- Department of Medicine, Institute of Medical Microbiology, University of Zürich, Zürich, Switzerland
The ubiquitous Gram-negative bacterium Legionella pneumophila parasitizes environ mental amoebae and, upon inhalation, replicates in alveolar macrophages, thus causing a life-threatening pneumonia called “Legionnaires’ disease.” The opportunistic pathogen employs a bi-phasic life cycle, alternating between a replicative, non-virulent phase and a stationary, transmissive/virulent phase. L. pneumophila employs the Lqs (Legionella quorum sensing) system as a major regulator of the growth phase switch. The Lqs system comprises the autoinducer synthase LqsA, the homologous sensor kinases LqsS and LqsT, as well as a prototypic response regulator termed LqsR. These components produce, detect, and respond to the α-hydroxyketone signaling molecule LAI-1 (Legionella autoinducer-1, 3-hydroxypentadecane-4-one). LAI-1-mediated signal transduction through the sensor kinases converges on LqsR, which dimerizes upon phosphorylation. The Lqs system regulates the bacterial growth phase switch, pathogen-host cell interactions, motility, natural competence, filament production, and expression of a chromosomal “fitness island.” Yet, LAI-1 not only mediates bacterial intra-species signaling, but also modulates the motility of eukaryotic cells through the small GTPase Cdc42 and thus promotes inter-kingdom signaling. Taken together, the low molecular weight compound LAI-1 produced by L. pneumophila and sensed by the bacteria as well as by eukaryotic cells plays a major role in pathogen-host cell interactions.
Inter-Bacterial and Inter-Kingdom Small Molecule Signaling
Bacteria communicate with each other through small diffusible organic molecules and thus coordinate their group behavior. This phenomenon is termed “quorum sensing” and triggered upon reaching a threshold concentration (the “quorum”) of the signaling molecule. Quorum sensing is mediated by various chemical classes of small molecules called “autoinducers” (Fuqua and Greenberg, 2002; Ng and Bassler, 2009; Shank and Kolter, 2009). Prominent examples of low molecular weight compounds promoting quorum sensing of Gram-negative bacteria include N-acyl-homoserine lactone (HSL) autoinducers, diffusible signal factor (DSF), autoinducer-2 (AI-2) and its precursor 4,5-dihydroxy-2,3-pentanedione (DPD), 2-heptyl-3-hydroxy-4-quinolone (PQS), as well as dialkylresorcinol (DAR) and α-hydroxyketone (AHK) molecules (Papenfort and Bassler, 2016).
The bacterial response to autoinducer classes is multi-faceted and versatile. The same compound can be detected by vastly different bacterial genera, e.g., AI-2 appears to be an almost universal signal (Pereira et al., 2013). Slightly modified molecules of the same chemical class can trigger distinct responses among different species of the same genus, e.g., AHK derivatives confer signaling specificity among Vibrio species (Ng and Bassler, 2009; Ng et al., 2010, 2011). Also, a number of chemically different quorum sensing molecules co-operate in a single organism, e.g., Vibrio harveyi responds to and integrates the signaling of N-(3-hydroxybutyryl)-HSL (HAI-1), autoinducer-2 (AI-2, a furanosyl borate diester), and (Z)-3-aminoundec-2-en-4-one (CAI-1; Ng and Bassler, 2009). Thus, in a complex environmental niche, such as a mixed-species biofilm, a plethora of chemical “languages” is spoken, and a single bacterial cell needs to selectively respond to distinct “idioms.”
Small molecule communication is not restricted to prokaryotes. Rather, reciprocal signaling also occurs between prokaryotes and eukaryotes, either of which can produce the corresponding low molecular weight molecule(s). This process is termed inter-kingdom signaling (Shiner et al., 2005; Pacheco and Sperandio, 2009). Prominent examples of autoinducers mediating inter-kingdom signaling are bacteria-produced AHLs, which modulate eukaryotic cell migration and chemotaxis (Karlsson et al., 2012; Holm and Vikstrom, 2014), cell death (Tateda et al., 2003; Schwarzer et al., 2012; Kravchenko et al., 2013; Valentine et al., 2013), inflammatory responses (Kravchenko et al., 2008; Kravchenko and Kaufmann, 2013), as well as plant development and immunity (Schikora et al., 2016). Furthermore, AHLs as well as AHKs are chemo-attractants for the nematode Caenorhabditis elegans (Werner et al., 2014), and AHKs play a role in bacteria-fungi interactions (Haack et al., 2016).
Host cell-produced inter-kingdom signaling molecules include adrenergic compounds (catecholamines), which are detected by a number of bacterial genera through the QseBC two-component system (TCS; Kendall and Sperandio, 2016). Phagocytes such as neutrophils and macrophages (Flierl et al., 2007, 2009), as well as amoebae (Coppi et al., 2002) synthesize and respond to the catecholamines adrenaline and noradrenaline. In turn, these hormones activate macrophages and thus restrict intracellular growth of, e.g., Mycobacterium spp. (Weatherby et al., 2003). In this review, we will highlight recent research on small molecule signaling underlying the intra-species and inter-kingdom signaling of the amoebae-resistant, opportunistic pathogen Legionella pneumophila.
Legionella pneumophila: Environmental Niches and Human Infection
Legionella pneumophila is a ubiquitous Gram-negative bacterium that colonizes complex aquatic biofilm communities (Declerck, 2010; Abdel-Nour et al., 2013) and also forms single species biofilms in rich and minimal artificial media (Mampel et al., 2006; Piao et al., 2006; Pécastaings et al., 2010). In the environment, L. pneumophila preferentially parasitizes free-living protozoa (amoebae and ciliates), wherein the bacteria naturally replicate (Fields, 1996; Hoffmann et al., 2014b). Another possible niche of L. pneumophila is the intestinal tract of nematodes, e.g., C. elegans, which under laboratory conditions can be infected with the pathogen (Brassinga et al., 2010; Komura et al., 2010). The bacteria grow best at ambient temperatures (25–42°C) with an optimal growth temperature of around 35°C (Fields et al., 2002). However, Legionella spp. can persist at temperatures above 60°C in association with thermo-tolerant amoebae such as Acanthamoeba, Naegleria, Hartmannella, and Vahlkampfia spp. (Taylor et al., 2009).
Upon inhalation of Legionella-contaminated aerosols, the opportunistic pathogens reach the lung and replicate in alveolar macrophages, thus causing a severe pneumonia called Legionnaires’ disease (McDade et al., 1977; Horwitz and Silverstein, 1980), reviewed by (Newton et al., 2010; Hilbi et al., 2011). About half of the more than 55 Legionella spp. currently identified, have been associated with human disease; yet the clinically most relevant species are L. pneumophila and Legionella longbeachae. L. pneumophila serogroup (sg) 1 causes about 85% of all clinical cases in most parts of the world, while L. longbeachae accounts for about 30% of the reported cases in Australia and New Zealand (Fields et al., 2002; Newton et al., 2010).
Legionnaires’ disease mainly affects elderly or immuno-compromised persons and can spread in outbreaks comprising as many as 450 cases (Garcia-Fulgueiras et al., 2003). Since the identification of L. pneumophila 40 years ago, it was believed that the “accidental” pathogen is transmitted to humans only from environmental sources (Hilbi et al., 2010; Newton et al., 2010). However, after a recent outbreak of Legionnaires’ disease (Shivaji et al., 2014), the first case of a probable person-to-person transmission has been reported (Correia et al., 2016). The genome sequence of this outbreak strain revealed a phylogenetic divergence from most other outbreak-associated L. pneumophila sg1 strains studied (Borges et al., 2016). Of note, the strain harbors a mosaic genome carrying eight different horizontally acquired regions, some of which are also found in other L. pneumophila isolates.
Legionella pneumophila Replicates Intracellularly in a Distinct Pathogen Vacuole
Legionella pneumophila is taken up by phagocytes through macropinocytosis, evades the canonical bactericidal endocytic pathway and instead forms a replication-permissive membrane-bound compartment, the LCV (Isberg et al., 2009; Hilbi and Haas, 2012). Using an evolutionarily seemingly conserved mechanism, the nascent LCV avoids fusion with lysosomes, but communicates with the endosomal, secretory, and retrograde vesicle trafficking pathways (Personnic et al., 2016) and finally associates with the endoplasmic reticulum (ER) in a tight manner (Swanson and Isberg, 1995; Lu and Clarke, 2005; Robinson and Roy, 2006).
