
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 12 January 2017
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.02173
This article is part of the Research TopicAntifungals used against animal and plant pathogenic fungiView all 12 articles
 Sarah G. Whaley1
Sarah G. Whaley1 Elizabeth L. Berkow1†
Elizabeth L. Berkow1† Jeffrey M. Rybak1
Jeffrey M. Rybak1 Andrew T. Nishimoto1
Andrew T. Nishimoto1 Katherine S. Barker1
Katherine S. Barker1 P. David Rogers1,2*
P. David Rogers1,2*Within the limited antifungal armamentarium, the azole antifungals are the most frequent class used to treat Candida infections. Azole antifungals such as fluconazole are often preferred treatment for many Candida infections as they are inexpensive, exhibit limited toxicity, and are available for oral administration. There is, however, extensive documentation of intrinsic and developed resistance to azole antifungals among several Candida species. As the frequency of azole resistant Candida isolates in the clinical setting increases, it is essential to elucidate the mechanisms of such resistance in order to both preserve and improve upon the azole class of antifungals for the treatment of Candida infections. This review examines azole resistance in infections caused by C. albicans as well as the emerging non-albicans Candida species C. parapsilosis, C. tropicalis, C. krusei, and C. glabrata and in particular, describes the current understanding of molecular basis of azole resistance in these fungal species.
Candida albicans and emerging non-albicans Candida (NAC) species such as C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei can cause superficial infections of the oral and vaginal mucosa as well as disseminated bloodstream and deep-tissue infections. Species involvement varies by infection site and by geography. Candida infections are most often caused by C. albicans as evidenced by epidemiological studies in the United States (Cleveland et al., 2015), Europe (Klingspor et al., 2015), and the Middle East (Sharifzadeh et al., 2013). Of all the NAC species, C. glabrata is the most commonly isolated from patients with candidemia in North America (Sobel, 2010; Azie et al., 2012; Pfaller et al., 2014b), and Northern Europe (Lortholary et al., 2014; Milazzo et al., 2014), as well as other geographic areas studied with the exception of Latin America (Pfaller et al., 2010). C. glabrata is also the most common NAC species found to be the causative agent in vulvovaginal candidiasis (VVC) (Corsello et al., 2003; Holland et al., 2003; Richter et al., 2005; Vermitsky et al., 2008; Mahmoudi Rad et al., 2012) and candiduria (Sobel et al., 2000; Kauffman, 2005). In some patient populations, for example, candidemia in patients with hematologic malignancy and VVC in diabetic patients, C. glabrata is even more common than C. albicans (Goswami et al., 2006; Ray et al., 2007; Hachem et al., 2008). C. parapsilosis is well known for its threat to the pediatric population, as it is responsible for 17–50% of all fungemia in infants and neonates (Abi-Said et al., 1997; Krcmery et al., 1999). C. parapsilosis is also second only to C. albicans in incidence as a cause of Candida endocarditis with mortality rates between 42 and 65% (Weems, 1992; Garzoni et al., 2007). In the Asia-Pacific region, C. tropicalis has been reported to constitute 20–45% of Candida isolates (Kothari and Sagar, 2009; Pfaller et al., 2010). C. tropicalis infections are commonly associated with malignancy, with some studies reporting higher prevalence among patients with hematologic diseases such as acute myeloid leukemia (Weinberger et al., 2005; Nucci and Colombo, 2007; Tang et al., 2014, 2015; Cornely et al., 2015). Mortality associated with C. tropicalis candidemia in these populations unfortunately remains high, ranging from 30 to 70%, with the highest rates most commonly observed among the elderly (Weinberger et al., 2005; Nucci and Colombo, 2007; Morii et al., 2014; Cornely et al., 2015; Wang et al., 2015). C. krusei is the fourth most common NAC species associated with invasive candidiasis and candidemia, accounting for approximately 2.7% of NAC species isolated across the United States (Pfaller et al., 2014b). Moreover, the number of C. krusei isolates implicated in these types of infections has increased over time (Pfaller et al., 2014a,b). In particular, patients with hematologic malignancies and bone marrow transplants have been shown to be at increased risk of C. krusei infection (Merz et al., 1986; Wingard et al., 1991; Pfaller et al., 2008).
There are several classes of compounds that comprise the arsenal used to treat Candida infections. The polyenes, azoles, echinocandins, nucleoside analogs, and allylamines are used with varying efficacy depending on the type and site of infection and the sensitivity of the Candida species (Pfaller et al., 2010; Pfaller and Diekema, 2012b; Pfaller et al., 2013; Pappas et al., 2016). The most commonly prescribed antifungal used for most C. albicans infections is fluconazole, a member of the azole class of antifungals (Pfaller et al., 2010). Azoles inhibit 14-α-sterol demethylase, encoded by the ERG11 gene, which is an enzyme involved in the biosynthesis of the fungal-specific membrane sterol ergosterol. As some NAC species exhibit intrinsic resistance to azoles, their use is likely a contributing factor to the more frequent incidence of infections caused by these NAC species (Oxman et al., 2010; Lortholary et al., 2011; Fothergill et al., 2014). Moreover, many studies have documented the ability of Candida to develop high-level resistance to azole antifungals (Oxman et al., 2010; Lortholary et al., 2011). A compilation of fluconazole MIC ranges and epidemiological cutoff values for Candida species is presented in Table 1.
Infections caused by C. albicans are associated with varying levels of fluconazole resistance depending on the type of infection. C. albicans isolates from candidemic patients have the lowest incidence of azole resistance (0–5%) (Diekema et al., 2012; Pfaller et al., 2015; Ying et al., 2015). The incidence of fluconazole resistance in C. albicans isolates from oropharyngeal candidiasis (OPC) is higher and depends upon previous fluconazole treatment and prior OPC infections (Enwuru et al., 2008; Berberi et al., 2015). C. glabrata has the highest incidence of azole resistance among Candida clinical isolates and exhibits intrinsic decreased susceptibility to the azole class of antifungals (Oxman et al., 2010; Pfaller et al., 2014b), including the newest addition to the class, isavuconazole (Castanheira et al., 2014). C. glabrata is also able to develop high-level resistance after exposure to azole antifungals (Fidel et al., 1999; Lee et al., 2009) and is one of the most frequent species isolated in breakthrough infections from patients receiving azole prophylaxis (Bennett et al., 2004; Imhof et al., 2004; Hachem et al., 2008). Of increasing concern are the number of multidrug resistant isolates of C. glabrata that are being recovered clinically (Manzano-Gayosso et al., 2003; Chapeland-Leclerc et al., 2010; Hull et al., 2012; Pfaller et al., 2012a; Cho et al., 2015). In the Asia-Pacific region, fluconazole resistance in C. tropicalis ranges from 0 to as high as 83% (Yang et al., 2004, 2008; Yoo et al., 2009). The worldwide incidence of fluconazole resistance in C. parapsilosis disseminated infections ranges between 2 and 5% (Chen et al., 2006; Martí-Carrizosa et al., 2014; Pfaller et al., 2015). As C. krusei exhibits intrinsic resistance to fluconazole, there is some controversy whether its increased infection rate is related to fluconazole prophylaxis or previous treatment (Hope et al., 2002; Lin et al., 2005; Blot et al., 2006; Gong et al., 2016). Clearly, an understanding of molecular mechanisms driving intrinsic and development of high-level azole resistance is warranted.
Resistance to azole antifungals in Candida (summarized in Figure 1) has been most extensively studied in C. albicans. One mechanism of resistance identified in this species is the presence of point mutations in ERG11. Previous studies have identified amino acid substitutions that result in decreased fluconazole susceptibility and have noted that several of these critical allelic variations cluster in three “hot spot” regions within Erg11p (Marichal et al., 1999). Recently, 63 fluconazole-resistant C. albicans clinical isolates were examined for mutations within their ERG11 alleles, and 55 were found to carry at least one mutation that resulted in amino acid substitutions, with nine such predicted amino acid substitutions being novel (Flowers et al., 2015). Molecular modeling of the substitutions that resulted in decreased fluconazole susceptibility when expressed in a susceptible background revealed that the mutations clustered in either the predicted catalytic site, the fungus-specific external loop, or on the proximal surface potentially interacting with the loop or near the heme. Additionally, a study involving site-directed mutagenesis of wild-type ERG11 to introduce mutations identified in 23 C. albicans clinical isolates demonstrated nine of these mutations result in increased fluconazole resistance (Xiang et al., 2013). Five of the amino acid substitutions were predicted to be at or near the active site of Erg11p.
