
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 June 2017
Sec. Aquatic Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.01094
This article is part of the Research Topic The Responses of Marine Microorganisms, Communities and Ecofunctions to Environmental Gradients View all 22 articles
 Shan-Hua Yang1†
Shan-Hua Yang1† Ching-Hung Tseng2†
Ching-Hung Tseng2† Chang-Rung Huang1‡
Chang-Rung Huang1‡ Chung-Pin Chen3
Chung-Pin Chen3 Kshitij Tandon1,4,5
Kshitij Tandon1,4,5 Sonny T. M. Lee6
Sonny T. M. Lee6 Pei-Wen Chiang1
Pei-Wen Chiang1 Jia-Ho Shiu1,7,8
Jia-Ho Shiu1,7,8 Chaolun A. Chen1
Chaolun A. Chen1 Sen-Lin Tang1*
Sen-Lin Tang1*The coral holobiont is the assemblage of coral host and its microbial symbionts, which functions as a unit and is responsive to host species and environmental factors. Although monitoring surveys have been done to determine bacteria associated with coral, none have persisted for >1 year. Therefore, potential variations in minor or dominant community members that occur over extended intervals have not been characterized. In this study, 16S rRNA gene amplicon pyrosequencing was used to investigate the relationship between bacterial communities in healthy Stylophora pistillata in tropical and subtropical Taiwan over 2 years, apparently one of the longest surveys of coral-associated microbes. Dominant bacterial genera in S. pistillata had disparate changes in different geographical setups, whereas the constitution of minor bacteria fluctuated in abundance over time. We concluded that dominant bacteria (Acinetobacter, Propionibacterium, and Pseudomonas) were stable in composition, regardless of seasonal and geographical variations, whereas Endozoicomonas had a geographical preference. In addition, by combining current data with previous studies, we concluded that a minor bacteria symbiont, Ralstonia, was a keystone species in coral. Finally, we concluded that long-term surveys for coral microbial communities were necessary to detect compositional shifts, especially for minor bacterial members in corals.
Corals harbor a variety of microorganisms, including algae, fungi, bacteria, archaea, and viruses (Knowlton and Rohwer, 2003; Wegley et al., 2007; Marhaver et al., 2008). Coral-associated microbes, which live in coral mucus, tissue, and skeleton, have various interactions with their host and maintain coral holobiont function, including antimicrobial defense and nutrient acquisition (Bourne et al., 2016). With advances in high-throughput sequencing technologies, there is growing research interest in microbial ecology in rare biospheres (Lynch and Neufeld, 2015), characterizing genetic and functional diversity supporting global ecosystem health (Yachi and Loreau, 1999; Elshahed et al., 2008).
Composition of coral-associated microorganisms is affected by coral species (Littman et al., 2009; Reis et al., 2009; Kvennefors et al., 2010; Lee et al., 2012; Lema et al., 2012; McKew et al., 2012; Morrow et al., 2012; Carlos et al., 2013), environmental factors (Hong et al., 2009; Ceh et al., 2011; Chen et al., 2011), geographical differences (Hong et al., 2009; Littman et al., 2009; Kvennefors et al., 2010; Lee et al., 2012; Morrow et al., 2012; Lema et al., 2014), and human activities (Klaus et al., 2007). According to previous studies, some microbes had host specificity, regardless of geographical difference (Rohwer et al., 2001, 2002), whereas others had regional specificity rather than host types (Littman et al., 2009).
Various coral-associated microbial species reportedly shared similar functional roles in coral ecology (Li et al., 2013). However, bacterial succession in corals has not been well characterized, as the longest monitoring survey lasted only 1 year (Rohwer et al., 2002; Hong et al., 2009; Ceh et al., 2011; Carlos et al., 2013; Lema et al., 2014; Hernandez-Agreda et al., 2016). Furthermore, although some microbes were consistently detected in coral (Rohwer et al., 2002; Carlos et al., 2013), it is still unknown whether these common microbes change their abundance over longer intervals. A 2-years survey characterizing dynamics of Symbiodinium rare biosphere provided evidence for symbiont switching in reef-building corals after environmental changes (Boulotte et al., 2016), but dynamics of the bacterial rare biosphere in coral, and bacterial shifting/switching within corals, are not established.
The objective was to determine whether a long-term survey was necessary to detect compositional dynamics of coral-associated bacteria in various locations. Based on previous short-term studies, we hypothesized that coral-associated bacteria require varying time scales to manifest detectable variations in different environments. Cumulative stressors, e.g., nutrient availability and elevated sea surface temperature, have been postulated to change coral reef ecosystems; therefore, coral-associated microbes might either attenuate or exacerbate effects from stressors through their roles in coral holobiont fitness (Bourne et al., 2008, 2016). Since cumulative stressors vary from place to place, duration of detectable changes in coral-associated microbes is also likely to vary, which should be apparent in a long-term survey (Mouchka et al., 2010; Ceh et al., 2011; Chen et al., 2011; Lee et al., 2012).
To determine dynamics of coral-associated microbial communities among locations and to study temporal variations of dominant and minor bacterial groups, 16S rRNA gene amplicon pyrosequencing was used in a 2-years observational study investigating bacterial communities in coral Stylophora pistillata, which is widely distributed in Taiwan and around the globe. That Taiwan lies across the Tropic of Cancer, S. pistillata in tropical (Kenting and Lyudao) and subtropical (Yehliu) Taiwan were collected to provide a comparative basis for latitude influence on bacterial composition in corals. This 2-years survey was to assess coral-associated microbial differences among various climate zones, and determine importance of long-term observation to detect bacterial dynamics in coral hosts.
Coral and seawater samples were collected from three locations, Kenting, Yehliu, and Lyudao, in Taiwan (Figure 1) from February 2008 to January 2010. In each location and at each sampling time, three colonies (replicates) of S. pistillata at depths of 3–7 m were selected and tagged for sampling. For each colony, a coral fragment approximately 2 cm long and 2 cm in diameter was collected. Samples were washed twice with filtered seawater and stored at 4°C until coral tissue was processed. In addition, 1 L of seawater surrounding the coral colony was simultaneously collected to analyze the microbial community in reef water. A total of 88 S. pistillata and seawater samples were collected (Supplementary Table S1). Environmental data (precipitation, daylight duration and air and water temperatures) were obtained from the Central Weather Bureau in Taiwan (Supplementary Table S1).
