- 1School of Science, Monash University Malaysia, Bandar Sunway, Malaysia
- 2Tropical Medicine and Biology Multidisciplinary Platform, Monash University Malaysia, Bandar Sunway, Malaysia
- 3Novel Bacteria and Drug Discovery Research Group, School of Pharmacy, Monash University Malaysia, Bandar Sunway, Malaysia
- 4Center of Health Outcomes Research and Therapeutic Safety (Cohorts), School of Pharmaceutical Sciences, University Phayao, Phayao, Thailand
A novel Gram negative rod-shaped bacterium, designated strain MSh1T, was isolated from Southeast Pahang tropical peat swamp forest soil in Malaysia and characterized using a polyphasic taxonomy approach. The predominant cellular fatty acids (>10.0%) were C16:0 (31.7%), C17:0 cyclo (26.6%), and C19:0 cyclo ω8c (16.1%). The polar lipids detected were phosphatidylglycerol, phosphatidylethanolamine, and diphosphatidylglycerol. The predominant ubiquinone was Q-8. This revealed that strain MSh1T belongs to the genus Burkholderia. The type strain MSh1T can be differentiated from other Burkholderia cepacia complex (Bcc) species by phylogenetic analysis of 16S rRNA gene sequence, multilocus sequence analysis (MLSA), average nucleotide identity (ANI) and biochemical tests. DNA–DNA relatedness values between strain MSh1T and closely related type strains were below the 70% threshold value. Based on this polyphasic study of MSh1T, it can be concluded that this strain represents a novel species within the Bcc, for which the name Burkholderia paludis sp. nov. is proposed. The type strain is MSh1T (= DSM 100703T = MCCC 1K01245T). The dichloromethane extract of MSh1T exhibited antimicrobial activity against four Gram positive bacteria (Enterococcus faecalis ATCC 29212, E. faecalis ATCC 700802, Staphylococcus aureus ATCC 29213, S. aureus ATCC 700699) and a Gram negative bacteria (Escherichia coli ATCC 25922). Further purification work has led to the isolation of Compound 1, pyochelin. Pyochelin demonstrated antimicrobial activity against four S. aureus strains and three E. faecalis strains with MIC-values of 3.13 μg/ml and 6.26 μg/ml, respectively. SEM analysis showed that the cellular morphology of E. faecalis ATCC 700802 was not affected by pyochelin; suggesting that it might target the intracellular components. Pyochelin, a siderophore with antimicrobial activity might be useful in treating bacterial infections caused by S. aureus and E. faecalis, however further work has to be done.
Introduction
The misuse of antimicrobial compounds to treat infections has led to an increase in the prevalence of antimicrobial resistant bacteria (ARB), often associated with nosocomial (hospital acquired) infections (Mishra et al., 2012). It was reported that nosocomial infections constitute the sixth leading cause of death in the United States and more than 70% of these nosocomial pathogens are resistant to at least one of the common antimicrobial drugs used to treat them (Iowa Department of Public Health). This is an alarming issue because ARB infections often result in increased mortality rates, limit treatment options and cause increased healthcare costs (Ammerlaan et al., 2016). Thus, there is a need for new antimicrobial compounds to combat ARB infections.
One strategy to discover new antimicrobials is bioprospecting—the exploration for new compounds in unique ecological niches (Imhoff et al., 2011). The discovery of abyssomicins, novel antibiotics produced by Verrucosispora sp. from South China Sea sediments is a successful example of bioprospecting (Wang et al., 2013). Bacteria producing antibiotics may have an advantage in competing for resources and colonization of new habitats in natural environments. Novel microorganisms thriving in extreme and isolated environments have the potential to develop unique biosynthesis gene clusters giving rise to novel compounds (Bull and Stach, 2007; Hibbing et al., 2010). Consequently, tropical peat swamp forests in Malaysia were chosen as potential bioprospecting locations for novel antimicrobial compounds. Tropical peat swamp forests are unique wetland ecosystems characterized by their acidic (pH range of 2.9 to 4.5) and waterlogged conditions. They are ombotrophic hence receiving nutrients solely from rain and atmospheric deposition which results in an extremely nutrient poor environment (Yule, 2010). Despite being such a harsh environment, we successfully isolated an antimicrobial-producing bacterium, belonging to the Burkholderia cepacia complex (Bcc) in a previous study (Ong et al., 2015).
The Bcc consists of a group of diverse, ubiquitous bacteria that occur in aquatic environments, plant rhizospheres and animals, including humans (Coenye and Vandamme, 2003; Vanlaere et al., 2009; Peeters et al., 2013). Certain Bcc species, for example B. cepacia, can be used for biocontrol as they can produce antifungal compounds to repress soil borne pathogens (Caballero-Mellado et al., 2004). An example of one such compound is pyrrolnitrin which targets the electron transport chain of both Gram positive bacteria and fungi (El-Banna and Winkelmann, 1998). Bcc species share a high degree of 16S rRNA (98–100%) and recA (94–95%) gene sequence similarity, and moderate levels of DNA–DNA hybridization (30–50%) (Coenye et al., 2001). In order to differentiate different species of Bcc, multi-locus sequence analysis (MLSA) are usually adopted as these taxonomic techniques provide the discriminatory power needed for both identification and differentiation of Bcc species (Spilker et al., 2009; Vanlaere et al., 2009; Peeters et al., 2013).
In this paper, we report a novel antimicrobial-producing Bcc species, isolated from the peat soil of the Southeast Pahang tropical peat swamp forest reserve in Malaysia. This polyphasic taxonomic study revealed that strain MSh1T represents a novel Bcc species, for which the name B. paludis is proposed. Based on current literature, Burkholderia species are known to produce narrow-spectrum antimicrobial compounds such as iminopyrrolidines (produced by B. plantari) and occidiofungin (B. contaminans) which only targets Gram negative Erwinia amylovora and fungi, respectively (Lu et al., 2009; Tawfik et al., 2010). In this study, strain MSh1T exhibit a different spectrum of antimicrobial activity, which targets Gram positive bacteria only, particularly strains of S. aureus and E. faecalis.
Materials and Methods
Test Microorganism Strains and Culture Conditions
Test microorganism strains that were used in this study include Bacillus cereus ATCC 14579, Bacilus subtilis ATCC 8188, Enterococcus faecalis ATCC 700802, Enterococcus faecalis ATCC 29212, Enterococcus faecalis JH-22, Staphylococcus aureus ATCC 700699, Staphylococcus aureus ATCC 43300, Staphylococcus aureus ATCC 6538P, Staphylococcus aureus ATCC 29213, Aeromonas hydrophila ATCC 49140, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 10031, Proteus mirabilis ATCC 49140, Proteus vulgaris (Institute of Medical Research, Malaysia), Pseudomonas aeruginosa ATCC 10145, Pseudomonas aeruginosa ATCC BAA-47, Salmonella Typhimurium ATCC 14028, and Shigella flexneri ATCC 12022. Strains were cultured on Muller Hinton borth (MHB) (Oxoid, UK) at 37°C and maintained at −80°C in MHB with 25% (v/v) glycerol.
Isolation and Maintenance of Isolate
Strain MSh1T was previously isolated from surface peat collected from the Southeast Pahang tropical peat swamp forest reserve (3° 01′ 19.56″ N; 103° 39′ 29.67″ E) in Malaysia on March 2013. Strain MSh1T was maintained on nutrient agar (NA) at 30°C and in 25% (v/v) glycerol in nutrient broth (NB) (Merck, Germany) at −80°C for long term preservation.