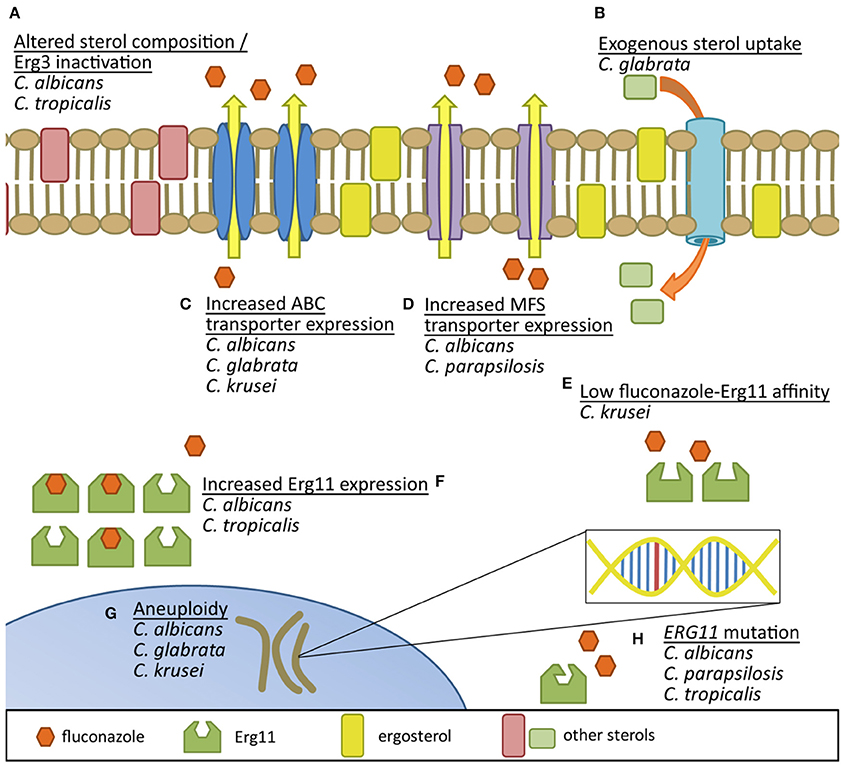
Figure 1. Comparison of documented fluconazole resistance mechanisms in Candida species. (A) Erg3 inactivation results in utilization of alternative sterols in the yeast membrane. (B) Uptake of exogenous sterols helps circumvent endogenous sterol production inhibition by fluconazole. Increased production of both (C) ATP-binding cassette efflux pumps and (D) major facilitator superfamily transporters reduces intracellular accumulation of azoles. (E) Inherently low affinity of fluconazole binding to species-specific Erg11 may decrease fluconazole's potential to inhibit the protein. (F) Increased expression of Erg11 protein can help overcome azole activity and (G) aneuploidy may promote genetic adaptation to azole exposure. (H) Mutations in ERG11 can also result in proteins with reduced affinity for fluconazole binding.
Another mechanism of fluconazole resistance in C. albicans is the increased expression of ERG11 due to activating mutations in the gene encoding the zinc-cluster transcriptional regulator Upc2p. C. albicans Upc2 is a homolog of the Saccharomyces cerevisiae ERG gene regulator pair Upc2/Ecm22. Initially, Upc2 involvement in fluconazole resistance in C. albicans was demonstrated when Δupc2 C. albicans strains were shown to be highly susceptible to azoles while those over-expressing Upc2 had increased fluconazole resistance (MacPherson et al., 2005). Further studies examining a matched set of fluconazole-susceptible and—resistant C. albicans clinical isolates in which fluconazole resistance was not associated with overexpression of drug efflux pumps revealed SNPs in one UPC2 allele and overexpression of several ERG genes and UPC2 in the resistant isolate (Dunkel et al., 2008a). Expression of UPC2 alleles in fluconazole-susceptible strains resulted in increased fluconazole resistance (Dunkel et al., 2008a; Heilmann et al., 2010; Hoot et al., 2011). Interestingly, three additional matched sets of ERG11-overexpressing clinical C. albicans isolates have been described which have no sequence differences in UPC2 between the susceptible and resistant isolates in each pair (Heilmann et al., 2010), indicating that other mechanisms of ERG11 upregulation exist. While these studies were important in establishing Upc2p as a regulator of ERG11 expression in the context of fluconazole resistance, it was assumed that UPC2-mediated fluconazole resistance is a rare occurrence. However, a large study involving 63 fluconazole-resistant C. albicans clinical isolates demonstrated 47 of these isolates overexpressed ERG11 by at least 2-fold (Flowers et al., 2012). Twenty-nine of these ERG11-overexpressing isolates contained a missense mutation in UPC2, and eight single amino acid substitutions were elucidated from their UPC2 alleles. Seven of these alleles were found to be associated with increased ERG11 expression, increased ergosterol production, and decreased fluconazole susceptibility.
Two other mechanisms of fluconazole resistance in C. albicans involve the overexpression of drug efflux pumps Mdr1p and Cdr1p/Cdr2p. TAC1 (transcriptional activator of CDR genes) is a zinc-cluster transcription factor whose regulon is hallmarked by the ATP-binding cassette (ABC) transporter-encoding genes CDR1 and CDR2 (Coste et al., 2004). Activation of expression of the TAC1 regulon is through binding of TAC1 to the DRE (drug response element) present in the promoters of TAC1-regulated genes (Coste et al., 2004; Liu et al., 2007). At least nine hyperactive TAC1 alleles have been identified (Coste et al., 2007), and fluconazole minimum inhibitory concentrations (MIC) associated with the isolates from which these alleles have been discovered have revealed that TAC1 demonstrates codominance resulting in intermediate fluconazole MIC in TAC1-heterologous strains and high fluconazole MIC upon loss of heterozygosity (Coste et al., 2006, 2007). Because TAC1 resides on the left arm of Chr5 with ERG11, such loss of heterozygosity in the presence of hyperactive TAC1 and mutated ERG11 results in high-level azole resistance (Coste et al., 2007; Selmecki et al., 2008).
Mdr1p is a major facilitator superfamily (MFS) efflux pump usually expressed at non-detectable levels in wildtype C. albicans strains, induced in the presence of benomyl, diamide, and hydrogen peroxide, and constitutively overexpressed in some fluconazole-resistant C. albicans isolates (Alarco and Raymond, 1999). MRR1, multidrug resistance regulator 1, was identified by comparing the transcriptomes of sets of matched isolates in which the fluconazole-resistant isolates overexpressed MDR1 (Morschhäuser et al., 2007). Disruption of MRR1 in these resistant isolates led to a decrease in fluconazole MIC, while introduction of each of the mutant alleles individually into a wildtype fluconazole-susceptible background in the native MRR1 locus conferred fluconazole resistance to the constructed strain. Another study examined additional MRR1 allelic variations in MDR1-mediated fluconazole resistance (Dunkel et al., 2008b). In most cases the resistant isolates/strains were homozygous for the MRR1 allele containing the gain-of-function mutations due to mitotic recombination and chromosome loss.
A less common mechanism of azole resistance in C. albicans is inactivation of the ERG3 gene, which encodes the ergosterol biosynthesis enzyme sterol Δ5,6 desaturase. Erg3p catalyzes one of the final steps in the pathway and also converts nontoxic 14α-methylated sterol intermediates, that accumulate during azole treatment, into the toxic sterol 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Inactivation or deletion of the ERG3 gene, therefore, prevents such toxic sterols from being synthesized. Only a handful of clinical C. albicans isolates have documented azole resistance due to ERG3 inactivation (Kelly et al., 1997; Nolte et al., 1997; Miyazaki et al., 1999; Chau et al., 2005; Martel et al., 2010; Morio et al., 2012).
Aneuploidy plays a role in azole resistance in C. albicans as demonstrated by comparative genome hybridization (Selmecki et al., 2006). As alluded to earlier, a common aneuploidy found in azole-resistant strains involves Chr5. Similarly, loss of heterozygosity (LOH) has been shown to occur in azole-resistant C. albicans (Coste et al., 2006). Examination of TAC1 in a matched set of azole-susceptible and—resistant C. albicans isolates revealed that the susceptible isolate harbored two wildtype alleles of TAC1, while the resistant isolate contained only one of those alleles in which a single nucleotide polymorphism (SNP) translated into an activating amino acid substitution (N977D).
Because azole resistance has been extensively studied in C. albicans, attempts to elucidate mechanisms of azole resistance in C. parapsilosis have involved examining orthologous genes and yielded mixed results. A study of a series of six isolates from a single patient found a single SNP in MRR1 present in the two fluconazole-resistant isolates (Zhang et al., 2015). Nine fluconazole-resistant isolates were obtained from candidemia patients in a Brazilian hospital and examined for CDR1, MDR1, and ERG11 overexpression as well as the presence of SNPs in the ERG11 gene (Souza et al., 2015). Each of the resistant isolates possessed a single homozygous SNP (A395T) which corresponds to a Y132F amino acid substitution. In addition, while none of the isolates overexpressed MDR1 as compared to the C. parapsilosis reference strain ATCC22019, CDR1 expression was between 3.3- and 9.2-fold higher in these isolates as compared to the reference strain, and ERG11 was overexpressed between 1.5- and 7.4-fold. While this study indicated an association between CDR1 and ERG11 and fluconazole resistance in C. parapsilosis, a causal link was not definitively proven.
In a larger-scale study, 30 resistant isolates, 37 susceptible-dose-dependent isolates, and 55 susceptible isolates were collected from hospitals in four U.S. cities, and their ERG11 genes were sequenced (Grossman et al., 2015). Five SNPs were identified in 54 of the isolates; amino acid substitution Y132F, found in 17 resistant isolates, was the only one found exclusively in resistant isolates. Twenty-three isolates harbored SNPs in MRR1. Of the nine SNPs identified, only three were found exclusively in resistant isolates. Quantitative PCR measuring relative MDR1 expression revealed nine isolates (six with a SNP in MRR1, three without) with at least 5-fold increase in MDR1 expression compared to a composite expression level from a subset of susceptible isolates. However, the expression levels were a fraction of the levels achieved by MDR1-mediated azole resistance in C. albicans. Without definitive experiments in which introduction of a mutated ERG11 allele confers azole resistance in a susceptible isolate, these results remain suggestive.
