- 1State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Office of Emergency Response, Chenzhou Center for Disease Control and Prevention, Chenzhou, China
- 3School of Public Health, University of South China, Hengyang, China
Bacterial mammalian cell entry (Mce) proteins have been implicated in pathogen invasion of mammalian host cells. The aim of this study was to examine the invasion-conferring ability of mce1E operon-encoded proteins, in vivo expression of Mce1E in cells from infected mice and rabbits, and Mce1E immunogenicity. Nocardia farcinica mce1E was cloned into pet30a(+) vectors, expressed in Escherichia coli, and purified. Invasion assays, transmission electron microscopy (TEM), immunoblots, and enzyme-linked immunosorbent assay (ELISA) detection of cytokines were conducted. TEM confirmed the invasion of HeLa cells by Mce1E-coated beads. The antigenicity of E. coli-expressed recombinant Mce1E was confirmed in immunoblots with sera from N. farcinica-infected mouse and rabbit sera. Co-incubation of Mce1E with splenocytes of N. farcinica-infected mice demonstrated upregulation of interferon (IFN-γ), but not interleukin (IL)-4 or IL-10, in the cultural supernatant. These findings demonstrate that Mce1E may facilitate N. farcinica interactions with and invasion of mammalian cells. Notably, Mce1E are expressed and elicited antibody responses in mice and rabbits during infection. Besides, it may play a role in cell-mediated immune reactions and cause host inflammation responses to N. farcinica infection.
Introduction
Nocardia genus bacteria are Gram-positive filamentous rod, aerobic pathogens found in soil and water worldwide (Scharfen et al., 2010). They are considered opportunistic pathogens, affecting predominantly immunocompromised patients, including patients with AIDS and transplant recipients (Kim et al., 2016). Pulmonary disease is the most common presentation of Nocardia in immunosuppressed patients and approximately one-third of affected patients have a disseminated disease (Ambrosioni et al., 2010; Kandi, 2015; Scorey and Daniel, 2016). Infection of traumatic wounds produces chronic inflammation that may lead to fistulas, abscesses, cellulitis, ulcerations, and mycetoma (Smego and Gallis, 1984; Salinas-Carmona, 2000; Salinas-Carmona et al., 2009), and may extend into muscles, bones, the brain, kidneys, the prostate, cornea, heart, and adjacent organs (De Nardo et al., 2013; Sirijatuphat et al., 2013; Kumar et al., 2014; Park et al., 2014; Sharma and O’Hagan, 2016). Nocordia infection of the central nervous system may be acquired by cutaneous or respiratory routes (Smego and Gallis, 1984; Beaman and Beaman, 1994; Inamadar and Palit, 2003; Zakaria et al., 2008; Chen et al., 2016).
The incidence of Nocardia infection cases has been increasing in recent years. Thus far, some 25 Nocordia species have been found to infect human patients, including Nocordia brasiliensis, N. asteroides, N. farcinica, N. abscessus, N. nova, and N. transvalensis complex; among them, N. farcinica is the most commonly encountered species (Kandi, 2015). Clinically, untreated pulmonary nocardiosis resembles tuberculosis and thus represents a risk for misdiagnosis (Ekrami et al., 2014).
The mechanisms of Nocardia-macrophage interaction have not been resolved. To achieve infection, bacteria must first escape host defenses. Bacteria can enter human macrophages through interactions with cell-surface receptors (Glickman and Jacobs, 2001). The mammalian cell entry (Mce) proteins, encoded by mce genes, are a family of invasion proteins expressed by Mycobacteria. They have putative signal sequences at the N-terminus and are thought to be localized to the mycobacterial cell surface (Ahmad et al., 2005). Mce protein expression in non-pathogenic Escherichia coli has been shown to enable the bacteria to enter and survive within HeLa cells and macrophages (Arruda et al., 1993; El-Shazly et al., 2007; Saini et al., 2008). Six mce operons were identified in N. farcinica (Ishikawa et al., 2004). However, no prior study has clarified whether the Mce1E protein in N. farcinica enables host cell invasion. Furthermore, the immunological reactivity of Mce1E has not been described in the literature.
In the present study, we examined whether expression of purified recombinant N. farcinica Mce1E protein can promote N. farcinica invasion of mammalian cells. Additionally, we assessed expression of Mce1E in N. farcinica infections. Finally, we explored Mce1E immunogenicity in murine splenocytes infected with N. farcinica.
Materials and Methods
Ethics Statement
Laboratory animal care and experimentation were performed in accordance with animal ethics guidelines and approved protocols. The animal experiments were approved by the Ethics Review Committee of the National Institute for Communicable Disease Control and Prevention at the Chinese Center for Disease Control and Prevention.
Bacterial Strains, Plasmid, and Anti-N. farcinica Sera
Standard DSM43131 strain N. farcinica bacteria were purchased from the German Resource Centre for Biological Materials and grown in brain-heart-infusion medium at 37°C (Difco Laboratories Inc., Detroit, MI, USA). The pET30a(+) plasmid was used as an expression vector and E. coli BL21 (DE3) were used a host for the vector, as recommended by the manufacturer. E. coli colonies (TransGen Biotech, China) were grown in Luria-Bertani (LB) medium at 37°C. Anti-N. farcinica sera were prepared from BALB/c mice and New Zealand rabbits in our laboratory.
