
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 02 March 2017
Sec. Food Microbiology
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00316
This article is part of the Research TopicRecent Discoveries in Human Serious Foodborne Pathogenic Bacteria: Resurgence, Pathogenesis, and Control StrategiesView all 22 articles
This study evaluated the antimicrobial and desiccation resistance of Cronobacter sakazakii and Cronobacter malonaticus isolates from powdered infant formula and processing environments. The antimicrobial susceptibility tests showed that the 70 Cronobacter strains, representing 19 sequence types, were susceptible to the most of the antibiotics except for amoxicillin-clavulanate, ampicillin, and cefazolin. Furthermore, the growth of six C. sakazakii and two C. malonaticus strains from different sequence types (STs) in hyperosmotic media was measured. The growth of the two C. sakazakii strains (CE1 and CE13) from the neonatal pathovars ST4 and ST8, were significantly higher (p < 0.05) than that of other strains. C. malonaticus strain CM35 (ST201) was the slowest grower in all strains, and most could not grow in more than 8% NaCl solution. Also the survival of these strains under desiccation conditions was followed for 1 year. The viable count of Cronobacter spp. under desiccation conditions was reduced on average by 3.02 log cycles during 1 year, with CE13 (ST8) being the most desiccation resistant strain. These results will improve our understanding of the persistence of the two closely related species C. sakazakii and C. malonaticus which are of concern for neonatal and adult health.
Cronobacter spp. (formerly Enterobacter sakazakii) is an emerging opportunistic bacterial pathogen comprised of seven species: Cronobacter sakazakii, Cronobacter malonaticus, Cronobacter turicensis, Cronobacter muytjensii, Cronobacter condimenti, Cronobacter universalis, and Cronobacter dublinensis (Joseph et al., 2012b; Lu et al., 2014). This organism can cause necrotizing enterocolitis, bacteremia, and meningitis in neonates and infants, with a 40–80% mortality rate (Forsythe et al., 2014; Holy and Forsythe, 2014; Li et al., 2015). To date, only the three species C. sakazakii, C. malonaticus, and C. turicensis have been isolated from neonatal infections (Sonbol et al., 2013). Cronobacter spp. has been recovered from a wide variety of foods, with powdered infant formula (PIF) being of particular concern as it is the most significant source of Cronobacter strains resulting in neonatal infections (Iversen and Forsythe, 2004; Craven et al., 2010). Cronobacter strains cannot survive after the standard pasteurization procedures, therefore, the addition of non-heat treated materials and environmental contamination during filling and packaging are the plausible causes of Cronobacter contamination of PIF (Nazarowec-White and Farber, 1997). Besides, the strong resistant ability of the organism in dry PIF factory environments can increase the likelihood of post-pasteurization contamination (Riedel and Lehner, 2007).
Currently, antibiotic therapy is considered to be the common and preferred way to prevent the Cronobacter infection in humans (Depardieu et al., 2007). Many studies have confirmed that Cronobacter strains can be effectively eliminated by antibiotics, however, prolonged use of antibiotics is undesirable as it may result in the development of Cronobacter antibiotic resistance (Yoneyama and Katsumata, 2006; McMahon et al., 2007). Hochel et al. (2012) reported that all of 53 Cronobacter strains isolated from 399 retail food samples were resistant to erythromycin, and two of them were resistant to both erythromycin and tetracycline. Chon et al. (2012) found that 77.8% Cronobacter strains isolated from desiccated foods in Korea were not susceptible to cephalothin. The Cronobacter strains resistance to cefotaxime and streptomycin have also been isolated from various foods in Korea and commercial PIF in China (Lee et al., 2012). Caubilla-Barron and co-workers reported that the C. sakazakii strains isolated from two fatal neonatal infections expressed β–lactamase activity (Caubilla-Barron et al., 2007). In addition, the effective dose of antibiotics for treatment may change after long-term antibiotic use, therefore screening environmental and PIF isolates of Cronobacter for antibiotic resistance would be a useful comparison with clinical isolates.
