- 1Division of Genetics and Molecular Biology, Institute of Biological Sciences, Faculty of Science, University of Malaya, Kuala Lumpur, Malaysia
- 2Novel Bacteria and Drug Discovery Research Group, School of Pharmacy, Monash University Malaysia, Selangor, Malaysia
- 3Department of Pharmacy, Abasyn University, Peshawar, Pakistan
- 4Department of Pharmaceutics, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 5UKM Medical Molecular Biology Institute, UKM Medical Centre, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
- 6Center of Health Outcomes Research and Therapeutic Safety (Cohorts), School of Pharmaceutical Sciences, University of Phayao, Phayao, Thailand
Bacteria must develop resistance to various inhospitable conditions in order to survive in the human gastrointestinal tract. Bile, which is secreted by the liver, and plays an important role in food digestion also has antimicrobial properties and is able to disrupt cellular homeostasis. Paradoxically, although bile is one of the guts defenses, many studies have reported that bacteria such as Vibrio parahaemolyticus can sense bile and use its presence as an environmental cue to upregulate virulence genes during infection. This article aims to discuss how bile is detected by V. parahaemolyticus and its role in regulating type III secretion system 2 leading to human infection. This bile–bacteria interaction pathway gives us a clearer understanding of the biochemical and structural analysis of the bacterial receptors involved in mediating a response to bile salts which appear to be a significant environmental cue during initiation of an infection.
Introduction
Humans have a complex digestive system that not only aids in digestion of food but also has a role in self-defense against microorganisms in the body. Microorganisms such as bacteria have to tolerate various extreme environments in order to survive in the human gastrointestinal tract (Begley et al., 2005). Bile is an alkaline substance that is continuously secreted by liver and stored in the gall bladder in humans; the presence of bile plays an important role in the digestive system process. During the digestive process, the lipids are emulsified and solubilized by bile. In addition, bile has the capability to affect the cell membranes proteins and phospholipid structures and cause cellular homeostasis. Bile aids in the emulsification and solubilization of lipids in the gastrointestinal tract. In addition, it has the capability to affect the phospholipids and proteins of cell membranes and disrupt cellular homeostasis. Hence the ability to overcome the potentially lethal effects of bile is important for bacteria in order to survive and subsequently colonize the gastrointestinal tract (Begley et al., 2005; Hung and Mekalanos, 2005; Edwards and Slater, 2009). Recently, there has been increased evidence showing bile is been used as a signaling cue by enteric bacteria to initiate virulence genes in host infection (Pope et al., 1995; Krukonis and DiRita, 2003; Prouty et al., 2004). Bile acids are a major component of crude bile that triggers the expression of bacterial virulence in the body. In this article, we aim to discuss how Vibrio parahaemolyticus senses bile in the human GI tract to regulate type III secretion system 2. This bile–bacteria interaction pathway gives us a clearer understanding of the biochemical and structural analysis of the bacterial receptors that takes action upon sensing the bile salts during an infection.
Virulence Factors of Vibrio parahaemolyticus
The bacterial protagonist in this story is V. parahaemolyticus, a Gram-negative, halophilic bacterium which naturally inhabits marine and estuarine environments worldwide (Zhang and Orth, 2013; Letchumanan et al., 2014). V. parahaemolyticus is recognized as the causative agent of foodborne gastroenteritis, a disease often associated with consumption of raw or undercooked seafood (Raghunath, 2015). Global climate change and rising ocean temperatures have led to the increase in the distribution of this pathogen worldwide (O’Boyle and Boyd, 2014). This is of concern as approximately half the reported foodborne cases in Asian countries are caused by V. parahaemolyticus (Alam et al., 2002; Bhuiyan et al., 2002). Frequent outbreaks of V. parahaemolyticus cases have also been reported in the United States and coastal countries of Europe such as Spain, Italy, and Norway (Caburlotto et al., 2008; Scallan et al., 2011; Ottaviani et al., 2013).