Expression and Purification of Recombinant Mce1E
The mce1E gene was amplified from N. farcinica genomic DNA by polymerase chain reaction with the following specific primers: forward 5′-GTA TCA TAT GAT GAG ACG CGC GCG TCG CAC-3′ and reverse 5′-GAT CAA GCT TTC GGC CCT GTC CCC CCT CGA-3′. Polymerase chain reaction products were digested by Nde I and Hind III and then introduced into the pET-30a(+) prokaryotic expression vector. The recombinant plasmids were sequenced and then transformed into E. coli BL21 cells for fusion protein expression. The E. coli BL21 cells were cultured at 37°C with agitation in LB medium containing 50 μg/ml kanamycin until their optical density at 600 nm reached 0.8. Subsequently, the cells were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 30°C for 6 h.
After sonication and centrifugation, Mce1E protein molecules were solubilized in binding buffer containing 6 M urea. The solubilized proteins were chromatographed on a His column in accordance with the manufacturer’s instructions (Novagen, Germany). Purified protein were dialyzed in a concentration gradient of urea (6, 4, 2, and 1 M) to allow renaturing at 4°C for 24 h. The renatured proteins were placed in phosphate-buffered saline (PBS) overnight at 4°C. The recombinant Mce1E proteins were examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and their concentrations were determined with a BCA protein assay kit (Thermo Scientific, USA).
Coating of Beads with Recombinant Proteins and HeLa Cell Culture
A 5-μl sample of stock latex bead suspension (4%w/v, 0.3-μm diameter, Thermo Fisher) was mixed with 1 ml PBS containing 60 μg Mce1E protein; uncoated latex beads served as the control treatment. Samples from all groups were incubated for 2 h at 37°C. HeLa cells were cultured with Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal calf serum (FCS; Gibco) at 37°C.
Invasion Assays and Transmission Electron Microscopy (TEM) Analysis
HeLa cells were harvested, washed, and re-suspended in Dulbecco’s modified Eagle’s medium. Subsequently, they were seeded in a 24-well polystyrenetissue culture plates and incubated to form monolayers of cells. MceIE-coated latex beads and uncoated latex beads (200 μl per aliquot) were added to near-confluent HeLa cell monolayers grown in 24-well plates. The cells were incubated at 37°C for 24 h in a CO2 incubator, and then washed three times with PBS. The washed cells were fixed in 2% glutaraldehyde, postfixed in 1% osmium tetroxide, and dehydrated through increasing grades of ethanol solution. Samples of cells were embedded, and then cut into ultrathin sections. The ultrathin sections were stained and examined by transmission electron microscopy (TEM) with a microscope HT7700 (Japan).
Western Blot
To confirm recombinant protein expression, proteins were separated by SDS-PAGE (5–12%) and transferred onto polyvinylidene fluoride membranes with 100 V for 1 h. The membranes were then blocked with blocking buffer (5% skim milk in PBS, pH 7.4, with 0.05% Tween 20) overnight at 4°C. A 1:4000 dilution of a horse-radish peroxidase (HRP)-conjugated monoclonal anti-pentahistidine (His) antibody (New England Biolabs Inc., USA) was applied to the membranes for 1 h to detectthe His tag. Mouse and rabbit antisera (1:2000) were used as primary antibodies and an IgG-HRP goat antibody (Sigma) was used as secondary antibody to detect recombinant Mce1E. Detection was performed with a diaminobenzidine kit (DAB, Tiangen).
Spleen Cell Preparation and Cytokine Detection
A unicellular suspension containing 108 colony forming units per ml of N. farcinica in the log phase of growth was injected subcutaneously (500 μl) into BALB/c mice (6–8 weeks). Spleens were harvested 1, 3, 7, and 14 days after infection (N = 3 per time point). The red blood cells were removed from the spleen samples and the spleen lymphocytes were washed and then suspended at a concentration of 1 × 106 cells per ml of RPMI-1640 medium with 10% FCS. Cultures containing 1 × 106 cells were stimulated with 1 μg/ml of recombinant Mce1E or phytohemagglutinin A (PHA). The stimulated cultures were incubated for 24 h in 5% CO2 at 37°C with constant humidity (95%). Subsequently, the cultures were centrifuged at 10000 rpm for 5 min to remove the cells. Interferon (IFN)-γ, interleukin (IL)-10, and IL-4 concentrations in the supernatant were determined by quantitative enzyme-linked immunosorbent assay (ELISA; IFN-γ ELISA kit from RD, IL-10 and IL-4 ELISA kits from BD, USA). All cultures were processed in triplicate.
Statistical Analysis
Group means and standard deviations (SDs) were compared with Student’s t-tests. Differences were considered statistically significant when p-values were below 0.05.
Results
Expression and Purification of Recombinant Mce1E
Nucleotide sequencing analysis confirmed that mce1E had been inserted correctly into the pet30a vector. As shown in Figure 1, SDS-PAGE results showed that after cells were induced with 1 mM IPTG (30°C for 6 h), they exhibited increased expression of a 48-kDa protein. It was further demonstrated that purification removed, to a large extent, other proteins, leaving a predominant band at 48 kDa.