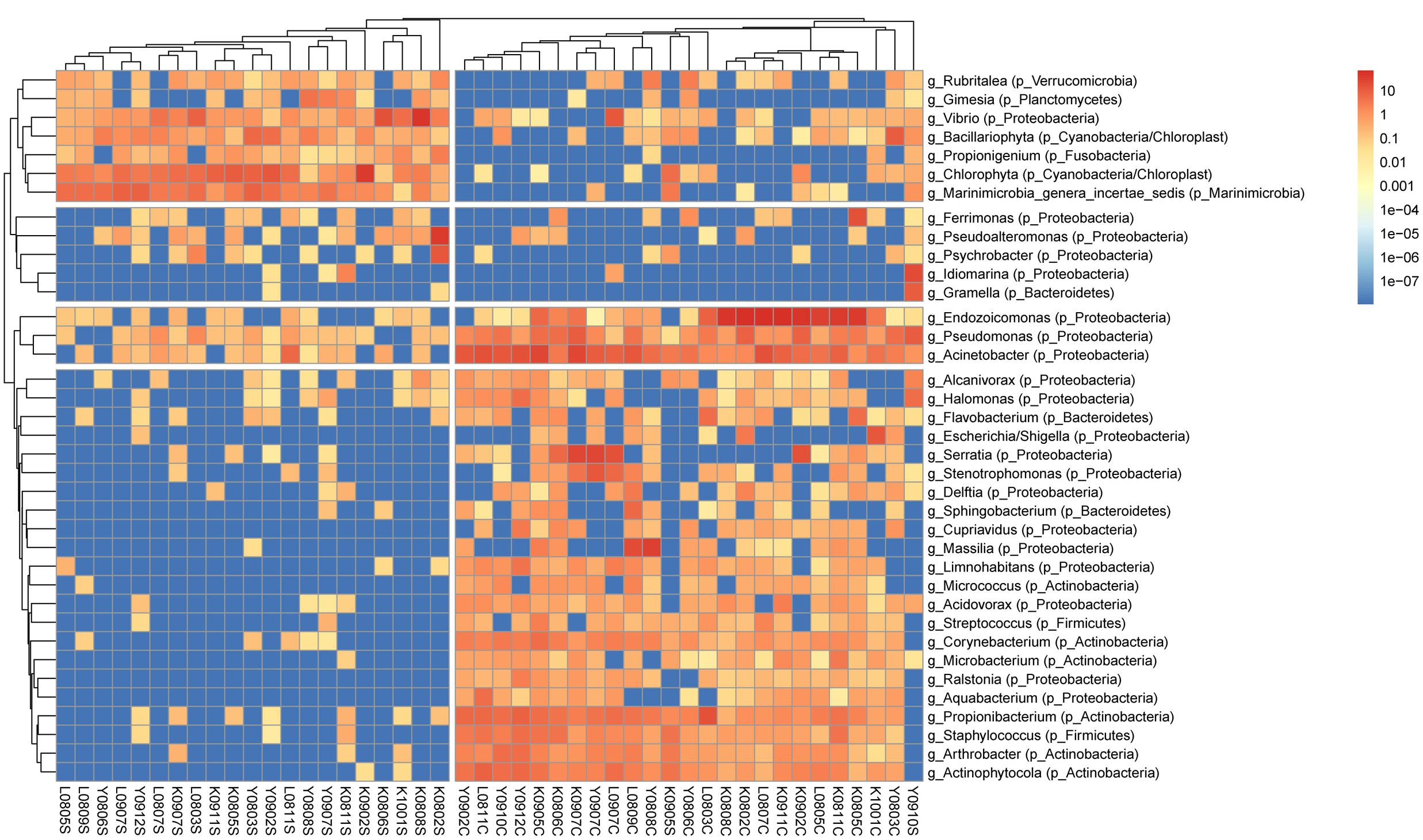
FIGURE 1. Clustering analysis of bacterial genera in S. pistillata and seawater. A total of 37 genera remained in the analysis after excluding genera of unknown taxonomy or <0.2% relative abundance. Trees were cut at 2 and 4 groups in columns and rows, respectively. Within each sample, the color scale indicates relative abundance (percentage). Genus names are prefixed with “g_,” and phylum names with “p_” in parenthesis. Columns are clustered by Bray–Curtis distance and rows by Euclidean distance of their abundance profile.
Coral tissues were sprayed (using an airbrush) with 2–5 mL 10× TE buffer (10 mM Tris–HCl, pH 8.5, 1 mM EDTA, pH 8.0) and collected in a sterilized bag. Coral tissues were transferred to a 15 mL sterilized tube and stored at -20°C pending DNA extraction.
Coral tissue was recovered for genomic DNA extraction by centrifugation (10,000 × g, 15 min). After discarding the supernatant, tissue pellets were put into 600 μL of 10× TE buffer and liquid nitrogen, homogenized (mortar and pestle), and the resulting solution transferred to a clean tube. Seawater samples were filtered through membrane filters (0.2 μm pore size; Adventec, Tokyo, Japan) to collect microbial particulate. Total genomic DNA of coral tissue and seawater were extracted as outlined in the UltraClean Soil DNA Kit (MioBio, Solana Beach, CA, United States).
The PCR was performed using two bacterial universal primers: 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 341R (5′-CTGCTGCCTCCCGTAGG-3′), which were designed for the V1–V2 hypervariable region of the bacterial 16S rRNA gene (Hamp et al., 2009). The reaction mixture contained 1 μL of 5 U TaKaRa Ex Taq HS (Takara Bio, Otsu, Japan), 5 μL of 10×Ex Taq buffer, 4 μL of 2.5 mM deoxynucleotide triphosphate mixture, 1 μL of each primer (10 μM), and 1–5 μL (10–20 ng) template DNA in a volume of 50 μL. The temperature program for 30 PCR cycles was the initial step of 94°C for 3 min, 94°C for 20 s, 52°C for 20 s, 72°C for 20 s, and 72°C for 2 min (final extension after the last cycle).
The PCR amplicons of the bacterial 16S rRNA gene V1–V2 region were verified by DNA agarose gel electrophoresis with 1.5% agarose gel and 10× TE buffer. The expected DNA band (∼320 bp) was cut from the gel and DNA was recovered with a QIAEX II Agarose Gel Extraction kit (QIAGEN, Hilden Germany) and quality verified with a Scandrop spectrophotometer (Thermo Scientific, Vantaa, Finland).
A DNA tagging PCR was used to tag each PCR product of the bacterial 16S rRNA gene V1–V2 region (Chen et al., 2011). According to Chen et al. (2011), the tag primer was designed with four overhanging nucleotides; this arrangement ensured 256 distinct tags, at the 5′ end of the 27F and 341R primers for bacterial DNA. The tagging PCR conditions consisted of an initial step of 94°C for 3 min, 5 cycles of 94°C for 20 s, 60°C for 15 s, 72°C for 20 s, and a final step at 72°C for 2 min.
Pooled 40 ng lots of each tagged bacterial 16S rRNA gene V1–V2 region DNA samples (88 samples in total) were used for massively parallel pyrosequencing using a Roche 454 Genome Sequencer FLX System (Mission Biotech, Taipei, Taiwan). Raw sequencing reads were sorted according to unique tags using an in-house script, and deposited in NCBI Sequence Read Archive database, under accession number SRP098878, after the primer was removed. Reads from replicate colonies, sampled at the same time and location, were combined into one sample.
On a per-sample basis, bacterial reads were quality-filtered using Mothur v1.36.1 (Schloss et al., 2009) to retain reads of lengths from 290 to 340 base pairs (bp), and an average quality score >27. Reads containing any ambiguous base or homopolymer >8 bp were excluded. Chimeric reads were detected and removed using USEARCH v8.1.1861 (Edgar, 2010) with reference mode (3% minimum divergence). Quality-filtered and non-chimeric reads were analyzed using UPARSE pipeline (Edgar, 2013) to generate OTUs per sample (97% identity level), which were classified with RDP classifier (v2.12) (Wang et al., 2007) with a pseudo-bootstrap threshold of 80%.
The rarefied diversity measures (e.g., Shannon, Simpson, Chao 1, ACE, and Good’s coverage) were estimated using Mothur with 1000 iterations. The sample size (N) varied from 567 to 29819; therefore, 567 was chosen for rarefaction analysis.
