- 1National Reference Laboratory of Veterinary Drug Residues and MOA Key Laboratory for Detection of Veterinary Drug Residues, Huazhong Agricultural University, Wuhan, China
- 2MOA Laboratory for Risk Assessment of Quality and Safety of Livestock and Poultry Products, Huazhong Agricultural University, Wuhan, China
- 3Division of Microbiology, National Center for Toxicological Research, US Food and Drug Administration, Jefferson, AR, USA
Campylobacter jejuni is one of the most common foodborne pathogen worldwide. A putative transcriptional regulator, Cj0440c, was up-regulated in the erythromycin-resistant C. jejuni, however, the precise role of Cj0440c is yet to be determined. The aim of this study was to determine the biological functions of Cj0440c. The Cj0440c isogenic mutants were constructed from erythromycin-susceptible C. jejuni NCTC 11168 (S) and -resistant C. jejuni 68-ER (R), designating as SM and RM, respectively. The isogenic Cj0440c mutants (SM and RM) and parental strains (S and R) were subjected to microarray and qRT-PCR analysis to examine the transcriptional profile changes contributed by Cj0440c. The antimicrobial susceptibility, flagellar morphology, in vitro growth and in vivo colonization in chickens were carried out to analyze the biological function of Cj0440c. The results showed that 17 genes were down-regulated in SM compared to S, while 9 genes were down-regulated in RM compared to R. The genes with transcriptional change were mainly involved in flagella biosynthesis and assembly. Using transmission electron microscopy, we found that the filaments were impaired in SM and lost in RM. The chicken colonization experiments showed that Cj0440c mutants (SM and RM) had reduced colonization ability in chickens when compared with corresponding parental strains (S and R). In conclusion, Cj0440c regulates flagella biosynthesis and assembly, and consequently affect the in vivo colonization of erythromycin-susceptible and -resistant C. jejuni.
Introduction
Campylobacter jejuni has been recognized as one of the most important pathogens, which can cause infectious diarrhea and severe forms of disease such as Guillain-Barre Syndrome or Miller Fischer Syndrome (Samuel et al., 2004; Hughes and Cornblath, 2005; Riddle et al., 2006). The CDC estimated that in 2009 the number of Campylobacter infection was 13.02 per 100,000 people (Silva et al., 2011). The cost of human Campylobacteriosis in the United States is estimated at $1.3 to 6.8 billion dollars annually (Scharff, 2012; Epps et al., 2013). Macrolides (e.g., erythromycin) are the most important drugs of choice for clinical treatment of Campylobacter infections (Gibreel et al., 2005). Unfortunately, macrolides-resistant Campylobacter have emerged and impose a global public health concerns (Gibreel and Taylor, 2006; ECDC et al., 2009). In earlier study we demonstrated that the transcription level of Cj0440c was increased in high-level erythromycin-resistant C. jejuni (Hao et al., 2013).
Bioinformatic analyses suggested that Cj0440c is a putative transcriptional regulator and encodes a TENA/THI-4 family protein, however, the molecular function of this family is yet to be determined. The gene Cj0440c is located downstream of the Cj0437–Cj0439 operon (mfr, methylmenaquinol:fumarate reductase) which plays an important role in the susceptibility to hydrogen peroxide (H2O2) (Parkhill et al., 2000; Weingarten et al., 2009; Kassem et al., 2012) and upstream of Cj0441 (acpP, acyl carrier protein) which is a universal and highly conserved acyl donor for synthesis of fatty acid, endotoxin and acylated homoserine lactones for the quorum sensing in C. jejuni (Byers and Gong, 2007). Both the downstream and upstream genes of Cj0440c were essential for the growth, survival, colonization and pathogenesis in Campylobacter. Although Cj0440c is located on the opposite DNA coding strand, it may divergently transcribed with its up-and-downstream genes, and likely to act as a transcriptional regulator and play an important role in gene regulation and the biological function in C. jejuni. The biological functions of Cj0440c in C. jejuni are largely unknown.
In the present study, Cj0440c-inactivated mutation was constructed in both erythromycin-susceptible (S) and -resistant C. jejuni (R), the transcriptional profile and relative in vitro and in vivo phenotype determination were implemented to decipher the function and regulation mechanism of Cj0440c.
Materials and Methods
Plasmids, Bacterial Strains, and Growth Conditions
The C. jejuni NCTC11168 (designated as S) was kindly provided by Chinese Center for Disease Control and Prevention. C. jejuni strains were routinely cultured in Mueller-Hinton (MH) medium at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) in the anaerobic incubator (YQX-II, Shanghai, China) (Mace et al., 2015). The Escherichia coli DH5α was grown aerobically in Luria-Bertani medium at 37°C. The erythromycin-resistant C. jejuni strain 68-ER (designated as R) was descendant of C. jejuni NCTC11168 resulting from in vitro step-wise selection by erythromycin. Plasmids pGEM-T (Promega, Madison, WI, USA) and pMW10 was kindly provided by China Agricultural University and used for mutant vector construction.
Construction of Isogenic ΔCj0440c Mutants
The DNA fragment containing Cj0440c gene and its flanking regions was amplified from C. jejuni NCTC 11168 genome using Pfu polymerase (Promega) with primers of Cj0440cF2 and Cj0440cR2 (Table 1) and was cloned into pGEM-T easy vector (Promega,) to generate plasmid pCJ0440c. Primers pCj0440cU and pCj0440cL (Table 1) carrying endonuclease restriction sites of KpnI and XbaI were used to inversely amplify DNA fragment from the vector of pCJ0440c using Taq and Pfu polymerase (8:1). A kanamycin resistance cassette (kan) was amplified from pMW10 plasmid with Pfu polymerase (Promega) using primers KanF and KanR (Table 1) which have the same restriction sites of KpnI and XbaI. The amplified DNA fragments of inverse PCR and kan were digested with KpnI and XbaI and purified with a PCR clean-up kit (Generay, Shanghai, China). The digested inverse PCR product was ligated to the kan cassette using T4 DNA ligase (Takara, Dalian, China) to obtain the construct plasmid pCJ0440c-Kan, which was then transformed into E. coli DH5α. The purified plasmid of pCJ0440c-Kan was introduced into S and R via electroporation according to the method described previously (Jeon et al., 2011). Insertional mutants, named SM and RM, respectively, were selected on MH agar plates with 25 μg/ml kanamycin and 50 μg/ml ampicillin. Both PCR and sequencing analysis of the Cj0440c mutants (SM and RM) confirmed that the mutation resulted in deletion of 200 bp of coding sequence in Cj0440c and simultaneous insertion of the kan gene into the same location.
RNA Microarray and Data Analysis
The transcriptional difference between Cj0440c mutants and their parental strains (SM&S and RM&R) was examined by microarray (CapitalBio Corporation, Shanghai, China). Briefly, the strains were separately grown in MH broth for 24 h at 42°C under microaerophilic conditions with shaking. Immediately after the incubation, twice volume of RNA protective reagent (Qiagen, Germantown, MD, USA) was added to the culture (with same OD600 of 0.3) to stabilize mRNA. The total RNA from each sample was extracted using RNeasy Protect Mini Kit (Qiagen) and purified using Nucleo®Spin RNA clean-up kit (Macherey-nagel, Germany). The RNA quality and quantity was determined by formaldehyde denatured gel electrophoresis and NanodropTM 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The cDNA was synthesized from the extracted RNA using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). The cDNA was labeled by Cy5 or Cy3 dye and co-hybridized onto one microarray slide (NimbleGen 4 K × 72K), scanned by Axon Instruments Gene Pix 4000B (Union City, CA, USA) and read by Gene Pix Pro 6.0 (Axon Instruments). Microarray data were analyzed using Array Star software. The genes with False Discovery Rate (FDR)-corrected q-values < 0.01 and fold change >2 were selected as differentially expressed genes and subjected to NCBI gene annotation, KEGG pathway analysis and STRING 9.05 protein network analysis.