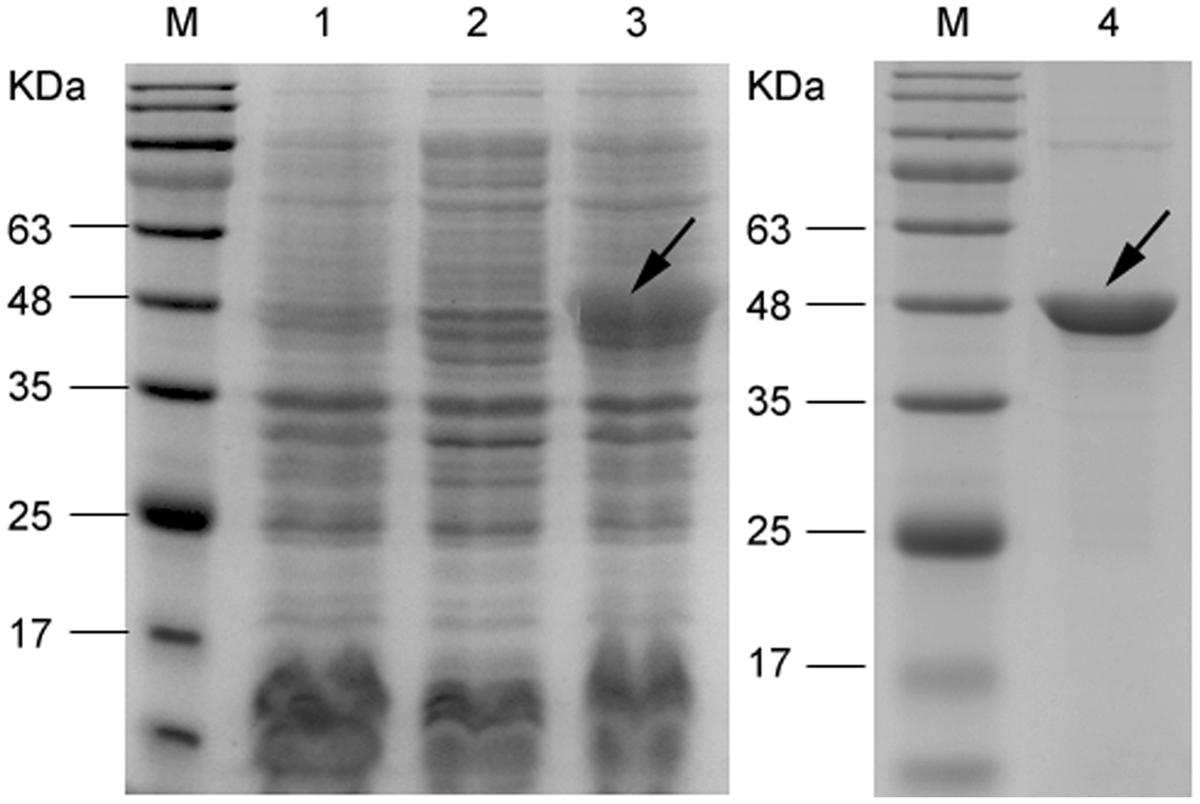
FIGURE 1. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of recombinant Mce1E expressed in Escherichia coli BL21. Lane M, marker; lane 1, vector control (whole-cell protein); lane 2, uninduced control (whole-cell protein); lane 3, induced (30°C for 6 h; whole-cell protein); and lane 4, purified Mce1E protein.
Invasion of HeLa Cells by mce1E-Coated Latex Beads
As shown in Figure 2, TEM confirmed that beads coated with Mce1E entered HeLa cells. Internalized coated-beads were observed within vacuolar compartments after 24 h of incubation; non-coated beads were not observed within vacuolar compartments after 24 h of incubation.