V. parahaemolyticus possess wide range of virulence factors that enables them to cause a gastrointestinal infection including adhesin (Liu and Chen, 2015), toxins, and secreted effectors (Zhang and Orth, 2013). These virulence factors play a vital role in the pathogenesis of the disease. During the initial host cell binding, adhesion is the first important step in bacterial pathogenesis (Liu and Chen, 2015). This factor is present on the surface of all V. parahaemolyticus to form a platform for them to attach onto host cell and secrete toxins during an infection (Broberg et al., 2011; Zhang and Orth, 2013; Letchumanan et al., 2014). The thermostable direct hemolysin (tdh) and TDH related hemolysin (trh) are the two major toxins found in V. parahaemolyticus (Honda et al., 1988; Nishibuchi et al., 1992; Okada et al., 2009). These two virulence toxins are believed to cause hemolysis and cytotoxic activity in a host cell (Broberg et al., 2011; Ceccarelli et al., 2013). The tdh is a pore-forming toxin which forms pores in the erythrocyte’s membrane (Matsuda et al., 2010). The large pore size enables both water and ions to flow through the membrane (Honda and Iida, 1993). The subsequent alterations in ion flux in the intestine causes the diarrhea which is observed during an infection (Raghunath, 2015). Similar to the tdh gene, the trh gene also triggers cl- channels resulting in altered ion flux during an infection (Takahashi et al., 2000). Both the tdh and trh are correlated with pathogenic V. parahaemolyticus strains, however, these genes do not completely account for the pathogenicity of V. parahaemolyticus (Lynch et al., 2005). There are several studies have reported that even in the absence of tdh and/or trh genes, V. parahaemolyticus strains remain virulent indicating the existence of other virulence factors (Jones et al., 2012; Pazhani et al., 2014). The thermolabile hemolysin (tlh), a type of phospholipase is another virulence toxin found in V. parahaemlyticus (DePaola et al., 2003; Zhang and Austin, 2005). Although the specific function of this gene in human infection remains unclear, tlh gene expression is upregulated under conditions mimic the intestinal environmental of human (Broberg et al., 2011; West et al., 2013). Hence, in the process of infection, tlh gene may be equally important as the tdh and trh genes.
The type III secretion system (T3SSs) is another important virulence factor of V. parahaemolyticus which is responsible for its pathogenicity (Broberg et al., 2011). This protein like structure has a secretion apparatus consisting of three main parts: the basal body that extends into the inner and outer membranes; a needle like structure that allows toxins to travel; and the translocon which is a pore injected into a target cell membrane (Izore et al., 2011). The T3SS1 and T3SS2 are the two main T3SSs encoded by V. parahaemolyticus. The cytotoxic T3SS1 is reported to be present in all V. parahaemolyticus and causes mouse lethality and possible initiation of autophagy (Park et al., 2004; Burdette et al., 2009; Hiyoshi et al., 2010). The enterotoxin T3SS2, on the other hand plays a vital part in determining the environmental fitness of strains (Hiyoshi et al., 2010; Matz et al., 2011). The T3SS2, tdh and trh are also known to be encoded on the pathogenicity island (Vp-PAI), signifying that V. parahaemolyticus acquires virulence determines through horizontal gene transfer (Okada et al., 2009; Matz et al., 2011). It is believed that the progression and severity of infection in humans are effected by the V. parahaemolyticus T3SS toxins (Ono et al., 2006). The strains that possess this needle-like T3SSs have the advantage of being able to secrete bacterial protein effectors directly into the host cell membrane and cytoplasm without facing the extracellular environment (Cornelis, 2006). In addition, the T3SS2 is suggested to be associated with tdh- and/or trh-positive V. parahaemolyticus strains (Raghunath, 2015). There are two distinct lineages of T3SS2 that have been described and associations were demonstrated of tdh with T3SSα and trh with T3SSβ (Park et al., 2004; Noriea et al., 2010). This could suggest that V. parahaemolyticus strains with the tdh and/or trh genes and T3SSs system have better ability to overcome host defenses in humans, conferring virulence that facilitates the development of infection.
Further analysis on the virulence properties has led to the discovery of type VI secretion systems encoded by T6SS1 and T6SS2 in V. parahaemolyticus. The T6SS1 is located on chromosome 1 where else, T6SS2 is located on chromosome 2 on V. parahaemolyticus RIMD 2210633 (Boyd et al., 2008; Izutsu et al., 2008). Salomon et al. (2013) proposed the role of T6SSs in V. parahaemolyticus. The T6SS1 is very active under warm marine-like conditions where else, T6SS2 is active under low salt conditions. It is also noted that surface sensing and quorum sensing differentially regulate both systems (Salomon et al., 2013). The T6SS2 and T3SS2 co-exist, suggesting the both systems may cooperate during an infection. T6SS2 takes the first step of infection as a role of adhesion where else T3SS2 exports effectors by inducing enterocytotoxicity (Park et al., 2004; Yu et al., 2012).
