- 1State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2College of Animal Science and Technology, Jilin Agricultural University, Changchun, China
- 3College of Animal Science and Technology, Changchun Sci-Tech University, Shuangyang, China
Enterocytozoon bieneusi is an important zoonotic parasite. It can infect virtually all animal species and has a global distribution. However, the prevalence of E. bieneusi in donkeys (Equus asinus) has only been reported in Algeria and Spain, and no information is available concerning genotypes of E. bieneusi in donkeys worldwide. In the present study, a total of 301 donkey fecal samples (48 from Jilin Province, 224 from Shandong Province and 29 from Liaoning Province) were collected and examined by PCR amplification of the internal transcribed spacer (ITS) region. The overall E. bieneusi prevalence was 5.3% (16/301), with 6.3% (3/48) in Jilin Province, 4.9% (11/224) in Shandong Province, and 6.9% (2/29) in Liaoning Province. Prevalence in different age groups ranged from 4.2 to 5.5%. E. bieneusi prevalence in donkeys sampled in different seasons varied from 4.2 to 6.5%. Altogether, four E. bieneusi genotypes were identified in this study, with two known genotypes (J and D) and two novel genotypes (NCD-1and NCD-2). Phylogenetic analysis revealed that genotypes D, NCD-1 and NCD-2 belonged to group 1, while the remaining genotype J was clustered into group 2. These findings revealed the occurrence of E. bieneusi in donkeys in China for the first time. Moreover, the present study also firstly genotyped the E. bieneusi in donkeys worldwide. These findings extend the distribution of E. bieneusi genotypes and provide baseline data for controlling E. bieneusi infection in donkeys, other animals and humans.
Introduction
Microsporidiosis is of increasing concern because Microsporidia can infect virtually all animals (Santín and Fayer, 2011; Abu-Akkada et al., 2015; Santin and Fayer, 2015; Zhang et al., 2016a; Zhao et al., 2016). Fecal-oral routes, such as ingestion of contaminated water and food (Zhao et al., 2014a) are the major route to transmit Microsporidia which consist of 1300 named species (Karim et al., 2014c). Of these, Enterocytozoon bieneusi is considered as the most important species and responsible for more than 90% of human microsporidiosis (Desportes et al., 1985). Although E. bieneusi was firstly detected in HIV patients in 1985 (Desportes et al., 1985), as research continues, more and more animals were also considered as susceptible hosts for E. bieneusi. To date, more than 200 distinct genotypes have been reported on the basis of sequences of the internal transcribed spacer (ITS) region (Karim et al., 2014b; Tian et al., 2015). These genotypes were divided into several groups: the zoonotic groups (syn. Group 1), host-adapted groups (groups 2–5 and an outlier genotypes in dogs), and some other small groups (groups 6–9). However, surprisingly, some of the genotypes in Group 2 (the so-called host-adapted group) were found in both animals and humans, which should also be considered as a zoonotic agent (Hu et al., 2014; Karim et al., 2014a; Ma et al., 2015a,b).
To our knowledge, limited data has been published on genotypes of E. bieneusi from China (Tian et al., 2015; Zhang et al., 2015, 2016a,b; Qi et al., 2016) and no data is available for donkeys. The objectives of the present study were to investigate the E. bieneusi prevalence and identify their genotypes in donkeys in Jilin, Liaoning and Shandong Provinces, eastern and northeastern China.
Materials and Methods
Ethics Approval and Consent to Participate
This study was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Donkeys used for the study were handled in accordance with good animal practices required by the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Collection and Preparation of Donkey Fecal Samples
Between May 2015 and October 2016, a total of 301 fecal samples were collected from donkeys from Jilin Province (n = 48), Liaoning Province (n = 29), and Shandong Province (n = 224). At the time of the sampling, all the donkeys were in apparently good health status. Fecal samples were collected from each animal after defecation onto the ground, and then were taken to the laboratory. All the detailed information of investigated donkeys was obtained and listed in Tables 1, 2.
