- Department of Microbiology, School of Life Sciences, Sikkim University, Gangtok, India
Dahi and datshi are common naturally fermented milk (NFM) products of Bhutan. Population of lactic acid bacteria (LAB) in dahi (pH 3.7) and datshi (pH 5.2) was 1.4 × 107 and 3.9 × 108 cfu/ml, respectively. Based on 16S rRNA gene sequencing isolates of LAB from dahi and datshi were identified as Enterococcus faecalis, E. faecium, Lactococcus lactis subsp. lactis. LAB strains were tested for some technological properties. All LAB strains except E. faecalis CH2:17 caused coagulation of milk at both 30°C for 48 h. Only E. faecium DH4:05 strain was resistant to pH 3. No significant difference (P > 0.05) of viable counts was observed in MRS broth with and without lysozyme. All LAB strains grew well in 0.3% bile showing their ability to tolerate bile salt. None of the LAB strains showed >70% hydrophobicity. This study, being the first of its microbiological analysis of the NFM of Bhutan, has opened up to an extent of research work that gives a new insight to the products.
Introduction
Naturally fermented milk (NFM) products are prepared by the practice of one of the oldest techniques of milk fermentation known as the ‘back-sloping’ method in which a previous batch of a fermented product is used to inoculate the new batch (Josephsen and Jespersen, 2004; Tamang et al., 2016b). NFM products are prepared and consumed daily in Bhutan. Some NFM products of Bhutan are dahi, datshi, mohi, gheu, hard-chhurpi (chugo/churkam) and hitpa. Dahi (Figure 1A) is a yogurt-like NFM product of Bhutan, which is traditionally prepared by allowing the boiled milk to undergo spontaneous fermentation at room temperature for 2–3 days with the inoculation of the previous dahi sample. Dahi is drunk as a refreshing non-alcoholic beverage in Bhutan. Datshi (Figure 1B) is a cottage cheese like product, which is prepared by churning dahi for 10–15 min until a clumping product; butter (locally called gheu) is extracted. The butter is collected in another vessel and the buttermilk, locally called mohi is then heated for 15–20 min for the curdling of the product, called datshi, which is made into round small balls. It is consumed as curry in main meals in Bhutan. Most of these NFM products are occasionally used for religious ceremonies in Bhutan. Some people are economically dependent upon these NFM products where they sell at local markets. Some NFM products of other countries were well studied such as dahi, misti dahi, shrikhand, chhu, chhurpi, philu and somar of India, Nepal, Pakistan, and Bangladesh (Tamang et al., 2000; Dewan and Tamang, 2006, 2007; Harun-ur-Rashid et al., 2007; Sarkar, 2008; Patil et al., 2010; Tamang, 2010), kurut of China (Sun et al., 2010), aaruul, airag, byasulag, chigee, tarag, and khoormog of Mongolia (Watanabe et al., 2008; Takeda et al., 2011; Oki et al., 2014), ergo of Ethiopia, lben, rayeb, zabady, and zeer of Morocco and Northern African and Middle East countries, rob (from camel milk), biruni, mish (cow/camel milk) of Sudan, amasi (hodzeko, mukaka wakakora) of Zimbabwe, nunu of Ghana (Akabanda et al., 2013), filmjölk and långfil of Sweden (Mayo et al., 2010), and koumiss or kumis or kumys or kymys of the Caucasian area (Wu et al., 2009). Among species of lactic acid bacteria (LAB), Lactococcus lactis subsp. cremoris, and Lc. lactis subsp. lactis are the dominant microbiota along with other mesophilic lactobacilli (Lactobacillus casei/Lb. paracasei, Lb. fermentum, Lb. helveticus, Lb. plantarum, and/or Lb. acidophilus), Enterococcus faecium, species of Leuconostoc and Pediococcus in NFMs (Tamang et al., 2000, 2016b; Mathara et al., 2004; Dewan and Tamang, 2006, 2007; Patrignani et al., 2006; Watanabe et al., 2008; Wu et al., 2009; Hao et al., 2010; Yu et al., 2011; Akabanda et al., 2013; Oki et al., 2014). Technological properties including probiotics characters have been extensively studied in some NFM products of the world (Patrignani et al., 2006; Dewan and Tamang, 2007; Harun-ur-Rashid et al., 2007; Wu et al., 2009; Tamang et al., 2016a). Till date, there has been no report on the microbiological analysis and technological properties of the NFM from Bhutan, making this research the first of this kind. This paper is aimed to determine some technological properties of the LAB isolates from two popular NFM products of Bhutan- dahi and datshi such as acidification and coagulation, resistance to low pH, tolerance against bile, lysozyme tolerance and hydrophobicity assay, and also to isolate and identify LAB species by 16S rRNA sequencing.