The Sensing of Bile
V. parahaemolyticus with the virulence factors described are able to launch an attack on and cause illness to humans. Even with its arsenal of virulence factors, this bacterium still has to first survive the harsh conditions in the human gastrointestinal tract. It is suggested that exposure to harsh environmental conditions enables bacteria to be able to withstand the effects of bile in humans. The various pH conditions, temperatures and growth harden the bacteria toward the antimicrobial effects of bile in human. This will eventually increase their tolerance toward bile and the bacteria is able to survive in the human gastrointestinal tract. In addition, the bile levels in the human intestine are not constant and particularly in the presence of food, the bacteria would be less affected by the bile (Begley et al., 2005). Therefore, with these added advantages, it could be suggested that V. parahaemolyticus can indeed survive in the human gastrointestinal tract and regulate virulence during infections.
Bile is a bactericidal agent that are made up from various proteins, ions, pigments, cholesterol and bile salts. In an infection, the bile salts is believe to provide protection against bacteria (Merritt and Donaldson, 2009). When there is high amount of bile acids in the small intestine, the bacterial growth is inhibited (Inagaki et al., 2006). Where else, the growth of bacteria increases in the small intestine when bile is secreted in low amount, such seen in liver cirrhosis patients (Slocum et al., 1992). However, enteric pathogens including Vibrio species have now developed a mechanism to resist the action off bile.
Gotoh et al. (2010) discovered the production of T3SS2 proteins are induced by bile under osmotic conditions similarly to the environments in gastrointestinal tract. They identified that the T3SS2 system is encoded in the pathogenicity island (Vp-PAI) and causes enterotoxicity effects to host cell. The VtrA and VtrB are the two transcriptional regulators that regulates encoded genes. Based on the study, V. parahaemolyticus initially recognizes its location in the human gastrointestinal tract by detecting bile acids. The transcription of Vp-PAI will be induced by bile acids via two main proteins, the VtrA and VtrB. The virulence genes then are regulated by the transduction of signals in the human intestinal tract (Gotoh et al., 2010). It was revealed that crude bile is a potent host derived inducer of tdh gene and T3SS2 under osmotic conditions corresponding to those in the intestinal tract.
Recently, Li et al. (2016) reported how V. parahaemolyticus has the ability to sense bile as an environmental cue to regulate its virulence mainly the T3SS2 during an infection. The study utilized bioinformatics tools to identify the proteins that are responsible for bile salt sensing and T3SS2 activation. Figure 1 illustrates how the bacteria-bile sensing mechanism happens in the human body. VtrA and VtrC are two gene encoded proteins that is identified to be responsible for bile salt sensing. These two genes interact to form a protein complex on the surface of the membrane that surrounds the bacterial cell. The two proteins then create a barrel like structure that binds to bile salts and triggers the cell to produce toxins. Upon binding of bile salts to the hydrophobic chamber in the VtrA/VtrC complex, the cytoplasmic DNA binding domain of VtrA is activated which in turn induces VtrB to activate the T3SS2 virulence system. The VtrA/VtrC complex is described to be highly conserved in a group of diverse Vibrionaceae family (Li et al., 2016). Additionally, the study also found a family of monomeric lipid binding calycin domain proteins that has expanded to include an obligate heterodimer which binds to bile salts and can be utilized to transmit a signal. This increases the ability of V. parahaemolyticus to sense bile salt as an environmental cue to regulate virulence.
