- 1State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Biotechnology Program, Department of Environmental Sciences, COMSATS Institute of Information Technology, Abbottabad, Pakistan
- 3Institute of Biochemistry and NCHU Agricultural Biotechnology Center, National Chung Hsing University, Taichung, Taiwan
Kdp-ATPase is an inducible high affinity potassium uptake system that is widely distributed in bacteria, and is generally regulated by the KdpD/KdpE two-component system (TCS). In this study, conducted on Mycobacterium smegmatis, the kdpFABC (encoding Kdp-ATPase) expression was found to be affected by low concentration of K+, high concentrations of Na+, and/or of the medium. The KdpE was found to be a transcriptional regulator that bound to a specific 22-bp sequence in the promoter region of kdpFABC operon to positively regulate kdpFABC expression. The KdpE binding motif was highly conserved in the promoters of kdpFABC among the mycobacterial species. 5′-RACE data indicated a transcriptional start site (TSS) of the kdpFABC operon within the coding sequence of MSMEG_5391, which comprised a 120-bp long 5′-UTR and an open reading frame of the 87-bp kdpF gene. The kdpE deletion resulted in altered growth rate under normal and low K+ conditions. Furthermore, under K+ limiting conditions, a single transcript (kdpFABCDE) spanning kdpFABC and kdpDE operons was observed. This study provided the first insight into the regulation of kdpFABC operon by the KdpD/KdpE TCS in M. smegmatis.
Introduction
Potassium ion (K+) is a ubiquitous monovalent cation that is essential for all living cells. It plays important roles in a wide variety of cellular functions such as turgor pressure maintenance, osmo-regulation, acid-base homeostasis, membrane potential adjustment, enzyme activation, and gene expression regulation (Booth, 1985; Epstein, 1986, 2003; Dinnbier et al., 1988; Stumpe et al., 1996; Follmann et al., 2009). Generally, a higher intracellular concentration of K+ (150–500 mM) is required than that of extracellular environment (Epstein and Schultz, 1965; Dinnbier et al., 1988) to efficiently carry out these functions. Hence, cell has to precisely regulate the uptake of K+ for its growth and survival in various hostile conditions (Stumpe et al., 1996; Kuo et al., 2005). For this purpose, animal cells are usually equipped with Na+/K+ ATP pumps that are generally missing in bacterial cells (Heitzmann and Warth, 2008; Gumz et al., 2015), whereas several K+ channels and active transporters are present in the prokaryotic cells to maintain potassium homeostasis. These bacterial K+ uptake systems vary a lot in their structures and affinities for the cation, which ultimately help bacteria to precisely maintain intracellular K+ concentrations under diverse environmental conditions (Altendorf et al., 1992, 1998; Epstein, 2003; Nanatani et al., 2015). In prokaryotes, three major families of K+ uptake transporters, Trk/Ktr/HKT, Kup/HAK/KT, and Kdp, have been discovered (Buurman et al., 1995; Ballal et al., 2002, 2007; Berry et al., 2003; Sato et al., 2014). Besides these, some K+ channels have also been reported (Epstein, 2003). Among these K+ uptake transporters, Trk and Kup are constitutively expressed in Escherichia coli, with the uptake rate of Trk system nearly 10 times higher than that of Kup transporter (Rhoads et al., 1976). On the contrary, the Kdp-ATPase is an inducible system, exhibiting high affinity for K+ uptake that is specific to bacterial and archeal kingdoms (Diskowski et al., 2015). This system becomes operational under low K+ concentrations (~2 μM as estimated from the Km-value; Epstein, 2016) when other K+ uptake systems such as Trk and Kup fail to function (Epstein, 1992). Generally, Kdp-ATPase complex is consisted of four subunits, encoded in the kdpFABC operon. Among these subunits, KdpF is required for stabilizing the complex (Altendorf et al., 1998; Gassel et al., 1999), KdpA is involved in the binding and translocation of K+ (Altendorf et al., 1992; Greie, 2011), while KdpB is associated with KdpC that is essential for the ATP hydrolysis (Haupt et al., 2005; Greie and Altendorf, 2007). In most cases, kdpFABC is found adjacent to kdpDE, which encodes proteins that are involved in the regulation of Kdp-ATPase system. The inducible Kdp-ATPase is regulated by histidine kinase KdpD and response regulator KdpE, which constitute a typical two-component system (TCS) (Walderhaug et al., 1992), with KdpD responding to a multitude of signals such as low K+ concentrations, osmotic pressure upshift, different intracellular ATP levels, and pH of the medium (Asha and Gowrishankar, 1993; Jung and Altendorf, 2002; Hamann et al., 2008; Epstein, 2016). Previously, it was suggested that turgor pressure was another stimulus for kdpDE induction (Laimins et al., 1981), however recent observations did not support this idea (Epstein, 2016). Other than its role in K+ homeostasis, KdpD/KdpE TCS is also well-reported for its role in the regulation of bacterial virulence in diverse bacterial species including Mycobacterium tuberculosis, Staphylococcus aureus, Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa (Parish et al., 2003; Alegado et al., 2011; Xue et al., 2011; Feinbaum et al., 2012; Freeman et al., 2013). Moreover, as a regulator, KdpE was found to regulate many essential genes involved in key metabolisms, stress tolerance and virulence (Njoroge et al., 2013; Burda et al., 2014; Samir et al., 2016; Parker et al., 2017).
The Kdp-ATPase and its cognate TCS (KdpD/KdpE) have been well-characterized in E. coli (Walderhaug et al., 1992; Gassel et al., 1998; Brandon et al., 2000; Heermann and Jung, 2010; Laermann et al., 2013; Epstein, 2016). Cytoplasmic stimuli such as K+ concentration, ionic strength and ATP content are perceived by histidine kinase KdpD (Jung and Altendorf, 2002; Heermann and Jung, 2010). After detecting these stimuli, KdpD experiences auto-phosphorylation, with its phosphoryl group transferred to the transcriptional regulator KdpE (Voelkner et al., 1993). The phosphorylated KdpE (KdpE~P) binds to a 23-bp T-rich sequence in the promoter of kdpFABC with stronger affinity as compared to un-phosphorylated KdpE (Nakashima et al., 1992, 1993; Sugiura et al., 1992). Moreover, some accessory proteins such as UspC (Heermann et al., 2009) and enzyme IIANtr (Lüettmann et al., 2009) have also been reported for their interactions with KdpD for signal transduction and cross talk. Among pathogenic bacteria, KdpD/KdpE TCS has been well-studied in S. aureus (Xue et al., 2011). The kdpFABC genes were significantly up-regulated (up to 100-fold) under salt stress in this osmo-tolerant bacterial species (Price-Whelan et al., 2013). In S. aureus, KdpE also regulates the expression of many other genes. Among these are the capsular polysaccharide genes (cap) that play a vital role in the pathogenicity of S. aureus. It was found that KdpE specifically bound to the cap promoter region and deletion of kdpDE resulted in the decreased transcription of cap genes (Zhao et al., 2010). Expression of other virulence factors such as spa, hla, aur, geh, and hlgB was also regulated by KdpE, and direct binding of KdpE to the promoter regions of many of these genes, has been observed (Xue et al., 2011).
Based on sequence homology to E. coli, Kdp-ATPase was found to be widely distributed among different bacterial and archeal genera (Ballal et al., 2007; Heermann and Jung, 2010). In case of genus Mycobacterium, the Kdp system has been found in M. tuberculosis, M. avium, M. bovis, M. smegmatis, M. marinum, and others, except M. leprae and M. ulcerans (Cholo et al., 2008; Bretl et al., 2011). Among these species, M. smegmatis is a fast growing and comparatively safe bacterial species to work with, which is now commonly used as a surrogate for M. tuberculosis. Generally, M. smegmatis is perceived as a non-pathogenic species. However, some investigations have demonstrated its pathogenic potential (Wallace et al., 1988; Alvarado-Esquivel et al., 2009; Best and Best, 2009; Driks et al., 2011; Xu et al., 2013). Up to now, KdpD/KdpE TCS in M. smegmatis has not been well-characterized. Due to the different organizations of kdpFABC and kdpDE in various mycobacterial species, investigation of this system in M. smegmatis would be valuable. As KdpD/KdpE is involved in regulating K+ homeostasis, elucidation of its detailed function and regulatory mechanism for its downstream target genes would provide better options to cope with mycobacterial infections. Moreover, due to the structural differences between the K+ uptake systems of prokaryotes and eukaryotes (Cholo et al., 2008), the current system may present a potential target for antibiotic development against mycobacterial infection. In the current study, the promoter of kdpFABC was clearly identified and the regulation of kdpFABC expression by KdpD/KdpE TCS under different potential stimuli was investigated in detail. To the best of our knowledge, for the first time this study provided strong evidences that KdpE bound to the promoter region of kdpFABC operon (PkdpF) at a specific site in M. smegmatis. In addition, we also predicted the coding sequence of KdpF in M. smegmatis as well as in many other mycobacterial species.
Materials and Methods
Bacterial Strains and Growth Conditions
Strains and plasmids used in this study are listed in Table 1. E. coli strains DH5α and BL21 (DE3) were grown in Lysogeny broth (LB) media. M. smegmatis wild type strain MC2155 and its mutants were grown at 37°C in Middlebrook 7H9 broth (Difco Becton Dickinson, USA) supplemented with 0.2% (v/v) glycerol and 0.05% (v/v) Tween80 or on Middlebrook 7H10 agar (Difco Becton Dickinson, USA) supplemented with 0.5% (v/v) glycerol (Tang et al., 2014). For potassium limitation study, Hartmans-de Bont (HdB) medium [35 μM EDTA, 490 μM MgCl2. 6H2O, 7 μM CaCl2, 0.8 μM NaMoO4. 2H2O, 5.49 μM MnCl2. 2H2O, 6.95 μM ZnSO4. 7H2O, 1.68 μM CoCl2. 6H2O, 20 μM FeSO4. 7H2O, 0.8 μM CuSO4. 5H2O, 7.08 mM NaH2PO4, 8.9 mM K2HPO4, or 8.9 mM Na2HPO4, 15 mM (NH4)2SO4, supplemented with 0.2% (v/v) glycerol and 0.05% (v/v) Tween 80, pH 7.0] (Smeulders et al., 1999; Hartmans et al., 2006) was used. Kanamycin (30 μg/mL), hygromycin B (100 μg/mL), 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) (50 μg/mL) and 10% sucrose (w/v) were added in media when required.
Construction of ΔkdpD and ΔkdpE Mutants and Functional Complementation of ΔkdpE
The mutant strains of M. smegmatis were constructed by the method of homologous recombination as described earlier (Yang et al., 2012). The detailed procedures used to construct kdpD and kdpE mutants (ΔkdpD and ΔkdpE) and the kdpE complementary strain (CΔkdpE) are provided in Supplementary Material (Figures S1–S3).
Purification of KdpE Protein
The kdpE gene was amplified and cloned into pET28a(+) (Novagen) to construct expression plasmid pET28-kdpE. The 6-His-tagged KdpE protein was heterologously expressed in E. coli BL21(DE3)-pET28-kdpE (Table 1) and purified with Ni-NTA affinity column (GenScript). Purified KdpE protein was concentrated and desalted by MWCO 3000 Amicon ultra centrifugal filters (Millipore). The purity of KdpE was analyzed on 12% SDS-PAGE (Figure S4) and quantified by Bradford assay (Bradford, 1976).
Electrophoretic Mobility Shift Assay (EMSA)
DNA probes were amplified with and without 6-FAM-labeled primers (Table S1) and the probes were purified with a gel purification kit (Bioteke). Labeled probes were incubated with different amounts of KdpE in EMSA binding buffer [10 mM Tris (pH 7.5 at 20°C), 100 mM KCl, 1 mM EDTA, 0.1 mM DTT, 5% v/v glycerol, 0.010 mg/mL BSA] (Hellman and Fried, 2007) with the addition of 0.5 mM Na2ATP to activate phosphorylation of KdpE. Unlabeled probes were used for competition experiments. The reaction mixture was incubated at 25°C for 1 h and then subjected to 6% native polyacrylamide gel electrophoresis at 150 V for 1 h in a 0.25 × Tris-borate-EDTA (TBE) running buffer. The gel image was obtained using Typhoon Scanner (GE Healthcare).