Sequence and Phylogenetic Analysis
The genomic DNA of strain MSh1T was isolated using GF-1 nucleic acid extraction kit (Vivantis, Malaysia). The 16S rRNA gene sequence of strain MSh1T was amplified using the universal primers 27f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492r (5′-ACG GCT ACC TTG TTA CGA CTT 3′) (Kane et al., 1993). PCR were set up as follows: 150 ng (5 μL) of DNA extract, 10 μL of 5x MyTaq Red Reduction Buffer, 5 μM of forward primer, 5 μM of reverse primer, and 1.25 U of MyTaq DNA polymerase. The reaction volume was made up to 50 μL using sterile milliQ water (Millipore, Germany). The PCR included an initial denaturation step at 95°C for 1 min, followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 45 s and elongation at 72°C for 45 s. PCR products were separated on a 1.5% (w/v) agarose gel and the bands were visualized with 1x GelRed. Amplification products were purified and cloned into pJET1.2 vector (FirstBase, Malaysia). The 16S rRNA gene sequence of strain MSh1T was aligned with sequences of closely related type strains of the genus Burkholderia retrieved from the GenBank/EMBL/DDBJ databases using CLUSTAL-X software (Thompson et al., 1997). The alignment was manually verified and adjusted prior to the construction of phylogenetic tree using the neighbor-joining (Saitou and Nei, 1987) algorithm with the MEGA version 6.0 software (Tamura et al., 2011). The stability of the resultant tree topologies were evaluated by using the bootstrap resampling method (Felsenstein, 1985). The evolutionary distances for the neighbor-joining algorithm were computed using the Kimura's two-parameter model (Kimura, 1980).
DNA–DNA Hybridization (DDH)
DDH-tests were carried out by the Identification Service of Deutsche Sammlung von Mikroorganisen und Zellkulturen (DSMZ, Germany) to evaluate the DNA–DNA relatedness between strain MSh1T, B. arboris R-24201T, B. cenocepacia J2315T, and B. lata 383T using the optical renaturation rate method as described by De Ley et al. (1970) under consideration of the modifications described by Huss et al. (1983).
Multilocus Sequence Analysis (MLSA)
MLSA was performed on strain MSh1T based on the method described by Spilker et al. (2009). A phylogenetic tree of the concatenated sequence (2773 bp) of seven housekeeping gene fragments [atpD (443 bp), gltB (400 bp), gyrB (454 bp), recA (393 bp), lepA (397 bp), phaC (385 bp) and trpB (301 bp)] was constructed using MEGA6.0 (Tamura et al., 2011). The mean number of nucleotide substitution per site (i.e., the percentage of divergence of concatenated allele sequences) between established Bcc type strains and strain MSh1T was calculated using the DnaSP v5.10 (Librado and Rozas, 2009) based on the Jukes-Cantor method (Jukes and Cantor, 1969). Nucleotide sequences of each allele, allelic profiles and sequence types of strain MSh1T from the present study are available on the Bcc PubMLST database (http://pubmlst.org/bcc).
PCR Amplification of prnD (Pyrrolnitrin) Gene
The synthesis of pyrrolnitrin is regulated mainly by four genes prnA, prnB, prnC and prnD, where prnD being the most crucial gene. The gene product of prnD catalyzes the oxidation of the amino group of aminopyrrolnitrin to a nitro group to form pyrrolnitrin (Souza and Raaijmakers, 2003). As pyrrolnitrin is the only reported compound with antimicrobial activity against Gram positive bacteria known to be produced by Bcc (El-Banna and Winkelmann, 1998), this assay was carried out to avoid the rediscovery of a known compound (pyrrolnitrin). Hence in order to assess the ability of strain MSh1T to produce pyrrolnitrin, the prnD gene was amplified with a forward primer PRND1 (5′-GGG GCG GGC CGT GGT GAT GGA-3′) and a reverse primer PRND2 (5′-YCC CGC SGC CTG YCT GGT CTG-3′) (Souza and Raaijmakers, 2003). PCR were set up as follows: 5 μL of DNA extract, 10 μL of 5x MyTaq Red Reduction Buffer, 5 μM of forward primer, 5 μM of reverse primer, and 1.25 U of MyTaq DNA polymerase. The reaction volume was made up to 50 μL using sterile milliQ water (Millipore, Germany). The PCR included an initial denaturation step at 95°C for 1 min, followed by 30 cycles of denaturation at 95°C for 60 s, annealing at 68°C for 30 s, and elongation at 72°C for 30 s. Genomic DNA of pyrrolnitrin producer B. lata 383T was used as a positive control and non-pyrrolnitrin producer B. cenocepacia J2315T was used as a negative control. PCR products were separated on a 1.5% (w/v) agarose gel and the bands were visualized with 1x GelRed.
Phenotypic and Biochemical Characterization
Cellular morphological characteristics of strain MSh1T were observed by using a Hitachi S-4800 field-emission scanning electron microscopy (FE-SEM) at 25,000 × magnification after 2 days of growth in nutrient broth (NB) at 30°C. Colony morphology of strain MSh1T was examined after 2 days of growth on NA at 30°C. Cell motility was measured using hanging drop method after incubation for 2 days at 30°C in NB (Robbie, 1945). The optimal temperature for growth was measured at 4, 15, 28, 30, 37, 40, and 45°C in NB for 2 days. The pH range for growth was tested with in NB adjusted with HCl or NaOH to pH 3–11, at intervals of 0.5 pH units. NaCl tolerance at 0–5% (w/v) at interval of 0.5% (w/v) was determined in NB for 2 days. Anaerobic growth was tested in NB for up to 5 days in a 2.5 l jar containing an AnaeroPack-Anaero (Thermo Scientific, USA). Phenotypic characteristics including Gram staining, catalase, and oxidase activity were examined using the methods described by Buck (1982) and Cappuccino and Sherman (2002) after 2 days of growth on NA at 30°C. Physiological and biochemical properties were further determined using API 50CH, API 20NE, and API ZYM strips (bioMerieux, France) according to the manufacturer's instructions. All tests were conducted in duplicate. The API 50CH and API 20NE-tests were read after 24–48 h at 30°C, while the API ZYM-tests were read after 4 h of incubation at 30°C.
Chemotaxonomic Characterization
The cellular fatty acids analysis of strain MSh1T, B. arboris R-24201T, B. cenocepacia J2315T, and B. lata 383T was carried out by the Identification Service of DSMZ (Braunschweig, Germany). The cell mass of strain MSh1T and closely related type strains were harvested from NB after incubation at 30°C for 2 days. Extraction and analysis of the cellular fatty acids were performed according to the standard protocols of the Sherlock Microbial Identification System (MIDI) (Miller, 1982), analyzed using an Agilent 6890N gas chromatograph fitted with a 5% phenyl-methyl silicone capillary column. Peaks were integrated and identified using the peak-naming table TSBA40 at the DSMZ. Polar lipids extracted from 200 mg freeze-dried cell material using chloroform: methanol: 0.3% (w/v) aqueous NaCl mixture 1:2:0.8 (v/v/v) as described by Tindall (1990). The extracted polar lipids were then separated by two dimensional silica gel thin layer chromatography with chloroform: methanol: water (65:25:4, v/v/v) as mobile phase for the first direction, followed by chloroform: methanol: acetic acid: water (80:12:15:4, v/v/v/v) as the mobile phase for the second direction (Tindall et al., 2007). Cellular ubiquinones were extracted and purified as described previously by Tindall (1990). The different quinone classes were firstly separated using thin layer chromatography on silica gel using hexane-tert-butylmethylether (9:1, v/v) as a solvent and then further purified with reversed-phase HPLC using methanol:heptane (9:1, v/v) as the eluent. The purified quionones were compared to standards at the DSMZ.
Genome-To-Genome Distance Calculator (GGDC) and Average Nucleotide Identity (ANI)
GGDC was performed to predict the DNA–DNA hybridization value between strain MSh1T with related Burkholderia species. GGDC was performed at http://ggdc.dsmz.de/distcalc2.php using the standard parameters (GGDC 2.0 BLAST+) (Meier-Kolthoff et al., 2013). The genome sequence of strain MSh1T (DDBJ/EMBL/GenBank JPGL00000000) (Ong et al., 2014) was queried with related Bcc species. Results were expressed as similarity percentage ± confidence interval. ANI was performed to estimate the mean values between homologous genomic regions shared by strain MSh1T with related Bcc species using the whole genome sequence obtained. ANI was performed at http://enve-omics.ce.gatech.edu/ani using the standard parameters (Goris et al., 2007). Results were expressed as ANI percentage ± standard deviation.
Genome Analysis of Secondary Metabolites Gene Clusters Using Antismash
The whole genome of strain MSh1T was screened for biosynthetic gene clusters responsible for the synthesis of secondary metabolites using Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) 2.0) (Blin et al., 2013). The antiSMASH 2.0 program analyses the whole genome sequence for homologs to known secondary metabolites via BLAST search and annotates them based on different biosynthetic gene clusters, for example polyketide synthase (PKS), non-ribosomal peptide synthase (NRPS), terpene and bacteriocins.