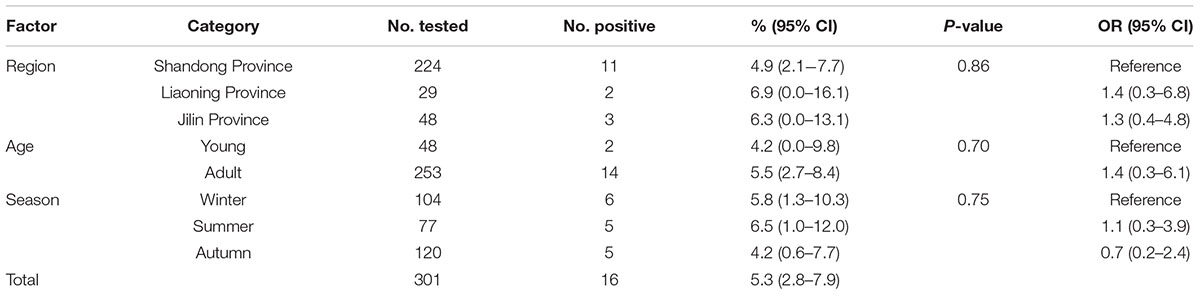
TABLE 1. Factors associated with prevalence of Enterocytozoon bieneusi in donkeys in Northern China.
DNA Extraction and PCR Amplification
Genomic DNA from each fecal sample was obtained by using a DNA extraction kit (OMEGA, USA). The extracted DNA samples were stored at -20°C until PCR amplification.
The E. bieneusi genotypes were determined by the nested PCR of the ITS region of the ribosomal RNA (rRNA) gene using primers F1 [5′-GGTCATAGGGATGAAGAG-3′] and R1 [5′-TTCGAGTTCTTTCGCGCTC-3′] for primary amplification, and primers F2 [5′-GCTCTGAATATCTATGGCT-3′] and R2 [5′-ATCGCCGACGGATCCAAGTG-3′] for secondary amplification. 25 μl of PCR reaction composed of 200 μM each deoxy-ribonucleoside triphosphate (dNTP), 1 × Ex Taq buffer (Mg2+ free), 0.4 μM of each primer, 0.625 U of Ex Taq DNA polymerase (TAKARA, Japan), 2 mM MgCl2, and 2 μl of DNA template. Both positive and negative controls were included in each test. All the PCR products were detected under UV light after electrophoresis on a 2% agarose gels containing GoldViewTM (Solarbio, China).
Sequence and Phylogenetic Analyses
Positive secondary PCR products were sequenced using bi-directional sequencing. Genotypes that produced sequences with mutations, including single nucleotide substitutions, deletions or insertion which were confirmed by the DNA sequencing of at least two PCR products, were identified as novel genotypes. The obtained sequences were aligned with reference sequences to determine the E. bieneusi genotypes/subtypes using the BLAST1. Phylogenetic trees were reconstructed using neighbor-joining (NJ) method in Mega 5.0 software (Kimura 2-parameter model, 1000 replicates).
Statistical Analysis
The variation in E. bieneusi prevalence (y) of donkeys of different geographical location (x1), season (x2) and age (x3) were analyzed by χ2 test using SPSS V20.0 (IBM, Chicago, IL, USA) (Letchumanan et al., 2015; Cabal et al., 2016). Using multivariable regression analysis, each of these variables was included in the binary logit model as an independent variable. The best model was judged by Fisher’s scoring algorithm. All tests were two-sided. When the values of P < 0.05, results were considered statistically significant. 95% confidence intervals (95% CIs) were also calculated based on the formula of X ± Z × {X(1 - X) / N}½ (Z = 1.96; X represents the prevalence; N represents the sample sizes).