DNase-I Foot Printing
To determine the binding sequence of KdpE in the PkdpF, the fluorescence labeled probes and the reaction system were the same as used in EMSA. The mixtures were treated with 0.02 U DNase-I for 2 min. Further procedures of purification and analysis of samples were same as described previously (Tang et al., 2014).
RNA Extraction from M. smegmatis and Synthesis of cDNA
To study gene expression, bacterial cells were grown in HdB media (with and without K+) to the late logarithmic phase. Total RNA was extracted by TRIzol method (Schmittgen and Livak, 2008) and the samples were analyzed on 1% agarose gel for qualitative assessment and NanoDrop 2000 (Thermo Scientific, USA) spectrophotometer was used for the quantitative analysis. First-strand cDNA was constructed with the commercially available PrimeScript RT reagent kit with gDNA Eraser (Takara Biotechnology, Japan) according to manufacturer's instructions.
Real Time qPCR
For real time qPCR reaction (RT-qPCR), gene specific primers were used (Table S1). Appropriate dilutions of cDNA were prepared and used in RT-qPCR. The reaction was performed in real-time PCR machine (Lightcycler 480 instrument II, Roche) with following conditions: 45 cycles of 95°C for 10 s, 63°C for 10 s, and 72°C for 10 s. Relative quantification of the gene expression was determined by 2−ΔΔCT method (Livak and Schmittgen, 2001). The sigA (MSMEG_2758) was used as a reference gene for the determination of relative expression.
5′-Rapid Amplification of cDNA Ends (5′-Race)
To identify the transcriptional start site (TSS) of kdpFABC operon, 5′-RACE analysis was performed with RNA extracted from M. smegmatis cells grown in HdB media (with and without K+) using 5′-Full RACE kit (Takara biotechnology, Japan) according to manufacturer's instructions and as reported in our previous studies (Tang et al., 2014, 2016). The PkdpF region was amplified by reverse transcriptase (RT) PCR from RACE cDNA libraries using 5′-RACE outer primers and PkdpF specific outer primers (Table S1). The resulting amplified product was purified from the agarose gel and used as template in next amplification step with 5′-RACE inner primers along with PkdpF specific inner primers (Table S1). Finally, 250 bp sequence obtained was purified and ligated into pMD19-T (Takara biotechnology, Japan) and mobilized into E. coli DH5α. Clones were sequenced and the first nucleotide next to 5′-RACE adaptor was considered to be the TSS of the operon.
β-Galactosidase Assay
All strains were grown in HdB (with and without K+) and 7H9 broth with different concentrations of salts (NaCl, NH4Cl and KCl), sucrose and with different pH-values containing kanamycin (30 μg/mL) at 37°C for 20 h (late-logarithmic phase). The collected culture was assayed for β-galactosidase activity as previously described (Bharati et al., 2013).
Effects of Different Stress Conditions on the Growth of M. smegmatis Strains
M. smegmatis MC2155 and its mutant derivatives were subjected to various stress conditions such as temperature, pH, and salt stress. Previously described methodology was used to introduce stress conditions to the bacterial strains (Gebhard et al., 2008). For each stress experiment, bacterial cells were grown to mid-logarithmic phase in the 7H9 medium supplemented with 0.2% glycerol and 0.05% Tween 80. The specific stimulation conditions are described in Table S2.
Bioinformatics Analysis
Promoter sequences of kdpFABC in various mycobacterial species were found by NCBI blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and KEGG server (http://www.genome.jp/kegg/). The multiple sequence alignments of KdpF proteins and KdpE binding motifs were conducted using BioEdit program, and final output was prepared by ESPript 3.0 server (http://espript.ibcp.fr/ESPript/ESPript/) (Robert and Gouet, 2014). DNA logo was generated using program WebLogo (http://weblogo.berkeley.edu/) (Crooks et al., 2004).
Results
Expression of kdpFABC Operon under K+ Limiting Condition and Its Regulation by the KdpD/KdpE TCS
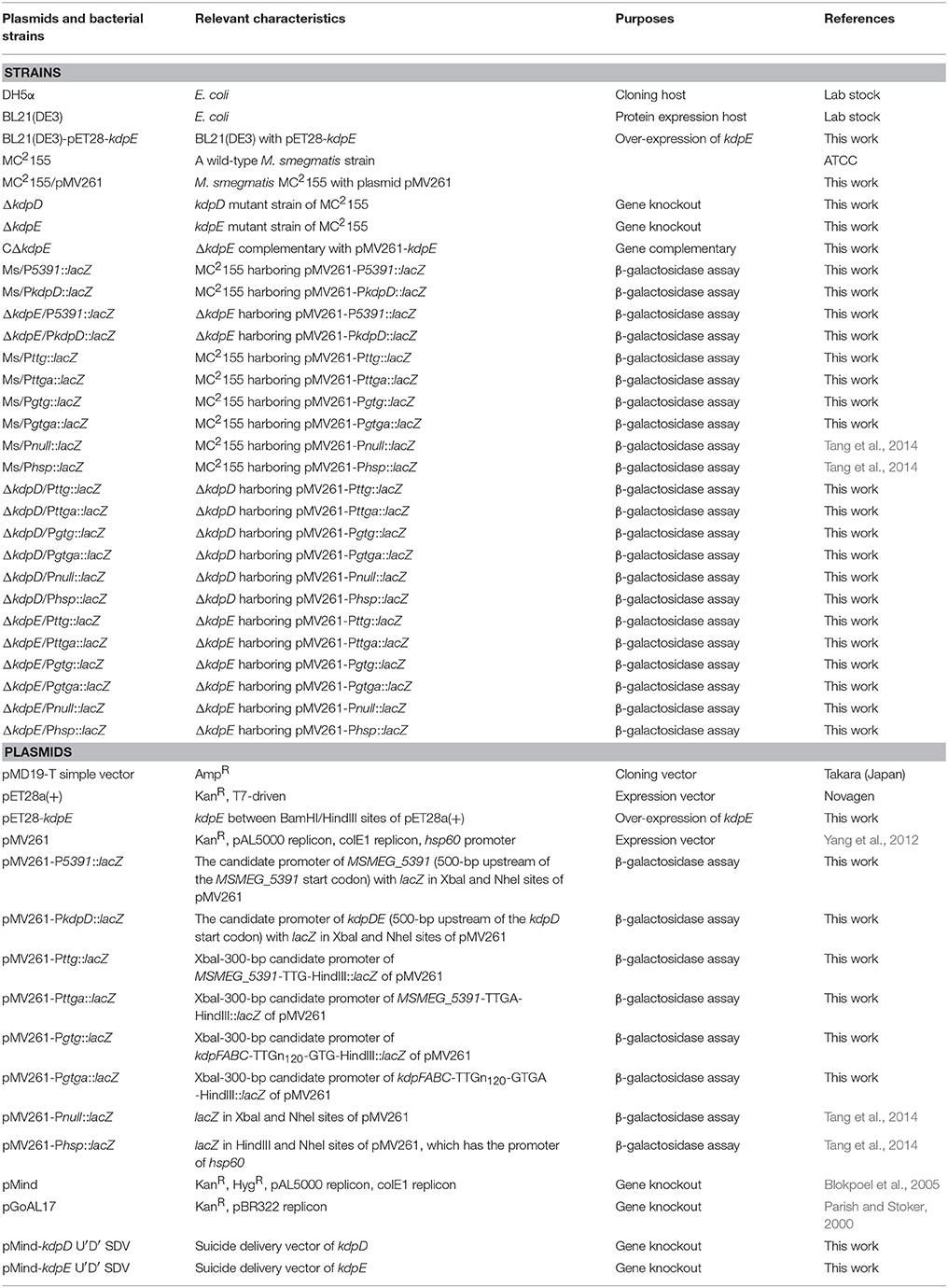
Initially, the relative expression levels of kdpFABC and kdpDE genes were studied in bacterial strains grown in the presence and absence of K+ in the medium. In wild type MC2155, the expression of above genes occurred at a very basal level under normal K+ condition whilst considerably higher levels were observed in 0 mM K+ condition (Figure 1). In both ΔkdpD and ΔkdpE mutants, no expression levels of kdpFABC genes were detected compared to the wild type MC2155 growing under normal K+ condition (Figure 1). This indicated that expression levels of kdpFABC were controlled by KdpD/KdpE TCS and were up-regulated under K+ limiting condition.
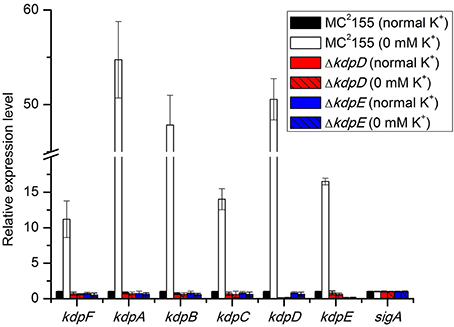
Figure 1. Relative expression levels of kdp loci in response to growth in K+ limiting and non-limiting conditions. Cultures of wild type, as well as ΔkdpD and ΔkdpE mutants were grown to late exponential phase in the presence and absence of K+ in HdB medium. Total RNA was extracted and cDNA was used for quantitative analysis. Data represent the averages of biological triplicates. Error bars indicate standard deviation. sigA (MSMEG-2758) gene was used as reference gene for the determination of relative expression levels of kdpFABC and kdpDE genes in K+ limiting condition with respect to normal K+ condition (wild type strain). Normal K+ stands for HdB medium where K+ concentration was similar to that defined earlier (Smeulders et al., 1999), which was ~17.8 mM as measured by flame photometry, whereas 0 mM K+ stands for HdB medium in which K+ salt was replaced by similar concentration of Na+ salt and the residual K+ concentration was ~23 μM as measured by flame photometry.
Co-Transcription of kdpFABC and kdpDE Operons under Low k+ Condition
Next, we sought to investigate the transcription patterns of kdpFABC and kdpDE operons. RT-PCR analysis showed an obvious amplification band in the region spanning kdpC (last gene of the kdpFABC operon) and kdpD (first gene of the kdpDE operon) when cDNA from wild type strain grown under K+ limiting condition was used as a template (Figure S5). In contrast, no amplification band was detected with cDNA from wild type strain grown under normal K+ condition in HdB medium (data not shown). These results showed that in M. smegmatis, the kdpFABC and kdpDE operons were co-transcribed as a single transcript (kdpFABCDE) from PkdpF under K+ limiting condition but not under normal K+ condition.
Binding of KdpE Protein to PkdpF
To gain further insights into the regulation of kdpFABC operon, we determined the binding of KdpE to the promoter PkdpF. The in vitro EMSA of KdpE with the candidate promoter of kdpF was carried out in the presence of ATP, and a dramatic shift of the labeled DNA probe in the presence of KdpE was observed (Figure 2A). It was noteworthy that such binding was relatively weaker in the absence of ATP (data not shown), indicating phosphorylation of KdpE was required for efficient promoter binding. The EMSA was also carried out in a competition way to reveal the specific binding of KdpE with the PkdpF; when unlabeled PkdpF probe was added to the reaction mixture in increasing proportions, the unlabeled probe clearly competed with the labeled probe, causing labeled probe to move to the position of no protein binding (Figure 2A). In S. aureus, KdpE was reported to bind not only to the promoter region of kdpFABC but also to that of kdpDE (Xue et al., 2011). However, no such binding was observed when we performed the EMSA experiment with the promoter of kdpDE operon (PkdpD) (data not shown).
