- 1Key Laboratory of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2School of Environmental Sciences, University of Guelph, Guelph, ON, Canada
False smut of rice, caused by Ustilaginoidea virens (Cooke) Takahashi (teleomorph: Villosiclava virens), is one of the most important diseases affecting rice worldwide. Agrobacterium tumefaciens-mediated transformation was used to identify functional genes in U. virens. In this study, we selected a single-copy insertion mutant T133 with deficiency in producing conidia by screening the T-DNA insertion mutant library of U. virens. The UvPRO1-deletion mutant was successfully obtained after cloning the targeted gene by analysis of the T-DNA insert site of mutant T133. Further research showed that the UvPRO1 mutant was reduced in growth rate and could not produce conidia in PSB medium, while sensitivities to sodium dodecyl sulfate, Congo red, and hyperosmotic stress increased. Moreover, the UvPRO1 deletion mutant hyphae could extend along the surface of spikelets at 1–3 dpi, but mycelia became shriveled and completely lost the ability to infect spikelets at 4 dpi. The relative expression level of UvPRO1 at 8 dpi was more than twice as high as that at 1–2 dpi. These results suggest that UvPRO1 plays a critical role in hyphal growth and conidiation, as well as in stress response and pathogenesis. These findings provide a novel mode of action for the PRO1 protein in fungi and improve the understanding of the function of UvPRO1 in the life cycle of U. virens.
Introduction
False smut of rice, caused by Ustilaginoidea virens (Cooke) Takahashi (teleomorph: Villosiclava virens), is a minor disease that has been present in the major rice-growing areas of Asia, Africa, and America for some time (Deng, 1989; Savary et al., 2000; Ashizawa et al., 2010). Since the beginning of this century, it has become one of the most devastating grain diseases that threatens rice production worldwide, due to the widespread cultivation of susceptible high-yield hybrid rice varieties, intensive application of chemical fertilizers, and an apparent change in global climates (Rush et al., 2000; Wang et al., 2004; Singh and Pophaly, 2010; Guo et al., 2012). Occurrence of rice false smut not only affects yield, but creates a health issue by producing ustiloxins, which are microtubule inhibitors toxic to humans and animals (Koiso et al., 1994; Miyazaki et al., 2009).
Prior research on Ustilaginoidea virens has concentrated on the biology of the organism, including its distribution and detection, toxin production, and disease cycle and management (Zhou et al., 2003; Brooks et al., 2009; Tang et al., 2013). Compared with other important diseases such as rice blast and bacterial leaf blight, studies on the interaction of the false smut pathogen and the rice host at the molecular level are few. Sun et al. (2013) reported the genome sequence of U. virens and predicted possible effectors. Zhang et al. (2008) characterized the first MAPK protein from U. virens and verified that UVMK1 is a homolog of Magnaporthe grisea PMK1. Rao et al. (2014) cloned a homolog of HOG1 from U. virens and measured transcript levels of UvHog1 under salinity conditions, suggesting that UvHog1 may be involved in the specific response to salt stress. Fan et al. (2015) used time-course microscopic and transcriptional approaches to investigate host responses to U. virens infection, and the results implied that U. virens may hijack rice nutrient reservoir systems to successfully colonize rice floral organs and to form false smut balls.
In recent years, generation of random mutant collections via Agrobacterium tumefaciens-mediated transformation (ATMT) has been widely used in different fungal species to study gene functions (Mullins and Kang, 2001; Mullins et al., 2001; Sugui et al., 2005; Frandsen, 2011). Zhang et al. (2006) first reported the transformation of U. virens by the ATMT method. Yu et al. (2013) cloned the spo76 gene in the T-DNA insertion mutant A2588, which is a high-yield mutant of rice germ, and found that reduced levels of spo76 gene expression may enhance conidiation of U. virens. Yu et al. (2015) obtained 37 mutants with reproducible pathogenic defects and cloned the UvSUN2 gene from mutant B20; their morphophysiological characterization analysis suggested that UvSUN2 was required for hyphal growth, cell wall construction, stress response, and virulence. Wang et al. (2015) selected an avirulent T-DNA insertion mutant, B1464, and obtained a C2H2-type zinc finger protein gene, which might be related to sporulation and pathogenicity. Bo et al. (2016) found a GH18 family gene in U. virens by screening of a T-DNA insertional library, which is most likely related to hyphal growth, sporulation, and pathogenicity. Zheng M.T. et al. (2016) cloned and analyzed Uvt3277, which is a low-affinity iron transport protein, verifying the relationship with pathogenicity by RNAi.
Although previous research studies have reported many genes which might be related to hyphal growth, sporulation, or pathogenicity, few studies of deletion targeted genes by homologous recombination have been reported in U. virens. It may be possible that U. virens has a relatively low homologous recombination frequency, as so far only Zheng D. et al. (2016) obtained the UvHOG1 deletion mutant and demonstrated that UvHOG1 likely has a conserved role in regulation stress responses, hyphal growth, and possibly secondary metabolism.
In this study, we selected four strains of sporulation defect mutants and one strain that does not produce a conidia by screening the T-DNA insertion mutant library, and we successfully obtained a UvPRO1 deletion mutant after cloning the target gene by analysis of the T-DNA insert site of mutant T133. Further research showed the UvPRO1 mutant was reduced in for growth rate and conidiation, and had increased sensitivity to sodium dodecyl sulfate (SDS), Congo red (CR) and hyperosmotic stress, and significantly reduced virulence. However, the PRO1 gene has not been reported in U. virens; it was first identified in Sordaria macrospora in a genetic screen for mutations defective in perithecia development (Masloff et al., 1999, 2002).
In Cryphonectria parasitica, disruption of the PRO1 gene resulted in a significant reduction in asexual sporulation and loss of female fertility (Sun et al., 2009). Tanaka et al. (2013) identified a mutant with an insertion in PRO1 in a forward genetic screen to identify Epichloe festucae symbiosis genes, and demonstrated that PRO1 is a central regulator for in planta specific growth of E. festucae. Compared with the role of PRO1 in other fungi, UvPRO1 not only regulated hyphal growth and conidiation, but was also involved in stress response and pathogenesis. Functional elucidation can provide a novel mode of action of PRO1 in fungi and improve our understanding of the function of UvPRO1 in the life cycle of U. virens.
Materials and Methods
Strains, Plasmids, and Plants
The wild-type strain HWD2 and all the transformants of U. virens generated in this study were routinely cultured on potato sucrose agar (PSA, 2% sucrose plus extract from boiled peeled potato) at 28°C, and stored in the form of mycelial-colonized filter paper at -20°C. The A. tumefaciens strain EHA105 and binary vector pTFCM were used for U. virens transformation. Plasmids KS1004 and pneoP3300III were used for gene disruption or complementation vector construction.
The susceptible rice cultivar Wanxian 98 was used in virulence assays. The seeds were kept for 24 h at 30°C before planting. After 10 days, four seedlings were placed into pots (25 cm × 20 cm × 30 cm, length × width × height) each containing 5 kg of autoclaved paddy soil. In the greenhouse, pots were fertilized twice (4 g carbamide per bucket): once at tillering (after 45 days of growth) and just before inoculation at the at the booting stage (after 90 days of growth; Jia et al., 2015).
Agrobacterium-Mediated Transformation of U. virens
Agrobacterium-mediated transformation was carried out following the protocols described Yu et al. (2015) with minor modifications. The wild-type strain HWD2 was cultured in a 250 mL flask containing 150 mL liquid potato sucrose broth (PSB). The flask was placed in a shaking incubator at 28°C in the dark. After shaking at 160 rpm for 7 days, the cultures were filtered through multiple layers of cheese cloth, and conidia were obtained from the filtrate by centrifugation (3,000 rpm for 5 min). The conidial suspension was adjusted to 1 × 106 conidia per mL using a haemocytometer.
The A. tumefaciens strain EH105 was grown at 28°C with shaking at 180 rpm for 48 h in minimal medium supplemented with kanamycin (50 μg/mL). Then, A. tumefaciens cells were grown in induction medium supplemented with 200 μM acetosyringone. After shaking at 180 rpm for an additional 10 h at 28°C, bacterial cultures were diluted to an optical density of 0.5 OD units at 600 nm and were mixed 1:1 with a conidial suspension from HWD2 (106 spores/mL). The mix was plated onto co-cultivation medium with a layer of nitrocellulose filter. After co-cultivation at 24°C for 4 days, the membrane was removed, and placed mycelium-side down onto PSA containing 500 μg/mL of cefotaxime to counter-select bacteria, and 200 μg/mL of hygromycin to select for U. virens transformants. After incubation at 28°C for 5–7 days, transformant colonies were transferred to PSA plates containing 200 μg/mL of hygromycin for a second round of selection.
To test for the mitotic stability of the integrated hygromycin resistance cassette, 20 randomly chosen transformants were cultivated on PSA without hygromycin. After weekly transfer to new plates for four passages by subculturing of hyphal tips, transformants were grown on PSA plates containing hygromycin (200 mg/mL).
Conidiation Test of ATMT Transformants
The fungus was propagated in PSA plates for 14 days at 28°C. Then, 3-mm-diameter mycelia dishes were cut from the edge of a colony and inoculated in a 50 mL flask containing 30 mL PSB which was placed in a shaking incubator. After shaking at 180 rpm for 7 days, the cultures were filtered through three layers of gauze, and conidial production was measured using a haemocytometer. The experiment was repeated three times with three replicates each time.
Amplification and Analysis of T-DNA Flanking Sequences
Genomic DNA sequences of the transformants flanking T-DNA insertions were amplified by TAIL-PCR (thermal asymmetric interlaced-polymerase chain reaction) and inverse PCR with primer sequences shown in Supplementary Table 1. For TAIL-PCR, genomic DNA was used as a template in successive reactions with nested left border primers (LB1, 2, and 3) and right border primers (RB1, 2, and 3) together with the degenerate primers (AD1, 2, 3, or 4). PCR settings for TAIL-PCR followed Liu et al. (2013). For inverse PCR, genomic DNA was digested with SacI and circularized with T4 DNA ligase (Invitrogen, Karlsruhe, Germany). The product was purified using a Nucleic Acid Purification kit (Axygen, Union City, CA, USA). The reaction conditions for first round PCR were: 1 cycle at 95°C for 5 min, 30 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 4 min and a final cycle at 72°C for 5 min. The second round nested PCR was performed with the same PCR program using 1 ml of the first round PCR product (diluted 1: 50) as a template together with nested primers (Liu et al., 2013). Flanking sequences recovered by TAIL-PCR and inverse PCR were analyzed with the BLAST tool hosted by the National Center for Biotechnology Information1 against the GenBank database and the genome sequences of U. virens (NCBI, JHTR00000000.1). Nucleotide sequences were compared with known protein sequences using BLASTX (NCBI2). Open reading frames (ORFs) were analyzed using FGENESH (Softberry Inc., Mount Kisco, NY, USA), conserved domains were detected by comparison to the Conserved Domain Database of NCBI3.
