
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 10 May 2017
Sec. Food Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00827
Over thousands of years, modernization could be predicted for the use of microorganisms in the production of foods and beverages. However, the current accelerated pace of new food production is due to the rapid incorporation of biotechnological techniques that allow the rapid identification of new molecules and microorganisms or even the genetic improvement of known species. At no other time in history have microorganisms been so present in areas such as agriculture and medicine, except as recognized villains. Currently, however, beneficial microorganisms such as plant growth promoters and phytopathogen controllers are required by various agricultural crops, and many species are being used as biofactories of important pharmacological molecules. The use of biofactories does not end there: microorganisms have been explored for the synthesis of diverse chemicals, fuel molecules, and industrial polymers, and strains environmentally important due to their biodecomposing or biosorption capacity have gained interest in research laboratories and in industrial activities. We call this new microbiology Technological Microbiology, and we believe that complex techniques, such as heterologous expression and metabolic engineering, can be increasingly incorporated into this applied science, allowing the generation of new and improved products and services.
The history of the use of biotechnological techniques by humanity is confounded by the history of the establishment of microbiology as a science. The first indication of the use of microorganisms for cereal grain fermentation to produce an alcoholic beverage was obtained from molecular evidence from the Neolithic village of Jiahu in China and dates to 7000 BC (McGovern et al., 2004). Similar evidence was found in the Zagros Mountains of northern Mesopotamia, dating to 5400–5000 BC (McGovern et al., 1996). The first indication of wine production comes from the presence of tartaric acid in an old jar, also dated to 5400–5000 BC, at the Neolithic site of Tepe in Mesopotamia (McGovern et al., 1996) and from grape juice residues, found in Dikili Tash in Greece and dated to 5000 BC (Valamoti et al., 2007). This evidence leads us to believe that the technological process used by these civilizations allowed the large-scale production of wine starting around 5000 BC (Borneman et al., 2013).
The Egyptians, who already used yeast to brew beer, began to employ this microorganism to make bread. Samples were found in different archeological sites dating to 2000–1200 BC (Samuel, 1996). The establishment and dissemination of fermentation practices throughout Asia, Mesopotamia, Egypt, and the Old World are traits of the empirical domestication of yeasts (Sicard and Legras, 2011), which later stimulated the interest of Louis Pasteur in explaining the true cause of fermentation (Pasteur, 1857). Evidence suggests that in 1856, Pasteur was approached by a beetroot-based alcohol producer from the Lille agricultural-industrial region, who faced production problems. Thus began the pioneering studies of Pasteur on lactic acid and alcoholic fermentations (Gal, 2008). He then became an admirer of the microscopic universe, describing the association of microorganisms with diseases and proposing vaccination methods such as used against anthrax (1881) and human rabies (1885) (Pasteur, 2002; Plotkin and Plotkin, 2011). Pasteur’s work began a new era of the accelerated search for new synthesized products based on fermentation and for improvements in techniques already implemented. His studies also provided support for the establishment of microbiology as a science, which had as its initial interest the sanitary control of diseases.
Technological Microbiology, however, started to draw the attention of the market when products originating from microbial activity began to be required on an industrial scale. This occurred with the glycerol demand for the manufacture of explosives during World War I (Wang et al., 2001) and the large-scale production of penicillin, discovered by Fleming, in the 1940s (Neushul, 1993).
The American economic expansion brought on by the end of World War II and known as the Golden Age of Capitalism (Stephen et al., 1991), as well as the knowledge of microbial genetics that was emerging at that time (e.g., Zinder and Lederberg, 1952; Jacob et al., 1960; Ames and Martin, 1964; Holloway, 1969; Sussman, 1970; Bagdasarian and Timmis, 1982), stimulated the emergence of microorganism-based industrial processes, triggering modern Technological Microbiology. However, Technological Microbiology is considered to have begun in the 1980s, following a decision made by the United States Supreme Court that allowed the patenting of a Pseudomonas putida variant that is effective in the organic digestion of compounds found in crude oil spills (Robinson and Medlock, 2005). The patent for a genetically modified microorganism, requested by Ananda Chakrabarty, contributed to a revolution in biotechnology that resulted in the issuance of thousands of patents, the founding of hundreds of new companies, and the development of thousands of bioengineering and food plants (Holloway, 2014).
The studies of Warner Arber, Hamilton Smith, and Daniel Nathans on bacterial endonucleases that hydrolyze the DNA of the viruses invading these microorganisms (Smith and Nathans, 1973; Arber, 1974) in the early 1970s earned these researchers the Nobel Prize in Physiology or Medicine in 1978. These enzymes, also known as restriction enzymes for “breaking” the DNA and providing gene fragments, became frequently used in biotechnological processes such as cloning, hybridization, fingerprinting, gene identification, and other genetic manipulations for the production of transgenic animals and plants.
The genetic modification of Escherichia coli in the 1970s allowed the production of artificial insulin, which was the first product obtained from recombinant DNA technology (Walsh, 2012) and was approved by the United States Food and Drug Administration in 1982 (Johnson, 1983). Over time, the selection of improved microbial strains became frequent, as did the manipulation of other microorganisms to obtain products to meet human demands, and as a result, Technological Microbiology has become a science essentially applied to several branches of production, including food, chemical, agricultural, and pharmacological.
Alternatively, the success of the genetic transformation carried out by Herbert Boyer and Stanley Cohen in California from the construction of chimeric E. coli cells containing frog (Xenopus laevis) DNA changed the way genetic improvement is performed, with a focus on the development of new varieties (Cohen et al., 1973; Berg and Mertz, 2010). In 1976, a thermostable DNA polymerase was isolated from the bacterium Thermus aquaticus. Kary Mullis and others contributors found that this enzyme could be used in the polymerase chain reaction (PCR) to amplify DNA fragments (Chien et al., 1976; Saiki et al., 1988). The development of that technique, as well as the enhancement of molecular cloning techniques using plasmids as vectors, expanded the possibilities of microorganism manipulation (Simon and Chopin, 1988; Olsen, 2016) and the large-scale production of microbial products, including those from modified microorganisms.
In addition, in the 1970s, Carl Woese and colleagues used the 16S rRNA molecule, a universally conserved sequence, as a taxonomic marker and revealed that our ignorance about microbial diversity was enormous, capable of hiding the existence of a new prokaryotic domain, the Archaea (Woese and Fox, 1977; Woese et al., 1990). Recent genomic discoveries have shown that the tree of life seems to be even more complex, as the existence of two extraordinarily complex and poorly studied groups has been revealed: the bacterial group Candidate Phyla Radiation (CPR) and the Archaea superphylum DPANN (Spang and Ettema, 2016). CPR bacteria have small genomes and unusual ribosomal compositions, in addition to lacking numerous biosynthetic pathways (Brown et al., 2015), whereas DPANN has been defined as a function of the metabolic capacity (Rinke et al., 2013; Castelle et al., 2015). These studies indicate that the tree of life may continue to grow in the future. Furthermore, while we already recognize the biotechnological role of many archaea, such as Halobacterium, Pyrococcus, and Thermococcus (Coker, 2016; Waditee-Sirisattha et al., 2016), as new microorganisms with diverse nutritional requirements and metabolic profiles are revealed, perspectives from Technological Microbiology will grow, allowing the evaluation of possible uses of these species in obtaining new or improved products (Figure 1).
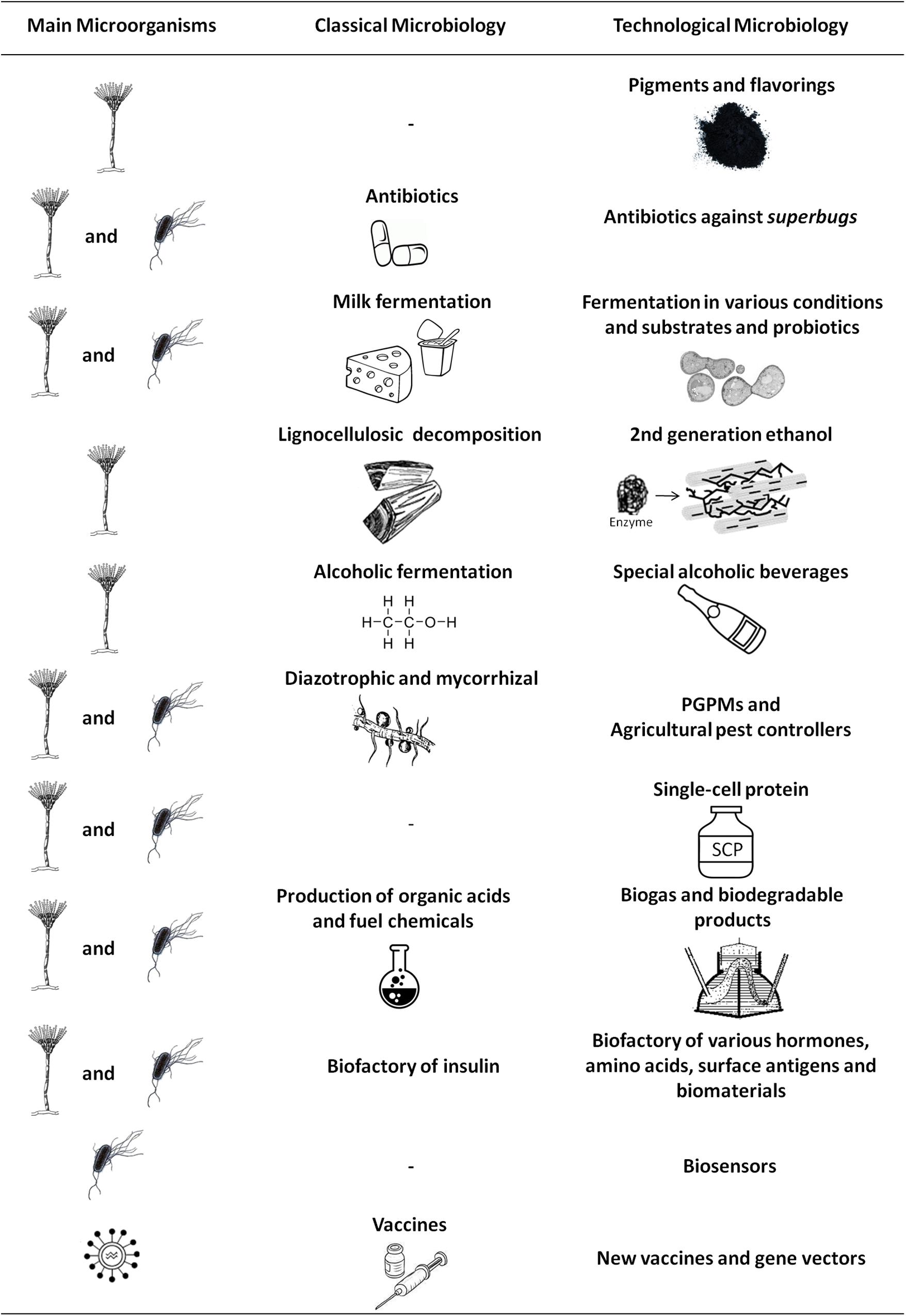
FIGURE 1. The main applications of fungi, bacteria, and viruses for obtaining new or improved products. Comparison between the possibilities generated by Classical Microbiology and Technological Microbiology, where the incorporation of techniques has led to market novelties, as well as to the improvement of commonly used products and services. PGPMs, plant growth-promoting microorganisms.
Therefore, we have seen a contemporary improvement in Classical Microbiology through the discovery of new species, selection and improvement of known strains until the introduction of non-native genes for the acquisition of expressed products or new functional traits. We have decided to call this complex and applied microbiology Technological Microbiology, and although many of its fields overlap, to facilitate our discussion, we chose to divide it into six areas: Food Technological Microbiology, Agricultural Technological Microbiology, Chemical and Fuel Technological Microbiology, Environmental Technological Microbiology, Medical Technological Microbiology, and Materials Technological Microbiology, as follows.
Despite the application of biotechnological techniques to the food-processing industry and the agroindustry, which occurred prior to the technological advances of the 1970s, the current trend incorporates the use of genetically modified microorganisms or even the use of enzymes, dyes, and other compounds obtained from microbial metabolism with the aim of improving productivity, enhancing organoleptic characteristics, or even attributing new nutritional functions to certain foods. Microorganisms, therefore, may have two different roles in current food production. First, acting as starters in fermentations (in this case, GMOs or engineered microorganisms are not allowed). Secondly, they are used as factories for the production of food ingredients. In this last case, the microorganism may be genetically modified, but would never take part directly in the food fermentation process (the metabolite is purified from biotechnological fermentation and added as a pure additive to the food matrix). The participation of the microorganisms in this case is indirect.
Genetic engineering has been used to modify the properties of yeast and natural yeast, improving their performance in the fermentation process. In the future, breads and pastas of the better quality can be obtained in less time. Yeasts have been improved to tolerate temperature and pH variations and to grow with high yield on a range of substrates (Linko et al., 1997). Compounds involved in stress tolerance in yeasts, such as proline and trehalose, are promising for the development of resistant strains (Takagi and Shima, 2015). Thus, yeasts subjected to new processes such as UV radiation have allowed foods with new nutritional attributes to be developed, such as foods with increased vitamin D levels (Degré et al., 2008; Lipkie et al., 2016).
Selection of β-lyase-producing yeasts improves aromatic thiol release and, consequently, the sensory properties of wines (Belda et al., 2016), whereas the selection of yeasts specializing in certain processes such as flocculation may improve the fermentation of special wines such as sparkling wines (Tofalo et al., 2016). In turn, the current trend of using non-Saccharomyces strains, which in the past were considered yeasts of secondary importance or yeasts that produced undesirable changes, has positively impacted the vinification process, given the ability of the strains to produce enzymes, secondary metabolites, glycerol, ethanol, and other compounds that can increase the organoleptic complexity of wines (Padilla et al., 2016).
The prospection of lactic acid bacteria present in products fermented with various cultures (e.g., Satish et al., 2013; Mokoena et al., 2016) has created new sources of probiotics and the discovery of strains that can improve the quality of fermented products. Probiotics are living microorganisms that have been linked to host health benefits (Gawkowski and Chikindas, 2016). Currently, the best-known probiotic microorganisms are those belonging to the genera Lactobacillus and Bifidobacterium (Prasad et al., 2000). Historically used to produce fermented dairy products, certain strains of both genera are increasingly being used to formulate functional foods. The result of this phenomenon is the increase in the number of probiotic foods available on the market, including a rapidly emerging variety of non-dairy probiotic beverages (Enujiugha and Badejo, 2017).
A number of enzyme preparations of microbial origin have been evaluated in food processing. Amylases obtained from cultures of Aspergillus niger (e.g., Omemu et al., 2005; Djekrif-Dakhmouche et al., 2006; Adejuwon et al., 2015) or Bacillus subtilis (e.g., Ploss et al., 2016; Salman et al., 2016), for example, have been used in place of chemical additives in the treatment of wheat flour (e.g., Bueno et al., 2016), improving the preparation of dough for baking and allowing the acquisition of pre-cooked foods. A. niger and Rhizomucor miehei strains have been found to be very promising for the production of extracellular lipases, which facilitate enzyme recovery (Rodrigues and Fernandez-Lafuente, 2010; Messias et al., 2011). These microbial lipases are being employed in the hydrolysis of milk fat, improving the aromatization of dairy products. They can also enhance the aroma of beverages and the quality of margarine and mayonnaise (Sharma et al., 2001). Cellulases and pectinases, used especially in juice clarification and viscosity reduction, have also been easily recovered from cultures of filamentous fungi that are efficient in the degradation of plant biomass, such as Cladosporium sphaerospermum, Penicillium chrysogenum (Andersen et al., 2016), and Trichoderma viride (Ismail et al., 2016). In addition, the technology for immobilization of these enzymes in prefabricated supports or polymer matrices improves their stability, activity, and selectivity, favoring their application and reuse for long periods in industrial reactors (Mateo et al., 2007; Sheldon, 2007).
In addition, microbial enzymes have been used to obtain natural scents and flavorings for foods (Carroll et al., 2016), although these compounds can often be directly obtained from the general metabolism of filamentous fungi, such as A. niger and Pycnoporus cinnabarinus, which can act together in a process that leads to the synthesis of vanillin, an important food flavoring, from autoclaved maize bran (Lesage-Meessen et al., 2002). Additionally, yeasts of the genus Pichia have been added to coffee fermentation to improve the quality and flavor of the beverage because they increase the production of the natural flavoring isoamyl acetate (Saerens and Swiegers, 2016). Microbial biosynthetic pathways have been explored mainly because they can enzymatically convert inexpensive precursors, such as glucose or glycerol, into expensive aromatic compounds. One example is the synthesis by E. coli of acetoin, which is responsible in part for a buttery aroma, using glucose as a substrate (Nielsen et al., 2010).
High rates of population growth have increased the demand for new foods around the world. Protein extracted from cultivated microbial biomass (single-cell protein – SCP) can be used for protein supplementation in basic diets, replacing expensive conventional sources and alleviating the problem of protein shortages (Anupama and Ravindra, 2000). SCP has been widely used as a source of protein in animal and human food (Adedayo et al., 2011). Many bacterial strains of Bacillus, Hydrogenomonas, Methanomonas, Methylomonas, and Pseudomonas have been used as substrate for the production of S on an industrial scale because these bacteria can contain approximately 80% crude protein in the total dry weight. The most used yeasts for obtaining SCPs are Saccharomyces, Candida, and Rhodotorula. Cultivation of yeasts is more practical because these microorganisms are able to use a wide variety of substrates (Patelski et al., 2015); however, the SCPs obtained are insufficient in sulfur-containing amino acids. The most commonly used filamentous fungi are Fusarium, Aspergillus, and Penicillium, and among the prokaryotic algae, the most used belong to the genus Spirulina, with approximately 65% of their dry weight consisting of protein (Nalage et al., 2016). The possibility of using microorganisms to obtain food, food additives, or even microbial biomass for food has reinvigorated the food-processing industry, which sees new possibilities for conventional foods, such as flavors, textures, and aromas, or even the discovery of new foods.
Recently, the interest in microorganisms has focused on compounds with pesticidal activity, mainly herbicidal, insecticidal, and nematicidal. The first commercially registered mycoherbicide consisted of a suspension of chlamydospores of Phytophthora palmivora to control Morrenia odorata (McRae, 1988), and since then, many other plant parasite and phytotoxin-producing microbial species have been identified. Colletotrichum gloeosporioides (Penz) Sacc. f. sp. aeschynomene can induce symptoms of anthracnose in Aeschynomene virginica, thus controlling this legume, which is a rice and soybean weed. On the other hand, Puccinia canaliculata can control yellow nutsedge (Cyperus esculentus L) by completely inhibiting flowering and reducing tuber formation (Duke et al., 2015). Bioherbicides, however, have not been widely applied in agronomic and horticultural crops for weed management because they have a number of requirements, such as ideal humidity conditions, which diminish their effectiveness when compared to chemical herbicides. In the future, biotechnological advances will likely reverse this situation and improve the performance of bioherbicides.