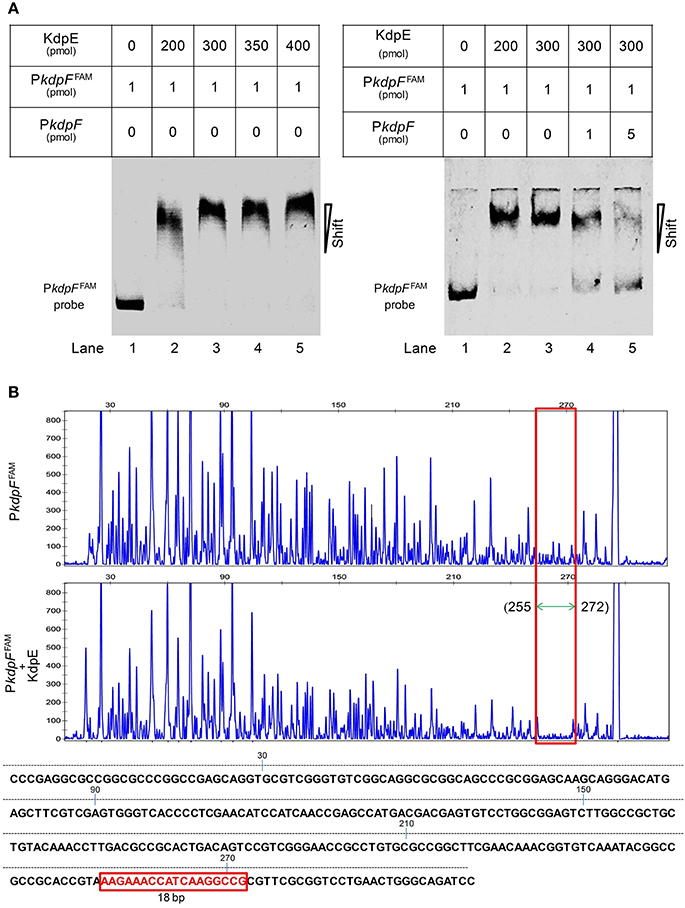
Figure 2. Binding of purified KdpE protein to specific DNA motif in the PkdpF. (A) EMSA of KdpE with PkdpF. Left lane 1: 6-FAM-labeled probe of PkdpF, left lanes 2–5: complex formation with increasing concentrations of KdpE. Right lane 1: 6-FAM-labeled probe of PkdpF, right lanes 2–3: complex formation with increasing concentrations of KdpE, right lanes 4–5: competition with increasing concentration of unlabeled probe. (B) Electropherograms indicating protected DNA region in PkdpF after DNase-I digestion of 6-FAM-labeled probe in the absence of KdpE (upper panel) and in the presence of KdpE (lower panel). The fluorescence signals of 6-FAM-labeled fragments are plotted against probe length. The protected region in electropherograms and the 18-bp DNA motif of KdpE are boxed in red. The 300-bp DNA probe used in the assay is shown below with numerical values along the DNA sequence representing the positions of nucleotides with reference to the above graphical scale.
In order to further verify the role of KdpE as a transcriptional regulator, a β-galactosidase assay was performed using lacZ as the reporter gene. Surprisingly, no β-galactosidase activity was obtained when 500-bp upstream of MSMEG_5391 (P5391) (MSMEG_5391 which is present upstream of kdpA gene and it has been annotated to translate hypothetical protein) was selected as the candidate promoter (Figure S6A). On the other hand, very basal level activities were detected with the PkdpD without any significant difference between the wild type and ΔkdpE under normal and limiting K+ conditions (Figure S6B).
Minimum KdpE Binding Motif in the PkdpF
Above EMSA data provided clear evidences for the in vitro binding between KdpE and candidate PkdpF. To map the precise binding sequence of KdpE, a DNase-I foot printing assay was performed using the 300-bp 6-FAM-labeled promoter fragment of the kdpFABC operon. It is clear from the electropherograms that a short 18-bp sequence (5′-AAGAAACCATCAAGGCCG-3′) appeared to be protected from DNase-I digestion as a result of KdpE binding (Figure 2B). To validate this result, the EMSA experiment was further performed using essentially the same 18-bp sequence (mentioned above). Unexpectedly, we failed to obtain binding between the 18-bp probe identified from DNase-I foot printing and KdpE protein (data not shown). We thus extended the 18-bp probe with few base pairs flanking the central 18-bp probe to make a longer 33-bp probe and repeated the EMSA experiment, again KdpE was found to be able to bind successfully with this longer probe (Figure 3). Furthermore, a competition EMSA was carried out with increasing concentrations of unlabeled 33-bp probe along with 300-bp 6-FAM-labeled probe in the reaction mixture. The unlabeled 33-bp probe showed specific binding with KdpE and competed well with the labeled probe as revealed in the gel (Figure 3A).
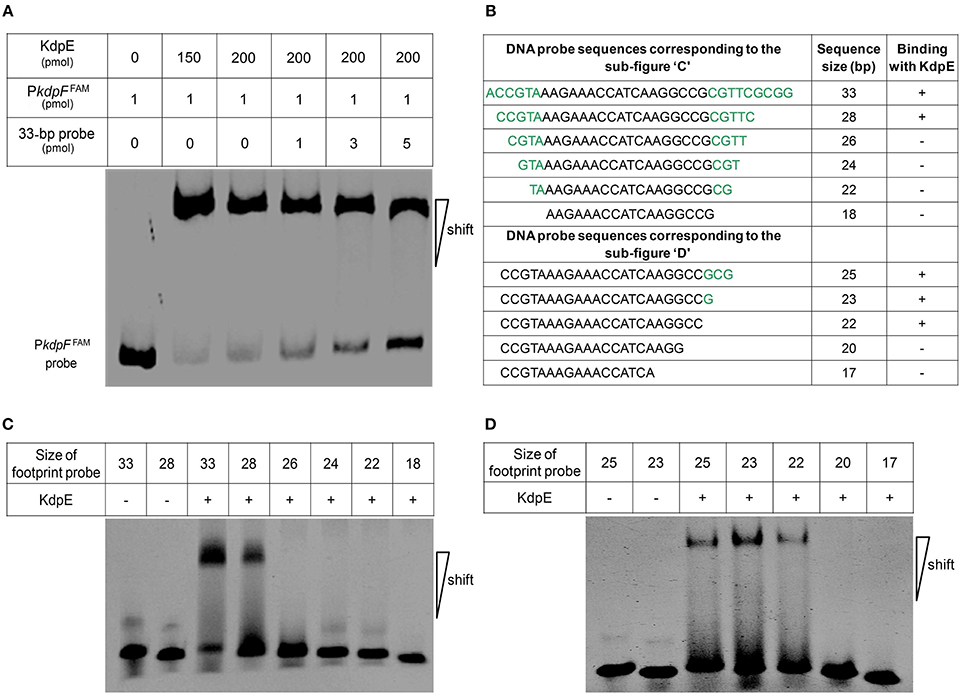
Figure 3. Determination of minimum DNA sequence required for successful binding of KdpE protein with PkdpF. (A) EMSA competition assay performed with labeled (300-bp 6-FAM-labeled probe) and unlabeled 33-bp DNA probes. With increasing concentration of unlabeled 33-bp probe, the band intensity of labeled probe with KdpE protein decreased while that of unlabeled probe increased, indicating that unlabeled 33-bp probes could effectively compete with the 300-bp 6-FAM-labeled probe in binding KdpE. (B) The sizes and sequences of different DNA probes used for the determination of exact foot print sequence. (C) Shorter double stranded DNA probes used (based on DNase-I foot printing results) in EMSA to determine the minimum DNA sequence required for KdpE binding with PkdpF. Sizes of DNA probes were decreased by removing few nucleotides on both ends of the 33-bp probe to determine minimum nucleotides required for binding. (D) Unnecessary nucleotides (based on DNA sequence homology with other mycobacterial species) were trimmed down from the 3′ end of the 28-bp probe to determine the minimum sequence required for KdpE-DNA interaction.
To attain the minimum sequence required for the KdpE binding, few nucleotides were sequentially removed from both ends of the effective 33-bp probe (Figure 3B) to see if shorter fragments still retained the binding ability with KdpE. In this aspect, a sequence of 28-bp but not any other shorter sequence was found to retain successful binding with KdpE (Figure 3C). We then further removed nucleotides from 3′-end of the 28-bp sequence to generate a series of shorter fragments (Figure 3B). Finally, a minimum of 22-bp sequence (5′-CCGTAAAGAAACCATCAAGGCC-3′) with strong binding ability with KdpE was identified (Figure 3D). This 22-bp sequence contained five additional nucleotides at the 5′-end and one nucleotide less from the 3′-end of the original 18-bp foot print sequence. This newly determined 22-bp motif was found to be highly conserved in many mycobacterial species (Figure 4).
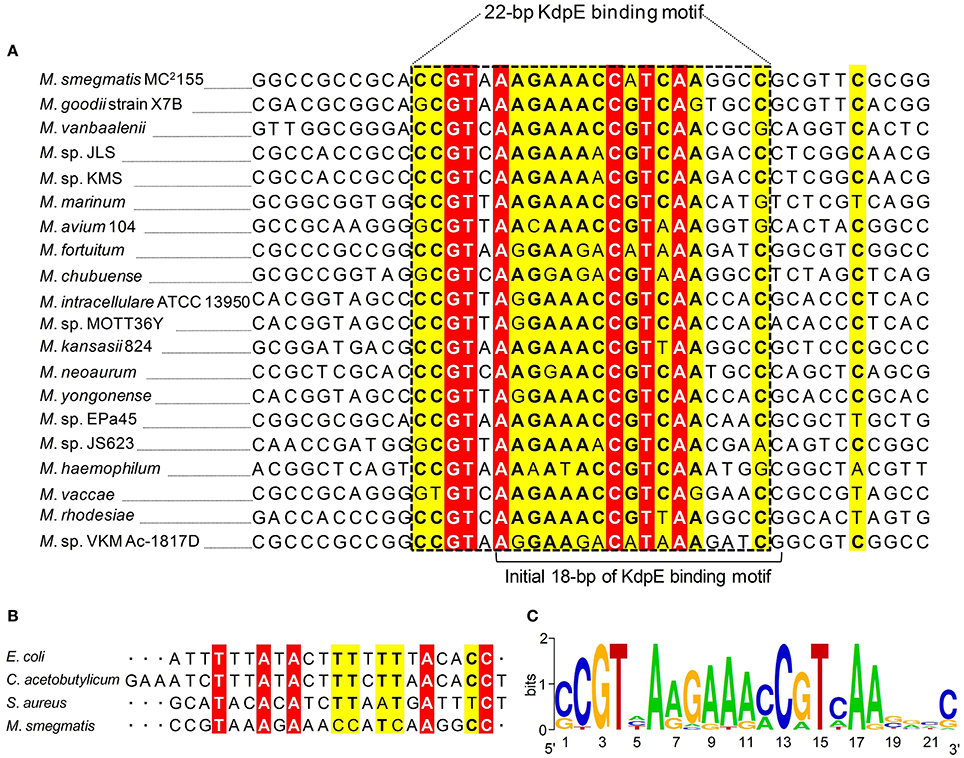
Figure 4. Multiple DNA sequences alignments of KdpE binding motifs from different mycobacterial species. (A) The homology of minimum 22-bp binding sequence was searched in different species of genus Mycobacterium. Highly similar nucleotides are shown in white letters highlighted in red background, less similar nucleotides in black bold letters highlighted in yellow, while dissimilar nucleotides in black letters in white background. (B) KdpE binding sequences found in different bacterial species; E. coli (Sugiura et al., 1992), C. acetobutylicum (Behrens and Duerre, 2000), S. aureus (Xue et al., 2011) and M. smegmatis (present study). (C) The DNA logo of DNA motifs required for efficient binding of KdpE protein to the PkdpF based on DNA homology among different mycobacterial species. The logo was prepared using WebLogo (Crooks et al., 2004).