Microarray Data Accession Number
The microarray data obtained in this study have been deposited in the NCBI Gene Expression Omnibus database1 and assigned accession number GSE49255 and GSE49256.
qRT-PCR
The same batches cDNA of Cj0440c mutants (SM and RM) and their parental strains (S and R) used in microarray were subjected to qRT-PCR analysis to confirm the transcriptional difference of some respective genes identified by microarray following method described in previous study (Hao et al., 2010). Briefly, the PCR amplification was performed in IQ5 Multicolor Real-time PCR Detection System (Bio-Rad). The cycling conditions were as follows: 3 min of pre-incubation at 95°C, followed by 30 cycles of 10 s at 95°C and 40 s at 60°C. The primer sets used for specific genes are listed in Table 1. 16S rDNA was used as an internal control for normalization. The experiment was done in triplicate to obtain the average value of fold change. The student’s t-test was performed to analyze the significant difference between mutants and their parental stains.
Antimicrobial Susceptibility Test
Minimum inhibitory concentrations (MICs) of nine antimicrobial agents (azithromycin, erythromycin, tylosin, ciprofloxacin, olaquindox, chloromycetin, tetracycline, gentamicin, and ceftriaxone) were determined using agar dilution method recommended by Clinical and Laboratory Standards Institute (CLSI). C. jejuni ATCC 33560 was used as a quality control strain.
Transmission Electron Microscopy
The presence and length of flagella on the four C. jejuni strains (S, SM, R and RM) were examined using transmission electron microscopy according to a previously described method (Barrero-Tobon and Hendrixson, 2014; Matsunami et al., 2016). Briefly, bacterial suspensions were obtained after washing plate with 2 ml sterile phosphate-buffered saline and spotted on carbon-coated copper grids. The cells were stained with 2% phosphotungstic acid (pH 6.7) for 1 min. Samples were observed employing a HITACHI H-7650 transmission electron microscope (Hitachi, Japan).
In Vitro Growth Determination
To compare the growth kinetics of the mutants with that of the parental strains, a fresh culture (100 μL) of each C. jejuni strain (0.5 McFarland) was inoculated into 100 mL MH broth and the cultures were incubated at 42°C under microaerobic conditions for 36 h with shaking. The growth kinetics was determined by measuring the absorbance in 600 nm (OD600) at 0, 4, 8, 12, 20, 22, 24, 27, 31, 33, and 36 h post-inoculation.
Single and Competitive Colonization in Chicken
Newly hatched broiler chickens (White Leghorns) were purchased from Zhengda Limited Company (Wuhan, China). All the broiler chickens were examined for C. jejuni to ascertain that birds are free of C. jejuni prior to infection all the chickens (Hao et al., 2015).
These chickens were randomly assigned to seven groups with 6 to 10 chickens per group. One group was used as a control. Four groups were used for single colonization test in which 109 CFU C. jejuni strains (S, SM, R and RM) were individually inoculated via oral gavage into each group. Another two groups were used for pairwise competition test in which 109 CFU C. jejuni pairwise mixtures (S&SM or R&RM) were inoculated via oral gavage to each group. Fecal samples were collected from each bird at 3, 6, 9, and 12 days’ post-infection. The CFU of S, SM, R and RM were determined using Campylobacter selective CCDA agar (Oxoid, Thermo Fisher Scientific, Waltham, MA, USA) with or without 25 μg/ml Kanamycin or 50 μg/ml erythromycin. Each sample was spread onto three respective selective plates to obtain the average CFU.
The significance of differences between mutant and parental strain in colonization at each sampling time point was determined by using Student’s t-test, Welch’s t-test to allow for non-constant variation across treatment groups, and the Wilcoxon rank-sum test to allow for non-normality (Guo et al., 2008; Luangtongkum et al., 2012; Xia et al., 2013). Differences were considered significant at a P-value of <0.01.
Ethics Statement
The animal study was approved by Animal Ethics Committee of Huazhong Agricultural University (HZAUCH 2013-002) and the Animal Care Center, Hubei Science and Technology Agency in China (SYXK 2013-0044). All experimental procedures in this study were performed according to the guidelines of the committee on the use and care of the laboratory animals in Hubei Province, China. All the animals were monitored throughout the study for any signs of adverse effects.
Results
Differentially Expressed Genes in SM and RM
The target gene Cj0440c was down-regulated in the Cj0440c-inactivated mutants (SM and RM). The other differentially expressed genes in Cj0440c mutants (SM and RM) compared to their parental strains (S and R) were shown in Tables 2, 3. The relationship of these different genes was summarized in Figure 1.
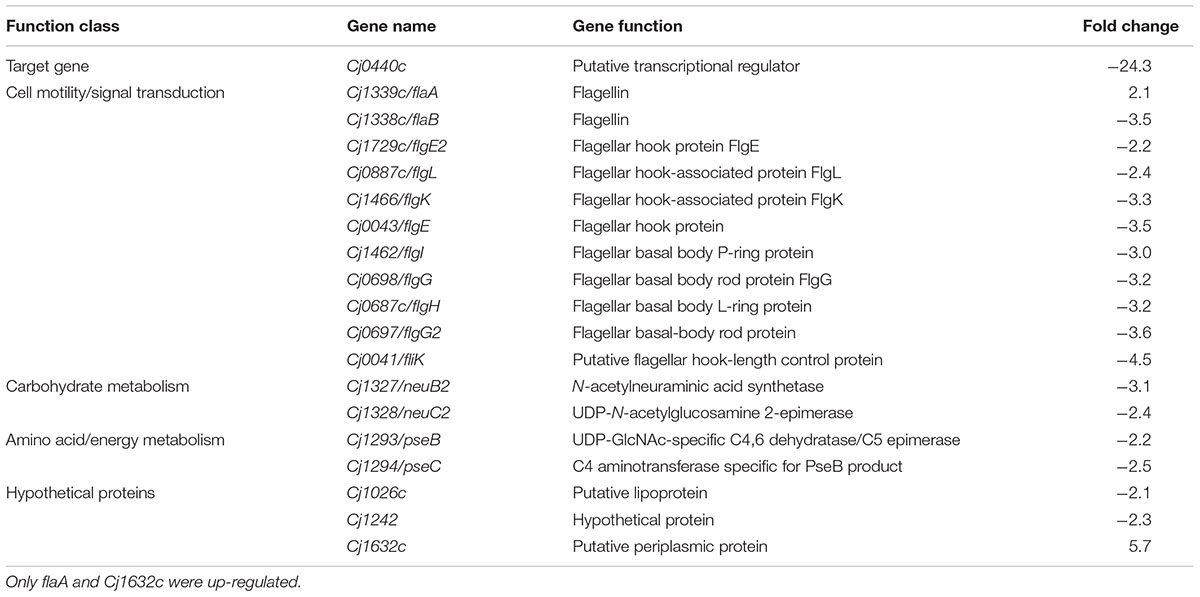
TABLE 2. Transcriptional difference in the mutant SM comparing to its parental strain S determined by microarray.
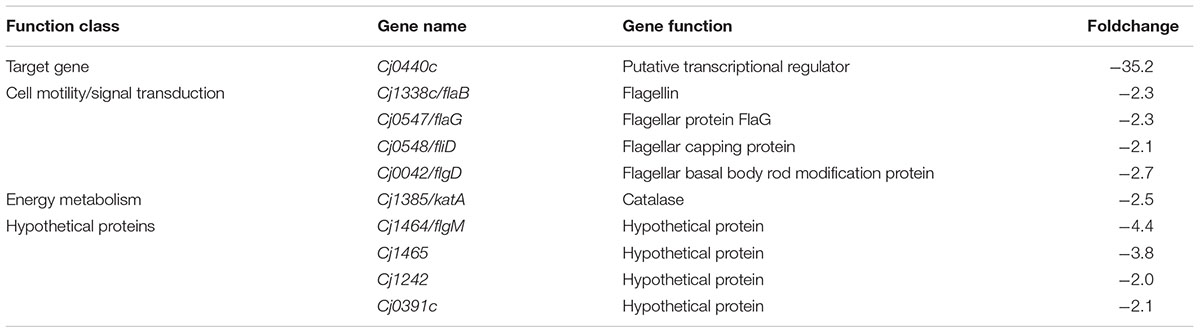
TABLE 3. Transcriptional difference in the mutant RM comparing to its parental strain R determined by microarray.
