate change have resulted in mass coral bleaching (loss of dinoflagellate photosymbionts, Symbiodinium spp., from coral tissues) and subsequent coral mortality, devastating reefs worldwide. Genetic variation within and between Symbiodinium species strongly influences the bleaching tolerance of corals, thus recent papers have called for genetic engineering of Symbiodinium to elucidate the genetic basis of bleaching-relevant Symbiodinium traits. However, while Symbiodinium has been intensively studied for over 50 years, genetic transformation of Symbiodinium has seen little success likely due to the large evolutionary divergence between Symbiodinium and other model eukaryotes rendering standard transformation systems incompatible. Here, we integrate the growing wealth of Symbiodinium next-generation sequencing data to design tailored genetic engineering strategies. Specifically, we develop a testable expression construct model that incorporates endogenous Symbiodinium promoters, terminators, and genes of interest, as well as an internal ribosomal entry site from a Symbiodinium virus. Furthermore, we assess the potential for CRISPR/Cas9 genome editing through new analyses of the three currently available Symbiodinium genomes. Finally, we discuss how genetic engineering could be applied to enhance the stress tolerance of Symbiodinium, and in turn, coral reefs.
Photosynthetic dinoflagellates are critical primary producers in the aquatic environment, yet, their functional genomics are largely unexplored (Leggat et al., 2011; Murray et al., 2016). Symbiodinium is considered one of the most important dinoflagellate genera given its role as the essential photosymbiont of many tropical reef invertebrates, notably reef-building corals (Trench and Blank, 1987). Provision of photosynthetically derived metabolites from Symbiodinium to the coral host drives coral calcification and growth that forms the foundation of coral reef ecosystems (Muscatine and Porter, 1977; Muscatine, 1990; Kirk and Weis, 2016). Thermal and light stress cause photosynthetic dysfunction of Symbiodinium and increased leakage of harmful reactive oxygen species from their cells, a process considered largely responsible for the dissociation of Symbiodinium from corals characterized as “coral bleaching” (Warner et al., 1999; Suggett et al., 2008; Weis, 2008; Levin et al., 2016). Symbiodinium has therefore become established as a major focus for research globally, and in effect, a model genus for dinoflagellates.
Dinoflagellates evolved an estimated 520 million years ago (Moldowan and Talyzina, 1998) and exhibit substantial evolutionary divergence from model eukaryotic organisms including other microalgae such as Chlamydomonas and diatoms (Shoguchi et al., 2013). Consequently, dinoflagellates possess unusual biological features that have hindered research progress, such as some of the largest known nuclear genomes (1.5–112 Gbp, typically exceeding the size of the human haploid genome), permanently condensed liquid-crystalline chromosomes, trans-splicing of polycistronic mRNAs, and plastid genomes that are divided up into minicircles (Shoguchi et al., 2013; Zhang et al., 2013; Lin et al., 2015; Murray et al., 2016). The Symbiodinium genus evolved an estimated 50 million years ago and is highly diverse, containing nine major evolutionary lineages or “clades” (A–I; Coffroth and Santos, 2005; Pochon et al., 2006; Pochon and Gates, 2010) with hundreds of genetically distinct “types/sub-clades” considered to be different species1 (Tonk et al., 2013). Genetic factors that promote differences in stress tolerance between Symbiodinium variants (both inter- and intra-specific) strongly influence coral gene expression and bleaching susceptibility (Berkelmans and van Oppen, 2006; DeSalvo et al., 2010; Yuyama et al., 2012; Levin et al., 2016). However, the capacity to fully explore Symbiodinium genetics is currently restricted by a lack of genetic engineering capability. Genetic engineering has been central to the study of gene function and phenotypic enhancement in organisms ranging from microbes to mammals and a key platform for socioeconomic industries and biotechnologies; yet only two cases of transgene expression in Symbiodinium have ever been validated (ten Lohuis and Miller, 1998; Ortiz-Matamoros et al., 2015a).
In 1998, a type A1 strain was transformed at very low efficiencies using silicon carbide whiskers with plasmids encoding expression constructs with plant, plant-viral, and agrobacterial promoters (nos, CaMV 35S, and p1′2′) to drive transcription of antibiotic resistance genes (nptII and hptII) and a reporter gene (GUS) (ten Lohuis and Miller, 1998); however, these results have yet to be reproduced. It was not until 2015 that another case of transgene expression in Symbiodinium was reported (Ortiz-Matamoros et al., 2015b). Plasmids encoding expression constructs with plant and plant-viral promoters (nos and double CaMV 35S) to drive transcription of a herbicide resistance gene (bar) and a reporter gene (GFP) were introduced to type A1, B1, and F1 strains using glass beads. Whilst cells transiently exhibited improved herbicide resistance and suggestive GFP signal, transformations were not validated through DNA, RNA, or protein analysis (Ortiz-Matamoros et al., 2015b). Further transformation of these strains was attempted using Agrobacterium carrying plasmids with the same expression constructs, but the transformants were transient and unable to divide (Ortiz-Matamoros et al., 2015a). Of these studies, none attempted manipulation of ecologically relevant genes thereby limiting new insight gained into Symbiodinium biology.
Therefore, in an attempt to overcome the bottleneck that has become established in transforming Symbiodinium (and other dinoflagellates), we recommend a new approach that capitalizes on the recent surge in “omics” breakthroughs (Figure 1). By evaluating the rapidly increasing supply of next-generation sequencing (NGS) data, we propose a genetic engineering framework for Symbiodinium that may markedly advance our understanding of these important dinoflagellates. Furthermore, genetic manipulation of Symbiodinium in order to reduce coral bleaching has been hypothesized as a strategy to facilitate coral management as reefs continue to rapidly deteriorate under climate change (van Oppen et al., 2017). Combatting the impacts of climate change and conserving marine organisms are both key goals for sustainable development set forth by the United Nations2. Thus, we believe genetic engineering of Symbiodinium may open a novel avenue to achieve these goals by protecting corals from climate change.
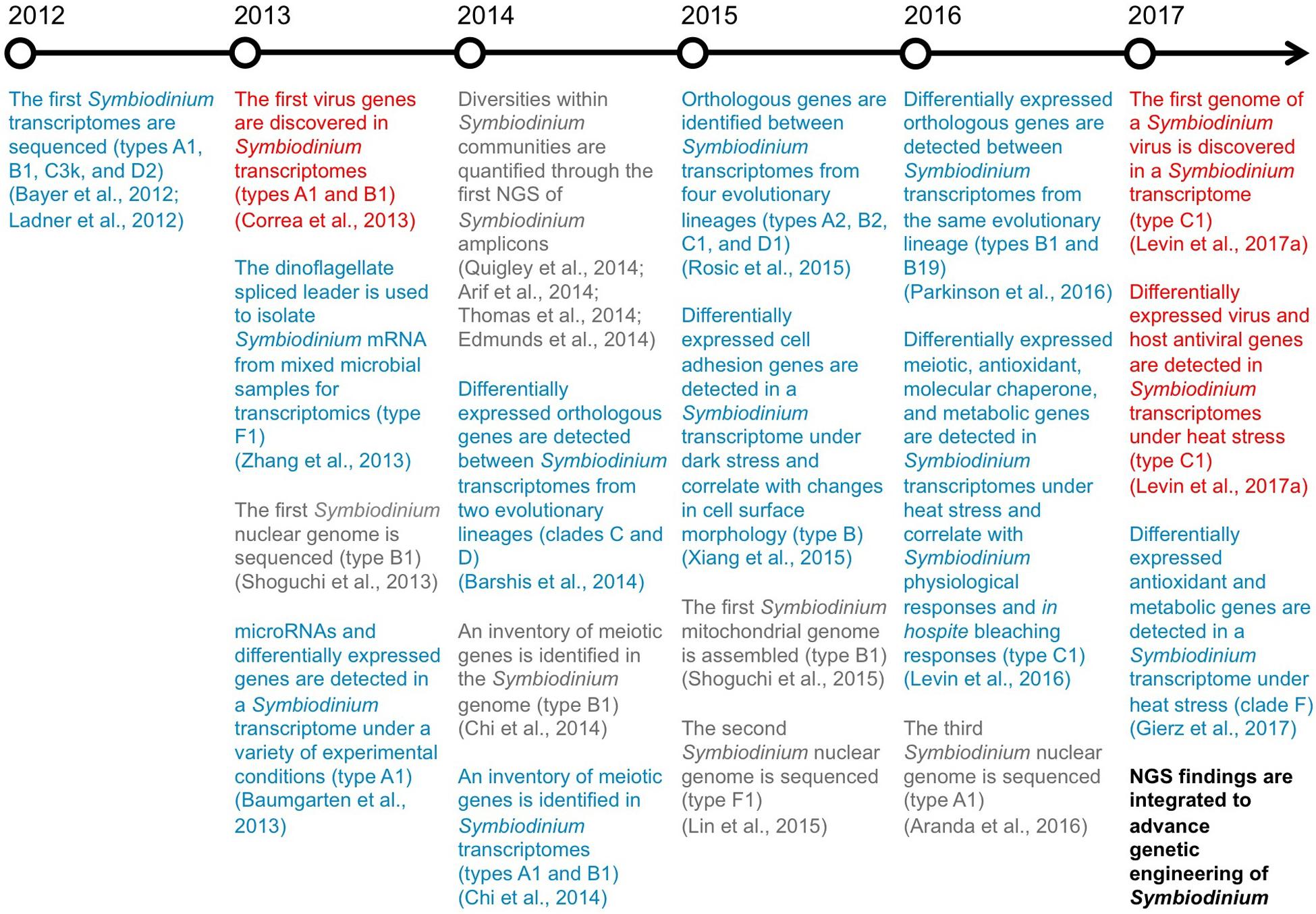
FIGURE 1. Breakthroughs in NGS of Symbiodinium. A timeline highlighting the key genomic (gray), transcriptomic (blue), and virus RNA (red) findings from recent NGS studies of Symbiodinium.