The endotoxin proteins Cry and Cyt are currently best known as pesticides. These endotoxins are synthesized by the soil bacterium B. thuringiensis (Bt) and have an entomopathogenic action, controlling the pests present in cabbage, potato, and grains (Sarwar, 2015a). Several transgenic species expressing Bt protein crystals, such as tomato, tobacco, and corn, have been cultivated worldwide because they have been successful in preventing the spread of caterpillars, especially Lepidoptera (Khan et al., 2016). Caterpillars and eggs of pests such as Spodoptera frugiperda can also be infected by baculovirus, thus reducing the agricultural losses caused by this caterpillar, especially in corn. In addition, the progress achieved by the genetic improvement of this virus has increased its effectiveness as an insecticide (Popham et al., 2016). Several fungi pathogenic of insects are also being used as control agents, including Beauveria, Metarhizium, and Paecilomyces. These are most frequently used against leaf caterpillars in greenhouses or other places where the humidity is relatively high (Sarwar, 2015b).
In recent years, much progress has been achieved in the development and commercialization of bionematicides (Wilson and Jackson, 2013). Examples of this are the products of the bacterium Streptomyces avermitilis, which are metabolites known as avermectins. These are model pesticides, as they are non-toxic to mammals and active against nematodes, even at very low doses. Thus, filtrates of B. firmus cultures induce paralysis and mortality of adult nematodes and larvae, including Radopholus similis, Meloidogyne incognita, and Ditylenchus dipsaci, which suggests that the synthesis of toxic metabolites (Mendoza et al., 2008) is involved in the control of these pests. Toxic metabolites are also produced by Myrothecium verrucaria when grown in bioreactors, and when in contact with adult nematodes, the metabolites in suspension kill the adults, in addition to inhibiting egg development and hatching (Twomey et al., 2000). By contrast, the endospores of the bacterium Pasteuria sp. use parasitism as a method of control. When these endospores come into contact with nematodes such as Meloidogyne spp., Heterodera spp., Globodera spp., and Belonolaimus spp., they germinate, become parasitic, and strongly decrease host fecundity (Davies et al., 2011).
Among the microorganisms that act in the biological control of pests, the most widely disseminated species are the fungi belonging to the genus Trichoderma. These fungi are saprophytes, mycoparasite decomposers, and plant symbionts, usually associated with soil ecosystems, and have a global geographical distribution (Druzhinina et al., 2011). This range of lifestyles within the genus explains why Trichoderma is the source of many strains commercially used in biological control (Howell, 2003). Trichoderma spp. parasitize and successfully control phytopathogenic fungal species such as Sclerotinia (Jones et al., 2014, 2016), Fusarium (Saravanakumar et al., 2016), Verticillium (Carrero-Carrón et al., 2016), and Macrophomina (Khaledi and Taheri, 2016), among others, and have nematicidal effect on the gall-forming Meloidogyne (Sahebani and Hadavi, 2008; Feyisa et al., 2016; Sokhandani et al., 2016). This functional characteristic of Trichoderma and other biocontrol species responds to the increasing call for practices that minimize the side effects left by pesticides, such as resistance in pest populations, reduction of soil and water quality, and the generation of residues with harmful effects on non-target organisms.
Sustainable agriculture, however, provides not only the control of phytopathogens but also the use of functional microbial characteristics related to the promotion of plant growth. Symbiotic microorganisms such as mycorrhizal fungi and rhizobacteria develop activities that can improve plant fitness, facilitating nutrient acquisition by the plant. Mycorrhizal fungi and roots are complementary in plant foraging within nutrient patches (Cheng et al., 2016) and facilitate the acquisition of phosphorus by the plant, through the expression of genes that code for inorganic transporters of this ion (Walder et al., 2016). Likewise, PGPRs (plant growth-promoting rhizobacteria) act through direct and indirect mechanisms to promote plant growth. Direct mechanisms include mainly biofertilization, with nitrogen synthesis by strains belonging to the genera Rhizobium, Sinorhizobium, Mesorhizobium, Bradyrhizobium, Azorhizobium, and Allorhizobium, and the stimulation of root growth through the synthesis of auxins, cytokinins, and gibberellins. Indirect mechanisms are related to the reduction of susceptibility to diseases, including antibiosis, induction of systemic resistance and competition for nutrients and niches (Lugtenberg and Kamilova, 2009).
On the other hand, endophytic microorganisms colonize plant tissues without triggering any disease symptoms, establishing a stable long-term interaction with the host plant. During the interaction, endophytes synthesize bioactive metabolites that may confer greater fitness to the plant. This promotion of growth by endophytic action may be a consequence of nitrogen fixation, synthesis of phytohormones, biocontrol of phytopathogens through the synthesis of antibiotics or siderophores, competition for nutrients, and the induction of systemic disease resistance (Ahmad et al., 2016). However, the bioprospection and characterization of these microorganisms, associated with the most diverse plant species, is aimed not only at obtaining strains of agronomic importance but also at the identification of species that produce metabolites with potential for the synthesis of antibiotics (e.g., Ma et al., 2016), as well as potential for obtaining biotechnologically important chemicals.
Obtaining chemicals such as organic acids via microbial activity is very promising, especially if it is thought to occur from renewable carbon sources. Most organic acids are natural products or intermediates of the microbial metabolism present in important metabolic pathways (Sauer et al., 2008). Due to their functional groups, these acids, such as acetic, citric, lactic, and succinic acid, are extremely useful as raw materials for the chemical or food industry. Citric acid, for example, has been required on the market for use as a food additive, and all annual worldwide industrial-scale production occurs via the fermentation of glucose, beet molasses, cane molasses, or corn starch using A. niger (Adham, 2002; Ikram-ul et al., 2004; Wang et al., 2016). On the other hand, all the annual world production of lactic acid also comes from the fermentation performed by microorganisms. This acid and its derivatives are widely used in the food, pharmaceutical, leather, and textile industries. In addition, lactic acid fermentation processes have recently received more attention because of the growing demand for new biomaterials, such as biodegradable products and biocompatible polylactics (Gao et al., 2011). The most common method for obtaining this acid is using Lactobacillus spp. cultivated in whey (Hofvendahl and Hahn-Hägerdal, 2000). However, it may also be obtained via the activity of Rhizopus sp. under aerobic conditions in glucose-rich medium and with limited amounts of nitrogen (Papagianni, 2004; Fu et al., 2014), and even via the fermentation of Saccharomyces cerevisiae in glucose- and cane juice-based medium (Saitoh et al., 2005; Valli et al., 2006). In the future, the microbiological processes for obtaining a variety of organic acids are expected to will be competitive, become established in the market, and allow for an annual increase in the production of these compounds.
The microbial production of acetone and butanol, efficiently performed by the genus Clostridium, was one of the first large-scale industrial fermentation processes to gain global importance, but this production has been losing ground to chemical synthesis. Similarly, the centennial microbial synthesis of glycerol was impacted by the inability to compete with chemical synthesis from petrochemical feedstocks. However, in a scenario where the cost of propylene increased because its availability decreased, especially in developing countries, glycerol became an important raw material for the production of various chemicals, which made its alternative synthesis by fermentation more attractive (Wang et al., 2001).
Likewise, the production of 1,3-propanediol (1,3-PDO), which occurs through the fermentation of glycerol by bacteria of the genus Clostridium or Enterobacteriaceae, a technique described in 1881, lost status front to chemical synthesis through petroleum products, remaining forgotten for more than a century. In the last decade, however, research related to the synthesis of microbial 1,3-PDO expanded considerably (Biebl et al., 1999) because this diol began to be used in the synthesis of biodegradable polymers and for obtaining solvents, films, adhesives, antifreezes, and polyesters. Currently, a potentially viable alternative for the synthesis of 1,3-PDO is the use of genetically modified microorganisms. Genes from pathogenic bacteria, such as Citrobacter freundii and Klebsiella pneumoniae, were recently introduced in E. coli to allow the efficient synthesis of 1,3-PDO from waste glycerol (Przystałowska et al., 2015).
This synthesis of chemicals through microbial metabolic processes meets an urgent need to reduce dependence on fossil fuels for energy generation. In modern biorefineries, renewable resources such as biomass or waste products are converted into substrates susceptible to microbial action (Sauer, 2016), and thus, interest in bio-based chemicals has recently been renewed because increasing climate change and environmental problems have pushed the industry, moving it away from fossil fuel consumption and toward renewable raw materials (Moon et al., 2016). Microorganisms have also been potentially explored for the production of a new generation of biofuels (Liao et al., 2016). The production of second-generation ethanol, for example, obtained from lignocellulosic biomass, already occurs in some countries, although improvements are still needed to make the technology economically competitive. Recent developments such as the discovery of functional xylose isomerases (Kuyper et al., 2005) resulted in the creation of new yeasts capable of fermenting 5-carbon (C5) sugars, as well as 6-carbon (C6) sugars. Co-fermentation of C5 sugars with cane juice can produce up to 37% more ethanol in first-generation fermenters (Losordo et al., 2016). Another problem to be overcome for the effective production of second-generation ethanol is the tolerance to acetic acid. This acid is one of the main inhibitors of lignocellulose hydrolysates. The polygenic basis of the high acetic acid tolerance present in some strains of S. cerevisiae is still unknown, but its identification may lead to greater efficacy in improving acetic acid tolerance in strains without negatively affecting other industrially important yeast properties (Meijnen et al., 2016). However, in addition to yeast genetic improvements, the prospection of new cellulose sources, such as forestry and crop residues (eucalyptus bark, corn, and rice husks), and the development of pretreatment techniques (e.g., McIntosh et al., 2016) can leverage the production of second-generation ethanol.
In addition to bioethanol, other energy molecules such as biogas can be obtained from the microbial conversion of biomass. Biogas is a combination of methane, CO2, nitrogen, H2S, and traces of other gasses produced by anaerobic digestion (AD) (Appels et al., 2008). Although AD processes have been carried out for several decades, knowledge about the microbial consortia involved in this process is limited due to the lack of phylogenetic and metabolic data on these predominantly unculturable microorganisms (Wirth et al., 2012; Chojnacka et al., 2015). Studies carried out to isolate and identify the microbial community associated with the production of biogas revealed the presence of Proteobacteria, Chloroflexi, Firmicutes, Bacteroidetes, Actinobacteria, Bacteroides, Acidobacteria, and Spirochetes (Chouari et al., 2005; Chojnacka et al., 2015). Methanogenic Archaea, such as Methanosarcina barkeri, M. frisius, and Methanobacterium formicicum, were also identified in anaerobic digestions (Godon et al., 1997; Satpathy et al., 2016). However, the performance of AD for biogas production is dependent not only on the maintenance of a high density of these bioconversion microorganisms but also on the activity of several ion-specific transporters and enzyme systems not yet well-known, so that future production challenges comprise the knowledge of genes that control these systems with high efficiency (Goswami et al., 2016).
The expectation is that, in the future, at least 25% of all bioenergy can originate from biogas (Holm-Nielsen et al., 2009), and therefore, studies that seek to optimize the methanogenesis process or describe the structure of microbial communities have been encouraged (e.g., Ennouri et al., 2016; Mulat et al., 2016; Suksong et al., 2016). Metagenomic approaches associated with next-generation sequencing (NGS) techniques will help to unravel the diversity of natural communities and in biogas fermenters communities (e.g., Schlüter et al., 2008). However, studies have shown that most of the microorganisms isolated from the reactors are still unexplored (e.g., Krause et al., 2008; Figure 2) and may be a source for new products and services in the future.
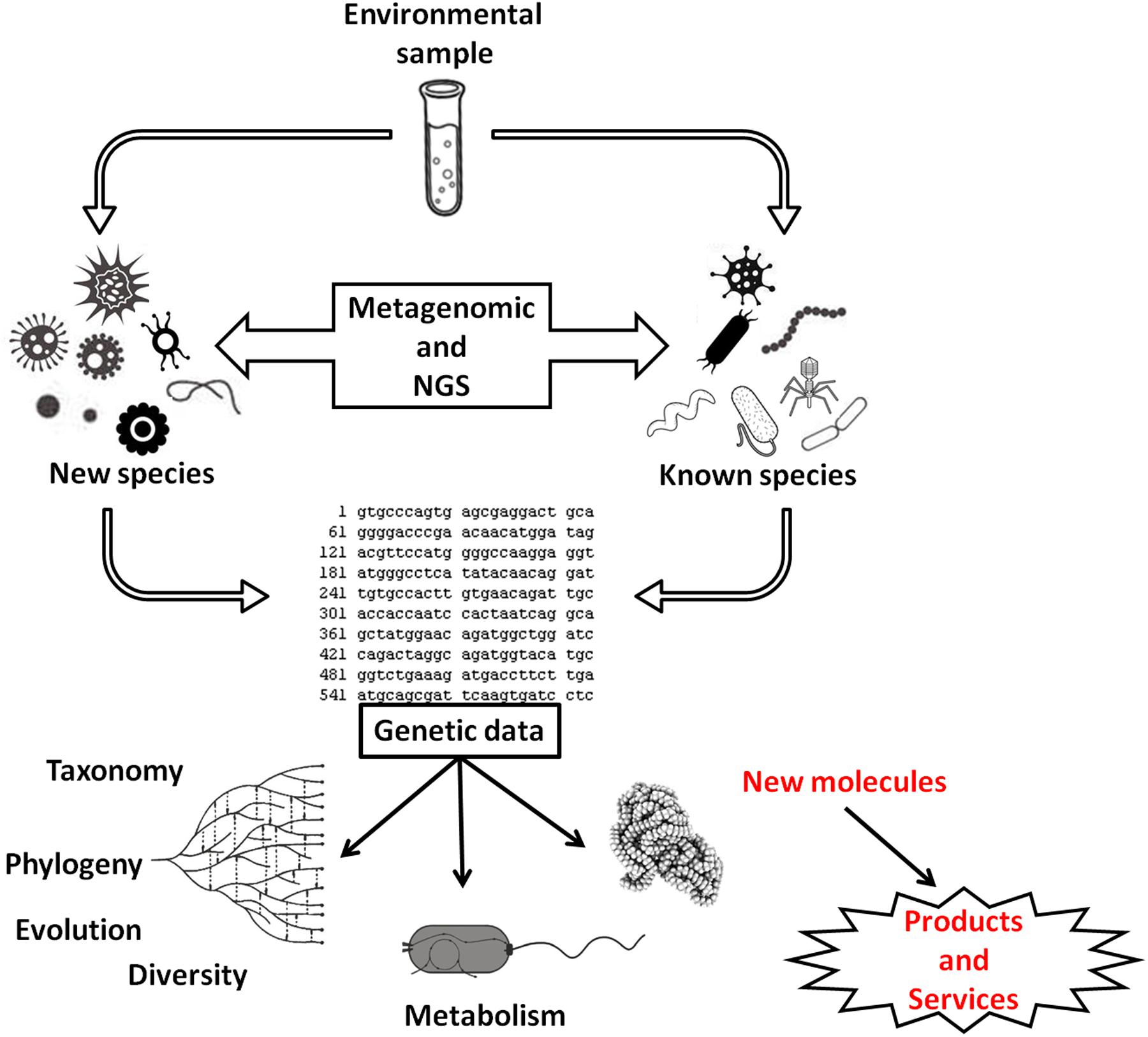
FIGURE 2. Use of metagenomics and next-generation sequencing in the study of microbial communities to obtain genetic data. These data can be used in studies of microbial diversity, group phylogeny, species diversification, and microbial metabolism. These data have also allowed the discovery of new or modified molecules used to obtain improved products, new products, or services.
A large variety of microorganisms, including heterotrophic or autotrophic aerobic bacteria, actinomycetes, fecal coliforms, and thermophiles, as well as yeasts and other fungi, have been reported in solid waste composting processes (Beffa et al., 1996; Tiquia et al., 2002). Many factors determine the microbial community present during composting, but under aerobic conditions, temperature is the main factor determining not only the microbial types but also the species diversity and metabolic rate (Hassen et al., 2001). On the other hand, the direct use of microbial enzymes in the treatment of effluents, especially industrial effluents, has been encouraged because the enzymatic action is faster, dispensing with the conditions necessary for the fermentative process. Lipases, for example, are used in the treatment of wastewater containing mainly triglycerides (Jamie et al., 2016). The presence of these enzymes in activated sludge and in other aerobic degradation processes is important for the continuous removal of the fat layers formed on the surface of aerated tanks to allow oxygen transport (Hasan et al., 2006). Peroxidases, phenoloxidases, dioxygenases, and phenoloxidase-like compounds have also been used for the removal of contaminants present in wastewater (Durán and Esposito, 2000). Peroxidases, polyphenol oxidases, and tyrosinases obtained from microorganisms such as P. syringae, Arthromyces ramosus, and Agaricus bisporus may be applied to the removal of phenols, biphenols, and chlorophenols (Tatsumi et al., 1996; Tong et al., 1998; Akay et al., 2002; Kampmann et al., 2014). Laccases of P. cinnabarinus were found to be efficient for the degradation of benzopyrene (Rama et al., 1998), while manganese peroxidases of Phanerochaete chrysosporium, Nematoloma frowardii, and Phlebia radiata can be applied to the elimination of lignin in wastewater (Hofrichter et al., 1999; Kunz et al., 2001).
Currently, research efforts have focused on integrating the treatment of solid wastes or even wastewater with the use of microbial fuel cells (MFC), i.e., microbial cells that use electrons donated by low-value organic substrates, contained in the waste, to generate energy (Xu et al., 2016). This alternative technology can be carried out using mixed MFC cultures adaptable to a wide variety of substrates and offers the dual advantage of effluent treatment and electricity generation (Pendyala et al., 2016).
Research efforts have also been directed at improving the purification of drinking water. A recent biotechnological process called biologically active carbon (BAC) has been found to be very efficient in removing water contaminants. In this process, microbial cells colonize the surface of the granular activated carbon (GAC) used in the filtering mechanism. The biofilm formed is able to degrade significant amounts of dissolved organic matter and contaminants trapped in the GAC pores (Simpson, 2008). In addition, the BAC biofilm can also biodegrade the cyanotoxins and organic substances that can change the taste and odor of potable water (Brown and Lauderdale, 2006).
Waste treatment based on enzymatic processes tends to be less expensive; however, the enzymes are biodegradable, and further studies and prospection of microbial enzymes that are thermostable or resilient to large pH variations are needed. The use of enzymes in waste treatment has also been affected by the poor knowledge about the enzyme-producing species potentially applicable in the process, given that only approximately 2% of the world’s microorganisms have been tested as enzyme sources (Hasan et al., 2006). Genetic improvement, as well as the genetic manipulation of cells and the heterologous expression of genes, is expected to help increase the enzymatic biosynthesis in microorganisms of interest, or even to contribute to the development of microorganism biofactories for important enzymes not only for food or industry but also for environmental applications, thereby expanding alternatives for the elimination of the wastes that have historically accumulated in soils and watercourses.