Legionella-containing vacuole formation is a complex and robust process, which on the pathogen side requires as an essential virulence factor the Icm/Dot T4SS (Kubori and Nagai, 2016). The Icm/Dot T4SS is a multi-component molecular apparatus that translocates the stunning number of up to 300 different putative “effector” proteins into eukaryotic host cells (Hubber and Roy, 2010; Finsel and Hilbi, 2015). The effector proteins subvert crucial cellular processes, such as signal transduction, vesicle trafficking, motility, death pathways, gene expression, and protein production. Some of these effectors target host components implicated in antibacterial defense or membrane dynamics, including the chelator phytate (Weber et al., 2014a), small GTPases (Itzen and Goody, 2011; Sherwood and Roy, 2013; Hoffmann et al., 2014a), phosphoinositide (PI) lipids (Weber et al., 2006, 2014b; Ragaz et al., 2008; Brombacher et al., 2009; Haneburger and Hilbi, 2013; Dolinsky et al., 2014), the PI phosphatase OCRL (Weber et al., 2009), the retromer complex (Finsel et al., 2013), microtubules (Rothmeier et al., 2013; Simon et al., 2014), or the actin cytoskeleton (Franco et al., 2012; Guo et al., 2014; Michard et al., 2015). In summary, the plethora of Icm/Dot-translocated effector proteins subverts the host cell’s physiology in a highly sophisticated and customized manner to ensure intracellular survival and growth of the pathogen.
The Bi-Phasic Life Cycle of L. pneumophila
Legionella pneumophila employs a bi-partite metabolism, where serine serves as major energy supply, while glycerol and carbohydrates like glucose are mainly fed into anabolic processes (Häuslein et al., 2016). The facultative intracellular pathogen survives and replicates in extracellular as well as intracellular niches. The transfer of the bacteria between different niches is facilitated by a bi-phasic life cycle, comprising a replicative, non-virulent phase and a transmissive, virulent phase (Molofsky and Swanson, 2004; Manske and Hilbi, 2014). The cycle is controlled by the bacterial growth phase and nutritional conditions (Byrne and Swanson, 1998), such as amino acid availability (Byrne and Swanson, 1998; Sauer et al., 2005) or fatty acid biosynthesis activity (Dalebroux et al., 2009).
In the post-exponential phase L. pneumophila up-regulates virulence, motility, and stress resistance, while in the exponential phase these traits are repressed, and metabolic pathways are up-regulated (Brüggemann et al., 2006; Faucher et al., 2011). The transition from exponential to post-exponential phase upon growth of the bacteria in broth is considered to reflect the transmission from the replicative to the transmissive phase in host cells. Collectively, the transmissive and virulence traits enable L. pneumophila to evade protozoan predators, survive in the environment as motile planktonic cells and re-establish a replicative niche in biofilms, protozoa or – perhaps – nematodes.
The master regulator of L. pneumophila’s bi-phasic life cycle is CsrA (carbon storage regulator A), a conserved and essential global activator of replication and repressor of transmission traits (Fettes et al., 2001; Molofsky and Swanson, 2003; Forsbach-Birk et al., 2004). Accordingly, the overproduction of CsrA in L. pneumophila leads to a reduction of flagellation (Fettes et al., 2001; Suzuki et al., 2006). CsrA is an RNA-binding regulatory protein, which is sequestered by the small non-coding RNAs (snRNAs) RsmY and RsmZ, thus relieving the repression of virulence and transmissive traits (Rasis and Segal, 2009; Sahr et al., 2009).
Distribution of AHK-Based Quorum Sensing Systems
As a major regulator of the growth phase switch L. pneumophila employs the Lqs system (Tiaden et al., 2007), which produces, detects and responds to the AHK molecule LAI-1 (Legionella autoinducer-1; 3-hydroxypentadecane-4-one; Spirig et al., 2008) (Figure 1). The system is encoded by genes arranged in a genomic cluster (lqsA–lqsR–hdeD–lqsS; Tiaden et al., 2008) and an orphan gene (lqsT; Kessler et al., 2013). All of these genes are expressed from individual promoters (Sahr et al., 2012). L. pneumophila but not L. longbeachae harbors the Lqs system, and thus, the system is not conserved among Legionella spp. (Tiaden et al., 2010a). L. pneumophila apparently lacks AI-2- and AHL-based sensing circuits, leaving Lqs the only known quorum sensing system of this species (Tiaden et al., 2010a).
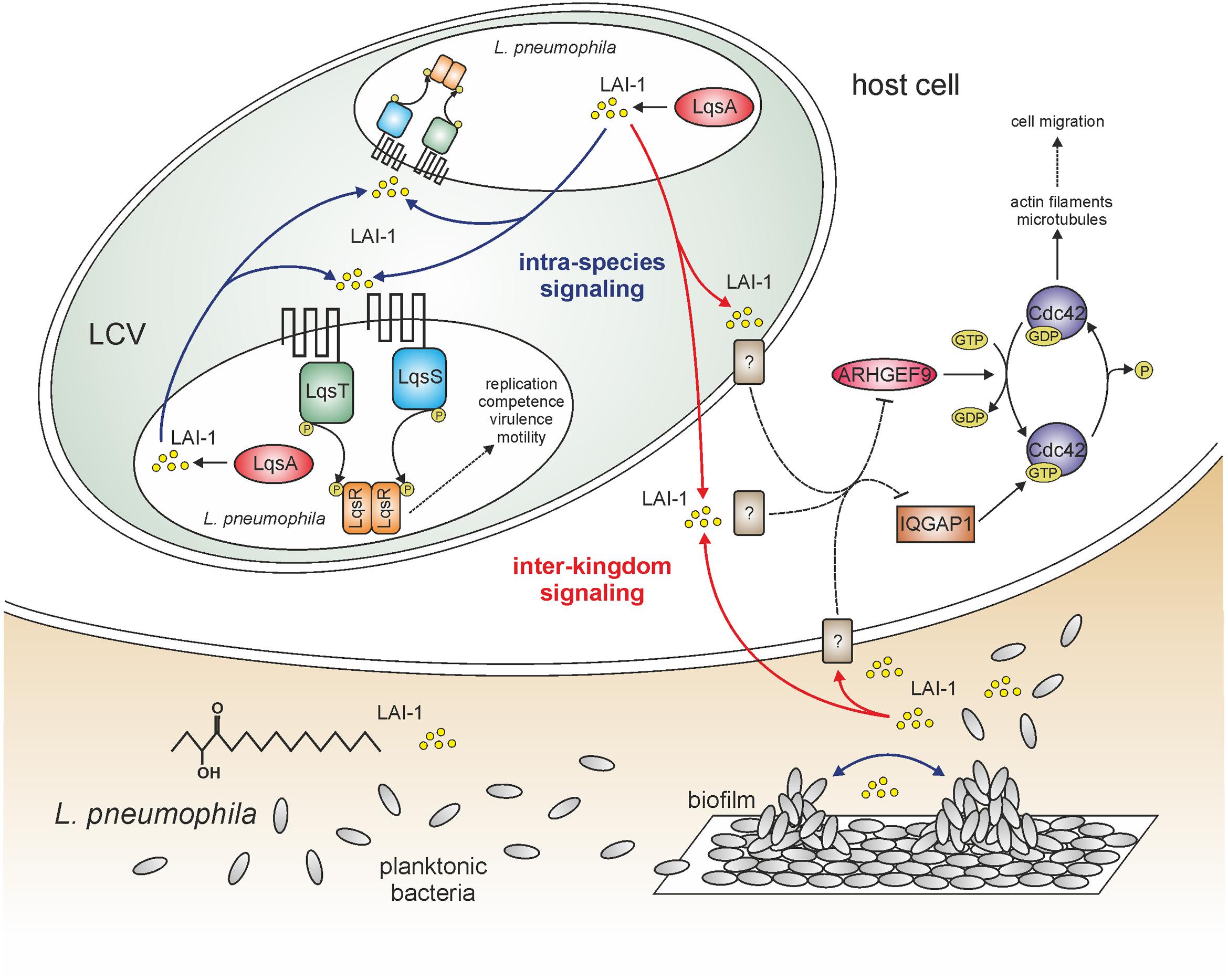
FIGURE 1. LAI-1-mediated intra-species and inter-kingdom signaling of Legionella pneumophila. The amoebae-resistant opportunistic pathogen L. pneumophila colonizes different niches in the environment, including biofilms and protozoa. L. pneumophila employs a bi-phasic life cycle, alternating between a replicative and a stationary/transmissive phase. As a major regulator of the growth phase switch L. pneumophila employs the Lqs system, which produces, detects, and responds to the AHK (α-hydroxyketone) signaling molecule LAI-1 (Legionella autoinducer-1; 3-hydroxypentadecane-4-one). The Lqs system comprises the autoinducer synthase LqsA, the sensor kinases LqsS and LqsT, as well as the response regulator LqsR. The system regulates the bacterial growth phase switch, virulence, motility, filaments, and competence. LAI-1 not only promotes bacterial cell–cell communication and quorum sensing, but also modulates the migration of eukaryotic cells through a pathway requiring the scaffold protein IQGAP1, the small GTPase Cdc42 and its activator ARHGEF9. The eukaryotic LAI-1 receptor(s) is/are unknown [?]. Solid and dashed lines represent known/direct or hypothetical/indirect pathways, respectively.