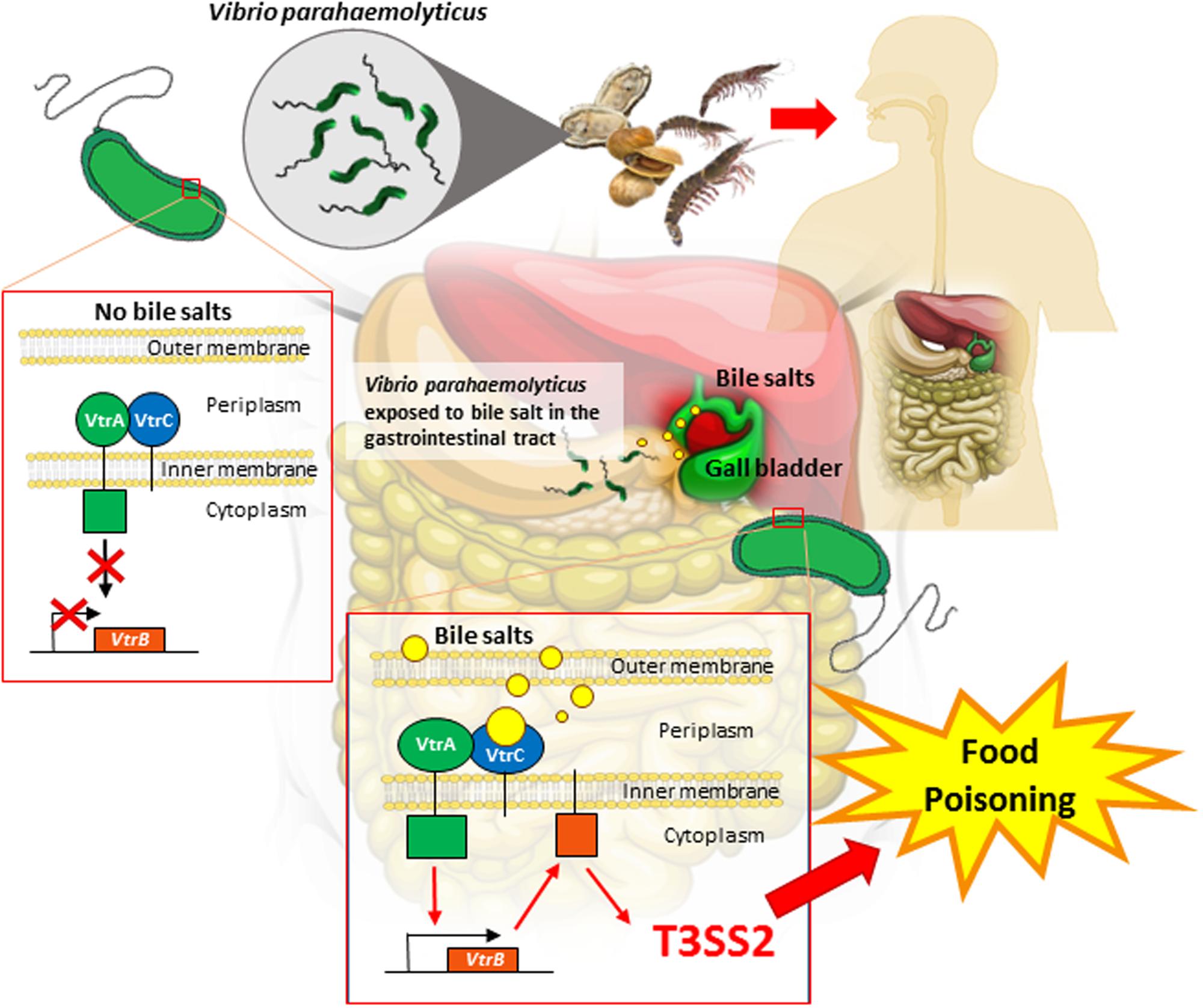
FIGURE 1. Illustration on how Vibrio parahaemolyticus sense bile salts in the human gastrointestinal tract. Human ingest V. parahaemolyticus from contaminated seafood such as clams, shrimps and oysters. Upon ingestion, V. parahaemolyticus is exposed to bile salts in the gastrointestinal tract. VtrA and VtrC are two gene encoded proteins that is identified to be responsible for bile salt sensing. VtrA and VtrC will interact and form a protein complex on the surface of the membrane of the bacterial cell. This complex structure then binds to bile salts and triggers the cell to produce toxins. Upon binding of bile salts to the VtrA/VtrC complex, the cytoplasmic DNA binding domain of VtrA is activated which in turn induces VtrB to activate, resulting in the T3SS2 expression. T3SS2 virulence is secreted thus causing illness to human.