The increasing market demand for biodegradable polymers has also stimulated the prospection of microorganisms that act in the synthesis of these compounds. The class of biodegradable biopolymers of major interest is the polyhydroxyalkanoates (PHAs), and the best known among these are poly(beta-hydroxybutyrate; PHB), poly(beta-hydroxyvalerate; PHV), and poly(hydroxybutyrate-co-valerate; PHB-V), the latter being commercially known as Biopol. These biopolymers are accumulated intracellularly by bacteria as a carbon and/or energy reserve under the limitation of a nutrient essential for their growth, such as nitrogen, phosphorus, sulfur, or oxygen (Philip et al., 2007). The species Cupriavidus necator is one of those responding more favorably to the conditions for industrial production. This bacterium can accumulate approximately 80% of its dry mass in polymer and uses different types of substrates, such as glucose, fructose, and crude glycerin (Figueiredo et al., 2014). Biopolymers offer a possible solution for eliminating the problem of the residuals associated with petroleum-based plastic, but for researchers, the real challenge lies not in obtaining these molecules, which are the products of microbial metabolism, but in finding applications that consume large amounts of these materials, promoting price reductions and allowing biopolymers to compete economically in the market (Mohanty et al., 2000).
Another aspect of environmental technological microbiology is advances in the knowledge and use of the symbiotic relationship between plants and mycorrhizal fungi as a strategy to increase plant biomass or increase the yields of products of agricultural or pharmacological interest (e.g., Boyer et al., 2016; Gabriele et al., 2016; Köhl et al., 2016). Such benefits are the product of the positive and multifunctional roles of mycorrhizal fungi in plant nutrition, pathogen protection, stress tolerance, and soil structure supply (Smith and Read, 2008). Due to their unique biological traits, which include obligatory biotrophy and intracellular development within plant tissues, and multiple ecological functions, arbuscular mycorrhizal fungi (AMFs; which are also thought to have helped plants conquer terrestrial environments) have aroused agronomic interests with the aim of sustainable production with low chemical consumption (Lanfranco et al., 2016). In sustainable agriculture, AMFs are known as biofertilizers. In the plant-microorganism relationship, the mineral nutrients (mainly phosphorus, nitrogen and water) are extracted from the soil through the extensive network of hyphae and transferred to the plant, and organic compounds of carbon are transferred from the plant to the AMF. These microorganisms therefore reduce the need for application of chemical fertilizers in soils (e.g., Abdel-Fattah et al., 2016; Oliveira et al., 2016a,b). In the current context, “mycorrhizal technology” aims to increase the abundance and diversity of AMFs in soils in general to enhance the function of the mycorrhizal community (Rillig et al., 2016) and, consequently, crop production efficiency.
The participation of microorganisms in the generation of medical products or services involves four distinct aspects: (1) biocontrol of diseases, (2) production of vaccines, (3) production of antibiotics, and (4) production of biotherapeutics (hormones, biomaterials, and others). These aspects will be discussed throughout this session.
A problem commonly encountered in developing countries is the difficulty of implementing public policies to control the spread of parasitic vectors such as those of the genera Aedes and Anopheles. However, recent epidemic outbreaks of emerging and reemerging diseases have stimulated the development of biotechnological techniques that can not only assist diagnosis but also serve as alternatives for controlling transmission. An example is the potential presented by the introduction of the bacterium Wolbachia as an endosymbiont of the mosquito Aedes aegypti, which transmits diseases such as dengue, yellow fever, chikungunya, and the more recently detected Zika virus (Walker et al., 2011). The focus of this approach is on the reduction of mosquito longevity and not on abundance. The presence of the bacteria reduces the mosquito life span, thus decreasing the possibility of dengue virus transmission, since only adult females are able to transmit (Cook et al., 2008; Turley et al., 2009; Bian et al., 2010). Mosquitoes containing the Wolbachia wMelPop-CLA strain showed an approximately 50% reduction of the survival of females compared to mosquitoes without the strain (McMeniman et al., 2009). These bacteria are transmitted vertically from the female to the offspring. To guarantee this transmission, the bacterium manipulates its host in diverse ways such as feminization, death of males, parthenogenesis, and cytoplasmic incompatibility. In cytoplasmic incompatibility, fertilization of females not infected with Wolbachia by infected males results in embryonic mortality. By contrast, females infected with the bacterium will produce the highest number of viable offspring, increasing the number of infected individuals in the population (Figure 3A). Cytoplasmic incompatibility facilitates the propagation of Wolbachia in natural populations and their persistence over time (McMeniman et al., 2009).
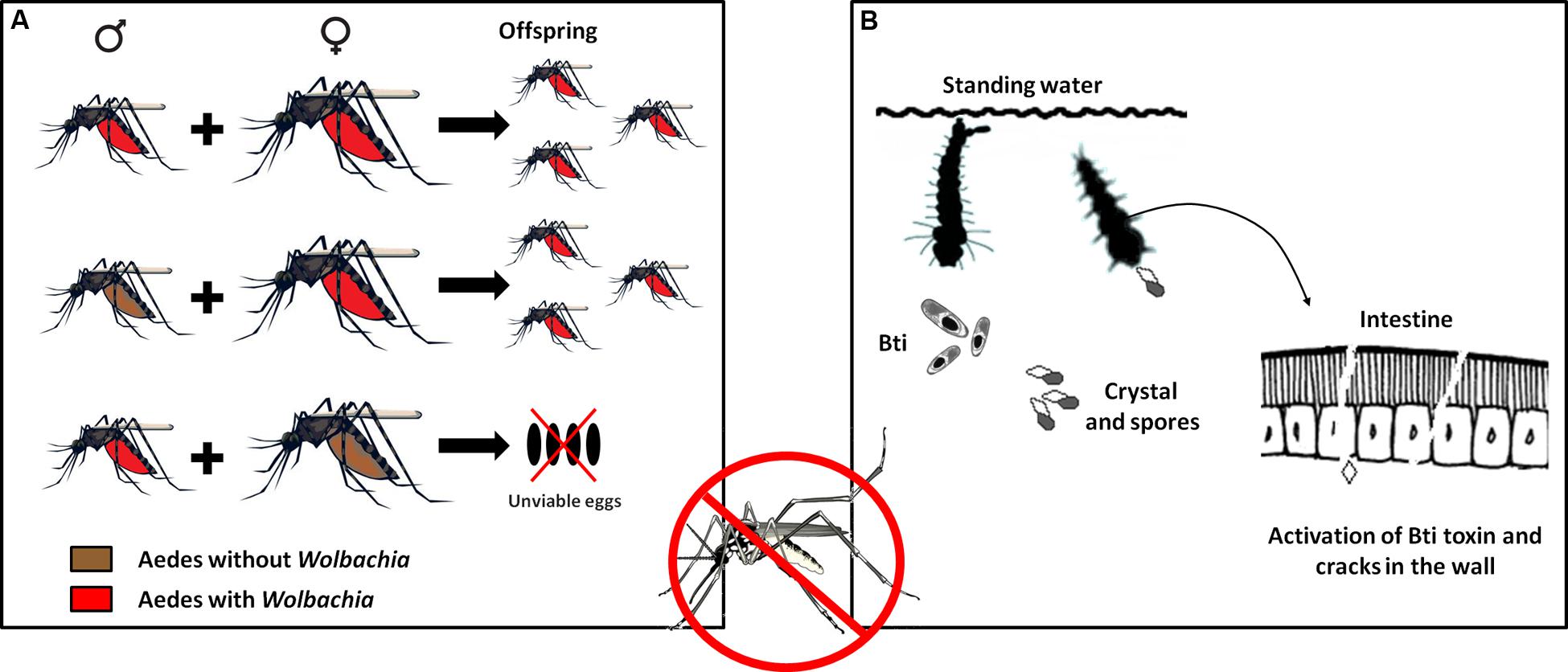
FIGURE 3. Use of Technological Microbiology to prevent the proliferation of the Aedes aegypti vector and the DENV virus. (A) Mechanisms of transmission of the bacterium Wolbachia to the offspring of the vector. Cytoplasmic incompatibility causes females with Wolbachia to always breed offspring with Wolbachia, whether mating with males with or without the bacterium. When females without Wolbachia mate with males with Wolbachia, the fertilized eggs die. With successive generations, the number of male and female mosquitoes with the bacterium tends to increase until the entire mosquito population bears this characteristic. (B) Use of Bacillus thuringiensis israelensis bioinsecticide to fight dengue. This bacterium synthesizes protein crystals that, when consumed by Aedes larvae, solubilize in the mosquito’s intestine and are transformed into efficient toxins that damage the intestinal wall, allowing the attack of pathogenic bacteria that cause the death of the larva.
Another alternative, also focused on preventing the proliferation of the dengue virus vector, is the use of a biological insecticide. The action of this insecticide is based on the activation of endotoxins (Cry and Cyt) produced naturally by the bacterium B. thuringiensis serotype israelensis (Bti) (Figure 3B). These toxins, previously mentioned in this text, are inactive crystals that, when ingested by Aedes larvae, are solubilized by intestinal proteases and converted into active toxins that interact with the cell membrane of the midgut, leading to pore formation, cell lysis, septicemia, and finally death (Gill et al., 1992; de Maagd et al., 2001). This alternative has been successfully applied in several regions of the world (e.g., Mohiddin et al., 2016; Setha et al., 2016), and many Bti-based commercial products have been introduced to the market, although some strains of A. aegypti have already shown resistance to Bt toxins (Paris et al., 2011, 2012; Wu et al., 2016).
At the same time, the technological race for the development of a vaccine against dengue continues. Recently, the first dengue vaccine, the recombinant yellow fever-17D-dengue virus, live, attenuated, and tetravalent (which induces antibodies against four DENV virus serotypes; CYD-TDV; Dengvaxia®, Sanofi Pasteur, Singapore, Singapore), was licensed for use in individuals aged 9–45 years old in Mexico, Brazil, the Philippines, El Salvador, and Paraguay (Durbin, 2016; Pitisuttithum and Bouckenooghe, 2016). This represents an important complement to the other techniques that focus mainly on vector control. This vaccine, however, is currently only available under private health care systems at very high prices.
The HPV (human papilloma virus) vaccine is another example of a newly developed vaccine that has been widely used in many countries. The focus of campaigns in developing countries has been on women who are not yet sexually active, although being sexually active does not contraindicate the vaccine, which can be tetravalent (protects against types 6, 11, 16, and 18) or bivalent (protects against types 16 and 18). This virus has been frequently linked to cases of cervical cancer, with types 16 and 18 accounting for approximately 70% of cases (Cutts et al., 2007). However, these same viral types account for 86–95% of cases of non-cervical cancer, i.e., anal, oropharynx, vulval, and vaginal cancers in women, as well as anal, oropharyngeal, and penile cancers in men (Gillison et al., 2008). Thus, prophylaxis should also extend to men as a way to prevent potential non-cervical cancers; however, affordable prices, funding mechanisms, and multidisciplinary partnerships are essential for the HPV vaccine to reach most populations in need, especially considering that cervical cancer is the second leading cause of death of cancer in women and is more worrying in populations that do not have screening programs to detect precursor lesions (Roden and Wu, 2006).
For the production of vaccines, microorganisms do not function as biofactories but are instead only used (whole or fractionated) to stimulate the synthesis of specific antibodies. Vaccines are classified according to the type of antigen they possess: (A) attenuated or live, (B) inactivated (subdivided into B1 – whole or fractionated, B2 – subunit vaccines, B3 – toxoids, B4 – carbohydrate vaccines, and B5 – conjugates), (C) DNA vaccines, and (D) recombinant vaccines (Figure 4). In attenuated vaccines, the pathogens (virus or bacteria) are alive and induce immune reactions similar to those resulting from a real infection (Plotkin et al., 2008). Attenuated vaccines have been efficiently developed for a range of diseases: mumps, polio (Sabin), rubella, measles, smallpox, chickenpox, tuberculosis, yellow fever, and dengue. These vaccines are considered highly immunogenic and efficiently stimulate humoral immunity, such that only one dose is capable of conferring immunity for decades. Recent studies have revealed strategies for the development of attenuated strains of the influenza virus that trigger robust immune responses (e.g., Si et al., 2016; Wang et al., 2017). These studies not only provide perspectives for producing more effective vaccines against influenza but also suggest innovative approaches for the generation of live attenuated strains for viral pathogens, which remains a challenge (Bournazos and Ravetch, 2017). Studies have also shown that live vaccines induce non-specific immunity, conferring protection beyond the target pathogen (e.g., Aaby et al., 2010; Higgins et al., 2014; Sørup et al., 2014), and that revaccination may be indicated to reduce general mortality caused by all diseases (Benn et al., 2016).
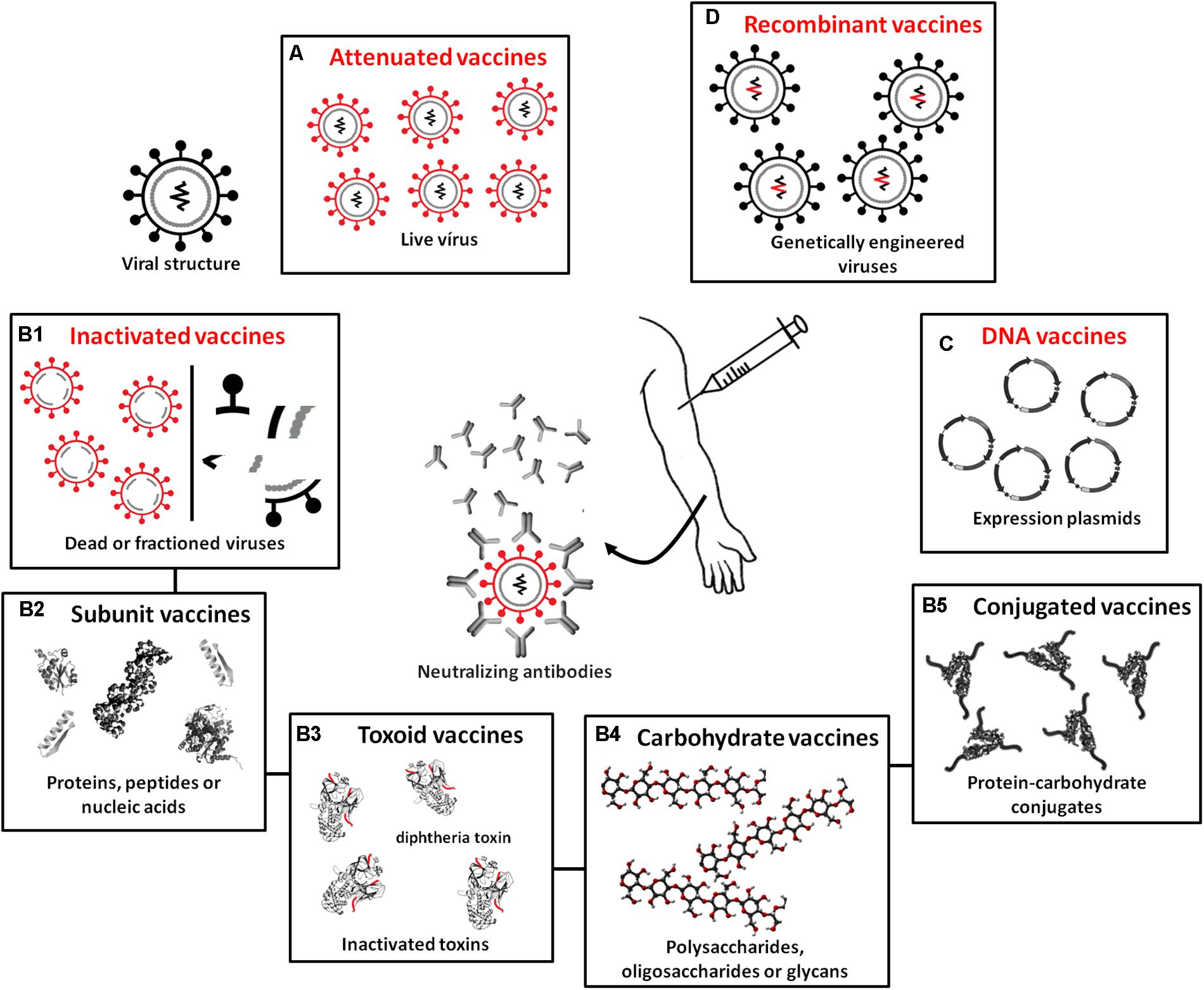
FIGURE 4. Different biotechnological techniques used in the production of currently available vaccine types. (A) Attenuated or live vaccines, which use attenuated pathogens. (B) Inactivated vaccines containing completely inactivated or fractionated pathogens or only antigenic components of these pathogens, subdivided into (B1) whole or fractioned; (B2) subunit vaccines, which use proteins, peptides, or nucleic acids as antigens; (B3) toxoids, which use inactivated pathogen toxins as antigens; (B4) carbohydrate vaccines produced from polysaccharides, oligosaccharides, and glycans; and (B5) conjugate vaccines, which have polysaccharides combined with transport proteins. (C) DNA vaccines carrying plasmids containing genes encoding immunogenic antigens. (D) Recombinant vaccines containing viruses engineered to carry genes encoding antigens from other disease-causing viruses.
Inactivated vaccines, which are classified as whole or fractionated, contain completely inactivated or fractionated pathogens or only antigenic components of these pathogens (including subunit vaccines, toxoid vaccines, carbohydrate vaccines, and conjugate vaccines). In general, these vaccines are effective stimulators of humoral immune responses, but in many cases, multiple doses are required for long-term immunity since they do not stimulate the production of immunoglobulin A (IgA) or cytotoxic T cell responses because the virus does not replicate. The advantage of inactivated vaccines is that they are safer because the virulence of dead organisms cannot be reversed. Inactivated vaccines are currently available for hepatitis A, rabies, cholera, influenza, poliomyelitis (Salk), typhoid fever, and pertussis. Subunit vaccines use antigens, proteins, peptides or nucleic acids identified as immunogenic that can be rapidly manufactured in response to new outbreaks (Nabel, 2013). These antigens are purified from microorganisms, produced by recombinant DNA techniques, or chemically synthesized. They are poorly reactogenic, which is an advantage in terms of adverse effects but a disadvantage in terms of stimulating potent and long-lasting immune responses (Bobbala and Hook, 2016). Thus, these vaccines often require the co-administration of efficient adjuvants to activate and modulate immune responses (Reed et al., 2009; Schmidt et al., 2016). Despite this poor reactogenicity, subunit vaccines have been developed against a variety of pathogens, including Streptococcus pneumoniae, hepatitis B virus, and HPV. By contrast, toxoid vaccines stimulate the immune response by using inactivated pathogen toxins as antigens. Vaccines of this type are available for tetanus, diphtheria, and anthrax. Carbohydrate vaccines are developed based on the knowledge that the vast majority of pathogens have dense distributions of polysaccharides, oligosaccharides, and complex glycans on their cell surface (glycocalyx) and that infected organisms detect the presence of pathogens through glycocalyx recognition and pattern recognition receptors (PRRs) that stimulate host defense responses (Astronomo and Burton, 2010; Pifferi et al., 2017). Low immunogenicity is a major obstacle to the manufacture of carbohydrate vaccines, and because of the lack of long-term immune protection by these antigens, few such vaccines are commercially available today. These include vaccines against Salmonella typhi and Neisseria meningitides (Keitel et al., 1994; King et al., 1996). Traditional covalent linkages between carbohydrates and carrier proteins have been used to increase immune responses to polysaccharide antigens (Nishat and Andreana, 2016), thereby obtaining conjugate vaccines. In this case, proteins function as carriers for capsular polysaccharides, which induce T-cell dependent antibody responses and B-cell differentiation in plasma cells and long-term memory. As a result, protein-polysaccharide vaccines induce immunological memory, a protection that is longer lasting than that induced by the use of simple polysaccharide antigens (Knuf et al., 2011; Frenck and Yeh, 2012; Pichichero, 2013). Conjugate vaccines are already currently used in the control of Haemophilus influenza type B and S. pneumoniae.
