
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 23 December 2016
Sec. Microbial Symbioses
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.02102
This article is part of the Research Topic Experimental models in animal-associated microbiota View all 36 articles
Sponges have a significant impact on marine benthic communities, they are of biotechnological interest owing to their production of bioactive natural compounds, and they promise to provide insights into conserved mechanisms of host–microbe interactions in basal metazoans. The natural variability of sponge-microbe associations across species and environments provides a meaningful ecological and evolutionary framework to investigate animal-microbial symbiosis through experimentation in the field and also in aquaria. In addition, next-generation sequencing technologies have shed light on the genomic repertoire of the sponge host and revealed metabolic capacities and symbiotic lifestyle features of their microbiota. However, our understanding of symbiotic mechanisms is still in its infancy. Here, we discuss the potential and limitations of the sponge-microbe symbiosis as emerging models for animal-associated microbiota.
Each model of animal-associated microbiota offers unique opportunities to address questions related to symbiosis (Ruby, 2008; Bosch and McFall-Ngai, 2011; Kostic et al., 2013). Existing laboratory models, such as fly (Drosophila melanogaster), worm (Caenorhabditis elegans), zebra fish or mice, enable to test the influence of genetic or environmental factors on symbioses. However, it remains unclear to which extent the findings in lab models apply to natural systems and to other taxa. Natural models provide a relevant ecological and evolutionary framework, but are frequently restricted to systems in which the number of players and interactions is reduced (e.g., the squid Euprymna scolopes and the bacterium Vibrio fischeri, or the mussels Bathymodiolus spp. and their microbiota). However, owing to deep-sequencing technologies, it has become clear that many animals are associated with complex microbial consortia and the implications of such symbioses are just beginning to be unraveled (McFall-Ngai et al., 2013). The study of complex consortia is challenging because multiple interactions take place simultaneously, making it difficult to decipher the specific roles of each symbiont. Additionally, methodologies are limiting and microbes are frequently recalcitrant to cultivation. Enhancing the tractability of the symbiosis within different animal phyla would contribute to our understanding on animal–microbe interactions.
Marine sponges (phylum Porifera) represent prominent examples for such complex symbioses. Many sponge species contain diverse microbial consortia within their mesohyl matrix that can reach densities of up to 109 microbial cells/cm3 of sponge (Hentschel et al., 2006). Sponges belong to a phylum that originated ca. 600 million years ago (Li et al., 1998). Their porous body plan contains a highly ramified aquiferous canal system through which seawater is pumped. Specific cells lining the choanocyte chambers (termed “choanocytes”) take up particles, such as bacterioplankton, from the seawater and transfer them into the mesohyl interior where these are digested by phagocytosis. Sponges lack organs, muscles, and a nervous system. In spite of their simple anatomy, recent studies have revealed an unexpected genomic complexity in sponges (Srivastava et al., 2010; Riesgo et al., 2014a). For example, they express homologs of genes involved in the animal nervous system (Ludeman et al., 2014) and possess central elements of the Toll-like receptor signaling cascade, potentially involved in innate immunity (Hentschel et al., 2012; Riesgo et al., 2014a). In addition, sponges and their associated microorganisms yield secondary metabolites that are of relevance to biotechnological and medical applications (Mehbub et al., 2014; Indraningrat et al., 2016).
The sponge-associated microbiota is exceedingly complex with thousands of symbiont lineages reported per sponge individual (Thomas et al., 2016). The most abundant phyla are Proteobacteria, Chloroflexi, and Crenarchaeota, among others. Altogether, more than 40 microbial phyla, including several candidate phyla (e.g., Tectomicrobia, Poribacteria) were recovered from sponges. In contrast, most animal-associated microbiota belong within 3–5 phyla and diversification is found at species and strain level (Kostic et al., 2013). Some members of the sponge-associated consortia are vertically transmitted to the next generation by larval stages (De Caralt et al., 2007b; Schmitt et al., 2008), but horizontal acquisition also appears likely (Taylor et al., 2007). Sponge symbiont OTUs have also been recovered from seawater, albeit at very low abundances, and their activity in the free-living state still needs to be explored (Webster et al., 2010). The diversity patterns, metabolic repertoire and genomic features of sponge-associated microbiota have been reviewed in detail elsewhere (Taylor et al., 2007; Hentschel et al., 2012; Webster and Thomas, 2016). Despite a species-specific composition, similar functions are detected in the microbiomes of distantly related sponge species, suggesting convergent evolution (Fan et al., 2012; Ribes et al., 2012; Thomas et al., 2016; Horn et al., 2016). These functions relate to nutritional interactions (e.g., nitrification, vitamin B synthesis), host–microbe recognition [e.g., eukaryotic-like proteins (ELPs)] and adaptation to host’s internal environment (e.g., CRISPR-Cas defense system).
A significant body of information has been accrued from analyzing the natural variability of sponge microbiomes in different host species and environments. Host species appears to be the main factor driving microbial diversity (Erwin et al., 2012; Easson and Thacker, 2014), although environmental factors (e.g., intertidal vs. subtidal habitat, Weigel and Erwin, 2016; depth, Steinert et al., 2016; location, Pita et al., 2013b) can also cause intraspecific variability. Also, differences in symbiont density within the mesohyl (i.e., high microbial abundance HMA sponges vs. low microbial abundance LMA sponges; Gloeckner et al., 2014) have an impact on microbial community composition as well as on the host pumping rate and other metabolic parameters (Weisz et al., 2008; Ribes et al., 2012). Monitoring microbiota changes over seasons, bleaching episodes, natural gradients, or upon transplantation showed the plasticity of the symbiosis at scales that are difficult to mimic in laboratory (Steindler et al., 2007; López-Legentil et al., 2010; Erwin et al., 2015; Morrow et al., 2015). As similar past and present environment has been also faced by other benthic invertebrates, the features and mechanisms of sponge-microbe symbioses contribute to a more comprehensive view of marine symbiotic systems.
