
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 January 2017
Sec. Extreme Microbiology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.02132
This article is part of the Research Topic Recent Advances in Acidophile Microbiology: Fundamentals and Applications View all 11 articles
 Mark Dopson1
Mark Dopson1 David S. Holmes2,3
David S. Holmes2,3 Marcelo Lazcano2,3
Marcelo Lazcano2,3 Timothy J. McCredden4
Timothy J. McCredden4 Christopher G. Bryan4
Christopher G. Bryan4 Kieran T. Mulroney4
Kieran T. Mulroney4 Robert Steuart4
Robert Steuart4 Connie Jackaman4
Connie Jackaman4 Elizabeth L. J. Watkin4*
Elizabeth L. J. Watkin4*Extremely acidophilic microorganisms (pH optima for growth of ≤3) are utilized for the extraction of metals from sulfide minerals in the industrial biotechnology of “biomining.” A long term goal for biomining has been development of microbial consortia able to withstand increased chloride concentrations for use in regions where freshwater is scarce. However, when challenged by elevated salt, acidophiles experience both osmotic stress and an acidification of the cytoplasm due to a collapse of the inside positive membrane potential, leading to an influx of protons. In this study, we tested the ability of the halotolerant acidophile Acidihalobacter prosperus to grow and catalyze sulfide mineral dissolution in elevated concentrations of salt and identified chloride tolerance mechanisms in Ac. prosperus as well as the chloride susceptible species, Acidithiobacillus ferrooxidans. Ac. prosperus had optimum iron oxidation at 20 g L−1 NaCl while At. ferrooxidans iron oxidation was inhibited in the presence of 6 g L−1 NaCl. The tolerance to chloride in Ac. prosperus was consistent with electron microscopy, determination of cell viability, and bioleaching capability. The Ac. prosperus proteomic response to elevated chloride concentrations included the production of osmotic stress regulators that potentially induced production of the compatible solute, ectoine uptake protein, and increased iron oxidation resulting in heightened electron flow to drive proton export by the F0F1 ATPase. In contrast, At. ferrooxidans responded to low levels of Cl− with a generalized stress response, decreased iron oxidation, and an increase in central carbon metabolism. One potential adaptation to high chloride in the Ac. prosperus Rus protein involved in ferrous iron oxidation was an increase in the negativity of the surface potential of Rus Form I (and Form II) that could help explain how it can be active under elevated chloride concentrations. These data have been used to create a model of chloride tolerance in the salt tolerant and susceptible species Ac. prosperus and At. ferrooxidans, respectively.
Acidophilic and extremely acidophilic microorganisms have pH optima for growth of ≤5 and ≤3, respectively, and comprise a phylogenetically and phenotypically diverse group of microorganisms from all three domains of life (reviewed in Aguilera et al., 2016; Dopson, 2016; Golyshina et al., 2016). As a group, they comprise species across wide temperature ranges for growth (from eurypsychrophilic to thermophiles), with the ability to exploit organic and/or inorganic electron donors and carbon sources, and can utilize molecular oxygen, ferric iron, and sulfate as electron acceptors. Acidophilic microorganisms have generated considerable interest as: (i) they catalyze the dissolution of sulfide minerals for recovery of valuable metals, termed “biomining” or “bioleaching” (Vera et al., 2013); (ii) they can cause uncontrolled sulfide mineral oxidation leading to the release of toxic, acidic and metal containing waters, called “acid mine drainage” (Mendez-Garcia et al., 2015); (iii) they are a source of extremozymes for use in biotechnologies (Elleuche et al., 2014); (iv) liposomes from these species have been investigated as a method for drug delivery (Jensen et al., 2015); and (v) these microorganisms may be analogs for early life on earth and potential life on other planets (Bauermeister et al., 2014).
Acidithiobacillus ferrooxidans was the first microorganism recognized to generate acid mine drainage (Colmer and Hinkle, 1947) and has since been identified in many acidic environments playing an important role during heap bioleaching of sulfide minerals. At. ferrooxidans fixes carbon dioxide for cellular carbon and couples ferrous iron, inorganic sulfur compound, and hydrogen oxidation to the reduction of either molecular oxygen or ferric iron. The type strain genome sequence is available (Valdes et al., 2008) and the genetic basis of many aspects of its metabolism has been elucidated (Osorio et al., 2008, 2013; Quatrini et al., 2009; Esparza et al., 2010; Ponce et al., 2012). Acidihalobacter prosperus (originally described as “Thiobacillus prosperus”) is another autotrophic and acidophilic species capable of growth via oxidation of ferrous iron and inorganic sulfur compounds (Huber and Stetter, 1989; Cardenas et al., 2015). The Ac. prosperus type strain was isolated from a volcanic marine environment and is halotolerant, being able to grow in chloride concentrations from 0.04 to 0.6 M (2.3–35 g L−1; Nicolle et al., 2009). The underlying mechanisms for Ac. prosperus growth are far less well-understood than for At. ferrooxidans, although the recent publication of its genome sequence (Ossandon et al., 2014) now aids investigation of this species.
Acidophiles employ a number of methods to maintain their intracellular pH near to neutral despite a proton ion gradient of up to 10,000 fold across the cytoplasmic membrane (reviewed in Slonczewski et al., 2009; Zammit and Watkin, 2016). These mechanisms include: (i) primary proton pumps that export protons during electron transport; (ii) cytoplasmic membranes that are highly resistant to the influx of protons; (iii) an inside positive membrane potential that creates a chemiosmotic gradient that reduces proton influx by electrostatic repulsion; (iv) alterations in cytoplasmic membrane structure; (v) proton-consuming reactions such as carboxylases; and (vi) cytoplasmic buffering. Osmotic stress occurs when the soluble extracellular salts differ from the concentration within the cell that either leads to cellular dehydration or lysis (Zammit and Watkin, 2016). Typical acidophile biomining strains are highly sensitive to anions and in particular chloride that have been demonstrated to inhibit ferrous iron oxidation by a Leptospirillum ferriphilum-dominated culture (Gahan et al., 2010) and the bioleaching ability of an undefined mixed acidophile consortium (Shiers et al., 2005). One exception is the salt tolerant industrial isolate, L. ferriphilum Sp-Cl and its genome sequence will aid in discovering adaptations to high salt concentrations in acidophiles (Issotta et al., 2016). The greater sensitivity to the membrane permeable anion chloride is due to its ability to cross the cell membrane. This reduces the transmembrane potential resulting in an influx of protons and acidification of the cytoplasm (Suzuki et al., 1999). Other anions such as , and cations such as Na+, have little effect beyond their impact on osmotic potential (Blight and Ralph, 2004; Shiers et al., 2005; Davis-Belmar et al., 2008; Rea et al., 2015; Boxall et al., 2016).
The isolation and investigation of halotolerant/halophilic acidophiles has long been a goal due to their potential use for biomining in countries where freshwater is limited and the use of seawater would be beneficial (Zammit et al., 2012; Rea et al., 2015). The major constituents of standard seawater are; Cl− (0.56M) and Na+ (0.48M) with at much lower concentrations (0.03M; Millero et al., 2008). Given the sensitivity of acidophiles to Cl−, those to be utilized in biomining with seawater must be able to tolerate the dual stresses of low pH and high Cl− concentrations. Adaptations to high salt concentrations exhibited by halophilic/halotolerant microorganisms include: (i) accumulation of cytoplasmic potassium; production of osmo-protectants in the cytoplasm to maintain an even turgor pressure inside and outside of the cell; (ii) alterations in the cell membrane, and (iii) an increase in acidic amino acids on the surface of proteins resulting in an elevated negative potential that aids in keeping the protein in solution (Shivanand and Mugeraya, 2011; Oren, 2013; Graziano and Merlino, 2014). In addition, changes in the surface electrostatic potential of a halophilic/halotolerant electron transport proteins is likely to affect their interactions with redox partners as has been shown for the interaction of the blue copper protein amicyanin with methylamine dehydrogenase (Ma et al., 2007; Choi et al., 2011). The combined effect of low pH and an anion such as chloride is to collapse the inside positive membrane potential involved in pH homeostasis (Alexander et al., 1987; Suzuki et al., 1999). However, the mechanisms halo-acidophiles utilize to combat these combined stresses are poorly understood and the majority of the studies to date have focused on species susceptible to increased salt while halotolerant acidophiles have been neglected.
Acidophilic bacteria have proven to be recalcitrant to the development of genetic methods, such as the creation of knockout mutants, and such techniques are only recently becoming more common (Wen et al., 2014; Yu et al., 2014). As a result, many acidophile studies have utilized “omics” techniques, including proteomics to investigate not only whole communities (Belnap et al., 2011; Mueller et al., 2011; Goltsman et al., 2013) but also specific responses in a single species (Baker-Austin et al., 2010; Mykytczuk et al., 2011; Potrykus et al., 2011; Mangold et al., 2013). In this study, iron oxidation and biomining studies along with a proteomic strategy are used to investigate the differing responses to chloride by the salt susceptible and tolerant acidophiles At. ferrooxidans and Ac. prosperus, respectively.
At. ferrooxidans ATCC 23270T and Ac. prosperus DSM 5130T were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) and grown under the following conditions. At. ferrooxidansT was cultured in pH 1.8 basal salts medium (BSM) (Plumb et al., 2002) and Ac. prosperusT in DSMZ media 477 with 12.5 g/L NaCl (pH 2.5). Filter sterilized (0.2 μm Minisart, Sartorius Stedim) substrates (50 mM FeSO4·7 H2O and 5 mM K2S4O6) were added to both media. Cultures were incubated on a rotary shaker at 120 rpm at 30°C. The effect of NaCl on the iron oxidizing activity of At. ferrooxidansT and Ac. prosperusT was investigated by addition of NaCl to the media to achieve the required Cl− concentrations. Cells were counted using a Helber bacteria counting chamber (Hawksley) at 400-fold magnification. Iron(II) oxidation was determined in At. ferrooxidansT using spectrophotometry (Govender et al., 2012) and in Ac. prosperusT experiments by titration against CeSO4 (Dopson and Lindström, 1999).