The greater resistance of Cronobacter spp. to desiccation compared with other Enterobacteriaceae will enable their long term survival under a low water activity (aw) condition such as in PIF (aw: 0.2–0.5) which has a long shelf life of up to 1 year (Gurtler and Beuchat, 2007). In general, the viability of Cronobacter spp. in PIF decreases by 0.5–0.6 log cycles per month (Edelson-Mammel et al., 2005; Gurtler and Beuchat, 2007). Caubilla-Barron et al. (2007) found there was considerable reduction in the viable count of Cronobacter strains (about 3.34 log10 cycles) in the first 6 months, whereas after the following 24 months, the average reduction in viability decreased by 1.88 log10 cycles. It is important to note that some capsulated C. sakazakii strains were still recovered after 2.5 years, by contrast, both Salmonella enteritidis, and Escherichia coli were undetectable after 15 months (Barron and Forsythe, 2007). Some difference in desiccation resistance can be found among the different Cronobacter strains, for example, C. sakazakii NCTC11467T has atypical growth characteristics compared with most of Cronobacter strains, and cannot be recovered from desiccated condition after 1 year (Iversen et al., 2004). Cronobacter spp. produce highly mucoid colonies on milk agar plates and the capsular material could be linked to virulence traits such as macrophage survival as well as desiccation resistance (Ogrodzki and Forsythe, 2015). Particular capsule profiles correlate with the meningitic C. sakazakii pathovar clonal complex (CC) 4 (Ogrodzki and Forsythe, 2015).
Cronobacter is a diverse genus and has been extensively studied using multilocus sequence typing for over 1,000 strains (Forsythe et al., 2014). This has revealed the high clonality within the genus as well as identified particular pathovars. C. sakazakii sequence type (ST) four identified using multi-locus sequence typing (MLST) was the dominant ST, and was associated with neonatal meningitis (Joseph and Forsythe, 2011). In addition, Cronobacter pathovars, associated with clonal complex are now recognized. Of particular relevance is the C. sakazakii CC4 pathovar which is associated with neonatal meningitis (Hariri et al., 2013; Forsythe et al., 2014). C. sakazakii ST8 and C. sakazakii ST1 were mainly isolated from clinical sources and PIF, respectively (Forsythe et al. 2014). C. sakazakii CC4 was also the genotype of ~25% of isolates recovered from the processing environment of PIF manufacturing plants (Sonbol et al., 2013). Why this CC predominates in PIF and neonatal infections is uncertain since no particular virulence genes have been detected (Joseph et al., 2012a). Therefore, a better understanding of the environmental fitness and antibiotic resistance of Cronobacter isolates with significant sequence type from PIF production areas is warranted.
In previous studies, our group isolated 66 Cronobacter sakazakii and four C. malonaticus strains from PIF and processing environments from 2009 to 2012, and identified their sequence type by MLST (Fei et al., 2015). In this study, we further characterized the strains according to their antibiograms and desiccation tolerance, and considered whether the differences corresponded with their sequence type.
A total of 70 Cronobacter strains (66 C. sakazakii and four C. malonaticus strains) were studied. All strains had been isolated from PIF and processing environments, as previously reported (Fei et al., 2015). All 70 Cronobacter strains were used for the antimicrobial assays, and six C. sakazakii (CE21, CE1, CE13, CE38, CE52, and CE25) and two C. malonaticus (CM3 and CM35) strains with different sequence types were selected for studying the survivals under desiccation condition.
Strains were recovered from storage at −80°C in 40% (v/v) glycerol, by inoculating 0.1 mL portions of thawed cultures into 10 mL Luria-Bertani (LB) broth, followed by cultivation at 37°C for 12 h. The cultures were streaked onto Tryptic Soy Agar (TSA) plates and incubated at 37°C for 24 h for single colony isolation. A single colony of each strain was inoculated into the LB and incubated at 37°C for 18 h for following study.
The antimicrobial susceptibility determination was performed using the BD PhoenixTM100 Automated Microbiology System (BD Diagnostic Systems, Sparks, MD) as according to the manufacturer's instructions. Twenty-one antibiotics were selected for the susceptibility test, including amikacin, amoxicillin-clavulanate, ampicillin, ampicillin-sulbactam, aztreonam, cefotaxime, ceftazidime, cefazolin, cefepime, chloramphenicol, ciprofloxacin, colistin, gentamicin, imipenem, levofloxacin, meropenem, moxifloxacin, piperacillin, piperacillin-tazobactam, tetracycline, and trimethoprim-sulfamethoxazole. The results were expressed as sensitive (S), intermediate (I), and resistant (R), and resistant according to the PhoenixTM100 guidelines. E. coli ATCC 25922 and E. coli ATCC 35218 were used as the quality control organisms, and were included in each run according to the manufacturer's recommendations.