Fundamental components of Symbiodinium biology have recently been uncovered through a boom in NGS (Figure 1), particularly the assembly of the first Symbiodinium genomes and transcriptomes, direct correlation between Symbiodinium transcriptional and physiological states, and discovery of genes from viruses actively infecting Symbiodinium cells. Furthermore, NGS of Symbiodinium has revealed genetic elements that may allow for transformation of Symbiodinium. In the following sections, we detail how unique Symbiodinium promoters, specific Symbiodinium genes underpinning important phenotypes, and a viral internal ribosomal entry site recognized by Symbiodinium ribosomes could be integrated to build expression constructs for Symbiodinium.
Currently, dinoflagellate nuclear genome assemblies are all from the genus Symbiodinium (types A1, B1, and F1; Shoguchi et al., 2013; Lin et al., 2015; Aranda et al., 2016), emphasizing the importance of Symbiodinium to dinoflagellate research. The assemblies have revealed the immense size of Symbiodinium genomes with 36,850–49,109 genes, unidirectional gene orientation, prevalent gene tandem arrays, microRNAs along with putative gene targets, and unique promoter architecture (Shoguchi et al., 2013; Lin et al., 2015; Aranda et al., 2016). Rather than the traditional TATA-box of eukaryotic promoters, Symbiodinium promoters appear to have a TTTT-box that is followed by a unique transcription start site (YYANWYY), branch point (YTNAY), and acceptor for the dinoflagellate spliced leader (AG) (Lin et al., 2015). Additionally, instead of the typical eukaryotic polyadenylation signal AAUAAA, dinoflagellate terminators use AAAAG/C (Bachvaroff and Place, 2008). Hence, utilization of endogenous Symbiodinium promoters and terminators (as opposed to promoters and terminators from other organisms) would likely improve expression and stability of transgenes introduced into Symbiodinium. By chance, the CAMV 35S (plant-viral) promoter happens to contain all of the described Symbiodinium promoter elements, and the CAMV 35S (plant-viral) and nos (plant) terminators both contain the dinoflagellate polyadenylation signal; this may have contributed to their ability to drive transgene expression in Symbiodinium previously (ten Lohuis and Miller, 1998; Ortiz-Matamoros et al., 2015a).
Recent transcriptomic studies have identified highly expressed Symbiodinium nuclear genes that can be genome-mapped to uncover strong, endogenous promoters and their corresponding terminators. These promoters and terminators can be isolated from purified genomic DNA (gDNA) through PCR and incorporated into custom DNA expression constructs for Symbiodinium (Figure 2). Among the most highly expressed transcripts in Symbiodinium transcriptomes are genes for peridinin-chlorophyll a-binding protein, caroteno-chlorophyll a-c-binding protein, major basic nuclear protein 2, dinoflagellate viral nucleoprotein, and glyceraldehyde-3-phosphate dehydrogenase (Baumgarten et al., 2013; Levin et al., 2016; Parkinson et al., 2016); though all are multi-copy genes (Shoguchi et al., 2013; Lin et al., 2015; Aranda et al., 2016). Ideally, highly expressed nuclear genes chosen for promoter selection should not have high copy numbers, as their expression levels may largely be due to prevalence in the genome rather than strong promoters. Constitutively expressed nuclear genes are also desirable for selection of promoters that drive consistent transcription regardless of experimental conditions, and thus, drive reliable transgene expression.
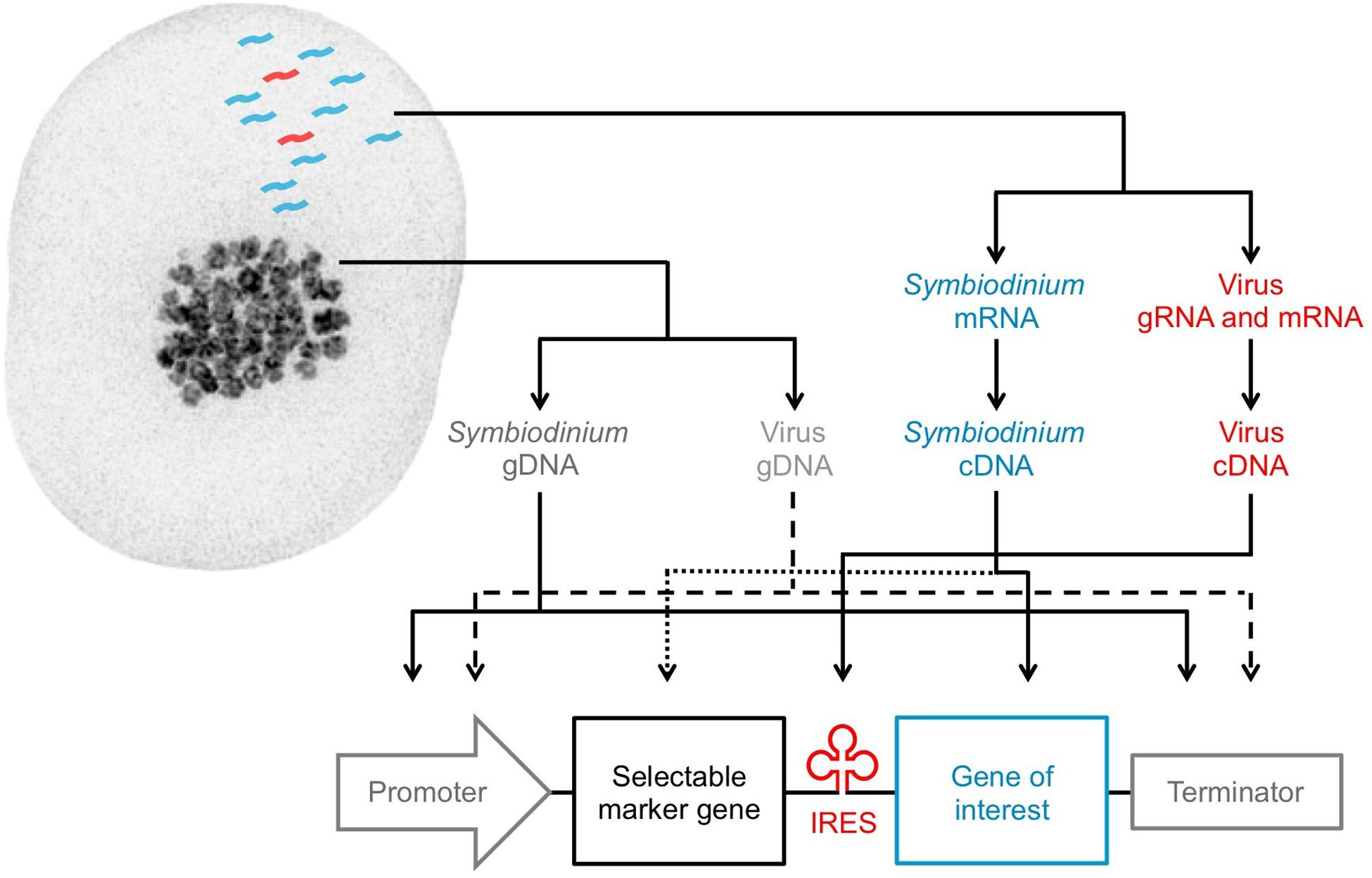
FIGURE 2. Design of a tailored expression construct for Symbiodinium. Genetic elements that can be isolated from Symbiodinium cells: Symbiodinium genomic DNA (dark gray), Symbiodinium messenger RNA (blue), resident virus genomic DNA (light gray), resident virus genomic or messenger RNA (red). Solid lines (identified elements) and dashed or dotted lines (unidentified elements) are used to arrange the elements into a Symbiodinium expression construct. The pictured Symbiodinium cell (type C1) was stained with DAPI for imaging on a DeltaVision OMX Blaze microscope (excitation/emission: 405 nm/419–465 nm).
