
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 May 2017
Sec. Systems Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00904
This article is part of the Research Topic Microorganisms, pesticides, antibiotics, and organic pollutants: exploring the interactions and biotechnological applications View all 24 articles
The development of bacterial biofilms in natural environments may alter important functions, such as pollutant bioremediation by modifying both the degraders' physiology and/or interactions within the matrix. The present study focuses on the influence of biofilm formation on the metabolism of a pesticide, 2,4-dichlorophenoxyacetic acid (2,4-D), by Cupriavidus necator JMP134. Pure cultures were established in a liquid medium with 2,4-D as a sole carbon source with or without sand grains for 10 days. Bacterial numbers and 2,4-D concentrations in solution were followed by spectrophotometry, the respiration rate by gas chromatography and the surface colonization by electron microscopy. In addition, isotopic techniques coupled with Fatty Acid Methyl Ester (FAME) profiling were used to determine possible metabolic changes. After only 3 days, approximately 80% of the cells were attached to the sand grains and microscopy images showed that the porous medium was totally clogged by the development of a biofilm. After 10 days, there was 25% less 2,4-D in the solution in samples with sand than in control samples. This difference was due to (1) a higher (+8%) mineralization of 2,4-D by sessile bacteria and (2) a retention (15%) of 2,4-D in the biofilm matrix. Besides, the amount of carbohydrates, presumably constituting the biofilm polysaccharides, increased by 63%. Compound-specific isotope analysis revealed that the FAME isotopic signature was less affected by the biofilm lifestyle than was the FAME composition. These results suggest that sessile bacteria differ more in their anabolism than in their catabolism compared to their planktonic counterparts. This study stresses the importance of considering interactions between microorganisms and their habitat when studying pollutant dynamics in porous media.
The functioning of microorganisms is closely linked to their distribution in a structured media, such as soil. A review by Kuzyakov and Blagodatskaya (2015) stressed the importance of considering the spatial and temporal heterogeneity of microbial processes in soils. They defined microbial hotspots as small soil volumes with much faster process rates and much more intensive interactions compared to the average soil conditions. Considering microbial hotspots is particularly important when studying the fate of pesticides in soils because of the very high spatial variability of their degradation. For example, Monard et al. (2012) showed that 2,4-D mineralisation variability was higher than that of a simple molecule, such as glucose, due to the spatial heterogeneity of specific 2,4-D degraders. Dechesne et al. (2014) reported that the presence and activity of pesticide degraders frequently displays non-random spatial patterns with coefficients of variation often exceeding 50%. Soil microorganism localization is indeed restricted to very small microhabitats comprising much <1% of total soil volume (Young et al., 2008) and covering <10−6% of the soil surface area (Young and Crawford, 2004). These habitats are composed of diverse microbial assemblages ranging from single colonies to biofilms (Hodge et al., 1998; Ekschmitt et al., 2005). In situ evidence of the role of such microhabitats on pesticide degradation is often difficult to obtain. Therefore, biofilms should be considered as relevant microbial hotspots for studying processes in vitro.
Biofilm research can be traced back to the 1930s when bacterial attachment and growth on solid supports was first noted and the term “surface effect” proposed (see Zobell and Anderson, 1936; Zobell, 1943). This effect describes the ability of solid surfaces to adsorb minute but demonstrable quantities of organic matter, thus retarding exocellular enzymes and hydrolysates diffusion away from surface-localized bacterial cells (Vanloosdrecht et al., 1990). Most of the batch studies on pesticide degradation have been done without taking into account this surface effect, using shaken-liquid medium, which favors bacteria that grow well in suspension. However, biofilms are considered to be a major mode of bacterial life in nature (Costerton et al., 1995). The use of liquid media might introduce a significant bias when studying pesticide degradation. Compared to their planktonic counterparts, sessile bacteria often benefit from physical and physiological interactions that enhance nutrient availability and, potentially, toxic metabolite removal (Davey and O'Toole, 2000). Cooperative and competitive cell–cell interactions that define biofilm form and function are influenced by the spatial arrangement of genotypes within a community (Nadell et al., 2016). The extracellular polysaccharide (EPS) matrix also provides a protection against a variety of environmental stresses, including pesticides exposure, which could explain the tolerance of microbial communities to such xenobiotics (Imfeld and Vuilleumier, 2012). While recent studies have demonstrated that biofilms induced changes in pesticide degradation rate, due to the modifications in the microbial communities (e.g., Verhagen et al., 2011; Tien et al., 2013), very few studies have focused on the consequences for the metabolism of pesticides of biofilm formation by a single strain of degrader. The first objective of the present study is to address this question.
The second objective of this study is to evaluate the potential change in membrane lipid profiles between planktonic and sessile bacteria. Indeed, phospholipid fatty acids (PLFA) are commonly used as biomarkers in soil microbial ecology. Such molecules provide information about the whole community structure of a sample, but also allow the targeting of microbial groups specifically involved in the degradation of a given pesticide when the latter has been labeled with 13C labeling. This method is called Stable Isotope Probing (SIP). PLFA were the first to be analyzed by SIP (Boschker et al., 1998) and then the technique was extended to nucleic acids (NA) a few years later (Radajewski et al., 2000). Although less taxonomically precise, lipid based method are much more sensitive than NA based SIP. This method has been successfully used to assess microbial community of 2,4-D degraders on long term incubation experiment (e.g., Lerch et al., 2009a) and even bound residues when the dilution of the labeling is high (Lerch et al., 2009b). In order to provide taxonomic information on the targeted communities, isotopically enriched lipid profiles have to be compared to existing fatty acid databases of cultured microorganisms. Thus, pure culture studies are still necessary not only to increase phylogenetic resolution of lipid biomarkers, but also to ensure their accuracy. Indeed, the fatty acid composition of the cell membrane is known to be affected by many stresses, such as high temperature (Heipieper et al., 1996), low pH (Baath and Anderson, 2003), heavy metals (Frostegard et al., 1993), organic pollutants (Rosas et al., 1980). More recently, the biofilm formation has also been showed to induce changes in membrane lipids (Benamara et al., 2011).
Here, we compared the biodegradation of 2,4-D by a pure strain of Cupriavidus necator JMP134 in the presence or absence of a solid phase. A previous study based on the same microbial model showed that C. necator JMP134 preferentially used C originating from the 2,4-D acetic chain for energy while C originating from the benzenic ring was rather used as C source (Lerch et al., 2007). In the present study, we hypothesize that adding sand grains to a culture will induce the development of a microbial biofilm that might change the bacterial metabolic activity and/or the retention of 2,4-D. We combined classical optical density measurements, respirometry, microscopy and isotopic analysis. The fatty acid composition of C. necator JMP134 has been reported to be modified by the nature of the growth substrate (Lerch et al., 2011). We expect that such potential change in fatty acid profiles and isotopic signature could also occur with the biofilm formation.
Unlabelled 2,4-D (chemical purity > 99%, δ13C = −29.1‰) was purchased from Sigma-Aldrich Co., Ltd. and ring-U-labeled 13C-2,4-D (99% of chemical purity, isotopic enrichment > 98%) was obtained from Dislab'system (France). Before any experiments, the 13C-2,4-D level of enrichment was checked by GC-IRMS (see description below).