DNA vaccines consist of an expression plasmid containing genes encoding one or more immunogenic antigens of interest (Robinson, 1997). The use of viral promoters enhances gene expression and improves mRNA stability related to antigen synthesis. In addition, the incorporation of immunological adjuvants and new methods of insertion of this material into cells of the host organism beyond the intramuscular and dermal routes have increased the popularity as well as the immunogenicity of DNA vaccines, since the action of nucleases can inactivate plasmid DNA. The other insertion pathways with potential applications in humans include (1) bioballistics, which requires further technical enhancement for use in humans (Fynan et al., 1993; Brouillette et al., 2016); (2) intradermal needleless administration of the DNA plasmid: in tests conducted with non-human primates, these intradermal needleless devices did not increase immunogenicity compared to conventional syringes (Rao et al., 2006); (3) intradermal tattoo, in which the DNA plasmid is delivered to the epidermal layer using thousands of injections; and (4) through the mucosa, which simulates the entry of pathogens by this route. One approach for this last route uses bacteria as carriers of a DNA plasmid to target specific cells and activate receptors of pathogen-associated molecular patterns (Becker et al., 2008). These vaccines are still under clinical testing, and microbiological research efforts have focused on HIV, hepatitis B, hepatitis C, influenza, and HPV.
Recombinant (gene) vaccines are prepared from viruses engineered to carry genes encoding antigens from other disease-causing viruses for expression in the host after inoculation. This expression induces antibody production and immunization. The immunity induced by recombinant vaccines is usually attributed to the ability of the recombinant virus to express the gene of interest at high levels within the host cells. The viral vectors used for this purpose are attenuated to the host and are therefore intrinsically safe. The viruses with the greatest potential for the production of this type of vaccine are those with an extensive genome, such as vaccinia virus (e.g., He et al., 2000). In the manufacture of such vaccines, all genes that are not essential for replication are first eliminated from the virus. Subsequently, the genes of the other virus are introduced. Recombinant vaccines are not yet available clinically but are quite promising.
Recently, the incorporation of biotechnological techniques has allowed wide access to numerous monoclonal antibodies (mAbs). Human virus-neutralizing MAbs have already been isolated from non-immune and immune sources using a range of newly developed antibody isolation technologies. One such technology employs microorganisms, such as phages, yeasts, bacteria, and viruses, to display repertoires of single-chain variable-domain antibody fragments (ScFvs), antigen-binding fragments (Fab), or domain antibodies (Dabs) on their surfaces (Carter, 2006). These antibodies can also be obtained directly from memory B cells of viral-infected patients or even from mouse lymphocyte cells (Marasco and Sui, 2007). These antibodies have also been used for the treatment of infectious diseases. Recently, two antibodies were approved for this purpose: palivizumab, a human respiratory syncytial virus (RSV)-neutralizing monoclonal antibody that blocks virus replication; and raxibacumab, which prevents binding of the protective antigen of the anthrax toxin to its receptors in host cells (The IMpact-RSV Study Group, 1998; Kummerfeldt, 2014; Baldo, 2016). MAbs represent one of the largest classes of drugs in development, and between 2010 and 2014, 17 of the 54 protein drugs approved were mAbs (31.5%). These drugs therefore provide a new and promising way of thinking about the treatment of diseases caused by microorganisms.
In addition to increasing progress in the production of vaccines and MAbs, technological advances have also increased the availability of new drugs, such as antibiotics and hormones. The boom of antibiotic discovery occurred between 1950 and 1960. However, despite the need for new antibiotics, only two new classes of antibiotics have been introduced in medicine since 1963, both of which are based on nalidixic acid (Brito and Cordeiro, 2012). Limited research results, inappropriate prescription of antibiotics, and misuse of antibiotics by the general population have threatened antibiotic potency and increased the occurrence of superbugs, i.e., microorganisms that appear at an alarming rate and are resistant to most or all clinical antibiotics in use (Srivastava et al., 2011).
In the United States, the Centers for Disease Control and Prevention estimate that antibiotic-resistant bacteria infect more than two million people annually (Nizet, 2015). In this country alone, methicillin-resistant Staphylococcus aureus (MRSA) accounts for approximately 10,000 cases of hospital-acquired bloodstream infections, whereas Clostridium difficile, associated with diarrhea, is the most common infection in the United States, with more than 80,000 estimated annual cases (Magill et al., 2014). Many multi-antibiotic-resistant gram-negative bacilli also fit the description of superbugs, such as P. aeruginosa, K. pneumoniae, E. coli, Acinetobacter baumannii, and Stenotrophomonas maltophilia, and polymyxins have emerged as the major last-line of defense against these gram-negative superbugs (Velkov et al., 2016). Recently, Ling et al. (2015) developed an important method that allowed the in situ growth of soil microorganisms not cultivable under laboratory conditions. Hence, chemicals produced naturally by the microorganisms could be tested, such as teixobactin, the first compound of a new important class of antibiotics. Teixobactin is capable of eliminating MRSA, and bacteria are believed not to be susceptible to developing resistance to teixobactin, which targets the lipids essential for the maintenance of the bacterial cell wall (Borghesi and Stronati, 2015). Thus, biotechnological alternatives have been developed to circumvent the problem of the low rate of discovery of new antibiotic molecules. Selenium nanoparticles (SeNPs) combined with the synergistic properties of quercetin and acetylcholine showed inhibitory effect on MRSA. A study showed that SeNPs attach to the bacterial cell wall, causing irreversible damage to the membrane, thus achieving a remarkable synergistic antibacterial effect that inhibits MRSA (Huang X. et al., 2016).
The use of recombinant microbial cells has allowed large-scale production of a large number of products of pharmaceutical interest, such as hormones, anticoagulants, high-value proteins, antibodies or antigens, and others. This has been crucial in determining the structure-function relation of proteins, as well as for developing a better understanding of immune system reactions, cell biology, and signaling events. The major microorganisms explored as biofactories are the bacterium E. coli, followed by the yeast S. cerevisiae; both prokaryotic and eukaryotic systems are constantly evolving and competing to improve their properties and intensify as platforms of choice for the production of biopharmaceuticals (Chumnanpuen et al., 2016; Sanchez-Garcia et al., 2016). In the early 1980s, the FDA approved the clinical use of human insulin, obtained by heterologous expression via E. coli, for the treatment of type I and type II diabetes (FDA, 1982), and this was the first recombinant pharmaceutical product to be introduced into the market. Since then, the improvement of new heterologous protein production systems via E. coli enabled the commercial approval of several other products, including hormones (calcitonin, parathyroid hormone, human growth hormone, glucagon, and somatropin), interferons, and interleukins (Ferrer-Miralles et al., 2009). For example, approximately 30% of commercially available recombinant proteins are currently produced in prokaryotic systems (Overton, 2014). This production method is due to the unusual physiology of the cells as well as the ease of genetically manipulating them, but the understanding is that it is possible, by adding heterologous reactions, to synthesize 1,777 non-native products from E. coli, of which 279 have commercial applications. Among the latter are 4-hydroxybenzoate, tyrosine, and phenylalanine, which are precursors common to a large number of non-native commercial products (Zhang et al., 2016).
By contrast, the products of heterologous proteins obtained from S. cerevisiae comprise hormones (insulin, insulin analogs, and glucagon), vaccines (hepatitis B virus surface antigen), and virus-like particles (VLPs) (Ferrer-Miralles et al., 2009). Prokaryotic production systems are required whenever the recombinant proteins are smaller or do not require post-translational modifications (PTMs), such as glycosylation, phosphorylation, or proteolytic cleavage. However, production in yeasts, such as S. cerevisiae and Pichia pastoris, is generally required when the target protein cannot be produced in a soluble form in a prokaryotic system, when it is rich in disulfide bonds, or when a specific PTM, essential for the biological activity of the protein, cannot be artificially created from the purified product (Jenkins, 2007; Demain and Vaishnav, 2009).
Modified strains of S. carnosus, Corynebacterium glutamicum, B. subtilis, and Lactococcus lactis, for example, have been used in the controlled biological synthesis of calcitonin, amino acids (glutamate and lysine), proinsulin (Olmos-Soto and Contreras-Flores, 2003; Sandgathe et al., 2003; Liu et al., 2016), and protein nanoparticles (Cano-Garrido et al., 2016), respectively. Further manipulations of these species are expected to create strains capable of producing a wide variety of non-native commercial products.
Filamentous fungi, used for centuries in traditional Chinese medicine, have also been evaluated for the potential production of biopharmaceuticals. Polysaccharides of secondary metabolites can be obtained using Ganoderma lucidum, Cordyceps sinensis, and C. militaris (Paterson, 2006, 2008; Wadt et al., 2015). Endophytic fungi such as Metarhizium anisopliae and C. gloeosporioides (Gangadevi and Muthumary, 2008; Liu et al., 2009) were found to be efficient in the synthesis of Taxol, being a viable alternative to obtain this antineoplastic. The great diversity of molecules produced by filamentous fungi justifies the exploration of these microorganisms, and therefore, the development of production systems in bioreactors has been encouraged.
The metabolic engineering of molecules from microorganisms has been stimulated mainly by the need for new functional biomaterials in emerging drugs (nanostructured or not) (Vázquez and Villaverde, 2013). A large number of substances with nanomedical application have emerged, including polymers, metallic nanoparticles, magnetic nanoparticles, VLPs, virions or virion components, and a growing diversity of self-organized protein materials, some with adjustable biomechanical properties such as stiffness, elasticity, adhesion or controllable disintegration or release of incorporated functional blocks or conventional chemical drugs (Rodriguez-Carmona and Villaverde, 2010). Obviously, the success of the use of microbial nanoparticles in nanotechnology and nanomedicine depends on the identification of new species of microorganisms and on the knowledge of the microbial interactions that occur in natural environments that can lead to the discovery of new molecules (Bajaj et al., 2012; Avendaño et al., 2016; Borghese et al., 2016).
Recently, the term ‘biobetter’ has been used to refer to next-generation therapeutic macromolecules, which have a more effective drug delivery system. These macromolecules are modified by chemical and/or engineering methods using molecular biology techniques to display better pharmacological properties, such as higher activity, greater stability, fewer side effects, and lower immunogenicity (Beck, 2011; Jozala et al., 2016). Because they require original research and development, and because they propose alternative methods of administration such as dermatological applications and inhaled formulations in order to minimize biological instability, biobetters still have significantly higher costs compared to reference biopharmaceutical versions (Mitragotri et al., 2014; Sandeep et al., 2016). However, the future popularization of protein engineering techniques, especially site-directed mutagenesis (SDM), which allows the substitution, elimination or insertion of one or more amino acids in the sequence of a protein, is expected to enable the availability of less expensive biobetters, which are the main growing class of biopharmaceuticals (Courtois et al., 2016; Jozala et al., 2016).
The application of biotechnological techniques to microbiology has also made it possible to obtain a great diversity of biomaterials and biosensors. Biomaterials are artificial or natural products, usually synthesized by microorganisms in different environmental conditions, that can act in biological systems (tissues or organs). Biosensors integrate microorganisms with a physical transducer to generate a measurable signal proportional to the concentration of analytes, allowing rapid and accurate detection of analysis targets in diverse fields such as medicine, environmental monitoring, food processing, and others (D’Souza, 2001; Paitan et al., 2003; Lei et al., 2006; Su et al., 2011).
An important family of biomaterials includes the bioplastics. Bioplastics are polyesters that accumulate intracellularly in microorganisms in the form of storage granules, with physicochemical properties similar to petrochemical plastics. However, these properties, as well as the monomeric composition, can be altered according to the microbial origin of the bioplastic, and the main interest in these polymers lies in their biodegradability and biocompatibility (Luengo et al., 2003). Bioplastic can also be produced as a byproduct of biorefinery using acidogenic fermentation or pyrolysis of lignocellulosic biomass, as well as a by-product of the biotreatment of solid or liquid wastes (Ivanov and Stabnikov, 2016).
Bioplastics are being used in the manufacture of high-value-added medical materials, such as films that function as vehicles for drug delivery (Awadhiya et al., 2016). For use as medical materials, the high purity of PHAs is enhanced by using bioceramics and/or bioactive glasses that improve their biomechanical properties and increase their bioactivity (Radecka et al., 2016). Bioplastic formulations were recently tested for seed coating of agronomic species. These coatings, which contain spores of growth promoters such as T. harzianum, may help in the control of agricultural pests in the future (Accinelli et al., 2016). Bioplastics also have the potential to lead to the rise, within civil construction, of materials that have low incorporated energy, contributing to energy efficiency (Ivanov and Stabnikov, 2016).
Polysaccharides of microbial origin, such as chitosan, alginate, xanthan gum, and cellulose, are another class of biomaterials that have gained considerable interest for medical use because of their properties, including that they are renewable, biodegradable, and mimic the components of the extracellular matrix, which make them key elements in biological processes (Pires and Moraes, 2015; Pires et al., 2015). Chitosan can be easily recovered from the cell wall of fungi such as A. niger, Rhizopus oryzae, Cunninghamella elegans, and Mucor indicus, among others (Pochanavanich and Suntornsuk, 2002; Franco et al., 2004; Ruholahi et al., 2016; Abdel-Gawad et al., 2017). Specific interactions with extracellular matrix components allow the use of chitosan in the field of tissue engineering to repair skin, bone, and cartilage (Khor and Lim, 2003). The potential of fungal chitosan, when present in bioactive filters, to chelate heavy metals and inhibit pathogenic microbial agents in contaminated water was recently evaluated (e.g., Ruholahi et al., 2016; Tayel et al., 2016a) for the development of a renewable, ecofriendly, and cost-effective polymer that can help overcome the current problems of chemical and microbial water pollution (Tayel et al., 2016b).
Alginate is a polysaccharide synthesized by several genera of brown algae and two genera of bacteria: Pseudomonas and Azotobacter (Hay et al., 2013; Maleki et al., 2016). Its properties have broadened its use in the encapsulation or controlled release of drugs, enzymes, or cells, or as a matrix for tissue engineering (Andersen et al., 2012; Lee and Mooney, 2012), similarly to chitosan. When mixed in the aqueous phase, alginate and chitosan combine spontaneously by strong electrostatic attraction, forming a polyelectrolyte complex (PEC) that may be employed for the production of thin, transparent membranes that allow good absorption of physiological fluids, as well as the incorporation of several bioactive compounds (Pires and Moraes, 2015). Xanthan gum is also a good alternative for combination with chitosan, forming a complex used in the immobilization of enzymes and in the production of microparticles and membranes (Bejenariu et al., 2008). This exopolysaccharide is commercially synthesized by the bacterium Xanthomonas campestris, using different carbon sources (Barua et al., 2016; Li et al., 2016; Velu et al., 2016).
Cellulose synthesized (in abundance) by bacteria such as Gluconacetobacter xylinus (e.g., Huang C. et al., 2016) displays the same polymeric structure of cellulose from plants but is superior in its mechanical properties, purity, and uniformity, allowing the production of higher-quality devices (Pires et al., 2015). These devices include dialysis membranes and scaffolds for tissue engineering (Svensson et al., 2005). Microbial cellulose has great potential for the treatment of skin lesions and replacement of small-diameter blood vessels (Czaja et al., 2006).
The focus on microorganisms as an alternative in the production of biosensors is mainly due to the ability to produce them massively through cell culture (Su et al., 2011). In addition, the recombinant DNA technique has facilitated the availability of microbial biosensors in the market, providing a new direction to manipulate their selectivity and sensitivity at the DNA level. This technique consists of the construction of recombinant microbial strains that contain a reporter gene (lux, GFP, or lacZ), i.e., a gene that generates a signal when the biological reaction between a microorganism and analyte occurs (Bechor et al., 2002; Lei et al., 2006). An example of a microbial biosensor currently being used for pollutant monitoring purposes consists of immobilized recombinant E. coli cells expressing organophosphorus hydrolase (OPH). The OPH catalyzes the hydrolysis of organophosphorus pesticides releasing protons, whose concentration is proportional to the amount of substrate analyzed (Mulchandani et al., 1998; Kim H. J. et al., 2016). Recently, E. coli cells were developed to function as CadC-T7 biosensors, which are based on synthetic genetic circuits that combine a fluorescence reporter gene and heavy metal-responsive proteins. These biosensors showed high specificity for the detection of heavy metals (Kim K. R. et al., 2016). For environmental monitoring of cadmium, a biosensor in which cells express β-galactosidase in the presence of this metal is also available (Shin, 2016). Bioluminescent E. coli have also been used to signal DNA damage, superoxide radical production, and membrane damage caused by potentially toxic liquids (Bharadwaj et al., 2017).
In addition to the use of E. coli in biosensors, other microorganisms have already been evaluated. P. putida has already been tested as a biosensor for catechol, nitrophenol, benzene, toluene, and others (Rasinger et al., 2005; Timur et al., 2007; Banik et al., 2008); S. cerevisiae, for Cu2++ (Tag et al., 2007); Acidithiobacillus ferrooxidans and Leptospirillum ferrooxidans, for Fe2+, S2O32-, Cr2O72-, and others (Zlatev et al., 2006; Stoytcheva et al., 2009); and Gluconobacter oxydans, for propanediol and ethanol (Katrlik et al., 2007; Valach et al., 2009), among others. Despite the great leap forward made by biotechnology in the area of microbial biosensor development, many challenges still need to be overcome. New microorganisms still need to be evaluated for efficiency, more precise methods for immobilizing microbial cells still need to be developed, and the induction techniques need to be continuously evaluated because they may vary in terms of their efficiency depending on the analyte.