Identification and Disruption of the U. virens PRO1 Gene
The full sequence of UvPRO1 was obtained from the genome sequence of U. virens (NCBI, JHTR00000000.1). To confirm sequence presence, the primers UvPRO1F and UvPRO1R (Supplementary Table 1) were designed and used for the amplification of the UvPRO1 gene from HWD2 isolates. Primers were all designed using Primer Premier 5.04, and ORFs were analyzed using FGENESH. Protein domain and motif predictions were performed with SMART software5.
The PRO1 protein sequences from different organisms were obtained from the GenBank database, using the BLAST algorithm with the UvPRO1 sequence. Sequence alignments were performed using the Clustal X (version 2.06), and a phylogenetic tree was generated with Mega software (version 7.07) using the Neighbor-Joining method.
To assess the function of UvPRO1, which was potentially mutated in T133, a vector was constructed for the targeted disruption of UvPRO1 by means of homologous recombination. Vector KS1004 was constructed by cloning a 1.9 kb PtrpC-hph cassette into the SmaI site of pBluescriptII KS, and the hygromycin resistance was used as the first selectable marker for screening of disruption transformants. Vector pneoP3300III was generated by cloning a 2.1 kb neomycin resistant gene cassette into the XbaI site of pCAMBIA3300, and the neomycin resistance was used as the second selectable marker.
A pair of gene-specific primers, UvPRO1F1F and UvPRO1F1R (Supplementary Table 1), was used to amplify the 900 bp fragment (Figure 4A) in the 5′ coding region of UvPRO1. Another pair of gene-specific primers, UvPRO1F2F and UvPRO1F2R (Supplementary Table 1), was used to amplify the 978 bp fragment, containing part of the 3′ coding region of UvPRO1 (Figure 4A). The 900 bp HindIII/SalI-fragment (5′ region of UvPRO1) and the 978 bp XbaI/KpnI-fragment (3′ region of UvPRO1) were cloned into the corresponding restriction sites of the vector KS1004, resulting in the preliminary vector KS1004-UvPRO1. The hph-UvPRO1 cassette (with a 900 bp HindIII/SalI-fragment, a 1909 bp hph-fragment, and a 978 bp XbaI/KpnI-fragment) was cloned into pneoP3300III, resulting in the gene disruption vector p3300neoUvPRO1.
This vector, p3300neoUvPRO1, was transformed into A. tumefaciens EHA105 by electroporation, and then hyphae were transformed with the ATMT protocol. To find UvPRO1 disruption transformants, cultures were grown on PSA amended with hygromycin (200 mg/mL), and then subcultured onto PSA amended with 800 μg/mL of antibiotic G418 (Amresco, Solon, OH, USA). Gene disruption transformants were subjected to PCR with two pairs of primers, UvPRO1KF/UvPRO1KR and HphF/HphR (Supplementary Table 1), and amplicons were detected by PCR and Southern blot analysis.
Complementation of UvPRO1 Disruption Mutant
To confirm targeted gene disruption, the disruption mutant ΔUvPRO1-27 was complemented with a full length sequence of UvPRO1. Because UvPRO1 disruption mutants were unable to grow on the PSA supplemented with G418, the neomycin resistance cassette was chosen as a selectable marker for the complementation transformation. The complementation plasmid p3300neoUvPRO1-Com was based on pneoP3300III. The 3,315 bp UvPRO1 fragment (UvPRO1 ORF plus 574 bp 5′-flanking and 905 bp 3′-flanking sequences) was amplified from genomic DNA of the wild-type with the primer pair UvPRO1ComF and UvPRO1ComR (Supplementary Table 1), and cloned into the BamHI site of pneoP3300III to generate the complementation plasmid p3300neoUvPRO1-Com. To obtain the UvPRO1 complementation transformants, ΔUvPRO1-27 was transformed with vector p3300neoUvPRO1-Com by the ATMT method. The complementation transformants were screened on PSA containing 800 μg/mL G418, and gene fragments were detected by RT-PCR.
DNA Manipulation and Southern Blot Analysis
Genomic DNA was extracted using CTAB (Sambrook et al., 1989). For Southern blot analysis of T-DNA insertion in U. virens, PCR was used to confirm the presence of T-DNA insertions by using primers HphF and HphR (Supplementary Table 1) to amplify an 887 bp internal region of the hygromycin resistance gene (hph). DNA from the wild-type and the transformants was completely digested with SacI, which has only one recognition site in the binary vector pTFCM, and then size-fractionated through a 0.8% agarose gel and mounted onto a positively charged nylon membrane (Figure 2A). The hph gene was excised from the pTFCM vector and labeled with digoxigenin (DIG)-dUTP using the PCR DIG probe synthesis kit (Roche, Mannheim, Germany) following manufacturer’s instructions. Hybridization was detected using a DIG luminescence detection kit. For Southern blot analysis of UvPRO1 disruption mutants, genomic DNA from the wild-type and the putative UvPRO1 disruption mutants were digested with SacI at 37°C for 24 h. The nylon membrane was hybridized with probe P (Figure 4A).
RNA Isolation and qRT-PCR Analysis
Hyphae harvested from PSB medium were collected at different points in time (3, 4, 5, 6, 7, 8, and 9 days), as well as inoculated spikelets at different points in time (1, 2, 3, 4, 6, 8, 10, and 12 days). These were frozen in liquid nitrogen and stored at 80°C until required. RNA was extracted using a TRIzol Plus RNA purification kit (Invitrogen, Carlsbad, CA, USA). DNA contamination was removed by DNaseI treatment (RNase free; TaKaRa, Dalian, China). First-strand cDNA was synthesized by using a RevertAidTM first strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany). Expression of UvPRO1 in disruption mutants and the complementation strain were examined by RT-PCR, and a 1560-bp fragment was amplified with gene-specific primers qRT-UvPRO1F and qRT-UvPRO1R (Supplementary Table 1). PCR conditions used 25 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min.
Expression of UvPRO1 at different developmental stages of the fungus in vitro or in planta was analyzed by qRT-PCR with UvPRO1 gene-specific primers qRT-PRO1F/qRT-PRO1R (Supplementary Table 1). PCR conditions were 40 cycles of 94°C for 15 s, 55°C for 20 s, and 72°C for 15 s, and with a final extension from 65°C to 95°C (0.5°C/5 s; Gu et al., 2012). The U. virensα-tubulin2, as the reference gene, was amplified with primers α-tubulin2F and α-tubulin2R (Supplementary Table 1). PCR reactions were run on a PTC-200 DNA Engine Peltier thermal cycler (BioRad, Hercules, CA, USA). The whole experiment was repeated three times.
Phenotypic Analysis
For mycelial growth, mycelial plugs (5 mm in diameter) were transferred from 12-day-old PSA plates and grown on fresh PSA medium at 28°C. After 6 and 12 days of being cultured, the radial growth of vegetative mycelia was measured. For conidial production, strains were grown in PSB medium at 28°C. After shaking at 180 rpm for different lengths of time (4, 5, 6, 7, 8, and 9 days), the cultures were filtered through three layers of gauze, and conidial production was measured using the haemocytometer. For testing the sensitivity to various stress chemicals, the strains exposed to CM medium containing either exogenous 0.1–0.5 M NaCl, 0.01–0.05% SDS, or 30–70 mg/L CR were assessed also by measuring colony diameter of 14-day cultures. Each treatment was repeated three times.
Pathogenicity Assay
For pathogenicity analysis, mycelial plugs of the wild-type, UvPRO1 knock out and complementation strains were transferred from 12-day-old PSA plates and grown in PSB medium at 28°C. After shaking at 180 rpm for 5 days, the cultures were homogenized in a blender, and rice plants were inoculated with 2 mL of mycelial suspension using a syringe in the middle section of distal internodes at the eight stage of panicle development. The rice plants were placed in a plant growth chamber (Wuhan Ruihua Instrument and Equipment Co., Ltd., Wuhan, China) equipped with a high pressure sodium lamp (12 h light/dark cycle) with conditions set at a RH of 95 ± 5% and a temperature of 25 ± 1°C. After a post-inoculation surface wetness period of 120 h, plants were transferred to a greenhouse equipped with an automatic climate control system set at 28 ± 2°C and 75 ± 7% RH. This experiment was repeated three times (Jia et al., 2015). Five of the injected panicles were sampled at each time point (1, 2, 3, 4, 6, 8, 10, and 12 days after inoculation) and others were used to count the severity of false smut infection 15 days after inoculation.
Scanning Electron Microscopy
The samples for scanning electron microscopy were first fixed with 2.5% (v/v) glutaraldehyde in 50 mM phosphate buffer (pH 7.2) for 6–8 h at 4°C, before a rinse with the same buffer for 2 h. They were then fixed in 1% (w/v) osmium tetroxide in 50 mM phosphate buffer for 1 h. After dehydration in a graded acetone series, the samples were critical-point dried, mounted on stubs, sputter coated with gold-palladium, and viewed using a JEOL JSM-6390LV scanning electron microscope operating at 10 kV (Hu et al., 2014).
Statistical Analysis
The quantitative data were analyzed with DPS statistical analysis software (version 3.01, China Agric. Press, Beijing, China), using ANOVA. When significant treatment effects were found (P < 0.05), separation of means was done using Fisher’s Least Significant Difference test.
Results
Screening and Analysis of Sporulation Deficiency Mutants
Using the modified protocol for ATMT, a total of 3,016 hygromycin-resistant transformants of U. virens were obtained. The mitotic stability of the integrated T-DNA was tested by analysis of 20 randomly selected transformants. Transformants were serially subcultured for five time on PSA medium not containing hygromycin. Transformants retained the integrated T-DNA, as indicated the ability to grow on PSA containing hygromycin.
