
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 June 2017
Sec. Aquatic Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01133
 Zhaojun Chen1†
Zhaojun Chen1† Daojun Yu2†
Daojun Yu2† Songzhe He3
Songzhe He3 Hui Ye4
Hui Ye4 Lei Zhang5
Lei Zhang5 Yanping Wen6
Yanping Wen6 Wenhui Zhang1
Wenhui Zhang1 Liping Shu1
Liping Shu1 Shuchang Chen1*
Shuchang Chen1*This study investigated the distribution of antibiotic resistant Escherichia coli (E. coli) and examined the possible relationship between water quality parameters and antibiotic resistance from two different drinking water sources (the Qiantang River and the Dongtiao Stream) in Hangzhou city of China. E. coli isolates were tested for their susceptibility to 18 antibiotics. Most of the isolates were resistant to tetracycline (TE), followed by ampicillin (AM), piperacillin (PIP), trimethoprim/sulfamethoxazole (SXT), and chloramphenicol (C). The antibiotic resistance rate of E. coli isolates from two water sources was similar; For E. coli isolates from the Qiantang River, their antibiotic resistance rates decreased from up- to downstream. Seasonally, the dry and wet season had little impact on antibiotic resistance. Spearman's rank correlation revealed significant correlation between resistance to TE and phenicols or ciprofloxacin (CIP), as well as quinolones (ciprofloxacin and levofloxacin) and cephalosporins or gentamicin (GM). Pearson's chi-square tests found certain water parameters such as nutrient concentration were strongly associated with resistance to some of the antibiotics. In addition, tet genes were detected from all 82 TE-resistant E. coli isolates, and most of the isolates (81.87%) contained multiple tet genes, which displayed 14 different combinations. Collectively, this study provided baseline data on antibiotic resistance of drinking water sources in Hangzhou city, which indicates drinking water sources could be the reservoir of antibiotic resistance, potentially presenting a public health risk.
There is a growing concern regarding the occurrence of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in aquatic environments (Kummerer, 2009; Diwan et al., 2010). As the main receptacle for pollution from industry, agriculture, or domestic life, aquatic environment provides an ideal setting for the acquisition and dissemination of antibiotic resistance (Pereira et al., 2013; Marti et al., 2014). Natural bodies of water, commonly used for irrigation, aquaculture, or recreation activities, are closely associated with human life. Direct or indirect contact with water (for drinking, or recreational use) contaminated by ARB could harm and infecte the human population with antibiotic resistant pathogens, and/or ARGs carried by bacteria may transfer to microorganisms in humans as a consequence of horizontal gene transfer (Chen et al., 2011; Heuer et al., 2011; Ribeiro et al., 2012; Jiang et al., 2013). Such events would undermine our ability to prevent and control disease, and thus expose a great threat to public health.
Most of the studies focusing on antibiotic resistance are done in aquatic environments with serious pollution or waters that have been strongly influenced by anthropogenic activities, such as agricultural watershed and rivers near wastewater treatment plant outflows (Maal-Bared et al., 2013; Middleton and Salierno, 2013; Zhang et al., 2014). However, the presence and distribution of antibiotic resistance in a special aquatic environment, such as a drinking water source, is always neglected due to strict legal protection and less anthropogenic activities. According to literatures, the context of antibiotic resistance in drinking water sources is of serious grave concern (Jiang et al., 2013; Flores Ribeiro et al., 2014; Guo et al., 2014; Machado and Bordalo, 2014; Mohanta and Goel, 2014). For example, high antibiotic resistance rates of 72 and 59% were found in a water source in Guinea-Bissau (West Africa) during both the dry and wet seasons, (Machado and Bordalo, 2014). High antibiotic resistance levels were also observed in the source waters of the Huangpu River of China (Jiang et al., 2013). Considering the source water is directly related with human activity and health, understanding the prevalence of antibiotic resistance in human drinking water sources is of great importance.
Hangzhou, the capital of Zhejiang Province in China, is one of the most economically developed cities in China. The Qiantang River and the Dongtiao Stream both serve this city of 1.96 million residents as the drinking water sources. Unfortunately, the Qiantang River and the Dongtiao Stream both face serious pollution from the upstream cities' discharge. For instance, the Fuchun River is upstream to the Qiantang River, with many potential pollution sources existing in its downstream, such as wastewater treatment plants (WWTPs), large and small industrial plants, sand excavation operation sites, and livestock farmland. With this, various pollutants including antibiotics, pesticides, and insecticides likely flow into the Qiantang River promoting the emergence and spread of antibiotic resistance in drinking water sources (Zhao and Dang, 2011). Nevertheless, limited information about antibiotic resistance in these water sources has been made available.
Escherichia coli (E. coli) is an opportunistic pathogen that can survive well in aquatic environments, and E. coli is highly adept at horizontal gene transfer, which is deemed as the vector for antibiotic resistance dissemination (Maal-Bared et al., 2013; Pereira et al., 2013). For the present study, as the situation of drinking water source can be best represented by water collected from the intake sites of water plants, we collected samples from five intake sites of water plants whose water sources are the Qiangtang River and the Dongtiao Stream. We investigated the presence of antibiotic resistant E. coli from samples to 18 antibiotics that are commonly used in clinical and farming practices. We further examined the effects of water quality parameters and seasonal variations on antibiotic resistance. The working hypothesis was that (1) E. coli isolates from different water sources and different seasons may have different antibiotic resistance levels; (2) With good protection and less new pollution, the antibiotic resistance levels of E. coli may be decreased; (3) Some water quality parameters could have an impact on the levels of antibiotic resistance.
The Qiantang River (Q) supplies water to three water plants while the Dongtiao Stream (D) is the source for one additional water plant, respectively. Sampling sites (Q1, Q2, Q3, and D1) were selected at the corresponding water intakes of the four water plants. The Tiesha River (site Q4) is one tributary of the Qiantang River, which serves as a water source with an urban backup. All of the sampling sites (Figure 1) are routinely used for water quality monitoring by the local environmental authorities.