At. ferrooxidansT and Ac. prosperusT cells (80 mL) were removed from log phase planktonic grown cultures, filtered (8.0 μm pore size nitrocellulose filters; Miltex™) to remove iron hydroxysulfate precipitate, and concentrated by centrifugation for 20 min at 48,000 × g at 4°C. Cell pellets were washed and resuspended with growth media and then 5.0 × 106 cells were collected by centrifugation for 20 min at 48,000 × g at 4°C and resuspended in 100 μL of the appropriate growth media. Of this concentrated culture, 30 μL was pipetted onto an A1 SEM aluminum stub and incubated at 37°C for 10–40 min (until the surface appeared barely dry). Twenty Five microliters of 2.5% gluteraldehyde in BSM pH 2.5 was pipetted onto the surface of the stubs and then incubated at 4°C for 3 h. The sample stub was then washed by gently pipetting Invitrogen Gibco Ultrapure Distilled Water over the surface. Samples were dehydrated by sequential 30 min incubations with 70, 90, and 100% ethanol at 37°C before being transferred to a desiccator for 24 h. Stubs were coated with a 5 nm layer of platinum and imaged using a Zeiss Neon 40ESB Crossbeam Electron Microscope. Cell debris was differentiated from inorganic precipitates using SEM-EDX Spectra.
A single dye viability assay was developed for cells with an internal positive membrane potential. SYTO 9 is natively fluorescent although the fluorescence increases by a factor of ten when bound to DNA (Ankarcrona et al., 1995; Knowles et al., 1996). SYTO9 will cross the membrane of cells with an inside negative membrane potential via passive diffusion. However, the inside positive membrane potential of live acidophiles will exclude SYTO9. Cells of non-viable acidophiles will lose their membrane potential and SYTO 9 will be able to cross the membrane by passive diffusion, binding to the DNA, and fluorescing brighter. The difference in fluorescence intensity between live and dead cells is 10-fold (Supplemental File 1). The cell viability assay was carried out by first removing ferric iron precipitates from cultures by filtration using 8.0 μm Miltex™ nitrocellulose filters followed by centrifugation at 700 × g for 1 min at 4°C. Cells were harvested from the supernatant by centrifugation at 48,000 × g for 20 min at 4°C and re-suspended in either BSM, pH 1.8 (At. ferrooxidansT) or DSMZ 477 media, pH 2.5 (Ac. prosperusT). Cell suspensions were adjusted to a density of 5.0 × 105 cells/mL. SYTO 9 (ThermoFisher, Eugene, OR) was added to a concentration of 5 μM and the samples were incubated, protected from light, for 15 min. Three controls were prepared: (i) “no stain,” a 1 mL aliquot of cells was incubated at 4°C until time of acquisition with no further handling; (ii) “untreated,” a 1 mL aliquot of cells was incubated at 4°C for 15 min prior to acquisition, at which time it was stained with 5 μM SYTO 9, and (iii) “heat treated,” a 1 mL aliquot was heated to 60°C for 120 min and then incubated in 80% vol/vol molecular biology grade ethanol at room temperature for 60 min and then stained with 5 μM SYTO 9. To confirm the non-viability of this sample, a 200 μL aliquot was inoculated into 800 μL of appropriate growth media with 50 mM FeSO4.7 H2O and 5 mM K2S4O6 and incubated for 48 h at 30°C.
Samples were acquired in technical triplicates on an Attune Acoustic Flow Cytometer (ThermoFisher) using Attune Cytometric software version 1.2.5. on high sensitivity mode at a flow rate of 25 μL/min. Acquisition was terminated once 10,000 events on all gates were collected or 3 min had elapsed. Photomultiplier tube (PMT) voltages for forward scatter and side scatter were adjusted such that the bacterial population was clearly visible, with no truncation of relevant populations. PMT voltages were set on an unstained aliquot of cells with mean fluorescence intensity (MFI) of ~102 arbitrary fluorescence units excited at 488 nm with a 515–545 nm filter. Gating strategies and the determination of the fluorescence properties of populations of interest were established using FlowJo v10.0 (FlowJo LLC) and an unpaired t-test between conditions was performed using the GraphPad Prism v6 Software Suite (Graphpad Software, Inc.).
Milled concentrates (<0.75 μm) of pyrite (FeS2), chalcopyrite (CuFeS2), and pentlandite [(Ni,Fe)9S8] were sterilized by gamma irradiation (50 kGray). The elemental compositions of the concentrates were determined using inductively coupled plasma—atom emission spectroscopy (ICP-AES) after borax flux and re-dissolution in 5% (vol/vol) HNO3. The pyrite concentrate contained (wt/wt) 36.6% Fe, 0.24% Cu, 0.04% Ni, and 39.8% S; the chalcopyrite contained 26.6% Fe, 26.8% Cu, and 29.8% S; and the pentlandite contained 40.7% Fe, 0.73% Cu, 7.01% Ni, and 35.4% S. Ac. prosperusT was incubated in 100 mL of DSM 144 media containing increasing concentrations of NaCl and 0.5% (wt/vol) of the respective sulfide mineral concentrates. The cultures were incubated at 30°C with shaking at 130 rpm and evaporation was compensated for by the addition of sterile deionized water acidified to pH 2.5 with sulfuric acid. Leachates were assayed for pH and ORP (vs. Ag/AgCl) using Ionode pH (IJ44A) and ORP (IJ64) electrodes connected to a TPS SmartCHEM pH reader; Fe(III) concentration using the method of Govender et al. (2012); and Cu and Ni concentrations using flame atomic absorption spectrophotometry (Avanta Σ) with standards supplied by FLUKA chemicals.
Two liter cultures of the isolates were grown as described above with low and high concentrations of NaCl (0 and 8 g/L for At. ferrooxidansT and 3.5 and 30 g/L for Ac. prosperusT). To avoid alterations within the proteome as a result of differences in the growth phase, cultures were harvested by centrifugation for 20 min at 48,000 × g and 4°C during mid-exponential phase as determined by Fe3+ concentrations. Cell pellets were washed in acidified ultrapure water (HpH2O, pH 1.8 or 2.5), re-pelleted by centrifugation for 20 min at 48,000 × g and 4°C, and stored at −80°C.
The At. ferrooxidansT total soluble proteome was analyzed by 2D-PAGE as described by Mangold et al. (2011) except the initially extracted proteins were concentrated through methanol precipitation and 400 μg of protein was added to each IPG strip. Two dimensional gels were run in duplicate for cells grown in high salt and four gels were run for cells grown at low salt. Images of gels were taken on a PerkinElmer ProXPRESS and analyzed using Progenesis Same Spots program (Non-Linear Dynamics, USA). The two stained gels, of proteins isolated from cells grown in high NaCl concentrations, were aligned to the four stained gels, of proteins isolated from cells grown in low NaCl concentrations. Protein spots that showed change in abundance >1.8-fold and p < 0.05 were included in the identification process. Protein spots were analyzed at the Proteomics Node of the Lotterywest State Biomedical Facility within the Western Australian Institute for Medical Research. Protein samples were trypsin digested and analyzed by tandem mass spectrometry using a 5800 Proteomics Analyser (AB Sciex, USA) and peptides identified using Mascot sequence matching software with Ludwig NR Database and taxonomy set to bacteria.
Differential expression of the Ac. prosperusT proteome was analyzed by isobaric tags for relative and absolute quantification (iTRAQ). The cell pellet was suspended in lysis buffer [0.2% vol/vol IGEPAL, 0.2% vol/vol Triton X, 0.2% wt/vol CHAPS, 75 mM NaCl, 1 mM EDTA, protease inhibitors; in phosphate buffered saline (PBS)] and sonicated using Misonix Ultrasonic Liquid Processor S-4000 (Sonica LLC) with the following parameters; Amplitude 30% and 5 s cycles (pulse on and off) for a total of 2 min. Cellular debris was removed by centrifugation at 13,000 × g for 10 min at 4°C and the supernatant stored at −80°C. iTraq analysis was performed by Proteomics International as follows: protein samples were acetone diafiltrated, reduced, alkylated, and trypsin digested according to the iTRAQ protocol (Applied Biosystems) and labeled using the iTRAQ reagents. Peptides were desalted on a Strata-X 33 μm polymeric reversed phase column (Phenomenex) and dissolved in a buffer containing 10 mM KH2PO4 pH 3 in 10% vol/vol acetonitrile before separation by strong cation exchange liquid chromatography (SCX) on an Agilent 1100 High Performance Liquid Chromatography system using a PolySulfoethyl column (4.6 × 100 mm; 5 μm; 300 A). Peptides were eluted with a linear gradient of 0 to 400 mM KCl. Eight fractions containing the peptides were collected and after desalting on Strata-X columns were loaded onto a Agilent Zorbax 300SB-C18, 3.5 μm column (Agilent Technologies) running on an Shimadzu Prominence nano HPLC system (Shimadzu). Peptides were resolved with a gradient of 10–40% vol/vol acetonitrile (0.1% vol/vol trifluoroacetic acid) over 160 min. The resultant spots were analyzed on a 5600 Triple Time of Flight mass spectrometer (AB Sciex). Spectral data were analyzed against a protein sequence database for the whole genome (Ossandon et al., 2014) using ProteinPilot™ 4.5 Software (AB Sciex).
The amino acid sequence of the rusticyanin protein of At. ferrooxidansT (locus tag: AFE_3146) was used as a query in a BlastP search (Altschul et al., 1997) to predict similar proteins (genes) in the genome of Ac. prosperusT (Ossandon et al., 2014). Predicted protein sequences were aligned with Clustal Omega (Sievers et al., 2011) using the server at http://www.ebi.ac.uk/Tools/msa/clustalo/. Predictions of peptide signal and subcellular location were carried out using SignalP 4.1 (Petersen et al., 2011) and Cello (Yu et al., 2006) using the servers at http://www.cbs.dtu.dk/services/SignalP/ and http://cello.life.nctu.edu.tw/, respectively.