Methanol Extraction of MSh1T
Strain MSh1T was first grown on nutrient agar supplemented with 5 g/l of glycerol for 5 days at 30°C. The whole agar including the bacteria was extracted three times with 100% methanol (Merck, Germany) (Isnansetyo and Kamei, 2003). The crude methanol extract was lyophilized using a Freezone 4.5 Plusfreeze Dryer (Labconco, USA).
Sequential Solvent Fractionation and Purification of the Crude Methanol Extract
Sequential solvent fractionation was performed on the crude methanol extract to fractionate the extract into different fractions with different polarity. Five grams of crude methanol extract was first dissolved in 500 ml of distilled water, and then extracted with hexane (HEX), dichloromethane (DCM) and ethyl acetate (EtOAc) sequentially. Each extraction step was performed three times and combined into one fraction before lyophylization in a Freezone 4.5 Plusfreeze dryer (Labconco, USA). The lyophilized active fraction was dissolved in methanol and partially purified on an open C18 column (Merck, Germany), followed by reversed-phase HPLC using a Cosmosil 5C18-MS-II, 20 × 250 mm, 5 μm column (Nacalai, USA), to yield compound 1. All processes were monitored by bioassay.
Identification of the Antimicrobial Compound(s) from Strain MSh1T
Structural determination of the antimicrobial compound was performed by spectroscopic techniques and literature comparison. The antimicrobial compound was analyzed by thin-layer chromatography on a silica 60 plates (Merck, Germany) with chloroform-acetic acid-ethanol at 95:5:2.5 (v/v) as the mobile phase, followed by spraying of an iron reagent (0.1 M FeCl3 in 0.1 M HCl). LC-MS was performed with an Agilent 1290 Infinity LC system coupled to Agilent 6520 Accurate-Mass Q-TOF mass spectrometer (dual ESI source) equipped with an Agilent Zorbax Eclipse XDB-C18 column. The ultraviolet/visible absorption spectrum was recorded with the photodiode array detector equipped with the above-mentioned HPLC. The mobile phase was composed of water (A, 0.5% formic acid) and acetonitrile (B, 0.5% formic acid), the gradient program of which was 0–12.00 min 90% A and 10% B and 12.00–15.00 min 100% B. The follow rate of the mobile phase was 0.3 ml/min, and the column temperature was set to 25°C. The injection volume was 10 μl.
Antimicrobial Testing Via Broth Microdilution
The antimicrobial activity of each fraction was evaluated using a broth microdilution assay to determine the minimum inhibitory concentration (MIC) of an antimicrobial compound as described by the Clinical and Laboratory Standard Institute (CLSI). MIC is defined as the lowest concentration of an antimicrobial to inhibit the visible growth of a microorganism after overnight incubation. Briefly, the test microorganisms were grown in MHB at 37°C for 24 h and adjusted to 0.5 McFarland standard (OD625 0.08–0.11). The adjusted cultures were then diluted 100 times in MHB and used as inocula. The extracts were serially diluted using sterile MHB in a 96-well flat bottomed microtiter plate. One hundred micro liters of test microorganisms corresponding to approximately 105 colony forming units (CFU) was added to each well. Determination of MIC was performed in triplicate. The positive control for bacteria was 100 μg/ml chloramphenicol. The negative control contained MHB with test microorganisms. The blank control consisted only of MHB. The microtiter plate was incubated at 37°C aerobically for 24 h and the MIC was determined by the concentration of extract (μg/ml) where no visible growth was observed.
Scanning Electron Microscopy (SEM)
SEM was performed based on the method described by Pilsczek et al. (2010) with modification, to determine the effect of the extract on the cellular morphology of the bacteria. E. faecalis ATCC 700802 was grown in MHB at 37°C for 24 h and the turbidity was adjusted to 0.5 McFarland standard. The MIC of the extract was added to the adjusted bacterial culture. An untreated control was used as a negative control, while treatment with 100 μg/mL chloramphenicol was used as the positive control. All samples were incubated at 37°C for 4 h. The cultures were then centrifuged at 5000 × g for 3 min and the supernatant was discarded. The bacterial pellet was washed with phosphate buffered saline (PBS) and subjected to centrifugation at 5000 × g for 3 min. This washing process was repeated three times. The washed pellets were reconstituted in minimal volume of PBS, placed onto glass slides (5 × 5 mm) and allowed to air dry for 30 min. The slides were fixed using 2.5% (v/v) gluteraldehyde in PBS for 4 h and washed three times with PBS to remove excess glutaraldehyde. The slides were then serially dehydrated with increasing concentration of ethanol and kept in a desiccator overnight. The slide was spur-coated with platinum using Q150R rotary-pumped sputter coater before observed using SU8010 FE-SEM (Hitachi, Japan).
Nucleotide Sequence Accession Number
The 16s rRNA gene sequence of strain MSh1T has been deposited in GenBank/EMBL/DDBJ under the accession number KT159931. The gene sequences of each MLSA loci have been deposited at the Bcc PubMLST database with sequence typing (ST) 1043; and GenBank/EMBL/DDBJ under the accession number KU301866–301872.
Results
16S rRNA Gene Sequence Analysis
The 16S rRNA gene sequence was obtained for strain MSh1T (1497 bp; GenBank/EMBL/DDBJ accession number KT159931) and a phylogenetic tree was constructed (Figure 1). Phylogenetic analysis demonstrated that strain MSh1T is closely related to B. arboris R-24201T and B. lata 383T, as they formed a distinct clade (Figure 1). Pairwise comparison of the 16S rRNA gene sequence of strain MSh1T with those Bcc type strains revealed similarity levels between 97.1 and 99.9% (data not shown).
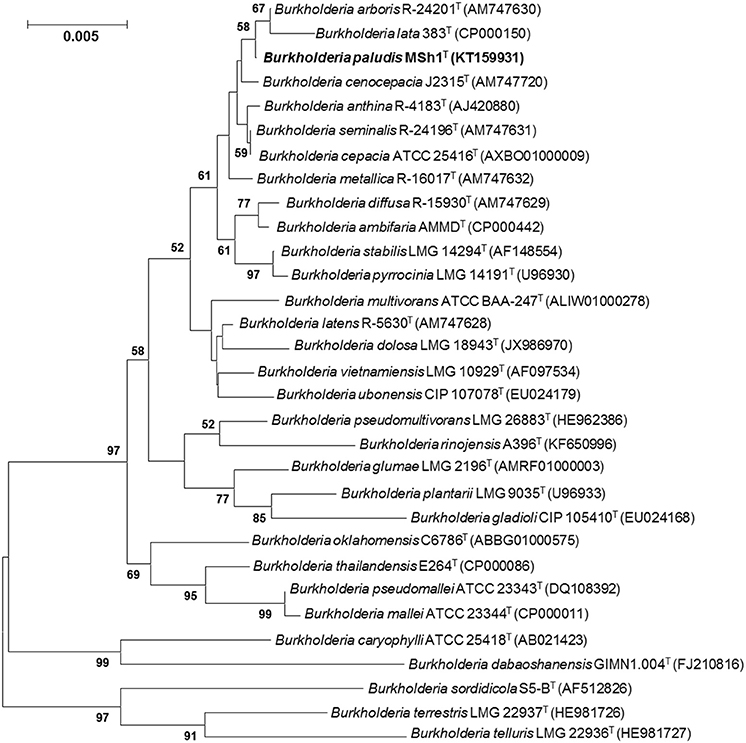
Figure 1. Neighbor-joining tree based on 16S rRNA sequences showing relationship between strain MSh1T and representatives of some other related taxa. Bootstrap values (>50%) based on 1000 resampled datasets are shown at branch nodes. Bar, 5 substitutions per 1000 nucleotide positions.
DDH
The DNA–DNA relatedness values between strain MSh1T with its close neighbor based on 16S rRNA phylogenetic analysis: B. arboris R-24201T (29.0 ± 3.7%), B. cenocepacia J2315T (32.8 ± 2.2%) and B. lata 383T (19.5 ± 1.8%) were significantly below the 70% threshold value for species delineation (Wayne et al., 1987).
