- Programa de Genómica Funcional de Procariotes, Centro de Ciencias Genómicas, Universidad Nacional Autónoma de México, Cuernavaca, Mexico
Background
Rhizobia are soil bacteria that can interact with legumes through the formation of root nodules, where they fix nitrogen symbiotically. Rhizobia are also present in other legume tissues, including the interiors of roots and leaves, as well as in other plants and trees (Sturz et al., 1997; López-López et al., 2010; Rozahon et al., 2014). We recently reported the isolation and characterization of strains from common bean seeds (Phaseolus vulgaris) capable of nodulating and fixing nitrogen when inoculated onto bean (Mora et al., 2014). The seed-borne strain CCGM7 was characterized by screening for genes that allow the bacteria to survive in seeds (Peralta et al., 2016). To extend this research, we tested nodules formed by CCGM7 (or a strain from soil, CFNEI73) and isolated two strains, named CCGM10 and CCGM11, capable of growing in LB medium, a phenotype reported for certain strains of agrobacteria (Tanaka et al., 2009).
Genus Agrobacterium includes several species of bacteria commonly found in soil capable of forming tumors on the stems and roots of plants and trees (Singh and Prasad, 2016). Consistent with this phenotype, introduction of Agrobacterium T-DNA into the plant genome results in tumor formation (Bourras et al., 2015). Agrobacterium fabrum (formerly A. tumefaciens) has been widely used as a natural genetic engineer to transform plants of agricultural interest (Lassalle et al., 2011); however, A. radiobacter does not form tumors.
The newly defined species Agrobacterium pusense includes the type strain NRCPB10T, isolated from Cicer arietinum roots (Panday et al., 2011; Mousavi et al., 2015), as well as other root and nodule bacteria generally incapable of forming nodules or fixing nitrogen. Strain H13-3 has been studied for many years as a model for chemotaxis and motility, but it is non-symbiotic and was isolated from the rhizosphere of Lupinus luteus (Wibberg et al., 2011). Strain HPC(L) was isolated from desert soil (Agarwal and Purohit, 2013). Other strains closely related to A. fabrum have been found in nodules of legumes, including common bean, but cannot fix nitrogen (De Lajudie et al., 1999; Mhamdi et al., 2002, 2005; Aserse et al., 2012). IRBG74, the only strain with a symbiotic plasmid, can fix nitrogen in Sesbania cannabina and infect rice endophytically (Tan et al., 2001; Crook et al., 2013). Intriguingly, several isolates of A. pusense have been found in human wounds and body fluids (Aujoulat et al., 2015). Although genus Agrobacterium is to some extent phylogenetically entangled with rhizobia (see a recent review on Rhizobium taxonomy, Mousavi et al., 2015), one means for clear identification is the linear chromosome present in several Agrobacterium strains, which is accompanied by a protelomerase gene (telA) (Ramírez-Bahena et al., 2014).
Here, we report the genomic sequences of the A. pusense strains CCGM10 and CCGM11 isolated from bean nodules, a genomic analysis of the homology of their chromosomes and plasmids relative to other agrobacteria, and a functional profile of their genes.
Methods
Strains and Culture Media
Strains were maintained on solid rich medium LB (1% peptone, 0.5% yeast extract, 1% NaCl), supplemented with appropriate antibiotics. Liquid cultures for inoculation were grown in LB at 30°C with shaking at 200 rpm (Encarnación et al., 1995). Antibiotics were added as follows: nalidixic acid, 20 μg/mL; streptomycin, 200 μg/mL; and fosfomycin, 200 μg/mL.
Isolation of A. pusense Strains CCGM10 and CCGM11
P. vulgaris Negro Jamapa plantlets were inoculated with Sinorhizobium americanum strain CFNEI73 or CCGM7, both of which form nodules and fix nitrogen (Toledo et al., 2003; Mora et al., 2014). When the nodules were crushed and applied to LB plates, where symbiotic rhizobia normally do not grow, single colonies appeared after 72 h of incubation at 29°C. Three colonies from each spot were washed exhaustively in 10 mM MgSO4/0.01% Tween 40 and plated on LB medium, followed by incubation for 72 h at 29°C. To purify the strains, bacteria were grown overnight in liquid LB, serially diluted in 10 mM MgSO4-0.01% Tween 40, plated on LB plates, and incubated as above. Individual colonies were isolated from these plates.
Visualization of Plasmids
Plasmids were visualized by the Eckhardt technique with modifications, as previously reported (Hynes and McGregor, 1990).