In an effort to identify potential mechanisms of azole resistance on a genome-wide scale in C. parapsilosis, fluconazole-, voriconazole-, and posaconazole-resistant strains were developed experimentally by serial passage in liquid culture containing either fluconazole, voriconazole, or posaconazole (Silva et al., 2011). The fluconazole- and voriconazole-resistant strains were cross-resistant to both fluconazole and voriconazole and possessed similar transcriptional profiles as assessed by microarray analysis; however, the posaconazole-resistant strain was not cross-resistant to the other azoles and had a distinct transcriptional profile. Among the genes differentially expressed in fluconazole- and voriconazole-resistant strains were the stress response gene GRP2, as well as MDR1 and MRR1. ERG11 was not differentially expressed in these strains. However, in the posaconazole-resistant strain, the ergosterol biosynthesis genes ERG11 and ERG6, as well as ERG gene regulator UPC2 were among the genes differentially expressed.
In a study using laboratory strains of C. parapsilosis in which previously-determined gain-of-function alleles of CpMRR1 were introduced into the native locus, strains containing Mrr1p with a G583R amino acid substitution from a fluconazole-resistant C. parapsilosis isolate led to resistant fluconazole and voriconazole MIC compared to strains harboring the wildtype allele (Branco et al., 2015). Similarly, strains with single SNP-containing MRR1 alleles had a ~5-fold increase in MRR1 gene expression and ~70-fold increase in MDR1 gene expression.
In another study, 35 unrelated fluconazole-resistant and four unrelated susceptible isolates of C. parapsilosis were examined to elucidate mechanisms of fluconazole resistance in C. parapsilosis (Berkow et al., 2015). Sixteen resistant isolates overexpressed CDR1, three other resistant isolates exhibited MDR1 overexpression, and eight resistant isolates demonstrated overexpression of ERG11 as compared to the susceptible isolates. When sequencing orthologues of UPC2, MRR1, and TAC1 in order to identify putative gain-of-function mutations that would lead to overexpression of ERG11, MDR1, and CDR1, only one heterozygous mutation in UPC2 was recovered from one isolate, suggesting that ERG11 overexpression in fluconazole-resistant C. parapsilosis is not mediated by UPC2. TAC1 mutations that were recovered did not fully correspond with CDR1 overexpression and those recovered were not analogous to those found in gain-of-function CaTAC1 alleles. Similarly, MRR1 mutations recovered did not correspond to any mutations found in gain-of-function alleles of CaMRR1. Subsequently, CDR1 was deleted from three of the CDR1-overexpressing isolates which only resulted in a one-dilution decrease in fluconazole MIC. MDR1 deletion in three MDR1-overexpressing isolates revealed a one-dilution decrease in fluconazole MIC in two isolates and no change in fluconazole MIC in the third. To address the role of alterations in the ergosterol biosynthesis pathway in azole resistance in C. parapsilosis, ERG11, and ERG3 were sequenced. No ERG3 mutations were recovered, which was supported by the sterol profiles of the isolates. A single ERG11 mutation (Y132F) was recovered in one resistant isolate and a combination of Y132F and R398I mutations was found in an additional ten isolates. In nine of these eleven isolates there was a change in the sterol profile indicative of a change in Erg11 functionality. This study indicates that while differential expression of efflux pumps is commonly found in azole-resistant C. parapsilosis isolates, the resistant phenotype is not solely due to their overexpression but instead is multifactorial and involves ERG11 mutation and/or overexpression.
As compared with other species of Candida, relatively little is known about the mechanisms of azole resistance in C. tropicalis. An analysis of 52 clinical C. tropicalis isolates from China found the average ERG11 expression level more than 4-fold higher among fluconazole-resistant isolates than -susceptible isolates (Jiang et al., 2013). Moreover, ERG11 expression was even higher among a subset of fluconazole-resistant isolates also resistant to itraconazole and voriconazole. These results were recently echoed by a similar study characterizing 35 C. tropicalis isolates from Korean university hospitals, nine of which were fluconazole-non-susceptible (Choi et al., 2016). While considerable variability in ERG11 expression (~150-fold) was observed in the highly fluconazole-susceptible group, ERG11 expression was significantly higher among both less fluconazole-susceptible (MIC 1–2 μg/ml) and fluconazole-non-susceptible (MIC ≥ 4 μg/ml) isolates. This study also sequenced the C. tropicalis UPC2 gene and found several heterozygous and homozygous mutations. However, many of these mutations have been observed in fluconazole-susceptible isolates not found to overexpress ERG11, and further characterization of their impact on the regulatory function of UPC2 is needed.
Molecular characterization of azole-resistant clinical C. tropicalis isolates has also revealed alterations in the ergosterol biosynthetic pathway (Vandeputte et al., 2005; Eddouzi et al., 2013; Jiang et al., 2013; Choi et al., 2016). A fluconazole-resistant C. tropicalis isolate recovered from a clinical blood specimen from Tunisia was found to have mutations in both ERG3 and ERG11 which were individually observed to be detrimental to ergosterol biosynthesis when heterologously expressed in S. cerevisiae (Eddouzi et al., 2013). Notably, the ERG11 mutation in this isolate consisted of a deletion of 132 nucleotides resulting in a D275V amino acid substitution and the loss of 44 amino acids near the N-terminus of Erg11p. Homozygous replacement of the wild-type C. tropicalis ERG11 with the truncated clinical variant, with or without the associated clinical ERG3 mutation, resulted in high-level fluconazole resistance in a fluconazole-susceptible reference strain of C. tropicalis. Additionally, an ERG11 mutation resulting in decreased fluconazole susceptibility due to the amino acid substitution Y132F, has been well characterized in C. albicans and was recently observed in a fluconazole-resistant C. tropicalis isolate from a patient with candidemia (Tan et al., 2015).
One of the first studies to associate the overexpression of efflux pumps with azole resistance in C. tropicalis utilized serial passaging of a reference C. tropicalis isolate on media containing various concentrations of fluconazole to produce genetically-related isolates with reduced fluconazole susceptibility (Barchiesi et al., 2000). After passaging, all isolates with reduced susceptibility to fluconazole demonstrated increased expression of both C. tropicalis MDR1 and a gene with high homology to C. albicans CDR1. In both cases, the increased expression was found to then be diminished in fluconazole-susceptible revertants obtained from further passaging on fluconazole-free media. The role of efflux pump overexpression in azole resistance among clinical C. tropicalis isolates has been less clearly defined. When the expression of MDR1 and CDR1 was examined in the aforementioned 52 clinical C. tropicalis isolates from China, no significant difference was observed between fluconazole-susceptible and -resistant isolates (Jiang et al., 2013). In contrast, among the 35 clinical isolates from Korean university hospitals, expression of both MDR1 and CDR1 was observed to be significantly higher among both less-fluconazole-susceptible and fluconazole-non-susceptible isolates. However, it is important to note the large degree of variability in the expression of MDR1 and CDR1 observed in the highly fluconazole-susceptible control group, ~50-fold and ~30-fold respectively (Wang et al., 2015). To date, experiments to directly delineate the potential role of these efflux pumps has yet to be performed in C. tropicalis, and the homologs of C. albicans MRR1 and TAC1 have not been examined.
C. krusei is intrinsically resistant to fluconazole, though the precise mechanism is not completely understood. Several studies have attributed C. krusei's innate azole resistance to efflux pump activity, namely through the ATP-binding cassette transporter Abc1p, and reduced drug accumulation (Marichal et al., 1995; Katiyar and Edlind, 2001; Lamping et al., 2009) in combination with reduced azole affinity for Erg11p (Marichal et al., 1995; Venkateswarlu et al., 1997; Orozco et al., 1998; Guinea et al., 2006; Lamping et al., 2009). Changes in the cell membrane affecting membrane fluidity may be implicated in azole resistance as well since there is evidence to suggest that intracellular azole accumulation occurs through one or possibly both mechanisms of passive and facilitated diffusion (Mansfield et al., 2010; Kolaczkowska and Kolaczkowski, 2016). Additionally, the discovery of a trisomy in the ERG11-containing chromosome in a C. krusei strain suggests aneuploidy may not be uncommon in this species, though the effects as it relates to azole resistance are not yet known (Lamping et al., 2009).
