
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 06 December 2016
Sec. Evolutionary and Genomic Microbiology
Volume 7 - 2016 | https://doi.org/10.3389/fmicb.2016.01964
This article is part of the Research Topic Planctomycetes-Verrucomicrobia-Chlamydiae bacterial superphylum: New model organisms! View all 14 articles
Bacteria from the Planctomycetes, Verrucomicrobia, and Chlamydiae (PVC) superphylum are exceptions to the otherwise dominant mode of division by binary fission, which is based on the interaction between the FtsZ protein and the peptidoglycan (PG) biosynthesis machinery. Some PVC bacteria are deprived of the FtsZ protein and were also thought to lack PG. How these bacteria divide is still one of the major mysteries of microbiology. The presence of PG has recently been revealed in Planctomycetes and Chlamydiae, and proteins related to PG synthesis have been shown to be implicated in the division process in Chlamydiae, providing important insights into PVC mechanisms of division. Here, we review the historical lack of observation of PG in PVC bacteria, its recent detection in two phyla and its involvement in chlamydial cell division. Based on the detection of PG-related proteins in PVC proteomes, we consider the possible evolution of the diverse division mechanisms in these bacteria. We conclude by summarizing what is known and what remains to be understood about the evolutionary cell biology of PVC division modes.
Most bacteria divide by binary fission using a mechanism centered on the interaction between the FtsZ protein and the peptidoglycan (PG) biosynthesis machinery. With limited exceptions, both FtsZ and PG are ubiquitous in bacteria. Amongst those exceptions are the members of the Planctomycetes, Verrucomicrobia, and Chlamydiae (PVC) superphylum, which encompasses these three phyla as well as the Lentisphaerae and some uncultured candidate phyla, such as the Candidatus Omnitrophica (previously known as OP3; Wagner and Horn, 2006; Fuerst, 2013a; Devos and Ward, 2014) (Figure 1).
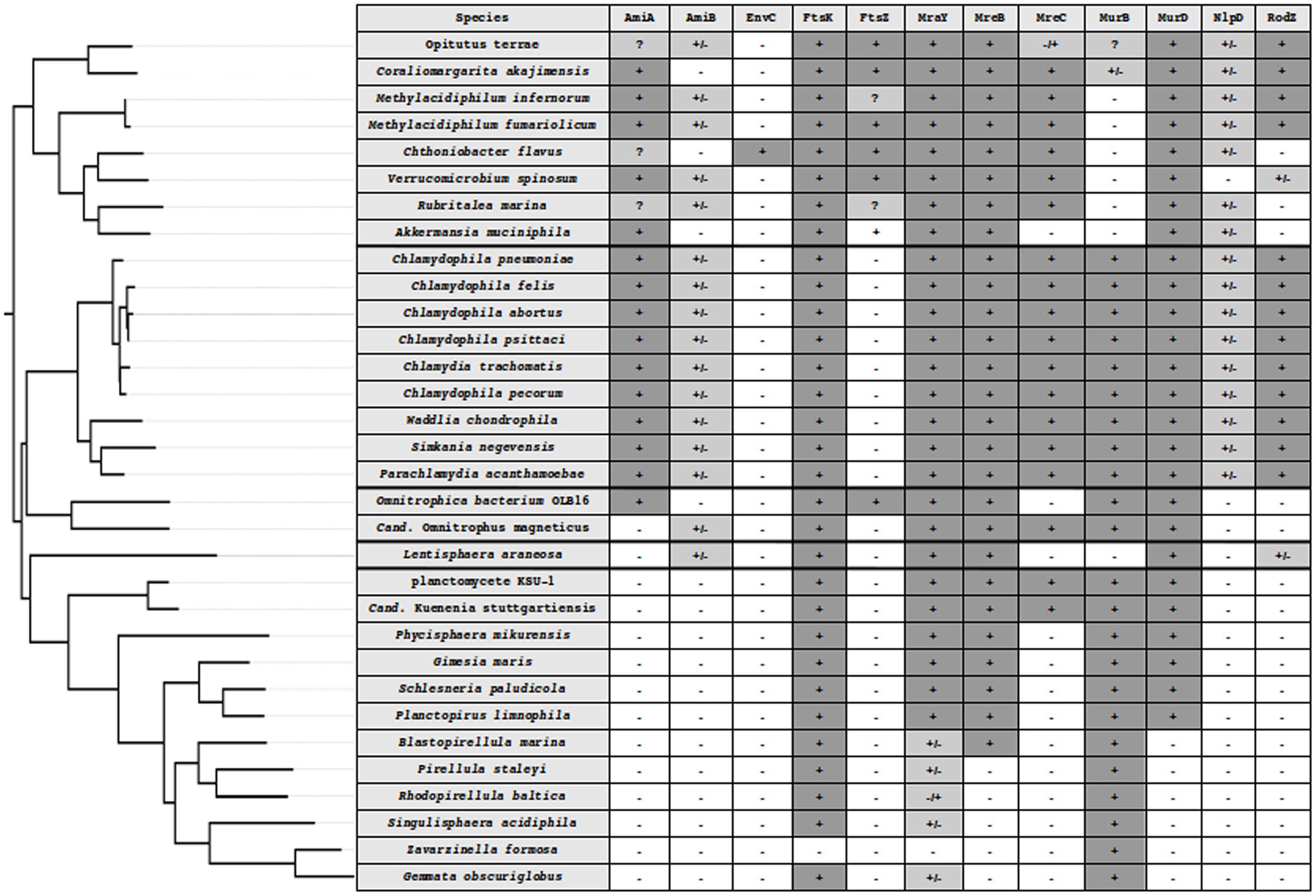
FIGURE 1. Phylogenetic tree of the PVC superphylum and detection of division and PG synthesis-related proteins in PVC. PVC species from five well-supported monophyletic groups, Verrucomicrobia, Lentisphaera, Chlamydiae, Planctomycetes, and Cand. Omnitrophica. The tree was built using 16S rRNA sequences in PhyML 3.1 with custom parameters and the HKY85 matrix (Guindon et al., 2010) and rooted on E. coli. In silico protein detection in the PVC proteomes is denoted by the following symbols: -, not found; +, found from both searches; +/-, found in the search starting from the W. chondrophila sequence but not from the one starting from E. coli; -/+, found in the search starting from the E. coli sequence but not from the one starting from W. chondrophila; ?, indicating that a candidate could be found using an alternative query (as in the case of Methylacidiphilum infernorum FtsZ), that only a fragment of the protein could be found (incomplete data, as in the case of Rubritalea marina FtsZ) or that both searches found different proteins that were the reciprocal best hit of both queries (as in the case of AmiA in Verrucomicrobia).
The Chlamydiae phylum was initially restricted to members of the family Chlamydiaceae due to their human pathogenicity, but now includes the families Parachlamydiaceae, Waddliaceae, Criblamydiaceae, and Simkaniaceae of the order Chlamydiales (Bertelli and Greub, 2013). These newly described families of chlamydia-related bacteria diverged from the Chlamydiaceae more than 700 million years ago (Horn et al., 2004). The phyla Lentisphaerae and Verrucomicrobia form a separate group within the PVC superphylum, with the latter containing the orders Opitutales, Puniceicoccales, and Verrucomicrobiales, amongst others. Within the Planctomycetes, three orders are recognized: Cand. Brocadiales, Phycisphaerales, and Planctomycetales. The Cand. Brocadiales is a deep-branching order responsible for anaerobic ammonium oxidation (anammox; van Niftrik, 2013). The Phycisphaerales only contain a few species that are still uncultured and for which limited description is available (Fukunaga et al., 2009). The order Planctomycetales is formed by the Gemmata, Blastopirellula, and Pirellula genera, amongst others. The Cand. Omnitrophica phylum was recently added to the PVC group with scarce information available, including genomic one.
Bacteria belonging to the PVC superphylum are fascinating for their peculiar biology. Their lifestyles range from the free-living soil and aquatic Planctomycetes, Verrucomicrobia, and Lentisphaerae, through the commensal and mutualistic Verrucomicrobia and Lentisphaerae, to the obligate pathogens of the Chlamydiae. Cand. Omnitrophica are mostly found in fresh water. Cell division is one of the particularities of this superphylum: some PVC species, such as the members of the order Planctomycetales, divide by budding, whereas most others divide by binary fission (Figure 2). Although Chlamydiae were previously described as dividing by binary fission, new data have revealed an asymmetric division in Chlamydia trachomatis (Abdelrahman et al., 2016). With the exception of the Verrucomicrobia and Cand. Omnitrophica, PVC bacteria also lack the FtsZ protein, the central player of bacterial division. How bacteria deprived of FtsZ divide without this landmark protein is unknown. Some members of the PVC superphylum have also been described as lacking PG. The lack of both PG and FtsZ, and the different mode of division observed in Planctomycetes and Chlamydiae, once represented one of the biggest mysteries of microbiology – as well as an excellent breeding ground for hypotheses on the evolution of new modes of cell division.