For determining bacterial growth in a hyperosmotic environment, cultures (0.1 mL) were inoculated into brain–heart infusion (BHI) broth containing molar equivalents of either sodium chloride (NaCl) (4, 6, 8, 10% w/v) or sorbitol (12.5, 19, 25, 31% w/v). The above cultures were incubated at 37°C for 24 h, and subsequent growth was measured the growth according to the absorption values at 600 nm (OD 600) using an ultraviolet spectrophotometer (Biochrom Ltd., Cambridge, England). The “time to detection” (TTD), defined as the time (in h) to reach an OD 600 of 0.2, and “growth rate” (in h−1) were used to determine the growth of each strain under the different treatments, as previously reported (Alvarez-Ordonez et al., 2014).
C. sakazakii and C. malonaticus were grown in LB for 18 h at 37°C before analysis. For dry stresses, the cultures were harvested by centrifugation at 8,000 g for 10 min, then the cell pellets were diluted in 0.85% sterile normal saline (NS) to obtain a cell density of 8.00 log10 cfu/mL.
The C. sakazakii and C. malonaticus cell suspensions were desiccated as previously described with minor modifications (Breeuwer et al., 2003). Unstressed cultures (50 μL) were transferred to sterile petri dishes without their lids, which were placed in a constant temperature humidity chamber, and incubated at 25°C with 20.7% air relative humidity (RH) for air-drying. After drying for 1.5 h, the petri dishes were covered and kept at 21°C with 20.7% RH for 1 year. Periodically, the bacterial survival was determined by conventional colony counting.
Mean values and standard deviations were obtained from three replicate experiments with duplicated plating (n = 6). Statistical analysis was performed by Analysis of Variance (ANOVA) with the SPSS 20.0 software. Tukey's multiple range test was used to determine the significant differences (p < 0.05) between treatments.
The antimicrobial susceptibility pattern and Minimal Inhibitory Concentration (MIC) of the 70 Cronobacter strains are shown in Tables 1, 2. All Cronobacter strains were susceptible to most antibiotics, including amikacin, ampicillin-sulbactam, aztreonam, cefepime, cefotaxime, ceftazidime, chloramphenicol, ciprofloxacin, colistin, gentamicin, imipenem, levofloxacin, meropenem, moxifloxacin, piperacillin, piperacillin- tazobactam, tetracycline, and trimethoprim-sulfamethoxazole. However, they were resistant to amoxicillin-clavulanate, ampicillin, and cefazolin. The MIC of antibiotics, except cefazolin, did not vary across the 70 Cronobacter strains. Ciprofloxacin and trimethoprim-sulfamethoxazole were considered to be the most effective antibiotics against the 70 Cronobacter strains at MIC of ≤ 0.5 and ≤ 0.5/0.95 μg/mL. In contrast, there were some differences in their resistance to cefazolin; Table 2. The 70 Cronobacter strains were divided into four groups according to their cefazolin MIC-values (≤4, >4, ≤8, >8, ≤16, >16 μg/mL). A majority of Cronobacter isolates with the same ST had a same MIC-value.
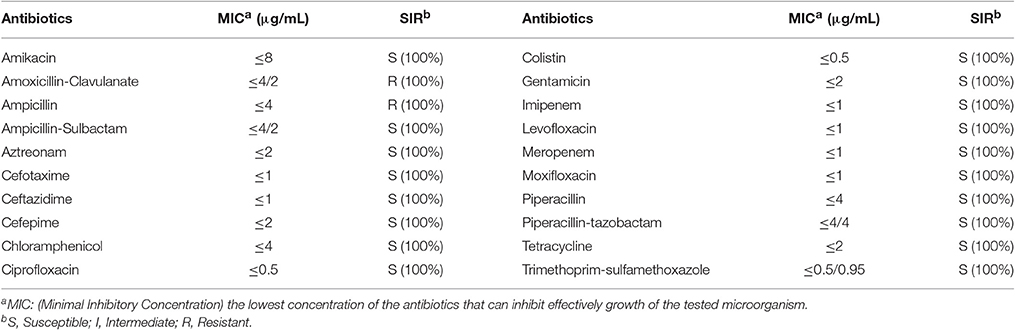
Table 1. Antimicrobial susceptibility and MIC of 70 Cronobacter strains isolated from PIF and processing environments.
