
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 19 April 2017
Sec. Aquatic Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00637
This article is part of the Research Topic Molecular Ecology and Genetic Diversity of the Roseobacter Clade View all 11 articles
The elucidation of the pathways for dimethylsulfoniopropionate (DMSP) synthesis and metabolism and the ecological impact of DMSP have been studied for nearly 70 years. Much of this interest stems from the fact that DMSP metabolism produces the climatically active gas dimethyl sulfide (DMS), the primary natural source of sulfur to the atmosphere. DMSP plays many important roles for marine life, including use as an osmolyte, antioxidant, predator deterrent, and cryoprotectant for phytoplankton and as a reduced carbon and sulfur source for marine bacteria. DMSP is hypothesized to have become abundant in oceans approximately 250 million years ago with the diversification of the strong DMSP producers, the dinoflagellates. This event coincides with the first genome expansion of the Roseobacter clade, known DMSP degraders. Structural and mechanistic studies of the enzymes of the bacterial DMSP demethylation and cleavage pathways suggest that exposure to DMSP led to the recruitment of enzymes from preexisting metabolic pathways. In some cases, such as DmdA, DmdD, and DddP, these enzymes appear to have evolved to become more specific for DMSP metabolism. By contrast, many of the other enzymes, DmdB, DmdC, and the acrylate utilization hydratase AcuH, have maintained broad functionality and substrate specificities, allowing them to carry out a range of reactions within the cell. This review will cover the experimental evidence supporting the hypothesis that, as DMSP became more readily available in the marine environment, marine bacteria adapted enzymes already encoded in their genomes to utilize this new compound.
Dimethylsulfoniopropionate (DMSP) was first identified in 1948 and has since been found to be not only abundant in marine surface waters but also a valuable resource for many marine organisms and an integral part of the global sulfur cycle (Challenger and Simpson, 1948; van Duyl et al., 1998; Stefels et al., 2007). DMSP is the precursor of the climate-active gas dimethyl sulfide (DMS), which upon release into the atmosphere aids in the formation of cloud condensation nuclei (Lovelock et al., 1972; Hatakeyama et al., 1982). Additionally, DMS is the largest natural source of sulfur to the atmosphere, comparable in magnitude to the sulfur dioxide formed during the burning of coal. As DMS oxidation products display a longer residence time in the atmosphere than anthropogenic sulfur dioxide, their contribution to the global sulfur burden is also greater (Lovelock et al., 1972; Chin and Jacob, 1996).
From an organismal viewpoint, DMSP is equally important. The ability to produce and metabolize DMSP is concentrated into specific classes of life. The main producers of DMSP are phytoplankton, mostly the classes Dinophyceae (dinoflagellates) and Prymnesiophycaea (coccolithophores) (Keller, 1989). DMSP production has also been noted in diatoms (Lyon et al., 2011; Kettles et al., 2014), the green algae Ulva intestinalis (Gage et al., 1997), corals (Raina et al., 2013), and certain higher plants like sugarcane (Paquet et al., 1994), and the coastal angiosperms Spartina alterniflora (Kocsis et al., 1998) and Wollastonia biflora (Hanson et al., 1994). Recently, DMSP biosynthesis was detected in several marine Alphaproteobacteria (Curson et al., 2017). The basis of the need for DMSP is not entirely understood. Several physiological functions for DMSP in phytoplankton and green algae have been demonstrated, including roles as an osmolyte, antioxidant, predator deterrent, and cryoprotectant (Kirst et al., 1990; Karsten et al., 1996; Wolfe and Steinke, 1997; Sunda et al., 2002). At present, each of the proposed pathways for DMSP biosynthesis begins with methionine, although subsequent steps vary (Figure 1). The pathways proposed for phytoplankton, algae, corals, and perhaps the DMSP-producing Alphaproteobacteria share similar reactions and intermediates which differ distinctly from those predicted in the coastal angiosperms (Hanson et al., 1994; Gage et al., 1997; Kocsis et al., 1998; Lyon et al., 2011; Curson et al., 2017). These variations indicate that the ability to synthesize DMSP has evolved at least twice (Stefels, 2000).
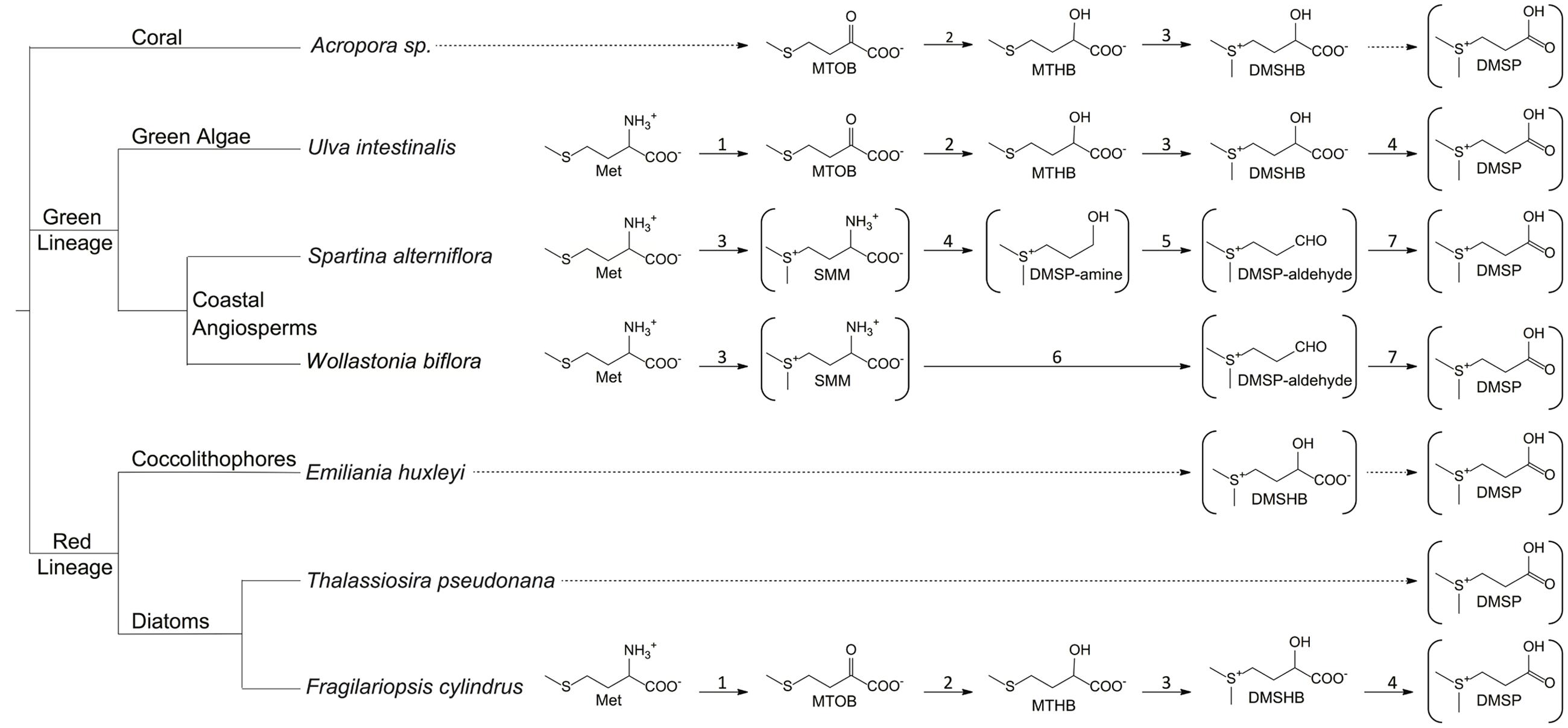
FIGURE 1. Proposed DMSP biosynthetic pathways. Structures in brackets have been detected and verified. Complete arrows signify reactions that are identified or predicted based on the observed intermediates. Dotted arrows signify unknown reactions. 1, aminotransferase; 2, NADPH-reductase; 3, methyltransferase; 4, decarboxylase; 5, oxidase; 6, decarboxylase/transaminase; 7, dehydrogenase. MTOB, 4-methylthio-2-oxobutyrate; MTHB, 4-methylthio-2-hydroxybutyrate; DMSHB, 4-dimethylsulfonio-2-hydroxybutyrate; SMM, S-methyl-L-methionine. (Hanson et al., 1994; James et al., 1995; Kocsis et al., 1998; Summers et al., 1998; Kocsis and Hanson, 2000; Lyon et al., 2011; Raina et al., 2013).
Bacteria may metabolize DMSP via two pathways, the cleavage or the demethylation pathway (Figure 2). The cleavage pathway results in the formation of DMS, while the demethylation pathway produces methanethiol (MeSH). The DMSP demethylation and cleavage pathway enzymes are hypothesized to be adapted versions of enzymes that were already contained within bacterial genomes and developed in response to the availability of this substrate (Reisch et al., 2011a,b). In this review, we investigate the likely evolutionary path that led to the development of DMSP biosynthesis and subsequently the specialized DMSP catabolic pathways. The members of the Alphaproteobacteria, specifically members of the Roseobacter clade, appear to be uniquely adapted to utilize this valuable source of reduced carbon and sulfur. Bacteria within the Roseobacter and SAR11 clades possess enzymes that specifically and efficiently catalyze reactions of the demethylation pathway (Reisch et al., 2008, 2011b; Curson et al., 2011b; Tan et al., 2013; Bullock et al., 2014; Johnston et al., 2016; Sun et al., 2016). Bacteria are also responsible for the majority of DMSP catabolism via the cleavage pathway (Figure 2). There is additional evidence suggesting the use of DMSP as an osmolyte and antioxidant in marine bacteria (Kiene et al., 2000; Simo et al., 2002; Lesser, 2006; Reisch et al., 2011b; Salgado et al., 2014). Many microorganisms encode enzymes that share a great deal of similarity to the demethylation pathway enzymes (Figure 3), demonstrating their adaptability and plasticity. The many roles of DMSP may have helped to drive the adaptation of existing enzymes for DMSP metabolism.
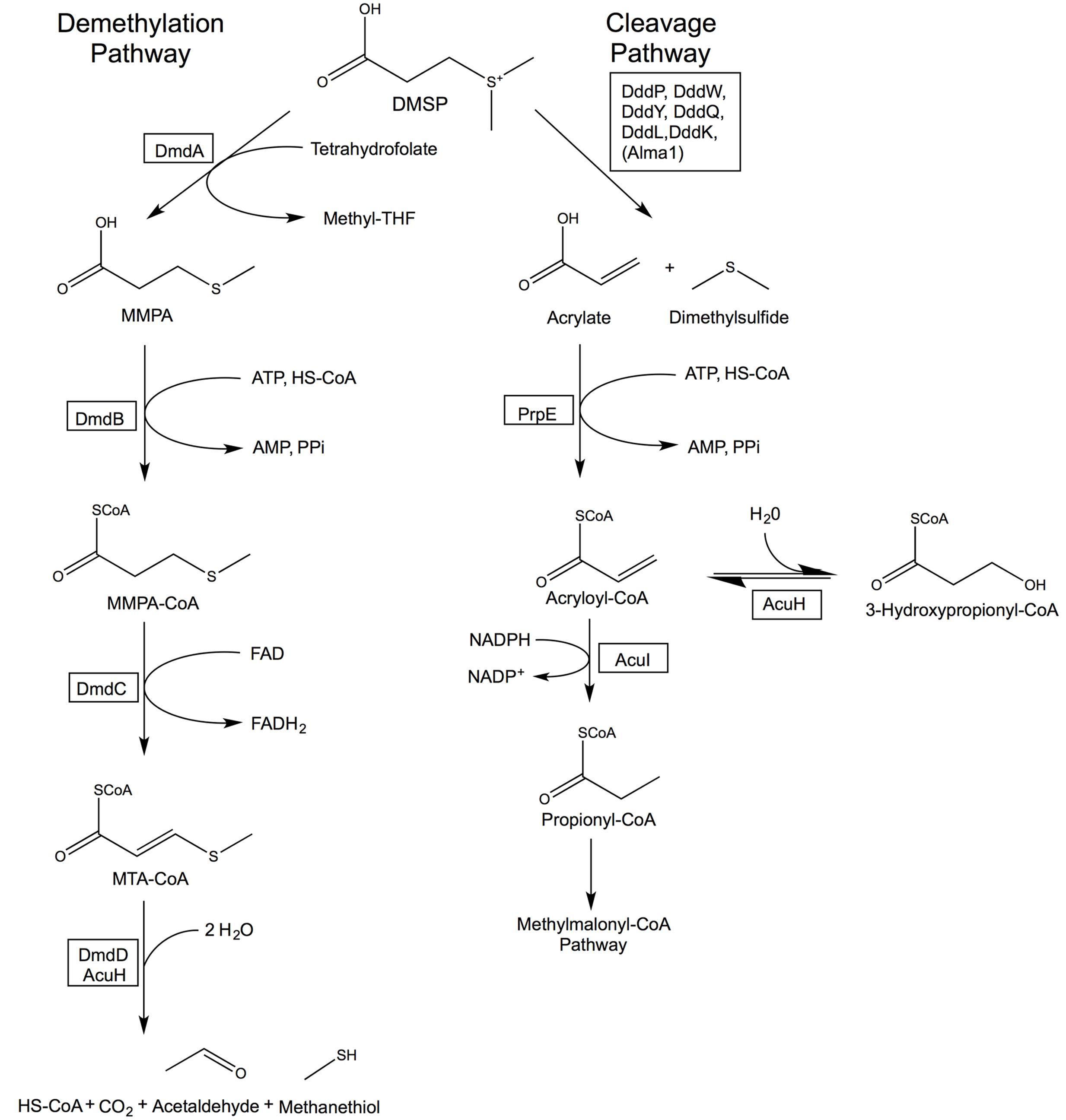
FIGURE 2. Bacterial DMSP demethylation and cleavage pathways. The DMSP demethylation pathway is catalyzed by the DMSP demethylase (DmdA), MMPA-CoA ligase (DmdB), MMPA-CoA dehydrogenase (DmdC), and either the MTA-CoA hydratase (DmdD) or acrylate utilization hydratase (AcuH). The cleavage pathway is catalyzed by a DMSP lyase (DddP, DddW, DddY, DddQ, DddL, DddK, or the algal Alma1), an acrylate-CoA ligase (PrpE), and an acryloyl-CoA reductase (AcuI). AcuH catalyzes a side reaction forming 3-hydroxypropionyl-CoA in the cleavage pathway. Revised from Reisch et al. (2011b, 2013).
