- 1National and Regional Joint Engineering Laboratory for Medicament of Zoonosis Prevention and Control, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2Key Laboratory of Zoonosis, Key Laboratory of Animal Vaccine Development, Ministry of Agriculture, Guangzhou, China
- 3Key Laboratory of Zoonosis Prevention and Control of Guangdong Province, Ministry of Agriculture, Guangzhou, China
The H5 subtype virus of Highly Pathogenic Avian Influenza Virus has caused huge economic losses to the poultry industry and is a threat to human health. Until 2010, H5N1 subtype virus was the major genotype in China. Since 2011, reassortant H5N2, H5N6, and H5N8 viruses were identified in domestic poultry in China. The clade 2.3.4.4 H5N6 and H5N8 AIV has now spread to most of China. Clade 2.3.4.4 H5N6 virus has caused 17 human deaths. However, the prevalence, pathogenicity, and transmissibility of the distinct NA reassortment with H5 subtypes viruses (H5Nx) is unknown. We constructed five clade 2.3.4.4 reassortant H5Nx viruses that shared the same HA and six internal gene segments. The NA gene segment was replaced with N1, N2, N6, ΔN6 (with an 11 amino acid deletion at the 58th to 68th of NA stalk region), and N8 strains, respectively. The reassortant viruses with distinct NAs of clade 2.3.4.4 H5 subtype had different degrees of fitness. All reassortant H5Nx viruses formed plaques on MDCK cell monolayers, but the ΔH5N6 grew more efficiently in mammalian and avian cells. The reassortant H5Nx viruses were more virulent in mice as compared to the H5N2 virus. The H5N6 and H5N8 reassortant viruses exhibited enhanced pathogenicity and transmissibility in chickens as compared to the H5N1 reassortant virus. We suggest that comprehensive surveillance work should be undertaken to monitor the H5Nx viruses.
Introduction
Influenza A virus (IAV) belongs to the Orthomyxoviridae family of RNA viruses that have negative-sense, single-stranded, and segmented RNA genomes. IAVs are classified into subtypes based on antibody responses to the two major viral surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (Yoon et al., 2014). Sixteen HA subtypes (H1–H16) and nine NA subtypes (N1–N9) are circulating in birds (Fouchier et al., 2005). The A/goose/Guangdong/1/1996 (H5N1) subtype virus was first isolated in Guangdong Province of China in 1996 and has posed threat to public health (Duan et al., 2007). To date, 856 people have been infected with the H5N1 subtype virus and 452 people have died since 20031. IAV continues to evolve and clade 2.3.4.4 IAVs with other NA subtypes (N2, N6, and N8) are now prevalent (Lee et al., 2015; Pasick et al., 2015; Su et al., 2015; Claes et al., 2016). H5N2 and H5N8 viruses spread from migratory birds to poultry in multiple continents in 2014 (Verhagen et al., 2015; Lycett et al., 2016). Humans can be infected with the H5N6 subtype avian influenza virus (Chen and Zhang, 2015; Yang et al., 2015). These H5Nx viruses (H5N1, H5N2, H5N6, and H5N8) seriously threaten human health and the poultry industry.
Influenza A virus attaches to cellular membranes via HA interacting with sialic acids expressed at the terminal position of carbohydrate chains of cell surface glycoproteins and glycolipids (Matrosovich et al., 1997). NA activity catalyzes the removal of sialic acids from viral and cellular components, promotes the release of viral progeny, and prevents virion aggregation (Ruangrung et al., 2016). NA reassortment with H5 subtype viruses in the evolution of H5N1 avian influenza virus leads to novel combinations of H5Nx (H5N2, H5N6, and H5N8) subtypes (Claes et al., 2016). The balance between HA and NA proteins contributes to expression of the HPAI phenotype (Diederich et al., 2015). Furthermore, the NA gene has been shown to play an important role in influenza virus enzyme activity, transmission, and pathogenicity (Lv et al., 2015; Stech et al., 2015; Ranadheera et al., 2016).
Structural analyses of the H5Nx NAs (N1, N2, N6, and N8) indicated that the overall NA structures are very similar (Yang et al., 2016). Using next-generation sequencing (NGS), virus isolation methods and genetic evolution, H5N6 has been confirmed to have replaced H5N1 as one of the dominant subtypes among poultry in southern China (Bi et al., 2016; Li et al., 2017). In addition, an analysis of how 2.3.4.4 H5Nx viruses had spread in the geographical and environmental space was carried out using Boosted Regression Tree (BRT) models (Dhingra et al., 2016). However, the biological characteristics of distinct NAs and functional comparisons with the 2.3.4.4 H5 HA subtype viruses are not well clarified. To study the pandemic potential, NA biological characteristics, and genetic interactions between HA and NA of the clade 2.3.4.4 H5Nx virus, we constructed and rescued five reassortant H5Nx viruses. A clade 2.3.4.4 H5N1 virus, A/goose/Guangdong/SH7/2013 was used as the genetic backbone and the N1 gene segment was replaced with N2, N6, ΔN6 (with an 11 amino acid deletion at the 58th to 68th in NA stalk region), and N8 NAs using reverse genetics. We then characterized the biology of these viruses in mammalian and avian cells, determined their virulence in mice and chickens, and characterized their horizontal transmission in chickens.
Materials and Methods
H5Nx Influenza Virus Analyses
All sequences of H5Nx viruses (H5N1, H5N2, H5N6, and H5N8) from 1996 to 2016 in China that were available from the GISAID and GenBank databases were downloaded. And the sequences of H5N6 virus aligned using the MegAlign 6.06 program (DNASTAR) with the ClustalW method.
Viruses and Cells
The clade 2.3.4.4 H5 subtype avian influenza viruses, including A/goose/Guangdong/SH7/2013 (H5N1) (SH7, EPI_ISL_259927), A/duck/Guangdong/673/2014 (H5N6) (673, EPI_ISL_259923), A/duck/Guangdong/674/2014 (H5N6) (674, EPI_ISL_259924), and A/goose/Jiangsu/1306/2014 (H5N8) (Js1306, EPI_ISL_259926) were isolated from sick ducks or geese. The H5N2 avian influenza virus, A/chicken/Hebei/LZF/2014 (H5N2) (LZF, EPI_ISL_259925) was isolated from a sick chicken. All viruses were propagated in 10-day-old SPF embryonated eggs at 37°C and frozen at -80°C. Human embryonic kidney cells (HEK293T), Madin-Darby canine kidney (MDCK), and Chicken Embryo Fibroblast (CEF) cells were grown in Dulbecco’s modified essential medium (DMEM; Gibco) with 10% fetal bovine serum (FBS; BI) and 1% penicillin/streptomycin (Gibco).