α-Hydroxyketone signaling molecules and the corresponding quorum sensing system have been first identified in Vibrio spp. and termed CAI-1 (cholerae autoinducer-1; 3-hydroxytridecane-4-one; Higgins et al., 2007) and CqsAS, respectively, (Miller et al., 2002; Henke and Bassler, 2004), reviewed in (Ng and Bassler, 2009; Tiaden and Hilbi, 2012). The system appears to be wide-spread, as homologs of the cqsA and cqsS genes are found not only in members of the family Legionellaceae (Legionella spp.) and Vibrionaceae (Vibrio spp., Photobacterium spp.), but also in the Burkholderiaceae (Burkholderia spp., Cupriavidus spp.), Chlorobiaceae (Chlorobium spp., Prosthecochloris spp.; Tiaden et al., 2010a), and Oxalobacteraceae (Janthinobacterium spp., Duganella spp.; Hornung et al., 2013; Haack et al., 2016). Photobacterium angustum indeed produces CqsAS orthologs and employs AHK-dependent quorum sensing (Ke et al., 2014). Given its broad distribution, AHK signaling might be commonly used for inter-species and inter-genera crosstalk among environmental bacteria.
Functional Characterization of L. pneumophila Lqs System Components
Components of the L. pneumophila Lqs system comprise the pyridoxal-5′-phosphate-dependent autoinducer synthase LqsA (Spirig et al., 2008), the cognate membrane-bound sensor kinase LqsS (Tiaden et al., 2010b) and its homolog LqsT (Kessler et al., 2013), as well as the prototypic response regulator LqsR (Figure 1) (Tiaden et al., 2007). The hdeD gene also present in the lqs cluster encodes a protein of unknown function (Tiaden et al., 2008).
The biochemical characterization of LqsS and LqsT revealed that the putative sensor histidine kinases are indeed autophosphorylated by [γ-32P]-ATP at their conserved histidine residues (His200 or His204, respectively), located in the cytoplasmic C-terminal histidine kinase domain (Schell et al., 2014). LqsS and LqsT are both bound by LqsR or phospho-LqsR, and dependent on its conserved receiver domain aspartate (Asp108), the response regulator abrogated autophosphorylation of the sensor kinases by catalyzing the dephosphorylation of phospho-LqsS or phospho-LqsT. LqsR forms dimers upon phosphorylation at Asp108 by either acetyl-phosphate or phospho-LqsT. Upon heterologous production in Escherichia coli LqsT (but not LqsS) is autophosphorylated by ATP, and LqsR prevents phosphorylation of the sensor kinase under these conditions by catalyzing the dephosphorylation of phospho-LqsT. Taken together, phosphorylation signaling through the sensor histidine kinases LqsS and LqsT converges on LqsR, which forms dimers upon phosphorylation (Schell et al., 2014).
Synthetic LAI-1 inhibits autophosphorylation of LqsS or LqsT by [γ-32P]-ATP in a dose-dependent manner (Schell et al., 2016). LAI-1 does neither affect the stability of phospho-LqsS or phospho-LqsT, nor the dephosphorylation by LqsR, suggesting that the AHK compound inhibits the kinase reaction. In contrast, the Vibrio cholerae autoinducer CAI-1 (3-hydroxytridecane-4-one) promotes the phosphorylation of LqsS (but not LqsT). Moreover, synthetic LAI-1 promotes the motility of L. pneumophila in an LqsS/LqsT- and LqsR-dependent manner. Transcriptome analysis of L. pneumophila treated with LAI-1 revealed that the signaling molecule negatively regulates the RNA-binding global regulator crsA and positively regulates a number of genes, including the snRNAs rsmY and rsmZ. In summary, these findings indicate that LAI-1 regulates motility and the switch from the replicative to the transmissive growth phase of L. pneumophila by phosphorylation signaling through LqsS, LqsT and LqsR (Schell et al., 2016).
Legionella pneumophila Traits Regulated by the Lqs System
To assess the function of the lqs genes genetically, the individual genes or the entire lqs gene cluster (lqsA–lqsR–hdeD–lqsS) were deleted from the L. pneumophila chromosome by double homologous recombination (Tiaden et al., 2007, 2008, 2010b; Kessler et al., 2013). These studies revealed that the Lqs system regulates a number of processes in L. pneumophila, including the entry into replicative growth phase, pathogen-phagocyte interactions, bacterial motility, the formation of extracellular filaments, natural competence for DNA uptake, and the expression of a bona fide genomic “fitness island.”
Legionella pneumophila lacking lqsA is only mildly defective for pathogen-host cell interactions (Tiaden et al., 2010b), but outcompeted by the parental strain upon co-infection of Acanthamoeba castellanii (Kessler et al., 2013). Moreover, the ΔlqsA mutant strain shows reduced motility, expression of the flagellin promoter PflaA, and flagellin production (Schell et al., 2016). Strikingly, L. pneumophila ΔlqsA takes up external DNA 3–4 orders of magnitude more efficiently, and the expression of the PcomEA promoter is up-regulated. The promoter controls the expression of comEA encoding a small periplasmic DNA-binding protein essential for competence (Charpentier et al., 2011). These results revealed that the Lqs system is a major negative regulator of natural competence of L. pneumophila.
Legionella pneumophila lacking lqsS is severely defective for intracellular replication (Tiaden et al., 2010b), impaired for motility (Schell et al., 2016), outcompeted by the parental strain upon co-infection of amoebae and, similar to the ΔlqsA strain, more competent for DNA uptake (Kessler et al., 2013). L. pneumophila ΔlqsS also sediments slower than wild-type or ΔlqsA mutant bacteria, due to the formation of extracellular filaments (Tiaden et al., 2010b). Finally, in the ΔlqsS mutant strain 52 genes located in a 133 kb high plasticity genomic “fitness island” are up-regulated at least two-fold (Tiaden et al., 2010b). The fitness island (lpg0973-lpg1096) shows a higher G+C content than the L. pneumophila core genome, is located adjacent to the tRNAThr gene lpg0972 and flanked by putative DNA-mobilizing genes such as integrases, transposases, and phage-like genes. Two regions can be discriminated: region I (lpg0973-lpg1003, 26 kb) harbors many (conserved) unknown genes, some of which encode putative pili components (PilE, PilT), and region II (lpg1006-lpg1096, 107 kb) encodes the subunits of a FoF1 ATP synthase and several metal ion resistance transporters. The region lpg1008-lpg1035 has been characterized previously as a 40 kb efflux pump genomic island, which is induced upon (but not required for) L. pneumophila infection of macrophages (McClain et al., 1996; Rankin et al., 2002). Taken together, the L. pneumophila 133 kb genomic region fulfills the criteria of a canonical genomic “fitness island” (Dobrindt et al., 2004).
Legionella pneumophila lacking lqsT (or both lqs sensor kinase genes) is also severely defective for intracellular replication and outcompeted by the parental strain upon co-infection of amoebae (Kessler et al., 2013), impaired for motility (Schell et al., 2016), as well as – similar to the ΔlqsS and ΔlqsA strains – more competent for DNA uptake (Kessler et al., 2013). However, in contrast to lqsS, lqsT does not regulate the production of extracellular filaments. The lqsT and lqsS genes are divergently expressed in the post-exponential growth phase, and transcriptome studies reveal that 90% of the genes down-regulated in absence of lqsT, are up-regulated in absence of lqsS. Reciprocally regulated genes encode constituents of the 133 kb genomic island or Icm/Dot-translocated effectors. The phenotypes of a mutant strain lacking lqsS and lqsT are partially complemented by either lqsT or lqsS, but are not reversed by overexpression of lqsA, as the single mutants are. This suggests that LqsT and LqsS are the sole LAI-1-responsive sensor kinases in L. pneumophila. Collectively, these results indicate that the Lqs system comprises two partially antagonistic LAI-1-responsive sensor kinases, which regulate distinct pools of genes implicated in various physiological and pathogenic processes of L. pneumophila.
Legionella pneumophila lacking lqsR shows a reduced lag phase before initiating growth in broth, and thus, the response regulator LqsR regulates the switch between the transmissive and the replicative phase (Tiaden et al., 2007). The ΔlqsR mutant strain is defective for uptake by and intracellular replication in phagocytes (Tiaden et al., 2007), impaired for motility (Schell et al., 2016), outcompeted by the parental strain upon co-infection of amoebae, and more competent for DNA uptake (Kessler et al., 2013). The virulence phenotypes of ΔlqsR are stronger than those of the other lqs single mutant strains. Yet, L. pneumophila lacking the entire lqs cluster showed even more severe and pleiotropic phenotypes, suggesting that the lqs genes act synergistically (Tiaden et al., 2008).