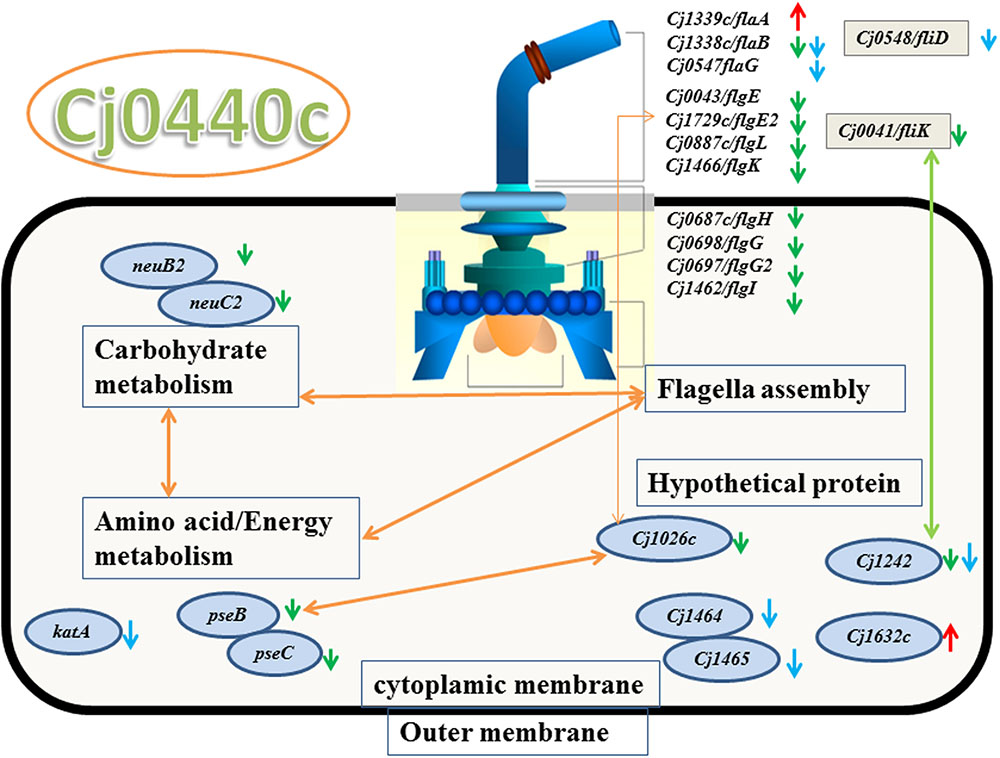
FIGURE 1. Significant genes in Cj0440c-inactivated mutants and their relationship based on KEGG and STRING protein network analysis. Green arrow and red arrow were genes down-regulated and up-regulated in SM, respectively. Genes with blue arrow were down-regulated in RM. The yellow double sided arrow means positive relationship between these genes and green double sided arrow means negative relationship between these genes.
A flagellin gene (flaA) and a gene (Cj1632c) encoding a putative periplasmic protein were up-regulated in SM as compared to S (indicated by red arrows in Figure 1). Among the down-regulated genes in SM (indicated by green arrows in Figure 1), 10 genes (flaB, flgE, flgE2, flgG, flgG2, flgH, flgI, flgK, flgL, and fliK) are possible involve in flagellar assembly; 2 genes (pseB and pseC) in carbohydrate metabolism; 2 genes (neuB2 and neuC) in surface glycoprotein metabolism.
None of the genes were up-regulated genes were found in RM when compared the expression of different genes with R. Eleven down regulated genes in RM (indicated by blue arrows in Figure 1) are flagellar associated genes (flgD, fliD, flaG and flaB), a catalase encoding gene (katA) and four genes with unknown function (flgM, Cj1465, Cj0391, and Cj1242).
When submitted to STRING 9.05 and KEGG pathway analysis, the result showed that 10 flagellar genes were interacted with other down-regulated genes (pseB/C, neuB2/C2 and Cj1026c) (Figure 1).
The transcriptional change of several representative genes detected in microarray was validated by qRT-PCR. The similar change of the selected genes was found both in microarray and qRT-PCR (Figure 2).
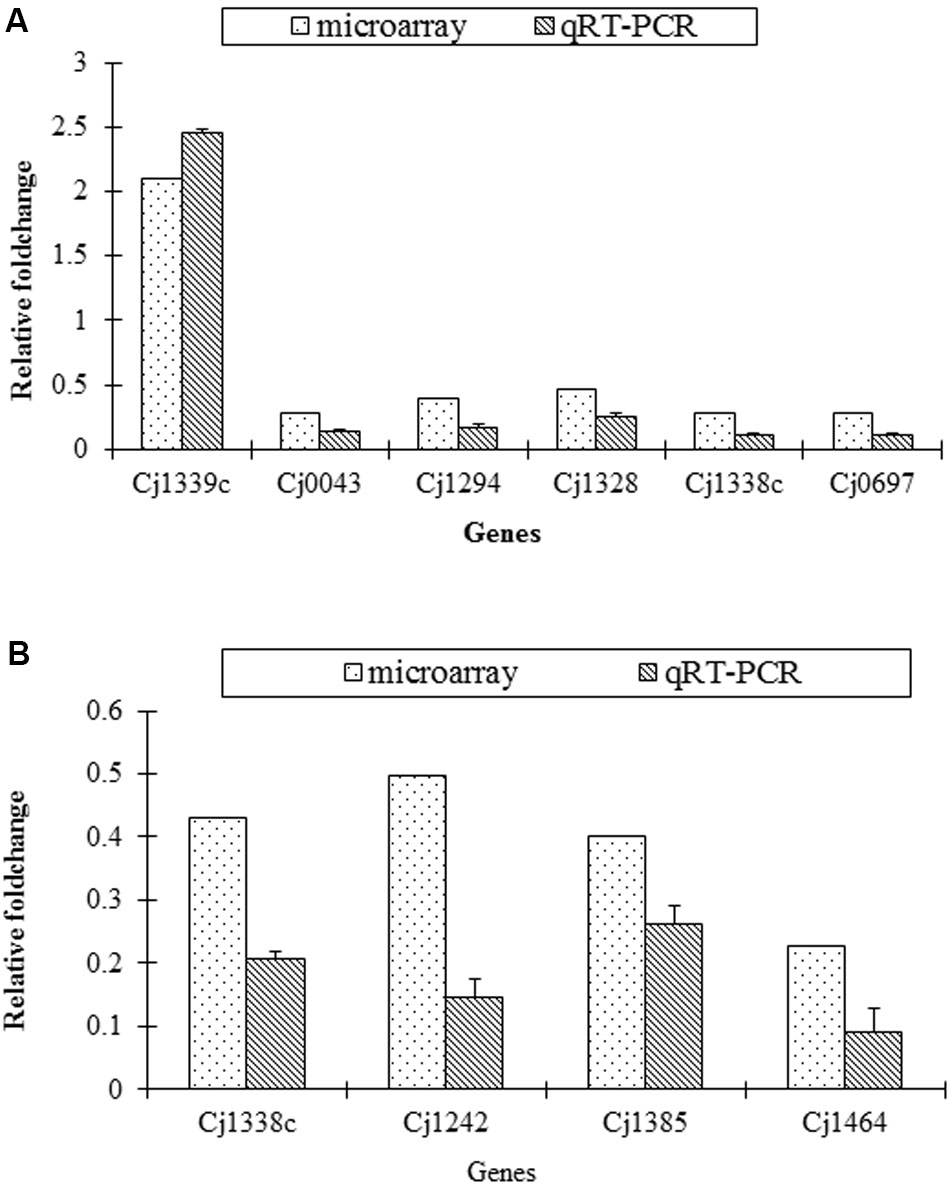
FIGURE 2. Confirmation of transcriptional change of target genes in the mutants and the parental strains by qRT-PCR. (A) Fold change of six genes in SM versus S; (B) Fold change of four genes in RM versus R.