Microorganisms are the most biodiverse class, leading us to believe that the emergence and spread of new human and/or agricultural pathogens may shift from a current critical situation, aggravated by globalization, to a recurrent situation at various points in the future. Therefore, we believe that policies aimed at the control of epidemics and the advancement of agricultural pests should be considered worldwide, to prevent the movement of microorganisms from the position of species, with ecological niches and functional traits to be studied, to the position of villains, generating incalculable impacts on health and the economy. We must face the resurgence of diseases such as Zika and the appearance of superbugs as public health alerts, requiring emergency decision-making. These decisions must be made while considering the entire technological framework currently available, including transgenics and recombinant DNA, such that incentives exist so that microbiology research can be converted into products and services for society (Figure 5).
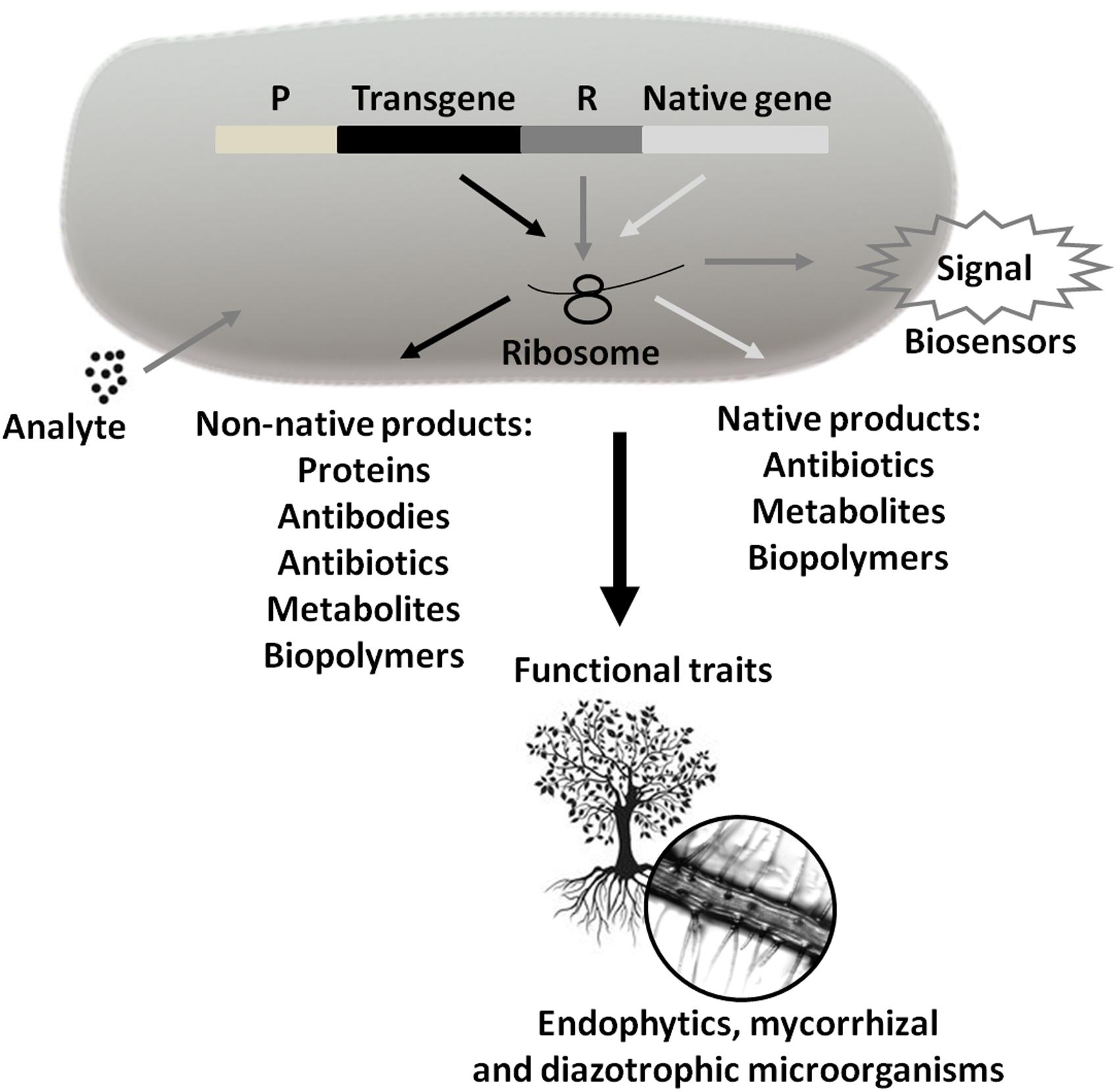
FIGURE 5. Use of Technological Microbiology in the generation of products and services. These products and services can be obtained from the expression of transgenes or native microbial genes (P = promoter and R = reporter). Marker expression generates signals that may indicate the presence and concentration of analytes (biosensors). In turn, the symbiotic interaction between plant species and endophytic, mycorrhizal, and/or diazotrophic microorganisms can help plant growth and development through N2 uptake, immobilized phosphate solubilization, siderophore production, competition with phytopathogenic species, etc. Arrows of the same color inside the bacterium signal the same pathway.
Not only biodiversity but also the unique nature and the biosynthetic capacities under specific environmental conditions make microorganisms the probable candidates to solve particularly complex environmental problems, such as the biodegradation of xenobiotics, or even recurrent problems, such as the decomposition of garbage and waste piles produced daily in urban environments. Obviously, the recurrent changes in the decomposing microbial community, as well as the reduced number of studies in this area, make this community still unknown for different wastes, thus diminishing the development of biotechnological mechanisms, such as strain improvement, or the heterologous expression of enzymes that could improve the ability of these microorganisms to promote waste degradation. This is a challenge to be faced by Technological Microbiology in the coming years.
The group’s biodiversity, however, also means microorganisms offer the greatest potential for the exploration of molecules and processes, and the knowledge of unconventional species, especially within the Archaea group, has stimulated the research of genes of interest. These new genes may be incorporated by recombinant technology into biologically known species, such as E. coli and S. cerevisiae, for the large-scale synthesis of products. To date, molecular strategies have advanced by establishing heterologous expression systems for the production of valuable industrial compounds, such as biofuels, chemicals, pharmaceuticals, enzymes, and food ingredients. However, Technological Microbiology has obstacles to overcome, and these obstacles extend beyond the continuous search for unconventional microbial species with valuable metabolic properties. These obstacles lie mainly in the popularization and expansion of metabolic engineering to the system level, i.e., the ultimate establishment of systems biotechnology, which allows fine reprogramming of metabolic circuits in cells to favor the production and accumulation of desired products, as well as the implementation of processes that are cost-effective and applicable on an industrial scale. Most of the time, the products and processes generated by systems biotechnology are expensive and of little benefit when implemented on a large scale. Thus, only in-depth research in this area could result in more complex and efficient microbial factories.
LV conceived this article, reviewed the existing literature, participated in writing, and created the figures. LB participated in writing and critically reviewed all content.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aaby, P., Martins, C. L., Garly, M. L., Bale, C., Andersen, A., Rodrigues, A., et al. (2010). Non-specific effects of standard measles vaccine at 4.5 and 9 months of age on childhood mortality: randomised controlled trial. BMJ 341:c6469. doi: 10.1136/bmj.c6495
Abdel-Fattah, G. M., Shukry, W. M., Shokr, M. M. B., and Ahmed, M. A. (2016). Application of mycorrhizal technology for improving yield production of common bean plants. Int. J. Appl. Sci. Biotechnol. 4, 191–197. doi: 10.3126/ijasbt.v4i2.15103
Abdel-Gawad, K. M., Hifney, A. F., Fawzy, M. A., and Gomaa, M. (2017). Technology optimization of chitosan production from Aspergillus niger biomass and its functional activities. Food Hydrocoll. 63, 593–601. doi: 10.1016/j.foodhyd.2016.10.001
Accinelli, C., Abbas, H. K., Little, N. S., Kotowicz, J. K., Mencarelli, M., and Shier, W. T. (2016). A liquid bioplastic formulation for film coating of agronomic seeds. Crop Prot. 89, 123–128. doi: 10.1016/j.foodhyd.2016.10.001
Adedayo, M. R., Ajiboye, E. A., Akintunde, J. K., and Odaibo, A. (2011). Single cell proteins: as nutritional enhancer. Adv. Appl. Sci. Res. 2, 396–409.
Adejuwon, A. O., Oluduro, A. O., Agboola, F. K., Olutiola, P. O., Burkhardt, B. A., and Segal, S. J. (2015). Expression of α-amylase by Aspergillus niger: effect of nitrogen source of growth medium. Adv. Biosci. Bioeng. 3, 12–19.
Adham, N. Z. (2002). Attempts at improving citric acid fermentation by Aspergillus niger in beet-molasses medium. Bioresour. Technol. 84, 97–100. doi: 10.1016/S0960-8524(02)00007-X
Ahmad, S., Abbas, S. S., Prakash, R., Alam, A., and Husain, M. A. (2016). Applications of endophytic Actinomycetes and their role in protection. Imp. J. Interdiscip. Res. 2, 854–858.
Akay, G., Erhan, E., Keskinler, B., and Algur, O. F. (2002). Removal of phenol from wastewater using membrane-immobilized enzymes Part II. Cross-flow filtration. J. Membr. Sci. 206, 61–68. doi: 10.1016/S0376-7388(01)00626-3
Ames, B. N., and Martin, R. G. (1964). Biochemical aspects of genetics: the operon. Ann. Rev. Biochem. 33, 235–258. doi: 10.1146/annurev.bi.33.070164.001315
Andersen, B., Poulsen, R., and Hansen, G. H. (2016). Cellulolytic and xylanolytic activities of common indoor fungi. Int. Biodeter. Biodegr. 107, 111–116. doi: 10.1016/j.ibiod.2015.11.012
Andersen, T., Strand, B. L., Formo, K., Alsberg, E., and Christensen, B. E. (2012). “Alginates as biomaterials in tissue engineering,” in Carbohydrate Chemistry: Chemical and Biological Approaches, ed. A. P. Rauter (Cambridge: The Royal Society of Chemistry), 227–258. doi: 10.1039/9781849732765-00227
Anupama, and Ravindra, P. (2000). Value-added food: single cell protein. Biotechnol. Adv. 18, 459–479. doi: 10.1016/S0734-9750(00)00045-8
Appels, L., Baevens, J., Degrève, J., and Dewil, R. (2008). Principles and potential of the anaerobic digestion of waste-activated sludge. Prog. Energy Combust. Sci. 34, 755–781. doi: 10.1016/j.pecs.2008.06.002
Arber, W. (1974). DNA Modification and Restriction. Prog. Nucleic Acid Res. Mol. Biol. 14, 1–37. doi: 10.1016/S0079-6603(08)60204-4
Astronomo, R. D., and Burton, D. R. (2010). Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat. Rev. Drug. Discov. 9, 308–324. doi: 10.1038/nrd3012
Avendaño, R., Chaves, N., Fuentes, P., Sánchez, E., Jiménez, J. I., and Chavarría, M. (2016). Production of selenium nanoparticles in Pseudomonas putida KT2440. Sci. Rep. 6, 37155. doi: 10.1038/srep37155
Awadhiya, A., Tyeb, S., Rathore, K., and Verma, V. (2016). Agarose bioplastic-based drug delivery system for surgical and wound dressings. Eng. Life Sci. 17, 204–214. doi: 10.1002/elsc.201500116
Bagdasarian, M., and Timmis, K. N. (1982). Host: vector systems for gene cloning in Pseudomonas. Curr. Top. Microbiol. 96, 47–67.
Bajaj, M., Schmidt, S., and Winter, J. (2012). Formation of Se (0) nanoparticles by Duganella sp. and Agrobacterium sp. isolated from Se-laden soil of North-East Punjab, India. Microb. Cell Fact. 11:64. doi: 10.1186/1475-2859-11-64
Baldo, B. A. (2016). “Other approved therapeutic monoclonal antibodies,” in Safety of Biologics Therapy (New York City, NY: Springer International Publishing), 141–215. doi: 10.1007/978-3-319-30472-4_4
Banik, R. M., Mayank Prakash, R., and Upadhyay, S. N. (2008). Microbial biosensor based on whole cell of Pseudomonas sp. for Online Measurement of p-Nitrophenol. Sens. Actuatators B Chem. 131, 295–300. doi: 10.1016/j.snb.2007.11.022
Barua, R., Alam, M. J., Salim, M., and Ashrafee, T. S. (2016). Small scale production and characterization of xanthan gum synthesized by local isolates of Xanthomonas campestris. Indian J. Exp. Biol. 54, 151–155.
Bechor, O., Smulski, D. R., Van Dyk, T. K., LaRossa, R. A., and Belkin, S. (2002). Recombinant microorganisms as environmental biosensors: pollutants detection by Escherichia coli bearing fabA’::lux fusions. J. Biotechnol. 94, 125–132. doi: 10.1016/S0168-1656(01)00423-0
Beck, A. (2011). Biosimilar, Biobetter and next generation therapeutic antibodies. MAbs 3, 107–110. doi: 10.4161/mabs.3.2.14785
Becker, P. D., Noerder, M., and Guzmán, C. A. (2008). Genetic immunization: bacteria as DNA vaccine delivery vehicles. Hum. Vaccin. 4, 189–202. doi: 10.4161/hv.4.3.6314
Beffa, T., Blanc, M., Marilley, L., Fischer, J. L., Lyon, P.-F., and Aragno, M. (1996). “Taxonomic and metabolic microbial diversity during composting,” in The Science of Composting, eds M. Bertoldi, P. Sequi, B. Lemmes, and T. Papi (Dordrecht: Springer), 149–161. doi: 10.1007/978-94-009-1569-5-16
Bejenariu, A., Popa, M., Cerf, D. L., and Picton, L. (2008). Stiffness xanthan hydrogels: synthesis, swelling characteristics and controlled release properties. Polym. Bull. 61, 631–641. doi: 10.1007/s00289-008-0987-6
Belda, I., Ruiz, J., Navascués, E., Marquina, D., and Santos, A. (2016). Improvement of aromatic thiol release through the selection of yeasts with increased β-lyase activity. Int. J. Food. Microbiol. 225, 1–8. doi: 10.1016/j.ijfoodmicro.2016.03.001
Benn, C. S., Fisker, A. B., Whittle, H. C., and Aaby, P. (2016). Revaccination with live attenuated vaccines confer additional beneficial nonspecific effects on overall survival: a review. EBioMedicine 10, 312–317. doi: 10.1016/j.ebiom.2016.07.016
Berg, P., and Mertz, J. E. (2010). Personal reflections on the origins and emergence of recombinant DNA technology. Genetics 184, 9–17. doi: 10.1534/genetics.109.112144
Bharadwaj, S., Mitchell, R. J., Qureshi, A., and Niazi, J. H. (2017). Toxicity evaluation of e-juice and its soluble aerosols generated by electronic cigarettes using recombinant bioluminescent bacteria responsive to specific cellular damages. ıBiosens. Bioelectron. 90, 53–60. doi: 10.1016/j.bios.2016.11.026
Bian, G., Xu, Y., Lu, P., Xie, Y., and Xi, Z. (2010). The Endosymbiotic bacterium Wolbachia induces resistance to Dengue virus in Aedes aegypti. PLoS Pathog. 6:e1000833. doi: 10.1371/journal.ppat.1000833
Biebl, H., Menzel, K., Zeng, A.-P., and Deckwer, W.-D. (1999). Microbial production of 1,3-propanediol. Appl. Microbiol. Biotechnol. 52, 289–297. doi: 10.1007/s002530051523
Bobbala, S., and Hook, S. (2016). Is there an optimal formulation and delivery strategy for subunit vaccines? Pharm. Res. 33, 2078–2097. doi: 10.1007/s11095-016-1979-0
Borghese, R., Brucale, M., Fortunato, G., Lanzi, M., Mezzi, A., Valle, F., et al. (2016). Extracellular production of tellurium nanoparticles by the photosynthetic bacterium Rhodobacter capsulatus. J. Hazard. Mater. 309, 202–209. doi: 10.1016/j.jhazmat.2016.02.011
Borghesi, A., and Stronati, M. (2015). Superbugs and antibiotics in the newborn. J. Pediatr. Neonat. Individ. Med. 4:e040253. doi: 10.7363/040253
Borneman, A. R., Schmidt, S. A., and Pretorius, I. S. (2013). At the cutting-edge of grape and wine biotechnology. Trends Genet. 29, 263–271. doi: 10.1016/j.tig.2012.10.014
Bournazos, S., and Ravetch, J. V. (2017). Attenuated vaccines for augmented immunity. Cell Host Microbe 21, 314–315. doi: 10.1016/j.chom.2017.02.016
Boyer, L. R., Feng, W., Gulbis, N., Hadju, K., Harrison, R. J., Jeffries, P., et al. (2016). The use of arbuscular mycorrhizal fungi to improve strawberry production in coir substrate. Front. Plant Sci. 7:1237. doi: 10.3389/fpls.2016.01237
Brito, M. A., and Cordeiro, B. C. (2012). The need of new antibiotics. J. Bras. Patol. Med. Lab. 48, 247–249. doi: 10.1590/S1676-24442012000400002
Brouillette, M., Doré, M., Hébert, C., Spooner, M.-F., Marchand, S., Côté, J., et al. (2016). A new biolistic intradermal injector. Shock Waves 26, 25–37. doi: 10.1007/s00193-013-0464-5
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Brown, J., and Lauderdale, C. (2006). Efficient, simultaneous destruction of multiple drinking water contaminants using biological filtration. Florida Water Resour. J. 3, 28–30.
Bueno, M. M., Thys, R. C. S., and Rodrigues, R. C. (2016). Microbial enzymes as substitutes of chemical additives in baking wheat flour-part II: combined effects of nine enzymes on dough rheology. Food Bioprocess Technol. 9, 1598–1611. doi: 10.1007/s11947-016-1744-1748
Cano-Garrido, O., Céspedes, M. V., Unzueta, U., Saccardo, P., Roldán, M., Sánchez-Chardi, A., et al. (2016). CXCR4+-targeted protein nanoparticles produced in the food-grade bacterium Lactococcus lactis. Nanomedicine 11, 2387–2398. doi: 10.2217/nnm-2016-0200
Carrero-Carrón, I., Trapero-Casas, J. L., Olivares-García, C., Monte, E., Hermosa, R., and Jiménez-Díaz, R. M. (2016). Trichoderma asperellum is effective for biocontrol of Verticillium wilt in olive caused by the defoliating pathotype of Verticillium dahliae. Crop Prot. 88, 45–52. doi: 10.1016/j.cropro.2016.05.009
Carroll, A. L., Desai, S. H., and Atsumi, S. (2016). Microbial production of scent and flavour compounds. Curr. Opin. Biotechnol. 37, 8–15. doi: 10.1016/j.copbio.2015.09.003
Carter, P. J. (2006). Potent antibody therapeutics by design. Nat. Rev. Immunol. 6, 343–357. doi: 10.1038/nri1837
Castelle, C. J., Wrighton, K. C., Thomas, B. C., Hug, L. A., Brown, C. T., Wilkins, M. J., et al. (2015). Genomic expansion of domain Archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr. Biol. 25, 690–701. doi: 10.1016/j.cub.2015.01.014
Cheng, L., Chen, W., Adams, T. S., Wei, X., Li, L., McCormack, M. L., et al. (2016). Mycorrhizal fungi and roots are complementary in foraging within nutrient patches. Ecology 97, 2815–2823. doi: 10.1002/ecy.1514
Chien, A., Edgar, D. B., and Trela, J. M. (1976). Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J. Bacteriol. 127, 1550–1557.
