- 1Servei de Microbiologia, Hospital de la Santa Creu i Sant Pau, Institut d’Investigació Biomèdica Sant Pau, Barcelona, Spain
- 2Department of Microbiology, University of Barcelona, Barcelona, Spain
Bacteriophages, viruses that infect bacteria, have re-emerged as powerful regulators of bacterial populations in natural ecosystems. Phages invade the human body, just as they do other natural environments, to such an extent that they are the most numerous group in the human virome. This was only revealed in recent metagenomic studies, despite the fact that the presence of phages in the human body was reported decades ago. The influence of the presence of phages in humans has yet to be evaluated; but as in marine environments, a clear role in the regulation of bacterial populations could be envisaged, that might have an impact on human health. Moreover, phages are excellent vehicles of genetic transfer, and they contribute to the evolution of bacterial cells in the human body by spreading and acquiring DNA horizontally. The abundance of phages in the human body does not pass unnoticed and the immune system reacts to them, although it is not clear to what extent. Finally, the presence of phages in human samples, which most of the time is not considered, can influence and bias microbiological and molecular results; and, in view of the evidences, some studies suggest that more attention needs to be paid to their interference.
Introduction
Bacteriophages were discovered in the second decade of the 20th century (Twort, 1915; D’Herelle, 1917). It was initially suggested the idea they could be used to lyse pathogenic bacteria as a treatment of infectious diseases. However, the idea was rapidly abandoned in western countries due to the introduction of antibiotics. For decades, phages have been the most common model entities for the study of viruses and their replication cycles. Studies of certain model phages have contributed significantly to the advancement of molecular biology, for example in identifying the basis of genetic material, as the code of nucleotide triplets of individual amino acids (Crick et al., 1961), and the restriction enzymes (Dussoix and Arber, 1962). Moreover, the first sequenced genome was that of an Escherichia coli phage: ϕX174 (Sanger et al., 1977). For some years, the interest for phages was limited to ecological studies and proposals for their use as indicators of fecal pollution (IAWPRC Study Group on Health Related Water Microbiology, 1991; Jofre et al., 2016); in general, bacteriophages have deserved less interest in comparison to their bacterial hosts or to animal viruses.
Nevertheless, the remarkable estimated number of 1031 phages on the Earth (Suttle, 2005) is commonly used by researchers to highlight the importance of phages, which are believed to outnumber any other class of biological entity on the planet. Phages have recently re-emerged as powerful regulators of the bacterial populations in natural ecosystems (Fuhrman, 1999). Moreover, because of the appearance of resistances to different antimicrobial agents, their potential use as antimicrobials has been revisited (Reardon, 2014). Most significantly, recent metagenomes describe the abundance of viral sequences both outside and inside bacterial cells. This highlights their ubiquity as mobile genetic elements that contribute and affect bacterial evolution, causing the emergence of new bacterial pathogens, mobilizing genes outside the cells, and other functions. Interest in metagenomes includes the study of human microbiomes, where phages again appear as extremely abundant and diverse elements. Researchers are only now starting to suspect that phages actively contribute to the homeostasis of the bacterial flora (De Paepe et al., 2014). Because many studies focus on the role of the human symbiotic microbiota in our wellness, phages thus appear as contributing actors that are directly related with human health (Manrique et al., 2016), and therefore the interest in them is rising.
Phages as a Part of Human and Animal Microbiota
Many metagenomic analyses of human microbiomes show the abundance of phages, which is generally greater than that of eukaryotic viruses. This has been shown in metagenomic analysis of lung, vaginal, skin, oral or intestinal microbiota (Breitbart et al., 2003; Colomer-Lluch et al., 2011a; Minot et al., 2011; Oh et al., 2014; Virgin, 2014). More recently, infectious phages have been found in different clinical samples such as ascitic fluid and urine (Brown-Jaque et al., 2016). It was suggested that they could reach the peritoneal cavity after translocation from the intestine (Górski et al., 2006), where they are present (Figure 1) and abundant. They are also present in voided urine (Brown-Jaque et al., 2016), probably coming from the periurethral area. In animals, phages infecting Bacteroides were found in serum (Keller and Traub, 1974), confirming their presence in the blood stream. Translocation of phages from blood to mouse fetal tissues has also been demonstrated in pregnant mice (Srivastava et al., 2004a).