Triplicate samples were collected on April 3, 2014 (dry season) and July 4, 2014 (wet season). These samples were then separated into two parts: (1) 500 ml water samples were collected using sterile glass bottles for the evaluation of microbiological parameters and the isolation of E. coli; (2) 5 L water samples were obtained using plastic barrels for the analysis of physicochemical parameters of the water. All samples were preserved in cold boxes, transported to the laboratory within 4 h and maintained at 4°C until use.
All water quality parameters were measured according to Chinese National Standard Methods. Briefly, water temperature (Thermometer method), pH (Glass electrode method), and dissolved oxygen (Electrochemical probe method) were measured in situ using a digital display thermometer (Ruiming, Changzhou, China), a pH meter (METTLER, Switzerland) and an YSI-58 instrument (Yellow Spring, USA) respectively. Other water quality parameters analyzed in the lab were ammonia nitrogen, total phosphorus, total nitrogen, nitrate-N, biological oxygen demand, chemical oxygen demand, fecal coliforms, and E. coli. Ammonia nitrogen (Nessler's reagent spectrophotometry method) and total phosphorus (Ammonium molybdate spectrophotometric method) were measured using the visible spectrophotometer (UNICO, Shanghai, China). Total nitrogen (Potassium persulfate digestion UV spectrophotometric method nitrate-N) and nitrate-N (Spectrophotometric method with phenol disulfonic acid method) were measured using a double beam UV-visible spectrophotometer (PERSEE, Beijing, China). Biological oxygen demand was measured using the dilution and seeding method. COD was measured using the dichromate method. Fecal coliforms were measured using the manifold zymotechnics method. And E. coli was measured using the membrane filter method.
The number of E. coli was determined in water samples by membrane filtration method (Talukdar et al., 2013). Briefly, a 50 ml aliquot of water sample was filtered through a 0.22 μm membrane filter (Millipore, Carrightwohill, IRL). The membranes were placed on E. coli-Coliforms Chromogenic Mediums (Nissui, Qingdao, China) and incubated at 37°C for 24 h. After incubation, blue colonies represented E. coli. Typical E. coli were picked and purified on the MacConkey Agar (Nissui, Qingdao, China), and then they were confirmed using the automated identification and susceptibility testing system (BD Phoenix NMIC/IDNMIC/ID-4 panel, Sparks, USA) Confirmed isolates were stored at −80°C in tryptic soy broth with 20% (vol/vol) glycerol.
Antibiotic susceptibility testing for confirmed E. coli was conducted using the automated identification and susceptibility testing system (BD Phoenix NMIC/IDNMIC/ID-4 panel, Sparks, USA). Eighteen antibiotics were contained: amikacin (30 μg), gentamicin (10 μg), imipenem (10 μg), meropenem(30 μg), cefazolin (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), aztreonam (30 μg), ampicillin (10 μg), piperacillin (100 μg), amoxicillin/clavulanate (20/10 μg), ampicillin/sulbactam (10/10 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), tetracycline (30 μg). “Sensitive” “intermediate resistant” or “resistant” patterns of E. coli were judged using antibiotic minimum inhibitory concentration(MIC) provided by the guidelines of the Clinical and Laboratory Standards Institute (CLSI). E. coli strain ATCC 25922 was sensitive to all the antibiotics tested and so was used as the quality control strain. All isolates showing “resistant” or “intermediate resistant” patterns were classified as “resistant,” whereas all other isolates were classified as “sensitive”. E. coli isolates resistant to three or more classes of antibiotics were multiple resistance E. coli isolates, which were classified as R3-6 subpopulation. And isolates resistant to one or two classes of antibiotics were classified as R1-2 subpopulation.
The antibiotic resistance index (ARI) is used for analyzing the prevalence of E. coli isolated from drinking water sources. ARI was determined using the following formula: ARI = a/(b × c), where “a” is the total antibiotic resistance score of all E. coli from specified location, “b” is the number of tested antibiotics, and “c” is the number of E. coli isolates from specified location (Mohanta and Goel, 2014; Zhang et al., 2014).
Genomic DNA was extracted from E. coli isolates by the boiling method (Dei-Tutuwa and Rahman, 2014). Briefly, colonies of E. coli were suspended in 100 μl of distilled water and heated at 100°C for 10 min followed by cooling for 10 min. The lysed cell suspension was centrifuged at 12,000 g for 10 min, and the recovered supernatant was frozen at −20°C until use.
PCR assays were used to determine the presence of tetracycline resistance genes in tetracycline-resistant E. coli. We designed the primers using Primer Primiers 5.0 software. Primers with the best specificity and similar melting temperatures (Tm) were selected based on a blast sequence comparison. The primers were synthesized by Lift Technology, and their sequences are listed in Table 1.
Amplification of the DNA was performed in a PCR apparatus with TaKaRa Ex Taq buffer (TaKaRa, Japan). A positive control, a negative control and a blank control were employed in this assay.
PCR products of tetracycline resistance genes obtained from the resistant isolates were cloned, sequenced, and searched against GenBank using the BLAST alignment tool (http://www.ncbi.nlm.nih.gov/blast/).
All statistical analyses were conducted using SPSS15.0. Rank-sum tests were performed to evaluate the difference of water quality parameters in different drinking water sources or seasons. Chi-square tests were used to determine the effect of different water quality parameters or different seasons on the resistance patterns of E. coli. To describe the relationships between antibiotic resistance rates and water quality parameters, Spearman's rank correlations were used for each antibiotic. Person's chi-square tests were used to evaluate the association between different antibiotic resistance phenotypes of E. coli isolates from drinking water sources.
As shown in Table 2, some physicochemical and bacteriological parameters of water samples were measured, including temperature, dissolved oxygen (DO), pH, chemical oxygen demand (COD), Nitrate -N(-N), and fecal coliforms. Except for DO and fecal coliforms at site Q4 in the wet season, all values of water quality parameters tested met Surface Water Environmental Quality Standard for grade III (GB3838-2002), which was the standard for evaluating the water quality of centralized drinking water source supplies in China. It should be noted that, however, the values of total phosphorus (TP) and fecal coliforms at some sampling sites were close to the upper limit (0.2 mg/l and 1*104 CFU/l, respectively), and the values of total nitrogen (TN) were high in all samples which ranged from 1.91 to 3.54 mg/l, indicating that some degree of organic pollution and fecal pollution existed in drinking water sources.