Chojnacka, A., Szczêsny, P., Błaszczyk, M. K., Zielenkiewicz, U., Detman, A., Salamon, A., et al. (2015). Noteworthy facts about a methane-producing microbial community processing acidic effluent from sugar beet molasses fermentation. PLoS ONE 10:e0128008. doi: 10.1371/journal.pone.0128008
Chouari, R., Le, P. D., Daegelen, P., Ginestet, P., Weissenbach, J., and Sghir, A. (2005). Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7, 1104–1115. doi: 10.1111/j.1462-2920.2005.00795.x
Chumnanpuen, P., Kocharin, K., and Vongsangnak, W. (2016). “Yeast expression systems for industrial biotechnology,” in Gene Expression Systems in Fungi: Advancements and Applications, Part of the Series Fungal Biology, eds M. Schmoll and C. Dattenböck (New York City, NY: Springer International Publishing), 227–237. doi: 10.1007/978-3-319-27951-0-9
Cohen, S. N., Chang, A. C., Boyer, H. W., and Helling, R. B. (1973). Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U.S.A. 70, 3240–3244. doi: 10.1073/pnas.70.11.3240
Coker, J. A. (2016). Extremophiles and biotechnology: current uses and prospects. F1000 Res. 5, 396. doi: 10.12688/f1000research.7432.1
Cook, P. E., McMeniman, C. J., and O’Neill, S. L. (2008). Modifying insect population age structure to control vector-borne disease. Adv. Exp. Med. Biol. 627, 126–140. doi: 10.1007/978-0-387-78225-6-11
Courtois, F., Agrawal, N. J., Lauer, T. M., and Trout, B. L. (2016). Rational design of therapeutic mAbs against aggregation through protein engineering and incorporation of glycosylation motifs applied to bevacizumab. MAbs 8, 99–112. doi: 10.1080/19420862.2015.1112477
Cutts, F. T., Franceschi, S., Goldie, S., Castellsague, X., de Sanjose, S., Garnett, G., et al. (2007). Human papillomavirus and HPV vaccines: a review. Bull. World Health Organ. 85, 719–726. doi: 10.1590/S0042-96862007000900018
Czaja, W., Krystynowics, A., Bielecki, S., and Brown, R. M. Jr. (2006). Microbial cellulose-the natural power to heal wounds. Biomaterials 27, 145–151. doi: 10.1016/j.biomaterials.2005.07.035
Davies, K. G., Rowe, J., Manzanilla-López, R., and Opperman, C. H. (2011). Re-evaluation of the life-cycle of the nematode-parasitic bacterium Pasteuria penetrans in root-knot nematodes. Meloidogyne spp. Nematologica 13, 825–835. doi: 10.1163/138855410X552670
de Maagd, R. A., Bravo, A., and Crickmore, N. (2001). How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 17, 193–199. doi: 10.1016/S0168-9525(01)02237-5
Degré, R., Edwards, G., and Zhang, Z. (2008). Novel Vitamin D2 Yeast Preparation, a Method for Producing the Same, and the use thereof. US 2008/0138469.
Demain, A. L., and Vaishnav, P. (2009). Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 27, 297–306. doi: 10.1016/j.biotechadv.2009.01.008
Djekrif-Dakhmouche, S., Gheribi-Aoulmi, Z., Meraihi, Z., and Bennamoun, L. (2006). Application of a statistical design to the optimization of culture medium for α-amylase production by Aspergillus niger ATCC 16404 grown on orange waste powder. J. Food Eng. 73, 190–197. doi: 10.1016/j.jfoodeng.2005.01.021
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B. A., Kenerley, C. M., Monet, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
D’Souza, S. F. (2001). Microbial biosensors. Biosens. Bioelectron. 16, 337–353. doi: 10.1016/S0956-5663(01)00125-7
Duke, S. O., Scheffler, B. E., Boyette, C. D., and Dayan, F. E. (2015). Biotechnology in Weed Control. Kirk-Othmer Encyclopedia of Chemical Technology. New York, NY: John Wiley & Sons, Inc. doi: 10.1002/0471238961.herbduke.a01.pub2
Durán, N., and Esposito, E. (2000). Potential applications of oxidative enzymes and phenoloxidase-like compounds in wastewater and soil treatment: a review. Appl. Catal. B Environ. 28, 83–99. doi: 10.1016/S0926-3373(00)00168-5
Ennouri, H., Miladi, B., Diaz, S. Z., Guelfo, L. A. F., Solera, R., Hamdi, M., et al. (2016). Effect of thermal pretreatment on the biogas production and microbial communities balance during anaerobic digestion of urban and industrial waste activated sludge. Bioresour. Technol. 214, 184–191. doi: 10.1016/j.biortech.2016.04.076
Enujiugha, V. N., and Badejo, A. A. (2017). Probiotic potentials of cereal-based beverages. Crit. Rev. Food Sci. Nutr. 57, 790–804. doi: 10.1080/10408398.2014.930018
Ferrer-Miralles, N., Domingo-Espín, J., Corchero, J. L., Vázquez, E., and Villaverde, A. (2009). Microbial factories for recombinant pharmaceuticals. Microb. Cell Fact. 8:17. doi: 10.1186/1475-2859-8-17
Feyisa, B., Lencho, A., Selvaraj, T., and Getaneh, G. (2016). Evaluation of some botanicals and Trichoderma harzianum against root-knot nematode (Meloidogyne incognita (Kofoid and White) Chit wood) in tomato. J. Entomol. Nematol. 8, 11–18. doi: 10.5897/JEN2015.0145
Figueiredo, T. V. B., Campos, M. I., Sousa, L. S., Silva, J. R., and Druzian, J. I. (2014). Produção e caracterização de polihidroxialcanoatos obtidos por fermentação da glicerina bruta residual do biodiesel. Quím. Nova 37, 1111–1117. doi: 10.5935/0100-4042.20140183
Franco, L. O., Maia, R. C. C., Porto, A. L. F., Messias, A. S., Fukushima, K., and Campostakaki, G. M. (2004). Heavy metal biosorption by chitin and chitosan isolated from Cunninghamella elegans (IFM 46109). Braz. J. Microbiol. 35, 2004. doi: 10.1590/S1517-83822004000200013
Frenck, R. W., and Yeh, S. (2012). The development of 13-valent pneumococcal conjugate vaccine and its possible use in adults. Expert Opin. Biol. Ther. 12, 63–77. doi: 10.1517/14712598.2012.636348
Fu, Y. Q., Yin, L. F., Zhu, H. Y., Jiang, R., Li, S., and Xu, Q. (2014). Effects of pellet characteristics on L-lactic acid fermentation by R. oryzae: pellet morphology, diameter, density, and interior structure. Appl. Biochem. Biotechnol. 174, 2019–2030. doi: 10.1007/s12010-014-1146-1
Fynan, E. F., Wwbster, R. G., Fuller, D. H., Haynes, J. R., Santoro, J. C., and Robinson, H. L. (1993). DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc. Natl. Acad. Sci. U.S.A. 90, 11478–11482. doi: 10.1073/pnas.90.24.11478
Gabriele, M., Gerardi, C., Longo, V., Lucejko, J., Degano, I., Pucci, L., et al. (2016). The impact of mycorrhizal fungi on Sangiovese red wine production: phenolic compounds and antioxidant properties. LWT Food Sci. Technol. 72, 310–316. doi: 10.1016/j.lwt.2016.04.044
Gal, J. (2008). The discovery of biological enantioselectivity: louis Pasteur and the fermentation of tartaric acid, 1857-A review and analysis 150 yr later. Chirality 20, 5–19. doi: 10.1002/chir.20494
Gangadevi, V., and Muthumary, J. (2008). Isolation of Colletotrichum gloeosporioides, a novel endophytic taxol-producing fungus from the leaves of a medicinal plant, Justicia gendarussa. Mycol. Balc. 5, 1–4.
Gao, C., Ma, C., and Xu, P. (2011). Biotechnological routes based on lactic acid production from biomass. Biotechnol. Adv. 29, 930–939. doi: 10.1016/j.biotechadv.2011.07.022
Gawkowski, D., and Chikindas, M. L. (2016). Non-dairy probiotic beverages: the next step into human health. Benef. Microbes 4, 127–142. doi: 10.3920/BM2012.0030
Gill, S. S., Cowles, E. A., and Pietrantonio, P. V. (1992). The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 37, 615–636. doi: 10.1146/annurev.en.37.010192.003151
Gillison, M. L., Chaturvedi, A. K., and Lowy, D. R. (2008). HPV prophylactic vaccines and the potential prevention of noncervical cancers in both men and women. Cancer 113, 3036–3046. doi: 10.1002/cncr.23764
Godon, J. J., Zumstein, E., Dabert, P., Habouzit, F., and Moletta, R. (1997). Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63, 2802–2813.
Goswami, R., Chattopadhyay, P., Shome, A., Banerjee, S. N., Chakraborty, A. K., Mathew, A. K., et al. (2016). An overview of physico-chemical mechanisms of biogas production by microbial communities: a step towards sustainable waste management. 3 Biotech 6, 72. doi: 10.1007/s13205-016-0395-9
Hasan, F., Shah, A. A., and Hameed, A. (2006). Industrial applications of microbial lipases. ıEnzyme Microb. Technol. 39, 235–251. doi: 10.1016/j.enzmictec.2005.10.016
Hassen, A., Belguith, K., Jedidi, N., Cherif, A., Cherif, M., and Boudabous, A. (2001). Microbial characterization during composting of municipal solid waste. Bioresour. Technol. 80, 217–225. doi: 10.1016/S0960-8524(01)00065-7
Hay, I. D., Rehman, Z. U., Moradali, M. F., Wang, Y., and Rehm, B. H. A. (2013). Microbial alginate production, modification and its applications. Microb. Biotechnol. 6, 637–650. doi: 10.1111/1751-7915.12076
He, Z., Wlazlo, A. P., Kowalczyk, D. W., Cheng, J., Xiang, Z. Q., Giles-Daves, W., et al. (2000). Viral recombinant vaccines to the E6 and E7 antigens of HPV-16. Virology 270, 146–161. doi: 10.1006/viro.2000.0271
Higgins, J. P. T., Soares-Weiser, K., and Reingold, A. (2014). Systematic Review of the Non-Specific Effects of BCG, DTP and Measles Containing Vaccines. Available at: http://www.who.int/immunization/sage/meetings/2014/april [accessed June 1, 2014].
Hofrichter, M., Vares, K., Scheibner, K., Galkin, S., Sipila, J., and Hatakka, A. (1999). Mineralization and solubilization of synthetic lignin by manganese peroxidases from Nematoloma frowardii and Phlebia radiata. J. Biotechnol. 67, 217–228. doi: 10.1016/S0168-1656(98)00180-1
Hofvendahl, K., and Hahn-Hägerdal, B. (2000). Factors affecting the fermentative lactic acid production from renewable resources. Enzyme Microb. Technol. 26, 87–107. doi: 10.1016/S0141-0229(99)00155-6
Holloway, B. W. (2014). “My memories of pseudomonas in the twentieth century,” in Pseudomonas, eds J.-L. Ramos, J. B. Goldberg, and A. Filloux (Dordrecht: Springer), 289–314. doi: 10.1007/978-94-017-9555-5-11
Holm-Nielsen, J. B., Al Seadi, T., and Oleskowicz-Popiel, P. (2009). The future of anaerobic digestion and biogas utilization. Bioresour. Technol. 100, 5478–5484. doi: 10.1016/j.biortech.2008.12.046
Howell, C. R. (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87, 4–10. doi: 10.1094/PDIS.2003.87.1.4
Huang, C., Guo, H.-J., Xiong, L., Wang, B., Shi, S.-L., Chen, X.-F., et al. (2016). Using wastewater after lipid fermentation as substrate for bacterial cellulose production by Gluconacetobacter xylinus. Carbohydr. Polym. 136, 198–202. doi: 10.1016/j.carbpol.2015.09.043
Huang, X., Chen, X., Chen, Q., Yu, Q., Sun, D., and Liu, J. (2016). Investigation of functional selenium nanoparticles as potent antimicrobial agents against superbugs. Acta Biomater. 30, 397–407. doi: 10.1016/j.actbio.2015.10.041
Ikram-ul, H., Ali, S., Qadeer, M. A., and Iqbal, J. (2004). Citric acid production by selected mutants of Aspergillus niger from cane molasses. Bioresour. Technol. 93, 125–130. doi: 10.1016/j.biortech.2003.10.018
Ismail, A.-M., Abo-Elmagd, H., and Housseiny, M. M. (2016). A safe potential juice clarifying pectinase from Trichoderma viride EF-8 utilizing Egyptian onion skins. J. Genet. Eng. Biotechnol. 14, 153–159. doi: 10.1016/j.jgeb.2016.05.001
Ivanov, V., and Stabnikov, V. (2016). “Construction biotechnological plastics,” in Construction Biotechnology, Part of the Series Green Energy and Technology (Singapore: Springer), 51–75. doi: 10.1007/978-981-10-1445-1_4
Jacob, F., Perrin, D., Sanchez, C., and Monod, J. (1960). L’operon: groupe de génes á expression coordonné par un opérateur. Comp. Rend. Acad. Sci. Paris 250, 1727–1729.
Jamie, A., Alshami, A. S., Maliabari, Z. O., Ateih, M. A., and Al Hamouz, O. C. S. (2016). Immobilization and enhanced catalytic activity of lipase on modified MWCNT for oily wastewater treatment. Environ. Prog. Sustain. Energy 35, 1441–1449. doi: 10.1002/ep.12375
Jenkins, N. (2007). Modifications of therapeutic proteins: challenges and prospects. Cytotechnology 53, 121–125. doi: 10.1007/s10616-007-9075-2
Johnson, I. S. (1983). Human insulin from recombinant DNA technology. Science 219, 632–637. doi: 10.1126/science.6337396
Jones, E. E., Bienkowski, D. A., and Stewart, A. (2016). The importance of water potential range tolerance as a limiting factor on Trichoderma spp. biocontrol of Sclerotinia sclerotiorum. Ann. Appl. Biol. 168, 41–51. doi: 10.1111/aab.12240
Jones, E. E., Rabeendran, N., and Stewart, A. (2014). Biocontrol of Sclerotinia sclerotiorum infection of cabbage by C. minitans and Trichoderma spp. Biocontrol. Sci. Technol. 24, 1363–1382. doi: 10.1080/09583157.2014.940847
Jozala, A. F., Geraldes, D. C., Tundisi, L. L., Feitosa, V. A., Breyer, C. A., Cardoso, S. L., et al. (2016). Biopharmaceuticals from microorganisms: from production to purification. Braz. J. Microbiol. 47, 51–63. doi: 10.1016/j.bjm.2016.10.007
Kampmann, M., Boll, S., Kossuch, J., Bielecki, J., Uhl, S., Kleiner, B., et al. (2014). Efficient immobilization of mushroom tyrosinase utilizing whole cells from Agaricus bisporus and its application for degradation of bisphenol A. Water Res. 57, 295–303. doi: 10.1016/j.watres.2014.03.054
Katrlik, J., Vostiar, I., Sefcovicova, J., Tkac, J., Mastihuba, V., Valach, M., et al. (2007). A novel microbial biosensor based on cells of Gluconobacter oxydans for the selective determination of 1,3-propanediol in the presence of glycerol and its application to bioprocess monitoring. Anal. Bioanal. Chem. 388, 287–295. doi: 10.1007/s00216-007-1211-5
Keitel, W. A., Bond, N. L., Zahradnik, J. M., Cramton, T. A., and Robbins, J. B. (1994). Clinical and serological responses following primary and booster immunization with Salmonella typhi Vi capsular polysaccharide vaccines. Vaccine 12, 195–199. doi: 10.1016/0264-410X(94)90194-5
Khaledi, N., and Taheri, P. (2016). Biocontrol mechanisms of Trichoderma harzianum against soybean charcoal rot caused by Macrophomina phaseolina. J. Plant Prot. Res. 56, 21–31. doi: 10.1515/jppr-2016-0004
Khan, M. A., Paul, B., Ahmad, W., Paul, S., Aggarwal, C., Khan, Z., et al. (2016). “Potential of Bacillus thuringiensis in the management of pernicious lepidopteran pests,” in Plant, Soil and Microbes, eds K. R. Hakeem and M. S. Akhtar (New York City, NY: Springer International Publishing), 277–301. doi: 10.1007/978-3-319-29573-2_13
Khor, E., and Lim, L. Y. (2003). Implantable applications of chitin and chitosan. Biomaterials 24, 2339–2349. doi: 10.1016/S0142-9612(03)00026-7
Kim, H. J., Lim, J. W., Jeong, H., Lee, S.-J., Lee, D.-W., Kim, T., et al. (2016). Development of a highly specific and sensitive cadmium and lead microbial biosensor using synthetic CadC-T7 genetic circuitry. Biosens. Bioelectron. 79, 701–708. doi: 10.1016/j.bios.2015.12.101
Kim, K. R., Song, Y. H., Seo, J. H., and Kang, D. G. (2016). Effect of culture conditions on the whole cell activity of recombinant Escherichia coli expressing periplasmic organophosphorus hydrolase and cytosolic GroEL/ES chaperone. ıBiotechnol. Bioprocess Eng. 21, 502–507. doi: 10.1007/s12257-016-0342-y
King, W. J., MacDonald, N. E., Wells, G., Huang, J., Allen, U., Chan, F., et al. (1996). Total and functional antibody response to a quadrivalent meningococcal polysaccharide vaccine among children. J. Pediatr. 128, 196–202. doi: 10.1016/S0022-3476(96)70389-X
Knuf, M., Kowalzik, F., and Kieninger, D. (2011). Comparative effects of carrier proteins on the immunological response induced by the vaccine. Vaccine 29, 4881–4890. doi: 10.1016/j.vaccine.2011.04.053
Köhl, L., Lukasiewicz, C. E., and van der Heijden, M. G. A. (2016). Establishment and effectiveness of inoculated arbuscular mycorrhizal fungi in agricultural soils. Plant Cell Environ. 39, 136–146. doi: 10.1111/pce.12600
Krause, L., Diaz, N. N., Edwards, R. A., Gartemann, K.-H., Krömeke, H., Neuweger, H., et al. (2008). Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotechnol. 136, 91–101. doi: 10.1016/j.jbiotec.2008.06.003
Kummerfeldt, C. E. (2014). Raxibacumab for the treatment of inhalational Anthrax. Infect. Drug Resist. 7, 101–109. doi: 10.2147/IDR.S47305
Kunz, A., Reginatto, V., and Durán, N. (2001). Combined treatment of textile effluent using the sequence Phanerochaete chrysosporium-ozone. Chemosphere 44, 281–287. doi: 10.1016/S0045-6535(00)00165-X
Kuyper, M., Hartog, M. M. P., Toirkens, M. J., Almering, M. J. H., Winkler, A. A., van Dijken, J. P., et al. (2005). Metabolic engineering of a xylose-isomerase-expressing strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 5, 399–409. doi: 10.1016/j.femsyr.2004.09.010
Lanfranco, L., Bonfante, P., and Genre, A. (2016). The mutualistic interaction between plants and arbuscular mycorrhizal fungi. Microbiol. Spectr. 4:FUNK-0012-2016. doi: 10.1128/microbiolspec.FUNK-0012-2016
Lee, K. Y., and Mooney, D. J. (2012). Alginate: properties and biomedical applications. Prog. Polym. Sci. 37, 106–126. doi: 10.1016/j.progpolymsci.2011.06.003
Lei, Y., Chen, W., and Mulchandani, A. (2006). Microbial biosensors. Anal. Chim. Acta 568, 200–210. doi: 10.1016/j.aca.2005.11.065
Lesage-Meessen, L., Lomascolo, A., Bonnin, E., Thibault, J.-F., Buleon, A., Roller, M., et al. (2002). A biotechnological process involving filamentous fungi to produce natural crystalline vanillin from maize bran. Appl. Biochem. Biotechnol. 102, 141–153. doi: 10.1385/ABAB:102-103:1-6:141
Li, P., Li, T., Zeng, Y., Li, X., Jiang, X., Wang, Y., et al. (2016). Biosynthesis of xanthan gum by Xanthomonas campestris LRELP-1 using kitchen waste as the sole substrate. Carbohydr. Polym. 151, 684–691. doi: 10.1016/j.carbpol.2016.06.017
Liao, J. C., Mi, L., Pontrelli, S., and Luo, S. (2016). Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14, 288–304. doi: 10.1038/nrmicro.2016.32
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. doi: 10.1038/nature14098
Linko, Y.-Y., Javanainen, P., and Linko, S. (1997). Biotechnology of bread baking. Trends Food Sci. Technol. 81, 339–344. doi: 10.1016/S0924-2244(97)01066-2
Lipkie, T. E., Ferruzzi, M., and Weaver, C. M. (2016). Bioaccessibility of vitamin D from bread fortified with UV-treated yeast is lower than bread fortified with crystalline vitamin D2 and bovine milk. FASEB J 30, 918.