Materials and Methods
Samples
A total number of eight fresh samples of dahi (4) and datshi (4) were collected from Tabthangbu village, Bhutan in pre-sterilized sampling bags and were transported to the laboratory in an icebox carrier, stored at 4°C and analyzed within a week.
Microbiological Analysis
Samples (10 ml) were homogenized with sterile physiological saline (90 ml) in a stomacher lab-blender (400, Seward, London, UK) for 1 min, and were serially diluted in the same diluent. LAB were enumerated on MRS agar (M641, HiMedia, Mumbai, India) plates under anaerobic conditions in an anaerobic gas-pack system (LE002, HiMedia, Mumbai, India) and incubated at 30°C for 48–72 h (Dewan and Tamang, 2007). Colonies were selected randomly from the plates which contained less than 10 colonies, according to Leisner et al. (1997). Purity of the isolates was checked by streaking again and sub-culturing on fresh agar plates of the isolation media, followed by microscopic examinations. LAB isolates were preserved at -20°C in MRS broth (M369, HiMedia, Mumbai, India) mixed with 20% (v/v) glycerol.
Determination of pH
The pH of samples was determined using a pH meter (Crison basic 20, Barcelona, Spain) calibrated with standard buffers.
Phenotypic Characterization
Cell morphology of all isolates and their motility was determined using a phase contrast microscope (Olympus CH3-BH-PC, Japan). Isolates were Gram-stained and tested for catalase production, and were preliminarily identified based on the phenotypic properties including sugar fermentations, following the methods of Schillinger and Lücke (1987) and Dykes et al. (1994).
Molecular Identification
DNA Extraction
Based on similar sugar fermentation and other phenotypic characteristics criteria, six representative strains of LAB were randomly selected from 44 strains of LAB. Total genomic DNA of six representative strains of LAB was extracted from 2-ml samples of overnight cultures grown in MRS broth at 30°C according to the methods of Martín-Platero et al. (2007). DNA was quantified using fluorometer (Qubitol® 3.0, Fisher Scientific, USA).
16S rRNA Gene Sequencing
The 16S rRNA gene was amplified by PCR mixtures (25 μL) contained approximately 30–50 ng template DNA, 1 μM forward primer 27F and 1 μM reverse primer 1492R (Lane, 1991) using a PCR Master Mix (Promega, Canada) performed under the standard PCR amplification procedure in a SimpliAmpTM Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA). The PCR amplicons were checked for their purity on 1% agarose gel electrophoresis in the presence of ethidium bromide (10 mg/mL), which was later analyzed by the Gel Doc System (Ultra-Violet Products Ltd, UK). Sequencing service was outsourced.
Phylogenetic Analysis
The BLAST (Basic Phylogenetic Local Alignment Search Tool) program was used for comparing DNA databases for sequence similarities available in the NCBI database. Five different strains/species from each BLAST results were chosen for phylogenetic analysis using Molecular Evolutionary genetics Analysis software (MEGA version 6).
Technological Properties
Activation of LAB Strains
Enterococcus faecalis CH1:14, E. faecalis CH2:02, E. faecalis CH2:17, E. durans CH3:03, Lactococcus lactis subsp. cremoris CH4:01 and E. faecium DH4:05, isolated from dahi and datshi, were grown in MRS broth for 16-24 h at 30°C, and were used for determinations of acidification and coagulation, tolerance against bile, and lysozyme tolerance. Activation of LAB strains for resistance to pH 3 and hydrophobicity were mentioned below.