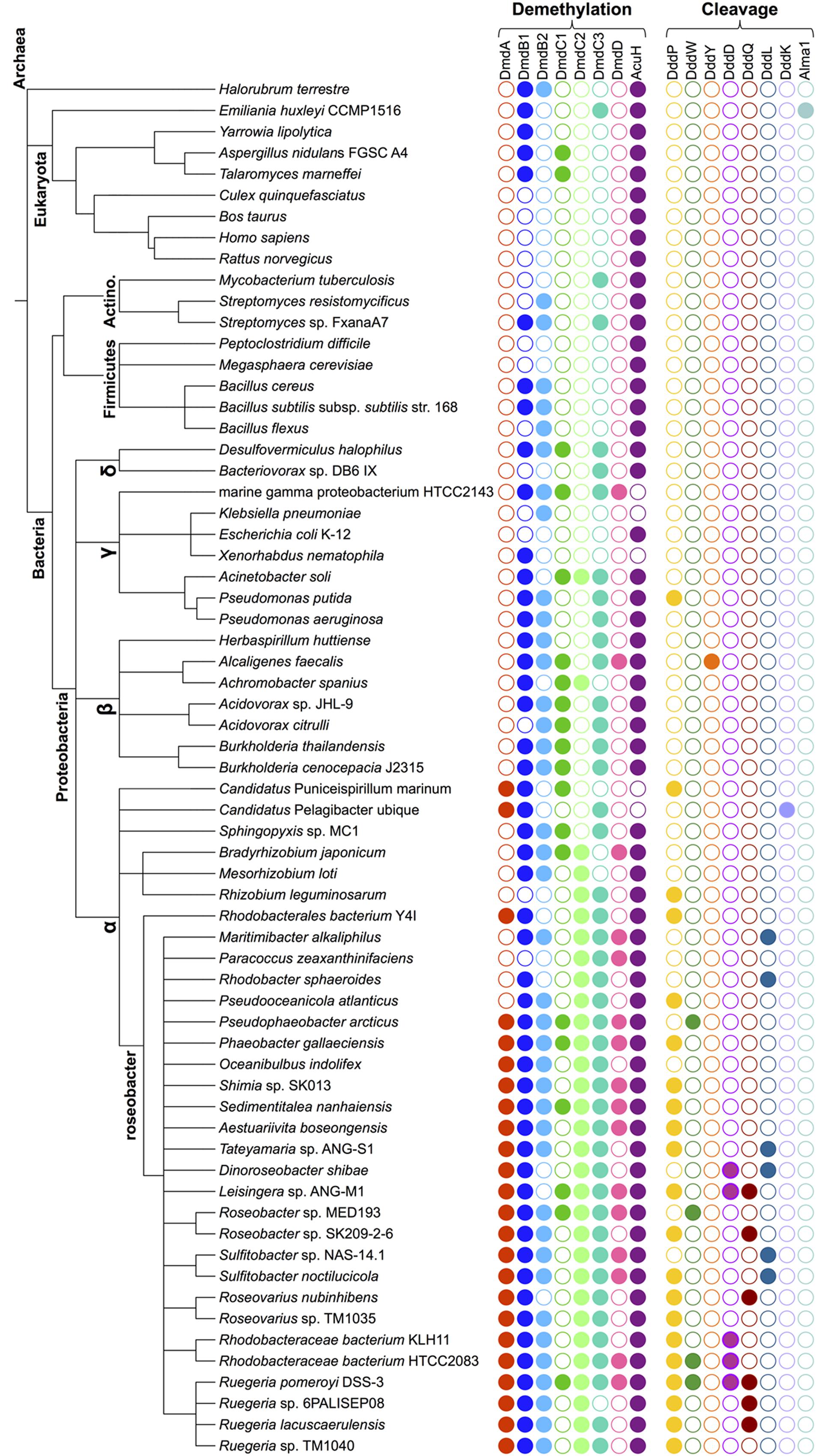
FIGURE 3. Phylogenetic species tree representing the diversity of organisms that possess enzymes from the DMSP demethylation pathway and the DMSP lyases. The relatedness of representative lineages is indicated schematically on the left. Colored filled circles represent the presence of the indicated protein-encoding gene. Protein designations and query sequences are as follows: From R. pomeroyi, DmdA (SPO1913); DmdB1 (SPO0677); DmdB2 (SPO2045); DmdC1 (SPO3805); DmdC2 (SPO0298); DmdC3 (SPO2915); DmdD (SPO3804); and AcuH (SL1157_0807) from R. lacuscaerulensis. DMSP lyase protein designations and query sequences are R. pomeroyi DddP (SPO2299), DddW (SPO0453), DddQ (SPO1596), DddD (SPO1703); A. faecalis DddY (ADT64689.1); R. sphaeroides DddL (RSP1433), P. ubique DddK (SAR11_0394); E. huxleyi Alma1 (XP_005784450). The e value cut off used in all cases was < e 10-70 with the exception of DddW (cut off of < e 10-40) and DddQ (cut off of < e 10-30). See Figure 2 for the names of the enzymes.
The first photosynthetic eukaryotes developed as the result of the acquisition of a cyanobacterium endosymbiont by a eukaryotic host, creating a membrane bound plastid (Bhattacharya and Medlin, 1998; Palmer, 2003; Yoon et al., 2004). Further diversification led to the formation of three clades from this original photosynthetic eukaryote, the green algae (green plastid lineage), the red algae (red plastid lineage), and the microbial algae glaucophytes (Delwiche, 1999). These lineages are distinguished by the chlorophyll present in their plastids. All the plastids contain chlorophyll a, but the green plastids also contain chlorophyll b, and the red plastids contain phycobilin (Keeling, 2010). Members of the Charophyta branch of the green plastid lineage colonized the land approximately 430 million year ago (mya). The Chlorophyta branch evolved into the green algae species seen today, including the Euglenoids and Chlorarachniophytes (Sanderson, 2003; Lewis and McCourt, 2004; McCourt et al., 2004). Meanwhile, today’s marine phytoplankton are largely descended from the red plastid lineage. The red plastid lineage phytoplankton, including coccolithophores, diatoms, and most dinoflagellates, first began to increase in abundance after the end-Permian extinction about 250 mya (Falkowski et al., 2004a,b).
The coccolithophores and dinoflagellates both began appearing in the fossil record about 250 mya in the Mesozoic period, while diatoms first appeared during the Early Cretaceous. All three groups saw extensive subsequent diversification in the Mesozoic period (250–65 mya) (Harwood and Nikolaev, 1995; Moldowan et al., 1996; Stover et al., 1996; Moldowan and Talyzina, 1998; Moldowan and Jacobson, 2000; Bown et al., 2004). The red lineage first began to proliferate in the benthic coastal regions, which were the first consistently oxic marine habitats. The breakup of Pangea increased sea levels and the total length of coastal area available for phytoplankton to colonize. This event also allowed nutrients that had been locked in the interior portions of continents to reach coastal waters (Vail et al., 1977; Haq et al., 1987). Changes in ocean redox chemistry from more reducing conditions that favored the green plastid lineage prior to the end-Permian extinction to the higher oxidation states of the Mesozoic ocean further contributed to the success of the red plastid lineage (Whitfield, 2001). Quigg et al. (2003) present evidence for the role of trace element availability in the proliferation of the red plastid lineage based on differences in the trace element composition between members of the red and green plastid lineages (Quigg et al., 2003). Members of the green plastid lineage have much higher requirements for iron, zinc, and copper while members of the red plastid lineage have high requirements for manganese, cobalt, and cadmium. It has been predicted that these differences in trace element requirements reflect differences in green vs. red plastid biochemistry (Quigg et al., 2003; Falkowski et al., 2004a).
The dominance of the red plastid lineage is such that all but one of the eight major taxa of eukaryotic phytoplankton in the present day oceans contains the red plastid (Falkowski et al., 2004a). The diversity of the red plastid lineage also greatly expanded as a result of secondary and tertiary endosymbiotic events, which are evident from the presence of multiple membranes surrounding some plastids of modern day phytoplankton. These events involved the engulfment of an algal cell by another eukaryote via endocytosis (Delwiche, 1999; Archibald and Keeling, 2002; Palmer, 2003; Keeling, 2010). The majority of the phytoplankton present today are the result of secondary and sometimes tertiary endosymbiotic events (Archibald, 2009). Today’s phytoplankton play key roles in global nutrient cycles and particularly in the global sulfur cycle as producers of DMSP and DMS (Lovelock et al., 1972).
Marine phytoplankton and algae live in an environment that is continually changing based on shifts in ocean currents. Living in this dynamic environment requires that these organisms adapt continually to varying temperatures, light, and nutrient availability (Cortes et al., 2001; Wagner et al., 2005; Allen et al., 2006). These changes may have been even more extreme in the Paleozoic and Mesozoic oceans. Abiotic forces have been found to have a large impact on the population variability of Emiliania huxleyi and Florisphaera profunda (Cortes et al., 2001). Phytoplankton in general, however, adapt quickly and relatively readily to environmental changes due to their rapid cell division rates and large population sizes (Simo, 2001; Allen, 2005; Wagner et al., 2005; Allen et al., 2006).
One specific adaptation that may help phytoplankton deal with their ever changing environment is the ability to synthesize and utilize DMSP and DMS. DMSP makes up about 90% of the reduced sulfur found in algae, but much about the regulation of its biosynthesis and uptake is still not well understood (Gage et al., 1997). Nevertheless, many of the proposed roles for these compounds would be beneficial to phytoplankton trying to survive in an ever-changing environment. DMSP is proposed to have roles as an osmolyte (Kirst et al., 1990), an antioxidant (Sunda et al., 2002), and as a means of balancing excess cellular energy (Stefels, 2000; Allen, 2005). Additionally, polar diatoms and algae are thought to produce DMSP as a cryoprotectant (Karsten et al., 1996). This hypothesis is supported by the higher levels of DMSP in sea ice diatoms compared with those from more temperate climates (Lyon et al., 2011; Kettles et al., 2014). DMSP is also a predator/grazing deterrent owing to its cleavage to acrylate (Stefels, 2000). New studies of the coral genus Acropora have generated still more uses for DMSP. Reef building coral juveniles increase DMSP production when subject to thermal stress and may also use DMSP as a bacterial signaling molecule, attracting particular microbial communities that are necessary for coral health (Raina et al., 2013).
The role of DMSP and DMS as antioxidants could be particularly useful for phytoplankton as plastids are typically hyperoxic and produce reactive oxygen species (ROS) during oxygenic photosynthesis. Other stresses such as exposure to ultraviolet radiation (UVR) and thermal stress can further increase ROS production (Asada and Takahashi, 1987; Fridovich, 1998; Lesser, 2006). The production of ROS by plastids might explain why DMSP and DMS production are observed in both phytoplankton and land plants. There is also evidence to suggest that the final step of DMSP synthesis in the flowering plant W. biflora takes place in the plastid (chloroplast) (Trossat et al., 1996). DMSP, DMS, and acrylate are all able to quench HO∙ radicals, although acrylate and DMS are more efficient than DMSP. The resultant product of HO∙ quenching is dimethylsulfoxide (DMSO), which subsequently reacts with additional HO∙ radicals to form methane sulfinic acid and then methane sulfonic acid. In contrast to DMSP and acrylate, DMS is uncharged and can diffuse through biological membranes, acting as an antioxidant nearly anywhere in the cell (Sunda et al., 2002; Lesser, 2006; Husband et al., 2012).