Liu, K., Ding, X., Deng, B., and Chen, W. (2009). Isolation and characterization of endophytic taxol-producing fungi from Taxus chinensis. J. Ind. Microbiol. Biotechnol. 36, 1171–1177. doi: 10.1007/s10295-009-0598-8
Liu, X., Zhang, W., Zhao, Z., Dai, X., Yang, Y., and Bai, Z. (2016). Protein secretion in Corynebacterium glutamicum. Crit. Rev. Biotechnol. 37, 541–551. doi: 10.1080/07388551.2016.1206059
Losordo, Z., McBride, J., Van Rooyen, J., Wenger, K., Willies, D., Froehlich, A., et al. (2016). Cost competitive second-generation ethanol production from hemicellulose in a Brazilian sugarcane biorefinery. Biofuel Bioprod. Bior. 10, 589–602. doi: 10.1002/bbb.1663
Luengo, J. M., Garcia, B., Sandoval, A., Naharro, G., and Oliveira, E. R. (2003). Bioplastics from microorganisms. Curr. Opin. Microbiol. 6, 251–260. doi: 10.1016/S1369-5274(03)00040-7
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Ma, Y.-M., Liang, X.-A., Zhang, H.-C., and Liu, R. (2016). Cytotoxic and antibiotic cyclic pentapeptide from an endophytic Aspergillus tamarii of Ficus carica. J. Agric. Food Chem. 64, 3789–3793. doi: 10.1021/acs.jafc.6b01051
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Maleki, S., Almaas, E., Zotchev, S., Valla, S., and Ertesvåg, H. (2016). Alginate biosynthesis factories in Pseudomonas fluorescens: localization and correlation with alginate production level. Appl. Environ. Microbiol. 82, 41227–41236. doi: 10.1128/AEM.03114-15
Marasco, W. A., and Sui, J. (2007). The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 25, 1421–1434. doi: 10.1038/nbt1363
Mateo, C., Palomo, J. M., Fernandez-Lorente, G., Guisan, J. M., and Fernandez-Lafuente, R. (2007). Improvement of enzyme activity, stability and selectivity via immobilization techniques. ıEnzyme Microb. Technol. 40, 1451–1463. doi: 10.1016/j.enzmictec.2007.01.018
McGovern, P. E., Glusker, D. L., and Exner, L. J. (1996). Neolithic resinated wine. Nature 381, 480–481. doi: 10.1038/381480a0
McGovern, P. E., Zhang, J., Tang, J., Zhang, Z., Hall, G. R., Moreau, R. A., et al. (2004). Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. U.S.A. 101, 17593–17598. doi: 10.1073/pnas.0407921102
McIntosh, S., Zhang, Z., Palmer, J., Wong, H.-H., Doherty, W. O. S., and Vancov, T. (2016). Pilot-scale cellulosic ethanol production using eucalyptus biomass pre-treated by dilute acid and steam explosion. Biofuel Bioprod. Bior. 10, 346–358. doi: 10.1002/bbb.1651
McMeniman, C. J., Lane, R. V., Cass, B. N., Fong, A. W. C., Sidhu, M., Wang, Y.-F., et al. (2009). Stable introduction of a Life-Shortening Wolbachia Infection into the Mosquito Aedes aegypti. Science 323, 141–144. doi: 10.1126/science.1165326
McRae, C. F. (1988). Classical and inundative approaches to biological weed control compared. Plant Prot. Quart. 3, 124–127.
Meijnen, J.-P., Randazzo, P., Foulquié-Moreno, M. R., Brink, J. V. D., Vandecruys, P., Stojiljkovic, M., et al. (2016). Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae. Biotechnol. Biofuels 9, 5. doi: 10.1186/s13068-015-0421-x
Mendoza, A. R., Kiewnick, S., and Sikora, R. A. (2008). In vitro activity of Bacillus firmus against the burrowing nematode Radopholus similis the root-knot nematode Meloidogyne incognita and the stem nematode Ditylenchus dipsaci. Biocontrol. Sci. Technol. 18, 377–389. doi: 10.1080/09583150801952143
Messias, J. M., da Costa, B. Z., Lima, V. M. G., Giese, E. C., Dekker, R. F. H., and Barbosa, A. M. (2011). Lipases microbianas: produção, propriedades e aplicações biotecnológicas. Ciênc. Exatas Tecnol. 32, 213–234. doi: 10.5433/1679-0375.2011v32n2p213
Mitragotri, S., Burke, P. A., and Langer, R. (2014). Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat. Rev. Drug Discov. 13, 655–672. doi: 10.1038/nrd4363
Mohanty, A. K., Misra, M., and Hinrichsen, G. (2000). Biofibres, biodegradable polymers and biocomposites: an overview. Macromol. Mater. Eng. 276-277, 1–24. doi: 10.1002/(SICI)1439-2054(20000301)276:1
Mohiddin, A., Lasim, A. M., and Zuharah, W. F. (2016). Susceptibility of Aedes albopictus from dengue outbreak areas to temephos and Bacillus thuringiensis subsp. israelensis. Asian Pac. J. Trop. Biomed. 6, 295–300. doi: 10.1016/j.apjtb.2016.01.006
Mokoena, M. P., Mutanda, T., and Olaniran, A. O. (2016). Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr. Res. 60:29630. doi: 10.3402/fnr.v60.29630
Moon, H. G., Jang, Y.-S., Cho, C., Lee, J., Binkley, R., and Lee, S. Y. (2016). One hundred years of clostridial butanol fermentation. FEMS Microbiol. Lett. 363, fnw001. doi: 10.1093/femsle/fnw001
Mulat, D. G., Jacobi, H. F., Feilberg, A., Adamsen, A. P. S., Richnow, H.-H., and Nikolausz, M. (2016). Changing feeding regimes to demonstrate flexible biogas production: effects on process performance, microbial community structure, and methanogenesis pathways. Appl. Environ. Microbiol. 82, 438–449. doi: 10.1128/AEM.02320-15
Mulchandani, A., Mulchandani, P., Kaneva, I., and Chen, W. (1998). Biosensor for direct determination of organophosphate nerve agents using recombinant Escherichia coli with surface-expressed organophosphorus hydrolase. 1. Potentiometric microbial electrode. Anal. Chem. 70, 4140–4145. doi: 10.1021/ac9805201
Nabel, G. J. (2013). Designing tomorrow’s vaccines. N. Engl. J. Med. 368, 551–560. doi: 10.1056/NEJMra1204186
Nalage, D. N., Khedkar, G. D., Kalyankar, A. D., Sarkate, A. P., Ghodke, S. R., Bedre, V. B., et al. (2016). “Single cell proteins,” in The Encyclopedia of Food and Health, eds B. Caballero, P. Finglas, and F. Toldrá (Oxford: Academic Press), 790–794. doi: 10.1016/B978-0-12-384947-2.00628-0
Neushul, P. (1993). Science, government and the mass production of penicillin. J. Hist. Med. Allied Sci. 48, 371–395. doi: 10.1093/jhmas/48.4.371
Nielsen, D. R., Yoon, S. H., Yuan, C. J., and Prather, K. L. (2010). Metabolic engineering of acetoin and meso-2,3-butanediol biosynthesis in E. coli. Biotechnol. J. 5, 274–284. doi: 10.1002/biot.200900279
Nishat, S., and Andreana, P. R. (2016). Entirely carbohydrate-based vaccines: an emerging field for specific and selective immune responses. Pharmaceutics 8, 7. doi: 10.3390/pharmaceutics8010007
Nizet, V. (2015). Stopping superbugs, maintaining the microbiota. Sci. Transl. Med. 7, 295ed8. doi: 10.1126/scitranslmed.aab2373
Oliveira, R. S., Ma, Y., Rocha, I., Carvalho, M. F., Vosátka, M., and Freitas, H. (2016a). Arbuscular mycorrhizal fungi are an alternative to the application of chemical fertilizer in the production of the medicinal and aromatic plant Coriandrum sativum L. J. Toxicol. Environ. Health 79, 320–328. doi: 10.1080/15287394.2016.1153447
Oliveira, R. S., Rocha, I., Ma, Y., Vosátka, M., and Freitas, H. (2016b). Seed coating with arbuscular mycorrhizal fungi as an ecotechnological approach for sustainable agricultural production of common wheat (Triticum aestivum L.). J. Toxicol. Environ. Health 79, 329–337. doi: 10.1080/15287394.2016.1153448
Olmos-Soto, J., and Contreras-Flores, R. (2003). Genetic system constructed to overproduce and secrete proinsulin in Bacillus subtilis. Appl. Microbiol. Biotechnol. 62, 369–373. doi: 10.1007/s00253-003-1289-4
Olsen, J. L. (2016). “Polymerase chain reaction,” in Encyclopedia of Immunotoxicology, ed. H.-W. Vohr (Berlin: Springer), 715–720. doi: 10.1007/978-3-642-54596-2-1193
Omemu, A. M., Akpan, I., Bankole, M. O., and Teniola, O. D. (2005). Hydrolysis of raw tuber starches by amylase of Aspergillus niger AM07 isolated from the soil. Afr. J. Biotechnol. 4, 19–25.
Overton, T. W. (2014). Recombinant protein production in bacterial hosts. Drug Discov. Today 19, 590–601. doi: 10.1016/j.drudis.2013.11.008
Padilla, B., Gil, J. V., and Manzanares, P. (2016). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7:411. doi: 10.3389/fmicb.2016.00411
Paitan, Y., Biran, D., Biran, I., Shechter, N., Babai, R., Rishpon, J., et al. (2003). On-line and in situ biosensors for monitoring environmental pollution. Biotechnol. Adv. 22, 27–33. doi: 10.1016/j.biotechadv.2003.08.014
Papagianni, M. (2004). Fungal morphology and metabolite production in submerged mycelia processes. Biotechnol. Adv. 22, 189–259. doi: 10.1016/j.biotechadv.2003.09.005
Paris, M., Melodelima, C., Coissac, E., Tetreau, G., Reynaud, S., David, J.-P., et al. (2012). Transcription profiling of resistance to Bti toxins in the mosquito Aedes aegypti using next-generation sequencing. J. Invertebr. Pathol. 109, 201–208. doi: 10.1016/j.jip.2011.11.004
Paris, M., Tetreau, G., Laurent, F., Lelu, M., Despres, L., and David, J.-P. (2011). Persistence of Bacillus thuringiensis israelensis (Bti) in the environment induces resistance to multiple Bti toxins in mosquitoes. Pest Manag. Sci. 67, 122–128. doi: 10.1002/ps.2046
Pasteur, L. (2002). Summary report of the experiments conducted at Pouilly-le-Fort, near Melun, on the anthrax vaccination, 1881. Yale J. Biol. Med. 75, 59–62.
Patelski, P., Berlowska, J., Dziugan, P., Pielech-Przybylska, K., Balcerek, M., Dziekonska, U., et al. (2015). Utilisation of sugar beet bagasse for the biosynthesis of yeast SCP. J. Food Eng. 67, 32–37. doi: 10.1016/j.jfoodeng.2015.03.031
Paterson, R. R. M. (2006). Ganoderma–A therapeutic fungal biofactory. Phytochemistry 67, 1985–2001. doi: 10.1016/j.phytochem.2006.07.004
Paterson, R. R. M. (2008). Cordyceps–A traditional Chinese medicine and another fungal therapeutic biofactory? Phytochemistry 69, 1469–1495. doi: 10.1016/j.phytochem.2008.01.027
Pendyala, B., Chaganti, S. R., Lalman, J. A., and Heath, D. D. (2016). Optimizing the performance of microbial fuel cells fed a combination of different synthetic organic fractions in municipal solid waste. Wast Manag. 49, 73–82. doi: 10.1016/j.wasman.2015.12.032
Philip, S., Keshavarz, T., and Roy, I. (2007). Polyhydroxyalkanoates: biodegradable polymers with a range of applications. J. Chem. Technol. Biotechnol. 82, 233–247. doi: 10.1002/jctb.1667
Pichichero, M. E. (2013). Protein carriers of conjugate vaccines: characteristics, development, and clinical trials. Hum. Vaccin. Immunother. 9, 2505–2523. doi: 10.4161/hv.26109
Pifferi, C., Berthet, N., and Renaudet, O. (2017). Cyclopeptide scaffolds in carbohydrate-based synthetic vaccines. Biomater. Sci. 5, 953–965. doi: 10.1039/C7BM00072C
Pires, A. L. R., Bierhalz, A. C. K., and Moraes, A. M. (2015). Biomateriais: tipos, aplicações e mercado. Quim. Nova 38, 957–971. doi: 10.5935/0100-4042.20150094
Pires, A. L. R., and Moraes, A. M. (2015). Improvement of the mechanical properties of chitosan-alginate wound dressings containing silver through the addition of a biocompatible silicone rubber. J. Appl. Polym. Sci. 132:41686. doi: 10.1002/APP.41686
Pitisuttithum, P., and Bouckenooghe, A. (2016). The first licensed dengue vaccine: an important tool for integrated preventive strategies against dengue virus infection. Expert Rev. Vaccines. 15, 795–798. doi: 10.1080/14760584.2016.1189331
Ploss, T. N., Reilman, E., Monteferrante, C. G., Denham, E. L., Piersma, S., Lingner, A., et al. (2016). Homogeneity and heterogeneity in amylase production by Bacillus subtilis under different growth conditions. Microb. Cell Fact. 15, 57. doi: 10.1186/s12934-016-0455-1
Plotkin, S. A., Orenstein, W. A., and Offit, P. A. (2008). Vaccines, 5th Edn. Philadelphia, PA: Saunders/Elsevier, 399–434.
Plotkin, S. L., and Plotkin, S. A. (2011). “A short history of vaccination,” in Vaccines, eds S. A. Plotkin and W. A. Orenstein (Philadelphia, PA: Elsevier), 1–16.
Pochanavanich, P., and Suntornsuk, W. (2002). Fungal chitosan production and its characterization. Lett. Appl. Microbiol. 35, 17–21. doi: 10.1046/j.1472-765X.2002.01118.x
Popham, H. J. R., Nusawardani, T., and Bonning, B. C. (2016). “Introduction to the use of baculoviruses as biological insecticides,” in Baculovirus and Insect Cell Expression Protocols, ed. D. W. Murhammer (New York, NY: Springer), 383–392. doi: 10.1007/978-1-4939-3043-2_19
Prasad, J., Gill, H., Smart, J., and Gopal, P. K. (2000). Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int. Dairy J. 8, 993–1002. doi: 10.1016/S0958-6946(99)00024-2
Przystałowska, H., Zeyland, J., Szymanowska-Powałowska, D., Szalata, M., Słomski, R., and Lipiñski, D. (2015). 1,3-Propanediol production by new recombinant Escherichia coli containing genes from pathogenic bacteria. Microbiol. Res. 171, 1–7. doi: 10.1016/j.micres.2014.12.007
Radecka, I. K., Guozhan, J., Hill, D. J., and Kowalczuk, M. M. (2016). “Poly(Hydroxyalkanoates) composites and their applications,” in Green Polymer Composites Technology, ed. Inamuddin (New York, NY: Taylor & Francis Group), 163–176.
Rama, R., Mougin, C., Boyer, F. D., Kollmann, A., Malosse, C., and Sigoillot, J. C. (1998). Biotransformation of benzo[a] pyrene in bench scale reactor using laccase of Pycnoporus cinnabarinus. Biotechnol. Lett. 20, 1101–1104. doi: 10.1023/A:1005387016390
Rao, S. S., Gomez, P., Mascola, J. R., Dang, V., Krivulka, G. R., Yu, F., et al. (2006). Comparative evaluation of three different intramuscular delivery methods for DNA immunization in a nonhuman primate animal model. Vaccine 24, 367–373. doi: 10.1016/j.vaccine.2005.07.072
Rasinger, J. D., Marrazza, G., Briganti, F., Scozzafava, A., Mascini, M., and Turner, A. P. F. (2005). Evaluation of an FIA operated amperometric bacterial biosensor, based on Pseudomonas putida F1 for the detection of benzene, toluene, ethylbenzene, and xylenes (BTEX). Anal. Lett. 38, 1531–1547. doi: 10.1081/AL-200065793
Reed, S. G., Bertholet, S., Coler, R. N., and Friede, M. (2009). New horizons in adjuvants for vaccine development. Trends Immunol. 30, 23–32. doi: 10.1016/j.it.2008.09.006
Rillig, M. C., Sosa-Hernández, M. A., Roy, J., Aguilar-Trigueros, C. A., Vályi, K., and Lehmann, A. (2016). Towards an integrated mycorrhizal technology: harnessing mycorrhiza for sustainable intensification in agriculture. Front. Plant Sci. 7:1625. doi: 10.3389/fpls.2016.01625
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J.-F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Robinson, D., and Medlock, N. (2005). Diamond v. Chakrabarty: a retrospective on 25 years of biotech patents. Intell. Prop. Technol. Law J. 17, 12–15.