Three dimensional models of the structures of Rus Forms I and II from Ac. prosperusT were constructed using the experimentally determined structure of rusticyanin from At. ferrooxidansT (PDB “1RCY”) as a template (Walter et al., 1996). Electrostatic potentials of all three rusticyanins were determined using SWISS MODEL and visualized in Swiss-PDBViewer using the Swiss Model server at https://swissmodel.expasy.org/ (Bordoli et al., 2008). Default parameters were used [dielectric constant (solvent) 80.000, using only charged residues] using the Coulomb computational method with a dielectric constant (protein) 1.000 and solvent ionic strength (mol/L) 0.0.
Ac. prosperusT maintained activity (as defined by iron oxidation) at a higher concentration of NaCl compared to At. ferrooxidansT (Figure 1). At. ferrooxidansT ferrous iron oxidation was ~25% inhibited in the presence of 8 g L−1 NaCl and ~65% inhibited with the addition of 10 g L−1 NaCl (Figure 1A). In contrast, Ac. prosperusT had the highest rate of ferric iron generation in the presence of 20 g L−1 NaCl. In addition, while Ac. prosperusT ferrous iron oxidation in the presence of 50 g L−1 NaCl was less rapid, the ferrous iron was completely oxidized within 96 h (Figure 1B). Scanning electron micrographs indicate that At. ferrooxidansT was healthier at 0 g L−1 NaCl compared to 3.5 g L−1 NaCl with many of the At. ferrooxidansT cells lysed at the higher concentration. The lysed cells were confirmed as organic in nature by SEM-EDX Spectra (data not shown). In comparison Ac. prosperusT cells appeared more healthy in the presence of 30 g L−1 NaCl compared with 12.5 g L−1 NaCl (Figure 2). An optimum NaCl concentration of 20 g L−1 (342 mM) suggests Ac. prosperusT is a “slight halophile” (Ollivier et al., 1994).
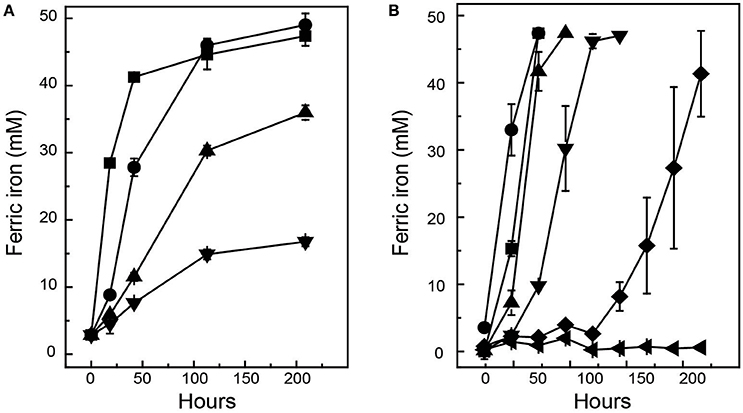
Figure 1. Effect of NaCl upon of ferric iron generation during growth of At. ferrooxidansT (A) and Ac. prosperusT (B). Symbols for At. ferrooxidansT: ■, 0 g L−1; ●, 6 g L−1; ▲, 8 g L−1; and ▼, 10 g L−1 while the symbols for Ac. prosperusT are: ■, 3.8 g L−1; ●, 20 g L−1; ▲, 35 g L−1; ▼, 50 g L−1; ♦, 60 g L−1; and ◄, 75 g L−1. Data are averages ± SD of duplicate biological replicates and one to three technical replicates.
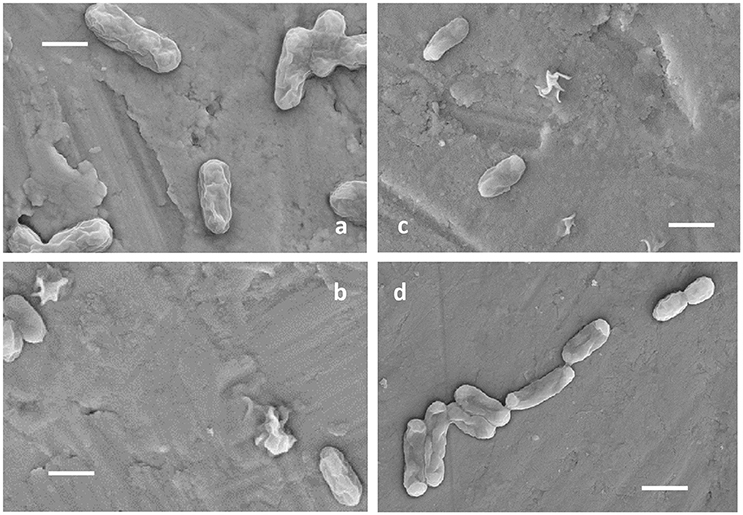
Figure 2. Electron micrographs of At. ferrooxidansT grown in the presence of 0 g L−1 (A) and 3.5 g L−1 NaCl (B) and Ac. prosperusT in the presence of 12.5 g L−1 (C), and 30 g L−1 NaCl (D). All scale bars are 1 μm.
A single dye viability assay using SYTO9 was developed based on positively charged SYTO 9 being excluded by live cells with an inside positive membrane potential. The cell viability of At. ferrooxidansT grown at 3.5 g L−1 NaCl decreased by 50% (P < 0.01) compared to 0 g L−1 NaCl whereas Ac. prosperusT had a 30% increase (P < 0.001) in viable cells when grown at 30 g L−1 NaCl compared to 12.5 g L−1 NaCl (Supplemental File 1).
Previous studies have indicated that the ability of At. ferrooxidansT to leach metal sulfides in the presence of chloride is completely inhibited in the presence of ~3.5 g/L NaCl (Deveci, 2002; Deveci et al., 2008; Zammit et al., 2012; Bevilaqua et al., 2013). Due to the potential use of halo-acidophiles to carry out biomining in arid environments where saline groundwater is used (Zammit et al., 2012; Rea et al., 2015), the ability of Ac. prosperusT to catalyze metal release from sulfide mineral concentrates was evaluated (Figure 3). The generation of ferric iron during Ac. prosperusT catalyzed pyrite bioleaching was more rapid in the presence of 15 and 30 g L−1 NaCl compared to either 3.8 or 50 g L−1 NaCl. The pyrite bioleaching in the presence of NaCl confirmed that Ac. prosperusT is a slight halophile. Ferric iron generation from pentlandite was similar in the presence of 15, 30, and 50 g L−1 NaCl and more rapid than observed at 3.5 and 75 g L−1 NaCl. Nickel release was greatest at 30 g L−1 NaCl (Figure 3). However, this trend was not supported for ferric iron generation and copper release from chalcopyrite where the leaching rates were very low and there was no statistically significant difference between 3.5 and 75 g L−1 NaCl. This lack of difference in leaching rates was potentially due to the advantages of chalcopyrite bioleaching in chloride systems as opposed to sulfate systems (reviewed in Watling, 2014). However, not all studies find an advantage with higher chloride ions at temperatures below 50°C (Dutrizac and Macdonald, 1971).
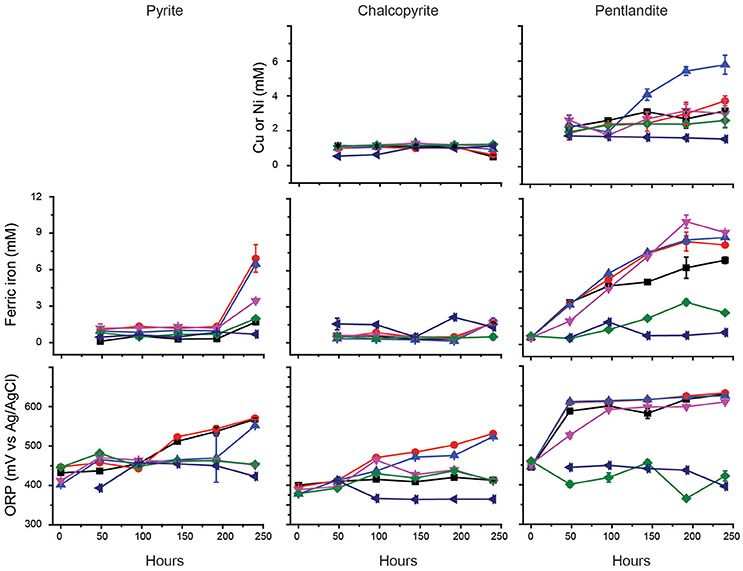
Figure 3. Effect of NaCl on the bioleaching of pyrite, chalcopyrite, and pentlandite by Ac. prosperusT: ■, 3.8 g L−1; ●, 15 g L−1; ▲, 30 g L−1; ▼, 50 g L−1; ♦, 75 g L−1; and ◄ abiotic control in the presence of 15 g L−1 NaCl (chosen as closest to the optimal NaCl concentration for Ac. prosperusT growth). The figure shows the copper and nickel concentrations during chalcopyrite and pentlandite leaching, respectively. Data are averages ± SD of triplicate biological replicates and three technical replicates.
Differential expression of the total soluble proteome from Ac. prosperusT cultures grown in the presence of 3.5 and 30 g L−1 NaCl identified 617 proteins in each of the proteomes of which 125 were differentially expressed (P < 0.05; Table 1 plus the complete data set in Supplemental File 2). The COG classifications with the highest number of differentially expressed proteins were cell envelope integrity, protein synthesis, energy acquisition, central carbon metabolism, and protein fate (Supplemental File 3). This likely reflected the need to adjust the cell envelope to maintain cellular integrity and the increased energy required to maintain pH and osmotic balance (reviewed in Slonczewski et al., 2009; Zammit and Watkin, 2016).