TSS of the kdpFABC Operon
Two 5′-RACE cDNA libraries (in the presence and absence of K+) were constructed from the wild type strain. Amplification of expected RACE inner fragment was only possible when RT-PCR was performed using the K+ limiting library, which revealed a sharp band on agarose gel, whereas no such band was detected using the normal K+ library (data not shown). First nucleotide base identified next to the 5′-RACE adaptor was an adenosine residue (A) that was considered as the TSS of the kdpFABC operon (Figure 5A). Unexpectedly, this TSS was found within the coding sequence of MSMEG_5391 at the +4-bp position. Hence, the ORF of the candidate kdpF was supposed to be present within this region. DNA sequence analysis revealed another possible start codon GTG located at the +120-bp position of TSS (Figure 5B). The candidate kdpF gene (87-bp) shared significant homology to the kdpF of M. tuberculosis (93-bp). Furthermore, this candidate kdpF was aligned to the upstream regions of kdpA in various mycobacterial species. Almost all species in which KdpE binding sequence was conserved, contained a kdpF gene which shares significant identity to the kdpF of M. smegmatis and M. tuberculosis (Figure 6). Further, β-galactosidase assay (with and without frame shift) was conducted to verify candidate kdpF as a functional gene of kdpFABC operon. Together, all data showed that the 87-bp ORF is the part of kdpFABC operon which was successfully induced under low K+ conditions (Figure 7). Since the start codon of MSMEG_5391 was not functional, this confirmed that the previously identified hypothetical protein (MSMEG_5391) was very likely to be annotated incorrectly. In contrast, this genomic locus (MSMEG_5391) was believed to contain a long 5′-UTR and a functional kdpF gene.
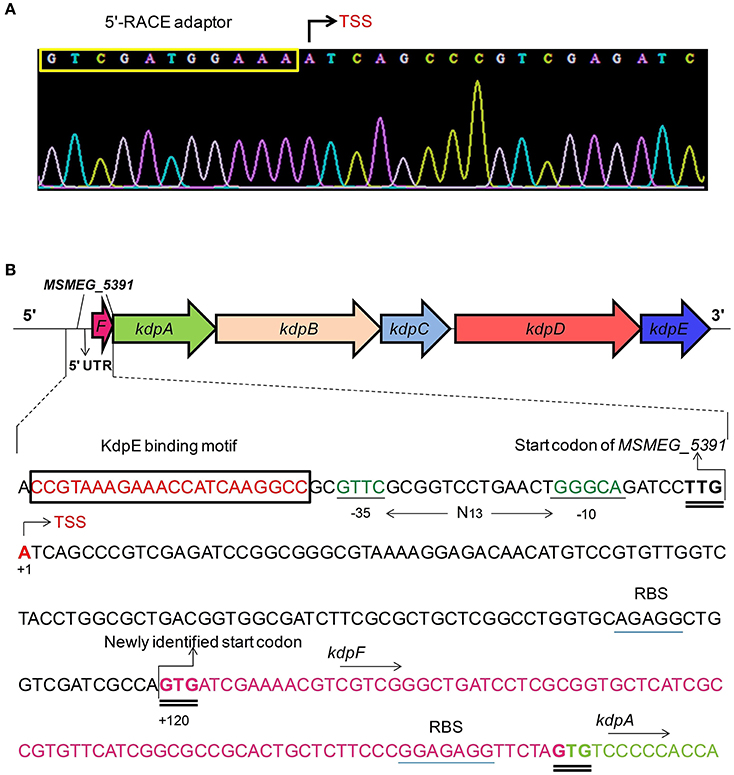
Figure 5. Mapping of TSS and characterization of the PkdpF. (A) The 5′-RACE adaptor sequence (boxed in yellow) along with the transcript sequence after DNA sequencing. The TSS is marked with a bent arrow. (B) Fully characterized PkdpF sequence, where the KdpE binding sequence is shown in red nucleotides boxed in black, the SigF-dependent promoter consensus −10 and −35 regions shown in dark green, the TSS nucleotide shown with red letter marked by a bent arrow. All potential start codons are double underlined, with the newly identified start codon indicated at the +120 position. The candidate kdpF gene is shown in magenta letters. The sequence of MSMEG_5391 containing a 5′-UTR region (120 bp) of kdpFABC and the newly determined kdpF coding sequence (87-bp) is shown overlapped with the start codon of kdpA. The possible Shine-Dalgarno sequences in RBS are underlined with blue line.
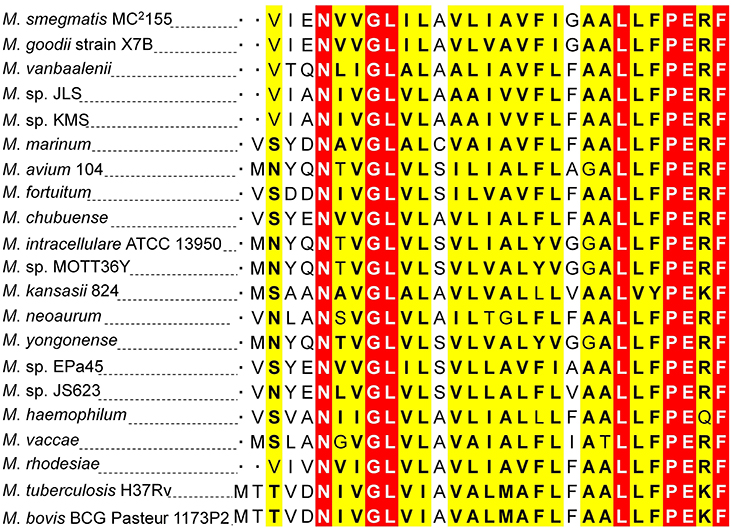
Figure 6. Protein sequence homology of KdpF of M. smegmatis with other mycobacterial species. The DNA sequence of kdpF in M. smegmatis was determined based on 5′-RACE and subsequent β-galactosidase assays. Its homologs were searched in other mycobacterial species in the upstream region of kdpA. The KdpF protein sequences of high similarity are shown in white letters highlighted in red background, low similarity in black bold letters highlighted in yellow background, and dissimilar residues in black letters in white background.
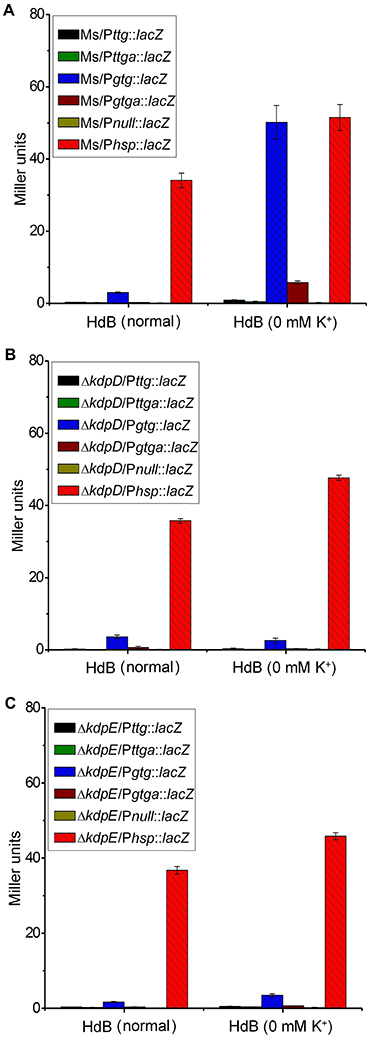
Figure 7. Induction of kdpF under K+ limiting conditions. β-galactosidase assay was performed to determine functionality of PkdpF, translation start codon of MSMEG_5391 (TTG) as well as newly identified translation start codon of kdpF (GTG). Promoter regions were fused with promoter less-lacZ gene (in frame and out of frame) in pMV261 to generate the Pttg::lacZ, Pttga::lacZ, Pgtg::lacZ, and Pgtga::lacZ plasmids. Plasmids were separately transferred into wild type (A), ΔkdpD (B), and ΔkdpE (C) to determine regulation of PkdpF by KdpD/KdpE TCS. For positive control, Phsp was fused with the lacZ gene (Phsp::lacZ) and for the negative control promoter less-lacZ plasmid (Pnull::lacZ) was used. Strains were grown in HdB broth under K+ normal and limiting conditions (0 mM K+) and β-galactosidase activity was determined to investigate promoter functionality.
Effects of the Salts on the Expression of kdpFABC
A series of β-galactosidase assays was performed to investigate potential stimuli for the transcriptional induction of the kdpFABC operon and its regulation by the KdpD/KdpE TCS. For this purpose, different promoter sequences were fused with lacZ (Figure S7). Under K+ non-limiting condition, no β-galactosidase activity was observed in wild type strains, except for some basal level activity in the Ms/Pgtg::lacZ, whereas under K+ limiting condition, very high β-galactosidase activity was observed for Ms/Pgtg::lacZ compared to all other strains (Figure 7A). There was no β-galactosidase activity observed in the ΔkdpD and ΔkdpE in both, normal and 0 mM K+ HdB (Figures 7B,C). These results also supported our previous RT-qPCR results in which deletion of kdpD/kdpE genes resulted in silencing of the kdpFABC operon.
In order to further explore the potential signals other than low K+ condition that could turn on kdpFABC transcription in M. smegmatis, all strains previously constructed for β-galactosidase activity (Table 1) were grown in 7H9 medium with different concentrations of NaCl, NH4Cl, KCl, and sucrose as well as under different pH-values. In case of 7H9 medium alone, none of the wild type derivatives exhibited noticeable β-galactosidase activity. However, with increasing concentrations of NaCl and NH4Cl, β-galactosidase activity of Ms/Pgtg::lacZ was increased. On the other hand, minor and no activity was observed in the case of increasing concentrations of sucrose and KCl, respectively. Moreover, no activity change was observed under different pH-values. The derivatives of ΔkdpD showed no β-galactosidase activity in all above mentioned conditions, while in the ΔkdpE derivatives, only ΔkdpE/Pgtg::lacZ showed slight activity with increasing NaCl or NH4Cl concentrations that was considerably lower than Ms/Pgtg::lacZ under the same conditions (Figure 8).
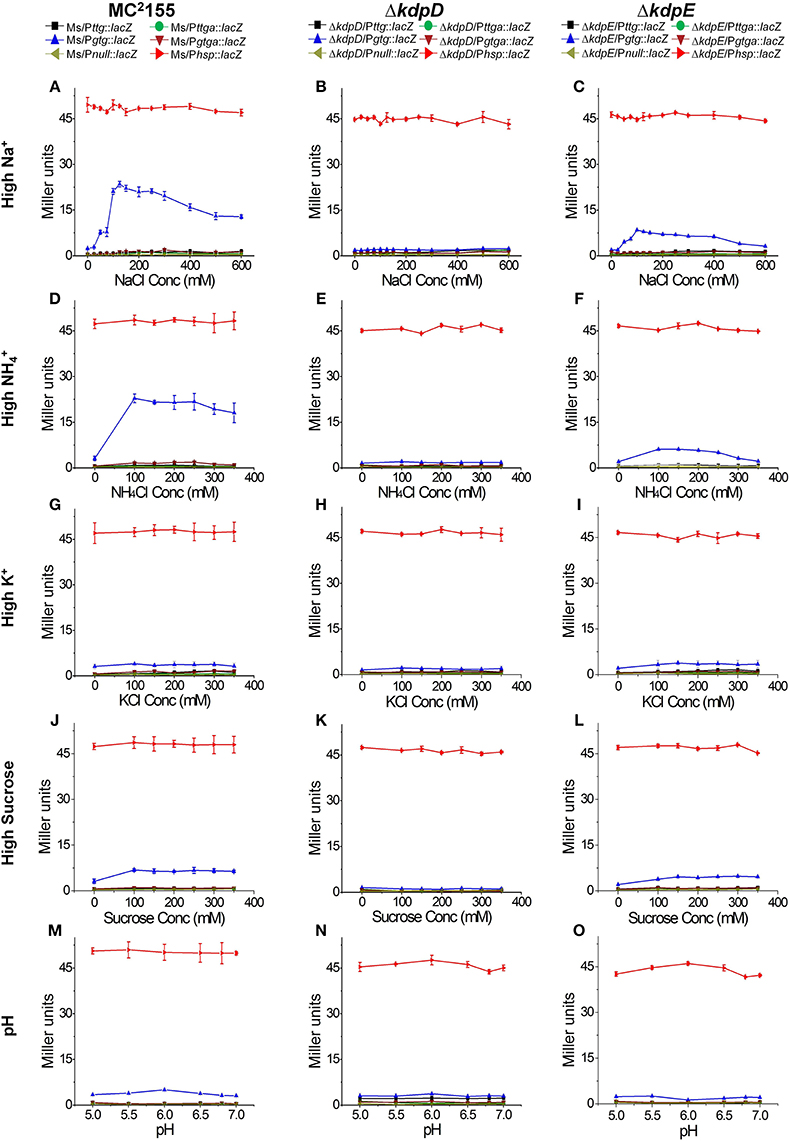
Figure 8. Promoter functionality and effect of different stimuli on kdpFABC transcription. The strains used are described in Figure 7. Strains were grown in 7H9 broth under different concentrations of NaCl (A–C), NH4Cl (D–F), KCl (G–I), sucrose (J–L), and pH-values (M–O). β-galactosidase activities were determined to investigate promoter functionality and expression of kdpFABC.