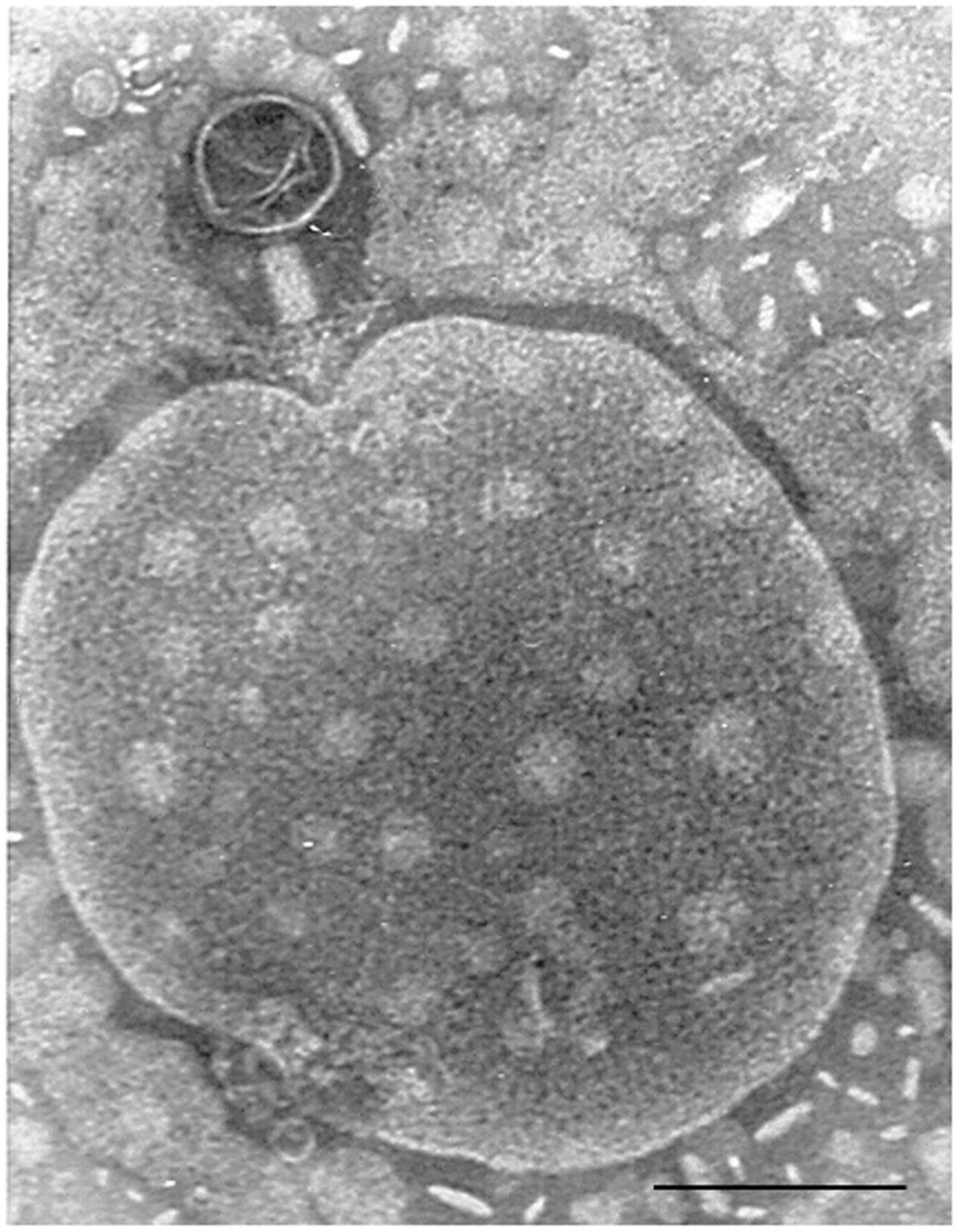
FIGURE 1. Bacteriophage of Myoviridae morphology isolated from a fecal sample, attached to an unidentified particle. Bar 100 nm.
In the light of these results, and as a second level of study, some researchers have analyzed solely the virome fraction of these microbiomes. To do this, they have devised methods that allow discrimination of the viral fraction, while discarding bacterial and free DNA. Those studies have yielded some surprising results; many viral particles in fact carry sequences identified as bacterial DNA. Shared genetic content is observed when analyzing the phage and bacterial DNA fractions of the same sample (Breitbart et al., 2003; Minot et al., 2011; Colombo et al., 2016; Howe et al., 2016), including sequences belonging to CRISPR-Cas systems (Dutilh et al., 2014).
CRISPR-Cas systems constitute a immune system that protect bacteria against bacteriophages and foreign DNA (Mojica and Rodriguez-Valera, 2016), that has later been applied for genome engineering in bacteria and eukaryotes. The different activity of the CRISPR-Cas systems influences the allowance of bacterial cells to foreign DNA or their immunity to phage infection, and this can shape the evolution of human microbiomes. Besides the use of CRISPR-Cas systems in genome engineering, the analysis of CRISPR sequences from raw metagenomic data has revealed unidentified phages, as crAssphage phage, that is claimed to be present in the majority of human fecal microbiomes, although it has never been isolated (Dutilh et al., 2014).
Phages as Mobile Genetic Elements
Transduction, the process by which the DNA is mobilized between cell by a virus or viral vector was reported the last century (Zinder and Lederberg, 1952), although the rates of this mobilization has never been well defined. For this reason, the detection of an important proportion of bacterial DNA in phage particles observed in metagenomic analysis was indeed a surprise, and it initially prompted the belief that the methods for segregating phage and bacterial particles were not accurate enough, and either bacterial or free DNA contaminated the phage samples. However, the protocols have been optimized allowing specific extraction of packaged DNA. Another suspicion is that the bioinformatic analysis failed to identify phage DNA sequences correctly and they were mistaken for bacterial DNA. Nevertheless, subsequent repetitions and more accurate approaches have shown that despite some of these problems occurring, a relevant fraction of the virome is actually mobilizing bacterial DNA. This has led to the suspicion that bacterial cells use the numerous capsid genes that they possess, probably inherited from ancient prophage remnants, to build protein capsids that pack and spread their DNA content (Asadulghani et al., 2009; Lang et al., 2012; Penades et al., 2015).