Genome Sequencing, Assembly, and Annotation
Total DNA from strains was extracted from cultures grown in liquid LB medium; the cells were centrifuged, resuspended in 50 mM Tris/20 mM EDTA pH 8.0, and then lysed with proteinase K (2.5 mg/mL) and 10% SDS. After treatment with a mix of 1:1 phenol-chloroform, DNA was precipitated with absolute ethanol. Genomes were sequenced by Macrogen (Seoul, South Korea). For each strain, libraries of 3 kilobase pairs (kb) were prepared and run on an Illumina HiSeq2000 to obtain 100–base pair (bp) mated pair reads. Total read counts were 13,969,230 and 17,424,244 for CCGM10 and CCGM11, respectively; filtered read counts were 6,951,885 and 8,690,792, with genome coverage of 121 × and 152 ×. Assembly was performed with SOAPdenovo2 (Luo et al., 2012). Annotation was performed with RAST v4.0 (Aziz et al., 2008) with manual curation. Functions were assigned using the extended annotation of clusters of orthologous groups (COG) of COG 2014 database update (Galperin et al., 2015). Phages were detected using the PHASTER server (Arndt et al., 2016).
Comparative Genomic Analysis
The rpoB genes of strains CCGM10 and CCGM11 were sent to BLAST server (http://www.ncbi.nlm.nih.gov/Blast.cgi) to detect closely related strains. The genomes of selected strains were compared with genomes of CCGM10 and CCGM11 using Proteinortho version 5, with BLAST parameters E < 1E-5; 30% identity, and 70% coverage. The organisms selected for comparison were A. pusense IRBG74; A. pusense HPC(L), also known as R. pusense; and A. fabrum C58, also designated A. tumefaciens.
Nucleotide Accession Numbers
The genomes of A. pusense strains CCGM10 and CCGM11 were registered at GenBank with the provisional accessions LNZW00000000 and MAPG00000000, respectively.
Results
Homology and Chromosomal Conservation
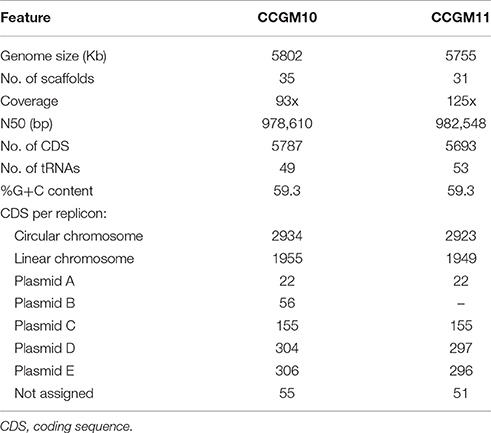
The main characteristics of the genomes of A. pusense strains CCGM10 and CCGM11 are listed in Table 1. The similarity between the strains was remarkable, with 93 and 92% of shared orthologs in the circular and linear chromosomes, respectively (Supplementary Table 1). Interestingly, some portions of the chromosomes were not shared with strain IRBG74; these segments mainly corresponded to genes with hypothetical functions (Supplementary Table 2). The sequence identity of shared orthologs in both strains was surprisingly high, 99.97%, and 5096 products had 100% identity at the amino acid level. Only about 142–205 genes in each chromosome were strain-specific. In addition, the plasmids were almost the same size (Figure 1A) and exhibited very high proportion of shared orthologs, ranging from 82 to 95%. These features indicate that the strains diverged very recently, mainly by insertion or deletion of genes.
Figure 1. Genome comparison of Agrobacterium pusense strains. (A) Eckhardt plasmid profile of strains CCGM10, CCGM11, and R. etli CFN42 (for which the pattern was known). Estimated molecular weights are in kb. (B) Schematic representation of the chromosomes of A. pusense strains. The chromosomes of A. pusense IRBG74 were used as a reference. Top, comparison of circular chromosome. From the innermost circle: GC skew, %GC content, CDS prediction with direction of transcription of the IRBG74 chromosome (gray boxes), comparison with CCGM10 (red bars), comparison with CCGM11 (orange), comparison with HPC(L) (turquoise), and comparison with A. fabrum C58 (green). Bottom, comparison of linear chromosome. From bottom, CDS prediction with direction of transcription of the IRBG74 chromosome (gray boxes), comparison with CCGM10 (red bars), comparison with CCGM11 (orange), comparison with HPC(L) (turquoise), and comparison with A. fabrum C58 (green). (C) Homology of plasmid genes of strain CCGM10.