Requirement of KdpE for the Normal Growth of M. smegmatis
The growth of wild type as well as ΔkdpD and ΔkdpE mutant strains was investigated in two different growth media (7H9 and HdB). The growth patterns of wild type and ΔkdpD were mostly similar without much difference, whereas ΔkdpE showed a slightly faster growth rate (Figures 9A,B).
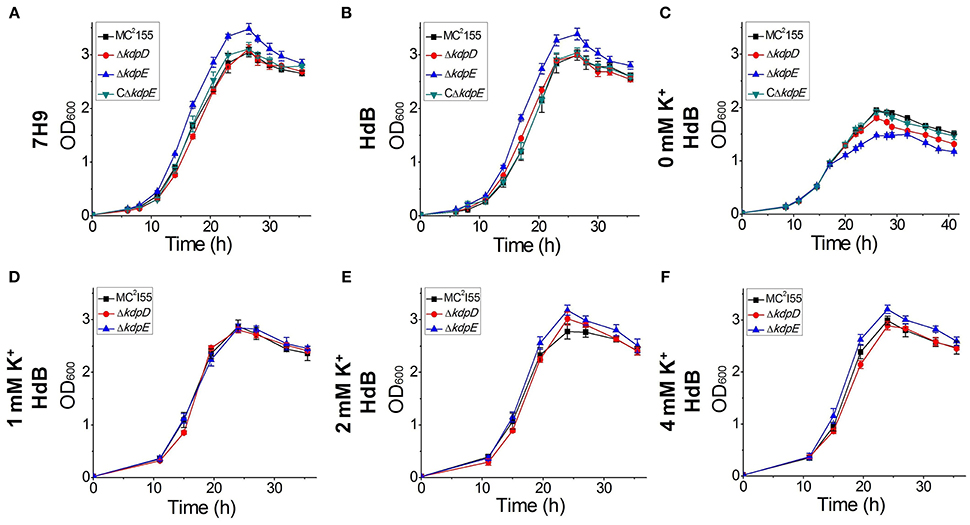
Figure 9. Growth curves of wild type MC2155 and its derivative strains under different concentrations of K+ in HdB medium. Bacterial growth curves were generated by growing all bacterial strains including wild type MC2155, ΔkdpD, ΔkdpE, and CΔkdpE, in 7H9 and HdB broth (with and without K+). Overnight cultures of all the strains were used to inoculate fresh media at a starting OD600 of 0.02. Each experiment was performed in triplicate with mean values shown in the graph. Cultures were incubated at 37°C under stirring condition for 40 h. OD600 was determined by sampling cultures in every 3 h. (A) Growth experiment for M. smegmatis and its derivatives was studied in 7H9 broth. Similarly, growth curves of M. smegmatis and its derivatives were also generated in HdB medium under different concentrations of K+ (B = normal, C = 0 mM, D = 1 mM, E = 2 mM, F = 4 mM).
Medium K+ concentrations influenced the growth rates of the wild type and mutant strains. At 0 mM K+ in HdB medium, the growth of all strains started with a relatively longer lag phase but shorter logarithmic phase in comparison to the growth in normal K+ condition. Besides, a noticeable defect was observed for the growth of ΔkdpE (Figure 9C). At 1 mM K+ concentration, all strains grew equally well with rates similar to those of the wild type in 7H9 and HdB media (Figure 9D). Increasing K+ up to 2 mM or above, the growth rates of ΔkdpE slightly raised as compared to the wild type and ΔkdpD (Figures 9E,F). Furthermore, to confirm whether this altered growth behavior of ΔkdpE strain was solely associated with kdpE deletion, we repeated the growth experiments in 7H9, HdB media (with normal K+) and HdB (with 0 mM K+) using the complementary strain of ΔkdpE. The growth curve of the CΔkdpE strain was found to be similar to the wild type strain (Figures 9A–C), confirming that the altered growth behavior of ΔkdpE was attributed to kdpE deletion.
Attempts were also made to detect responses of ΔkdpD and ΔkdpE to different stress conditions. For this, bacterial strains were exposed to various stress conditions for different time intervals and then plated onto 7H10 medium. No significant differences were observed in the growth patterns of wild type and mutant strains under most of the stresses studied. However, under the heat shock stress of 50°C for 5 h, ΔkdpE appeared to be slightly resistant. On the contrary, under hypo-osmotic stress condition in H2O and acidic pH 4.0 in citrate phosphate buffer for 5 h, ΔkdpE appeared to be slightly sensitive (Table S2). Overall these data demonstrated that KdpE was required for the normal growth of M. smegmatis.
Discussion
K+ plays many physiological roles in bacteria and its sufficient supply is essential for the pathogenicity of various species including M. tuberculosis (Salina et al., 2014). Upon K+ deficiency, bacterial cells of M. tuberculosis entered into a dormant non-culturable state but could be revitalized to culturable state once the availability of K+ was restored. Moreover, those non-culturable cells acquired resistance to the cell wall-targeting antimicrobials. Hence this resistance dormancy may raise the probability of latent tuberculosis (Salina et al., 2014). Regarding the important roles of K+ in mycobacterial physiology and pathogenicity, elucidation about K+ uptake and its regulatory mechanism would enable us to control mycobacterial infections in a more efficient way.
The species of genus Mycobacterium relies mainly on the Trk and Kdp systems for K+ uptake (Cholo et al., 2008). The Kdp-ATPase is regulated by KdpD/KdpE TCS, which was found to be functional in M. tuberculosis (Agrawal and Saini, 2014) and the bacterial strain became hyper-virulent upon its deletion (Parish et al., 2003). However, noticeable variations in the genetic organization of kdpFABC and kdpDE operons were observed within the genus Mycobacterium (Steyn et al., 2003; Cholo et al., 2008; Agrawal and Saini, 2014) (Figure S8). Some of the pathogenic species of genus Mycobacterium such as M. leprae and M. ulcerans are even deprived of Kdp-ATPase system (Cholo et al., 2008). Since different species of the genus Mycobacterium pose serious threats to a wide variety of organisms, the mechanism of K+ uptake within this genus needs to be elucidated in detail in order to cope with their pathogenic potential. To date, regulation of Kdp-ATPase has not been thoroughly investigated in M. smegmatis, a model species that has been used as a surrogate for pathogenic mycobacterial species. This manuscript focused on the regulation of kdpFABC operon by KdpD/KdpE TCS and, for the first time, proposed a detailed regulatory mechanism in M. smegmatis (Figure 10). These findings shall provide a foundation to further explore the mechanism of K+ uptake by Kdp-ATPase in other pathogenic species of genus Mycobacterium.
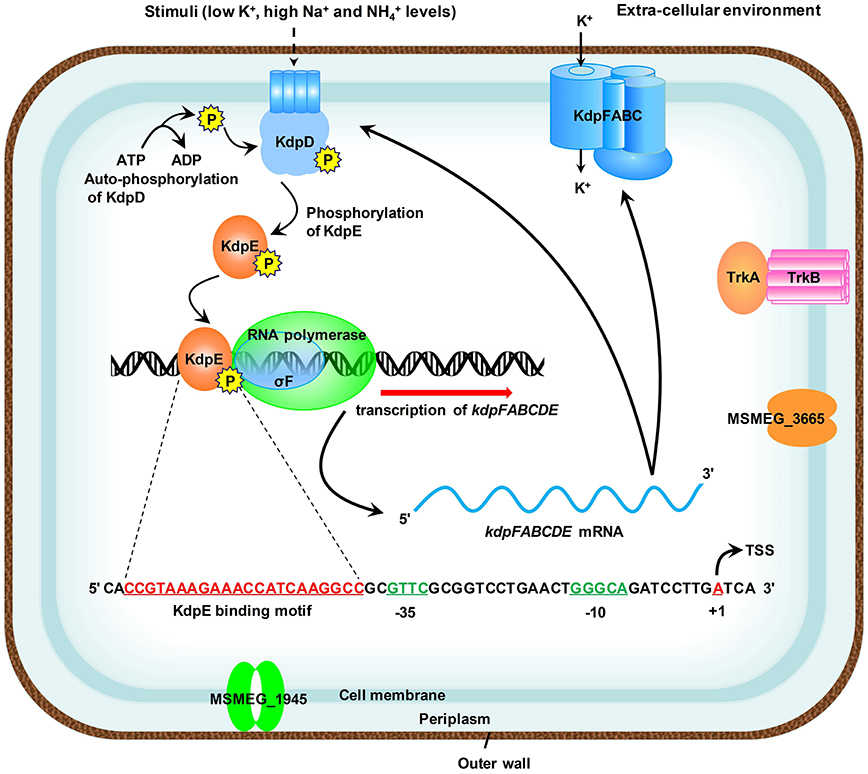
Figure 10. Proposed schematic representation of the induction and transcription of kdpFABC operon in M. smegmatis. Membrane bound KdpD senses stimuli such as low K+, high Na+, and concentrations of the medium. KdpE acts as a transcriptional regulator and is phosphorylated by KdpD to positively regulate the transcription of kdpFABC operon along with SigF-dependent RNA polymerase in a cooperative way. The 22-bp KdpE binding sequence is shown in red and SigF-dependent promoter consensus −35 and −10 regions are shown in green. The TSS nucleotide is shown in red with bend arrow. The Trk K+ uptake system and other possible channels are also shown in the diagram.
In M. smegmatis, it was found that the expression of kdpFABC operon was regulated by a KdpD/KdpE TCS that was induced under K+ limiting condition. Similar phenomenon was also reported in other bacteria (Altendorf et al., 1992; Frymier et al., 1997; Treuner-Lange et al., 1997). The KdpE binding sequence in PkdpF was found to be well-conserved in various mycobacterial species (Figure 4). However, no such sequence similarity could be detected in M. tuberculosis. Our results support the previous study that attempted to reveal binding of KdpE~P to the 192-bp intergenic region between kdpFABC and kdpDE in M. tuberculosis, although no such binding was observed (Agrawal and Saini, 2014). Contrary to M. tuberculosis, the direct binding of KdpE in PkdpF was confidentially observed in M. smegmatis in the present study.
Initially, candidate PkdpF (upstream of MSMEG_5391) was used for EMSA and DNase-I foot printing studies, considering MSMEG_5391 as a part of the kdp operon. However, the newly identified TSS was the fourth nucleotide of MSMEG_5391 hence, we believed that the original annotation of ORF (MSMEG_5391) might be inaccurate since the first three nucleotides of this frame were not the part of mRNA. The downward sequence of MSMEG_5391 was therefore searched for another potential ORF of kdpF gene. The sequence similarity of this newly identified kdpF of M. smegmatis with those of other species in genus Mycobacterium was noteworthy, although in many mycobacterial species, kdpF is still un-annotated. Our findings provide a strong foundation to identify and characterize kdpF in other species of genus Mycobacterium. Previously, KdpF was reported as a regulatory peptide (Gannoun-Zaki et al., 2013, 2014), hence its role under stress conditions needs further investigations in M. smegmatis.