The need for sponge models for symbiosis has been debated within the sponge microbiology community (i.e., at the 1st International Symposium of Sponge Microbiology, Taylor et al., 2011). While the benefits of pooling resources, developing standardized protocols and limiting redundancy were clearly acknowledged, the dangers of developing too narrow a view of the natural diversity were also voiced (Taylor et al., 2011; Webster and Taylor, 2012). An experimental sponge model would, however, be immensely useful to put the large amount of sequence data into functional context. Even though some advances toward an experimental sponge model have been made, such as the generation of protocols and procedures for sponge aquaculture (Schippers et al., 2012) as well as the silencing of sponge genes for functional studies (Rivera et al., 2011), the overall efforts are still in its infancy. Here we discuss the current status and future directions toward establishing an experimental sponge model for symbiosis.
Disclosing the mechanistic bases of sponge–microbe interactions requires experimental tractability. The desired features of an experimental sponge model are depicted in Table 1. First, species that are accessible to as many laboratories as possible would be useful candidates. Most studied sponge species thrive in shallow temperate or tropical waters, where they are easily collected by snorkeling or scuba diving. Sponges from remote or deep environments in need of ocean-going equipment, expertise, and resources are naturally less amenable to experimental manipulation. Sponge species inhabiting a wide geographic range should be prioritized; however, many species are limited to distinct geographic regions. Consequently, most research groups have focused on the abundant species at their local sites (e.g., the Great Barrier Reef species Amphimedon queenslandica, Rhopaloides odorabile, Cymbastela concentrica, the West-Atlantic/Caribbean/East-Pacific sponges Xestospongia muta, Mycale laxissima, Axinella mexicana, or the Mediterranean/East-Atlantic species Aplysina aerophoba, Petrosia ficiformis, Dysidea avara, Ircinia spp.), thus making an overall comparison across geographic boundaries difficult. On the genus level, some sponges show an almost global distribution within temperate and/or tropical latitudes (e.g., Aplysina, Halichondria, Ircinia, and Xestospongia) and laboratories in different geographic areas would benefit from protocols applicable to these sponges. In addition, when choosing a model system, the parameters of morphology and size should be taken into account. Smaller, compact species are easier to collect and maintain than massive or fragile species. For example, while Xestospongia muta satisfies the criteria of wide distribution and ecological relevance, it will hardly be amenable to aquarium maintenance owing to its large size and fragility.
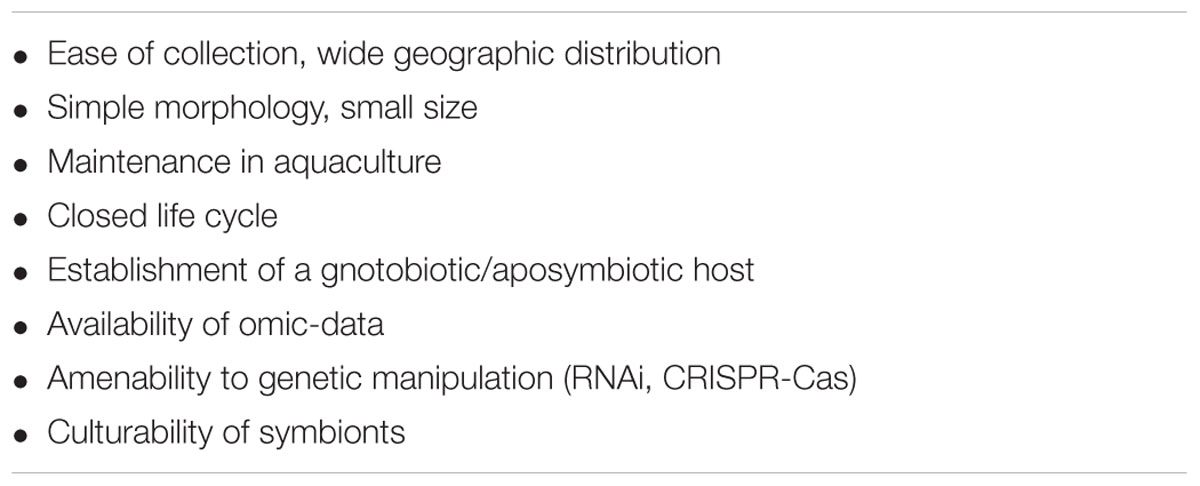
TABLE 1. Desired properties for an experimental sponge model for symbiosis (adapted after Ruby, 2008).
An experimental model requires the long-term controlled maintenance of the holobiont (here defined as the host plus its symbionts, Margulis, 1991). Different laboratories have kept sponges in aquaria for days/weeks (e.g., Fan et al., 2013; Pita et al., 2013a), several months (e.g., Aplysina aerophoba, Gerçe et al., 2009; Sacristán-Soriano et al., 2016), and up to 2 years (Ircinia strobilina, Mohamed et al., 2008b; Mycale laxissima, Mohamed et al., 2008a). The aquaculture setups vary from closed systems with external food supply (i.e., bacteria or algae) to open flow-through systems with direct seawater uptake, which are closer to in situ conditions but demand advanced infrastructure. Depending on the sponge species and/or the aquaculture system, the microbiota changes upon transfer and long-term maintenance in aquaria with respect to community composition and abundance, although the core microbiome seems to persist (e.g., Mohamed et al., 2008a; Webster et al., 2011; Ribes et al., 2016). This plasticity requires careful monitoring of the microbial diversity during aquaculture by use of standardized protocols (Earth Microbiome Project, Thomas et al., 2016).
