- 1Warwick Crop Centre, School of Life Sciences, University Warwick, Warwick, UK
- 2Eden Project, Bodelva, UK
- 3Institute of Integrative Biology, University of Liverpool, Liverpool, UK
- 4Centre for Crop and Disease Management, Curtin University, Bentley, WA, Australia
- 5Faculty of Science, School of Agriculture and Environment, University of Western Australia, Crawley, WA, Australia
- 6Division of Biotechnology and Plant Health, Norwegian Institute of Bioeconomy Research, Ås, Norway
Sclerotinia species are important fungal pathogens of a wide range of crops and wild host plants. While the biology and population structure of Sclerotinia sclerotiorum has been well-studied, little information is available for the related species S. subarctica. In this study, Sclerotinia isolates were collected from different crop plants and the wild host Ranuculus ficaria (meadow buttercup) in England, Scotland, and Norway to determine the incidence of Sclerotinia subarctica and examine the population structure of this pathogen for the first time. Incidence was very low in England, comprising only 4.3% of isolates while moderate and high incidence of S. subarctica was identified in Scotland and Norway, comprising 18.3 and 48.0% of isolates respectively. Characterization with eight microsatellite markers identified 75 haplotypes within a total of 157 isolates over the three countries with a few haplotypes in Scotland and Norway sampled at a higher frequency than the rest across multiple locations and host plants. In total, eight microsatellite haplotypes were shared between Scotland and Norway while none were shared with England. Bayesian and principal component analyses revealed common ancestry and clustering of Scottish and Norwegian S. subarctica isolates while English isolates were assigned to a separate population cluster and exhibited low diversity indicative of isolation. Population structure was also examined for S. sclerotiorum isolates from England, Scotland, Norway, and Australia using microsatellite data, including some from a previous study in England. In total, 484 haplotypes were identified within 800 S. sclerotiorum isolates with just 15 shared between England and Scotland and none shared between any other countries. Bayesian and principal component analyses revealed a common ancestry and clustering of the English and Scottish isolates while Norwegian and Australian isolates were assigned to separate clusters. Furthermore, sequencing part of the intergenic spacer (IGS) region of the rRNA gene resulted in 26 IGS haplotypes within 870 S. sclerotiorum isolates, nine of which had not been previously identified and two of which were also widely distributed across different countries. S. subarctica therefore has a multiclonal population structure similar to S. sclerotiorum, but has a different ancestry and distribution across England, Scotland, and Norway.
Introduction
Sclerotinia species are important pathogens of a wide range of crop plants as well as many wild hosts. Of these, S. sclerotiorum (Lib.) de Bary is probably the best studied with a worldwide distribution and a wide host range of more than 400 plants including many important dicotyledonous crops and wild species (Boland and Hall, 1994). Some of the major crops affected include oilseed rape, soybean, sunflower, lettuce, carrot, potatoes, beans, and peas (Bolton et al., 2006). Infection of the majority of host plants is by ascospores released from apothecia produced through carpogenic germination of soilborne sclerotia, although direct infection by myceliogenic germination can occasionally occur (Hao et al., 2003). Apothecia are formed through sexual reproduction, and as S. sclerotiorum is predominantly homothallic, a multiclonal population structure has generally been observed in studies carried out on a variety of crop plants in Alaska, Australia, Brazil Canada, China, Iran, New Zealand, Turkey, UK, and USA using DNA fingerprinting (Kohn et al., 1991; Kohn, 1995; Cubeta et al., 1997; Carbone et al., 1999; Carpenter et al., 1999; Carbone and Kohn, 2001b; Hambleton et al., 2002; Phillips et al., 2002) or microsatellite genotyping (Sexton and Howlett, 2004; Sexton et al., 2006; Winton et al., 2006; Mert-Turk et al., 2007; Hemmati et al., 2009; Gomes et al., 2011; Attanayake et al., 2013; Clarkson et al., 2013; Aldrich-Wolfe et al., 2015; Lehner et al., 2015). In these studies, the typical population structure is such that one or a small number of clones is sampled at high frequency, with the remainder sampled only once or a few times (Kohn, 1995). The high frequency S. sclerotiorum clones found at a local scale can sometimes be sampled repeatedly over several years in the same locality and in some cases over a wider geographic area (Hambleton et al., 2002; Clarkson et al., 2013). There is, however, a limit to the geographic distribution of S. sclerotiorum clones; for instance, none of the S. sclerotiorum clones from oilseed rape and soybean identified by DNA fingerprinting in Canada (Kohn et al., 1991; Kohli et al., 1992, 1995; Hambleton et al., 2002) were found in various crops from different locations in the USA (Cubeta et al., 1997; Malvárez et al., 2007). The distribution of most S. sclerotiorum clones is therefore restricted geographically with little or no sharing of genotypes between different locations in the same country, resulting in genetically distinct subdivided populations as identified in Australia (Sexton and Howlett, 2004), UK (Clarkson et al., 2013) and USA (Malvárez et al., 2007). Although there is overwhelming support for homothallism and clonal reproduction in S. sclerotiorum, there has been some evidence for outcrossing and genetic exchange based on linkage disequilibrium measures (Atallah et al., 2004; Sexton and Howlett, 2004; Malvárez et al., 2007; Hemmati et al., 2009; Clarkson et al., 2013) and lack of association of molecular markers with mycelial compatibility group (MCG) (Atallah et al., 2004). More direct evidence of outcrossing has been through rare observations of sibling ascospores derived from a single apothecium belonging to more than one MCG (Atallah et al., 2004; Malvárez et al., 2007) and ascospore dimorphism (Ekins et al., 2006).
Although the population structure of S. sclerotiorum has been well-studied, there are fewer reports for related species such as Sclerotinia minor Jagger (Wu and Subbarao, 2006) S. trifoliorum Erikss. (Njambere et al., 2014) and none for S. subarctica nom. prov. S. minor has a reported host range of just over 90 species (Melzer et al., 1997) and like S. sclerotiorum is a major pathogen of lettuce (Wu and Subbarao, 2006). In one of the few population studies, Wu and Subbarao (2006) reported much lower levels of genetic diversity in S. minor compared with S. sclerotiorum based on MCGs for isolates from lettuce in California. In contrast to S. sclerotiorum and S. minor, S. trifoliorum is bipolar heterothallic (Uhm and Fujii, 1983) and has a more limited host range, being found mainly on cool-season forage and vegetable legumes (Willetts et al., 1980). A recent population study of S. trifoliorum on chickpea in California identified high levels of diversity based on MCGs and microsatellites (Njambere et al., 2014). Compared to the other Sclerotinia spp., S. subarctica was only identified relatively recently on the wild hosts yellow marsh marigold (Caltha palustris), dandelion (Taraxacum sp.), and northern wolfsbane (Aconitum septentrionale) in Norway (Holst-Jensen et al., 1998). It was first reported on horticultural crop hosts in Alaska, often in sympatry with S. sclerotiorum (Winton et al., 2006). A possible reason for this is that S. subarctica is difficult to distinguish from S. sclerotiorum as symptoms on plants are identical, and the two species look very similar in culture although S. subarctica generally forms larger sclerotia (Clarkson et al., 2010). Identification and designation as a new species was therefore based on three nucleotide substitutions in the ITS region and the absence of a 304 base group I intron in the large subunit (LSU) of the ribosomal RNA gene (Holst-Jensen et al., 1998). Since then, S. subarctica was first reported in the UK on meadow buttercup (Ranunculus acris) at a single location in England and pathogenicity demonstrated on oilseed rape (Clarkson et al., 2010). More recently, the pathogen has also been identified on a turnip rape crop (Brassica rapa subsp. oleifera) in Norway (Brodal et al., 2017). Little is known about the biology and epidemiology of S. subarctica, but one hypothesis is that it is more endemic to Northern latitudes (Winton et al., 2006). In addition, there have been no studies so far on S. subarctica population structure, although microsatellite markers have been published (Winton et al., 2007).
In a previous study, the population structure of S. sclerotiorum was examined in England and Wales (UK) for the first time using microsatellites and sequencing the intergenic spacer (IGS) region of the rRNA gene. (Clarkson et al., 2013). In total, 228 microsatellite haplotypes were identified within 384 isolates with one found at high frequency across different crop types and meadow buttercup. Of 14 IGS haplotypes identified, six were unique to buttercup and three were found at high frequency and were also present in S. sclerotiorum populations from Canada, the USA, and New Zealand published previously.
To date, S. subarctica has only been found on meadow buttercup at one location in England in sympatry with S. sclerotiorum, but it was hypothesized that S. subarctica may be more prevalent in the north of the UK and is likely to be found on crop plants (Clarkson et al., 2013). Hence one of the main aims of the present study was to sample and identify the species in further Sclerotinia populations from both crop plants and buttercup in England and Scotland. In addition, we sought to confirm that S. subarctica could still be isolated from the single location in Herefordshire (England) where it was first identified in samples collected in 2009 (Clarkson et al., 2010). For comparison, the relative incidence of S. subarctica and S. sclerotiorum was also examined for crop plants in Norway, a northern “neighbour” of the UK, where a high incidence of S. subarctica might be expected. Following identification of S. subarctica, a further aim was to genotype these isolates using microsatellites, hence providing a population structure analysis of this pathogen for the first time. Microsatellite and IGS data were also generated for isolates collected and identified as S. sclerotiorum, allowing population structure at a country scale to be examined for England, Scotland, and Norway. This added to the existing information generated in the previous study that focussed only on S. sclerotiorum populations from different locations in England. Finally, S. sclerotiorum isolates collected from Western Australia and genotyped using the same microsatellite markers and/or by IGS sequencing were also used as a geographically distant comparison with the UK and Norwegian populations.