Another impetus for the production of DMSP may be the need for an osmolyte that does not contain nitrogen. Nitrogen is often limiting in ocean surface waters, which may in turn limit the production of the nitrogen-containing osmolyte glycine betaine. Ito et al. (2011) observed that under conditions where sulfate limited growth of the marine algae Ulva pertusa, the sulfur from methionine was used primarily for the synthesis of S-adenosyl methionine and methionyl-tRNA, rather than for DMSP synthesis. However, when the salinity and abundance of sulfate increased; the sulfur from methionine was increasingly used for DMSP biosynthesis, and the intracellular DMSP levels increased (Ito et al., 2011).
One additional hypothesis for the origin of DMSP biosynthesis proposes that it developed as a means of dispelling excess energy, carbon and reducing equivalents when growth becomes unbalanced due to nutrient limitation (Stefels, 2000). Rapid changes in the ocean environment can require phytoplankton to have an equally rapid response to imbalances between photosynthesis and growth (Allen, 2005; Wagner et al., 2005; Allen et al., 2006). Since photon capture cannot be quickly stopped, production of nitrogen or phosphorous poor molecules when growth is limited by these nutrients is a means of consuming extra carbon, energy, and reducing equivalents that cannot be used for protein biosynthesis or cell division (Stefels, 2000; Simo, 2001; Allen, 2005). Further, the continued production of DMSP may also serve to regenerate and redistribute nitrogen for the production of new amino acids and to stimulate continued sulfate assimilation by keeping the cellular concentration of methionine and cysteine low (Gage et al., 1997; Stefels, 2000). Thus, DMSP may have originally been produced as a means of dissipating excess energy and carbon and was then adapted for other functions.
The main producers of DMSP are phytoplankton, mostly in the classes Dinophyceae (dinoflagellates) and the Prymnesiophycaea (which includes the coccolithophores). Certain members of the Chryosphyceae and Bacillariophyceae (diatoms) can also produce DMSP (Keller, 1989). DMSP likely first became abundant in ocean environments about 250 mya in conjunction with the increasing abundance of dinoflagellates and coccolithophores. Based on a comparison of literature reports for 95 DMSP-producing species (Table 1), it was determined that dinoflagellates produced the highest amounts of DMSP, with intracellular levels ranging from 0.00011 to 14.7 pmol/cell (Keller, 1989; Wiesemeier and Pohnert, 2007; Caruana, 2010; Caruana and Malin, 2014). In particular, Alexandrium minutum and Protoperidinium pellucidum produced 14.2 and 14.7 pmol DMSP/cell, respectively. Diatoms have intracellular DMSP levels ranging from 0.0006 to 0.257 pmol/cell, while haptophytes (coccolithophores) contained from 0.00037 to 0.148 pmol DMSP/cell (Keller, 1989; Caruana and Malin, 2014). DMSP production is less common among the higher plants, although it has been observed in Spartina species (Kocsis et al., 1998), certain sugarcanes (Paquet et al., 1994), and the flowering plant W. biflora (Hanson et al., 1994; James et al., 1995). DMSP production has also been observed in members of the coral genus Acropora in the absence of their algal endosymbiont Symbiodinium, also a known DMSP producer (Raina et al., 2013). Thus, while DMSP is widely distributed in a large number of phototrophs, only a few groups produce very high amounts, and it is likely that DMSP only became widely available as a nutrient in marine environments following the evolution of these groups.
Little is understood about the biosynthetic pathways for DMSP in marine phytoplankton and corals. The first complete DMSP biosynthetic pathways were described in the green algae U. intestinalis (Gage et al., 1997), the marine cordgrass S. alterniflora (Kocsis et al., 1998), and coastal plant W. biflora (Hanson et al., 1994; James et al., 1995) (Figure 1). Each pathway identified thus far begins with methionine and includes a deamination reaction, supporting the hypothesis that DMSP biosynthesis is used by these organisms to regenerate nitrogen from methionine. The DMSP biosynthetic pathways of S. alterniflora and W. biflora are more similar to each other than they are to the pathway in U. intestinalis, suggesting that the plant pathways evolved independently from those in marine algae, corals, and phytoplankton. If true, this would indicate that there was selective pressure for the evolution of DMSP biosynthetic pathways even in very different organisms.
The DMSP biosynthetic pathways of the major producers in the marine environment are still largely unknown, but they are likely similar to the pathway described in U. intestinalis. The U. intestinalis pathway begins with methionine and utilizes an aminotransferase, a NADPH-linked reductase, a methyltransferase, and an oxidative decarboxylase to produce DMSP (Gage et al., 1997; Summers et al., 1998). The commitment step is hypothesized to be the third step, the conversion of 4-methylthio-2-hydroxybutyrate (MTHB) to 4-dimethylsulfonio-2-hydroxybutyrate (DMSHB) by a methyltransferase (Figure 1). The key intermediate DMSHB has been identified in U. intestinalis, U. pertusa, E. huxleyi, Tetraselmis sp., and Melosira nummuliodes, indicating that this pathway is present in a range of phytoplankton (Gage et al., 1997; Stefels, 2000; Ito et al., 2011). Lyon et al. (2011) identified candidate proteins and genes for this four-step pathway in the sea-ice diatom Fragilariopsis cylindrus. Proteins from the same enzyme classes proposed in the U. intestinalis pathway were more abundant when F. cylindrus was exposed to conditions that increased DMSP production. However, the activities of these proteins still need to be verified (Lyon et al., 2011). Orthologs for the genes encoding a NADPH-reductase and an AdoMet-dependent methyltransferase have also been found in the corals Acropora millepora and Acropora digitifera and in the coral dinoflagellate symbiont Symbiodinium, all known DMSP producers. Based on the collective data, Raina et al. (2013) hypothesized that the enzymes of the DMSP biosynthetic pathway are conserved between diatoms, alveolates, green algae, and corals. Interestingly, a study of the diatom Thalassiosira pseudonana did not identify any of the same proteins proposed for the F. cylindrus biosynthetic pathway under conditions that increased intracellular DMSP levels, suggesting that it may contain an alternative pathway (Kettles et al., 2014).
A recent study has reported the biosynthesis of DMSP by marine bacteria. DMSP production was observed from Oceanicola batsensis HTCC2597, Pelagibaca bermudensis HTCC2601, Sediminimonas qiaohouensis DSM21189, Amorphus coralli DSM18348, Sagittula stellata E-37, Labrenzia aggregata LZB033, Labrenzia aggregata IAM12614, and Thalassobaculum salexigens DSM19539 (Curson et al., 2017). DMSP biosynthesis in marine bacteria proceeds in a similar manner to that observed in algae and phytoplankton, via the methionine transamination based pathway. A methyltransferase gene, dysB, was identified in marine Alphaproteobacteria and appears to be the key enzyme for DMSP biosynthesis in these microorganisms. When dysB was cloned into the non-DMSP producer Rhizobium leguminosarum, the ability to synthesize DMSP was conferred. Thus, the addition of a single gene, in certain cases, is sufficient to enable the production of DMSP. Further, dysB expression from L. aggregata LZB033 is up-regulated during increased salinity, nitrogen limitation, and at low temperatures, conditions already predicted to stimulate DMSP production in marine phytoplankton and algae. Selective pressures, like changes in salinity or nitrogen limitation, could result in the acquisition of dysB by marine bacteria to enable DMSP biosynthesis and gain a competitive advantage in their environment (Curson et al., 2017).
While the demethylation pathway appears to be unique to marine bacteria, several marine phytoplankton lyse DMSP into DMS. Multiple studies have reported significant DMSP lyase activity within phytoplankton blooms and among individual phytoplankton, including Phaeocystis sp., Heterocapsa triquetra, Scrippsiella trochoidea, and several Symbiodinium strains (Stefels et al., 1995; Niki et al., 1997, 2000; Yoch, 2002). To date, while several marine phytoplankton have been observed to produce DMS from DMSP, the genes responsible for this activity have not been identified in most cases. It has been known for many years that E. huxleyi cleaves DMSP into DMS and acrylate (Yoch, 2002), but only recently was the responsible gene, Alma1, identified (Alcolombri et al., 2015). Alma1 is a member of the aspartate racemase superfamily. Based on sequence similarity, Alma1 and its paralogs from E. huxleyi are present in a wide range of phytoplankton as well as certain bacteria, highlighting the diversity of this protein (Yost and Mitchelmore, 2009; Alcolombri et al., 2015). There are seven Alma1 paralogs within the E. huxleyi genome. Alma1 paralogs from E. huxleyi, Phaeocystis Antarctica, A. millepora (coral), and Symbiodinium sp. were synthesized and tested for activity toward DMSP. Of those tested, however, only one E. huxleyi paralog, Alma2, and a Symbiodinium paralog had DMSP lyase activity, indicating that there is still much to learn about the phytoplankton DMSP lyases (Alcolombri et al., 2015).
Marine bacteria have developed many uses for DMSP, from a source of reduced sulfur and carbon (Kiene et al., 1999, 2000), to use as an osmolyte (Sunda et al., 2002; Salgado et al., 2014), and potentially a cryoprotectant (Karsten et al., 1996). The details of the bacterial catabolism of DMSP have only recently come to light (Figure 2). The characterization of the enzymes involved in the DMSP demethylation pathway as well as the identification of several DMSP lyases from the DMSP cleavage pathway have provided new insights into the evolution of these enzymatic activities. Some of the enzymes of the demethylation pathway have likely roots in fatty acid ß-oxidation (Reisch et al., 2011a,b; Bullock et al., 2014). The DMSP lyases are widely distributed and varied in sequence, structure, and activity (Curson et al., 2011b; Johnston et al., 2016). Many of the enzymes involved in the microbial DMSP catabolic pathways are widespread, particularly among the Proteobacteria (Figure 3). Even those Roseobacters with reduced genomes, such as the lineages SAG-O19, DC5-80-3, and NAC11-7, have been found to encode dmdA and at least one DMSP lyase (Zhang et al., 2016). Presumably, the relatively modern evolution of phytoplankton producing high levels of DMSP provided the impetus for developing and maintaining these functions. To learn more about how the degradation pathways evolved, the structural and functional characteristics of the DMSP catabolic enzymes were examined to posit how they may have been adapted from existing enzymes.
The enzymatic cleavage of DMSP produces DMS and acrylate. To date, eight DMSP lyases have been identified (Table 2). The lyases were recently reviewed (Johnston et al., 2016). Except DddD which produces 3-hydroxypropionate, these enzymes all carry out the same reaction to form DMS and acrylate despite differing drastically in sequence and size (Todd et al., 2010; Curson et al., 2011b). Based upon a survey of lyase encoding genes in representative genomes of marine bacteria, dddP is the most widely distributed (Figure 3). However, dddD, dddW, dddQ, and dddL are also relatively common. In contrast, dddY, dddK and Alma1 are rare in marine bacteria. There are now several reports of DMSP lyase activity being induced by the presence of DMSP. In Ruegeria pomeroyi DSS-3 and Roseovarius nubinhibens, dddP and dddQ expression was induced when cells were pre-grown with DMSP as compared to cells not exposed to DMSP. Likewise, expression of dddY increased following growth of Alcaligenes faecalis with DMSP (Todd et al., 2007, 2009). Further, a field study in Monterey Bay, California, found that expression of dddP increased during mixed-community DMSP-producing phytoplankton blooms (Varaljay et al., 2015). Expression of R. pomeroyi dddW also increased after exposure to DMSP in growth medium (Todd et al., 2012b). These observations are consistent with a role of these enzymes in DMSP cleavage.
Evidence for the physiological relevance of the two best studied lyases, DddP and DddQ, has been mounting. The dddP and dddQ genes are the most abundant of the bacterial DMSP lyase genes in the marine metagenome as determined by the Global Ocean Sampling Expedition (GOS) (Rusch et al., 2007; Todd et al., 2011). The role of DddP and DddQ from R. pomeroyi DSS-3 and R. nubinhibens in DMSP cleavage has been clearly demonstrated. Studies using 14C or 13C labeled DMSP show that Escherichia coli extracts expressing dddP and dddQ are able to produce DMS and acrylate from DMSP (Kirkwood et al., 2010; Todd et al., 2011). Additionally, dddP and dddQ mutants in R. pomeroyi produce significantly less DMS when compared with wild-type cells, 50% less in the case of dddP and 97% less in the case of dddQ. A dddQ mutant from R. nubinhibens produced 20% less DMS from DMSP, while a dddP mutant produced only 10% of the wild-type levels (Todd et al., 2009, 2011; Kirkwood et al., 2010).