Thus far, sponge experimentation in aquaria depends on the collection of specimens from the field and their transfer to the lab. The limitations to in vitro reproduction of sponges are mainly due to their life cycle as the reproductive period typically occurs once a year, because reproductive cues vary among species (Riesgo and Maldonado, 2008), and because juveniles have higher mortality rates in aquaria (De Caralt et al., 2007a). It is therefore noteworthy, that the life cycles of the Mediterranean sponges Dysidea avara and Crambe crambe and of the Australian sponge Amphimedon queenslandica were successfully closed in vitro (De Caralt et al., 2007a; Conaco et al., 2012). In addition to field collections, clonal populations of sponges can be generated and maintained owing to sponge reproduction by gemmulation or budding (Adams et al., 2010; Di Bari et al., 2015). The quick regeneration capacity after fragmentation can be used to generate “explants,” which are clonal pieces of sponge that will continue to be metabolically active in aquaria. Such efforts were undertaken with the species Halichondria panicea (Barthel and Theede, 1986), Corticium candelabrum (De Caralt et al., 2003), Geodia barretti (Hoffmann et al., 2003), and Rhopaloides odorabile (Webster et al., 2011) and have provided new insights into primary metabolism of the sponge holobiont. The generation of sponge fragments may be useful in the future to rear sponge individuals of similar/identical genetic background at small sizes and with a high number of replicates.
Aposymbiotic states (i.e., deprivation of a particular group of symbionts) and germ-free hosts are crucial tools to dissect the animal-microbiota crosstalk. The most common method to rear germ-free animals is the application of antibiotics. However, the sponge microbiota appears to be resistant to antibiotics (Friedrich et al., 2001; De Caralt et al., 2003), even when applied to sponge cell aggregates (Richardson et al., 2012). Interestingly, aposymbiotic sponges occur in the field. The Mediterranean sponge Petrosia ficiformis harbors the cyanobacterium Synechococcus feldmannii; however, individuals in caves appear photosymbiont-free (Burgsdorf et al., 2014). Also, the Caribbean sponge Xestospongia muta undergoes cyclic bleaching – density loss of symbiont Synechococcus spp. –, from which the sponge can recover (López-Legentil et al., 2008). If the cue for aposymbiosis is discerned, it could potentially be simulated in the lab. In a different approach, Riesgo et al. (2014b) created dinoflagellate-free Cliona varians specimens by removing the outer layer of the sponges, where the photosymbionts thrive. Aposymbiotic Cliona varians sponges were able to recapture photosymbionts. Unfortunately, the available examples are still restricted to few species and the photosynthetic symbionts. Alternative approaches based on phage therapy, a clinical strategy to clean infections and tumoral cells (e.g., Cattaneo et al., 2008; Nobrega et al., 2015), remain unexplored but are promising tools to selectively remove symbionts.
In the last 5 years, the amount of genetic information available on sponge hosts in the context of symbiosis has increased exponentially (Table 2). The first published genome – from A. queenslandica, with a size of 166.7 Mb and mean GC percentage of 37.5% (Srivastava et al., 2010)– is still the reference for Porifera. Recently, the enhanced annotation of this genome raised the total number of genes from 28898 (first version, Srivastava et al., 2010) to 40122 (Fernandez-Valverde et al., 2015). This finding suggests a more compacted genome than previous thought, but also reflects the difficulties of identifying the structure and function of genes in species that are phylogenetically distant from traditional model organisms. Even the role of genes with known homologs in other phyla needs to be confirmed in experimental studies in this group. As Porifera is a highly diversified phylum, the genetic information of different species is necessary to gain a more comprehensive view of the sponge genetic toolkit (Riesgo et al., 2014a).
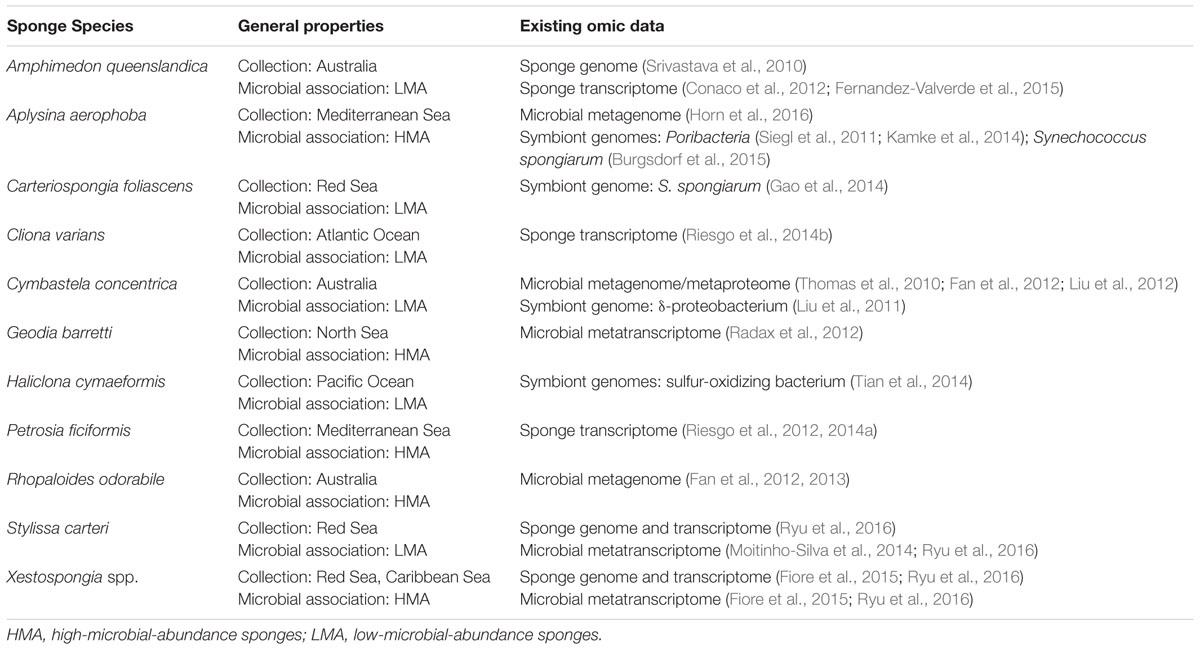
TABLE 2. A compilation of sponge species for which published omic-data in the context of the sponge symbiosis are available.