The fact that phage capsids can mobilize bacterial DNA has multiple consequences, such as, for example, the fact that they can mobilize and transduce virulence genes (O’Brien et al., 1984; Griffiths et al., 2000; Allué-Guardia et al., 2011; Penades et al., 2015), antibiotic resistances (Muniesa et al., 2004; Colomer-Lluch et al., 2011b; Ross and Topp, 2015; Haaber et al., 2016) or genes related to fitness (Lindell et al., 2004; Müller et al., 2013) to new bacterial hosts. This causes horizontal genetic exchange and leads to the evolution of bacterial populations.
Phages as Regulators of Populations
Bacterial populations can change and evolve through acquisition of new genes transferred by phages, but also by predation and lysis caused by phages. Experimental evidence from chemostats and observations of phages/hosts in open systems has shown that for some bacterial species, populations of phages and hosts oscillate over time, following a “Red Queen/kill-the-winner” dynamics,” which describes prey–predator variations (Rodriguez-Brito et al., 2010; Jover et al., 2013; Lim et al., 2015). However, phage–host dynamics can change in accordance with the homogeneity and structure of the environment, and also depending on the conditions that facilitate phage–cell encounters (De Paepe et al., 2014).
Changes or a total replacement of the microbiome by a fecal transplant in diseases without a well-defined etiological agent, such as inflammatory bowel diseases (Crohn’s disease or ulcerative colitis), can result in different disease outcomes (Loh and Blaut, 2012; Moayyedi et al., 2015). Comparison of the viromes of individuals suffering from Crohn’s disease and healthy relatives revealed differences in composition and variability (Pérez-Brocal et al., 2013; Wagner et al., 2013). Whether changes in the phagome of human biomes is a cause or a consequence of dysbiosis in such diseases has not yet been established. Considering that the phagome could influence bacterial populations, two options are plausible: changes in bacteria could cause variations in the distribution of phage groups; or changes in the phagome could be responsible of dysbacteriosis (Norman et al., 2015; Pérez-Brocal et al., 2015).
Similarly, phages have been detected in the metagenomes of sputum of patients suffering cystic fibrosis (Willner et al., 2009); and both the phage diversity and relative abundances were reported to be different from those of non-cystic fibrosis patients. It is hard, however, to conclude from these results what the cause of these differences is. Some variations in bacteria are caused by phages and those variations could be harmful to the patients. For example, mucoid isolates of Pseudomonas fluorescens are more virulent than their non-mucoid isogenic variants. This mucoid overproduction is a virulence factor contributing to more persistent infections in cystic fibrosis patients (Scanlan and Buckling, 2012). This phenotypic characteristic is favorably selected in the presence of phages, because it confers protection against phage infection. Accordingly, the mucoid isolates became resistant to the phages with the corresponding detrimental consequence for the patients (Scanlan and Buckling, 2012).
A different example of the regulation of human bacterial populations by phages is observed when we look at the competition between Streptococcus pneumoniae and Staphylococcus aureus. The former produces hydrogen peroxide; an agent that induces the bacterial SOS response and can induce temperate prophages. Meanwhile, the vast majority of S. aureus strains carry prophages that could be induced in the presence of the concentrations of H2O2 produced by S. pneumoniae. S. pneumoniae prophages, in turn, are not induced at these concentrations. The result is that S. pneumoniae prevails by killing S. aureus lysogenic strains via induction of prophages that cause the subsequent lysis of the cell (Selva et al., 2009).
Yet another example of how bacteriophages can impact the dynamics of bacterial populations has been observed in Enterococcus faecalis V583. This strain produces a composite phage ΦV1/7, derived from two distinct chromosomally encoded prophage elements. Prophage ΦV1 produces the capsids, while prophage ΦV7 is in charge of infection of susceptible hosts and V583 can produce infectious ΦV1/7. The induction of ΦV1/7 is highly enhanced by the availability of free amino acids in the medium. The strain producing ΦV1/7 has an advantage over other E. faecalis strains in the intestine, because these are lysed by ΦV1/7, while V583 is resistant to superinfection, enhancing the success of E. faecalis V583 during competitive growth (Duerkop et al., 2012).
Interactions With the Immune System
It is not clear whether phages can easily be detected by the immune system, or whether they interact with it. Because the size of phage particles is usually bigger than eukaryotic viruses, activation of the immune system might occur as for other viruses. The desire to use phages to treat bacterial infections has led to explorations of the responses that phages might cause within the human immune system.
Very soon after the discovery of phages, it was observed that antibodies against bacteriophages in humans or animals were produced (Jerne, 1952, 1956); and it is easy to generate phage antiserums by immunization of humans or animals with phage lysates (Puig et al., 2001; Gorski et al., 2012; Bacon et al., 2017). The sera of non-immunized individuals (humans or animals) present antibodies against phages, although at low levels; the so-called “natural antibodies.” For instance, antibodies against T4 phages are naturally present in human serum (Dabrowska et al., 2014) presumably as a consequence of the confirmed constant presence of phages in human biomes (Górski et al., 2006; Brown-Jaque et al., 2016). However, the origin of natural antibodies, generally of IgM class, with broad cross reactivity and low affinity, is not clear in the majority of cases.
