
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 March 2017
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00344
This article is part of the Research Topic Molecular Self-assembly Inspired Advanced Materials Against Antibiotic-resistant Pathogens View all 6 articles
Copper is one of the most abundant heavy metals present in swine manure. In this study, a laboratory-scale aerobic composting system was amended with Cu at three levels (0, 200, and 2000 mg kg-1, i.e., control, Cu200, and Cu2000 treatments, respectively) to determine its effect on the fate of copper resistance genes [copper resistance genes (CRGs): pcoA, cusA, copA, and tcrB], antibiotic resistance genes [antibiotic resistance genes (ARGs): erm(A) and erm(B)], and intl1. The results showed that the absolute abundances of pcoA, tcrB, erm(A), erm(B), and intl1 were reduced, whereas those of copA and cusA increased after swine manure composting. Redundancy analysis showed that temperature significantly affected the variations in CRGs, ARGs, and intl1. The decreases in CRGs, ARGs, and intI1 were positively correlated with the exchangeable Cu levels. The bacterial community could be grouped according to the composting time under different treatments, where the high concentration of copper had a more persistent effect on the bacterial community. Network analysis determined that the co-occurrence of CRGs, ARGs, and intI1, and the bacterial community were the main contributors to the changes in CRGs, ARG, and intl1. Thus, temperature, copper, and changes in the bacterial community composition had important effects on the variations in CRGs, ARGs, and intl1 during manure composting in the presence of added copper.
Poultry and livestock feeds are frequently supplemented with trace elements such as copper to promote optimum growth due to their antimicrobial properties (Xiong et al., 2010). However, poultry and livestock absorb these additives at low rates and they excrete up to 95% in their dung and urine, which often contain high concentrations of heavy metals such as copper (Xiong et al., 2010). High concentrations of copper have been found in manure (22–3388 mg kg-1; Ji et al., 2012), and large amounts of copper may enter the environment via the application of manure. Thus, the ecological environment may be damaged by copper, which could perturb the structure and function of the microbial community.
Composting is a common and effective approach for manure treatment before its application to land. In the composting process, microbes play a key role in the biotransformation (i.e., biosynthesis or biodegradation) of compost materiel (Guo et al., 2012). In addition, copper can impose selective pressures on the bacterial community’s tolerance of copper during composting (Li et al., 2014), which may be attributable to bacteria carrying copper resistance gene (CRGs) in the bacterial community. CRGs may reflect the actual responses of bacteria to the selective pressure imposed by copper, but the effects of copper on the evolution of CRGs during swine composting are still unclear. The main mechanisms that mediate copper resistance in microorganisms include the following: (1) the efflux ATPase pump encoded by copA can extrude copper ions from the cytoplasm into the periplasmic space (Rensing and Grass, 2003); (2) the cus system where the cusA gene encodes a resistance nodulation cell protein with an antiport system (Outten et al., 2001); (3) the pco system for Cu resistance encodes a multicopper oxidase protein, which is responsible for the oxidation of copper (I) to copper (II) in the periplasmic space (Brown et al., 1997); (4) the cue system is the main mechanism responsible for copper resistance in E. coli, where cueO encodes a periplasmic multicopper oxidase (Outten et al., 2001; Rensing and Grass, 2003); and (5) tcrB in Enterococcus faecium belongs to the CPX-type ATPase family of heavy metal transporters (Henrik and Frankm, 2002). The bacteria that carry these genes may be transferred from human and animal sources into the environment, thereby leading to disease. For example, Mourão et al. (2015a) found that pcoA-pcoD were chromosomally located in Salmonella enterica serotype 4,[5],12:i:, which is a new epidemic multidrug-resistant serotype in Europe that is associated with human infections. In E. faecium, tcrB belongs to the CPx-type ATPase family, and human homologs of the human CPx-type ATPase are related to Menkes and Wilson disease (Trenor et al., 1994).
Recently, pollution-induced community tolerance (PICT) was elucidated based on the rate of microbial metabolism and several researchers have represented the copper resistance of the microbial community as PICT (Fernández-Calviño and Bååth, 2013; Li et al., 2014). However, this has only been shown for the culturable fraction of bacteria (Huysman et al., 1994). Screening resistant bacteria by culture only samples microbes that are culturable and that can express their CRGs under these conditions. Recently, culture-independent methods have been investigated for quantifying the abundances of CRGs using quantitative PCR (qPCR; Besaury et al., 2013; Xiong et al., 2015; Zhang et al., 2016a). In addition, heavy metal and antibiotic resistance are frequently linked on the same plasmid (Baker-Austin et al., 2006; Laffite et al., 2016), and increased mobilization under metal-selective conditions might increase the risk of antibiotic resistance genes (ARGs) spreading via horizontal gene transfer to bacteria that cause human infections (Zhu et al., 2013; Martínez et al., 2014). The integron platforms are defective for self-transposition, but they are often associated with transposons and/or conjugative plasmids, which can serve as vehicles for the intra- and interspecies transmission of genetic material (Mazel, 2006). Gillings et al. (2015) proposed that the class 1 integron-integrase gene (intI1) can be used as a generic marker for anthropogenic pollutants. However, Pal et al. (2015) showed that no CRGs co-occur with intI1 or any other ARGs on plasmids based on large-scale genomic analyses of bacterial genomes and plasmids. Thus, it is necessary to obtain a better understanding of the changes in CRGs as well as the relationship between CRGs and the ARGs and intl1 during swine manure composting in the presence of added copper.
In the present study, we investigated the changes in CRGs, ARGs, and intl1 using qPCR, and we employed 16S rRNA high-throughput sequencing to determine the changes in the bacterial community. The results of this study should help to determine the effects of copper exposure on CRGs, ARGs, intl1, and the bacterial community during the manure composting process, thereby providing a better understanding of how and why the abundances of CRGs/ARGs change during composting under copper exposure.
The manure was collected from a medium-sized swine farm in Yangling, Shaanxi, China (i.e., producing swines > 500 head/year), where the diet comprised corn, bran, and soybean meal. The pre-mix feed included tetracycline, macrolide, CuSO4, and ZnSO4. In order to replicate the actual responses of resistance genes at different copper concentrations, the feed given to swine supplemented with trace elements and antibiotics was stopped 2 weeks before collecting the manure. The fresh manure was mixed well, air dried to obtain a water content <30%, crushed, and sieved through a 5-mm mesh. Wheat straw was cut into pieces that measured <1 cm long. The swine manure had an organic nitrogen content of 26 g kg-1, organic carbon content of 380.2 g kg-1, pH of 7.6, and total copper content of 121 mg kg-1. The wheat straw had an organic nitrogen content of 6.5 g kg-1 and an organic carbon content of 496.3 g kg-1.