Bacterial OTU representative sequences of all samples affiliated with a given taxonomy were collectively clustered using USEARCH, with options –cluster_smallmem and –id 0.97. Abundance of each centroid (or cross-sample OTU) was represented by average relative abundance of its cluster members. Phylogenetic analysis was conducted in MEGA7 (Kumar et al., 2016), with maximum-likelihood trees generated with 1000 bootstraps for aligned representative sequences.
The present study defined six resident types of bacteria in S. pistillata and seawater, based on relative abundance and detection frequency. Type I (prevalent abundant group), type II (moderately prevalent abundant group), and type III (non-prevalent abundant group) each had ≥1% average relative abundance and were detected in ≥80, 80–60, and <60% samples, respectively. Type IV (prevalent minor group), type V (moderately prevalent minor group), and type VI (non-prevalent minor group) had <1% average relative abundance and were detected in ≥80, 80–60, and <60% samples, respectively. Coral and seawater samples were separately considered (in terms of detection frequency).
Relative abundance of each classified bacterial genera in individual samples was incorporated into a matrix to estimate the Bray–Curtis distance. Results were presented by non-metric multidimensional scaling (nMDS) analysis using Primer 6 (PRIMER-E, Lutton, Lvybridge, United Kingdom; Clarke and Warwick, 2001) to determine relationships of bacterial communities among times and locations. Analysis of similarity (ANOSIM) via Primer 6 was used to test bacterial diversity indices and compositions in spatiotemporal variations.
For analysis of abundant and minor bacterial members, abundant groups (type I–III) and minor groups (type IV–VI) were separately incorporated into two matrixes. These matrices were used to perform ANOSIM in a Primer 6 based Bray–Curtis distance derived from relative abundance transformed by three methods, including no transformation, square root transformation, and binary transformation (i.e., presence and absence analysis).
Bacterial compositions between sampling times in each location were compared with Pearson correlations using R package Hmisc1. In addition, Pearson correlations were used to compare fluctuations of Endozoicomonas and Acinetobacter.
Hierarchical clustering analysis was performed using the average linkage via R package pheatmap (Kolde, 2015) and visualized on a heat map. The distance matrix used for clustering was calculated using the R package vegan (Dixon, 2003) based on a base-10 logarithmic transformation of relative abundance (in percentage), plus a pseudo-count of 1 × 10-8 (Costea et al., 2014). Genera of unknown taxonomy or <0.2% relative abundance were excluded.
Seawater sample K1001S yielded the most operational taxonomic units (OTUs), 406.15 (in rarefaction), whereas coral sample K0808C had the fewest (58.68 OTUs; Supplementary Table S2). According to ANOSIM, bacterial diversity indices in corals and seawater samples differed (richness: R = 0.634, p = 0.001; Shannon: R = 0.298, p = 0.001) and bacterial community in seawater samples had greater richness and Shannon diversity than those in corals (Supplementary Table S2).
Overall, bacterial compositions in S. pistillata from Kenting, Yehliu, and Lyudao were similar at a class level, similar to seawater samples (Supplementary Figure S1). However, bacterial communities in S. pistillata and seawater differed from each other. Dominant bacterial classes in seawater were Flavobacteria, unclassified Marinimicrobia, Gammaproteobacteria, and Alphaproteobacteria. Among these, most dominant genera were unclassified, such as unclassified Marinimicrobia, unclassified Gammaproteobacteria, and unclassified Flavobacteriaceae. In addition, the genus Vibrio, which belongs to Gammaproteobacteria, was higher than those in corals.
In S. pistillata, Actinobacteria, Betaproteobacteria, and Gammaproteobacteria were dominant, and their relative abundances higher than those in seawater. In Gammaproteobacteria, dominant genera were Endozoicomonas, Acinetobacter, Pseudomonas, and Serratia (Supplementary Figure S1), among which Endozoicomonas was particularly more dominant in Kenting and Lyudao compared to Yehliu. Propionibacterium and Actinophytocola were the two dominant Actinobacteria genera, and Massilia Betaproteobacteria in coral.
Bacterial community structure in S. pistillata and seawater at various sampling times is shown (Figure 1). Some genera, such as Ralstonia, Cupriavidus, and Aquabacterium, were only detected in coral samples, whereas Pseudoalteromonas was common only in seawater, and Gramella was exclusively retrieved from seawater samples. In addition, among coral samples, although some bacterial taxa were prevalent at every collection, their relative abundance varied from time to time (Figure 1). For example, Endozoicimonas and Acinetobacter fluctuated in abundance throughout this survey.
There were differences among sampling years in bacterial community structure (ANOSIM: R = 0.301, p = 0.002; Supplementary Figure S2). Among all sites, based on nMDS, the bacterial community changed along the sampling time (Figure 2). Within each location, bacterial composition also differed among sampling years or times (Supplementary Figure S3), especially in samples from Kenting (ANOSIM: R = 0.370, p = 0.028). In Lyudao and Yehliu, bacterial compositions were clustered according to sampling years (difference not significant), but the Pearson correlation value between each sampling time in Lyudao and Yehliu decreased significantly (in Lyudao, from 1.00 to 0.24; in Yehliu, from 1.00 to 0.16) with increasing time span (Supplementary Figure S4), which indicated changes in bacterial composition during sampling times. In addition, there was no difference between seasons (ANOSIM: R = -0.072, p = 0.791).