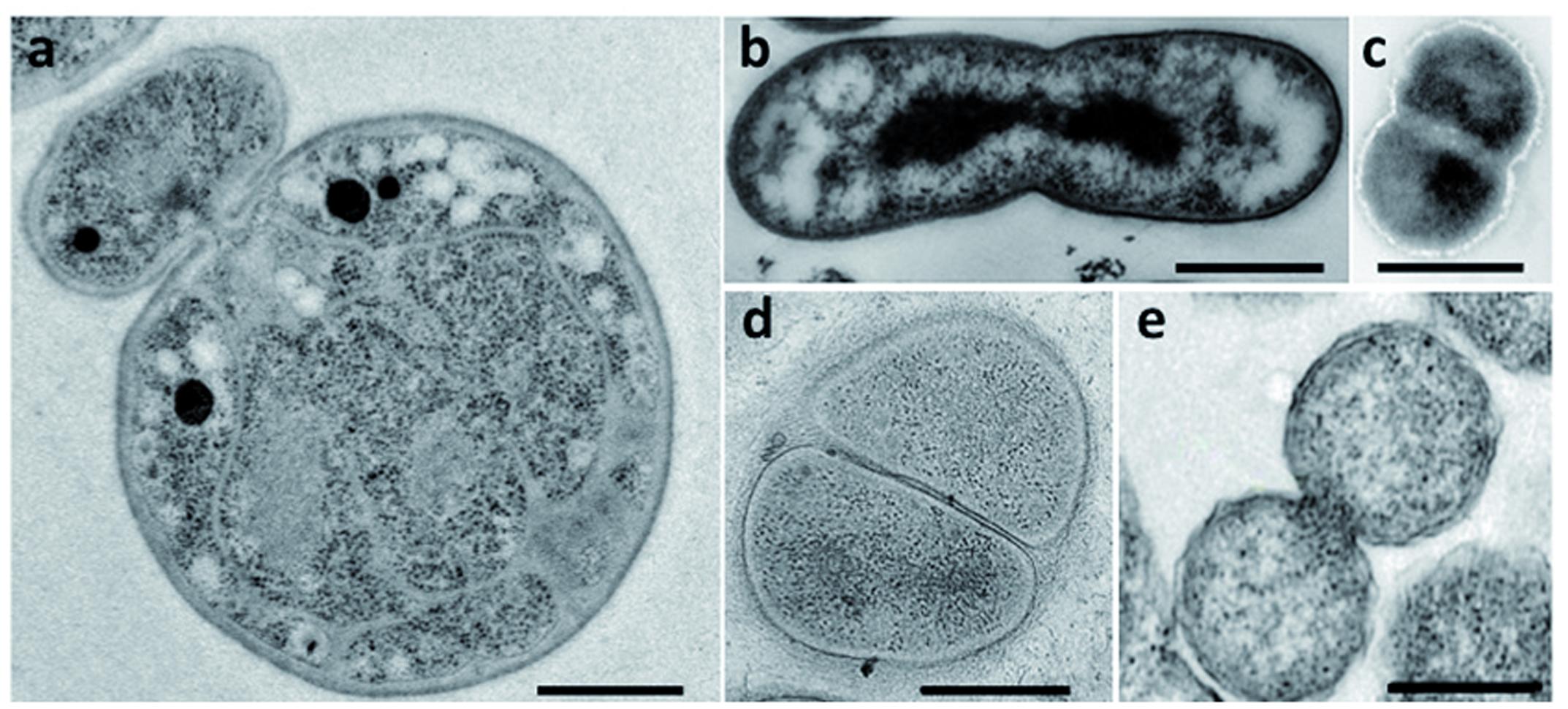
FIGURE 2. Division modes in the PVC superphylum. Transmission electron micrographs of thin sections of dividing cells from (a) G. obscuriglobus (Santarella-Mellwig, personal communication), (b) Chthoniobacter flavus [adapted with permission from Sangwan et al. (2004), License number 3662961133966], (c) L. araneosa [adapted with permission from Cho et al. (2004), License number 3662980203389], (d) P. mikurensis [reprinted with permission from Fukunaga et al. (2009)], and (e) C. trachomatis [adapted with permission from Lewis et al. (2014)]. Scale bars, 0.5 μm.
Evolutionary cell biology is a recent field that aims to meld an understanding of evolutionary processes with variation in intracellular structure, based on the comparison of mechanisms between species (Lynch et al., 2014). The development of a new division mode is one of the most important evolutionary transitions. How a binary fission mechanism based on FtsZ evolved into an FtsZ-independent mechanism of division by budding is a major question in this field.
Various publications have recently revealed the presence of PG in some planctomycetal and chlamydial species, suggesting that it is also present in other species in these phyla and in other PVC phyla (Pilhofer et al., 2013; Liechti et al., 2014, 2016; Jeske et al., 2015; van Teeseling et al., 2015). Furthermore, a link between PG synthesis at the septum and division in Chlamydiae is emerging, offering clues about the mechanism of bacterial division without FtsZ (Liechti et al., 2016).
Here, we review the most striking features of the PVC bacteria that are linked to cell division without FtsZ and the involvement of PG in this process. We first summarize the historical analyses that led to the conclusion that PG was absent from some of these bacteria. We then recapitulate the recent detection of PG in some chlamydias and planctomycetes, as well as the role of PG, PG-related and division proteins in chlamydial division. Based on the detection of PG-related proteins in PVC proteomes, we evaluate the possible evolution of division mechanisms in these bacteria. Finally, we discuss the implications of these findings for division in Chlamydiae and their extrapolation to the other PVC phyla.
In the majority of bacteria, cell division is performed by binary fission. There are a few exceptions: a small proportion of bacteria use mechanisms such as intracellular offspring production or multiple fission (Angert, 2005). Budding is an alternative mode that has been reported in many bacterial lineages, including some members of Cyanobacteria, Firmicutes, Planctomycetes, and the prosthecate proteobacteria. Recent research also reveals that C. trachomatis divides asymmetrically, which is reminiscent of budding (Abdelrahman et al., 2016; Liechti et al., 2016). In model organisms, such as Escherichia coli, division by binary fission is realized using a molecular machinery assembled around the FtsZ protein, a homolog of the eukaryotic tubulin (Figure 3) (den Blaauwen et al., 2008; Erickson et al., 2010). FtsZ is one of the first proteins to localize at the division site in the middle of the cell. It forms a ring while recruiting the components of the division machinery, the divisome, including FtsA, FtsI, FtsK, FtsL, and FtsW, as well as PBP1b, EnvC, NlpD, and the Tol-Pal system (Lutkenhaus et al., 2012). During division, the FtsZ ring and the associated divisome contract, pulling the membranes toward the inside of the cell. As a consequence of its central role in division, FtsZ is present in almost all bacteria. Only a few groups, including Chlamydiae, Planctomycetes, and some of the Mollicutes, are unusual in that they lack a recognizable homolog of FtsZ (Figure 1; Supplementary Table S1).