All of the 3,016 transformants were screened for sporulation deficiency and five transformants with sporulation deficiency were found. Four transformants (T420, T896, T1296, T2328) were found to have significantly (P < 0.05) lower conidial production, and one transformant (T133) was found to have no conidia (Figure 1). Southern blot analysis showed that, among several mutants with sporulation deficiency, four (T133, T420, T896, T2328) contained a single T-DNA insertion and T1296 contained two T-DNA copies (Figure 2B). T-DNA flanking sequences were recovered from these mutants by amplifying genomic DNA sequences flanking T-DNA insertions of transformants with TAIL-PCR and inverse PCR (Supplementary Table 1). These sequences were used to screen the GenBank database and the genome sequences of U. virens (NCBI, JHTR00000000.1). FGENESH was used to identify ORFs around the T-DNA insertion site. ORF sequences were compared against protein sequences from NCBI with BLASTX. Details on affected genes and disruption sites are shown in Table 1.
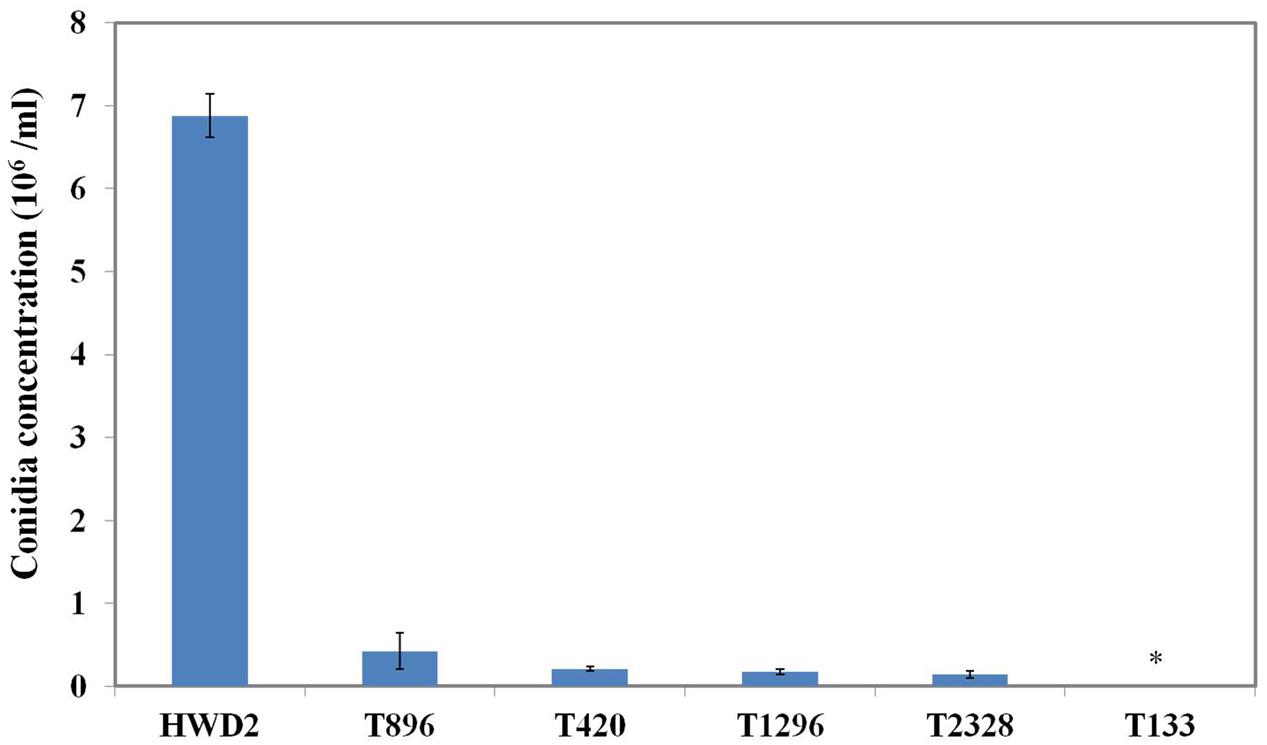
FIGURE 1. Sporulation by conidiation defect mutants. Quantitative analysis of the conidiation of Ustilaginoidea virens wild-type HWD2 and five mutants following growth for 7 days at 28°C in potato sucrose broth (PSB) medium. Asterisks indicate no conidia were observed in T133.
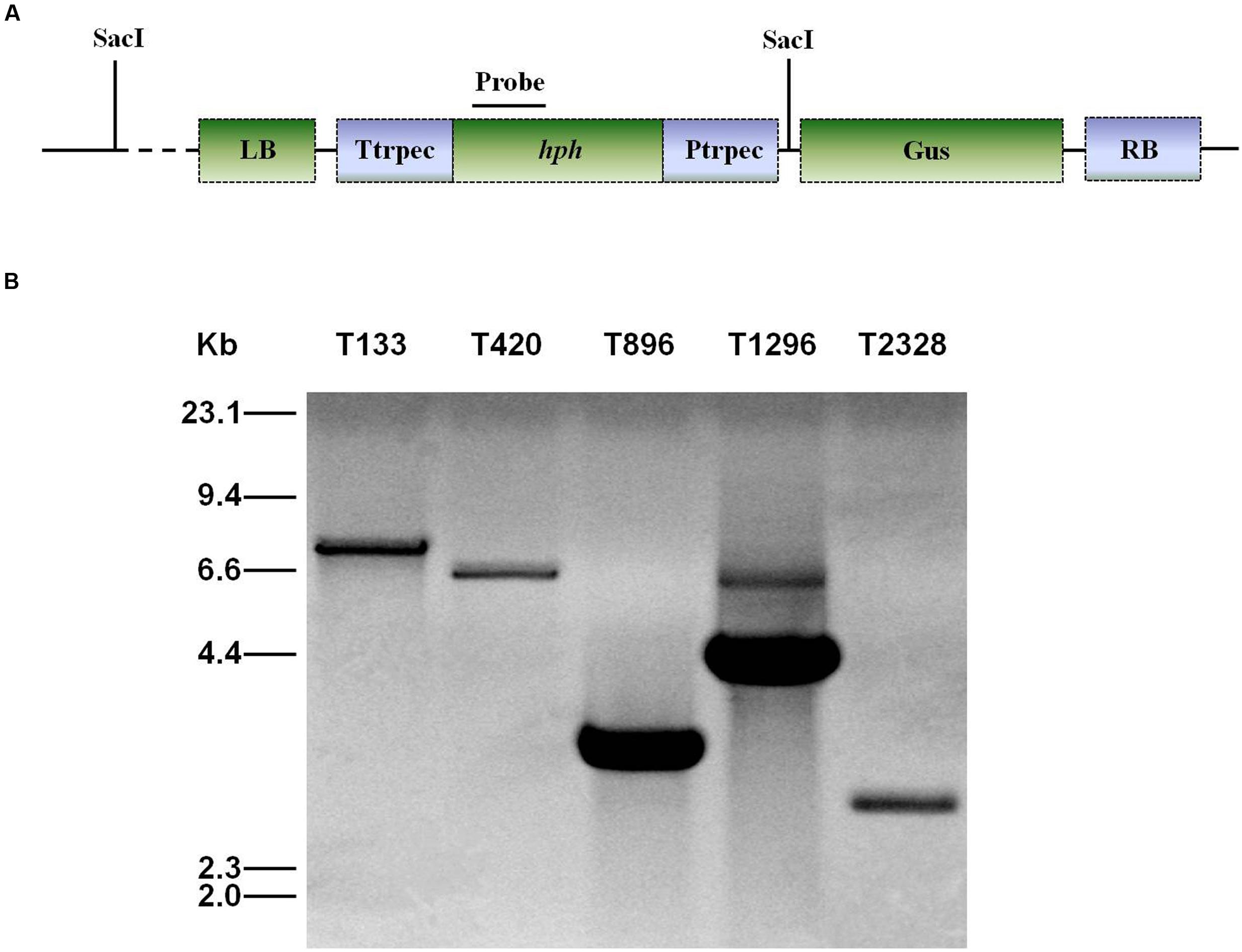
FIGURE 2. Southern blot analysis of genomic DNA of U. virens mutants. (A) The top section shows the ATMT vector pTFCM. The region for the probe used for Southern blot hybridization and SacI restriction sites are indicated. (B) The bottom section shows Southern blot analysis of conidiation mutants of U. virens. Genomic DNA of transformants was digested with SacI.
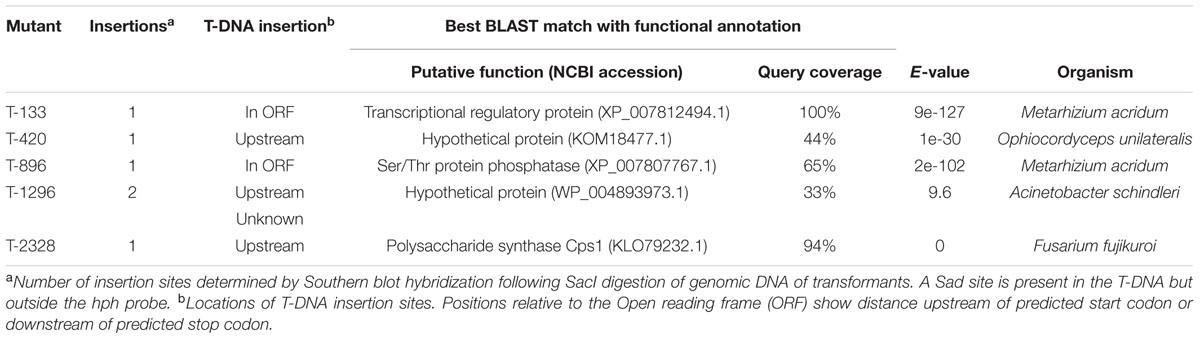
TABLE 1. Summary of Ustilaginoidea virens genes identified from T-DNA flanking sequences with the best BLAST matches.
