
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 16 March 2017
Sec. Virology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00390
This article is part of the Research Topic Highly mutable animal RNA viruses: adaptation and evolution View all 37 articles
Anti-retroviral therapy (ART) is crucial for controlling human immunodeficiency virus type-1 (HIV-1) infection. Recently, progress in identifying and characterizing highly potent broadly neutralizing antibodies has provided valuable templates for HIV-1 therapy and vaccine design. Nevertheless, HIV-1, like many RNA viruses, exhibits genetically diverse populations known as quasispecies. Evolution of quasispecies can occur rapidly in response to selective pressures, such as that exerted by ART and the immune system. Hence, rapid viral evolution leading to drug resistance and/or immune evasion is a significant barrier to the development of effective HIV-1 treatments and vaccines. Here, we describe our recent investigations into evolutionary pressure exerted by anti-retroviral drugs and monoclonal neutralizing antibodies (NAbs) on HIV-1 envelope sequences. We also discuss sensitivities of HIV-1 escape mutants to maraviroc, a CCR5 inhibitor, and HIV-1 sensitized to NAbs by small-molecule CD4-mimetic compounds. These studies help to develop an understanding of viral evolution and escape from both anti-retroviral drugs and the immune system, and also provide fundamental insights into the combined use of NAbs and entry inhibitors. These findings of the adaptation and evolution of HIV in response to drug and immune pressure will inform the development of more effective antiviral therapeutic strategies.
Human immunodeficiency virus type-1 (HIV-1) exhibits extremely high genetic diversity (Rambaut et al., 2004) indicating that rapidly changing genetic variation can confer on the virus the capacity to escape the immune system and anti-retroviral therapy (ART). The HIV-1 components presenting the highest degree of sequence diversity are the surface-expressed viral envelope glycoproteins (Env), which are prime targets for both entry inhibitors and neutralizing antibodies (NAbs) (Goulder and Watkins, 2008).
The function of Env is to facilitate the entry of HIV-1 into the target cell, a process mediated by recognition of the CD4 receptor and coreceptor (usually CCR5 or CXCR4) on the cellular membrane (Dalgleish et al., 1984; Klatzmann et al., 1984; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996; Dragic et al., 1996; Feng et al., 1996). Env is composed of the surface glycoprotein, gp120, and the transmembrane glycoprotein, gp41, which associate as a non-covalent complex to form a single subunit of a trimeric viral envelope spike (Wyatt and Sodroski, 1998). Gp120 is responsible for interactions with CD4 and the coreceptor, whereas gp41 anchors the Env machinery at the viral membrane and induces membrane fusion during viral entry (Freed and Martin, 1995; Bazan et al., 1998).
Many entry inhibitors have been developed to block the interaction of Env with the CD4 receptor, the coreceptor, or the fusion reaction. Currently, two entry inhibitors have been approved for clinical use, the fusion inhibitor, enfuvirtide (T-20) (Robertson, 2003), and the CCR5 inhibitor, maraviroc (MVC) (Dorr et al., 2005; Gulick et al., 2008). As with any anti-retroviral drug, HIV can develop resistance to T-20 and MVC. The major mechanism of resistance to T-20 is caused by mutations within the binding site on the HR1 region of gp41 (Greenberg and Cammack, 2004) (Figure 1C). On the other hand, clinical resistance to MVC involves different genetic alterations in env giving rise to highly divergent Env phenotypes (Roche et al., 2013). Potential molecular mechanisms of resistance to MVC include tropism switching to CXCR4-using (X4) viruses (Westby et al., 2006; Raymond et al., 2015), increased kinetics of the entry step (Reeves et al., 2002; Putcharoen et al., 2012), increased affinity for CD4 and/or CCR5 (Agrawal-Gamse et al., 2009; Pugach et al., 2009; Pfaff et al., 2010; Ratcliff et al., 2013), and utilization of MVC-bound CCR5 for entry (Pugach et al., 2007; Westby et al., 2007; Tilton et al., 2010; Roche et al., 2011).
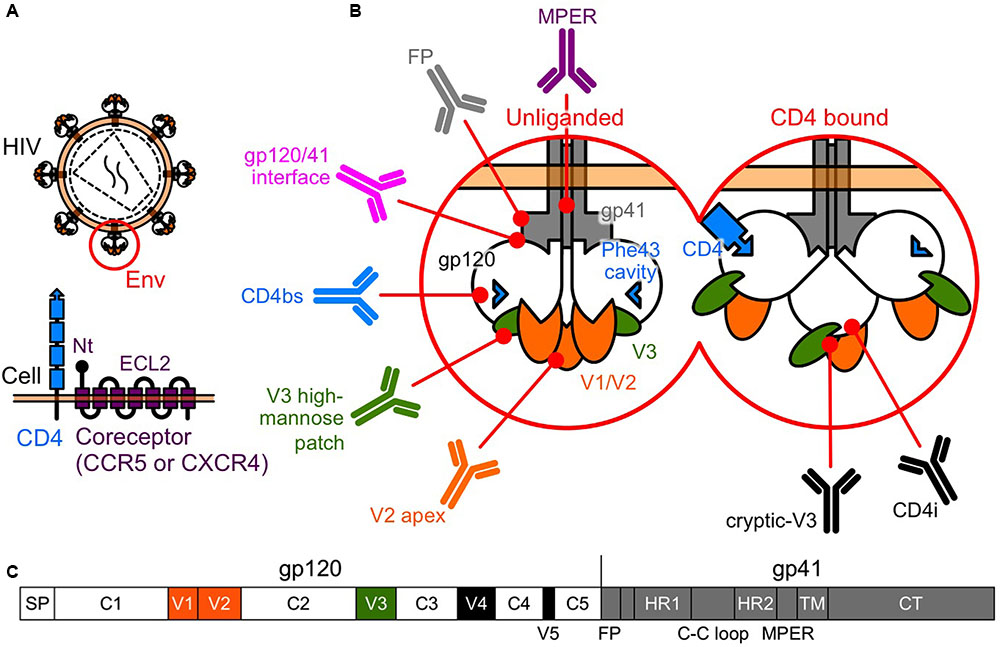
FIGURE 1. Human immunodeficiency virus type-1 (HIV-1) Env. (A) Entry of HIV-1 into a host cell involves interactions between the Env and the two-receptor mechanism of CD4 and the coreceptor. (B) Tertiary schematic view of HIV-1 Env. Following the binding of CD4 and gp120, gp120 undergoes conformational changes, moving from a rigid (unliganded) to a flexible state, allowing a subsequent interaction with the coreceptors. bNAbs have been identified that target the V2 apex, the V3 high-mannose patch, the CD4bs, the gp120/41 interface, the FP, and the MPER of gp41. In the CD4-bound state, a larger area is uncovered and potentially available for recognition by NAbs, such as V3-directed or CD4i, which recognize the conserved coreceptor-binding site. (C) Linear schematic view of HIV-1 Env. Gp120 is composed of five conserved regions (C1 to C5) that are interspersed with five variable regions (V1 to V5).