MLSA
Phylogenetic analysis of concatenated allele sequences demonstrated that strain MSh1T diverged from the other closely related Bcc type strains, supported by a bootstrap value of 86% (Figure 2). MLSA data were used to assign allele types and allelic profiles. The allelic profile were as follows: atpD, 302; gltB, 11; gyrB, 50; recA, 350; lepA, 288; phaC, 249; and trpB, 345 (GenBank/EMBL/DDBJ accession number KU301866–301872). Strain MSh1T showed distinct allelic profile when compared with those closely related type strains (Table S1). For each established Bcc species, all allele types and allelic profiles were exported from the Bcc MLST database. The average concatenated allele sequence divergence of strain MSh1T toward its nearest neighbor B. stabilis LMG 14294T (5.23%), B. pyrrocinia LMG 14191T (5.81%) and B. cenocepacia IIIC 19230T (6.87%) were above the 3% cut-off value hence indicating that strain MSh1T is a novel species within the Bcc (Table S2) (Vanlaere et al., 2009; Peeters et al., 2013). Moreover, these data further substantiated the results obtained from DDH which confirmed that strain MSh1T is a novel species within the Bcc as the concatenated allele divergences between strain MSh1T and its close neighbor, B. arboris R-24201T, B. cenocepacia J2315T and B. lata 383T were more than 3%.

Figure 2. Phylogenetic tree based on the concatenated sequences of seven housekeeping gene fragments of established Bcc species and strain MSh1T. Bootstrap values (>50%) based on 1000 resampled datasets are shown at branch nodes. Bar, 2 substitutions per 100 nucleotide positions.
Phenotypic and Biochemical Characterization
Strain MSh1T was Gram negative, facultative anaerobic, motile, rod-shaped, 0.6–0.8 × 1.6–2.1 μm bacterium (Figure 3). Colonies produced by the strain MSh1T on NA were round, yellow colored with a smooth surface and 2–3 mm in diameter. Growth of strain MSh1T occurs at 15–40°C (optimum 30°C) and pH 4.0–10.0 (optimum pH 7.0) in NB. Growth occurs with 0–2.5% NaCl (optimum without NaCl). Table 1 shows that strain MSh1T can be differentiated biochemically from the closely related members of the genus Burkholderia. It was shown that strain MSh1T differs from the other Bcc species by the ability to acidify adonitol, having arginine dihydrolase activity and inability to assimilate N-acetyl-glucosamine (Vandamme et al., 1997, 2000, 2002; Coenye et al., 2001; Henry et al., 2001; Vanlaere et al., 2008, 2009; Peeters et al., 2013).

Figure 3. SEM images of Burkholderia paludis MSh1T. Images were taken under 25,000 × magnification at 1 kV. The bacterial cells sizes are approximately 0.6–0.8 × 1.6–2.1 μm.
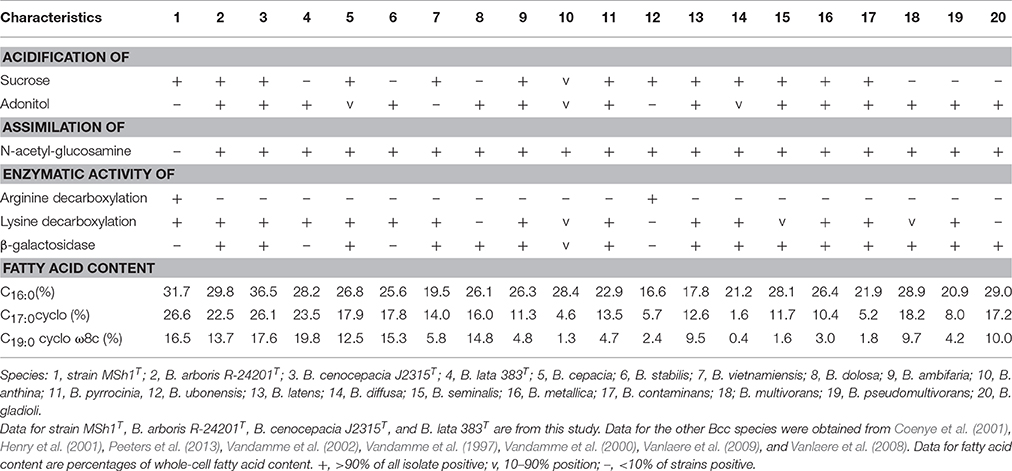
Table 1. Differentiation characteristics of strain MSh1T and type strains of closely related species of Bcc.
Chemotaxonomic Characterization
Chemotaxonomic analysis revealed that the major cellular fatty acids were C16:0 (31.7%), C17:0 cyclo (26.6%) and C19:0 cyclo ω8c (16.1%). The fatty acid profile of MSh1T was consistent with these of closely related type strains such as B. arboris R-24201T, B. cenocepecia J2315T, and B. lata 383T which contained fatty acid C16:0 (28.2–36.5%), C17:0 cyclo (22.5–26.1%) and C19:0 cyclo ω8c (13.7–19.8%) (Table 1). The ubiquinone Q-8 was detected. The polar lipids consisted phosphatidylglycerol (PG), phosphatidylethanolamine (PE) and diphosphatidylglycerol (DPG), aminolipid and aminophospholipid (Figure 4). The fatty acid profile, polar lipid profile and major isoprenoid quinone of strain MSh1T were consistent with Burkholderia type strains (Gillis et al., 1995).
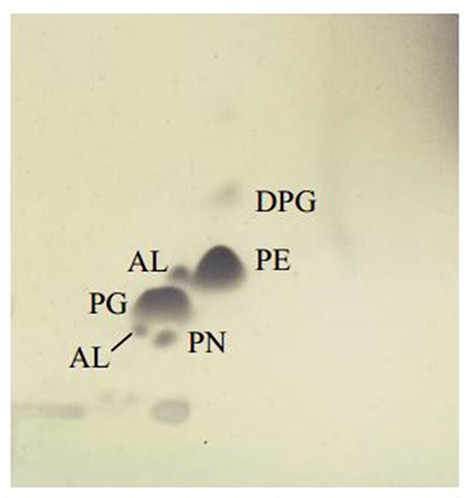
Figure 4. Thin layer chromatogram polar lipid detected in Burkholderia paludis MSh1T. AL, aminolipid; PN, aminophospholipid; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; DPG, diphosphatidylglycerol.
Genome-To-Genome Distance Calculator (GGDC) and Average Nucleotide Identity (ANI)
GGDC is an in silico genome-to-genome comparison tool used to calculate the intergenomic distances and relatedness of strain MSh1T with known type strains (Meier-Kolthoff et al., 2013). The data obtained is converted to similarity values analogous to DNA–DNA hybridization (DDH) hence similarity value of 70% is generally regarded as a threshold for new species determination (Meier-Kolthoff et al., 2013). It was found that the similarity values between strain MSh1T with related known Bcc species are less than 70%, thus suggesting that this isolate might represent new species. This is further supported by the ANI results which revealed that strain MSh1T had ANI-value lower than the 96% threshold for new species (Goris et al., 2007; Table S3).
Genome Analysis of Secondary Metabolites Gene Clusters Using Antismash
Antimicrobial compounds produced by bacteria are usually secondary metabolites which are regulated by biosynthetic gene clusters. These gene clusters can be annotated using antiSMASH 2.0 (Blin et al., 2013, 2014). Analysis showed that the genome of strain MSh1T had 43 gene clusters responsible for the biosynthesis of secondary metabolites which include the PKS, NRPS, bacteriocin, and terpene biosynthetic genes. Besides that, it was found that strain MSh1T (43 gene clusters) possess a higher number of biosynthetic gene clusters compared to other related Burkholderia species (9 to 25 gene clusters) despite having similar genome size (Table 2). This might indicate that some of the putative biosynthetic gene clusters might be new as antiSMASH was unable to annotate due to the lack of similarity between query sequences from the database. These results revealed that the antimicrobial compounds produced by strain MSh1T might either be new or under-studied.