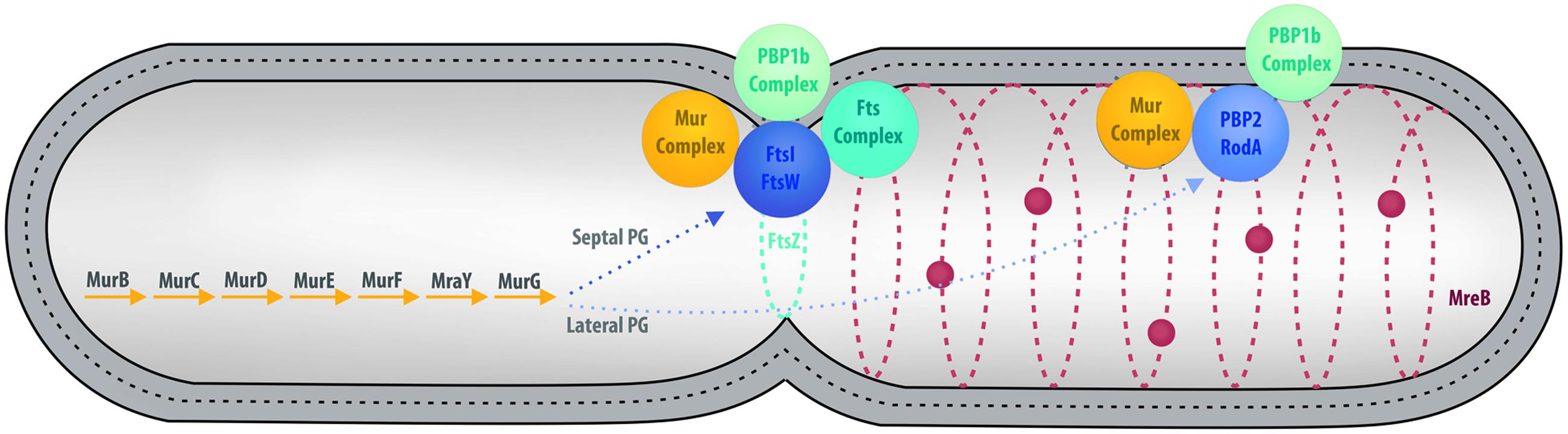
FIGURE 3. Overview of cell division in model organisms. Major cell division and PG synthesis complexes are represented. Cytoplasmic steps of PG synthesis enzymes (Left), major cell division and septal PG synthesis complexes (Center), and elongation of lateral PG complexes (Right) are represented by colored spheres. PG is represented by a dotted line between the inner and outer membranes.
Peptidoglycan also plays an important role in division by binary fission. PG is a net-like heteropolymer comprising linear glycan strands that are assembled from two sugar precursors, N-acetyl-glucosamine acid (GlcNAc) and N-acetyl-muramic acid (MurNAc), through β-(1,4)-glycosidic bonds and stabilized through peptide bridges containing D-amino acids. PG has a structural role: it surrounds the bacterial cell, forming an exoskeleton, the sacculus, that amongst other functions, protects cells from osmotic stress. The sacculus is so stable that it can be purified as a single entity (den Blaauwen et al., 2008). The presence of PG is one of the characteristics that differentiates bacteria from the other two domains of life, archaea and eukaryotes. There are, however, a few exceptions to the PG universality in bacteria, which are now restricted to the Mollicutes but until recently also included the Chlamydiae and Planctomycetes.
Peptidoglycan has historically been thought to be absent from the periplasm of most PVC bacteria. Early attempts failed to detect PG in Chlamydiae (Garrett et al., 1974; Fox et al., 1990), efforts to purify chlamydial sacculi were unsuccessful (Caldwell et al., 1981; Barbour et al., 1982), and periplasmic density layers between the inner and outer membranes were not detected by electron microscopy (Tamura et al., 1971; Huang et al., 2010). Surprisingly, bacteria from the Chlamydiae are sensitive to β-lactam antibiotics, which inhibit PG synthesis (Matsumoto and Manire, 1970). In addition, most available chlamydial genomes encode functional enzymes for PG synthesis, and the functionality of a nearly complete biosynthesis pathway in several species is well supported (Hesse et al., 2003; McCoy et al., 2003; McCoy and Maurelli, 2005, 2006; Patin et al., 2009, 2012). These contradictory observations have been referred to as the “chlamydial anomaly” (Moulder, 1993). By contrast, Planctomycetes are resistant to all PG-targeting antibiotics (Cayrou et al., 2010), and early biochemical analyses were unable to detect PG components in isolated cell envelopes of various strains (König et al., 1984; Stackebrandt et al., 1986). Thus, PG was though to be absent from Planctomycetes. Nevertheless, the growth of the anammox planctomycetes Cand. Kuenenia stuttgartiensis has been shown to be inhibited by lysozyme and penicillin, which both act on PG (Hu et al., 2013). PG had also been detected in some, but not all, verrucomicrobial species investigated (Yoon et al., 2007a,b, 2008a,b, 2010). In agreement, most PG synthesis genes were detected in Verrucomicrobium spinosum and the functionality of at least one of them has been confirmed (McGroty et al., 2013). Recently, PG has been detected in subdivision 5 of Verrucomicrobia (Spring et al., 2016).
In addition, and of chief interest here, is the observation that PVC bacteria encode a diversity of division proteins. Verrucomicrobia, Lentisphaera araneosa, and Omnitrophica bacterium OLB16 encode the FtsZ protein and synthesize PG, whereas the Planctomycetes and Chlamydiae lack FtsZ as well as other division proteins (Figure 1; Supplementary Table S1). Whereas the Verrucomicrobia and Lentisphaerae, as well as the lowest branching Planctomycetes orders, Cand. Brocadiales and Phycisphaerales, divide by binary fission, members of the order Planctomycetales divide by budding. Chlamydiae were previously described as dividing by binary fission, however, asymmetric polarized cell division that might resemble budding has recently been reported in C. trachomatis (Abdelrahman et al., 2016; Liechti et al., 2016). This intracellular pathogen has differential localization of major cell components, such as the major outer-membrane protein and lipopolysaccharide present on opposite poles of the cell (Abdelrahman et al., 2016).
Thus, the Verrucomicrobia were classified as Gram-negative bacteria dividing by binary fission with FtsZ and PG (Krieg, 2011); the Chlamydiae as PG-deprived Gram-negative bacteria dividing by binary fission without FtsZ; and the Planctomycetes as dividing by budding without FtsZ and PG, but also as defining a “third cell plan” (Fuerst, 2013b).
The first analysis to address the conservation of the division and cell wall (dcw) cluster in PVC genomes failed to detect some key PG synthesis genes and concluded that the last common ancestor of the PVC superphylum possessed a ‘classical’ dcw cluster (Pilhofer et al., 2008). Major modifications, mostly losses, were found in the Planctomycetes. The presence of genes was recently re-addressed using a profile-based method more appropriate for detecting remote relationships between proteins (Jeske et al., 2015; van Teeseling et al., 2015). Contrary to previous analyses, these studies revealed that all planctomycetal and chlamydial species investigated harbored the genes essential for PG synthesis.
Given the detection of PG synthesis enzymes in the chlamydial and planctomycetal genomes, the presence of PG in their cell walls was re-evaluated (Pilhofer et al., 2013; Liechti et al., 2014; Jeske et al., 2015; van Teeseling et al., 2015). Using a combination of methods, PG was observed in four planctomycetes strains: Gemmata obscuriglobus, Planctopirus limnophila, Rhodopirellula baltica, and strain L21-Rpul-D3 (Jeske et al., 2015). Those methods included a radioactive kinase assay to detect the transfer of a radioactive γ-phosphoryl group from [γ-32P]-ATP to GlcNAc and MurNAc monomers, gas chromatography and mass spectrometry to reveal the presence of the PG component 2,6-diaminopimelic acid, visualization of cell wall sacculi disruption after lysozyme treatment, and direct visualization of a thin PG layer by electron microscopy in vitrified cells. The presence of PG was simultaneously reported for the anammox planctomycete Cand. Kuenenia stuttgartiensis (van Teeseling et al., 2015). A similar set of methods was used, including direct observation of sacculi digestion by lysozyme, incorporation of PG specific probes, chemical analyses using ultra-performance liquid chromatography and mass spectrometry, and direct visualization of the cell wall layer by cryo-electron microscopy of vitreous sections. Similarly, the use of mass spectrometry and fluorescent labeling dyes combined with electron cryotomography and a PG synthesis inhibitor revealed the presence of a PG sacculus in Protochlamydia amoebophila, a symbiotic member of the Chlamydiae (Pilhofer et al., 2013). However, the same technique failed to reveal PG in Simkania negevensis, a pathogenic chlamydia-like bacterium. This lack of detection is in agreement with the resistance of this bacterium to penicillin and phosphomycin (Pilhofer et al., 2013), but is at odds with the presence of genes coding for most PG synthesis enzymes in its genome (Supplementary Table S1). In addition, two of the S. negevensis genes, amiA and nlpD, have been shown to encode functional proteins that are active in E. coli (Frandi et al., 2014). A new metabolic cell-wall labeling method based on D-amino acid dipeptide and click chemistry revealed the presence of PG in C. trachomatis (Liechti et al., 2014). Remarkably, whereas a typical, cell-surrounding sacculus is formed in P. amoebophila, only a ring-like band is observed at mid-cell in C. trachomatis (Liechti et al., 2014).