The innate immune system, particularly by the components of the reticuloendothelial system (RES), could be a mechanism for removing phages that are circulating in the human body (Gorski et al., 2012). Certainly, this system was credited with the rapid removal of administered wild-type phage λ from the circulatory system in humans (Geier et al., 1973). Moreover, different phage λ mutants could induce different host responses. When using certain phage λ mutants that were capable of circumventing the RES immune response, these mutants prevailed for longer periods in the blood stream than the wild-type phage (Merril et al., 1996).
Data on anti-phage cellular responses are very scarce in comparison with data on phage–humoral responses. The only study we are aware of, evaluated the cellular response to MS2 phage that was intradermally administrated in guinea pigs. The presence of the phage produced erythema and induration, that are signs of cell-mediated immunity (Langbeheim et al., 1978). In contrast, another study showed that the permanence of phages in blood is the same when comparing immunocompetent mice or those deficient in T-cells, indicating no specific role of T-cell response in phage inactivation (Srivastava et al., 2004b).
When administered together with the host bacteria, some studies showed that phages seem to stimulate bacterial phagocytosis, and this is attributed to certain “opsonization” of the bacterial cells by phages. In addition, phages can remain active and infective when adsorbed onto the bacteria on intake by granulocytes. Therefore, some authors have suggested that during phagocytosis, phages continue lysing the phagocytosed bacteria, helping the activity of phagocytic cells. This process is limited in time and phages are no longer active after the completion of phagocytosis (Gorski et al., 2012). Despite these descriptions, there is no definitive evidence that phages activate phagocytosis by themselves, and some years ago, a contrary outcome was reported (Kantoch et al., 1958). In those studies, when used at very high doses (1010/ml), phages inhibited phagocytosis of their host bacteria, and this inactivation was observed using either infectious or heat-inactivated phages (Kantoch et al., 1958). Inhibition was greater when using antibody-treated phages, and therefore the authors suggested that the immunocomplexes phage–antibody would be inactivating factors particularly active (Kantoch et al., 1958). Moreover, purified phages have anti-inflammatory effects via suppression of ROS (reactive oxygen species) production and inhibition of NF-κβ activity, affecting the production of cytokines [for a review, see (Gorski et al., 2012)]. Despite this evidence, it should be borne in mind that many experiments have been conducted with phage lysates, which on many occasions could contain remnants of bacteria lysed by the phages (e.g., lipopolysaccharide) or perhaps fragments of the host bacterial cell wall adhered to the phage tails. This makes it extremely difficult to determine the components truly responsible for the modulation of the immune response.
Interference With Clinical Diagnoses
Assuming the relative occurrence and distribution of phages throughout the human body described above, coincident with the location of their bacterial hosts, and highly plausible translocation of phage particles to other areas of the body, some reports indicate that the neglected presence of phages in human samples could have an important influence by interfering in clinical practice (Brown-Jaque et al., 2016).
It has been shown that the presence of phages could interfere with many protocols intended to isolate bacteria by enrichment broth, since the phages in the sample destroy the bacterial cells during the enrichment procedure (Muniesa et al., 2005; Quiros et al., 2015). Phages might also interfere in clinical settings during bacterial isolation. As indicated above, there is evidence of a lack of, or reduced, bacterial isolation in clinical samples (ascitic fluid and urine) carrying a high titer of phages, because the lytic activity of the phages disturbs the isolation of the target bacteria (Brown-Jaque et al., 2016).
In addition, some results obtained using molecular methods when targeting some virulence genes present in pathogenic bacteria could be confusing. This is because some genes are located in temperate phages and DNA extraction methods do not distinguish between bacterial and phage DNA. This is the case for phages encoding the Shiga toxin gene, which can be detected in the absence of Shiga toxin-producing bacteria (Martínez-Castillo and Muniesa, 2014). Detection of certain bacterial groups by 16SrDNA qPCR or by genomic sequencing in mixed samples might also be confusing if the sample contains phages and the DNA in the phages is actually what is amplified in the absence of intact bacterial cells. This might be an explanation of the mismatch between the high number of gene copies of 16SrDNA obtained by qPCR amplification and the lack of bacterial isolation observed sometimes (Esparcia et al., 2011). Among others, one hypothesis could be that the positive results were due to amplification of bacterial DNA within phage particles or of bacterial DNA released in the sample after phage-mediated lysis.
Use of Phages Against Human Bacterial Pathogens
The problems of fighting antibiotic resistance in bacteria are continually increasing and severely undermine our capacity to control bacterial infectious diseases. After the increased incidence of bacterial resistance to antibiotics over recent decades, phages have surfaced again as alternative or complementary therapies to control bacterial infections (Fischetti et al., 2006; Doyle and Erickson, 2012; Hertwig et al., 2013; Reardon, 2014; Schmelcher and Loessner, 2014; Górski et al., 2016).