The sigF loci in M. smegmatis encoded an alternative stress-response sigma factor F (SigF) (Gebhard et al., 2008). Previously, SigF-dependent promoters were first identified in M. tuberculosis (Rodrigue et al., 2007), and also characterized in M. smegmatis (Gebhard et al., 2008) in later studies. A GTTT-N(15−17)-GGGTA motif was predicted as SigF-dependent promoter consensus after inspecting promoter sequences of numerous SigF-regulated genes through bioinformatics analysis by the same group (Hümpel et al., 2010). In our study, we found that the −10 and −35 consensus sequences of PkdpF were highly similar to numerous SigF-regulated sequences so far identified, but with a smaller N numbers (GTTC-N(13)-GGGCA). In the previous study, PkdpF was not identified as a SigF-regulated promoter (Hümpel et al., 2010). This difference might be due to two reasons. First, the previous conclusion was based on different growth phases, whereas extremely low K+ concentration or high salt stress are required for the activation of kdpFABC transcription. Since their microarray study was performed under a non-inducible condition, kdpFABC might not be expressed at all. Second, different criteria were adopted in choosing spacing between the −10 and −35 regions of the SigF-dependent promoter consensus.
Ionic strength has been reported as stimulus for the induction of Kdp system. For instance, the Kdp system was found to be stimulated at high Na+ and concentrations in E. coli (Jung et al., 2001; Hamann et al., 2008; Epstein, 2016). Similar to these findings, Kdp-ATPase of M. smegmatis was also induced under high concentrations of Na+ and salts. Results of our RT-qPCR data further verified the induction of Kdp system under Na+ and salts stresses (Figure S9). In the case of M. tuberculosis, bacterial systems related to K+ uptake were induced at acidic pH (Cholo et al., 2015). Similarly, in E. coli, the expression of kdpFABC also altered due to the change in the medium pH (Asha and Gowrishankar, 1993). However, we found that the Kdp-ATPase system of M. smegmatis was not induced by medium pH change.
In our study on M. smegmatis, a single transcript of kdpFABCDE was observed under K+ limiting condition. In E. coli, however, it was proposed that some long transcripts were made to contain transcripts of both operons but most of the transcripts stopped before the start of kdpD (Altendorf et al., 1992). The single transcript (kdpZYABCXDE) in K+ limiting condition was reported in Clostridium acetobutylicum (Treuner-Lange et al., 1997) and (kdpABC-DE) in Acinetobacter baumannii (Samir et al., 2016). In case of M. smegmatis, it could be proposed that under low K+ concentration, both operons (kdpFABC and kdpDE) were co-transcribed at least to some extent from PkdpF, whereas under normal K+ condition, kdpDE was transcribed to very basal level and this transcription was initiated from PkdpD.
Kdp-ATPase and its regulatory KdpD/KdpE TCS have been reported in many bacterial species including E. coli, S. aureus, C. acetobutylicum, M. tuberculosis, S. typhimurium, Leptospira interrogans, Alicyclobacillus acidocaldarius, and A. baumannii for its role in K+ uptake (Ballal et al., 2007; Xue et al., 2011; Matsunaga and Coutinho, 2012; Samir et al., 2016). In bacterial species such as E. coli, L. interrogans, and A. baumannii, KdpE was highly expressed in order to positively regulate expression of kdpFABC operon under low K+ concentrations (Polarek et al., 1992; Matsunaga and Coutinho, 2012; Samir et al., 2016) except in S. aureus where KdpE negatively regulated kdpFABC expression (Xue et al., 2011). In M. tuberculosis, KdpD/KdpE TCS was found to be functional (Agrawal and Saini, 2014) and it was reported that KdpD interacted with two membrane lipoproteins (LprF and LprJ) to modulate expression of kdpFABC controlled by KdpE under low K+ concentrations (Steyn et al., 2003). At present, KdpE binding sequence in the PkdpF has only been identified in four bacterial species including E. coli (Sugiura et al., 1992), C. acetobutylicum (Behrens and Duerre, 2000), S. aureus (Xue et al., 2011), and M. smegmatis (current study). The C. acetobutylicum KdpE binding motif shared significant sequence similarity with that of E. coli (23 bp T-rich). However, M. smegmatis KdpE binding motif (22 bp A-rich) did not show noticeable sequence similarity with any other previously reported KdpE binding motif (Figure 4B).
In M. smegmatis, bioinformatics study revealed another constitutively expressed system for K+ uptake, namely the Trk system comprised TrkA (MSMEG_2771) and TrkB (MSMEG_2769) subunits. In addition, TrkA domain-containing protein (MSMEG_3665) and ion channel membrane protein (MSMEG_1945) were found as possible K+ channels. Expression of these genes was down-regulated in K+ limiting (0 mM K+) condition when compared to K+ non-limiting conditions (normal K+) in wild type and mutant strains (Figure S10). These results indicated that under K+ limiting condition, the Kdp-ATPase appeared to be functional to scavenge K+ as compared to other K+ systems.
It can be concluded that in M. smegmatis, Kdp-ATPase plays a vital role in the bacterial physiology during K+ limitation. The system is positively regulated by KdpD/KdpE TCS and triggered for osmo-regulation during low K+ and salt stress. The regulation of kdpFABC operon via KdpD/KdpE TCS appeared to be dependent upon SigF. Based on growth discrepancies of ΔkdpD and ΔkdpE, it can be proposed that some other histidine kinases may also activate KdpE, which in return regulates genes other than kdpFABC. However, a detailed study is required to verify this hypothesis.
For the first time, DNA sequence required for successful binding of KdpE with PkdpF was identified in genus Mycobacterium. Based on sequence similarity of the DNA motif, the binding of KdpE protein to positively regulate Kdp-ATPase can be expected in other mycobacterial species such as M. avium, M. fortuitum, M. marinum, and M. chubuense. On the other hand, a different mechanism might be speculated for the regulation of Kdp-ATPase in M. tuberculosis and M. bovis, which lack this KdpE-binding DNA motif and disclose different genomic organization for the kdpFABC and kdpDE operons.
In the present manuscript, we also observed a comparatively long 5′-UTR region in the transcript of kdpFABC for M. smegmatis. It would be worthy of further studies to see whether this 5′-UTR region exhibits some kind of regulatory role for the translation of Kdp-ATPase transporter.
Author Contributions
MKA performed most of the experiments and made most of the data evaluation. XLi, QT, XLiu, FC, JX and MA participated in partial experiments and interpretation of the data. MKA, QT, and JH conceived the study and drafted the manuscript. JH, SC, and MA revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jamshaid Hussain for critically reading the manuscript. This study was supported by the National Natural Science Foundation of China (grant 31270105), the Fundamental Research Funds for the Central Universities (grant 2662015PY175) the National Basic Research Program of China (973 Program, grant 2010CB126105), and the National High-tech R&D Program of China (grant 2011AA10A205) to JH, and in part by the Ministry of Education, Taiwan, under the ATU plan to SC.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00570/full#supplementary-material
Figure S1. Schematic diagram of kdpD gene knock-out procedure in M. smegmatis MC2155. The kdpD mutation was carried out by the method of homologous recombination as described earlier (Yang et al., 2012). The 800-bp upstream and downstream regions flanking the target kdpD gene were amplified by PCR, purified and transferred into pMD19-T for sequence verification. These fragments were cloned into pMind vector (containing a hygromycin marker) at specific cloning sites. For ΔkdpD, hygromycin marker was excised from vector to create unmarked mutation. The resulting plasmid containing upstream and downstream regions was digested with PacI enzyme to insert the sacB-lacZ cassette from pGOAL17. This final suicide delivery vector was then electroporated into M. smegmatis MC2155 competent cells after 5 min direct exposure to UV radiation, and plated on 7H10 medium containing kanamycin (30 μg/mL) and X-gal (50 μg/mL) for single crossover (SCO). The obtained SCO colonies were allowed to grow in 7H9 broth without antibiotics to facilitate double crossover (DCO). One hundred microliters of culture was then plated on 7H10 medium containing 10% sucrose and X-gal (50 μg/mL). Resulting white colonies were tested for kanamycin susceptibility.
Figure S2. Schematic diagram of kdpE gene knock-out procedure in M. smegmatis MC2155. The kdpE knock-out was performed by the method of homologous recombination as described earlier (Yang et al., 2012). The 800-bp upstream and downstream regions flanking the target kdpE gene were amplified by PCR, purified and transferred into pMD19-T for sequence verification. These fragments were cloned into pMind vector (containing a hygromycin marker) at specific cloning sites. For kdpE gene, marked mutation method was used. The resulting plasmid containing upstream and downstream regions was digested with PacI enzyme to insert the sacB-lacZ cassette from pGOAL17. This final suicide delivery vector was then electroporated into M. smegmatis MC2155 competent cells after 5 min direct exposure to UV radiation and plated on 7H10 medium containing kanamycin (30 μg/mL), X-gal (50 μg/mL), and hygromycin B (100 μg/mL) for SCO. The obtained SCO colonies were allowed to grow in 7H9 broth without antibiotics to facilitate DCO and 100 μl culture was spread over 7H10 medium containing 10% sucrose, X-gal (50 μg/mL) and hygromycin B (100 μg/mL). Resulting white colonies were tested for kanamycin susceptibility.
Figure S3. Construction of complementary strain of kdpE. Promoter region (PkdpD) was amplified by PCR using CP-D-F and CP-D-R primers (Table S1) and confirmed by sequencing. The hsp60 promoter (Phsp) in expression plasmid pMV261 (Yang et al., 2012) was replaced by PkdpD followed by insertion of kdpE gene that was amplified by primers C-kdpE-F and C-kdpE-R (Table S1). The resulting recombinant plasmid was confirmed by restriction endonucleases and PCR amplification. Plasmid was then electroporated into ΔkdpE competent cells. Resulting colonies were plated on 7H10 with kanamycin (30 μg/mL), and confirmed by RT-PCR for the expression of kdpE gene in CΔkdpE strain.
Figure S4. Heterologous expression of KdpE in E. coli BL21(DE3)-pET28-kdpE. KdpE protein from M. smegmatis genome was amplified and cloned into pET28a(+) to obtain expression plasmid pET28-kdpE that was transferred to E. coli BL21(DE3) to obtain the over-expression strain BL21(DE3)-pET28-kdpE. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the culture when OD600 reached to 0.5 and incubated at 28°C for 12 h for protein expression. Cells were harvested and re-suspended in lysis buffer (20 mM imidazole, 20 mM tris-base, 1 M NaCl) and lysed by ultrasonication. Crude cell lysate was centrifuged to remove debris and supernatant was used to purify 6 × His KdpE by using affinity column (Ni-NTA, GenScript). Cell lysate and purified KdpE protein were mixed with 2 × SDS loading buffer, boiled and centrifuged to run on 12% SDS PAGE. Lane 1: cell lysate, lane 2: protein marker, lane 3: purified KdpE protein (28.5 kDa).
Figure S5. Co-transcription of kdpFABC and kdpDE operons under K+ limiting conditions. Genomic organization of the kdpFABC and kdpDE genes and small regions amplified in RT-PCR are shown by numbered short lines. Amplified PCR products obtained from K+ limiting cDNA were separated on 1% agarose gel. CK+ denotes amplification of a segment of sigA which was used as positive control, while CK− denotes PCR mixture without cDNA which was used as negative control.
Figure S6. Promoter functionality β-galactosidase assays. (A) About 500 bp upstream region of MSMEG_5391 was taken as candidate promoter (P5391) and fused with promoter less lacZ gene to construct plasmid pMV261-P5391::lacZ. Newly constructed plasmid pMV261-P5391::lacZ was transformed into wild type and ΔkdpE. β-galactosidase activity was determined under normal and 0 mM K+ condition. (B) To study induction of promoter of kdpDE (PkdpD) under normal and 0 mM K+, about 500 bp upstream region of kdpDE operon was fused with promoter less lacZ and the newly constructed plasmid pMV261-PkdpD::lacZ was transformed into wild type and ΔkdpE. β-galactosidase activity was determined under normal and 0 mM K+ condition. Data represents the averages of biological triplicates. Error bars indicate standard deviation.