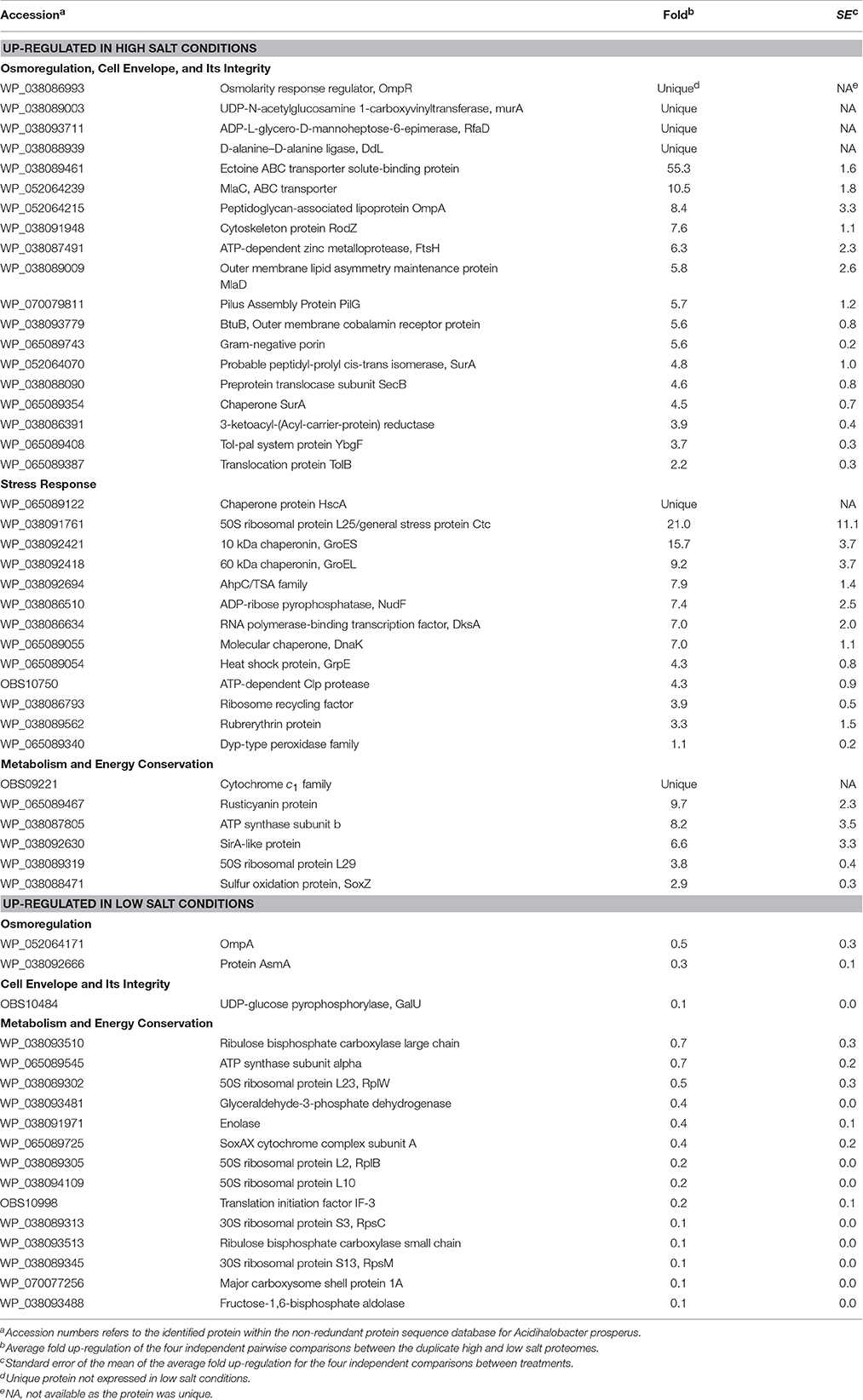
Table 1. Up- and down-regulated Ac. prosperusT proteins in the presence of high (30 g L−1) and low (3.5 g L−1) concentrations of sodium chloride.
Growth in high salt compared to low salt resulted in the unique identification of the osmolarity response regulator, OmpR. This regulator senses alterations in the membrane surface tension as a result of changes in the medium osmolarity (Cai and Inouye, 2002). A further regulatory protein, PilG which acts to control the transcription of many genes including osmotically inducible (Bouvier et al., 1998) and osmotic control (Lucht et al., 1994) genes, was 5.7 ± 1.2 fold up-regulated. On exposure to osmotic stress microorganisms will accumulate compatible solutes of which the most common is ectoine (Empadinhas and da Costa, 2008) and its ABC transporter was 55.3 ± 1.6 fold up-regulated in high salt conditions.
Another known response to osmotic stress is the production of proteins involved in the maintenance of the cell membrane integrity (reviewed: Zammit and Watkin, 2016). Growth in high salt conditions resulted in up-regulation of many Ac. prosperusT membrane integrity proteins. These included cytoskeleton protein RodZ (7.6 ± 1.1 fold) that is linked to maintaining cell shape (Bendezu et al., 2009); the cell membrane integrity Tol-Pal system (Lloubes et al., 2001; Zakharov et al., 2004) proteins BtuB (5.6 ± 0.8 fold), SecB (4.6 ± 0.8 fold), YbgF (3.7 ± 0.3 fold), and TolB (2.2 ± 0.3 fold); the MlaD outer membrane lipid asymmetry maintenance protein (5.8 ± 2.6 fold) and MlaC phospholipid ABC transporter (10.5 ± 1.8 fold) that maintain outer membrane integrity (Malinverni and Silhavy, 2009); a Gram-negative porin (5.6 ± 0.2 fold) and SurA (4.8 ± 1.0 and 4.5 ± 0.7 fold) involved in outer membrane protein folding (Vertommen et al., 2009). Additionally, several proteins that form the cell membrane had higher levels of abundance including MurA (unique in high salt conditions); RfaD and DdL (both unique) involved in lipopolysaccharide and peptidoglycan biosynthesis, respectively; and a peptidoglycan-associated lipoprotein (8.4 ± 3.3 fold). An increase in membrane biosynthesis proteins in the presence of chloride has also been observed in Acidithiobacillus caldus (Guo et al., 2014).
A second group of Ac. prosperusT proteins with increased concentrations in response to high salt conditions were related to the stress response. These proteins included protein folding chaperones such as DnaK (7.0 ± 1.1 fold) and GrpE (4.3 ± 0.8 fold) that form a homolog of the eukaryotic Hsp70 chaperone (Mayer and Bukau, 2005); HscA (unique) that forms part of a chaperone similar to DnaK (Silberg et al., 1998); and GroL (9.2 ± 3.7 fold) that acts under stress conditions (Chuang and Blattner, 1993). A further group of up-regulated proteins were involved in oxidative stress and included a AhpC/TSA family protein (7.9 ± 1.4 fold), ruberythrin (3.3 ± 1.5 fold), and a Dyp-type peroxidase family protein (1.1 ± 0.2 fold). These proteins may have been produced due to the increased metabolic and electron transport rate (see below) necessary to remove protons from the cytoplasm. This response has previously been observed in the acidophiles At. caldus (Zammit et al., 2012; Guo et al., 2014), Acidimicrobium ferrooxidans (Zammit et al., 2012), and Leptospirillum ferrooxidans (Parro et al., 2007). The final up-regulated proteins involved in the stress response to chloride were an ATP-dependent Clp protease (4.3 ± 0.9 fold) that degrades proteins (Katayama et al., 1988); GroS (15.7 ± 3.7 fold) that acts in concert with GroE in the response to DNA mutation (Al Mamun et al., 2012); ADP-ribose pyrophosphatase, NudF (7.4 ± 2.5 fold) that if deleted increases sensitivity to heat stress (Krisko et al., 2014); and RNA polymerase-binding transcription factor, DksA (7.0 ± 2.0 fold) that is induced at low pH (Stancik et al., 2002).
Metabolic and electron transport proteins with a higher concentration in high salt conditions included rusticyanin (9.7 ± 2.3 fold) and cytochrome c1 (unique) involved in ferrous iron oxidation (Quatrini et al., 2009). As mentioned above, these proteins were likely used during proton exclusion from the cytoplasm. In addition, ATP synthase subunit b had an 8.2 ± 3.5 fold higher concentration in high salt conditions. In addition to synthesizing ATP, the Enterococcus hirae ATPase extrudes protons from the cytoplasm to regulate pH (Shibata et al., 1992) and increasing the concentration of subunit b may result in the same function.
Proteins with a statistically higher concentration in low salt conditions generally had much lower fold differences (Table 1). These proteins included OmpA (0.5 ± 0.3 fold) and AsmA (0.3 ± 0.1 fold) involved in OMP assembly that were likely decreased in high salt conditions to reduce pores in the outer membrane that allow influx of chloride, as has been reported for OmpC and OmpF in E. coli (Csonka and Hanson, 1991). In addition, the ATPase α-subunit (0.7 ± 0.2 fold) had a higher concentration in low salt conditions, potentially as the complex was being used to produce ATP rather than extrude protons (Shibata et al., 1992). In a similar vein, several central carbon metabolism (e.g., enolase; 0.4 ± 0.1 fold), Calvin-Benson-Bassham cycle (e.g., ribulose bisphosphate carboxylase large chain; 0.7 ± 0.3 fold), and ribosomal (e.g., 50S ribosomal protein L23, RplW; 0.5 ± 0.3 fold) proteins had higher concentrations as energy production via ferrous iron oxidation was likely utilized for cellular growth rather than as a response to osmotic and pH stress.
At. ferrooxidansT response to growth in high (8 g/L) and low (0 g/L) salt conditions was investigated by two-dimensional polyacrylamide gel based proteomics (Supplemental File 4) that identified a total of 24 statistically valid up-regulated proteins during growth in high salt conditions (Supplemental File 5 with proteins discussed in the text in Table 2). At. ferrooxidansT growth in high salt exhibited several similar strategies as employed by Ac. prosperusT such as the increased abundance of peptidyl-prolyl cis-trans isomerase (two protein spots that were 2.8 and 2.5 fold up-regulated in 8 vs. 0 g/L salt) which is involved in outer membrane protein folding (Vertommen et al., 2009). Another three protein spots with increased abundance were identified as periplasmic solute binding proteins that are involved in the maintenance of the cell envelope integrity (2.8, 2.6, and 2.3 fold). However, the periplasmic solute binding protein also had a 3.1 higher concentration in low salt conditions suggesting that it had undergone regulation via post-translational modification. Several At. ferrooxidansT stress proteins with higher concentrations in 8 g/L NaCl included heat shock protein Hsp20 (2.5 fold) that aids in reducing protein denaturation (Lindquist and Craig, 1988); ribosome recycling factor (4.0 fold) also observed when Ac. prosperusT was cultured in high salt conditions; and a serine protease, DO/DeqQ family protein (2.0 fold) that has a chaperone function and also has a higher concentration in the At. ferrooxidans response to heat stress (Ribeiro et al., 2011). Finally, the major outer membrane protein 40 had 1.8 fold lower concentration in high salt conditions, potentially to reduce the influx of chloride (Csonka and Hanson, 1991).