Individually, these reports provide compelling evidence for the presence of PG in the cell wall of the investigated strains. Taken together, the direct and indirect evidence accumulated from orthogonal sources constitutes irrefutable proof of PG synthesis in phylogenetically dispersed strains of the PVC superphylum. In addition, PG has been detected in strains belonging to the genus Prosthecobacter and in strain L21-Fru-ABT from the Verrucomicrobia (Hedlund et al., 1996; Spring et al., 2016). Combined with the genomic data, these detections strongly suggest that PG synthesis takes place in most, if not all, members of the PVC superphylum. To the best of our knowledge, the synthesis of PG has not yet been tested in Lentisphaerae, although the only genomic sequence information available so far suggests that L. araneosa is capable of PG synthesis (Figure 1; Supplementary Table S1). These data strongly supports that PVC bacteria are a variation of, not an exception to, the Gram-negative cell plan, putting a clear end to the controversy about the planctomycetal cell plan (Fuerst, 2013b; Devos, 2014a,b; Sagulenko et al., 2014).
Considerable modifications of the chlamydial PG structure and timing of its synthesis have recently been revealed in four pathogenic strains: C. trachomatis, C. muridarum, Chlamydophila psittaci, and C. caviae (Liechti et al., 2016). The use of chemical probes with super-resolution microscopy demonstrated that PG in these species is limited to a narrow ring at the division plane only during the replicative stage (Liechti et al., 2014, 2016). Because the host immune system recognizes and responds to PG, the authors suggest that members of the family Chlamydiaceae, all of which are pathogens, may limit the synthesis of PG to the place and time that are absolutely necessary, i.e., the division septum of replicating cells (Liechti et al., 2016). Thus, PG synthesis is crucial to chlamydial division, even in pathogenic strains, which have reduced it to a minimum. This spatiotemporally limited synthesis of PG explains why it was not detected before and also why Chlamydiae are susceptible to antibiotics, thus resolving the ‘chlamydial anomaly.’
The detection of PG in Planctomycetes and Chlamydiae poses the question of why it remained undetected for so long. In the case of Chlamydiae, it was due mostly to technical limitations, combined with the restricted presence of PG in time and space in pathogenic strains (Pilhofer et al., 2013; Liechti et al., 2014, 2016). The case of Planctomycetes appears to be different. Since early analyses (König et al., 1984; Liesack et al., 1986; Stackebrandt et al., 1986), the absence of PG in Planctomycetes cell walls was not reconsidered, despite several investigations into these bacteria and their structure (Fuerst, 2005, 2013b; Fuerst and Sagulenko, 2013; Sagulenko et al., 2014). Planctomycetal sacculi was purified in the presence of SDS and high temperature, but instead of PG, a proteinaceous cell wall was found (Stackebrandt et al., 1986). However, protein recovery was only 51%, with no indication of what comprised the remaining 49% of the envelope. It is unlikely that the sacculi consisted exclusively of proteins, as proteins of mesophilic organisms cannot resist hours of boiling in SDS and would be denatured under those conditions. Attempts to determine the presence of PG in Planctomycetes never involved lysozyme treatment, until recently (Giovannoni et al., 1987; Hu et al., 2013; Jeske et al., 2015; van Teeseling et al., 2015).
Given that PG is present in Planctomycetes, their resistance to β-lactam antibiotics might appear confusing (Cayrou et al., 2010). This resistance is possibly due to the presence of putative β-lactamases encoded in their genomes (Kahane et al., 1993). At least one putative β-lactamase gene is present in each Planctomycetes genome, often in multiple copies. For instance, there are 13 putative β-lactamases encoded in the P. limnophila genome (Labutti et al., 2010). By contrast, Chlamydiae are sensitive to β-lactam antibiotics despite the presence of β-lactamases in their genomes (Bertelli et al., 2010).
The detection of PG in PVC species and its interaction with division proteins are important, as they provide potential clues about PVC division mechanisms. The few PVC species for which division mechanisms have been investigated so far belong to the Chlamydiae (Jacquier et al., 2015). Chlamydial division was previously believed to resemble FtsZ-dependent binary fission (Brown and Rockey, 2000; Abdelrahman and Belland, 2005). However, an asymmetric polarized division mode has recently been revealed in Chlamydiaceae (Abdelrahman et al., 2016) and progress on understanding the link between chlamydial PG and division has been reported (Jacquier et al., 2015). This new knowledge allows us to draw a preliminary model of chlamydial division that reveals important modifications to the mechanisms of division in model organisms. In particular, the central role of MreB (a bacterial homolog of actin) and its interaction with PG synthesis enzymes at the septum is emerging.
Division is tightly regulated in Chlamydiales, as only one specific cell type, the reticulate body, is metabolically active and able to divide. Expression analyses of several division and PG biosynthesis genes in C. trachomatis and C. pneumoniae show that these genes are over-expressed in the dividing forms compared with the non-dividing forms (Albrecht et al., 2010, 2011). Typical peptide components of PG accumulate at the chlamydial division site, as in most other bacteria (Jacquier et al., 2014). These PG precursors are required for proper localization of PG- and MreB-binding proteins at the division septum (Jacquier et al., 2014). The activity of MurA, MurF, and MurG, which are involved in the early and mid-phase stages of PG synthesis, are required to organize the chlamydial septum, further supporting the understanding that PG synthesis is required for division (Liechti et al., 2014).
The cytoskeletal proteins MreB and RodZ, which are responsible for cell-shape determination in model organisms, seem to play an important role in this process in Chlamydiae, as they may bring together the PG biosynthesis and remodeling enzymes to the divisome (Jacquier et al., 2015). Indeed, it appears that FtsZ-less Chlamydiae uses MreB to define the division plane by interaction with FtsK, which in turn may recruit PBP2, FtsI (also called PBP3) and likely other unidentified proteins (Ouellette et al., 2012). Co-localization results suggest that MreB polymerization is required to guide new PG incorporation along the PG ring in Chlamydiales (Gaballah et al., 2011; Liechti et al., 2016), supporting the proposal that MreB acts as the division plane organizer in pathogenic chlamydias. However, MreB does not seem to be an early cell division protein like FtsZ, because it is recruited late at the septum (Gaballah et al., 2011). Like FtsZ, MreB appears to be a central coordinator of the large multi-protein PG synthesizing complex that cooperates to direct cell elongation. This complex includes PBP2, RodA and many additional partners, such as MreC, MreD, and RodZ, that are involved in the stabilization and/or regulation of this machinery. C. pneumoniae MreB has been shown to interact with MurF, MraY, and MurG, three key components of lipid II biosynthesis (Gaballah et al., 2011). In W. chondrophila, RodZ is recruited early to the septal site in a process that is dependent on the presence and dispersal of cell wall precursors (Jacquier et al., 2014). The endopeptidase NlpD localizes at the division plane in Chlamydiae and its septal sequestration depends on prior cell wall synthesis (Frandi et al., 2014). Thus, the division mechanisms of Chlamydiae share similarities but also have important differences with model organisms.
The diversity of characteristics of related bacteria makes the PVC superphylum particularly attractive for the field of evolutionary cell biology. To paraphrase Theodosius Dobzhansky, it could be said that “nothing in PVC cell division makes sense except in the light of evolutionary cell biology.” Some clues about PVC division can be derived from the limited knowledge available about division in the Chlamydiae and from gene conservation in other species. We thus revisited the presence of the main genes involved in PG synthesis and division in the PVC proteomes for a representative set of genomes across the superphylum (Figure 1; Supplementary Table S1). It is unclear if the inability to detect some proteins is due to their absence or represents a false negative result. The latter nevertheless indicates a profound modification of the protein sequences and thus most likely of the molecular mechanisms associated with it.
The diversity of division modes and proteins observed in PVC species is due to divergent evolution from a common ancestor (Figure 4). The most parsimonious explanation for the gene distribution of the dcw cluster is that the last PVC common ancestor was a Gram-negative bacterium that divided by binary fission using PG and FtsZ, and that had a mostly classical dcw cluster, as suggested by others (Pilhofer et al., 2008; Jogler et al., 2012). Within the PVC superphylum, species have diverged from this common ancestor, accumulating differences including gene losses but also modifications of the function of existing genes (Figure 1; Supplementary Table S1). How gene losses are related to modifications of this division mechanism is one of the main questions remaining in this field. Even in the PVCs containing FtsZ, modifications of the division modes are to be expected, as demonstrated by the losses of molecular components. Due to the diversity of phenotypes and genotypes encountered in the PVC superphylum, generalization of their features is a difficult exercise.