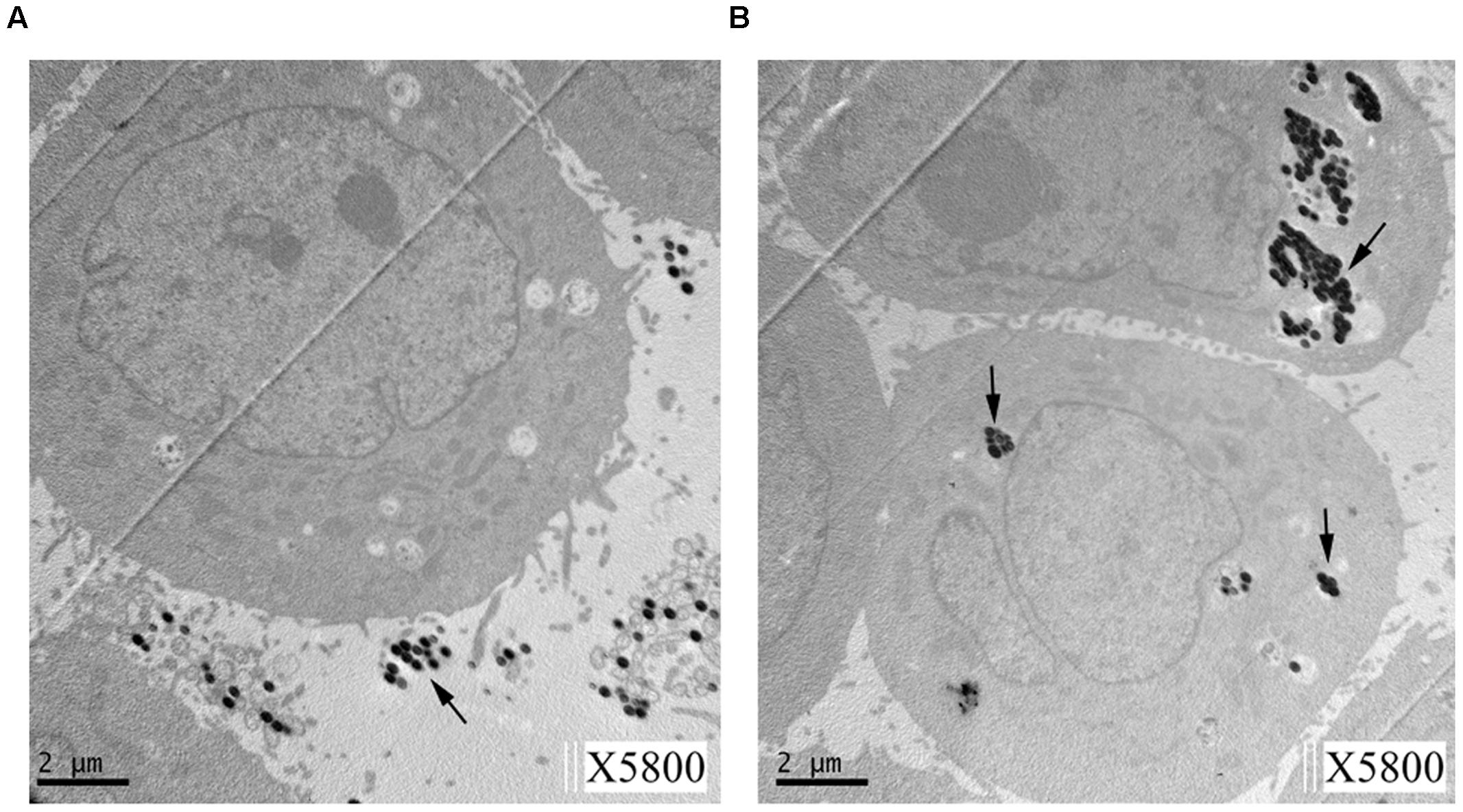
FIGURE 2. Transmission electron microscopy (TEM) of HeLa cells showing internalization of Mce1E-coated beads. (A) Non-coated beads were not observed in HeLa cells. (B) Mce1E-coated beads were completely internalized by HeLa cells. The beads were present in HeLa cells either alone or in clusters.
Putative Mce1E Protein Band Bound Specifically by Sera from N. farcinica-Infected Animals
As shown in Figure 3, expressed 48-kDa fusion proteins were detected in immunoblots with a major band of reactivity at the expected migration positions for the anti-His antibody employed. Importantly, the 48-kDa immunopositive band was also observed when purified Mce1E was immunoblotted with sera from N. farcinica-infected mice and rabbits. No bands were detected at 48 kDa when purified Mce1E was immunoblotted with control mouse or rabbit sera.
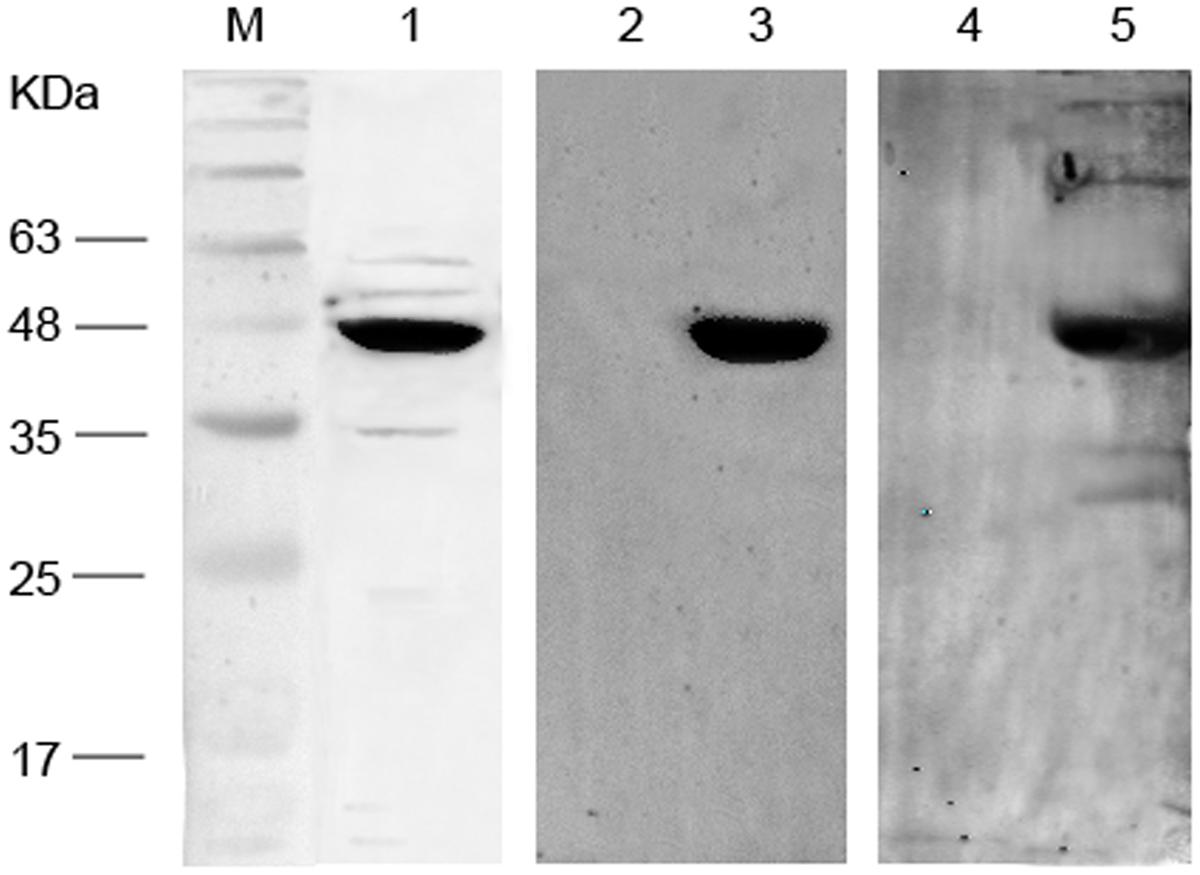
FIGURE 3. Western blot analysis of recombinant Mce1E. Lane M: Marker; lane1, purified Mce1E immunoblotted with anti-His antibodies; lane 2, purified Mce1E immunoblotted