Plasmids Construction and Generation of Reassortant Viruses
N1 gene segments were replaced with NAs from LZF, 673, 674, and Js1306, respectively, and were cloned into a pHW2000 plasmid system according to a previous study (Hoffmann et al., 2000; Xiao et al., 2016). HEK293T cells monolayers in 6-well plates were transfected at 80–90% confluency with 4 μg of the eight plasmids (500 ng of each plasmid) by using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. Six hours later, the mixture was replaced with Opti-MEM (Gibco) containing 0.2% bovine serum albumin and 1 μg/ml TPCK trypsin (Sigma). After 48 h, the supernatant was harvested and injected into SPF embryonated eggs for virus propagation. Viruses were titrated in embryonated eggs using hemagglutination assays and sequenced. The five reassortant H5Nx viruses were named rH5N1, rH5N2, rH5N6, ΔrH5N6, and rH5N8.
Virus Plaque Assay
The plaque assay was adapted from previously described procedure (Tian et al., 2012). MDCK cells were grown in DMEM with 1 mM L-glutamine and 10% FBS and seeded onto 6-well plates. Confluent monolayers were washed twice with phosphate-buffered saline (PBS) and infected with serial 10-fold dilutions of the virus at 37°C. After 2 h of incubation, the cells were washed twice with PBS and then overlaid with MEM containing 1% agarose. After 48–72 h of incubation at 37°C, the agarose was removed and cells were stained with 0.5% crystal violet in 10% formaldehyde solution. The plaques were visualized and manually counted.
Hemagglutination and Hemagglutination-Elution Assays
The balance between HA binding and NA cleavage was measured by performing hemagglutination assays as previously described (Jean-Sébastien Casalegno, 2014). Viruses were prepared using serial twofold dilutions in 50 μl of PBS in 96-well plates and incubated with 50 μl of 0.8% guinea pig erythrocytes at 4°C for 75 min and then at 37°C for 2 h. HA titers were recorded at both temperatures, respectively. Each sample was assayed in triplicate.
Neuraminidase Activity Assay
A fluorescent substrate 2′ (4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA; Sigma) was used to measure NA enzymatic activity, according to the manufacturer’s recommendations. Viruses were prepared using serial twofold dilutions in 50 μl of calcium saline buffer in 96-well plates and 50 μl of 200 μmol MUN was added to each well, and then incubated for 60 min at 37°C in darkness. One hour later, stop solution was added and NA activity was quantified using a Synergy HT Multi-Detection microplate reader (BioTek, Winooski, VT, United States) with excitation and emission wavelength of 360 and 440 nm.
Virus Growth Kinetics
Confluent MDCK cells were infected at a multiplicity of infection (MOI) of 0.001 TCID50/cell, CEF cells were infected at MOI of 0.0001 TCID50/cell for 1 h at 37°C on 6-well plates, as described previously (Xiao et al., 2016). One hour later, plates were washed twice with PBS, and then incubated with DMEM containing 0.2% BSA at 37°C or 39°C with 5% CO2. Culture supernatants were collected at 12, 24, 36, and 48 hour post-inoculation (h.p.i.). The virus titers were determined by performing 50% tissue culture infective dose (TCID50) assays in MDCK cells.
Animal Experiments
Groups of eight female 4- to 6-week-old BALB/c mice (Vitalriver Company, Beijing, China) were anesthetized with CO2 and inoculated intranasally (i.n.) with 105 EID50/50 μl of the five reassortant H5Nx viruses (rH5N1, rH5N2, rH5N6, ΔrH5N6, and rH5N8). Body weight and clinical symptoms were monitored daily for 14 days after infection. The mice were euthanized if they lost more than 25% of their initial body weight. Three mice from each group were euthanized on 4 days post-inoculation (d.p.i.), and the lung, brain and turbinate tissues were collected to determine the virus titers by EID50 assay. The 50% mouse lethal dose (MLD50) were determined by inoculating groups of five mice with serial 10-fold dilutions of virus ranging from 101 to 105 EID50/50 μl. The MLD50 values were calculated using the Reed and Muench method.