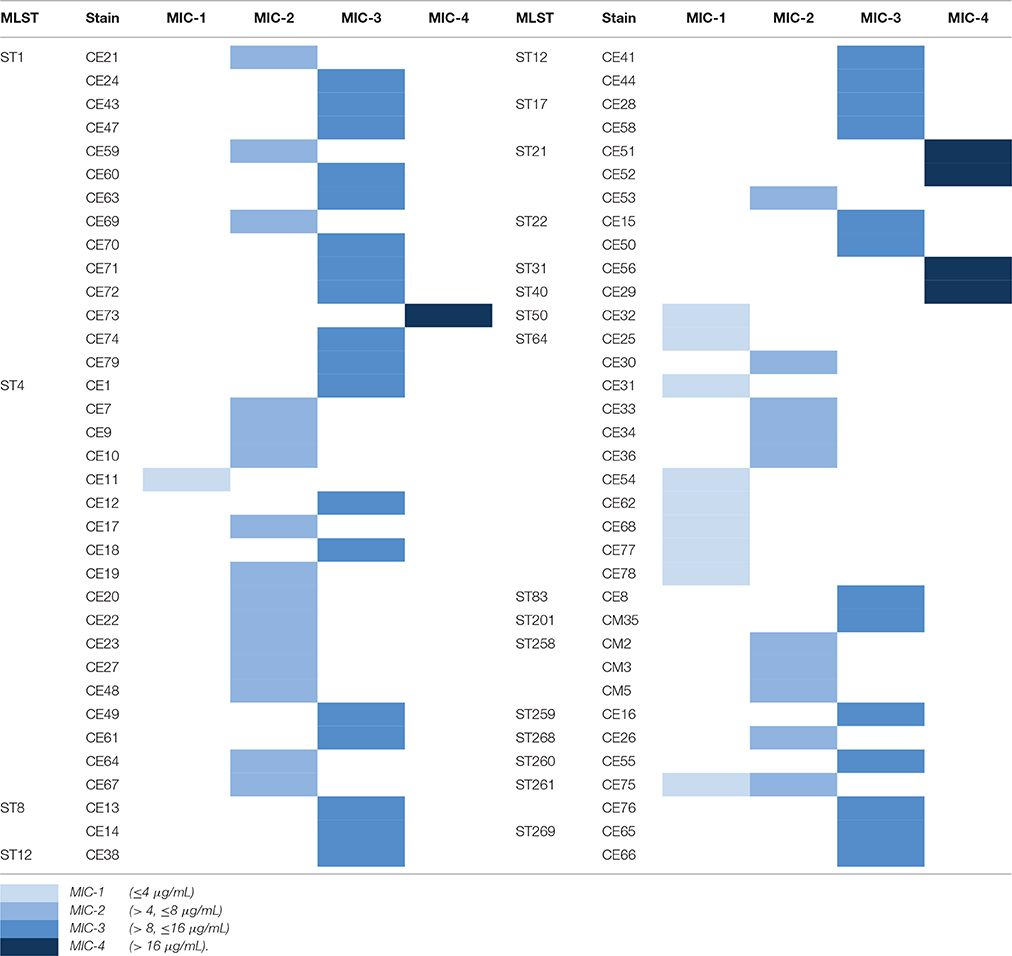
Table 2. The differences of resistance to cefazolin in 70 Cronobacter strains isolated from PIF and processing environments.
Six C. sakazakii and two C. malonaticus strains were incubated in BHI with various molar equivalents of either NaCl or sorbitol at 37°C for 24 h. The TTD values and growth rates of eight Cronobacter strains are shown in Table 3. Compared with growth in BHI without high concentration solutes, the growth of all eight strains in BHI with different concentrations of NaCl and sorbitol was significantly delayed and slower. The TTD-values of all eight strains growing at >8% NaCl were >24 h, and the growth rates of those ranged from 0.0004/h to 0.0035/h. Meanwhile, in BHI with sorbitol, all strains reached an OD 600 of 0.2 within 12 h. This result showed that the growth of strains was not completely inhibited in <31% sorbitol, whereas there was no growth in the molar equivalent NaCl. In addition, there was variation between strains in their resistance to osmotic stress. Among the eight strains in this study, the growth of CE25 (ST64) was the most significantly delayed (p < 0.05), while CE13 (ST8) had more resistance to the delayed growth compared with other strains. The growth rates of CE13 (ST8) and CE1 (ST4) were significantly higher (p < 0.05) than that of other strains, and CM35 (ST201) was the slowest grower (p < 0.05) in all strains.