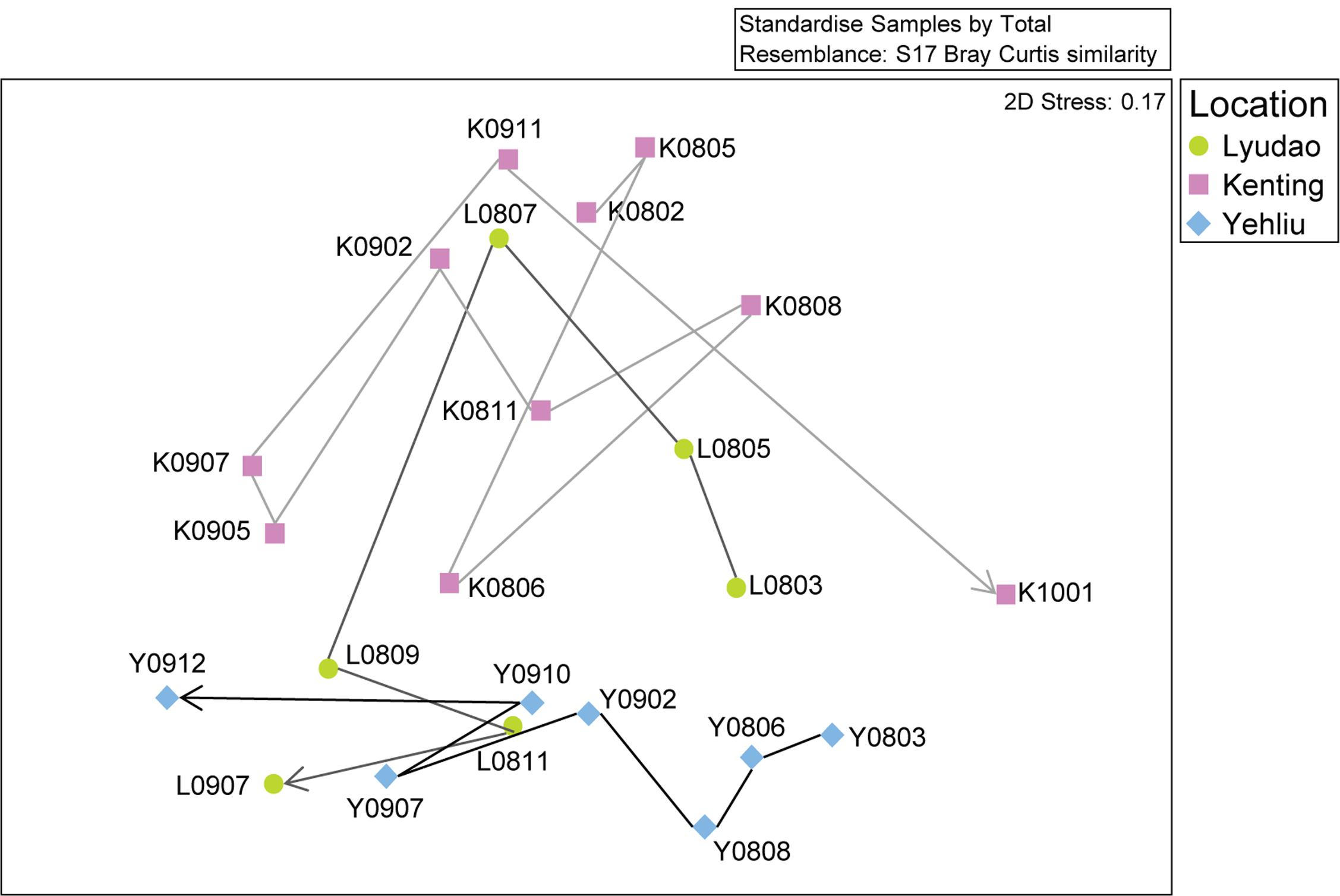
FIGURE 2. Non-metric multidimensional scaling (nMDS) analysis of the changing S. pistillata-associated bacterial communities in three locations at various times (2008–2010). Symbols indicate samples from different locations; circles are for Lyudao, squares for Kenting, and diamonds for Yehliu. The lines and arrow indicate the change of bacterial communities from the first to the last sampling time and colors of lines distinguish locations.
To determine differential effects of short- versus long-term observations, consecutive patterns of correlations between bacterial communities were analyzed (Supplementary Figure S5). Correlations fluctuated over five consecutive samplings (Supplementary Figures S5A–F). However, a longer time span revealed a more holistic view of variation in bacterial composition (Supplementary Figure S5G), not apparent in short-term observations. The longer time span also revealed that changes followed a pattern. For example, two peaks (times 4–6 and time 9) had different duration, indicating different resilience status (Supplementary Figure S5G). Both abundant and minor (<1% relative abundance) genera had a significant differences between sampling years, and only minor genera did so by considering presence/absence data (binary transformation; Supplementary Table S3). In Yehliu, these abundant genera (except Endozoicomonas) retained similar abundance and moderately increased in 2009, although they had more variable fluctuations in abundance in Lyudao and Kenting, especially Endozoicomonas and Acinetobacter (Figure 3). In Lyudao, Acinetobacter increased during July to November in 2008, and Endozoicomonas increased during May and July in 2008, but decreased thereafter. In Kenting, abundance of Acinetobacter was low during 2008, increased during May and July in 2009, and decreased from November 2009 to January 2010. However, Endozoicomonas bloomed in most sampling times during 2008 and decreased during May and July in 2009, followed by an increase in November 2009. Intriguingly, both in Lyudao and Kenting, the times of Endozoicomonas and Acinetobacter blooms were staggered (Figure 3). Furthermore, in Kenting, there was a negative correlation (Pearson correlation: R = -0.899, p = 0.038) between Endozoicomonas and Acinetobacter abundance fluctuations from November 2008 to November 2009.
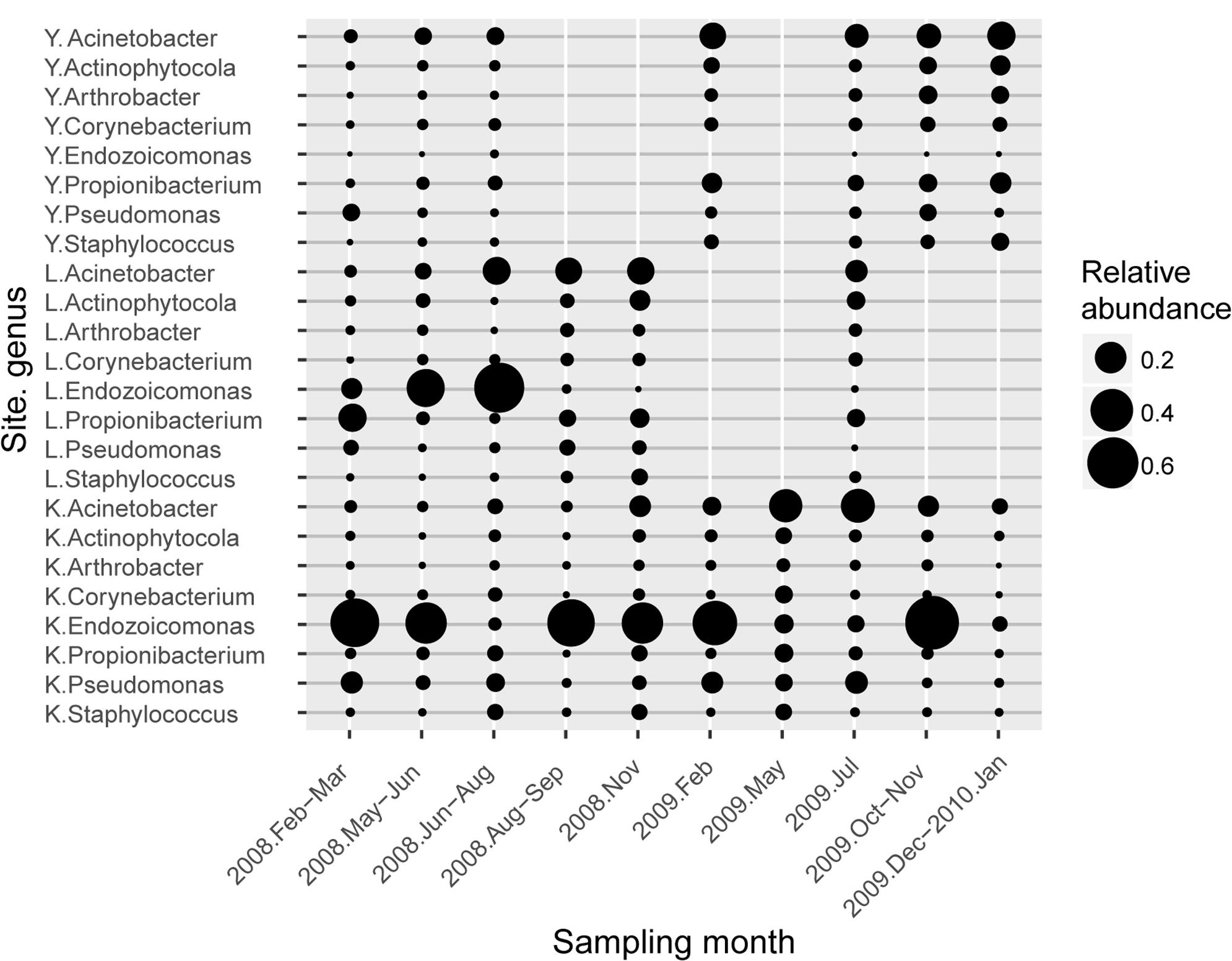
FIGURE 3. Average abundance profile of resident type-I genera associated with S. pistillata. Circle size represents the average relative abundance of each genus. Each genus name was prefixed with a one-letter abbreviation for sampling sites (K is for Kenting, L for Lyudao, and Y for Yehliu).