Concluding Remarks
Phages, the most abundant entities on the planet, are also present in human biomes. This presence is known and recognized, but sometimes neglected; and it has a strong influence on the distribution and dynamics of different bacterial populations. Considering the influence of these populations in human health, as their reported ability to improve digestive health, it is clear that phages can be directly related with human well-being (Figure 2). The influence of phages in different mechanisms of our immune system suggests a long-term relationship that we are just starting to elucidate. Moreover, considering our interest in isolating and identifying bacterial pathogens, the presence of phages could certainly interfere with that analysis if it is not considered. A One Health multidisciplinary approach, not restricted to academic or clinical settings and not limited either to microbiological studies, is advisable to evaluate the real extent of and the role played by the phagome in human bodies.
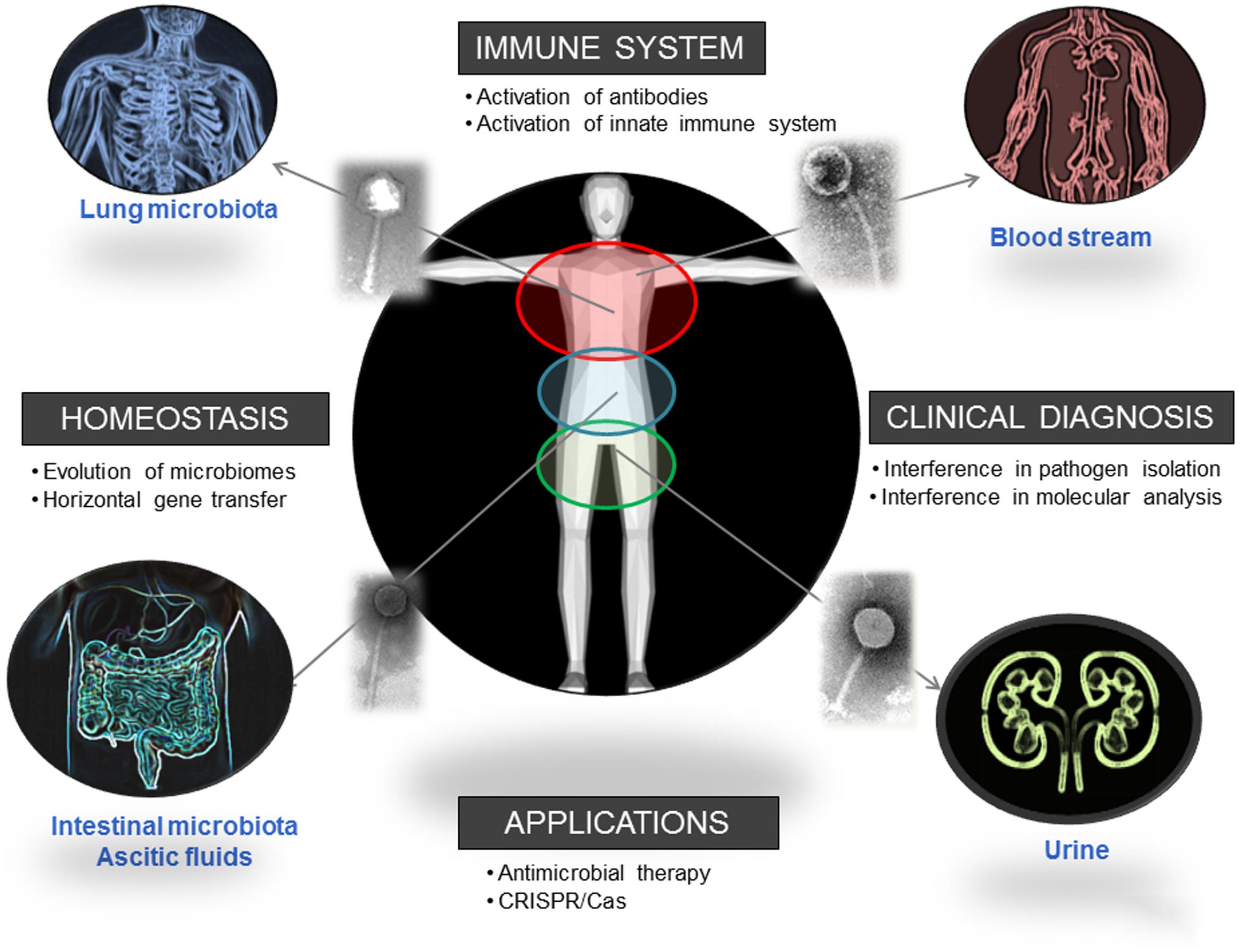
FIGURE 2. Biomes where the presence of phages has been reported and direct and indirect ways in which phages influence human health. Pictures has been adapted from Pixabay.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This study was supported by the Generalitat de Catalunya (2009SGR1043), the Centre de Referència en Biotecnologia (XeRBa), the Sira Carrasco Foundation Grant and by the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III, cofinanced by the European Development Regional Fund, A Way To Achieve Europe, ERDF; the Fondo de Investigación Sanitaria (grant PI16/00158) and project MINECO AGL2016-75536-P (AEI/FEDER, EU).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Authors thank Prof. P. Coll and Prof. J. Jofre for useful comments on the manuscript.