The compost reactors comprised 54 identical 500 mL plastic containers, where 150 g dry weight (DW) of the raw material was placed in each container. The swine manure and wheat straw were mixed (3:1 DW) to prepare the compost mixture. The composting experiment was designed as described by Li et al. (2014) and determined based on the Cu residue levels in swine manure (Xiong et al., 2010). A stock solution of Cu was prepared and the diluted Cu solution was then mixed thoroughly with each sample to spike them with additional Cu concentrations of 200 and 2000 mg Cu kg-1 (DW), i.e., Cu200 and Cu2000, respectively. A similar volume of sterilized distilled water was added to the control (without the addition of CuSO4). Before use in the composting experiment, CuSO4 was stabilized for 6 h in swine manure. The moisture content was adjusted to 55%. To provide the optimal conditions for composting, the temperature was artificially controlled in an incubator in the following four phases: mesophilic phase (20–55°C, 0–5 days), thermophilic phase (55°C, 6–10 days; 55–50°C, 11–13 days; 50°C, 14–16 days), cooling phase (50–40°C, 17–21 days), and maturity (decreased to 20°C, 22–35 days), as described previously (Li et al., 2014). The composts were turned fully and mixed every 2 days during the composting process.
Three plastic containers for each treatment were then sampled as triplicates on days 0, 2, 7, 14, 21, and 35, and these samples were mixed well to form a test subsample. The sample was divided into two parts, where one was used to determine the physical and chemical properties, and the second was freeze-dried using a vacuum freeze dryer (Songyuan, China), milled to 1 mm by an ultra-centrifugal mill (Retsch Z200, Germany), and stored at –80°C.
The moisture contents of fresh samples were measured by drying at 105°C for 24 h. The pH values were determined based on fresh samples in water suspension at 1:10 (w/w) using a Thermo Orion 3-star pH-meter (San Diego, CA, USA). NH4+ and NO3- were extracted with 2 M KCl and determined by flow injection analysis (Systea, Italy).
Cu was sequentially extracted, as described by Tessier et al. (1979), where the extracts were then analyzed using a Model Z-2000 Series Polarized Zeeman atomic absorption spectrophotometer (Hitachi, Japan). For the data shown in Supplementary Figure S1 and Miaomiao et al. (2009), exchangeable Cu is defined as the bioavailable Cu in swine manure compost.
DNA was extracted from the freeze-dried compost samples using a Fast DNA Kit for Soil (MP Biomedicals, USA) according to the manufacturer’s instructions. Five CRGs (copA, cusA, cusO, pcoA, and tcrB), five macrolide resistance genes [erm(A), erm(B), erm(C), erm(T), and erm(X)], and the integrase gene of class 1 integrons (intI1) were determined by standard PCR. In addition, four CRGs (copA, cusA, pcoA, and tcrB), two macrolide resistance genes [erm(A) and erm(B)], and the integrase gene of class 1 integrons (intI1) were detected by standard PCR. These genes were quantified by real-time qPCR, where the conditions and primer sequences are described in the Supporting Information (Supplementary Material S1 and Tables S1a,b). The samples were quantified by qPCR using the Bio-Rad CFX connectTM Real-Time PCR Detection System (Bio-Rad). The absolute abundances of the CRGs and ARGs were expressed as copies g-1 of dry compost. The relative abundances of the CRGs and ARGs were calculated as: copy number of a CRG or ARG/copy number of 16S rRNA.
The V4 region of the 16S rRNA gene was analyzed by high-throughput sequencing using the Illumina HiSeq platform at Novogene (Beijing, China). The raw data were analyzed using QIIME software and the UPARSE pipeline (Caporaso et al., 2010). The UPARSE pipeline was employed for taxonomic assignment based on ≥97% similarity. Taxonomic classification was performed using the RDP classifier (Wang et al., 2007). All the raw sequence data have been deposited in the NCBI SRA database (Accession number: SRP093470).
All of the statistical analyses were performed using SPSS 19.0 (SPSS Inc., USA). Significant differences in the relative abundances of CRGs and ARGs (at P < 0.05) between treatments were detected using a one-way ANOVA/LSD test. Principal coordinates analysis (PCoA) was conducted using R (Version 2.15.3). Network analysis based on Spearman’s rank analysis was performed using the relative abundances of CRGs, ARGs, the bacterial community (based on OTU), and intI1 with the Gephi platform. Redundancy analysis was conducted with CANOCO 4.5.
As shown in Supplementary Table S2, the pH of the control increased gradually during the first week and the maximum value (pH 9.14) was reached on day 7. The pH then declined to 8.51 in the final compost. The increase in the pH during the initial stage of composting was due mainly to the release of ammonia and the degradation of organic acids (Li et al., 2014), whereas the decrease in the pH during the later phase was probably caused by the synthesis of phenolic compounds, or the nitrifying process (Zhang et al., 2015). In addition, the pH of Cu2000 was slightly lower than that of the control and Cu200 after 21 days, which may have been related to the adverse effects of Cu on the urease activity (Li et al., 2014), thereby inhibiting the release of ammonia. As the pile temperature increased, the NH4+ value increased during the first 7 days, but then decreased until the end of composting. The NO3- value increased on day 2 and decreased gradually during days 7–21, before increasing again from days 21 to 35. However, compared with the control, the NO3- values were lower in Cu200 (62.9% lower) and Cu2000 (76.4% lower) on day 35, which may have been related to inhibition of the activity of nitrifying microorganisms by copper.
During composting, the distribution of metals can be influenced by physicochemical changes within the mixture, such as organic matter mineralization, decreases in pH, and the formation of humic substances (Hsu and Lo, 2001; Farrell and Jones, 2009). The exchangeable Cu concentrations with different treatments are shown in Supplementary Table S2, which indicate that the exchangeable Cu concentration was 3.10–129.57 mg kg-1 during composting, where there was a decrease from early in the first week, although there was a transient increase in the control during the first 2 days. In the cooling phase, the exchangeable Cu contents remained fairly constant. This indicates that the forms of metals were more stable after composting, as shown in previous studies (Tandy et al., 2009; Li et al., 2014).
After composting for 35 days, the absolute abundances of pcoA and tcrB decreased (Supplementary Tables S3a,b), which might have been due to cell death. Burch et al. (2013) found that death of the bacterial host led to reductions in ARGs. In addition, composting provides aerobic conditions, which might have been unfavorable for the maintenance of pcoA and tcrB, e.g., pcoA and tcrB may function better under anaerobic conditions than aerobic conditions (Mourão et al., 2015a,b). However, the abundances of copA and cusA increased during the composting process (Supplementary Figure S2 and Table S3b). Outten et al. (2001) and Toes et al. (2008) found that copA and cusA were expressed under copper stress in aerobic and anaerobic conditions. Furthermore, the abundance of copA was higher than that that of cusA at the end of composting, especially in the Cu200 and Cu2000 treatments (Supplementary Tables S3a,b), which indicates that copA dominated when copper was added compared with cusA. By contrast, Besaury et al. (2014) reported that cusA was dominant compared with copA in copper-contaminated sediment, where this difference may be explained by variations in the evolution of the bacterial communities in the swine compost and sediment environments.
The variations in the relative abundances of CRGs are shown in Figure 1, Cu2000 enhanced the abundance of pcoA by 1.7–6.0 times compared with the control during days 2–14 (except on day 7), so it is probable that most of the microbes carrying the pcoA gene were enhanced by the high concentration of copper in the early composting phase. However, Cu2000 increased the abundance of tcrB by 1.3–8.9 times and 4.7–15.2 times compared with the control and Cu200, respectively, during days 14–35, which indicates that the bacterial hosts carrying tcrB may have increased under a high concentration of copper, as reported previously (Amachawadi et al., 2015). In addition, Cu200 and Cu2000 enhanced the abundances of cusA (1.4–2.4 times) and copA (1.7–2.9 times) compared with the control throughout the entire composting process, but there were no significant differences between Cu200 and Cu2000. This suggests that the abundances of the bacteria carrying copA and cusA increased during composting. Based on these results, we conclude that Cu2000 slowed down the loss of CRGs during composting.