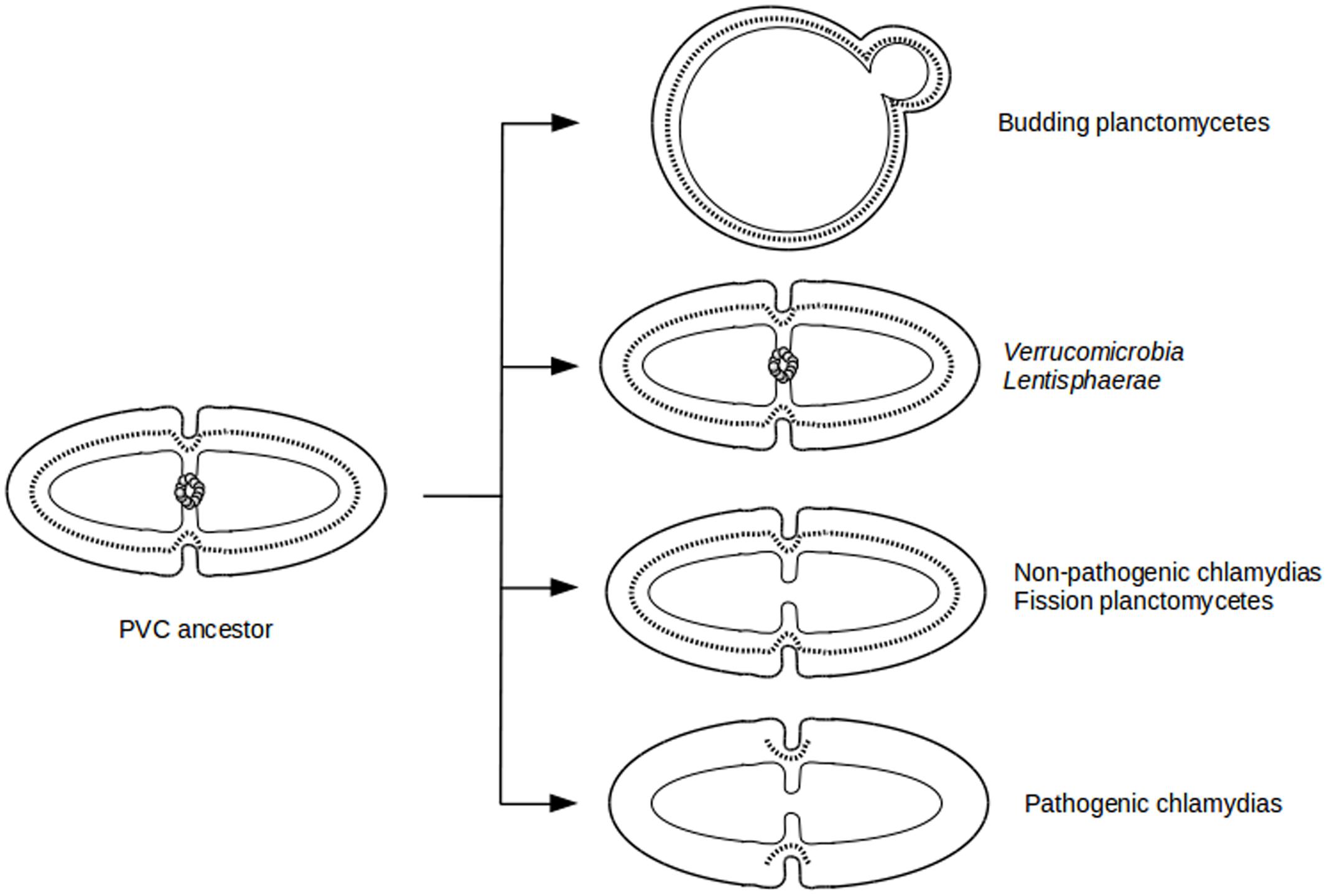
FIGURE 4. Evolutionary cell biology of division modes in PVCs. Schematic representation of cell division modes in the last PVC common ancestor (Left) and in current PVC species (Right). Outer and inner membranes are in thick and thin lines, respectively. PG layer is in dotted line. FtsZ proteins are represented as a ring of gray spheres.
First and foremost, FtsZ, the landmark protein of binary fission that is conserved in almost all bacteria, is not found in Chlamydiae or in Planctomycetes. It is detected only in Omnitrophica bacterium OLB16, and it is not clear if its absence in the other two members of the Cand. Omnitrophica is real or merely a result of incomplete data. On the other side of the spectrum of conservation, a putative FtsK protein, the DNA translocase, is likely to be present in all genomes (its absence in Zavarzinella formosa is likely due to incomplete data). The same applies to MraY, an integral membrane enzyme that catalyzes the transfer of the PG precursor to the lipid carrier undecaprenyl phosphate, forming the lipid I.
In between those two extremes, interesting intermediary cases are found. MreB is found in almost all PVC proteomes, with the exception of some of the budding planctomycetes. This conservation supports the central role of MreB in the absence of FtsZ. In model organisms, MreB requires the membrane proteins MreC and MreD for the organization of the PG (White et al., 2010). Interestingly, MreD is not found in any PVC proteomes, while MreC shows a patchy distribution: it is found in verrucomicrobias and chlamydias, and in two out of three binary fission planctomycetes, but not in L. araneosa or in the budding planctomycetes (with the possible exception of Planctomyces brasiliensis, which is probably a false positive). The protein RodZ might form the link between MreB and the PG-modifying penicillin-binding proteins (PBPs), and it locates early at the septum in Chlamydiae (Jacquier et al., 2014). Interestingly, RodZ is only found in Chlamydiae and Verrucomicrobia but not in Planctomycetes or in L. araneosa. Thus, important modifications of division mode due to the lack of RodZ are expected. This is in agreement with the fact that the chlamydial RodZ is truncated, lacking its C-terminal periplasmic domain, and cannot complement an E. coli RodZ mutant (Ouellette et al., 2014).
The peptidase NlpD is another protein that localizes at the septum in chlamydial pathogens (Frandi et al., 2014). NlpD, like RodZ, is found only in Chlamydiae and Verrucomicrobia, and not in Planctomycetes or Lentisphaera. The amidase activator EnvC is not found in any PVC proteomes (with a single exception, which is probably a false positive). In model organisms, EnvC is a regulator of the cell wall amidases AmiA and AmiB. Like NlpD, AmiA is found in Chlamydiae and Verrucomicrobia, but not in Planctomycetes, Omnitrophica, or Lentisphaera. AmiB is detected in Chlamydiae, L. araneosa, and only one Omnitrophica, but only in five members of the Verrucomicrobia (out of nine) and not at all in Planctomycetes. Most PG synthesis enzymes are found in PVC members, with the exception of some budding planctomycetes.
Hence, this pattern of undetected proteins illustrates important modifications of the cell division modes in the various branches of the PVC superphylum. The diverse division modes in PVC bacteria offer a fascinating field of study for the years to come.
Great progress has been made in uncovering the division modes in members of the PVC superphylum. Many relevant discoveries reveal new questions that require further research to answer. The data presented here calls for a few remarks and questions.
First, PG is almost universal in bacteria, with a few exceptions limited to obligate intracellular bacteria, and is one of the true features defining bacteria (including bacteria-derived organelles such as plastids). The recent detection of PG in PVC members, combined with genomic information, suggests that it is present in most PVC species. The only bacterial group that is possibly devoid of a PG cell wall is the Mollicutes. In addition, to the best of our knowledge, the mollicutes Phytoplasma sp., Ureaplasma sp., and Mycoplasma mobile are the only species deprived of both FtsZ and PG (Bernander and Ettema, 2010).
Second, PVC bacteria are variations of, but not exceptions to, the Gram-negative cell plan (Devos, 2014a). The presence of PG in PVC species definitively resolves this controversy.