The structures of the DMSP lyases provide insights into their evolutionary roots. The crystal structures of DddP and DddQ from Ruegeria lacuscaerulensis and DddP from Roseobacter denitrificans have been solved (Li et al., 2013; Wang et al., 2015). Data gathered from the available structures suggests that subtle changes in the active sites of these lyases make sulfur containing substrates, like DMSP, the preferred substrates for these enzymes. The sequence and structure of DddP most closely resembles that of M24 peptidase. Typically, an M24 peptidase hydrolyzes C-N bonds. DddP, however, cleaves C-S bonds (Todd et al., 2009; Wang et al., 2015). Wang and coworkers expressed the recombinant R. lacuscaerulensis dddP in E. coli and found it displayed no measurable activity toward the M24 peptidase substrate valine-proline, but it did exhibit DMSP lyase activity, producing acrylate and DMS (Wang et al., 2015). DddP is a homodimeric protein in which one monomer has a metal center containing Fe, while the other monomer generally contains Fe, but may also contain Ni, Zn, or Cu instead (Hehemann et al., 2014; Wang et al., 2015). The explanation for the change in substrate preference and activity appears to be due to the change of the active ion from Co or Mn coordinated by five residues in the M24 peptidases to Fe coordinated by six residues in DddP. The two metal ions in DddP are coordinated with three aspartates, two glutamates, and a histidine residue, which are conserved in the known functional DddPs (Hehemann et al., 2014; Wang et al., 2015). The substitution of any of the active site residues for alanine in DddP results in the elimination of DMSP lyase activity, indicating that all six are necessary for activity (Kirkwood et al., 2010; Wang et al., 2015). Additionally, two conserved histidine residues in M24 peptidases that help to bind and stabilize substrates are exchanged for aspartate and phenylalanine in DddP (Hehemann et al., 2014; Wang et al., 2015). Wang et al. (2015) suggest that this change abolishes the peptidase activity of DddP and allows the active site aspartate to act as a nucleophilic base for DMSP cleavage (Wang et al., 2015). It is further proposed that DddP is a case of divergent evolution from the M24 peptidases as DddPs cluster in a separate clade in phylogenetic analyses. In support of this hypothesis, the M24 peptidase conserved C-domain has up to 31% sequence identity with the C-domain of the R. lacuscaerulensis DddP. The N-domain of DddP, by contrast is structurally different than the N-domains of M24 peptidases and allows for the formation of a compact dimer and a smaller catalytic cavity for DMSP binding (Wang et al., 2015). In conclusion, DddP appears to have acquired specific adaptations for DMSP lyase activity, supporting the assertion that this is its major role.
A structure for DddQ from R. lacuscaerulensis has recently been solved (Li et al., 2013). DddQ is one of the cupin motif containing DMSP lyases, along with DddW and DddL (Curson et al., 2011b; Johnston et al., 2016). DddQs have been identified in a number of Roseobacters, but they display substantial amino acid sequence variation, even when multiple copies are present in the same organism (Todd et al., 2011). Despite this variation, certain amino acids in the cupin motifs, two histidines and a glutamate in cupin motif 1 and a histidine in cupin motif 2 are conserved in DddQ, DddW, and DddL. In addition to these conserved amino acids, two tyrosines in motif 1 are highly conserved in all the cupin protein DMSP lyases but not among other cupin proteins. These conserved active site residues are predicted to play a role in DMSP cleavage as substitution at any of these residues decreased activity toward DMSP (Li et al., 2013).
The formation of DMS and acrylate from DMSP is proposed to be the result of a β-elimination reaction (Li et al., 2013; Wang et al., 2015). The DMSP lyases appear to have developed different catalytic mechanisms for carrying out the same reaction, indicting separate evolutionary paths to this activity. DddP is proposed to implement an ion shift. When DMSP enters the active site, a moveable Fe binds to the carboxyl group of DMSP, stabilizing the molecule in the active site, while two other conserved residues, tryptophan and tyrosine, bind to the sulfur in DMSP. This orientation allows for the abstraction of a proton by aspartate from the alpha carbon of the DMSP carboxyl group, cleavage of the C-S bond, and the subsequent formation of a double bond between the alpha and beta carbons of DMSP to produce acrylate (Hehemann et al., 2014; Wang et al., 2015). In DddQ, it has been proposed that binding of DMSP to the metal cofactor causes a conserved tyrosine residue to shift closer to the DMSP molecule. This shift allows the oxygen atom of one of the conserved tyrosine residues to interact with the alpha carbon of DMSP. The resultant conformational change enables the abstraction of a proton from the DMSP carboxyl group by the oxygen atom of tyrosine (Li et al., 2013). The algal DMSP lyase, Alma1, is proposed to function in a similar manner, abstracting a proton from the carbon adjacent to the carboxylate to cause β-elimination and the subsequent release of DMS and acrylate (Alcolombri et al., 2015). Further investigations into the structures and mechanisms of the other DMSP lyases, like the algal Alma1 or DddY, may yield still more variability in reaction mechanisms.
DddY from A. faecalis M3A was the first identified DMSP lyase (de Souza and Yoch, 1995). It is the only DMSP lyase that is a periplasmic protein and has no similarity to any other enzyme of known function. DddYs have been identified in A. faecalis M3A and Desulfovibrio acrylicus, as well as in several Shewanella species and Arcobacter nitrofigilis DSM7299. DMS production from DMSP was observed in Shewanella halifaxensis HAW-EB4, Shewanella putrefaciens CN-32, and A. nitrofigilis DSM7299 (Curson et al., 2011a). S. halifaxensis and S. putrefaciens are found in marine sediments and shale sandstone, respectively, while A. nitrofigilis can be found in sediment around Spartina roots. It is likely that dddY was spread via horizontal gene transfer (HGT) among these distantly related bacteria. In addition to dddY, A. faecalis also has acrylate utilization (acu) genes that resemble those used for DMSP and acrylate metabolism in other DMSP-utilizing bacteria (Curson et al., 2011a). More in depth studies of DddY have not been undertaken.
Despite convincing evidence for the physiological role of the DMSP lyases, the affinities for DMSP of the currently known lyases are lower than expected for a natural substrate, displaying Kms for DMSP in the millimolar range (Table 2) (Johnston et al., 2016). The Kms for the most widely distributed lyases, DddP and DddQ, are among the highest (Rusch et al., 2007; Todd et al., 2011). The lowest Km for DMSP observed thus far is for DddY. The DddYs from A. faecalis and D. acrylicus have Kms for DMSP of 1.4 and 0.4 mM, respectively (Table 2) (de Souza and Yoch, 1996; van der Maarel et al., 1996). Both of these organisms are found in coastal marine sediments and likely obtain DMSP from Spartina spp. (Curson et al., 2011a). High Km values for DMSP are also shared with the DMSP demethylases (see DmdA below), which is the first committed step of the demethylation pathway. Thus, the low affinities of the lyases may simply reflect the requirement for high intracellular concentrations of DMSP to initiate its metabolism. If DMSP serves as an osmolyte in bacterioplankton, cells should maintain high concentrations in the cell. For instance, during growth on DMSP, a concentration of 70 mM has been observed in R. pomeroyi. Under these conditions, low Kms for DMSP are not necessary for DMSP lyases to function effectively in vivo (Reisch et al., 2008). Concentrations of DMSP in ocean surface waters range from less than 1 nM in the open ocean to micromolar levels within phytoplankton blooms (van Duyl et al., 1998). Senescence and autolysis of DMSP producers like Spartina or phytoplankton can also produce microenvironments with high concentrations of DMSP (de Souza and Yoch, 1995). Provided a bacterium has the necessary transporters for the uptake of DMSP, intracellular concentrations of DMSP have the potential to reach to millimolar levels (Kiene, 1998; Kiene and Williams, 1998; Kiene et al., 1998; Todd et al., 2010).
In conclusion, the sequence and structural variability of the DMSP lyases that have been identified so far indicates that they likely evolved independently. For this to happen, the new activity must be readily acquired in evolution from multiple ancestral enzymes, Moreover, there must be strong selective pressures to maintain this function in very different groups of organisms. In addition, some bacteria contain multiple DMSP lyases, and it is possible that their physiological functions are somewhat different. This would allow cells to maintain lyases with the same catalytic activity but different regulatory or other functional properties.
The DMSP demethylation pathway consists of a series of reactions that convert DMSP into methanethiol (MeSH), HS-CoA, CO2, and acetaldehyde (Reisch et al., 2011a,b). While DMS production from phytoplankton has been observed, there is no indication that these organisms possess the demethylation pathway. Instead, the demethylation pathway is restricted to the Alphaproteobacteria (Figure 3). Based on the current evidence, it seems likely that the individual steps of the demethylation pathway may have evolved independently.
The initial step of the demethylation pathway is mediated by the DMSP demethylase DmdA (Figure 2). This step also commits DMSP to the demethylation pathway because demethylation precludes the formation of DMS (Howard et al., 2006; Reisch et al., 2008, 2011b). As with the DMSP lyases, the Kms for DMSP of the two characterized DmdAs from R. pomeroyi and Pelagibacter ubique are relatively high, 5.4 and 13.2 mM, respectively. The deletion of dmdA from R. pomeroyi, however, results in a mutant incapable of producing MeSH, indicating that this gene encodes the only protein in R. pomeroyi able to perform this reaction (Howard et al., 2006; Reisch et al., 2008). Additionally, field measurements indicate that dmdA expression is upregulated during blooms of DMSP-producing phytoplankton (Varaljay et al., 2015). DmdA in R. pomeroyi was initially annotated as a glycine cleavage T-protein (GcvT) (Reisch et al., 2011a). However, when analyzed phylogenetically, DmdA-like proteins share sequence identity ranging from 22 to 26% with GcvT, dimethylglycine oxidase and sarcosine oxidase, but form a separate clade from known GcvTs.
The crystal structure of DmdA from P. ubique provides further evidence supporting a common ancestry for DmdA and GcvT (Figure 4). Schuller et al. (2012) described the structure of DmdA, noting that while DmdA is structurally similar to GcvT, the low sequence similarity between the two indicated that the enzymes are evolutionarily distant (Schuller et al., 2012). Both proteins possess a very similar tri-domain structure (Figure 4) with the conserved residues between the proteins being mainly involved with tetrahydrofolate (THF) binding. Specifically, the residues that interact with the folate moiety and those involved in the ring stacking of THF are highly conserved (Lee et al., 2004; Reisch et al., 2008; Schuller et al., 2012). In contrast, DmdA possesses high substrate specificity for DMSP and closely related compounds, so the binding site for this substrate must differ from that of GcvT. Despite structural similarity, DmdA and GcvT are mechanistically distinct. DmdA produces 5-methyl-THF from DMSP as the result of a redox-neutral methyl transfer while GcvT coverts glycine to 5,10-methylene-THF (Howard et al., 2006; Reisch et al., 2008; Schuller et al., 2012). Small changes to the THF- binding fold in DmdA allow for hydrogen bond formation between amino acid residues in the fold and THF, enabling DmdA to carry out a redox-neutral methyl transfer to produce 5-methyl-THF. Overall, the mechanism of DmdA catalysis appears to be more similar to the S-adenosylmethionine SAM-dependent N-methyltransferases than the more closely related GcvTs (Schuller et al., 2012).
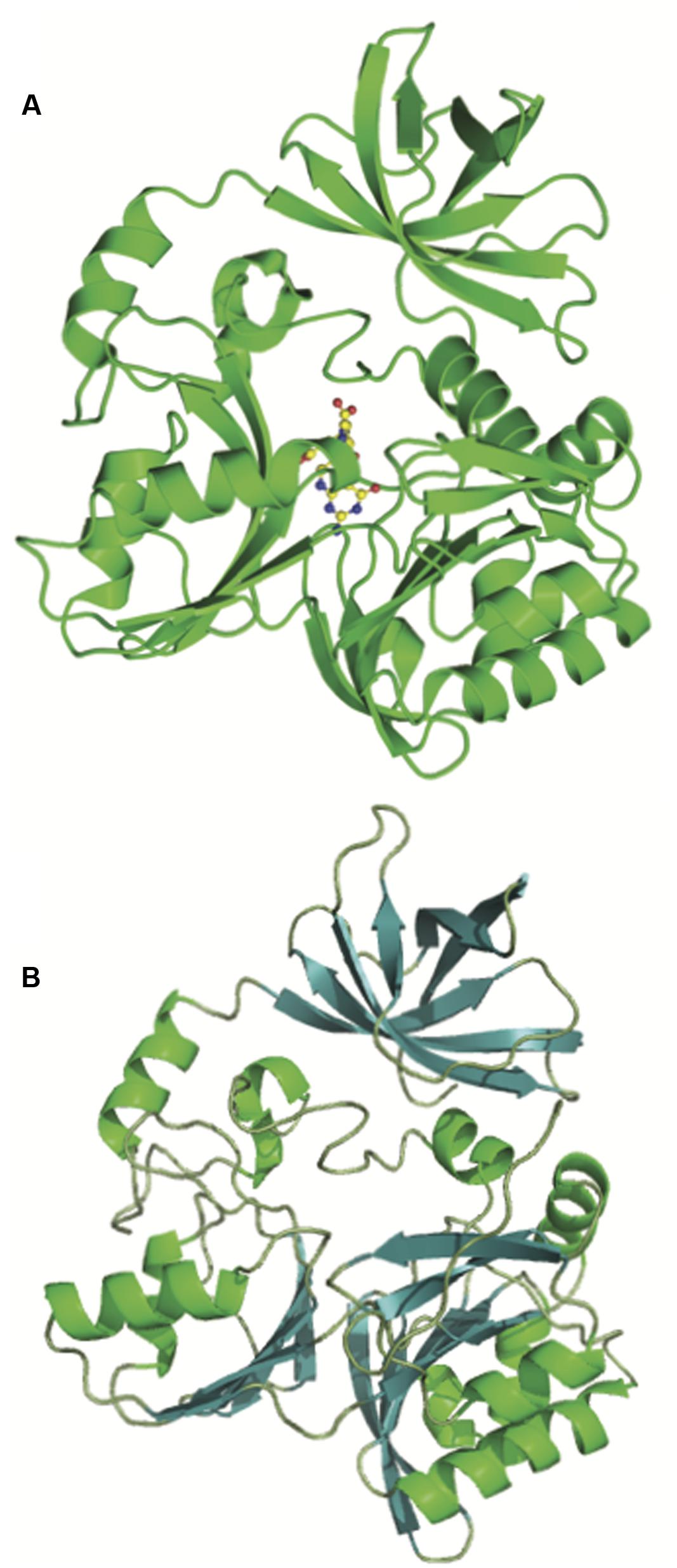
FIGURE 4. Structural similarity of the glycine cleavage T-protein and the DMSP demethylase (DmdA). Crystal structures of the (A) glycine cleavage T-protein from Thermotoga maritima (Lee et al., 2004) (PBD ID:1WOO) and (B) the DmdA monomer from P. ubique (Schuller et al., 2012) showing the shared tri-domain structure.