To illustrate this approach of Symbiodinium promoter selection, we examined NGS data from a type A1 Symbiodinium strain for which the nuclear genome has been recently sequenced (Aranda et al., 2016) and the transcriptional responses to various conditions (temperatures, ionic stress, dark stress, and contrasting circadian rhythm time points) have been determined (Baumgarten et al., 2013). Locus 144 and Locus 1768 in the type A1 transcriptome, a subunit of a large neutral amino acids transporter and a putative ATP-binding cassette transporter gene, both show high expression across all conditions (average expression in the top 2% of all genes; Baumgarten et al., 2013) and map tightly to the type A1 genome scaffolds 710 and 484, respectively. No significant open reading frames are found >5 kb up- or down-stream of either gene, confirming that they are not part of tandem arrays. For each gene, all Symbiodinium promoter elements are within 1 kb of the start codon, and the dinoflagellate polyadenylation signal is found ∼300 bp after the stop codon. These promoter and terminator regions could therefore be isolated and utilized to drive high and consistent expression of transgenes in a Symbiodinium expression construct.
Recent transcriptomic studies have been fundamental in the discovery of Symbiodinium nuclear genes that underpin phenotypic traits, such as those related to cell adhesion (e.g., GspB, Svep1, Slap1; Xiang et al., 2015), sexual reproduction (e.g., Msh4, Msh5, Spo11-2; Chi et al., 2014; Levin et al., 2016; Gierz et al., 2017), antiviral response (e.g., Birc3, Ns1bp, Ifih1; Levin et al., 2017a), and antioxidant activity/thermal tolerance (e.g., Fe-sod, Mn-sod, Pxrd, Hsp70; Levin et al., 2016; Gierz et al., 2017). Symbiodinium antioxidant genes are of particular interests because of their potential role in defining bleaching susceptibility of the coral host (Krueger et al., 2015; Levin et al., 2016). For instance, iron-type superoxide dismutase (Fe-sod) genes are believed to minimize thermally induced oxidative damage to photosynthetic apparatuses and leakage of harmful reactive oxygen species from type C1 Symbiodinium cells—determinants of coral bleaching (Weis, 2008); however, these genes are not expressed at detectable levels in all Symbiodinium variants (Krueger et al., 2015; Levin et al., 2016). A Fe-sod gene could therefore be inserted after a strong Symbiodinium promoter in an expression construct to drive its over-expression for evaluation of its phenotypic influence on Symbiodinium. Endogenous genes of interest should be isolated through PCR of complementary DNA (cDNA) reverse transcribed from purified mRNA, since gDNA introns may prevent proper expression in constructs (Figure 2).
Expression of exogenous genes of interest in Symbiodinium could also greatly advance investigations of ecological processes central to coral reef health. For instance, documenting competition between Symbiodinium types, transmission and acquisition of Symbiodinium types by the coral host, and shuffling of Symbiodinium types within host tissues (Toller et al., 2001; van Oppen et al., 2001; Little et al., 2004; Berkelmans and van Oppen, 2006; Byler et al., 2013; Boulotte et al., 2016) is currently reliant upon sequencing since it is not possible to visually differentiate many types. As a result, studies have been restricted to low temporal and spatial resolution relative to real-time imaging. Instead, the ability to color-code Symbiodinium types through genetic transformation with various fluorescent proteins could illuminate these phenomena by enabling real-time imaging for visually differentiating types. Additionally, tagging endogenous genes of interest through fluorescent protein fusions would permit imaging of protein localization within Symbiodinium cells and potential protein secretion out of Symbiodinium cells (Xiang et al., 2015). When selecting appropriate fluorescent proteins, it will be imperative to consider the extreme autofluorescence of Symbiodinium (Shaner et al., 2005); for example, venus (excitation/emission: 515/528 nm), tdTomato (excitation/emission: 554/581 nm), and mCherry (excitation/emission: 587/610 nm) are promising candidates as their fluorescence properties are off-peak of the Symbiodinium excitation and emission spectra (Hennige et al., 2009; Jiang et al., 2012). Finally, codon optimization may be necessary for optimal exogenous gene expression in Symbiodinium since codon usage of Symbiodinium genes can be divergent from foreign genes (Levin et al., 2017a) and even between Symbiodinium nuclear and minicircle genes (Bayer et al., 2012).
Although antibiotics have previously been used to select transformed Symbiodinium (ten Lohuis and Miller, 1998; Ortiz-Matamoros et al., 2015a), their use is problematic for two main reasons. Firstly, eliminating wild-type Symbiodinium in culture requires high concentrations of antibiotics (e.g., 3 mg/ml of G418 or hygromycin; ten Lohuis and Miller, 1998), making experimentation and long-term maintenance of transformed cell lines extremely costly. It is also important to note that natural antibiotic resistances are not uniform across all strains (Supplementary Table 1), so dosage curves are necessary before conducting transformation trials. Secondly, dinoflagellates including Symbiodinium require symbiotic bacteria to grow optimally (Alavi et al., 2001; Croft et al., 2005; Miller and Belas, 2006; Ritchie, 2012). Since eukaryotic antibiotics can also be toxic to prokaryotes (Gonzalez et al., 1978; Colanduoni and Villafranca, 1986; Pline et al., 2001; Vicens and Westhof, 2003), bacterial communities in Symbiodinium cultures are removed during antibiotic selection.
To preserve symbiotic bacteria, alternatives to antibiotic selection markers should be considered, such as genes that provide growth advantages under specific conditions by increasing pathogen resistance, increasing thermal tolerance, or allowing for utilization of non-metabolized carbohydrates (Breyer et al., 2014). The precise functions of these alternative marker genes (e.g., phosphomannose isomerase) are well defined and shown to be applicable to many photosynthetic species (Stoykova and Stoeva-Popova, 2011), though their compatibilities with dinoflagellates are unknown. Discovery of endogenous selectable markers should therefore also be pursued. Recent Symbiodinium transcriptomic studies have uncovered genes involved in selection-relevant phenotypes like photosynthetic ability at unique light regimes (Parkinson et al., 2016) or tolerance to increased temperature regimes (Levin et al., 2016). These Symbiodinium genes could first be expressed in more easily transformed microalgae like Chlamydomonas and diatoms to gauge the potential for their up-regulation to grant a significant selectable advantage under specific conditions.
Viral promoters and terminators, internal ribosome entry sites (IRES), and 2A peptides are staple regulatory elements incorporated in expression constructs since they have evolved to be recognized by eukaryotic machinery for efficient and stable foreign gene expression (Benfey and Chua, 1990; Martínez-Salas, 1999; Levin et al., 2014). Symbiodinium transcriptomics have led to the discovery of genes, as well as an entire RNA genome, from novel eukaryotic viruses that infect Symbiodinium (Correa et al., 2013; Levin et al., 2017a). A putative viral IRES, which allows cap-independent translation to produce separate proteins from one mRNA transcript, was found between the two open reading frames in the RNA genome of the +ssRNA virus infecting type C1 Symbiodinium (GenBank accession: KX538960 and KX787934; Levin et al., 2017a). The +ssRNA virus transcripts were extremely abundant in a type C1 Symbiodinium transcriptome (Levin et al., 2017a), and such rampant +ssRNA virus replication indicates that Symbiodinium ribosomes have high affinity to this IRES.