Despite the increase of genetic data on sponge holobionts, functional studies are missing. Research on gene expression (e.g., transcriptomics) is helpful to fill this gap, especially if applied from an experimental approach (e.g., Riesgo et al., 2014b). Also, RNA interference (RNAi) provides a valuable tool to temporary silence specific genes (e.g., Timmons et al., 2001; Franzenburg et al., 2012). In sponges, promising results from RNAi have been obtained in the freshwater sponge Ephydatia muelleri and the marine sponge Tethya wilhelma, for genes involved in animal development (Rivera et al., 2011). The authors performed RNAi in marine sponges by feeding them with bacteria expressing the double-stranded RNA of the target gene. Emerging methods to generate targeted genome editing -e.g., clustered regularly interspaced short palindromic repeat associated proteins (CRISPR)/Cas9 mediated genome editing (Gaj et al., 2013)- could allow sponge genome modification. CRISPR, together with Cas proteins, is a nucleic-acid based adaptive immunity found in prokaryotes (Wiedenheft et al., 2012) leading to sequence-specific cleavage of viral invading nucleic acids (Barrangou, 2015) by Cas endonucleases (Jinek et al., 2012). The sequence-specific cleavage of invading DNA by CRISPR-associated endonuclease Cas9 has been adopted to edit genomes in bilaterian model systems (Friedland et al., 2013; Gratz et al., 2013; Hwang et al., 2013; Wang et al., 2013). Since then, this method has revolutionized the possibilities to alter the genome of non-model organisms with unprecedented ease and specificity. Recently, CRISPR/Cas9 genome editing was established in two marine organisms: the cnidaria Nematostella vectensis (Ikmi et al., 2014) and the echinoderm Strongylocentrotus purpuratus (Lin and Su, 2016). Thus, CRISPR/Cas9 mediated genome-editing presents a promising tool to study mechanisms of symbiosis in sponges, but further effort and investment are required.
The availability of genetically tractable sponges would further contribute to revealing host mechanisms of symbiosis. In other models, host immunity seems to mediate microbial interactions. In particular, pattern-recognition receptors (PRRs) sense microbes (both pathogens and symbionts) and either initiate the defense against pathogenic infection or promote microbiota homeostasis (reviewed in Chu and Mazmanian, 2013). Studies in sponge genomic repertoires have identified a collection of extracellular (i.e., scavenger receptor cysteine-rich, SRCR, domain), membrane-bounded (immunoglobulin-like domains), and intracellular (NOD-like receptor, NLR, domains) PRRs (Hentschel et al., 2012). Some results suggest that immunity may mediate microbial recognition in sponges: (i) high expansion of NLR (in A. queenslandica, Degnan, 2015), and SRCR (in Stylissa carteri, Ryu et al., 2016) in sponge genomes may correspond to the need for a diverse array of PRRs for effective discrimination upon microbial encounter; (ii) a gene codifying for a SRCR-containing protein was overexpressed in individuals of Petrosia ficiformis in symbiosis with cyanobacteria vs. aposymbiotic individuals (Steindler et al., 2007); (iii) Ryu et al. (2016) detected different enrichment in immune domains depending on symbiont densities within the mesohyl (i.e., the genomes of LMA sponges A. queenslandica and S. carteri vs. the HMA sponge, X. testudinaria). The picture of sponge immunity is still incomplete and the development of experimental sponge models would allow validating the function of the predicted genes and the role of immunity in mediating symbiosis.
As for the host, the ability to culture and manipulate symbionts is necessary to further understand the mechanisms of host–microbe interactions. But sponge-associated symbionts, revealed by molecular techniques, remain recalcitrant to culture (Schippers et al., 2012; Hardoim et al., 2014). Alternative cultivation efforts are based on diffusion growth chambers (Steinert et al., 2014), longer incubations, and/or on adjusting culture conditions to match the symbiotic metabolic properties revealed from genomic data (Lavy et al., 2014). In recent years, genomic features of the microbiota from several sponge species have been published (Table 2). Also, state-of-the-art sequencing and bioinformatics methods allowed the characterization of the genomes of key sponge-associated symbionts: Cenarchaeum symbiosum (Hallam et al., 2006), representatives of the Candidate phylum Poribacteria (Siegl et al., 2011), a sponge-associated sulfur-oxidizing bacterium (Tian et al., 2014), and different clades of Ca. Synechococcus spongiarum (Gao et al., 2014; Burgsdorf et al., 2015). Sponge symbionts show a wide variety of metabolisms, and some hint at physiological interactions with the sponge host (reviewed in Webster and Thomas, 2016). Metagenomes of sponge-associated microbiota also showed an enrichment on ELPs, suggested to be involved in avoiding host consumption (Thomas et al., 2010). In the absence of tractable models, researchers tested their hypothesis by feeding amoebas with recombinant E. coli cells expressing sponge-symbiont-derived ELPs; as a result, the recombinant cells escaped digestion (Nguyen et al., 2014). Further sequencing efforts and cultivation of target symbionts will allow further insight into environment-regulated response, chemical characterization or colonization mechanisms and may become a system amenable to genetic manipulation.
Research on sponge-associated microbiota adds valuable insights to our understanding of animal-microbiota symbiosis, mainly because of the natural range of symbiosis within this early-divergent phylum. However, the field needs to move from exploratory to mechanistic projects, where state-of-the-art techniques are applied to meaningful experimental design and the symbiosis is manipulated. Although certain infrastructure is required, aquaculture conditions are described for several species and the regeneration capacity of sponges could serve for keeping clone lines in laboratory. Further research and resources should focus on the physiology and microbiology of cultured sponges over the long term. The new techniques for targeted genome editing appear to be the most promising method for investigating the sponge-microbiota symbiosis through host manipulation.