Results and Discussion
A total of 16 (5.3%) out of 301 donkeys were PCR-positive for E. bieneusi (Table 1). Donkeys in Shandong Province (11/224, 4.9%) showed the higher E. bieneusi prevalence (Table 1). E. bieneusi prevalence in different season groups ranged from 4.2 to 6.5% (Table 1). Prevalence of E. bieneusi in young donkeys and adult donkeys was 4.2 and 5.5%, respectively (Table 1). Moreover, E. bieneusi prevalence in different farm groups varied from 4.2 to 6.9% (Table 2).
In this study, the overall prevalence was 5.3% (16/301) which was lower than that in horses from the Czech Republic (6.9%) (Wagnerová et al., 2012), Colombia (10.8%) (Santín et al., 2010), and Algeria (6.8%). However, it is higher than that in donkeys in Algeria (1.6%) (Laatamna et al., 2015), and Spain (0%) (Lores et al., 2002). Moreover, it is also considerably higher than that in horses in Spain (0%) (Lores et al., 2002). The different infection rates may be due to the different susceptibility between donkeys and horses. Moreover, animal welfare, sample sizes, age distribution of the samples and geo-ecological conditions can also influence the E. bieneusi prevalence.
In the present study, although donkeys from Liaoning Province has relatively high rates of E. bieneusi infection compared with donkeys from Jilin Province and Shandong Province, the difference was not statistically significant (χ2 = 0.30, df = 2, P = 0.86) (Table 1). This may be due to the similar climates during sampling times in Jilin, Liaoning and Shandong Provinces, and also relate to the same management mode in these farms. Moreover, the present study also showed that E. bieneusi prevalence in donkeys increased gradually with age, which is consistent with previous reports in that E. bieneusi may accumulate throughout the life time (Thellier and Breton, 2008; Zhao et al., 2014a), but the difference was not statistically significant (χ2 = 0.15, df = 1, P = 0.70) (Table 1).
To our knowledge, although two studies of E. bieneusi prevalence in donkeys have been reported previously, these isolates were not genotyped successfully (Lores et al., 2002; Laatamna et al., 2015). Previous studies have identified sixteen E. bieneusi ITS genotypes, namely Horse 1 – Horse 11, G, WL15, D, EpbA and CZ3 in horses who is a close relative to donkeys (Santín et al., 2010; Wagnerová et al., 2012; Laatamna et al., 2015). But probably due to the smaller sample sizes and different collection times, only four E. bieneusi genotypes were observed in donkeys in the present study, including two known genotypes J and D, and two novel genotypes NCD-1 and NCD-2 (Figure 1). Thus, four E. bieneusi genotypes were endemic in donkeys in eastern and northeastern China. This study also indicated that genotype J was the most prevalent in donkeys, which were different from the results reported in horses in Colombia (Santín et al., 2010) and Czech Republic (Wagnerová et al., 2012), where the predominance of genotype D was found, and in horses in Algeria, where predominance of horse1 was identified (Laatamna et al., 2015). Furthermore, a total of 10 polymorphic sites were observed among these genotypes (Table 3), which suggested the high genetic diversity of E. bieneusi in the investigated donkeys.