Based on nMDS, there was distinct clustering of coral-associated bacterial composition in Yehliu, Lyudao, and Kenting (ANOSIM: R = 0.224, p = 0.014; Figure 2). Kenting and Yehliu had differences not only in bacterial composition (ANOSIM: R = 0.437, p = 0.004) but also in community richness (ANOSIM: R = 0.218, p = 0.045). There was a compositional difference among sampling sites in dominant genera (ANOSIM: R = 0.240, p = 0.008), but not in minor genera (Supplementary Table S3).
Endozoicomonas and Acinetobacter were the two frequently detected genera of high abundance (Figure 3), of which both had disparate inhabiting preferences on S. pistillata in different locations (Figure 4). Endozoicomonas were mostly present in corals from Lyudao and Kenting, and composed of distinct genotypes in each locations (i.e., OTU02, 03, 04, 07, 08, and 10 in Kenting; OTU05, 06, and 09 in Lyudao). These Endozoicomonas OTUs were merely detected in both Kenting and Lyudao, indicating that geographical setups determined their inhabiting specificity (Figure 4A). Based on phylogenetic analysis, these OTUs were highly associated with E. elysicola and E. atrinae (Figure 4C). In contrast to exclusive detection of Endozoicomonas in coral, Acinetobacter were present in both S. pistillata and seawater (Figure 4B). Furthermore, most Acinetobacter OTUs were shared by three sampling sites, with less geographical preference than Endozoicomonas (Figure 4B). These Acinetobacter OTUs were also more diverse compared to Endozoicomonas OTUs in terms of phylogenetic affiliation (Figure 4D).
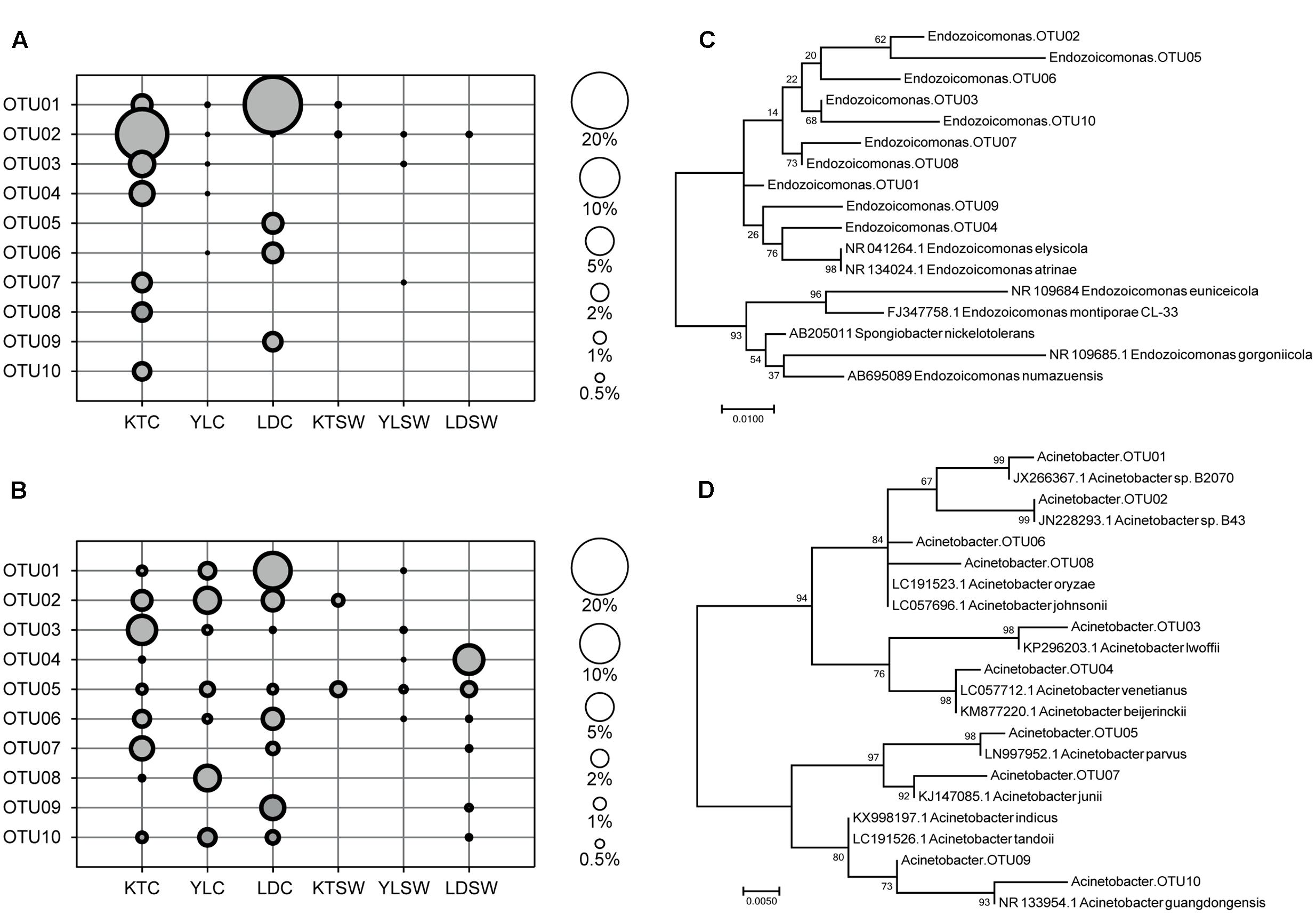
FIGURE 4. Top-10 abundant OTU profiles and molecular phylogenetic analysis of Endozoicomonas and Acinetobacter in S. pistillata and seawater. The abundance profile of top-10 OTUs of (A) Endozoicomonas and (B) Acinetobacter were organized in a descendent order according to their average relative abundance (scaled by circle size). Maximum-likelihood trees (with 1000 bootstraps) among OTUs of (C) Endozoicomonas and (D) Acinetobacter were also displayed. KT, Kenting; YL, Yehlui; LD, Lyudao; C, colony; SW, seawater.
According to relative abundance and detection frequency, bacteria in S. pistillata and seawater were categorized into six resident types (Section Resident Types of Bacterial Lineages in Materials and Methods and Table 1). In S. pistillata, type I group (i.e., with >1% relative abundance and detected in ≥80% samples) had eight genera, including Acinetobacter, Endozoicomonas, Propionibacterium, Corynebacterium, Staphylococcus, Arthrobacter, Actinophytocola, and Pseudomonas. However, there were only Vibrio and unclassified Marinimicrobia as type I in seawater. Among these abundant genera (resident type I–III), coral and seawater shared no genus.