All cultures were grown in a minimum medium (MM) containing K2HPO4 (1.5 g.L−1), KH2PO4 (0.5 g L−1), (NH4)2SO4 (1 g.L−1), MgSO4 (H2O)7 (207 mg.L−1), ZnSO4 (H2O)7 (200 μg.L−1), MgCl2 (H2O)4 (10 μg.L−1), H3BO3 (5 μg.L−1), CoCl2 (H2O)6 (25 μg.L−1), CuSO4 (100 μg.L−1), NiCl2 (H2O)6 (5 μg.L−1), FeSO4 (H2O)7 (250 μg.L−1), and EDTA (ED4S) (125 μg.L−1). 2,4-D (250 mg.L−1) was added as the sole carbon source (the amount of C from EDTA was considered negligible compared to that in 2,4-D, 2,000 times higher). Two types of incubations were performed. Incubations with unlabelled 2,4-D were performed in Erlenmeyer's containing 200 mL of MM with or without (Control) 200 g of fine sterilized sand (sable de Fontainebleau, 100–200 μm, Prolabo). Incubations with 13C-labeled 2,4-D (δ13C = 134‰) were performed in serum bottles (120 mL) with Teflon® rubber stoppers crimped on with aluminum seals containing 50 mL of MM with or without (Control) 50 g of sand. There were 4 replicates per treatment in the unlabelled and 13C-labeled 2,4-D experiments. The sand was completely saturated and there was approximately 80 mL of free water phase. So, it was possible to shake and sample the liquid medium in the same way as for the control samples (without sand).
The inoculation of the pure C. necator JMP134 strain (courtesy of F. Martin-Laurent, INRA Dijon, France), collected at the end of the exponential growth phase, was done at a level of 105 CFU.mL−1. Pre-cultures were performed in a TY liquid medium: 5 g.L−1 of tryptone (δ13C = −23.1‰), 3 g.L−1 of yeast extract (δ13C = −26.1‰) and 50 mg.L−1 of 2,4-D. According to the C content of each substrate, the average isotopic enrichment of the preculture was −23.9‰. The inoculum was rinsed once with phosphate buffer to limit the addition of C from the TY medium in the MM. All batch cultures were performed at 25°C in the dark on a rotary shaker at 150 rpm.
For all experiments (unlabelled and 13C-labeled 2,4-D experiments) we estimated 2,4-D (% of initial amount) and bacterial concentrations [Log(CFU).mL−1] after 1, 2, 3, 5, and 10 days of incubation by measuring the optical density in the liquid medium using a Lambda 5 (Perkin-Elmer) spectrophotometer (λ2,4−D = 282 nm and λBacteria = 600 nm). At each sampling date, 4 replicates of each treatment were destructively sampled for microscopy and carbohydrates measurements (unlabeled 2,4-D experiments) or for Fatty Acid Methyl Ester (FAME) profiling (13C-labeled 2,4-D experiments).
Scanning Electron Microscopy (SEM) of sand surface was performed on the unlabelled 2,4-D experiments at each sampling date. In order to preserve the original structure of the sample we used a SEM equipped with a low temperature system (Chenu and Tessier, 1995). Sand samples of about 1 mm3 were mounted on the metal stubs of a specimen holder (Hexland cryostrans system CT 1,500) and were cryofixed by immersion in a nitrogen slush at −200°C, transferred to the microscope preservation chamber at −180°C where they were fractured with a cold blade. Ice was partially sublimated under vacuum at about −80°C for 20 min, and the disappearance of ice in the pores of the samples was controlled by observation under the microscope. The surface of the sample was coated at −180°C with gold, and the samples were then introduced in the refrigerated columns of the microscope (Philips SM 525 SEM) to be observed at a temperature < −160°C. For each case date of sampling three replicates were observed.
Measurements of the amount and the 13C enrichment of the CO2 evolved were performed during the 13C-labeled 2,4-D experiments. At each sampling date, 3 gas samples were taken from the serum bottle headspace and the CO2 was directly quantified with a micro-GC (Agilent 3,000A) equipped with a HP-Plot-U column with helium as the carrier gas. The gas sample was introduced into the micro GC via a built-in vacuum pump. The column and the detector were maintained at 80°C. The isotopic content of the CO2 was measured with a GC-IRMS (Isochrom Optima, Micromass) on 3 other gas samples injected with a syringe into the GC. The GC was equipped with a Haysep Q column (Valco Instruments Co. Inc.) and helium as the carrier gas. The injector and detector were maintained at 180°C. The column temperature was programmed at 80°C then ramped at 30°C per min to 160°C. The δ13C standard deviations were 0.1‰or better. At the beginning of the experiment and after measuring CO2, all the flasks were flushed with reconstituted air (19% O2, 81% N2).
Carbohydrate concentrations of each sample were measured as hexoses by the phenol-sulfuric method as previously described by Dubois et al. (1956). Briefly, 2.5 μl of phenol (80%), and 250 μl of concentrated sulfuric acid were added to 100 μl of each cell wall fraction. Following incubation at 30°C for 30 min, color was detected by an automated plate reader at 490 nm. The carbohydrate concentration was determined using glucose controls (37.5, 75, 150, and 300 g.mL−1). Three analytical replicates were performed for each experimental replicate and each date of sampling.
At each sampling date, cells from control samples were harvested by centrifugation (3,500 g for 20 min at 4°C) using a Beckman-Coulter Adventi J20 XP1 and then lyophilised before lipid extraction (see protocols described below). Samples containing sand were first filtered (GF/F, < 0.7 μm, Whatmann) and then lyophilised with the filter. For 13C-labeled 2,4-D experiments, the 4 replicates samples were put together to obtain a sufficient amount of biomass. The total lipid fraction was preferred to the PLFA method because it may also contain information about 2,4-D and its metabolites, as these are extractable in the FAME extractants (Lerch et al., 2007). Briefly, samples were extracted with a mixture of dichloromethane and methanol by using the modified Bligh/Dyer method as described previously by White et al. (1979). The total lipids obtained were transesterified with BF3/Methanol to recover the FAME (Morrison and Smith, 1964) and 2,4-D as methyl-2,4-D (Cochrane et al., 1981) in hexane/ether.
The FAME were separated and quantified by GC with flame ionization detection (GC-FID), identified by mass spectrometry (GC-MS) and their individual isotopic analysis was done by isotopic ratio mass spectrometry (GC-IRMS) as follows. The fatty acid methyl esters were analyzed by capillary GC-FID on a Hewlett-Packard model 5890 series II chromatograph with a 60 m SGE BPX5 column (0.32 mm inside diameter and 0.25 μm film thickness) with helium as the carrier gas. The injector and detector were maintained at 280°C. The column temperature was programmed at 50°C for 1 min, then ramped at 5°C per min to 300°C, followed by an isothermal period of 10 min. Samples were injected in splitless mode. The preliminary peak identification was done by comparison of retention times with commercial standards for FAME (BAME Mix, Supelco) and some molecules, such as 2,4-D, 2,4-DCP (2,4-dichlorophenol) and 3,5-CC (3,5-dichlorocatechol). Results were expressed as equivalent peak responses to these standards. Peak identification was completed by GC-MS of selected samples with a Hewlett-Packard model 6890N gas chromatograph interfaced to a HP 5373 quadrupole mass spectrometer by using the same column and temperature program as previously described. The electron impact ion source was maintained at 250°C, and the quadrupole temperature was 150°C. Mass spectra were obtained by electron impact at 70 eV.