Materials and Methods
Sclerotinia Isolates from Different Host Plants
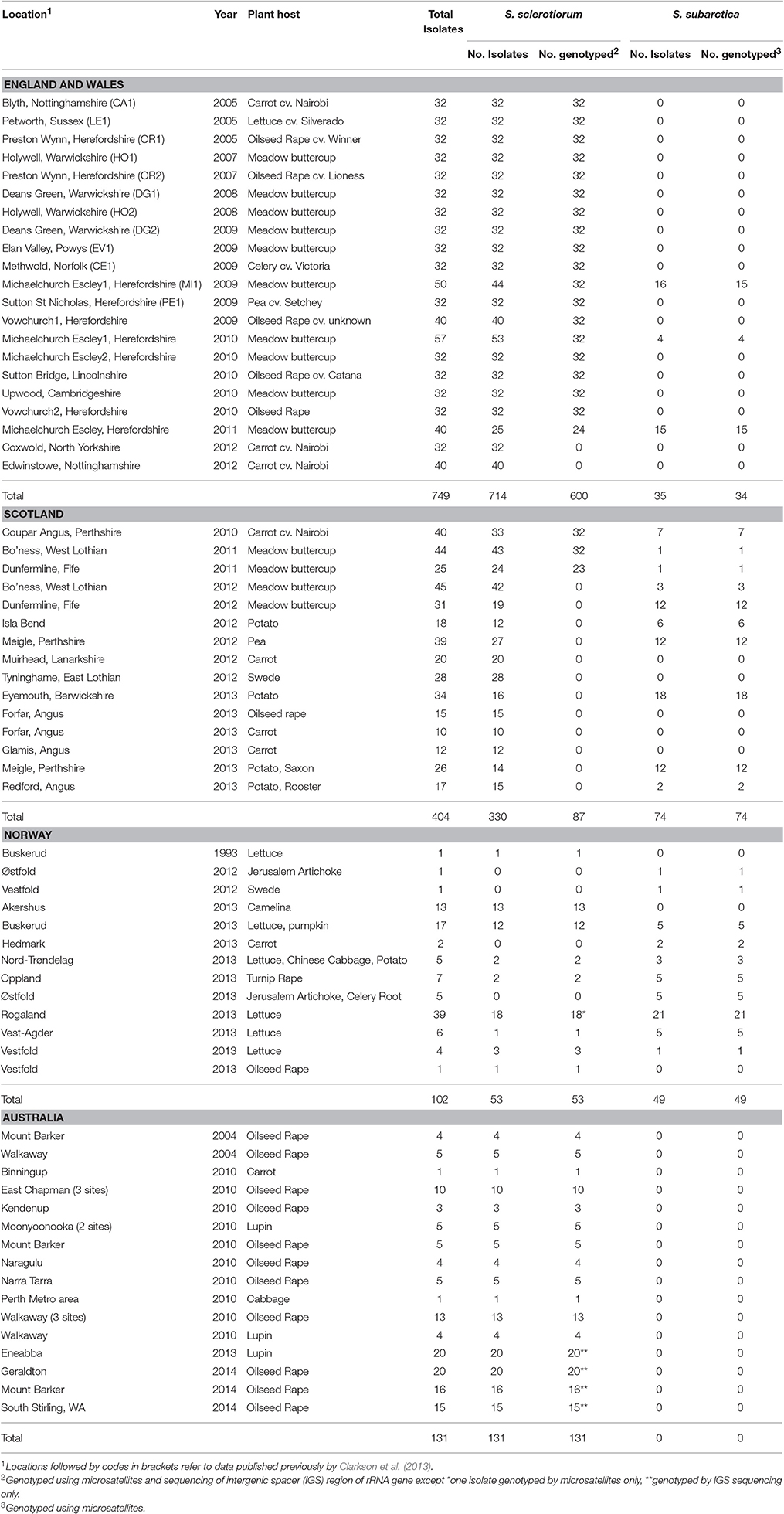
Sclerotinia species sclerotia were collected from a range of diseased crop plants comprising carrot (Daucus carota), cabbage (Brassica oleracea), celery (Apium graveolens), chinese cabbage (Brassica rapa subsp. pekinensis), camelina (Camelina sativa), Jerusalem artichoke (Helianthus tuberosus), lettuce (Lactuca sativa), oilseed rape (Brassica napus subsp. napus), potato (Solanum tuberosum), pumpkin (Cucurbita pepo), swede (B. napus), and turnip rape (B. rapa subsp. oleifera) from different locations in England, Scotland, and Norway between 2009 and 2013 (Table 1). For some crops, structured sampling was carried out whereby sclerotia were collected from infected plants at points at least 8 m apart along transects, with sclerotia collected from different points stored separately. For others, low levels of disease meant that this was not possible and sclerotia were collected from individual infected plants where found. Cultures of Sclerotinia were obtained from individual sclerotia by surface sterilizing them in a solution of 50% sodium hypochlorite (11–14% available chlorine, VWR International Ltd, UK) and 50% ethanol (v/v) for 4 min with agitation followed by two washes in sterile distilled water (SDW) for 1 min. The sclerotia were then bisected, placed on potato dextrose agar (PDA; Oxoid) and incubated at 20°C. After 2–3 days, agar plugs from the leading edge of actively growing mycelium were sub-cultured onto PDA and after ~6 weeks the mature sclerotia formed were stored both dry at 5°C and submerged in potato dextrose broth (PDB; Formedium, UK) amended with 10% glycerol (Sigma-Aldrich Company Ltd, UK) at −20°C. These stock sclerotia were used to initiate new cultures as required.
Isolates of Sclerotinia spp. were also isolated from meadow buttercup in England and Scotland (Table 1) following the method described by Clarkson et al. (2013). Briefly, this was done by sampling flowers from five plants showing symptoms of infection, which were collected at up to 40 points at 10 m intervals along transects, with flowers from each plant stored separately. The flowers were then incubated on damp tissue paper in sealed plastic boxes at room temperature (~22°C) for 4 weeks. Sclerotia formed on the damp tissue paper were then picked off and cultured as described previously.
Isolates of S. sclerotiorum collected between 2009 and 2014 were also obtained from sclerotia collected from infected lupin and oilseed rape from the northern and southern agricultural regions of Western Australia in addition to “standard” isolates from oilseed rape (collected 2004), cabbage, and carrot (collected 2010) (Ge et al., 2012). These were cultured and stored as described previously.
DNA Extraction and Molecular Identification of S. sclerotiorum and S. subarctica
Sclerotinia spp. cultures were initiated from stock sclerotia and incubated on PDA at 20°C for 3–4 days to produce actively growing colonies. Three agar plugs were taken from the leading edge, placed into Petri dishes containing half strength PDB, and incubated at 20°C for 3 days. The agar plugs were then removed and the mycelial mat washed twice in sterilized reverse osmosis (RO) water and blotted dry on tissue (KimTech; Kimberly-Clark Ltd, UK) before being freeze-dried overnight. Genomic DNA was extracted from the freeze-dried mycelium using a DNeasy Plant Mini Kit (Qiagen Ltd, UK) following the manufacturer's protocol.
S. subarctica and S. sclerotiorum isolates were distinguished by PCR amplification of the large subunit of the ribosomal DNA (LSU), where a large (304 bp) intron is absent in S. subarctica compared to S. sclerotiorum (Holst-Jensen et al., 1998). The 25 μl PCR reaction mixture consisted of 1 x REDTaq ReadyMix PCR reaction mix (Sigma-Aldrich, UK), LR5 and LROR primers (0.4 μmol L−1; (Vilgalys and Hester, 1990) and ~10 ng DNA template. Thermal cycling parameters were 94°C for 2 min; 35 cycles of 94°C for 60 s, 52°C for 60 s, 72°C for 60 s; 72°C for 10 min and then a hold at 12°C. PCR products were visualized on a 1.5% agarose gel with a DNA ladder (EasyLadder I, Bioline Reagents Ltd, UK). Isolates associated with the smaller sized amplicons were identified as S. subarctica and this was further confirmed by PCR amplification and sequencing of the rRNA ITS region. Here, the PCR reaction mixture of 25 μl consisted of 1 x REDTaq ReadyMix PCR reaction mix (Sigma-Aldrich, UK), modified standard ITS primers (White et al., 1990) for S. sclerotiorum ITS2AF (TCGTAACAAGGTTTCCGTAGG) and ITS2AR (CGCCGTTACTGAGGTAATCC; 0.4 μmol L−1) and approximately 10 ng DNA template. Thermal cycling parameters were 94°C for 2 min; 40 cycles of 94°C for 15 s, 59°C for 15 s, 72°C for 30 s; 72°C for 10 min and then a hold at 12°C. PCR products were visualized on a 1.5% agarose gel to confirm amplification, purified using the QIAquick PCR purification kit (Qiagen, UK), and sequenced (ITS2AF/ITS2AR primers) by GATC Biotech (Germany). ITS sequences obtained for all the S. subarctica isolates were aligned using the ClustalW algorithm implemented in MEGA v6 (Tamura et al., 2013) and sequence identity was confirmed by BLASTn analysis.
Molecular Characterization of Sclerotinia subarctica Isolates Using Microsatellites
Isolates identified as S. subarctica were characterized using eight microsatellite markers in two multiplexed PCR reactions (loci MS01, MS03, MS06, MS08 and MS02, MS04, MS05, MS07) with fluorescent-labeled primer pairs (Applied Biosystems, UK) as developed by Winton et al. (2007). Each PCR reaction mixture of 20 μl consisted of 1 x QIAGEN Multiplex PCR Master Mix, 0.5 x Q solution, primer mix (0.4 μmol L−1) and ~10 ng DNA template. Thermal cycling parameters were 95°C for 15 min; 35 cycles of 94°C for 30 s, 55°C for 90 s, 69°C for 75 s; 60°C for 30 min and then a hold at 12°C. PCR products were visualized on a 1.5% agarose gel to confirm amplification and two separate PCR amplifications per locus were carried out for each isolate to ensure reproducibility of results. The size of all PCR amplicons was determined by Eurofins (Germany) using an ABI 3130xl genetic analyser and allele sizes assigned using GENEMARKER (Version 1.6; SoftGenetics, USA). FLEXIBIN (Amos et al., 2007) was then used to bin allele sizes and estimate the relative number of repeats at each locus.
Molecular Characterization of Sclerotinia Sclerotiorum Isolates Using Microsatellites and IGS sequencing
Isolates identified as S. sclerotiorum were characterized using eight microsatellite markers (Sirjusingh and Kohn, 2001) in two multiplexed PCR reactions (loci 13-2, 17-3, 55-4, 110-4, 114-4 and 7-2, 8-3, 92-4) using fluorescent-labeled primer pairs (Applied Biosystems, UK). The PCR reaction mixture of 10 μl consisted of 1 x QIAGEN Multiplex PCR Master Mix, 0.5 x Q solution, forward and reverse primer pairs (0.2 μmol L−1) and ~10 ng DNA template. Thermal cycling parameters were 95°C for 15 min; 35 cycles of 94°C for 30 s, 55°C for 90 s, 69°C for 75 s; 69°C for 75 s and then a hold at 12°C. PCR products were visualized on a 1.5% agarose gel to confirm amplification and two separate PCR amplifications per locus were carried out for each isolate to ensure reproducibility of results. The size of all PCR amplicons was determined by Eurofins (Germany) using an ABI 3130xl genetic analyser and allele sizes assigned using GENEMARKER (Version 1.6; SoftGenetics, USA). FLEXIBIN (Amos et al., 2007) was then used to bin allele sizes and estimate the number of repeats for each locus.