The microarray data obtained in this study have been deposited in the NCBI Gene Expression Omnibus database2 and assigned accession numbers GSE49255 (RM&R) and GSE49255(SM&S).
Antimicrobial Susceptibility of Cj0440c Mutants
As shown in Table 4, there was no significant difference between MIC of nine antimicrobial agents in Cj0440c mutants (SM and RM) comparing to their parental strains (S and R). Inactivation of Cj0440c did not affect antimicrobial susceptibility of C. jejuni.
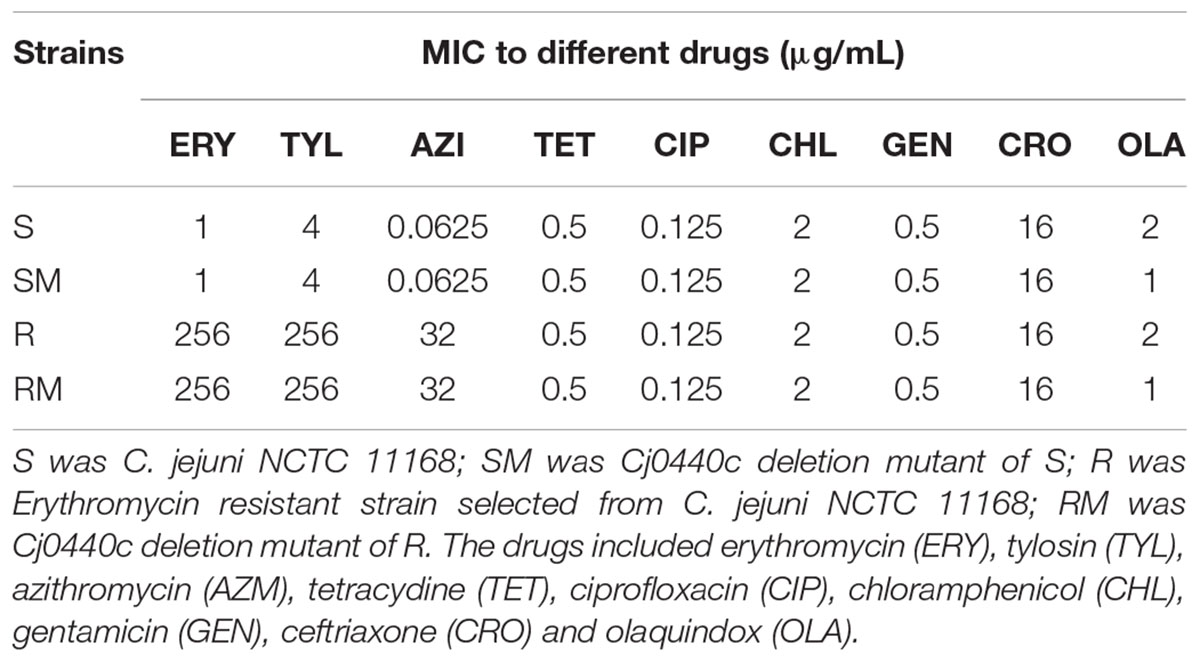
TABLE 4. Minimum inhibitory concentration (MIC) of Cj0440c mutant strains and parental strains to different drugs.
Flagella Presence and Length
The electron micrographs of all tested strains were shown in Figure 3. The results showed that parental strains (S and R) had long, spiral and complete flagella filaments in two sides (Figures 3A,C). However, SM had shorter filaments in only one side (Figure 3B). No filaments of RM were detected in RM (Figure 3D). These findings indicated that Cj0440c may affect the formation of flagella in C. jejuni.
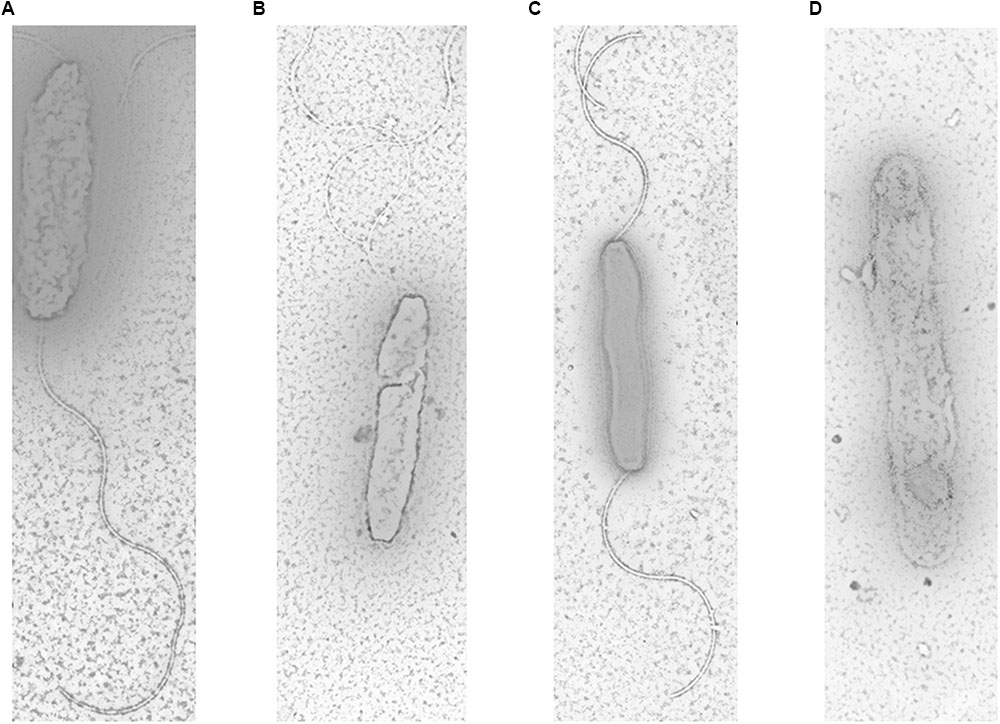
FIGURE 3. Flagella morphology of S (A), SM (B), R (C), and RM (D) under transmission electron microscope. The magnification used for TEM images in the captionis are 1 μm.