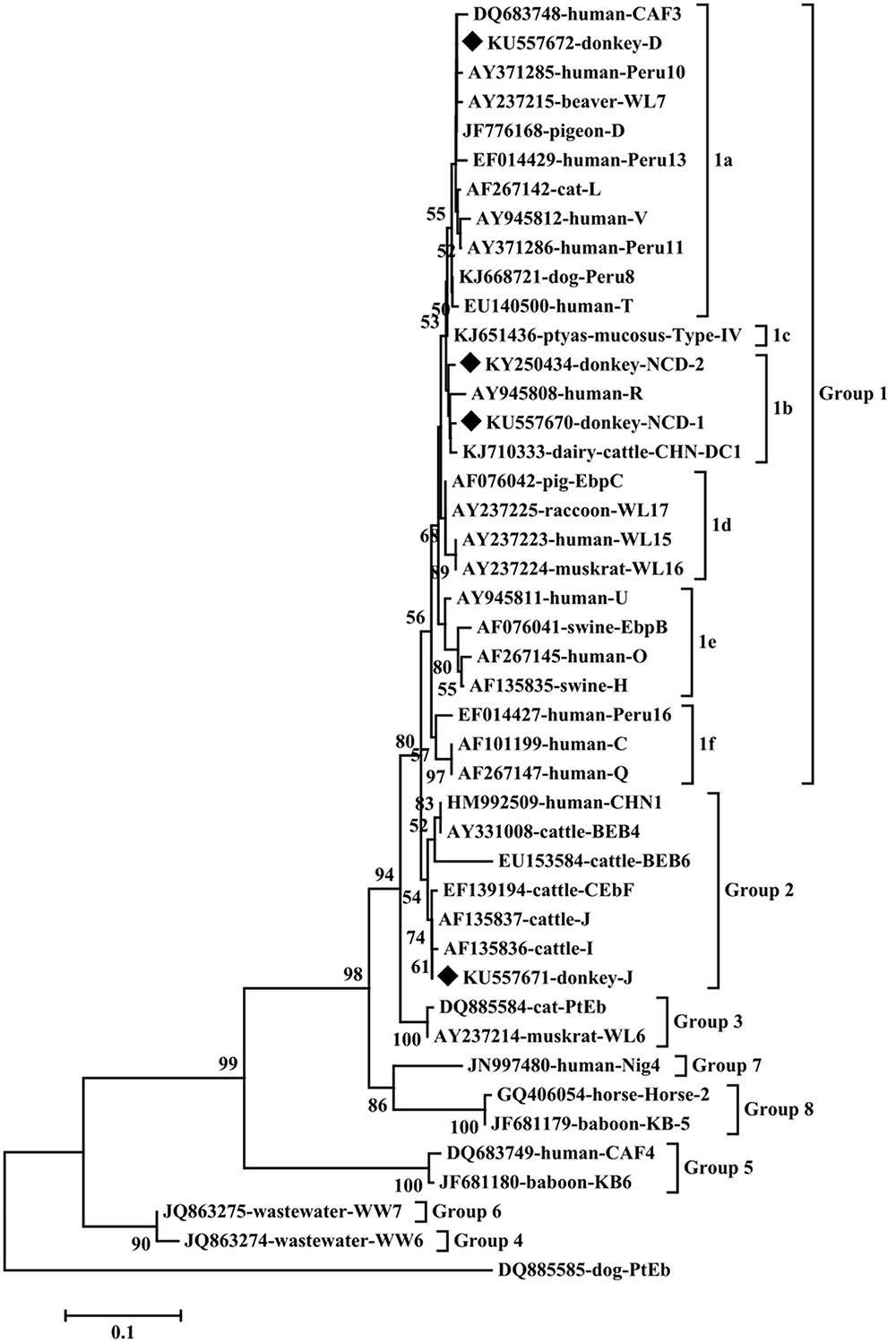
FIGURE 1. Phylogenetic analyses of Enterocytozoon bieneusi using neighbor-joining (NJ) method (Kimura 2-parameter model). Bootstrapping (1000 replicates) was performed. The values below 50% are not shown. E. bieneusi isolates identified in the present study are indicated by solid diamond.
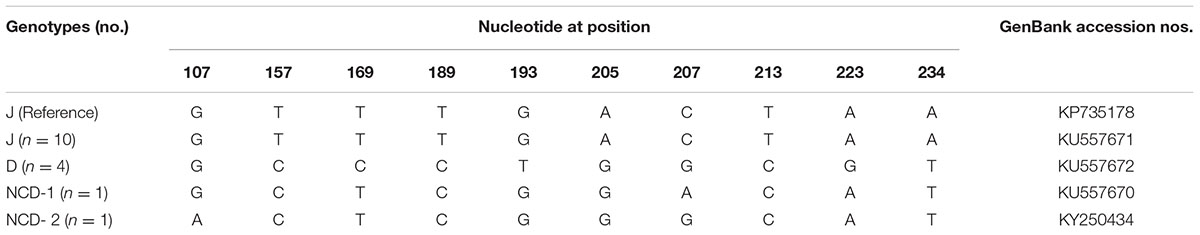
TABLE 3. Variations in the ITS nucleotide sequences among genotypes of the Enterocytozoon bieneusi in donkeys in Northern China.