In recent years, progress in identifying and characterizing highly potent broadly NAbs (bNAbs), has provided valuable templates for HIV-1 therapy and vaccine design (Kwong and Mascola, 2012; Kwong et al., 2013; Burton and Mascola, 2015; Burton and Hangartner, 2016). However, attempts to elicit such highly potent bNAbs by immunization have not been successful, due in part to the high genetic diversity of Env and the complex escape mechanisms employed by Env (Seaman et al., 2010).
Moreover, the replication capacity of HIV-1 is largely related to the efficiency of viral entry (Arts and Quinones-Mateu, 2003; Rangel et al., 2003). In this respect, evolutionary patterns of Env are important, and selective pressures exerted by NAbs and anti-retroviral drugs can contribute to its evolution. Thus, elucidation of these patterns would inform the development of more effective antiviral therapeutic strategies.
Recently, we investigated dynamic features of selective pressure on Env by assessing NAb sensitivities of HIV-1 escape mutants from MVC, and small-molecule CD4-mimetic compounds (CD4mc) that sensitize HIV-1 to NAbs. Thus, we summarize these recent advances and discuss the application of these findings to the development of more effective combinations of NAbs and anti-retroviral drugs.
Entry of HIV-1 into a target cell involves interactions between Env and the two-receptor mechanism involving CD4 and the coreceptor. This interaction activates conformational changes in Env that lead to the membrane fusion reaction (Sattentau and Moore, 1995) (Figure 1B).
Gp120 is composed of five conserved regions (C1 to C5) that are interspersed with five variable regions (V1 to V5) (Starcich et al., 1986) (Figure 1C). The CD4 binding site (CD4bs) and especially the Phe 43 cavity, where Phe 43 of CD4 contacts gp120, are highly conserved among the different subtypes (Kwong et al., 1998). Following the binding of CD4 and gp120, the gp120 core undergoes conformational changes, moving from a rigid (unliganded) to a flexible state, allowing a subsequent interaction with the coreceptor (Myszka et al., 2000) (Figure 1B). Binding of gp120 to the coreceptor triggers further conformational changes in Env that fuse the viral membrane with the target cell membrane (Chan and Kim, 1998). Current models suggest the V3 tip interacts with the coreceptor second extracellular loop (ECL2), whereas the gp120 bridging sheet and the V3 stem interact with the coreceptor N terminus (Brelot et al., 1999; Farzan et al., 1999; Cormier and Dragic, 2002; Huang et al., 2005) (Figure 1A).
Recently, bNAbs have been isolated from HIV-1-infected individuals. Most major target specificities of these bNAbs have been mapped to various sites on Env, and include the V2 N160 glycan (V2 apex), the V3 N332 glycan (high-mannose patch), the CD4bs, the gp120/41 interface region, the fusion peptide (FP), and the membrane proximal external region (MPER) of gp41 (Burton and Mascola, 2015; Burton and Hangartner, 2016; Kong et al., 2016; van Gils et al., 2016). In addition, CD4 binding exposes highly conserved cryptic epitopes recognized by V3-directed or CD4-induced (CD4i) NAbs, which recognize the coreceptor binding site (Kwong and Mascola, 2012) (Figure 1B).
The V3-directed NAb, KD-247, is a humanized NAb with potent neutralizing activity. The epitope recognized by KD-247 was mapped to the IGPGR sequence of the V3-tip, which covers about half of subtype B. A phase-1b clinical study indicates that KD-247 reduces viral load in patients with chronic HIV-1 infection (Matsushita et al., 2015). However, HIV-1 can escape from the adaptive immune responses, and can become resistant to all anti-retroviral drugs. Therefore, in our previous in vitro study, we induced resistant variants against KD-247 using the JR-FL strain (Yoshimura et al., 2006). Resistance against KD-247 was associated with G314E substitution in the epitope on the V3-tip. Unexpectedly, the KD-247-resistant variant exhibited higher sensitivity to CCR5 inhibitors (TAK-779, aplaviroc and SCH-C) compared with the parental virus. Furthermore, our data showed strong synergistic interactions between KD-247 and CCR5 inhibitors (Yoshimura et al., 2006).
In addition to our studies, recent investigations of passive NAb therapy in HIV-infected individuals demonstrated that particular bNAbs could reduce levels of plasma viremia and suppress neutralization-sensitive viruses (Caskey et al., 2015; Lynch et al., 2015; Matsushita et al., 2015; Bar et al., 2016). However, a single use of NAbs could not suppress HIV completely and poses the danger of inducing escape variants in vivo. These findings suggest that combination strategies containing NAbs are needed to maintain virus suppression and prevent appearance of NAb-escape variants. Therefore, in the near future, combinations of NAbs and CCR5 inhibitors are likely to be efficient weapons against HIV-1.
The main mechanism of resistance to MVC appears to be related to changes in the V3 region, which enables the virus to utilize MVC-bound CCR5 coreceptors. The resistance is characterized by reductions in the maximal percent inhibition (MPI) value rather than shifts in the IC50 value (Pugach et al., 2007; Roche et al., 2011).
Pugach et al. noted that resistant variants against two CCR5 inhibitors (vicriviroc and AD101) were more sensitive to several types of NAbs compared with the parental virus (Pugach et al., 2008; Berro et al., 2009). Subsequently, we have reported the resistance induction of the primary KP-5P virus (subtype B, R5) against MVC in vitro (Yoshimura et al., 2014). Resistance to MVC was associated with V200I, T297I, K305R, and M434I substitutions near the CCR5 binding site. This MVC-resistant variant also exhibited extremely high sensitivity to three NAbs: b12 (CD4bs), 4E9C (CD4i), and KD-247. These results indicated that the MVC-resistance mutations might improve the accessibility of epitopes for the NAbs and, therefore, be incompatible with resistance to the NAbs (Yoshimura et al., 2014). More recently, Kuwata et al. (2015) showed that resistant variants against a CCR5 inhibitor, cenicriviroc, also became sensitive to three NAbs: VRC01 (CD4bs), 4E9C, and 0.5γ (V3).