with sera from BALB/c mice infected with normal saline; lane 3, purified Mce1E immunoblotted with sera from BALB/c mice infected with Nocardia farcinica; lane 4, purified Mce1E immunoblotted with sera from New Zealand white rabbits infected with normal saline; lane5, purified Mce1E immunoblotted with sera from New Zealand white rabbits infected with N. farcinica.
Cytokine Detection
Spleen cell IFN-γ production was increased by Mce1E stimulation 7 and 14 days after N. farcinica infection (both p < 0.05) and this IFN-γ production increased with the progression of time (Figure 4). Spleen cell production of IL-4 and IL-10 remained low following stimulation with recombinant Mce1E protein, with no statistically significant differences among the time-point groups (data not shown).
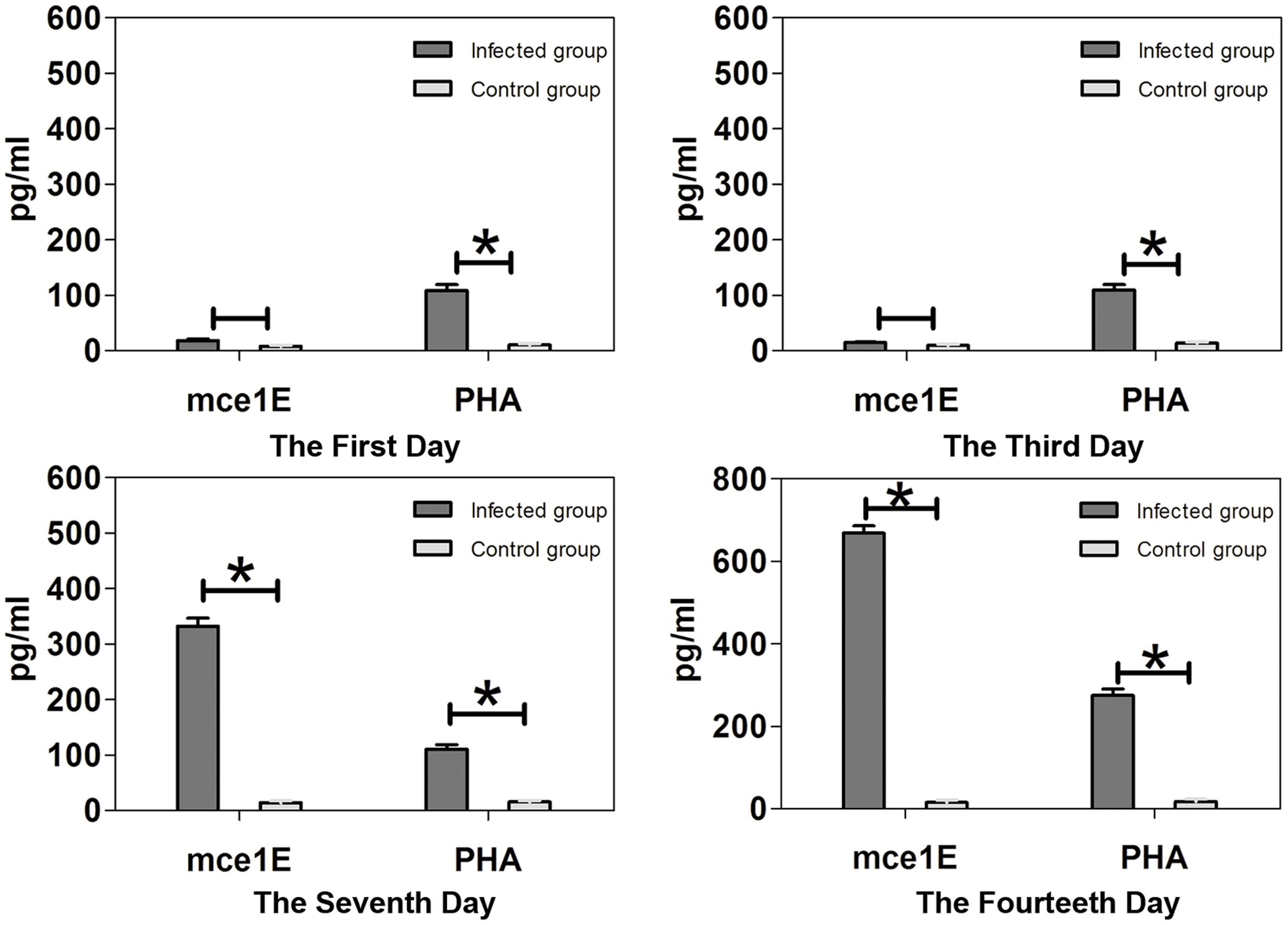
FIGURE 4. Interferon-γ production by splenocytes stimulated with Mce1E or PHA. Upregulated production of IFN-γ was detected in Mce1E-stimulated cultures of cells from N. farcinica-infected mice 7 and 14 days, but not 1 or 3 days, after infection. ∗P < 0.05.