Acidification and Coagulation
Acidification and coagulation ability of LAB strains were assayed by inoculating 10% skim milk (RM1254, HiMedia, Mumbai, India) at 1% level and incubated at 30°C for 72 h. Observation was made for commencement of clotting, followed by pH measurement (Olasupo et al., 2001).
Tolerance against Bile
MRS broth containing 0.3% bile was inoculated with active cultures for 4 h (Prasad et al., 1998) and viable cells were enumerated in MRS agar plates after 24 h incubation and growth was recorded.
Lysozyme Tolerance
10 mL of MRS broth with lysozyme (MB098-1G, HiMedia, India) and without lysozyme, respectively, was inoculated with 1 mL of both culture suspensions of 108 cfu/ml cell concentration and incubated at 30°C for 24 h and viable cells were enumerated in MRS agar plates after 24 h incubation (Brennan et al., 1986).
Resistance to Low pH
Active cultures were harvested by centrifugation and pellets were washed once in phosphate-saline buffer (PBS, pH 7.2), re-suspended in PBS (pH 3) and incubated in MRS agar plates at 30°C for 24 h, and growth was recorded (Prasad et al., 1998).
Hydrophobicity Assay
Bacterial affinity to hydrocarbons was determined and results were expressed according to Perez et al. (1998), modified by Tamang et al. (2009) as follows. Fresh cultures were grown in MRS broth at 30°C for 24 h and centrifuged at 8,000 g for 5 min. The pellet was washed with 9 ml of Ringer solution (Merck, Germany) and thoroughly mixed. Suspension (1 ml) was taken and the absorbance at 580 nm was measured. Then, 1.5 ml of suspension was mixed with equal volume of n-hexadecane (RM 2238, HiMedia, Mumbai, India) in duplicates and mixed thoroughly. Phases were allowed to separate for 30 min at room temperature, after which aqueous phase was carefully transferred to a new tube and absorbance at 580 nm was measured. The percentage hydrophobicity was expressed as follows:
where A0 and A are the absorbance values of the aqueous phase before and after contact with n-hexadecane.
Results and Discussion
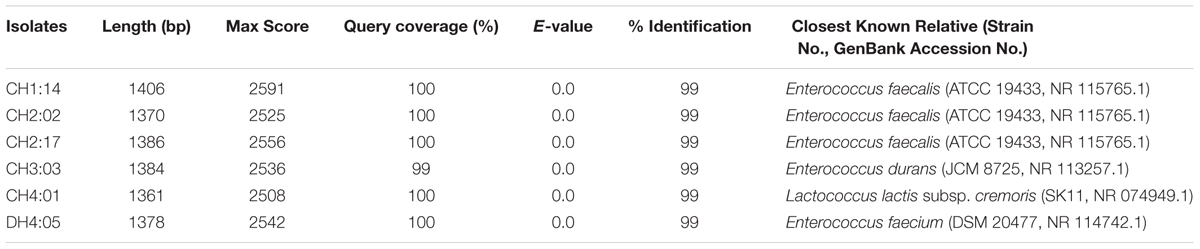
Dahi and datshi are acidic fermented milk products showing an average pH of 3.7 ± 0.17 and 5.2 ± 0.12, respectively. Isolation of LAB was performed on the classical media i.e., Lactobacillus MRS Agar media under anaerobic conditions at 30°C incubation for 48 h. The microbial load of LAB in dahi was 1.4 × 107 cfu/ml and in datshi was 3.9 × 108 cfu/mL, respectively. A total of 44 LAB isolates were isolated from dahi and datshi and phenotypically characterized and were randomly grouped into six representative strains based on similar sugar fermentation and other phenotypic characteristics (Table 1). These isolates were tentatively identified as Enterococcus and Lactococcus (Table 1).