Third, most PVC bacteria have a well developed endomembrane system. This system can take the form of invaginations of the cytoplasmic membrane toward the inside of the cell (as observed in most planctomycetes), of evaginations of the outer membrane (in some verrucomicrobias), or of a tubulo-vesicular network in the periplasm of the cell (in the planctomycete G. obscuriglobus; Santarella-Mellwig et al., 2013; Acehan et al., 2014; Devos, 2014a). The anammox planctomycetes even appear to display the first prokaryotic organelle with a compartment apparently separated from the inner membrane (Neumann et al., 2014). How the PG organizes within or around this endomembrane system, including the periplasmic tubulo-vesicular network, is another intriguing feature that requires deeper investigation. How PG is synthesized and organized in the bud is yet another interesting area of research.
Fourth, during evolutionary divergence from their common ancestor with a classical FtsZ-based binary fission mode of division, PVC members have evolved different modifications, including the loss of FtsZ and the development of alternative division mechanisms and modes, raising various questions for evolutionary cell biology. What forces and constraints acted on which proteins and processes? What has been the evolutionary pressure behind those innovations? Given the degradation of the dcw cluster, the total conservation of some genes, such as mreB or ftsK, is intriguing.
Fifth, recent progress has been made in deciphering the chlamydial division mechanism, which requires the active synthesis of PG, suggesting a role at the septum related to that of PG in classical Gram-negative species. This opens up more important questions. How much of the ancestral division mechanism is conserved in current species? And what are the differences? What is the role of PG in division without FtsZ? Has MreB replaced FtsZ, as has been suggested (Jacquier et al., 2015)? A key question is how to transfer the chlamydial knowledge to other PVCs.
Sixth, recent advances in the field of PVC genetic manipulation are providing us with better tools to answer some of these questions. Genetic tools have been described recently in the three initial PVC phyla Planctomycetes, Verrucomicrobia, and Chlamydiae (Domman et al., 2011; Jogler et al., 2011; Kari et al., 2011; Wang et al., 2011; Gérard et al., 2013; Erbilgin et al., 2014; Kokes et al., 2015; Rivas-Marín et al., 2016). The analyses reported here and elsewhere constitute a very good place not only to continue deciphering the modes of division of Chlamydiae, but also to start exploring those of the other PVC members. The isolation and characterization of new PVC members is important, as it will provide more information about the diversity of division modes in PVC bacteria. Is there an intermediary organism with a mode of division between binary fission and budding? Genome information for additional PVC species is needed to evaluate the conservation of division proteins (and mechanisms) in this superphylum.
The PVC bacterial superphylum presents an unparalleled opportunity for the study of evolutionary cell biology of division modes. We are looking forward to progress in this fascinating field.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
ER-M and DD are supported by the Spanish Ministry of Economy and Competitiveness (Grant BFU2013-40866-P) and the Junta de Andalucía (CEIC Grant C2A program to DD). IC is supported by the Spanish Ministry of Economy and Competitiveness (Grant BIO2014-57545-R).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Rachel Santarella-Mellwig (EMBL, Heidelberg, Germany) for helpful comments on the manuscript, Eduardo Santero (CABD, Seville, Spain) for support, and Nicola Bordin (CABD) for building the phylogenetic tree.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01964/full#supplementary-material
TABLE S1 | Division and cell wall cluster and PG synthesis protein detection in PVC. Due to the divergence of the PVC species from model organisms and for the sake of completeness, two complementary searches were combined (Sheet 1). We first performed two iterations of Psi-Blast using the W. chondrophila (Sheet 2) and E. coli (Sheet 3) proteins as a query on the non-redundant GenBank (nr) database, using default parameters and accepting hits with an e-value below 10-3. One additional iteration was run on the PVC proteomes with the obtained profile as query. A reciprocal search was then used to verify that the starting proteins were found when using the hit as a query. The PVC hit was only accepted if the reciprocal search found the starting sequence. Each cell contains two identifiers, if the reciprocal best hits are different, the first one corresponding to the hit from the W. chondrophila search (left), followed by the hit from the E. coli search (right). In most cases, the two approaches hit the same proteins, reinforcing their results. Only one identifier is indicated if the reciprocal best hits are the sames. Sheets 2 and 3 show the e-values of detection corresponding to the searches from the W. chondrophila and E. coli proteins, respectively. NA, not applicable; PF, binary fission planctomycetes; PB, budding planctomycetes.
Abdelrahman, Y., Ouellette, S. P., Belland, R. J., and Cox, J. V. (2016). Polarized cell division of Chlamydia trachomatis. PLOS Pathog. 12:e1005822. doi: 10.1371/journal.ppat.1005822
Abdelrahman, Y. M., and Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959. doi: 10.1016/j.femsre.2005.03.002
Acehan, D., Santarella-Mellwig, R., and Devos, D. P. (2014). A bacterial tubulovesicular network. J. Cell Sci. 127, 277–280. doi: 10.1242/jcs.137596
Albrecht, M., Sharma, C. M., Dittrich, M. T., Müller, T., Reinhardt, R., Vogel, J., et al. (2011). The transcriptional landscape of Chlamydia pneumoniae. Genome Biol. 12:R98. doi: 10.1186/gb-2011-12-10-r98
Albrecht, M., Sharma, C. M., Reinhardt, R., Vogel, J., and Rudel, T. (2010). Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 38, 868–877. doi: 10.1093/nar/gkp1032
Angert, E. R. (2005). Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3, 214–224. doi: 10.1038/nrmicro1096
Barbour, A. G., Amano, K., Hackstadt, T., Perry, L., and Caldwell, H. D. (1982). Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151, 420–428.
Bernander, R., and Ettema, T. J. (2010). FtsZ-less cell division in archaea and bacteria. Curr. Opin. Microbiol. 13, 747–752. doi: 10.1016/j.mib.2010.10.005
Bertelli, C., Collyn, F., Croxatto, A., Rückert, C., Polkinghorne, A., Kebbi-Beghdadi, C., et al. (2010). The Waddlia genome: a window into chlamydial biology. PLoS ONE 5:e10890. doi: 10.1371/journal.pone.0010890
Bertelli, C., and Greub, G. (2013). “Phyla related to planctomycetes: members of phylum Chlamydiae and their implications for planctomycetes cell biology,” in Planctomycetes: Cell Structure, Origins and Biology, ed. J. A. Fuerst (New York, NY: Humana Press), 229–241. doi: 10.1007/978-1-62703-502-6_10
Brown, W. J., and Rockey, D. D. (2000). Identification of an antigen localized to an apparent septum within dividing chlamydiae. Infect. Immun. 68, 708–715. doi: 10.1128/IAI.68.2.708-715.2000
Caldwell, H. D., Kromhout, J., and Schachter, J. (1981). Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31, 1161–1176.
Cayrou, C., Raoult, D., and Drancourt, M. (2010). Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J. Antimicrob. Chemother. 65, 2119–2122. doi: 10.1093/jac/dkq290
Cho, J.-C., Vergin, K. L., Morris, R. M., and Giovannoni, S. J. (2004). Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum. Lentisphaerae. Environ. Microbiol. 6, 611–621. doi: 10.1111/j.1462-2920.2004.00614.x
den Blaauwen, T., de Pedro, M. A., Nguyen-Distèche, M., and Ayala, J. A. (2008). Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32, 321–344. doi: 10.1111/j.1574-6976.2007.00090.x
Devos, D. P. (2014a). PVC bacteria: variation of, but not exception to, the gram-negative cell plan. Trends Microbiol. 22, 14–20. doi: 10.1016/j.tim.2013.10.008
Devos, D. P. (2014b). Re-interpretation of the evidence for the PVC cell plan supports a gram-negative origin. Antonie Van Leeuwenhoek 105, 271–274. doi: 10.1007/s10482-013-0087-y
Devos, D. P., and Ward, N. L. (2014). Mind the PVCs. Environ. Microbiol. 16, 1217–1221. doi: 10.1111/1462-2920.12349
Domman, D. B., Steven, B. T., and Ward, N. L. (2011). Random transposon mutagenesis of Verrucomicrobium spinosum DSM 4136(T). Arch. Microbiol. 193, 307–312. doi: 10.1007/s00203-010-0666-5
Erbilgin, O., McDonald, K. L., and Kerfeld, C. A. (2014). Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl. Environ. Microbiol. 80, 2193–2205. doi: 10.1128/AEM.03887-13
Erickson, H. P., Anderson, D. E., and Osawa, M. (2010). FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. MMBR 74, 504–528. doi: 10.1128/MMBR.00021-10
Fox, A., Rogers, J. C., Gilbart, J., Morgan, S., Davis, C. H., Knight, S., et al. (1990). Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect. Immun. 58, 835–837.