In mutant T420, the targeted gene encodes a hypothetical protein, showing similarity to a protein of unknown function from Ophiocordyceps unilateralis (GenBank KOM18477.1). In mutant T896, a single insertion was located inside a predicted ORF of a gene with significant similarity to the Ser/Thr protein phosphatase gene of Metarhizium acridum (GenBank XP_007807767.1). In mutant T1296, one insertion was located upstream of a gene that showed high sequence similarity to a hypothetical protein from Acinetobacter schindleri (GenBank WP_004893973.1), and the other failed during cloning. In mutant T2328, T-DNA targeted upstream of a gene with significant similarity to a polysaccharide synthase Cps1 gene of Fusarium fujikuroi (GenBank KLO79232.1). In mutant T133, T-DNA was inserted into a predicted ORF of a PRO1 gene encoding C6 transcription factor that showed high sequence similarity to a PRO1 gene of M. acridum (GenBank XP_007812494.1; Table 1). The mutant T133 was characterized by no sporulation and contained a single T-DNA insertion. Therefore, our subsequent work focused on the gene UvPRO1 in the mutant.
Identification and Characterization of UvPRO1
The aligned sequences of overlapping DNA fragments of the PRO1 gene amplified by PCR from U. virens genomic DNA and from corresponding mRNA revealed a 2,440-bp ORF. The coding domain was predicted to encode a polypeptide consisting of 611 amino acids and a high level of sequence identity (84%) with transcriptional regulatory protein PRO1 of M. acridum CQMa 102. Sequence analysis with SMART revealed that UvPRO1 contained a Fungal_trans_2 conserved domain (Figure 3A).
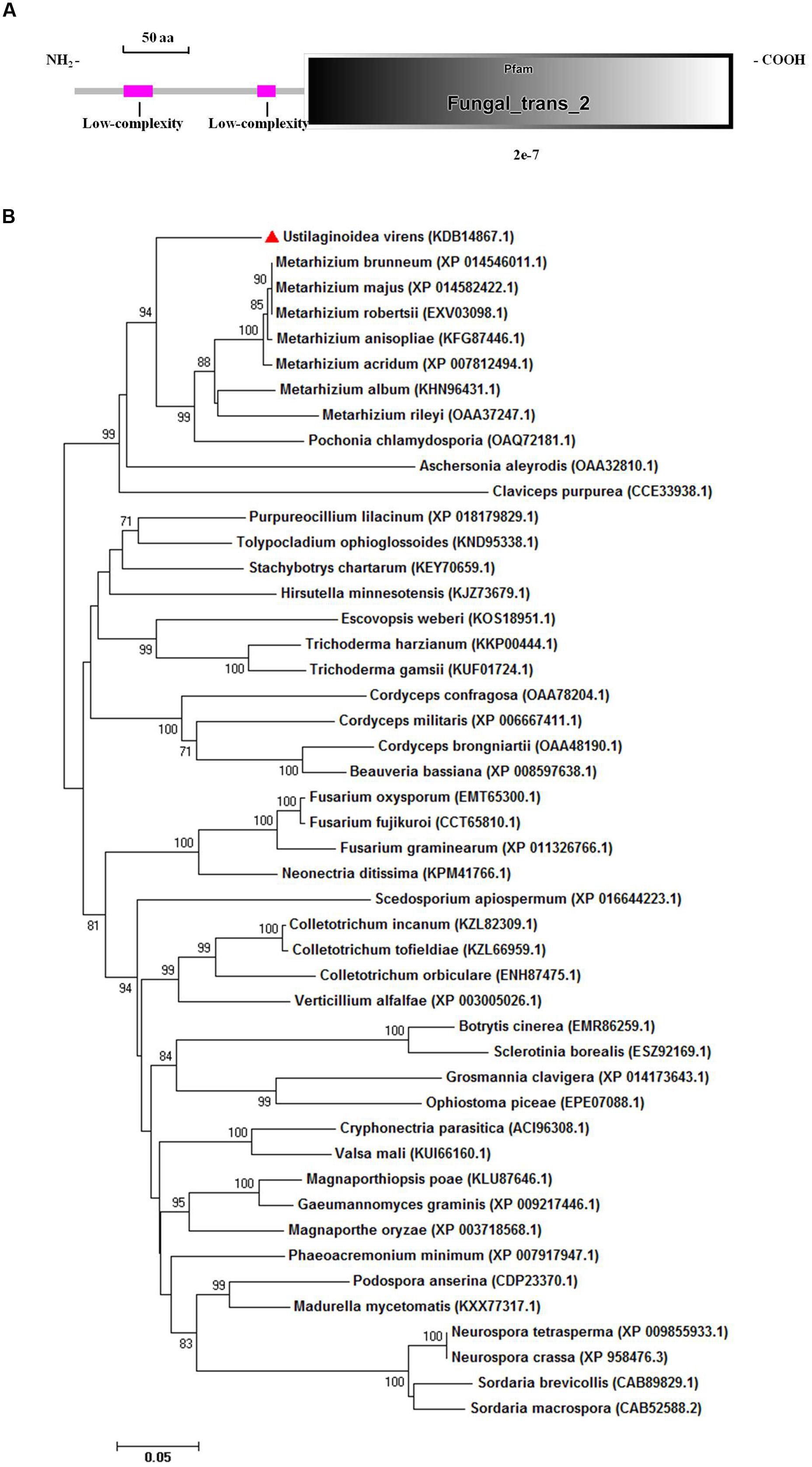
FIGURE 3. Functional domain identification and phylogenetic tree. (A) A conserved Fungal specific transcription factor domain (Fungal_trans_2 domain) and two low-complexity regions in UvPRO1 were predicted using SMART website. (B) Neighbor-Joining analysis of UvPRO1 with 46 homologs from other fungal species. Sequence alignments were performed using the Clustal X 2.0 program and the tree was generated using Mega 7.0 program with 1,000 bootstrap replicates. All 47 protein sequences of the PRO1 homologs were downloaded from the NCBI database.
Phylogenetic analysis of UvPRO1 (GenBank KDB14867.1) to other PRO1 proteins (Figure 3B) revealed that UvPRO1 was most similar to PRO1 proteins of Pochonia chlamydosporia and species of Metarhizium (with identities above 81%), and more distant from those of other fungi (with identities above 58%). This result indicates that PRO1 proteins are conserved among fungi tested.
Disruption and Complementation of UvPRO1
A gene disruption vector, p3300neoUvPRO1, containing the hph gene and both the 3′ and 5′ flanking regions of UvPRO1, was constructed with two vectors, KS1004 and pneoP3300III (Figure 4A). Vector p3300neoUvPRO1 was transformed into the wild-type, and transformants were selected on hygromycin-containing medium and on G418-containing medium. Among 628 hygromycin-resistant transformants, three without resistance to G418 were obtained.
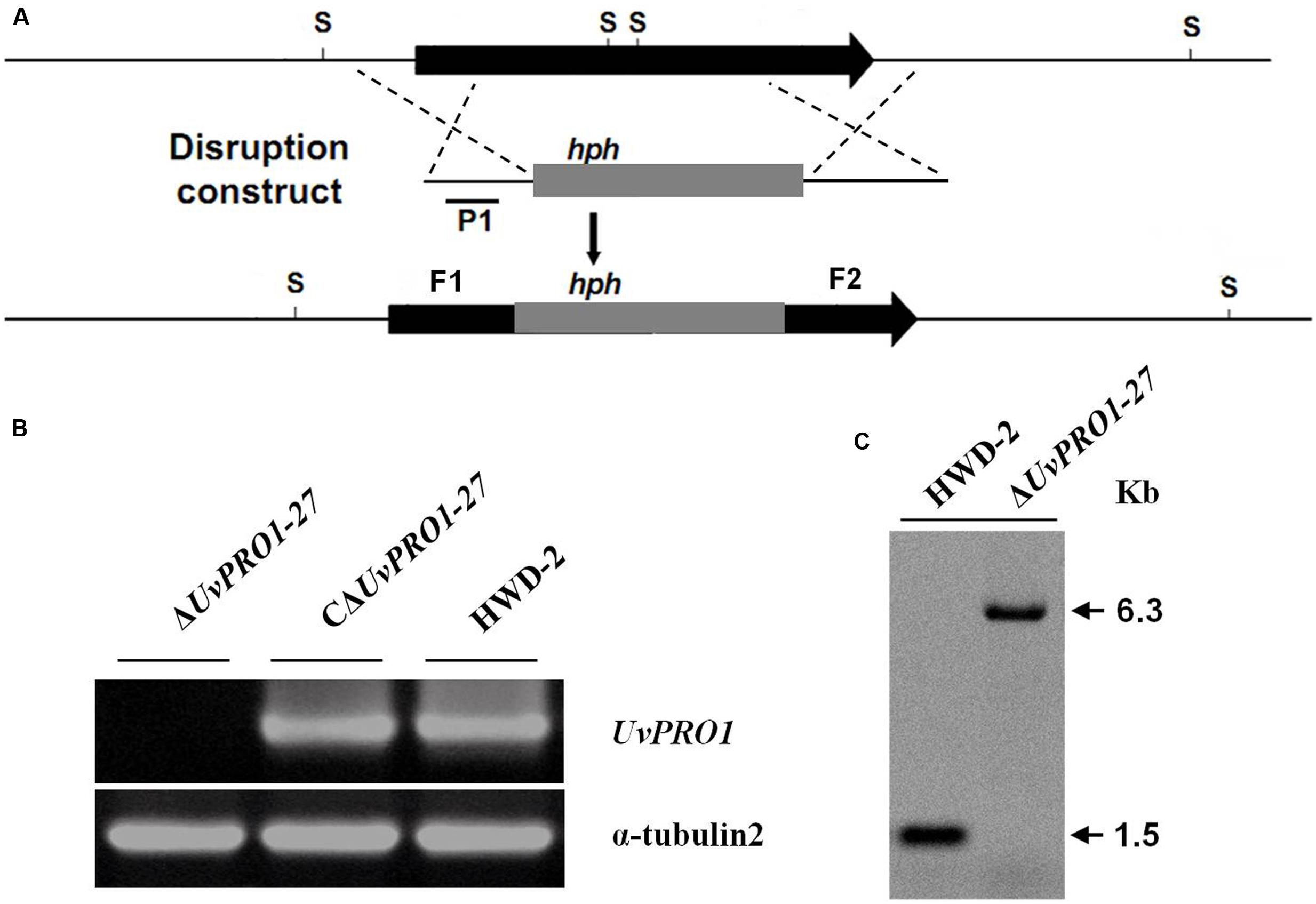
FIGURE 4. Targeted disruption of UvPRO1 gene. (A) Strategic map of gene disruption vector construction and restriction map of UvPRO1 genomic region. (B) RT-PCR analysis of the transcription of UvPRO1 disruption mutant, complementation mutant, and wild-type strain with gene-specific primers UvPRO1F and UvPRO1R. (C) Southern blotting of the SacI-digested genomic DNA from the wild-type and UvPRO1 disruption mutant, which was hybridized with probe P1.