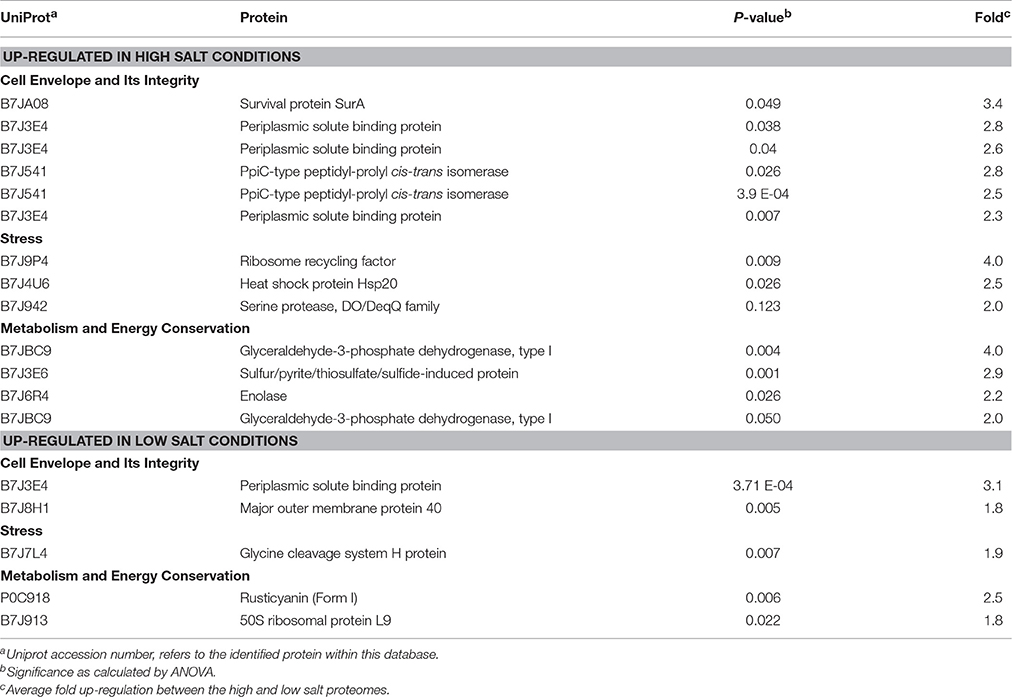
Table 2. At. ferrooxidansT proteins with statistically supported altered abundance when grown in high or low NaCl concentration.
In contrast to the increase in rusticyanin seen in Ac. prosperusT when cultured in high salt conditions, At. ferrooxidansT had a 2.5 fold decrease implying a reduction in iron oxidation (Quatrini et al., 2009) as was demonstrated in the growth experiments, where a reduction of iron oxidation by 25% was observed.
Iron oxidation in the well-studied acidophile At. ferrooxidansT involves a protein complex that transfers electrons from iron to oxygen (Castelle et al., 2008; Li et al., 2015) and includes the copper protein rusticyanin encoded in the rus operon (Levicán et al., 2002). Rusticyanin is located in the periplasmic space where the pH is low. A cluster of genes has been detected in Ac. prosperus V6 (DSM 14174) that has similarity to the rus operon of At. ferrooxidansT (Nicolle et al., 2009) and it is hypothesized that expression of the rusticyanin gene is actively involved in Fe oxidation, presumably in a similar way to that described for At. ferrooxidansT. However, a major difference in the two systems is that iron oxidation in At. ferrooxidansT is inhibited by low concentrations of chloride (Blake et al., 1991; Harahuc et al., 2000), whereas chloride is required for expression of rusticyanin in Ac. prosperus V6 (Nicolle et al., 2009) and maximum iron oxidation in Ac. prosperusT was seen at 20 g/L NaCl.
Using the rusticyanin gene of At. ferrooxidansT (locus tag: AFE_3146) as a query, two rusticyanin genes, termed Form I and Form II (locus tags: Thpro_021557 and Thpro_020703, respectively) were predicted in the genome of Ac. prosperusT (Ossandon et al., 2014). Relative to the rusticyanin of At. ferrooxidansT, Form I was detected with a score of 142, a query coverage of 100%, an E-value of 2e-48, and an identity of 46%. The same parameters for Form II were 116, 89%, 2e-38, and 43%. The extent of sequence similarity and coverage suggest that the two forms of Rus in Ac. prosperusT are encoded by genes that are orthologs of rus from At. ferrooxidansT. Both Rus Forms I and II are predicted to contain signal peptides and to reside in the periplasm. If this is true, then they are most likely to be subjected to the low pH and high salt conditions typical for Ac. prosperus. However, the genetic contexts of the two Forms differ (Supplemental File 6). Form I is embedded in a gene cluster very similar to the classical rus operon of At. ferrooxidansT (Valdes et al., 2008). This supports the hypothesis that Form I Rus is involved in iron oxidation in a manner similar to that described for At. ferrooxidansT. In contrast, the gene encoding Form II Rus is found as a singleton gene with no other known genes involved in iron oxidation in the gene neighborhood (Supplemental File 6). The function of this Rus remains unknown. However, because of its sequence similarity to Rus from At. ferrooxidansT, it can be speculated that it is also involved in iron oxidation, perhaps under different growth conditions from Form I Rus.
As Form I Rus increases in abundance (9.7 ± 2.3 fold) when Ac. prosperusT is subjected to high salt conditions, both its primary amino acid sequence and its predicted tertiary structure were interrogated for clues that might suggest how it maintains activity in high salt conditions. Form II Rus (no change in abundance with increasing salt concentration) and Rus from At. ferrooxidansT (2.5 fold decrease) were included for comparison (Figure 4). Four critical amino acids (two histidines, one cysteine, and one methionine) have been shown to be ligands in the inner sphere coordinating the copper ion in Rus in At. ferrooxidansT and many other members of the family of small blue copper proteins (Gray et al., 2000). The ligands Cys, Met, and one of the histidines are close to each other at the C terminal end in the primary sequence whereas the other histidine is far away from them in the amino acid chain. The loop length that connects these ligands has been shown to be important for coordination of the copper in related blue copper proteins (Gough and Chothia, 2004). Also, as observed in other small blue copper proteins including Rus from At. ferrooxidansT, both Form I and Form II Rus from Ac. prosperusT are predicted to contain the so-called Greek key β-barrel (data not shown). This is a rigid structure formed by an extended network of hydrogen bonds and tertiary interactions between amino acid side chains (Gray et al., 2000). This rigidity is transmitted to the metal ion and is essential for electron transfer. As shown in Figure 4, these ligands, their relative positions in the primary amino acid sequence, and the length of the connecting loop are conserved in Forms I and II Rus from Ac. prosperus and in Rus from At. ferrooxidansT. Due to the conservation of these properties between the acidophilic At. ferrooxidansT and the haloacidophilic Ac. prosperusT, it is unlikely that they contribute to salt tolerance in Rus Form I (and perhaps Form II).

Figure 4. Clustal Omega multiple alignment of rusticyanins from At. ferrooxidansT (locus tag: AFE_3146), Ac. prosperusT Form I (locus tag: Thpro_021557) and Ac. prosperusT Form II (locus tag: Thpro_020703). An * (asterisk) indicates positions which have a single, fully conserved residue. (A), (colon) indicates conservation between groups of strongly similar properties, a. (period) indicates conservation between groups of weakly similar properties. A Δ (triangle) indicates the position of cutting of the peptide signal using At. ferrooxidansT as reference. The meaning of the colors is described in (Sievers et al. (2011), Mol Sys Bio 7:539). A † (dagger) indicates the four conserved amino acids that bind the copper ion.
The number and distribution of positively (His, Lys, and Arg) and negatively charged (Asp and Glu) amino acids in Rus Forms I and II differ from that observed for Rus of At. ferrooxidansT (Figure 4). This is in agreement with an observation made earlier for Rus from Ac. prosperus V6 (Nicolle et al., 2009). In order to examine whether these differences in charged amino acids could affect the surface electrostatic potential of the different Rus, three dimensional models of the structures of Rus Forms I and II were constructed using the experimentally determined structure of rusticyanin from At. ferrooxidansT (PDB “1RCY”) as a template (Walter et al., 1996). The predicted surface electrostatic potentials of Rus Forms I and II of Ac. prosperusT (Figures 5B,C) are significantly more negative compared to that of Rus from At. ferrooxidansT (Figure 5A). In the case of Form I Rus, this negative electrostatic potential is widely distributed over the surface of the entire molecule, including around the copper ion. In contrast, in Form II, it is principally distributed around the copper ion. It has been well-established that the electrostatic field directly influences the electrostatic properties of the metal-binding site of blue copper proteins, being a major determinant of the redox potential of the copper ion (Olsson et al., 2003). It is possible that the noticeable negative shift in surface electrostatic potential of Rus Forms I and II could help stabilize them in high salt conditions and assist in the maintenance of an appropriate redox potential of the copper ion. It could also help to repel negatively charged chloride ions in the immediate environment of the proteins.