Discussion
In the present study, we obtained data confirming the ability of N. farcinicaon Mce1E to enable invasion of mammalian cells. We further confirmed that the recombinant Mce1E that produced internationalization of associated beads was immunologically reactive. These data suggest that N. farcinicaon Mce1E is functionally similar to Mycobacterium tuberculosis Mce proteins, which enable M. tuberculosis mammalian-cell invasion, and thus pathogenesis, leading to long-term survival and proliferation of the pathogenic bacteria in host cells (Gioffre et al., 2005). The present work extends the work of Arruda et al. (1993) who showed that Mce1E can confer upon non-pathogenic E. coli the ability to invade HeLa cells, escape host defenses, augment macrophage phagocytosis, and survive for at least 24 h in human macrophages.
In Mycobacteria, as well as five other Actinomycetales genera and some Gram-negative bacteria, mce operons are widely distributed but structurally identical (Casali and Riley, 2007). The pathogenicity of these factors might be determined by their expression (Haile et al., 2002; Shimono et al., 2003; Casali and Riley, 2007). Mce3A, Mce3D, and Mce3E—encoded by the mce3 operon—are expressed by M. tuberculosis and elicit antibody responses in a majority of naturally infected human patients (Ahmad et al., 2004, 2005). Indeed, Mce1A and Mce1E have been found in the sera of tuberculosis patients (Ahmad et al., 1999). Meanwhile, it was reported recently that N. brasiliensis HUJEG-1 possesses 33 mce genes distributed in six operons (Vera-Cabrera et al., 2013). Moreover, N. nova SH22a was found to have significantly more mce clusters than N. farcinica IFM 10152 or N. brasiliensis HUJEG-1 (Luo et al., 2014). Notwithstanding, Nocardia Mce proteins have been given scant attention despite the fact that the corresponding proteins are considered to be an important virulence factor in M. tuberculosis (Arruda et al., 1993).
Recombinant M. bovis Mce4A and Mce4E have been shown previously to stimulate alveolar macrophages, there by upregulating the expression of tumor necrosis factor-alpha, inducible nitric oxide synthase, and IL-6, without affecting IL-12 (Xu et al., 2007, 2008), suggesting that these proteins may cause host inflammation responses to M. bovis infection. Conversely, Li et al. (2015) showed that M. tuberculosis Mce3E can suppress host innate immune responses by inhibiting activation of the extracellular signal-regulated kinase 1/2 signaling pathway and suppressing expression of tumor necrosis factor and IL-6. Here, we found that our recombinant N. farcinica Mce1E can stimulate spleen lymphocytes of N. farcinica-infected mice to express IFN-γ, while IL-4 and IL-10 were not detected. The steady increase in lymphocyte production of IFN-γ from 3 days post-infection onward may be explained by N. farcinica achieving entry of macrophages 3 days after infection.
Conclusion
The present data showed that Mce1E encoded by the mce1 operon of the N. farcinica genome facilitated internalization of latex beads by non-phagocytic mammalian (HeLa) cells. Furthermore, we found that Mce1E was expressed by N. farcinica in the process of infection in animals, suggesting it may also be expressed in humans with N. farcinica infections. Our results showing that Mce1E can stimulate spleen lymphocytes of N. farcinica-infected mice to express IFN-γ suggest that mce1E play a role in cell-mediated immune reactions.
Author Contributions
XJ, XT, ZL, and XY conceived and designed the experiments. XJ and XT wrote the manuscript. XJ and XT performed the experiments. XJ and XT analyzed the data. XH, CS, SX, and LT contributed reagents/materials/analysis tools. ZL supported financially and administratively, final approval of manuscript.
Funding
This work was supported by the Special Key Project of Biosafety Technologies (2016YFC1202603, 2016YFC1200701) for the National Major Research & Development Program of China and the National High Research and Development Program of China (“863” Program, grant no. 2014AA021404).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We are grateful to Lina Sun for critical reading and helpful comments on our manuscript.