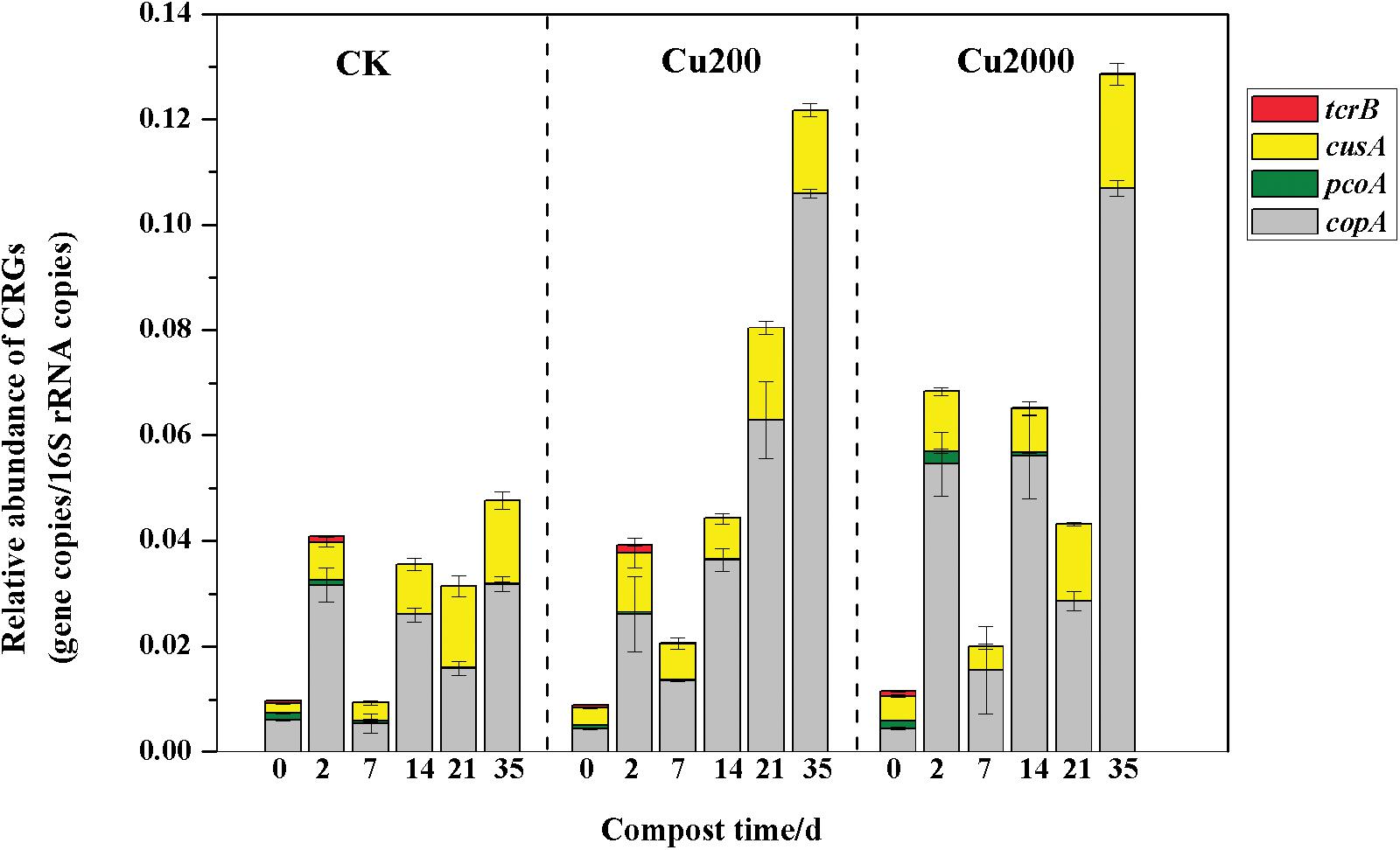
FIGURE 1. Changes in the relative abundances of copper resistance genes (CRGs) under different manure composting treatments.
Copper resistance genes and ARGs are commonly found on plasmids, and copper can potentially co-select for resistance to multiple antibiotics, especially the erm macrolide resistance gene (Amachawadi et al., 2013). As shown in Supplementary Figure S3, there were marked decreases in the absolute abundances of erm(A) (0.4–1.2 logs) and erm(B) (1.2–1.6 logs) after composting for 35 days. erm(B) had the highest relative abundance during the composting process (Figure 2). The relative abundance of erm(B) was significantly higher in Cu200 than that in the control (2.0–8.5 times higher) and in Cu2000 (1.7–4.2 times higher) during days 0–7. The relative abundance of erm(B) in Cu2000 was significantly higher than that in the control (1.1–6.1 times) and in Cu200 (1.2–5.9 times) during days 14–35. Compared with the control and Cu 200, Cu2000 enhanced the relative abundance of erm(A) by 0.9–2.0 times throughout the entire composting process. These results suggest that the relative abundances of erm(A) and erm(B) were increased by copper during swine manure composting. Previous studies also found that copper could increase the abundances of macrolide-resistant bacteria in compost (Li et al., 2014) and soil (Fernández-Calviño and Bååth, 2013).
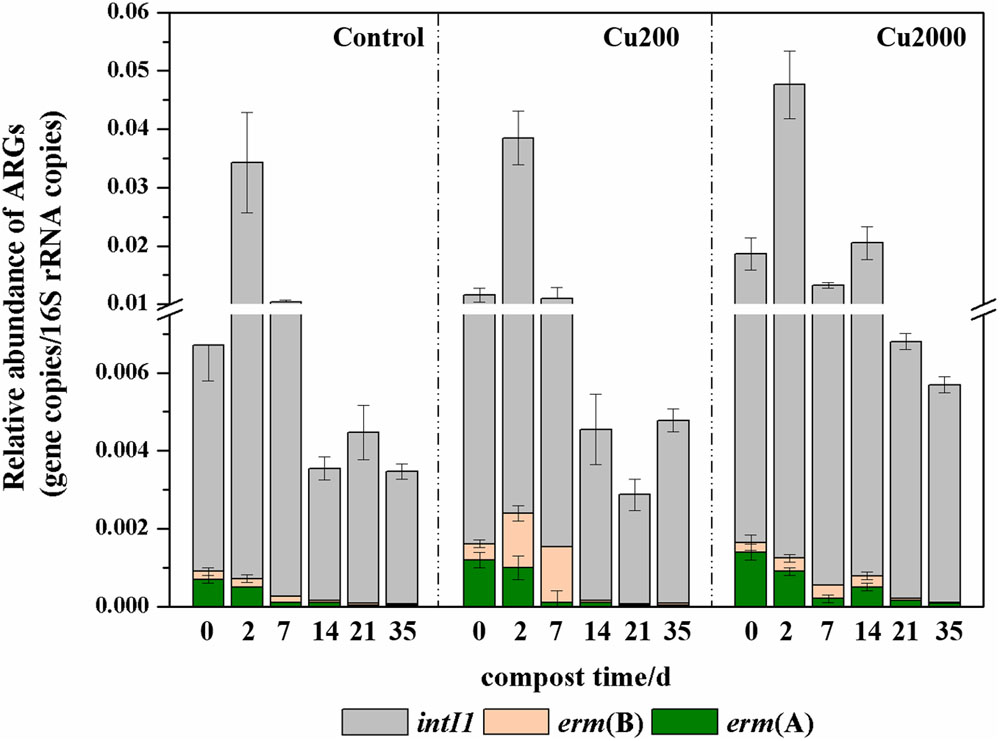
FIGURE 2. Changes in the relative abundances of antibiotic resistance genes (ARGs) and intI1 under different manure composting treatments.