S. sclerotiorum isolates were also characterized by sequencing part of the IGS region of the rRNA gene where PCR primers IGS2F (TTACAAAGATCCTCTTTCCATTCT) and IGS2R (GCCTTTACAGGCTGACTCTTC) (Clarkson et al., 2013) were used to amplify an 834 bp (approx.) fragment. The PCR reaction mixture of 25 μl consisted of 0.5 x REDTaq ReadyMix PCR reaction mix (Sigma-Aldrich, UK), IGS2F and IGS2R primers (4 μmol L−1) and ~10 ng DNA template. Thermal cycling parameters were 94°C for 2 min, 40 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 2 min followed by 72°C for 10 min and a hold of 12°C. PCR products were visualized on a 1.5% agarose gel to confirm amplification, purified using the QIAquick PCR purification kit (Qiagen, UK), and sequenced (IGS2F/IGS2R primers) by GATC Biotech (Germany). IGS primers did not consistently amplify an initial selection of S. subarctica isolates and hence sequences were not generated for this species.
Analysis of Sclerotinia sclerotiorum and Sclerotinia subarctica Microsatellite Data
ARLEQUIN (Excoffier et al., 2005) was used to determine the haplotype frequency of S. subarctica /S. sclerotiorum isolates for each country based on the relative number of repeats at each microsatellite locus and to identify shared haplotypes. Genodive (Meirmans and van Tienderen, 2004) was used to calculate gene diversity (Hs) for each locus (Nei, 1987) and the average across all loci and furthermore generate clonal (haplotype) diversity statistics for each Sclerotinia species in the different countries. These comprised haplotype diversity (div) (Nei, 1987) and a corrected form of the Shannon-Wiener Index (shc). The former is based on frequencies of haplotypes in each population while the latter is based on the abundance and evenness of haplotypes. While div is independent of sample size (Nei, 1987) the Shannon index is prone to bias when comparing unequal sample sizes. However, the corrected form calculated in Genodive accounts for this through a non-parametric approach which uses unequal probability sampling theory (Chao and Shen, 2003). Calculations of both div and shc as implemented in Genodive also included a jackknife approach to estimate the relationship between sample size and diversity and in all cases, the variance in diversity decreased with increasing population size and leveled off below the population size sampled. A bootstrap test (1,000 permutations) also implemented in Genodive allowed us to test if Sclerotinia populations from different countries differed in their haplotype diversity as measured by div and shc. POPPR (Kamvar et al., 2014) was used to calculate multilocus indices of disequilibrium; the index of association IA and the index rd which accounts for the number of loci (Agapow and Burt, 2001). ARLEQUIN was used to test for subdivision for both S. sclerotiorum and S. subarctica populations from different countries and was estimated through pairwise comparisons of RST (Slatkin, 1995), a statistic which uses a stepwise mutation model which has been widely implemented for microsatellite data [including for S. sclerotiorum e.g., Aldrich-Wolfe et al. (2015)], with significance tested by permuting (1,023) haplotypes between populations. The analysis for S. sclerotiorum included data sets for the 12 English S. sclerotiorum populations (384 isolates, Table 1) published previously (Clarkson et al., 2013).
S. subarctica and S. sclerotiorum microsatellite data were subjected to Bayesian population structure analyses using STRUCTURE v2.3.3 (Pritchard et al., 2000), an approach used previously for Sclerotinia spp. (Attanayake et al., 2013, 2014; Njambere et al., 2014). The markers used in this study map to different chromosomes of the S. sclerotiorum reference genome (Amselem et al., 2011) with the exception of 7-2 and 114-4 which both map to chromosome 4 and 13-2 and 110-4 which both map to chromosome 6. These pairs that map to the same chromosomes were shown by Attanayake et al. (2014) to be of sufficient distance for LD to decay (Bastien et al., 2014) and therefore satisfy the assumptions of STRUCTURE. A burn-in period of 300,000 Markov Chain Monte Carlo iterations and a 300,000 run-length was implemented using an admixture model and correlated allele frequencies for K values between 1 and 6. For each simulated cluster for K = 1–6, four runs were repeated independently for consistency. For the S. sclerotiorum data, this was followed by a second analysis using a burn-in period of 500,000 Markov Chain Monte Carlo iterations and a 500,000 run-length implemented using an admixture model and correlated allele frequencies for K values between 3 and 5. Again, for each simulated cluster for K = 3–5, four runs were repeated independently. For both Sclerotinia species, the python script structure Harvester.py v0.6.92 (Earl and vonHoldt, 2012) was then used to summarize the STRUCTURE output, producing ΔK values using the Evanno method (Evanno et al., 2005) to estimate the most likely underlying K. Replicate simulations of cluster membership (q-matrices) at K = 4 for S. sclerotiorum isolates and K = 2 for S. subarctica isolates were used as input for CLUMPP_OSX.1.1.2 (Jakobsson and Rosenberg, 2007) using the Fullsearch algorithm, with weighted H and the G similarity statistic. Summarized cluster membership matrices (q-values) for both individuals and populations were then visualized using distruct_OSX1.1 (Rosenberg, 2004).
The microsatellite data represented a multivariate dataset and to reduce the complexity of the data, principal component analyses (PCA) were used to complement the STRUCTURE analyses. For both Sclerotinia species, analyses were performed first on the microsatellite repeat size data, and then on an allele score matrix constructed by subdividing each microsatellite into repeat size categories, then scoring if the repeat size is present or absent in each isolate, forming a binary score data matrix. Principal components were estimated using the singular value decomposition method implemented in R v3.2.3 (R-Development-Core-Team, 2015) using the built in “prcomp” function and the package FactoMineR v1.31.4 (Lê et al., 2008). Scatter plots of the component scores were produced using ggplot2 (Wickham, 2016), with ellipses representing Euclidean distance from the center (confidence level = 0.95) of each cluster.
Analysis of IGS Sequence Data for Sclerotinia sclerotiorum
S. sclerotiorum IGS sequences were aligned using the ClustalW algorithm implemented in MEGA v6 (Tamura et al., 2013) and DNASP v. 5 (Librado and Rozas, 2009) was used to identify haplotypes based on sequence differences (omitting indels), calculate haplotype diversity and also used to examine subdivision between populations from different countries using pairwise comparisons of the nearest neighbor statistic (Snn) (Hudson, 2000) with significance calculated with 1,000 permutations. Again, data sets for the 12 English S. sclerotiorum populations (384 isolates) published previously (Clarkson et al., 2013) were also included in the analysis. A median joining network of IGS haplotypes (Bandelt et al., 1999) which included sequence data from Canada, New Zealand, Norway, and the USA (Carbone and Kohn, 2001a) was constructed using NETWORK v. 4.6 (Fluxus Technology, USA) for all the datasets from England, Scotland, Norway, and Australia.
Results
Molecular Identification and Frequency of Sclerotinia subarctica
A total of 843 Sclerotinia isolates were collected from crop plants and buttercup in England (337), Scotland (404), and Norway (102). Identification through amplification of the LSU rDNA showed that 142 of these were S. subarctica (Table 1) with the proportion rising to 157 of 1,255 isolates when previous data from England (412 isolates) was included (Table 1). S. subarctica was detected on a wide range of host plants and there was increased incidence in samples collected from Scotland and Norway. In England, only 35 of 749 Sclerotinia isolates (4.7%) were S. subarctica and these were all isolated from a single buttercup meadow in Herefordshire over 3 years (2009–2011). In Scotland however, 74 of 404 isolates (18.3%) were S. subarctica and were found in the majority of crops, locations and years between 2010 and 2013. In Norway, 49 of 102 Sclerotinia isolates (48.0%) collected in 2012/2013, again from a wide range of crop types and locations were identified as S. subarctica. Identity of all S. subarctica isolates was further confirmed by sequencing of the rRNA ITS region and all sequences were identical to that previously deposited in Genbank (GU018183) (Clarkson et al., 2010).
Molecular Characterization of Sclerotinia subarctica Isolates Using Microsatellites
Microsatellite analysis of the S. subarctica isolates resulted in 5 to 10 polymorphic alleles per locus, with loci MS01, MS02, MS03, MS04, MS05, MS06, MS07, and MS08 having 10, 6, 5, 10, 5, 7, 7, and 6 alleles respectively (Table 2). Two of the loci (MS02 and MS04) were monomorphic for the isolates from England. Overall, 75 microsatellite haplotypes were identified within the 157 S. subarctica isolates genotyped from England, Scotland, and Norway (Figure 1A). The number of different haplotypes as a proportion of the total number of isolates differed between countries, England having fewer haplotypes (14%; 5 haplotypes within 34 isolates) compared to Scotland (51%; 38 haplotypes within 74 isolates) and Norway (82%, 40 haplotypes within 49 isolates). Over all the 75 S. subarctica haplotypes, 18 were represented by more than one isolate with eight shared between Scotland and Norway but none shared between England and Scotland or England and Norway (Figure 1A). A few haplotypes were sampled more frequently than the rest. The most prevalent haplotypes in England were haplotypes 1 and 4 (19 and 7 isolates respectively; Table 3) and both were found in each of the 3 years sampling at the buttercup meadow in Herefordshire. Haplotypes 2 and 3 were most prevalent for both Scotland and Norway (19 and 15 isolates respectively; Table 3) and were represented in samples from potato, buttercup and swede (Scotland) and carrot, lettuce and swede (Norway). Haplotype diversity measures div and shc were significantly lower for the S. subarctica isolates from England compared to Scotland and Norway (Table 4; P < 0.001) as was the diversity in Scotland compared to Norway (P < 0.05). The index of association IA for S. subarctica microsatellite data ranged between 0.57 (Norway) and 2.9 (England) while rd was between 0.08 (Norway) and 0.58 (England). Significance testing showed that the hypothesis of random mating was rejected in all cases (P < 0.001; Table 4). This was also true when clone-corrected data was used in the analysis (Table 4). There was also evidence of subdivision between the S. subarctica populations from the three different countries with the RST fixation index statistic highly significant (P < 0.0001) for pairwise combinations of England/Scotland (RST = 0.637) and England/Norway (RST = 0.591). However, this was less significant (P < 0.05) for the Scotland/Norway combination (RST = 0.030) as might be expected given some sharing of haplotypes.
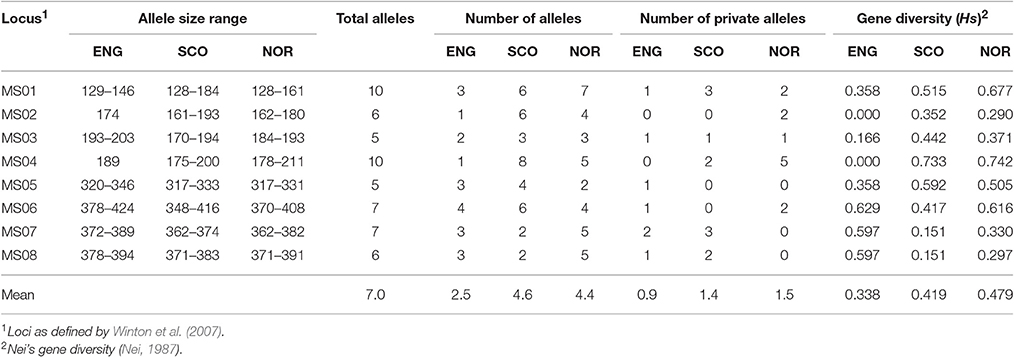
Table 2. Summary of microsatellite data for S. subarctica isolates from England, Scotland, and Norway.