References
Ahmad, S., Akbar, P. K., Wiker, H. G., Harboe, M., and Mustafa, A. S. (1999). Cloning, expression and immunological reactivity of two mammalian cell entry proteins encoded by the mce1 operon of Mycobacterium tuberculosis. Scand. J. Immunol. 50, 510–518. doi: 10.1046/j.1365-3083.1999.00631.x
Ahmad, S., El-Shazly, S., Mustafa, A. S., and Al-Attiyah, R. (2004). Mammalian cell-entry proteins encoded by the mce3 operon of Mycobacterium tuberculosis are expressed during natural infection in humans. Scand. J. Immunol. 60, 382–391. doi: 10.1111/j.0300-9475.2004.01490.x
Ahmad, S., El-Shazly, S., Mustafa, A. S., and Al-Attiyah, R. (2005). The six mammalian cell entry proteins (Mce3A-F) encoded by the mce3 operon are expressed during in vitro growth of Mycobacterium tuberculosis. Scand. J. Immunol. 62, 16–24. doi: 10.1111/j.1365-3083.2005.01639.x
Ambrosioni, J., Lew, D., and Garbino, J. (2010). Nocardiosis: updated clinical review and experience at a tertiary center. Infection 38, 89–97. doi: 10.1007/s15010-009-9193-9
Arruda, S., Bomfim, G., Knights, R., Huima-Byron, T., and Riley, L. W. (1993). Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261, 1454–1457. doi: 10.1126/science.8367727
Beaman, B. L., and Beaman, L. (1994). Nocardia species: host-parasite relationships. Clin. Microbiol. Rev. 7, 213–264. doi: 10.1128/CMR.7.2.213
Casali, N., and Riley, L. W. (2007). A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8:60. doi: 10.1186/1471-2164-8-60
Chen, B., Tang, J., Lu, Z., Wang, N., Gao, X., and Wang, F. (2016). Primary cutaneous nocardiosis in a patient with nephrotic syndrome: a case report and review of the literature. Medicine 95:e2490. doi: 10.1097/md.0000000000002490
De Nardo, P., Giancola, M. L., Noto, S., Gentilotti, E., Ghirga, P., Tommasi, C., et al. (2013). Left thigh phlegmon caused by Nocardia farcinica identified by 16S rRNA sequencing in a patient with leprosy: a case report. BMC Infect. Dis. 13:162. doi: 10.1186/1471-2334-13-162
Ekrami, A., Khosravi, A. D., Samarbaf Zadeh, A. R., and Hashemzadeh, M. (2014). Nocardia co-infection in patients with pulmonary tuberculosis. Jundishapur J. Microbiol. 7:e12495. doi: 10.5812/jjm.12495
El-Shazly, S., Ahmad, S., Mustafa, A. S., Al-Attiyah, R., and Krajci, D. (2007). Internalization by HeLa cells of latex beads coated with mammalian cell entry (Mce) proteins encoded by the mce3 operon of Mycobacterium tuberculosis. J. Med. Microbiol. 56(Pt 9), 1145–1151. doi: 10.1099/jmm.0.47095-0
Gioffre, A., Infante, E., Aguilar, D., Santangelo, M. P., Klepp, L., Amadio, A., et al. (2005). Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 7, 325–334. doi: 10.1016/j.micinf.2004.11.007
Glickman, M. S., and Jacobs, W. R. Jr. (2001). Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell 104, 477–485. doi: 10.1016/S0092-8674(01)00236-7
Haile, Y., Caugant, D. A., Bjune, G., and Wiker, H. G. (2002). Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT). FEMS Immunol. Med. Microbiol. 33, 125–132. doi: 10.1111/j.1574-695X.2002.tb00581.x
Inamadar, A. C., and Palit, A. (2003). Primary cutaneous nocardiosis: a case study and review. Indian J. Dermatol. Venereol. Leprol. 69, 386–391.