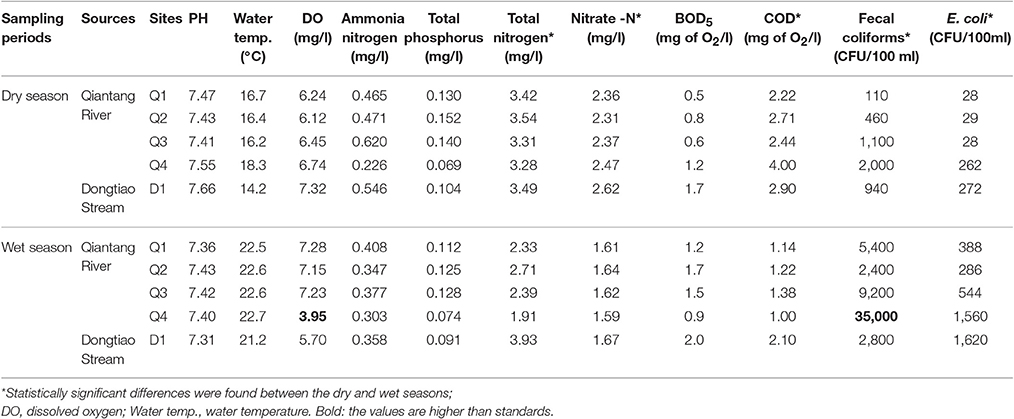
Table 2. Water quality parameters of the Qiantang River and the Dongtiao Stream between the dry and wet seasons.
Rank-sum tests were used to determine the statistical significance of organic pollution and fecal pollution in different drinking water sources and seasons. The results showed significant seasonal differences for -N, TN, COD, fecal coliforms, and E. coli, with p-values of 0.009, 0.009, 0.021, 0.009, and 0.009, respectively, no significant difference was found between the two drinking water sources for organic pollution and fecal pollution. These data suggest that the level of organic pollution was higher in the dry season, while the pollution with fecal materials was more serious in the wet season.
A total of 200 E. coli isolated from drinking water sources were examined. Among them, 99 isolates (49.50%) were resistant to at least one of the 18 antibiotics tested, and the antibiotic resistance index was 0.13. As shown in Figure 2, the most frequently resisted antibiotic was tetracycline (TE) (42.00%), followed by ampicillin (AM) (29.00%), piperacillin (PIP) (27.00%), trimethoprim/sulfamethoxazole (SXT) (25.50%), and chloramphenicol (C) (19.00%). Isolates were more susceptible to aztreonam (ATM) (94.00%), amoxicillin/clavulanate (AMC) (96.00%), ceftazidime (CAZ) (98.00%), and amikacin (AN) (99.00%). No isolates were resistant to imipenem (IPM) or meropenem (MEM). Additionally, 48 isolates (24.00%) exhibited multiple resistances, and six isolates were found to be resistant to all classes of antibiotics. Notably, 75.51% multiple resistant E. coli (37/49) in this study was resistant to tetracycline and β-lactams concurrently. The antibiotic resistance profiles of 200 coli isolates were shown in Table S1.
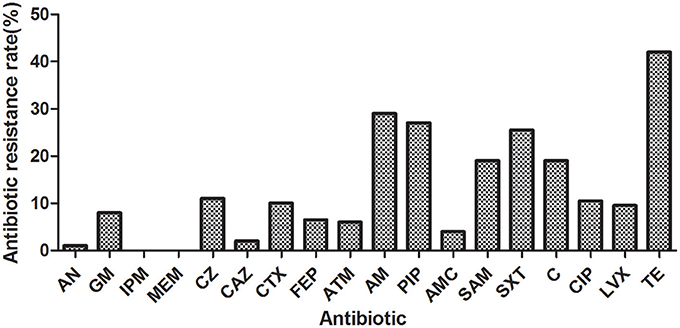
Figure 2. Antibiotic resistance rates of the E. coli isolated from drinking water sources (the Qiantang River and the Dongtiao Stream). AN, amikacin; GM: gentamicin; IPM, imipenem; MEM, meropenem; CZ, cefazolin; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; AMP, ampicillin; PIP, piperacillin; AMC, amoxicillin/clavulanate; SAM, ampicillin/sulbactam; C, chloramphenicol; CIP, ciprofloxacin; LVX, levofloxacin; TE, tetracycline.
The antibiotic resistance level in different seasons and different water sources was presented in Table 3 and Figure 3. In general, E.coli isolated from the Qiantang River and the Dongtiao Stream had similar antibiotic resistant rates. And the dry/wet seasons had minor effects on the resistance rate of E. coli. Even for the individual antibiotics tested, still no significant differences can be observed between the dry/wet seasons or between the Qiantang River and the Dongtiao Stream. However, it should be noted that 83.33% isolates (5/6,) resistant to six classes of antibiotics were found in the dry season.
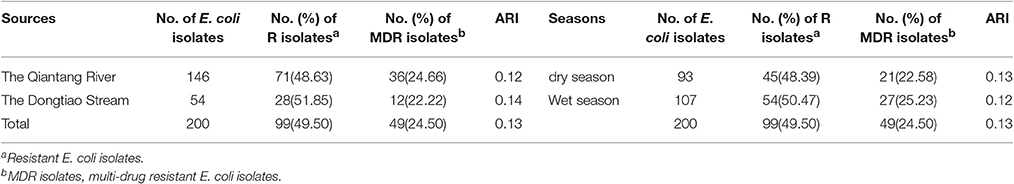
Table 3. Antibiotic resistance of E. coli isolated from the Qiantang River and the Dongtiao Stream over two seasons.