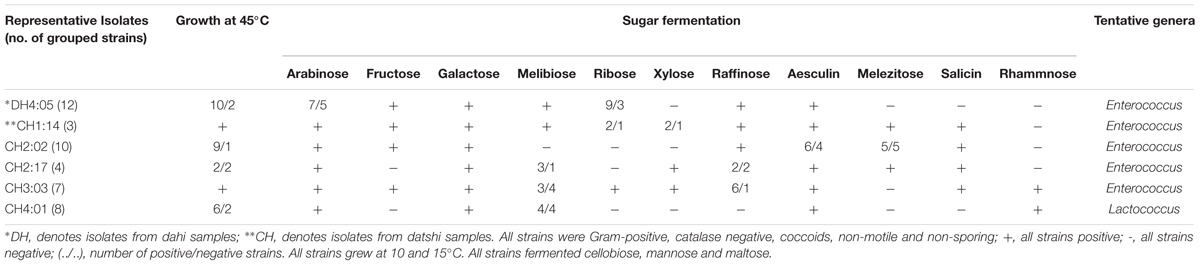
TABLE 1. Phenotypic characteristics of the lactic acid bacteria (LAB) isolated from dahi and datshi of Bhutan.
Total genomic DNA of 6 representative strains of LAB was extracted and amplified and were identified by partial 16S rRNA gene sequencing which were compared to the NCBI database for their phylogenetic relationship by using the software MEGA 6 (Figure 2). On the basis of molecular identification, the following species of LAB were identified from dahi and datshi of Bhutan with percentage similarity of LAB: E. faecalis CH1:14 (99%), E. faecalis CH2:02 (99%), E. faecalis CH2:17 (99%), E. durans CH3:03 (99%), Lactococcus lactis subsp. cremoris CH4:01 (99%), and E. faecium DH4:05 (99%; Table 2).
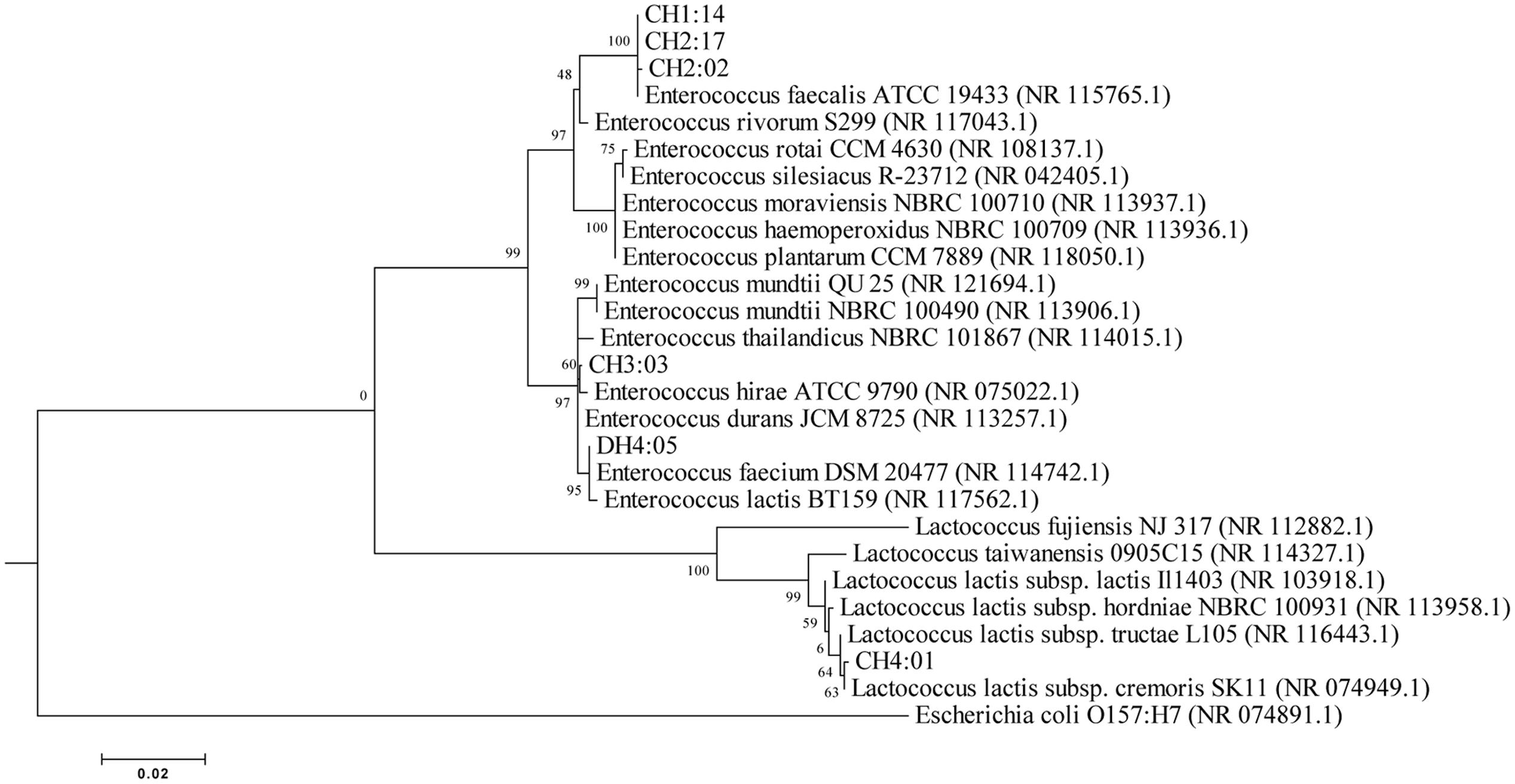
FIGURE 2. Phylogenetic tree based upon the Neighbor-Joining of 16S rDNA sequences (E. coli 84 to 1437) derived by PCR with the primer 27F and 1492R.
Lactococcus lactis subsp. lactis, Lc. lactis subsp. cremoris, E. faecium, E. faecalis, Leuconostoc mesenteroides and Pediococcus and lactobacilli (Lactobacillus casei, Lb. fermentum, Lb. helveticus, Lb. plantarum, and/or Lb. acidophilus), were reported from many NFM products of different countries (Tamang et al., 2000; Mathara et al., 2004; Dewan and Tamang, 2006, 2007; Patrignani et al., 2006; Watanabe et al., 2008; Wu et al., 2009; Hao et al., 2010; Yu et al., 2011; Akabanda et al., 2013).