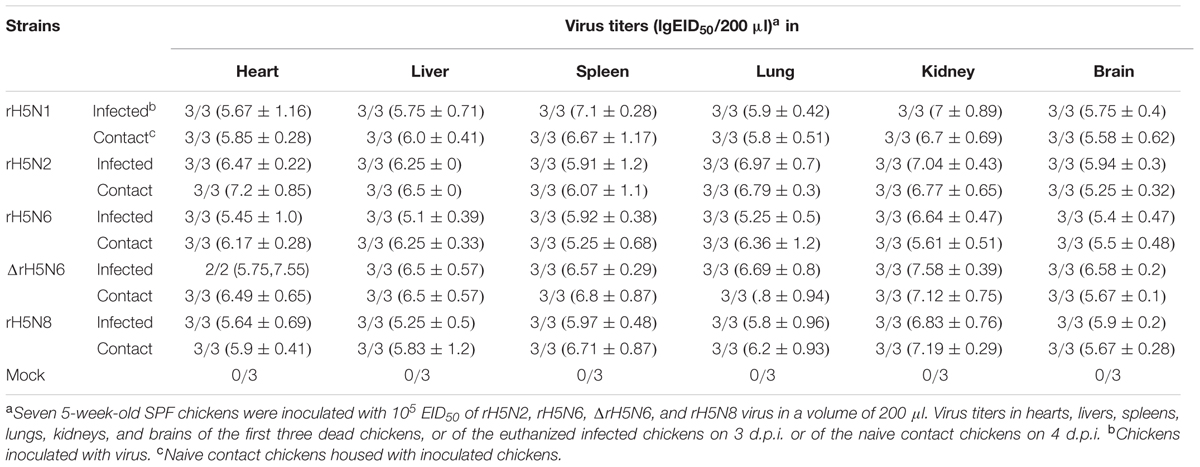
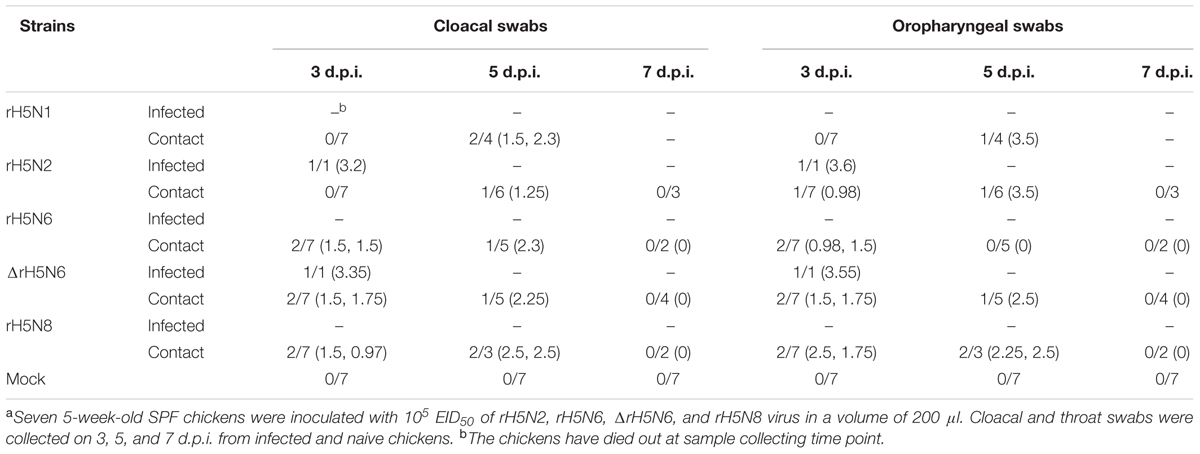
Groups of seven 5-week-old SPF chickens (Merial Vital Company, Beijing, China) were inoculated intranasally with 105 EID50/200 μl of five reassortant H5Nx viruses, respectively. Seven uninfected chickens served as negative controls. All chickens were observed for clinical symptoms for 14 days. On 1 d.p.i., seven naive chickens (contact) were mixed with treatment group chickens. Cloacal and throat swabs were collected on 3, 5, and 7 d.p.i. from infected and naive chickens. Three inoculated chickens in each group were euthanized on 3 d.p.i. to test the virus replication in organs, including heart, lung, kidney, brain, spleen, and liver. Similar actions were performed on chickens if they died within 3 days. For the chickens in the naive contact group, the virus replication in heart, lung, kidney, brain, spleen, and liver were detected after the chickens died.
Ethics Statement and Biosafety
All experiments were carried out in ABSL-3 facilities in compliance with the biosafety committee of South China Agriculture University (SCAU) protocols. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at SCAU and were carried out in accordance with the approved guidelines.
Results
Evolution and Epidemiology of H5Nx Influenza Viruses in China
Until 2010, H5N1 subtype virus was the major genotype in China (Figure 1A). The H5N2 and H5N8 viruses were sporadically reported in China after 2011. The H5N6 virus was isolated in 2013 and became a major genotype in China. The clade 2.3.4.4 viruses were detected in most regions of China from 2013 to 2016 (Figures 1B,C). In addition, H5N2 virus was mainly isolated in northern China. The NA6 stalk region can be divided into two groups according to the presence of an amino acid deletion in the positions 58–68 of NA stalk region. The percentage of 11 amino acids deletion in NA stalk region of H5N6 viruses were 63% (24 of 38 viruses), 37% (69 of 186 viruses), 87% (359 of 408 viruses), and 100% (23 of 23 viruses) respectively, from 2013 to 2016 (Figure 1C). Seventeen H5N6 strains of clade 2.3.4.4 have caused human infections2. Sixteen viruses had the 11 amino acid deletion at the 58th to 68th in the NA stalk region. The H5N8 virus has become the pandemic genotype in 2016 (Figure 1C).
FIGURE 1. Epidemiology and distribution of H5Nx avian influenza viruses in China. (A) Distribution of H5Nx AIVs in China 1996–2010. (B) Distribution of H5Nx AIVs in China 2011–2016. (C) The isolation rate of H5Nx viruses isolated in China. Data are from NCBI (https://www.ncbi.nlm.nih.gov/genome/viruses/variation/flu/) and GISAID (http://platform.gisaid.org/epi3/frontend). Colors depict different virus subtypes. H5N1, H5N2, H5N6, ΔrH5N6, and H5N8 are shown in red, purple, green, gray, and black, respectively.
HA and NA Properties of H5Nx Reassortant Viruses
To directly measure the balance between HA and NA, the loss of HA titer at 37°C was compared to the erythrocyte agglutination rate at 4°C. The binding and release of five reassortant viruses were quantitatively measured via guinea pig erythrocytes. At 4°C, the NA is inactive while HA can bind to its receptors and agglutinate the erythrocytes. At 37°C, the NA becomes active; thus, the loss of HA titer evaluates the equilibrium between HA affinity and NA activity. HA titers were reduced by five- and sixfold for the rH5N6 and rH5N8 viruses, respectively. The rH5N1, rH5N2, and ΔrH5N6 viruses did not show any change of HA titer at either temperature (Figure 2A). 2′ (4-methylumbelliferyl)-α-D-N-acetylneuraminic acid was used to evaluate the NA activities of five H5Nx reassortant virus (Potier et al., 1979). The rH5N2 and rH5N8 viruses had RFU values, and the rH5N6 and ΔrH5N6 viruses had lower activities, as compared to rH5N1 (Figure 2B).