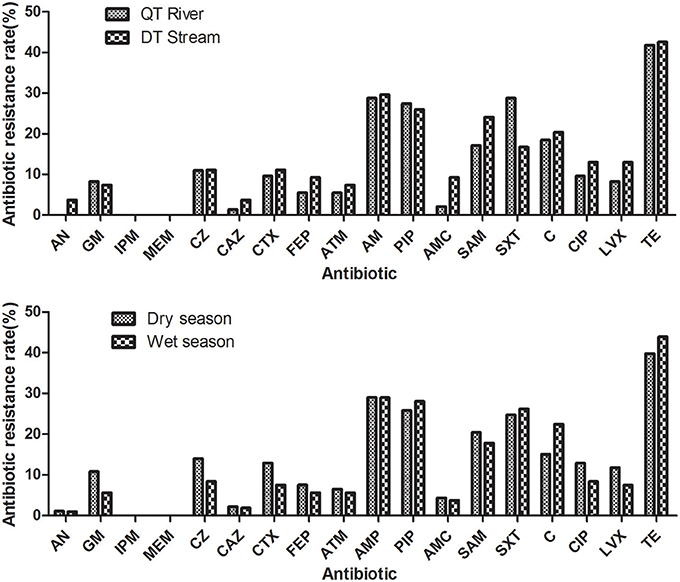
Figure 3. Antibiotic resistance rates of the E. coli isolated from the Qiantang River and the Dongtiao Stream over two seasons. AN, amikacin; GM, gentamicin; IPM, imipenem; MEM, meropenem; CZ, cefazolin; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; AMP, ampicillin; PIP, piperacillin; AMC, amoxicillin/clavulanate; SAM, ampicillin/sulbactam; C, chloramphenicol; CIP, ciprofloxacin; LVX, levofloxacin; TE, tetracycline.
As shown in Table 4, the resistance rate of E. coli noticeably decreased (except R1-2 subpopulation in the wet season) from up- to downstream of the Qiantang River. This finding indicates that antibiotic resistance level of E. coli can be reduced gradually in source water protection area.
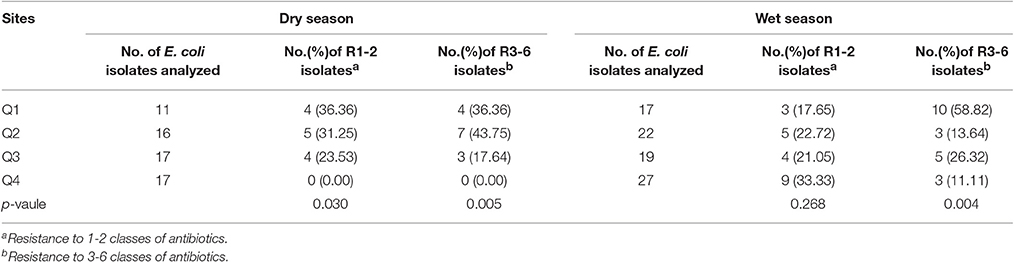
Table 4. The distribution of antibiotic resistance rate for R1-2 and R3-6 subpopulation isolated from the Qiantang River in two seasons.
Spearman's rank correlation analyses were conducted to reveal the correlations between the resistance rates of E. coli and water quality parameters in drinking water sources. As shown in Table 5, the results demonstrated that water quality parameters, including the density of E. coli and fecal coliforms, did not have correlation with antibiotic resistance rates and multi-drug resistance rates. However, ammonia nitrogen correlated significantly with the resistance rates for gentamicin (GM), cefazolin (CZ), ampicillin/sulbactam (SAM), levofloxacin (LVX) (p < 0.05), also a strong but statistically insignificant trend with cefotaxime (CTX), C, and CIP resistance (0.05 < p < 0.1). Total phosphorus was closely correlated with the resistance rates for GM (p < 0.001), CZ, CTX, and SXT (p < 0.05). Total nitrogen was significantly correlated with cefepime (FEP) resistance (p < 0.05), while biological oxygen demand (BOD5) was correlated with the resistance rate for amikacin (AN) (p < 0.05).
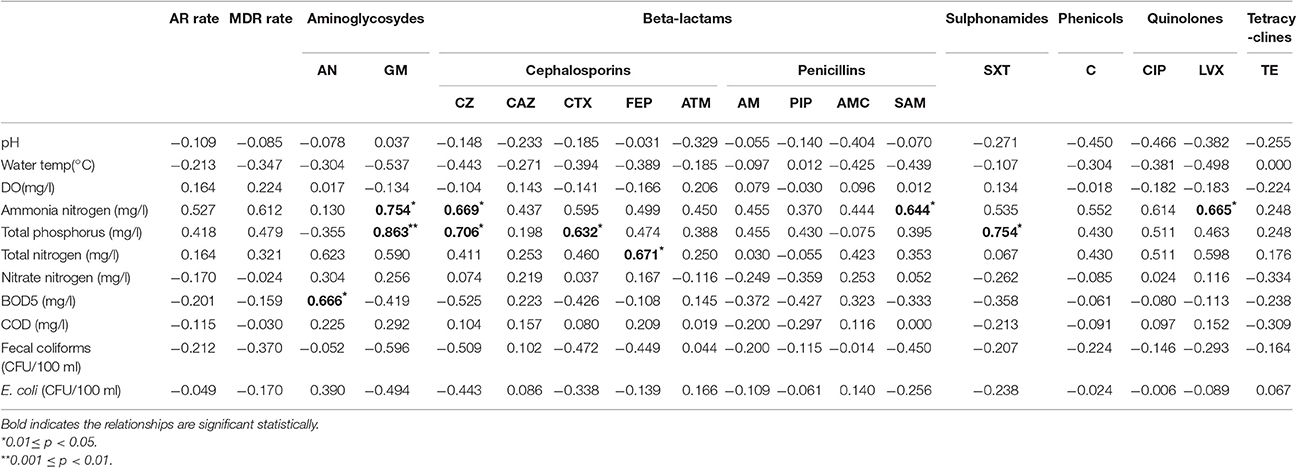
Table 5. Spearman's rank correlation coefficients between antibiotic resistance rates of E. coli and water quality parameters.
The association between antibiotic resistance phenotypes of E. coli isolates from drinking water sources were analyzed by Pearson's chi-square tests (Table 6). Besides the co-resistance to the same class of antibiotics, resistance to quinolones (CIP and LVX) was closely associated with cephalosporins or GM (p < 0.001). In addition, resistance to tetracyclines was associated with CIP resistance (0.001 ≤ p < 0.01), and to a relatively weaker extent with phenicols (0.01 ≤ p < 0.05).