Phylogenetic analysis reveals that GcvTs and other similar proteins are nearly universally distributed among the prokaryotes, while DmdA proteins cluster separately. DmdA appears to be most prevalent among members of the Alphaproteobacteria (Figure 3) (Reisch et al., 2008; Moran et al., 2012). DmdA may have originally been a GcvT, but the development of a new activity and substrate preference has uniquely adapted this enzyme for DMSP metabolism (Reisch et al., 2008). Other organisms without DmdA may simply maintain DMSP as an osmolyte or utilize one of the many DMSP lyases identified so far to metabolize it.
Once methylmercaptopropionate (MMPA) is produced by DmdA, it is converted first to MMPA-CoA by the MMPA-CoA ligase or DmdB and then to methylthioacryloyl (MTA)-CoA by the MMPA-CoA dehydrogenase DmdC (Reisch et al., 2011a). In contrast to the narrower distribution of DmdA, DmdB and DmdC are found in up to 60% of surface ocean bacteria, assuming one copy per cell, as well as in bacteria from terrestrial and other environments (Figure 3) (Reisch et al., 2011b). The DmdB and DmdC enzymes characterized thus far show activity with a wide range of substrates, mostly with small to medium chain length fatty acids and their CoA derivatives (Reisch et al., 2011b; Bullock et al., 2014). These enzymes probably did not originate specifically for DMSP metabolism, potentially having been recruited from the pathways of methionine degradation and ß-fatty acid oxidation (Reisch et al., 2011a,b). The ability of DmdB and DmdC to act upon MMPA and MMPA-CoA is a demonstration of the plasticity and flexibility of these enzymes.
R. pomeroyi possesses more than 20 CoA ligases, but not all are predicted to have activity with MMPA. R. pomeroyi has two DmdB isozymes, RPO_DmdB1 and RPO_DmdB2 (Table 3). RPO_DmdB1 has a Km of 0.08 mM for MMPA but even lower Kms for butyrate and propionate, 0.02 and 0.04 mM, respectively. RPO_DmdB2 has a Km for MMPA similar to that of RPO_DmdB1, 0.07 mM, but this was the lowest Km it displayed with any of the substrates tested (Table 3) (Bullock et al., 2014). There are distinct differences between the DmdB enzymes from marine and non-marine microorganisms. Particularly, only the DmdBs from marine microorganisms are inhibited by concentrations of DMSP likely to be present in the cell. The R. pomeroyi DmdB isozymes exhibit different regulatory mechanisms to reverse this inhibition. RPO_DmdB1 responds to changes in cellular energy charge, while RPO_DmdB2 responds to increases in MMPA concentration (Bullock et al., 2014). These regulatory mechanisms may have developed during the specialization of the DmdB isozymes for DMSP rather than fatty acid metabolism. Because they are not found in the DmdBs from terrestrial bacteria, they appear to be specific adaptations to the importance of DMSP as a nutrient for marine bacteria.
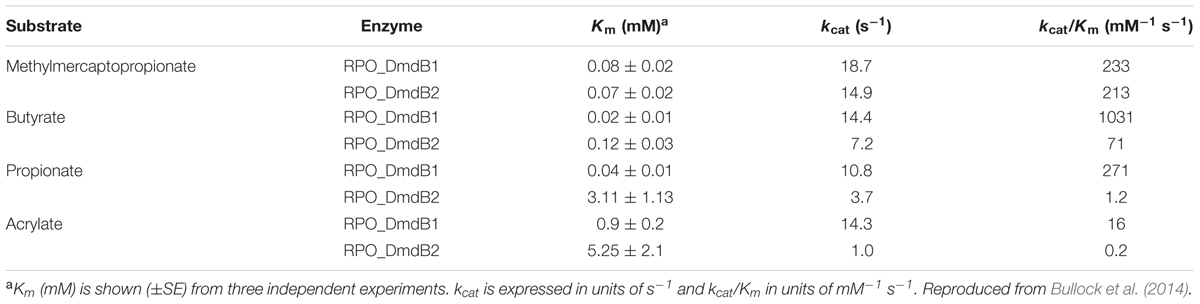
TABLE 3. Apparent kinetic constants for R. pomeroyi DSS-3 MMPA-CoA ligases RPO_DmdB1 and RPO_DmdB2a.
Three DmdC isozymes were identified in R. pomeroyi and verified to have activity toward MMPA-CoA (Reisch et al., 2011b). The Km of one of the DmdC isozymes (SPO3804; DmdC1) from R. pomeroyi for MMPA-CoA is low at 0.03 mM. However, lower Kms were observed for this enzyme with caproyl-CoA, valeryl-CoA, and butyrl-CoA. Thus, MMPA-CoA is not necessarily the preferred substrate for this enzyme. Instead, the substrate specificity of DmdC appears, like DmdB, to be based primarily on the length of the carbon chain of a substrate.
DmdB and DmdC isozymes are more widely distributed than DmdA, suggesting that these enzymes may be important in organisms that either metabolize only MMPA but not DMSP or possess pathways that form MMPA from substrates other than DMSP. Methionine degradation is one potential source of MMPA (Steele and Benevenga, 1979), a side reaction of the methionine salvage pathway can also produce MMPA (Sekowska et al., 2004; Albers, 2009). Xanthomonas campestris produces MMPA to induce bacterial blight in cassava (Perreaux et al., 1982; Ewbank and Maraite, 1990), and many plants, particularly fruiting plants, produce sulfur volatiles which closely resemble MMPA in structure (e.g., 3-methylthio-propanol, 3-methylthio-propanal, and ethyl-3-methylthio-propionate) (Gonda et al., 2013). These compounds might also be substrates of DmdB. Alternatively, the primary function of DmdB and DmdC in many bacteria may be fatty acid oxidation, and MMPA may only be an occasional substrate.
DmdD, a member of the crotonase superfamily, appears to be uniquely adapted for the metabolism of DMSP. DmdD has a crystal structure largely similar to that of other crotonases, a hexamer made up of a dimer of trimers. Figure 5 shows an overlay of one DmdD monomer with a monomer of rat liver enoyl-CoA hydratase (ECH). DmdD is similar to the rat liver ECH, sharing 32% amino acid identity. The main difference is that in DmdD the C-terminal loops of one of the trimers is oriented so that it can interact with the phosphate groups of CoA (Figure 5) (Tan et al., 2013). The same glutamate residues that are conserved and important for catalysis in the rat liver ECHs are also conserved in DmdD. However, DmdD is not nearly as efficient as an ECH at catalyzing the hydration of crotonyl-CoA, with a catalytic efficiency of 2100 mM-1s-1 compared with the typical values of 45000–119000 mM-1s-1 of other crotonases. DmdD instead displays a Km of 0.008 mM for MTA-CoA and a high catalytic efficiency, 5400 mM-1s-1 (Kiema et al., 1999; Feng et al., 2002; Tan et al., 2013). This greater catalytic efficiency only applies to reactions with MTA-CoA and appears to be due, at least in part, to the structure of MTA-CoA. The combination of the double bond and sulfur atom in MTA-CoA appear to be key for high rates DmdD hydrolysis activity as reactions with MMPA-CoA and crotonyl-CoA occur at lower rates (Tan et al., 2013).
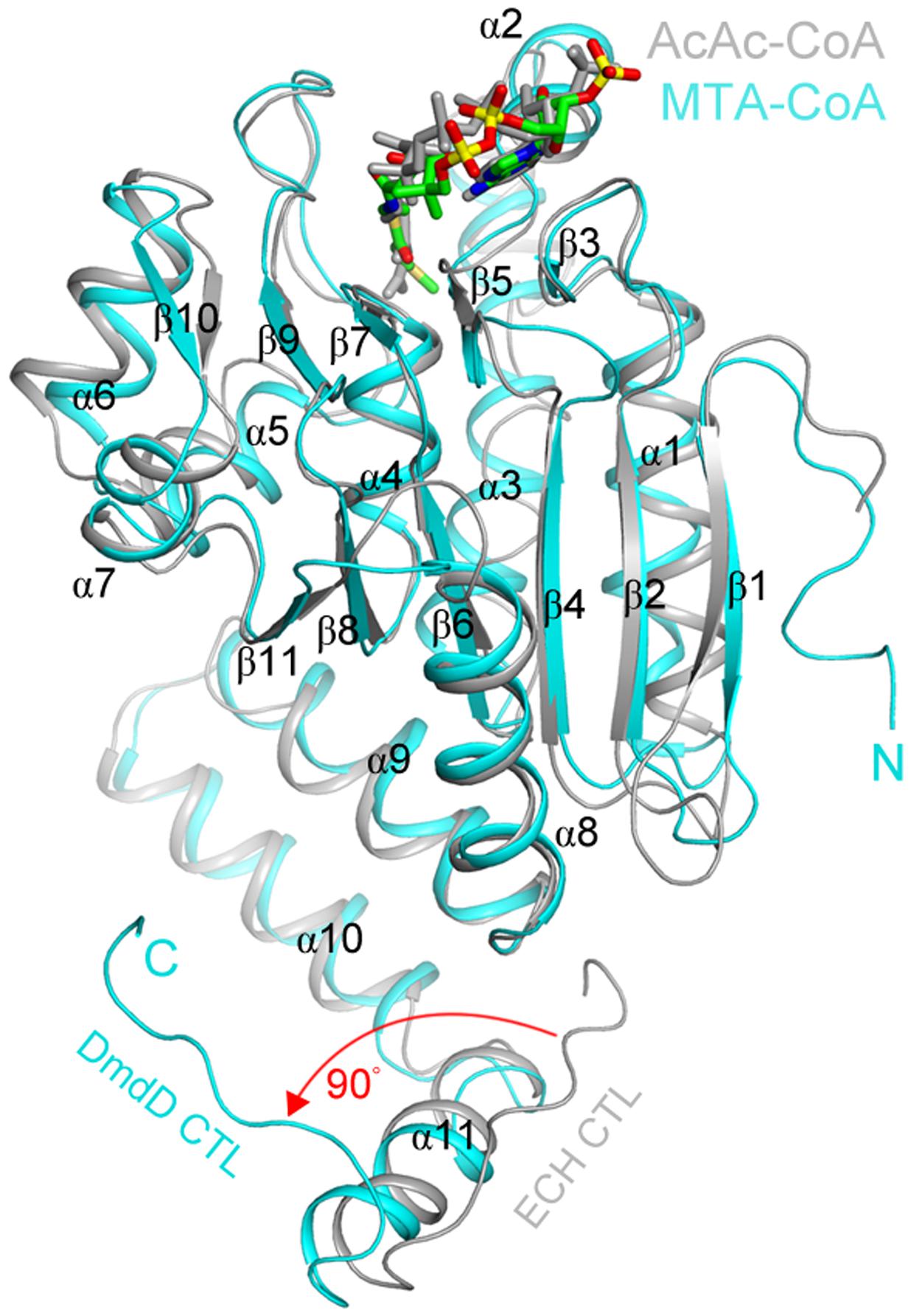
FIGURE 5. Structural similarities of the MTA-CoA hydratase and the enoyl-CoA hydratase from rat liver. Comparison of the structures of the R. pomeroyi MTA-CoA hydratase (DmdD, E121A mutant) monomer (cyan) in complex with MTA-CoA (green) and the rat liver ECH monomer (gray) in complex with acetoacetyl-CoA (gray) (Engel et al., 1996). The red arrow indicates a difference in the conformation of the C-terminal loop between the two structures. Figure reproduced from Tan et al. (2013).