For 13C-labeled 2,4-D experiments, the individual δ13C measurement of each FAME was performed in triplicate on an Isochrom III Isotopic mass spectrometer (Micromass-GVI Optima) coupled to a GC HP5890 with the same column and conditions as those previously described for GC-MS analysis. The column effluent was combusted on-line in an oxidation oven (copper-nickel-platinum catalyst at 980°C) and passed through a reactor with elemental copper (600°C) to reducing NOx and remove surplus O2. Combustion gases were purified in a cryogenic trap containing liquid N2 (−100°C).
The standard notation for expressing high-precision gas IRMS results in δ is defined as follows:
where RSample and RVPDB are the 13C/12C isotope ratios corresponding respectively to the sample and to the international internal standard Vienna PeeDee Belemnite (RVPDB = 0.0112372 ± 0.0000090).
The derivatization of the fatty acids introduces one additional carbon which is not present in the parent compound and which alters the original isotope ratio of the fatty acids. However, the derivatization process introduces a distinct reproducible fraction that is constant for each fatty acid (Goodman and Brenna, 1992). The measured isotope ratios of the FAME were corrected for the isotope ratio of the methyl moiety to obtain the isotope ratios of the fatty acids. This was done by using the Formula (2):
where δ13CFA is the δ13C of the fatty acid, Cn is the number of carbons in the fatty acid, δ13CFAME is the δ13C of the FAME, and δ13CMeOH is the δ13C of the methanol used for the methylating reaction (−63.2‰) to calculate the isotope ratios of the fatty acids.
Considering the 2,4-D as the sole carbon source in the medium, we used the mass conservation Equation (3) and the mixing isotopic mass Equation (4) in order to calculate the amount of ring-C and chain-C in a considered fraction:
where CFraction is the amount of total C, CRing is the amount of C coming from the 2,4-D benzenic ring (ring-C) and CChain is the amount of C coming from the 2,4-D acetate chain (chain-C). By simplifying equations (3) and (4), the equations giving CRing and CChain in a given fraction are the following:
where δ13CFraction is the measured value, δ13CChain (−32.1‰) and δ13CRing (+177‰) are the isotopic signatures of the acetate chain and benzene ring previously determined by Lerch et al. (2007). Equations 5 and 6 were used for every FAME analyzed and the CO2 evolved. It is important to notice that the C of the inoculum was neglected since it was at least 100 fold lower than the amount of biomass C after 1 day of incubation.
Significant differences among treatments (with or without sand) were tested by ANOVA followed by Tukey's HSD test. Principal component analysis (PCA) was performed both on total FAME profiles and on the percentage of CRing incorporated into each FAME. All multivariate analyses were performed using the “ade4TkGUI” package in R while graphical representations were performed with SigmaPlot 11.0. All statistical analyses were performed with R software (version 2.12.0 R Development Core Team, 2008).
Electron microscopy observations revealed that C. necator JMP134 was able to form a biofilm in a minimum medium containing only 2,4-D as C source and sand as a solid phase. After just 1 day of incubation, bacteria (about 1 μm diameter) and exopolymers (thin white filament) were detected on the surface (Figure S1). After 3 days, the density of sessile bacteria increased slightly as well as the amount of polymers between sand grains. After 5 days, sand grains were totally recovered by a film of polymers of approximately 1 μm thick. When analyzing sand grains at a larger scale (Figure 1), we observed that the porous media became totally clogged by biopolymers after 5 days of incubation.
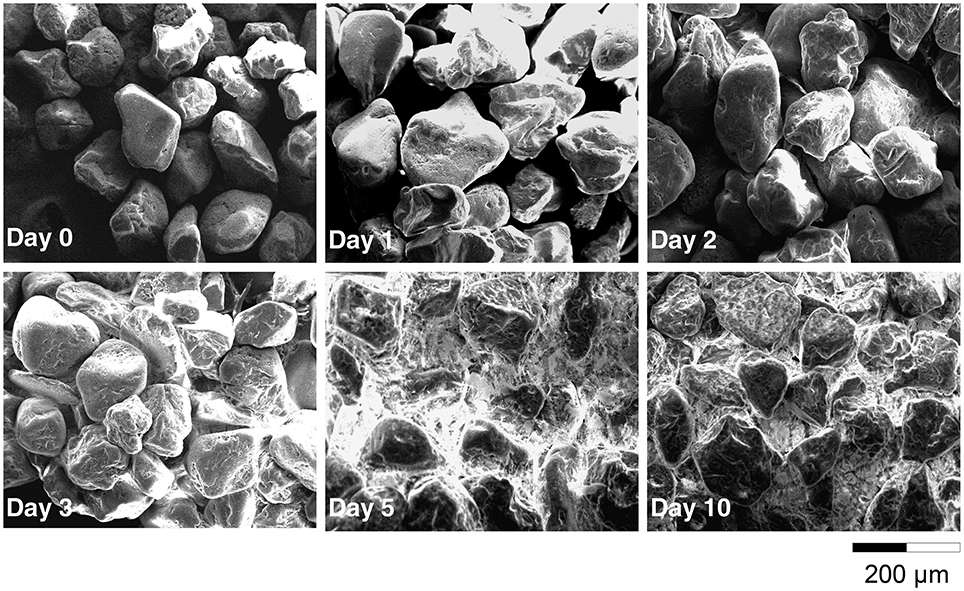
Figure 1. Scanning Electron Microscopy of the sand grains surface after 0, 1, 2, 3, 5, and 10 days of incubation.
The concentration of 2,4-D in solution was always (P < 0.001) lower in samples containing sand than in control samples (Figure 2A). After 1 day of incubation, the amount of 2,4-D in samples with sand was already significantly lower than for control samples. For both curves, a minimum was reached after 3 days of incubation. At the end of the experiment, the 2,4-D concentrations in solution were 5 ± 3% and 30 ± 3% of the initial amount in samples with or without sand, respectively. The amount of 2,4-D mineralized throughout the experiment was always higher (P < 0.001) in samples containing sand (Figure 2B). After 10 days of incubation, the cumulative amounts of CO2 evolved were 68 ± 3% and 60 ± 2% of the initial amount of C in the medium in samples with or without sand, respectively.
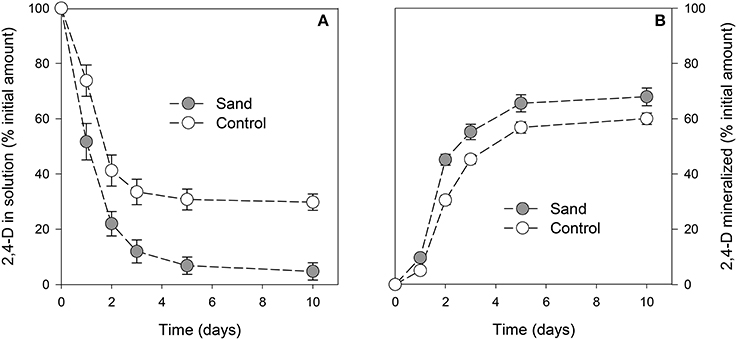
Figure 2. Proportion of 2,4-D in solution (A) or mineralized (B) with time. The gray circles and white circles represent incubation with or without addition of sand, respectively. Error bars correspond to the standard deviation calculated for 3 replicates.