Another mechanism of resistance to MVC appears to be by a change in coreceptor tropism from CCR5 to CXCR4, or by the selection of minority variants of X4 or dual/mixed viruses (Westby et al., 2006). Indeed, Raymond et al. (2015) has reported that half of MVC-treated patients who experienced virological failure harbored X4 viruses at failure. Remarkably, previous studies have shown that early X4 variants are more sensitive to NAbs compared with their coexisting R5 variants (Ganesh et al., 2004; Lusso et al., 2005; Margolis and Shattock, 2006; Bunnik et al., 2007). In addition, increased CCR5 affinity is also a potential resistance mechanism, but we have shown that low-CCR5 affinity-adapted variants also became sensitive to CD4bs and CD4i NAbs (Yoshimura et al., 2014). Thus, several studies have demonstrated diverse resistance mechanisms against MVC, but all these resistance pathways might drive viral evolution into a corner, escape from which would require high sensitivity to NAbs. Moreover, these observations indicate that MVC and NAbs might limit the emergence of mutants that are resistant to each other, supporting the clinical use of combination therapy (Figure 2A).
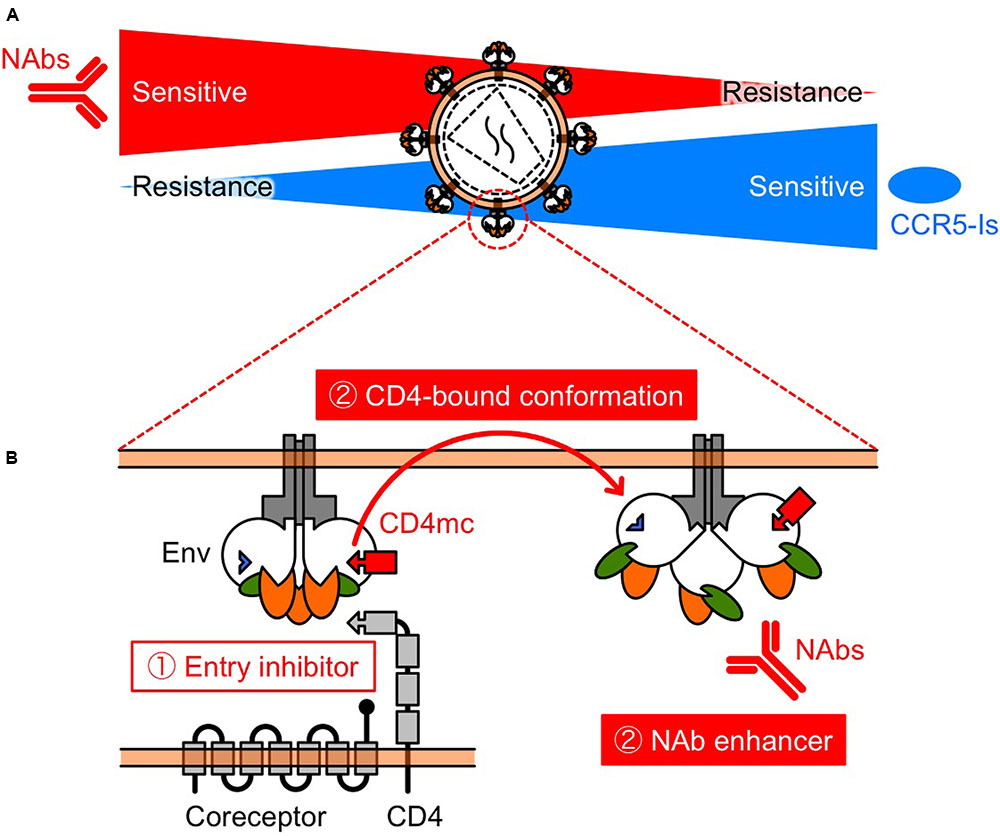
FIGURE 2. Schematic view of the relationship between NAb resistance and CCR5 inhibitor resistance (A), and the function of CD4mc (B). (A) Selection pressures on HIV-1 Env by NAbs and/or CCR5 inhibitors might turn the tide in the fight against HIV-1. The relationship indicates that NAbs and CCR5 inhibitors may restrict the emergence of variants that are resistant to each other. (B) CD4mc is a bifunctional entry inhibitor. The use of bifunctional entry inhibitors that display direct blockade of viral entry and exposure of epitopes to NAbs, should be effective in passive NAb immunization.
However, it is not clear whether patients’ plasma IgG under MVC treatment can induce mutations in Env to enhance neutralizing activity. We are currently investigating the relationship between NAb responses and MVC treatment using patients’ plasma IgGs before and after MVC-containing combination ART (cART). Moreover, we think treatment with particular entry inhibitors and/or CD4mc can induce bNAbs in vivo; however, we await results for this prediction. Thus, we will perform experiments in animal models to induce or enhance NAbs using novel entry inhibitors and/or CD4mc treatment.
Evolution of HIV-1 helps it to evade NAbs (Moore et al., 2012; Liao et al., 2013; Bouvin-Pley et al., 2014). cART, however, results in a reduction in the virus population size, which creates a genetic bottleneck. In vivo studies indicate that the bottleneck affects not only drug-target regions (e.g., reverse transcriptase), but also other regions of the viral genome, including the Env region (Zhang et al., 1994; Sheehy et al., 1996; Delwart et al., 1998; Nijhuis et al., 1998; Ibanez et al., 2000; Kitrinos et al., 2005; Charpentier et al., 2006; Nora et al., 2007). The population dynamics of the Env region might be important when bNAbs and novel entry inhibitors become available in the near future. However, it is hard to observe effects of an anti-retroviral drug-induced bottleneck on the Env region in vivo.