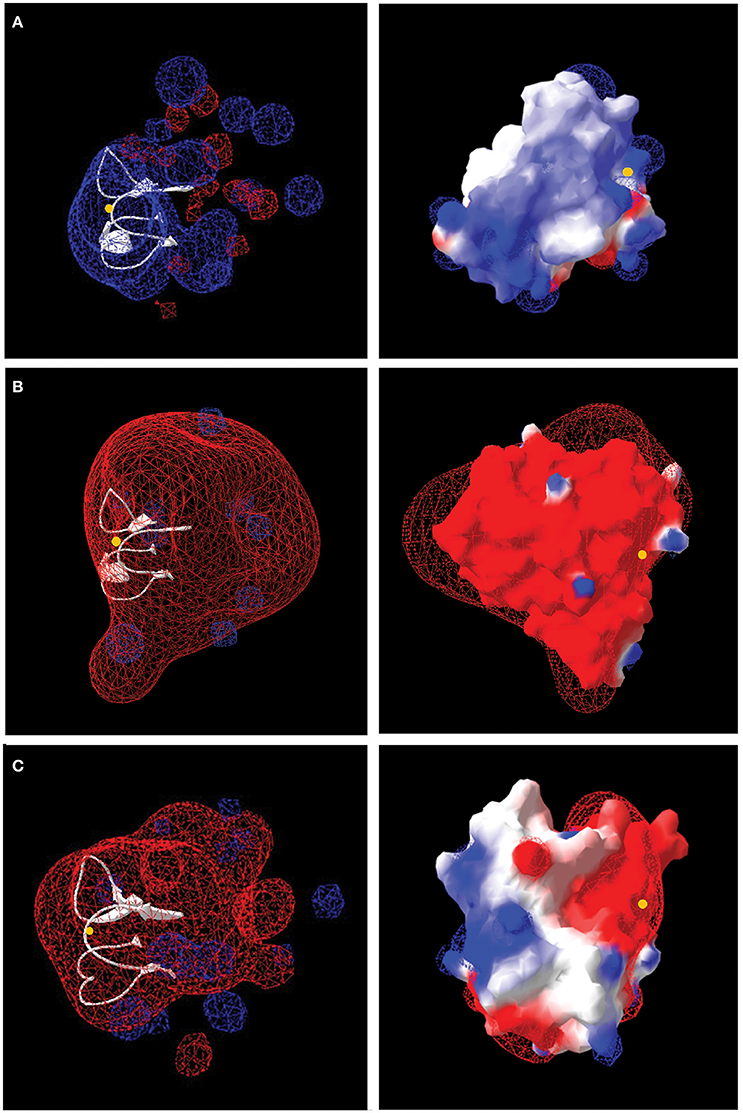
Figure 5. Models of the electrostatic surface potential of rusticyanin of: (A) At. ferrooxidansT; (B) Ac. prosperusT Form I, and (C) Ac. prosperusT Form II. The surface is colored according to the protein electrostatic potential from red (negative) to blue (positive); the copper ion is shown as a yellow dot. The models on the left hand side are rendered transparent to show (in white) the critical protein folds that binds the copper ion. The models on the right have been rotated 180° (y-axis) compared with the models on the left to provide a different perspective.
The increased negative surface electrostatic potential of Form I rusticyanin of Ac. prosperusT likely affects its interactions with its redox partners that, based upon amino acid sequence similarities and gene neighborhood conservation, are predicted to be the same as in At. ferrooxidansT [i.e., a high molecular weight c-type cytochrome Cyc2 located in the external membrane, a periplasmic diheme cytochrome c Cyc1, and a periplasmic diheme cytochrome Cyc42 (Cyc1A; Bruscella et al., 2007; Castelle et al., 2008)]. Although the potential changes evidently still permit electron transfer, their nature requires experimental verification. The redox partners of Rus Form II are not known.
Other changes in amino acid sequence between Rus of At. ferrooxidansT and Ac. prosperusT might reveal clues regarding stabilization and activity of Rus at high salt concentrations such as changes in the outer coordination sphere (Cascella et al., 2006; Warren et al., 2012), but these await discovery and investigation. Although a reasonable argument can be made that an increase in the negativity of the surface potential of Rus Form I (and Form II) could help explain salt tolerance perhaps by modulating the environment of copper ion and very likely by affecting its interaction with redox partners, significant effort is still required to understand and experimentally validate these ideas. However, the current suggestions do lead to testable hypotheses and can be used a basis for guiding future research.
When challenged by elevated salt concentration, acidophiles experience both osmotic stress and an acidification of the intracellular pH (reviewed in Zammit and Watkin, 2016). This is due to a collapse of the inside positive membrane potential as a result of Cl− crossing the cell membrane, leading to an influx of protons. Notwithstanding the caveat that the iTRAQ analysis of Ac. prosperusT in high salt will identify many more proteins than the 2D-PAGE analysis of At. ferrooxidansT, the response of the two species were distinct (Figure 6). At. ferrooxidansT responded to even low levels of Cl− with a generalized stress response and decreased iron oxidation which was confirmed by a reduced abundance of the protein rusticyanin. However, despite the reduced ability to generate energy there was an increase in central carbon metabolism and carbon fixation. The most significant responses to increased salt concentration by Ac. prosperusT were an increase in abundances of osmotic stress regulators; uptake of the compatible solute ectoine protein and increased iron oxidation as confirmed by the raised abundance of the proteins rusticyanin and cytochrome c1 that consumes cytoplasmic protons and/or provides reducing power for the stress response. Both central carbon metabolism and carbon fixation decreased suggesting the increased ability to generate energy is utilized for the potential efflux of protons via the F0F1 ATPase at the expense of ATP suggested by the greater abundance of the ATP synthase subunit b.
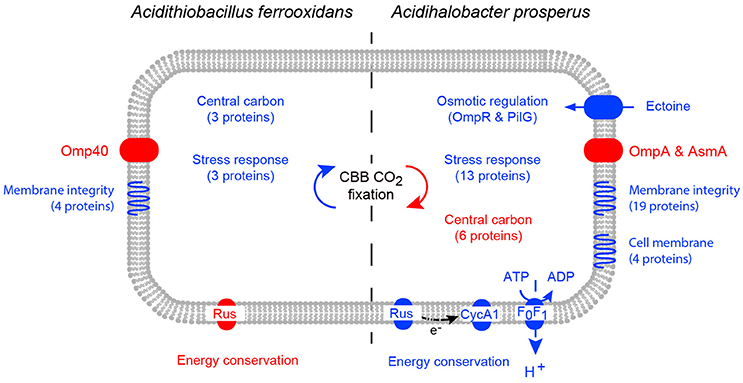
Figure 6. A model of the cellular response of At. ferrooxidansT (left) and Ac. prosperusT (right) to increased NaCl levels. Increases in protein abundance in high NaCl conditions are represented by blue and decreases in protein abundance in high NaCl conditions are represented by red. Differential protein expression was determined by 2D-PAGE for At. ferrooxidansT and iTraq for Ac. prosperusT.
EW, MD, CB, and DH conceived and designed the experiments. EW, TM, KM, and ML performed the experiments. MD, EW, DH analyzed the data. EW, MD, DH, RS, and CJ contributed to the reagents/materials/analysis tools. MD, EW, and DH wrote the paper. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CSD declared a past co-authorship with one of the authors DH to the handling Editor, who ensured that the process met the standards of a fair and objective review. The reviewer SH and the handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
EW was funded by an Ian Potter Foundation Travel Grant. This project was partially funded by a Bioplatforms Australia Omics grant. DH and ML were funded by Conicyt Basal CCTE PFB16 and Fondecyt 1130683.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02132/full#supplementary-material
Aguilera, A., Olsson, S., and Puente-Sánchez, F. (2016). “Physiological and phylogenetic diversity of acidophilic eukaryotes,” in Acidophiles: Life in Extremely Acidic Environments, eds R. Quatrini and D. B. Johnson (Poole, UK: Caister Academic Press), 107–118.
Alexander, B., Leach, S., and Ingledew, W. J. (1987). The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. J. Gen. Microbiol. 133, 1171–1179. doi: 10.1099/00221287-133-5-1171
Al Mamun, A. A., Lombardo, M. J., Shee, C., Lisewski, A. M., Gonzalez, C., Lin, D., et al. (2012). Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338, 1344–1348. doi: 10.1126/science.1226683
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ankarcrona, M., Dypbukt, J. M., Bonfoco, E., Zhivotovsky, B., Orrenius, S., Lipton, S. A., et al. (1995). Glutamate-induced neuronal death:a succession of necrosis or apoptosis depending on mitochondrial function. Neuron 15, 961–973. doi: 10.1016/0896-6273(95)90186-8
Baker-Austin, C., Potrykus, J., Wexler, M., Bond, P. L., and Dopson, M. (2010). Biofilm development in the extremely acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 14, 485–491. doi: 10.1007/s00792-010-0328-1
Bauermeister, A., Rettberg, P., and Flemming, H. C. (2014). Growth of the acidophilic iron-sulfur bacterium Acidithiobacillus ferrooxidans under Mars-like geochemical conditions. Planet. Space Sci. 98, 205–215. doi: 10.1016/j.pss.2013.09.009
Belnap, C. P., Pan, C., Denef, V. J., Samatova, N. F., Hettich, R. L., and Banfield, J. F. (2011). Quantitative proteomic analyses of the response of acidophilic microbial communities to different pH conditions. ISME J. 5, 1152–1161. doi: 10.1038/ismej.2010.200
Bendezu, F. O., Hale, C. A., Bernhardt, T. G., and De Boer, P. A. (2009). RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28, 193–204. doi: 10.1038/emboj.2008.264
Bevilaqua, D., Lahti, H., Suegama, P. H., Garcia, O. Jr., Benedetti, A. V., Puhakka, J. A., et al. (2013). Effect of Na-chloride on the bioleaching of a chalcopyrite concentrate in shake flasks and stirred tank bioreactors. Hydrometallurgy 138, 1–13. doi: 10.1016/j.hydromet.2013.06.008
Blake, R. C. II, White, K. J., and Shute, E. A. (1991). Effect of divers anions on the electron-transfer reaction between iron and rusticyanin from Thiobacillus ferrooxidans. Biochemistry 30, 9443–9449. doi: 10.1021/bi00103a008
Blight, K. R., and Ralph, D. E. (2004). Effect of ionic strength on iron oxidation with batch cultures of chemolithotrophic bacteria. Hydrometallurgy 73, 325–334. doi: 10.1016/j.hydromet.2003.12.006
Bordoli, L., Kiefer, F., Arnold, K., Benkert, P., Battey, J., and Schwede, T. (2008). Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13. doi: 10.1038/nprot.2008.197
Bouvier, J., Gordia, S., Kampmann, G., Lange, R., Hengge-Aronis, R., and Gutierrez, C. (1998). Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28, 971–980. doi: 10.1046/j.1365-2958.1998.00855.x
Boxall, N. J., Rea, S. M., Li, J., Morris, C., and Kaksonen, A. H. (2016). Effect of high sulfate concentrations on chalcopyrite bioleaching and molecular characterisation of the bioleaching microbial community. Hydrometallurgy. doi: 10.1016/j.hydromet.2016.07.006. [Epub ahead of print].