It should be mentioned that whatever the sampling date, the lipid fraction of control samples did not contain any residues of 2,4-D or its metabolites (data not shown). In contrast, the analysis of the lipid fraction extracted from samples containing sand revealed the presence of 2,4-D (Figure S2). Other 2,4-D metabolites were not detected. The quantity of 2,4-D in this fraction corresponded to 9 ± 3% of the initial amount on the first day and then remained stable between 13 ± 4 and 15 ± 4% of the initial amount from day 2 onwards.
The concentration of suspended cells in solution was always (P < 0.001) lower in samples containing sand than in control samples (Figure 3A). The planktonic bacterial concentration reached a plateau after 3 days for control (approximately 9 × 107 cells.mL−1) and 2 days for sand samples (2 × 107 cells.mL−1). It should be noted that no significant differences were observed between unlabelled and 13C-labeled experiments, indicating that both incubations were similar. The total amount of FAME (detailed below) was similar in both treatments throughout the incubation (Figure 3B), indicating similar microbial biomass in planktonic and biofilm cultures. The maximum amount of FAME was reached after 5 days of incubation with 243 ± 14 μg and 241 ± 17 μg per replicate for sample with or without sand, respectively. The amount of total carbohydrates (Figure 3C) was also significantly (P < 0.001) higher in the biofilm culture than in the suspended cell cultures from 3 to 10 days of incubation (approximately 47 to 63% more).
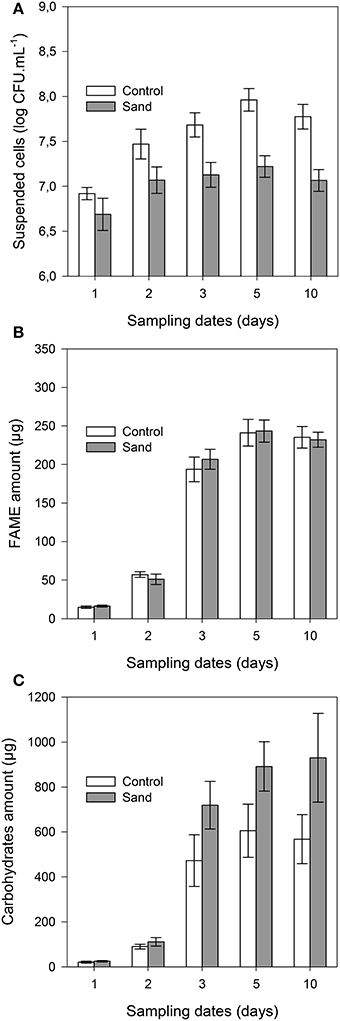
Figure 3. Cell concentration in solution (A), total amounts of FAME (B), and carbohydrates (C) per sample with time. The gray bars and white bars represent incubation with or without addition of sand, respectively. Error bars correspond to the standard deviation calculated for 3 replicates.
The 10 most concentrated FAME identified by GC analyses are expressed in percentage of their relative amount (Figure S3). Five other FAME (iC16:0, iC17:0, C17:0, C18:0, C18:1ω9t and C19:0) were present at trace levels (relative amount < 0.5%) and were not represented. The ANOVA revealed significant differences among sampling dates, but, no significant differences between sessile bacteria (control) and sessile + planktonic bacteria (+ sand), except at day 10. At this sampling date, the relative amount of C16:0 was significantly lower and that of the cycC17:0 was significantly higher for samples containing sand grains. In both treatments, four major FAME (C16:1ω9c, C16:0, cycC17:0, and C18:1ω9c) accounted for approximately 90% of total FAME. Six minor fatty acids (C14:0, iC5:0, aC15:0, C15:0, C18:0, and cycC19:0) were found at a level of approximately 10%. The proportion of three of the major FAME (C16:1ω9c, cycC17:0, and C18:1ω9c) clearly evolved with time (P < 0.001). The evolution of C16:0 proportion was less significant (P < 0.05). It should be mentioned that the cyclopropyl/precursor ratio (cycC17:0+cycC19:0/C16:1ω9c + C18:1ω9c) increased significantly (P < 0.001) for both treatments (Table S1). This ratio was significantly higher (P < 0.01) for sessile bacteria, especially from day 3 onwards. PCA performed on FAME profiles (Figure 4) showed change with time (F1 = 39%) and a distinction between planktonic and sessile cells (F2 = 37%). A redundancy analysis showed that 95% of the variability was explained by these two factors.
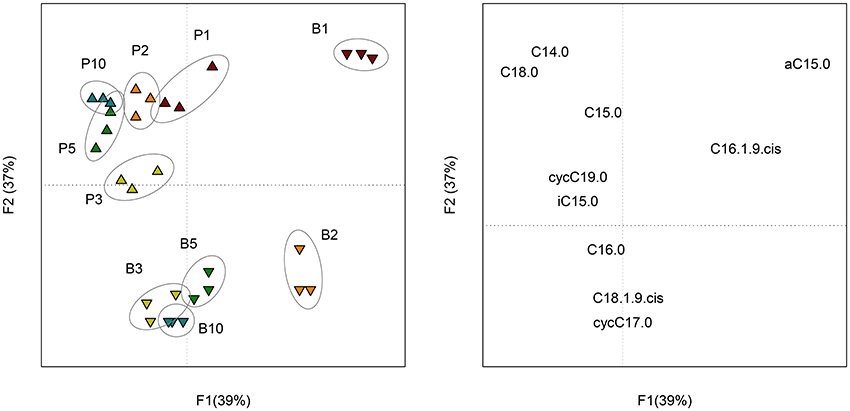
Figure 4. Scores (left) of the PCA performed on the composition (molar percentage) of FAME profiles of planktonic (P) and biofilm (B) cells at each sampling date (1, 2, 3, 5, and 10 days of incubation). Loadings (right) of the PCA representing the 10 FAME detected in the profiles.
The isotopic analysis of the CO2 did not show significant differences in the origin of C respired between samples with or without sand (Figure S4). In both treatments, the proportion of C originating from the benzenic ring (CRing) was slightly lower in the CO2 (≈ 73%) than in the 2,4-D (75%), indicating that C. necator JMP134 preferentially used the C originating from the acetic chain (CChain) for energy.
The isotopic analysis of individual FAME did not reveal significant differences between samples containing sand and control samples. As a result, the proportion of C originating from the benzenic ring (CRing) and the acetic chain (CChain) were similarly incorporated in sessile and planktonic bacteria. For both treatments, the proportion of CRing in FAME increased with time (Figure S5). PCA performed on the proportion of CRing in FAME (Figure 5) showed a small change with time (F1 = 81%) and a little distinction between planktonic and sessile cells during the first 2 days of incubation (F2 = 7%).