FIGURE 2. Hemagglutinin (HA)-neuraminidase (NA) properties and growth kinetics of H5Nx reassortant viruses. (A) Elution assays of guinea pig RBCs. H5Nxreassortant viruses were serially diluted in phosphate-buffered saline (PBS) and incubated with 0.8% RBC suspensions at 4°C for 75 min, then 37°C for 2 h. And then the loss of HA activity was recorded. Each virus was analyzed in triplicate wells. (B) NA activity was measured by a solution assay using the fluorescence substrate 4-MU-NANA. NA activity was represented by the fluorescence intensity represent the NA activity (∗∗P < 0.01, ∗∗∗P < 0.001). (C) Madin-Darby canine kidney (MDCK) cells were inoculated with viruses at an multiplicity of infection (MOI) of 0.001. (D) Chicken Embryo Fibroblast (CEF) was inoculated with viruses at an MOI of 0.0001. Samples were collected at 12, 24,36, and 48 hour post-inoculation (h.p.i.). Virus titers were determined by using the 50% tissue culture infectious dose (TCID50) assay. The virus titers are means ± standard deviations (SD) (n = 3), Statistical significance was analyzed using an unpaired t-test: ∗P < 0.05, ∗∗P < 0.01.
Plaque-Forming Ability and Replication Kinetics of Reassortant H5Nx Viruses
We then characterized the cleavage properties of the HA protein combined with different NAs by performing plaque assays. All reassortant H5Nx viruses formed plaques on MDCK cells without TPCK trypsin. MDCK cells infected with rH5N8 virus grew the largest plaques (1.80 mm) when compared to rH5N6 and ΔrH5N6 viruses. The rH5N1 virus and rH5N2 virus formed pin-point plaques in MDCK cells (from 0.48 to 0.73 mm) (Figure 3).
FIGURE 3. Plaque-forming ability of H5Nx reassortant viruses in MDCK cell monolayers. Cells were infected with rH5N1, rH5N2, rH5N6, ΔrH5N6, and rH5N8 virus. At 2 h.p.i., cells were washed twice with PBS and then overlaid with MEM containing 1% agarose. After 48–72 h of incubation at 37°C, plaques were counted.
The growth kinetics of reassortant H5Nx viruses on mammalian and avian cells (MDCK and CEF) were determined. In MDCK cells, the rH5N1 and ΔrH5N6 viruses replicated more efficiently and had the highest virus titers at 12, 24, and 36 h. By contrast, rH5N2 had the lowest virus titer at each time point. The rH5N6 and rH5N8 reassortant viruses exhibited an intermediate level of virus titers among H5Nx reassortant viruses. Growth kinetic of rH5N8 was performed using a significantly higher titer when compared to other reassortant H5Nx viruses at 48 h. And the H5N2 virus also appeared to surpass the rH5N6 virus at 48 h (Figure 2C). In avian CEF cells, ΔrH5N6 replicated more efficiently than the other reassortant H5Nx viruses. rH5N2 had the lowest virus titers at each investigated time point, except at 12 h. rH5N8 growth kinetics were similar to rH5N1 but higher than rH5N6 (Figure 2D). These results showed that the ΔrH5N6 grew efficiently in MDCK and CEF cells. In contract, rH5N2 virus had lower titer in these cells.