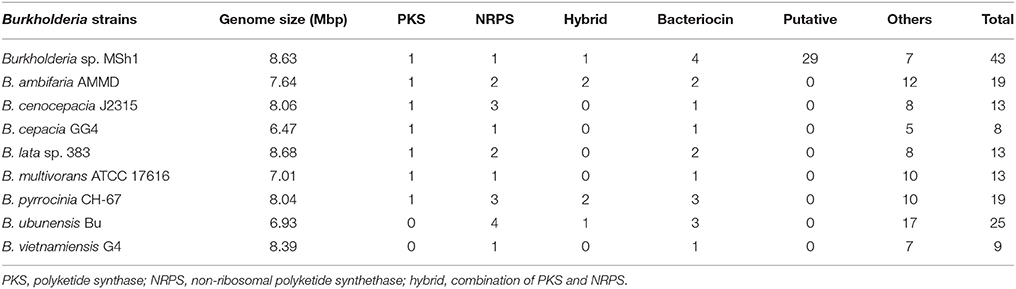
Table 2. Comparison of gene clusters responsible for secondary metabolites biosynthesis between strain MSh1 with related Burkholderia species.
Antimicrobial Activity of MSh1T
The methanol extract of MSh1T demonstrated antimicrobial activity four Gram positive bacteria and a Gram negative bacterium (Table 3). It was found that the Gram positive bacteria had lower MIC-value when compared to the Gram negative bacteria. Sequential solvent fractionation of the methanol crude extract yielded four different fractions with different polarities (HEX fraction, DCM fraction, EtOAc fraction and water fraction). The DCM fraction showed similar antimicrobial activity (MIC 0.020 mg/ml) to the crude methanol extract, indicating that the antimicrobial compounds had successfully been fractionated from the original extract. Further purification of the DCM extract has led to the isolation of Compound 1, pyochelin. Antimicrobial testing of Compound 1 was performed with 13 other test microorganisms via broth microdilution. Compound 1 demonstrated antimicrobial activity against four Staphylococcus strains and three Enterococcus strains; but not Bacillus subtilis ATCC 8188, B. cereus ATCC 14579 and all other Gram negative bacteria tested. SEM analysis showed that the cellular morphology of E. faecalis ATCC 700802 was not affected by pyochelin.
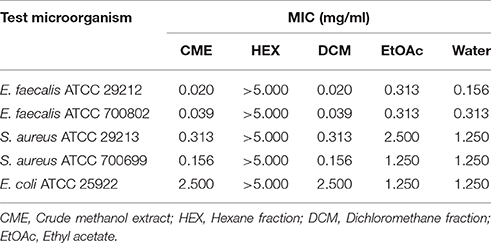
Table 3. The minimum inhibitory concentration (MIC) of different fractions and crude methanol extract against the five test microorganism strains.
Discussion
The taxonomic position of strain MSh1T was determined and characterized using a polyphasic approach. The 16S rRNA sequence of strain MSh1T revealed that it belonged to the genus Burkholderia and was closely related to the Bcc. Phylogenetic analysis further showed that strain MSh1T clades closely to B. arboris, B. lata and B. cenocepacia. Despite having high 16S rRNA gene sequence similarity (data not shown), phylogenetic analysis indicated that strain MSh1T is situated on a different branch in the tree, signifying the possibility of being a novel species. As members of Bcc are known to have high similarity in terms of 16S rRNA gene sequence, other molecular taxonomy techniques have to be used to differentiate and identify them (Coenye and Vandamme, 2003; Spilker et al., 2009; Peeters et al., 2013). Hence, DDH and MLSA were performed in this study. The DDH results which showed that the DNA–DNA relatedness value between strain MSh1T and its closely related neighbors (based on 16S rRNA phylogenetic analysis) were below the 70% threshold value for species delineation (Wayne et al., 1987). This was further supported by the average concatenated allele sequence divergence (obtained from the MLSA analysis) of strain MSh1T toward other Bcc species which were above the 3% threshold (Librado and Rozas, 2009). Moreover, based on the biochemical profile of strain MSh1T, it can be distinguished from most Bcc by its inability to assimilate N-acetyl-glucosamine. Furthermore, this study has used a combination of both polyphasic and genome comparison (via next generation sequencing) to determine the species delineation of strain MSh1T. For every technique used, each result revealed that strain MSh1T is indeed a novel species within the Bcc.
The genus Burkholderia is well known for producing a wide range of secondary metabolites which include siderophores (Asghar et al., 2011), antifungal (Lu et al., 2009; Tawfik et al., 2010), antibacterial (El-Banna and Winkelmann, 1998; Mitchell and Teh, 2005; Mitchell et al., 2008), antitumor (Klausmeyer et al., 2011; He et al., 2014), and exotoxins (Jeong et al., 2003; Partida-Martinez and Hertweck, 2007). Secondary metabolites are compounds which are non-essential for microbial growth or reproduction but provide diverse survival functions in nature (Martin et al., 2005). Many of these secondary metabolites are regulated by complex synthesis mechanisms within the bacteria itself which include the PKS and NRPS. The assembly of a polyketide via the PKS system begins by priming the starter molecule to a catalytic domain, followed by chain elongation with extender units by different kind of enzymes. As for the NRPS system, a NRP will firstly be generated and then linked with other molecules for instance a phosphate group, methyl group or fatty acids (Donadio et al., 2007; Amoutzias et al., 2008). In this present study, antiSMASH was used to annotate the secondary metabolite biosynthetic gene clusters present in the draft genome of strain MSh1T. It was found that strain MSh1T has a higher number of secondary metabolite biosynthetic gene clusters as compared to other known antimicrobial producing Bcc species, for example B. cepacia and B. pyroccinia (Souza and Raaijmakers, 2003). Both PKS and NRPS gene clusters are present in the genome, indicating the antimicrobial compounds produced might be assembled by either of these two gene clusters. Moreover, strain MSh1T might be able to produce a more complex antimicrobial compound as a hybrid PKS-NRPS was annotated by antiSMASH. One example of such an antimicrobial compound is a lipopeptide class compound (Sorensen et al., 2014). Lipopeptides are antimicrobial compounds, known to disrupt the surface membrane charges of its target. Thus far, there have been only two reported lipopeptides (burkholdine and occidiofungin) produced by Bcc species and they only possess antifungal activity (Lu et al., 2009; Tawfik et al., 2010). Bacteriocins are small peptides with narrow-spectrum antimicrobial properties (Cotter et al., 2013). Capistruin, is an example of antimicrobial peptide produced by a Burkholderia species. Nevertheless, the antimicrobial activity is only limited to several Gram negative bacteria which include Burkholderia species, Psuedomonas aeruginosa, and E. coli (Knappe et al., 2008). The data obtained from antiSMASH provided a preliminary insight into the secondary metabolite gene clusters present in the genome of strain MSh1T and it is certain that strain MSh1T can indeed produce antimicrobial compounds. As many secondary metabolite biosynthetic gene clusters were present in the genome, the antimicrobial activity observed might be due to the presence of multiple compounds. Therefore, extraction of the antimicrobial compounds produced by strain MSh1T was performed in this study.
Since the antimicrobial activity of strain MSh1T was determined with agar overlay assay in the previous study (Ong et al., 2015), this showed that the antimicrobial compounds produced could be extracted from the agar itself. Therefore, agar extraction (solid-liquid extraction) was carried out. This technique was adopted from a study conducted by Isnansetyo and Kamei (2003) in which they successfully extracted an antimicrobial compound produced by Pseudoalteromonas phenolica cultured on marine solid media. It was shown that E. faecalis ATCC 29212 and E. faecalis ATCC 700802 were highly sensitive to the antimicrobial compounds with MIC-value of 0.020 mg/ml and 0.039 mg/ml, respectively, as compared to S. aureus ATCC 29213 (0.313 mg/ml), S. aureus ATCC 700699 (0.156 mg/ml) and E. coli ATCC 25922 (2.500 mg/ml) (consistent with Ong et al., 2015). To the best of our knowledge, there has only been one antimicrobial compound with inhibitory effect against Gram positive bacteria and fungus (pyrrolnitrin), isolated from Burkholderia species, and based on the PCR performed in this study, the prnD gene was not found in strain MSh1T which further substantiate that it might be producing other types of antimicrobial compounds. Furthermore, the genes that code for prnA, prnB, prnC and prnD were absent from the draft genome of strain MSh1T.