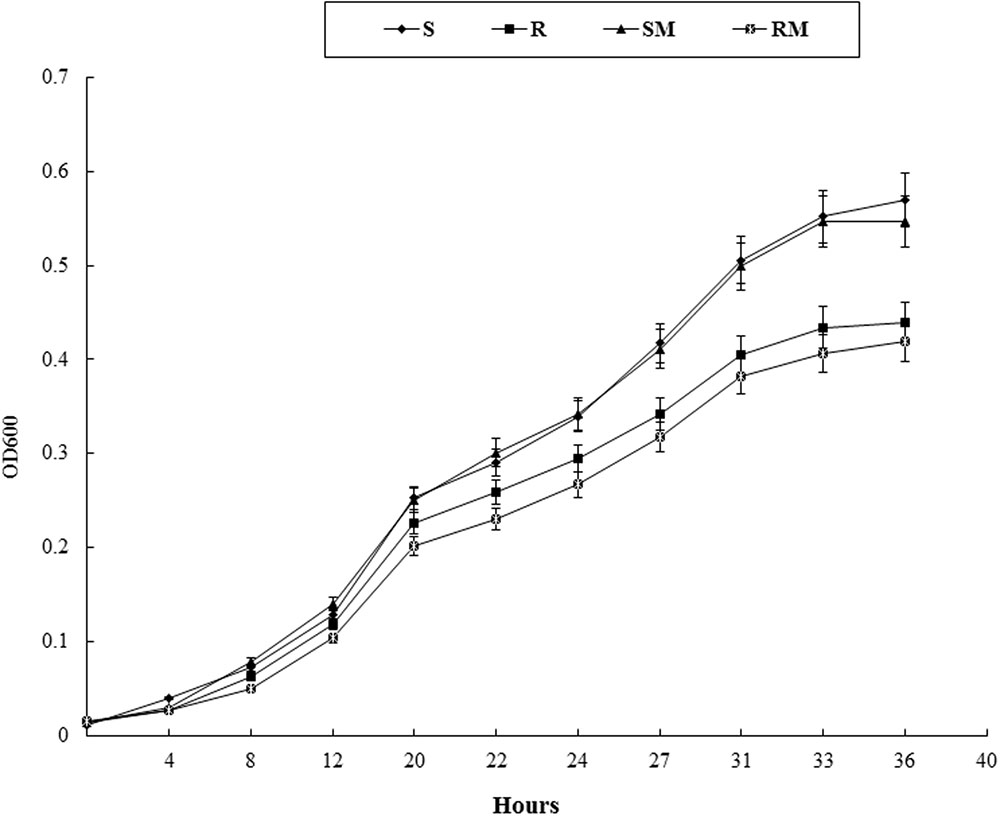
In Vitro Growth of Cj0440c Mutants
Growth kinetics of Cj0440c mutants (SM and RM) and their parental strains (S and R) were determined in MH broth. No significant difference in growth rate was observed between SM and S. The RM exhibited slower growth rate compared to its parental R, however, the difference was not significant (Figure 4).
In Vivo Colonization of Cj0440c Mutants
To determine the colonization capacity, broiler chickens were infected individually with four C. jejuni strains (S, R, SM and RM). All the strains were able to colonize in chicken intestinal tract, albeit at different rate. Comparing with the parental strains (S or R), the Cj0440c mutants (SM and RM) showed a significant reduction in colonization on 12 days’ post-inoculation (Figure 5).
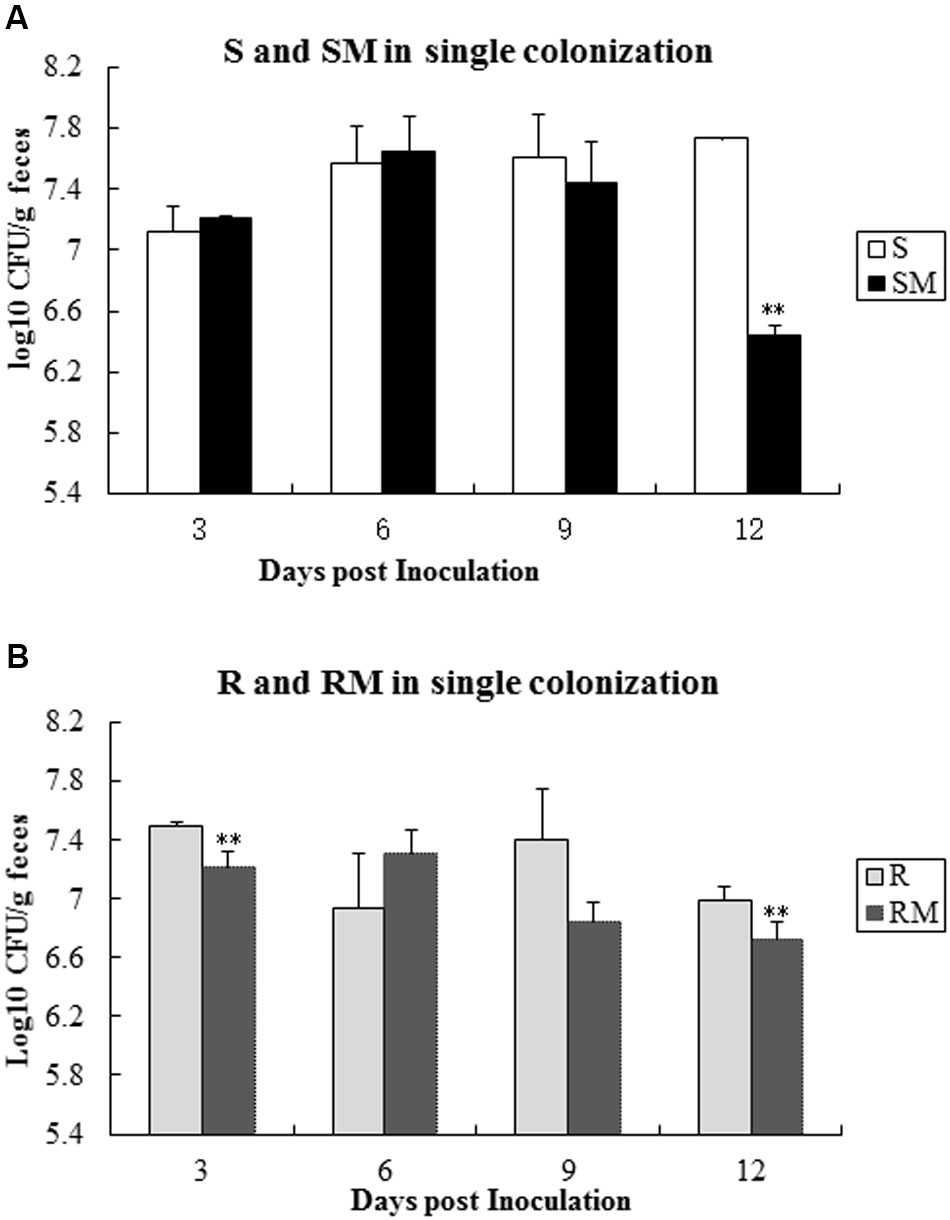
FIGURE 5. Single colonization of four C. jejuni strains in chickens after oral inoculation. (A) Colonization of S and SM; (B) Colonization of R and RM. The asterisk (∗∗) represent statistically significant difference with P ≤ 0.01 comparing with parental strains, respectively.
When the two pairs of C. jejuni strains (SM&S and RM&R) were infected chickens with one pair at a time, Cj0440c mutants (SM and RM) exhibited lower colonization level compared to their parental strains (S and R) after 9 days’ post-inoculation (Figure 6).