IRES sequences enable the creation of polycistronic constructs transcriptionally controlled by a single promoter (Martínez-Salas, 1999). By permitting simultaneous expression of two independent proteins from one mRNA, a bicistronic construct can achieve long-term expression of a gene of interest because the gene of interest is transcriptionally fused to the selectable marker gene (Gurtu et al., 1996; Figure 2). Conversely, in monocistronic constructs, the selectable marker gene often maintains expression, while the gene of interest becomes transcriptionally repressed over time if it does not increase fitness of the cell (Allera-Moreau et al., 2007). Therefore, the IRES from the Symbiodinium +ssRNA virus is a valuable viral element that is recognized by Symbiodinium ribosomes and may improve the stability of transgene expression in Symbiodinium. Moving forward, NGS data of viruses in Symbiodinium cultures (Weynberg et al., 2017) and the coral holobiont (Weynberg et al., 2014; Correa et al., 2016) should be mined for promoter, terminator, and other regulatory elements from Symbiodinium viruses, given the proven benefits of viral elements to genetic engineering. Once assembled, the Symbiodinium expression construct (Figure 2) can be combined with the backbone of a standard cloning plasmid; added into an artificial, replicating minicircle (Nehlsen et al., 2006; Karas et al., 2015); or serve as a repair template for CRISPR/Cas9 genome editing (Cong et al., 2013).
Within the past 5 years, CRISPR/Cas9 has revolutionized genome editing by allowing precise changes to be made to target sites in the genome (Cong et al., 2013; Baek et al., 2016; Nymark et al., 2016). In short, a single guide RNA (sgRNA) is designed to recruit the Cas9 endonuclease protein and to match a specific, desired target site in the genome that must be immediately followed by a protospacer adjacent motif (PAM) sequence (5′-NGG-3′). Once complexed with Cas9, the sgRNA guides Cas9 to the target genome site. Cas9 then interacts with the PAM sequence and creates a double-strand break in the target site. The cell can either repair the double stranded break through non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Ran et al., 2013). NHEJ genome editing arises from introduction of a random mutation/insertion/deletion when the broken ends of DNA are directly ligated, which can cause the target gene to be knocked out (i.e., non-functional). Gene knockout provides insight into the role and criticality of a gene by assessing the effect of its absence. Alternatively, HDR genome editing uses a repair template flanked by 5′ and 3′ homologous arm sequences that match the up- and down-stream regions of the double-strand break. The repair template can be designed for gene knockout, introduction of a specific mutation/insertion/deletion, or genomic integration of a transgene(s)/entire expression construct (Ran et al., 2013).
Symbiodinium exhibits an asexual haploid vegetative stage (Santos and Coffroth, 2003) with sister chromatids developing in S-phase of the cell cycle (Watrin and Legagneux, 2003), but HDR has yet to be directly observed in Symbiodinium. Therefore, CRISPR/Cas9 genome editing of Symbiodinium may be restricted to NHEJ. Ku70, Ku80, and DNA ligase IV (genes central to NHEJ; Chu et al., 2015) are all expressed in Symbiodinium transcriptomes (Levin et al., 2016). That said, some evidence does suggest Symbiodinium can enter a transient sexual diploid stage (Chi et al., 2014; Wilkinson et al., 2015; Levin et al., 2016), which has been documented in other dinoflagellates (Figueroa et al., 2015). In yeast, ploidy shifts the dominant double-stranded break repair mechanism—diploid cells favor HDR, while haploid cells favor NHEJ (Lee et al., 1999). Moreover, genes specific to meiosis, a process during which HDR occurs (Thacker and Keeney, 2016), have been found in Symbiodinium genomes and transcriptomes (Chi et al., 2014; Lin et al., 2015; Rosic et al., 2015; Levin et al., 2016). Msh4, Msh5, and Spo11-2 are all highly up-regulated at elevated temperatures (Levin et al., 2016), suggesting that HDR pathways in Symbiodinium are activated. Brca2, a gene that controls HDR (Holloman, 2011), is likewise up-regulated in heat stressed Symbiodinium (SM population: TR74441| c0_g1; MI population: TR63986| c0_g1; Levin et al., 2016). Hence, the potential for genomic integration of transgenes through HDR may improve if Symbiodinium are pre-stressed. HDR in Symbiodinium may also be increased by suppression of Ku70, Ku80, or DNA ligase IV (Chu et al., 2015).
The permanently condensed chromosomes of Symbiodinium could present an obstacle for CRISPR/Cas9 genome editing by possibly limiting access of sgRNAs to certain target sites. An additional challenge for genome editing is the abundance of multi-copy genes in the large Symbiodinium genomes. Gene redundancy can prevent knockout of gene function since the CRISPR/Cas9 system is not 100% efficient, meaning uncleaved functional gene copies can remain. Additionally, CRISPR/Cas9 targeting of genes with high copy numbers has been found to decrease cell proliferation and survival likely due to an increased frequency of DNA damage events (Aguirre et al., 2016). Also, design of sgRNAs requires a sequenced genome, but only three Symbiodinium genomes—each from a separate evolutionary lineage—are currently available.
As a first step to overcome some of these limitations, we analyzed the three published Symbiodinium genomes (types A1, B1, and F1; Shoguchi et al., 2013; Lin et al., 2015; Aranda et al., 2016) to identify conserved single copy genes. We then predicted a target site in each conserved gene with high sgRNA efficiency and specificity across the genomes (Supplementary Materials and Methods). Conserved target sites may permit CRISPR/Cas9 genome editing of Symbiodinium types that have yet to be sequenced. Our analysis revealed 1792 conserved single copy orthologs, 261 of which have an optimal target site compatible with all genomes (Supplementary Dataset 1a). The 261 single copy orthologs for CRISPR/Cas9 genome editing were enriched for a wide array of functional gene groups of interest, including cellular components for photosynthesis and biological pathways for oxidation-reduction and for response to UV-B (Supplementary Figure 1 and Supplementary Tables 2–4). Knockout of these genes would critically improve our understanding of Symbiodinium gene function, and if HDR is present in Symbiodinium, these sgRNA target sites could also be used to introduce genes of interest or entire Symbiodinium expression constructs into the genome. Furthermore, we identified sgRNA target sites in the type A1 genome scaffolds 710 and 484 (Aranda et al., 2016) immediately downstream from the potentially strong, constitutive Symbiodinium promoters discussed earlier (Supplementary Dataset 1b). Assuming HDR, reporter genes such as fluorescent proteins could be introduced at these sites to measure promoter activity.
The CRISPR/Cas9 system can be carried by plasmids that contain expression constructs for the Cas9, sgRNA, and in the case of HDR, the repair template with homologous arms. Target site cleavage is improved by increased CRISPR/Cas9 construct expression (Hsu et al., 2013), so strong endogenous promoters and terminators from Symbiodinium discussed earlier could be employed to drive transcription of Cas9 by Symbiodinium. However, transcription of sgRNAs requires RNA polymerase III (Pol III) rather than RNA polymerase II. Therefore, promoters specifically recognized by Pol III (e.g., promoter of the U6 snRNA gene) are needed. Such promoters have been isolated from other eukaryotes for sgRNA transcription; but, as discussed earlier, they contain motifs (e.g., TATA-box) that Symbiodinium lack (Goomer and Kunkel, 1992; Clarke et al., 2013). In Symbiodinium, 26 U6 snRNA gene copies have been identified (see Supplementary Table 5 in Shoguchi et al., 2013), one of which is unusually located in a cluster with U1, U2, U4, U5, 5S, and spliced leader snRNA genes (type B1 genome scaffold 8131; Shoguchi et al., 2013). Thus, genomic sequences found upstream and downstream of these Symbiodinium U6 snRNA genes could be isolated and trialed in sgRNA expression constructs as potential promoters and terminators recognized by Symbiodinium Pol III. Alternatively, the CRISPR/Cas9 system can be introduced to cells as pre-complexed sgRNA and purified Cas9 protein, which can achieve higher genome editing specificity by ∼10-fold compared to CRISPR/Cas9 plasmids and also removes the need to optimize Cas9 codon usage or to find appropriate promoters that will express Cas9 or sgRNAs (Zuris et al., 2015).