Genotypes J and D have been widely identified in different hosts in northern and eastern China. For example, genotype D was identified in golden takins (Budorcas taxicolor bedfordi) in Shannxi (Karim et al., 2014a), non-human primates in Henan (Wang et al., 2013; Karim et al., 2014b), dairy cattle, sheep, goats, pig, raccoon dog, cats, blue foxes and dogs in Heilongjiang (Zhao et al., 2014a,b, 2015a,b; Li et al., 2015), and blue foxes in Jilin (Zhao et al., 2015c); genotype J was found in cattle in Heilongjiang, Jilin, Shandong, Henan, captive wildlife in Henan, yaks in Qinghai. In addition, genotypes D was also identified in HIV patients in Henan (Wang et al., 2013; Karim et al., 2014b) and J was found in a child in Jilin (Zhang et al., 2011). Moreover, two of the ITS sequences (accession numbers: KU557671 and KU557672) of the identified E. bieneusi isolates were identical to that of genotypes J (GenBank accession no. KP735178, from a Homo sapiens in Iran) and D (GenBank accession no. KP262379, from a goat in China) sequences available in GenBank, respectively. These results indicate cross-species transmission of these E. bieneusi genotypes in northern China.
Phylogenetic analysis indicated that genotypes D, NCD-1 and NCD-2 belonged to group 1, the most important zoonotic groups; J was grouped into group 2, a cattle-specific groups, but genotype J has also been identified in humans. These results suggest that donkeys are potential source of animal and human microsporidiosis.
Conclusion
This is the first study of E. bieneusi prevalence (5.3%, 16/301) in donkeys in China. Moreover, two known genotypes (genotypes D and J), and two novel genotypes (NCD-1 and NCD-2) were detected for the first time in donkeys. Donkeys should be considered as an important potential source of human microsporidiosis, and effective strategies should be performed to control E. bieneusi infection in donkeys, other animals and humans.
Data Availability Statement
Representative nucleotide sequences were deposited in GenBank with the following accession numbers: KU557670-KU557672 and KY250434.
Author Contributions
X-QZ and X-XZ conceived and designed the study, and critically revised the manuscript. D-MY, J-GM, and W-BZ performed the experiments. X-XZ and D-MY analyzed the data. D-MY and J-GM drafted the manuscript. F-CL, J-LH and QZ helped in study design, study implementation and manuscript preparation. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Fundamental Research Funds of Chinese Academy of Agricultural Sciences (Grant No. Y2016JC05) and the Agricultural Science and Technology Innovation Program (ASTIP) (Grant No. CAAS-ASTIP-2014-LVRI-03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abu-Akkada, S. S., Ashmawy, K. I., and Dweir, A. W. (2015). First detection of an ignored parasite, Encephalitozoon cuniculi, in different animal hosts in Egypt. Parasitol. Res. 114, 843–850. doi: 10.1007/s00436-014-4247-4
Cabal, A., García-Castillo, M., Cantón, R., Gortázar, C., Domínguez, L., and Álvarez, J. (2016). Prevalence of Escherichia coli virulence genes in patients with diarrhea and a subpopulation of healthy volunteers in madrid. Spain. Front. Microbiol. 7:641. doi: 10.3389/fmicb.2016.00641