Resistance mechanisms against other azoles are also not clearly defined. For example, analysis of itraconazole-resistant C. krusei isolates revealed that reduced intracellular content of the drug and not altered affinity for the drug target likely drives itraconazole resistance (Marichal et al., 1995; Venkateswarlu et al., 1996). However, more recently it has been suggested that overexpression of genes encoding both Erg11p and the efflux pump Abc2p may also play a role with itraconazole resistance (Tavakoli et al., 2010; He et al., 2015). Despite its fungicidal activity in C. krusei (Rubio et al., 2005), resistance to voriconazole has also emerged, and current research supports a theory where overexpression of the genes encoding the efflux pump Abc2 and Erg11 impart more transient resistance properties, while increased expression of Abc1p and point mutations in ERG11 predominate as time progresses to yield a stably resistant pathogen in the prolonged presence of voriconazole (Ricardo et al., 2014). Erg11p amino acid substitutions have been observed in azole-resistant C. krusei and, in the case of Y166S, have been predicted to interfere with Erg11p function (Ricardo et al., 2014; Silva et al., 2016). While the newer antifungal agents posaconazole and isavuconazole have shown good activity against C. krusei (Lee et al., 2000; Rybak et al., 2015), reports of resistance against these agents are relatively sparse (Espinel-Ingroff et al., 2014; Pfaller et al., 2015). However, in a recent analysis examining NAC strains in the U.S. by region, Candida krusei resistance to posaconazole was highest in the eastern United States, with posaconazole resistance occurring in 13–16.7% of isolates (Pfaller et al., 2014b). Nevertheless, the mechanisms of resistance in C. krusei against these agents remain to be investigated.
C. glabrata is unique among the Candida species discussed here as it is a haploid yeast more closely related to S. cerevisiae. Development of azole resistance in clinical isolates of C. glabrata has been almost exclusively linked to the presence of activating mutations in the zinc cluster transcription factor Pdr1 (Vermitsky and Edlind, 2004) that lead to differential expression of downstream targets. Nearly all clinical isolates have been found to have PDR1 mutations, with such mutations found in the inhibitory domain, activating domain, middle homology region, and xenobiotic binding region. The rapid acquisition of PDR1 mutations could be due to the high incidence of mutations in the mismatch repair gene MSH2, which results in a hypermutable phenotype (Healey et al., 2016). The activating mutations exhibit distinct expression patterns of the downstream effector genes, with the exception of increased expression of CDR1 and PUP1, and no correlation has been found between location of the mutation and altered gene expression (Tsai et al., 2006, 2010; Ferrari et al., 2009; Caudle et al., 2011; Paul et al., 2011). Among the genes whose pleiotropic drug response element (PDRE) is directly bound by Pdr1 (Paul et al., 2014), only three, the ABC transporters CDR1 (Sanglard et al., 1999), PDH1 (CDR2) (Miyazaki et al., 1998; Sanglard et al., 2001), and SNQ2 (Sanguinetti et al., 2005; Torelli et al., 2008), have been linked directly to azole resistance. Recent work has shown increased expression of four MFS transporters in clotrimazole resistant isolates compared to clotrimazole susceptible clinical isolates. Disruption of one of these, TPO3, moderately increased susceptibility to clotrimazole and fluconazole (Costa et al., 2016). These findings suggest MFS transporters may have a minor role in azole resistance in C. glabrata.
Surprisingly, ERG11 does not appear to play an important role in clinical azole resistance in C. glabrata (Sanglard et al., 1999; Vermitsky and Edlind, 2004; Sanguinetti et al., 2005). Increased expression of ERG11 has been observed in only two clinical isolates of C. glabrata (vanden Bossche et al., 1992; Redding et al., 2003). The upregulation in one isolate was later found to be due to duplication of the entire chromosome containing ERG11 and the phenotype was lost with subsequent passaging in azole-free media (Marichal et al., 1997). A single resistant clinical isolate of C. glabrata has been shown to have a nonfunctional 14-α-sterol demethylase due to a missense mutation in ERG11, which led to the complete absence of ergosterol in the cell membrane (Hull et al., 2012). No additional clinical isolates have been identified to have resistance mechanisms related to the azole target.
C. glabrata has the ability to grow with altered cell membrane sterols, which allows for evasion of azole treatment. C. glabrata is able to take up exogenous sterols (Nakayama et al., 2000), both when the ergosterol biosynthesis pathway is blocked and under normal conditions in wild type strains (Tsai et al., 2004; Bard et al., 2005). Aus1p has been identified as the sterol transporter responsible for tolerance to azoles in the presence of exogenous sterols (Nakayama et al., 2007). C. albicans has recently been shown to take up sterols under aerobic conditions; however, C. glabrata is more liberal in its ability to take up sterols and does so in both aerobic and anaerobic conditions and, in the presence of serum and fluconazole, enhances uptake under aerobic conditions (Zavrel et al., 2013).
Azole resistance in C. glabrata has also been attributed to the formation of petite mutants, which are cells that have lost mitochondrial function resulting in respiratory deficiency (Defontaine et al., 1999; Brun et al., 2003). Petite mutants can be generated in the laboratory by treatment with azoles or ethidium bromide. This mutant phenotype has been recovered clinically (Bouchara et al., 2000; Ferrari et al., 2011), but is not common among clinical isolates. Azole resistance in petite mutants has been attributed to upregulation of the ABC transporters CDR1, CDR2, and SNQ2 (Sanglard et al., 2001; Ferrari et al., 2011), which is dependent on Pdr1 (Tsai et al., 2006). Petite mutants exhibit altered sterol profiles with a disproportionate amount of ergosterol and very little of ergosterol intermediates; however, no changes in the sequence of ERG11 or its expression have been detected (Brun et al., 2004).
Candida species are responsible for a majority of superficial and disseminated fungal infections in humans. While azole antifungals have long provided effective treatment for such infections, recent epidemiological studies indicate that intrinsic azole resistance in some Candida species as well as development of high-level azole resistance is a problem of critical importance in the clinical setting. While extensive studies to elucidate molecular mechanisms of high-level azole resistance in C. albicans has uncovered the role of ergosterol biosynthesis gene mutation and ERG gene and drug efflux pump upregulation as key mediators of azole resistance, there are clearly other factors at play that contribute significantly to such resistance. Similarly, while NAC are closely related to C. albicans, that does not necessarily translate to analogous molecular mechanisms of azole resistance.
Of the NAC species highlighted in this review, C. parapsilosis, C. tropicalis, C. krusei, and C. glabrata all express ABC transporter and/or MFS genes orthologous to CaCDR1 and CaMDR1. However, as discussed, the altered expression of these genes in azole-resistant NAC appear to contribute differently to resistance in different species. Moreover, the transcriptional regulators and genetic mutations governing azole efflux and sterol biosynthesis in C. tropicalis, C. parapsilosis, and C. krusei have not been fully examined. Finally, there exist clear differences in the mutations in ERG11 that are found to influence azole resistance in clinical isolates among these species. As azole resistance continues to emerge in these species, a more complete understanding of the important differences among resistance mechanisms employed by these species will be needed in order to circumvent this important clinical problem.
SW wrote the section regarding _Candida glabrata_, contributed to the Introduction, and edited the overall text. EB wrote the section regarding _Candida parapsilosis_ and contributed to the Introduction. JR wrote the section regarding _Candida tropicalis_, contributed to the Introduction, and compiled the data in Table 1. AN wrote the section regarding _Candida krusei_, contributed to the Introduction, and designed Figure 1. KB wrote the section regarding _Candida albicans_, contributed to the Introduction, and edited the overall text. PDR oversaw the review, wrote the abstract and conclusion, contributed to the Introduction, and edited the overall text.
Research from the laboratory of PDR is funded through a grant from the National Institutes of Health (R01 AI058145).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abi-Said, D., Anaissie, E., Uzun, O., Raad, I., Pinzcowski, H., and Vartivarian, S. (1997). The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24, 1122–1128. doi: 10.1086/513663
Alarco, A. M., and Raymond, M. (1999). The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181, 700–708.
Azie, N., Neofytos, D., Pfaller, M., Meier-Kriesche, H. U., Quan, S. P., and Horn, D. (2012). The PATH (Prospective Antifungal Therapy) Alliance(R) registry and invasive fungal infections: update 2012. Diagn. Microbiol. Infect. Dis. 73, 293–300. doi: 10.1016/j.diagmicrobio.2012.06.012
Barchiesi, F., Calabrese, D., Sanglard, D., Falconi Di Francesco, L., Caselli, F., Giannini, D., et al. (2000). Experimental induction of fluconazole resistance in Candida tropicalis ATCC 750. Antimicrob. Agents Chemother. 44, 1578–1584. doi: 10.1128/AAC.44.6.1578-1584.2000
Bard, M., Sturm, A. M., Pierson, C. A., Brown, S., Rogers, K. M., Nabinger, S., et al. (2005). Sterol uptake in Candida glabrata: rescue of sterol auxotrophic strains. Diagn. Microbiol. Infect. Dis. 52, 285–293. doi: 10.1016/j.diagmicrobio.2005.03.001
Bennett, J. E., Izumikawa, K., and Marr, K. A. (2004). Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob. Agents Chemother. 48, 1773–1777. doi: 10.1128/AAC.48.5.1773-1777.2004
Berberi, A., Noujeim, Z., and Aoun, G. (2015). Epidemiology of oropharyngeal candidiasis in human immunodeficiency virus/acquired immune deficiency syndrome patients and CD4+ counts. J. Int. Oral. Health 7, 20–23.