Thus, we induced variants against anti-retroviral drugs using primary swarm isolates (Harada et al., 2013). As a result, the phylogenetic clustering of raltegravir (an integrase inhibitor)-, lamivudine (a reverse transcriptase inhibitor)- and saquinavir (a protease inhibitor)-induced variants was entirely distinct from that of non-drug-treated controls. Among these drug-induced variants, the variable regions of gp120 were very similar to each other. Conversely, the non-drug-treated variant was quite different from the drug-induced variants. These results imply that, under selective pressure of non-entry inhibitors, the virus may choose a representative Env sequence from the viral population to gain a growth advantage (Harada et al., 2013). In addition to our results, a supporting study by Mesplede et al. (2015) showed that treatment with dolutegravir (an integrase inhibitor) results in a reduction in viral genetic diversity. Further studies are needed to confirm our observations, but these results may provide a new paradigm for viral evolution in the novel NAb plus anti-retroviral drug combination therapy era.
Binding of CD4 to gp120, is the first essential step of the entry process. The multiple contacts made by Phe 43 and Arg 59 of CD4 with gp120 residues in CD4bs contribute significantly to CD4–gp120 binding (Kwong et al., 1998). The critical Phe 43 of CD4 becomes buried in a binding pocket of gp120, termed the Phe 43 cavity. This cavity is known to be highly conserved among the different subtypes and is therefore considered a particularly interesting target for inhibitors of CD4–gp120 interaction (Kwong et al., 1998).
Molecules that mimic the CD4 receptor, such as soluble CD4 (sCD4), CD4 immunoadhesin (CD4-Ig), sCD4 mini-proteins, and CD4mc have been developed (Vita et al., 1999; Grupping et al., 2012). sCD4, CD4-Ig, and sCD4 mini-protein have been studied as potential therapeutics (Smith et al., 1987; Fisher et al., 1988; Hussey et al., 1988; Jacobson et al., 2000; Fletcher et al., 2007; Dereuddre-Bosquet et al., 2012; Gardner et al., 2015). These studies in patients and non-human primate models have provided proof of principle that viral entry can be successfully blocked in vivo. In particular, Gardner et al. (2015) demonstrated that eCD4-Ig, a fusion of CD4-Ig with a small CCR5-mimetic peptide, was on average more potent, and much broader than bNAbs. Moreover, adeno-associated virus-delivered eCD4-Ig, provided durable protection for immunized monkeys against high-dose intravenous SHIV challenge (Gardner et al., 2015).
The prototype of CD4mc, NBD-556, was identified in a screen for inhibitors of the CD4–gp120 interaction (Zhao et al., 2005). We and others have been exploring the potential of NBD-556-derived CD4mc as a novel class of HIV entry inhibitor (Madani et al., 2004, 2008, 2014, 2016, 2017; Narumi et al., 2010, 2011, 2013; Yamada et al., 2010; Yoshimura et al., 2010; Lalonde et al., 2011, 2012, 2013; Courter et al., 2014; Richard et al., 2015; Melillo et al., 2016; Mizuguchi et al., 2016; Ohashi et al., 2016). The binding of CD4mc in the Phe 43 cavity blocks CD4-gp120 interaction and, induces conformational changes in gp120 similar to those observed upon sCD4 binding (Figure 2B) (Schon et al., 2006; Haim et al., 2009; Curreli et al., 2014; Kwon et al., 2014). sCD4 significantly enhance neutralization by CD4i (Thali et al., 1993) and some V3 NAbs (Lusso et al., 2005). Remarkably, CD4i and V3 NAbs are present in HIV-infected individuals during the early stage of infection (Decker et al., 2005). Consequently, we hypothesized that CD4mc can cause exposure of cryptic epitopes to antibodies, allowing virus neutralization. As a result, combinations of CD4mc (NBD-556 or YYA-021) with CD4i or V3 NAbs produced strong synergistic antiviral interactions (Yamada et al., 2010; Yoshimura et al., 2010) (Figures 1, 2B). Moreover, we found that CD4mc sensitized a clinical isolate to autologous plasma antibodies from the same time point (Yoshimura et al., 2010).
Recently, this approach has been extended to combining vaccine with CD4mc. In studies using prototypic CD4mc BNM compounds, Madani et al. (2014, 2016) demonstrated that CD4mc sensitized the virus to antibodies elicited by immunization of humans and monkeys. These studies establish the proof of concept that CD4mc can sensitize primary viruses to antibodies that are present in plasma of infected or vaccinated individuals. In addition, Richard et al. (2015) reported that CD4mc could efficiently sensitize primary CD4 T cells from HIV-1-infected individuals to antibody-dependent cell-mediated cytotoxicity (ADCC) mediated by autologous sera and effector cells.
Based on these results, further studies are needed to investigate the effectiveness of delivery methods of CD4mc. Small molecules such as CD4mc have many advantages over conventional immunotherapeutic agents, including ease of production and the potential for oral administration. Furthermore, the use of bifunctional entry inhibitors that display direct blockade of viral entry and exposure of epitopes to NAbs should be effective in passive NAb immunization.
Extensive genetic diversity in the Env region presents significant obstructions to the development of promising therapies and vaccines against HIV-1. However, selection pressures on the Env region by NAbs, entry inhibitors, and/or non-entry antiviral inhibitors, might turn the tide in the fight against HIV-1. Moreover, bifunctional entry inhibitors such as CD4mc might potentiate these selection pressures. Thus, by taking advantage of the adaptation and evolution of HIV resulting from drug and immune pressure, we might drive HIV-1 into a vulnerable corner.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (JSPS KAKENHI Grant Number 15K08125), and Japan Agency for Medical Research and Development (AMED).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Agrawal-Gamse, C., Lee, F. H., Haggarty, B., Jordan, A. P., Yi, Y., Lee, B., et al. (2009). Adaptive mutations in a human immunodeficiency virus type 1 envelope protein with a truncated V3 loop restore function by improving interactions with CD4. J. Virol. 83, 11005–11015. doi: 10.1128/JVI.01238-09
Arts, E. J., and Quinones-Mateu, M. E. (2003). Sorting out the complexities of HIV-1 fitness. AIDS 17, 780–781. doi: 10.1097/00002030-200303280-00026
Bar, K. J., Sneller, M. C., Harrison, L. J., Justement, J. S., Overton, E. T., Petrone, M. E., et al. (2016). Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med. 375, 2037–2050. doi: 10.1056/NEJMoa1608243
Bazan, H. A., Alkhatib, G., Broder, C. C., and Berger, E. A. (1998). Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J. Virol. 72, 4485–4491.