Only one candidate disruption transformant ΔUvPRO1-27 was found lacking the 1208-bp UvPRO1 fragment compared to the wild-type strain after PCR amplification with PRO1F/PRO1R (Supplementary Table 1); however, an 887-bp hph fragment was obtained by PCR amplification with hphF/hphR (Supplementary Table 1) in this candidate transformant. Furthermore, Southern blot analysis showed that single integration events had occurred in selected UvPRO1 knockout transformant ΔUvPRO1-27 (Figure 4C), which had the 6.3-kb SacI fragment, while the wild-type strain HWD2 had the 1.5-kb SacI fragment. Null mutation of the UvPRO1 gene was further confirmed by RT-PCR analysis, since the UvPRO1 transcript was not detected in the targeted disruption transformant. These results demonstrated that the UvPRO1 gene was deleted in the UvPRO1 disruption transformant ΔUvPRO1-27. To investigate whether altered growth phenotypes and the loss of virulence in UvPRO1 disruption transformants could be restored by reintroduction of a wild-type copy of UvPRO1, we transformed ΔUvPRO1-27 with plasmid pNeo3300IIIUvPRO1-Com. Subsequently, complementation transformant CΔUvPRO1-27 was confirmed by RT-PCR analysis (Figure 4B) and was selected for further studies.
UvPRO1 Affects Vegetative Growth and Conidiation
The morphology of the strains was monitored on PSA medium. The ΔUvPRO1-27 mutant produced white colonies with long and abundant aerial hyphae, in contrast with the colonies with a light yellow center surrounded by a white edge of the wild-type rescued strain CΔUvPRO1-27 (Figure 5A). Furthermore, the ΔUvPRO1-27 strain had a reduced apical extension rate (2.6 mm/d), producing smaller colonies than the wild-type (2.8 mm/d) and CΔUv-PRO1-27 (2.8 mm/d), and the mycelial growth rate measured at 6 days on PSA of the ΔUvPRO1-27 strain (2.20 mm/d) was significantly less than the wild-type (2.73 mm/d; Figure 5B).
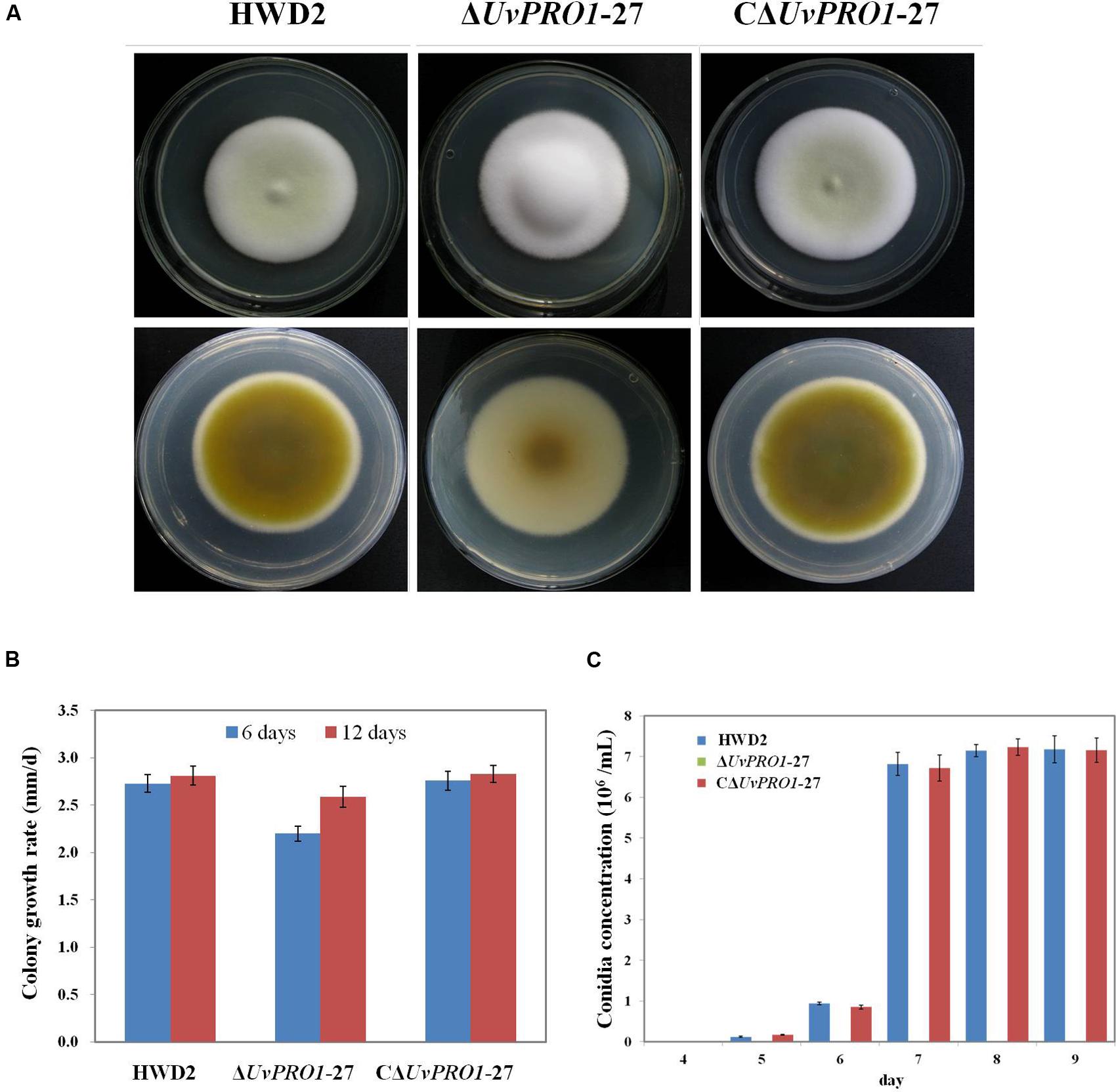
FIGURE 5. Growth phenotypes of wild-type HWD2, ΔUvPRO1-27, and CΔUvPRO1-27. (A) The morphology of the strains on PSA medium after incubation at 28°C for 14 days. (B) The growth rate of wild-type HWD2, ΔUvPRO1-27, and CΔUvPRO1-27 on potato sucrose agar (PSA) medium. (C) Sporulation of wild-type HWD2, ΔUvPRO1-27, and CΔUvPRO1-27 at different times in potato sucrose broth (PSB) medium. Asterisks indicate no conidia were observed in ΔUvPRO1-27, in contrast to wild-type HWD2 and CΔUvPRO1-27.
In PSB medium, mycelia from wild-type strains and the rescued strain CΔUvPRO1-27 produced hyphae with conidiophores at their tips after 5–6 days, and conidia were produced after 7 days at 6.7 or 6.8 × 106 conidia/mL, respectively. However, mycelia of the ΔUv-PRO1-27 produced hyphae without conidiophore formation at 6 days, and no conidia were observed up to 9 days (Figure 5C).
The Importance of UvPRO1 for Regulation Responses to Hyperosmotic and Cell Membrane Stresses
Because the mycelial growth of the UvPRO1 mutant was interrupted, we further monitored the effects of hyperosmotic and cell membrane stresses on CM medium with 0.1–0.5 M NaCl, 0.01–0.05% SDS, or 30–70 ug/mL CR. In the presence of 0.1–0.5 M NaCl, the growth rate of all strains decreased, and ΔUvPRO1-27 mutant displayed more sensitivity under salt stress compared to the wild-type and CΔUvPRO1-27, and the growth rate of ΔUvPRO1-27 mutant was reduced by 16–65%, respectively (Figure 6A). These results suggested that the UvPRO1 may play a role in regulation response to hyperosmotic conditions in U. virens.