Robinson, H. L. (1997). Nucleic acid vaccines: an overview. Vaccine 15, 785–787. doi: 10.1016/S0264-410X(96)00249-6
Roden, R., and Wu, T.-C. (2006). How will HPV vaccines affect cervical cancer? Nat. Rev. Cancer 6, 753–763. doi: 10.1038/nrc1973
Rodrigues, R. C., and Fernandez-Lafuente, R. (2010). Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J. Mol. Catal. B Enzym. 66, 15–32. doi: 10.1016/j.molcatb.2010.03.008
Rodriguez-Carmona, E., and Villaverde, A. (2010). Nanostructured bacterial materials for innovative medicines. Trends Microbiol. 18, 423–430. doi: 10.1016/j.tim.2010.06.007
Ruholahi, F., Mohammadi, M., Karimi, K., and Zamani, A. (2016). Nickel biosorption by fungal chitosan from Mucor Indicus. J. Chitin. Chitosan. 4, 69–73. doi: 10.1166/jcc.2016.1102
Saerens, S., and Swiegers, J. H. (2016). Enhancement of Coffee Quality and Flavor by Using Pichia kluyveri Yeast Starter Culture for Coffee Fermentation. US 20160058028 A1.
Sahebani, N., and Hadavi, N. (2008). Biological control of the root-knot nematode Meloidogyne javanica by Trichoderma harzianum. Soil Biol. Biochem. 40, 2016–2020. doi: 10.1016/j.soilbio.2008.03.011
Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., et al. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491. doi: 10.1126/science.2448875
Saitoh, S., Ishida, N., Onishi, T., Tokuhiro, K., Nagamori, E., Kitamoto, K., et al. (2005). Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl. Environ. Microbiol. 71, 2789–2792. doi: 10.1128/AEM.71.5.2789-2792
Salman, T., Kamal, M., Ahmed, M., Siddiqa, S. M., Khan, R. A., and Hassan, A. (2016). Medium optimization for the production of amylase by Bacillus subtilis RM16 in Shake-flask fermentation. Pak. J. Pharm. Sci. 29, 439–444.
Samuel, D. (1996). Investigation of ancient Egyptian baking and brewing methods by correlative microscopy. Science 273, 488–490. doi: 10.1126/science.273.5274.488
Sanchez-Garcia, L., Martín, L., Mangues, R., Ferrer-Miralles, N., Vázquez, E., and Villaverde, A. (2016). Recombinant pharmaceuticals from microbial cells: a 2015 update. Microb. Cell Fact. 15, 33. doi: 10.1186/s12934-016-0437-3
Sandeep, V., Parveen, J., and Chauhan, P. (2016). Biobetters: the better biologics and their regulatory overview. Int. J. Drug Regul. Aff. 4, 13–20.
Sandgathe, A., Tippe, D., Dilsen, S., Meens, J., Halfar, M., Weuster-Botz, D., et al. (2003). Production of a human calcitonin precursor with Staphylococcus carnosus: secretory expression and single-step recovery by expanded bed adsorption. Process Biochem. 38, 1351–1363. doi: 10.1016/S0032-9592(02)00332-1
Saravanakumar, K., Yu, C., Dou, K., Wang, M., Li, Y., and Chen, J. (2016). Synergistic effect of Trichoderma-derived antifungal metabolites and cell wall degrading enzymes on enhanced biocontrol of Fusarium oxysporum f. sp. Cucumerinum. Biol. Control. 94, 37–46. doi: 10.1016/j.biocontrol.2015.12.001
Sarwar, M. (2015a). Biopesticides: an effective and environmental friendly insect-pests inhibitor line of action. Int. J. Eng. Adv. Res. Technol. 1, 10–15.
Sarwar, M. (2015b). Microbial insecticides- an ecofriendly effective line of attack for insect pests management. Int. J. Eng. Adv. Res. Technol. 1, 4–9.
Satish, K. R., Kanmani, P., Yuvaraj, N., Paari, K. A., Pattukumar, V., and Arul, V. (2013). Traditional Indian fermented foods: a rich source of lactic acid bacteria. Int. J. Food. Sci. Nutr. 64, 415–428. doi: 10.3109/09637486.2012.746288
Satpathy, P., Steinigeweg, S., Cypionka, H., and Engelen, B. (2016). Different substrates and starter inocula govern microbial community structures in biogas reactors. Environ. Technol. 37, 1441–1450. doi: 10.1080/09593330.2015.1118559
Sauer, M. (2016). Industrial production of acetone and butanol by fermentation-100 years later. FEMS Microbiol. Lett. 363:fnw134. doi: 10.1093/femsle/fnw134
Sauer, M., Porro, D., Mattanovich, D., and Branduardi, P. (2008). Microbial production of organic acids: expanding the markets. Trends Biotechnol. 26, 100–108. doi: 10.1016/j.tibtech.2007.11.006
Schlüter, A., Bekel, T., Diaz, N. N., Dondrup, M., Eichenlaub, R., Gartemann, K.-H., et al. (2008). The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J. Biotechnol. 136, 77–90. doi: 10.1016/j.jbiotec.2008.05.008
Schmidt, S. T., Foged, C., Korsholm, K. S., Rades, T., and Christensen, D. (2016). Liposome-based adjuvants for subunit vaccines: formulation strategies for subunit antigens and immunostimulators. Vaccines 4:19. doi: 10.3390/vaccines4020019
Setha, T., Chantha, N., Benjamin, S., and Socheat, D. (2016). Bacterial larvicide, Bacillus thuringiensis israelensis strain AM 65-52 water dispersible granule formulation impacts both dengue vector, Aedes aegypti (L.) population density and disease transmission in Cambodia. PLoS Negl. Trop. Dis. 10:e0004973. doi: 10.1371/journal.pntd.0004973
Sharma, R., Chisti, Y., and Banerjee, U. C. (2001). Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 19, 627–662. doi: 10.1016/S0734-9750(01)00086-6
Sheldon, R. A. (2007). Enzyme immobilization: the quest for optimum performance. Adv. Synth. Catal. 349, 1289–1307. doi: 10.1002/adsc.200700082
Shin, H. J. A. (2016). Recombinant microbial biosensor for cadmium and lead detection. J. Life Sci. 26, 503–508. doi: 10.5352/JLS.2016.26.5.503
Si, L., Xu, H., Zhou, X., Zhang, Z., Tian, Z., Wang, Y., et al. (2016). Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 354, 1170–1173. doi: 10.1126/science.aah5869
Sicard, D., and Legras, J.-L. (2011). Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C. R. Biol. 334, 229–236. doi: 10.1016/j.crvi.2010.12.016
Simon, D., and Chopin, A. (1988). Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70, 559–566. doi: 10.1016/0300-9084(88)90093-4
Simpson, D. R. (2008). Biofilm processes in biologically active carbon water purification. Water Res. 42, 2839–2848. doi: 10.1016/j.watres.2008.02.025
Smith, H. O., and Nathans, D. (1973). A Suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J. Mol. Biol. 81, 419–423. doi: 10.1016/0022-2836(73)90152-6
Sokhandani, Z., Moosavi, M. R., and Basirnia, T. (2016). Optimum concentrations of Trichoderma longibrachiatum and cadusafos for controlling Meloidogyne javanica on Zucchini plants. J. Nematol. 48, 54–63.
Sørup, S., Benn, C. S., Poulsen, A., Krause, T., Aaby, P., and Ravn, H. (2014). Live vaccine against measles, mumps, and rubella and the risk of hospital admissions for nontargeted infections. JAMA 311, 826–835. doi: 10.1001/jama.2014.470
Spang, A., and Ettema, T. J. G. (2016). Microbial diversity: the tree of life comes of age. Nat. Microbiol. 1, 16056. doi: 10.1038/nmicrobiol.2016.56
Srivastava, R. K., Ichhpujani, R. I., Khare, S., Rai, A., and Chauhan, L. S. (2011). Superbug–the so-called NDM-1. Ind. J. Med. Res. 133, 458–460.
Stephen, A., Marglin, S. A., and Schor, J. B. (1991). The Golden Age of Capitalism. Oxford: Clarendon Press.
Stoytcheva, M., Zlatev, R., Magnin, J.-P., Ovalle, M., and Valdez, B. (2009). Leptospirillum ferrooxidans basead Fe 2+ sensor. Biosens. Bioelectron. 25, 482–487. doi: 10.1016/j.bios.2009.08.019
Su, L., Jia, W., Hou, C., and Lei, Y. (2011). Microbial biosensors: a review. Biosens. Bioelectron. 26, 1788–1799. doi: 10.1016/j.bios.2010.09.005
Suksong, W., Kongjan, P., Prasertsan, P., Imai, T., and O-Thong, S. (2016). Optimization and microbial community analysis for production of biogas from solid waste residues of palm oil mill industry by solid-state anaerobic digestion. Bioresour. Technol. 214, 166–174. doi: 10.1016/j.biortech.2016.04.077
Sussman, M. (1970). General bacterial genetics. Ann. Rev. Genet. 4, 135–176. doi: 10.1146/annurev.ge.04.120170.001031
Svensson, A., Nicklasson, E., Harrah, T., Panilaitis, B., Kaplan, D. L., Brittberg, M., et al. (2005). Bacterial cellulose as a potential scaffold for tissue engineering of cartilage. Biomaterials 26, 419–431. doi: 10.1016/j.biomaterials.2004.02.049
Tag, K., Riedel, K., Bauer, H. J., Hanke, G., Baronian, K. H. R., and Kunze, G. (2007). Amperometric detection of Cu2+ by yeast biosensors using flow injection analysis (FIA). Sens. Actuators B. 122, 403–409. doi: 10.1016/j.snb.2006.06.007
Takagi, H., and Shima, J. (2015). “Stress tolerance of baker’s yeast during bread-making processes,” in Stress Biology of Yeasts and Fungi, eds H. Takagi and H. Kitagaki (Tokyo: Springer), 23–42. doi: 10.1007/978-4-431-55248-2-2
Tatsumi, K., Wada, S., and Ichikawa, H. (1996). Removal of chlorophenols from wastewater by immobilized horseradish peroxidase. Biotechnol. Bioeng. 51, 126–130. doi: 10.1002/(SICI)1097-0290(19960705)51:1<126::AID-BIT15>3.0.CO;2-O
Tayel, A. A., El-Tras, W., and Elguindy, N. M. (2016a). The potentiality of cross-linked fungal chitosan to control water contamination through bioactive filtration. Int. J. Biol. Macromol. 88, 59–65. doi: 10.1016/j.ijbiomac.2016.03.018
Tayel, A. A., Gharieb, M. M., Zaki, H. R., and Elguindy, N. M. (2016b). Bio-clarification of water from heavy metals and microbial effluence using fungal chitosan. Int. J. Biol. Macromol. 83, 277–281. doi: 10.1016/j.ijbiomac.2015.11.072
The IMpact-RSV Study Group (1998). Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102, 3. doi: 10.1542/peds.102.3.531
Timur, S., Haghighi, B., Tkac, J., Pazarhoglu, N., Telefoncu, A., and Gorton, L. (2007). Electrical wiring of Pseudomonas putida and Pseudomonas fluorescens with osmium redox polymers. Bioelectrochemistry 71, 38–45. doi: 10.1016/j.bioelechem.2006.08.001
Tiquia, S. M., Wan, H. C., and Tam, N. F. Y. (2002). Microbial population dynamics and enzyme activities during composting. Compost Sci. Util. 10, 150–161. doi: 10.1080/1065657X.2002.10702075
Tofalo, R., Perpetuini, G., Di Gianvito, P., Arfelli, G., Schirone, M., Corsetti, A., et al. (2016). Characterization of specialized flocculent yeasts to improve sparkling wine fermentation. J. Appl. Microbiol. 120, 1574–1584. doi: 10.1111/jam.13113
Tong, Z., Qingxiang, Z., Hui, H., Qin, L., Yi, Z., and Min, Q. (1998). Kinetic study on the removal of toxic phenol and chlorophenol from waste water by horseradish peroxidise. Chemosphere 37, 1571–1577. doi: 10.1016/S0045-6535(98)00140-4
Turley, A. P., Moreira, L. A., O’Neill, S., and McGraw, E. A. (2009). Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl. Trop. Dis. 3:e516. doi: 10.1371/journal.pntd.0000516
Twomey, U., Warrior, P., Kerry, B. R., and Perry, R. N. (2000). Effects of the biological nematicide, DiTera, on hatching of Globodera rostochiensis and G. pallida. Nematology 2, 355–362. doi: 10.1163/156854100509114
Valach, M., Katrlik, J., Sturdik, E., and Gemeiner, P. (2009). Ethanol Gluconobacter biosensor designed for flow injection analysis: application in ethanol fermentation off-line monitoring. Sens. Actuators B 138, 581–586. doi: 10.1016/j.snb.2009.02.017
Valamoti, S. M., Mangafa, M., Koukouli-Chrysanthaki, C. H., and Malamidou, D. (2007). Grape-pressings from northern Greece: the earliest wine in the Aegean? Antiquity 81, 54–61. doi: 10.1017/S0003598X00094837
Valli, M., Sauer, M., Branduardi, P., Borth, N., Porro, D., and Mattanovich, D. (2006). Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl. Environ. Microbiol. 72, 85492–85499. doi: 10.1128/AEM.00683-06
Vázquez, E., and Villaverde, A. (2013). Microbial biofabrication for nanomedicine: biomaterials, nanoparticles and beyond. Nanomedicine 8, 1895–1898. doi: 10.2217/nnm.13.164
Velkov, T., Roberts, K. D., Thompson, P. E., and Li, J. (2016). Polymyxins: a new hope in combating Gram-negative superbugs? ıFuture Med. Chem. 8, 1017–1025. doi: 10.4155/fmc-2016-0091
Velu, S., Velayutham, V., and Manickkam, S. (2016). Optimization of fermentation media for xanthan gum production from Xanthomonas campestris using response surface methodology and artificial neural network techniques. Indian J. Chem. Technol. 23, 353–361.
Waditee-Sirisattha, R., Kageyama, H., and Takabe, T. (2016). Halophilic microorganism resources and their applications in industrial and environmental biotechnology. AIMS Microbiol. 2, 42–54. doi: 10.3934/microbial.2016.1.42
Wadt, N. S. Y., Okamoto, M. K. H., Bach, E. M., and Bach, E. E. (2015). Chemical, toxicological, anti-inflammatory and antimicrobial evaluation of Ganoderma lucidum extracts. Emir. J. Food Agric. 27, 577–584. doi: 10.9755/ejfa.2015.05.309
Walder, F., Boller, T., Wiemken, A., and Courty, P.-E. (2016). Regulation of plants’ phosphate uptake in common mycorrhizal networks: role of intraradical fungal phosphate transporters. Plant Signal. Behav. 11:e1131372. doi: 10.1080/15592324.2015.1131372
Walker, T., Johnson, P. H., Moreira, L. A., Iturbe-Ormaetxe, I., Frentiu, F. D., McMeniman, C. J., et al. (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476, 450–453. doi: 10.1038/nature10355
Wang, L., Cao, Z., Hou, L., Yin, L., Wang, D., Gao, Q., et al. (2016). The opposite roles of agdA and glaA on citric acid production in Aspergillus niger. Appl. Microbiol. Biotechnol. 100, 5791–5803. doi: 10.1007/s00253-016-7324-z
Wang, L., Liu, S.-Y., Chen, H.-W., Xu, J., Chapon, M., Zhang, T., et al. (2017). Generation of a live attenuated influenza vaccine that elicits broad protection in mice and ferrets. Cell Host Microbe 8, 334–343. doi: 10.1016/j.chom.2017.02.007
Wang, Z., Zhuge, J., Fang, H., and Prior, B. A. (2001). Glycerol production by microbial fermentation: a review. Biotechnol. Adv. 19, 201–223. doi: 10.1016/S0734-9750(01)00060-X
Wilson, M. J., and Jackson, T. A. (2013). Progress in the commercialisation of bionematicides. BioControl 58, 715–722. doi: 10.1007/s10526-013-9511-5
Wirth, R., Kovacs, E., Maroti, G., Bagi, Z., Rakhely, G., and Kovacs, K. L. (2012). Characterization of a biogas-producing microbial community by short-read next generation DNA sequencing. Biotechnol. Biofuels 5:41. doi: 10.1186/1754-6834-5-41
Woese, C. R., and Fox, G. E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. U.S.A. 74, 5088–5090. doi: 10.1073/pnas.74.11.5088
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. U.S.A. 87, 4576–4579. doi: 10.1073/pnas.87.12.4576
Wu, S., Wu, W., Zhu, X., Liu, Z., Rebeca, C.-L., Fu, T., et al. (2016). Physiological and biochemical response of Aedes aegypti tolerance to Bacillus thuringiensis. Biocontrol Sci. Technol. 26, 227–238. doi: 10.1080/09583157.2015.1089216
Xu, L., Zhao, Y., Doherty, L., Hu, Y., and Hao, X. (2016). The integrated processes for wastewater treatment based on the principle of microbial fuel cells: a review. Crit. Rev. Environ. Sci. Technol. 46, 60–91. doi: 10.1080/10643389.2015.1061884
Zhang, X., Tervo, C. J., and Reed, J. L. (2016). Metabolic assessment of E. coli as a biofactory for commercial products. Metab. Eng. 35, 64–74. doi: 10.1016/j.ymben.2016.01.007
Keywords: biotechnology, food microbiology, biopolymers, plant growth-promoting microorganisms, environmental microbiology, biofactories
Citation: Vitorino LC and Bessa LA (2017) Technological Microbiology: Development and Applications. Front. Microbiol. 8:827. doi: 10.3389/fmicb.2017.00827
Received: 22 February 2017; Accepted: 24 April 2017;
Published: 10 May 2017.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Daniel M. Linares, Teagasc – The Irish Agriculture and Food Development Authority, IrelandCopyright © 2017 Vitorino and Bessa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luciana C. Vitorino, bHVjaWFuYS52aXRvcmlub0BpZmdvaWFuby5lZHUuYnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.