Finally, the most suitable sponge species for experimental models will depend on the specific focus of the study. In terms of host manipulation, Tethya wilhelma and Ephydatia muelleri seem the most advanced model species, together with Amphimedon queenslandica, with a well annotated genome and possibility of aquaria maintenance. Based on adequate performance in aquaculture, Clathria prolifera, Dysidea avara, Halichondria panicea, Ianthella basta, Ircinia spp., or Mycale laxissima are valuable candidates but they still require comprehensive genomic data on the symbiosis. The cumulative genomic information on their symbiotic communities and possibilities for in situ manipulation or aquaculture of other species such as Aplysina aerophoba, R. odorabile, or Xestospongia sp., (Table 2), necessitate further studies to enhance their tractability. Developing more than one model species will produce a more comprehensive view of the mechanisms of symbiosis. The amenability of laboratory sponge models to manipulation would certainly help to identify key players and key functions of the interaction, and their relevance can be further validate by in situ studies in different species and environmental conditions. Thus, researchers take advantage of the insights from an experimentally tractable model in combination with the holistic view of the sponge symbiosis in its natural ecological context.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
UH and SF received financial support from the DFG (CRC1182-TPB1) and LP was awarded a postdoctoral fellowship from Alexander von Humboldt Foundation, which was sponsored by The Future Ocean Cluster.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful to L. Rix (GEOMAR Helmholtz Centre for Ocean Research) for English language and style corrections.
Adams, E. D. M., Goss, G. G., and Leys, S. P. (2010). Freshwater sponges have functional, sealing epithelia with high transepithelial resistance and negative transepithelial potential. PLoS ONE 5:e15040. doi: 10.1371/journal.pone.0015040
Barrangou, R. (2015). Diversity of CRISPR-Cas immune systems and molecular machines. Genome Biol. 16:247. doi: 10.1186/s13059-015-0816-9
Barthel, D., and Theede, H. (1986). A new method for the culture of marine sponges and its application for experimental studies. Ophelia 25, 75–82. doi: 10.1080/00785326.1986.10429715
Bosch, T. C. G., and McFall-Ngai, M. J. (2011). Metaorganisms as the new frontier. Zoology 114, 185–190. doi: 10.1016/j.zool.2011.04.001
Burgsdorf, I., Erwin, P. M., López-Legentil, S., Cerrano, C., Haber, M., Frenk, S., et al. (2014). Biogeography rather than association with cyanobacteria structures symbiotic microbial communities in the marine sponge Petrosia ficiformis. Front. Microbiol. 5:529. doi: 10.3389/fmicb.2014.00529
Burgsdorf, I., Slaby, B. M., Handley, K. M., Haber, M., Blom, J., Marshall, C. W., et al. (2015). Lifestyle evolution in cyanobacterial symbionts of sponges. MBio 6:e00391-15.
Cattaneo, R., Miest, T., Shashkova, E., and Barry, M. (2008). Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat. Rev. Microbiol. 6, 529–540. doi: 10.1016/j.pestbp.2011.02.012
Chu, H., and Mazmanian, S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Conaco, C., Neveu, P., Zhou, H., Arcila, M. L., Degnan, S. M., Degnan, B. M., et al. (2012). Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics 13:209. doi: 10.1186/1471-2164-13-209
De Caralt, S., Agell, G., and Uriz, M. J. (2003). Long-term culture of sponge explants: conditions enhancing survival and growth, and assessment of bioactivity. Biomol. Eng. 20, 339–347. doi: 10.1016/S1389-0344(03)00045-5
De Caralt, S., Otjens, H., Uriz, M. J., and Wijffels, R. H. (2007a). Cultivation of sponge larvae: settlement, survival, and growth of juveniles. Mar. Biotechnol. 9, 592–605. doi: 10.1007/s10126-007-9013-5
De Caralt, S., Uriz, M. J., and Wijffels, R. H. (2007b). Vertical transmission and successive location of symbiotic bacteria during embryo development and larva formation in Corticium candelabrum (Porifera: Demospongiae). J. Mar. Biol. Assoc. UK 87, 1693–1699. doi: 10.1017/S0025315407056846
Degnan, S. M. (2015). The surprisingly complex immune gene repertoire of a simple sponge, exemplified by the NLR genes: a capacity for specificity? Dev. Comp. Immunol. 48, 269–274. doi: 10.1016/j.dci.2014.07.012
Di Bari, G., Cardone, F., Gaino, E., Liuzzi, G. M., Nonnis Marzano, C., Scoccia, F., et al. (2015). Biological variations in a long-term ex situ cultivation: a Mediterranean demosponge as model system. Mediterr. Mar. Sci. 16, 73–81. doi: 10.12681/mms872
Easson, C. G., and Thacker, R. W. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5:532. doi: 10.3389/fmicb.2014.00532
Erwin, P. M., Coma, R., López-Sendino, P., Serrano, E., and Ribes, M. (2015). Stable symbionts across the HMA-LMA dichotomy: low seasonal and inter-annual variation in sponge-associated bacteria from taxonomically diverse hosts. FEMS Microbiol. 91, 1–11. doi: 10.1093/femsec/fiv115
Erwin, P. M., Pita, L., López-Legentil, S., and Turon, X. (2012). Stability of sponge-associated bacteria over large seasonal shifts in temperature and irradiance. Appl. Environ. Microbiol. 78, 7358–7368. doi: 10.1128/AEM.02035-12
Fan, L., Liu, M., Simister, R., Webster, N. S., and Thomas, T. (2013). Marine microbial symbiosis heats up: the phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 7, 991–1002. doi: 10.1038/ismej.2012.165
Fan, L., Reynolds, D., Liu, M., Stark, M., Kjelleberg, S., Webster, N. S., et al. (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U.S.A. 109, e1878–e1887. doi: 10.1073/pnas.12032871091073/pnas.1203287109
Fernandez-Valverde, S. L., Calcino, A. D., and Degnan, B. M. (2015). Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics 16:387. doi: 10.1186/s12864-015-1588-z
Fiore, C. L., Labrie, M., Jarett, J. K., and Lesser, M. P. (2015). Transcriptional activity of the giant barrel sponge, Xestospongia muta holobiont: molecular evidence for metabolic interchange. Front. Microbiol. 6:364. doi: 10.3389/fmicb.2015.00364