Sequential solvent fractionation was performed to fractionate and partially purify, at the same time removing impurities present in the crude methanol extract. A similar method was adopted from Tawfik et al. (2010) to isolate antifungal burkholdines produced by B. ambifaria 2.2N. In this study, four different solvents were used: hexane (HEX), dichloromethane (DCM), ethyl acetate (EtOAc) and water. HEX is a non-polar solvent commonly used to remove non-polar compounds from the crude extract. DCM and EtOAc both have middle-range polarity, thus is able to attract a wider range of compounds with different polarity (Moreau et al., 2003). Water being the most polar solvent will retain any compounds with highly polarity (Wang and Weller, 2006). It was shown that the HEX fraction had no antimicrobial activity against all test microorganisms up to 5 mg/mL. The DCM fraction had the best antimicrobial activity against all the test microorganism strains (when compared with the EtOAc and water fractions) as the MIC-values were comparable or similar to the crude methanol extract (Table 3). Thus, further purification was conducted on the DCM fraction using C18 which led to the isolation of Compound 1. Chromatographic analysis of Compound 1 on a TLC plate with chloroform-acetic acid-ethanol at 90:5:2.5 (v/v) as the mobile phase showed one spot (Rf 0.35) with yellow-green fluorescent band that turned red-brown after spraying with the iron reagent. Analysis of Compound 1 by analytical HPLC indicated that the compound contained one peak with three maxima at 210, 270, and 320 nm. The mass of Compound 1 (m/z 325, [M+H]+) was determined by liquid chromatography-electrospray ionization-mass spectrometer (LC-ESI-MS). The ESI-MS indicated that the molecular formula of the compound was C14H16N2O3S2. The interpretation of ESI-MS and UV spectrum of Compound 1 were found to be identical to pyochelin which was in good agreement with previous literature (Cox et al., 1981; Adler et al., 2012) (Figures S1, S2).
Pyochelin is a type of siderophore commonly produced by the genus Pseudomonas (Cox et al., 1981; Buysens et al., 1996; Lim et al., 2016). Siderophores can solubilize ferric ion, hence is an iron chelating growth factor for many bacteria. Thus far, only three Burkholderia species (B. arboris, B. cenocepacia and B. contaminans) have been known to produce pyochelin (Dang et al., 2011; Schwagner et al., 2012; Deng et al., 2015). As pyochelin is a siderophore, its antimicrobial properties were not extensively studied. Pyochelin can inhibit or kill bacteria by catalyzing the generation of reactive oxygen species (ROS) (Adler et al., 2012). However, this is bacteria dependent as shown in Table 4. The Gram negative bacteria (typically the Enterobacteriaceae) were found to be resistant to Compound 1 (pyochelin) and this is consistent with the study conducted by Adler et al. (2012). The resistant profile shown by the Enterobacteriaceae was due to the production of catecholate siderophores such as enterobactin. These siderophores can act as hydrogen atom donors and efficiently terminate radical chain reactions; hence rendering pyochelin ineffective. The inhibitory effect of pyochelin on the four S. aureus and three E. faecalis strains might due to presence of ROS generated by pyochelin. Nevertheless, no literature has reported on the downstream effect of the ROS produced by pyochelin, as it might target the DNA/RNA replication, electron transport chain or bacterial cell membrane. Moreover, the nutrient availability of these strains might be compromised as pyochelin can chelate the vital metal ions present in the environment. Consequently, these sensitive strains might be starved and ultimately led to death, as they lack the receptors to mediate the entry of metal-bounded pyochelin into the bacterial cells. Hence as a preliminary study, we investigated the effect of pyochelin on the cellular morphology of E. faecalis ATCC 700802 via SEM. Pyochelin did not affect the cellular morphology of E. faecalis ATCC 700802, when compared to the positive control in which the morphology of the bacteria were distorted when treated with 100 μg/ml of chloramphenicol (Figure 5). This result supports that the ROS generated by pyochelin might target the intracellular components of E. faecalis ATCC 700802. Further work on the effect of pyochelin on the DNA/RNA replication will be validated via qPCR in the future. Moreover, the antimicrobial activity of pyochelin was not only restricted to normal strains of S. aureus and E. faecalis, but it also affects the antimicrobial-resistant strains, for instance methicillin-resistant S. aureus ATCC 700699, methicillin-resistant S. aureus ATCC 43300 and vancomycin-resistant E. faecalis ATCC 700802. However, this has to be validated by increasing the number of test strains used. Additional work is on-going to further elucidate the downstream effect of pyochelin on these sensitive strains, and also to determine its potential synergistic action with antibiotics. Nevertheless, this study has provided an insight that the infamous pyochelin might potentially be used to treat infections caused by ARB.
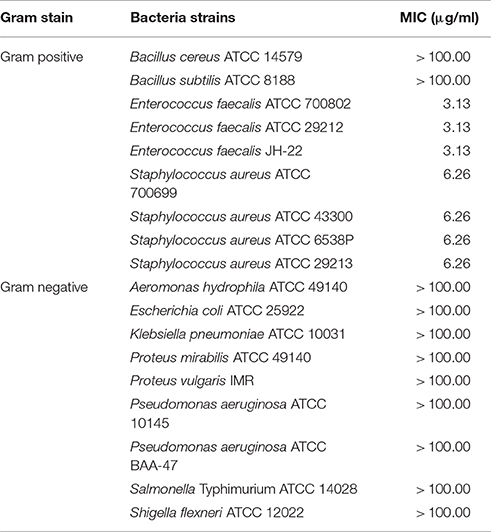
Table 4. The minimum inhibitory concentration (MIC) of Compound 1 (pyochelin) against 18 test microorganisms.

Figure 5. FE-SEM images of E. faecalis ATCC 700802 showing (A) negative control, (B) treated with MIC of Compound 1 and (C) treated with 100 μg/ml of chloramphenicol (positive control). Images were taken under 5000x magnification at 1 kV.
Based on the results of phylogenetic analysis, DDH, MLSA, GGDC, ANI, phenotypic, chemotaxonomic and biochemical characterization, strain MSh1T was assigned to a novel species in the genus Burkholderia, for which the name Burkholderia paludis sp. nov. is proposed.
Description of Burkholderia paludis sp. nov.
Burkholderia paludis (pa.lu.dis. L. gen. n. paludis of the swamp). Cells are Gram negative, facultative anaerobic, motile rods, about 0.6–0.8 μm wide and 1.6–2.1 μm long. Colonies are regular, circular, convex, translucent, moist and 1.0–3.0 mm in diameter after 2 days of cultivation at 30°C on NA. Grows at 15–40°C (optimum 30°C) and pH 4.0–10.0 (optimum pH 7.0) in NB. Grows at 0–2.5% NaCl (optimum without NaCl). Cells are positive for oxidase, glucose fermentation, arginine dihydrolase, urease, esculin hydrolysis and assimilation of glucose, but negative for nitrate/nitrite reduction, indole production, gelatin hydrolysis, β-galactosidase, assimilation of arabinose, mannose, mannitol, N-acetyl-glucosamine, maltose, potassium gluconate, capric acid, adipic acid, malate, trisodium citrate and phenylacetic acid (API 20NE).
In API ZYM-tests, positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-glucosidase and n-acetyl-β-glucosaminidase, but negative for valine arylamidase, cystine arylamidase, trypsin, a-chymotrypsin, α-galactosidase, β-galactosidase, β-glucoronidase, α-glucosidase, αmannosidase and α-fucosidase. The following carbon sources are utilized in the API 50CH: glycerol, erythritol, D-arabinose, L-arabinose, ribose, D-xylose, galactose, D-glucose, D-fructose, D-mannose, dulcitol, inositol, mannitol, sorbitol, α-methyl-D-glucoside, arbutin, esculin, salicin, cellobiose, maltose, lactose, saccharose, trehalose, D-turanose, D-xylose, D-tagatose, D-fucose, L-fucose, D-arabitol and L-arabitol; the other substrates, L-xylose, adonitol, α-methyl-D-xiloside, L-sorbose, rhamnose, α-methyl-D-mannoside, α-methyl-D-glucosamine, amygdalin, melibiose, inulin, melezitose, D-raffinose, amidon, glycogen, xylitol, β-gentiobiose, gluconate, 2-keto-gluconate and 5-keto-gluconate, are not utilized.
The fatty acid profile is composed mainly of C16:0 (31.7%), C17:0 cyclo (26.6%), and C19:0 cyclo ω8c (16.1%). The major compounds in the polar lipid profile are phosphatidylglycerol, phosphatidylethanolamine and diphosphatidylglycerol. The predominant ubiquinone is Q-8. Strain MSh1T is able to produce antimicrobial compounds active against four strains of S. aureus and three strains of E. faecalis.