While DmdD is highly efficient at catalyzing the hydration of MTA-CoA, it is not widely distributed (Reisch et al., 2011b). DmdD is absent from the majority of marine bacteria that utilize the demethylation pathway, i.e., possess DmdA (Figure 3). An ortholog of DmdD has been identified in the DmdD negative R. lacuscaerulensis as well as in R. pomeroyi (Reisch et al., 2011b). This enzyme, now designated AcuH for acrylate utilization hydratase, is an ECH with high activity toward acryloyl-CoA and crotonyl-CoA, but also displays activity toward MTA-CoA. The designation is similar to that of the acryloyl-CoA reductase AcuI. As a result of its activity toward acryloyl-CoA and MTA-CoA, AcuH is predicted to play an important role in the metabolism of acrylate formed from the cleavage pathway as well as MTA-CoA formed from the demethylation pathway (Figure 2) (Sullivan et al., 2011; Todd et al., 2012a; Reisch et al., 2013). By contrast, DmdD has no activity toward acryloyl-CoA. AcuH is less efficient than DmdD at hydrolyzing MTA-CoA, however, it is far more common, being found in a wide range of microorganisms, including those in the Roseobacter clade (Figure 3). AcuH appears to be a more versatile enzyme than DmdD and has maintained more of its functional similarity to other ECHs. Since AcuH likely functions in both the cleavage and the demethylation pathways, this strategy gives the cells increased metabolic flexibility and may also protect against acryloyl toxicity. DmdD, by contrast, has adapted specifically to function in the demethylation pathway, possibly allowing organisms which possess DmdD to utilize DMSP more efficiently.
Interactions between the cleavage pathway and demethylation pathways in organisms that contain both are an ongoing field of study. One proposal is that a ‘bacterial switch’ allows bacteria possessing both pathways to alternate between producing more or less DMS and MeSH (Kiene et al., 2000; Simo, 2001). While there is currently no consensus as to what signal controls the switch, the identification of the acrylate utilization enzymes AcuH and AcuI has begun to shed light on the topic. AcuH, as mentioned above, may function in both the cleavage and demethylation pathways (Reisch et al., 2011b, 2013). AcuI is an acryloyl-CoA reductase whose gene has been found immediately downstream of dmdA in many members of the Roseobacter clade. In R. pomeroyi, dmdA and acuI are co-regulated with acrylate acting as an inducer (Sullivan et al., 2011; Todd et al., 2012a). Since acrylate and acryloyl-CoA are inhibitory for bacterial growth, it has been proposed that AcuI maintains cellular acrylate concentrations below inhibitory levels. Thus, when acrylate concentrations increase as a result of DMSP lyase activity, AcuI and DmdA co-regulation results in increased activity of both enzymes. Elevated AcuI activity then alleviates inhibition caused by the build-up of acrylate, while increases in DmdA activity stimulate the demethylation pathway, allowing DMSP to be utilized in a manner that does not produce acrylate (Todd et al., 2012a). The activity of the demethylation pathway may also respond to carbon and energy limitation, with regulation resulting from changes in the energy charge of the cell (Bullock et al., 2014). Further research is still needed to investigate the physiological cues for balancing the demethylation and cleavage pathways.
It is unclear what the original impetus for the development of DMSP biosynthetic pathway may have been. The proposed roles for DMSP in marine phytoplankton as an osmolyte, antioxidant, predator deterrent, cryoprotectant, and as an energy overflow mechanism each could provide great benefits, particularly in consistently changing marine environments. If DMSP was originally produced as part of an overflow mechanism for dealing with unbalanced growth due to nutrient limitation, the other benefits provided by this compound may have selected for the maintenance of this pathway. The case has been made for the co-evolution of the marine Roseobacter and the DMSP producing-phytoplankton. Members of the Roseobacter clade are abundant in coastal waters and are one of the main bacterial groups enriched during DMSP-producing phytoplankton blooms (Gonzalez et al., 2000; Zubkov et al., 2001; Moran et al., 2007). Based on independent time estimates assisted by the cyanobacterial fossil calibration and estimates derived from the mutation rate clock method, the Roseobacter ancestor likely underwent a genome expansion, coincident with the increase in abundance and diversification of the dinoflagellates and coccolithophores around 250 mya (Luo et al., 2013; Luo and Moran, 2014; Sun et al., 2017). Thus, the radiation of dinoflagellates and coccolithophores may have provided new environments for members of the Roseobacter clade, in much the same way that the breakup of Pangea and changes in ocean redox chemistry created new environments for the proliferation of the red plastid lineage members (Whitfield, 2001; Quigg et al., 2003; Luo et al., 2013).
Research into the diversity of bacterial DMSP utilization enzymes and their regulation is still ongoing. New Roseobacter isolates showing adaptations to their particular environmental niches are continually being discovered. Recently, two new members of the Roseobacter clade were isolated from deep-sea water, Thiobacimonas profunda JLT2016 and Pelagibaca abyssi JLT2014. While these isolates did not possess DMSP metabolic genes, their genomes included genes for inorganic sulfur oxidation and CO2 fixation, further demonstrating the metabolic flexibility of this clade (Tang et al., 2016) and illustrating that members of the Roseobacter clade have exploited different routes to metabolically thrive in their environments (Luo et al., 2014). Since the first Roseobacter genome expansion 250 mya, many factors could have played a role in the development of metabolic pathways for the utilization of a specific carbon source like DMSP. There was not one path for these organisms to follow but many, allowing for a diversity of solutions to a single goal. As DMSP became more readily available in the environment, marine organisms, such as R. pomeroyi and other Roseobacters, likely adapted to utilize this compound as a source of carbon as well as reduced sulfur. Members of the Roseobacter clade were well poised for this task, being metabolically versatile bacteria and thus able to thrive in dynamic environments (Moran et al., 2007, 2012). Exposure to DMSP would be the driving force behind this evolution of function, putting pressure on organisms to adapt proteins already encoded in their genomes to utilize this new compound. Possible examples of this can be seen in the enzymes DmdA and DmdD from the demethylation pathway and DddP from the cleavage pathway. Each of these enzymes became more specialized to function in DMSP metabolism. Although structurally similar to their likely ancestral enzymes, they have undergone major changes in substrate specificities and, in some cases, enzymatic mechanism. AcuH seems to have adopted a different strategy and can function as both a MTA-CoA hydratase in the demethylation pathway and an acryloyl-CoA hydratase in acrylate metabolism. DmdB and DmdC also maintained activity with a wide range of substrates while adapting to function efficiently in DMSP metabolism as well. In these latter cases, there appears to have been minimal adaptations to DMSP metabolism, with changes in regulatory strategy as well as small changes in substrate specificity to accommodate a novel substrate.
Enzymes are known to diverge from a parental function to develop new substrate specificities, often via the duplication of genes encoding a multifunctional and multispecific enzymes that then undergo an alteration in substrate specificity (Noda-Garcıa et al., 2013). Although a certain core set of amino acid residues are required for functionality and structure, there is also room for variation and change, allowing evolution of new functions and substrate specificities (Perona and Craik, 1997). Thus, the amino acid sequences and structures of the enzymes catalyzing the DMSP catabolic reactions do not vary greatly from their non-DMSP degrading counterparts. From this perspective, the enzymes of the DMSP demethylation and cleavage pathways are examples of the various processes of enzyme adaptation and evolution that occurred within the Roseobacter clade in the last 250 million years.
WW and HB were responsible for the conceptualization and design of this manuscript. HB and HL collected data and analyzed the literature for this review. HB drafted the original manuscript. WW and HL reviewed and edited the manuscript. HB, WW, and HL provided final approval of the manuscript prior to submission.
This work was supported in part by grants from the National Science Foundation MCB1158037 and OCE1342694.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Norman Hassell for assistance with computational data analysis and Mak Yun Yi for assistance with literature collection.
Albers, E. (2009). Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5′-methylthioadenosine. IUBMB Life 61, 1132–1142. doi: 10.1002/iub.278
Alcolombri, U., Ben-Dor, S., Feldmesser, E., Levin, Y., Tawfik, D. S., and Vardi, A. (2015). MARINE SULFUR CYCLE. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348, 1466–1469. doi: 10.1126/science.aab1586
Alcolombri, U., Laurino, P., Lara-Astiaso, P., Vardi, A., and Tawfik, D. S. (2014). DddD is a CoA-transferase/lyase producing dimethyl sulfide in the marine environment. Biochemistry 53, 5473–5475. doi: 10.1021/bi500853s
Allen, A. E. (2005). Defining the moelcular basis for energy balance in marine diatoms under fluctuating environmental conditions. J. Phycol. 6, 1073–1076. doi: 10.1111/j.1529-8817.2005.00156.x
Allen, A. E., Vardi, A., and Bowler, C. (2006). An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr. Opin. Plant Biol. 9, 264–273. doi: 10.1016/j.pbi.2006.03.013
Archibald, J. M. (2009). The puzzle of plastid evolution. Curr. Biol. 19, R81–R88. doi: 10.1016/j.cub.2008.11.067
Archibald, J. M., and Keeling, P. J. (2002). Recycled plastids: a ’green movement’ in eukaryotic evolution. Trends Genet. 18, 577–584. doi: 10.1016/S0168-9525(02)02777-4
Asada, K., and Takahashi, M. (1987). “Production and scavenging of active oxygen in photosynthesis,” in Photoinhibition, eds D. J. Kyle, C. B. Osmond, and C. J. Arntzen (Amsterdam: Elsevier), 227–287.
Bhattacharya, D., and Medlin, L. (1998). Algal phylogeny and the origin of land plants. Plant Physiol. 116, 9–15. doi: 10.1104/pp.116.1.9
Bown, P. R., Lees, J. A., and Young, J. R. (2004). “Calcareous nannoplankton diversity and evolution through time,” in Coccolithophores, eds H. S. Thierstein and J. R. Young (Berlin: Springer-Verlag), 481–508.
Breckels, M. N., Boakes, D. E., Codling, E. A., Malin, G., Archer, S. D., and Steinke, M. (2010). Modelling the concentration of exuded dimethylsulfoniopropionate (DMSP) in the boundary layer surrounding phytoplankton cells. J. Plankton Res. 32, 253–257. doi: 10.1093/plankt/fbp116
Brummett, A. E., Schnicker, N. J., Crider, A., Todd, J. D., and Dey, M. (2015). Biochemical, kinetic, and spectroscopic characterization of Ruegeria pomeroyi DddW–A mononuclear iron-dependent DMSP lyase. PLoS ONE 10:e0127288. doi: 10.1371/journal.pone.0127288
Bullock, H. A., Reisch, C. R., Burns, A. S., Moran, M. A., and Whitman, W. B. (2014). Regulatory and functional diversity of methylmercaptopropionate coenzyme A ligases from the dimethylsulfoniopropionate demethylation pathway in Ruegeria pomeroyi DSS-3 and other proteobacteria. J. Bacteriol. 196, 1275–1285. doi: 10.1128/JB.00026-14
Caruana, A. (2010). DMS and DMSP Production by Marine Dinoflagellates. Ph.D. dissertation, University of East Anglia, Norwich.
Caruana, A., and Malin, G. (2014). The variability in DMSP content and DMSP lyase activity in mairne dinoflagellates. Prog. Oceanogr. 120, 410–424. doi: 10.1016/j.pocean.2013.10.014
Challenger, F., and Simpson, M. I. (1948). Studies on biological methylation; a precursor of the dimethyl sulphide evolved by Polysiphonia fastigiata; dimethyl-2-carboxyethylsulphonium hydroxide and its salts. J. Chem. Soc. 3, 1591–1597. doi: 10.1039/jr9480001591
Chin, M., and Jacob, D. J. (1996). Anthropogenic and natural contributions to tropospheric sulfate: a global model analysis. J. Geophys. Res. 101, 18691–18699. doi: 10.1029/96jd01222
Corn, M., Belviso, S., Partensky, F., Simon, N., and Christaki, U. (1996). “Origin and importance of picoplanktonic DMSP,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds, ed. R. Kiene (New York, NY: Plenum Press), 191–201.
Cortes, M. Y., Bollmann, J., and Thierstein, H. R. (2001). Coccolithophore ecology at the HOT station HOT. Hawaii. Deep Sea Res. II 48, 1957–1981. doi: 10.1016/S0967-0645(00)00165-X
Curson, A. R., Liu, J., Bermejo Martinez, A., Green, R. T., Chan, Y., Carrion, O., et al. (2017). Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat. Microbiol. 2, 17009. doi: 10.1038/nmicrobiol.2017.9
Curson, A. R., Rogers, R., Todd, J. D., Brearley, C. A., and Johnston, A. W. (2008). Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine alpha-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10, 757–767. doi: 10.1111/j.1462-2920.2007.01499.x
Curson, A. R., Sullivan, M. J., Todd, J. D., and Johnston, A. W. (2011a). DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 5, 1191–1200. doi: 10.1038/ismej.2010.203
Curson, A. R., Todd, J. D., Sullivan, M. J., and Johnston, A. W. (2011b). Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat. Rev. Microbiol. 9, 849–859. doi: 10.1038/nrmicro2653
de Souza, M. P., and Yoch, D. C. (1995). Comparative physiology of dimethyl sulfide production by dimethylsulfoniopropionate lyase in Pseudomonas doudoroffii and Alcaligenes sp. strain M3A. Appl. Environ. Microbiol. 61, 3986–3991.
de Souza, M. P., and Yoch, D. C. (1996). Differential metabolism of dimethylsulfoniopropionate and acrylate in saline and brackish intertidal sediments. Microb. Ecol. 31, 319–330. doi: 10.1007/BF00171575
Delwiche, C. F. (1999). Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 154, S164–S177. doi: 10.1086/303291
Engel, C. K., Mathieu, M., Zeelen, J. P., Hiltunen, J. K., and Wierenga, R. K. (1996). Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket. EMBO J. 15, 5135–5145.