Franzenburg, S., Fraune, S., Künzel, S., Baines, J. F., Domazet-lo, T., and Bosch, T. C. G. (2012). MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc. Natl. Acad. Sci. U.S.A. 109, 19374–19379. doi: 10.1073/pnas.1213110109
Friedland, A., Tzur, Y., Esvelt, K., Colaiácovo, M., Church, G., and Calarco, J. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743. doi: 10.1038/nmeth.2532
Friedrich, A. B., Fischer, I., Proksch, P., Hacker, J., and Hentschel, U. (2001). Temporal variation of the microbial community associated with the mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38, 105–115. doi: 10.1111/j.1574-6941.2001.tb00888.x
Gaj, T., Gersbach, C. A., and Barbas, C. F. I. I. I. (2013). ZFN, TALEN and CRISPR/Cas based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Gao, Z., Wang, Y., and Tian, R. (2014). Symbiotic adaptation drives genome streamlining of the cyanobacterial sponge symbiont “Candidatus Synechococcus spongiarum”. MBio 5:e00079-14. doi: 10.1128/mBio.00079-14
Gerçe, B., Schwartz, T., Voigt, M., Rühle, S., Kirchen, S., Putz, A., et al. (2009). Morphological, bacterial, and secondary metabolite changes of Aplysina aerophoba upon long-term maintenance under artificial conditions. Microb. Ecol. 58, 865–878. doi: 10.1007/s00248-009-9560-6
Gloeckner, V., Wehrl, M., Moitinho-Silva, L., Gernert, C., Schupp, P., Pawlik, J. R., et al. (2014). The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol. Bull. 227, 78–88. doi: 10.1086/BBLv227n1p78
Gratz, S. J., Cummings, A. M., Nguyen, J. N., Hamm, D. C., Donohue, L. K., Harrison, M. M., et al. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. doi: 10.1534/genetics.113.152710
Hallam, S. J., Konstantinidis, K. T., Putnam, N., Schleper, C., Watanabe, Y., Sugahara, J., et al. (2006). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U.S.A. 103, 18296–18301. doi: 10.1073/pnas.0608549103
Hardoim, C. C. P., Cardinale, M., Cúcio, A. C. B., Esteves, A. I. S., Berg, G., Xavier, J. R., et al. (2014). Effects of sample handling and cultivation bias on the specificity of bacterial communities in keratose marine sponges. Front. Microbiol. 5:611. doi: 10.3389/fmicb.2014.00611
Hentschel, U., Piel, J., Degnan, S. M., and Taylor, M. W. (2012). Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641–654. doi: 10.1038/nrmicro2839
Hentschel, U., Usher, K. M., and Taylor, M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55, 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Hoffmann, F., Rapp, H. T., Zöller, T., and Reitner, J. (2003). Growth and regeneration in cultivated fragments of the boreal deep water sponge Geodia barretti bowerbank, 1858 (Geodiidae, Tetractinellida, Demospongiae). J. Biotechnol. 100, 109–118. doi: 10.1016/S0168-1656(02)00258-4
Horn, H., Slaby, B., Jahn, M., Bayer, K., Moitinho-silva, L., Förster, F., et al. (2016). An enrichment of CRISPR and other defense-related features in marine sponge-associated microbial metagenomes. Front. Microbiol. 7:1751. doi: 10.3389/fmicb.2016.01751
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Shengdar, Q., Sander, J. D., et al. (2013). Efficient in vivo genome editing using RNA-guided nucleases. Nat. Biotechnol. 31, 227–229. doi: 10.1038/nbt.2501
Ikmi, A., McKinney, S. A., Delventhal, K. M., and Gibson, M. C. (2014). TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 5:5486. doi: 10.1038/ncomms6486
Indraningrat, A., Smidt, H., and Sipkema, D. (2016). Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 14:87. doi: 10.3390/md14050087
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclase in adaptative bacterial immunity. Science 337, 816–822. doi: 10.1126/science.1225829
Kamke, J., Rinke, C., Schwientek, P., Mavromatis, K., Ivanova, N., Sczyrba, A., et al. (2014). The candidate phylum Poribacteria by single-cell genomics: new insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS ONE 9:e87353. doi: 10.1371/journal.pone.0087353
Kostic, A. D., Howitt, M. R., and Garrett, W. S. (2013). Exploring host – microbiota interactions in animal models and humans. Genes Dev. 27, 701–718. doi: 10.1101/gad.212522.112
Lavy, A., Keren, R., Haber, M., Schwartz, I., and Ilan, M. (2014). Implementing sponge physiological and genomic information to enhance the diversity of its culturable associated bacteria. FEMS Microbiol. Ecol. 87, 486–502. doi: 10.1111/1574-6941.12240
Li, C., Chen, J., and Hua, T. (1998). Precambrian sponges with cellular structures. Science 279, 879–882. doi: 10.1126/science.279.5352.879
Lin, C.-Y., and Su, Y.-H. (2016). Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Dev. Biol. 409, 420–428. doi: 10.1016/j.ydbio.2015.11.018
Liu, M., Fan, L., Zhong, L., Kjelleberg, S., and Thomas, T. (2012). Metaproteogenomic analysis of a community of sponge symbionts. ISME J. 6, 1515–1525. doi: 10.1038/ismej.2012.1
Liu, M. Y., Kjelleberg, S., and Thomas, T. (2011). Functional genomic analysis of an uncultured δ-proteobacterium in the sponge Cymbastela concentrica. ISME J. 5, 427–435. doi: 10.1038/ismej.2010.139
López-Legentil, S., Erwin, P. M., Pawlik, J. R., and Song, B. (2010). Effects of sponge bleaching on ammonia-oxidizing Archaea: distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta. Microb. Ecol. 60, 561–571. doi: 10.1007/s00248-010-9662-1
López-Legentil, S., Song, B., McMurray, S. E., and Pawlik, J. R. (2008). Bleaching and stress in coral reef ecosystems: hsp70 expression by the giant barrel sponge Xestospongia muta. Mol. Ecol. 17, 1840–1849. doi: 10.1111/j.1365-294X.2008.03667.x
Ludeman, D. A., Farrar, N., Riesgo, A., Paps, J., and Leys, S. P. (2014). Evolutionary origins of sensation in metazoans: functional evidence for a new sensory organ in sponges. BMC Evol. Biol. 14:3. doi: 10.1186/1471-2148-14-3
Margulis, L. (1991). “Symbiogenesis and symbionticism,” in Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis, eds L. Margulis and R. Fester (Cambridge, MA: MIT Press), 1–14.