intI1 genes cannot mobilize and transfer themselves between microbes, but they are often associated with mobile genetic elements such as conjugative plasmids, transposons, and insertion sequences (Wang et al., 2015; Zhang et al., 2016a). In this study, the abundance of intI1 decreased in all of the treatments (Supplementary Figure S3). Thus, the abundance of intI1 decreased by 0.7–1.1 logs during the thermophilic phase, but its abundance was stable throughout the subsequent composting period. Cu2000 significantly improved the abundance of intI1, and a high absolute abundance of intI1 in compost can enhance the dissemination of ARGs in the soil environment (Su et al., 2015; Qian et al., 2016a).
This study used 16S rRNA gene sequencing to investigate the responses of the bacterial community to copper stress during composting, where the addition of copper had limited effects on the succession of the bacterial community throughout composting. According to PCoA, PC1, and PC2 accounted for 70.49% of the total variation in the bacterial community (Supplementary Figure S4). The bacterial community structure was separated significantly during different composting periods, thereby indicating that the composting period played an important role in shaping the microbial community, as also shown in previous studies (Qian et al., 2016a; Zhang et al., 2016b). However, Cu2000 was most distinct from the control beyond day 2, and it tended toward the days 0–2 clusters (Supplementary Figure S4). These results demonstrate that higher concentrations of copper had more persistent effects on the bacterial community (Liu et al., 2015; Sun et al., 2016).
Among the 34 bacterial phyla identified, the most frequently detected were Firmicutes (17.6–47.0% in all composting treatments), Proteobacteria (16.2–39.2%), Actinobacteria (6.2–23.2%), and Bacteroidetes (2.8–20.7%), which accounted for 68.1.4–94.5% of the total bacteria (Supplementary Figure S5). Firmicutes was the dominant phyla in the mesophilic and thermophilic phases (Supplementary Figure S5). The genus Clostridium_sensu_stricto_1 was the main contributor to the changes in Firmicutes, and Clostridium _sensu_stricto_1 strains, which are widely distributed in soil, composts, and the gastrointestinal tracts of animals and humans (Rainisalo et al., 2011), were enhanced in Cu2000 after day 7 (Figure 3). Reed et al. (2013) found that Clostridium spp. accelerated the utilization of cellulose. Actinobacteria and Proteobacteria were dominant in the cooling and maturation phases. The abundance of Actinobacteria is considered a marker of compost maturity (Wang et al., 2013). Cu2000 decreased the abundance of these bacteria by 38.4% compared with the control on day 35, and the decreased abundance of this phylum after copper addition indicated the lower maturity of the compost obtained. Actinobacteria and Proteobacteria are important hosts of ARGs (Heuer et al., 2011; Stegmann et al., 2015) and previous studies have detected significant positive correlations between them and ARGs (Su et al., 2015; Zhang et al., 2016b). Compared with the control, Proteobacteria were enriched in Cu 2000 throughout composting (Supplementary Figure S5), whereas Actinobacteria were only enriched in Cu 200 and Cu2000 during the mesophilic stage. Pseudomonas and Corynebacterium_1 were the dominant Proteobacteria and Actinobacteria in the raw material (Figure 3), and they declined in all treatments by the end of composting. However, compared with the control, they were enriched in Cu2000 during composting. Pseudomonas species are highly resistant to and tolerant of copper (Liu et al., 2015), which may be attributed to the presence of the operon cop regulating the copper concentration in the cell as well as their ability to produce a large quantity of exopolymer to reduce the toxicity of copper in the soil (Kunito et al., 2001; Andreazza et al., 2012). The genus Corynebacterium includes potential human pathogens (Mohammadi et al., 2013), and Wang et al. (2016) claimed that Corynebacterium has a positive correlation with pco genes. In addition, Bacteroides were enriched in Cu200 and Cu2000 after 14 days (Figure 3). Liu and Pop (2009) demonstrated that macrolide resistance genes are often present in Bacteroides.
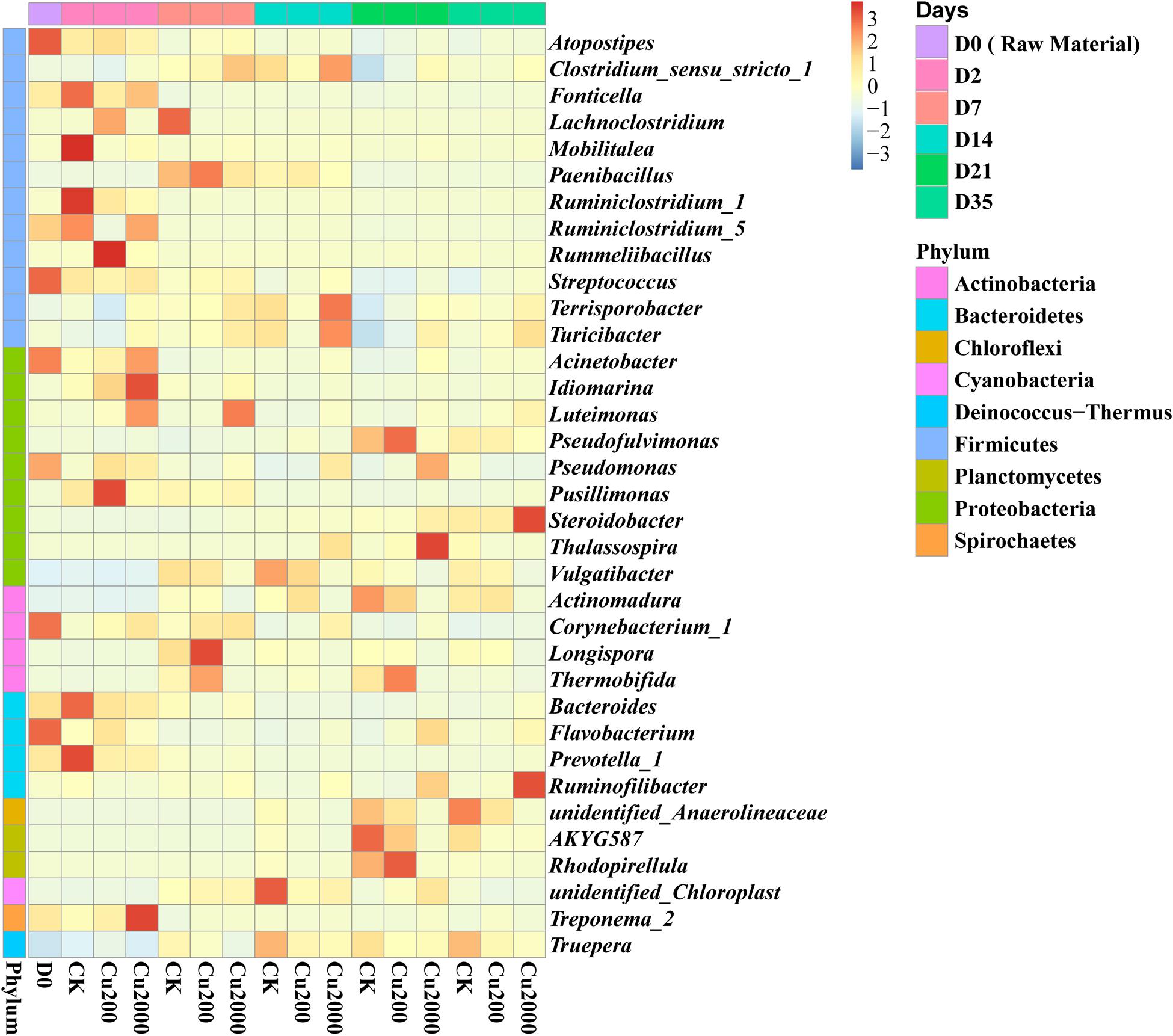
FIGURE 3. Heatmap of the top 35 genera identified in compost supplemented with 0, 200, and 2000 mg kg-1 copper.