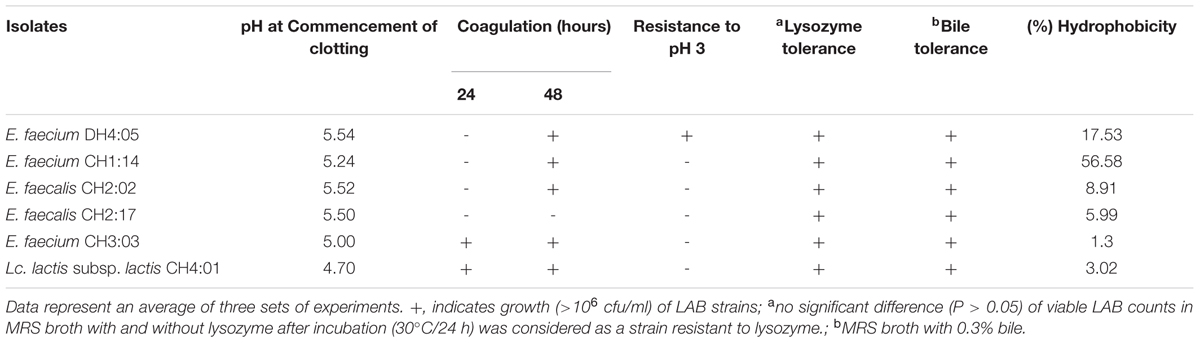
Lactic acid bacteria strains were tested for some technological properties (Table 3). All LAB strains except E. faecalis CH2:17 caused coagulation of milk at both 30°C for 48 h with a significant drop in pH (Table 3). Coagulation of milk by LAB strains reveals their potential as starters or adjunct cultures in the production of NFM of Bhutan. Only E. faecium DH4:05 strain showed positive result indicating its resistance to pH 3 in applied method (Table 3). Resistance to pH 3 is often used in vitro assays to determine the resistance to stomach pH (Prasad et al., 1998). Resistances to the lysozyme by all six strains of LAB were evaluated in MRS broth with and without lysosome at 30°C for 24 h (Table 3). Lysozyme is capable of lysing bacteria, but it doesn’t impair activities of LAB (Saran et al., 2012). Tolerance against bile was also tested and found that all LAB strains grew well in 0.3% bile showing their ability to tolerate bile salt. The mean intestinal bile concentration is 0.3% (w/v) and the staying time of food in small intestine is suggested to be 4 h (Prasad et al., 1998). The probiotic bacteria survival in the gastrointestinal transit is primordial, and implies in the ability of microorganisms to survive at the stomach acidity and bile, so that they can exert their beneficial effects on the host (Pozza et al., 2011).