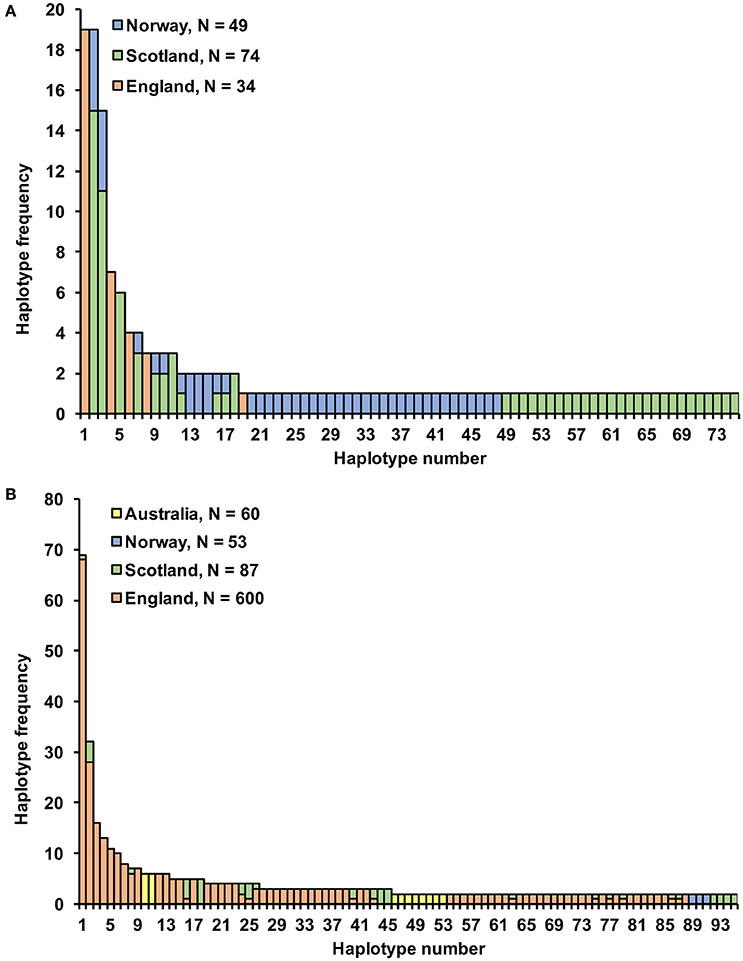
Figure 1. Microsatellite haplotype frequency for (A) 157 S. subarctica isolates from England, Scotland, and Norway; (B) 800 S. sclerotiorum isolates from England, Scotland, Norway, and Australia for 95 haplotypes comprising more than one isolate.
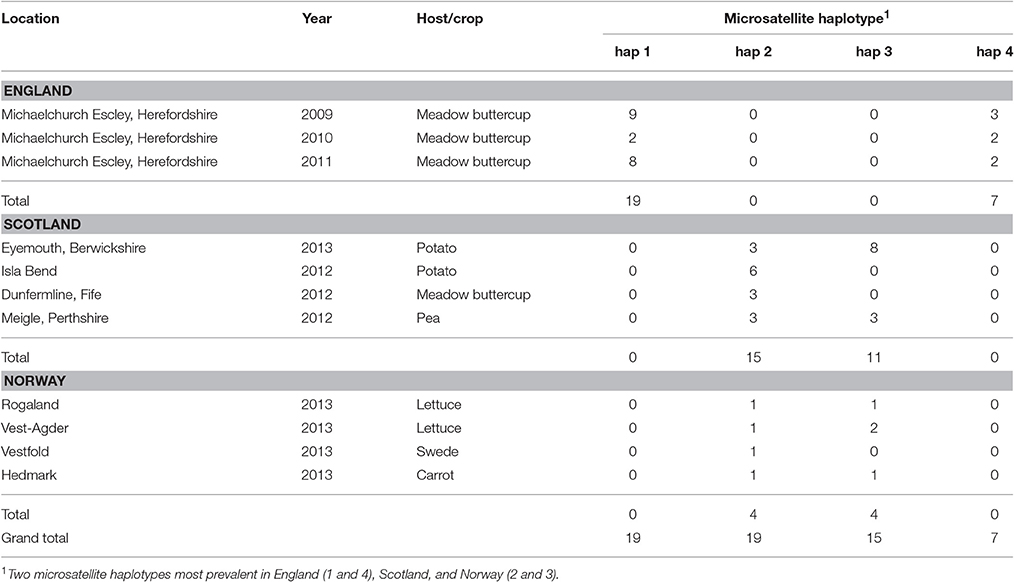
Table 3. Location, year, host, and frequency of most common S. subarctica microsatellite haplotypes in England, Scotland, and Norway.
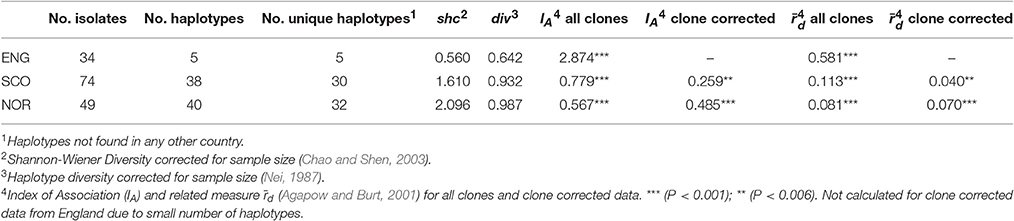
Table 4. Diversity statistics and disequilibrium measures for S. subarctica isolates from England (ENG), Scotland (SCO), and Norway (NOR) based on microsatellite data.
The Bayesian cluster analysis of the S. subarctica microsatellite data using STRUCTURE suggested two genetically distinct ancestral populations (K = 2, being the value associated with the highest value of ΔK). Examining the probability of each isolate belonging to either of the two sub-populations described by the membership coefficient matrix, all 34 of the isolates from buttercup collected from the single site in Herefordshire (Michaelchurch Escley) in England between 2009 and 2012 were assigned to cluster q1, whereas all the isolates from Norway (49) and 68 of the 74 (92%) isolates from Scotland were assigned to q2 (Figure 2A). This suggests that the majority of the isolates from Norway and Scotland share a common ancestry. However, six of the Scottish isolates (SC52-SC57), which were all collected from meadow buttercup at a site near Dunfermline (Fife) in 2012 were assigned to q1 and hence appear to share ancestry with the English isolates. The reduced genetic diversity in the English isolates and the shared ancestry of Scottish and Norwegian S. subarctica isolates was also evident in the principal component analysis using either microsatellite repeat number or the binary matrix. The first principal component clearly distinguished Scottish and Norwegian isolates from the English isolates and the reduced genetic variation of the English isolates was indicated by the reduced space they occupied in the PCA map, particularly in dimension 2 (Figures 3A,B).
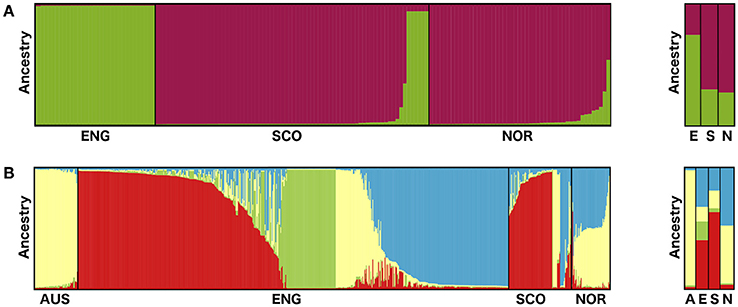
Figure 2. Bar plots showing the assignment of (A) S. subarctica isolates (N = 157) from England (ENG, N = 34), Scotland (SCO, N = 74), and Norway (NOR, N = 49) to two ancestral populations and (B) S. sclerotiorum isolates (N = 800) from Australia (AUS, N = 60), England (N = 600), Scotland (N = 87), and Norway (N = 53) to three ancestral populations from STRUCTURE analysis of microsatellite data. Assignment to populations is based on q values for each isolate (S. subarctica q1 green, q2 purple; S. sclerotiorum q1 red, q2 green, q3 yellow, q4 blue) and population q values for each country (A, Australia; E, England; S, Scotland; N, Norway.).
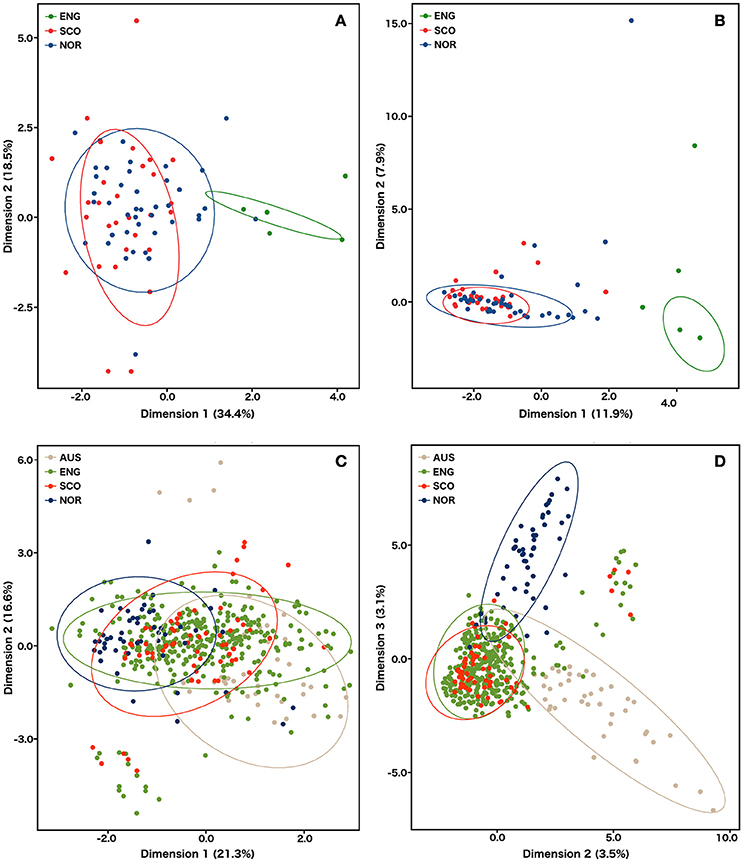
Figure 3. Principal component analysis of microsatellite data for S. subarctica isolates from England (N = 34), Scotland (N = 74), and Norway (N = 49) and S. sclerotiorum isolates from Australia (N = 60), England (N = 600), Scotland (N = 87), Norway (N = 53). (A) S. subarctica individuals factor map estimated using microsatellite repeat number; (B) S. subarctica individuals factor map estimated using an allele presence binary matrix; (C) S. sclerotiorum individuals factor map estimated using microsatellite repeat number; (D) S. sclerotiorum individuals factor map estimated using an allele presence binary matrix. Individual isolates are colored by geographic origin. Ellipses represent the Euclidean distance from the center (confidence level = 0.95) of each cluster.