Figure S7. Design of promoter::lacZ vectors constructed for the identification of exact promoter sequence and kdpF identification. (A) The organization of annotated MSMEG_5391 with nucleotide sequence is shown in green. The start and termination codons of annotated MSMEG_5391 are boxed in black. (B) The 300 bp candidate promoter region upstream of annotated MSMEG_5391 gene with the start codon of MSMEG_5391 (TTG) fused with lacZ. (C) The 300 bp candidate promoter region upstream of MSMEG_5391 with the start codon of MSMEG_5391 plus one extra nucleotide (TTGA) to induce a frame-shifted lacZ. (D) The organization of newly proposed kdpF with nucleotide sequence is shown in magenta. The start and termination codons of newly proposed kdpF are boxed in black. (E) The PkdpF plus 5′-UTR of newly proposed kdpF with start codon (GTG) fused with lacZ. (F) The PkdpF plus 5′-UTR of newly proposed kdpF with start codon plus one extra nucleotide (GTGA) to induce frame shifted lacZ.
Figure S8. Organization of kdpFABC and kdpDE operons in different mycobacterial species. The positioning of kdpFABC and kdpDE operons in different mycobacterial species are shown in gene locus tags. Colored arrows showed different genes, with kdpF in magenta, kdpA in light green, kdpB in light orange, kdpC in light blue, kdpD in red, and kdpE in dark blue. Arrow head represents gene direction. Most of the kdpF sequences shown are identified in this study by sequence homology, and locus tags of kdpF are designated only to those species in which kdpF was previously and correctly annotated by NCBI/KEGG. In all species of mycobacteria that are analyzed in this study, the kdpDE operon is located adjacent to kdpFABC operon with the exception of M. chubuense, in which two operons are distantly located in same orientation. In case of M. chubuense, two slanted red lines indicate that two operons are not adjacent to each other.
Figure S9. Induction of kdpF and kdpDE genes under osmotic stress. Wild type was grown under normal (7H9) and salt stress conditions (150 mM NaCl, and 150 mM NH4Cl). Total RNA was extracted to synthesize cDNA. Relative expressions of kdpF, kdpD, and kdpE genes were studied by RT-qPCR using the 2−ΔΔCt method. Data represent the averages of biological triplicates. Error bars indicate standard deviation. sigA was used as reference gene. Wild type strain grown under normal conditions (7H9 medium) was used as control to compare relative expressions.
Figure S10. Expression of other K+ uptake loci in wild type and kdp mutants. Genes of Trk and Trk related system such as trkA (MSMEG_2771), trkB (MSMEG_2769), TrkA domain-containing protein (MSMEG_3665), and ion channel membrane protein (MSMEG_1945) were studied in this experiment. Wild type, ΔkdpD and ΔkdpE were grown under normal and low K+ condition. Total RNA was extracted to synthesize cDNA. Relative expression was studied using the 2−ΔΔCt method. Data represents the averages of biological triplicates. Error bars indicate standard deviation. sigA was used as a reference gene. Relative expression of above genes under K+ limiting conditions was determined in comparison with normal K+ conditions.
Table S1. List of primers used in this study.
Table S2. Responses of ΔkdpE under various stress conditions.
References
Agrawal, R., and Saini, D. K. (2014). Rv1027c-Rv1028c encode functional KdpDE two-component system in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 446, 1172–1178. doi: 10.1016/j.bbrc.2014.03.066
Alegado, R. A., Chin, C.-Y., Monack, D. M., and Tan, M.-W. (2011). The two-component sensor kinase KdpD is required for Salmonella typhimurium colonization of Caenorhabditis elegans and survival in macrophages. Cell Microbiol. 13, 1618–1637. doi: 10.1111/j.1462-5822.2011.01645.x
Altendorf, K., Gassel, M., Puppe, W., Mollenkamp, T., Zeeck, A., Boddien, C., et al. (1998). Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. Suppl. 643, 137–146.
Altendorf, K., Siebers, A., and Epstein, W. (1992). The KDP ATPase of Escherichia coli. Ann. N. Y. Acad. Sci. 671, 228–243. doi: 10.1111/j.1749-6632.1992.tb43799.x
Alvarado-Esquivel, C., Garcia-Corral, N., Carrero-Dominguez, D., Enciso-Moreno, J. A., Gurrola-Morales, T., Portillo-Gomez, L., et al. (2009). Molecular analysis of Mycobacterium isolates from extrapulmonary specimens obtained from patients in Mexico. BMC Clin. Pathol. 9:1. doi: 10.1186/1472-6890-9-1
Asha, H., and Gowrishankar, J. (1993). Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J. Bacteriol. 175, 4528–4537. doi: 10.1128/jb.175.14.4528-4537.1993
Ballal, A., Basu, B., and Apte, S. K. (2007). The Kdp-ATPase system and its regulation. J. Biosci. 32, 559–568. doi: 10.1007/s12038-007-0055-7
Ballal, A., Heermann, R., Jung, K., Gassel, M., Apte, S. K., and Altendorf, K. (2002). A chimeric Anabaena/Escherichia coli KdpD protein (Anacoli KdpD) functionally interacts with E. coli KdpE and activates kdp expression in E. coli. Arch. Microbiol. 178, 141–148. doi: 10.1007/s00203-002-0435-1
Behrens, M., and Düerre, P. (2000). KdpE of Clostridium acetobutylicum is a highly specific response regulator controlling only the expression of the kdp operon. J. Mol. Microbiol. Biotechnol. 2, 45–52.
Berry, S., Esper, B., Karandashova, I., Teuber, M., Elanskaya, I., Rögner, M., et al. (2003). Potassium uptake in the unicellular cyanobacterium Synechocystis sp strain PCC 6803 mainly depends on a Ktr-like system encoded by slr1509 (ntpJ). FEBS Lett. 548, 53–58. doi: 10.1016/S0014-5793(03)00729-4
Best, C. A., and Best, T. J. (2009). Mycobacterium smegmatis infection of the hand. Hand 4, 165–166. doi: 10.1007/s11552-008-9147-6
Bharati, B. K., Swetha, R. K., and Chatterji, D. (2013). Identification and characterization of starvation induced msdgc-1 promoter involved in the c-di-GMP turnover. Gene 528, 99–108. doi: 10.1016/j.gene.2013.07.043
Blokpoel, M. C. J., Murphy, H. N., O'Toole, R., Wiles, S., Runn, E. S. C., Stewart, G. R., et al. (2005). Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 33:e22. doi: 10.1093/nar/gni023
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brandon, L., Dorus, S., Epstein, W., Altendorf, K., and Jung, K. (2000). Modulation of KdpD phosphatase implicated in the physiological expression of the Kdp ATPase of Escherichia coli. Mol. Microbiol. 38, 1086–1092. doi: 10.1046/j.1365-2958.2000.02219.x
Bretl, D. J., Demetriadou, C., and Zahrt, T. C. (2011). Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582. doi: 10.1128/MMBR.05004-11
Burda, W. N., Miller, H. K., Krute, C. N., Leighton, S. L., Carroll, R. K., and Shaw, L. N. (2014). Investigating the genetic regulation of the ECF sigma factor σS in Staphylococcus aureus. BMC Microbiol. 14:280. doi: 10.1186/s12866-014-0280-9
Buurman, E. T., Kim, K. T., and Epstein, W. (1995). Genetic evidence for two sequentially occupied K+ binding sites in the KDP transport ATPase. J. Biol. Chem. 270, 6678–6685. doi: 10.1074/jbc.270.12.6678
Cholo, M. C., Van Rensburg, E. J., and Anderson, R. (2008). Potassium uptake systems of Mycobacterium tuberculosis: genomic and protein organisation and potential roles in microbial pathogenesis and chemotherapy. South Afr. J. Epidemiol. Infect. 23, 13–16. doi: 10.1080/10158782.2008.11441327
Cholo, M. C., van Rensburg, E. J., Osman, A. G., and Anderson, R. (2015). Expression of the genes encoding the Trk and Kdp potassium transport systems of Mycobacterium tuberculosis during growth in vitro. Biomed Res. Int. 2015:608682. doi: 10.1155/2015/608682
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dinnbier, U., Limpinsel, E., Schmid, R., and Bakker, E. P. (1988). Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150, 348–357. doi: 10.1007/BF00408306
Diskowski, M., Mikusevic, V., Stock, C., and Hänel, I. (2015). Functional diversity of the superfamily of K+ transporters to meet various requirements. Biol. Chem. 396, 1003–1014. doi: 10.1515/hsz-2015-0123
Driks, M., Weinhold, F., and Cokingtin, Q. (2011). Pneumonia caused by Mycobacterium smegmatis in a patient with a previous gastrectomy. BMJ Case Rep. 2011:3281. doi: 10.1136/bcr.08.2010.3281
Epstein, W. (1986). Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Lett. 39, 73–78. doi: 10.1111/j.1574-6968.1986.tb01845.x
Epstein, W. (1992). Kdp, a bacterial P-type ATPase whose expression and activity are regulated by turgor pressure. Acta Physiol. Scand. Suppl. 607, 193–199.
Epstein, W. (2003). The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 75, 293–320. doi: 10.1016/S0079-6603(03)75008-9
Epstein, W. (2016). The KdpD sensor kinase of Escherichia coli responds to several distinct signals to turn on expression of the Kdp transport system. J. Bacteriol. 198, 212–220. doi: 10.1128/JB.00602-15
Epstein, W., and Schultz, S. G. (1965). Cation transport in Escherichia coli: V. Regulation of cation content. J. Gen. Physiol. 49, 221–234. doi: 10.1085/jgp.49.2.221
Feinbaum, R. L., Urbach, J. M., Liberati, N. T., Djonovic, S., Adonizio, A., Carvunis, A. R., et al. (2012). Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 8:e1002813. doi: 10.1371/journal.ppat.1002813
Follmann, M., Becker, M., Ochrombel, I., Ott, V., Krämer, R., and Marin, K. (2009). Potassium transport in Corynebacterium glutamicum is facilitated by the putative channel protein CglK, which is essential for pH homeostasis and growth at acidic pH. J. Bacteriol. 191, 2944–2952. doi: 10.1128/JB.00074-09
Freeman, Z. N., Dorus, S., and Waterfield, N. R. (2013). The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLoS Pathog. 9:e1003201. doi: 10.1371/journal.ppat.1003201
Frymier, J. S., Reed, T. D., Fletcher, S. A., and Csonka, L. N. (1997). Characterization of transcriptional regulation of the kdp operon of Salmonella typhimurium. J. Bacteriol. 179, 3061–3063. doi: 10.1128/jb.179.9.3061-3063.1997
Gannoun-Zaki, L., Alibaud, L., Carrère-Kremer, S., Kremer, L., and Blanc-Potard, A. B. (2013). Overexpression of the KdpF membrane peptide in Mycobacterium bovis BCG results in reduced intramacrophage growth and altered cording morphology. PLoS ONE 8:e60379. doi: 10.1371/journal.pone.0060379
Gannoun-Zaki, L., Belon, C., Dupont, C., Hilbert, F., Kremer, L., and Blanc-Potard, A. B. (2014). Overexpression of the Salmonella KdpF membrane peptide modulates expression of kdp genes and intramacrophage growth. FEMS Microbiol. Lett. 359, 34–41. doi: 10.1111/1574-6968.12559
Gassel, M., Möllenkamp, T., Puppe, W., and Altendorf, K. (1999). The KdpF subunit is part of the K+ translocating Kdp complex of Escherichia coli and is responsible for stabilization of the complex in vitro. J. Biol. Chem. 274, 37901–37907. doi: 10.1074/jbc.274.53.37901
Gassel, M., Siebers, A., Epstein, W., and Altendorf, K. (1998). Assembly of the Kdp complex, the multi-subunit K+-transport ATPase of Escherichia coli. Biochim. Biophys. Acta 1415, 77–84. doi: 10.1016/S0005-2736(98)00179-5
Gebhard, S., Hümpel, A., McLellan, A. D., and Cook, G. M. (2008). The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 154, 2786–2795. doi: 10.1099/mic.0.2008/018044-0
Greie, J. C. (2011). The KdpFABC complex from Escherichia coli: a chimeric K+ transporter merging ion pumps with ion channels. Eur. J. Cell Biol. 90, 705–710. doi: 10.1016/j.ejcb.2011.04.011
Greie, J. C., and Altendorf, K. (2007). The K+-translocating KdpFABC complex from Escherichia coli: a p-type ATPase with unique features. J. Bioenerg. Biomembr. 39, 397–402. doi: 10.1007/s10863-007-9111-0
Gumz, M. L., Rabinowitz, L., and Wingo, C. S. (2015). An integrated view of potassium homeostasis. N. Engl. J. Med. 373, 60–72. doi: 10.1056/NEJMra1313341
Hamann, K., Zimmann, P., and Altendorf, K. (2008). Reduction of turgor is not the stimulus for the sensor kinase KdpD of Escherichia coli. J. Bacteriol. 190, 2360–2367. doi: 10.1128/JB.01635-07
Hartmans, S., de Bont, J. A. M., and Stackebrandt, E. (2006). “The genus Mycobacterium-nonmedical,” in The Prokaryotes, Vol. 3, Archaea. Bacteria: Firmicutes, Actinomycetes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 889–918.