The type strain is MSh1T (= DSM 100703T = MCCC 1K01245T), isolated from surface peat from the Southeast Pahang tropical peat swamp forest reserve, Malaysia. The 16S rRNA and MLSA gene sequence of strain MSh1T has been deposited in GenBank/EMBL/DDBJ under the accession number KT159931 and KU301866–301872, respectively.
Accession Numbers
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain MSh1T is KT159931. The GenBank/EMBL/DDBJ accession number for the MLSA gene sequences of strain MSh1T are KU301866–301872.
Author Contributions
KSO performed the laboratory experiments, data analysis and the manuscript write-up. SML supervised the entire study. LHL and YLC co-supervised the study. SML, KSO and YKA contributed to the experimental designs. LHL and KSO contributed to the polyphasic taxonomy. CMY, SML and YKA apprehended the idea of bioprospecting in tropical peat swamp forest. All authors proofread and reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the School of Science and Tropical Medicine and Biology Multidisciplinary Platform, Monash University Malaysia for funding this project.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02046/full#supplementary-material
Abbreviations
Bcc, Burkholderia cepacia complex; MLSA, multilocus sequence analysis.
References
Adler, C., Corbalán, N. S., Seyedsayamdost, M. R., Pomares, M. F., de Creistóbal, R. E., Clardy, J., et al. (2012). Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS ONE 7:e46754. doi: 10.1371/journal.pone.0046754
Ammerlaan, H. S., Harbarth, S., Buiting, A. G., Crook, D. W., Fitzpatrick, F., Hanberger, H., et al. (2016). Secular trends in nosocomiam bloodstream infections: antibiotic-resistant bacteria increase the total burden of infection. Clin. Infect. Dis. 56, 798–805. doi: 10.1093/cid/cis1006
Amoutzias, G. D., Van de Peer, Y., and Mossialos, D. (2008). Evolution and taxanomic distribution of nonribosomal peptide and polyketide synthase. Future Microbiol. 3, 361–370. doi: 10.2217/17460913.3.3.361
Asghar, A. H., Shastri, S., Dave, E., Wowk, I., Agnoli, K., Cook, A. M., et al. (2011). The pobA gene of Burkholderia cenocepacia encodes a Group I Sfp-type phosphopantetheinyltransferase required for biosynthesis of the siderophores ornibactin and pyochelin. Microbiology 157, 349–361. doi: 10.1099/mic.0.045559-0
Blin, K., Kazempour, D., Wohlleben, W., and Weber, T. (2014). Improved lanthipeptide detection and prediction for antiSMASH. PLoS ONE 9:e89420. doi: 10.1371/journal.pone.0089420
Blin, K., Medema, M. H., Kazempour, D., Fischbach, M. A., Breitling, R., Takano, E., et al. (2013). antiSMASH 2.0: a versatile platform for genome mining for secondary metabolite producers. Nucleic Acids Res. 41, 204–212. doi: 10.1093/nar/gkt449
Buck, J. D. (1982). Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol. 44, 992–993.
Bull, A. T., and Stach, J. E. (2007). Marine actinobacteria: new opportunities for natural product search and discovery. Trends. Microbiol. 15, 491–499. doi: 10.1016/j.tim.2007.10.004
Buysens, S., Heuengens, K., Poppe, J., and Hofte, M. (1996). Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Appl. Environ. Microbiol. 62, 865–871.
Caballero-Mellado, J. Martínez-Aguilar L., Paredes-Valdez, G., and Estrada-de los Santos, P. (2004). Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophtic species. Int. J. Syst. Evol. Microbiol. 54, 1165–1172. doi: 10.1099/ijs.0.02951-0
Cappuccino, J. G., and Sherman, N. (2002). Microbiology: A Laboratory Manual, 6th Edn. Menlo Park, CA: Benjamin Cummings.
Coenye, T., LiPuma, J. J., Henry, D., Hoste, B., Vandemeulebroecke, K., Gillis, M., et al. (2001). Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51, 271–279. doi: 10.1099/00207713-51-2-271
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins: a viable alternatie to antibiotics? Nat. Rev. Micriobiol. 11, 95–105. doi: 10.1038/nrmicro2937
Cox, C. D., Rinehart, K. L. Jr., Moore, M. L., and Cook, C. (1981). Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 78, 4256–4250. doi: 10.1073/pnas.78.7.4256
Dang, L. D., Son, S. W., Cheon, H. M., Choi, G. J., Choi, Y. H., Jang, K. S., et al. (2011). Pyochelin isolated from Burkholderia arboris KRICT1 carried by pine wood nematodes exhibits phytotoxicity in pine callus. Nematology 13, 521–528. doi: 10.1163/138855410X528271
De Ley, J., Cattoir, H., and Reynaerts, A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12, 133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x
Deng, P., Wang, X., Baird, S. M., Showmaker, K. C., Smith, L., Peterson, D., et al. (2015). Comparative genome-wide analysis reveals that Burkholderia contaminans MS14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. Microbiol. Open 5, 353–369. doi: 10.1002/mbo3.333
Donadio, S., Monciardini, P., and Sosio, M. (2007). Polyketide synthase and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat. Prod. Rep. 24, 1073–1109. doi: 10.1039/b514050c
El-Banna, N., and Winkelmann, G. (1998). Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 85, 69–78. doi: 10.1046/j.1365-2672.1998.00473.x
Felsenstein, J. (1985). Phylogenies and the comparative method. Am. Nat. 125, 1–15. doi: 10.1086/284325
Gillis, M., Van Van, T., Bardin, R., Goor, M., Hebbar, P., Willems, A., et al. (1995). Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Evol. Microbiol. 45, 274–289. doi: 10.1099/00207713-45-2-274
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
He, H., Ratnayake, A. S., Janso, J. E., He, M., Yang, H. Y., Loganzo, F., et al. (2014). Cytotoxic spliceostatins from Burkholderia sp. and their semisynthetic analogues. J. Nat. Prod. 2014, 1864–1870. doi: 10.1021/np500342m
Henry, D., Mahenthiralingam, E., Vandamme, P., Coenye, T., and Speert, D. P. (2001). Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J. Clin. Microbiol. 39, 1073–1078. doi: 10.1128/JCM.39.3.1073-1078.2001
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Micriobiol. 8, 15–25. doi: 10.1038/nrmicro2259
Huss, V. A. R., Festl, H., and Schleifer, K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4, 184–192. doi: 10.1016/S0723-2020(83)80048-4
Imhoff, J. F., Labes, A., and Wiese, J. (2011). Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol. Adv. 29, 468–482. doi: 10.1016/j.biotechadv.2011.03.001
Isnansetyo, A., and Kamei, Y. (2003). MC21-a, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30T, against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47, 480–488. doi: 10.1128/AAC.47.2.480-488.2003
Jeong, Y., Kim, J., Kim, S., Kang, Y., Nagamatsu, T., and Hwang, I. (2003). Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing wilt in many field crops. Plant Dis. 87, 890–895. doi: 10.1094/PDIS.2003.87.8.890
Jukes, T., and Cantor, C. (1969). Evolution of protein molecules. Mamm. Protein. Metab. 3, 21–132. doi: 10.1016/B978-1-4832-3211-9.50009-7
Kane, M. D., Poulsen, L. K., and Sathl, D. A. (1993). Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59, 682–686.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutes through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Klausmeyer, P., Shipley, S. M., Zuck, K. M., and McCloud, T. G. (2011). Histone deacetylase inhibitors from Burkholderia thailandensis. J. Nat. Prod. 74, 2039–2044. doi: 10.1021/np200532d
Knappe, T. A., Linne, U., Zirah, S., Rebuffat, S., XIe, X., and Marahiel, M. A. (2008). Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J. Am. Chem. Soc. 130, 11446–11454. doi: 10.1021/ja802966g
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lim, C. K., Penesyan, A., Hassan, K. A., Loper, J. E., and Paulsen, I. T. (2016). Disruption of transporters affiliated with enantio-pyochelin biosynthesis gene cluster of Pseudomonas protegens Pf-5 has pleiotropic effects. PLoS ONE 11:e0159884. doi: 10.1371/journal.pone.0159884
Lu, S., Novak, J., Austin, F. W., Gu, G., Ellis, D., Kirk, M., et al. (2009). Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 48, 8312–8321. doi: 10.1021/bi900814c
Martin, J. F., Casqueiro, J., and Liras, P. (2005). Secretion system for secondary metabolites: how producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 8, 282–293. doi: 10.1016/j.mib.2005.04.009
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Miller, L. T. (1982). A single derivatization method for bacterial fatty acid methyl esters including hydroxy acids. J. Clin. Microbiol. 16, 584–586.