Molecular Characterization of Sclerotinia sclerotiorum Isolates Using Microsatellites
Microsatellite analysis of the S. sclerotiorum isolates resulted in 7 to 19 polymorphic alleles per locus, with loci 7-2, 8-3, 13-2, 17-3, 55-4, 92-4, 110-4, 114-4 having 11, 15, 19, 20, 20, 7, 8, and 2 alleles respectively (Table 5). Overall, 484 microsatellite haplotypes were identified within the 800 S. sclerotiorum isolates that were genotyped (Figure 1B). In England, 343 haplotypes were found within 600 isolates (57%), with 64 (74%), 50 (94%), and 42 haplotypes (70%) identified within 87, 53, and 60 isolates from Scotland, Norway, and Australia respectively. Over all 800 S. sclerotiorum haplotypes, 95 were represented by more than one isolate while 15 were shared between England and Scotland (Figure 1B). No haplotypes were shared between any other countries. In each country, there were a small number of haplotypes sampled more frequently than the rest. In England, 68 isolates representing the most prevalent haplotype 1 were found in the majority of hosts and locations sampled and a further single representative was identified in Scottish buttercup (Figure 1B, Table 6). The second most prevalent haplotype 2 in England comprised 25 isolates from different buttercup meadows but was only identified in a single crop location (lettuce, three isolates). Haplotype 2 was also found in two buttercup meadows (four isolates, 2011; Table 6) in Scotland. However, the most prevalent haplotype 18 in Scotland was represented by five isolates found in two different buttercup locations (2011) and in carrot (2010) (Table 6), but was not present in England. Haplotype 16 comprising four isolates from buttercup and carrot in Scotland (Table 6) was also found in one buttercup meadow in England (Holywell, 2008). In Norway, the most prevalent haplotypes were haplotype 89, 90, and 91 each of which was represented by just two isolates from oilseed rape, pumpkin, and turnip rape hosts while the two most prevalent haplotypes 10 and 11 in Australia both comprised six isolates each from oilseed rape and lupin from different locations (Table 6).
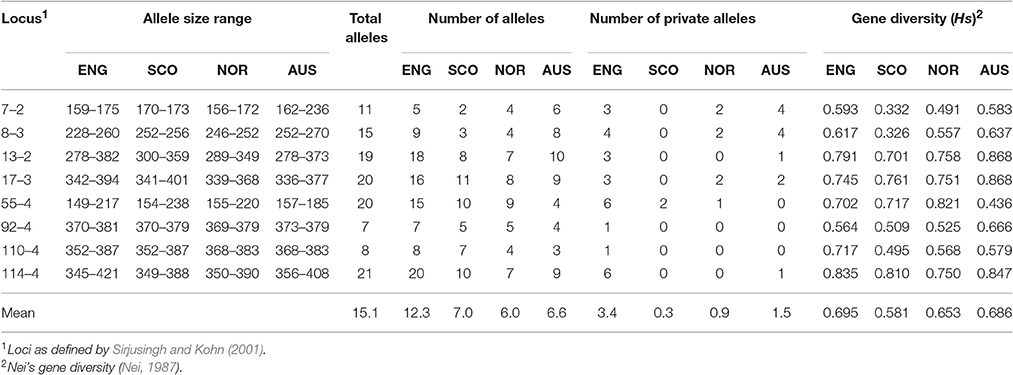
Table 5. Summary of microsatellite data for S. sclerotiorum isolates from England (ENG), Scotland (SCO), Norway (NOR), and Australia (AUS).
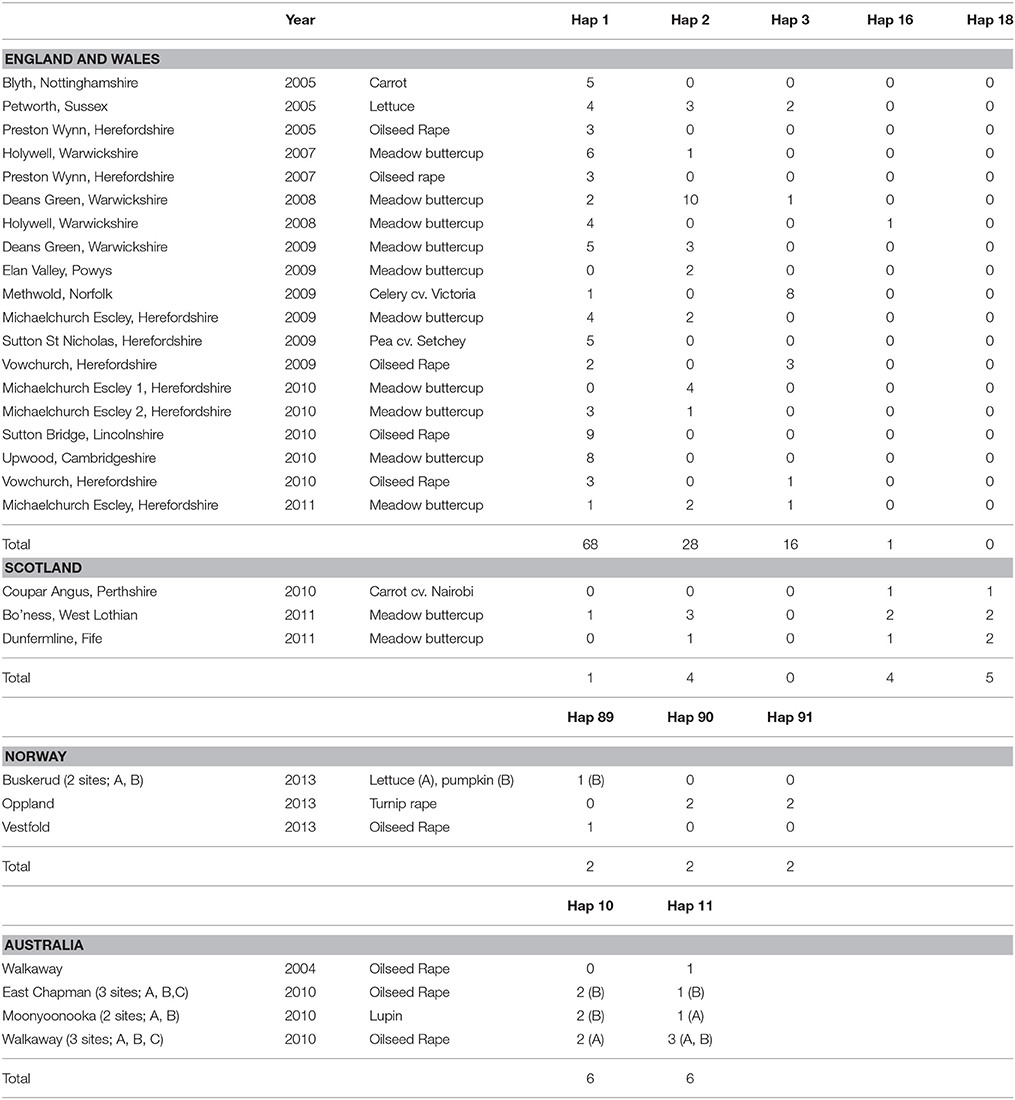
Table 6. Location, year, host, and frequency of the most prevalent S. sclerotiorum microsatellite haplotypes in England (Hap 1, 2, 3), Scotland (Hap 2, 16, 18), Norway (Hap 89, 90, 91), and Australia (Hap 10, 11).
Haplotype diversity measures of div and shc for the S. sclerotiorum microsatellite data were generally high for England Scotland and Norway but lower for the Australian isolates (Table 7) but this was only significant for the Norway/Australia pairwise combination for div (P < 0.05). However, pairwise comparisons showed that shc was significantly greater for England compared to Scotland (P < 0.05) and Australia (P < 0.01) while shc was significantly greater in Norway compared to all the other three countries (P < 0.001). There was also evidence of genetic differentiation between the S. sclerotiorum populations from the four different countries with the RST fixation index statistic highly significant (P < 0.0001) for pairwise combinations of England/Norway (RST = 0.094), England/Australia (RST = 0.186), Scotland/Norway (RST = 0.068), Scotland/Australia (RST = 0.152), and Norway/Australia (RST = 0.198). Differentiation between England/Scotland was less significant (P < 0.01, RST = 0.015). The index of association IA for S. sclerotiorum microsatellite data ranged between 0.26 (Norway) and 0.95 (Australia) while rd was between 0.04 (Norway) and 0.14 (Australia). Significance testing showed that the hypothesis of random mating was rejected in all cases (P < 0.001; Table 7). This was also the case when clone-corrected data was used in the analysis (Table 7).
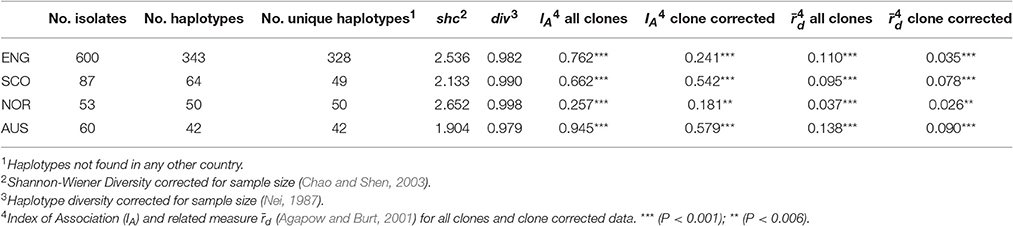
Table 7. Diversity statistics and disequilibrium measures for S. sclerotiorum isolates from England (ENG), Scotland (SCO), Norway (NOR), and Australia (AUS) based on microsatellite data.