Berro, R., Sanders, R. W., Lu, M., Klasse, P. J., and Moore, J. P. (2009). Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548. doi: 10.1371/journal.ppat.1000548
Bouvin-Pley, M., Morgand, M., Meyer, L., Goujard, C., Moreau, A., Mouquet, H., et al. (2014). Drift of the HIV-1 envelope glycoprotein gp120 toward increased neutralization resistance over the course of the epidemic: a comprehensive study using the most potent and broadly neutralizing monoclonal antibodies. J. Virol. 88, 13910–13917. doi: 10.1128/JVI.02083-14
Brelot, A., Heveker, N., Adema, K., Hosie, M. J., Willett, B., and Alizon, M. (1999). Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol. 73, 2576–2586.
Bunnik, E. M., Quakkelaar, E. D., Van Nuenen, A. C., Boeser-Nunnink, B., and Schuitemaker, H. (2007). Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J. Virol. 81, 525–531. doi: 10.1128/JVI.01983-06
Burton, D. R., and Hangartner, L. (2016). Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 34, 635–659. doi: 10.1146/annurev-immunol-041015-055515
Burton, D. R., and Mascola, J. R. (2015). Antibody responses to envelope glycoproteins in HIV-1 infection. Nat. Immunol. 16, 571–576. doi: 10.1038/ni.3158
Caskey, M., Klein, F., Lorenzi, J. C., Seaman, M. S., West, A. P. Jr., Buckley, N., et al. (2015). Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491. doi: 10.1038/nature14411
Chan, D. C., and Kim, P. S. (1998). HIV entry and its inhibition. Cell 93, 681–684. doi: 10.1016/S0092-8674(00)81430-0
Charpentier, C., Nora, T., Tenaillon, O., Clavel, F., and Hance, A. J. (2006). Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 80, 2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006
Choe, H., Farzan, M., Sun, Y., Sullivan, N., Rollins, B., Ponath, P. D., et al. (1996). The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85, 1135–1148. doi: 10.1016/S0092-8674(00)81313-6
Cormier, E. G., and Dragic, T. (2002). The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76, 8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002
Courter, J. R., Madani, N., Sodroski, J., Schon, A., Freire, E., Kwong, P. D., et al. (2014). Structure-based design, synthesis and validation of CD4-mimetic small molecule inhibitors of HIV-1 entry: conversion of a viral entry agonist to an antagonist. ACC Chem. Res. 47, 1228–1237. doi: 10.1021/ar4002735
Curreli, F., Kwon, Y. D., Zhang, H., Yang, Y., Scacalossi, D., Kwong, P. D., et al. (2014). Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study. Antimicrob. Agents Chemother. 58, 5478–5491. doi: 10.1128/AAC.03339-14
Dalgleish, A. G., Beverley, P. C., Clapham, P. R., Crawford, D. H., Greaves, M. F., and Weiss, R. A. (1984). The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312, 763–767. doi: 10.1038/312763a0
Decker, J. M., Bibollet-Ruche, F., Wei, X., Wang, S., Levy, D. N., Wang, W., et al. (2005). Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201, 1407–1419. doi: 10.1084/jem.20042510
Delwart, E. L., Pan, H., Neumann, A., and Markowitz, M. (1998). Rapid, transient changes at the env locus of plasma human immunodeficiency virus type 1 populations during the emergence of protease inhibitor resistance. J. Virol. 72, 2416–2421.
Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., et al. (1996). Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666. doi: 10.1038/381661a0
Dereuddre-Bosquet, N., Morellato-Castillo, L., Brouwers, J., Augustijns, P., Bouchemal, K., Ponchel, G., et al. (2012). MiniCD4 microbicide prevents HIV infection of human mucosal explants and vaginal transmission of SHIV(162P3) in cynomolgus macaques. PLoS Pathog. 8:e1003071. doi: 10.1371/journal.ppat.1003071
Doranz, B. J., Rucker, J., Yi, Y., Smyth, R. J., Samson, M., Peiper, S. C., et al. (1996). A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85, 1149–1158. doi: 10.1016/S0092-8674(00)81314-8
Dorr, P., Westby, M., Dobbs, S., Griffin, P., Irvine, B., Macartney, M., et al. (2005). Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49, 4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005
Dragic, T., Litwin, V., Allaway, G. P., Martin, S. R., Huang, Y., Nagashima, K. A., et al. (1996). HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381, 667–673. doi: 10.1038/381667a0
Farzan, M., Mirzabekov, T., Kolchinsky, P., Wyatt, R., Cayabyab, M., Gerard, N. P., et al. (1999). Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676. doi: 10.1016/S0092-8674(00)80577-2
Feng, Y., Broder, C. C., Kennedy, P. E., and Berger, E. A. (1996). HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872–877. doi: 10.1126/science.272.5263.872
Fisher, R. A., Bertonis, J. M., Meier, W., Johnson, V. A., Costopoulos, D. S., Liu, T., et al. (1988). HIV infection is blocked in vitro by recombinant soluble CD4. Nature 331, 76–78. doi: 10.1038/331076a0
Fletcher, C. V., Deville, J. G., Samson, P. M., Moye, J. H. Jr., Church, J. A., Spiegel, H. M., et al. (2007). Nonlinear pharmacokinetics of high-dose recombinant fusion protein CD4-IgG2 (PRO 542) observed in HIV-1-infected children. J. Allergy Clin. Immunol. 119, 747–750. doi: 10.1016/j.jaci.2006.10.045
Freed, E. O., and Martin, M. A. (1995). The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270, 23883–23886. doi: 10.1074/jbc.270.41.23883
Ganesh, L., Leung, K., Lore, K., Levin, R., Panet, A., Schwartz, O., et al. (2004). Infection of specific dendritic cells by CCR5-tropic human immunodeficiency virus type 1 promotes cell-mediated transmission of virus resistant to broadly neutralizing antibodies. J. Virol. 78, 11980–11987. doi: 10.1128/JVI.78.21.11980-11987.2004