Pathogenicity of H5Nx Reassortant Viruses in Mice
To investigate whether these five H5Nx reassortant virus also replicate efficiently in mammalian hosts, BALB/c mice were infected with 105 EID50/50 μl of these H5Nx reassortant viruses. All viruses were lethal to mice with 2 d.p.i., except for rH5N2 (at 5 d.p.i.). Mice infected with rH5N1, rH5N6, ΔrH5N6, and rH5N8 died within 6 d.p.i., whereas mice infected with rH5N2 showed delayed fatality on 8 d.p.i. (Figures 4A,B). All viruses replicated efficiently in lung and nasal tissues. As H5N1 IAV can cause systemic infection, neurotropism, and long-term effects on the central nervous system (CNS) (Shinya et al., 2011), the replication of H5Nx reassortant viruses in mice brain was tested. The rH5N1, rH5N6, and rH5N8 virus were found in all tested mice brains, while rH5N2 and ΔrH5N6 were only detected in 2/3 or 1/3 mice, respectively (Figures 4C–E). Subsequently, to study the virulence of H5Nx reassortant viruses, BALB/c mice were inoculated with serial 10-fold dilutions of the virus to determine viral MLD50. The MLD50 (lgEID50) of rH5N1, rH5N6, ΔrH5N6, and rH5N8 were 2.5, 3.3, 2.68, 3.16, and 2.5, respectively.
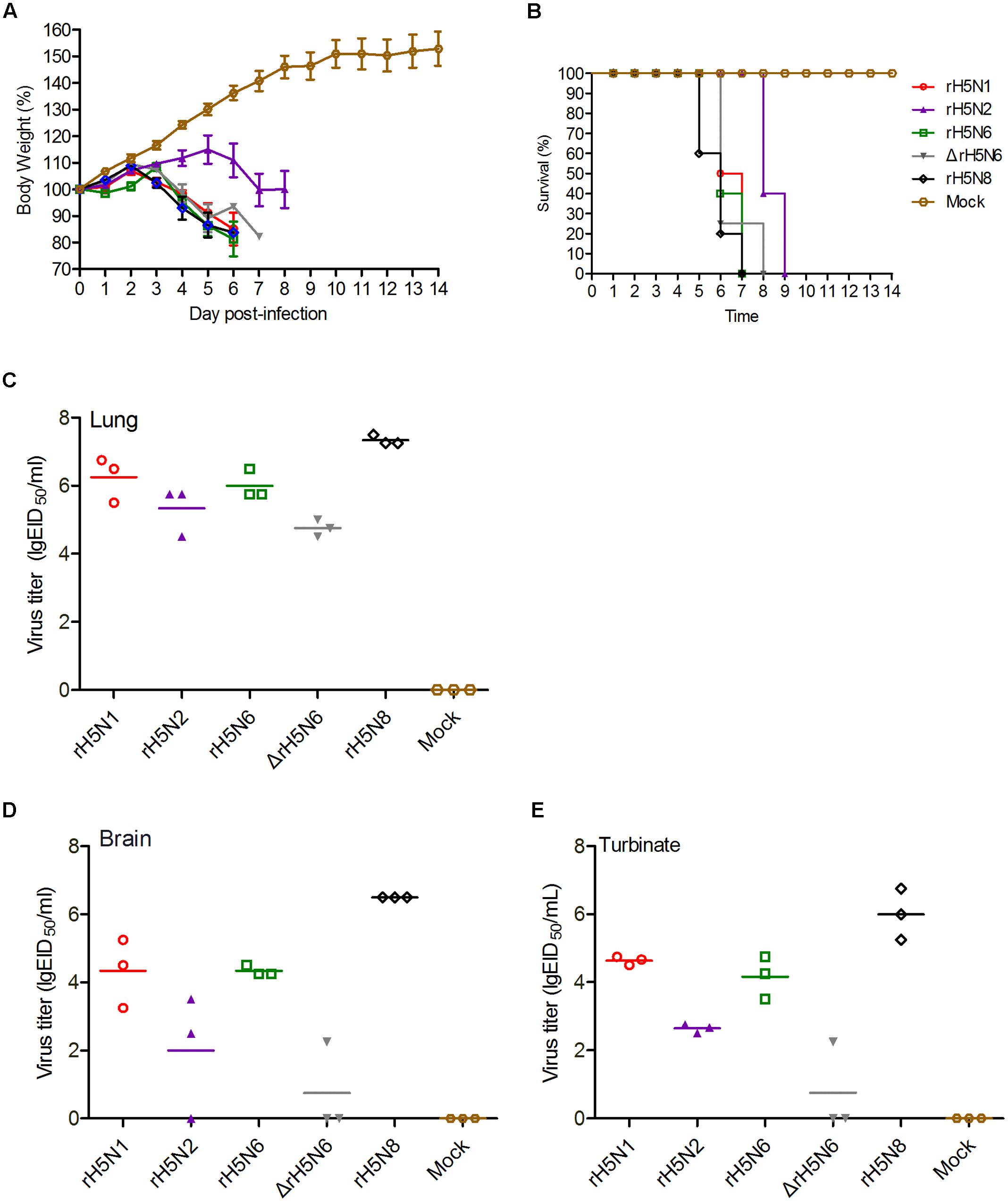
FIGURE 4. Pathogenicity of the of H5Nx reassortant viruses in mice. Groups of eight female BALB/c mice were intranasally inoculated with 105 EID50/50 μl of the indicated viruses or with PBS as a negative control. Mouse body weight (A) and survival (B) were monitored daily for 14 days. Mice that lost more than 25% of their initial weight were euthanized. The percentage weight from each group and each time point are presented as means ± SD. The titers of viruses were detected in organs lung (C), brain (D), turbinate (E). Three mice from each group were euthanized on 4 days post-inoculation (d.p.i.).
Pathogenicity and Transmission of H5Nx Reassortant Viruses in Chickens
Groups of seven 5-week-old SPF chickens were inoculated intranasally with 105 EID50/50 μl of the five H5Nx reassortants. Chickens infected with rH5N2 and ΔrH5N6 died within 4 d.p.i. ΔrH5N6, rH5N6, and rH5N8 viruses caused sudden death without symptoms of illness on 2 d.p.i. (Figure 5A). Reassortant viruses were isolated from lung, spleen, kidney, and brain samples at high virus titers. All H5Nx reassortant viruses replicated in lungs, with mean titers from 5.8 to 6.97 lgEID50. The ΔrH5N6 virus replicated highly in brains, with a mean titer of 6.58 lgEID50 (Table 1).
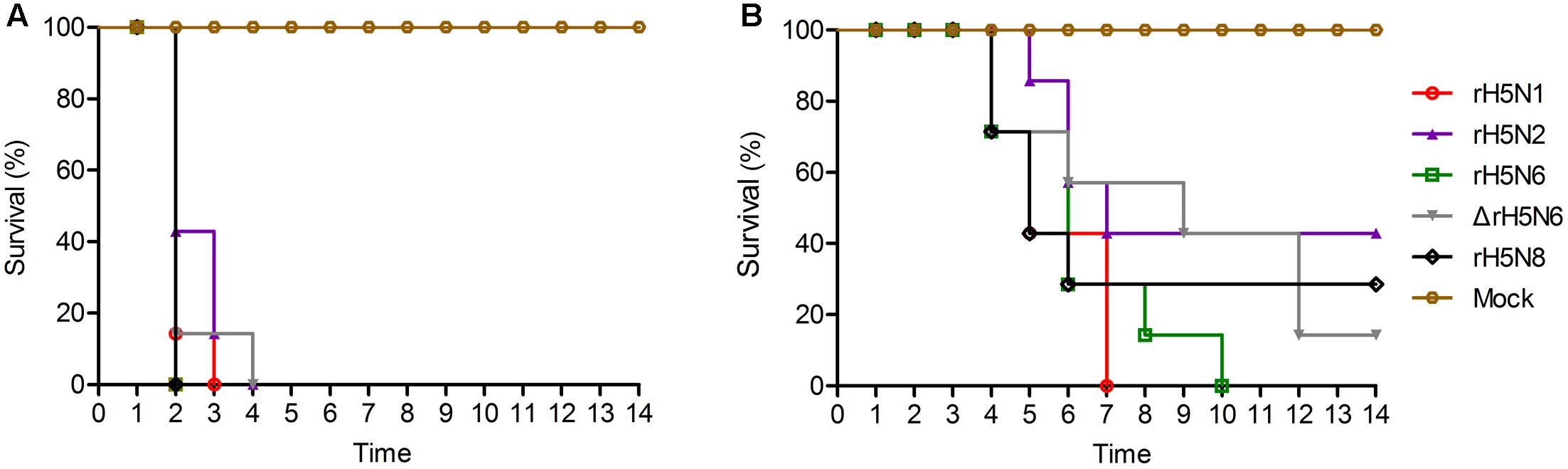
FIGURE 5. Pathogenicity and transmission of H5Nx reassortant viruses in chickens. Groups of seven 5-week-old SPF chickens were inoculated intranasally with 105 EID50/200 μl of the indicated viruses. Seven chickens were used as negative controls. (A) Survival rate of chickens infected with H5Nx reassortant viruses. (B) Survival rate of chickens co-housed with the animals infected indicated virus. All chickens were observed for clinical symptoms for 14 days.