Frandi, A., Jacquier, N., Théraulaz, L., Greub, G., and Viollier, P. H. (2014). FtsZ-independent septal recruitment and function of cell wall remodelling enzymes in chlamydial pathogens. Nat. Commun. 5:4200. doi: 10.1038/ncomms5200
Fuerst, J. A. (2005). Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 59, 299–328. doi: 10.1146/annurev.micro.59.030804.121258
Fuerst, J. A. (ed.) (2013a). Planctomycetes: Cell Structure, Origins and Biology. Totowa, NJ: Humana Press.
Fuerst, J. A. (2013b). The PVC superphylum: exceptions to the bacterial definition? Antonie Van Leeuwenhoek 104, 451–466. doi: 10.1007/s10482-013-9986-1
Fuerst, J. A., and Sagulenko, E. (2013). Nested bacterial boxes: nuclear and other intracellular compartments in Planctomycetes. J. Mol. Microbiol. Biotechnol. 23, 95–103. doi: 10.1159/000346544
Fukunaga, Y., Kurahashi, M., Sakiyama, Y., Ohuchi, M., Yokota, A., and Harayama, S. (2009). Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275. doi: 10.2323/jgam.55.267
Gaballah, A., Kloeckner, A., Otten, C., Sahl, H.-G., and Henrichfreise, B. (2011). Functional analysis of the cytoskeleton protein MreB from Chlamydophila pneumoniae. PLoS ONE 6:e25129. doi: 10.1371/journal.pone.0025129
Garrett, A. J., Harrison, M. J., and Manire, G. P. (1974). A search for the bacterial mucopeptide component, muramic acid, in Chlamydia. J. Gen. Microbiol. 80, 315–318. doi: 10.1099/00221287-80-1-315
Gérard, H. C., Mishra, M. K., Mao, G., Wang, S., Hali, M., Whittum-Hudson, J. A., et al. (2013). Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomed. Nanotechnol. Biol. Med. 9, 996–1008. doi: 10.1016/j.nano.2013.04.004
Giovannoni, S. J., Godchaux, W., Schabtach, E., and Castenholz, R. W. (1987). Cell wall and lipid composition of Isosphaera pallida, a budding eubacterium from hot springs. J. Bacteriol. 169, 2702–2707. doi: 10.1128/jb.169.6.2702-2707.1987
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hedlund, B. P., Gosink, J. J., and Staley, J. T. (1996). Phylogeny of Prosthecobacter, the fusiform caulobacters: members of a recently discovered division of the bacteria. Int. J. Syst. Bacteriol. 46, 960–966. doi: 10.1099/00207713-46-4-960
Hesse, L., Bostock, J., Dementin, S., Blanot, D., Mengin-Lecreulx, D., and Chopra, I. (2003). Functional and biochemical analysis of Chlamydia trachomatis MurC, an enzyme displaying UDP-N-acetylmuramate:amino acid ligase activity. J. Bacteriol. 185, 6507–6512. doi: 10.1128/JB.185.22.6507-6512.2003
Horn, M., Collingro, A., Schmitz-Esser, S., Beier, C. L., Purkhold, U., Fartmann, B., et al. (2004). Illuminating the evolutionary history of chlamydiae. Science 304, 728–730. doi: 10.1126/science.1096330
Hu, Z., van Alen, T., Jetten, M. S. M., and Kartal, B. (2013). Lysozyme and penicillin inhibit the growth of anaerobic ammonium-oxidizing planctomycetes. Appl. Environ. Microbiol. 79, 7763–7769. doi: 10.1128/AEM.02467-13
Huang, Z., Chen, M., Li, K., Dong, X., Han, J., and Zhang, Q. (2010). Cryo-electron tomography of Chlamydia trachomatis gives a clue to the mechanism of outer membrane changes. J. Electron Microsc. (Tokyo) 59, 237–241. doi: 10.1093/jmicro/dfp057
Jacquier, N., Frandi, A., Pillonel, T., Viollier, P. H., Viollier, P., and Greub, G. (2014). Cell wall precursors are required to organize the chlamydial division septum. Nat. Commun. 5:3578. doi: 10.1038/ncomms4578
Jacquier, N., Viollier, P. H., and Greub, G. (2015). The role of peptidoglycan in chlamydial cell division: towards resolving the chlamydial anomaly. FEMS Microbiol. Rev. 39, 262–275. doi: 10.1093/femsre/fuv001
Jeske, O., Schüler, M., Schumann, P., Schneider, A., Boedeker, C., Jogler, M., et al. (2015). Planctomycetes do possess a peptidoglycan cell wall. Nat. Commun. 6:7116. doi: 10.1038/ncomms8116
Jogler, C., Glöckner, F. O., and Kolter, R. (2011). Characterization of planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum planctomycetes. Appl. Environ. Microbiol. 77, 5826–5829. doi: 10.1128/AEM.05132-11
Jogler, C., Waldmann, J., Huang, X., Jogler, M., Glöckner, F. O., Mascher, T., et al. (2012). Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in planctomycetes by comparative genomics. J. Bacteriol. 194, 6419–6430. doi: 10.1128/JB.01325-12
Kahane, S., Gonen, R., Sayada, C., Elion, J., and Friedman, M. G. (1993). Description and partial characterization of a new chlamydia-like microorganism. FEMS Microbiol. Lett. 109, 329–333. doi: 10.1111/j.1574-6968.1993.tb06189.x
Kari, L., Goheen, M. M., Randall, L. B., Taylor, L. D., Carlson, J. H., Whitmire, W. M., et al. (2011). Generation of targeted Chlamydia trachomatis null mutants. Proc. Natl. Acad. Sci. U.S.A. 108, 7189–7193. doi: 10.1073/pnas.1102229108
Kokes, M., Dunn, J. D., Granek, J. A., Nguyen, B. D., Barker, J. R., Valdivia, R. H., et al. (2015). Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of chlamydia. Cell Host Microbe 17, 716–725. doi: 10.1016/j.chom.2015.03.014
König, E., Schlesner, H., and Hirsch, P. (1984). Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch. Microbiol. 138, 200–205. doi: 10.1007/BF00402120
Krieg, N. R. (2011). Bergey’s Manual of Systematic Bacteriology: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, Vol. 4. Berlin: Springer.
Labutti, K., Sikorski, J., Schneider, S., Nolan, M., Lucas, S., Glavina Del Rio, T., et al. (2010). Complete genome sequence of Planctomyces limnophilus type strain (Mü 290). Stand. Genomic Sci. 3, 47–56. doi: 10.4056/sigs.1052813
Lewis, M. E., Belland, R. J., AbdelRahman, Y. M., Beatty, W. L., Aiyar, A. A., Zea, A. H., et al. (2014). Morphologic and molecular evaluation of Chlamydia trachomatis growth in human endocervix reveals distinct growth patterns. Front. Cell. Infect. Microbiol. 4:71. doi: 10.3389/fcimb.2014.00071
Liechti, G., Kuru, E., Packiam, M., Hsu, Y.-P., Tekkam, S., Hall, E., et al. (2016). Pathogenic chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLOS Pathog. 12:e1005590. doi: 10.1371/journal.ppat.1005590
Liechti, G. W., Kuru, E., Hall, E., Kalinda, A., Brun, Y. V., VanNieuwenhze, M., et al. (2014). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510. doi: 10.1038/nature12892
Liesack, W., König, H., Schlesner, H., and Hirsch, P. (1986). Chemical composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch. Microbiol. 145, 361–366. doi: 10.1007/BF00470872
Lutkenhaus, J., Pichoff, S., and Du, S. (2012). Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton 69, 778–790. doi: 10.1002/cm.21054
Lynch, M., Field, M. C., Goodson, H. V., Malik, H. S., Pereira-Leal, J. B., Roos, D. S., et al. (2014). Evolutionary cell biology: two origins, one objective. Proc. Natl. Acad. Sci. U.S.A. 111, 16990–16994. doi: 10.1073/pnas.1415861111
Matsumoto, A., and Manire, G. P. (1970). Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 101, 278–285.