Berkow, E. L., Manigaba, K., Parker, J. E., Barker, K. S., Kelly, S. L., and Rogers, P. D. (2015). Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob. Agents Chemother. 59, 5942–5950. doi: 10.1128/AAC.01358-15
Blot, S., Janssens, R., Claeys, G., Hoste, E., Buyle, F., De Waele, J. J., et al. (2006). Effect of fluconazole consumption on long-term trends in candidal ecology. J. Antimicrob. Chemother. 58, 474–477. doi: 10.1093/jac/dkl241
Bouchara, J. P., Zouhair, R., Le Boudouil, S., Renier, G., Filmon, R., Chabasse, D., et al. (2000). In-vivo selection of an azole-resistant petite mutant of Candida glabrata. J. Med. Microbiol. 49, 977–984. doi: 10.1099/0022-1317-49-11-977
Branco, J., Silva, A. P., Silva, R. M., Silva-Dias, A., Pina-Vaz, C., Butler, G., et al. (2015). Fluconazole and voriconazole resistance in Candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob. Agents Chemother. 59, 6629–6633. doi: 10.1128/AAC.00842-15
Brun, S., Aubry, C., Lima, O., Filmon, R., Bergès, T., Chabasse, D., et al. (2003). Relationships between respiration and susceptibility to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 47, 847–853. doi: 10.1128/AAC.47.3.847-853.2003
Brun, S., Berges, T., Poupard, P., Vauzelle-Moreau, C., Renier, G., Chabasse, D., et al. (2004). Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48, 1788–1796. doi: 10.1128/AAC.48.5.1788-1796.2004
Castanheira, M., Messer, S. A., Rhomberg, P. R., Dietrich, R. R., Jones, R. N., and Pfaller, M. A. (2014). Isavuconazole and nine comparator antifungal susceptibility profiles for common and uncommon Candida species collected in 2012: application of new CLSI clinical breakpoints and epidemiological cutoff values. Mycopathologia 178, 1–9. doi: 10.1007/s11046-014-9772-2
Caudle, K. E., Barker, K. S., Wiederhold, N. P., Xu, L., Homayouni, R., and Rogers, P. D. (2011). Genomewide expression profile analysis of the Candida glabrata Pdr1 regulon. Eukaryot. Cell 10, 373–383. doi: 10.1128/EC.00073-10
Chapeland-Leclerc, F., Hennequin, C., Papon, N., Noël, T., Girard, A., Socie, G., et al. (2010). Acquisition of flucytosine, azole, and caspofungin resistance in Candida glabrata bloodstream isolates serially obtained from a hematopoietic stem cell transplant recipient. Antimicrob. Agents Chemother. 54, 1360–1362. doi: 10.1128/AAC.01138-09
Chau, A. S., Gurnani, M., Hawkinson, R., Laverdiere, M., Cacciapuoti, A., and McNicholas, P. M. (2005). Inactivation of sterol Δ5,6-desaturase attenuates virulence in Candida albicans. Antimicrob. Agents Chemother. 49, 3646–3651. doi: 10.1128/AAC.49.9.3646-3651.2005
Chen, K. H., Chang, C. T., Yu, C. C., Huang, J. Y., Yang, C. W., and Hung, C. C. (2006). Candida parapsilosis peritonitis has more complications than other Candida peritonitis in peritoneal dialysis patients. Ren. Fail. 28, 241–246. doi: 10.1080/08860220600580431
Cho, E. J., Shin, J. H., Kim, S. H., Kim, H. K., Park, J. S., Sung, H., et al. (2015). Emergence of multiple resistance profiles involving azoles, echinocandins and amphotericin B in Candida glabrata isolates from a neutropenia patient with prolonged fungaemia. J. Antimicrob. Chemother. 70, 1268–1270. doi: 10.1093/jac/dku518
Choi, M. J., Won, E. J., Shin, J. H., Kim, S. H., Lee, W. G., Kim, M. N., et al. (2016). Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with fluconazole-less-susceptible isolates. Antimicrob. Agents Chemother. 60, 3653–3661. doi: 10.1128/AAC.02652-15
Cleveland, A. A., Harrison, L. H., Farley, M. M., Hollick, R., Stein, B., Chiller, T. M., et al. (2015). Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008-2013: results from population-based surveillance. PLoS ONE 10:e0120452. doi: 10.1371/journal.pone.0120452
Clinical Laboratory Standards Institute (2012). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Fourth Informational Supplement, M27-S4. Wayne, PA: CLSI.
Cornely, O. A., Gachot, B., Akan, H., Bassetti, M., Uzun, O., Kibbler, C., et al. (2015). Epidemiology and outcome of fungemia in a cancer Cohort of the Infectious Diseases Group (IDG) of the European Organization for Research and Treatment of Cancer (EORTC 65031). Clin. Infect. Dis. 61, 324–331. doi: 10.1093/cid/civ293
Corsello, S., Spinillo, A., Osnengo, G., Penna, C., Guaschino, S., Beltrame, A., et al. (2003). An epidemiological survey of vulvovaginal candidiasis in Italy. Eur. J. Obstet. Gynecol. Reprod. Biol. 110, 66–72. doi: 10.1016/S0301-2115(03)00096-4
Costa, C., Ribeiro, J., Miranda, I. M., Silva-Dias, A., Cavalheiro, M., Costa-de-Oliveira, S., et al. (2016). Clotrimazole drug resistance in Candida glabrata clinical isolates correlates with increased expression of the drug:H(+) antiporters CgAqr1, CgTpo1, CgTpo3, and CgQdr2. Front. Microbiol. 7:526. doi: 10.3389/fmicb.2016.00526
Coste, A., Selmecki, A., Forche, A., Diogo, D., Bougnoux, M. E., d'Enfert, C., et al. (2007). Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryotic Cell 6, 1889–1904. doi: 10.1128/EC.00151-07
Coste, A. T., Karababa, M., Ischer, F., Bille, J., and Sanglard, D. (2004). TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3, 1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004
Coste, A., Turner, V., Ischer, F., Morschhäuser, J., Forche, A., Selmecki, A., et al. (2006). A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172, 2139–2156. doi: 10.1534/genetics.105.054767
Defontaine, A., Bouchara, J. P., Declerk, P., Planchenault, C., Chabasse, D., and Hallet, J. N. (1999). In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48, 663–670. doi: 10.1099/00222615-48-7-663
Diekema, D., Arbefeville, S., Boyken, L., Kroeger, J., and Pfaller, M. (2012). The changing epidemiology of healthcare-associated candidemia over three decades. Diagn. Microbiol. Infect. Dis. 73, 45–48. doi: 10.1016/j.diagmicrobio.2012.02.001
Dunkel, N., Blass, J., Rogers, P. D., and Morschhäuser, J. (2008b). Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69, 827–840. doi: 10.1111/j.1365-2958.2008.06309.x
Dunkel, N., Liu, T. T., Barker, K. S., Homayouni, R., Morschhäuser, J., and Rogers, P. D. (2008a). A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7, 1180–1190. doi: 10.1128/EC.00103-08
Eddouzi, J., Parker, J. E., Vale-Silva, L. A., Coste, A., Ischer, F., Kelly, S., et al. (2013). Molecular mechanisms of drug resistance in clinical Candida species isolated from Tunisian hospitals. Antimicrob. Agents Chemother. 57, 3182–3193. doi: 10.1128/AAC.00555-13
Enwuru, C. A., Ogunledun, A., Idika, N., Enwuru, N. V., Ogbonna, F., Aniedobe, M., et al. (2008). Fluconazole resistant opportunistic oropharyngeal Candida and non-Candida yeast-like isolates from HIV infected patients attending ARV clinics in Lagos, Nigeria. Afr. Health Sci. 8, 142–148.
Espinel-Ingroff, A., Pfaller, M. A., Bustamante, B., Canton, E., Fothergill, A., Fuller, J., et al. (2014). Multilaboratory study of epidemiological cutoff values for detection of resistance in eight Candida species to fluconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 58, 2006–2012. doi: 10.1128/AAC.02615-13
Ferrari, S., Ischer, F., Calabrese, D., Posteraro, B., Sanguinetti, M., Fadda, G., et al. (2009). Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. doi: 10.1371/journal.ppat.1000268
Ferrari, S., Sanguinetti, M., De Bernardis, F., Torelli, R., Posteraro, B., Vandeputte, P., et al. (2011). Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 55, 1852–1860. doi: 10.1128/AAC.01271-10
Fidel, P. L. Jr., Vazquez, J. A., and Sobel, J. D. (1999). Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12, 80–96.