To evaluate the transmission of five H5Nx reassortants in chickens, after 1 d.p.i., seven SPF chickens were co-housed with infected chickens. The naive contact chickens co-housed with chickens infected with rH5N1, rH5N6, ΔrH5N6, or rH5N8 viruses infection died at 4 d.p.i. Naïve chickens co-housed with chickens infected with the rH5N2 virus died on 5 d.p.i. (Figure 5B). No seroconversion was found in surviving contact animals. Oropharyngeal and cloacal swabs from contact chickens were tested at 3 d.p.i. or 5 d.p.i. No virus was detected in oropharyngeal among contact chickens at 7 d.p.i. (Table 2). The virus was detected in the lung, spleen, kidney, and brain of contact chickens (Table 1). Overall, the H5Nx reassortant viruses were highly pathogenic to chickens, and rH5N6, ΔrH5N6, and rH5N8 exhibited enhanced pathogenicity and greater transmissibility in chickens.
Discussion
The novel clade 2.3.4.4 H5Nx (H51N1, H5N2, H5N6, and H5N8) viruses present in migrating wild aquatic birds contributes to the circulation of avian influenza viruses. More recently, structural and biochemical analysis of clade 2.3.4.4 H5Nx virus as well as the emergence, spread, and persistence of H5Nx viruses have been well annotated (Claes et al., 2016; Sun et al., 2016; Yang et al., 2016). Our study focused on the pandemic H5Nx viruses of clade 2.3.4.4 embodying different NA genes and described the basic characteristics of viral virulence and transmission.
The NA activity balance HA activity was beneficial to viral entry and exit (Gen et al., 2013). NA with a stalk region that varies in length influences its enzymatic function and showed the different pathogenicity in chicken and mice (Zhou et al., 2009). In our study, ΔrH5N6 reassortant viruses was with the 11 amino acid deletion at the 58th to 68th in NA stalk region. In addition, previous studies indicated that the different NA genes with not the same pathogenicity of H9 influenza virus (Yan et al., 2016). The functional match between HA and NA is crucial to virus adaptation and evolution (Xu et al., 2012; Gaymard et al., 2016). rH5N2 and rH5N8 had higher NA activity as compared to the other H5Nx reassortants, while rH5N6 was inactive. Our results suggest that rH5N6 and rH5N8 may have a stable fitness between their HA and NA activities, which might explain why they have become prevalent viruses in China.
Hemagglutinin cleavage is a major determinant of avian influenza virus virulence (Horimoto and Kawaoka, 2001). Eurasian A(H5N8), North American A (H5N8), and North American A (H5N2) viruses were as susceptible to HA cleavage as HPAI H5N1 (Kaplan et al., 2016). The ability of rH5N2, rH5N6, ΔrH5N6, and rH5N8 viruses to form plaques were enhanced, as compared to rH5N1. Together, these data suggested that clade 2.3.4.4 HA of H5 subtypes combined with N2, N6, or N8 was beneficial to viral fitness. Furthermore, HA of H5 subtype combined with N6 and N8 virus grew more efficiently than N2, which demonstrated that the match of HA/NA enhanced virus replication, consistent with previous reports that HA and NA balance has a great impact on viral growth (Yen et al., 2011; Diederich et al., 2015).
H5N2 and H5N6 viruses are known to have increased virulence and replication efficiency in mice (Li et al., 2014, 2015). In our study, rH5N1, rH5N6, ΔrH5N6, and rH5N8 were highly virulent, while rH5N2 showed moderate virulence in mice (Katz et al., 2000). Virus titers were detected in the lungs of mice infected with the H5Nx reassortant viruses and organs from mice infected rH5N8 showed high titers. The ΔrH5N6 virus did not replicate in mice brain and nasal cavities. The H5N6 virus which has caused human infections has an 11 amino acids deletion at the 58th to 68th in the NA stalk region. However, the influence of this deletion in NA stalk region on virus transmission needs to be investigated further.
Our findings indicated that rH5N6, ΔrH5N6, and rH5N8 were more pathogenic than rH5N1, and rH5N2 was less efficiently transmitted to contact chickens. These data suggest that N2 is likely not matched to the clade 2.3.4.4. HA and thus may explain why 2.3.4.4 H5N2 subtype viruses are not prevalent in China.
The rH5N6 virus was highly pathogenic to contact chickens, which is consistent with previous studies (Jiao et al., 2016). In addition, a virus created with short stalk NA showed adaptation and virulence enhancement of waterfowl influenza viruses among chickens (Munier et al., 2010). The ΔrH5N6 virus also replicated effectively in MDCK and CEF cells. Thus, our results suggest a possibility that high mortality of H5N6 with long-stalk NA on hosts limits the transmission and epidemics in chickens. Chickens inoculated with rH5N8 had severe infections, yet this virus was only poorly transmitted to contact chickens. The H5N8 virus occurs sporadically in reassortment events, but does not circulate as widely as the H5N1 subtype virus in China. However, the clade 2.3.4.4 H5N8 virus has spread around the world along two main long-distance migration routes and genetic analyses have shown close phylogenetic relationships among the H5N8 viruses in Asia and North America. This suggests future outbreak possibilities for the 2.3.4.4 H5N8 (Lycett et al., 2016).
In summary, we show that distinct NAs reassortments with clade 2.3.4.4 H5N1 subtype viruses are important for virus replication, pathogenicity, and transmission. The N6 and N8 NAs possess a fitness balance between HA and NA activity, which contributes to the high pathogenicity of H5N8 and H5N6 in mice and chickens. H5N2 virus doses not efficiently infect chickens by direct contact. H5Nx viruses constitute a threat to poultry and public health. Although a reassortant vaccine matched with clade 2.3.4.4 has been developed and used in poultry, more comprehensive surveillance work also should conducted.
Author Contributions
YY, ZaZ, WQ, and ML conceived and designed the experiments; YY, ZaZ, and HL performed epidemiological investigation; YY, ZaZ, HL, XW, BL, XR, ZhZ, XZ, SL, and PH performed the animal experiments; YY, ZaZ, WQ, and ML contributed analysis; YY, ZaZ, WQ, and ML drafted the manuscript. All authors reviewed and revised the first and final drafts of this manuscript.
Funding
This work was partially supported by National Natural Science Foundation of China (31672586, U1501212, 31272563), Program for National Broiler Industry (CARS-42-G09), Natural Science Foundation of Guangdong Province (2015A030313422), Science and Technology Planning Project of Guangdong Province (2013B020224), and South China Co-innovation Center for Prevention and Control of Poultry Infectious Diseases and Product Safety, Guangzhou, 510642. WQ and ML is supported by “Special Support Plan of Guangdong Province in Science and Technology for Talents.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s and Genbank’s EpiFlu Database on which this research is based. We also thank Dr. Philip R. Hardwidge (Associated professor, College of Veterinary Medicine, Kansas State University, Manhattan, KS, United States) for editing the manuscript.