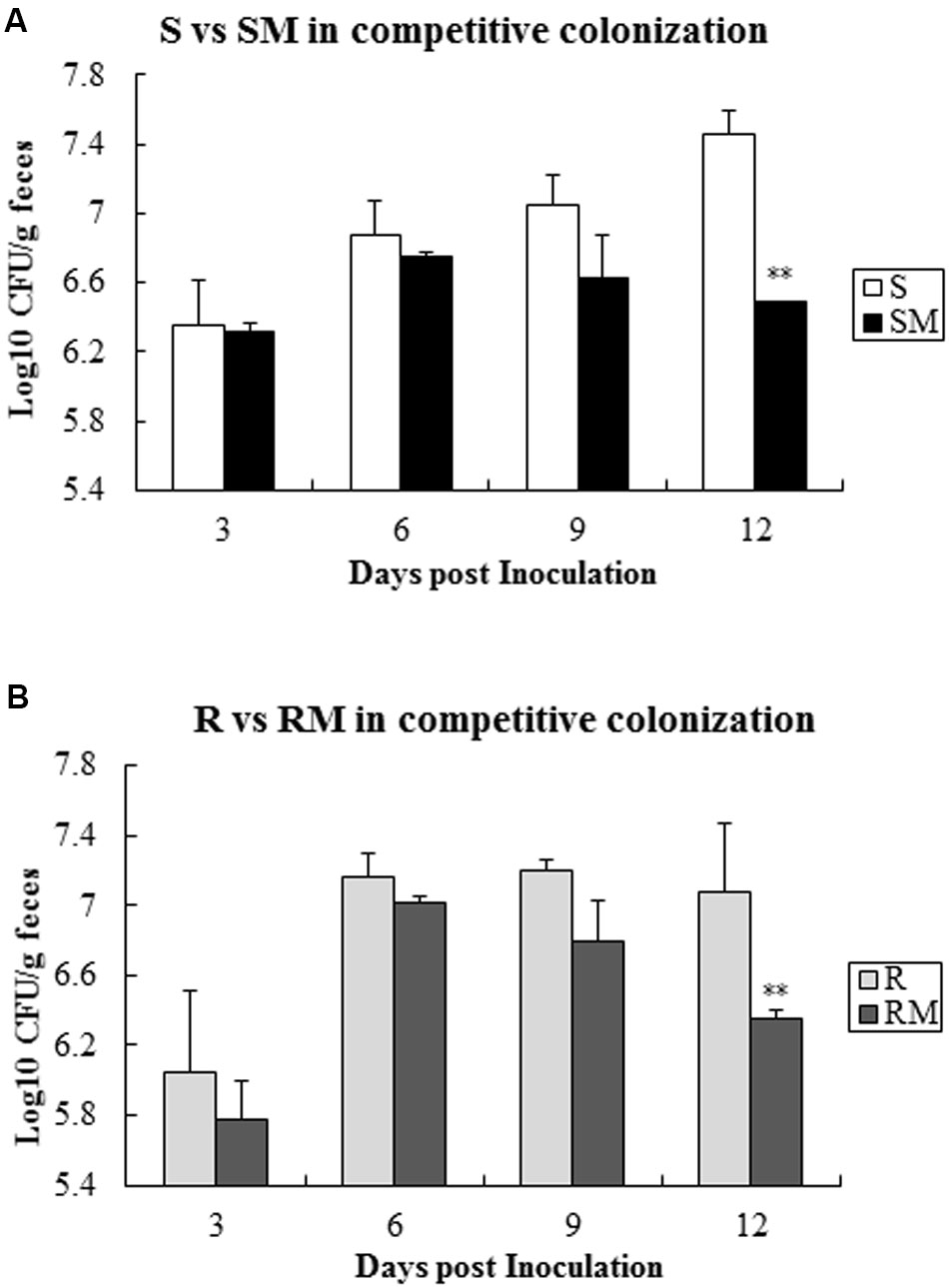
FIGURE 6. Competitive colonization of Cj0440c mutants with their patent strains (SM&S and RM&R). (A) Competitive colonization of the pair of SM&S; (B) Competitive colonization of the pair of RM&R. The asterisk (∗∗) represent statistically significant difference with P ≤ 0.01 comparing with parental strains, respectively.
Discussion
Campylobacter jejuni is a very common foodborne pathogen in the developed world. Its biology and pathogenicity is largely unknown (Young et al., 2007). Cj0440c is a putative transcriptional regulator and an increased transcriptional level expression was detected in the erythromycin-resistant C. jejuni (Hao et al., 2013). The gene may encode a TENA/THI-4/PQQC family protein. TENA is one of a number of proteins that enhance the expression of extracellular enzymes (e.g., alkaline protease, neutral protease and levansucrase) (Pang et al., 1991). The extracellular enzymes may be regulated by the master regulator of flagellar genes (e.g., flhDC) (Cui et al., 2008). THI-4 protein is involved in thiamine biosynthesis (Akiyama and Nakashima, 1996). This family also includes bacterial coenzyme pyrroloquinoline quinone (PQQ) synthesis protein C (PQQC) proteins. PQQ is the prosthetic group of several bacterial enzymes, including methanol dehydrogenase of methylotrophs and the glucose dehydrogenase (Toyama et al., 2002, 2007). In E. coli, PQQ biosynthesis may be affected by tldD gene which encodes a peptidase involved in processing of small peptides (Holscher and Gorisch, 2006). The tldD may lead to chromosomal DNA relaxation and subsequent derepression of cdtB and lgeR which may regulate the expression of some flagellar genes (Haghjoo and Galan, 2007). Therefore, the TENA/THI-4/PQQC family may have some indirect relationship with flagellar genes.
The flagella formation plays an important role in the pathogenesis of Campylobacter including motility, microcolony formation, biofilm formation, autoagglutination, protein secretion, adherence to host cell, and host invasion (Guerry et al., 2006; Guerry, 2007). The major groups of down-regulated genes in Cj0440c mutants (SM and RM) were involved in flagellar assembly, including 11 genes (flaB, flgE/E2/L/K/H/G/G2/I, flgK, flgL, fliK) in SM and 4 genes (flaB/G, flgD, fliD) in RM (Figure 1). The down-regulation of those flagella-associated genes in Cj0440c mutants can reasonably explain why SM and RM lose one or two sides of filament. The reduced colonization of Cj0440c mutants may result from the down-regulation of flagella genes.
A second group of genes (pseB, pseC, neuB2 and neuC2) down-regulated in SM were involved in O-linked glycosylation which was also essential for flagellin assembly. The pseB/C in C. jejuni contribute in glycosylation modifications of flagellin, often by non-specifically modifying the surface-exposed Thr, Ser, and Tyr residues of filament proteins FlaA and FlaB (Ewing et al., 2009). While neuB2/C2 requires in O-linked glycosylation which may contribute to flagella antigen diversity of Campylobacter (Linton et al., 2000; Sundaram et al., 2004; Tabei et al., 2009). The down-regulation of these glycosylation-associated genes in SM suggested that Cj0440c may play an important role in flagella assembly.
Several hypothetical genes (Cj1026c, Cj1242, Cj1464, Cj1465 and Cj0391c) were down-regulated in SM or RM. The Cj1026c (FlgP) was essential for motility of C. jejuni and possible encode the promoter of flaG (Sommerlad and Hendrixson, 2007). The Cj1464 (FlgM) may regulate temperature-dependent FlgM/FliA complex formation and flagella length of C. jejuni (Wösten et al., 2010). The Cj0391c generally co-expressed with flagella-associated genes and involved in biofilm formation of Campylobacter (Kalmokoff et al., 2006). The down-regulation of these genes suggested that Cj0440c may be closely associated with flagella biosynthesis and assembly.
All our data showed that Cj0440c may have close relationship with flagella biosynthesis and assembly, however, the precise role of Cj0440c in flagella formation pathway is yet to be determined. Flagellar biogenesis in C. jejuni requires three distinct sigma factors, including σ80, σ54 (or RpoN) and σ28 (or FliA) (McCarter, 2006; Anderson et al., 2010). The FlgS/FlgR two-component system is required for transcription of the RpoN regulon (Joslin and Hendrixson, 2009). The FliK likely part of a negative feedback loop that turns off expression of σ54-dependent genes (Ryan et al., 2005; Kamal et al., 2007). The FlgM (anti σ28) may negatively regulate the class III motility genes (Wang et al., 2005). The present study showed that the transcription of fliK was down-regulated in SM and the transcription of flgM (Cj1464) was down-regulated in RM. The down-regulation of fliK and flgM can influence the down-regulation of class II and class III motility. The roles of Cj0440c on flagellar genes are complex and further investigations are required.