Desportes, I., Le Charpentier, Y., Galian, A., Bernard, F., Cochand-Priollet, B., Lavergne, A., et al. (1985). Occurrence of a new microsporidan: Enterocytozoon bieneusi n.g., n. sp., in the enterocytes of a human patient with AIDS. J. Protozool. 32, 250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x
Hu, Y., Feng, Y., Huang, C., and Xiao, L. (2014). Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ. Sci. Technol. 48, 14219–14227. doi: 10.1021/es504464t
Karim, M. R., Dong, H., Yu, F., Jian, F., Zhang, L., Wang, R., et al. (2014a). Genetic diversity in Enterocytozoon bieneusi isolates from dogs and cats in China: host specificity and public health implications. J. Clin. Microbiol. 52, 3297–3302. doi: 10.1128/JCM.01352-14
Karim, M. R., Wang, R., Dong, H., Zhang, L., Li, J., Zhang, S., et al. (2014b). Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl. Environ. Microbiol. 80, 1893–1898. doi: 10.1128/AEM.03845-13
Karim, M. R., Wang, R., He, X., Zhang, L., Li, J., Rume, F. I., et al. (2014c). Multilocus sequence typing of Enterocytozoon bieneusi in nonhuman primates in China. Vet. Parasitol. 200, 13–23. doi: 10.1016/j.vetpar.2013.12.004
Laatamna, A. E., Wagnerová, P., Sak, B., Květoňová, D., Xiao, L., Rost, M., et al. (2015). Microsporidia and Cryptosporidium in horses and donkeys in Algeria: etection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet. Parasitol. 208, 135–142. doi: 10.1016/j.vetpar.2015.01.007
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Li, W., Li, Y., Song, M., Lu, Y., Yang, J., Tao, W., et al. (2015). Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet. Parasitol. 208, 125–134. doi: 10.1016/j.vetpar.2015.01.014
Lores, B., del Aguila, C., and Arias, C. (2002). Enterocytozoon bieneusi (microsporidia) in faecal samples from domestic animals from Galicia, Spain. Mem. Inst. Oswaldo Cruz 97, 941–945. doi: 10.1590/S0074-02762002000700003
Ma, J., Cai, J., Ma, J., Feng, Y., and Xiao, L. (2015a). Enterocytozoon bieneusi genotypes in yaks (Bos grunniens) and their public health potential. J. Eukaryot. Microbiol. 62, 21–25. doi: 10.1111/jeu.12141
Ma, J., Li, P., Zhao, X., Xu, H., Wu, W., Wang, Y., et al. (2015b). Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 207, 220–227. doi: 10.1016/j.vetpar.2014.10.011
Qi, M., Wang, R., Wang, H., Jian, F., Li, J., Zhao, J., et al. (2016). Enterocytozoon bieneusi genotypes in grazing horses in China and their zoonotic transmission potential. J. Eukaryot. Microbiol. 63, 591–597. doi: 10.1111/jeu.12308
Santín, M., and Fayer, R. (2011). Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 90, 363–371. doi: 10.1016/j.rvsc.2010.07.014
Santin, M., and Fayer, R. (2015). Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J. Eukaryot. Microbiol. 62, 34–43. doi: 10.1111/jeu.12155
Santín, M., Vecino, J. A., and Fayer, R. (2010). A zoonotic genotype of Enterocytozoon bieneusi in horses. J. Parasitol. 96, 157–161. doi: 10.1645/GE-2184.1
Thellier, M., and Breton, J. (2008). Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 15, 349–358. doi: 10.1051/parasite/2008153349
Tian, G. R., Zhao, G. H., Du, S. Z., Hu, X. F., Wang, H. B., Zhang, L. X., et al. (2015). First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect. Genet. Evol. 34, 32–35. doi: 10.1016/j.meegid.2015.06.015
Wagnerová, P., Sak, B., Květoňová, D., Buňatová, Z., Civišová, H., Maršálek, M., et al. (2012). Enterocytozoon bieneusi and Encephalitozoon cuniculi in horses kept under different management systems in the Czech Republic. Vet. Parasitol. 190, 573–577. doi: 10.1016/j.vetpar.2012.07.013