Flowers, S. A., Barker, K. S., Berkow, E. L., Toner, G., Chadwick, S. G., Gygax, S. E., et al. (2012). Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 11, 1289–1299. doi: 10.1128/EC.00215-12
Flowers, S. A., Colón, B., Whaley, S. G., Schuler, M. A., and Rogers, P. D. (2015). Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob. Agents Chemother. 59, 450–460. doi: 10.1128/AAC.03470-14
Fothergill, A. W., Sutton, D. A., McCarthy, D. I., and Wiederhold, N. P. (2014). Impact of new antifungal breakpoints on antifungal resistance in Candida species. J. Clin. Microbiol. 52, 994–997. doi: 10.1128/JCM.03044-13
Garzoni, C., Nobre, V. A., and Garbino, J. (2007). Candida parapsilosis endocarditis: a comparative review of the literature. Eur. J. Clin. Microbiol. Infect. Dis. 26, 915–926. doi: 10.1007/s10096-007-0386-1
Gong, X., Luan, T., Wu, X., Li, G., Qiu, H., Kang, Y., et al. (2016). Invasive candidiasis in intensive care units in China: risk factors and prognoses of Candida albicans and non-albicans Candida infections. Am. J. Infect. Control 44, e59–e63. doi: 10.1016/j.ajic.2015.11.028
Goswami, D., Goswami, R., Banerjee, U., Dadhwal, V., Miglani, S., Lattif, A. A., et al. (2006). Pattern of Candida species isolated from patients with diabetes mellitus and vulvovaginal candidiasis and their response to single dose oral fluconazole therapy. J. Infect. 52, 111–117. doi: 10.1016/j.jinf.2005.03.005
Grossman, N. T., Pham, C. D., Cleveland, A. A., and Lockhart, S. R. (2015). Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob Agents Chemother. 59, 1030–1037. doi: 10.1128/AAC.04613-14
Guinea, J., Sánchez-Somolinos, M., Cuevas, O., Peláez, T., and Bouza, E. (2006). Fluconazole resistance mechanisms in Candida krusei: the contribution of efflux-pumps. Med. Mycol. 44, 575–578. doi: 10.1080/13693780600561544
Hachem, R., Hanna, H., Kontoyiannis, D., Jiang, Y., and Raad, I. (2008). The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 112, 2493–2499. doi: 10.1002/cncr.23466
He, X., Zhao, M., Chen, J., Wu, R., Zhang, J., Cui, R., et al. (2015). Overexpression of both ERG11 and ABC2 genes might be responsible for itraconazole resistance in clinical isolates of Candida krusei. PLoS ONE 10:e0136185. doi: 10.1371/journal.pone.0136185
Healey, K. R., Zhao, Y., Perez, W. B., Lockhart, S. R., Sobel, J. D., Farmakiotis, D., et al. (2016). Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 7:11128. doi: 10.1038/ncomms11128
Heilmann, C. J., Schneider, S., Barker, K. S., Rogers, P. D., and Morschhäuser, J. (2010). An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob. Agents Chemother. 54, 353–359. doi: 10.1128/AAC.01102-09
Holland, J., Young, M. L., Lee, O., and C-A Chen, S. (2003). Vulvovaginal carriage of yeasts other than Candida albicans. Sex. Transm. Infect. 79, 249–250. doi: 10.1136/sti.79.3.249
Hoot, S. J., Smith, A. R., Brown, R. P., and White, T. C. (2011). An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob Agents Chemother. 55, 940–942. doi: 10.1128/AAC.00995-10
Hope, W., Morton, A., and Eisen, D. P. (2002). Increase in prevalence of nosocomial non-Candida albicans candidaemia and the association of Candida krusei with fluconazole use. J. Hosp. Infect. 50, 56–65. doi: 10.1053/jhin.2001.1131
Hull, C. M., Parker, J. E., Bader, O., Weig, M., Gross, U., Warrilow, A. G., et al. (2012). Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob. Agents Chemother. 56, 4223–4232. doi: 10.1128/AAC.06253-11
Imhof, A., Balajee, S. A., Fredricks, D. N., Englund, J. A., and Marr, K. A. (2004). Breakthrough fungal infections in stem cell transplant recipients receiving voriconazole. Clin. Infect. Dis. 39, 743–746. doi: 10.1086/423274
Jiang, C., Dong, D., Yu, B., Cai, G., Wang, X., Ji, Y., et al. (2013). Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 68, 778–785. doi: 10.1093/jac/dks481
Katiyar, S. K., and Edlind, T. D. (2001). Identification and expression of multidrug resistance-related ABC transporter genes in Candida krusei. Med. Mycol. 39, 109–116. doi: 10.1080/mmy.39.1.109.116
Kelly, S. L., Lamb, D. C., and Kelly, D. E. (1997). Sterol 22-desaturase, cytochrome P45061, possesses activity in xenobiotic metabolism. FEBS Lett. 412, 233–235. doi: 10.1016/S0014-5793(97)00785-0
Klingspor, L., Tortorano, A. M., Peman, J., Willinger, B., Hamal, P., Sendid, B., et al. (2015). Invasive Candida infections in surgical patients in intensive care units: a prospective, multicentre survey initiated by the European Confederation of Medical Mycology (ECMM) (2006-2008). Clin. Microbiol. Infect. 21, 87.e1–87.e10 doi: 10.1016/j.cmi.2014.08.011
Kolaczkowska, A., and Kolaczkowski, M. (2016). Drug resistance mechanisms and their regulation in non-albicans Candida species. J. Antimicrob. Chemother. 71, 1438–1450. doi: 10.1093/jac/dkv445
Kothari, A., and Sagar, V. (2009). Epidemiology of candida bloodstream infections in a tertiary care institute in India. Ind. J. Med. Microbiol. 27, 171–172. doi: 10.4103/0255-0857.49440
Krcmery, V. Jr., Mrazova, M., Kunova, A., Grey, E., Mardiak, J., Jurga, L., et al. (1999). Nosocomial candidaemias due to species other than Candida albicans in cancer patients. Aetiology, risk factors, and outcome of 45 episodes within 10 years in a single cancer institution. Support. Care Cancer 7, 428–431. doi: 10.1007/s005200050304
Lamping, E., Ranchod, A., Nakamura, K., Tyndall, J. D., Niimi, K., Holmes, A. R., et al. (2009). Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei. Antimicrob. Agents Chemother. 53, 354–369. doi: 10.1128/AAC.01095-08
Lee, I., Fishman, N. O., Zaoutis, T. E., Morales, K. H., Weiner, M. G., Synnestvedt, M., et al. (2009). Risk factors for fluconazole-resistant Candida glabrata bloodstream infections. Arch. Intern. Med. 169, 379–383. doi: 10.1001/archinte.169.4.379
Lee, J. K., Peters, D., Obias, A. A., Noskin, G. A., and Peterson, L. R. (2000). Activity of voriconazole against Candida albicans and Candida krusei isolated since 1984. Int. J. Antimicrob. Agents 16, 205–209. doi: 10.1016/S0924-8579(00)00191-6
Lin, M. Y., Carmeli, Y., Zumsteg, J., Flores, E. L., Tolentino, J., Sreeramoju, P., et al. (2005). Prior antimicrobial therapy and risk for hospital-acquired Candida glabrata and Candida krusei fungemia: a case-case-control study. Antimicrob. Agents Chemother. 49, 4555–4560. doi: 10.1128/AAC.49.11.4555-4560.2005
Liu, T. T., Znaidi, S., Barker, K. S., Xu, L., Homayouni, R., Saidane, S., et al. (2007). Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot. Cell 6, 2122–2138. doi: 10.1128/EC.00327-07
Lortholary, O., Desnos-Ollivier, M., Sitbon, K., Fontanet, A., Bretagne, S., and Dromer, F. (2011). Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob. Agents Chemother. 55, 532–538. doi: 10.1128/AAC.01128-10
Lortholary, O., Renaudat, C., Sitbon, K., Madec, Y., Denoeud-Ndam, L., Wolff, M., et al. (2014). Worrisome trends in incidence and mortality of candidemia in intensive care units (Paris area, 2002-2010). Intensive Care Med. 40, 1303–1312. doi: 10.1007/s00134-014-3408-3
MacPherson, S., Akache, B., Weber, S., De Deken, X., Raymond, M., and Turcotte, B. (2005). Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 49, 1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005
Mahmoudi Rad, M., Zafarghandi, A., Amel Zabihi, M., Tavallaee, M., and Mirdamadi, Y. (2012). Identification of Candida species associated with vulvovaginal candidiasis by multiplex PCR. Infect. Dis. Obstet. Gynecol. 2012:872169. doi: 10.1155/2012/872169
Mansfield, B. E., Oltean, H. N., Oliver, B. G., Hoot, S. J., Leyde, S. E., Hedstrom, L., et al. (2010). Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 6:e1001126. doi: 10.1371/journal.ppat.1001126
Manzano-Gayosso, P., Hernández-Hernández, F., Mendez-Tovar, L. J., González- Monroy, J., and López-Martínez, R. (2003). Fungal peritonitis in 15 patients on continuous ambulatory peritoneal dialysis (CAPD). Mycoses 46, 425–429. doi: 10.1046/j.0933-7407.2003.00922.x
Marichal, P., Gorrens, J., Coene, M. C., Le Jeune, L., and Vanden Bossche, H. (1995). Origin of differences in susceptibility of Candida krusei to azole antifungal agents. Mycoses 38, 111–117. doi: 10.1111/j.1439-0507.1995.tb00032.x
Marichal, P., Koymans, L., Willemsens, S., Bellens, D., Verhasselt, P., et al. (1999). Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145 (Pt 10), 2701–2713. doi: 10.1099/00221287-145-10-2701
Marichal, P., Vanden Bossche, H., Odds, F. C., Nobels, G., Warnock, D. W., Timmerman, V., et al. (1997). Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41, 2229–2237.
Martel, C. M., Parker, J. E., Bader, O., Weig, M., Gross, U., Warrilow, A. G., et al. (2010). Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54, 4527–4533. doi: 10.1128/AAC.00348-10
Martí-Carrizosa, M., Sánchez-Reus, F., March, F., and Coll, P. (2014). Fungemia in a Spanish hospital: the role of Candida parapsilosis over a 15-year period. Scand. J. Infect. Dis. 46, 454–461. doi: 10.3109/00365548.2014.900190
Merz, W. G., Karp, J. E., Schron, D., and Saral, R. (1986). Increased incidence of fungemia caused by Candida krusei. J. Clin. Microbiol. 24, 581–584.