Footnotes
- ^ http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/
- ^ http://www.who.int/en/
References
Bi, Y., Chen, Q., Wang, Q., Chen, J., Jin, T., Wong, G., et al. (2016). Genesis, evolution and prevalence of H5N6 avian influenza viruses in china. Cell Host Microbe 20, 810–821. doi: 10.1016/j.chom.2016.10.022
Chen, T., and Zhang, R. (2015). Symptoms seem to be mild in children infected with avian influenza A (H5N6) and other subtypes. J. Infect. 71, 702–703. doi: 10.1016/j.jinf.2015.09.004
Claes, F., Morzaria, S. P., and Donis, R. O. (2016). Emergence and dissemination of clade 2.3.4.4 H5Nx influenza viruses-how is the Asian HPAI H5 lineage maintained. Curr. Opin. Virol. 16, 158–163. doi: 10.1016/j.coviro.2016.02.005
Dhingra, M. S., Artois, J., Robinson, T. P., Linard, C., Chaiban, C., Xenarios, I., et al. (2016). Global mapping of highly pathogenic avian influenza H5N1 and H5Nx clade 2.3.4.4 viruses with spatial cross-validation. Elife 5:e19571. doi: 10.7554/eLife.19571
Diederich, S., Berhane, Y., Embury-Hyatt, C., Hisanaga, T., Handel, K., Cottam-Birt, C., et al. (2015). Hemagglutinin-neuraminidase balance influences the virulence phenotype of a recombinant H5N3 influenza A virus possessing a polybasic HA0 cleavage site. J. Virol. 89, 10724–10734. doi: 10.1128/jvi.01238-15
Duan, L., Campitelli, L., Fan, X. H., Leung, Y. H. C., Vijaykrishna, D., Zhang, J. X., et al. (2007). Characterization of low-pathogenic H5 subtype influenza viruses from eurasia: implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 81, 7529–7539. doi: 10.1128/JVI.00327-07
Fouchier, R. A. M., Munster, V., Wallensten, A., Bestebroer, T. M., Herfst, S., Smith, D., et al. (2005). Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79, 2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005
Gaymard, A., Le Briand, N., Frobert, E., Lina, B., and Escuret, V. (2016). Functional balance between neuraminidase and hemagglutinin in influenza viruses. Clin. Microbiol. Infect. 22, 975–983. doi: 10.1016/j.cmi.2016.07.007
Gen, F., Yamada, S., Kato, K., Akashi, H., Kawaoka, Y., and Horimoto, T. (2013). Attenuation of an influenza A virus due to alteration of its hemagglutinin-neuraminidase functional balance in mice. Arch. Virol. 158, 1003–1011. doi: 10.1007/s00705-012-1577-3
Hoffmann, E., Neumann, G., Kawaoka, Y., Hobom, G., and Webster, R. G. (2000). A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U.S.A. 97, 6108–6113. doi: 10.1073/pnas.100133697
Horimoto, T., and Kawaoka, Y. (2001). Pandemic threat posed by avian influenza A viruses. Clin. Microbiol. Rev. 14, 129–149. doi: 10.1128/CMR.14.1.129-149.2001
Jean-Sébastien Casalegno, O. F. V. E. (2014). Functional balance between the hemagglutinin and neuraminidase of influenza A(H1N1)pdm09 HA D222 variants. PLoS ONE 9:e104009. doi: 10.1371/journal.pone.0104009
Jiao, P., Cui, J., Song, Y., Song, H., Zhao, Z., Wu, S., et al. (2016). New reassortant H5N6 highly pathogenic avian influenza viruses in Southern China, 2014. Front. Microbiol. 7:754. doi: 10.3389/fmicb.2016.00754
Kaplan, B. S., Russier, M., Jeevan, T., Marathe, B., Govorkova, E. A., Russell, C. J., et al. (2016). Novel highly pathogenic avian A(H5N2) and A(H5N8) influenza viruses of clade 2.3.4.4 from north america have limited capacity for replication and transmission in mammals. mSphere 1:e00003-16. doi: 10.1128/mSphere.00003-16
Katz, J. M., Lu, X., Tumpey, T. M., Smith, C. B., Shaw, M. W., and Subbarao, K. (2000). Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 74, 10807–10810.
Lee, D.-H., Torchetti, M. K., Winker, K., Ip, H. S., Song, C.-S., and Swayne, D. E. (2015). Intercontinental spread of Asian-origin H5N8 to north america through beringia by migratory birds. J. Virol. 89, 6521–6524. doi: 10.1128/JVI.00728-15
Li, M., Zhao, N., Luo, J., Li, Y., Chen, L., Ma, J., et al. (2017). Genetic characterization of continually evolving highly pathogenic H5N6 influenza viruses in China, 2012-2016. Front. Microbiol. 8:260. doi: 10.3389/fmicb.2017.00260
Li, Q., Wang, X., Zhong, L., Wang, X., Sun, Z., Gao, Z., et al. (2014). Adaptation of a natural reassortant H5N2 avian influenza virus in mice. Vet. Microbiol. 172, 568–574. doi: 10.1016/j.vetmic.2014.06.018
Li, X., Fu, Y., Yang, J., Guo, J., He, J., Guo, J., et al. (2015). Genetic and biological characterization of two novel reassortant H5N6 swine influenza viruses in mice and chickens. Infect. Genet. Evol. 36, 462–466. doi: 10.1016/j.meegid.2015.08.017
Lv, J., Wei, L., Yang, Y., Wang, B., Liang, W., Gao, Y., et al. (2015). Amino acid substitutions in the neuraminidase protein of an H9N2 avian influenza virus affect its airborne transmission in chickens. Vet. Res. 46, 44. doi: 10.1186/s13567-014-0142-3
Lycett, S. J., Bodewes, R., Pohlmann, A., Banks, J., Bányai, K., Boni, M. F., et al. (2016). Role for migratory wild birds in the global spread of avian influenza H5N8. Science 354, 213–217. doi: 10.1126/science.aaf8852
Matrosovich, M. N., Gambaryan, A. S., Teneberg, S., Piskarev, V. E., Yamnikova, S. S., Lvov, D. K., et al. (1997). Avian influenza A viruses differ from human viruses by recognition of Sialyloligosaccharides and Gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233, 224–234. doi: 10.1006/viro.1997.8580
Munier, S., Larcher, T., Cormier-Aline, F., Soubieux, D., Su, B., Guigand, L., et al. (2010). A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J. Virol. 84, 940–952. doi: 10.1128/JVI.01581-09
Pasick, J., Berhane, Y., Joseph, T., Bowes, V., Hisanaga, T., Handel, K., et al. (2015). Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to eurasian H5N8 in British Columbia, Canada, 2014. Sci. Rep. 5:9484. doi: 10.1038/srep09484
Potier, M., Mameli, L., Belisle, M., Dallaire, L., and Melancon, S. B. (1979). Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal. Biochem. 94, 287–296.