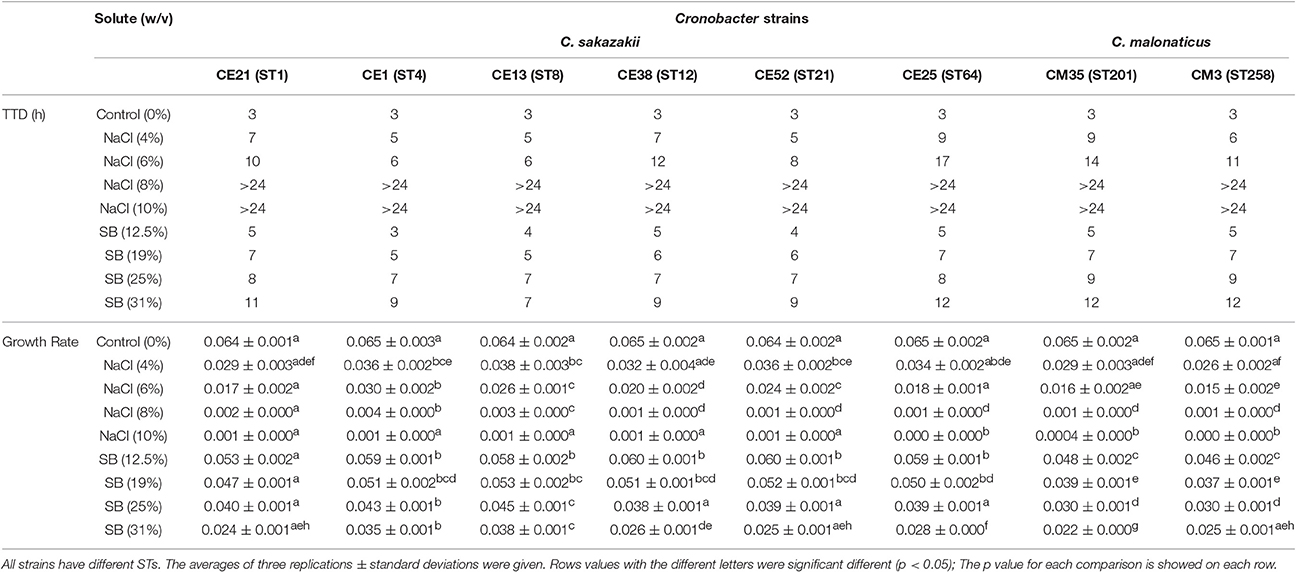
Table 3. Growth ability of the six C. sakazakii (CE21, CE1, CE13, CE38, CE52, and CE25) and two C. malonaticus (CM35 and CM3) strains under concentrations of NaCl (4, 6, 8, 10% w/v) and sorbitol (SB) (12.5, 19, 25, 31%w/v) conditions.
The survival of six C. sakazakii and two C. malonaticus strains stored under desiccation condition (21°C, RH = 20.7%) was monitored for up to 1 year (Table 4). In the first 60 days, the average recovery declined by about 1.55 log cycles, then, a smaller decrease was observed during the next 10 months, ranging from 1.07 log cycles (CE13, ST8) to 1.85 log cycles (CE25, ST64). Within 1 year, the reduction of all strains ranged from 2.43 log cycles (CE13, ST8) to 3.46 log cycles (CE25, ST64). The survival of C. malonaticus CM3 (ST258) and C. sakazakii CE25 (ST64) were significantly less (p < 0.05) than the other six Cronobacter strains, and C. sakazakii CE13(ST8) was the highest (p < 0.05) survival in all eight Cronobacter strains. In addition, during the different time-phase in 1 year, the CM3 (ST258) had significant lower (p < 0.05) survival value compared with other strains.
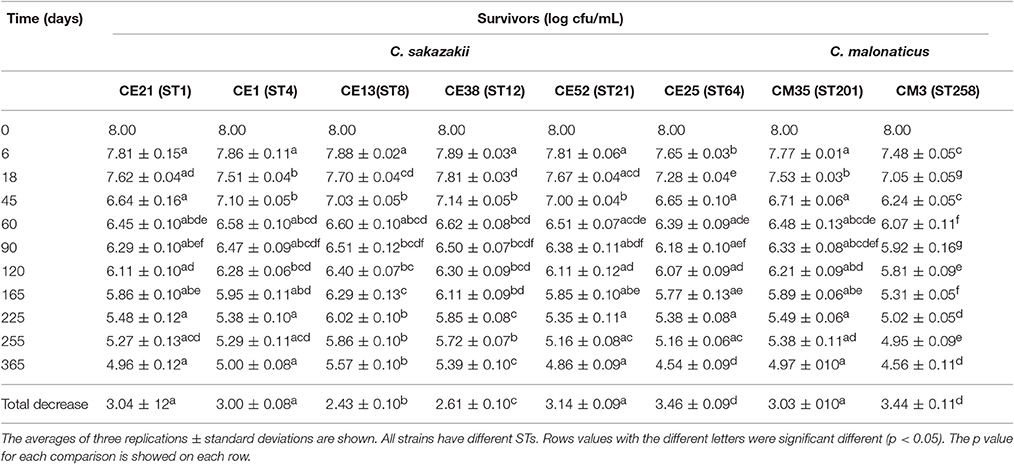
Table 4. Survivors (log cfu/mL) of the six C. sakazakii (CE21, CE1, CE13, CE38, CE52, and CE25) and two C. malonaticus (CM35 and CM3) after dehydration for up to 1 year.