In summary, these studies revealed a unique organization of the L. pneumophila Lqs system comprising two homologous, partially antagonistic LAI-1-responsive sensor kinases. The Lqs system and LAI-1 circuit regulate many distinct features of L. pneumophila, in particular transmissive traits, such as pathogen-host cell interactions and virulence, bacterial motility, natural competence, production of extracellular filaments, and expression of a genomic “fitness island.” Hence, the Lqs system is a major regulator of the bi-phasic life cycle of L. pneumophila.
Connection of the Lqs System With Other L. pneumophila Two-Component Systems
The production of LqsR is controlled by the stationary phase sigma factor RpoS, and, less stringently, by the response regulator LetA (Tiaden et al., 2007), as well as on a post-transcriptional level probably by CsrA (Sahr et al., 2009). Thus, the Lqs system represents an element of the stationary growth phase virulence regulatory network of L. pneumophila comprising several TCSs (Figure 2) (Segal, 2013). The network includes the LetAS TCS, which is homologous to Pseudomonas GacAS (Laville et al., 1992). Upon entering stationary phase, LetAS regulates L. pneumophila transmission and virulence by promoting motility, contact-dependent cytotoxicity, infectivity, and evasion of lysosomes in macrophages (Hammer et al., 2002), as well as intracellular replication in A. castellanii (Gal-Mor and Segal, 2003b; Lynch et al., 2003). The response regulator LetA, in concert with RpoS, directly up-regulates the snRNAs rsmY and rsmZ, which bind and sequester the global repressor CsrA (Rasis and Segal, 2009; Sahr et al., 2009).
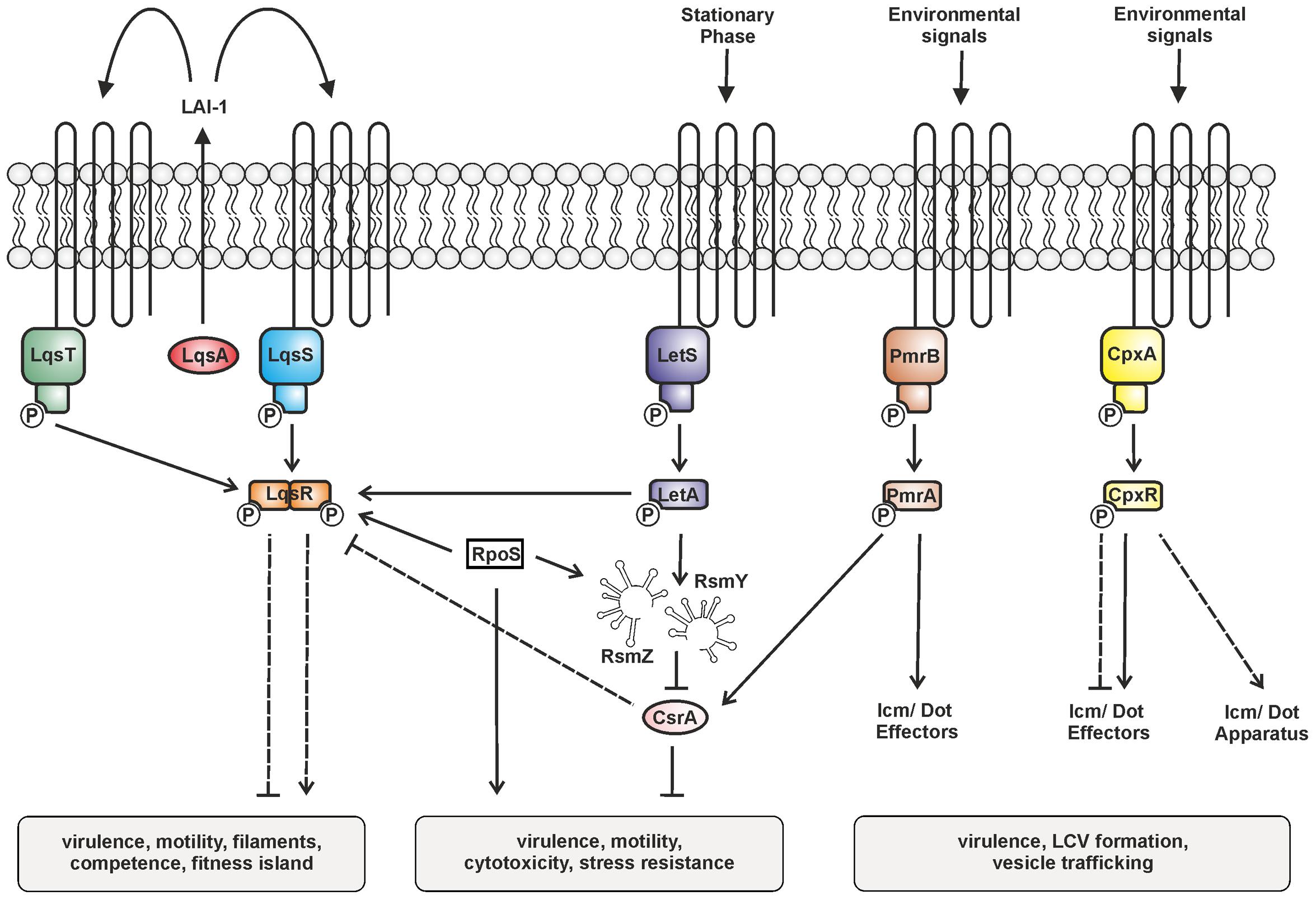
FIGURE 2. Two-component systems (TCSs) controlling the bi-phasic life cycle of L. pneumophila. The TCSs LqsRS, LetAS, PmrAB, and CpxRA, as well as the RsmYZ-CsrA regulatory unit are illustrated. Under conditions of nutrient abundance the RNA-binding carbon storage regulator A (CsrA) represses transmission traits and promotes replication. Upon nutrient depletion (at the onset of stationary phase), LetAS and the sigma factor RpoS up-regulate transmission traits. LetA induces the small non-coding snRNAs RsmY and RsmZ, which sequester CsrA and relieve post-transcriptional repression. LAI-1 produced by LqsA is detected by and prevents phosphorylation of the sensor kinases LqsS and LqsT. Thus, the cognate response regulator LqsR is dephosphorylated. LqsR is positively regulated by RpoS and negatively regulated by the LetA-RsmYZ-CsrA cascade. The response regulators PmrA and CpxR regulate the production of Icm/Dot components and substrates. The signals triggering the sensor histidine kinases LetS, PmrB, and CpxA are not known. Solid and dashed lines represent known/direct or hypothetical/indirect regulation, respectively.
The PmrAB TCS is a direct regulator of several Icm/Dot-translocated effector proteins and required for motility and intracellular replication of L. pneumophila in protozoa and macrophages (Zusman et al., 2007; Al-Khodor et al., 2009). The response regulator PmrA positively regulates metabolic genes including csrA and thus links the TCS with the LetA-RsmYZ-CsrA regulatory cascade (Rasis and Segal, 2009).
Finally, the CpxRA TCS plays a major role in L. pneumophila virulence gene regulation by controlling the expression of the icmR gene and at least 27 Icm/Dot substrates, as well as type II-secreted virulence factors (Gal-Mor and Segal, 2003a; Altman and Segal, 2008; Feldheim et al., 2016; Tanner et al., 2016). CpxR is a dual regulator, which acts as an activator or repressor, and can still self-interact in absence of phosphorylation to repress but not activate target genes (Feldheim et al., 2016).
Inter-Kingdom Signaling Through Lai-1
Small signaling molecules promote the communication not only between bacteria but also between prokaryotes and eukaryotes. To test the hypothesis that the Lqs system and LAI-1 affect eukaryotic cells, we used migration of eukaryotic cells as readout. L. pneumophila inhibits the chemotactic migration of amoebae, macrophages, and neutrophils in an Icm/Dot-dependent manner (Simon et al., 2014). Rather unexpectedly, however, L. pneumophila lacking lqsA no longer impeded the migration of infected amoebae or macrophages, and the defect was complemented by plasmid-borne lqsA (Simon et al., 2015). Interestingly, the overexpression of lqsA in an Icm/Dot deficient ΔicmT mutant strain abolished cell migration, indicating that the overproduction of LqsA and in consequence LAI-1 inhibits cell migration (in parallel to the Icm/Dot T4SS). In agreement with this notion, synthetic LAI-1 (as well as the Vibrio autoinducer CAI-1) dose-dependently inhibited cell migration (Figure 1). That is, LAI-1 reduced the forward migration index but not the velocity, suggesting that the directionality but not speed of the cells was affected.