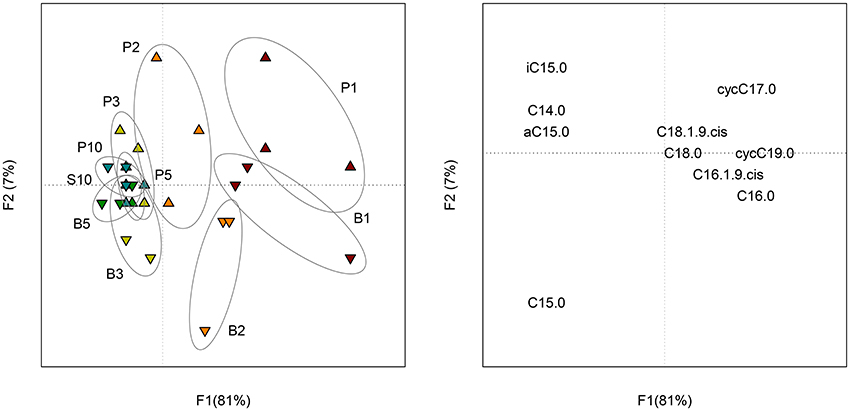
Figure 5. Scores (left) of the PCA performed on the proportion of CRing in each FAME of planktonic (P) and biofilm (B) cells at each sampling date (1, 2, 3, 5 and 10 days of incubation). Loadings (right) of the PCA representing the 10 FAME detected in the profiles.
The similarity of the total FAME amounts in both treatments (with or without sand) suggested that there was no difference in the total microbial biomass. Spectrophotometric measurements of the bacterial density in the liquid phase showed that C. necator JMP134 cultures in the presence of 2,4-D presented similar growth as reported previously (Lerch et al., 2007, 2011). For both treatments, the exponential phase reached its maximum after 3 days followed by a stationary phase that lasted for the next 2 days. Thereafter, there was a similar stage of cellular death with decreasing OD. While both treatments showed the same kinetics, the concentration of suspended bacteria in samples containing sand was approximately 25% that of control samples. In parallel, electron microscopy observations showed that bacterial cells were already attached to the sand grain surfaces after 1 day of incubation. Thus, we conclude that from the early growth stage, most of the cells were attached to the sand grains than being in suspension. By using mass conversion of cell density and with comparison to optical density measurements, we estimated that approximately 80% of the cells in samples containing sand were fixed to the mineral phase from 2 days onwards. Assuming sand grain were perfect spheres (specific area of 0.02 m2.g−1 and density of 2.7 g.cm−3), the surface density of sessile bacteria was estimated to be 0.02 to 0.1 cell.μm−2, which is consistent with SEM analyses.
Most biotreatment experiments using biofilms are enhanced by previous encapsulation and/or support material with high surface area in conjunction with co-substrate to enhance the immobilization of the cells (e.g., Yanez-Ocampo et al., 2009). Our study showed that C. necator JMP134 is capable to rapidly attach to sand grains to reach a density of 0.1 cell per μm2 and develop a thin biofilm using 2,4-D as a sole carbon source of a biofilm with the cell. The ability of this bacteria to form biofilm has been reported previously in the literature. Alcaligenes eutrophus AE1308, a transconjugant of A. eutrophus CH34 and A. eutrophus JMP134 (former name of C. necator JMP134) have been successfully used to form active biofilm into membrane reactor for wastewater treatment (Diels et al., 1995). Other studies carried out in different environments showed that strains similar to C. necator JMP134 were able to form biofilms: Ralstonia eutropha (Steinle et al., 1998; Simkus et al., 2004), Ralstonia solanacaerum (Tans-Kersten et al., 2001; Kang et al., 2002), A. eutrophus (Mergeay et al., 1985; VanRoy et al., 1997), Alcaligenes denitrificans (Elvers et al., 1998), Alcaligenes xylosoxydans (Meade et al., 2001). We did not measure exactly the surface area and thickness of the biofilm. However, electron microscopy images showed that after 3 days of incubation, the thickness of the biofilm was approximately that of the bacteria (c.a. 1 μm), while the surface area of sand grains was almost entirely covered with polymers. Karthikeyan et al. (1999) reported that the thickness/area ratio of a biofilm was correlated to physicochemical characteristics of the organic pollutants being degraded by the biofilm, such as the octanol/water partition coefficient (KOW). It is hypothesized that a close positioning of cells is a disadvantage when cells compete for a labile carbon source. The more recalcitrant and hydrophobic is a molecule, the higher thickness/surface area ratio of the biofilm. This finding is in line with the observation we did with 2,4-D, a very soluble molecule: the biofilm surrounding cells was relatively thin but covered a large part of the grain sand surface. We also observed that after 5 days, all pores were clogged, which certainly modified the diffusion of substrates and metabolites (Chenu and Roberson, 1996) as well as exchanges between the liquid and the solid phases.
Cell immobilization has been employed for biological removal of pesticides because it allows maintaining a catalytic activity over long periods of time. Advantages over cultures using suspended cells include greater cellular content in the support, enhanced cellular viability (weeks or months) and greater tolerance to high concentrations of pollutants. For example, a membrane reactor composed of a microbial consortium showed a better removal of 2,4-D than activated sludge (Gonzalez et al., 2006). Our results suggest that even a single population of bacteria is able to remove more 2,4-D when organized into a biofilm. Some studies using bacterial strains similar to C. necator JMP134 showed that the cell immobilization enhanced the removal of monochlorophenol (Balfanz and Rehm, 1991; Menke and Rehm, 1992) or dichlorophenol (Steinle et al., 1998). Here, we showed that the greater 2,4-D decrease in the liquid medium was partly due to a higher mineralisation. It could be hypothesized that metabolic activity of the cell is higher when immobilized than in planktonic form (Kinniment and Wimpenny, 1992). In the present study, the total amount of biomass was not affected by the bacterial attachment onto the surface. However, SEM observations and carbohydrate measurements suggested that higher exopolymer amounts where produced when C. necator JMP134 was grown with sand. This EPS production likely required more energy, which could also explain the relative increase in 2,4-D mineralisation. The better efficiency of 2,4-D uptake may be related to the exchange of metabolites between cells and/or the nutrient recycling after cell death (Laspidou and Rittmann, 2002, 2004). These possible advantages of sessile bacteria over their planktonic counterparts may explain results obtained in this study and also those previously published (Wolfaardt et al., 1998; Karthikeyan et al., 1999).
While the additional 2,4-D removal from the liquid medium was approximately 30% of added substrate in samples containing sand, only 10% more of added 2,4-D was mineralized. 2,4-D directly from the immobilized cells represented approximately 15% of the added substrate after 2 days of incubation (data not shown), which could explain the difference between the 2 treatments. Since 2,4-D is not adsorbed on the sand grains (preliminary experiments, data not shown) and the accumulation of 2,4-D into the cells is unlikely to happen (Lerch et al., 2007), we hypothesize that the retention of the pesticide was mainly due to the exopolysaccharides (EPS) matrix. It seems unlikely that EPS surrounding directly the immobilized cells may play a major role with regard to the low thickness (1 μm) of the biofilm in this area. However, SEM images revealed much more EPS in the porosity of the sand grains. In these specific area, the 2,4-D diffusion coefficient in the EPS matrix could be lower than in water, as shown already for glucose (Chenu and Roberson, 1996). Many authors have considered this phenomenon as a microbial process that enables to trap nutrients in various environments (Decho, 1990). The EPS matrix may also adsorb 2,4-D in soils as it has been reported for other molecules, such as dissolved organic matter (Sherr, 1988; Kaiser and Guggenberger, 2003), and organic pollutants (Flemming, 1995; Holden et al., 1997; Spath et al., 1998; Holden et al., 2002). An accumulation of pesticide diclofop in the EPS matrix was reported by Wolfaardt et al. (1994) with no consequence on its mineralisation rate. In our study, the accumulation of 2,4-D in the biofilm was accompanied by a higher mineralisation rate and higher carbohydrate amounts that are likely involved in EPS production. Considering that retention within the EPS matrix can allow further chemical transformation that would not happen in suspension (Confer and Logan, 1998), the increase in carbohydrate production could be a storage strategy to improve microbial survival since the production of biopolymers changed the habitat locally and improve substrate immobilization. In soils, the retention of 2,4-D on such biopolymers might play an important role in the formation of non-extractable residues (Lerch et al., 2009a) but also on their mineralisation (Lerch et al., 2009b).