Verified delivery of expression constructs into Symbiodinium was previously achieved using silicon carbide whiskers, which yielded very few transformants (ten Lohuis and Miller, 1998), and with Agrobacterium, which produced transient transformants that were unable to divide (Ortiz-Matamoros et al., 2015a). Low efficiency foreign DNA delivery may be due to obstruction by the thick, multilayer Symbiodinium cell covering comprised of an external polysaccharide or glycoprotein layer atop an internal cell wall (thecal plates and the pellicle) then finally the plasma membrane (Markell et al., 1992; Wakefield et al., 2000). To overcome this barrier, methods including high-voltage electroporation, bioballistics, microinjection, and viral transduction should be trialed. Continued exploration into Symbiodinium viruses may facilitate development of a compatible transduction system. Additionally, the first method to produce viable Symbiodinium protoplasts (cells with their cell wall removed) was developed (Levin et al., 2017b). Protoplasts have been instrumental in genetic manipulation of cell-walled organisms through somatic hybridization as well as by allowing for alternate DNA delivery methods (Davey et al., 2005). Protoplast-dependent methods such as polyethylene glycol-mediated transformation (Mathur and Koncz, 1998) and liposome-mediated transformation (Caboche, 1990) may improve efficiency of construct delivery into Symbiodinium. Cell walls also serve as a barrier to RNA/protein complexes like pre-complexed sgRNA and Cas9 protein. Thus, genome editing of Symbiodinium with pre-complexed sgRNA and Cas9 protein may require the use of protoplasts (Woo et al., 2015). Polyethylene glycol-mediated transformation (Woo et al., 2015), cationic lipid transformation (Zuris et al., 2015), and electroporation (Baek et al., 2016) have all been used to effectively deliver pre-complexed sgRNA and Cas9 protein through cell membranes of other eukaryotes that lacked cell walls.
Coral reefs are the most diverse marine habitat per unit area (Reaka-Kudla et al., 1996; Knowlton et al., 2010) and provide world economies with nearly US$30 billion in net benefits from goods and services annually (Cesar et al., 2003). Climate change impact models predict that most reefs will be severely damaged or lost in this century unless immediate protection efforts are made (Hoegh-Guldberg et al., 2007; Pandolfi et al., 2011; Mora et al., 2016; Hughes et al., 2017) prompting calls for the development of novel mitigation and restoration approaches (Rinkevich, 2014; van Oppen et al., 2015, 2017; Piaggio et al., 2016). Exceptional genetic variability naturally exists within the genus Symbiodinium, suggesting that seeding vulnerable corals with more climate-change tolerant Symbiodinium variants could provide a means to reduce bleaching susceptibility of corals (van Oppen et al., 2015). Although, uptake of non-native Symbiodinium variants by corals may not be widely achievable since many coral species only associate with specific Symbiodinium types (LaJeunesse et al., 2004). Furthermore, shifts from innately less stress tolerant Symbiodinium types to more stress tolerant Symbiodinium types (e.g., from type C2 to D) can have negative impacts on a number of coral fitness traits including growth and fecundity (Little et al., 2004; Jones and Berkelmans, 2011).
Environmental bioengineering is an alternative strategy to safeguard against climate change (Solé, 2015; Piaggio et al., 2016). Microalgae, such as Symbiodinium, are clear and promising candidates for genetic engineering with the aim of regaining and preserving ecosystem-climate homeostasis (Solé, 2015) because they can significantly influence the health of entire ecosystems (Berkelmans and van Oppen, 2006; Kirk and Weis, 2016; Murray et al., 2016). Genetic engineering to increase stress tolerance of the Symbiodinium variants that are naturally harbored by at-risk corals holds potential to reduce bleaching susceptibility without negatively impacting the fitness of the coral host since existing Symbiodinium-coral partnerships would be preserved. Fe-sod, Mn-sod Prxd, and Hsp70 genes from Symbiodinium (Levin et al., 2016; Gierz et al., 2017; Goyen et al., 2017) are standout candidates whose engineered up-regulation may enhance thermal and bleaching tolerance by reducing heat-induced oxidative damage, but thorough evaluation of how this artificial up-regulation contributes to long term fitness and the Symbiodinium-coral symbiosis would be mandatory.
Application of genetic engineering to support environmental management practices has been gaining momentum. Notably, sterile male mosquitoes have been engineered to control mosquito-borne diseases (Gabrieli et al., 2014). Field releases of the sterile males significantly reduced wild mosquito populations, supporting their value to disease control (Harris et al., 2012). Similarly, fungus-resistance has been engineered in American chestnut trees in order to restore the natural population that was nearly eradicated from the spread of a foreign fungus. Introduction of these transgenic trees into the wild may receive federal approval in just the next few years, which would make them the first threatened plant species to be restored through genetic engineering (Jacobs et al., 2013; Powell, 2014).
Considering the great promise shown by genetic engineering-based approaches to promote environmental health (Jacobs et al., 2013; Powell, 2014) and human health (Paine et al., 2005; Harris et al., 2012; Gabrieli et al., 2014), as well as to sustain food security (Schroeder et al., 2013), it is logical for genetic engineering to be proposed as an important component of the growing repertoire of forward-looking coral reef management approaches (van Oppen et al., 2015; Piaggio et al., 2016). Due to the urgent need to protect coral reefs from climate change, the Symbiodinium research community must commit to an all-hands-on-deck attitude to achieve and extensively test genetic enhancement of Symbiodinium and other novel reef restoration strategies in the laboratory setting. In parallel, comprehensive cost-benefit-risk evaluation of the potential ecological and socioeconomic impacts from implementation of such strategies in the natural environment must be exhaustive before field-based trials are initiated. Additionally, transparent dialogs with policy makers, coral reef managers, and the general public need to be initiated now to begin the process of education and public acceptance of genetic engineering approaches for coral reef mitigation and restoration.
As we have discussed here, recent NGS breakthroughs have revealed natural genetic elements of Symbiodinium and their viruses (Figure 1). Based on these discoveries, we have developed a tailored genetic engineering framework for Symbiodinium based on empirical data that may also be applicable to other dinoflagellate genera. In doing so, we have opened a new prospective avenue to decode Symbiodinium functional genomics that may ultimately allow for engineering increased stress tolerance of Symbiodinium to reduce coral bleaching.
RL conceived the manuscript concept, analyzed NGS data, and wrote the manuscript. CRV analyzed NGS data and critically edited the theory and writing of the manuscript. SA analyzed NGS data. PS, DS, and MvO critically edited the theory and writing of the manuscript.
Funding from the University of New South Wales and King Abdullah University of Science and Technology (KAUST) supported the analyses presented here.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AWL declared a shared affiliation, though no other collaboration, with one of the authors DS to the handling Editor, who ensured that the process nevertheless met the standards of a fair and objective review.