C. sakazakii and C. malonaticus are the dominant species of Cronobacter spp. isolated from PIF and processing environment, and can infect infants and adults, respectively (Fei et al., 2015). Given the long term shelf-life of PIF (up to 1 year) and repeated isolation from PIF production plants (Craven et al., 2010; Sonbol et al., 2013), environmental fitness of Cronobacter is very important trait to understand as it may lead to increased persistence and neonatal exposure.
The phylogenetic relationship of concatenated sequences (3,036 bp) based on conventional MLST (7 loci), ribosomal-MLST (53 loci), and core genome MLST (1865 loci) reflects the whole genome phylogeny of the Cronobacter genus (Joseph et al., 2012b; Forsythe et al., 2014). The application of MLST has also led to the recognition of stable clonal complexes and sequence types associated with certain clinical presentations. Subsequently the diversity and clonal stability within the Cronobacter genus needs to be taken into consideration when interpreting laboratory studies. Therefore, Cronobacter strains with different sequence types were chosen for detailed study here.
In this study, 70 Cronobacter strains, previously isolated from PIF and processing environments, were used. These represented 19 sequence types of C. sakazakii and C. malonaticus, detailed information can be obtained from Cronobacter MLST databases (http://pubmlst.org/cronobacter/) (Fei et al., 2015). The main STs from PIF and processing environments, were the neonatal pathovars C. sakazakii ST4, ST1, and ST8, as well as ST64, ST12, ST21, ST258, and ST201.
The isolates were susceptible to most antibiotics, except for amoxicillin-clavulanate, and ampicillin. Previous reports revealed that the occurrence of susceptibility to cefotaxime, chloramphenicol, ciprofloxacin, gentamicin, and tetracycline in Cronobacter strains isolated from different sources (Chon et al., 2012; Xu et al., 2014). Chon et al. (2012) reported that only 5.6% strains from desiccated foods in Korea were resistance to ampicillin whereas in our study, all isolates were resistant to this antibiotic. Furthermore, we found that the MIC values of cefazolin varied across different STs, but were similar within each ST. This may reflect a connection between the ST and antimicrobial resistance.
The ability of eight Cronobacter isolates with different STs to resist the osmotic stress was evaluated in this study. Compared with C. malonaticus isolates, C. sakazakii isolates had a higher capacity to grow in the hyperosmotic media, in agreement with previous studies (Caubilla-Barron et al., 2007; Avelino Alvarez-Ordóñez et al., 2014). C. sakazakii CE13 (ST8) was the most resistant to osmotic stress in the eight strains, interestingly, C. sakazakii ATCC 29544T reported to have atypical growth characteristics is also ST8 (Osaili and Forsythe, 2009). C. sakazakii CE1 (ST4) also showed a more osmotic stress resistance, which might be one of the major reasons why C. sakazakii CC4 is frequently isolated from infant food, ready-to-eat foods, potable water. This could result in higher infant exposure and therefore risk of infection (Forsythe et al., 2014).
Cronobacter spp. tend to persist more in PIF with a water activity (aw) 0.25–0.30, compared with aw 0.43–0.50, and the survival rate was not associated with whether the PIF was milk-based or soybean-based (Osaili and Forsythe, 2009). In general, this organism can persist in PIF for more than 1 year, therefore, monitoring of survival under desiccated conditions is warranted for the two main Cronobacter species, C. sakazakii and C. malonaticus. Breeuwer et al. (2003) reported that the reduction of Cronobacter strains (species undetermined) which had been dried in air and incubated for 46 days at 25°C ranged from 1.0 to 1.5 log cycles. Using similar desiccation conditions, the viable counts of Cronobacter strains maintained at 25°C for 45 days decreased by 0.86–1.76 log cycles in our study. Barron and Forsythe (2007) reported that the number of survivors of C. sakazakii, C. muytjensis, and C. turicensis under desiccated conditions in the first 30 days reduced by 0.58 log cycles on average, and decreased 3.34 log cycles during the first 6 months. While, under the similar desiccation conditions, the results in current study indicated that the reduction of Cronobacter strains declined by an average of 0.86 log cycles during the first month, and 2.25 log cycles during the first 6 months. The differences in reduction of Cronobacter strains between different reports indicated that a continuous assessment of resistance to dry stress was very necessary.