McCoy, A. J., and Maurelli, A. T. (2005). Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting D-alanyl-D-alanine ligase activity involved in peptidoglycan synthesis and D-cycloserine sensitivity. Mol. Microbiol. 57, 41–52. doi: 10.1111/j.1365-2958.2005.04661.x
McCoy, A. J., and Maurelli, A. T. (2006). Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 14, 70–77. doi: 10.1016/j.tim.2005.12.004
McCoy, A. J., Sandlin, R. C., and Maurelli, A. T. (2003). In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J. Bacteriol. 185, 1218–1228. doi: 10.1128/JB.185.4.1218-1228.2003
McGroty, S. E., Pattaniyil, D. T., Patin, D., Blanot, D., Ravichandran, A. C., Suzuki, H., et al. (2013). Biochemical characterization of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: meso-2,6-diaminopimelate ligase (MurE) from Verrucomicrobium spinosum DSM 4136(T.). PloS One 8:e66458. doi: 10.1371/journal.pone.0066458
Moulder, J. W. (1993). Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect. Agents Dis. 2, 87–99.
Neumann, S., Wessels, H. J. C. T., Rijpstra, W. I. C., Sinninghe Damsté, J. S., Kartal, B., Jetten, M. S. M., et al. (2014). Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol. Microbiol. 94, 794–802. doi: 10.1111/mmi.12816
Ouellette, S. P., Karimova, G., Subtil, A., and Ladant, D. (2012). Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol. Microbiol. 85, 164–178. doi: 10.1111/j.1365-2958.2012.08100.x
Ouellette, S. P., Rueden, K. J., Gauliard, E., Persons, L., de Boer, P. A., and Ladant, D. (2014). Analysis of MreB interactors in Chlamydia reveals a RodZ homolog but fails to detect an interaction with MraY. Front. Microbiol. 5:279. doi: 10.3389/fmicb.2014.00279
Patin, D., Bostock, J., Blanot, D., Mengin-Lecreulx, D., and Chopra, I. (2009). Functional and biochemical analysis of the Chlamydia trachomatis ligase MurE. J. Bacteriol. 191, 7430–7435. doi: 10.1128/JB.01029-09
Patin, D., Bostock, J., Chopra, I., Mengin-Lecreulx, D., and Blanot, D. (2012). Biochemical characterisation of the chlamydial MurF ligase, and possible sequence of the chlamydial peptidoglycan pentapeptide stem. Arch. Microbiol. 194, 505–512. doi: 10.1007/s00203-011-0784-8
Pilhofer, M., Aistleitner, K., Biboy, J., Gray, J., Kuru, E., Hall, E., et al. (2013). Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 4:2856. doi: 10.1038/ncomms3856
Pilhofer, M., Rappl, K., Eckl, C., Bauer, A. P., Ludwig, W., Schleifer, K.-H., et al. (2008). Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J. Bacteriol. 190, 3192–3202. doi: 10.1128/JB.01797-07
Rivas-Marín, E., Canosa, I., Santero, E., and Devos, D. P. (2016). Development of genetic tools for the manipulation of the planctomycetes. Front. Microbiol. 7:914. doi: 10.3389/fmicb.2016.00914
Sagulenko, E., Morgan, G. P., Webb, R. I., Yee, B., Lee, K.-C., and Fuerst, J. A. (2014). Structural studies of planctomycete gemmata obscuriglobus support cell compartmentalisation in a bacterium. PLoS ONE 9:e91344. doi: 10.1371/journal.pone.0091344
Sangwan, P., Chen, X., Hugenholtz, P., and Janssen, P. H. (2004). Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl. Environ. Microbiol. 70, 5875–5881. doi: 10.1128/AEM.70.10.5875-5881.2004
Santarella-Mellwig, R., Pruggnaller, S., Roos, N., Mattaj, I. W., and Devos, D. P. (2013). Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol. 11:e1001565. doi: 10.1371/journal.pbio.1001565
Spring, S., Bunk, B., Spröer, C., Schumann, P., Rohde, M., Tindall, B. J., et al. (2016). Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum. ISME J. doi: 10.1038/ismej.2016.84 [Epub ahead of print].
Stackebrandt, E., Wehmeyer, U., and Liesack, W. (1986). 16S ribosomal RNA- and cell wall analysis of gemmata obscuriglobus, a new member of the order Planctomycetales. FEMS Microbiol. Lett. 37, 289–292. doi: 10.1111/j.1574-6968.1986.tb01810.x
Tamura, A., Matsumoto, A., Manire, G. P., and Higashi, N. (1971). Electron microscopic observations on the structure of the envelopes of mature elementary bodies and developmental reticulate forms of Chlamydia psittaci. J. Bacteriol. 105, 355–360.
van Niftrik, L. (2013). Cell biology of unique anammox bacteria that contain an energy conserving prokaryotic organelle. Antonie Van Leeuwenhoek 104, 489–497. doi: 10.1007/s10482-013-9990-5
van Teeseling, M. C. F., Mesman, R. J., Kuru, E., Espaillat, A., Cava, F., Brun, Y. V., et al. (2015). Anammox Planctomycetes have a peptidoglycan cell wall. Nat. Commun. 6:6878. doi: 10.1038/ncomms7878
Wagner, M., and Horn, M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249. doi: 10.1016/j.copbio.2006.05.005
Wang, Y., Kahane, S., Cutcliffe, L. T., Skilton, R. J., Lambden, P. R., and Clarke, I. N. (2011). Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7:e1002258. doi: 10.1371/journal.ppat.1002258
White, C. L., Kitich, A., and Gober, J. W. (2010). Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76, 616–633. doi: 10.1111/j.1365-2958.2010.07108.x
Yoon, J., Matsuo, Y., Adachi, K., Nozawa, M., Matsuda, S., Kasai, H., et al. (2008a). Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luteolibacter pohnpeiensis gen. nov., sp. nov. and Luteolibacter algae sp. nov., six marine members of the phylum “Verrucomicrobia”, and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae. Int. J. Syst. Evol. Microbiol. 58, 998–1007. doi: 10.1099/ijs.0.65520-0
Yoon, J., Matsuo, Y., Katsuta, A., Jang, J.-H., Matsuda, S., Adachi, K., et al. (2008b). Haloferula rosea gen. nov., sp. nov., Haloferula harenae sp. nov., Haloferula phyci sp. nov., Haloferula helveola sp. nov. and Haloferula sargassicola sp. nov., five marine representatives of the family Verrucomicrobiaceae within the phylum “Verrucomicrobia.”. Int. J. Syst. Evol. Microbiol. 58, 2491–2500. doi: 10.1099/ijs.0.2008/000711-0
Yoon, J., Matsuo, Y., Matsuda, S., Adachi, K., Kasai, H., and Yokota, A. (2007a). Cerasicoccus arenae gen. nov., sp. nov., a carotenoid-producing marine representative of the family Puniceicoccaceae within the phylum “Verrucomicrobia”, isolated from marine sand. Int. J. Syst. Evol. Microbiol. 57, 2067–2072. doi: 10.1099/ijs.0.65102-0
Yoon, J., Matsuo, Y., Matsuda, S., Adachi, K., Kasai, H., and Yokota, A. (2007b). Rubritalea spongiae sp. nov. and Rubritalea tangerina sp. nov., two carotenoid- and squalene-producing marine bacteria of the family Verrucomicrobiaceae within the phylum “Verrucomicrobia”, isolated from marine animals. Int. J. Syst. Evol. Microbiol. 57, 2337–2343. doi: 10.1099/ijs.0.65243-0
Yoon, J., Matsuo, Y., Matsuda, S., Kasai, H., and Yokota, A. (2010). Cerasicoccus maritimus sp. nov. and Cerasicoccus frondis sp. nov., two peptidoglycan-less marine verrucomicrobial species, and description of Verrucomicrobia phyl. nov., nom. rev. J. Gen. Appl. Microbiol. 56, 213–222. doi: 10.2323/jgam.56.213
Keywords: PVC superphylum, peptidoglycan, cell division, budding, ftsZ, dcw cluster
Citation: Rivas-Marín E, Canosa I and Devos DP (2016) Evolutionary Cell Biology of Division Mode in the Bacterial Planctomycetes-Verrucomicrobia-Chlamydiae Superphylum. Front. Microbiol. 7:1964. doi: 10.3389/fmicb.2016.01964
Received: 05 September 2016; Accepted: 23 November 2016;
Published: 06 December 2016.
Edited by:
Boran Kartal, Max-Planck-Gesellschaft, GermanyReviewed by:
Christian Jogler, Deutsche Sammlung von Mikroorganismen und Zellkulturen, GermanyCopyright © 2016 Rivas-Marín, Canosa and Devos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Damien P. Devos, ZGFtaWVucGRldm9zQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.