The ANOVA performed on each FAME did not reveal significant differences between sessile and planktonic bacteria. However, the second principal component (37% of the variability) of the PCA performed on the whole FAME profiles was partly linked to the microbial lifestyle. When comparing the cyclopropyl/precursor ratios, we observed that sessile bacteria synthetized more cyclopropyl fatty acid than planktonic bacteria after 3 days of incubation. For both treatments, important changes in FAME profile were related to the sampling date (39% of the variability). These results suggest that the C. necator JMP134 FAME composition was influenced by the growth stage as much as by its attachment to a solid surface. Such growth stage influence on FAME composition is important to consider when interpreting FAME (or PLFA) profiles in complex environments. Variations in the lipid biomarker composition are not only due to the modification of the microbial composition but also to physiological changes. For example, the sensitivity of FAME profiles to change in growth substrate has been reported (Lerch et al., 2011). According to the present study, the bacterial sessile mode of life also alters the signature of lipid biomarkers.
It should be noted that we did not analyse hydroxy-fatty acids in this study since a sylilation is a prerequisite for GC analyses and this derivatisation involves the incorporation of many exogenous C atoms with their own isotopic signature. These lipopolysaccharides (LPS) components have been reported in the FAME profile of R. eutropha B8562, a strain similar to C. necator JMP134 (Volova et al., 2006). The LPS amount is likely to increase with the formation of biofilm (Brandenburg et al., 2003; Keinänen et al., 2003). Thus, it could be hypothesized that the proportion of hydroxy-fatty acids also increases during this phenomenon. However, most of the studies using lipid biomarkers for in situ assessment of the microbial communities do not consider hydroxy-fatty acids.
The analysis of FAME isotopic signatures gives a good estimate of the whole biomass isotopic signature (Lerch et al., 2007, 2009a). Here, we did not report significant changes in the FAME 13C enrichment between sessile and planktonic bacteria, suggesting that the formation of a biofilm by C. necator JMP134 did not change its catabolic pathway. As observed for the FAME profiles, the main changes in the isotopic signature were due to the growth stage. Consequently, the formation of biofilms in complex environments may not alter the isotopic signature of a mono-specific population.
This study first reports that the C. necator JMP134 is able to very rapidly form a biofilm in a minimum medium supplemented with 2,4-D as a sole carbon and energy source. This ability induced direct consequences both on the mineralization and the retention of 2,4-D, leading to a better removal (25%) of the molecule from the solution. The accumulation of 2,4-D by EPS may be an active mechanism which enables C. necator JMP134 to use this molecule afterwards (e.g., when environmental conditions are more favorable). Considering the importance of the EPS matrix in this possible storage strategy, the increase in carbohydrates that constitute this biopolymer may be considered as an investment of energy and carbon to change locally the microbial habitat. This phenomenon may play an important role in the attenuation of pesticides in soils and other porous media since this kind of molecule is added intermittently in the microbial ecosystem. Isotopic analysis revealed that no major change of 2,4-D metabolism occurred when bacteria formed a biofilm: C. necator JMP134 preferentially used C originating from the acetic chain for energy while C originating from the benzenic ring is rather used as C source. The individual molecular analysis confirmed that the origin of 2,4-D carbon used for the synthesis of fatty acids did not differ between planktonic and sessile cells. However, we observed that the formation of the biofilm partially changed FAME profiles, which should be considered when using the signature of lipid biomarkers for in situ analysis. This study showed that physiological changes subsequent to biofilm formation mainly involved anabolic and not catabolic pathways.
TL conceived the study, performed analyses and wrote the article. CC, MD, EB, and AM directed research and contributed substantially to the writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with the authors and states that the process nevertheless met the standards of a fair and objective review.
This study was supported by the French National Research Program ECCO-ECODYN. The authors thank Yolaine Goubard Delaunay (AgroParisTech, ECOSYS) and Samir Abbad-Andaloussi (UPEC, LEESU) for their technical advices. Naoise Nunan (CNRS, IEES-Paris) is also acknowledged for his internal review of the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00904/full#supplementary-material
Figure S1. Scanning Electron Microscopy of the sand grains surface at 0, 1, 2, and 3 days of incubation.
Figure S2. Example of chromatograms (GC-FID) of the lipid fraction obtained from the sand samples at each sampling date. 2,4-D but no metabolites were detected. The other peaks corresponded to the bacterial fatty acids.
Figure S3. Composition (molar percentage) of C.necator JMP134 FAME profiles in the control (upper) and sand (lower) samples at each sampling date. Error bars correspond to the standard deviation calculated for 3 replicates.
Figure S4. Proportion of CRing in the CO2 evolved during the 10 day incubation of C.necator JMP134 in the control (white circles) and sand (gray circles) microcosms. Error bars correspond to the standard deviation calculated for 3 replicates. Dash line represents the proportion of CRing in the substrate (2,4-D) used.
Figure S5. Proportion of CRing in each FAME of C.necator JMP134 for the control (A) and sand (B) microcosms at each sampling date. Error bars correspond to the standard deviation calculated for 3 replicates. The gray line represents the proportion of CRing in the substrate (2,4-D) used.
Table S1. Evolution of the cyclopropyl/precursor ratio (cycC17:0 + cycC19:0/C16:1ω9c + C18:1ω9c) with time.
Baath, E., and Anderson, T. H. (2003). Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35, 955–963. doi: 10.1016/S0038-0717(03)00154-8
Balfanz, J., and Rehm, H. J. (1991). Biodegradation of 4-chlorophenol by adsorptive immobilized Alcaligenes sp. A 7-2 in soil. Appl. Microbiol. Biotechnol. 35, 662–668. doi: 10.1007/bf00169634
Benamara, H., Rihouey, C., Jouenne, T., and Alexandre, S. (2011). Impact of the biofilm mode of growth on the inner membrane phospholipid composition and lipid domains in Pseudomonas aeruginosa. Biochimi. Biophys. Acta 1808, 98–105. doi: 10.1016/j.bbamem.2010.09.004
Boschker, H. T. S., Nold, S. C., Wellsbury, P., Bos, D., de Graaf, W., Pel, R., et al. (1998). Direct linking of microbial populations to specific biogeochemical processes by C-13-labelling of biomarkers. Nature 392, 801–805. doi: 10.1038/33900
Brandenburg, K., Andra, J., Muller, M., Koch, M. H. J., and Garidel, P. (2003). Physicochemical properties of bacterial glycopolymers in relation to bioactivity. Carbohydr. Res. 338, 2477–2489. doi: 10.1016/j.carres.2003.08.008
Chenu, C., and Roberson, E. B. (1996). Diffusion of glucose in microbial extracellular polysaccharide as affected by water potential. Soil Biol. Biochem. 28, 877–884. doi: 10.1016/0038-0717(96)00070-3
Chenu, C., and Tessier, D. (1995). Low temperature scanning electron microscopy of clay and organic constituents and their relevance to soil microstructures. Scanning Microsc. 9, 989–1010.