The transcriptional change of majority parts of the genes was similar in both SM and RM except for few differences. The flaA and Cj1632c were up-regulated and O-linked glycosylation was down-regulated only in SM, while katA, encoding a sole catalase, was down-regulated in RM but not in SM. The flagellar filaments of Campylobacter spp. were composed primarily by FlaA and FlaB flagellin (Guerry et al., 1991). The flaA was merely up-regulated in SM but flaB was down-regulated in both SM and RM. Findings of our study suggested that that the role Cj0440c on transcription of FlaA and FlaB flagellin are different in Erys and in Eryr C. jejuni. The katA involves in oxidative stress and ROS defense which was essential for intra-macrophage persistence and environmental stress survival of Campylobacter (Farr and Kogoma, 1991; Day et al., 2000; Vliet et al., 2002; Flint et al., 2012). The down-regulation of katA in RM suggested that Cj0440c may interact with katA to improve their survival capacity in environmental stress.
The macrolide-resistance in C. jejuni generally suffered a fitness cost, however, several other factors may compensate the adaptation weakness (Björkman and Andersson, 2000; Kugelberg et al., 2006; Nilsson et al., 2006; Hao et al., 2010, 2013; Luangtongkum et al., 2012). Our previous study showed that Cj0440c was over-expressed in the Eryr C. jejuni (Hao et al., 2013). The result from the present study suggests that Cj0440c plays a role in compensating the fitness cost of erythromycin resistance through the positive relationship with flagellar and other related genes.
Conclusion
Cj0440c regulates expression of genes involved in flagella biosynthesis and assembly which consequently affects the growth or colonization of C. jejuni in vitro and in vivo environment.
Author Contributions
Conceived and designed the experiments: XF, HH, YW, XW, and ZY. Performed the experiments: XF and HH. Analyzed the data: XF, HH, JH, SF, GC, LH, and ZY. Contributed reagents/materials/analysis tools: ZY, ZL, MD, and HH. Wrote the paper: HH, XF, JH, SF, and ZY.
Funding
This work was supported by National Key research and development program (2016YFD0501302), National Natural Science Foundation of China (31101856), National Basic Research Program of China (2013CB127200), Morning program of Wuhan in China (2015070404010191), Fundamental Research Funds for the Central Universities (2662015PY035), and National Program for Risk Assessment of Quality and Safety of Livestock and Poultry Products (GJFP2016008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Disclaimer: The opinions expressed in this manuscript are solely the responsibility of the authors and do not necessarily represent the official views and policy of the US Food and Drug Administration. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the Food and Drug Administration.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Qijing Zhang and Zhangqi Shen from Iowa State University in USA, Congming Wu and Yang Wang from China Agricultural University for kindly providing the vector plasmids including plasmids pGEM-T and pMW10; and we thank Maojun Zhang from Chinese Center for Disease Control and Prevention for kindly providing C. jejuni NCTC11168. We thank Drs. Kidon Sung and Bijay Khajanchi from US FDA National Center for Toxicological Research for their critical review of manuscript.
Footnotes
References
Akiyama, M., and Nakashima, H. (1996). Molecular cloning of thi-4, a gene necessary for the biosynthesis of thiamine in Neurospora crassa. Curr. Genet. 30, 62–67. doi: 10.1007/s002940050101
Anderson, J. K., Smith, T. G., and Hoover, T. R. (2010). Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 18, 30–37. doi: 10.1016/j.tim.2009.11.001
Barrero-Tobon, A. M., and Hendrixson, D. R. (2014). Flagellar biosynthesis exerts temporal regulation of secretion of specific Campylobacter jejuni colonization and virulence determinants. Mol. Microbiol. 93, 957–974. doi: 10.1111/mmi.12711
Björkman, J., and Andersson, D. I. (2000). The cost of antibiotic resistance from a bacterial perspective. Drug Resist. Updat. 3, 237–245. doi: 10.1054/drup.2000.0147
Byers, D. M., and Gong, H. (2007). Acyl carrier protein: structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 85, 649–662. doi: 10.1139/O07-109
Cui, Y., Chatterjee, A., Yang, H., and Chatterjee, A. K. (2008). Regulatory network controlling extracellular proteins in Erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmB regulatory RNA production by affecting gacA and hexA (lrhA) expression. J. Bacteriol. 190, 4610–4623. doi: 10.1128/JB.01828-07
Day, W. A., Sajecki, J. L., Pitts, T. M., and Joens, L. A. (2000). Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68, 6337–6345. doi: 10.1128/IAI.68.11.6337-6345.2000
ECDC, EFSA, EMEA and SCENIHR (2009). Joint opinion on antimicrobial resistance (AMR) focused on zoonotic infections. EFSA J. 7, 1372.
Epps, S. V., Harvey, R. B., Hume, M. E., Phillips, T. D., Anderson, R. C., and Nisbet, D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health 10, 6292–6304. doi: 10.3390/ijerph10126292
Ewing, C. P., Andreishcheva, E., and Guerry, P. (2009). Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191, 7086–7093. doi: 10.1128/JB.00378-09
Farr, S. B., and Kogoma, T. (1991). Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55, 561–585.
Flint, A., Sun, Y.-Q., and Stintzi, A. (2012). Cj1386 is an ankyrin-containing protein involved in heme trafficking to catalase in Campylobacter jejuni. J. Bacteriol. 194, 334–345. doi: 10.1128/JB.05740-11
Gibreel, A., Kos, V. N., Keelan, M., Trieber, C. A., Levesque, S., Michaud, S., et al. (2005). Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob. Agents Chemother. 49, 2753–2759. doi: 10.1128/AAC.49.7.2753-2759.2005
Gibreel, A., and Taylor, D. E. (2006). Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58, 243–255. doi: 10.1093/jac/dkl210
Guerry, P. (2007). Campylobacter flagella: not just for motility. Trends Microbiol. 15, 456–461. doi: 10.1016/j.tim.2007.09.006
Guerry, P., Alm, R., Power, M., Logan, S., and Trust, T. (1991). Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 173, 4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991
Guerry, P., Ewing, C. P., Schirm, M., Lorenzo, M., Kelly, J., Pattarini, D., et al. (2006). Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60, 299–311. doi: 10.1111/j.1365-2958.2006.05100.x
Guo, B., Wang, Y., Shi, F., Barton, Y. W., Plummer, P., Reynolds, D. L., et al. (2008). CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J. Bacteriol. 190, 1879–1890. doi: 10.1128/JB.01796-07
Haghjoo, E., and Galan, J. E. (2007). Identification of a transcriptional regulator that controls intracellular gene expression in Salmonella Typhi. Mol. Microbiol. 64, 1549–1561. doi: 10.1111/j.1365-2958.2007.05754.x
Hao, H., Dai, M., Wang, Y., Chen, D., and Yuan, Z. (2010). Quantification of mutated alleles of 23S rRNA in macrolide-resistant Campylobacter by TaqMan real-time polymerase chain reaction. Foodborne Pathog. Dis. 7, 43–49. doi: 10.1089/fpd.2009.0339
Hao, H., Liu, J., Kuang, X., Dai, M., Cheng, G., Wang, X., et al. (2015). Identification of Campylobacter jejuni and determination of point mutations associated with macrolide resistance using a multiplex TaqMan MGB real-time PCR. J. Appl. Microbiol. 118, 1418–1425. doi: 10.1111/jam.12793