We thank the Aranda and Voolstra lab for providing the type A1 (Symbiodinium microadriaticum) genome prior to its publication.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01220/full#supplementary-material
Aguirre, A. J., Meyers, R. M., Weir, B. A., Vazquez, F., Zhang, C.-Z., Ben-David, U., et al. (2016). Genomic copy number dictates a gene-independent cell response to CRISPR-Cas9 targeting. Cancer Discov. 6, 914–929. doi: 10.1158/2159-8290.CD-16-0154
Alavi, M., Miller, T., Erlandson, K., Schneider, R., and Belas, R. (2001). Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3, 380–396. doi: 10.1046/j.1462-2920.2001.00207.x
Allera-Moreau, C., Delluc-Clavières, A., Castano, C., Van den Berghe, L., Golzio, M., Moreau, M., et al. (2007). Long term expression of bicistronic vector driven by the FGF-1 IRES in mouse muscle. BMC Biotechnol. 7:74. doi: 10.1186/1472-6750-7-74
Aranda, M., Li, Y., Liew, Y., Baumgarten, S., Simakov, O., Wilson, M., et al. (2016). Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6:39734. doi: 10.1038/srep39734
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Bachvaroff, T. R., and Place, A. R. (2008). From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS ONE 3:e2929. doi: 10.1371/journal.pone.0002929
Baek, K., Kim, D. H., Jeong, J., Sim, S. J., Melis, A., Kim, J.-S., et al. (2016). DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 6:30620. doi: 10.1038/srep30620
Barshis, D. J., Ladner, J. T., Oliver, T. A., and Palumbi, S. R. (2014). Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 31, 1343–1352. doi: 10.1093/molbev/msu107
Baumgarten, S., Bayer, T., Aranda, M., Liew, Y. J., Carr, A., Micklem, G., et al. (2013). Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics 14:704. doi: 10.1186/1471-2164-14-704
Bayer, T., Aranda, M., Sunagawa, S., Yum, L. K., DeSalvo, M. K., Lindquist, E., et al. (2012). Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 7:e35269. doi: 10.1371/journal.pone.0035269
Benfey, P. N., and Chua, N.-H. (1990). The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250, 959–966. doi: 10.1126/science.250.4983.959
Berkelmans, R., and van Oppen, M. J. (2006). The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’for coral reefs in an era of climate change. Proc. R. Soc. Lond. B Biol. Sci. 273, 2305–2312. doi: 10.1098/rspb.2006.3567
Boulotte, N. M., Dalton, S. J., Carroll, A. G., Harrison, P. L., Putnam, H. M., Peplow, L. M., et al. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 10, 2693–2701. doi: 10.1038/ismej.2016.54
Breyer, D., Kopertekh, L., and Reheul, D. (2014). Alternatives to antibiotic resistance marker genes for in vitro selection of genetically modified plants–scientific developments, current use, operational access and biosafety considerations. Crit. Rev. Plant Sci. 33, 286–330. doi: 10.1080/07352689.2013.870422
Byler, K. A., Carmi-Veal, M., Fine, M., and Goulet, T. L. (2013). Multiple symbiont acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata. PLoS ONE 8:e59596. doi: 10.1371/journal.pone.0059596
Caboche, M. (1990). Liposome-mediated transfer of nucleic acids in plant protoplasts. Physiol. Plant. 79, 173–176. doi: 10.1111/j.1399-3054.1990.tb05882.x
Cesar, H., Burke, L., and Pet-Soede, L. (2003). The Economics of Worldwide Coral Reef Degradation. Arnhem: Cesar Environmental Economics Consulting (CEEC).
Chi, J., Parrow, M. W., and Dunthorn, M. (2014). Cryptic sex in Symbiodinium (Alveolata, Dinoflagellata) is supported by an inventory of meiotic genes. J. Eukaryot. Microbiol. 61, 322–327. doi: 10.1111/jeu.12110
Chu, V. T., Weber, T., Wefers, B., Wurst, W., Sander, S., Rajewsky, K., et al. (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548. doi: 10.1038/nbt.3198
Clarke, B. D., Cummins, D. M., McColl, K. A., Ward, A. C., and Doran, T. J. (2013). Characterization of zebrafish polymerase III promoters for the expression of short-hairpin RNA interference molecules. Zebrafish 10, 472–479. doi: 10.1089/zeb.2012.0782
Coffroth, M. A., and Santos, S. R. (2005). Genetic diversity of symbiotic dinoflagellates in the genus Symbiodinium. Protist 156, 19–34. doi: 10.1016/j.protis.2005.02.004
Colanduoni, J. A., and Villafranca, J. J. (1986). Inhibition of Escherichia coli glutamine synthetase by phosphinothricin. Bioorg. Chem. 14, 163–169. doi: 10.1016/0045-2068(86)90026-X
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Correa, A. M., Ainsworth, T. D., Rosales, S. M., Thurber, A. R., Butler, C. R., and Thurber, R. L. V. (2016). Viral outbreak in corals associated with an in situ bleaching event: atypical herpes-like viruses and a new megavirus infecting Symbiodinium. Front. Microbiol. 7:127. doi: 10.3389/fmicb.2016.00127
Correa, A. M., Welsh, R. M., and Thurber, R. L. V. (2013). Unique nucleocytoplasmic dsDNA and + ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 7, 13–27. doi: 10.1038/ismej.2012.75
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Davey, M. R., Anthony, P., Power, J. B., and Lowe, K. C. (2005). Plant protoplast technology: current status. Acta Physiol. Plant. 27, 117–130. doi: 10.1007/s11738-005-0044-0
DeSalvo, M. K., Sunagawa, S., Fisher, P. L., Voolstra, C. R., Iglesias-Prieto, R., and Medina, M. (2010). Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol. Ecol 19, 1174–1186. doi: 10.1111/j.1365-294X.2010.04534.x
Edmunds, P. J., Pochon, X., Levitan, D. R., Yost, D. M., Belcaid, M., Putnam, H. M., et al. (2014). Long-term changes in Symbiodinium communities in Orbicella annularis in St. John, US Virgin Islands. Mar. Ecol. Prog. Ser. 506, 129–144. doi: 10.3354/meps10808
Figueroa, R. I., Dapena, C., Bravo, I., and Cuadrado, A. (2015). The hidden sexuality of alexandrium minutum: an example of overlooked sex in Dinoflagellates. PLoS ONE 10:e0142667. doi: 10.1371/journal.pone.0142667
Gabrieli, P., Smidler, A., and Catteruccia, F. (2014). Engineering the control of mosquito-borne infectious diseases. Genome Biol. 15, 535. doi: 10.1186/s13059-014-0535-7
Gierz, S. L., Forêt, S., and Leggat, W. (2017). Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front. Plant Sci. 8:271. doi: 10.3389/fpls.2017.00271
Gonzalez, A., Jimenez, A., Vazquez, D., Davies, J., and Schindler, D. (1978). Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim. Biophys. Acta 521, 459–469. doi: 10.1016/0005-2787(78)90287-3
Goomer, R., and Kunkel, G. (1992). The transcriptional start site for a human U6 small nuclear RNA gene is dictated by a compound promoter element consisting of the PSE and the TATA box. Nucleic Acids Res. 20, 4903–4912. doi: 10.1093/nar/20.18.4903
Goyen, S., Pernice, M., Szabó, M., Warner, M. E., Ralph, P. J., and Suggett, D. J. (2017). A molecular physiology basis for functional diversity of hydrogen peroxide production amongst Symbiodinium spp.(Dinophyceae). Mar. Biol. 164, 46. doi: 10.1007/s00227-017-3073-5
Gurtu, V., Yan, G., and Zhang, G. (1996). IRES bicistronic expression vectors for efficient creation of stable mammalian cell lines. Biochem. Biophys. Res. Commun. 229, 295–298. doi: 10.1006/bbrc.1996.1795
Harris, A. F., McKemey, A. R., Nimmo, D., Curtis, Z., Black, I., Morgan, S. A., et al. (2012). Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes. Nat. Biotechnol. 30, 828–830. doi: 10.1038/nbt.2350
Hennige, S., Suggett, D. J., Warner, M. E., McDougall, K., and Smith, D. J. (2009). Photobiology of Symbiodinium revisited: bio-physical and bio-optical signatures. Coral Reefs 28, 179–195. doi: 10.1007/s00338-008-0444-x
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Holloman, W. K. (2011). Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 18, 748–754. doi: 10.1038/nsmb.2096
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
Jacobs, D. F., Dalgleish, H. J., and Nelson, C. D. (2013). A conceptual framework for restoration of threatened plants: the effective model of American chestnut (Castanea dentata) reintroduction. New Phytol. 197, 378–393. doi: 10.1111/nph.12020