We conducted redundancy analyses to investigate the relationships among environmental factors, CRGs, and ARGs (Figure 4), where the results showed that the selected variables accounted for 43.8% of the total variation in the changes in the CRGs and ARGs quantified in this study (P = 0.002). Among the environmental factors considered in this study, NH4+ and NO3- significantly explained the variations in the distributions of CRGs, ARGs, and intl1. It is possible that the hosts of these genes may have been influenced by the NH4+ and NO3- contents during composting. In addition, temperature significantly explained the decreases in CRGs, ARGs, and intl1. A previous study showed that normal thermophilic and continuous thermophilic composting can decrease the abundances of most ARGs and intl1 (Qian et al., 2016b). Thus, temperature could still reduce the abundances of CRGs, ARGs, and intl1 under the pressure of a high concentration of Cu. Furthermore, exchangeable copper was positively correlated with pcoA, tcrB, erm(A), erm(B), and intl1 (Supplementary Table S4), which indicates that copper supplementation may affect the abundance of bacteria that carry these genes. Heavy metals will be more persistent if they are present inside living bacteria (Ji et al., 2012). Li et al. (2014) claimed that a high concentration of copper increased the microbial PICT to copper and tylosin during swine manure composting.
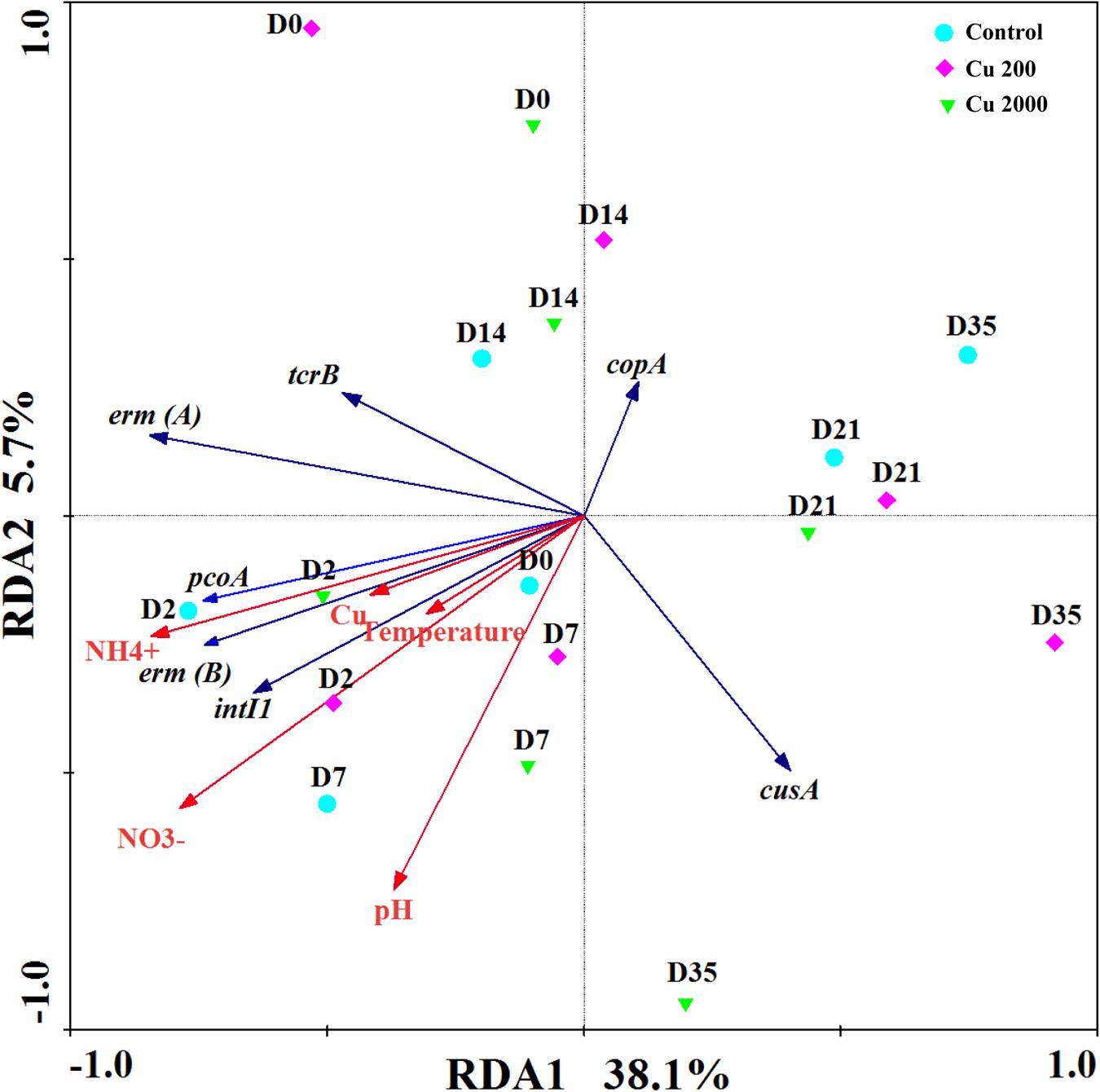
FIGURE 4. Redundancy analysis results illustrating the relationships between environmental factors, CRGs, intI1, and ARGs during manure composting in the control (A), Cu200 (B), and Cu2000 (C). Green triangles, purple diamonds, and blue circles represent samples from (A–C), respectively.