Ranadheera, C., Hagan, M. W., Leung, A., Collignon, B., Cutts, T., Theriault, S., et al. (2016). Reduction of neuraminidase activity exacerbates disease in 2009 pandemic influenza virus-infected mice. J. Virol. 90, 9931–9941.
Ruangrung, K., Suptawiwat, O., Maneechotesuwan, K., Boonarkart, C., Chakritbudsabong, W., Assawabhumi, J., et al. (2016). Neuraminidase activity and resistance of 2009 pandemic H1N1 influenza virus to antiviral activity in bronchoalveolar fluid. J. Virol. 90, 4637–4646. doi: 10.1128/jvi.00013-16
Shinya, K., Makino, A., Hatta, M., Watanabe, S., Kim, J. H., Hatta, Y., et al. (2011). Subclinical brain injury caused by H5N1 influenza virus infection. J. Virol. 85, 5202–5207. doi: 10.1128/JVI.00239-11
Stech, O., Veits, J., Abdelwhab el, S. M., Wessels, U., Mettenleiter, T. C., and Stech, J. (2015). The neuraminidase stalk deletion serves as major virulence determinant of H5N1 highly pathogenic avian influenza viruses in chicken. Sci. Rep. 5:13493. doi: 10.1038/srep13493
Su, S., Bi, Y., Wong, G., Gray, G. C., Gao, G. F., and Li, S. (2015). Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J. Virol. 89, 8671–8676. doi: 10.1128/jvi.01034-15
Sun, H., Pu, J., Hu, J., Liu, L., Xu, G., Gao, G. F., et al. (2016). Characterization of clade 2.3.4.4 highly pathogenic H5 avian influenza viruses in ducks and chickens. Vet. Microbiol. 182, 116–122. doi: 10.1016/j.vetmic.2015.11.001
Tian, J., Qi, W., Li, X., He, J., Jiao, P., Zhang, C., et al. (2012). A single E627K mutation in the PB2 protein of H9N2 avian influenza virus increases virulence by inducing higher glucocorticoids (GCs) level. PLoS ONE 7:e38233. doi: 10.1371/journal.pone.0038233
Verhagen, J. H., Herfst, S., and Fouchier, R. A. M. (2015). How a virus travels the world. Science 347:616.
Xiao, C., Ma, W., Sun, N., Huang, L., Li, Y., Zeng, Z., et al. (2016). PB2-588 V promotes the mammalian adaptation of H10N8, H7N9 and H9N2 avian influenza viruses. Sci. Rep. 6:19474. doi: 10.1038/srep19474
Xu, R., Zhu, X., McBride, R., Nycholat, C. M., Yu, W., Paulson, J. C., et al. (2012). Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J. Virol. 86, 9221–9232. doi: 10.1128/jvi.00697-12
Yan, L., Liu, Q., Su, X., Teng, Q., Bao, D., Che, G., et al. (2016). Pathogenicity of reassortant H9 influenza viruses with different NA genes in mice and chickens. Vet. Res. 47:67. doi: 10.1186/s13567-016-0352-y
Yang, H., Carney, P. J., Mishin, V. P., Guo, Z., Chang, J. C., Wentworth, D. E., et al. (2016). Molecular characterizations of surface proteins hemagglutinin and neuraminidase from recent H5Nx avian influenza viruses. J. Virol. 90, 5770–5784. doi: 10.1128/jvi.00180-16
Yang, Z. F., Mok, C. K., Peiris, J. S., and Zhong, N. S. (2015). Human infection with a novel avian influenza A(H5N6) virus. N. Engl. J. Med. 373, 487–489. doi: 10.1056/NEJMc1502983
Yen, H. L., Liang, C. H., Wu, C. Y., Forrest, H. L., Ferguson, A., Choy, K. T., et al. (2011). Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc. Natl. Acad. Sci. U.S.A. 108, 14264–14269. doi: 10.1073/pnas.1111000108
Yoon, S. W., Webby, R. J., and Webster, R. G. (2014). Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol. 385, 359–375. doi: 10.1007/82_2014_396
Keywords: H5Nx, highly pathogenic avian influenza virus, neuraminidase, evolution epidemic, pathogenicity
Citation: Yu Y, Zhang Z, Li H, Wang X, Li B, Ren X, Zeng Z, Zhang X, Liu S, Hu P, Qi W and Liao M (2017) Biological Characterizations of H5Nx Avian Influenza Viruses Embodying Different Neuraminidases. Front. Microbiol. 8:1084. doi: 10.3389/fmicb.2017.01084
Received: 15 March 2017; Accepted: 29 May 2017;
Published: 14 June 2017.
Edited by:
Dirk Dittmer, University of North Carolina at Chapel Hill, United StatesReviewed by:
Jianwei Wang, China Academy of Chinese Medical Sciences, ChinaThomas Paul Fabrizio, St. Jude Children’s Research Hospital, United States
Copyright © 2017 Yu, Zhang, Li, Wang, Li, Ren, Zeng, Zhang, Liu, Hu, Qi and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Liao, bWxpYW9Ac2NhdS5lZHUuY24= Wenbao Qi, cWl3ZW5iYW9Ac2NhdS5lZHUuY24=
†These authors have contributed equally to this study and have shared first authorship.
 Yuandi Yu1†
Yuandi Yu1† Zaoyue Zhang
Zaoyue Zhang Wenbao Qi
Wenbao Qi Ming Liao
Ming Liao