In order to determine host cell factors implicated in LAI-1-dependent cell migration inhibition, a scratch wound healing assay with A549 lung epithelial cells was used, alongside RNA interference (Simon et al., 2015). The depletion of any host factor relevant for LAI-1-dependent inter-kingdom signaling will abrogate the effects of synthetic LAI-1. This approach revealed that LAI-1-dependent inhibition of cell migration involves the scaffold protein IQGAP1, the small GTPase Cdc42 and the Cdc42-specific guanine nucleotide exchange factor (GEF) ARHGEF9 (Figure 1), but not other modulators of Cdc42, or the small GTPases RhoA, Rac1 or Ran. Furthermore, upon treatment with LAI-1, IQGAP1 redistributed to the cell cortex, and Cdc42 was inactivated. Taken together, the results indicate that LAI-1 modulates migration of eukaryotic cells through a signaling pathway involving IQGAP1, Cdc42, and ARHGEF9. The eukaryotic receptor(s) of L. pneumophila LAI-1 is/are currently not known.
Interestingly, Salmonella enterica serovar Typhimurium also modulates the actin cytoskeleton through Cdc42 and IQGAP1. The Salmonella type III-secreted GEF SopE directly activates Cdc42 (Hardt et al., 1998), and bacterial invasion of fibroblasts lacking IQGAP1 was decreased, while Cdc42 and Rac1 activation was abrogated (Brown et al., 2007).
Effects of Adrenergic Antagonists On L. pneumophila
Inter-kingdom communication not only occurs from prokaryotes to eukaryotes but also in a reciprocal manner. A prominent example of the latter is adrenergic signaling mediated by the catecholamines adrenaline (epinephrine) and noradrenaline (norepinephrine) produced by eukaryotic cells (Hughes and Sperandio, 2008). The adrenergic neurotransmitters adrenaline and noradrenaline moderate eukaryotic stress response, but are also agonists of virulence and motility gene expression of enterohemorrhagic Escherichia coli (EHEC) O157:H7 (Sperandio et al., 2003; Clarke et al., 2006; Njoroge and Sperandio, 2012). Accordingly, α- and β-adrenergic antagonists (targeting the α- or β-subgroup of adrenergic receptors) block the response of EHEC to these host hormones (Hughes and Sperandio, 2008).
Bacteria respond to adrenergic compounds through TCSs, in particular QseBC and QseEF (Kendall and Sperandio, 2016). The sensor kinase QseC has been identified as adrenergic receptor in EHEC (Clarke et al., 2006) as well as in uropathogenic Escherichia coli (UPEC; Kostakioti et al., 2009), and EHEC also employs QseE as a receptor (Reading et al., 2009). In S. enterica serovar Typhi the responses to adrenergic signaling appear to depend on the CpxRA TCS (Karavolos et al., 2011). The QseBC TCS is wide-spread among bacteria, including Enterobacteriaceae, Pseudomonas aeruginosa, and L. pneumophila (Rasko et al., 2008).
The genes apparently encoding the QseBC homolog in Legionella spp. have been named either pmrAB, qseBC, or lrpR, depending on the strain or species involved, and adrenergic compounds were assessed for effects on Legionella-phagocyte interactions (Harrison et al., 2015b). Adrenaline and noradrenaline had only mild, if any, effects on L. pneumophila growing in broth or intracellularly in phagocytes. However, the adrenergic receptor antagonists benoxathian, naftopidil, propranolol, and labetalol reduced the growth of L. pneumophila in broth or amoebae, while replication in macrophages was enhanced (Harrison et al., 2015b). Growth restriction was common to several members of the genus Legionella and also observed for Mycobacterium spp. The deletion of L. pneumophila pmrAB (qseBC) had only a minor effect on growth inhibition by adrenergic antagonists, implying a different mode of action and/or the presence of another adrenergic sensing system. Yet, regardless of their bacterial target, adrenergic antagonists might represent potential lead compounds in screens for novel anti-infective compounds against Legionella or Mycobacterium spp. using phagocytes as host cells (Harrison et al., 2013, 2015a; Kicka et al., 2014).
Conclusions
The causative agent of Legionnaires’ disease, L. pneumophila, is an amoebae-resistant opportunistic pathogen, which employs the AHK molecule LAI-1 for intra-species as well as inter-kingdom communication. Since AHK molecules are wide-spread in nature, environmental bacteria likely employ this class of signals for inter-genera signaling. L. pneumophila produces, detects and responds to LAI-1 through the Lqs system, which comprises the autoinducer synthase LqsA, the homologous sensor kinases LqsS and LqsT, as well as the response regulator LqsR. As part of the stationary phase regulatory network, the Lqs system regulates the growth phase switch, pathogen-host cell interactions, bacterial motility, natural competence, filament production and expression of a chromosomal “fitness island.” The responses of L. pneumophila to LAI-1 might be exploited for anti-virulence drug development, as has been described for Vibrio cholerae using agonist and antagonist derivatives of the corresponding signaling molecule CAI-1 (Bolitho et al., 2011). Furthermore, LAI-1 modulates the migration direction of eukaryotic cells and thus mediates inter-kingdom signaling. Adrenergic compounds synthesized by eukaryotic cells might also be sensed by L. pneumophila; however, the mere fact that the pathogen responds to adrenergic antagonists does not imply that this is the case. Future studies will address the question whether there are other classes of low molecular weight molecules produced and detected by Legionella spp. or by Legionella -infected host cells, and will explore the potential of small molecule signaling to interfere with Legionella virulence or eukaryote processes.
Author Contributions
RH and HH wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the two reviewers for insightful comments and suggestions. Aline Kessler is acknowledged for drafting Figure 2. This work was supported by the Institute of Medical Microbiology, a research grant from the University of Zürich awarded to RH, and the Swiss National Science Foundation (SNF; 31003A_153200).
Abbreviations
CAI-1, cholerae autoinducer-1; cqs, cholerae quorum sensing; icm/dot, intracellular multiplication/defective organelle trafficking; LAI-1, Legionella autoinducer-1; LCV, Legionella-containing vacuole; lqs, Legionella quorum sensing; T4SS, type IV secretion system.
References
Abdel-Nour, M., Duncan, C., Low, D. E., and Guyard, C. (2013). Biofilms: the stronghold of Legionella pneumophila. Int. J. Mol. Sci. 14, 21660–21675. doi: 10.3390/ijms141121660
Al-Khodor, S., Kalachikov, S., Morozova, I., Price, C. T., and Abu Kwaik, Y. (2009). The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect. Immun. 77, 374–386. doi: 10.1128/IAI.01081-08
Altman, E., and Segal, G. (2008). The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 190, 1985–1996. doi: 10.1128/JB.01493-07
Bolitho, M. E., Perez, L. J., Koch, M. J., Ng, W. L., Bassler, B. L., and Semmelhack, M. F. (2011). Small molecule probes of the receptor binding site in the Vibrio cholerae CAI-1 quorum sensing circuit. Bioorg. Med. Chem. 19, 6906–6918. doi: 10.1016/j.bmc.2011.09.021
Borges, V., Nunes, A., Sampaio, D. A., Vieira, L., Machado, J., Simoes, M. J., et al. (2016).Legionella pneumophila strain associated with the first evidence of person-to-person transmission of Legionnaires’ disease: a unique mosaic genetic backbone. Sci. Rep. 6:26261. doi: 10.1038/srep26261
Brassinga, A. K., Kinchen, J. M., Cupp, M. E., Day, S. R., Hoffman, P. S., and Sifri, C. D. (2010). Caenorhabditis is a metazoan host for Legionella. Cell Microbiol. 12, 343–361. doi: 10.1111/j.1462-5822.2009.01398.x
Brombacher, E., Urwyler, S., Ragaz, C., Weber, S. S., Kami, K., Overduin, M., et al. (2009). Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284, 4846–4856. doi: 10.1074/jbc.M807505200
Brown, M. D., Bry, L., Li, Z., and Sacks, D. B. (2007). IQGAP1 regulates Salmonella invasion through interactions with actin, Rac1, and Cdc42. J. Biol. Chem. 282, 30265–30272. doi: 10.1074/jbc.M702537200
Brüggemann, H., Hagman, A., Jules, M., Sismeiro, O., Dillies, M. A., Gouyette, C., et al. (2006). Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 8, 1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x
Byrne, B., and Swanson, M. S. (1998). Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66, 3029–3034.