The Bayesian cluster analysis of the S. sclerotiorum microsatellite data using STRUCTURE suggested the number of genetically distinct ancestral populations was best represented by K = 4 clusters, the value associated with the highest value of ΔK (Figure 2B) The majority of English isolates were assigned to cluster q1 (251, 42%) and q4 (189, 32%) followed by q2 (84, 14%) and to a lesser extent q3 (53 isolates, 9%). Similarly, most of the Scottish isolates were also assigned to cluster q1 (60 isolates, 69%) with the remainder approximately equally divided between q3 and q4. Norwegian isolates were predominantly assigned to either q3 or q4 populations, while the Australian isolates were all associated with cluster q3. Overall this suggests a shared ancestry for the English and Scottish isolates through q1 and q4, while the Norwegian isolates appear to share ancestry with England, Scotland, and Australia via q3 and q4 (Figure 2B).
Principal component analysis using the microsatellite repeat size data separated the isolates into broad clusters representing country of origin (Figure 3C). However, the extensive diversity captured in the English isolates led to a spread of these isolates across the first component space, overlapping with the Australian, Scottish, and Norwegian isolates and the second component failed to resolve the different populations into clear clusters. When the binary matrix values of the microsatellite data were used, the number of variables increased from 8 to 122 thereby enhancing the discriminative power of the principal component analysis. This resulted in the isolates being resolved into clear population clusters, with the English and Scottish isolates being grouped together, and the Norwegian and Australian isolates forming individual distinct clusters (Figure 3D), which confirmed the results of the STRUCTURE analysis.
Molecular Analysis of Sclerotinia sclerotiorum Isolates Using IGS Sequencing
In total, 26 IGS haplotypes were identified within the 870 S. sclerotiorum isolates (Table 8) from England (600 isolates), Scotland (87 isolates), Norway (52 isolates), and Australia (131 isolates). The most common haplotype was IGS3 (333 isolates), found in all three countries, closely followed by IGS2 (269 isolates) which was found in all countries except for Australia. In England, IGS3 (297 isolates) and IGS2 (179 isolates) were the most common haplotypes and were represented by isolates from every location and crop type as well as buttercup. In Scotland, the most prevalent haplotype was IGS2 (49 isolates) followed by IGS1 (16 isolates), both represented in all the buttercup and carrot populations genotyped. In Norway, the most prevalent haplotype was IGS2 (41 isolates) found in cabbage, camelina, lettuce, oilseed rape, and turnip rape from different locations. Conversely in Australia, the most common haplotype was IGS5 (58 isolates) followed by IGS7 (45 isolates) found in both lupin and oilseed rape across the majority of locations. A total of nine IGS haplotypes (IGS4, 8, 9, 10, 11, 12, 13, 20, 23) from England and Scotland were exclusively found in buttercup isolates. With these data, the haplotype network first published by Clarkson et al. (2013) was expanded considerably from 17 to 26 IGS haplotypes (Figure 4). The nine new haplotypes were from England (IGS18, Vowchurch oilseed rape 2009; IGS19, Sutton Bridge oilseed rape 2010; IGS20, Michaelchurch buttercup site 2, 2010), Australia (IGS21, oilseed rape different locations; IGS22, Mount Barker oilseed rape 2004), Scotland (IGS23, Dunfermline buttercup 2011), and Norway (IGS24, Buskerud lettuce 2013; IGS25, Nord-Trøndelag potato 2013; IGS26, Buskerud pumpkin 2013) (Genbank accession numbers KY798871-KY798879). The nearest neighbor statistic Snn values indicated that populations of S. sclerotiorum from different countries were all significantly differentiated from each other (Table 9).
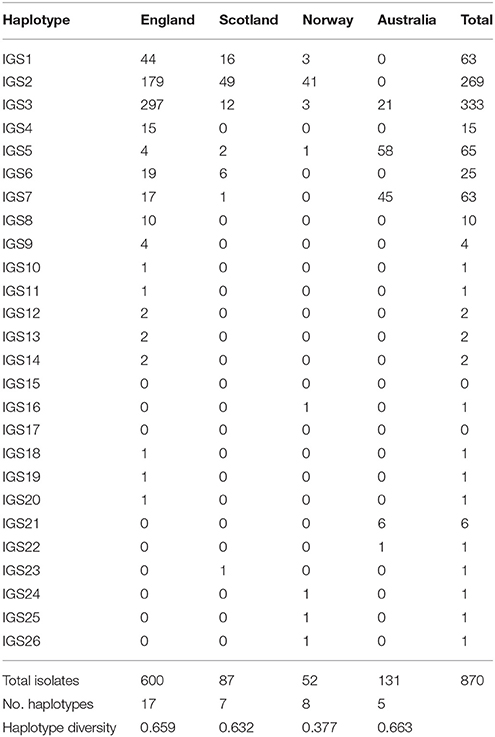
Table 8. IGS haplotype frequency and diversity for S. sclerotiorum isolates from England, Scotland, Norway, and Australia.
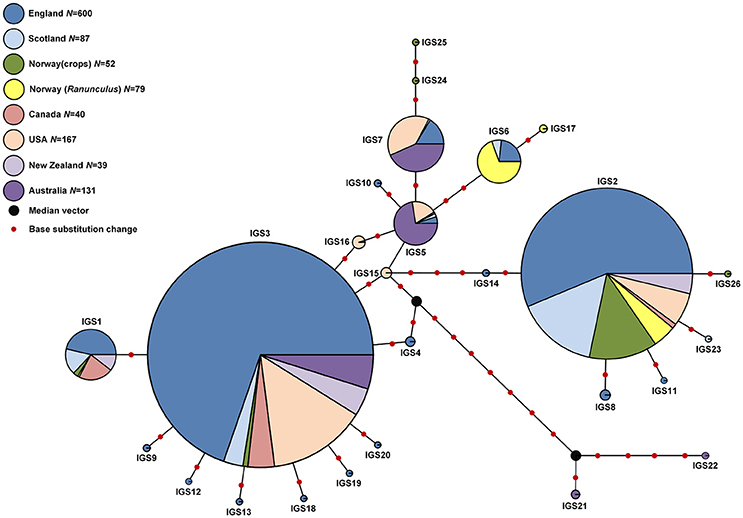
Figure 4. Median joining networks showing phylogenetic relationships between IGS haplotypes for S. sclerotiorum isolates from Australia, England, Scotland, and Norway (this study) and Canada, USA, New Zealand and Lesser Celandine (R. ficaria) in Norway (Carbone and Kohn, 2001a). The size of each circle is proportional to the corresponding haplotype frequency. Branch lengths are proportional to the number of base substitution changes. A median vector represents a hypothesized haplotype required to connect existing haplotypes within the network.
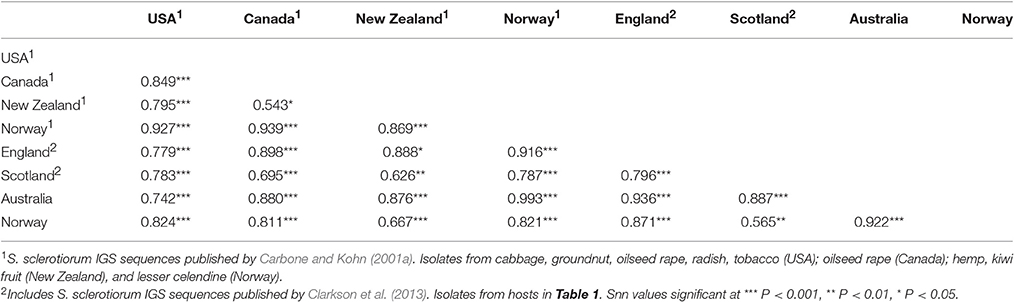
Table 9. Nearest neighbor statistic (Snn values) for S. sclerotiorum populations from USA, Canada, New Zealand, Norway (Carbone and Kohn, 2001a), and England, Scotland, Australia, and Norway (Clarkson et al., 2013, this study).
Discussion
There has been very little research concerning S. subarctica following the initial rDNA-based phylogeny of Sclerotinia species by Holst-Jensen et al. (1998) and the first report of the pathogen on crop plants in Alaska (Winton et al., 2006) where it was identified on potato, lettuce, cabbage, bean, and squash. This study shows for the first time a clear increase in the incidence of S. subarctica with increasing latitude from England, through to Scotland and Norway. In England, the pathogen appears to be insignificant compared to S. sclerotiorum, being identified in only one location (where originally first reported; Clarkson et al., 2010) on meadow buttercup despite widespread sampling. However, a much higher proportion of the Sclerotinia samples were identified as S. subarctica in Scotland (18%) and Norway (48%) with the latter very similar to the 46% reported in Alaska (Winton et al., 2006). Both Alaska and Norway occupy very similar latitude ranges. S. subarctica was also widely distributed across different crop plants and buttercup with new hosts reported here for the first time of carrot, celery root, Jerusalem artichoke, pea, and swede. This finding is therefore further evidence that S. subarctica has a broad host range similar to S. sclerotiorum or S. minor.
The reasons for the prevalence of S. subarctica in northern latitudes are unclear although it has been suggested that the larger sclerotia the pathogen generally produces may confer a survival advantage over S. sclerotiorum in harsh winters (Winton et al., 2006). Furthermore, we hypothesize that S. subarctica sclerotia may require an increased chilling requirement for rapid germination and apothecial production compared to S. sclerotiorum as observed in preliminary experiments (Warmington, 2014). If this is the case, then a lack of adequate cold temperature durations may therefore limit its reproductive ability, hence slowing down its spread further south. A requirement for cold “conditioning” for S. sclerotiorum sclerotia is well-documented for isolates from temperate regions (Phillips, 1987) although germination response to different temperature regimes varies between isolates from different geographic origins (Huang and Kozub, 1991). This, along with the extensive distribution of S. sclerotiorum (Anonymous, 2005), suggests adaptation to a much wider range of conditions for apothecial production than for S. subarctica.
This is the first study to extensively examine the population structure of S. subarctica and the finding that there are multiple clones (haplotypes) suggests a very similar reproductive strategy to S. sclerotiorum based on homothallic sexual reproduction through carpogenic germination of sclerotia. This is confirmed by observations of apothecial production by S. subarctica both in the laboratory (Warmington, 2014) and in the field (Winton et al., 2006). Furthermore, measures of linkage disequilibrium in the S. subarctica populations were significant which is again consistent with a selfing clonal population. This was also the case for the S. subarctica population from Alaska (Winton et al., 2007). This confirms the majority of studies with S. sclerotiorum where evidence of outcrossing is generally infrequent. However, more recent analyses using linkage disequilibrium decay with distance indicates that outcrossing can be much more common than is suggested solely by measurement of IA (Attanayake et al., 2014).