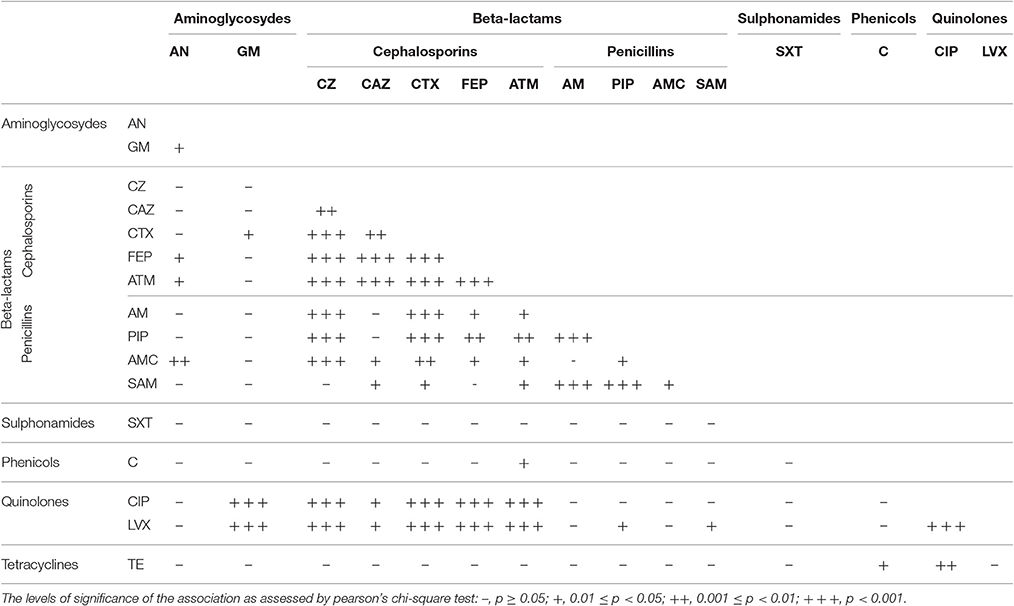
Table 6. Association between antibiotic resistance phenotypes of E. coli isolates from drinking water sources.
Tetracycline was the most prevalent antibiotic resistance in this study, thus five tetracycline resistance genes (tetA, tetC, tetD, tetE, and tetM) were analyzed in the 82 tetracycline positive E. coli isolates (Table S2). Of the five genes examined, tetA and tetC were the predominant genes. The detection ratios of these genes were: 89.02% for tetC (73/82), 70.73% for tetA (58/82), 39.02% for tetD (32/82), 14.63% for tetE (12/82), 9.76% for tetM (8/82), respectively. Most of the isolates (81.87%) contained two or more tet genes, while tetD and tetM were detected when other tet genes were present. There were 14 different combinations for the tet genes: the highest occurrence was tetA-tetC (34.15%), followed by tetA-tetC-tetD (15.85%), and tetC-tetD (7.32%). Some other combinations such as tetA-tetD and tetC-tetE, were only observed in one to three isolates.
The Polluting of drinking water sources and the presence of antibiotic resistant bacteria increase the risk to human health. It is important to have detailed information regarding such issues. In the present study, we assessed the water quality and the prevalence of antibiotic resistant E. coli in two drinking water sources (the Qiantang River and the Dongtiao Stream) in Hangzhou city. Although there was some degree of organic and fecal pollution found in these two sources, the situation was better than those reported at the other sites (Pereira et al., 2013; Ramirez Castillo et al., 2013), which could be the result of the good management of the special water function zone. The influx of fecal pollution deteriorated in the wet season, which was the opposite of organic pollution. This may have been due to rainfall. In the wet season, the feces of domestic animals and contaminated soils are easily washed into rivers by rain, and lead to the rise of bacteriological values. On the contrary, the absence of rain in the dry season reduces the dilution effect of water for pollution, which could cause a higher level of organic pollution. Except for DO and fecal coliforms at site Q4 during the wet season, the physicochemical and bacteriological parameters of all samples met the requirements for Surface Water Environmental Quality Standard for grade III (GB3838-2002).
In contrast to other types of pollution, antibiotic resistance is difficult to remove even if the release of antibiotic resistance determinants in the environment is discontinued (Martinez, 2009). In this study, 49.50% of the E. coli isolated was resistant to at least one of the tested antibiotics (with an ARI of 0.12) and 24.00% exhibited multiple resistances. The levels of antibiotic resistance are comparable to that in other aquatic environments (Chen et al., 2011; Ribeiro et al., 2012; Machado and Bordalo, 2014; Zhang et al., 2014). It indicates the issue of antibiotic resistance is also serious in qualitied source water. Most E. coli isolates detected here were resistant to tetracycline, followed by β-lactams (AMP and PIP), sulphonamides, and chloramphenicol, which is consistent with other studies (Maal-Bared et al., 2013; Pereira et al., 2013). This was expected as these antibiotics were old and they have been widely used in human and veterinary medicine (Jiang et al., 2013; Maal-Bared et al., 2013; Pereira et al., 2013). In Zhejiang province, tetracycline and chloramphenicol are the principal antibiotics used for livestock (Cui, 2011), and sulphonamides were also frequently detected in wastewater from piggeries and duck farms (Wu et al., 2012). Notably, 75.51% of multiple resistant E. coli (37/49) in this study was resistant to tetracycline and β-lactams concurrently. E. coli isolates from WWTP effluents or from the aquatic environments contaminated by these effluents, are present consistently resistant to tetracycline and β-lactams, and are associated with at least one other resistance, such as sulfamides, chloramphenicol, quinolones, in any combination (Ribeiro et al., 2012). Thus, a likely hypothesis for the origin of these multiresistant isolates is the WWTP effluents.
Thankfully, the resistance rate of E. coli noticeably decreased (with the exception of the R1-2 subpopulation in the wet season) from up- to downstream in the Qiantang River. This phenomenon, however, seems to contradict other reports (Ham et al., 2012; Zhang et al., 2014). Special water function zones with strict safeguards are possibly the main cause of such paradoxical phenomenon (Tao et al., 2010). In China, the range of secondary source water protection areas are not less than 3 km from intake upstream sites and not less than 300 m from the intake downstream locations. And Water Sources Protection Zones for Pollution Prevention and Control Regulations clearly point out that waste water, garbage, and any other activities that can cause water pollution are prohibited (not excluding unmanageably scattered pollutions) in water source protection areas. Thus, few new pollutants are discharged into such areas, and the level of antibiotic resistant E. coli from upstream decreases gradually due to gene loss or environmental self-purification.