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Mehbub, M. F., Lei, J., Franco, C., and Zhang, W. (2014). Marine sponge derived natural products between 2001 and 2010: trends and opportunities for discovery of bioactives. Mar. Drugs 12, 4539–4577. doi: 10.3390/md12084539
Mohamed, N. M., Enticknap, J. J., Lohr, J. E., McIntosh, S. M., and Hill, R. T. (2008a). Changes in bacterial communities of the marine sponge Mycale laxissima on transfer into aquaculture. Appl. Environ. Microbiol. 74, 1209–1222. doi: 10.1128/AEM.00454-08
Mohamed, N. M., Rao, V., Hamann, M. T., Kelly, M., and Hill, R. T. (2008b). Monitoring bacterial diversity of the marine sponge Ircinia strobilina upon transfer into aquaculture. Appl. Environ. Microbiol. 74, 4133–4143. doi: 10.1128/AEM.00454-08
Moitinho-Silva, L., Bayer, K., Cannistraci, C. V., Giles, E. C., Ryu, T., Seridi, L., et al. (2014). Specificity and transcriptional activity of microbiota associated with low and high microbial abundance sponges from the Red Sea. Mol. Ecol. 23, 1348–1363. doi: 10.1111/mec.12365
Morrow, K. M., Bourne, D. G., Humphrey, C., Botté, E. S., Laffy, P., Zaneveld, J., et al. (2015). Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 9, 894–908. doi: 10.1038/ismej.2014.188
Nguyen, M. T. H., Liu M., and Thomas, T. (2014). Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 23, 1635–1645. doi: 10.1111/mec.12384
Nobrega, F. L., Costa, A. R., Kluskens, L. D., and Azeredo, J. (2015). Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23, 185–191. doi: 10.1016/j.tim.2015.01.006.
Pita, L., Erwin, P. M., Turon, X., and López-Legentil, S. (2013a). Till death do us part: stable sponge-bacteria associations under thermal and food shortage stresses. PLoS ONE 8:e80307. doi: 10.1371/journal.pone.0080307
Pita, L., Turon, X., López-Legentil, S., and Erwin, P. M. (2013b). Host rules: spatial stability of bacterial communities associated with marine sponges (Ircinia spp.) in the Western Mediterranean Sea. FEMS Microbiol. Ecol. 86, 268–276. doi: 10.1111/1574-6941.12159
Radax, R., Rattei, T., Lanzen, A., Bayer, C., Rapp, H. T., Urich, T., et al. (2012). Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ. Microbiol. 14, 1308–1324. doi: 10.1111/j.1462-2920.2012.02714.x
Ribes, M., Calvo, E., Movilla, J., Logares, R., Coma, R., and Pelejero, C. (2016). Restructuring of the sponge microbiome favors tolerance to ocean acidification. Environ. Microbiol. Rep. 8, 536–544. doi: 10.1111/1758-2229.12430
Ribes, M., Jiménez, E., Yahel, G., López-Sendino, P., Diez, B., Massana, R., et al. (2012). Functional convergence of microbes associated with temperate marine sponges. Environ. Microbiol. 14, 1224–1239. doi: 10.1111/j.1462-2920.2012.02701.x
Richardson, C., Hill, M., Marks, C., Runyen-Janecky, L., and Hill, A. (2012). Experimental manipulation of sponge/bacterial symbiont community composition with antibiotics: sponge cell aggregates as a unique tool to study animal/microorganism symbiosis. FEMS Microbiol. Ecol. 81, 407–418. doi: 10.1111/j.1574-6941.2012.01365.x
Riesgo, A., Farrar, N., Windsor, P. J., Giribet, G., and Leys, S. P. (2014a). The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31, 1102–1120. doi: 10.1093/molbev/msu057
Riesgo, A., Peterson, K., Richardson, C., Heist, T., Strehlow, B., McCauley, M., et al. (2014b). Transcriptomic analysis of differential host gene expression upon uptake of symbionts: a case study with Symbiodinium and the major bioeroding sponge Cliona varians. BMC Genomics 15:376. doi: 10.1186/1471-2164-15-376
Riesgo, A., and Maldonado, M. (2008). Differences in reproductive timing among sponges sharing habitat and thermal regime. Invertebr. Biol. 127, 357–367. doi: 10.1111/j.1744-7410.2008.00128.x
Riesgo, A., Pérez-Porro, A. R., Carmona, S., Leys, S. P., and Giribet, G. (2012). Optimization of preservation and storage time of sponge tissues to obtain quality mRNA for next-generation sequencing. Mol. Ecol. Resour. 12, 312–322. doi: 10.1111/j.1755-0998.2011.03097.x
Rivera, A. S., Hammel, J. U., Haen, K. M., Danka, E. S., Cieniewicz, B., Winters, I. P., et al. (2011). RNA interference in marine and freshwater sponges: actin knockdown in Tethya wilhelma and Ephydatia muelleri by ingested dsRNA expressing bacteria. BMC Biotechnol. 11:67. doi: 10.1186/1472-6750-11-67
Ruby, E. G. (2008). Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol. 6, 752–762. doi: 10.1038/nrmicro1958
Ryu, T., Seridi, L., Moitinho-Silva, L., Oates, M., Liew, Y. J., Mavromatis, C., et al. (2016). Hologenome analysis of two marine sponges with different microbiomes. BMC Genomics 17:158. doi: 10.1186/s12864-016-2501-0