Compared to the preliminary study in Alaska (Winton et al., 2007) where only four microsatellite haplotypes were identified within 41 S. subarctica isolates (10% of maximum), we identified a more diverse range of haplotypes in Scotland and Norway with 38 and 40 haplotypes found within 74 and 49 isolates respectively (51 and 82% of maximum). Furthermore, only 2–3 alleles were identified per locus in the Alaskan work compared to 5–10 alleles in this study. One possible explanation for this is that the Alaskan isolates were all collected from a confined area in the Matanuska Valley which was only developed for agriculture in the early 1900s. Therefore, there has been less opportunity for immigration of different S. subarctica haplotypes via crop plant or soil-based introductions compared to Scotland or Norway.
Another significant finding in our study is that a few S. subarctica haplotypes were found more frequently than the rest across multiple crops and locations. For instance, two microsatellite haplotypes which were the most prevalent in both Scotland and Norway were identified in 13 different locations in crops of carrot, lettuce, pea, potato, swede as well as buttercup. Scotland and Norway also shared a further six microsatellite haplotypes. This therefore follows the same pattern of distribution as observed for S. sclerotiorum both in this study and in previous research (Kohli et al., 1992; Kohn, 1995; Hambleton et al., 2002). The common haplotypes between Scotland and Norway suggests either a common origin and/or exchange of isolates and admixture between these two S. subarctica populations. This was supported by the STRUCTURE and PCA analyses where the majority of isolates from each country were assigned to the same clusters. Therefore, as suggested for S. sclerotiorum (Kohn, 1995; Hambleton et al., 2002), it seems likely that certain S. subarctica haplotypes persist following initial immigration due to the longevity of sclerotia in the soil with new haplotypes arising locally through mutation and infrequent outcrossing. Haplotypes present across multiple hosts including both crop plants and buttercup for both S. subarctica and S. sclerotiorum in this study confirms previous reports that there is no evidence for host specialization (Bolton et al., 2006) in either pathogen and that wild hosts can also potentially act as a source of inoculum on crop plants as well as enabling the pathogen to survive in the absence of a susceptible crop host.
In England however, S. subarctica was restricted to a single buttercup meadow in Herefordshire where repeated detection of the same microsatellite haplotypes each year indicated continual survival and cycling of the same pathogen isolates. Although repeat sampling of this meadow makes direct comparisons with the populations from Scotland and Norway problematic, none of the English S. subarctica microsatellite haplotypes were found in Scotland or Norway and they were also clearly assigned to a different population cluster in both the STRUCTURE and PCA analyses. The lack of evidence for admixture in the English isolates suggests that the same ancestral population has endured in isolation without any influx of additional genetic diversity resulting in fixation of alleles. This may have been caused by a founder effect following an initial introduction, with the initial population being a skewed sampling of the alleles from an overall larger population. Evidence for the isolation of the English S. subarctica population was further supported by highly significant differentiation from Scottish and Norwegian populations and as well as low genetic diversity with only five microsatellite haplotypes identified. A similar situation was described for an isolated S. sclerotiorum population on R. ficaria in Norway which was characterized by low diversity and apparent localized inbreeding (Kohn, 1995). In addition, despite the availability of susceptible hosts in the area S. subarctica was not identified elsewhere locally. For instance, no S. subarctica was identified in an adjacent buttercup meadow (Michaelchurch Escley2, Herefordshire) approx. 100 m away (but separated by a road and hedges), although affected by S. sclerotiorum. Similarly, S. subarctica was not detected in two different oilseed rape fields 6 km away (Vowchurch 2009 and 2010), where S. sclerotiorum was again identified. This could suggest a limited ability of S. subarctica to spread, despite its potential ability to produce airborne ascospores via apothecia (Winton et al., 2006; Warmington, 2014). However, studies with S. sclerotiorum have shown that the majority of ascospores may only travel 40–60 m while long distance dispersal depends on wind speed and direction (Qandah and Del Rio Mendoza, 2012).
Populations of S. sclerotiorum from England, Scotland, Norway, and Australia were also analyzed in this study and in contrast to our previous work which only examined population structure at regional level in England (Clarkson et al., 2013), these additional isolates allowed us to examine populations at a different spatial (country) scale. Although clonal diversity measures were significantly lower for Australia compared to the other countries, especially for Hc, these estimates were comparable to other studies for S. sclerotiorum using microsatellites (e.g., Attanayake et al., 2013; Aldrich-Wolfe et al., 2015). The 15 shared microsatellite haplotypes between England and Scotland, assignment of the majority of isolates to two common clusters in the STRUCTURE analysis and a common cluster in the PCA analysis indicated admixture of these populations from a common origin which would be expected in adjacent countries. In contrast, the Australian S. sclerotiorum isolates were quite different from English/Scottish isolates as they were all assigned to different clusters in the STRUCTURE/PCA analyses suggesting a different ancestry which most likely reflects their geographic separation. However, despite the S. sclerotiorum isolates from Norway being separated from English and Scottish isolates in the PCA analysis, some appeared to potentially share some ancestry with both England/Scotland and Australia in the STRUCTURE analysis. Furthermore, in contrast to S. subarctica, there were no shared haplotypes between S. sclerotiorum populations in Norway and Scotland. This would therefore suggest a different pattern of initial distribution of S. sclerotiorum compared to S. subarctica.
The IGS sequence data for S. sclerotiorum provided a different level of phylogenetic resolution than the microsatellite data and a further nine new IGS haplotypes were identified, adding to the original 17 described by Clarkson et al. (2013), for S. sclerotiorum populations from the UK and previously published sequence data for populations from Canada, New Zealand, Norway, and the USA (Carbone and Kohn, 2001a). However, the new haplotypes were at low frequency while haplotype IGS 3, which was previously commonly found within S. sclerotiorum populations from all the above countries, was also identified in the Norwegian and Australian populations. The next most common haplotype IGS 2 was found in all countries except Australia. IGS sequencing therefore allows effective comparisons between different S. sclerotiorum populations globally and the frequency and relationship of the IGS haplotypes seen in the phylogenetic network continues to suggest the wide distribution of a small number of common haplotypes, with lower frequency haplotypes often emerging from these at the local scale as suggested by Clarkson et al. (2013).
Overall therefore, S. subarctica populations from Scotland and Norway appear admixed, with a common origin and shared microsatellite haplotypes. The lower incidence of S. subarctica in Scotland than in Norway and the rare occurrence of the pathogen in England may suggest a possible north-south migration and that the UK is at the limit of the pathogen's southerly distribution. However, further data and analysis would be required to test this theory. In contrast, for S. sclerotiorum, English and Scottish populations were similar, with shared microsatellite haplotypes and a common origin for many of the isolates. The Norwegian population showed only partial evidence of a common ancestry, and as for the Australian population, was clearly distinguished by PCA analysis. This therefore suggests limited admixture and geographic isolation between S. sclerotiorum populations from the UK, Norway, and Australia.
Author Contributions
JC conceived and obtained funding for the UK work, obtained isolates from diseased plants, carried out microsatellite genotyping and IGS sequencing of S. sclerotiorum and S. subarctica isolates and wrote paper. RW obtained isolates from diseased plants, carried out microsatellite genotyping and IGS sequencing of S. sclerotiorum and S. subarctica isolates and co-wrote paper. PW carried out Structure and PCA analyses of microsatellite data and edited paper. MD obtained isolates from diseased plants and carried out IGS genotyping of Australian S. sclerotiorum isolates. MB and GB obtained isolates from diseased plants and edited paper. BN conceived and obtained funding for the Norway work, obtained isolates from diseased plants and edited paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge funding from the UK Department for Food and Rural Affairs (Defra, project IF0188) and the Agriculture and Horticulture Development Board (AHDB, project CP80). The Norwegian contribution to this study was funded by Foundation for Research Levy on Agricultural Products/Agricultural Agreement Research Fund (research grant 207767), Regional Research Fund, Vestlandet (research grant 224787), with additional funding from agricultural industry partners and Norwegian lettuce growers. We also acknowledge funding from Australia's Grains Research & Development Corporation (research grant CUR00023).
References
Agapow, P.-M., and Burt, A. (2001). Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1, 101–102. doi: 10.1046/j.1471-8278.2000.00014.x
Aldrich-Wolfe, L., Travers, S., and Nelson, B. D. Jr. (2015). Genetic variation of Sclerotinia sclerotiorum from multiple crops in the North Central United States. PLoS ONE 10:e0139188. doi: 10.1371/journal.pone.0139188
Amos, W., Hoffman, J., Frodsham, A., Zhang, L., Best, S., and Hill, A. (2007). Automated binning of microsatellite alleles: problems and solutions. Mol. Ecol. Notes 7, 10–14. doi: 10.1111/j.1471-8286.2006.01560.x
Amselem, J., Cuomo, C. A., van Kan, J. A. L., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. doi: 10.1371/journal.pgen.1002230
Anonymous (2005). Distribution Maps of Plant Diseases (Edition 1). Sclerotinia sclerotiorum. Map 971.
Atallah, Z. K., Larget, B., Chen, X., and Johnson, D. A. (2004). High genetic diversity, phenotypic uniformity, and evidence of outcrossing in Sclerotinia sclerotiorum in the Columbia basin of Washington state. Phytopathology 94, 737–742. doi: 10.1094/PHYTO.2004.94.7.737
Attanayake, R. N., Carter, P. A., Jiang, D., Del Rio-Mendoza, L., and Chen, W. (2013). Sclerotinia sclerotiorum populations infecting canola from China and the United States are genetically and phenotypically distinct. Phytopathology 103, 750–761. doi: 10.1094/phyto-07-12-0159-r
Attanayake, R. N., Tennekoon, V., Johnson, D. A., Porter, L. D., del Rio-Mendoza, L., Jiang, D., et al. (2014). Inferring outcrossing in the homothallic fungus Sclerotinia sclerotiorum using linkage disequilibrium decay. Heredity 113, 353–363. doi: 10.1038/hdy.2014.37
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48.
Bastien, M., Sonah, H., and Belzile, F. (2014). Genome wide association mapping of resistance in soybean with a genotyping-by-sequencing approach. Plant Genome 7. doi: 10.3835/plantgenome2013.10.0030
Boland, G. J., and Hall, R. (1994). Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16, 93–108.
Bolton, M. D., Thomma, B. P., and Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/j.1364-3703.2005.00316.x
Brodal, G., Warmington, R., Grieu, C., Ficke, A., and Clarkson, J. P. (2017). First report of Sclerotinia subarctica nom. prov. (Sclerotinia sp. 1) causing stem rot on turnip rape (Brassica rapa subsp. oleifera) in Norway. Plant Dis. 101, 386. doi: 10.1094/PDIS-06-16-0785-PDN
Carbone, I., and Kohn, L. M. (2001a). A microbial population–species interface: nested cladistic and coalescent inference with multilocus data. Mol. Ecol. 10, 947–964. doi: 10.1046/j.1365-294X.2001.01244.x
Carbone, I., and Kohn, L. M. (2001b). Multilocus nested haplotype networks extended with DNA fingerprints show common origin and fine-scale, ongoing genetic divergence in a wild microbial metapopulation. Mol. Ecol. 10, 2409–2422. doi: 10.1046/j.0962-1083.2001.01380.x
Carbone, I., Anderson, J. B., and Kohn, L. M. (1999). Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. Evolution 53, 11–21.