Bacterial affinity to hydrocarbons, such as hexadecane, proved to be a simple method to determine cell surface hydrophobicity (van Loosdrecht et al., 1987). None of the LAB strains showed >70% hydrophobicity (Table 3). A percent hydrophobic index greater than 70% was classified as hydrophobic (Nostro et al., 2004). Hence, LAB strains from dahi and datshi do not show hydrophobic character in the applied method. However, these limited technological properties are not enough to validate the potential probiotic uses of these isolates.
Conclusion
Based on 16S rRNA gene sequencing isolates of LAB, isolated from dahi and datshi of Bhutan, were identified as E. faecalis, E. faecium, Lactococcus lactis subsp. lactis and some strains showed promising technological properties. This is the first report on NFM of Bhutan, which may be used as baseline data for further research on NFM products.
Author Contributions
HS: Molecular analysis of LAB isolates. SS: Isolation and phenotypic characterization. RR: Determination of technological properties of isolates. JT: Compilation of data and preparation of manuscript.
Funding
DST vide its sanction order: SB/EMEQ-367/2014, dated: 21.07.2014.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are grateful to Department of Science and Technology, New Delhi for research project. Authors are also thankful to Dr. K. Jeyaram and Dr. W. Romi of Institute of Bioresource and Sustainable Development, Imphal, India for analysis of the sequences in this paper.
References
Akabanda, F., Owusu-Kwarteng, J., Tano-Debrah, K., Glover, R. L. K., Nielsen, D. S., and Jespersen, L. (2013). Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 34, 277–283. doi: 10.1016/j.fm.2012.09.025
Brennan, M., Wanismail, B., Johnson, M. C., and Ray, B. (1986). Cellular damage in Dried Lactobacillus acidophilus. J. Food Prot. 49, 47–53. doi: 10.4315/0362-028X-49.1.47
Dewan, S., and Tamang, J. P. (2006). Microbial and analytical characterization of Chhu, a traditional fermented milk product of the Sikkim Himalayas. J. Sci. Indus. Res. 65, 747–752.
Dewan, S., and Tamang, J. P. (2007). Dominant lactic acid bacteria and their technological properties isolated from the Himalayan ethnic fermented milk products. Antonie Van Leeuwenhoek 92, 343–352. doi: 10.1007/s10482-007-9163-5
Dykes, G. A., Britz, T. J., and von Holy, A. (1994). Numerical taxonomy and identification of lactic acid bacteria from spoiled, vacuum packaged Vienna sausages. J. Appl. Bacteriol. 76, 246–252. doi: 10.1111/j.1365-2672.1994.tb01623.x
Hao, Y., Zhao, L., Zhang, H., Zhai, Z., Huang, Y., Liu, X., et al. (2010). Identification of the bacterial biodiversity in koumiss by denaturing gradient gel electrophoresis and species-specific polymerase chain reaction. J. Dairy Sci. 93, 1926–1933. doi: 10.3168/jds.2009-2822
Harun-ur-Rashid, M., Togo, K., Useda, M., and Miyamoto, T. (2007). Probiotic characteristics of lactic acid bacteria isolated from traditional fermented milk “Dahi” in Bangladesh. Pakistan J. Nutr. 6, 647–652. doi: 10.3923/pjn.2007.647.652
Josephsen, J., and Jespersen, L. (2004). “Handbook of food and beverage fermentation technology,” in Starter Cultures and Fermented Products, Vol. 3, eds Y. H. Hui, L. Meunier-Goddik, ÅS. Hansen, J. Josephsen, W. K. Nip, P. S. Stanfield, et al. (New York, NY: Marcel Dekker, Inc), 23–49.