Gardner, M. R., Kattenhorn, L. M., Kondur, H. R., Von Schaewen, M., Dorfman, T., Chiang, J. J., et al. (2015). AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519, 87–91. doi: 10.1038/nature14264
Goulder, P. J., and Watkins, D. I. (2008). Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8, 619–630. doi: 10.1038/nri2357
Greenberg, M. L., and Cammack, N. (2004). Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54, 333–340. doi: 10.1093/jac/dkh330
Grupping, K., Selhorst, P., Michiels, J., Vereecken, K., Heyndrickx, L., Kessler, P., et al. (2012). MiniCD4 protein resistance mutations affect binding to the HIV-1 gp120 CD4 binding site and decrease entry efficiency. Retrovirology 9:36. doi: 10.1186/1742-4690-9-36
Gulick, R. M., Lalezari, J., Goodrich, J., Clumeck, N., Dejesus, E., Horban, A., et al. (2008). Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359, 1429–1441. doi: 10.1056/NEJMoa0803152
Haim, H., Si, Z., Madani, N., Wang, L., Courter, J. R., Princiotto, A., et al. (2009). Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. doi: 10.1371/journal.ppat.1000360
Harada, S., Yoshimura, K., Yamaguchi, A., Boonchawalit, S., Yusa, K., and Matsushita, S. (2013). Impact of antiretroviral pressure on selection of primary human immunodeficiency virus type 1 envelope sequences in vitro. J. Gen. Virol. 94, 933–943. doi: 10.1099/vir.0.047167-0
Huang, C. C., Tang, M., Zhang, M. Y., Majeed, S., Montabana, E., Stanfield, R. L., et al. (2005). Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028. doi: 10.1126/science.1118398
Hussey, R. E., Richardson, N. E., Kowalski, M., Brown, N. R., Chang, H. C., Siliciano, R. F., et al. (1988). A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature 331, 78–81. doi: 10.1038/331078a0
Ibanez, A., Clotet, B., and Martinez, M. A. (2000). Human immunodeficiency virus type 1 population bottleneck during indinavir therapy causes a genetic drift in the env quasispecies. J. Gen. Virol. 81, 85–95. doi: 10.1099/0022-1317-81-1-85
Jacobson, J. M., Lowy, I., Fletcher, C. V., O’neill, T. J., Tran, D. N., Ketas, T. J., et al. (2000). Single-dose safety, pharmacology, and antiviral activity of the human immunodeficiency virus (HIV) type 1 entry inhibitor PRO 542 in HIV-infected adults. J. Infect. Dis. 182, 326–329. doi: 10.1086/315698
Kitrinos, K. M., Nelson, J. A., Resch, W., and Swanstrom, R. (2005). Effect of a protease inhibitor-induced genetic bottleneck on human immunodeficiency virus type 1 env gene populations. J. Virol. 79, 10627–10637. doi: 10.1128/JVI.79.16.10627-10637.2005
Klatzmann, D., Champagne, E., Chamaret, S., Gruest, J., Guetard, D., Hercend, T., et al. (1984). T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312, 767–768. doi: 10.1038/312767a0
Kong, R., Xu, K., Zhou, T., Acharya, P., Lemmin, T., Liu, K., et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. doi: 10.1126/science.aae0474
Kuwata, T., Enomoto, I., Baba, M., and Matsushita, S. (2015). Incompatible Natures of the HIV-1 envelope in resistance to the CCR5 antagonist cenicriviroc and to neutralizing antibodies. Antimicrob. Agents Chemother. 60, 437–450. doi: 10.1128/AAC.02285-15
Kwon, Y. D., Lalonde, J. M., Yang, Y., Elban, M. A., Sugawara, A., Courter, J. R., et al. (2014). Crystal structures of HIV-1 gp120 envelope glycoprotein in complex with NBD analogues that target the CD4-binding site. PLoS ONE 9:e85940. doi: 10.1371/journal.pone.0085940
Kwong, P. D., and Mascola, J. R. (2012). Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425. doi: 10.1016/j.immuni.2012.08.012
Kwong, P. D., Mascola, J. R., and Nabel, G. J. (2013). Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13, 693–701. doi: 10.1038/nri3516
Kwong, P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J., and Hendrickson, W. A. (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. doi: 10.1038/31405
Lalonde, J. M., Elban, M. A., Courter, J. R., Sugawara, A., Soeta, T., Madani, N., et al. (2011). Design, synthesis and biological evaluation of small molecule inhibitors of CD4-gp120 binding based on virtual screening. Bioorg. Med. Chem. 19, 91–101. doi: 10.1016/j.bmc.2010.11.049
Lalonde, J. M., Kwon, Y. D., Jones, D. M., Sun, A. W., Courter, J. R., Soeta, T., et al. (2012). Structure-based design, synthesis, and characterization of dual hotspot small-molecule HIV-1 entry inhibitors. J. Med. Chem. 55, 4382–4396. doi: 10.1021/jm300265j
Lalonde, J. M., Le-Khac, M., Jones, D. M., Courter, J. R., Park, J., Schon, A., et al. (2013). Structure-based design and synthesis of an HIV-1 entry inhibitor exploiting X-ray and thermodynamic characterization. ACS Med. Chem. Lett. 4, 338–343. doi: 10.1021/ml300407y
Liao, H. X., Lynch, R., Zhou, T., Gao, F., Alam, S. M., Boyd, S. D., et al. (2013). Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476. doi: 10.1038/nature12053
Lusso, P., Earl, P. L., Sironi, F., Santoro, F., Ripamonti, C., Scarlatti, G., et al. (2005). Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 79, 6957–6968. doi: 10.1128/JVI.79.11.6957-6968.2005
Lynch, R. M., Boritz, E., Coates, E. E., Dezure, A., Madden, P., Costner, P., et al. (2015). Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7:319ra206. doi: 10.1126/scitranslmed.aad5752
Madani, N., Perdigoto, A. L., Srinivasan, K., Cox, J. M., Chruma, J. J., Lalonde, J., et al. (2004). Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J. Virol. 78, 3742–3752. doi: 10.1128/JVI.78.7.3742-3752.2004
Madani, N., Princiotto, A. M., Easterhoff, D., Bradley, T., Luo, K., Williams, W. B., et al. (2016). Antibodies elicited by multiple envelope glycoprotein immunogens in primates neutralize primary Human Immunodeficiency Viruses (HIV-1) sensitized by CD4-mimetic compounds. J. Virol. 90, 5031–5046. doi: 10.1128/JVI.03211-15