New insights into the potential hosts and resistance genes present in environmental samples can be obtained by network analysis (Li et al., 2015; Zhang et al., 2016a). In this study, the CRGs, ARGs, and intl1 had significant (p < 0.05) positive correlations with the same bacterial taxa (Figure 5), and it has been demonstrated that these genes posses the same potential host bacteria according to network analysis (Zhang et al., 2016a). The genus Steroidobacter may be the potential host bacteria of copA and cusA, and the increases in these CRGs may have been due to the presence of Steroidobacter during composting. The abundance of Steroidobacter increased as composting continued, especially in the Cu200 and Cu2000 treatments (Figure 3). The co-occurrence of pcoA, erm(A), and intl1 was detected in Bacteroides and Corynebacterium_1, and the decreases in the abundances of Bacteroides and Corynebacterium_1 during composting may explain the declines in erm(A), pcoA, and intl1. Nesvera et al. (1998) and Nandi et al. (2004) also found intl1 in Gram-positive bacteria, including Corynebacterium. Whittle et al. (2002) demonstrated that Bacteroides can acquire resistance genes and transfer themselves and other elements into distantly related bacteria via conjugative transposons. Corynebacterium_1, a potential human pathogen (Mohammadi et al., 2013), mainly carries tcrB and erm(B). Forsberg et al. (2014) and Zhang et al. (2016a) showed that diverse and abundant ARGs are harbored by environmental bacteria and pathogens. In addition, tcrB is considered to be present on a conjugative plasmid and the co-localization of tcrB with erm(B) was demonstrated in a previous study (Amachawadi et al., 2013). Surprisingly, pcoA, which is common on plasmids (Besaury et al., 2014), co-occurred with erm(A), and intl1. This finding is not consistent with the results obtained Pal et al. (2015), who found that no CRGs (including pcoA) co-occurred with any ARGs on plasmids, whereas CRGs were connected with many ARGs in the genome. This may be explained by the high intensity of Cu exposure in the swine manure compost, which could differ from that found in other environment samples. Mourão et al. (2015a) found that pcoA were chromosomally located in Salmonella enterica serotype 4,[5],12:i:, but we did not find that pcoA co-occurred with ARGs in Salmonella. Thus, the co-occurrence relationships determined by network analysis in the present study need to be validated further using other approaches.
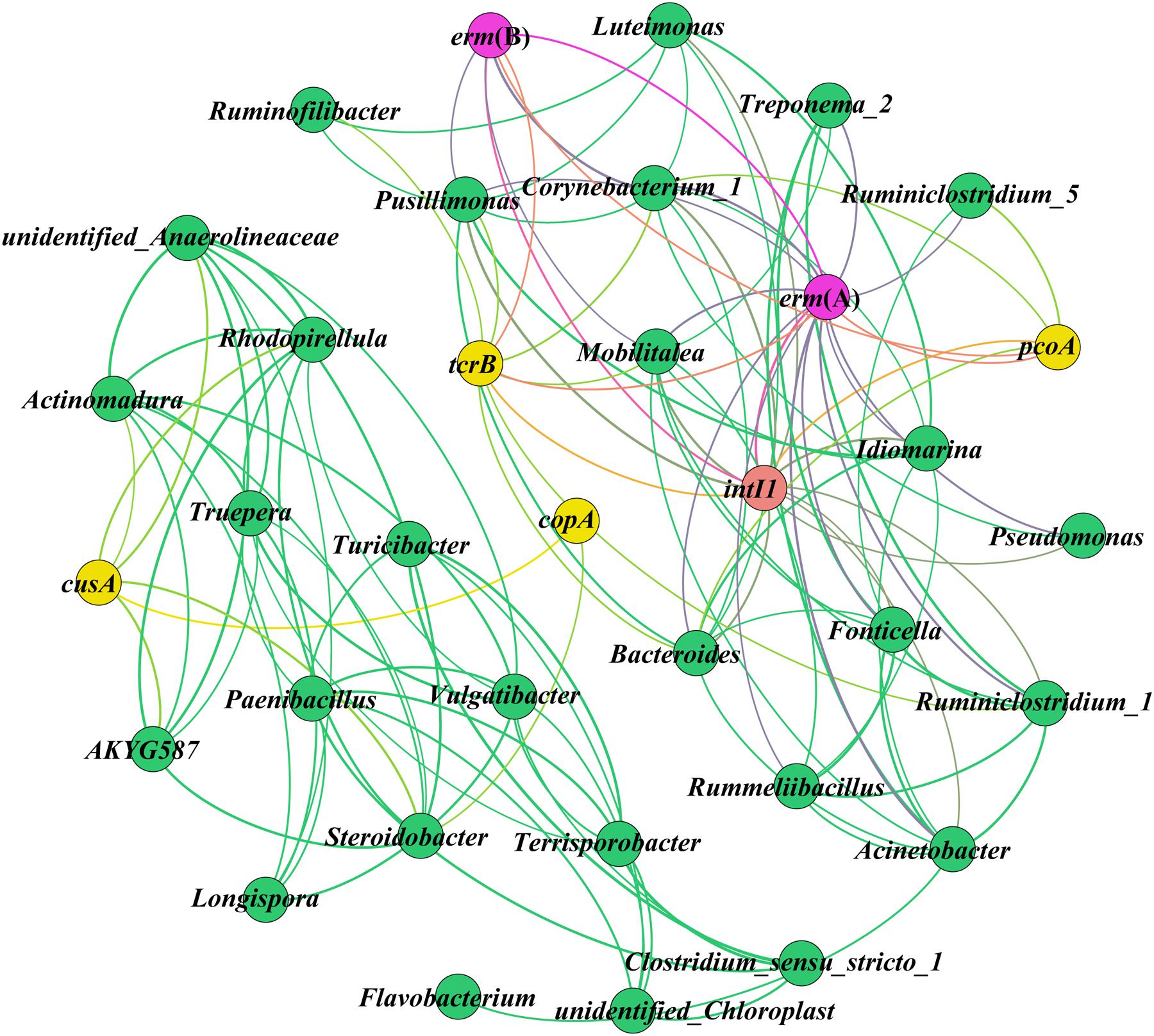
FIGURE 5. Network analysis showing the co-occurrence of ARGs, CRGs, and intl1 as well as their potential host bacteria. A connection represents a significant positive correlation (p < 0.05) according to Spearman’s rank analysis. The size of each node is proportional to the number of connections, i.e., the degree (details are given in the Supplementary Material S2).
This is the first study to investigate the fate of CRGs (pcoA, copA, cusA, and tcrB), ARGs [erm(A) and erm(B)], and intI1 during the composting of swine manure with added copper. The abundances of pcoA, tcrB, erm(A), erm(B), and intI1 decreased significantly, whereas those of copA and cusA increased significantly. The high concentration of copper slowed down the dissipation of CRGs, ARGs, and intI1 during composting. The composting period affected the succession of the bacterial community more greatly than the presence of copper, but a high concentration of Cu had more persistent effects on the bacterial community composition. The succession of the bacterial community, copper, and temperature are important factors that affect the fate of CRGs, ARGs, and intI1 during manure composting. In addition, we surprisingly detected the co-occurrence of CRGs, ARGs, and intI1 under copper exposure during swine manure composting, which should be validated using other approaches.