Hao, H., Yuan, Z., Shen, Z., Han, J., Sahin, O., Liu, P., et al. (2013). Mutational and transcriptomic changes involved in the development of macrolide resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 57, 1369–1378. doi: 10.1128/AAC.01927-12
Holscher, T., and Gorisch, H. (2006). Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. J. Bacteriol. 188, 7668–7676. doi: 10.1128/JB.01009-06
Hughes, R. A., and Cornblath, D. R. (2005). Guillain-Barré syndrome. Lancet 366, 1653–1666. doi: 10.1016/S0140-6736(05)67665-9
Jeon, B., Wang, Y., Hao, H., Barton, Y.-W., and Zhang, Q. (2011). Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J. Antimicrob. Chemother. 66, 79–85. doi: 10.1093/jac/dkq418
Joslin, S. N., and Hendrixson, D. R. (2009). Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191, 2656–2667. doi: 10.1128/JB.01689-08
Kalmokoff, M., Lanthier, P., Tremblay, T.-L., Foss, M., Lau, P. C., Sanders, G., et al. (2006). Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188, 4312–4320. doi: 10.1128/JB.01975-05
Kamal, N., Dorrell, N., Jagannathan, A., Turner, S. M., Constantinidou, C., Studholme, D. J., et al. (2007). Deletion of a previously uncharacterized flagellar-hook-length control gene fliK modulates the sigma54-dependent regulon in Campylobacter jejuni. Microbiology 153, 3099–3111. doi: 10.1099/mic.0.2007/007401-0
Kassem, I. I., Khatri, M., Esseili, M. A., Sanad, Y. M., Saif, Y. M., Olson, J. W., et al. (2012). Respiratory proteins contribute differentially to Campylobacter jejuni’s survival and in vitro interaction with hosts’ intestinal cells. BMC Microbiol. 12:258. doi: 10.1186/1471-2180-12-258
Kugelberg, E., Kofoid, E., Reams, A. B., Andersson, D. I., and Roth, J. R. (2006). Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. U.S.A. 103, 17319–17324. doi: 10.1073/pnas.0608309103
Linton, D., Karlyshev, A. V., Hitchen, P. G., Morris, H. R., Dell, A., Gregson, N. A., et al. (2000). Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 35, 1120–1134. doi: 10.1046/j.1365-2958.2000.01780.x
Luangtongkum, T., Shen, Z., Seng, V. W., Sahin, O., Jeon, B., Liu, P., et al. (2012). Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob. Agents Chemother. 56, 1300–1308. doi: 10.1128/AAC.05516-11
Mace, S., Haddad, N., Zagorec, M., and Tresse, O. (2015). Influence of measurement and control of microaerobic gaseous atmospheres in methods for Campylobacter growth studies. Food Microbiol. 52, 169–176. doi: 10.1016/j.fm.2015.07.014
Matsunami, H., Barker, C. S., Yoon, Y. H., Wolf, M., and Samatey, F. A. (2016). Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat. Commun. 7:13425. doi: 10.1038/ncomms13425
McCarter, L. L. (2006). Regulation of flagella. Curr. Opin. Microbiol. 9, 180–186. doi: 10.1016/j.mib.2006.02.001
Nilsson, A. I., Zorzet, A., Kanth, A., Dahlström, S., Berg, O. G., and Andersson, D. I. (2006). Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl. Acad. Sci. U.S.A. 103, 6976–6981. doi: 10.1073/pnas.0602171103
Pang, A. S., Nathoo, S., and Wong, S. L. (1991). Cloning and characterization of a pair of novel genes that regulate production of extracellular enzymes in Bacillus subtilis. J. Bacteriol. 173, 46–54. doi: 10.1128/jb.173.1.46-54.1991
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Riddle, M. S., Sanders, J. W., Putnam, S. D., and Tribble, D. R. (2006). Incidence, etiology, and impact of diarrhea among long-term travelers (US military and similar populations): a systematic review. Am. J. Trop. Med. Hyg. 74, 891–900.
Ryan, K. A., Karim, N., Worku, M., Penn, C. W., and O’Toole, P. W. (2005). Helicobacter pylori flagellar hook-filament transition is controlled by a FliK functional homolog encoded by the gene HP0906. J. Bacteriol. 187, 5742–5750. doi: 10.1128/JB.187.16.5742-5750.2005
Samuel, M. C., Vugia, D. J., Shallow, S., Marcus, R., Segler, S., McGivern, T., et al. (2004). Epidemiology of sporadic Campylobacter infection in the United States and declining trend in incidence, FoodNet 1996-1999. Clin. Infect. Dis. 38(Suppl. 3), S165–S174. doi: 10.1086/381583
Scharff, R. L. (2012). Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75, 123–131. doi: 10.4315/0362-028X.JFP-11-058
Silva, J., Leite, D., Fernandes, M., Mena, C., Gibbs, P. A., and Teixeira, P. (2011). Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2:200. doi: 10.3389/fmicb.2011.00200
Sommerlad, S. M., and Hendrixson, D. R. (2007). Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189, 179–186. doi: 10.1128/JB.01199-06
Sundaram, A. K., Pitts, L., Muhammad, K., Wu, J., Betenbaugh, M., Woodard, R. W., et al. (2004). Characterization of N-acetylneuraminic acid synthase isoenzyme 1 from Campylobacter jejuni. Biochem. J. 383, 83–89. doi: 10.1042/BJ20040218
Tabei, S. M., Hitchen, P. G., Day-Williams, M. J., Merino, S., Vart, R., Pang, P. C., et al. (2009). An Aeromonas caviae genomic island is required for both O-antigen lipopolysaccharide biosynthesis and flagellin glycosylation. J. Bacteriol. 191, 2851–2863. doi: 10.1128/JB.01406-08
Toyama, H., Fukumoto, H., Saeki, M., Matsushita, K., Adachi, O., and Lidstrom, M. E. (2002). PqqC/D, which converts a biosynthetic intermediate to pyrroloquinoline quinone. Biochem. Biophys. Res. Commun. 299, 268–272. doi: 10.1016/S0006-291X(02)02603-7
Toyama, H., Nishibayashi, E., Saeki, M., Adachi, O., and Matsushita, K. (2007). Factors required for the catalytic reaction of PqqC/D which produces pyrroloquinoline quinone. Biochem. Biophys. Res. Commun. 354, 290–295. doi: 10.1016/j.bbrc.2007.01.001
Vliet, A. H., Ketley, J. M., Park, S. F., and Penn, C. W. (2002). The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26, 173–186. doi: 10.1111/j.1574-6976.2002.tb00609.x
Wang, Q., Suzuki, A., Mariconda, S., Porwollik, S., and Harshey, R. M. (2005). Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24, 2034–2042. doi: 10.1038/sj.emboj.7600668
Weingarten, R. A., Taveirne, M. E., and Olson, J. W. (2009). The dual-functioning fumarate reductase is the sole succinate:quinone reductase in Campylobacter jejuni and is required for full host colonization. J. Bacteriol. 191, 5293–5300. doi: 10.1128/JB.00166-09
Wösten, M. M., Van Dijk, L., Veenendaal, A. K., De Zoete, M. R., Bleumink-Pluijm, N., and Van Putten, J. P. (2010). Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol. Microbiol. 75, 1577–1591. doi: 10.1111/j.1365-2958.2010.07079.x
Xia, Q., Muraoka, W. T., Shen, Z., Sahin, O., Wang, H., Wu, Z., et al. (2013). Adaptive mechanisms of Campylobacter jejuni to erythromycin treatment. BMC Microbiol. 13:133. doi: 10.1186/1471-2180-13-133
Keywords: Campylobacter jejuni, Cj0440c, flagella, colonization, erythromycin resistance
Citation: Hao H, Fang X, Han J, Foley SL, Wang Y, Cheng G, Wang X, Huang L, Dai M, Liu Z and Yuan Z (2017) Cj0440c Affects Flagella Formation and In Vivo Colonization of Erythromycin-Susceptible and -Resistant Campylobacter jejuni. Front. Microbiol. 8:729. doi: 10.3389/fmicb.2017.00729
Received: 01 August 2016; Accepted: 07 April 2017;
Published: 25 April 2017.
Edited by:
Avelino Alvarez-Ordóñez, Universidad de León, SpainCopyright © 2017 Hao, Fang, Han, Foley, Wang, Cheng, Wang, Huang, Dai, Liu and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zonghui Yuan, eXVhbjU4MDJAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Haihong Hao
Haihong Hao Xia Fang
Xia Fang Jing Han3
Jing Han3 Yulian Wang
Yulian Wang Guyue Cheng
Guyue Cheng Xu Wang
Xu Wang