It is well understood that enteric bacteria including V. parahaemolyticus has the ability to sense bile which helps them identify their immediate environments and virulence factors can be expressed. V. parahaemolyticus releases toxins and type III secretion systems (T3SS2) in order to trigger virulence during an infection. This mode of mechanism ensures the survival of pathogenic V. parahaemolyticus in the environments and increase in the bacterial infections. However, this mechanism will cause more harm to we humans in future. Our own body defense fails to protect us against bacterial infections and on the other hand helps bacteria to release virulence. This situation will be worsened by the emergence of antimicrobial resistant strains in the environment which has become a major therapeutic challenge. As the effectiveness of treating bacterial infections declines, interest has been renewed toward using bacteriophages as a non-antibiotic approach to control the spreading of evolutionary V. parahaemolyticus strains worldwide (Wittebole et al., 2014; Letchumanan et al., 2016). Bacteriophage belonging to the Siphoviridae family is suitable in controlling Vibrio species (Letchumanan et al., 2016). This bacteriophage is highly specific to the bacterial host cell, do not affect or alter the gut microbiota (Hagens and Offerhaus, 2008), and safe to be consumed by humans. The phages are able to perform as a bio-control agent to control and inhibit virulence of pathogenic Vibrio species from clinical and environmental samples (Jassim and Limoges, 2014). In addition, the application of bacteriophage in the aquaculture industry can reduce the dependency of antibiotics and control the spreading of antimicrobial resistant bacteria in the environment (Letchumanan et al., 2016). The listed advantages make bacteriophage therapy a promising tool to control bacterial infections.
Conclusion and Future Perspective
In summary, bile salts in human not only aid during digestion of food but possess antimicrobial activities as they have the ability to inhibit the survival of bacteria in the human gastrointestinal tract. However, certain conditions enable V. parahaemolyticus to develop resistance toward bile and eventually use bile as an environmental cue to regulate virulence. In order to treat infections, it is important to understand how V. parahaemolyticus senses bile salts and how this relates to their ability to regulate their virulence in the host during an infection. Given that there have been increasing numbers of multidrug resistant Vibrio strains from both clinical and environmental studies worldwide, drugs targeting suppression of bacterial virulence mechanisms should be designed instead of focusing on killing or inhibiting the growth of bacteria. Seen in this light, researchers will be able to design new drugs that may prevent the production of bacterial toxins and alleviate food poisoning symptoms. Future studies could focus on how other disease causing bacteria sense environmental cues to produce virulence during an infection.
Author Contributions
VL performed the literature review and manuscript writing. KG-C, TK, SB, N-SAM, B-HG, and L-HL provided vital guidance and insight to the writing. The project was conceptualized by L-HL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by PVC Award Grant (Project No. PVC-ECR-2016), External Industry Grant (Biotek Abadi–Vote No. GBA-808813), MOSTI eScience funds (Project No. 06-02-10-SF0300) awarded to L-HL, and University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027 and No. A000001-50001) awarded to K-GC, and a grant from “Research Center of the Female Scientific and Medical Colleges,” Deanship of Scientific Research, King Saud University awarded to SB.
References
Alam, M. J., Tomochika, K. I., Miyoshi, S. I., and Shinoda, S. (2002). Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208, 83–87. doi: 10.1111/j.1574-6968.2002.tb11064.x
Begley, M., Gahan, C. G., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Bhuiyan, N. A., Ansaruzzaman, M., Kamruzzaman, M., Alam, K., Chowdhury, N. R., Nishibuchi, M., et al. (2002). Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40, 284–286. doi: 10.1128/JCM.40.1.284-286.2002
Boyd, F. E., Cohen, L. V. A., Naughton, M. L., Ussery, W. D., Binnewies, T. T., Stine, C. O., et al. (2008). Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110. doi: 10.1186/1471-2180-8-110
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Burdette, D. L., Yarbrough, M. L., and Orth, K. (2009). Not without cause: Vibrio parahaemolyticus induces acute autophagy and cell death. Autophagy 5, 100–102. doi: 10.4161/auto.5.1.7264
Caburlotto, G., Ghidini, V., Gennari, M., Tafi, M. C., and Lleo, M. M. (2008). Isolation of a Vibrio parahaemolyticus pandemic strain from a marine water sample obtained in the northern Adriatic. Euro Surveill. 13, 1–2.