Seasonal variation is a possible factor for the change of antibiotic resistance (Mohanta and Goel, 2014). For example, the percentage of multi-drug resistant (MDR) bacteria from three different water sources in West Bengal followed the trend: post-monsoon>winter>summer (Mohanta and Goel, 2014). Similarly, the number of antibiotic resistant isolates from wells in West Africa was higher in the dry season than that in the wet season (Machado and Bordalo, 2014). However, our data indicates that no correlation was found between seasonality and the antibiotic resistance level of E. coli, which is consistent with the previous study (Maal-Bared et al., 2013). The discrepancy may be the result of the different major pollution sources and pollution types. For aquatic environments that mainly receives pollutants from scattered sources (such as farmlands and animal farms), leaching and runoff events, which are often seasonal, probably cause the change of antibiotic resistance in different seasons (Laroche et al., 2010). Point source pollution is the main pollution type in the Qiantang River and the Dongtiao Stream. Pollutants from wastewater treatment plants (WWTPs) or large/small enterprises in the upper streams of the rivers are relatively stable all year round, despite the existence of some small scattered pollution sources.
The relationships between certain water quality variables and antibiotic resistance of bacteria or ARGs have been reported by several studies, such as water depth, temperature, river flow, precipitation, ammonium, total dissolved solids concentrations, dissolved oxygen (Maal-Bared et al., 2013; Middleton and Salierno, 2013; Staley et al., 2015). In this study, we analyzed the links of some water quality parameters and the antibiotic resistance of E. coli. The resistance to different antibiotics had different associations with water quality parameters. The frequency of E. coli resistance to a part of the antibiotics had strong positive associations with nutrient concentrations, which was in accordance with the studies in British Columbia (Maal-Bared et al., 2013) and in northern China (Zhang et al., 2014). Blanco et al. explained that the addition of nutrient concentrations enhanced horizontal transfer of genetic resistance elements (Blanco et al., 2009). E. coli isolates carrying genetic resistance elements had much stronger viability than sensitive isolates under high nutrient concentrations, which led to increasing resistance. No relationships existed among water temperature, pH value, or DO with antibiotic resistance, which was different from other studies. Williams et al. (Williams et al., 1996) deemed that higher temperatures were also the stimulant for the increasing of natural transformation rates among microorganisms. Alkaline conditions easily made antibiotics degrade and hence decreased the resistance (Doi, 2000; Maal-Bared et al., 2013). The discrepancies with these studies are possibly due to the fact that all of the pH values in our study are neutral and the variable range (7.31–7.66) is negligible. For water temperature, even though the change range (14.2–22.7°C) is large, the lowest temperature is suitable for the growth and activity of E. coli. Water quality parameters were measured instantaneously, and hardly exhibit any direct effect on antibiotic resistance which is the result of prolonged period. Moreover, many important factors (such as suspended particles and biofilm) in aquatic environments, can not only influence the development of antibiotic resistance, but can also be influenced by water quality parameters (Dang and Lovell, 2016). All of these relative factors in the Qiantang River and the Dongtiao Stream have not been considered. Furthermore, natural resistance in the E. coli population present in water sources has also not been considered. Thus, these results should still be explained with caution.
Significant correlations were found frequently among the resistance rates of different antibiotics, and the correlations among tetracycline, chlortetracycline, and quinolone resistance were reported the most (Maal-Bared et al., 2013; Zhang et al., 2014). Similarly, in this study, we also found significant correlations between resistance to tetracycline and chloramphenicol or CIP. These strong correlations are probably the results of co-selection (Dang et al., 2006). In fact, tetracycline efflux proteins have similar amino acid and protein structure with other efflux proteins, including chloramphenicol and quinolone resistence, which result the concurrent resistance to multiple antibiotics (Chopra and Roberts, 2001). Besides, resistance to quinolone (CIP and LVX) also had significant correlations with resistance to cephalosporin or GM. They may indicate that an analogous mechanism of cross-selection and/or co-selection among these antibiotics had occured as well (Courvalin and Trieu-Cuot, 2001; Dang et al., 2006; Zhang et al., 2014). Moreover, many antibiotic resistance genes are often found on the same plasmid or mobile genetic elements (Chopra and Roberts, 2001; Roberts, 2005; Carattoli, 2013), which results in the correlations found among resistance to different antibiotics. The StrA-strB gene pair and the sul2 or tet genes were found in the same plasmid acquired in multiple resistant bacteria from the surface seawater of Jiaozhou Bay (Zhao and Dang, 2011). TetQ is often associated with a large conjugative transposon which carries the ermF (Chopra and Roberts, 2001). Thus, further studies should be carried out to illustrate the exact reasons of these correlations.
The frequent detection of tetracycline resistance genes have been reported widely in aquatic environments (Tao et al., 2010; Jia et al., 2014; Chen et al., 2015). In this study, tetracycline resistance also shows the highest resistance frequency among all of the antibiotics tested. Thus, we analyzed five tetracycline resistance genes (tetA, tetC, tetD, tetE, and tetM) from 82 tetracycline resistant E. coli isolates. As the representatives of efflux mechanism of tetracycline, tetA and tetC was widely found in gram-positive and gram-negative bacteria (Zhang et al., 2009). Similarly, our study also showed tetA and tetC were predominant genes (89.02 and 70.73%, respectively) in water sources. This may suggest the risk for increasing levels of multiple resistant bacteria, because these efflux genes are normally associated with large plasmids, and these plasmids often carry other antibiotic resistance genes (Chopra and Roberts, 2001). TetM, a gene for encoding ribosomal protection protein, had the lowest detectablerate. This result supports the fact that some gram-positive tet genes, such as tetK, tetL, tetO, tetM, are not often examined in gram-negative bacteria. In addition, we found it is common for most resistant E. coli isolates (81.87%) to carry two or more tet genes. Interestingly, all of the tetD and tetM were present with other tet genes, while tetA, tetC and tetE were sometimes the sole detectable tet genes. They are totally consistent with the study in Australia (Akinbowale et al., 2007).