YY and JG designed experiments; YY, WSo, XZ, HL, and YZ carried out experiments; YY, XW, WSu, and KZ analyzed experimental results. YY wrote the manuscript.
This study was supported by the National Nature Science Foundation of China (41671474 and 41601531), the Special Fund for Agro-scientific Research in the Public Interest (201303094), and the 948 Project of the Chinese Ministry of Agriculture under Grant number 2015-Z37.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Weier Hao of Duquesne University and Dr. Duncan E. Jackson for language editing.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00344/full#supplementary-material
Amachawadi, R. G., Scott, H. M., Alvarado, C. A., Mainini, T. R., Vinasco, J., Drouillard, J. S., et al. (2013). Occurrence of the transferable copper resistance gene tcrB among fecal enterococci of U.S. feedlot cattle fed copper-supplemented diets. Appl. Environ. Microbiol. 79, 4369–4375. doi: 10.1128/AEM.00503–13
Amachawadi, R. G., Scott, H. M., Aperce, C., Vinasco, J., Drouillard, J. S., and Nagaraja, T. G. (2015). Effects of in-feed copper and tylosin supplementations on copper and antimicrobial resistance in fecal enterococci of feedlot cattle. J. Appl. Microbiol. 118, 1287–1297. doi: 10.1111/jam.12790
Andreazza, R., Okeke, B. C., Pieniz, S., and Camargo, F. A. (2012). Characterization of copper-resistant rhizosphere bacteria from Avena sativa and Plantago lanceolata for copper bioreduction and biosorption. Biol. Trace Elem. Res. 146, 107–115. doi: 10.1007/s12011–011-9228–1
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and Mcarthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/j.tim.2006.02.006
Besaury, L., Bodilis, J., Delgas, F., Andrade, S., Iglesia, R. D. L., Ouddane, B., et al. (2013). Abundance and diversity of copper resistance genes cus A and cop A in microbial communities in relation to the impact of copper on Chilean marine sediments. Mar. Pollut. Bull. 67, 16–25. doi: 10.1016/j.marpolbul.2012.12.007
Besaury, L., Pawlak, B., and Quillet, L. (2014). Expression of copper-resistance genes in microbial communities under copper stress and oxic/anoxic conditions. Environ. Sci. Pollut. Res. Int. 23, 4013–4023. doi: 10.1007/s11356–014-3254–4
Brown, D. R., Qin, K., Herms, J. W., Madlung, A., Manson, J., Strome, R., et al. (1997). The cellular prion protein binds copper in vivo. Nature 390, 684–687. doi: 10.1038/37783
Burch, T. R., Sadowsky, M. J., and Lapara, T. M. (2013). Aerobic digestion reduces the quantity of antibiotic resistance genes in residual municipal wastewater solids. Front. Microbiol. 4:17. doi: 10.3389/fmicb.2013.00017
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Farrell, M., and Jones, D. L. (2009). Heavy metal contamination of a mixed waste compost: metal speciation and fate. Bioresour. Technol. 100, 4423–4432. doi: 10.1016/j.biortech.2009.04.023
Fernández-Calviño, D., and Bååth, E. (2013). Co-selection for antibiotic tolerance in Cu-polluted soil is detected at higher Cu-concentrations than increased Cu-tolerance. Soil Biol. Biochem. 57, 953–956. doi: 10.1016/j.soilbio.2012.08.017
Forsberg, K. J., Patel, S., Gibson, M. K., Lauber, C. L., Knight, R., Fierer, N., et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616. doi: 10.1038/nature13377
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Guo, X., Gu, J., Gao, H., Qin, Q., Chen, Z., Shao, L., et al. (2012). Effects of Cu on metabolisms and enzyme activities of microbial communities in the process of composting. Bioresour. Technol. 108, 140–148. doi: 10.1016/j.biortech.2011.12.087
Henrik, H., and Frankm, A. (2002). tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46, 1410–1416. doi: 10.1128/AAC.46.5.1410–1416.2002
Heuer, H., Schmitt, H., and Smalla, K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243. doi: 10.1016/j.mib.2011.04.009
Hsu, J. H., and Lo, S. L. (2001). Effect of composting on characterization and leaching of copper, manganese, and zinc from swine manure. Environ. Pollut. 114, 119–127. doi: 10.1016/S0269–7491(00)00198–6
Huysman, F., Verstraete, W., and Brookes, P. C. (1994). Effect of manuring practices and increased copper concentrations on soil microbial populations. Soil Biol. Biochem. 26, 103–110.
Ji, X., Shen, Q., Liu, F., Ma, J., Xu, G., Wang, Y., et al. (2012). Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai. China. J. Hazard. Mater. 23, 178–185. doi: 10.1016/j.jhazmat.2012.07.040
Kunito, T., Saeki, K., Nagaoka, K., Oyaizu, H., and Matsumoto, S. (2001). Characterization of copper-resistant bacterial community in rhizosphere of highly copper-contaminated soil. Eur. J. Soil Biol. 37, 95–102. doi: 10.1016/S1164–5563(01)01070–6
Laffite, A., Kilunga, P. I., Kayembe, J. M., Devarajan, N., Mulaji, C. K., Giuliani, G., et al. (2016). Hospital effluents are one of several sources of metal, antibiotic resistance genes, and bacterial markers disseminated in Sub-Saharan Urban Rivers. Front. Microbiol. 7:1128. doi: 10.3389/fmicb.2016.01128
Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J. M., et al. (2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 9, 2490–2502. doi: 10.1038/ismej.2015.59
Li, Y., Bei, L., Zhang, X., Min, G., and Jing, W. (2014). Effects of Cu exposure on enzyme activities and selection for microbial tolerances during swine-manure composting. J. Hazard. Mater. 283, 512–518. doi: 10.1016/j.jhazmat.2014.09.061
Liu, B., and Pop, M. (2009). ARDB—antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447. doi: 10.1093/nar/gkn656
Liu, J., He, X. X., Lin, X. R., Chen, W. C., Zhou, Q. X., Shu, W. S., et al. (2015). Ecological effects of combined pollution associated with e-waste recycling on the composition and diversity of soil microbial communities. Environ. Sci. Technol. 49, 6438–6447. doi: 10.1021/es5049804
Martínez, J. L., Coque, T. M., and Baquero, F. (2014). What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 13, 116–123. doi: 10.1038/nrmicro3399
Mazel, D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. doi: 10.1038/nrmicro1462
Miaomiao, H., Wenhong, L., Xinqiang, L., Donglei, W., and Guangming, T. (2009). Effect of composting process on phytotoxicity and speciation of copper, zinc and lead in sewage sludge and swine manure. Waste Manag. 29, 590–597. doi: 10.1016/j.wasman.2008.07.005
Mohammadi, N. S., Mafakheri, S., Abdali, N., Tauch, A., and Benz, R. (2013). Identification and characterization of the channel-forming protein in the cell wall of Corynebacterium amycolatum. Biochim. Biophys. Acta 1828, 2574–2582. doi: 10.1016/j.bbamem.2013.06.024
Mourão, J., Novais, C., Machado, J., Peixe, L., and Antunes, P. (2015a). Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- clones circulating in Europe. Int. J. Antimicrob. Agents 45, 610–616. doi: 10.1016/j.ijantimicag.2015.01.013
Mourão, J., Rae, J., Silveira, E., Freitas, A. R., Coque, T. M., Peixe, L., et al. (2015b). Relevance of tcrYAZB operon acquisition for Enterococcus survival at high copper concentrations under anaerobic conditions. J. Antimicrob. Chemother. 71, 560–563. doi: 10.1093/jac/dkv330
Nandi, S., Maurer, J. J., Hofacre, C., and Summers, A. O. (2004). Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. U.S.A. 101, 7118–7122. doi: 10.1073/pnas.0306466101
Nesvera, J., Hochmannova, J., and Patek, M. (1998). An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169, 391–395.