Bruscella, P., Appia-Ayme, C., Levicán, G., Ratouchniak, J., Jedlicki, E., Holmes, D. S., et al. (2007). Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology 153, 102–110. doi: 10.1099/mic.0.2006/000067-0
Cai, S. J., and Inouye, M. (2002). EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277, 24155–24161. doi: 10.1074/jbc.M110715200
Cardenas, J.-P., Ortiz, R., Norris, P. R., Watkin, E., and Holmes, D. S. (2015). Reclassification of ‘Thiobacillus prosperus’ (Huber and Stetter 1989) as Acidihalobacter prosperus gen. nov., sp. nov., a member of the family Ectothiorhodospiraceae. Int. J. Syst. Evol. Microbiol. 65, 3641–3644. doi: 10.1099/ijsem.0.000468
Cascella, M., Magistrato, A., Tavernelli, I., Carloni, P., and Rothlisberger, U. (2006). Role of protein frame and solvent for the redox properties of azurin from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 103, 19641–19646. doi: 10.1073/pnas.0607890103
Castelle, C., Guiral, M., Malarte, G., Ledgham, F., Leroy, G., Brugna, M., et al. (2008). A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 283, 25803–25811. doi: 10.1074/jbc.M802496200
Choi, M., Sukumar, N., Mathews, F. S., Liu, A., and Davidson, V. L. (2011). Proline 96 of the copper ligand loop of amicyanin regulates electron transfer from methylamine dehydrogenase by positioning other residues at the protein-protein interface. Biochemistry 50, 1265–1273. doi: 10.1021/bi101794y
Chuang, S. E., and Blattner, F. R. (1993). Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol. 175, 5242–5452. doi: 10.1128/jb.175.16.5242-5252.1993
Colmer, A. R., and Hinkle, M. E. (1947). The action of certain microorganisms in acid mine drainage: a preliminary report. Science 106, 253–256. doi: 10.1126/science.106.2751.253
Csonka, L. N., and Hanson, A. D. (1991). Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45, 569–606. doi: 10.1146/annurev.mi.45.100191.003033
Davis-Belmar, C. S., Nicolle, J. L. C., and Norris, P. R. (2008). Ferrous iron oxidation and leaching of copper ore with halotolerant bacteria in ore columns. Hydrometallurgy 94, 144–147. doi: 10.1016/j.hydromet.2008.05.030
Deveci, H. (2002). Effect of salinity on the oxidative activity of acidophilic bacteria during bioleaching of a complex Zn/Pb sulphide ore. Eur. J. Miner. Proc. Environ. Prot. 2, 141–150.
Deveci, H., Jordan, M. A., Powell, N., and Alp, I. (2008). Effect of salinity and acidity on bioleaching activity of mesophilic and extremely thermophilic bacteria. Trans. Nonferr. Met. Soc. China 18, 714–721. doi: 10.1016/S1003-6326(08)60123-5
Dopson, M. (2016). “Physiological and phylogenetic diversity of acidophilic bacteria,” in Acidophiles: Life in Extremely Acidic Environments, eds R. Quatrini and D. B. Johnson (Poole, UK: Caister Academic Press), 79–92.
Dopson, M., and Lindström, E. B. (1999). Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl. Environ. Microbiol. 65, 36–40.
Dutrizac, J. E., and Macdonald, R. J. C. (1971). The effect of sodium chloride on the dissolution of chalcopyrite under simulated dump leaching conditions. Metallurgical Mater. Trans. B 2, 2310–2312. doi: 10.1007/BF02917578
Elleuche, S., Schroder, C., Sahm, K., and Antranikian, G. (2014). Extremozymes - biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 29, 116–123. doi: 10.1016/j.copbio.2014.04.003
Empadinhas, N., and da Costa, M. S. (2008). Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int. Microbiol. 11, 151–161. doi: 10.2436/20.1501.01.55
Esparza, M., Cardenas, J. P., Bowien, B., Jedlicki, E., and Holmes, D. S. (2010). Genes and pathways for CO2 fixation in the obligate, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans. BMC Microbiol. 10:229. doi: 10.1186/1471-2180-10-229
Gahan, C. S., Sundkvist, J.-E., Dopson, M., and Sandström, Å. (2010). Effect of chloride on ferrous iron oxidation by a Leptospirillum ferriphilum-dominated chemostat culture. Biotechnol. Bioeng. 106, 422–431. doi: 10.1002/bit.22709
Goltsman, D. S., Dasari, M., Thomas, B. C., Shah, M. B., Verberkmoes, N. C., Hettich, R. L., et al. (2013). New group in the Leptospirillum clade: cultivation-independent community genomics, proteomics, and transcriptomics of the new species “Leptospirillum group IV UBA BS.” Appl. Environ. Microbiol. 79, 5384–5393. doi: 10.1128/AEM.00202-13
Golyshina, O., Ferrer, M., and Golyshin, P. N. (2016). “Diversity and physiologies of acidophilic archaea,” in Acidophiles: Life in Extremely Acidic Environments, eds R. Quatrini and D. B. Johnson (Poole, UK: Caister Academic Press), 93–106.
Gough, J., and Chothia, C. (2004). The linked conservation of structure and function in a family of high diversity: the monomeric cupredoxins. Structure 12, 917–925. doi: 10.1016/j.str.2004.03.029
Govender, E., Harrison, S. T. L., and Bryan, C. G. (2012). Modification of the ferric chloride assay for the spectrophotometric determination of ferric and total iron in acidic solutions containing high concentrations of copper. Miner. Eng. 35, 46–48. doi: 10.1016/j.mineng.2012.05.006
Gray, H. B., Malmstrom, B. G., and Williams, R. J. (2000). Copper coordination in blue proteins. J. Biol. Inorg. Chem. 5, 551–559. doi: 10.1007/s007750000146
Graziano, G., and Merlino, A. (2014). Molecular bases of protein halotolerance. Biochim. Biophys. Acta 1844, 850–858. doi: 10.1016/j.bbapap.2014.02.018
Guo, X., Jiang, C., Luo, Y., Zhang, M., Poetsch, A., and Liu, S. (2014). Proteomic and molecular investigations revealed that Acidithiobacillus caldus adopts multiple strategies for adaptation to NaCl stress. Chin. Sci. Bull. 59, 301–309. doi: 10.1007/s11434-013-0039-y
Harahuc, L., Lizama, H. M., and Suzuki, I. (2000). Selective inhibition of the oxidation of ferrous iron or sulfur in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 66, 1031–1037. doi: 10.1128/AEM.66.3.1031-1037.2000
Huber, H., and Stetter, K. O. (1989). Thiobacillus prosperus sp. nov., represents a new group of halotolerant metal-mobilizing bacteria isolated from a marine geothermal field. Arch. Microbiol. 151, 479–485. doi: 10.1007/BF00454862
Issotta, F., Galleguillos, P. A., Moya-Beltrán, A., Davis-Belmar, C. S., Rautenbach, G., Covarrubias, P. C., et al. (2016). Draft genome sequence of chloride-tolerant Leptospirillum ferriphilum Sp-Cl from industrial bioleaching operations in northern Chile. Stand. Genomic Sci. 11, 1–7. doi: 10.1186/s40793-016-0142-1
Jensen, S. M., Christensen, C. J., Petersen, J. M., Treusch, A. H., and Brandl, M. (2015). Liposomes containing lipids from Sulfolobus islandicus withstand intestinal bile salts: an approach for oral drug delivery? Int. J. Pharmaceut. 493, 63–69. doi: 10.1016/j.ijpharm.2015.07.026
Katayama, Y., Gottesman, S., Pumphrey, J., Rudikoff, S., Clark, W. P., and Maurizi, M. R. (1988). The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J. Biol. Chem. 263, 15226–15236.
Knowles, R. B., Sabry, J. H., Martone, M. E., Deerinck, T. J., Ellisman, M. H., Bassell, G. J., et al. (1996). Translocation of RNA granules in living neurons. J. Neurosci. 16, 7812–7820.
Krisko, A., Copic, T., Gabaldon, T., Lehner, B., and Supek, F. (2014). Inferring gene function from evolutionary change in signatures of translation efficiency. Genome Biol. 15:R44. doi: 10.1186/gb-2014-15-3-r44
Levicán, G., Bruscella, P., Guacunano, M., Inostroza, C., Bonnefoy, V., Holmes, D. S., et al. (2002). Characterization of the petI and res operons of Acidithiobacillus ferrooxidans. J. Bacteriol. 184, 1498–1501. doi: 10.1128/JB.184.5.1498-1501.2002
Li, T. F., Painter, R. G., Ban, B., and Blake, R. C. II. (2015). The multicenter aerobic iron respiratory chain of Acidithiobacillus ferrooxidans functions as an ensemble with a single macroscopic rate constant. J. Biol. Chem. 290, 18293–18303. doi: 10.1074/jbc.M115.657551
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Lloubes, R., Cascales, E., Walburger, A., Bouveret, E., Lazdunski, C., Bernadac, A., et al. (2001). The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152, 523–529. doi: 10.1016/S0923-2508(01)01226-8
Lucht, J. M., Dersch, P., Kempf, B., and Bremer, E. (1994). Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 269, 6578–6586.