Wang, L., Zhang, H., Zhao, X., Zhang, L., Zhang, G., Guo, M., et al. (2013). Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 51, 557–563. doi: 10.1128/JCM.02758-12
Zhang, X., Wang, Z., Su, Y., Liang, X., Sun, X., Peng, S., et al. (2011). Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 49, 2006–2008. doi: 10.1128/JCM.00372-11
Zhang, X. X., Cong, W., Lou, Z. L., Ma, J. G., Zheng, W. B., Yao, Q. X., et al. (2016a). Prevalence, risk factors and multilocus genotyping of Enterocytozoon bieneusi in farmed foxes (Vulpes lagopus), Northern China. Parasit Vectors 9, 72. doi: 10.1186/s13071-016-1356-1
Zhang, X. X., Jiang, J., Cai, Y. N., Wang, C. F., Xu, P., Yang, G. L., et al. (2016b). Molecular characterization of Enterocytozoon bieneusi in domestic rabbits (Oryctolagus cuniculus) in Northeastern China. Korean J. Parasitol. 54, 81–85. doi: 10.3347/kjp.2016.54.1.81
Zhang, X. X., Qin, S. Y., Zhang, Y., Meng, Q. F., Jiang, J., Yang, G. L., et al. (2015). First report of hepatitis E virus infection in sika deer in China. Biomed. Res. Int. 2015:502846. doi: 10.1155/2015/502846
Zhao, W., Yu, S., Yang, Z., Zhang, Y., Zhang, L., Wang, R., et al. (2016). Genotyping of Enterocytozoon bieneusi (Microsporidia) isolated from various birds in China. Infect. Genet. Evol. 40, 151–154. doi: 10.1016/j.meegid.2016.02.037
Zhao, W., Zhang, W., Wang, R., Liu, W., Liu, A., Yang, D., et al. (2014a). Enterocytozoon bieneusi in sika deer (Cervus nippon) and red deer (Cervus elaphus): deer specificity and zoonotic potential of ITS genotypes. Parasitol. Res. 113, 4243–4250. doi: 10.1007/s00436-014-4100-9
Zhao, W., Zhang, W., Yang, D., Zhang, L., Wang, R., and Liu, A. (2015a). Prevalence of Enterocytozoon bieneusi and genetic diversity of ITS genotypes in sheep and goats in China. Infect. Genet. Evol. 32, 265–270. doi: 10.1016/j.meegid.2015.03.026
Zhao, W., Zhang, W., Yang, F., Cao, J., Liu, H., Yang, D., et al. (2014b). High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Appl. Environ. Microbiol. 80, 3699–3707.
Zhao, W., Zhang, W., Yang, F., Zhang, L., Wang, R., Cao, J., et al. (2015b). Enterocytozoon bieneusi in dairy cattle in the Northeast of China: genetic diversity of ITS gene and evaluation of zoonotic transmission potential. J. Eukaryot. Microbiol. 62, 553–560. doi: 10.1111/jeu.12210
Keywords: Enterocytozoon bieneusi, Microsporidia, donkey, zoonotic disease, Internal transcribed spacer (ITS)
Citation: Yue D-M, Ma J-G, Li F-C, Hou J-L, Zheng W-B, Zhao Q, Zhang X-X and Zhu X-Q (2017) Occurrence of Enterocytozoon bieneusi in Donkeys (Equus asinus) in China: A Public Health Concern. Front. Microbiol. 8:565. doi: 10.3389/fmicb.2017.00565
Received: 25 January 2017; Accepted: 20 March 2017;
Published: 31 March 2017.
Edited by:
Paul J. Brindley, George Washington University, USAReviewed by:
Mario Santoro, Istituto Zooprofilattico Sperimentale del Mezzogiorno, ItalyAvi Peretz, The Baruch Padeh Medical Center, Poriya, Israel
Copyright © 2017 Yue, Ma, Li, Hou, Zheng, Zhao, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Xuan Zhang, zhangxiaoxuan1988@126.com Xing-Quan Zhu, xingquanzhu1@hotmail.com
†These authors have contributed equally to this work.
 Dong-Mei Yue1†
Dong-Mei Yue1† Xiao-Xuan Zhang
Xiao-Xuan Zhang