Charpentier, X., Kay, E., Schneider, D., and Shuman, H. A. (2011). Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J. Bacteriol. 193, 1114–1121. doi: 10.1128/JB.01146-10
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. doi: 10.1073/pnas.0604343103
Coppi, A., Merali, S., and Eichinger, D. (2002). The enteric parasite Entamoeba uses an autocrine catecholamine system during differentiation into the infectious cyst stage. J. Biol. Chem. 277, 8083–8090. doi: 10.1074/jbc.M111895200
Correia, A. M., Ferreira, J. S., Borges, V., Nunes, A., Gomes, B., Capucho, R., et al. (2016). Probable person-to-person transmission of Legionnaires’ disease. N. Engl. J. Med. 374, 497–498. doi: 10.1056/NEJMc1505356
Dalebroux, Z. D., Edwards, R. L., and Swanson, M. S. (2009). SpoT governs Legionella pneumophila differentiation in host macrophages. Mol. Microbiol. 71, 640–658. doi: 10.1111/j.1365-2958.2008.06555.x
Declerck, P. (2010). Biofilms: the environmental playground of Legionella pneumophila. Environ. Microbiol. 12, 557–566. doi: 10.1111/j.1462-2920.2009.02025.x
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi: 10.1038/nrmicro884
Dolinsky, S., Haneburger, I., Cichy, A., Hannemann, M., Itzen, A., and Hilbi, H. (2014). The Legionella longbeachae Icm/Dot substrate SidC selectively binds phosphatidylinositol 4-phosphate with nanomolar affinity and promotes pathogen vacuole-endoplasmic reticulum interactions. Infect. Immun. 82, 4021–4033. doi: 10.1128/IAI.01685-14
Faucher, S. P., Mueller, C. A., and Shuman, H. A. (2011). Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front. Microbiol. 2:60. doi: 10.3389/fmicb.2011.00060
Feldheim, Y. S., Zusman, T., Speiser, Y., and Segal, G. (2016). The Legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR’s function as a dual regulator and its connection to the effectors regulatory network. Mol. Microbiol. 99, 1059–1079. doi: 10.1111/mmi.13290
Fettes, P. S., Forsbach-Birk, V., Lynch, D., and Marre, R. (2001). Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291, 353–360. doi: 10.1078/1438-4221-00141
Fields, B. S. (1996). The molecular ecology of Legionellae. Trends Microbiol. 4, 286–290. doi: 10.1016/0966-842X(96)10041-X
Fields, B. S., Benson, R. F., and Besser, R. E. (2002). Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526. doi: 10.1128/CMR.15.3.506-526.2002
Finsel, I., and Hilbi, H. (2015). Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol. 17, 935–950. doi: 10.1111/cmi.12450
Finsel, I., Ragaz, C., Hoffmann, C., Harrison, C. F., Weber, S., Van Rahden, V. A., et al. (2013). The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe 14, 38–50. doi: 10.1016/j.chom.2013.06.001
Flierl, M. A., Rittirsch, D., Nadeau, B. A., Chen, A. J., Sarma, J. V., Zetoune, F. S., et al. (2007). Phagocyte-derived catecholamines enhance acute inflammatory injury. Nature 449, 721–725. doi: 10.1038/nature06185
Flierl, M. A., Rittirsch, D., Nadeau, B. A., Sarma, J. V., Day, D. E., Lentsch, A. B., et al. (2009). Upregulation of phagocyte-derived catecholamines augments the acute inflammatory response. PLoS ONE 4:e4414. doi: 10.1371/journal.pone.0004414
Forsbach-Birk, V., Mcnealy, T., Shi, C., Lynch, D., and Marre, R. (2004). Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int. J. Med. Microbiol. 294, 15–25. doi: 10.1016/j.ijmm.2003.12.003
Franco, I. S., Shohdy, N., and Shuman, H. A. (2012). The Legionella pneumophila effector VipA is an actin nucleator that alters host cell organelle trafficking. PLoS Pathog. 8:e1002546. doi: 10.1371/journal.ppat.1002546
Fuqua, C., and Greenberg, E. P. (2002). Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695. doi: 10.1038/nrm907
Gal-Mor, O., and Segal, G. (2003a). Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185, 4908–4919. doi: 10.1128/JB.185.16.4908-4919.2003
Gal-Mor, O., and Segal, G. (2003b). The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34, 187–194. doi: 10.1016/S0882-4010(03)00027-5
Garcia-Fulgueiras, A., Navarro, C., Fenoll, D., Garcia, J., Gonzalez-Diego, P., Jimenez-Bunuales, T., et al. (2003). Legionnaires’ disease outbreak in Murcia, Spain. Emerg. Infect. Dis. 9, 915–921. doi: 10.3201/eid0908.030337
Guo, Z., Stephenson, R., Qiu, J., Zheng, S., and Luo, Z. Q. (2014). A Legionella effector modulates host cytoskeletal structure by inhibiting actin polymerization. Microbes Infect. 16, 225–236. doi: 10.1016/j.micinf.2013.11.007
Haack, F. S., Poehlein, A., Kröger, C., Voigt, C. A., Piepenbring, M., Bode, H. B., et al. (2016). Molecular keys to the Janthinobacterium and Duganella spp. interaction with the plant pathogen Fusarium graminearum. Front. Microbiol. 7:1668. doi: 10.3389/fmicb.2016.01668
Hammer, B. K., Tateda, E. S., and Swanson, M. S. (2002). A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44, 107–118. doi: 10.1046/j.1365-2958.2002.02884.x
Haneburger, I., and Hilbi, H. (2013). Phosphoinositide lipids and the Legionella pathogen vacuole. Curr. Top. Microbiol. Immunol. 376, 155–173. doi: 10.1007/82_2013_341
Hardt, W. D., Chen, L. M., Schuebel, K. E., Bustelo, X. R., and Galan, J. E. (1998). S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815–826. doi: 10.1016/S0092-8674(00)81442-7
Harrison, C. F., Chiriano, G., Finsel, I., Manske, C., Hoffmann, C., Steiner, B., et al. (2015a). Amoebae-based screening reveals a novel family of compounds restricting intracellular Legionella pneumophila. ACS Inf. Dis. 1, 327–338. doi: 10.1021/acsinfecdis.5b00002
Harrison, C. F., Kicka, S., Kranjc, A., Finsel, I., Chiriano, G., Ouertatani-Sakouhi, H., et al. (2015b). Adrenergic antagonists restrict replication of Legionella. Microbiology 161, 1392–1406. doi: 10.1099/mic.0.000094
Harrison, C. F., Kicka, S., Trofimov, V., Berschl, K., Ouertatani-Sakouhi, H., Ackermann, N., et al. (2013). Exploring anti-bacterial compounds against intracellular Legionella. PLoS ONE 8:e74813. doi: 10.1371/journal.pone.0074813
Häuslein, I., Manske, C., Goebel, W., Eisenreich, W., and Hilbi, H. (2016). Pathway analysis using (13)C-glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Mol. Microbiol. 100, 229–246. doi: 10.1111/mmi.13313
Henke, J. M., and Bassler, B. L. (2004). Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186, 6902–6914. doi: 10.1128/JB.186.12.3794-3805.2004
Higgins, D. A., Pomianek, M. E., Kraml, C. M., Taylor, R. K., Semmelhack, M. F., and Bassler, B. L. (2007). The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886. doi: 10.1038/nature06284
Hilbi, H., and Haas, A. (2012). Secretive bacterial pathogens and the secretory pathway. Traffic 13, 1187–1197. doi: 10.1111/j.1600-0854.2012.01344.x
Hilbi, H., Hoffmann, C., and Harrison, C. F. (2011). Legionella spp. outdoors: colonization, communication and persistence. Environ. Microbiol. Rep. 3, 286–296. doi: 10.1111/j.1758-2229.2011.00247.x
Hilbi, H., Jarraud, S., Hartland, E., and Buchrieser, C. (2010). Update on Legionnaires’ disease: pathogenesis, epidemiology, detection and control. Mol. Microbiol. 76, 1–11. doi: 10.1111/j.1365-2958.2010.07086.x
Hoffmann, C., Finsel, I., Otto, A., Pfaffinger, G., Rothmeier, E., Hecker, M., et al. (2014a). Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell. Microbiol. 16, 1034–1052. doi: 10.1111/cmi.12256
Hoffmann, C., Harrison, C. F., and Hilbi, H. (2014b). The natural alternative: protozoa as cellular models for Legionella infection. Cell. Microbiol. 16, 15–26. doi: 10.1111/cmi.12235
Holm, A., and Vikstrom, E. (2014). Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front. Plant Sci. 5:309. doi: 10.3389/fpls.2014.00309
Hornung, C., Poehlein, A., Haack, F. S., Schmidt, M., Dierking, K., Pohlen, A., et al. (2013). The Janthinobacterium sp. HH01 genome encodes a homologue of the V. cholerae CqsA and L. pneumophila LqsA autoinducer synthases. PLoS ONE 8:e55045. doi: 10.1371/journal.pone.0055045