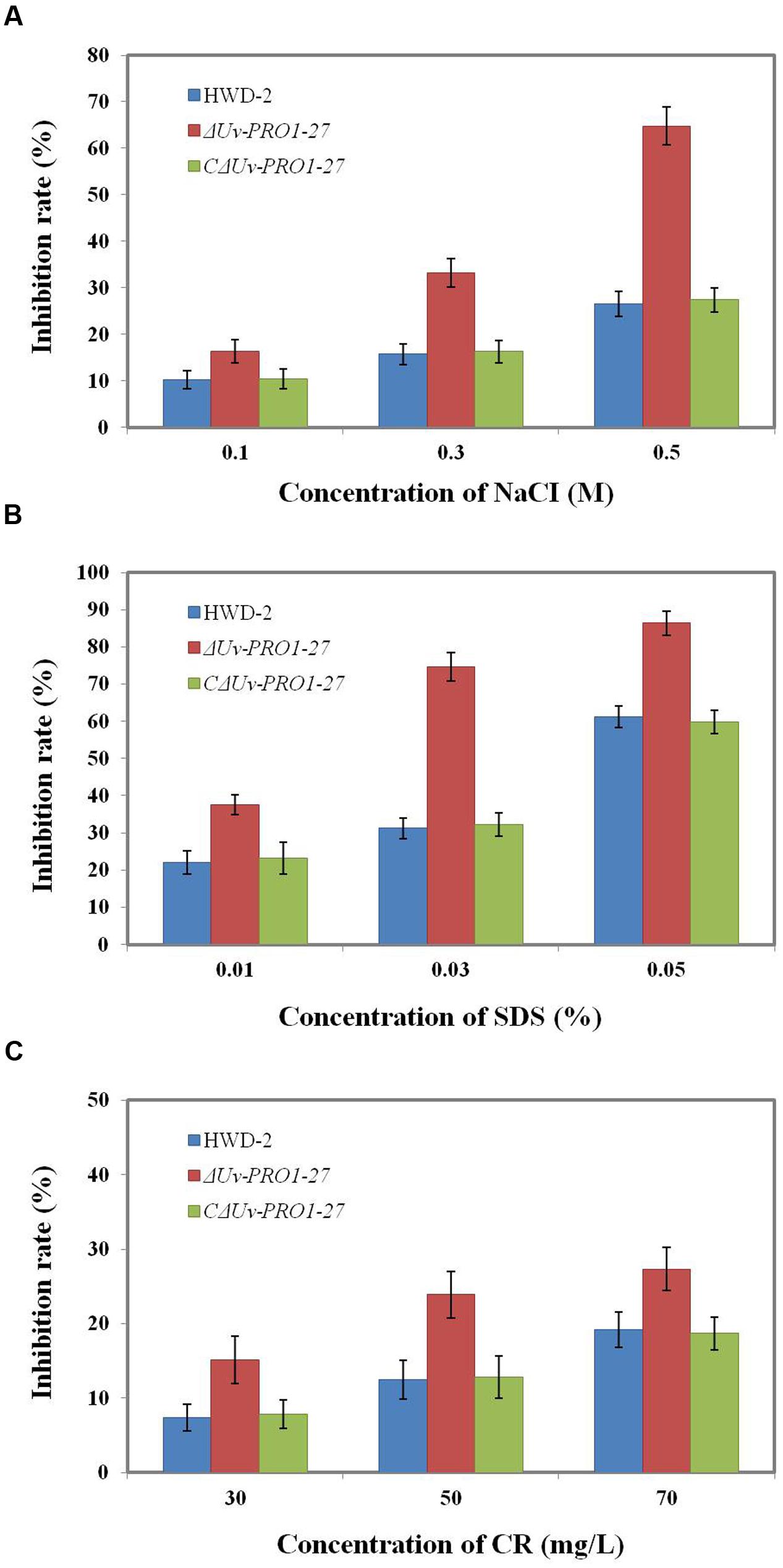
FIGURE 6. Growth of the UvPRO1 mutant in the presence of different stresses. (A) The wild-type strain HWD2, ΔUvPRO1-27, and CΔUvPRO1-27 were cultured on CM medium with 0.1–0.5 M NaCl after incubation at 28°C for 14 days. (B) The wild-type strain HWD2, ΔUvPRO1-27, and CΔUvPRO1-27 were cultured on CM medium amended with 0.01–0.03% sodium dodecyl sulfate (SDS) and incubated at 28°C for 14 days. (C) The wild-type strain HWD2, ΔUvPRO1-27, and CΔUvPRO1-27 were cultured on CM medium with 30–70 mg/L CR and incubated at 28°C for 14 days.
We also assayed the effects of SDS and CR treatments that mimic cytoplasm membrane and cell wall stresses, respectively. On CM with 0.01, 0.03, or 0.05% SDS, the growth rate of the ΔUvPRO1-27 mutant was, respectively, reduced by 37.5, 74.6, and 86.4%, while the decrease was 22.1, 31.5, and 61.3% in the wild-type and 23.2, 32.2, and 59.8% in CΔUvPRO1-27 (Figure 6B). In the presence of 30–70 mg/L CR, similar results to growth assays with SDS were obtained, in that ΔUvPRO1-27 mutant displayed a slower radial growth rate than the wild-type or CΔUvPRO1-27 (Figure 6C). These results suggested that the UvPRO1 mutant also had increased sensitivity to CR and SDS. Therefore, UvPRO1 may be involved in regulating responses to membrane and cell wall stresses in U. virens.
The Effect of UvPRO1 on the Pathogenicity of U. virens
Pathogenicity assays of the wild-type strain, ΔUvPRO1 mutant, and UvPRO1 complementary strain were performed on a susceptible host (Wanxian 98). Since the ΔUvPRO1 mutant produced no conidia, we also used a mycelial suspension of the wild-type strain and a UvPRO1 complementary strain for inoculation by injection as well as ΔUvPRO1 mutant. The inoculated plants were examined for colonization and infection by U. virens until 12 dpi. At 1–3 dpi, for the wild-type strain HWD2 and UvPRO1 complementary strain, many hyphal strands were observed to be elongated and extended along the surface of the spikelets (Figures 7D–F). At 4–6 dpi, hyphae were observed on the inner surfaces of spikelets, and filaments were infected by masses of hyphae (Figure 7G). At 7–8 dpi, the florets were covered profusely by hyphal growth with some wrapped around stamens and pistils (Figures 7H,I). At 9–12 dpi, the spaces in the spikelets were filled up by white mycelia (Figure 8A), and large mycelial masses grew out of the spikelets forming smut balls. After 15 dpi, 88.6% of the wild-type strain inoculated plants developed typical symptoms of false smut (Figure 8B), and similarly 86.2% in the UvPRO1 complementary strain (Table 2). For the ΔUvPRO1 mutant, at 1–3 dpi, hyphae were observed to be elongated and extended along the surface of spikelets (Figure 7A), which was similar to that of the wild-type strain and UvPRO1 complementary strain. At 4–6 dpi, mycelia became dehydrated and failed to grow further on spikelets (Figure 7B). No hyphae were observed inside of spikelets until 15 dpi (Figures 7C and 8A,B). This indicated that the ΔUvPRO1 mutant lost the ability for invasive growth on spikelets, and that the UvPRO1 is important for the pathogenicity of U. virens. Therefore, we conclude that the UvPRO1 plays an important role in virulence of U. virens.

FIGURE 7. Infection and colonization of rice spikelets by HWD2 and ΔUvPRO1-27at different time points. (A) Hyphae of ΔUvPRO1-27 growing and extending along spikelet surface at 1–3 days post-inoculation (dpi). (B) Mycelia of ΔUvPRO1-27 dehydrated and failed to thrive on spikelets at 4–6 dpi. (C) Floral organs were not infected by ΔUvPRO1-27 until 12 dpi. (D–F) Hyphae of HWD2 were observed to be elongated and extending along the surface of spikelets at 1–3 dpi. (G,H) Mycelia of wild-type strain HWD2 were observed on the inner surfaces of spikelets, and filaments were infected by masses of mycelia at 5 dpi. (I) The florets were covered with many hyphae with white mycelia of HWD2 wrapped around stamens and pistils at 7 dpi.

FIGURE 8. Comparison of the pathogenicity of HWD2, ΔUvPRO1-27, and CΔUvPRO1-27 on rice spikelets. (A) Floral organs were not infected by UvPRO1 at 9 dpi, but the spaces in the spikelets were filled by white mycelia of HWD2 and the UvPRO1 complementary strain at 9 dpi. (B) The pathogenicity phenotype of U. virens on rice spikelets at 15 dpi.
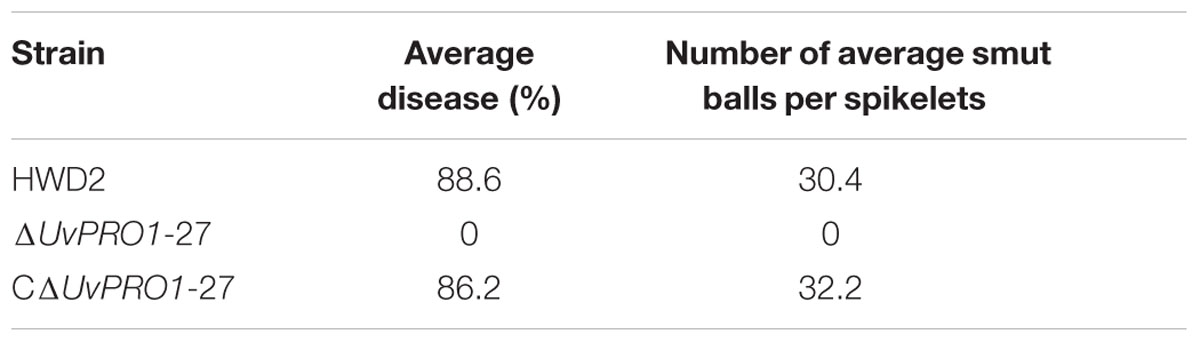
TABLE 2. Smut ball production by U. virens wild-type HWD2, and UvPRO1 deletion and complementation mutants.
Expression Dynamics of the UvPRO1 Gene
We first evaluated the UvPRO1 expression levels of U. virens in PSB medium using qRT-PCR. The results showed that lower expression levels were detected during early vegetative growth stages between 1 and 5 days, and were significantly increased during the conidiation stage between 6 and 9 days (Figure 9A). UvPRO1 expression during spikelet infection stage (4–12 dpi) was much higher than that in the early developmental stages (1–3 dpi), while the relative expression levels of UvPRO1 at 8 dpi was more than twofold higher than that at 1–2 dpi (Figure 9B).