Ma, J. K., Wang, Y., Carrell, C. J., Mathews, F. S., and Davidson, V. L. (2007). A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin. Biochemistry 46, 11137–11146. doi: 10.1021/bi7012307
Malinverni, J. C., and Silhavy, T. J. (2009). An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014. doi: 10.1073/pnas.0903229106
Mangold, S., Potrykus, J., Björn, E., Lövgren, L., and Dopson, M. (2013). Extreme zinc tolerance in acidophilic microorganisms from the bacterial and archaeal domains. Extremophiles 17, 75–85. doi: 10.1007/s00792-012-0495-3
Mangold, S., Valdes, J., Holmes, D. S., and Dopson, M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17. doi: 10.3389/fmicb.2011.00017
Millero, F. J., Feistel, R., Wright, D. G., and McDougall, T. J. (2008). The composition of standard seawater and the definition of the reference-composition salinity scale. Deep Sea Res. I 55, 50–72 doi: 10.1016/j.dsr.2007.10.001
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Méndez-García, C., Peláez, A. I., Mesa, V., Sánchez, J., Golyshina, O. V., and Ferrer, M. (2015). Microbial diversity and metabolic networks in acid mine drainage habitats. Front. Microbiol. 6:475. doi: 10.3389/fmicb.2015.00475
Mueller, R. S., Dill, B. D., Pan, C., Belnap, C. P., Thomas, B. C., Verberkmoes, N. C., et al. (2011). Proteome changes in the initial bacterial colonist during ecological succession in an acid mine drainage biofilm community. Environ. Microbiol. 13, 2279–2292. doi: 10.1111/j.1462-2920.2011.02486.x
Mykytczuk, N. C., Trevors, J. T., Foote, S. J., Leduc, L. G., Ferroni, G. D., and Twine, S. M. (2011). Proteomic insights into cold adaptation of psychrotrophic and mesophilic Acidithiobacillus ferrooxidans strains. Antonie van Leeuwen 100, 259–277. doi: 10.1007/s10482-011-9584-z
Nicolle, J. L. C., Simmons, S., Bathe, S., and Norris, P. R. (2009). Ferrous iron oxidation and rusticyanin in halotolerant, acidophilic ‘Thiobacillus prosperus’. Microbiology 155, 1302–1309. doi: 10.1099/mic.0.023192-0
Ollivier, B., Caumette, P., Garcia, J. L., and Mah, R. A. (1994). Anaerobic bacteria from hypersaline environments. Microbiol. Rev. 58, 27–38.
Olsson, M. H., Hong, G., and Warshel, A. (2003). Frozen density functional free energy simulations of redox proteins: computational studies of the reduction potential of plastocyanin and rusticyanin. J. Am. Chem. Soc. 125, 5025–5039. doi: 10.1021/ja0212157
Oren, A. (2013). Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4:315. doi: 10.3389/fmicb.2013.00315
Osorio, H., Mangold, S., Denis, Y., Nancucheo, I., Johnson, D. B., Bonnefoy, V., et al. (2013). Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79, 2172–2181. doi: 10.1128/AEM.03057-12
Osorio, H., Martinez, V., Nieto, P. A., Holmes, D. S., and Quatrini, R. (2008). Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility. BMC Microbiol. 8:203. doi: 10.1186/1471-2180-8-203
Ossandon, F. J., Cardenas, J. P., Corbett, M., Quatrini, R., Holmes, D. S., and Watkin, E. (2014). Draft genome sequence of the iron-oxidizing, acidophilic, and halotolerant “Thiobacillus prosperus” type strain DSM 5130. Genome Announce. 2:e01042–14. doi: 10.1128/genomeA.01042-01014
Parro, V., Moreno-Paz, M., and González-Toril, E. (2007). Analysis of environmental transcriptomes by DNA microarrays. Environ. Microbiol. 9, 453–464. doi: 10.1111/j.1462-2920.2006.01162.x
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Plumb, J. J., Gibbs, B., Stott, M. B., Robertson, W. J., Gibson, J. A. E., Nichols, P. D., et al. (2002). Enrichment and characterisation of thermophilic acidophiles for the bioleaching of mineral sulphides. Miner. Eng. 15, 787–794. doi: 10.1016/S0892-6875(02)00117-6
Ponce, J. S., Moinier, D., Byrne, D., Amouric, A., and Bonnefoy, V. (2012). Acidithiobacillus ferrooxidans oxidizes ferrous iron before sulfur likely through transcriptional regulation by the global redox responding RegBA signal transducing system. Hydrometallurgy 127–128, 187–194. doi: 10.1016/j.hydromet.2012.07.016
Potrykus, J., Rao Jonna, V., and Dopson, M. (2011). Iron homeostasis and responses to iron limitation in extreme acidophiles from the Ferroplasma genus. Proteomics 11, 52–63. doi: 10.1002/pmic.201000193
Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D., and Bonnefoy, V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. doi: 10.1186/1471-2164-10-394
Rea, S. M., Mcsweeney, N. J., Degens, B. P., Morris, C., Siebert, H. M., and Kaksonen, A. H. (2015). Salt-tolerant microorganisms potentially useful for bioleaching operations where fresh water is scarce. Miner. Eng. 75, 126–132. doi: 10.1016/j.mineng.2014.09.011
Ribeiro, D. A., Maretto, D. A., Nogueira, F. C., Silva, M. J., Campos, F. A., Domont, G. B., et al. (2011). Heat and phosphate starvation effects on the proteome, morphology and chemical composition of the biomining bacteria Acidithiobacillus ferrooxidans. World J. Microbiol. Biotechnol. 27, 1469–1479. doi: 10.1007/s11274-010-0599-9
Shibata, C., Ehara, T., Tomura, K., Igarashi, K., and Kobayashi, H. (1992). Gene structure of Enterococcus hirae (Streptococcus faecalis) F1F0-ATPase, which functions as a regulator of cytoplasmic pH. J. Bacteriol. 174, 6117–6124. doi: 10.1128/jb.174.19.6117-6124.1992
Shiers, D. W., Blight, K. R., and Ralph, D. E. (2005). Sodium sulphate and sodium chloride effects on batch culture of iron oxidising bacteria. Hydrometallurgy 80, 75–82. doi: 10.1016/j.hydromet.2005.07.001
Shivanand, P., and Mugeraya, G. (2011). Halophilic bacteria and their compatible solutes - osmoregulation and potential applications. Curr. Sci. 100, 1516–1521.
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Silberg, J. J., Hoff, K. G., and Vickery, L. E. (1998). The Hsc66-Hsc20 chaperone system in Escherichia coli: chaperone activity and interactions with the DnaK-DnaJ-grpE system. J. Bacteriol. 180, 6617–6624.
Slonczewski, J. L., Fujisawa, M., Dopson, M., and Krulwich, T. A. (2009). Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 55, 1–79. doi: 10.1016/S0065-2911(09)05501-5
Stancik, L. M., Stancik, D. M., Schmidt, B., Barnhart, D. M., Yoncheva, Y. N., and Slonczewski, J. L. (2002). pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184, 4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002
Suzuki, I., Lee, D., Mackay, B., Harahuc, L., and Oh, J. K. (1999). Effect of various ions, pH, and osmotic pressure on oxidation of elemental sulfur by Thiobacillus thiooxidans. Appl. Environ. Microbiol. 65, 5163–5168.
Valdes, J., Pedroso, I., Quatrini, R., Dodson, R. J., Tettelin, H., Blake, R., et al. (2008). Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. doi: 10.1186/1471-2164-9-597
Vera, M., Schippers, A., and Sand, W. (2013). Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation–part A. Appl. Microbiol. Biotechnol. 97, 7529–7541. doi: 10.1007/s00253-013-4954-2
Vertommen, D., Ruiz, N., Leverrier, P., Silhavy, T. J., and Collet, J. F. (2009). Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9, 2432–2443. doi: 10.1002/pmic.200800794
Walter, R. L., Ealick, S. E., Friedman, A. M., Blake, R. C. II, Proctor, P., and Shoham, M. (1996). Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability. J. Mol. Biol. 263, 730–751. doi: 10.1006/jmbi.1996.0612
Warren, J. J., Lancaster, K. M., Richards, J. H., and Gray, H. B. (2012). Inner- and outer-sphere metal coordination in blue copper proteins. J. Inorg. Biochem. 115, 119–126. doi: 10.1016/j.jinorgbio.2012.05.002
Watling, H. R. (2014). Chalcopyrite hydrometallurgy at atmospheric pressure: 2. Review of acidic chloride process options. Hydrometallurgy 146, 96–110. doi: 10.1016/j.hydromet.2014.03.013
Wen, Q., Liu, X. M., Wang, H. Y., and Lin, J. Q. (2014). A versatile and efficient markerless gene disruption system for Acidithiobacillus thiooxidans: application for characterizing a copper tolerance related multicopper oxidase gene. Environ. Microbiol. 16, 3499–3514. doi: 10.1111/1462-2920.12494
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Yu, Y. Y., Liu, X. M., Wang, H. Y., Li, X. T., and Lin, J. Q. (2014). Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 196, 2255–2264. doi: 10.1128/JB.01472-13
Zakharov, S. D., Eroukova, V. Y., Rokitskaya, T. I., Zhalnina, M. V., Sharma, O., Loll, P. J., et al. (2004). Colicin occlusion of OmpF and TolC channels: outer membrane translocons for colicin import. Biophys. J. 87, 3901–3911. doi: 10.1529/biophysj.104.046151
Keywords: salt, acidophile, biomining, bioleaching, proteomics, pyrite, chalcopyrite, environmental stress
Citation: Dopson M, Holmes DS, Lazcano M, McCredden TJ, Bryan CG, Mulroney KT, Steuart R, Jackaman C and Watkin ELJ (2017) Multiple Osmotic Stress Responses in Acidihalobacter prosperus Result in Tolerance to Chloride Ions. Front. Microbiol. 7:2132. doi: 10.3389/fmicb.2016.02132
Received: 29 September 2016; Accepted: 19 December 2016;
Published: 05 January 2017.
Edited by:
Axel Schippers, Federal Institute for Geosciences and Natural Resources, GermanyReviewed by:
Sabrina Hedrich, Federal Institute for Geosciences and Natural Resources, GermanyCopyright © 2017 Dopson, Holmes, Lazcano, McCredden, Bryan, Mulroney, Steuart, Jackaman and Watkin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth L. J. Watkin, ZS53YXRraW5AY3VydGluLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.