Ishikawa, J., Yamashita, A., Mikami, Y., Hoshino, Y., Kurita, H., Hotta, K., et al. (2004). The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U.S.A. 101, 14925–14930. doi: 10.1073/pnas.0406410101
Kandi, V. (2015). Human Nocardia infections: a review of pulmonary nocardiosis. Cureus 7:e304. doi: 10.7759/cureus.304
Kim, J., Kang, M., Kim, J., Jung, S., Park, J., Lee, D., et al. (2016). A case of Nocardia farcinica pneumonia and mediastinitis in an immunocompetent patient. Tuberc. Respir. Dis. 79, 101–103. doi: 10.4046/trd.2016.79.2.101
Kumar, V. A., Augustine, D., Panikar, D., Nandakumar, A., Dinesh, K. R., Karim, S., et al. (2014). Nocardia farcinica brain abscess: epidemiology, pathophysiology, and literature review. Surg. Infect. 15, 640–646. doi: 10.1089/sur.2012.205
Li, J., Chai, Q. Y., Zhang, Y., Li, B. X., Wang, J., Qiu, X. B., et al. (2015). Mycobacterium tuberculosis Mce3E suppresses host innate immune responses by targeting ERK1/2 signaling. J. Immunol. 194, 3756–3767. doi: 10.4049/jimmunol.1402679
Luo, Q., Hiessl, S., Poehlein, A., Daniel, R., and Steinbuchel, A. (2014). Insights into the microbial degradation of rubber and gutta-percha by analysis of the complete genome of Nocardia nova SH22a. Appl. Environ. Microbiol. 80, 3895–3907. doi: 10.1128/aem.00473-14
Park, S. D., Kim, H. J., Jang, I. H., Uh, Y., Kim, J., Yoon, K. J., et al. (2014). First report of Nocardia farcinica bursitis in a patient with diabetes mellitus. Ann. Lab. Med. 34, 252–255. doi: 10.3343/alm.2014.34.3.252
Saini, N. K., Sharma, M., Chandolia, A., Pasricha, R., Brahmachari, V., and Bose, M. (2008). Characterization of Mce4A protein of Mycobacterium tuberculosis: role in invasion and survival. BMC Microbiol. 8:200. doi: 10.1186/1471-2180-8-200
Salinas-Carmona, M. C. (2000). Nocardia brasiliensis: from microbe to human and experimental infections. Microbes Infect. 2, 1373–1381. doi: 10.1016/S1286-4579(00)01291-0
Salinas-Carmona, M. C., Zuniga, J. M., Perez-Rivera, L. I., Segoviano-Ramirez, J. C., and Vazquez-Marmolejo, A. V. (2009). Nocardia brasiliensis modulates IFN-gamma, IL-10, and IL-12 cytokine production by macrophages from BALB/c Mice. J. Interferon Cytokine Res. 29, 263–271. doi: 10.1089/jir.2008.0059
Scharfen, J., Buncek, M., Jezek, P., Urbaskova, P., Fridrichova, M., Kristufek, V., et al. (2010). Filamentous “contaminants” in the mycobacteriology laboratory; their culture, identification and clinical significance. Klin. Mikrobiol. Infekc. Lek. 16, 48–57.
Scorey, H., and Daniel, S. (2016). Nocardia farcinica bacteraemia presenting as a prostate abscess. IDCases 5, 24–26. doi: 10.1016/j.idcr.2016.06.001
Sharma, N., and O’Hagan, S. (2016). The role of oral co-trimoxazole in treating Nocardia farcinica keratitis-a case report. J. Ophthalmic Inflamm. Infect. 6:23. doi: 10.1186/s12348-016-0091-2
Shimono, N., Morici, L., Casali, N., Cantrell, S., Sidders, B., Ehrt, S., et al. (2003). Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. U.S.A. 100, 15918–15923. doi: 10.1073/pnas.2433882100
Sirijatuphat, R., Niltwat, S., Tiangtam, O., and Tungsubutra, W. (2013). Purulent pericarditis and cardiac tamponade caused by Nocardia farcinica in a nephrotic syndrome patient. Intern. Med. 52, 2231–2235. doi: 10.2169/internalmedicine.52.0453
Smego, R. A. Jr., and Gallis, H. A. (1984). The clinical spectrum of Nocardia brasiliensis infection in the United States. Rev. Infect. Dis. 6, 164–180. doi: 10.1093/clinids/6.2.164
Vera-Cabrera, L., Ortiz-Lopez, R., Elizondo-Gonzalez, R., and Ocampo-Candiani, J. (2013). Complete genome sequence analysis of Nocardia brasiliensis HUJEG-1 reveals a saprobic lifestyle and the genes needed for human pathogenesis. PLoS ONE 8:e65425. doi: 10.1371/journal.pone.0065425
Xu, G., Li, Y., Yang, J., Yin, X., Zhou, X., Liu, M., et al. (2008). Mycobacterium bovis Mce4E protein may play a role in modulating cytokine expression profile in alveolar macrophage. Int. J. Tuberc. Lung Dis. 12, 664–669.
Xu, G., Li, Y., Yang, J., Zhou, X., Yin, X., Liu, M., et al. (2007). Effect of recombinant Mce4A protein of Mycobacterium bovis on expression of TNF-alpha, iNOS, IL-6, and IL-12 in bovine alveolar macrophages. Mol. Cell. Biochem. 302, 1–7. doi: 10.1007/s11010-006-9395-0
Keywords: nocardiosis, Mce1E, entry protein, IFN-γ, recombinant protein expression
Citation: Ji X, Tan X, Hou X, Si C, Xu S, Tang L, Yuan X and Li Z (2017) Cloning, Expression, Invasion, and Immunological Reactivity of a Mammalian Cell Entry Protein Encoded by the mce1 Operon of Nocardia farcinica. Front. Microbiol. 8:281. doi: 10.3389/fmicb.2017.00281
Received: 16 December 2016; Accepted: 09 February 2017;
Published: 22 February 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Xunjia Cheng, Fudan University, ChinaHongbin Song, Military Medical Academy, China
Jun Han, National Institute for Viral Disease Control and Prevention (China CDC), China
Copyright © 2017 Ji, Tan, Hou, Si, Xu, Tang, Yuan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenjun Li, bGl6aGVuanVuQGljZGMuY24= Xiuqin Yuan, d3RqeXNoQDEyNi5jb20=
†These authors have contributed equally to this work.
 Xingzhao Ji
Xingzhao Ji Xiaoluo Tan2,3†
Xiaoluo Tan2,3†