Madani, N., Princiotto, A. M., Schon, A., Lalonde, J., Feng, Y., Freire, E., et al. (2014). CD4-mimetic small molecules sensitize human immunodeficiency virus to vaccine-elicited antibodies. J. Virol. 88, 6542–6555. doi: 10.1128/JVI.00540-14
Madani, N., Princiotto, A. M., Zhao, C., Jahanbakhshsefidi, F., Mertens, M., Herschhorn, A., et al. (2017). Activation and inactivation of primary human immunodeficiency virus envelope glycoprotein trimers by CD4-mimetic compounds. J. Virol. 91:e01880-16. doi: 10.1128/JVI.01880-16
Madani, N., Schon, A., Princiotto, A. M., Lalonde, J. M., Courter, J. R., Soeta, T., et al. (2008). Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16, 1689–1701. doi: 10.1016/j.str.2008.09.005
Margolis, L., and Shattock, R. (2006). Selective transmission of CCR5-utilizing HIV-1: the ’gatekeeper’ problem resolved? Nat. Rev. Microbiol. 4, 312–317. doi: 10.1038/nrmicro1427
Matsushita, S., Yoshimura, K., Ramirez, K. P., Pisupati, J., and Murakami, T. (2015). Passive transfer of neutralizing mAb KD-247 reduces plasma viral load in patients chronically infected with HIV-1. AIDS 29, 453–462. doi: 10.1097/QAD.0000000000000570
Melillo, B., Liang, S., Park, J., Schon, A., Courter, J. R., Lalonde, J. M., et al. (2016). Small-molecule CD4-mimics: structure-based optimization of HIV-1 entry inhibition. ACS Med. Chem. Lett. 7, 330–334. doi: 10.1021/acsmedchemlett.5b00471
Mesplede, T., Moisi, D., Oliveira, M., Ibanescu, I., Ohnona, F., Brenner, B., et al. (2015). Dolutegravir inhibits HIV-1 Env evolution in primary human cells. AIDS 29, 659–665. doi: 10.1097/QAD.0000000000000606
Mizuguchi, T., Harada, S., Miura, T., Ohashi, N., Narumi, T., Mori, H., et al. (2016). A minimally cytotoxic CD4 mimic as an HIV entry inhibitor. Bioorg. Med. Chem. Lett. 26, 397–400. doi: 10.1016/j.bmcl.2015.11.103
Moore, P. L., Gray, E. S., Wibmer, C. K., Bhiman, J. N., Nonyane, M., Sheward, D. J., et al. (2012). Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 18, 1688–1692. doi: 10.1038/nm.2985
Myszka, D. G., Sweet, R. W., Hensley, P., Brigham-Burke, M., Kwong, P. D., Hendrickson, W. A., et al. (2000). Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U.S.A. 97, 9026–9031. doi: 10.1073/pnas.97.16.9026
Narumi, T., Arai, H., Yoshimura, K., Harada, S., Hirota, Y., Ohashi, N., et al. (2013). CD4 mimics as HIV entry inhibitors: lead optimization studies of the aromatic substituents. Bioorg. Med. Chem. 21, 2518–2526. doi: 10.1016/j.bmc.2013.02.041
Narumi, T., Arai, H., Yoshimura, K., Harada, S., Nomura, W., Matsushita, S., et al. (2011). Small molecular CD4 mimics as HIV entry inhibitors. Bioorg. Med. Chem. 19, 6735–6742. doi: 10.1016/j.bmc.2011.09.045
Narumi, T., Ochiai, C., Yoshimura, K., Harada, S., Tanaka, T., Nomura, W., et al. (2010). CD4 mimics targeting the HIV entry mechanism and their hybrid molecules with a CXCR4 antagonist. Bioorg. Med. Chem. Lett. 20, 5853–5858. doi: 10.1016/j.bmcl.2010.07.106
Nijhuis, M., Boucher, C. A., Schipper, P., Leitner, T., Schuurman, R., and Albert, J. (1998). Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. U.S.A. 95, 14441–14446. doi: 10.1073/pnas.95.24.14441
Nora, T., Charpentier, C., Tenaillon, O., Hoede, C., Clavel, F., and Hance, A. J. (2007). Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J. Virol. 81, 7620–7628. doi: 10.1128/JVI.00083-07
Ohashi, N., Harada, S., Mizuguchi, T., Irahara, Y., Yamada, Y., Kotani, M., et al. (2016). Small-molecule CD4 mimics containing mono-cyclohexyl moieties as HIV entry inhibitors. ChemMedChem 11, 940–946. doi: 10.1002/cmdc.201500590
Pfaff, J. M., Wilen, C. B., Harrison, J. E., Demarest, J. F., Lee, B., Doms, R. W., et al. (2010). HIV-1 resistance to CCR5 antagonists associated with highly efficient use of CCR5 and altered tropism on primary CD4+ T cells. J. Virol. 84, 6505–6514. doi: 10.1128/JVI.00374-10
Pugach, P., Ketas, T. J., Michael, E., and Moore, J. P. (2008). Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology 377, 401–407. doi: 10.1016/j.virol.2008.04.032
Pugach, P., Marozsan, A. J., Ketas, T. J., Landes, E. L., Moore, J. P., and Kuhmann, S. E. (2007). HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361, 212–228. doi: 10.1016/j.virol.2006.11.004
Pugach, P., Ray, N., Klasse, P. J., Ketas, T. J., Michael, E., Doms, R. W., et al. (2009). Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor-CCR5 complex at low cell surface CCR5 densities. Virology 387, 296–302. doi: 10.1016/j.virol.2009.02.044
Putcharoen, O., Lee, S. H., Henrich, T. J., Hu, Z., Vanichanan, J., Coakley, E., et al. (2012). HIV-1 clinical isolates resistant to CCR5 antagonists exhibit delayed entry kinetics that are corrected in the presence of drug. J. Virol. 86, 1119–1128. doi: 10.1128/JVI.06421-11
Rambaut, A., Posada, D., Crandall, K. A., and Holmes, E. C. (2004). The causes and consequences of HIV evolution. Nat. Rev. Genet. 5, 52–61. doi: 10.1038/nrg1246
Rangel, H. R., Weber, J., Chakraborty, B., Gutierrez, A., Marotta, M. L., Mirza, M., et al. (2003). Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 77, 9069–9073. doi: 10.1128/JVI.77.16.9069-9073.2003
Ratcliff, A. N., Shi, W., and Arts, E. J. (2013). HIV-1 resistance to maraviroc conferred by a CD4 binding site mutation in the envelope glycoprotein gp120. J. Virol. 87, 923–934. doi: 10.1128/JVI.01863-12
Raymond, S., Maillard, A., Amiel, C., Peytavin, G., Trabaud, M. A., Desbois, D., et al. (2015). Virological failure of patients on maraviroc-based antiretroviral therapy. J. Antimicrob. Chemother. 70, 1858–1864. doi: 10.1093/jac/dkv026