Interestingly, after kept in the desiccation conditions for 1 year, the total decrease of CE13 (ST8) was the least, followed by CE38 (ST12) and CE1 (ST4). This trend was similar to osmotic stress resistance. Mechanisms of resistance to these environmental stressor have previously been reviewed by Osaili and Forsythe (2009). The higher resistance to both osmotic and desiccation stresses by C. sakazakii compared with C. malonaticus may in part account for the predominance of C. sakazakii in PIF and processing environments and subsequent greater infant exposure.
In summary, this study contributes to an improved understanding of the environmental persistence of C. sakazakii and C. malonaticus and subsequent risk of infant exposure through contaminated PIF. In addition, the relatively low antibiotic resistance is reassuring for the current treatment of Cronobacter infections.
Conceived and designed the experiments: CM and PF. Performed the experiments: PF, YJ, CM, JF, and YZ. Generated and analyzed the data: PF and RL. Wrote the paper: PF, CM, and SF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by the “Academic Backbone” Project of Northeast Agricultural University (15XG26) and Science Foundation for Distinguished Young Scholars of Heilongjiang Province (No. JC201415). We thank Lu Yan for her help in the antimicrobial resistance.
Alvarez-Ordonez, A., Begley, M., Clifford, T., Deasy, T., Collins, B., and Hill, C. (2014). Transposon mutagenesis reveals genes involved in osmotic stress and drying in Cronobacter sakazakii. Food Res. Int. 55, 45–54. doi: 10.1016/j.foodres.2013.10.037
Avelino Alvarez-Ordóñez, T. D., Clifford, T., Begley, M., and Hill, C. (2014). Acid stress management by Cronobacter sakazakii. Int. J. Food Microbiol. 178, 21–28. doi: 10.1016/j.ijfoodmicro.2014.03.001
Barron, J. C., and Forsythe, S. J. (2007). Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula. J. Food Prot. 70, 2111–2117. doi: 10.4315/0362-028X-70.9.2111
Breeuwer, P., Lardeau, A., Peterz, M., and Joosten, H. M. (2003). Desiccation and heat tolerance of Enterobacter sakazakii. J. Appl. Microbiol. 95, 967–973. doi: 10.1046/j.1365-2672.2003.02067.x
Caubilla-Barron, J., Hurrell, E., Townsend, S., Cheetham, P., Loc-Carrillo, C., Fayet, O., et al. (2007). Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J. Clin. Microbiol. 45, 3979–3985. doi: 10.1128/JCM.01075-07
Chon, J.-W., Song, K.-Y., Kim, S.-Y., Hyeon, J.-Y., and Seo, K.-H. (2012). Isolation and characterization of Cronobacter from desiccated foods in Korea. J. Food Sci. 77, 354–358. doi: 10.1111/j.1750-3841.2012.02750.x
Craven, H. M., McAuley, C. M., Duffy, L. L., and Fegan, N. (2010). Distribution, prevalence and persistence of Cronobacter (Enterobacter sakazakii) in the nonprocessing and processing environments of five milk powder factories. J. Appl. Microbiol. 109, 1044–1052. doi: 10.1111/j.1365-2672.2010.04733.x
Depardieu, F., Podglajen, I., Leclercq, R., Collatz, E., and Courvalin, P. (2007). Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20, 79–114. doi: 10.1128/CMR.00015-06
Edelson-Mammel, S. G., Porteous, M. K., and Buchanan, R. L. (2005). Survival of Enterobacter sakazakii in a dehydrated powdered infant formula. J. Food Prot. 68, 1900–1902. doi: 10.4315/0362-028X-68.9.1900
Fei, P., Man, C., Lou, B., Forsythe, S. J., Chai, Y., Li, R., et al. (2015). Genotyping and source tracking of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and an infant formula production factory in China. Appl. Environ. Microbiol. 81, 5430–5439. doi: 10.1128/AEM.01390-15
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15:1121. doi: 10.1186/1471-2164-15-1121
Gurtler, J. B., and Beuchat, L. R. (2007). Survival of Enterobacter sakazakii in powdered infant formula as affected by composition, water activity, and temperature. J. Food Prot. 70, 1579–1586. doi: 10.4315/0362-028X-70.7.1579
Hariri, S., Joseph, S., and Forsythe, S. J. (2013). Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerging Infect. Dis. 19, 175–177. doi: 10.3201/eid1901.120649