Mishra, R. P., Oviedo-Orta, E., Prachi, P., Rappuoli, R., and Bagnoli, F. (2012). Vaccines and antibiotic resistance. Curr. Opin. Microbiol. 15, 1–7. doi: 10.1016/j.mib.2012.08.002
Mitchell, R. E., Greenwood, D. R., and Sarojini, V. (2008). An antibacterial pyrazole derivative from Burkholderia glumae, a bacterial pathogen of rice. Phytochemistry 69, 2704–2707. doi: 10.1016/j.phytochem.2008.08.013
Mitchell, R. E., and Teh, K. L. (2005). Antibacterial iminopyrrolidines from Burkholderia plantarii, a bacterial pathogen of rice. Org. Biomol. Chem. 3, 3540–3543. doi: 10.1039/b509319h
Moreau, R. A., Powell, M. J., and Singh, V. (2003). Pressurized liquid extraction of polar and nonpolar lipids in corn and oats with hexane, methylene chloride, isopropanol and ethanol. J. Am. Oil Chem. Soc. 80, 1063–1067. doi: 10.1007/s11746-003-0821-y
Ong, K. S., Aw, Y. K., Gan, H. M., Yule, C. M., and Lee, S. M. (2014). Draft genome sequences of two antimicrobial-producing Burkholderia sp. strains, MSh1 and MSh2, isolated from Malaysian tropical peat swamp forest soil. Genome. Announc. 2, 1–2. doi: 10.1128/genomeA.01032-14
Ong, K. S., Yule, C. M., and Lee, S. M. (2015). Antimicrobial producing bacteria isolated from tropical peat swamp soil. Malays. J. Microbiol. 11, 170–175. doi: 10.21161/mjm.12914
Partida-Martinez, L. P., and Hertweck, C. (2007). A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochemistry 8, 41–45. doi: 10.1002/cbic.200600393
Peeters, C., Zlosnik, J. E., Spilker, T., Hird, T. J., LiPuma, J. J., and Vandamme, P. (2013). Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst. Appl. Microbiol. 36, 483–489. doi: 10.1016/j.syapm.2013.06.003
Pilsczek, F. H., Salina, D., Poon, K. K., Fahey, C., Yipp, B. G., Sibley, C. D., et al. (2010). A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J. Immunol. 185, 7413–7425. doi: 10.4049/jimmunol.1000675
Robbie, W. (1945). A hanging drop method for continuous observation of the activity of organisms in cyanide. Science 101, 649–650. doi: 10.1126/science.101.2634.649
Saitou, N., and Nei, M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Bio. Evol. 4, 406–425.
Schwagner, S., Agnoli, K., Kothe, M., Feldmann, F., Givskov, M., Carlier, A., et al. (2012). Identification of Burkholderia cenocepacia strain H111 virulence factors using nonmammalian infections hosts. Infect. Immun. 81, 143–153. doi: 10.1128/IAI.00768-12
Sorensen, J. L., Sondergaard, T. E., Covarelli, L., Feuertes, P. R., Hansen, F. T., Frandsen, R. J. N., et al. (2014). Identification of the biosynthetic gene clusters for the lipopeptides Fusaristatin, A., and W493 B in Fusarium graminearum and F. pseudograminearum. J. Nat. Prod. 77, 2619–2625. doi: 10.1021/np500436r
Souza, J. T., and Raaijmakers, J. M. (2003). Polymorphisms within the prnD and pltC gene from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 43, 21–34. doi: 10.1111/j.1574-6941.2003.tb01042.x
Spilker, T., Baldwin, A., Bumford, A., Dowson, C., Mahenthiralingam, E., and LiPuma, J. J. (2009). Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47, 2607–2610. doi: 10.1128/JCM.00770-09
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Bio. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tawfik, K., Jess, P., Bray, B., Dubay, G., Falkinham, J. O., Mesbah, M., et al. (2010). Bukholdines 1097 and 1229, potent antifungal peptides from Burkholderia ambifaria 2.2N. Org. Lett. 12, 664–666. doi: 10.1021/ol9029269
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tindall, B. J. (1990). A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 13, 128–130. doi: 10.1016/S0723-2020(11)80158-X
Tindall, B. J., Sikorski, J., Smibert, R. M., and Kreig, N. R. (2007). “Phenotypic characterization and the principles of comparative systematics,” in Methods for General and Molecular Microbiology, 3rd Edn., eds C. A. Reddy, T. J. Beveridge, J. A. Breznak, G. Marzluf, T. M. Schmidt, and L. R. Snyder (Washington, DC: ASM Press), 330–393.
Vandamme, P., Henry, D., Coenye, T., Nzula, S., Vancanneyt, M., LiPuma, J. J., et al. (2002). Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33, 143–149. doi: 10.1111/j.1574-695X.2002.tb00584.x
Vandamme, P., Holmes, B., Vancanneyt, M., Coenye, T., Hoste, B., Coopman, R., et al. (1997). Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Evol. Microbiol. 47, 1188–1200. doi: 10.1099/00207713-47-4-1188
Vandamme, P., Mahenthiralingam, E., Holmes, B., Coenye, T., Hoste, B., De Vos, P., et al. (2000). Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38, 1042–1047.
Vanlaere, E., Baldwin, A., Gevers, D., Henry, D., De Brandt, E., LiPuma, J. J., et al. (2009). Taxon, K., a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59, 102–111. doi: 10.1099/ijs.0.001123-0
Vanlaere, E., LiPuma, J. J., Baldwin, A., Henry, D., Brandt, E., Mahenthiralingam, E., et al. (2008). Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58, 1580–1590. doi: 10.1099/ijs.0.65634-0
Wang, L., and Weller, C. L. (2006). Recent advances in extraction of nutraceuticals from plants. Trends Food Sci. Tech. 17, 300–312. doi: 10.1016/j.tifs.2005.12.004
Wang, Q., Song, F., Xiao, X., Huang, P., Li, L., Monte, A., et al. (2013). Abyssomicins from the South China Sea deep-sea sediment Verrucosispora sp.: natural thioether Michael addition adducts as antitubercular prodrugs. Angew. Chem. Int. Microbiol. 78, 2039–2042. doi: 10.1002/anie.201208801
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 37, 463–464. doi: 10.1099/00207713-37-4-463
Keywords: antimicrobial, Burkholderia cepacia complex, multilocus sequence analysis (MLSA), polyphasic taxonomy, tropical peat swamp forest
Citation: Ong KS, Aw YK, Lee LH, Yule CM, Cheow YL and Lee SM (2016) Burkholderia paludis sp. nov., an Antibiotic-Siderophore Producing Novel Burkholderia cepacia Complex Species, Isolated from Malaysian Tropical Peat Swamp Soil. Front. Microbiol. 7:2046. doi: 10.3389/fmicb.2016.02046
Received: 07 September 2016; Accepted: 06 December 2016;
Published: 21 December 2016.
Edited by:
Tzi Bun Ng, The Chinese University of Hong Kong, ChinaReviewed by:
Vishvanath Tiwari, Central University of Rajasthan, IndiaDinesh Sriramulu, Shres Consultancy (Life Sciences), India
Sarah Victoria Harding, Defence Science and Technology Laboratory, UK
Sunil D. Saroj, Stockholm University, Sweden
Copyright © 2016 Ong, Aw, Lee, Yule, Cheow and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sui Mae Lee, bGVlLnN1aS5tYWVAbW9uYXNoLmVkdQ==
 Kuan Shion Ong
Kuan Shion Ong Yoong Kit Aw
Yoong Kit Aw Learn Han Lee
Learn Han Lee Catherine M. Yule
Catherine M. Yule Yuen Lin Cheow
Yuen Lin Cheow Sui Mae Lee
Sui Mae Lee