References
Allué-Guardia, A., García-Aljaro, C., and Muniesa, M. (2011). Bacteriophage-encoding cytolethal distending toxin type V gene induced from nonclinical Escherichia coli isolates. Infect. Immun. 79, 3262–3272. doi: 10.1128/IAI.05071-11
Asadulghani, M., Ogura, Y., Ooka, T., Itoh, T., Sawaguchi, A., Iguchi, A., et al. (2009). The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408. doi: 10.1371/journal.ppat.1000408
Bacon, E. J., Richmond, S. J., Wood, D. J., Stirling, P., Bevan, B. J., and Chalmers, W. S. (2017). Serological detection of phage infection in Chlamydia psittaci recovered from ducks. Vet. Rec. 119, 618–620.
Breitbart, M., Hewson, I., Felts, B., Mahaffy, J. M., Nulton, J., Salamon, P., et al. (2003). Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003
Brown-Jaque, M., Muniesa, M., and Navarro, F. (2016). Bacteriophages in clinical samples can interfere with microbiological diagnostic tools. Sci. Rep. 6:33000. doi: 10.1038/srep33000
Colombo, S., Arioli, S., Guglielmetti, S., Lunelli, F., and Mora, D. (2016). Virome-associated antibiotic-resistance genes in an experimental aquaculture facility. FEMS Microbiol. Ecol. 92:fiw003. doi: 10.1093/femsec/fiw003
Colomer-Lluch, M., Imamovic, L., Jofre, J., and Muniesa, M. (2011a). Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 55, 4908–4911. doi: 10.1128/AAC.00535-11
Colomer-Lluch, M., Jofre, J., and Muniesa, M. (2011b). Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 6:e17549. doi: 10.1371/journal.pone.0017549
Dutilh, B. E., Cassman, N., McNair, K., Sanchez, S. E., Silva, G. G. Z., Boling, L., et al. (2014). A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, 4498. doi: 10.1038/ncomms5498
Crick, F. H., Barnett, L., Brenner, S., and Watts-Tobin, R. J. (1961). General nature of the genetic code for proteins. Nature 192, 1227–1232. doi: 10.1038/1921227a0
Dabrowska, K., Miernikiewicz, P., Piotrowicz, A., Hodyra, K., Owczarek, B., Lecion, D., et al. (2014). Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 88, 12551–12557. doi: 10.1128/JVI.02043-14
De Paepe, M., Leclerc, M., Tinsley, C. R., and Petit, M.-A. (2014). Bacteriophages: an underestimated role in human and animal health? Front. Cell. Infect. Microbiol. 4:39. doi: 10.3389/fcimb.2014.00039
D’Herelle, F. (1917). sur un microbe invisible antagonist des bacilles disenterique. C. R. Acad. Sci. Ser. D 165, 373–375.
Doyle, M. P., and Erickson, M. C. (2012). Opportunities for mitigating pathogen contamination during on-farm food production. Int. J. Food Microbiol. 152, 54–74. doi: 10.1016/j.ijfoodmicro.2011.02.037
Duerkop, B. A., Clements, C. V., Rollins, D., Rodrigues, J. L. M., and Hooper, L. V. (2012). A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc. Natl. Acad. Sci. U.S.A. 109, 17621–17626. doi: 10.1073/pnas.1206136109
Dussoix, D., and Arber, W. (1962). Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J. Mol. Biol. 5, 37–49. doi: 10.1016/S0022-2836(62)80059-X
Esparcia, O., Montemayor, M., Ginovart, G., Pomar, V., Soriano, G., Pericas, R., et al. (2011). Diagnostic accuracy of a 16S ribosomal DNA gene-based molecular technique (RT-PCR, microarray, and sequencing) for bacterial meningitis, early-onset neonatal sepsis, and spontaneous bacterial peritonitis. Diagn. Microbiol. Infect. Dis. 69, 153–160. doi: 10.1016/j.diagmicrobio.2010.10.022
Fischetti, V. A., Nelson, D., and Schuch, R. (2006). Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24, 1508–1511. doi: 10.1038/nbt1206-1508
Fuhrman, J. A. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548. doi: 10.1038/21119
Geier, M. R., Trigg, M. E., and Merril, C. R. (1973). Fate of bacteriophage lambda in non-immune germ-free mice. Nature 246, 221–223. doi: 10.1038/246221a0
Gorski, A., Miedzybrodzk, R., Borysowski, J., Dabrowska, K., Wierzbicki, P., Ohams, M., et al. (2012). Chapter 2–“Phage as a modulator of immune responses: practical implications for phage therapy,” in Advances in Virus Research. Bacteriophages Part B, eds M. Lobocka and W. Szybalski (Sant Diego, CA: Academic Press), 41–72.
Górski, A., Miêdzybrodzki, R., Weber-Dąbrowska, B., Fortuna, W., Letkiewicz, S., Rogóż, P., et al. (2016). Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front. Microbiol. 7:1515. doi: 10.3389/fmicb.2016.01515
Górski, A., Wazna, E., Dabrowska, B.-W., Dabrowska, K., Switała-Jeleñ, K., and Miedzybrodzki, R. (2006). Bacteriophage translocation. FEMS Immunol. Med. Microbiol. 46, 313–319. doi: 10.1111/j.1574-695X.2006.00044.x
Griffiths, A. J., Miller, J. H., Suzuki, D. T., Lewontin, R. C., and Gelbart, W. M. (2000). “Gene transfer in bacteria and their viruses,” in An Introduction to Genetic Analysis, 7th Edn, ed. W. Freeman (New York, NY: W. H. Freeman).