Milazzo, L., Peri, A. M., Mazzali, C., Grande, R., Cazzani, C., Ricaboni, D., et al. (2014). Candidaemia observed at a university hospital in Milan (northern Italy) and review of published studies from 2010 to 2014. Mycopathologia 178, 227–241. doi: 10.1007/s11046-014-9786-9
Miyazaki, H., Miyazaki, Y., Geber, A., Parkinson, T., Hitchcock, C., Falconer, D. J., et al. (1998). Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42, 1695–1701.
Miyazaki, Y., Geber, A., Miyazaki, H., Falconer, D., Parkinson, T., Hitchcock, C., et al. (1999). Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene 236, 43–51. doi: 10.1016/S0378-1119(99)00263-2
Morii, D., Seki, M., Binongo, J. N., Ban, R., Kobayashi, A., Sata, M., et al. (2014). Distribution of Candida species isolated from blood cultures in hospitals in Osaka, Japan. J. Infect. Chemother. 20, 558–562. doi: 10.1016/j.jiac.2014.05.009
Morio, F., Pagniez, F., Lacroix, C., Miegeville, M., and Le Pape, P. (2012). Amino acid substitutions in the Candida albicans sterol Δ5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J. Antimicrob. Chemother. 67, 2131–2138. doi: 10.1093/jac/dks186
Morschhäuser, J., Barker, K. S., Liu, T. T., BlaB-Warmuth, J., Homayouni, R., and Rogers, P. D. (2007). The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. doi: 10.1371/journal.ppat.0030164
Nakayama, H., Izuta, M., Nakayama, N., Arisawa, M., and Aoki, Y. (2000). Depletion of the squalene synthase (ERG9) gene does not impair growth of Candida glabrata in mice. Antimicrob. Agents Chemother. 44, 2411–2418. doi: 10.1128/AAC.44.9.2411-2418.2000
Nakayama, H., Tanabe, K., Bard, M., Hodgson, W., Wu, S., Takemori, D., et al. (2007). The Candida glabrata putative sterol transporter gene CgAUS1 protects cells against azoles in the presence of serum. J. Antimicrob. Chemother. 60, 1264–1272. doi: 10.1093/jac/dkm321
Nolte, F. S., Parkinson, T., Falconer, D. J., Dix, S., Williams, J., Gilmore, C., et al. (1997). Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41, 196–199.
Nucci, M., and Colombo, A. L. (2007). Candidemia due to Candida tropicalis: clinical, epidemiologic, and microbiologic characteristics of 188 episodes occurring in tertiary care hospitals. Diagn. Microbiol. Infect. Dis. 58, 77–82. doi: 10.1016/j.diagmicrobio.2006.11.009
Orozco, A. S., Higginbotham, L. M., Hitchcock, C. A., Parkinson, T., Falconer, D., Ibrahim, A. S., et al. (1998). Mechanism of fluconazole resistance in Candida krusei. Antimicrob. Agents Chemother. 42, 2645–2649.
Oxman, D. A., Chow, J. K., Frendl, G., Hadley, S., Hershkovitz, S., Ireland, P., et al. (2010). Candidaemia associated with decreased in vitro fluconazole susceptibility: is Candida speciation predictive of the susceptibility pattern? J. Antimicrob. Chemother. 65, 1460–1465. doi: 10.1093/jac/dkq136
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 62, e1–e50. doi: 10.1093/cid/civ1194
Paul, S., Bair, T. B., and Moye-Rowley, W. S. (2014). Identification of genomic binding sites for Candida glabrata Pdr1 transcription factor in wild-type and rho0 cells. Antimicrob. Agents Chemother. 58, 6904–6912. doi: 10.1128/AAC.03921-14
Paul, S., Schmidt, J. A., and Moye-Rowley, W. S. (2011). Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata. Eukaryot. Cell 10, 187–197. doi: 10.1128/EC.00277-10
Pfaller, M. A., Andes, D. R., Diekema, D. J., Horn, D. L., Reboli, A. C., Rotstein, C., et al. (2014a). Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004-2008. PLoS ONE 9:e101510. doi: 10.1371/journal.pone.0101510
Pfaller, M. A., Castanheira, M., Lockhart, S. R., Ahlquist, A. M., Messer, S. A., and Jones, R. N. (2012a). Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J. Clin. Microbiol. 50, 1199–1203. doi: 10.1128/JCM.06112-11
Pfaller, M. A., and Diekema, D. J. (2012b). Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J. Clin. Microbiol. 50, 2846–2856. doi: 10.1128/JCM.00937-12
Pfaller, M. A., Diekema, D. J., Gibbs, D. L., Newell, V. A., Ellis, D., Tullio, V., et al. (2010). Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida Species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48, 1366–1377. doi: 10.1128/JCM.02117-09
Pfaller, M. A., Diekema, D. J., Gibbs, D. L., Newell, V. A., Nagy, E., Dobiasova, S., et al. (2008). Candida krusei, a multidrug-resistant opportunistic fungal pathogen: geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J. Clin. Microbiol. 46, 515–521. doi: 10.1128/JCM.01915-07
Pfaller, M. A., Jones, R. N., and Castanheira, M. (2014b). Regional data analysis of Candida non-albicans strains collected in United States medical sites over a 6-year period, 2006-2011. Mycoses 57, 602–611. doi: 10.1111/myc.12206
Pfaller, M. A., Messer, S. A., Woosley, L. N., Jones, R. N., and Castanheira, M. (2013). Echinocandin and triazole antifungal susceptibility profiles for clinical Opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J. Clin. Microbiol. 51, 2571–2581. doi: 10.1128/JCM.00308-13
Pfaller, M. A., Rhomberg, P. R., Messer, S. A., Jones, R. N., and Castanheira, M. (2015). Isavuconazole, micafungin, and 8 comparator antifungal agents' susceptibility profiles for common and uncommon opportunistic fungi collected in 2013: temporal analysis of antifungal drug resistance using CLSI species-specific clinical breakpoints and proposed epidemiological cutoff values. Diagn. Microbiol. Infect. Dis. 82, 303–313. doi: 10.1016/j.diagmicrobio.2015.04.008
Ray, D., Goswami, R., Banerjee, U., Dadhwal, V., Goswami, D., Mandal, P., et al. (2007). Prevalence of Candida glabrata and its response to boric acid vaginal suppositories in comparison with oral fluconazole in patients with diabetes and vulvovaginal candidiasis. Diabetes Care 30, 312–317. doi: 10.2337/dc06-1469
Redding, S. W., Kirkpatrick, W. R., Saville, S., Coco, B. J., White, W., Fothergill, A., et al. (2003). Multiple patterns of resistance to fluconazole in Candida glabrata isolates from a patient with oropharyngeal candidiasis receiving head and neck radiation. J. Clin. Microbiol. 41, 619–622. doi: 10.1128/JCM.41.2.619-622.2003
Ricardo, E., Miranda, I. M., Faria-Ramos, I., Silva, R. M., Rodrigues, A. G., and Pina-Vaz, C. (2014). In vivo and in vitro acquisition of resistance to voriconazole by Candida krusei. Antimicrob. Agents Chemother. 58, 4604–4611. doi: 10.1128/AAC.02603-14
Richter, S. S., Galask, R. P., Messer, S. A., Hollis, R. J., Diekema, D. J., and Pfaller, M. A. (2005). Antifungal susceptibilities of Candida species causing vulvovaginitis and epidemiology of recurrent cases. J. Clin. Microbiol. 43, 2155–2162. doi: 10.1128/JCM.43.5.2155-2162.2005
Rubio, M. C., de Ocáriz, I. R., Gil, J., Benito, R., and Rezusta, A. (2005). Potential fungicidal effect of voriconazole against Candida spp. Int. J. Antimicrob. Agents 25, 264–267. doi: 10.1016/j.ijantimicag.2004.11.007
Rybak, J. M., Marx, K. R., Nishimoto, A. T., and Rogers, P. D. (2015). Isavuconazole: pharmacology, pharmacodynamics, and current clinical experience with a new triazole antifungal agent. Pharmacotherapy 35, 1037–1051. doi: 10.1002/phar.1652
Sanglard, D., Ischer, F., and Bille, J. (2001). Role of ATP-binding- cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45, 1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001
Sanglard, D., Ischer, F., Calabrese, D., Majcherczyk, P. A., and Bille, J. (1999). The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43, 2753–2765.