Lane, D. J. (1991). “16S/23S rRNA Sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackenbrandt and M. Goodfellow (New York, NY: John Wiley & sons), 115–147.
Leisner, J. J., Rusul, G., Wee, B. W., Boo, H. C., and Mohammad, K. (1997). Microbiology of Chili Bo, a popular Malaysian food ingredient. J. Food Prot. 60, 1235–1240. doi: 10.4315/0362-028X-60.10.1235
Martín-Platero, A. M., Valdivia, E., Maqueda, M., and Martínez-Bueno, M. (2007). Fast, convenient, and economical method for isolating genomic DNA from lactic acid bacteria using a modification of the protein “salting-out” procedure. Anal. Biochem. 366, 102–104. doi: 10.1016/j.ab.2007.03.010
Mathara, J. M., Schillinger, U., Kutima, P. M., Mbugua, S. K., and Holzapfel, W. H. (2004). Isolation, identification and characterization of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int. J. Food Microbiol. 94, 269–278. doi: 10.1016/j.ijfoodmicro.2004.01.008
Mayo, B., Ammor, M. S., Delgado, S., and Alegría, A. (2010). “Fermented milk products,” in Fermented Foods and Beverages of the World, eds J. P. Tamang and K. Kailasapathy (Boca Raton, FL: CRC Press), 263–288.
Nostro, A., Cannatelli, M. A., Crisafi, G., Musolino, A. D., Procopio, F., and Alonzon, V. (2004). Modifications of hydrophobicity, in vitro adherence and cellular aggregation of Streptococcus mutans by Helichrysum italicum extract. Lett. Appl. Microbiol. 38, 423–427. doi: 10.1111/j.1472-765X.2004.01509.x
Oki, K., Dugersuren, J., Demberel, S., and Watanabe, K. (2014). Pyrosequencing analysis of the microbial diversity of airag, khoormog and tarag, traditional fermented dairy products of Mongolia. Biosci. Microbiol. Food Health 33, 53–64. doi: 10.12938/bmfh.33.53
Olasupo, N. A., Schillinger, U., and Holzpafel, W. H. (2001). Studies on some technological properties of predominant lactic acid bacteria isolated from Nigerian fermented foods. Food Biotechnol. 15, 157–167. doi: 10.1081/FBT-100107627
Patil, M. M., Pal, A., Anand, T., and Ramana, K. V. (2010). Isolation and characterization of lactic acid bacteria from curd and cucumber. Indian J. Biotechnol. 9, 166–172.
Patrignani, F., Lanciotti, R., Mathara, J. M., Guerzoni, M. E., and Holzapfel, W. H. (2006). Potential of functional strains, isolated from traditional Maasai milk, as starters for the production of fermented milks. Int. J. Food Microbiol. 107, 1–11. doi: 10.1016/j.ijfoodmicro.2005.08.004
Perez, P. F., Minnaard, Y., Disalvo, E. A., and De Antoni, G. L. (1998). Surface properties of bifidobacterial strains of human origin. Appl. Environ. Microbiol. 64, 21–26.
Pozza, M. S. S., Miglioranza, L. H. S., Garcia, J. E., Garcia, S., and Pozza, P. C. (2011). Human gastrointestinal tract resistance of Lactobacillus strains isolated from infant faeces. Ciec. Agraìrias 32, 1021–1032. doi: 10.5433/1679-0359.2011v32n3p1021
Prasad, J., Gill, H., Smart, J., and Gopal, P. K. (1998). Selection and characterization of Lactobacillus and Bifidobacterium strains for use as probiotic. Int. Dairy J. 8, 993–1002. doi: 10.1016/S0958-6946(99)00024-2
Saran, S., Bisht, M. S., Singh, K., and Toetia, U. S. (2012). Analyzing probiotic attributes to assess comparatively two isolates of Lactobacillus acidophilus in prebiotics, honey and inulin. DHR Int. J. Biomed. Life Sci. 2, 26–34.