Haaber, J., Leisner, J. J., Cohn, M. T., Catalan-Moreno, A., Nielsen, J. B., Westh, H., et al. (2016). Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat. Commun. 7:13333. doi: 10.1038/ncomms13333
Hertwig, S., Hammerl, J. A., Appel, B., and Alter, T. (2013). Post-harvest application of lytic bacteriophages for biocontrol of foodborne pathogens and spoilage bacteria. Berl. Münch. Tierärztl. Wochenschr. 126, 357–369.
Howe, A., Ringus, D. L., Williams, R. J., Choo, Z.-N., Greenwald, S. M., Owens, S. M., et al. (2016). Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J. 10, 1217–1227. doi: 10.1038/ismej.2015.183
IAWPRC Study Group on Health Related Water Microbiology (1991). Bacteriophages as model viruses in water quality control. Water Res 25, 529–545. doi: 10.1016/0043-1354(91)90126-B
Jerne, N. K. (1952). Bacteriophage inactivation by antiphage serum diluted in distilled water. Nature 169, 117–118. doi: 10.1038/169117b0
Jerne, N. K. (1956). The presence in normal serum of specific antibody against bacteriophage T4 and its increase during the earliest stages of immunization. J. Immunol. 76, 209–216.
Jofre, J., Lucena, F., Blanch, A., and Muniesa, M. (2016). Coliphages as model organisms in the characterization and management of water resources. Water 8, 199. doi: 10.3390/w8050199
Jover, L. F., Cortez, M. H., and Weitz, J. S. (2013). Mechanisms of multi-strain coexistence in host-phage systems with nested infection networks. J. Theor. Biol. 332, 65–77. doi: 10.1016/j.jtbi.2013.04.011
Kantoch, M., Skurski, A., and Wieczorek, Z. (1958). In vitro blockade of bacterial phagocytosis of leukocytes by means of bacterial viruses. Schweiz. Z. Pathol. Bakteriol. 21, 1106–1119. doi: 10.1159/000160571
Keller, R., and Traub, N. (1974). The characterization of Bacteroides fragilis bacteriophage recovered from animal sera: observations on the nature of Bacteroides phage carrier cultures. J. Gen. Virol. 24, 179–189. doi: 10.1099/0022-1317-24-1-179
Lang, A. S., Zhaxybayeva, O., and Beatty, J. T. (2012). Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482. doi: 10.1038/nrmicro2802
Langbeheim, H., Teitelbaum, D., and Arnon, R. (1978). Cellular immune response toward MS-2 phage and a synthetic fragment of its coat protein. Cell. Immunol. 38, 193–197. doi: 10.1016/0008-8749(78)90046-1
Lim, E. S., Zhou, Y., Zhao, G., Bauer, I. K., Droit, L., Ndao, I. M., et al. (2015). Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 21, 1228–1234. doi: 10.1038/nm.3950
Lindell, D., Sullivan, M. B., Johnson, Z. I., Tolonen, A. C., Rohwer, F., and Chisholm, S. W. (2004). Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. U.S.A. 101, 11013–11018. doi: 10.1073/pnas.0401526101
Loh, G., and Blaut, M. (2012). Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 3, 544–555. doi: 10.4161/gmic.22156
Manrique, P., Bolduc, B., Walk, S. T., van der Oost, J., de Vos, W. M., and Young, M. J. (2016). Healthy human gut phageome. Proc. Natl. Acad. Sci. U.S.A. 113, 10400–10405. doi: 10.1073/pnas.1601060113
Martínez-Castillo, A., and Muniesa, M. (2014). Implications of free Shiga toxin-converting bacteriophages occurring outside bacteria for the evolution and the detection of Shiga toxin-producing Escherichia coli. Front. Cell. Infect. Microbiol. 4:46. doi: 10.3389/fcimb.2014.00046
Merril, C. R., Biswas, B., Carlton, R., Jensen, N. C., Creed, G. J., Zullo, S., et al. (1996). Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. U.S.A. 93, 3188–3192. doi: 10.1073/pnas.93.8.3188
Minot, S., Sinha, R., Chen, J., Li, H., Keilbaugh, S. A., Wu, G. D., et al. (2011). The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. doi: 10.1101/gr.122705.111
Moayyedi, P., Surette, M. G., Kim, P. T., Libertucci, J., Wolfe, M., Onischi, C., et al. (2015). Fecal Microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6. doi: 10.1053/j.gastro.2015.04.001
Mojica, F. J. M., and Rodriguez-Valera, F. (2016). The discovery of CRISPR in archaea and bacteria. FEBS J. 283, 3162–3169. doi: 10.1111/febs.13766
Müller, M. G., Ing, J. Y., Cheng, M. K.-W., Flitter, B. A., and Moe, G. R. (2013). Identification of a phage-encoded Ig-binding protein from invasive Neisseria meningitidis. J. Immunol. 191, 3287–3296. doi: 10.4049/jimmunol.1301153
Muniesa, M., Blanch, A. R., Lucena, F., and Jofre, J. (2005). Bacteriophages may bias outcome of bacterial enrichment cultures. Appl. Environ. Microbiol. 71, 4269–4275. doi: 10.1128/AEM.71.8.4269-4275.2005