Carpenter, M. A., Frampton, C., and Stewart, A. (1999). Genetic variation in New Zealand populations of the plant pathogen Sclerotinia sclerotiorum. N. Z. J. Crop Hortic. Sci. 27, 13–21.
Chao, A., and Shen, T.-J. (2003). Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 10, 429–443. doi: 10.1023/A:1026096204727
Clarkson, J. P., Carter, H. E., and Coventry, E. (2010). First report of Sclerotinia subarctica nom. prov. (Sclerotinia species 1) in the UK on Ranunculus acris. Plant Pathol. 59 1173–1173. doi: 10.1111/j.1365-3059.2010.02271.x
Clarkson, J. P., Coventry, E., Kitchen, J., Carter, H. E., and Whipps, J. M. (2013). Population structure of Sclerotinia sclerotiorum in crop and wild hosts in the UK. Plant Pathol. 62, 309–324. doi: 10.1111/j.1365-3059.2012.02635.x
Cubeta, M. A., Cody, B. R., Kohli, Y., and Kohn, L. M. (1997). Clonality in Sclerotinia sclerotiorum on infected cabbage in eastern North Carolina. Phytopathology 87, 1000–1004.
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Ekins, M., Aitken, E. A., and Coulter, K. C. (2006). Homothallism in Sclerotinia minor. Mycol. Res. 110, 1193–1199. doi: 10.1016/j.mycres.2006.07.014
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47.
Ge, X. T., Li, Y. P., Wan, Z. J., You, M. P., Finnegan, P. M., Banga, S. S., et al. (2012). Delineation of Sclerotinia sclerotiorum pathotypes using differential resistance responses on Brassica napus and B. juncea genotypes enables identification of resistance to prevailing pathotypes. Field Crops Res. 127, 248–258. doi: 10.1016/j.fcr.2011.11.022
Gomes, E. V., Do Nascimento, L. B., De Freitas, M. A., Nasser, L. C. B., and Petrofeza, S. (2011). Microsatellite markers reveal genetic variation within Sclerotinia sclerotiorum populations in irrigated dry bean crops in Brazil. J. Phytopathol. 159, 94–99. doi: 10.1111/j.1439-0434.2010.01724.x
Hambleton, S., Walker, C., and Kohn, L. M. (2002). Clonal lineages of Sclerotinia sclerotiorum previously known from other crops predominate in 1999-2000 samples from Ontario and Quebec soybean. Can. J. Plant Pathol. 24, 309–315. doi: 10.1080/07060660209507014
Hao, J. J., Subbarao, K. V., and Duniway, J. M. (2003). Germination of Sclerotinia minor and S. sclerotiorum sclerotia under various soil moisture and temperature combinations. Phyopathology 93, 443–450. doi: 10.1094/PHYTO.2003.93.4.443
Hemmati, R., Javan-Nikkhah, M., and Linde, C. C. (2009). Population genetic structure of Sclerotinia sclerotiorum on canola in Iran. Eur. J. Plant Pathol. 125, 617–628. doi: 10.1007/s10658-009-9510-7
Holst-Jensen, A., Vaage, M., and Schumacher, T. (1998). An approximation to the phylogeny of Sclerotinia and related genera. Nord. J. Bot. 18, 705–719. doi: 10.1111/j.1756-1051.1998.tb01553.x
Huang, H., and Kozub, G. (1991). Temperature requirements for carpogenic germination of sclerotia of Sclerotinia sclerotiorum isolates of different geographic origin. Bot. Bull. Acad. Sin. 32, 279–286.
Hudson, R. R. (2000). A new statistic for detecting genetic differentiation. Genetics 155, 2011–2014.
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281
Kohli, Y., Brunner, L. J., Yoell, H., Milgroom, M. G., Anderson, J. B., Morrall, R. A. A., et al. (1995). Clonal dispersal and spatial mixing in populations of the plant pathogenic fungus, Sclerotinia sclerotiorum. Mol. Ecol. 4, 69–77.
Kohli, Y., Morrall, R. A. A., Anderson, J. B., and Kohn, L. M. (1992). Local and trans-Canadian clonal distribution of Sclerotinia sclerotiorum on canola. Phytopathology 82, 875–880.
Kohn, L. M. (1995). The clonal dynamic in wild and agricultural plant pathogen populations. Can. J. Bot. 73, S1231–S1240.
Kohn, L. M., Stasovski, E., Carbone, I., Royer, J., and Anderson, J. B. (1991). Mycelial incompatibility and molecular markers identify genetic variability in field populations of Sclerotinia sclerotiorum. Phytopathology 81, 480–485.
Lê, S., Josse, J., and Husson, F. (2008). FactoMineR: an R Package for multivariate analysis. 25:18. doi: 10.18637/jss.v025.i01
Lehner, M. S., Paula Júnior, T. J., Hora Júnior, B. T., Teixeira, H., Vieira, R. F., Carneiro, J. E. S., et al. (2015). Low genetic variability in Sclerotinia sclerotiorum populations from common bean fields in Minas Gerais State, Brazil, at regional, local and micro-scales. Plant Pathol. 64, 921–931. doi: 10.1111/ppa.12322
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Malvárez, G. M., Carbone, I., Grünwald, N. J., Subbarao, K. V., Schafer, M., and Kohn, L. M. (2007). New populations of Sclerotinia sclerotiorum from lettuce in California and lentils in Washington. Phytopathology 97, 470–483. doi: 10.1094/PHYTO-97-4-0470
Meirmans, P. G., and van Tienderen, P. H. (2004). genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794. doi: 10.1111/j.1471-8286.2004.00770.x
Melzer, M. S., Smith, E. A., and Boland, G. J. (1997). Index of plant hosts of Sclerotinia minor. Can. J. Plant Pathol. 19, 272–280. doi: 10.1080/07060669709500523
Mert-Turk, F., Ipek, M., Mermer, D., and Nicholson, P. (2007). Microsatellite and morphological markers reveal genetic variation within a population of Sclerotinia sclerotiorum from oilseed rape in the Canakkale Province of Turkey. J. Phytopathol. 155, 182–187. doi: 10.1111/j.1439-0434.2007.01223.x
Njambere, E. N., Peever, T. L., Vandemark, G., and Chen, W. (2014). Genotypic variation and population structure of Sclerotinia trifoliorum infecting chickpea in California. Plant Pathol. 63, 994–1004. doi: 10.1111/ppa.12176
Phillips, A. (1987). Carpogenic germination of sclerotia of Sclerolinia sclerotiorum: a review. Phytophylactica 19, 279–284.
Phillips, D. V., Carbone, I., Gold, S. E., and Kohn, L. M. (2002). Phylogeography and genotype-symptom associations in early and late season infections of canola by Sclerotinia sclerotiorum. Phytopathology 92, 785–793. doi: 10.1094/PHYTO.2002.92.7.785
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Qandah, I. S., and Del Rio Mendoza, L. E. (2012). Modelling inoculum dispersal and Sclerotinia stem rot gradients in canola fields. Can. J. Plant Pathol. 34, 390–400. doi: 10.1080/07060661.2012.705328
R-Development-Core-Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rosenberg, N. A. (2004). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Sexton, A. C., and Howlett, B. J. (2004). Microsatellite markers reveal genetic differentiation among populations of Sclerotinia sclerotiorum from Australian canola fields. Curr. Genet. 46, 357–365. doi: 10.1007/s00294-004-0543-3
Sexton, A. C., Whitten, A. R., and Howlett, B. J. (2006). Population structure of Sclerotinia sclerotiorum in an Australian canola field at flowering and stem-infection stages of the disease cycle. Genome 49, 1408–1415. doi: 10.1139/g06-101
Sirjusingh, C., and Kohn, L. M. (2001). Characterization of microsatellites in the fungal plant pathogen, Sclerotinia sclerotiorum. Mol. Ecol. Notes 1, 267–269. doi: 10.1046/j.1471-8278.2001.00102.x
Slatkin, M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 118, 705–711.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Uhm, J., and Fujii, H. (1983). Heterothallism and mating type mutation in Sclerotinia trifoliorum. Phytopathology 73, 569–572.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246.
Warmington, R. (2014). Pathogen Diversity, Epidemiology and Control of Sclerotinia Disease in Vegetable crops. PhD, University of Warwick.
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Willetts, H. J., Wong, J. A. L., and Kirst, G. D. (1980). The biology of Sclerotinia sclerotiorum, S. trifoliorum, and S. minor with emphasis on specific nomenclature. Bot. Rev. 46, 101–165.
Winton, L. M., Krohn, A. L., and Leiner, R. H. (2006). Genetic diversity of Sclerotinia species from Alaskan vegetable crops. Can. J. Plant Pathol. 28, 426–434. doi: 10.1080/07060660609507316
Winton, L. M., Krohn, A. L., and Leiner, R. H. (2007). Microsatellite markers for Sclerotinia subarctica nom. prov., a new vegetable pathogen of the High North. Mol. Ecol. Notes 7, 1077–1079. doi: 10.1111/j.1471-8286.2007.01782.x
Keywords: sclerotinia, sclerotiorum, subarctica, population, diversity, microsatellites, intergenic spacer region
Citation: Clarkson JP, Warmington RJ, Walley PG, Denton-Giles M, Barbetti MJ, Brodal G and Nordskog B (2017) Population Structure of Sclerotinia subarctica and Sclerotinia sclerotiorum in England, Scotland and Norway. Front. Microbiol. 8:490. doi: 10.3389/fmicb.2017.00490
Received: 12 October 2016; Accepted: 09 March 2017;
Published: 04 April 2017.
Edited by:
Martin G. Klotz, Queens College (CUNY), USAReviewed by:
Celesate Linde, Australian National University, AustraliaHelen L. Hayden, Department of Economic Development, Jobs, Transport and Resources, Australia
Merrick Ekins, Queensland Museum, Australia
Copyright © 2017 Clarkson, Warmington, Walley, Denton-Giles, Barbetti, Brodal and Nordskog. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John P. Clarkson, am9obi5jbGFya3NvbkB3YXJ3aWNrLmFjLnVr
 John P. Clarkson
John P. Clarkson Rachel J. Warmington
Rachel J. Warmington Peter G. Walley
Peter G. Walley Matthew Denton-Giles
Matthew Denton-Giles Martin J. Barbetti
Martin J. Barbetti Guro Brodal
Guro Brodal Berit Nordskog
Berit Nordskog