Horwitz, M. A., and Silverstein, S. C. (1980). Legionnaires’ disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66, 441–450. doi: 10.1172/JCI109874
Hubber, A., and Roy, C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283. doi: 10.1146/annurev-cellbio-100109-104034
Hughes, D. T., and Sperandio, V. (2008). Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. doi: 10.1038/nrmicro1836
Isberg, R. R., O’Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24. doi: 10.1038/nrmicro1967
Itzen, A., and Goody, R. S. (2011). Covalent coercion by Legionella pneumophila. Cell Host Microbe 10, 89–91. doi: 10.1016/j.chom.2011.08.002
Karavolos, M. H., Bulmer, D. M., Spencer, H., Rampioni, G., Schmalen, I., Baker, S., et al. (2011). Salmonella typhi sense host neuroendocrine stress hormones and release the toxin haemolysin E. EMBO Rep. 12, 252–258. doi: 10.1038/embor.2011.4
Karlsson, T., Turkina, M. V., Yakymenko, O., Magnusson, K. E., and Vikstrom, E. (2012). The Pseudomonas aeruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 8:e1002953. doi: 10.1371/journal.ppat.1002953
Ke, X., Miller, L. C., Ng, W. L., and Bassler, B. L. (2014). CqsA-CqsS quorum-sensing signal-receptor specificity in Photobacterium angustum. Mol. Microbiol. 91, 821–833. doi: 10.1111/mmi.12502
Kendall, M. M., and Sperandio, V. (2016). What a dinner party! Mechanisms and functions of interkingdom signaling in host-pathogen associations. MBio. 7:e01748. doi: 10.1128/mBio.01748-15
Kessler, A., Schell, U., Sahr, T., Tiaden, A., Harrison, C., Buchrieser, C., et al. (2013). The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ. Microbiol. 15, 646–662. doi: 10.1111/j.1462-2920.2012.02889.x
Kicka, S., Trofimov, V., Harrison, C., Ouertatani-Sakouhi, H., Mckinney, J., Scapozza, L., et al. (2014). Establishment and validation of whole-cell based fluorescence assays to identify anti-mycobacterial compounds using the Acanthamoeba castellanii-Mycobacterium marinum host-pathogen system. PLoS ONE 9:e87834. doi: 10.1371/journal.pone.0087834
Komura, T., Yasui, C., Miyamoto, H., and Nishikawa, Y. (2010). Caenorhabditis elegans as an alternative model host for Legionella pneumophila, and protective effects of Bifidobacterium infantis. Appl. Environ. Microbiol. 76, 4105–4108. doi: 10.1128/AEM.03021-09
Kostakioti, M., Hadjifrangiskou, M., Pinkner, J. S., and Hultgren, S. J. (2009). QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol. Microbiol. 73, 1020–1031. doi: 10.1111/j.1365-2958.2009.06826.x
Kravchenko, V., Garner, A. L., Mathison, J., Seit-Nebi, A., Yu, J., Gileva, I. P., et al. (2013). Facilitating cytokine-mediated cancer cell death by proteobacterial N-acylhomoserine lactones. ACS Chem. Biol. 8, 1117–1120. doi: 10.1021/cb4000184
Kravchenko, V. V., and Kaufmann, G. F. (2013). Bacterial inhibition of inflammatory responses via TLR-independent mechanisms. Cell Microbiol. 15, 527–536. doi: 10.1111/cmi.12109
Kravchenko, V. V., Kaufmann, G. F., Mathison, J. C., Scott, D. A., Katz, A. Z., Grauer, D. C., et al. (2008). Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science 321, 259–263. doi: 10.1126/science.1156499
Kubori, T., and Nagai, H. (2016). The type IVB secretion system: an enigmatic chimera. Curr. Opin. Microbiol. 29, 22–29. doi: 10.1016/j.mib.2015.10.001
Laville, J., Voisard, C., Keel, C., Maurhofer, M., Défago, G., and Haas, D. (1992). Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. U.S.A. 89, 1562–1566. doi: 10.1073/pnas.89.5.1562
Lu, H., and Clarke, M. (2005). Dynamic properties of Legionella-containing phagosomes in Dictyostelium amoebae. Cell. Microbiol. 7, 995–1007. doi: 10.1111/j.1462-5822.2005.00528.x
Lynch, D., Fieser, N., Gloggler, K., Forsbach-Birk, V., and Marre, R. (2003). The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219, 241–248. doi: 10.1016/S0378-1097(03)00050-8
Mampel, J., Spirig, T., Weber, S. S., Haagensen, J. A. J., Molin, S., and Hilbi, H. (2006). Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72, 2885–2895. doi: 10.1128/AEM.72.4.2885-2895.2006
Manske, C., and Hilbi, H. (2014). Metabolism of the vacuolar pathogen Legionella and implications for virulence. Front. Cell Infect. Microbiol. 4:125. doi: 10.3389/fcimb.2014.00125
McClain, M. S., Hurley, M. C., Brieland, J. K., and Engleberg, N. C. (1996). The Legionella pneumophila hel locus encodes intracellularly induced homologs of heavy-metal ion transporters of Alcaligenes spp. Infect. Immun. 64, 1532–1540.
McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A., and Dowdle, W. R. (1977). Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203. doi: 10.1056/NEJM197712012972202
Michard, C., Sperandio, D., Bailo, N., Pizarro-Cerda, J., Leclaire, L., Chadeau-Argaud, E., et al. (2015). The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. MBio. 6:e00354–15. doi: 10.1128/mBio.00354-15
Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K., and Bassler, B. L. (2002). Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314. doi: 10.1016/S0092-8674(02)00829-2
Molofsky, A. B., and Swanson, M. S. (2003). Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50, 445–461. doi: 10.1046/j.1365-2958.2003.03706.x
Molofsky, A. B., and Swanson, M. S. (2004). Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53, 29–40. doi: 10.1111/j.1365-2958.2004.04129.x
Newton, H. J., Ang, D. K., Van Driel, I. R., and Hartland, E. L. (2010). Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23, 274–298. doi: 10.1128/CMR.00052-09
Ng, W. L., and Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Ng, W. L., Perez, L. J., Wei, Y., Kraml, C., Semmelhack, M. F., and Bassler, B. L. (2011). Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 79, 1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x
Ng, W. L., Wei, Y., Perez, L. J., Cong, J., Long, T., Koch, M., et al. (2010). Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. Proc. Natl. Acad. Sci. U.S.A. 107, 5575–5580. doi: 10.1073/pnas.1001392107
Njoroge, J., and Sperandio, V. (2012). Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect. Immun. 80, 688–703. doi: 10.1128/IAI.05921-11
Pacheco, A. R., and Sperandio, V. (2009). Inter-kingdom signaling: chemical language between bacteria and host. Curr. Opin. Microbiol. 12, 192–198. doi: 10.1016/j.mib.2009.01.006
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Pécastaings, S., Berge, M., Dubourg, K. M., and Roques, C. (2010). Sessile Legionella pneumophila is able to grow on surfaces and generate structured monospecies biofilms. Biofouling 26, 809–819. doi: 10.1080/08927014.2010.520159
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Personnic, N., Bärlocher, K., Finsel, I., and Hilbi, H. (2016). Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol. 24, 450–462. doi: 10.1016/j.tim.2016.02.003
Piao, Z., Sze, C. C., Barysheva, O., Iida, K., and Yoshida, S. (2006). Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl. Environ. Microbiol. 72, 1613–1622. doi: 10.1128/AEM.72.2.1613-1622.2006
Ragaz, C., Pietsch, H., Urwyler, S., Tiaden, A., Weber, S. S., and Hilbi, H. (2008). The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell. Microbiol. 10, 2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x
Rankin, S., Li, Z., and Isberg, R. R. (2002). Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect. Immun. 70, 3637–3648. doi: 10.1128/IAI.70.7.3637-3648.2002
Rasis, M., and Segal, G. (2009). The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72, 995–1010. doi: 10.1111/j.1365-2958.2009.06705.x
Rasko, D. A., Moreira, C. G., Li De, R., Reading, N. C., Ritchie, J. M., Waldor, M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080. doi: 10.1126/science.1160354
Reading, N. C., Rasko, D. A., Torres, A. G., and Sperandio, V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 5889–5894. doi: 10.1073/pnas.0811409106