Reeves, J. D., Gallo, S. A., Ahmad, N., Miamidian, J. L., Harvey, P. E., Sharron, M., et al. (2002). Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U.S.A. 99, 16249–16254. doi: 10.1073/pnas.252469399
Richard, J., Veillette, M., Brassard, N., Iyer, S. S., Roger, M., Martin, L., et al. (2015). CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc. Natl. Acad. Sci. U.S.A. 112, E2687–E2694. doi: 10.1073/pnas.1506755112
Robertson, D. (2003). US FDA approves new class of HIV therapeutics. Nat. Biotechnol. 21, 470–471. doi: 10.1038/nbt0503-470
Roche, M., Jakobsen, M. R., Ellett, A., Salimiseyedabad, H., Jubb, B., Westby, M., et al. (2011). HIV-1 predisposed to acquiring resistance to maraviroc (MVC) and other CCR5 antagonists in vitro has an inherent, low-level ability to utilize MVC-bound CCR5 for entry. Retrovirology 8:89. doi: 10.1186/1742-4690-8-89
Roche, M., Salimi, H., Duncan, R., Wilkinson, B. L., Chikere, K., Moore, M. S., et al. (2013). A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology 10:43. doi: 10.1186/1742-4690-10-43
Sattentau, Q. J., and Moore, J. P. (1995). Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182, 185–196. doi: 10.1084/jem.182.1.185
Schon, A., Madani, N., Klein, J. C., Hubicki, A., Ng, D., Yang, X., et al. (2006). Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45, 10973–10980. doi: 10.1021/bi061193r
Seaman, M. S., Janes, H., Hawkins, N., Grandpre, L. E., Devoy, C., Giri, A., et al. (2010). Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84, 1439–1452. doi: 10.1128/JVI.02108-09
Sheehy, N., Desselberger, U., Whitwell, H., and Ball, J. K. (1996). Concurrent evolution of regions of the envelope and polymerase genes of human immunodeficiency virus type 1 during zidovudine (AZT) therapy. J. Gen. Virol. 77(Pt 5), 1071–1081. doi: 10.1099/0022-1317-77-5-1071
Smith, D. H., Byrn, R. A., Marsters, S. A., Gregory, T., Groopman, J. E., and Capon, D. J. (1987). Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science 238, 1704–1707. doi: 10.1126/science.3500514
Starcich, B. R., Hahn, B. H., Shaw, G. M., Mcneely, P. D., Modrow, S., Wolf, H., et al. (1986). Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45, 637–648. doi: 10.1016/0092-8674(86)90778-6
Thali, M., Moore, J. P., Furman, C., Charles, M., Ho, D. D., Robinson, J., et al. (1993). Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67, 3978–3988.
Tilton, J. C., Wilen, C. B., Didigu, C. A., Sinha, R., Harrison, J. E., Agrawal-Gamse, C., et al. (2010). A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5. J. Virol. 84, 10863–10876. doi: 10.1128/JVI.01109-10
van Gils, M. J., Van Den Kerkhof, T. L., Ozorowski, G., Cottrell, C. A., Sok, D., Pauthner, M., et al. (2016). An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2:16199. doi: 10.1038/nmicrobiol.2016.199
Vita, C., Drakopoulou, E., Vizzavona, J., Rochette, S., Martin, L., Menez, A., et al. (1999). Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 96, 13091–13096. doi: 10.1073/pnas.96.23.13091
Westby, M., Lewis, M., Whitcomb, J., Youle, M., Pozniak, A. L., James, I. T., et al. (2006). Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80, 4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006
Westby, M., Smith-Burchnell, C., Mori, J., Lewis, M., Mosley, M., Stockdale, M., et al. (2007). Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81, 2359–2371. doi: 10.1128/JVI.02006-06
Wyatt, R., and Sodroski, J. (1998). The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888. doi: 10.1126/science.280.5371.1884
Yamada, Y., Ochiai, C., Yoshimura, K., Tanaka, T., Ohashi, N., Narumi, T., et al. (2010). CD4 mimics targeting the mechanism of HIV entry. Bioorg. Med. Chem. Lett. 20, 354–358. doi: 10.1016/j.bmcl.2009.10.098
Yoshimura, K., Harada, S., Boonchawalit, S., Kawanami, Y., and Matsushita, S. (2014). Impact of maraviroc-resistant and low-CCR5-adapted mutations induced by in vitro passage on sensitivity to anti-envelope neutralizing antibodies. J. Gen. Virol. 95, 1816–1826. doi: 10.1099/vir.0.062885-0
Yoshimura, K., Harada, S., Shibata, J., Hatada, M., Yamada, Y., Ochiai, C., et al. (2010). Enhanced exposure of human immunodeficiency virus type 1 primary isolate neutralization epitopes through binding of CD4 mimetic compounds. J. Virol. 84, 7558–7568. doi: 10.1128/JVI.00227-10
Yoshimura, K., Shibata, J., Kimura, T., Honda, A., Maeda, Y., Koito, A., et al. (2006). Resistance profile of a neutralizing anti-HIV monoclonal antibody, KD-247, that shows favourable synergism with anti-CCR5 inhibitors. AIDS 20, 2065–2073. doi: 10.1097/01.aids.0000247587.31320.fe
Zhang, Y. M., Dawson, S. C., Landsman, D., Lane, H. C., and Salzman, N. P. (1994). Persistence of four related human immunodeficiency virus subtypes during the course of zidovudine therapy: relationship between virion RNA and proviral DNA. J. Virol. 68, 425–432.
Keywords: HIV-1, antiretroviral therapy, neutralizing antibody, evolution, escape
Citation: Harada S and Yoshimura K (2017) Driving HIV-1 into a Vulnerable Corner by Taking Advantage of Viral Adaptation and Evolution. Front. Microbiol. 8:390. doi: 10.3389/fmicb.2017.00390
Received: 31 October 2016; Accepted: 24 February 2017;
Published: 16 March 2017.
Edited by:
Akio Adachi, University of Tokushima, JapanReviewed by:
Takamasa Ueno, Kumamoto University, JapanCopyright © 2017 Harada and Yoshimura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuhisa Yoshimura, eWthenVAbmloLmdvLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.