Sanguinetti, M., Posteraro, B., Fiori, B., Ranno, S., Torelli, R., and Fadda, G. (2005). Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob. Agents Chemother. 49, 668–679. doi: 10.1128/AAC.49.2.668-679.2005
Selmecki, A., Forche, A., and Berman, J. (2006). Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313, 367–370. doi: 10.1126/science.1128242
Selmecki, A., Gerami-Nejad, M., Paulson, C., Forsche, A., and Berman, J. (2008). An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68, 624–641. doi: 10.1111/j.1365-2958.2008.06176.x
Sharifzadeh, A., Khosravi, A. R., Shokri, H., Asadi Jamnani, F., Hajiabdolbaghi, M., and Ashrafi Tamami, I. (2013). Oral microflora and their relation to risk factors in HIV+ patients with oropharyngeal candidiasis. J. Mycol. Med. 23, 105–112. doi: 10.1016/j.mycmed.2013.02.001
Silva, A. P., Miranda, I. M., Guida, A., Synnott, J., Rocha, R., Silva, R., et al. (2011). Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrob. Agents Chemother. 55, 3546–3556. doi: 10.1128/AAC.01127-10
Silva, D. B., Rodrigues, L. M., Almeida, A. A., Oliveira, K. M., and Grisolia, A. B. (2016). Novel point mutations in the ERG11 gene in clinical isolates of azole resistant Candida species. Mem. Inst. Oswaldo Cruz 111, 192–199. doi: 10.1590/0074-02760150400
Sobel, J. D. (2010). Changing trends in the epidemiology of Candida blood stream infections: a matter for concern? Crit. Care Med. 38, 990–992. doi: 10.1097/CCM.0b013e3181d16866
Sobel, J. D., Kauffman, C. A., McKinsey, D., Zervos, M., Vazquez, J. A., Karchmer, A. W., et al. (2000). Candiduria: a randomized, double-blind study of treatment with fluconazole and placebo. National Institute of Allergy and Infectious Diseases (NIAID) Mycoses Study Group. Clin. Infect. Dis. 30, 19–24. doi: 10.1086/313580
Souza, A. C., Fuchs, B. B., Pinhati, H. M., Sigueira, R. A., Hagen, F., Meis, J. F., et al. (2015). Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella larvae. Antimicrob. Agents Chemother. 59, 6581–6587. doi: 10.1128/AAC.01177-15
Tan, J., Zhang, J., Chen, W., Sun, Y., Wan, Z., Li, R., et al. (2015). The A395T mutation in ERG11 gene confers fluconazole resistance in Candida tropicalis causing candidemia. Mycopathologia 179, 213–218. doi: 10.1007/s11046-014-9831-8
Tang, H. J., Liu, W. L., Lin, H. L., and Lai, C. C. (2014). Epidemiology and prognostic factors of candidemia in cancer patients. PLoS ONE 9:e99103. doi: 10.1371/journal.pone.0099103
Tang, J. L., Kung, H. C., Lei, W. C., Yao, M., Wu, U. I., Hsu, S. C., et al. (2015). High incidences of invasive fungal infections in acute myeloid leukemia patients receiving induction chemotherapy without systemic antifungal prophylaxis: a prospective observational study in Taiwan. PLoS ONE 10:e0128410. doi: 10.1371/journal.pone.0128410
Tavakoli, M., Zaini, F., Kordbacheh, M., Safara, M., Raoofian, R., and Heidari, M. (2010). Upregulation of the ERG11 gene in Candida krusei by azoles. Daru 18, 276–280.
Torelli, R., Posteraro, B., Ferrari, S., La Sorda, M., Fadda, G., Sanglard, D., et al. (2008). The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol. Microbiol. 68, 186–201. doi: 10.1111/j.1365-2958.2008.06143.x
Tsai, H. F., Bard, M., Izumikawa, K., Krol, A. A., Sturm, A. M., Culbertson, N. T., et al. (2004). Candida glabrata erg1 mutant with increased sensitivity to azoles and to low oxygen tension. Antimicrob. Agents Chemother. 48, 2483–2489. doi: 10.1128/AAC.48.7.2483-2489.2004
Tsai, H. F., Krol, A. A., Sarti, K. E., and Bennett, J. E. (2006). Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50, 1384–1392. doi: 10.1128/AAC.50.4.1384-1392.2006
Tsai, H. F., Sammons, L. R., Zhang, X., Suffis, S. D., Su, Q., Myers, T. G., et al. (2010). Microarray and molecular analyses of the azole resistance mechanism in Candida glabrata oropharyngeal isolates. Antimicrob. Agents Chemother. 54, 3308–3317. doi: 10.1128/AAC.00535-10
vanden Bossche, H., Marichal, P., Odds, F. C., Le Jeune, L., and Coene, M. C. (1992). Characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 36, 2602–2610. doi: 10.1128/AAC.36.12.2602
Vandeputte, P., Larcher, G., Bergès, T., Renier, G., Chabasse, D., and Bouchara, J. P. (2005). Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob. Agents Chemother. 49, 4608–4615. doi: 10.1128/AAC.49.11.4608-4615.2005
Venkateswarlu, K., Denning, D. W., and Kelly, S. L. (1997). Inhibition and interaction of cytochrome P450 of Candida krusei with azole antifungal drugs. J. Med. Vet. Mycol. 35, 19–25. doi: 10.1080/02681219780000821
Venkateswarlu, K., Denning, D. W., Manning, N. J., and Kelly, S. L. (1996). Reduced accumulation of drug in Candida krusei accounts for itraconazole resistance. Antimicrob. Agents Chemother. 40, 2443–2446.
Vermitsky, J. P., and Edlind, T. D. (2004). Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48, 3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004
Vermitsky, J. P., Self, M. J., Chadwick, S. G., Trama, J. P., Adelson, M. E., Mordechai, E., et al. (2008). Survey of vaginal-flora Candida species isolates from women of different age groups by use of species-specific PCR detection. J. Clin. Microbiol. 46, 1501–1503. doi: 10.1128/JCM.02485-07
Wang, E., Farmakiotis, D., Yang, D., McCue, D. A., Kantarjian, H. M., Kontoyiannis, D. P., et al. (2015). The ever-evolving landscape of candidaemia in patients with acute leukaemia: non-susceptibility to caspofungin and multidrug resistance are associated with increased mortality. J. Antimicrob. Chemother. 70, 2362–2368. doi: 10.1093/jac/dkv087
Weems, J. J. Jr. (1992). Candida parapsilosis: epidemiology, pathogenicity, clinical manifestations, and antimicrobial susceptibility. Clin. Infect. Dis. 14, 756–766. doi: 10.1093/clinids/14.3.756
Weinberger, M., Leibovici, L., Perez, S., Samra, Z., Ostfeld, I., Levi, I., et al. (2005). Characteristics of candidaemia with Candida albicans compared with non-albicans Candida species and predictors of mortality. J. Hosp. Infect. 61, 146–154. doi: 10.1016/j.jhin.2005.02.009
Wingard, J. R., Merz, W. G., Rinaldi, M. G., Johnson, T. R., Karp, J. E., and Saral, R. (1991). Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N.Engl. J. Med. 325, 1274–1277. doi: 10.1056/NEJM199110313251803
Xiang, M. J., Liu, J. Y., Ni, P. H., Wang, S., Shi, C., Wei, B., et al. (2013). Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 13, 386–393. doi: 10.1111/1567-1364.12042
Yang, Y. L., Ho, Y. A., Cheng, H. H., Ho, M., and Lo, H. J. (2004). Susceptibilities of Candida species to amphotericin B and fluconazole: the emergence of fluconazole resistance in Candida tropicalis. Infect. Control Hosp. Epidemiol. 25, 60–64. doi: 10.1086/502294
Yang, Y. L., Wang, A. H., Wang, C. W., Cheng, W. T., Li, S. Y., Lo, H. J., et al. (2008). Susceptibilities to amphotericin B and fluconazole of Candida species in Taiwan Surveillance of Antimicrobial Resistance of Yeasts 2006. Diagn. Microbiol. Infect. Dis. 61, 175–180. doi: 10.1016/j.diagmicrobio.2008.01.011
Ying, Y., Zhang, J., Huang, S. B., Liu, F. D., Liu, J. H., Zhang, J., et al. (2015). Fluconazole susceptibility of 3,056 clinical isolates of Candida species from 2005 to 2009 in a tertiary-care hospital. Indian J. Med. Microbiol. 33, 413–415. doi: 10.4103/0255-0857.158569
Yoo, J. I., Choi, C. W., Lee, K. M., Kim, Y. K., Kim, T. U., Kim, E. C., et al. (2009). National surveillance of antifungal susceptibility of Candida species in South Korean hospitals. Med. Mycol. 47, 554–558. doi: 10.1080/13693780802354037
Zavrel, M., Hoot, S. J., and White, T. C. (2013). Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot. Cell 12, 725–738. doi: 10.1128/EC.00345-12
Keywords: antifungal, azole, resistance, Candida albicans, Candida parapsilosis, Candida glabrata, Candida tropicalis, Candida krusei
Citation: Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS and Rogers PD (2017) Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 7:2173. doi: 10.3389/fmicb.2016.02173
Received: 21 September 2016; Accepted: 28 December 2016;
Published: 12 January 2017.
Edited by:
Dominique Sanglard, University of Lausanne, SwitzerlandReviewed by:
Miguel Cacho Teixeira, Universidade de Lisboa, PortugalCopyright © 2017 Whaley, Berkow, Rybak, Nishimoto, Barker and Rogers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: P. David Rogers, cHJvZ2VyczNAdXRoc2MuZWR1
†Present Address: Elizabeth Berkow, Fungal Service Team, Mycotic Diseases Branch, Centers for Disease Control and Prevention, Atlanta, GA, USA
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.