Haupt, M., Bramkamp, M., Coles, M., Kessler, H., and Altendorf, K. (2005). Prokaryotic Kdp-ATPase: recent insights into the structure and function of KdpB. J. Mol. Microbiol. Biotechnol. 10, 120–131. doi: 10.1159/000091559
Heermann, R., and Jung, K. (2010). The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS Microbiol. Lett. 304, 97–106. doi: 10.1111/j.1574-6968.2010.01906.x
Heermann, R., Weber, A., Mayer, B., Ott, M., Hauser, E., Gabriel, G., et al. (2009). The universal stress protein UspC scaffolds the KdpD/KdpE signaling cascade of Escherichia coli under salt stress. J. Mol. Biol. 386, 134–148. doi: 10.1016/j.jmb.2008.12.007
Heitzmann, D., and Warth, R. (2008). Physiology and pathophysiology of potassium channels in gastrointestinal epithelia. Physiol. Rev. 88, 1119–1182. doi: 10.1152/physrev.00020.2007
Hellman, L. M., and Fried, M. G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861. doi: 10.1038/nprot.2007.249
Hümpel, A., Gebhard, S., Cook, G. M., and Berney, M. (2010). The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J. Bacteriol. 192, 2491–2502. doi: 10.1128/JB.00035-10
Jung, K., and Altendorf, K. (2002). Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli. J. Mol. Microbiol. Biotechnol. 4, 223–228.
Jung, K., Krabusch, M., and Altendorf, K. (2001). Cs+ induces the kdp operon of Escherichia coli by lowering the intracellular K+ concentration. J. Bacteriol. 183, 3800–3803. doi: 10.1128/JB.183.12.3800-3803.2001
Kuo, M. M. C., Haynes, W. J., Loukin, S. H., Kung, C., and Saimi, Y. (2005). Prokaryotic K+ channels: from crystal structures to diversity. FEMS Microbiol. Rev. 29, 961–985. doi: 10.1016/j.femsre.2005.03.003
Laermann, V., Cudic, E., Kipschull, K., Zimmann, P., and Altendorf, K. (2013). The sensor kinase KdpD of Escherichia coli senses external K+. Mol. Microbiol. 88, 1194–1204. doi: 10.1111/mmi.12251
Laimins, L. A., Rhoads, D. B., and Epstein, W. (1981). Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 78, 464–468. doi: 10.1073/pnas.78.1.464
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lüettmann, D., Heermann, R., Zimmer, B., Hillmann, A., Rampp, I. S., Jung, K., et al. (2009). Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIA(Ntr) in Escherichia coli. Mol. Microbiol. 72, 978–994. doi: 10.1111/j.1365-2958.2009.06704.x
Matsunaga, J., and Coutinho, M. L. (2012). Positive regulation of Leptospira interrogans kdp expression by KdpE as demonstrated with a novel beta-galactosidase reporter in Leptospira biflexa. Appl. Environ. Microbiol. 78, 5699–5707. doi: 10.1128/AEM.00713-12
Nakashima, K., Sugiura, A., Kanamaru, K., and Mizuno, T. (1993). Signal transduction between the two regulatory components involved in the regulation of the kdpABC operon in Escherichia coli: phosphorylation-dependent functioning of the positive regulator, KdpE. Mol. Microbiol. 7, 109–116. doi: 10.1111/j.1365-2958.1993.tb01102.x
Nakashima, K., Sugiura, A., Momoi, H., and Mizuno, T. (1992). Phosphotransfer signal transduction between two regulatory factors involved in the osmoregulated kdp operon in Escherichia coli. Mol. Microbiol. 6, 1777–1784. doi: 10.1111/j.1365-2958.1992.tb01350.x
Nanatani, K., Shijuku, T., Takano, Y., Zulkifli, L., Yamazaki, T., Tominaga, A., et al. (2015). Comparative analysis of kdp and ktr mutants reveals distinct roles of the potassium transporters in the model Cyanobacterium synechocystis sp strain PCC 6803. J. Bacteriol. 197, 676–687. doi: 10.1128/JB.02276-14
Njoroge, J. W., Gruber, C., and Sperandio, V. (2013). The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli. J. Bacteriol. 195, 2499–2508. doi: 10.1128/JB.02252-12
Parish, T., Smith, D. A., Kendall, S., Casali, N., Bancroft, G. J., and Stoker, N. G. (2003). Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71, 1134–1140. doi: 10.1128/IAI.71.3.1134-1140.2003
Parish, T., and Stoker, N. G. (2000). Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975. doi: 10.1099/00221287-146-8-1969
Parker, C. T., Russell, R., Njoroge, J. W., Jimenez, A. G., Taussig, R., and Sperandio, V. (2017). Genetic and mechanistic analyses of the periplasmic domain of the enterohemorrhagic E. coli (EHEC) QseC histidine sensor kinase. J. Bacteriol. JB. 199:e00861-16. doi: 10.1128/JB.00861-16
Polarek, J. W., Williams, G., and Epstein, W. (1992). The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J. Bacteriol. 174, 2145–2151. doi: 10.1128/jb.174.7.2145-2151.1992
Price-Whelan, A., Poon, C. K., Benson, M. A., Eidem, T. T., Roux, C. M., Boyd, J. M., et al. (2013). Transcriptional profiling of Staphylococcus aureus during growth in 2 M NaCl leads to clarification of physiological roles for Kdp and Ktr K+ uptake systems. MBio 4, e00407–e00413. doi: 10.1128/mBio.00407-13
Rhoads, D. B., Waters, F. B., and Epstein, W. (1976). Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67, 325–341. doi: 10.1085/jgp.67.3.325
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rodrigue, S., Brodeur, J., Jacques, P. E., Gervais, A. L., Brzezinski, R., and Gaudreau, L. (2007). Identification of mycobacterial σ factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189, 1505–1513. doi: 10.1128/JB.01371-06
Salina, E. G., Waddell, S. J., Hoffmann, N., Rosenkrands, I., Butcher, P. D., and Kaprelyants, A. S. (2014). Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biol. 4:140106. doi: 10.1098/rsob.140106
Samir, R., Hussein, S. H., Elhosseiny, N. M., Khattab, M. S., Shawky, A. E., and Attia, A. S. (2016). Adaptation to potassium limitation is essential for Acinetobacter baumannii pneumonia pathogenesis. J. Infect. Dis. 214, 2006–2013. doi: 10.1093/infdis/jiw476
Sato, Y., Nanatani, K., Hamamoto, S., Shimizu, M., Takahashi, M., Tabuchi-Kobayashi, M., et al. (2014). Defining membrane spanning domains and crucial membrane-localized acidic amino acid residues for K+ transport of a Kup/HAK/KT-type Escherichia coli potassium transporter. J. Biochem. 155, 315–323. doi: 10.1093/jb/mvu007
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Smeulders, M. J., Keer, J., Speight, R. A., and Williams, H. D. (1999). Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181, 270–283.
Steyn, A. J. C., Joseph, J., and Bloom, B. R. (2003). Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol. Microbiol. 47, 1075–1089. doi: 10.1046/j.1365-2958.2003.03356.x
Stumpe, S., Schlösser, A., Schleyer, M., and Bakker, E. P. (1996). “K+ circulation across the prokaryotic cell membrane: K+ uptake systems,” in Handbook of Biological Physics, eds. W. N. Konings, H. R. Kaback, and J. S. Lolkema (Amsterdam: Elsevier Science B. V.), 473–499.
Sugiura, A., Nakashima, K., Tanaka, K., and Mizuno, T. (1992). Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 6, 1769–1776. doi: 10.1111/j.1365-2958.1992.tb01349.x
Tang, Q., Li, X. F., Zou, T. T., Zhang, H. M., Wang, Y. Y., Gao, R. S., et al. (2014). Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol. Microbiol. 94, 1006–1023. doi: 10.1111/mmi.12817
Tang, Q., Yin, K., Qian, H. L., Zhao, Y. W., Wang, W., Chou, S. H., et al. (2016). Cyclic di-GMP contributes to adaption and virulence of Bacillus thuringiensis through a riboswitch-regulated collagen adhesion protein. Sci. Rep. 6:28807. doi: 10.1038/srep28807
Treuner-Lange, A., Kuhn, A., and Durre, P. (1997). The kdp system of Clostridium acetobutylicum: cloning, sequencing, and transcriptional regulation in response to potassium concentration. J. Bacteriol. 179, 4501–4512. doi: 10.1128/jb.179.14.4501-4512.1997
Voelkner, P., Puppe, W., and Altendorf, K. (1993). Characterization of the KdpD protein, the sensor kinase of the K+ translocating Kdp system of Escherichia coli. Eur. J. Biochem. 217, 1019–1026. doi: 10.1111/j.1432-1033.1993.tb18333.x
Walderhaug, M. O., Polarek, J. W., Voelkner, P., Daniel, J. M., Hesse, J. E., Altendorf, K., et al. (1992). KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174, 2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992
Wallace, R. J. Jr., Nash, D. R., Tsukamura, M., Blacklock, Z. M., and Silcox, V. A. (1988). Human disease due to Mycobacterium smegmatis. J. Infect. Dis. 158, 52–59. doi: 10.1093/infdis/158.1.52
Xu, Z., Lu, D., Zhang, X., Li, H. J., Meng, S. F., Pan, Y. S., et al. (2013). Mycobacterium smegmatis in skin biopsy specimens from patients with suppurative granulomatous inflammation. J. Clin. Microbiol. 51, 1028–1030. doi: 10.1128/JCM.03421-12
Xue, T., You, Y. B., Hong, D., Sun, H. P., and Sun, B. L. (2011). The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect. Immun. 79, 2154–2167. doi: 10.1128/IAI.01180-10
Yang, M., Gao, C. H., Cui, T., An, J. N., and He, Z. G. (2012). A TetR-like regulator broadly affects the expressions of diverse genes in Mycobacterium smegmatis. Nucleic Acids Res. 40, 1009–1020. doi: 10.1093/nar/gkr830
Keywords: Mycobacterium smegmatis, two-component system (TCS), KdpD/KdpE, KdpFABC, Kdp-ATPase, potassium transporter, K+ limitation
Citation: Ali MK, Li X, Tang Q, Liu X, Chen F, Xiao J, Ali M, Chou S-H and He J (2017) Regulation of Inducible Potassium Transporter KdpFABC by the KdpD/KdpE Two-Component System in Mycobacterium smegmatis. Front. Microbiol. 8:570. doi: 10.3389/fmicb.2017.00570
Received: 26 January 2017; Accepted: 20 March 2017;
Published: 24 April 2017.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Hyunwoo Lee, University of Illinois at Chicago, USAAndreas Burkovski, University of Erlangen-Nuremberg, Germany
Copyright © 2017 Ali, Li, Tang, Liu, Chen, Xiao, Ali, Chou and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin He, hejin@mail.hzau.edu.cn
 Maria K. Ali
Maria K. Ali Xinfeng Li1
Xinfeng Li1 Muhammad Ali
Muhammad Ali Shan-Ho Chou
Shan-Ho Chou Jin He
Jin He