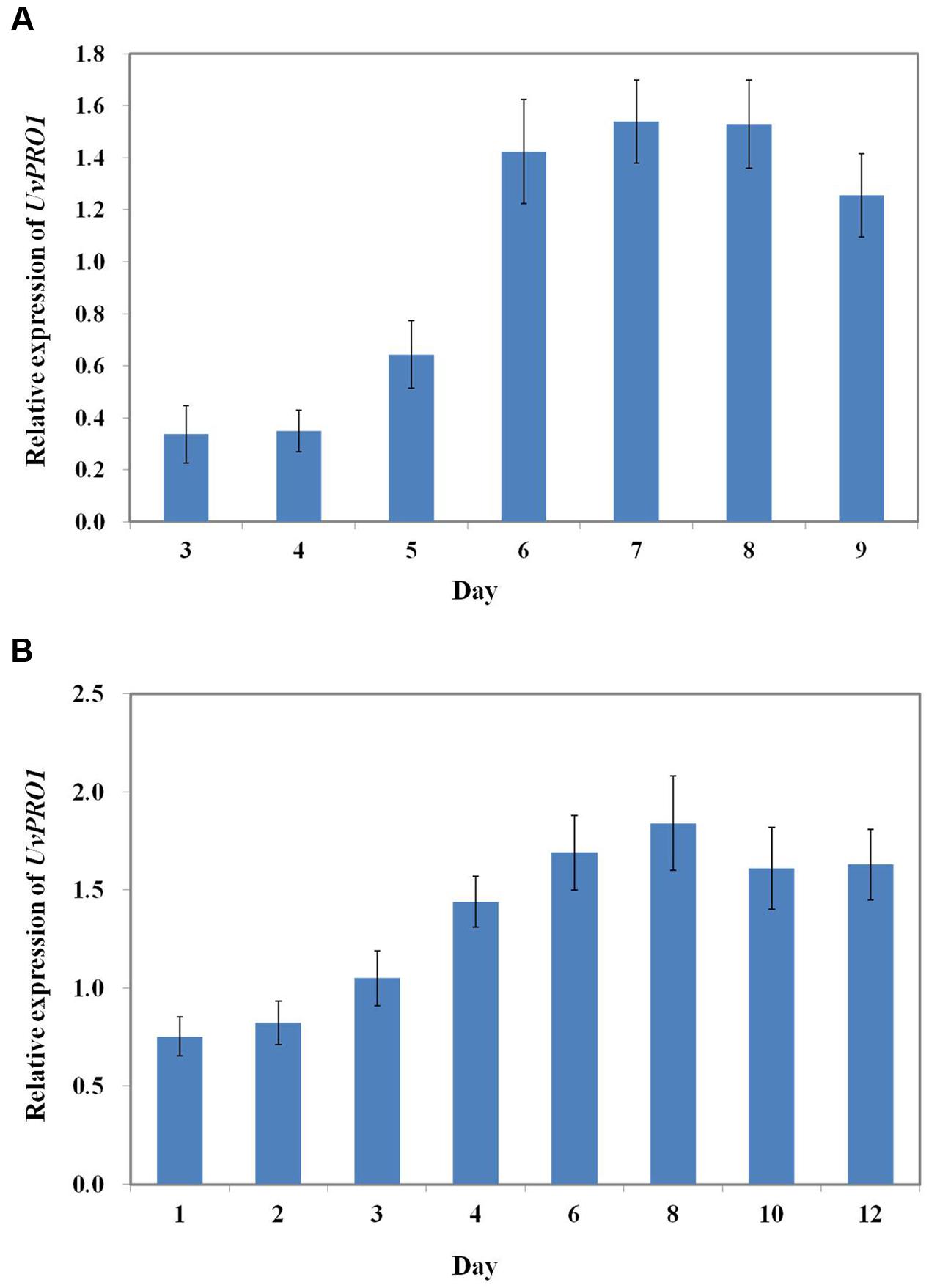
FIGURE 9. Expression dynamics of the UvPRO1 gene. (A) Expression of UvPRO1 during conidiation. An agar plug (3 mm in diam.) of wild-type strain placed into PSB medium. Values are relative to tubulin gene expression in RNA isolated from mycelia 3–9 dpi. (B) Expression of UvPRO1 during infection of rice spikelets. Rice spikelets were inoculated with mycelia of either the wild-type or the UvPRO1 deletion mutant. Values are relative to tubulin gene expression in RNA isolated from spikelets 1–12 dpi. Boxes and bars represent averages and standard error, respectively, of three independent biological replicates.
Discussion
The ATMT system has been used as an effective tool for insertional mutagenesis and homologous replacement in many phylogenetically diverse fungi (Mullins and Kang, 2001; Khang et al., 2005; Frandsen, 2011; Paz et al., 2011). Many target genes have been identified by screening phenotype and pathogenicity defective mutants from fungal T-DNA random insertion mutant library with homologous replacement and complementary methods (Munch et al., 2011; Giesbert et al., 2012; Xu and Chen, 2013; López-Pérez et al., 2015). In this study, we obtained five sporulation defective mutants by screening 3,016 strains of U. virens T-DNA insertion mutants. Southern blot analysis revealed that 80% of U. virens transformants contained single-copy T-DNA insertions, which is greater than the frequency described in a previous study (Yu et al., 2015). Therefore, the ATMT system used in this study was stable and reliable, and it could provide appropriate experimental material for screening targeted genes (Maruthachalam et al., 2011; Cai et al., 2013).
Among the conidiation defective mutants, T133 showed a decrease in mycelial growth and complete loss of conidiation. Sequence analysis showed that the mutant T133 has a T-DNA insertion in a predicted ORF encoding the amino acid sequence with high similarity (84%) to PRO1 of M. acridum which included the typical GAL4-like Zn(II)2Cys6 binuclear cluster DNA-binding domain.
Transcription factors of the Zn(II)2Cys6 binuclear cluster DNA-binding domain class, to which PRO1 belongs, are the most abundant class of transcription factors in fungal genomes (Borkovich et al., 2004). Most of the characterized members of this family participate in regulation of the primary and secondary metabolic pathways, but several have been shown to regulate fungal developmental processes (Vienken et al., 2005). PRO1 was first identified in S. macrosporea in a genetic screen for mutations defective in perithecial development, and gene deletion and complementation studies showed that SmPRO1 is required for sexual development. In C. parasitica, deletion of PRO1 resulted in a significant reduction in asexual sporulation and loss of female fertility (Sun et al., 2009). In E. festucae, Tanaka et al. (2013) identified a mutant with an insertion in PRO1, and disruption of targeted gene increased asexual sporulation and reduced cell fusion.
In this study, morphological observation of the UvPRO1 deletion mutant showed that UvPRO1 deficiency led to a decline in the hyphal growth rate, an increase in aerial hyphae, and a complete loss of sporulation. In contrast, the deletion PRO1 gene of Alternaria brassicae led to a similar effect on mycelial growth in that the mycelial growth rate of the AbPRO1 deletion mutant declined by 25% (Cho et al., 2009). Moreover, in C. parasitica, PRO1 gene deletion also resulted in production of few or no conidia and increased aerial hyphae, but the radial growth rate was not influenced (Sun et al., 2009). Therefore, by comparing the previous research findings, the PRO1 gene can be seen to participate in regulation of pathogen growth and development, but the role of PRO1 in different fungi was visibly different. In addition, the PRO1 gene has not been reported to be regulated in response to various environmental stresses such as oxidative and cell wall stresses. However, in our study, the PRO1 deletion mutant showed increased sensitivities to hyperosmotic and cell wall stresses, which provide a novel regulation of PRO1 in among pathogenic fungi.
In previous studies, PRO1 has been verified to be important for pathogen virulence in A. brassicicola and stable maintenance of hypovirus infection C. parasitica. To assess the role of UvPRO1 in virulence, we observed the infection process of the UvPRO1 mutant and wild-type after inoculation at rice booting stage. The results showed that hyphae of the PRO1 deletion mutant could extend along the surface of spikelets at 1–3 dpi, but mycelia became dehydrated and completely lost the ability to infect spikelets after 4 dpi. The qRT-PCR analysis of UvPRO1 showed that the expression levels of UvPRO1 in the infection stage (4–12 dpi) were much higher than that in early developmental stages (1–3 dpi), while the relative expression levels of UvPRO1 at 8 dpi was more than twice as high as that at 1–2 dpi. Therefore, we conclude that the UvPRO1 plays an important role in virulence of U. virens.
Conclusion
The UvPRO1 gene in U. virens was characterized as a Zn(II)2Cys6 transcription factor required for fungal developmental processes. The results of this study suggest that UvPRO1 plays a critical role in hyphal growth and conidiation, and is also involved in stress responses and pathogenesis, which provided novel actions of PRO1 in fungi and has improved the understanding of the function of UvPRO1 during the life cycle of U. virens.
Author Contributions
Conceived and designed the experiments: BL and JH. Performed the experiments: BL. Analyzed the experiment data: BL, LZ, HL, and JT. Contributed reagents/materials/analysis tools: BL and TH. Wrote the paper: BL. All authors have read and approve the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (Grant No. 2662015QC022) and the Natural Science Foundation of Hubei Provincial (Grant No. 2014CFB952).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02086/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/BLAST/
- ^ http://www.ncbi.nlm.nih.gov
- ^ http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
- ^ http://www.premierbiosoft.com/primerdesign/
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.clustal.org/clustal2/
- ^ http://www.megasoftware.net/index.php
References
Ashizawa, T., Takahashi, M., Moriwaki, J., and Hirayae, K. (2010). Quantification of the rice false smut pathogen Ustilaginoidea virens from soil in Japan using real-time PCR. Eur. J. Plant. Pathol. 128, 221–232. doi: 10.1007/s10658-010-9647-4
Bo, H., Yu, M., Yu, J., Yi, X., Ding, H., Wang, Y., et al. (2016). Molecular cloning flanking sequences of T-DNA insertion from the Ustilaginoidea virens mutant strain B1241. Chin. Agric. Sci. 49, 1685–1695. doi: 10.3864/j.issn.0578-1752.2016.09.005
Borkovich, K. A., Alex, L. A., Yarden, O., Freitag, M., Turner, G. E., Read, N. D., et al. (2004). Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68, 1–108. doi: 10.1128/MMBR.68.1.1-108.2004
Brooks, S. A., Anders, M. M., and Yeater, K. M. (2009). Effect of cultural management practices on the severity of false smut of rice. Plant Dis. 93, 1202–1208. doi: 10.1094/PDIS-93-11-1202
Cai, Z., Li, G., Lin, C., Shi, T., Zhai, L., Chen, Y., et al. (2013). Identifying pathogenicity genes in the rubber tree anthracnose fungus Colletotrichum gloeosporioides through random insertional mutagenesis. Microbiol. Res. 168, 340–350. doi: 10.1016/j.micres.2013.01.005
Cho, Y., Kim, K. H., La Rota, M., Scott, D., Santopietro, G., Callihan, M., et al. (2009). Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol. Microbiol. 72, 1316–1333. doi: 10.1111/j.1365-2958.2009.06689.x
Deng, G. S. (1989). Present status of research on false smut in China. Plant. Prot. China 15, 39–40.
Fan, J., Guo, X., Li, L., Huang, F., Sun, W. X., Li, Y., et al. (2015). Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain filling related genes. J. Integr. Plant. Biol. 57, 577–590. doi: 10.1111/jipb.12299
Frandsen, R. J. N. (2011). A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens-mediated transformation. J. Microbiol. Methods 87, 247–262. doi: 10.1016/j.mimet.2011.09.004
Giesbert, S., Schumacher, J., Kupas, V., Espino, J., Segmuller, N., Haeuser-Hahn, I., et al. (2012). Identification of pathogenesis-associated genes by T-DNA-mediated insertional mutagenesis in Botrytis cinerea: a type 2A phosphoprotein phosphatase and an SPT3 transcription factor have significant impact on virulence. Mol. Plant Microbe Interact. 25, 481–495. doi: 10.1094/MPMI-07-11-0199
Gu, Z., Ding, Z., Chen, X., Guo, L., Zeng, D., Qian, Q., et al. (2012). Reference Genes Selection of Ustilaginoidea virens by real-time quantitative polymerase chain reaction (PCR). Chin. Rice Sci. 26, 615–618.