Jiang, J., Zhang, H., Kang, Y., Bina, D., Lo, C. S., and Blankenship, R. E. (2012). Characterization of the peridinin–chlorophyll a-protein complex in the dinoflagellate Symbiodinium. Biochim. Biophys. Acta 1817, 983–989. doi: 10.1016/j.bbabio.2012.03.027
Jones, A. M., and Berkelmans, R. (2011). Tradeoffs to thermal acclimation: energetics and reproduction of a reef coral with heat tolerant Symbiodinium type-D. J. Mar. Biol. 2011, 12. doi: 10.1371/journal.pone.0010437
Karas, B. J., Diner, R. E., Lefebvre, S. C., McQuaid, J., Phillips, A. P., Noddings, C. M., et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6, 1–10. doi: 10.1038/ncomms7925
Kirk, N. L., and Weis, V. M. (2016). “Animal–Symbiodinium symbioses: foundations of coral reef ecosystems,” in The Mechanistic Benefits of Microbial Symbionts, ed. C. J. Hurst (Cham: Springer), 269–294. doi: 10.1007/978-3-319-28068-4_10
Knowlton, N., Brainard, R. E., Fisher, R., Moews, M., Plaisance, L., and Caley, M. J. (2010). “Coral reef biodiversity,” in Life in the World’s Oceans: Diversity Distribution and Abundance, ed. A. D. Mcintyre (Hoboken, NJ: John Wiley & Sons), 65–74. doi: 10.1002/9781444325508.ch4
Krueger, T., Fisher, P. L., Becker, S., Pontasch, S., Dove, S., Hoegh-Guldberg, O., et al. (2015). Transcriptomic characterization of the enzymatic antioxidants FeSOD, MnSOD, APX and KatG in the dinoflagellate genus Symbiodinium. BMC Evol. Biol. 15:48. doi: 10.1186/s12862-015-0326-0
Ladner, J. T., Barshis, D. J., and Palumbi, S. R. (2012). Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol. Biol. 12:217. doi: 10.1186/1471-2148-12-217
LaJeunesse, T. C., Thornhill, D. J., Cox, E. F., Stanton, F. G., Fitt, W. K., and Schmidt, G. W. (2004). High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs 23, 596–603. doi: 10.1007/s00338-004-0428-4
Lee, S. E., Pâques, F., Sylvan, J., and Haber, J. E. (1999). Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9, 767–770. doi: 10.1016/S0960-9822(99)80339-X
Leggat, W., Yellowlees, D., and Medina, M. (2011). Recent progress in Symbiodinium transcriptomics. J. Exp. Mar. Biol. Ecol. 408, 120–125. doi: 10.1016/j.jembe.2011.07.032
Levin, R. A., Beltran, V. H., Hill, R., Kjelleberg, S., McDougald, D., Steinberg, P. D., et al. (2016). Sex, scavengers, and chaperones: transcriptome secrets of divergent symbiodinium thermal tolerances. Mol. Biol. Evol. 33, 2201–2215. doi: 10.1093/molbev/msw119
Levin, R. A., Felsen, C. N., Yang, J., Lin, J. Y., Whitney, M. A., Nguyen, Q. T., et al. (2014). An optimized triple modality reporter for quantitative in vivo tumor imaging and therapy evaluation. PLoS ONE 9:e97415. doi: 10.1371/journal.pone.0097415
Levin, R. A., Suggett, D. J., Nitschke, M. R., van Oppen, M. J., and Steinberg, P. D. (2017b). Expanding the Symbiodinium (Dinophyceae, Suessiales) toolkit through protoplast technology. J. Eukaryot. Microbiol. doi: 10.1111/jeu.12393 [Epub ahead of print].
Levin, R. A., Voolstra, C. R., Weynberg, K. D., and van Oppen, M. J. H. (2017a). Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 11, 808–812. doi: 10.1038/ismej.2016.154
Lin, S., Cheng, S., Song, B., Zhong, X., Lin, X., Li, W., et al. (2015). The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350, 691–694. doi: 10.1126/science.aad0408
Little, A. F., Van Oppen, M. J., and Willis, B. L. (2004). Flexibility in algal endosymbioses shapes growth in reef corals. Science 304, 1492–1494. doi: 10.1126/science.1095733
Markell, D., Trench, R., and Iglesias-Prieto, R. (1992). Macromolecules associated with the cell walls of symbiotic dinoflagellates. Symbiosis 12, 19–31.
Martínez-Salas, E. (1999). Internal ribosome entry site biology and its use in expression vectors. Curr. Opin. Biotechnol. 10, 458–464. doi: 10.1016/S0958-1669(99)00010-5
Mathur, J., and Koncz, C. (1998). PEG-mediated protoplast transformation with naked DNA. Methods Mol. Biol. 82, 267–276. doi: 10.1385/0-89603-391-0:267
Miller, T. R., and Belas, R. (2006). Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8, 1648–1659. doi: 10.1111/j.1462-2920.2006.01071.x
Moldowan, J. M., and Talyzina, N. M. (1998). Biogeochemical evidence for dinoflagellate ancestors in the Early Cambrian. Science 281, 1168–1170. doi: 10.1126/science.281.5380.1168
Mora, C., Graham, N. A., and Nyström, M. (2016). Ecological limitations to the resilience of coral reefs. Coral Reefs 35, 1271–1280. doi: 10.1007/s00338-016-1479-z
Murray, S. A., Suggett, D. J., Doblin, M. A., Kohli, G. S., Seymour, J. R., Fabris, M., et al. (2016). Unravelling the functional genetics of dinoflagellates: a review of approaches and opportunities. Perspect. Phycol. 3, 37–52. doi: 10.1127/pip/2016/0039
Muscatine, L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals. Ecosyst. World 25, 75–87.
Muscatine, L., and Porter, J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Nehlsen, K., Broll, S., and Bode, J. (2006). Replicating minicircles: generation of nonviral episomes for the efficient modification of dividing cells. Gene Ther. Mol. Biol. 10, 233–244.
Nymark, M., Sharma, A. K., Sparstad, T., Bones, A. M., and Winge, P. (2016). A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6:24951. doi: 10.1038/srep24951
Ortiz-Matamoros, M. F., Islas-Flores, T., Voigt, B., Menzel, D., Baluška, F., and Villanueva, M. A. (2015a). Heterologous DNA uptake in cultured Symbiodinium spp. Aided by Agrobacterium tumefaciens. PLoS ONE 10:e0132693. doi: 10.1371/journal.pone.0132693
Ortiz-Matamoros, M. F., Villanueva, M. A., and Islas-Flores, T. (2015b). Transient transformation of cultured photosynthetic dinoflagellates (Symbiodinium spp.) with plant-targeted vectors Transformación de dinoflagelados fotosintéticos del género Symbiodinium en cultivo con vectores diseñados para plantas. Ciencias Marinas 41, 21–32. doi: 10.7773/cm.v41i1.2449
Paine, J. A., Shipton, C. A., Chaggar, S., Howells, R. M., Kennedy, M. J., Vernon, G., et al. (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487. doi: 10.1038/nbt1082
Pandolfi, J. M., Connolly, S. R., Marshall, D. J., and Cohen, A. L. (2011). Projecting coral reef futures under global warming and ocean acidification. Science 333, 418–422. doi: 10.1126/science.1204794
Parkinson, J. E., Baumgarten, S., Michell, C. T., Baums, I. B., LaJeunesse, T. C., and Voolstra, C. R. (2016). Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol. Evol. 8, 665–680. doi: 10.1093/gbe/evw019
Piaggio, A. J., Segelbacher, G., Seddon, P. J., Alphey, L., Bennett, E. L., Carlson, R. H., et al. (2016). Is it time for synthetic biodiversity conservation? Trends Ecol. Evol. 32, 97–107. doi: 10.1016/j.tree.2016.10.016
Pline, W. A., Lacy, G. H., Stromberg, V., and Hatzios, K. K. (2001). Antibacterial activity of the herbicide glufosinate on Pseudomonas syringae pathovar glycinea. Pestic. Biochem. Physiol. 71, 48–55. doi: 10.1006/pest.2001.2556
Pochon, X., and Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol. Phylogenet. Evol. 56, 492–497. doi: 10.1016/j.ympev.2010.03.040
Pochon, X., Montoya-Burgos, J. I., Stadelmann, B., and Pawlowski, J. (2006). Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol. Phylogenet. Evol. 38, 20–30. doi: 10.1016/j.ympev.2005.04.028
Powell, W. (2014). The american chestnut’s genetic rebirth. Sci. Am. 310, 68–73. doi: 10.1038/scientificamerican0314-68
Quigley, K. M., Davies, S. W., Kenkel, C. D., Willis, B. L., Matz, M. V., and Bay, L. K. (2014). Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS ONE 9:e94297. doi: 10.1371/journal.pone.0094297
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi: 10.1038/nprot.2013.143
Reaka-Kudla, M. L., Wilson, D. E., and Wilson, E. O. (1996). Biodiversity II: Understanding and Protecting our Biological Resources. Washington, DC: Joseph Henry Press.