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Cornelis, G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. doi: 10.1038/nrmicro1526
DePaola, A., Nordstrom, J. L., Bowers, J. C., Wells, J. G., and Cook, D. W. (2003). Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 69, 1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003
Edwards, A. D., and Slater, N. K. (2009). Protection of live bacteria from bile acid toxicity using bile acid adsorbing resins. Vaccine 27, 3897–3903. doi: 10.1016/j.vaccine.2009.04.006
Gotoh, K., Kodama, T., Hiyoshi, H., Izutsu, K., Park, K. S., Dryselius, R., et al. (2010). Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants.PLoS ONE 5:13365. doi: 10.1371/journal.pone.0013365
Hagens, S., and Offerhaus, M. L. (2008). Bacteriophages – new weapons for food safety. Food Technol. 62, 46–54.
Hiyoshi, H., Kodama, T., Iida, T., and Honda, T. (2010). Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 78, 1772–1780. doi: 10.1128/IAI.01051-09
Honda, T., and Iida, T. (1993). The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct hemoloysin and related hemolysin. Rev. Med. Microbiol. 4, 106–113. doi: 10.1097/00013542-199304000-00006
Honda, T., Ni, Y. X., and Miwatani, T. (1988). Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56, 961–965.
Hung, D. T., and Mekalanos, J. J. (2005). Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. U.S.A. 102, 3028–3033. doi: 10.1073/pnas.0409559102
Inagaki, T., Moschetta, A., Lee, Y.-K., Peng, L., Zhao, G., Downes, M., et al. (2006). Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 3920–3925. doi: 10.1073/pnas.0509592103
Izore, T., Job, V., and Dessen, A. (2011). Biogenesis, regulation, and targeting of the type III secretion system. Structure 13, 603–612. doi: 10.1016/j.str.2011.03.015
Izutsu, K., Kurokawa, K., Tashiro, K., Kuhara, S., Hayashi, T., Honda, T., et al. (2008). Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilo base pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect. Immun. 76, 1016–1023. doi: 10.1128/IAI.01535-07
Jassim, S. A. A., and Limoges, R. G. (2014). Natural solution to antibiotic resistance: bacteriophages “The Living Drugs”. World J. Microbiol. Biotechnol. 30,2153–2170. doi: 10.1007/s11274-014-1655-7
Jones, J. L., Ludeke, C. H. M., Bowers, J. C., Garrett, N., Fischer, M., Parsons, M. B., et al. (2012). Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 50, 2343–2352. doi: 10.1128/JCM.00196-12
Krukonis, E. S., and DiRita, V. J. (2003). From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6, 186–190. doi: 10.1016/S1369-5274(03)00032-8
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Chan, K.-G., Pusparajah, P., Saokaew, S., Duangjai, A., Goh, B.-H., et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. doi: 10.3389/fmicb.2016.01114
Li, P., Rivera-Cancel, G., Kinch, L. N., Salomon, D., Tomchick, D. R., Grishin, N. V., et al. (2016). Bile salt receptor complex activates a pathogenic type III secretion system. Elife 5:15718. doi: 10.7554/elife.15718
Liu, M., and Chen, S. (2015). A novel adhesive factor contributing to the virulence of Vibrio parahaemolyticus. Sci. Rep. 5:14449. doi: 10.1038/srep14449
Lynch, T., Livingstone, S., Buenaventura, E., Lutter, E., Fedwick, J., Buret, A. G., et al. (2005). Vibrio parahaemolyticus disruption of epithelial cell tight junctions occurs independently of toxin production. Infect. Immun. 73, 1275–1283. doi: 10.1128/IAI.73.3.1275-1283.2005
Matsuda, S., Kodama, T., Okada, N., Okayama, K., Honda, T., and Iida, T. (2010). Association of Vibrio parahaemolyticus thermostable direct hemolysin with lipid rafts is essential for cytotoxicity but not hemolytic activity. Infect. Immun. 78, 603–610. doi: 10.1128/IAI.00946-09
Matz, C., Nouri, B., McCarter, L., and Martinez-Urtaza, J. (2011). Acquired type III secretion system determines environmental fitness of epidemic Vibrio parahaemolyticus in the interaction with bacterivorous protists.PLoS ONE 6:e20275. doi: 10.1371/journal.pone.0020275
Merritt, M. E., and Donaldson, J. R. (2009). Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microb. 58, 1533–1541. doi: 10.1099/jmm.0.014092-0
Nishibuchi, M., Fasano, A., Russell, R. G., and Kaper, J. B. (1992). Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60, 3539–3545.