Outten, F. W., Huffman, D. L., Hale, J. A., and O’Halloran, T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677. doi: 10.1074/jbc.M104122200
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides, and metals reveals novel insights into their co-selection potential. BMC Genomics 16:964. doi: 10.1186/s12864–015-2153–5
Qian, X., Sun, W., Gu, J., Wang, X. J., Sun, J. J., Yin, Y. N., et al. (2016a). Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure. J. Hazard. Mater. 315, 61–69. doi: 10.1016/j.jhazmat.2016.05.002
Qian, X., Sun, W., Gu, J., Wang, X. J., Zhang, Y. J., Duan, M. L., et al. (2016b). Reducing antibiotic resistance genes, integrons, and pathogens in dairy manure by continuous thermophilic composting. Bioresour. Technol. 220, 425–432. doi: 10.1016/j.biortech.2016.08.101
Rainisalo, A., Romantschuk, M., and Kontro, M. H. (2011). Evolution of clostridia and streptomycetes in full-scale composting facilities and pilot drums equipped with on-line temperature monitoring and aeration. Bioresour. Technol. 102, 7975–7983. doi: 10.1016/j.biortech.2011.05.087
Reed, P. T., Izquierdo, J. A., and Lynd, L. R. (2013). Cellulose fermentation by Clostridium thermocellum and a mixed consortium in an automated repetitive batch reactor. Bioresour. Technol. 155, 50–56. doi: 10.1016/j.biortech.2013.12.051
Rensing, C., and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213. doi: 10.1016/S0168–6445(03)00049–4
Stegmann, E., Frasch, H. J., Kilian, R., and Pozzi, R. (2015). Self-resistance mechanisms of actinomycetes producing lipid II-targeting antibiotics. Int. J. Med. Microbiol. 305, 190–195. doi: 10.1016/j.ijmm.2014.12.015
Su, J., Wei, B., Ou-Yang, W., Huang, F., Zhao, Y., Xu, H., et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 49, 7356–7363. doi: 10.1021/acs.est.5b01012
Sun, F. L., Fan, L. L., and Xie, G. J. (2016). Effect of copper on the performance and bacterial communities of activated sludge using Illumina MiSeq platforms. Chemosphere 156, 212–219. doi: 10.1016/j.chemosphere.2016.04.117
Tandy, S., Healey, J. R., Nason, M. A., Williamsona, J. C., and Jonesa, D. L. (2009). Heavy metal fractionation during the co-composting of biosolids, deinking paper fibre and green waste. Bioresour. Technol. 100, 4220–4226. doi: 10.1016/j.biortech.2009.02.046
Tessier, A., Campbell, P. G. C., and Bisson, M. (1979). Sequential extraction procedure for the speciation of particulate trace metals. Anal. Chem. 51, 844–851.
Toes, A. C., Daleke, M. H., Kuenen, J. G., and Muyzer, G. (2008). Expression of copA and cusA in Shewanella during copper stress. Microbiology 154, 2709–2718. doi: 10.1099/mic.0.2008/016857–0
Trenor, C., Lin, W., and Andrews, N. C. (1994). Novel bacterial P-type ATPases with histidine-rich heavy-metal-associated sequences. Biochem. Biophys. Res. Commun. 205, 1644–1650.
Wang, C., Guo, X., Deng, H., Dong, D., Tu, Q., and Wu, W. (2013). New insights into the structure and dynamics of actinomycetal community during manure composting. Appl. Microbiol. Biotechnol. 98, 3327–3337. doi: 10.1007/s00253–013-5424–6
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062–07
Wang, Q., Lu, Q., Mao, D., Cui, Y., and Luo, Y. (2015). The horizontal transfer of antibiotic resistance genes is enhanced by ionic liquid with different structure of varying alkyl chain length. Front. Microbiol. 6:864. doi: 10.3389/fmicb.2015.00864
Wang, R., Zhang, J., Sui, Q., Wan, H., Tong, J., Chen, M., et al. (2016). Effect of red mud addition on tetracycline and copper resistance genes and microbial community during the full scale swine manure composting. Bioresour. Technol. 216, 1049–1057. doi: 10.1016/j.biortech.2016.06.012
Whittle, G., Shoemaker, N. B., and Salyers, A. A. (2002). The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59, 2044–2054.
Xiong, W., Zeng, Z., Zhang, Y., Ding, X., and Sun, Y. (2015). Fate of metal resistance genes in arable soil after manure application in a microcosm study. Ecotoxicol. Environ. Saf. 113, 59–63. doi: 10.1016/j.ecoenv.2014.11.026
Xiong, X., Li, Y., Li, W., Lin, C., Han, W., and Yang, M. (2010). Copper content in animal manures and potential risk of soil copper pollution with animal manure use in agriculture. Resour. Conserv. Recycl. 54, 985–990. doi: 10.1016/j.resconrec.2010.02.005
Zhang, J., Chen, M., Sui, Q., Rui, W., Tong, J., and Wei, Y. (2016a). Fate of antibiotic resistance genes and its drivers during anaerobic co-digestion of food waste and sewage sludge based on microwave pretreatment. Bioresour. Technol. 217, 28–36. doi: 10.1016/j.biortech.2016.02.140
Zhang, J., Chen, M., Sui, Q., Tong, J., Jiang, C., Lu, X., et al. (2016b). Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res. 91, 339–349. doi: 10.1016/j.watres.2016.01.010
Zhang, L., Zeng, G., Zhang, J., Chen, Y., Yu, M., Lu, L., et al. (2015). Response of denitrifying genes coding for nitrite (nirK or nirS) and nitrous oxide (nosZ) reductases to different physico-chemical parameters during agricultural waste composting. Appl. Microbiol. Biotechnol. 99, 4059–4070. doi: 10.1007/s00253–014-6293–3
Keywords: antibiotic resistance gene, bacterial community, composting, copper resistance gene, Cu
Citation: Yin Y, Gu J, Wang X, Song W, Zhang K, Sun W, Zhang X, Zhang Y and Li H (2017) Effects of Copper Addition on Copper Resistance, Antibiotic Resistance Genes, and intl1 during Swine Manure Composting. Front. Microbiol. 8:344. doi: 10.3389/fmicb.2017.00344
Received: 18 September 2016; Accepted: 20 February 2017;
Published: 03 March 2017.
Edited by:
Octavio Luiz Franco, Universidade Católica de Brasília, BrazilReviewed by:
Krassimira Hristova, Marquette University, USACopyright © 2017 Yin, Gu, Wang, Song, Zhang, Sun, Zhang, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Gu, Z3VqaWUyMDVAc2luYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.