Rinkevich, B. (2014). Rebuilding coral reefs: does active reef restoration lead to sustainable reefs? Curr. Opin. Environ. Sustain. 7, 28–36. doi: 10.1016/j.cosust.2013.11.018
Ritchie, K. B. (2012). “Bacterial symbionts of corals and Symbiodinium,” in Beneficial Microorganisms in Multicellular Life Forms, eds E. Rosenberg and U. Gophna (Berlin: Springer), 139–150. doi: 10.1007/978-3-642-21680-0_9
Rosic, N., Ling, E. Y. S., Chan, C.-K. K., Lee, H. C., Kaniewska, P., Edwards, D., et al. (2015). Unfolding the secrets of coral–algal symbiosis. ISME J. 9, 844–856. doi: 10.1038/ismej.2014.182
Santos, S. R., and Coffroth, M. A. (2003). Molecular genetic evidence that dinoflagellates belonging to the genus Symbiodinium Freudenthal are haploid. Biol. Bull. 204, 10–20. doi: 10.2307/1543491
Schroeder, J. I., Delhaize, E., Frommer, W. B., Guerinot, M. L., Harrison, M. J., Herrera-Estrella, L., et al. (2013). Using membrane transporters to improve crops for sustainable food production. Nature 497, 60–66. doi: 10.1038/nature11909
Shaner, N. C., Steinbach, P. A., and Tsien, R. Y. (2005). A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909. doi: 10.1038/nmeth819
Shoguchi, E., Shinzato, C., Hisata, K., Satoh, N., and Mungpakdee, S. (2015). The large mitochondrial genome of Symbiodinium minutum reveals conserved noncoding sequences between Dinoflagellates and Apicomplexans. Genome Biol. Evol. 7, 2237–2244. doi: 10.1093/gbe/evv137
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. doi: 10.1016/j.cub.2013.05.062
Solé, R. (2015). Bioengineering the biosphere? Ecol. Complex. 22, 40–49. doi: 10.1016/j.ecocom.2015.01.005
Stoykova, P., and Stoeva-Popova, P. (2011). PMI (manA) as a nonantibiotic selectable marker gene in plant biotechnology. Plant Cell Tissue Organ Cult. (PCTOC) 105, 141–148. doi: 10.1007/s11240-010-9858-6
Suggett, D. J., Warner, M. E., Smith, D. J., Davey, P., Hennige, S., and Baker, N. R. (2008). Photosynthesis and production of hydrogen peroxide by symbiodinium (Pyrrophyta) phylotypes with different thermal tolerances. J Phycol. 44, 948–956. doi: 10.1111/j.1529-8817.2008.00537.x
ten Lohuis, M. R., and Miller, D. J. (1998). Genetic transformation of dinoflagellates (Amphidinium and Symbiodinium): expression of GUS in microalgae using heterologous promoter constructs. Plant J. 13, 427–435. doi: 10.1046/j.1365-313X.1998.00040.x
Thacker, D., and Keeney, S. (2016). “Homologous recombination during meiosis,” in DNA Replication, Recombination, and Repair, eds F. Hanaoka and K. Sugasawa (Tokyo: Springer), 131–151. doi: 10.1007/978-4-431-55873-6_6
Thomas, L., Kendrick, G., Kennington, W., Richards, Z., and Stat, M. (2014). Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Mol. Ecol. 23, 3113–3126. doi: 10.1111/mec.12801
Toller, W. W., Rowan, R., and Knowlton, N. (2001). Repopulation of zooxanthellae in the Caribbean corals Montastraea annularis and M. faveolata following experimental and disease-associated bleaching. Biol. Bull. 201, 360–373. doi: 10.2307/1543614
Tonk, L., Bongaerts, P., Sampayo, E. M., and Hoegh-Guldberg, O. (2013). SymbioGBR: a web-based database of Symbiodinium associated with cnidarian hosts on the Great Barrier Reef. BMC Ecol. 13:7. doi: 10.1186/1472-6785-13-7
Trench, R. K., and Blank, R. J. (1987). Symbiodinium Microadriaticum Freudenthal, S. Goreauii Sp. Nov., S. Kawagutii Sp. Nov. and S. Pilosum Sp. Nov.: Gymnodinioid Dinoflagellate symbionts of marine invertebrates 1. J. Phycol. 23, 469–481. doi: 10.1111/j.1529-8817.1987.tb02534.x
van Oppen, M. J., Gates, R. D., Blackall, L. L., Cantin, N., Chakravarti, L. J., Chan, W. Y., et al. (2017). Shifting paradigms in restoration of the world’s coral reefs. Glob. Chang Biol. doi: 10.1111/gcb.13647 [Epub ahead of print].
van Oppen, M. J. H., Oliver, J. K., Putnam, H. M., and Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313. doi: 10.1073/pnas.1422301112
van Oppen, M. J. H., Palstra, F. P., Piquet, A. M. T., and Miller, D. J. (2001). Patterns of coral–dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host–symbiont selectivity. Proc. R. Soc. Lond. B. Biol. Sci. 268, 1759–1767. doi: 10.1098/rspb.2001.1733
Vicens, Q., and Westhof, E. (2003). Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J. Mol. Biol. 326, 1175–1188. doi: 10.1016/S0022-2836(02)01435-3
Wakefield, T. S., Farmer, M. A., and Kempf, S. C. (2000). Revised description of the fine structure of in situ” zooxanthellae” genus Symbiodinium. Biol. Bull. 199, 76–84. doi: 10.2307/1542709
Warner, M. E., Fitt, W. K., and Schmidt, G. W. (1999). Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc. Natl. Acad. Sci. U.S.A. 96, 8007–8012. doi: 10.1073/pnas.96.14.8007
Watrin, E., and Legagneux, V. (2003). Introduction to chromosome dynamics in mitosis. Biol. Cell 95, 507–513. doi: 10.1016/j.biolcel.2003.08.003
Weis, V. M. (2008). Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066. doi: 10.1242/jeb.009597
Weynberg, K. D., Neave, M., Clode, P. L., Voolstra, C. R., Brownlee, C., Laffy, P., et al. (2017). Prevalent and persistent viral infection in cultures of the coral algal endosymbiont Symbiodinium. Coral Reefs 2017, 1–12. doi: 10.1007/s00338-017-1568-7
Weynberg, K. D., Wood-Charlson, E. M., Suttle, C. A., and van Oppen, M. J. (2014). Generating viral metagenomes from the coral holobiont. Front. Microbiol. 5:206. doi: 10.3389/fmicb.2014.00206
Wilkinson, S. P., Fisher, P. L., van Oppen, M. J. H., and Davy, S. K. (2015). Intra-genomic variation in symbiotic dinoflagellates: recent divergence or recombination between lineages? BMC Evol. Biol. 15:46. doi: 10.1186/s12862-015-0325-1
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Xiang, T., Nelson, W., Rodriguez, J., Tolleter, D., and Grossman, A. R. (2015). Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 82, 67–80. doi: 10.1111/tpj.12789
Yuyama, I., Harii, S., and Hidaka, M. (2012). Algal symbiont type affects gene expression in juveniles of the coral Acropora tenuis exposed to thermal stress. Mar. Environ. Res. 76, 41–47. doi: 10.1016/j.marenvres.2011.09.004
Zhang, H., Zhuang, Y., Gill, J., and Lin, S. (2013). Proof that dinoflagellate spliced leader (DinoSL) is a useful hook for fishing dinoflagellate transcripts from mixed microbial samples: Symbiodinium kawagutii as a case study. Protist 164, 510–527. doi: 10.1016/j.protis.2013.04.002
Keywords: synthetic biology, genetic engineering, dinoflagellate, Symbiodinium, zooxanthellae, coral bleaching
Citation: Levin RA, Voolstra CR, Agrawal S, Steinberg PD, Suggett DJ and van Oppen MJH (2017) Engineering Strategies to Decode and Enhance the Genomes of Coral Symbionts. Front. Microbiol. 8:1220. doi: 10.3389/fmicb.2017.01220
Received: 16 March 2017; Accepted: 16 June 2017;
Published: 30 June 2017.
Edited by:
Diana Elizabeth Marco, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Michael Sweet, University of Derby, United KingdomCopyright © 2017 Levin, Voolstra, Agrawal, Steinberg, Suggett and van Oppen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachel A. Levin, cmFjaHlsZXZpbkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.