Noriea, N. F. III, Johnson, C. N., Griffitt, K. J., and Grimes, D. J. (2010). Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J. Appl. Microbiol. 109, 953–962. doi: 10.1111/j.1365-2672.2010.04722.x
O’Boyle, N., and Boyd, A. (2014). Manipulation of intestinal epithelial cell function by the cell contact-dependent type III secretion systems of Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 3:114. doi: 10.3389/fcimb.2013.00114
Okada, N., Iida, T., Park, K. S., Goto, N., Yasunaga, T., Hiyoshi, H., et al. (2009). Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect. Immun. 77, 904–913. doi: 10.1128/IAI.01184-08
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system. 1. Infect. Immun. 74, 1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006
Ottaviani, D., Leoni, F., Rocchegiani, E., Mioni, R., Costa, A., Virgilio, S., et al. (2013). An extensive investigation into the prevalence and the genetic and serological diversity of toxigenic Vibrio parahaemolyticus in Italian marine coastal waters. Environ. Microbiol. 15, 1377–1386. doi: 10.1111/j.1462-2920.2012.02839.x
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T., et al. (2004). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004
Pazhani, G. P., Bhowmik, S. K., Ghosh, S., Guin, S., Dutta, S., Rajendran, K., et al. (2014). Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 8:2815. doi: 10.1371/journal.pntd.0002815
Pope, L. M., Reed, K. E., and Payne, S. M. (1995). Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63, 3642–3648.
Prouty, A. M., Brodsky, I. E., Manos, J., Belas, R., Falkow, S., and Gunn, J. S. (2004). Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41, 177–185. doi: 10.1016/j.femsim.2004.03.002
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 8:61086. doi: 10.1371/journal.pone.0061086
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.091101
Slocum, M. M., Sittig, K. M., Specian, R. D., and Deitch, E. A. (1992). Absence of intestinal bile promotes bacterial translocation. Am. Surg. 58, 305–310.
Takahashi, A., Kenjyo, N., Imura, K., Myonsun, Y., and Honda, T. (2000). Cl-secretion in colonic epithelial cells induced by the Vibrio parahaemolyticus hemolytic toxin related to thermostable direct hemolysin. Infect. Immun. 68, 5435–5438. doi: 10.1128/IAI.68.9.5435-5438.2000
West, C. K. G., Klein, S. L., and Lovell, C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a Pristine Estuary. Appl. Environ. Microbiol. 79, 2247–2252. doi: 10.1128/AEM.03792-12
Wittebole, X., Rock, S. D., and Opal, S. M. (2014). A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5, 226–235. doi: 10.4161/viru.25991
Yu, Y., Yang, H., Li, J., Zhang, P., Wu, B., Zhu, B., et al. (2012). Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 194, 827–835. doi: 10.1007/s00203-012-0816-z
Zhang, H. Z., and Austin, B. (2005). Haemolysins in Vibrio species. J. Appl. Microbiol. 98, 1011–1019. doi: 10.1111/j.1365-2672.2005.02583.x
Keywords: bacteria, human gastrointestinal tract, bile, Vibrio parahaemolyticus, type III secretion system 2
Citation: Letchumanan V, Chan K-G, Khan TM, Bukhari SI, Ab Mutalib N-S, Goh B-H and Lee L-H (2017) Bile Sensing: The Activation of Vibrio parahaemolyticus Virulence. Front. Microbiol. 8:728. doi: 10.3389/fmicb.2017.00728
Received: 19 January 2017; Accepted: 07 April 2017;
Published: 21 April 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Adrian Canizalez-Roman, Autonomous University of Sinaloa, MexicoShihua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2017 Letchumanan, Chan, Khan, Bukhari, Ab Mutalib, Goh and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Learn-Han Lee, bGVlLmxlYXJuLmhhbkBtb25hc2guZWR1; bGVlbGVhcm5oYW5AeWFob28uY29t Bey-Hing Goh, Z29oLmJleS5oaW5nQG1vbmFzaC5lZHU=
 Vengadesh Letchumanan
Vengadesh Letchumanan Kok-Gan Chan
Kok-Gan Chan Tahir M. Khan
Tahir M. Khan Sarah I. Bukhari4
Sarah I. Bukhari4 Nurul-Syakima Ab Mutalib
Nurul-Syakima Ab Mutalib Bey-Hing Goh
Bey-Hing Goh Learn-Han Lee
Learn-Han Lee