Guo, X., Li, Y., Fan, J., Li, L., Huang, F., and Wang, W. (2012). Progress in the study of false smut disease in rice. J. Agric. Sci. Technol. A. 2, 1211–1217.
Hu, M., Luo, L., Wang, S., Liu, Y., and Li, J. (2014). Infection processes of Ustilaginoidea virens during artificial inoculation of rice panicles. Eur. J. Plant. Pathol. 139, 67–77. doi: 10.1007/s10658-013-0364-7
Jia, Q., Lv, B., Guo, M. Y., Luo, C. X., Zheng, L., Hsiang, T., et al. (2015). Effect of rice growth stage, temperature, relative humidity and wetness duration on infection of rice panicles by Villosiclava virens. Eur. J. Plant Pathol. 141, 15–25. doi: 10.1007/s10658-014-0516-4
Khang, C., Park, S., Lee, Y., and Kang, S. (2005). A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet. Biol. 42, 483–492. doi: 10.1016/j.fgb.2005.03.004
Koiso, Y., Li, Y., Iwasaki, S., Hanaka, K., Kobayashi, T., Sonoda, R., et al. (1994). Ustiloxins, antimitotic cydic peptides from false smut balls on rice panicles caused by Ustilaginoidea virens. J. Antibiot. 47, 765–773. doi: 10.7164/antibiotics.47.765
Liu, L., Zhao, D., Zheng, L., Hsiang, T., Wei, Y., Fu, Y., et al. (2013). Identification of virulence genes in the crucifer anthracnose fungus Colletotrichum higginsianum by insertional mutagenesis. Microb. Pathog. 64, 6–17. doi: 10.1016/j.micpath.2013.06.001
López-Pérez, M., Ballester, A. R., and González-Candelas, L. (2015). Identification and functional analysis of Penicillium digitatum genes putatively involved in virulence towards citrus fruit. Mol. Plant. Pathol. 16, 262–275. doi: 10.1111/mpp.1217
Maruthachalam, K., Klosterman, S. J., Kang, S., Hayes, R. J., and Subbarao, K. V. (2011). Identification of pathogenicity-related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens-mediated T-DNA insertional mutagenesis. Mol. Biotechnol. 49, 209–221. doi: 10.1007/s12033-011-9392-8
Masloff, S., Jacobsen, S., Pöggeler, S., and Kück, U. (2002). Functional analysis of the C6 zinc finger gene pro1 involved in fungal sexual development. Fungal Genet. Biol. 36, 107–116. doi: 10.1016/S1087-1845(02)00010-5
Masloff, S., Pöggeler, S., and Kück, U. (1999). The pro1(+) gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152, 191–199.
Miyazaki, S., Matsumoto, Y., Uchihara, T., and Morimoto, K. (2009). High-performance liquid chromatographic determination of ustiloxin A in forage rice silage. J. Vet. Med. Sci. 71, 239–241. doi: 10.1292/jvms.71.239
Mullins, E. D., Chen, X., Romaine, P., Raina, R., Geiser, D. M., and Kang, S. (2001). Agrobacterium tumefaciens-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91, 173–180. doi: 10.1094/phyto.2001.91.2.173
Mullins, E. D., and Kang, S. (2001). Transformation: a tool for studying fungal pathogens of plants. Cell. Mol. Life Sci. 58, 2043–2052. doi: 10.1007/PL00000835
Munch, S., Ludwig, N., Floss, D. S., Sugui, J. A., Koszucka, A. M., Voll, L. M., et al. (2011). Identification of virulence genes in the corn pathogen Colletotrichum graminicola by Agrobacterium tumefaciens-mediated transformation. Mol. Plant. Pathol. 12, 43–55. doi: 10.1111/j.1364-3703.2010.00651.x
Paz, Z., García-Pedrajas, M. D., Andrews, D. L., Klosterman, S. J., Baeza-Montañez, L., and Gold, S. E. (2011). One step construction of Agrobacterium-recombination-ready-plasmids (OSCAR), an efficient and robust tool for ATMT based gene deletion construction in fungi. Fungal Genet. Biol. 48, 677–684. doi: 10.1016/j.fgb.2011.02.003
Rao, Y., Ding, Z., Chen, X., Zeng, D., Ma, B., and Gu, Z. (2014). Cloning and expression an analysis of UvHog1 gene in Ustilaginoidea virens. Chin. J. Rice Sci. 28, 9–14. doi: 10.3969/j.issn.1001-7216.2014.01.002
Rush, M. C., Shahjahan, A. K. M., Jones, J. P., and Groth, D. E. (2000). Outbreak of false smut of rice in Louisiana. Plant. Dis. 84:100. doi: 10.1094/PDIS.2000.84.1.100D
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning, Vol. 2. New York, NY: Cold spring harbor laboratory press, 14–19.
Savary, S., Willocquet, L., Elazegui, F. A., Teng, P. S., Van Du, P., Zhu, D., et al. (2000). Rice pest constraints in tropical Asia: characterization of injury profiles in relation to production situations. Plant. Dis. 84, 341–356. doi: 10.1094/PDIS.2000.84.3.341
Singh, A. K., and Pophaly, D. J. (2010). An unusual rice false smut epidemic reported in Raigarh District, Chhattisgarh, India. Int. Rice Res. Notes 35, 1–3.
Sugui, J. A., Chang, Y. C., and Kwon-Chung, K. J. (2005). Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microb. 71, 1798–1802. doi: 10.1128/AEM.71.4.1798-1802.2005
Sun, Q., Choi, G. H., and Nuss, D. L. (2009). Hypovirusresponsive transcription factor gene pro1 of the chestnut blight fungus Cryphonectria parasitica is required for female fertility, asexual spore development, and stable maintenance of hypovirus infection. Eukaryot. Cell. 8, 262–270. doi: 10.1128/EC.00338-08
Sun, X., Kang, S., Zhang, Y., Tan, X., Yu, Y., He, H., et al. (2013). Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS ONE 8:e76879. doi: 10.1371/journal.pone.0076879
Tanaka, A., Cartwright, G. M., Saikia, S., Kayano, Y., Takemoto, D., Kato, M., et al. (2013). ProA, a transcriptional regulator of fungal fruiting body development, regulates leaf hyphal network development in the Epichloë festucae–Lolium perenne symbiosis. Mol. Microbiol. 90, 551–568. doi: 10.1111/mmi.12385
Tang, Y. X., Jin, J., Hu, D. W., Yong, M. L., and Xu, Y. (2013). Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 62, 1–8. doi: 10.1111/j.1365-3059.2012.02629.x
Vienken, K., Scherer, M., and Fischer, R. (2005). The Zn (II) 2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics 169, 619–630. doi: 10.1534/genetics.104.030767
Wang, D., Wang, S., and Fu, J. (2004). Research advance on false smut of rice. Liaoning Agri. Sci. 1, 21–24.
Wang, Y., Liu, Y., Lu, F., Yu, M., Huang, L., Zheng, M., et al. (2015). Molecular characterization of T-DNA integration into Ustilaginoidea virens mutant B1464. Chin. J. Rice Sci. 29, 311–318. doi: 10.3969/j.issn.1001-7216.2015.03.011
Xu, L., and Chen, W. (2013). Random T-DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 26, 431–441. doi: 10.1094/MPMI-07-12-0177-R
Yu, J., Nie, Y., Yu, M., Yi, X., Hu, J., Huang, L., et al. (2013). Characterization of T-DNA insertion flanking genes of enhanced-conidiation Ustilaginoidea virens mutant A2588. Chin. Agric. Sci. 46, 5132–5141. doi: 10.3864/j.issn.0578-1752.2013.24.007
Yu, M., Yu, J., Hu, J., Huang, L., Wang, Y., Yin, X., et al. (2015). Identification of pathogenicity-related genes in the rice pathogen Ustilaginoidea virens through random insertional mutagenesis. Fungal Genet. Biol. 76, 10–19. doi: 10.1016/j.fgb.2015.01.004
Zhang, Z., Du, X., Cai, R., Mao, X., Qiu, H., Wang, L., et al. (2006). Agrobacterium tumefaciens-mediated transformation of the pathogen of Ustilaginoidea virens. Chin. J. Rice Sci. 20, 440–442.
Zhang, Z., Du, X., Chai, R., Wang, J., Qiu, H., Mao, X., et al. (2008). Cloning of a homologous gene of Magnaporthe grisea PMK1 type MAPK from Ustilaginoidea virens and functional identification by complement in Magnaporthe grisea corresponding mutant. Acta Microbiol. Sin. 48, 1473–1478.
Zheng, D., Wang, Y., Han, Y., Xu, J. R., and Wang, C. (2016). UvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens. Sci. Rep. 6:24824. doi: 10.1038/srep24824
Zheng, M. T., Ding, H., Huang, L., Wang, Y. H., Yu, M. N., Zheng, R., et al. (2016). Low-affinity iron transport protein Uvt3277 is important for pathogenesis in the rice false smut fungus Ustilaginoidea virens. Curr. Genet. doi: 10.1007/s00294-016-0620-4 [Epub ahead of print].
Keywords: Ustilaginoidea virens, false smut, UvPRO1, conidiation, stress response, pathogenicity
Citation: Lv B, Zheng L, Liu H, Tang J, Hsiang T and Huang J (2016) Use of Random T-DNA Mutagenesis in Identification of Gene UvPRO1, A Regulator of Conidiation, Stress Response, and Virulence in Ustilaginoidea virens. Front. Microbiol. 7:2086. doi: 10.3389/fmicb.2016.02086
Received: 09 September 2016; Accepted: 09 December 2016;
Published: 27 December 2016.
Edited by:
Thorsten Lumbsch, The Field Museum, USAReviewed by:
Christian P. Kubicek, Vienna University of Technology, AustriaGerardo Díaz-Godínez, Autonomous University of Tlaxcala, Mexico
Copyright © 2016 Lv, Zheng, Liu, Tang, Hsiang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbin Huang, anVuYmluaHVhbmdAbWFpbC5oemF1LmVkdS5jbg==
 Bo Lv
Bo Lv Lu Zheng
Lu Zheng Hao Liu1
Hao Liu1