Hochel, I., Ruzickova, H., Krasny, L., and Demnerova, K. (2012). Occurrence of Cronobacter spp. in retail foods. J. Appl. Microbiol. 112, 1257–1265. doi: 10.1111/j.1365-2672.2012.05292.x
Holy, O., and Forsythe, S. (2014). Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 86, 169–177. doi: 10.1016/j.jhin.2013.09.011
Iversen, C., and Forsythe, S. J. (2004). Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 21, 771–776. doi: 10.1016/j.fm.2004.01.009
Iversen, C., Lane, M., and Forsythe, S. J. (2004). The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38, 378–382. doi: 10.1111/j.1472-765X.2004.01507.x
Joseph, S., Cetinkaya, E., Drahovska, H., Levican, A., Figueras, M. J., and Forsythe, S. J. (2012a). Cronobacter condimenti sp nov., isolated from spiced meat, and Cronobacter universalis sp nov., a species designation for Cronobacter sp genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 62, 1277–1283. doi: 10.1099/ijs.0.032292-0
Joseph, S., Desai, P., Ji, Y., Cummings, C. A., Shih, R., Degoricija, L., et al. (2012b). Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS ONE 7:e49455. doi: 10.1371/journal.pone.0049455
Joseph, S., and Forsythe, S. J. (2011). Predominance of Cronobacter sakazakii Sequence Type 4 in Neonatal Infections. Emerging Infect. Dis. 17, 1713–1715. doi: 10.3201/eid1709.110260
Lee, Y.-D., Park, J.-H., and Chang, H. (2012). Detection, antibiotic susceptibility and biofilm formation of Cronobacter spp. from various foods in Korea. Food Control 24, 225–230. doi: 10.1016/j.foodcont.2011.09.023
Li, R., Fei, P., Man, C. X., Lou, B. B., Niu, J. T., Feng, J., et al. (2015). Tea polyphenols inactivate Cronobacter sakazakii isolated from powdered infant formula. J. Dairy Sci. 99, 1019–1028. doi: 10.3168/jds.2015-10039
Lu, Y., Chen, Y., Lu, X. A., Lv, J., Man, C. X., Chai, Y. L., et al. (2014). Comparison of methods for the microbiological identification and typing of Cronobacter species in infant formula. J. Dairy Sci. 97, 632–641. doi: 10.3168/jds.2013-7147
McMahon, M. A. S., Xu, J., Moore, J. E., Blair, I. S., and McDowell, D. A. (2007). Environmental stress and antibiotic resistance in food-related pathogens. Appl. Environ. Microbiol. 73, 211–217. doi: 10.1128/AEM.00578-06
Nazarowec-White, M., and Farber, J. M. (1997). Incidence, survival, and growth of Enterobacter sakazakii in infant formula. J. Food Prot. 60, 226–230. doi: 10.4315/0362-028X-60.3.226
Ogrodzki, P., and Forsythe, S. (2015). Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16:758. doi: 10.1186/s12864-015-1960-z
Osaili, T., and Forsythe, S. (2009). Desiccation resistance and persistence of Cronobacter species in infant formula. Int. J. Food Microbiol. 136, 214–220. doi: 10.1016/j.ijfoodmicro.2009.08.006
Riedel, K., and Lehner, A. (2007). Identification of proteins involved in osmotic stress response in Enterobacter sakazakii by proteomics. Proteomics 7, 1217–1231. doi: 10.1002/pmic.200600536
Sonbol, H., Joseph, S., McAuley, C. M., Craven, H. M., and Forsythe, S. J. (2013). Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 30, 1–7. doi: 10.1016/j.idairyj.2012.11.004
Xu, X., Wu, Q., Zhang, J., Ye, Y., Yang, X., and Dong, X. (2014). Occurrence and characterization of Cronobacter spp. in powdered formula from Chinese retail markets. Foodborne Pathog. Dis. 11, 307–312. doi: 10.1089/fpd.2013.1657
Keywords: C. sakazakii, C. malonaticus, desiccation, antibiotic susceptibility, powdered infant formula
Citation: Fei P, Jiang Y, Feng J, Forsythe SJ, Li R, Zhou Y and Man C (2017) Antibiotic and Desiccation Resistance of Cronobacter sakazakii and C. malonaticus Isolates from Powdered Infant Formula and Processing Environments. Front. Microbiol. 8:316. doi: 10.3389/fmicb.2017.00316
Received: 17 October 2016; Accepted: 15 February 2017;
Published: 02 March 2017.
Edited by:
Lanming Chen, Shanghai Ocean University, ChinaReviewed by:
Séamus Fanning, University College Dublin, IrelandCopyright © 2017 Fei, Jiang, Feng, Forsythe, Li, Zhou and Man. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaoxin Man, bWN4d2gyMDA2QHFxLmNvbQ==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.