The quality of drinking water sources detected in Hangzhou city conformed to the Surface Water Environmental Quality Standard of China, which deems these water sources safe enough to supply to water plants. Despite this, antibiotic resistant E. coli isolates were still heavily seen in these source waters. As antibiotic resistance is hard to eliminate and E. coli is proficient at horizontal transfer, the prevalence of antibiotic resistance E. coli in source water reveals a potential public health risk. Thanks to the special water function zones that offer protection reducing new pollutants, the resistance rates of E. coli decreased gradually from upstream to downstream in Qiantang River. It indicates controlling pollutant emissions and anthropogenic activities could be an effective method to lower the level of antibiotic resistance. Seasonality (dry and wet seasons) had no effect on the resistance rates of E. coli isolates. While some water quality parameters (ammonia nitrogen, total phosphorus, total nitrogen, and BOD5) showed strong relationships with antibiotic resistance that is worth studying further. In addition, significant correlations were found among the resistance rates of different antibiotics. The specific reasons for this should also be explored further.
Because of the highest resistance to tetracycline, five tet genes (tetA, tetC, tetD, tetE, and tetM) were detected from tetracycline resistant positive isolates. We found that tetA and tetC were the predominant genes in source water. Considering tetracycline efflux genes are usually associated with large plasmids, and these plasmids often carry other antibiotic resistance genes, the potential risk for increasing multiple resistance bacteria in drinking water sources should be attached great importance.
ZC performed the isolation of E. coli and part of antibiotic susceptibility test, all of data analysis, and wrote the manuscript. DY supervised all the experiments. SH performed part of antibiotic susceptibility test. HY drew the map and measured part of water quality parameters. YW detected all tetracycline resistance genes. WZ and LS collected water samples and measured part of water quality parameters. LZ and SC designed the study.
This study was funded by Science Technology Bureau of Hangzhou Municipality (Grant No.20130533B22, 20140533B16, 20140733Q01) and Health and Family Planning Commission of Zhejiang Province (Grant No.2014KYB214).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01133/full#supplementary-material
ARB, antibiotic resistant bacteria; ARGs, antibiotic resistance genes; E. coli, Escherichia coli; MDR, multi-drug resistant; ARI, Antibiotic resistance index; AN, amikacin; GM, gentamicin; IPM, imipenem; MEM, meropenem; CZ, cefazolin; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; AMP, ampicillin; PIP, piperacillin; AMC, amoxicillin/clavulanate; SAM, ampicillin/sulbactam; C, chloramphenicol; CIP, ciprofloxacin; LVX, levofloxacin; TE, tetracycline; DO, dissolved oxygen; Water temp., water temperature; COD, Chemical oxygen demand; N03−-N, Nitrate-N; TP, total phosphorus; TN, total nitrogen.
Akinbowale, O. L., Peng, H., and Barton, M. D. (2007). Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J. Appl. Microbiol. 103, 2016–2025. doi: 10.1111/j.1365-2672.2007.03445.x
Blanco, G., Lemus, J. A., and Grande, J. (2009). Microbial pollution in wildlife: linking agricultural manuring and bacterial antibiotic resistance in red-billed choughs. Environ. Res. 109, 405–412. doi: 10.1016/j.envres.2009.01.007
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Chen, B., Hao, L., Guo, X., Wang, N., and Ye, B. (2015). Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ. Sci. Pollut. Res. Int. 22, 13950–13959. doi: 10.1007/s11356-015-4636-y
Chen, B., Zheng, W., Yu, Y., Huang, W., Zheng, S., Zhang, Y., et al. (2011). Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the Minjiang River, Fujian Province, China. Appl. Environ. Microbiol. 77, 148–155. doi: 10.1128/AEM.01676-10
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Courvalin, P., and Trieu-Cuot, P. (2001). Minimizing potential resistance: the molecular view. Clin. Infect. Dis. 33(Suppl. 3), S138–S146. doi: 10.1086/321840
Cui, H. H. (2011). The investigation of tetracycline and chloramphenicol residue in sterillized milk and unsterillized milk from part of zhejiang province. Guangxi J. Light Indus. 151, 3–4. doi: 10.3969/j.issn.1003-2673.2011.06.002
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Dang, H., Song, L., Chen, M., and Chang, Y. (2006). Concurrence of cat and tet genes in multiple antibiotic-resistant bacteria isolated from a sea cucumber and sea urchin mariculture farm in China. Microb. Ecol. 52, 634–643. doi: 10.1007/s00248-006-9091-3
Dei-Tutuwa, D. A. P., and Rahman, M. A. (2014). Rapid detection of microbial contamination in Ghanaian herbal medicines by PCR analysis. Ghana Med. J. 48, 106–111. doi: 10.4314/gmj.v48i2.9
Diwan, V., Tamhankar, A. J., Khandal, R. K., Sen, S., Aggarwal, M., Marothi, Y., et al. (2010). Antibiotics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health 10:414 doi: 10.1186/1471-2458-10-414
Doi, A. M. S. M. K. (2000). The kinetics of oxytetracycline degradation in deionized water under varying temperature, PH, light, substrate, and organic matter. J. Aquat. Anim. Health 12, 246–253. doi: 10.1577/1548-8667(2000)012<0246:TKOODI>2.0.CO;2
Flores Ribeiro, A., Bodilis, J., Alonso, L., Buquet, S., Feuilloley, M., Dupont, J. P., et al. (2014). Occurrence of multi-antibiotic resistant Pseudomonas spp. in drinking water produced from karstic hydrosystems. Sci. Total. Environ. 490, 370–378. doi: 10.1016/j.scitotenv.2014.05.012