Cochrane, W. P., Singh, J., Miles, W., and Wakeford, B. (1981). Determination of chlorinated dibenzo-p-dioxin contaminants in 2,4-D products by gas chromatography-mass spectrometric techniques. J. Chromatogr. 217, 289–299. doi: 10.1016/S0021-9673(00)88083-8
Confer, D. R., and Logan, B. E. (1998). Location of protein and polysaccharide hydrolytic activity in suspended and biofilm wastewater cultures. Water Res. 32, 31–38. doi: 10.1016/S0043-1354(97)00194-2
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., and Lappinscott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Davey, M. E., and O'Toole, G. A. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867. doi: 10.1128/MMBR.64.4.847-867.2000
Dechesne, A., Badawi, N., Aamand, J., and Smets, B. F. (2014). Fine scale spatial variability of microbial pesticide degradation in soil: scales, controlling factors, and implications. Front. Microbiol. 5:667. doi: 10.3389/fmicb.2014.00667
Decho, A. W. (1990). Microbial exopolymer secretions in ocean environments: their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. Ann. Rev. 28, 73–153.
Diels, L., Vanroy, S., Somers, K., Willems, I., Doyen, W., Mergeay, M., et al. (1995). The use of bacteria immobilized in tubular membrane reactors for heavy metal recovery and degradation of chlorinated aromatics. J. Memb. Sci. 100, 249–258. doi: 10.1016/0376-7388(94)00253-U
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
Ekschmitt, K., Liu, M. Q., Vetter, S., Fox, O., and Wolters, V. (2005). Strategies used by soil biota to overcome soil organic matter stability-why is dead organic matter left over in the soil? Geoderma 128, 167–176. doi: 10.1016/j.geoderma.2004.12.024
Elvers, K. T., Leeming, K., Moore, C. P., and Lappin-Scott, H. M. (1998). Bacterial-fungal biofilms in flowing water photo-processing tanks. J. Appl. Microbiol. 84, 607–618. doi: 10.1046/j.1365-2672.1998.00388.x
Flemming, H. C. (1995). Sorption sites in biofilms. Water Sci. Technol. 32, 27–33. doi: 10.1016/0273-1223(96)00004-2
Frostegard, A., Baath, E., and Tunlid, A. (1993). Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 25, 723–730. doi: 10.1016/0038-0717(93)90113-P
Gonzalez, S., Muller, J., Petrovic, M., Barcelo, D., and Knepper, T. P. (2006). Biodegradation studies of selected priority acidic pesticides and diclofenac in different bioreactors. Environ. Pollut. 144, 926–932. doi: 10.1016/j.envpol.2006.02.021
Goodman, K. J., and Brenna, J. T. (1992). High sensitivity tracer detection using high-precision gas chromatography-combustion isotope ratio mass spectrometry and highly enriched [U-13C]-labeled precursors. Anal. Chem. 64, 1088–1095. doi: 10.1021/ac00034a004
Heipieper, H. J., Meulenbeld, G., VanOirschot, Q., and DeBont, J. A. M. (1996). Effect of environmental factors on the trans/cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl. Environ. Microbiol. 62, 2773–2777.
Hodge, A., Paterson, E., Grayston, S. J., Campbell, C. D., Ord, B. G., and Killham, K. (1998). Characterisation and microbial utilisation of exudate material from the rhizosphere of Lolium perenne grown under CO2 enrichment. Soil Biol. Biochem. 30, 1033–1043. doi: 10.1016/S0038-0717(97)00269-1
Holden, P. A., Hunt, J. R., and Firestone, M. K. (1997). Toluene diffusion and reaction in unsaturated Pseudomonas putida biofilms. Biotechnol. Bioeng. 56, 656–670. doi: 10.1002/(SICI)1097-0290(19971220)56:6<656::AID-BIT9>3.0.CO;2-M
Holden, P. A., LaMontagne, M. G., Bruce, A. K., Miller, W. G., and Lindow, S. E. (2002). Assessing the role of Pseudomonas aeruginosa surface-active gene expression in hexadecane biodegradation in sand. Appl. Environ. Microbiol. 68, 2509–2518. doi: 10.1128/AEM.68.5.2509-2518.2002
Imfeld, G., and Vuilleumier, S. (2012). Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur. J. Soil Biol. 49, 22–30. doi: 10.1016/j.ejsobi.2011.11.010
Kaiser, K., and Guggenberger, G. (2003). Mineral surfaces and soil organic matter. Eur. J. Soil Sci. 54, 219–236. doi: 10.1046/j.1365-2389.2003.00544.x
Kang, Y. W., Liu, H. L., Genin, S., Schell, M. A., and Denny, T. P. (2002). Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46, 427–437. doi: 10.1046/j.1365-2958.2002.03187.x
Karthikeyan, S., Wolfaardt, G. M., Korber, D. R., and Caldwell, D. E. (1999). Functional and structural responses of a degradative microbial community to substrates with varying degrees of complexity in chemical structure. Microb. Ecol. 38, 215–224. doi: 10.1007/s002489900171
Keinänen, M. M., Korhonen, L. L., Martikainen, P. J., Vartiainen, T., Miettinen, I. T., Lehtola, M. J., et al. (2003). Gas chromatographic–mass spectrometric detection of 2- and 3-hydroxy fatty acids as methyl esters from soil, sediment and biofilm. J. Chromatogr. A. 783, 443–451.
Kinniment, S. L., and Wimpenny, J. W. T. (1992). Measurements of the distribution of adenylate concentrations and adenylate energy charge across Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 58, 1629–1635.
Kuzyakov, Y., and Blagodatskaya, E. (2015). Microbial hotspots and hot moments in soil: concept & review. Soil Biol. Biochem. 83, 184–199. doi: 10.1016/j.soilbio.2015.01.025
Laspidou, C. S., and Rittmann, B. E. (2002). A unified theory for extracellular polymeric substances, soluble microbial products, and active and inert biomass. Water Res. 36, 2711–2720. doi: 10.1016/S0043-1354(01)00413-4
Laspidou, C. S., and Rittmann, B. E. (2004). Modeling the development of biofilm density including active bacteria, inert biomass, and extracellular polymeric substances. Water Res. 38, 3349–3361. doi: 10.1016/j.watres.2004.04.037
Lerch, T. Z., Dignac, M. F., Barriuso, E., Bardoux, G., and Mariotti, A. (2007). Tracing 2,4-D metabolism in Cupriavidus necator JMP134 with C-13-labelling technique and fatty acid profiling. J. Microbiol. Methods 71, 162–174. doi: 10.1016/j.mimet.2007.08.003
Lerch, T. Z., Dignac, M. F., Barriuso, E., and Mariotti, A. (2011). Effect of glucose on the fatty acid composition of Cupriavidus necator JMP134 during 2,4-dichlorophenoxyacetic acid degradation: implications for lipid-based stable isotope probing methods. Appl. Environ. Microbiol. 77, 7296–7306. doi: 10.1128/AEM.06438-11
Lerch, T. Z., Dignac, M. F., Nunan, N., Bardoux, G., Barriuso, E., and Mariotti, A. (2009a). Dynamics of soil microbial populations involved in 2,4-D biodegradation revealed by FAME-based stable isotope probing. Soil Biol. Biochem. 41, 77–85. doi: 10.1016/j.soilbio.2008.09.020
Lerch, T. Z., Dignac, M. F., Nunan, N., Barriuso, E., and Mariotti, A. (2009b). Ageing processes and soil microbial community effects on the biodegradation of soil C-13-2,4-D nonextractable residues. Environ. Pollut. 157, 2985–2993. doi: 10.1016/j.envpol.2009.05.058
Meade, M. J., Waddell, R. L., and Callahan, T. M. (2001). Soil bacteria Pseudomonas putida and Alcaligenes xylosoxidans subsp denitrificans inactivate triclosan in liquid and solid substrates. FEMS Microbiol. Lett. 204, 45–48. doi: 10.1111/j.1574-6968.2001.tb10860.x
Menke, B., and Rehm, H. J. (1992). Degradation of mixtures of monochlorophenols and phenol as substrates for free and immobilized cells of Alcaligenes sp. A7-2. Appl. Microbiol. Biotechnol. 37, 655–661. doi: 10.1007/bf00240744
Mergeay, M., Nies, D., Schlegel, H. G., Gerits, J., Charles, P., and Vangijsegem, F. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162, 328–334.