Muniesa, M., García, A., Miró, E., Mirelis, B., Prats, G., Jofre, J., et al. (2004). Bacteriophages and diffusion of beta-lactamase genes. Emerg. Infect. Dis. 10, 1134–1137. doi: 10.3201/eid1006.030472
Norman, J. M., Handley, S. A., Baldridge, M. T., Droit, L., Liu, C. Y., Keller, B. C., et al. (2015). Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460. doi: 10.1016/j.cell.2015.01.002
O’Brien, A. D., Newland, J. W., Miller, S. F., Holmes, R. K., Smith, H. W., and Formal, S. B. (1984). Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226, 694–696. doi: 10.1126/science.6387911
Oh, J., Byrd, A. L., Deming, C., Conlan, S., Kong, H. H., and Segre, J. A. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64. doi: 10.1038/nature13786
Penades, J. R., Chen, J., Quiles-Puchalt, N., Carpena, N., and Novick, R. P. (2015). Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 23, 171–178. doi: 10.1016/j.mib.2014.11.019
Pérez-Brocal, V., García-López, R., Nos, P., Beltrán, B., Moret, I., and Moya, A. (2015). Metagenomic analysis of Crohn’s disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm. Bowel Dis. 21, 2515–2532. doi: 10.1097/MIB.0000000000000549
Pérez-Brocal, V., García-López, R., Vázquez-Castellanos, J. F., Nos, P., Beltrán, B., Latorre, A., et al. (2013). Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin. Transl. Gastroenterol. 4, e36. doi: 10.1038/ctg.2013.9
Puig, A., Araujo, R., Jofre, J., and Frias-Lopez, J. (2001). Identification of cell wall proteins of Bacteroides fragilis to which bacteriophage B40-8 binds specifically. Microbiology 147, 281–288. doi: 10.1099/00221287-147-2-281
Quiros, P., Martinez-Castillo, A., and Muniesa, M. (2015). Improving detection of Shiga toxin-producing Escherichia coli by molecular methods by reducing the interference of free Shiga toxin-encoding bacteriophages. Appl. Environ. Microbiol. 81, 415–421. doi: 10.1128/AEM.02941-14
Rodriguez-Brito, B., Li, L., Wegley, L., Furlan, M., Angly, F., Breitbart, M., et al. (2010). Viral and microbial community dynamics in four aquatic environments. ISME J. 4, 739–751. doi: 10.1038/ismej.2010.1
Ross, J., and Topp, E. (2015). Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microbiol. 81, 7905–7913. doi: 10.1128/AEM.02363-15
Sanger, F., Air, G. M., Barrell, B. G., Brown, N. L., Coulson, A. R., Fiddes, C. A., et al. (1977). Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265, 687–695. doi: 10.1038/265687a0
Scanlan, P. D., and Buckling, A. (2012). Co-evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J. 6, 1148–1158. doi: 10.1038/ismej.2011.174
Schmelcher, M., and Loessner, M. J. (2014). Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 4:e28137. doi: 10.4161/bact.28137
Selva, L., Viana, D., Regev-Yochay, G., Trzcinski, K., Corpa, J. M., Lasa, I., et al. (2009). Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. U.S.A. 106, 1234–1238. doi: 10.1073/pnas.0809600106
Srivastava, A. S., Chauhan, D. P., and Carrier, E. (2004a). In utero detection of T7 phage after systemic administration to pregnant mice. Biotechniques 37, 81–83.
Srivastava, A. S., Kaido, T., and Carrier, E. (2004b). Immunological factors that affect the in vivo fate of T7 phage in the mouse. J. Virol. Methods 115, 99–104.
Twort, F. W. (1915). An investigation on the nature of ultra-microscopic viruses. Lancet 186, 1241–1243. doi: 10.1016/S0140-6736(01)20383-3
Virgin, H. W. (2014). The virome in mammalian physiology and disease. Cell 157, 142–150. doi: 10.1016/j.cell.2014.02.032
Wagner, J., Maksimovic, J., Farries, G., Sim, W. H., Bishop, R. F., Cameron, D. J., et al. (2013). Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm. Bowel Dis. 19, 1598–1608. doi: 10.1097/MIB.0b013e318292477c
Willner, D., Furlan, M., Haynes, M., Schmieder, R., Angly, F. E., Silva, J., et al. (2009). Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 4:e7370. doi: 10.1371/journal.pone.0007370
Keywords: bacteriophages, human biomes, homeostasis, metagenomics, diagnosis, virome
Citation: Navarro F and Muniesa M (2017) Phages in the Human Body. Front. Microbiol. 8:566. doi: 10.3389/fmicb.2017.00566
Received: 13 February 2017; Accepted: 20 March 2017;
Published: 04 April 2017.
Edited by:
Manuel Espinosa, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Guillem Prats, Autonomous University of Barcelona, SpainSteven P. T. Hooton, University of Nottingham, UK
Radoslaw Pluta, International Institute of Molecular and Cell Biology in Warsaw, Poland
Copyright © 2017 Navarro and Muniesa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maite Muniesa, mmuniesa@ub.edu
 Ferran Navarro
Ferran Navarro Maite Muniesa
Maite Muniesa