Ewbank, E., and Maraite, H. (1990). Conversion of methionine to phytotoxic 3-methylthiopropionic acid by Xanthomonas campestris pv. manihotis. J. Gen. Microbiol. 136, 1185–1189. doi: 10.1099/00221287-136-7-1185
Falkowski, P. G., Katz, M. E., Knoll, A. H., Quigg, A., Raven, J. A., Schofield, O., et al. (2004a). The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. doi: 10.1126/science.1095964
Falkowski, P. G., Schofield, O., Katz, M. E., Van De Schootbrugge, B., and Knoll, A. H. (2004b). “Why is the land green and the ocean red?,” in Coccolithophores, eds H. R. Thierstein and J. R. Young (Amsterdam: Elsevier), 429–453.
Feng, Y., Hofstein, H. A., Zwahlen, J., and Tonge, P. J. (2002). Effect of mutagenesis on the stereochemistry of enoyl-CoA hydratase. Biochemistry 41, 12883–12890. doi: 10.1021/bi020382g
Gage, D. A., Rhodes, D., Nolte, K. D., Hicks, W. A., Leustek, T., Cooper, A. J., et al. (1997). A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 387, 891–894. doi: 10.1038/43160
Gonda, I., Lev, S., Bar, E., Sikron, N., Portnoy, V., Davidovich-Rikanati, R., et al. (2013). Catabolism of L-methionine in the formation of sulfur and other volatiles in melon (Cucumis melo L.) fruit. Plant J. 74, 458–472. doi: 10.1111/tpj.12149
Gonzalez, J. M., Simo, R., Massana, R., Covert, J. S., Casamayor, E. O., Pedros-Alio, C., et al. (2000). Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66, 4237–4246. doi: 10.1128/AEM.66.10.4237-4246.2000
Hanson, A. D., Rivoal, J., Paquet, L., and Gage, D. A. (1994). Biosynthesis of 3-dimethylsulfoniopropionate in Wollastonia biflora (L.) DC. Evidence that S-methylmethionine is an intermediate. Plant Physiol. 105, 103–110. doi: 10.1104/pp.105.1.103
Haq, B. U., Hardenbol, J., and Vail, P. R. (1987). Chronology of fluctuating sea levels since the triassic. Science 235, 1156–1167. doi: 10.1126/science.235.4793.1156
Harwood, D. M., and Nikolaev, V. A. (1995). “Cretaceous diatoms: morphology, taxonomy, biostratigraphy,” in Siliceousmicrofossils, eds C. D. Blome, P. M. Whalen, and K. M. Reed (Lawrence, KS: Paleontological Society), 81–106.
Hatakeyama, S., Okuda, M., and Akimoto, H. (1982). Formation of sulfur-dioxide and methanesulfonic-acid in the photo-oxidation of dimethyl sulfide in the air. Geophys. Res. Lett. 9, 583–586. doi: 10.1029/GL009i005p00583
Hehemann, J. H., Law, A., Redecke, L., and Boraston, A. B. (2014). The structure of RdDddP from Roseobacter denitrificans reveals that DMSP lyases in the DddP-family are metalloenzymes. PLoS ONE 9:e103128. doi: 10.1371/journal.pone.0103128
Howard, E. C., Henriksen, J. R., Buchan, A., Reisch, C. R., Burgmann, H., Welsh, R., et al. (2006). Bacterial taxa that limit sulfur flux from the ocean. Science 314, 649–652. doi: 10.1126/science.1130657
Husband, J. D., Kiene, R. P., and Sherman, T. D. (2012). Oxidation of dimethylsulfoniopropionate (DMSP) in response to oxidative stress in Spartina alterniflora and protection of a non-DMSP producing grass by exogenous DMSP plus acrylate. Environ. Expl Bot. 79, 44–48. doi: 10.1016/j.envexpbot.2012.01.006
Ito, T., Asano, Y., Tanaka, Y., and Takabe, T. (2011). Regulation of biosynthesis of dimethylsulfoniopropionate and its uptake in sterile mutant of Ulva pertusa (Chlorophyta). J. Phycol. 47, 517–523. doi: 10.1111/j.1529-8817.2011.00977.x
James, F., Paquet, L., Sparace, S. A., Gage, D. A., and Hanson, A. D. (1995). Evidence implicating dimethylsulfoniopropionaldehyde as an intermediate in dimethylsulfoniopropionate biosynthesis. Plant Physiol. 108, 1439–1448. doi: 10.1104/pp.108.4.1439
Johnston, A. W., Green, R. T., and Todd, J. D. (2016). Enzymatic breakage of dimethylsulfoniopropionate-a signature molecule for life at sea. Curr. Opin. Chem. Biol. 31, 58–65. doi: 10.1016/j.cbpa.2016.01.011
Karsten, U., Kuck, K., Vogt, C., and Kirst, G. O. (1996). “Dimethylsulphoniopropioante production in phototrophic organisms and its physiological function as a cryoprotectant,” in Biological and Environmental Chemstry of DMSP and Related Sulphonium Compounds, eds R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (New York, NY: Plenum Press), 143–153.
Keeling, P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748. doi: 10.1098/rstb.2009.0103
Keller, M. (1989). Dimethyl sulfide production and marine phytoplankton. Biol. Oceanogr. 6, 375–382. doi: 10.1021/bk-1989-0393.ch011
Kettles, N. L., Kopriva, S., and Malin, G. (2014). Insights into the regulation of DMSP synthesis in the diatom Thalassiosira pseudonana through APR activity, proteomics and gene expression analyses on cells acclimating to changes in salinity, light and nitrogen. PLoS ONE 9:e94795. doi: 10.1371/journal.pone.0094795
Kiema, T. R., Engel, C. K., Schmitz, W., Filppula, S. A., Wierenga, R. K., and Hiltunen, J. K. (1999). Mutagenic and enzymological studies of the hydratase and isomerase activities of 2-enoyl-CoA hydratase-1. Biochemistry 38, 2991–2999. doi: 10.1021/bi981646v
Kiene, R., Linn, L. J., and Bruton, J. A. (2000). New and important roles for DMSP in marine microbial communities. J. Sea Res. 43, 209–224. doi: 10.1016/S1385-1101(00)00023-X
Kiene, R. P. (1998). Uptake of choline and its conversion to glycine betaine by bacteria in estuarine waters. Appl. Environ. Microbiol. 64, 1045–1051.
Kiene, R. P., Linn, L. J., Gonzalez, J., Moran, M. A., and Bruton, J. A. (1999). Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65, 4549–4558.
Kiene, R. P., and Williams, L. P. H. (1998). Glycine betaine uptake, retention, and degradation by microorganisms in seawater. Limnol. Oceanogr. 43, 1592–1603. doi: 10.4319/lo.1998.43.7.1592
Kiene, R. P., Williams, L. P. H., and Walker, J. E. (1998). Seawater microorganisms have a high affinity glycine betaine uptake system which also recognizes dimethylsulfoniopropionate. Aquat. Microb. Ecol. 15, 39–51. doi: 10.3354/ame015039
Kirkwood, M., Le Brun, N. E., Todd, J. D., and Johnston, A. W. (2010). The dddP gene of Roseovarius nubinhibens encodes a novel lyase that cleaves dimethylsulfoniopropionate into acrylate plus dimethyl sulfide. Microbiology 156(Pt 6), 1900–1906. doi: 10.1099/mic.0.038927-0
Kirst, G. O., Thiel, C., Wolff, H., Nothnagel, J., Wanzek, M., and Ulmke, R. (1990). Dimethylsulfoniopropionate (DMSP) in icealgae and its possibe biological role. Mar. Chem. 35, 381–388. doi: 10.1016/S0304-4203(09)90030-5
Kocsis, M. G., and Hanson, A. D. (2000). Biochemical evidence for two novel enzymes in the biosynthesis of 3-dimethylsulfoniopropionate in Spartina alterniflora. Plant Physiol. 123, 1153–1161. doi: 10.1104/pp.123.3.1153
Kocsis, M. G., Nolte, K. D., Rhodes, D., Shen, T. L., Gage, D. A., and Hanson, A. D. (1998). Dimethylsulfoniopropionate biosynthesis in Spartina alterniflora. Evidence that S-methylmethionine and dimethylsulfoniopropylamine are intermediates. Plant Physiol. 117, 273–281. doi: 10.1104/pp.117.1.273
Lee, H. H., Kim, D. J., Ahn, H. J., Ha, J. Y., and Suh, S. W. (2004). Crystal structure of T-protein of the glycine cleavage system. Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia. J. Biol. Chem. 279, 50514–50523. doi: 10.1074/jbc.M409672200
Lesser, M. P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Lewis, L. A., and McCourt, R. M. (2004). Green algae and the origin of land plants. Am. J. Bot. 91, 1535–1556. doi: 10.3732/ajb.91.10.1535
Li, C., Wei, T., Zhang, S., Chen, X., Gao, X., Wang, P., et al. (2013). Molecular insight into bacterial cleavage of oceanic dimethylsulfoniopropionate into demethyl sulfide. Proc. Natl. Acad. Sci. U.S.A. 111, 1026–1031. doi: 10.1073/pnas.1312354111
Lovelock, J. E., Maggs, R. J., and Rasmusse, R. A. (1972). Atmospheric dimethyl sulfide and natural sulfur cycle. Nature 237, 452–453. doi: 10.1038/237452a0
Luo, H., Csuros, M., Hughes, A. L., and Moran, M. A. (2013). Evolution of divergent life history strategies in marine alphaproteobacteria. MBio 4:e00373-13. doi: 10.1128/mBio.00373-13
Luo, H., and Moran, M. A. (2014). Evolutionary ecology of the marine Roseobacter clade. Microbiol. Mol. Biol. Rev. 78, 573–587. doi: 10.1128/MMBR.00020-14
Luo, H., Swan, B. K., Stepanauskas, R., Hughes, A. L., and Moran, M. A. (2014). Evolutionary analysis of a streamlined lineage of surface ocean Roseobacters. ISME J. 8, 1428–1439. doi: 10.1038/ismej.2013.248
Lyon, B. R., Lee, P. A., Bennett, J. M., Ditullio, G. R., and Janech, M. G. (2011). Proteomic analysis of a sea-ice diatom: salinity acclimation provides new insight into the dimethylsulfoniopropionate production pathway. Plant Physiol. 157, 1926–1941. doi: 10.1104/pp.111.185025
McCourt, R. M., Delwiche, C. F., and Karol, K. G. (2004). Charophyte algae and land plant origins. Trends Ecol. Evol. 19, 661–666. doi: 10.1016/j.tree.2004.09.013
Moldowan, J. M., Dahl, J., Jacobson, S. R., Huizinga, B. J., Fago, F. J., Shetty, R., et al. (1996). Chemostratigraphic reconstruction of biofacies: molecular evidence linking cyst-forming dinoflagellates with pre-Triassic ancestors. Geology 24, 159–162. doi: 10.1130/0091-7613(1996)024<0159:CROBME>2.3.CO;2
Moldowan, J. M., and Jacobson, S. R. (2000). Chemical signals for early evolution of major taxa: biosignatures and taxon-specific biomarkers. Int. Geol. Rev. 42, 805–812. doi: 10.1080/00206810009465112
Moldowan, J. M., and Talyzina, N. M. (1998). Biogeochemical evidence for dinoflagellate ancestors in the Early Cambrian. Science 281, 1168–1170. doi: 10.1126/science.281.5380.1168
Moran, M. A., Belas, R., Schell, M. A., Gonzalez, J. M., Sun, F., Sun, S., et al. (2007). Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 73, 4559–4569. doi: 10.1128/AEM.02580-06
Moran, M. A., Reisch, C. R., Kiene, R. P., and Whitman, W. B. (2012). Genomic insights into bacterial DMSP transformations. Annu. Rev. Mar. Sci. 4, 523–542. doi: 10.1146/annurev-marine-120710-100827
Niki, T., Kohata, K., and Otsuki, A. (2000). DMSP-lyase activity in five marine phytoplankton species: its potential importance in DMS production. Mar. Biol. 136, 759–764. doi: 10.1007/s002279900235
Niki, T., Kunugi, M., Kohata, K., and Otsuki, A. (1997). Annual monitoring of DMS-producing bacteria in Tokyo Bay, Japan in relation to DMSP. Mar. Ecol. Prog. Ser. 156, 17–24. doi: 10.3354/meps156017
Noda-Garcıa, L., Camacho-Zarco, A., Medina-Ruız, S., Gaytan, P., Carrillo-Tripp, M., Fulop, V., et al. (2013). Evolution of substrate specificity in a recipient’s enzyme following horizontal gene transfer. Mol. Biol. Evol. 30, 2024–2034. doi: 10.1093/molbev/mst115
Palmer, J. D. (2003). The symbiotic birth and spread of plastids: how many time and whodunit. J. Phycol. 39, 4–11. doi: 10.1046/j.1529-8817.2003.02185.x
Paquet, L., Rathinasabapathi, B., Saini, H., Zamir, L., Gage, D. A., Huang, Z. H., et al. (1994). Accumulation of the compatible solute 3-dimethylsylfoniopropionate in sugarcane and its relatives, but not other gramineous crops. Aust. J. Plant Physiol. 21, 37–48. doi: 10.1071/PP9940037