Monard, C., McHergui, C., Nunan, N., Martin-Laurent, F., and Vieuble-Gonod, L. (2012). Impact of soil matric potential on the fine-scale spatial distribution and activity of specific microbial degrader communities. FEMS Microbiol. Ecol. 81, 673–683. doi: 10.1111/j.1574-6941.2012.01398.x
Morrison, W. R., and Smith, L. M. (1964). Preparation of fatty acids methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J. Lipid Res. 5, 600–608.
Nadell, C. D., Drescher, K., and Foster, K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600. doi: 10.1038/nrmicro.2016.84
Radajewski, S., Ineson, P., Parekh, N. R., and Murrell, J. C. (2000). Stable-isotope probing as a tool in microbial ecology. Nature 403, 646–649. doi: 10.1038/35001054
Rosas, S. B., Secco, M. D. C., and Ghittoni, N. E. (1980). Effects of pesticides on the fatty acid and phospholipid composition of Escherichia coli. Appl. Environ. Microbiol. 40, 231–234.
Sherr, E. B. (1988). Direct use of high molecular weight polysaccharide by heterotrophic flagellates. Nature 335, 348–351. doi: 10.1038/335348a0
Simkus, R., Meskiene, R., Leth, S., and Csoregi, E. (2004). Adaptive responses to static conditions in nutrient-rich cultures of luminous Ralstonia eutropha. Biotechnol. Lett. 26, 559–562. doi: 10.1023/B:BILE.0000021955.27361.4b
Spath, R., Flemming, H. C., and Wuertz, S. (1998). Sorption properties of biofilms. Water Sci. Technol. 37, 207–210. doi: 10.1016/S0273-1223(98)00107-3
Steinle, P., Stucki, G., Stettler, R., and Hanselmann, K. W. (1998). Aerobic mineralization of 2,6-dichlorophenol by Ralstonia sp. strain RK1. Appl. Environ. Microbiol. 64, 2566–2571.
Tans-Kersten, J., Huang, H. Y., and Allen, C. (2001). Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183, 3597–3605. doi: 10.1128/JB.183.12.3597-3605.2001
Tien, C. J., Lin, M. C., Chiu, W. H., and Chen, C. S. (2013). Biodegradation of carbamate pesticides by natural river biofilms in different seasons and their effects on biofilm community structure. Environ. Pollut. 179, 95–104. doi: 10.1016/j.envpol.2013.04.009
Vanloosdrecht, M. C. M., Lyklema, J., Norde, W., and Zehnder, A. J. B. (1990). Influence of interfaces on microbial activity. Microbiol. Rev. 54, 75–87.
VanRoy, S., Peys, K., Dresselaers, T., and Diels, L. (1997). The use of an Alcaligenes eutrophus biofilm in a membrane bioreactor for heavy metal recovery. Res. Microbiol. 148, 526–528. doi: 10.1016/S0923-2508(97)88356-8
Verhagen, P., De Gelder, L., Hoefman, S., De Vos, P., and Boon, N. (2011). Planktonic versus biofilm catabolic communities: importance of the biofilm for species selection and pesticide degradation. Appl. Environ. Microbiol. 77, 4728–4735. doi: 10.1128/AEM.05188-11
Volova, T. G., Trusova, M. Y., Kalacheva, G. S., and Kozhevnicov, I. V. (2006). Physiological-biochemical properties and the ability to synthesize polyhydroxyalkanoates of the glucose-utilizing strain of the hydrogen bacterium Ralstonia eutropha B8562. Appl. Microbiol. Biotechnol. 73, 429–433. doi: 10.1007/s00253-006-0460-0
White, D. C., Davis, W. M., Nickels, J. S., King, J. D., and Bobbie, R. J. (1979). Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 40, 51–62. doi: 10.1007/BF00388810
Wolfaardt, G. M., Lawrence, J. R., Headley, J. V., Robarts, R. D., and Caldwell, D. E. (1994). Microbial exopolymers provide a mechanism for bioaccumulation of contaminants. Microb. Ecol. 27, 279–291. doi: 10.1007/BF00182411
Wolfaardt, G. M., Lawrence, J. R., Robarts, R. D., and Caldwell, D. E. (1998). In situ characterization of biofilm exopolymers involved in the accumulation of chlorinated organics. Microb. Ecol. 35, 213–223. doi: 10.1007/s002489900077
Yanez-Ocampo, G., Sanchez-Salinas, E., Jimenez-Tobon, G. A., Penninckx, M., and Ortiz-Hernandez, M. L. (2009). Removal of two organophosphate pesticides by a bacterial consortium immobilized in alginate or tezontle. J. Hazard. Mater. 168, 1554–1561. doi: 10.1016/j.jhazmat.2009.03.047
Young, I. M., and Crawford, J. W. (2004). Interactions and self-organization in the soil-microbe complex. Science 304, 1634–1637. doi: 10.1126/science.1097394
Young, I. M., Crawford, J. W., Nunan, N., Otten, W., and Spiers, A. (2008). “Microbial distribution in soils: physics and scaling,” in Advances in Agronomy, Vol. 100, ed D. L. Sparks (Burlington, NJ: Academic Press), 81–121.
Zobell, C. E. (1943). The effect of solid surfaces upon bacterial activity. J. Bacteriol. 46, 39–56.
Keywords: biofilm, 2,4-D, Cupriavidus necator JMP134, PLFA-SIP
Citation: Lerch TZ, Chenu C, Dignac MF, Barriuso E and Mariotti A (2017) Biofilm vs. Planktonic Lifestyle: Consequences for Pesticide 2,4-D Metabolism by Cupriavidus necator JMP134. Front. Microbiol. 8:904. doi: 10.3389/fmicb.2017.00904
Received: 31 January 2017; Accepted: 03 May 2017;
Published: 23 May 2017.
Edited by:
Fabrice Martin-Laurent, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Arnaud Dechesne, Technical University of Denmark, DenmarkCopyright © 2017 Lerch, Chenu, Dignac, Barriuso and Mariotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Z. Lerch, dGhvbWFzLmxlcmNoQHUtcGVjLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.