Guo, X., Li, J., Yang, F., Yang, J., and Yin, D. (2014). Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 493, 626–631. doi: 10.1016/j.scitotenv.2014.06.035
Ham, Y. S., Kobori, H., Kang, J. H., Matsuzaki, T., Iino, M., and Nomura, H. (2012). Distribution of antibiotic resistance in urban watershed in Japan. Environ. Pollut. 162, 98–103. doi: 10.1016/j.envpol.2011.11.002
Heuer, H., Schmitt, H., and Smalla, K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243. doi: 10.1016/j.mib.2011.04.009
Jia, S., He, X., Bu, Y., Shi, P., Miao, Y., Zhou, H., et al. (2014). Environmental fate of tetracycline resistance genes originating from swine feedlots in river water. J. Environ. Sci. Health B 49, 624–631. doi: 10.1080/03601234.2014.911594
Jiang, L., Hu, X., Xu, T., Zhang, H., Sheng, D., and Yin, D. (2013). Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci. Total Environ. 458–460, 267–272. doi: 10.1016/j.scitotenv.2013.04.038
Kummerer, K. (2009). Antibiotics in the aquatic environment–a review–part II. Chemosphere 75, 435–441. doi: 10.1016/j.chemosphere.2008.12.006
Laroche, E. P. F., Fournier, M., and Pawlak, B. (2010). Transport of antibiotic-resistant Escherichia coli in a public rural karst water supply. J. Hydrol. 39:21. doi: 10.1016/jjhydrol.2010.07.022
Maal-Bared, R., Bartlett, K. H., Bowie, W. R., and Hall, E. R. (2013). Phenotypic antibiotic resistance of Escherichia coli and E. coli O157 isolated from water, sediment and biofilms in an agricultural watershed in British Columbia. Sci. Tot. Environ. 443, 315–323. doi: 10.1016/j.scitotenv.2012.10.106
Machado, A., and Bordalo, A. A. (2014). Prevalence of antibiotic resistance in bacteria isolated from drinking well water available in Guinea-Bissau (West Africa). Ecotoxicol. Environ. Saf. 106, 188–194. doi: 10.1016/j.ecoenv.2014.04.037
Marti, E., Variatza, E., and Balcazar, J. L. (2014). The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 22, 36–41. doi: 10.1016/j.tim.2013.11.001
Martinez, J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902. doi: 10.1016/j.envpol.2009.05.051
Middleton, J. H., and Salierno, J. D. (2013). Antibiotic resistance in triclosan tolerant fecal coliforms isolated from surface waters near wastewater treatment plant outflows (Morris County, NJ, USA). Ecotoxicol. Environ. Saf. 88, 79–88. doi: 10.1016/j.ecoenv.2012.10.025
Mohanta, T., and Goel, S. (2014). Prevalence of antibiotic-resistant bacteria in three different aquatic environments over three seasons. Environ. Monit. Assess. 186, 5089–5100. doi: 10.1007/s10661-014-3762-1
Ng, L. K., Martin, I., Alfa, M. J., and Mulvey, M. R. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15, 209–215. doi: 10.1006/mcpr.2001.0363
Pereira, A., Santos, A., Tacao, M., Alves, A., Henriques, I., and Correia, A. (2013). Genetic diversity and antimicrobial resistance of Escherichia coli from Tagus estuary (Portugal). Sci. Tot. Environ. 461–462, 65–71. doi: 10.1016/j.scitotenv.2013.04.067
Ramirez Castillo, F. Y., Avelar Gonzalez, F. J., Garneau, P., Marquez Diaz, F., Guerrero Barrera, A. L., and Harel, J. (2013). Presence of multi-drug resistant pathogenic Escherichia coli in the San Pedro River located in the State of Aguascalientes, Mexico. Front. Microbiol. 4:147. doi: 10.3389/fmicb.2013.00147
Ribeiro, A. F., Laroche, E., Hanin, G., Fournier, M., Quillet, L., Dupont, J. P., et al. (2012). Antibiotic-resistant Escherichia coli in karstic systems: a biological indicator of the origin of fecal contamination? FEMS Microbiol. Ecol. 81, 267–280. doi: 10.1111/j.1574-6941.2012.01382.x
Roberts, M. C. (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203. doi: 10.1016/j.femsle.2005.02.034
Staley, C., Gould, T. J., Wang, P., Phillips, J., Cotner, J. B., and Sadowsky, M. J. (2015). High-throughput functional screening reveals low frequency of antibiotic resistance genes in DNA recovered from the Upper Mississippi River. J. Water Health 13, 693–703. doi: 10.2166/wh.2014.215
Talukdar, P. K., Rahman, M., Nabi, A., Islam, Z., Hoque, M. M., Endtz, H. P., et al. (2013). Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE 8:e61090 doi: 10.1371/journal.pone.0061090
Tao, R., Ying, G. G., Su, H. C., Zhou, H. W., and Sidhu, J. P. (2010). Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ. Pollut. 158, 2101–2109. doi: 10.1016/j.envpol.2010.03.004
Wu, L. Q., Qian, M. R., Lu, L. Z., and Tao, Z. R. (2012). Antibiotic residues in waste waters from main livestock farms in Zhejiang Province. Acta Agric. Zhejiangen. 24, 699–705. doi: 10.3969/j.issn.1004-1524.2012.04.031
Williams, H. G., Day, M. J., Fry, J. C., and Stewart, G. J. (1996). Natural transformation in river epilithon. Appl. Environ. Microbiol. 62, 2994–2998.
Zhang, X. X., Zhang, T., and Fang, H. H. (2009). Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 82, 397–414. doi: 10.1007/s00253-008-1829-z
Zhang, X., Li, Y., Liu, B., Wang, J., Feng, C., Gao, M., et al. (2014). Prevalence of veterinary antibiotics and antibiotic-resistant Escherichia coli in the surface water of a livestock production region in northern China. PLoS ONE 9:e111026 doi: 10.1371/journal.pone.0111026
Keywords: antibiotic resistance, drinking water source, Escherichia coli, water quality, tet genes
Citation: Chen Z, Yu D, He S, Ye H, Zhang L, Wen Y, Zhang W, Shu L and Chen S (2017) Prevalence of Antibiotic-Resistant Escherichia coli in Drinking Water Sources in Hangzhou City. Front. Microbiol. 8:1133. doi: 10.3389/fmicb.2017.01133
Received: 19 March 2017; Accepted: 02 June 2017;
Published: 16 June 2017.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
Lorenzo Proia, Free University of Brussels, BelgiumCopyright © 2017 Chen, Yu, He, Ye, Zhang, Wen, Zhang, Shu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuchang Chen, aHpjZGNjaGVuQDEyNi5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.