Robinson, C. G., and Roy, C. R. (2006). Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol. 8, 793–805. doi: 10.1111/j.1462-5822.2005.00666.x
Rothmeier, E., Pfaffinger, G., Hoffmann, C., Harrison, C. F., Grabmayr, H., Repnik, U., et al. (2013). Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog. 9:e1003598. doi: 10.1371/journal.ppat.1003598
Sahr, T., Brüggemann, H., Jules, M., Lomma, M., Albert-Weissenberger, C., Cazalet, C., et al. (2009). Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72, 741–762. doi: 10.1111/j.1365-2958.2009.06677.x
Sahr, T., Rusniok, C., Dervins-Ravault, D., Sismeiro, O., Coppee, J. Y., and Buchrieser, C. (2012). Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 9, 503–519. doi: 10.4161/rna.20270
Sauer, J. D., Bachman, M. A., and Swanson, M. S. (2005). The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 9924–9929. doi: 10.1073/pnas.0502767102
Schell, U., Kessler, A., and Hilbi, H. (2014). Phosphorylation signalling through the Legionella quorum sensing histidine kinases LqsS and LqsT converges on the response regulator LqsR. Mol. Microbiol. 92, 1039–1055. doi: 10.1111/mmi.12612
Schell, U., Simon, S., Sahr, T., Hager, D., Albers, M. F., Kessler, A., et al. (2016). The a-hydroxyketone LAI-1 regulates motility, Lqs-dependent phosphorylation signalling and gene expression of Legionella pneumophila. Mol. Microbiol. 99, 778–793. doi: 10.1111/mmi.13265
Schikora, A., Schenk, S. T., and Hartmann, A. (2016). Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Mol. Biol. 90, 605–612. doi: 10.1007/s11103-016-0457-8
Schwarzer, C., Fu, Z., Patanwala, M., Hum, L., Lopez-Guzman, M., Illek, B., et al. (2012). Pseudomonas aeruginosa biofilm-associated homoserine lactone C12 rapidly activates apoptosis in airway epithelia. Cell. Microbiol. 14, 698–709. doi: 10.1111/j.1462-5822.2012.01753.x
Segal, G. (2013). The Legionella pneumophila two-component regulatory systems that participate in the regulation of Icm/Dot effectors. Curr. Top. Microbiol. Immunol. 376, 35–52. doi: 10.1007/82_2013_346
Shank, E. A., and Kolter, R. (2009). New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 12, 205–214. doi: 10.1016/j.mib.2009.01.003
Sherwood, R. K., and Roy, C. R. (2013). A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe 14, 256–268. doi: 10.1016/j.chom.2013.08.010
Shiner, E. K., Rumbaugh, K. P., and Williams, S. C. (2005). Inter-kingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol. Rev. 29, 935–947. doi: 10.1016/j.femsre.2005.03.001
Shivaji, T., Sousa Pinto, C., San-Bento, A., Oliveira Serra, L. A., Valente, J., Machado, J., et al. (2014). A large community outbreak of Legionnaires disease in Vila Franca de Xira, Portugal, October to November 2014. Euro. Surveill. 19:20991. doi: 10.2807/1560-7917.ES2014.19.50.20991
Simon, S., Schell, U., Heuer, N., Hager, D., Albers, M. F., Matthias, J., et al. (2015). Inter-kingdom signaling by the Legionella quorum sensing molecule LAI-1 modulates cell migration through an IQGAP1-Cdc42-ARHGEF9-dependent pathway. PLoS Pathog. 11:e1005307. doi: 10.1371/journal.ppat.1005307
Simon, S., Wagner, M. A., Rothmeier, E., Müller-Taubenberger, A., and Hilbi, H. (2014). Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell. Microbiol. 16, 977–992. doi: 10.1111/cmi.12258
Sperandio, V., Torres, A. G., Jarvis, B., Nataro, J. P., and Kaper, J. B. (2003). Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U.S.A. 100, 8951–8956. doi: 10.1073/pnas.1537100100
Spirig, T., Tiaden, A., Kiefer, P., Buchrieser, C., Vorholt, J. A., and Hilbi, H. (2008). The Legionella autoinducer synthase LqsA produces an α-hydroxyketone signaling molecule. J. Biol. Chem. 283, 18113–18123. doi: 10.1074/jbc.M801929200
Suzuki, K., Babitzke, P., Kushner, S. R., and Romeo, T. (2006). Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20, 2605–2617. doi: 10.1101/gad.1461606
Swanson, M. S., and Isberg, R. R. (1995). Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63, 3609–3620.
Tanner, J. R., Li, L., Faucher, S. P., and Brassinga, A. K. (2016). The CpxRA two-component system contributes to Legionella pneumophila virulence. Mol. Microbiol. 100, 1017–1038. doi: 10.1111/mmi.13365
Tateda, K., Ishii, Y., Horikawa, M., Matsumoto, T., Miyairi, S., Pechere, J. C., et al. (2003). The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71, 5785–5793. doi: 10.1128/IAI.71.10.5785-5793.2003
Taylor, M., Ross, K., and Bentham, R. (2009). Legionella, protozoa, and biofilms: interactions within complex microbial systems. Microb. Ecol. 58, 538–547. doi: 10.1007/s00248-009-9514-z
Tiaden, A., and Hilbi, H. (2012). α-Hydroxyketone synthesis and sensing by Legionella and Vibrio. Sensors 12, 2899–2919. doi: 10.3390/s120302899
Tiaden, A., Spirig, T., Carranza, P., Brüggemann, H., Riedel, K., Eberl, L., et al. (2008). Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions. J. Bacteriol. 190, 7532–7547. doi: 10.1128/JB.01002-08
Tiaden, A., Spirig, T., and Hilbi, H. (2010a). Bacterial gene regulation by α-hydroxyketone signaling. Trends Microbiol. 18, 288–297. doi: 10.1016/j.tim.2010.03.004
Tiaden, A., Spirig, T., Sahr, T., Wälti, M. A., Boucke, K., Buchrieser, C., et al. (2010b). The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ. Microbiol. 12, 1243–1259. doi: 10.1111/j.1462-2920.2010.02167.x
Tiaden, A., Spirig, T., Weber, S. S., Brüggemann, H., Bosshard, R., Buchrieser, C., et al. (2007). The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell. Microbiol. 9, 2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x
Valentine, C. D., Anderson, M. O., Papa, F. R., and Haggie, P. M. (2013). X-box binding protein 1 (XBP1s) is a critical determinant of Pseudomonas aeruginosa homoserine lactone-mediated apoptosis. PLoS Pathog. 9:e1003576. doi: 10.1371/journal.ppat.1003576
Weatherby, K. E., Zwilling, B. S., and Lafuse, W. P. (2003). Resistance of macrophages to Mycobacterium avium is induced by α2-adrenergic stimulation. Infect. Immun. 71, 22–29. doi: 10.1128/IAI.71.1.22-29.2003
Weber, S., Stirnimann, C. U., Wieser, M., Frey, D., Meier, R., Engelhardt, S., et al. (2014a). A type IV translocated Legionella cysteine phytase counteracts intracellular growth restriction by phytate. J. Biol. Chem. 289, 34175–34188. doi: 10.1074/jbc.M114.592568
Weber, S., Wagner, M., and Hilbi, H. (2014b). Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. MBio. 5:e00839. doi: 10.1128/mBio.00839-13
Weber, S. S., Ragaz, C., and Hilbi, H. (2009). The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell. Microbiol. 11, 442–460. doi: 10.1111/j.1462-5822.2008.01266.x
Weber, S. S., Ragaz, C., Reus, K., Nyfeler, Y., and Hilbi, H. (2006). Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2:e46. doi: 10.1371/journal.ppat.0020046
Werner, K. M., Perez, L. J., Ghosh, R., Semmelhack, M. F., and Bassler, B. L. (2014). Caenorhabditis elegans recognizes a bacterial quorum-sensing signal molecule through the AWCON neuron. J. Biol. Chem. 289, 26566–26573. doi: 10.1074/jbc.M114.573832
Keywords: α-hydroxyketone, amoeba, autoinducer, bacterial pathogenesis, cell–cell communication, Dictyostelium, Legionella, macrophage, small molecule signaling, phospho-transfer, response regulator, sensor kinase, quorum sensing
Citation: Hochstrasser R and Hilbi H (2017) Intra-Species and Inter-Kingdom Signaling of Legionella pneumophila. Front. Microbiol. 8:79. doi: 10.3389/fmicb.2017.00079
Received: 16 November 2016; Accepted: 11 January 2017;
Published: 03 February 2017.
Edited by:
Wolfgang R. Streit, University of Hamburg, GermanyReviewed by:
Mika Tapio Tarkka, Helmholtz Centre for Environmental Research (HZ), GermanyXavier Charpentier, Centre de Recherche en Infectiologie – INSERM U1111, France
Copyright © 2017 Hochstrasser and Hilbi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hubert Hilbi, aGlsYmlAaW1tLnV6aC5jaA==
 Ramon Hochstrasser
Ramon Hochstrasser Hubert Hilbi
Hubert Hilbi