Sacristán-Soriano, O., Banaigs, B., and Becerro, M. A. (2016). Can light intensity cause shifts in natural product and bacterial profiles of the sponge Aplysina aerophoba? Mar. Ecol. 37, 88–105. doi: 10.1111/maec.12252
Schippers, K. J., Sipkema, D., Osinga, R., Smidt, H., Pomponi, S. A., Martens, D. E., et al. (2012). Cultivation of sponges, sponge cells and symbionts. Achievements and future prospects. Adv. Mar. Biol. 62, 273–337. doi: 10.1016/B978-0-12-394283-8.00006-0
Schmitt, S., Angermeier, H., Schiller, R., Lindquist, N., and Hentschel, U. (2008). Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74, 7694–7708. doi: 10.1128/AEM.00878-08
Siegl, A., Kamke, J., Hochmuth, T., Piel, J., Richter, M., Liang, C., et al. (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5, 61–70. doi: 10.1038/ismej.2010.95
Srivastava, M., Simakov, O., Chapman, J., Fahey, B., Gauthier, M. E. A., Mitros, T., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. doi: 10.1038/nature09201
Steindler, L., Schuster, S., Ilan, M., Avni, A., Cerrano, C., and Beer, S. (2007). Differential gene expression in a marine sponge in relation to its symbiotic state. Mar. Biotechnol. 9, 543–549. doi: 10.1007/s10126-007-9024-22
Steinert, G., Taylor, M. W., Deines, P., Simister, R. L., de Voogd, N. J., Hoggard, M., et al. (2016). In four shallow and mesophotic tropical reef sponges from Guam the microbial community largely depends on host identity. PeerJ. 4:e1936. doi: 10.7717/peerj.1936
Steinert, G., Whitfield, S., Taylor, M. W., Thoms, C., and Schupp, P. J. (2014). Application of diffusion growth chambers for the cultivation of marine sponge-associated bacteria. Mar. Biotechnol. 16, 594–603. doi: 10.1007/s10126-014-9575-y
Taylor, M. W., Hill, R. T., and Hentschel, U. (2011). Meeting report: 1st international symposium on sponge microbiology. Mar. Biotechnol. 13, 1057–1061. doi: 10.1007/s10126-011-9397-0
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/MMBR.00040-06
Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J. R., Easson, C., Astudillo-García, C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7:11870. doi: 10.1038/ncomms11870
Thomas, T., Rusch, D., DeMaere, M. Z., Yung, P. Y., Lewis, M., Halpern, A., et al. (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4, 1557–1567. doi: 10.1038/ismej.2010.74
Tian, R.-M., Wang, Y., Bougouffa, S., Gao, Z.-M., Cai, L., Bajic, V., et al. (2014). Genomic analysis reveals versatile heterotrophic capacity of a potentially symbiotic sulfur-oxidizing bacterium in sponge. Environ. Microbiol. 16, 1–44. doi: 10.1111/1462-2920.12586
Timmons, L., Court, D. L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. doi: 10.1016/S0378-1119(00)00579-5
Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. doi: 10.1016/j.cell.2013.04.025
Webster, N. S., Cobb, R. E., Soo, R., Anthony, S. L., Battershill, C. N., Whalan, S., et al. (2011). Bacterial community dynamics in the marine sponge Rhopaloeides odorabile under in situ and ex situ cultivation. Mar. Biotechnol. 13, 296–304. doi: 10.1007/s10126-010-9300-4
Webster, N. S., and Taylor, M. W. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346. doi: 10.1111/j.1462-2920.2011.02460.x
Webster, N. S., Taylor, M. W., Behnam, F., Lücker, S., Rattei, T., Whalan, S., et al. (2010). Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x
Webster, N. S., and Thomas, T. (2016). The sponge hologenome. MBio 7:e00135-16. doi: 10.1128/mBio.00135-16
Weigel, B. L., and Erwin, P. M. (2016). Intraspecific variation in microbial symbiont communities of the sun sponge, Hymeniacidon heliophila, from intertidal and subtidal habitats. Appl. Environ. Microbiol. 82, 650–658. doi: 10.1128/AEM.02980-15
Weisz, J. B., Lindquist, N., and Martens, C. S. (2008). Do associated microbial abundances impact marine demosponge pumping rates and tissue densities? Oecologia 155, 367–376. doi: 10.1007/s00442-007-0910-0
Keywords: Porifera, holobiont, microbiota, aquaculture, metazoan symbiosis, RNAi, CRISPR-Cas
Citation: Pita L, Fraune S and Hentschel U (2016) Emerging Sponge Models of Animal-Microbe Symbioses. Front. Microbiol. 7:2102. doi: 10.3389/fmicb.2016.02102
Received: 21 September 2016; Accepted: 12 December 2016;
Published: 23 December 2016.
Edited by:
Mike Taylor, University of Auckland, New ZealandReviewed by:
Robert W. Thacker, Stony Brook University, USACopyright © 2016 Pita, Fraune and Hentschel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucia Pita, bHBpdGFAZ2VvbWFyLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.