The present study demonstrated that long-term observation of coral-associated microbial communities provided a baseline to elucidate coral–microbe and microbe–microbe interactions. Abundant and minor bacteria in S. pistillata had different inclinations to geographical and temporal changes. Based on relative abundance and detection frequency, prevalent and non-prevalent S. pistillata-associated bacterial genera were allotted into six putative resident types.
Shifts in bacterial communities in healthy coral tissues were only apparent with a sufficient duration of survey (∼2 years). Although changes in bacterial composition in Lyudao and Yehliu were not confirmed (samples were lost due to typhoons and human activities), based on the correlation between sampling times, community resilience may be present over a longer sampling interval. In the present study, the long and complete observation in Kenting successfully captured compositional resilience that was not apparent in short-term surveys. A small group of bacteria (that comprised the core microbiome) were ubiquitously associated with coral, regardless of abiotic environmental parameters (Hernandez-Agreda et al., 2016). However, based on our results, prolonged observation enabled characterizing their succession in coral (Lynch and Neufeld, 2015). In this study, changes in prevalent abundant genera Acinetobacter and Endozoicomonas had disparate fluctuation patterns among locations, with abundances constant in Yehliu but variable in Kenting in 2009. Negative abundance correlations between these two genera were not detected before November 2008 but afterward until November 2009; therefore, this successional pattern was far from conclusive. Apart from abundant ones, minor bacterial genera in S. pistillata also changed in abundance throughout the study, which emphasized the need for longer surveys of coral-associated bacteria. Although Pantos et al. (2003) proposed some explanations for observed fluctuations in coral-associated bacterial communities, their hypotheses were only based on short-term observations. It has been suggested that coral-associated microbes can attenuate or intensify cumulative stressors that may disrupt coral reef ecosystems (Bourne et al., 2016). Therefore, we believe that these stressors, which varied in times and degrees from place to place, also contributed to spatial and temporal dynamics in coral-associated bacterial composition and abundance.
Different coral reproduction strategies (i.e., broadcast spawning and brooding) affect symbiotic bacterial composition differently, and S. pistillata, a brooder coral, harbored distinguishable bacterial communities strongly clustered by geographical location (Neave et al., 2017b), implying, in S. pistillata, vertical transfer of microbes from parents to offspring, thereby restricting the microbial structure and development and leading to the observed geographical grouping. As some bacteria have inhabiting specificity to coral species (Hong et al., 2009; Reis et al., 2009), discrepant microbial compositions among corals reported by different studies were plausible.
Among abundant genera, Acinetobacter, Propionibacterium, and Pseudomonas were more constantly detected in S. pistillata regardless of seasonal and geographical differences, whereas Endozoicomonas had geographical variations. Endozoicomonas spp. have diverse roles in different hosts as being symbiotic (La Rivière et al., 2013) and parasitic (Mendoza et al., 2013). The documented high proportion of repeats and insertion sequences in E. montiporae (Ding et al., 2016) and pathogenic strain Candidatus Endozoicomonas cretensis (Katharios et al., 2015) corresponded to their high genome plasticity for wide adaptation to various hosts (reviewed by Neave et al., 2017a). Since Endozoicomonas was abundant in Lyudao and Kenting but not in Yehliu, their adaptation appeared more favorable for tropical versus subtropical oceans.
Based on the presence/absence transformation of abundance, minor genera had significant differences among sampling years, reflecting their fluctuating nature in S. pistillata over time. Ralstonia and Propionibacterium were two acknowledged minor symbiotic genera in corals and intimately associated with dinoflagellate endosymbionts (Ainsworth et al., 2015). However, in our study, Propionibacterium was abundantly detected among three locations during sampling, whereas Ralstonia had constantly low abundance among sampling sites. Lynch and Neufeld (2015) suggested that prevalent, minor taxa were just conditionally minor in abundance, and would opportunistically grow with abundance under optimal conditions. This accounted for the discrepant abundance of Propionibacterium in the current versus previous studies. In contrast, consistent detection of Ralstonia’s low abundance described its potential of being a keystone taxon permanently holding rare abundance in coral (Giovannoni and Stingl, 2005; Lynch and Neufeld, 2015), although the functional role of Ralstonia in S. pistillata remains unclarified.
Many bacterial taxa were reported as abundant members in coral (Littman et al., 2009; Liu et al., 2012; McKew et al., 2012; Morrow et al., 2012; Carlos et al., 2013), and mostly on Spongiobacter and Endozoicomonas (Lee et al., 2012; Morrow et al., 2012; Bayer et al., 2013; Carlos et al., 2013; Lema et al., 2014). These abundantly, frequently detected bacteria seemed adapted to live in coral tissues and acting as coral nutrition, pathogens, probiont, or purely commensal bacteria in the coral holobiont (Klaus et al., 2005). In the present study, we categorized S. pistillata-associated bacteria into six resident types, based on their abundance and detection frequency, with Acinetobacter (in type I) being the prevalent genera. Acinetobacter had been detected in stony corals from various regions (Littman et al., 2009; McKew et al., 2012; Morrow et al., 2012; Carlos et al., 2013; Li et al., 2014), and potentially played either beneficial or detrimental roles in corals. For example, Shnit-Orland and Kushmaro (2009) considered Acinetobacter sp. as a first-line defender that assisted the coral holobiont against pathogens resistant to multiple antibiotics. However, in a study of Dark Spot Syndrome in coral Stephanocoenia intersepta, Acinetobacter was regarded as a potential pathogen (Sweet et al., 2013). Given the common presence in stony corals, the uncertain role of Acinetobacter in S. pistillata needs to be clarified.
Among the eight genera in resident type I, four (i.e., Propionibacterium, Corynebacterium, Arthrobacter, and Actinophytocola) belong to the phylum Actinobacteria, consistent with other reports regarding Actinobacteria as a dominant genus associated with corals. Propionibacterium was present in coral Cirrhipathes lutkeni (Santiago-Vázquez et al., 2007), Mussismilia hispida (de Castro et al., 2010) and Acropora digitifera (Nithyanand et al., 2011). Using Actinobacteria-specific primers, Yang et al. (2013) reported 19 actinobacterial genera in two corals, suggesting high diversity of Actinobacteria in both hard and soft corals. Coral-associated actinobacteria reportedly had antimicrobial activity (Sweet et al., 2013; Mahmoud and Kalendar, 2016) attributed to production of bioactive substances (Yang et al., 2013). Therefore, these four type-I actinobacterial genera warrant careful consideration in future studies, considering their broad distribution in corals and antimicrobial properties.
In the ocean, heterotrophic bacterial communities (in terms of species composition, spatiotemporal variations, and community structure) are largely affected by the availability, composition, and spatiotemporal distribution of organic substrates. Similarly, variations in coral-associated bacterial community are also controlled, to a great extent, by organic matter secreted by coral, similar to the situation of algae-associated microbial communities (Dang and Lovell, 2016). For example, the genome of Endozoicomonas has a wide spectrum of genes related to generic transport for uptake of extracellular organic compounds and carbohydrates (Ding et al., 2016; Neave et al., 2017a). Numbers of transport molecules also implied interactions between Endozoicomonas and compounds produced by corals (Neave et al., 2014; Ding et al., 2016). Similarly, Acinetobacter was reportedly able to metabolize dimethyl sulfide (Horinouchi et al., 1997) produced by symbiotic algae in corals; furthermore, its precursor (dimethylsulfoniopropionate) has served as a chemoattractant to heterotrophic bacteria in oceans (Seymour et al., 2010). Therefore, availability of extracellular compounds in coral holobiont is likely associated with fluctuations in the abundance of Endozoicomonas and Acinetobacter.