Perona, J. J., and Craik, C. S. (1997). Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem. 272, 29987–29990. doi: 10.1074/jbc.272.48.29987
Perreaux, D., Maraite, H., and Meyer, J. A. (1982). Identification of 3(methylthio) propionic acid as a blight inducing toxin produced by Xanthomonas campestris pv. manihotis in vitro. Physiol. Plant Path. 20, 313–319. doi: 10.1016/0048-4059(82)90056-X
Quigg, A., Finkel, Z. V., Irwin, A. J., Rosenthal, Y., Ho, T. Y., Reinfelder, J. R., et al. (2003). The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature 425, 291–294. doi: 10.1038/nature01953
Raina, J. B., Tapiolas, D. M., Foret, S., Lutz, A., Abrego, D., Ceh, J., et al. (2013). DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502, 677–680. doi: 10.1038/nature12677
Reisch, C. R., Crabb, W. M., Gifford, S. M., Teng, Q., Stoudemayer, M. J., Moran, M. A., et al. (2013). Metabolism of dimethylsulphoniopropionate by Ruegeria pomeroyi DSS-3. Mol. Microbiol. 89, 774–791. doi: 10.1111/mmi.12314
Reisch, C. R., Moran, M. A., and Whitman, W. B. (2008). Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190, 8018–8024. doi: 10.1128/JB.00770-08
Reisch, C. R., Moran, M. A., and Whitman, W. B. (2011a). Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2:172. doi: 10.3389/fmicb.2011.00172
Reisch, C. R., Stoudemayer, M. J., Varaljay, V. A., Amster, I. J., Moran, M. A., and Whitman, W. B. (2011b). Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473, 208–211. doi: 10.1038/nature10078
Rusch, D. B., Halpern, A. L., Sutton, G., Heidelberg, K. B., Williamson, S., Yooseph, S., et al. (2007). The sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. doi: 10.1371/journal.pbio.0050077
Salgado, P., Kiene, R., Wiebe, W., and Magalhaes, C. (2014). Salinity as a regulator of DMSP degradation in Ruegeria pomeroyi DSS-3. J. Microbiol. 52, 948–954. doi: 10.1007/s12275-014-4409-1
Sanderson, M. J. (2003). Molecular data from 27 proteins do not support a Precambrian origin of land plants. Am. J. Bot. 90, 954–956. doi: 10.3732/ajb.90.6.954
Schuller, D. J., Reisch, C. R., Moran, M. A., Whitman, W. B., and Lanzilotta, W. N. (2012). Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique. Protein Sci. 21, 289–298. doi: 10.1002/pro.2015
Sekowska, A., Denervaud, V., Ashida, H., Michoud, K., Haas, D., Yokota, A., et al. (2004). Bacterial variations on the methionine salvage pathway. BMC Microbiol. 4:9. doi: 10.1186/1471-2180-4-9
Simo, R. (2001). Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16, 287–294. doi: 10.1016/S0169-5347(01)02152-8
Simo, R., Archer, S., Pedros-Ali, C., Gilpin, L., and Stelfox-Widdicombe, C. (2002). Coupled dynamics of dimethylsulfoniopropionate and dimethyl sulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47, 53–61. doi: 10.4319/lo.2002.47.1.0053
Steele, R. D., and Benevenga, N. J. (1979). The metabolism of 3-methylthiopropionate in rat liver homogenates. J. Biol. Chem. 254, 8885–8890.
Stefels, J. (2000). Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43, 183–197. doi: 10.1016/S1385-1101(00)00030-7
Stefels, J., Dijkhuizen, L., and Gieskes, W. C. (1995). DMSP-lyase activity in a spring phytoplankton bloom off the Dutch coast, related to Phaeocystis sp. abundance. Mar. Ecol. Prog. Ser. 123, 235–243. doi: 10.3354/meps123235
Stefels, J., Steinke, M., Turner, S., Malin, G., and Belviso, S. (2007). Environmental contraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modeling. Biogeochemistry 83, 245–275. doi: 10.1007/s10533-007-9091-5
Stover, L. E., Brinkhuis, H., Damassa, S. P., De Verteuil, L., and Helby, R. J. (1996). “Mesozoic-Tertiary dinoflagellates, acritarchs and prasinophyte,” in Palynology: Principles and Applications, eds D. C. Jansonius and J. McGregor (Washington DC: American Association of Stratigraphic Palynologists Foundation), 641–750.
Sullivan, M. J., Curson, A. R., Shearer, N., Todd, J. D., Green, R. T., and Johnston, A. W. (2011). Unusual regulation of a leaderless operon involved in the catabolism of dimethylsulfoniopropionate in Rhodobacter sphaeroides. PLoS ONE 6:e15972. doi: 10.1371/journal.pone.0015972
Summers, P. S., Nolte, K. D., Cooper, A. J. L., Borgeas, H., Leustek, T., Rhodes, D., et al. (1998). Identification and stereospecificity of the first three enzymes of 3-dimethylsulfoniopropionate biosynthesis in a chlorophyte alga. Plant Physiol. 116, 369–378. doi: 10.1104/pp.116.1.369
Sun, J., Todd, J. D., Thrash, J. C., Qian, Y., Qian, M. C., Temperton, B., et al. (2016). The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat. Microbiol. 1, 16065. doi: 10.1038/nmicrobiol.2016.65
Sun, Y., Powell, K. E., Sung, W., Lynch, M., Moran, M. A., and Luo, H. (2017). Spontaneous mutations of a model heterotrophic marine bacterium. ISME J. doi: 10.1038/ismej.2017.20 [Epub ahead of print].
Sunda, W., Kieber, D. J., Kiene, R. P., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320. doi: 10.1038/nature00851
Tan, D., Crabb, W. M., Whitman, W. B., and Tong, L. (2013). Crystal structure of DmdD, a crotonase superfamily enzyme that catalyzes the hydration and hydrolysis of methylthioacryloyl-CoA. PLoS ONE 8:e63870. doi: 10.1371/journal.pone.0063870
Tang, K., Yang, Y., Lin, D., Li, S., Zhou, W., Han, Y., et al. (2016). Genomic, physiologic, and proteomic insights into metabolic versatility in Roseobacter clade bacteria isolated from deep-sea water. Sci. Rep. 6:35528. doi: 10.1038/srep35528
Todd, J. D., Curson, A. R., Dupont, C. L., Nicholson, P., and Johnston, A. W. (2009). The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ. Microbiol. 11, 1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x
Todd, J. D., Curson, A. R., Kirkwood, M., Sullivan, M. J., Green, R. T., and Johnston, A. W. (2011). DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ. Microbiol. 13, 427–438. doi: 10.1111/j.1462-2920.2010.02348.x
Todd, J. D., Curson, A. R., Nikolaidou-Katsaraidou, N., Brearley, C. A., Watmough, N. J., Chan, Y., et al. (2010). Molecular dissection of bacterial acrylate catabolism–unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ. Microbiol. 12, 327–343. doi: 10.1111/j.1462-2920.2009.02071.x
Todd, J. D., Curson, A. R., Sullivan, M. J., Kirkwood, M., and Johnston, A. W. (2012a). The Ruegeria pomeroyi acuI gene has a role in DMSP catabolism and resembles yhdH of Escherichia coli and other bacteria in conferring resistance to acrylate. PLoS ONE 7:e35947. doi: 10.1371/journal.pone.0035947
Todd, J. D., Kirkwood, M., Newton-Payne, S., and Johnston, A. W. (2012b). DddW, a third DMSP lyase in a model Roseobacter marine bacterium, Ruegeria pomeroyi DSS-3. ISME J. 6, 223–226. doi: 10.1038/ismej.2011.79
Todd, J. D., Rogers, R., Li, Y. G., Wexler, M., Bond, P. L., Sun, L., et al. (2007). Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315, 666–669. doi: 10.1126/science.1135370
Trossat, C., Nolte, K. D., and Hanson, A. D. (1996). Evidence that the pathway of dimethylsulfoniopropionate biosynthesis begins in the cytosol and ends in the chloroplast. Plant Physiol. 111, 965–973. doi: 10.1104/pp.111.4.965
Vail, P. R., Mitchum, R. M., and Thompson, I. I. I. S. (1977). “Seismic stratigraphy and global changes of sea level, part 4: global cycles of relative changes of sea level,” in Seismic Stratigraphy-Applications to Hydrocarbon Exploration, ed. C. E. Payton (Tusla, OK: American Association of Petroleum Geologists Memoirs), 83–97.
van der Maarel, M., Van Bergeijk, S., Van Werkhoven, A., Laverman, A., Meijer, W., Stam, W., et al. (1996). Cleavage of dimethylsulfoniopropionate and reduction of acrylate by Desulfovibrio acrylicus sp. nov. Arch. Microbiol. 166, 109–115. doi: 10.1007/s002030050363
van Duyl, F. C., Gieskes, W. C., Kop, A. J., and Lewis, W. E. (1998). Biological control of short-term variations in the concentration of DMSP and DMS during Phaeocystis spring bloom. J. Sea Res. 40, 221–231. doi: 10.1016/S1385-1101(98)00024-0
Varaljay, V. A., Robidart, J., Preston, C. M., Gifford, S. M., Durham, B. P., Burns, A. S., et al. (2015). Single-taxon field measurements of bacterial gene regulation controlling DMSP fate. ISME J. 9, 1692. doi: 10.1038/ismej.2015.94
Wagner, H., Jakob, T., and Wilhelm, C. (2005). Balancing the energy flow from captured light into biomass under fluctuating light conditions. New Phytol. 169, 95–108. doi: 10.1111/j.1469-8137.2005.01550.x
Wang, P., Chen, X. L., Li, C. Y., Gao, X., Zhu, D. Y., Xie, B. B., et al. (2015). Structural and molecular basis for the novel catalytic mechanism and evolution of DddP, an abundant peptidase-like bacterial dimethylsulfoniopropionate lyase: a new enzyme from an old fold. Mol. Microbiol. 98, 289–301. doi: 10.1111/mmi.13119
Whitfield, M. (2001). Interactions between phytoplankton and trace metals in the ocean. Adv. Mar. Biol. 41, 3–128. doi: 10.1016/s0065-2881(01)41002-9
Wiesemeier, T., and Pohnert, G. (2007). Direct quantification of dimethylsulfoniopropionate (DMSP) in marine micro- and macroalgae using HPLC or UPLC/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 850, 493–498. doi: 10.1016/j.jchromb.2006.12.023
Wolfe, G. V., and Steinke, M. (1997). Grazing-activated chemical defense in a unicellular marine alga. Nature 387, 894–897. doi: 10.1038/43168
Yoch, D. C. (2002). Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68, 5804–5815. doi: 10.1128/AEM.68.12.5804-5815.2002
Yoon, H. S., Hackett, J. D., Ciniglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818. doi: 10.1093/molbev/msh075
Yost, D. M., and Mitchelmore, C. L. (2009). Dimethylsulfoniopropionate (DMSP) lyase activity in different strains of the symbiotic alga Symbiodinium microadriaticum. Mar. Ecol. Prog. Ser. 386, 61–70. doi: 10.3354/meps08031
Zhang, Y., Sun, Y., Jiao, N., Stepanauskas, R., and Luo, H. (2016). Ecological genomics of the uncultivated marine Roseobacter lineage CHAB-I-5. Appl. Environ. Microbiol. 82, 2100–2111. doi: 10.1128/AEM.03678-15
Zubkov, M. V., Fuchs, B. M., Archer, S. D., Kiene, R. P., Amann, R., and Burkill, P. H. (2001). Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3, 304–311. doi: 10.1046/j.1462-2920.2001.00196.x
Keywords: DMSP, dimethylsulfoniopropionate, evolution, phytoplankton, Roseobacter
Citation: Bullock HA, Luo H and Whitman WB (2017) Evolution of Dimethylsulfoniopropionate Metabolism in Marine Phytoplankton and Bacteria. Front. Microbiol. 8:637. doi: 10.3389/fmicb.2017.00637
Received: 14 January 2017; Accepted: 28 March 2017;
Published: 19 April 2017.
Edited by:
Bernd Wemheuer, University of New South Wales, AustraliaCopyright © 2017 Bullock, Luo and Whitman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannah A. Bullock, YnVsbG9ja2hAdWdhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.