Sarkar, S. (2008). Innovations in Indian fermented milk products-a review. Food Biotechnol. 22, 78–97. doi: 10.1080/08905430701864025
Schillinger, U., and Lücke, F. K. (1987). Identification of lactobacilli from meat and meat products. Food Microbiol. 4, 199–208. doi: 10.1016/0740-0020(87)90002-5
Sun, Z., Liu, W., Gao, W., Yang, M., Zhang, J., Wang, J., et al. (2010). Identification and characterization of the dominant lactic acid bacteria from kurut: the naturally fermented yak milk in Qinghai. China. J. Gen. Appl. Microbiol. 56, 1–10. doi: 10.2323/jgam.56.1
Takeda, S., Yamasaki, K., Takeshita, M., Kikuchi, Y., Tsend-Ayush, C., Dashnyam, B., et al. (2011). The investigation of probiotic potential of lactic acid bacteria isolated from traditional Mongolian dairy products. Animal Sci. J. 82, 571–579. doi: 10.1111/j.1740-0929.2011.00874.x
Tamang, J. P. (2010). Himalayan Fermented Foods: Microbiology, Nutrition, and Ethnic Values. New York, NY: CRC Press.
Tamang, J. P., Dewan, S., Thapa, S., Olasupo, N. A., Schillinger, U., and Holzapfel, W. H. (2000). Identification and enzymatic profiles of predominant lactic acid bacteria isolated from soft variety chhurpi, a traditional cheese typical of the Sikkim Himalayas. Food Biotechnol. 14, 99–112. doi: 10.1080/08905430009549982
Tamang, J. P., Shin, D. H., Jung, S. J., and Chae, S. W. (2016a). Functional properties of microorganisms in fermented foods. Front. Microbiol. 7:578. doi: 10.3389/fmicb.2016.00578
Tamang, J. P., Tamang, B., Schillinger, U., Guigas, C., and Holzapfel, W. H. (2009). Functional properties of lactic acid bacteria isolated from ethnic fermented vegetables of the Himalayas. Int. J. Food Microbiol. 135, 28–33. doi: 10.1016/j.ijfoodmicro.2009.07.016
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016b). Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
van Loosdrecht, M. C. M., Lyklema, J., Norde, W., Schraa, G., and Zehnder, A. J. B. (1987). The role of bacterial cell wall hydrophobicity in adhesion. Appl. Environ. Microbiol. 53, 1893–1897.
Watanabe, K., Fujimoto, J., Sasamoto, M., Dugersuren, J., Tumursuh, T., and Demberel, S. (2008). Diversity of lactic acid bacteria and yeasts in Airag and Tarag, traditional fermented milk products of Mongolia. World J. Microbiol. Biotechnol. 24, 1313–1325. doi: 10.1007/s11274-007-9604-3
Wu, R., Wang, L., Wang, J., Li, H., Menghe, B., Wu, J., et al. (2009). Isolation and preliminary probiotic selection of lactobacilli from Koumiss in Inner Mongolia. J. Basic Microbiol. 49, 318–326. doi: 10.1002/jobm.200800047
Keywords: technological properties, lactic acid bacteria, dahi, datshi, naturally fermented milk products
Citation: Shangpliang HNJ, Sharma S, Rai R and Tamang JP (2017) Some Technological Properties of Lactic Acid Bacteria Isolated from Dahi and Datshi, Naturally Fermented Milk Products of Bhutan. Front. Microbiol. 8:116. doi: 10.3389/fmicb.2017.00116
Received: 29 June 2016; Accepted: 17 January 2017;
Published: 01 February 2017.
Edited by:
Andrea Gomez-Zavaglia, Center for Research and Development in Food Cryotechnology (CONICET), ArgentinaReviewed by:
Bernadette Dora Gombossy De Melo Franco, University of São Paulo, BrazilPrakash M. Halami, Central Food Technological Research Institute (CSIR), India
Copyright © 2017 Shangpliang, Sharma, Rai and Tamang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jyoti P. Tamang, anlvdGlfdGFtYW5nQGhvdG1haWwuY29t
 H. N. J. Shangpliang
H. N. J. Shangpliang Sharmila Sharma
Sharmila Sharma Ranjita Rai
Ranjita Rai Jyoti P. Tamang
Jyoti P. Tamang