This long-term survey revealed dynamics of S. pistillata-associated bacterial community (e.g., compositional resilience), with fluctuating patterns of abundant and minor genera stratified by sampling times and geographical locations. Although Endozoicomonas and Acinetobacter were detected with high abundance and frequency, this long-term observation identified differential inhabiting inclination, defined by Endozoicomonas being more acclimated to tropical oceans, whereas Acinetobacter was merely confined by climatic zones. Regarding minor genera, despite the lack of functional evidence clarifying their opportunistic or keystone roles in S. pistillata, a long-term survey could legitimately distinguish them from somewhat transient colonizers, and also identify microbial candidates for future functional studies. Lastly, abundance-shuffling and species-switching of symbiotic algae improved environmental adaptation of corals (Buddemeier and Fautin, 1993; Fautin and Buddemeier, 2004; Boulotte et al., 2016), although some bacteria had beneficial effects relating to holobiont resilience without abundance fluctuations under short-term heat stress (Ziegler et al., 2017). Therefore, we inferred that large-scale and long-term time series observations are required to characterize fluctuating and steady microbial components in coral holobiont.
S-HY substantially analyzed data and wrote the manuscript. C-HT performed bioinformatics analysis, provided detailed methods and results for the analysis, and wrote the manuscript. C-RH performed sampling, DNA extraction, PCR, and data analysis. C-PC acquired samples and performed DNA extraction. KT performed statistical test and participated in the manuscript editing. SL participated in the manuscript writing and editing. P-WC performed sampling, DNA extraction, and PCR. J-HS and CC acquired samples. S-LT planned the experimental design and participated in manuscript editing.
Funding for this study was provided by the Biodiversity Research Center, Academia Sinica, Taiwan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank CC’s team for coral sampling. We also thank our dear lab member, the late C-RH for his efforts devoted to this project; may he rest in peace.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01094/full#supplementary-material
Ainsworth, T. D., Krause, L., Bridge, T., Torda, G., Raina, J. B., Zakrzewski, M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Bayer, T., Neave, M. J., Alsheikh-Hussain, A., Aranda, M., Yum, L. K., Mincer, T., et al. (2013). The microbiome of the red sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13
Boulotte, N. M., Dalton, S. J., Carroll, A. G., Harrison, P. L., Putnam, H. M., Peplow, L. M., et al. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J. 10, 2693–2701. doi: 10.1038/ismej.2016.54
Bourne, D., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Buddemeier, R. W., and Fautin, D. G. (1993). Coral bleaching as an adaptive mechanism: a testable hypothesis. Bioscience 43, 320–327. doi: 10.2307/1312064
Carlos, C., Torres, T. T., and Ottoboni, L. M. (2013). Bacterial communities and species-specific associations with the mucus of Brazilian coral species. Sci. Rep. 3:1624. doi: 10.1038/srep01624
Ceh, J., Van Keulen, M., and Bourne, D. G. (2011). Coral-associated bacterial communities on Ningaloo Reef, Western Australia. FEMS Microbiol. Ecol. 75, 134–144. doi: 10.1111/j.1574-6941.2010.00986.x
Chen, C. P., Tseng, C. H., Chen, C. A., and Tang, S. L. (2011). The dynamics of microbial partnerships in the coral Isopora palifera. ISME J. 5, 728–740. doi: 10.1038/ismej.2010.151
Clarke, K. R., and Warwick, R. M. (2001). Change in Marine Communities: an Approach to Statistical Analysis and Interpretation. Plymouth: Plymouth Press.
Costea, P. I., Zeller, G., Sunagawa, S., and Bork, P. (2014). A fair comparison. Nat. Methods 11, 359–359. doi: 10.1038/nmeth.2897
Dang, H., and Lovell, C. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
de Castro, A. P., Araújo, S. D. Jr., Reis, A. M., Moura, R. L., Francini-Filho, R. B., Pappas, G. Jr., et al. (2010). Bacterial community associated with healthy and diseased reef coral Mussismilia hispida from eastern Brazil. Microb. Ecol. 59, 658–667. doi: 10.1007/s00248-010-9646-1
Ding, J. Y., Shiu, J. H., Chen, W. M., Chiang, Y. R., and Tang, S. L. (2016). Genomic insight into the host-endosymbiont relationship of Endozoicomonas montiporae CL-33(T) with its coral host. Front. Microbiol. 7:251. doi: 10.3389/fmicb.2016.00251
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elshahed, M. S., Youssef, N. H., Spain, A. M., Sheik, C., Najar, F. Z., Sukharnikov, L. O., et al. (2008). Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74, 5422–5428. doi: 10.1128/AEM.00410-08
Fautin, D. G., and Buddemeier, R. W. (2004). Adaptive bleaching: a general phenomenon. Hydrobiology 530, 459. doi: 10.1007/s10750-004-2642-z
Giovannoni, S. J., and Stingl, U. (2005). Molecular diversity and ecology of microbial plankton. Nature 437, 343–348. doi: 10.1038/nature04158
Hamp, T. J., Jones, W. J., and Fodor, A. A. (2009). Effects of experimental choices and analysis noise on surveys of the “rare biosphere”. Appl. Environ. Microbiol. 75, 3263–3270. doi: 10.1128/AEM.01931-08
Hernandez-Agreda, A., Gates, R. D., and Ainsworth, T. D. (2016). Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 25, 125–140. doi: 10.1016/j.tim.2016.11.003
Hong, M. J., Yu, Y. T., Chen, C. A., Chiang, P. W., and Tang, S. L. (2009). Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Appl. Environ. Microbiol. 75, 7797–7806. doi: 10.1128/AEM.01418-09
Horinouchi, M., Kasuga, K., Nojiri, H., Yamane, H., and Omori, T. (1997). Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol. Lett. 155, 99–105. doi: 10.1111/j.1574-6968.1997.tb12692.x
Katharios, P., Seth-Smith, H. M. B., Fehr, A., Mateos, J. M., Qi, W., Richter, D., et al. (2015). Environmental marine pathogen isolation using mesocosm culture of sharpsnout seabream: striking genomic and morphological features of novel Endozoicomonas sp. Sci. Rep. 5:17609. doi: 10.1038/srep17609
Klaus, J. S., Frias-Lopez, J., Bonheyo, G. T., Heikoop, J. M., and Fouke, B. W. (2005). Bacterial communities inhabiting the healthy tissues of two Caribbean reef corals: inter-specific and spatial variation. Coral Reefs 24, 129–137. doi: 10.1007/s00338-004-0447-1
Klaus, J. S., Janse, I., Heikoop, J. M., Sanford, R. A., and Fouke, B. W. (2007). Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9, 1291–1305. doi: 10.1111/j.1462-2920.2007.01249.x
Knowlton, N., and Rohwer, F. (2003). Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 162, S51–S62. doi: 10.1086/378684
Kolde, R. (2015). Pheatmap: Pretty Heatmaps. R Package Version 1.0. 2. Available at: https://CRAN.R-project.org/package=pheatmap.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kvennefors, E. C., Sampayo, E., Ridgway, T., Barnes, A. C., and Hoegh-Guldberg, O. (2010). Bacterial communities of two ubiquitous great barrier reef corals reveals both site and species-specificity of common bacterial associates. PLoS ONE 5:e10401. doi: 10.1371/journal.pone.0010401
La Rivière, M., Roumagnac, M., Garrabou, J., and Bally, M. (2013). Transient shifts in cacterial communities associated with the temperate gorgonian Paramuricea clavata in the Northwestern Mediterranean Sea. PLoS ONE 8:e57385. doi: 10.1371/journal.pone.0057385
Lee, O. O., Yang, J., Bougouffa, S., Wang, Y., Batang, Z., Tian, R., et al. (2012). Spatial and species variations in bacterial communities associated with corals from the Red Sea as revealed by pyrosequencing. Appl. Environ. Microbiol. 78, 7173–7184. doi: 10.1128/AEM.01111-12
Lema, K. A., Bourne, D. G., and Willis, B. L. (2014). Onset and the establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23, 4682–4695. doi: 10.1111/mec.12899
Lema, K. A., Willis, B. L., and Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Li, J., Chen, Q., Long, L. J., Dong, J. D., Yang, J., and Zhang, S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 4:7320. doi: 10.1038/srep07320
Li, J., Chen, Q., Zhang, S., Huang, H., Yang, J., Tian, X. P., et al. (2013). Highly heterogeneous bacterial communities associated with the South China Sea reef corals Porites lutea, Galaxea fascicularis and Acropora millepora. PLoS ONE 8:e71301. doi: 10.1371/journal.pone.0071301
Littman, R. A., Willis, B. L., Pfeffer, C., and Bourne, D. G. (2009). Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68, 152–163. doi: 10.1111/j.1574-6941.2009.00666
Liu, Z. H., Chen, W., Gao, L., Zhao, J. J., Ren, C. H., Hu, C. Q., et al. (2012). Analysis of bacteria associated with Acropora solitaryensis by culture-dependent and -independent methods. Afr. J. Microbiol. Res. 6, 5205–5209.
Lynch, M. D., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Mahmoud, H. M., and Kalendar, A. A. (2016). Coral-associated actinobacteria: diversity, abundance, and biotechnological potentials. Front. Microbiol. 7:204. doi: 10.3389/fmicb.2016.00204
Marhaver, K. L., Edwards, R. A., and Rohwer, F. (2008). Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10, 2277–2286. doi: 10.1111/j.1462-2920.2008.01652
McKew, B. A., Dumbrell, A. J., Daud, S. D., Hepburn, L., Thorpe, E., Mogensen, L., et al. (2012). Characterization of geographically distinct bacterial communities associated with coral mucus produced by Acropora spp. and Porites spp. Appl. Environ. Microbiol. 78, 5229–5237. doi: 10.1128/AEM.07764-11
Mendoza, M., Güiza, L., Martinez, X., Caraballo, X., Rojas, J., Aranguren, L. F., et al. (2013). A novel agent (Endozoicomonas elysicola) responsible for epitheliocystis in cobia Rachycentrum canadum larvae. Dis. Aquat. Organ. 106, 31–37. doi: 10.3354/dao02636
Morrow, K. M., Moss, A. G., Chadwick, N. E., and Liles, M. R. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. doi: 10.1128/AEM.01162-12
Mouchka, M. E., Hewson, I., and Harvell, C. D. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 50, 662–674. doi: 10.1093/icb/icq061
Neave, M. J., Michell, C. T., Apprill, A., and Voolstra, C. R. (2014). Whole-genome sequences of three symbiotic endozoicomonas strains. Genome Announc. 2, e00802–14. doi: 10.1128/genomeA.00802-14
Neave, M. J., Michell, C. T., Apprill, A., and Voolstra, C. R. (2017a). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7:40579. doi: 10.1038/srep40579
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2017b). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Nithyanand, P., Manju, S., and Pandian, S. K. (2011). Phylogenetic characterization of culturable actinomycetes associated with the mucus of the coral Acropora digitifera from Gulf of Mannar. FEMS Microbiol. Lett. 314, 112–118. doi: 10.1111/j.1574-6968.2010.02149.x
Pantos, O., Cooney, R. P., Le Tissier, M. D., Barer, M. R., O’Donnell, A. G., and Bythell, J. C. (2003). The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5, 370–382. doi: 10.1046/j.1462-2920.2003.00427.x
Reis, A. M., Araújo, S. D. Jr., Moura, R. L., Francini-Filho, R. B., Pappas, G. Jr., Coelho, A. M., et al. (2009). Bacterial diversity associated with the Brazilian endemic reef coral Mussismilia braziliensis. J. Appl. Microbiol. 106, 1378–1387. doi: 10.1111/j.1365-2672.2008.04106.x
Rohwer, F., Breitbart, M., Jara, J., Azam, F., and Knowlton, N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20, 85–91. doi: 10.1007/s003380100138
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Santiago-Vázquez, L. Z., Brück, T. B., Brück, W. M., Duque-Alarcón, A. P., McCarthy, P. J., and Kerr, R. G. (2007). The diversity of the bacterial communities associated with the azooxanthellate hexacoral Cirrhipathes lutkeni. ISME J. 1, 654–659. doi: 10.1038/ismej.2007.77
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Seymour, J. R., Simó, R., Ahmed, T., and Stocker, R. (2010). Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345. doi: 10.1126/science.1188418
Shnit-Orland, M., and Kushmaro, A. (2009). Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol. Ecol. 67, 371–380. doi: 10.1111/j.1574-6941.2008.00644.x
Sweet, M., Burn, D., Croquer, A., and Leary, P. (2013). Characterisation of the bacterial and fungal communities associated with different lesion sizes of dark spot syndrome occurring in the coral Stephanocoenia intersepta. PLoS ONE 8:e62580. doi: 10.1371/journal.pone.0062580
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the wew cacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wegley, L., Edwards, R., Rodriguez-Brito, B., Liu, H., and Rohwer, F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x
Yachi, S., and Loreau, M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl Acad. Sci. U.S.A. 96, 1463–1468. doi: 10.1073/pnas.96.4.1463
Yang, S., Sun, W., Tang, C., Jin, L., Zhang, F., and Li, Z. (2013). Phylogenetic diversity of actinobacteria associated with soft coral Alcyonium gracllimum and stony coral Tubastraea coccinea in the East China Sea. Microb. Ecol. 661, 189–199. doi: 10.1007/s00248-013-0205-4
Keywords: long-term survey, microbial composition, corals, Stylophora pistillata, shifts
Citation: Yang S-H, Tseng C-H, Huang C-R, Chen C-P, Tandon K, Lee STM, Chiang P-W, Shiu J-H, Chen CA and Tang S-L (2017) Long-Term Survey Is Necessary to Reveal Various Shifts of Microbial Composition in Corals. Front. Microbiol. 8:1094. doi: 10.3389/fmicb.2017.01094
Received: 06 March 2017; Accepted: 30 May 2017;
Published: 13 June 2017.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
Luisa I. Falcon, National Autonomous University of Mexico, MexicoCopyright © 2017 Yang, Tseng, Huang, Chen, Tandon, Lee, Chiang, Shiu, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sen-Lin Tang, c2x0YW5nQGdhdGUuc2luaWNhLmVkdS50dw==
†These authors have contributed equally to this work and have shared co-first authorship.
‡Deceased 17 June, 2016
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.