A structural analysis of the genomes of these strains revealed very high conservation of both the linear and circular chromosomes, as shown in Figure 1B. The genomes were compared with that of the strains' closest relative, A. pusense IRBG74. The chromosomes were highly conserved, but the symbiotic plasmid of strain IRBG74 was not shared (not shown), and some regions were exclusive to the chromosomes of the reference strain. The genomes of A. pusense HPC(L) and that of A. fabrum C58, the model organism of the genus, were also included in the comparison.
Homologs of Plasmid Genes
Homology analysis of plasmid genes revealed that many had homologs in rhizobia and agrobacteria, but also with more distant organisms (Figure 1C). Most of genes in plasmid E were related to those from Agrobacterium and Rhizobium. Plasmid D harbored numerous genes with homologs in Roseomonas mucosa (with relatives isolated from human blood; Han et al., 2003) and Ochrobactrum anthropi, a member of the Brucellaceae that is found in natural ecosystems but also able to infect humans (Daxboeck et al., 2002). More than half of the genes on plasmid C were related to those from Rhizobium sp. H41, isolated from eroding rocks (Xi et al., 2014). Plasmid B, which is absent from strain CCGM11, is an intact temperate prophage of about 37 kb, harboring 47 genes, some shared with a prophage present in the circular chromosome of A. tumefaciens C58 and related to phage AmM-1 from Aurantimonas sp., a psychrotolerant bacterium belonging to the order Rhizobiales found in the deep ocean (Goldstein et al., 1982; Yoshida et al., 2015). Some genes assigned to plasmid A had homologs in Rhizobium sp. H41.
Other Prophages in the Genome
In light of our detection of the prophage in plasmid B, we analyzed the remaining replicons and identified additional intact prophages (Supplementary Table 3). The circular chromosome of strains CCGM10 and CCGM11 harbored three prophages: two related to phages RR1 and 16-3, also present in rhizobia, and vB_PmaS from the marine bacterium Paracoccus marcusii. In the putative linear chromosome of both strains, we found an intact prophage related to RC1 from Rhodobacter capsulatus. All prophages belonged to the family Caudovirales.
Gene Functions
Next, we analyzed the functions of genes in the plasmids of strain CCGM10 (Table 1 and Supplementary Table 4). Plasmid E was enriched in genes encoding transcriptional regulators and factors involved in signal transduction and amino acid or ion transport. Among these genes were those encoding proteins related to DNA uptake and folding (as HU), dnaE and ligD; central metabolism (e.g., fabG, ilvB, and arcCD), and ubiquinone synthesis. Plasmid D was especially enriched with genes involved in transcription, replication, and intracellular trafficking. Other notable genes encoded factors associated with secretion system IV, chemotaxis (cheBRW and methyl-accepting proteins mcp), transcriptional regulators and sensors, and genes for the redox response involving flavodoxin and glutathione. In addition, the plasmid contained a 500 bp segment homologous to the T-DNA segment of plasmid pTi.
Plasmid C was rich in genes involved in trafficking and ion transport, and it also harbored genes related to defense (toxin-antitoxin pairs) and central metabolism, such as arcACD, argF, speD, and the universal stress protein uspA. Plasmid A contained some genes involved in chemotaxis and glycerol utilization. In addition, we identified clusters of genes involved in conjugal plasmid transfer and replication (repABC family) in plasmids pC, pD, and pE.
In the circular chromosome, we identified the following genes involved in nitrogen fixation: fixGHS, fixNOQP, nifURS, nodLN, and fixR. A. fabrum C58 harbored homologs of these genes. However, some other genes not present in the C58 genome, such as the fixLJK cluster, were present in plasmid D. FixLJ are transcriptional regulators of nifA, the general activator of nitrogen fixation; FixK is regulator of the fixNOQP operon (encoding Cbb3, the cytochrome used by symbiotic rhizobia for respiration inside the nodules). These genes may represent the remnant of a complete gene cluster related to nitrogen fixation.
Bean Seeds As the Origin of A. pusense Strains
Several clues point to bean seeds as the origin of this pair of A. pusense strains, which were isolated from nodules but lack genes for nodulation. First, we have isolated 10 other rhizobial strains from nodules of non-inoculated plants; seeds from plants inoculated with two selected strains, CCGM1 or CCGM7, yielded positive signals for specific rhizobial gene markers (Mora et al., 2014). Second, some of the seed rhizobia strains did not have nodulation genes, but they are nonetheless able to inhabit nodules. Third, seed rhizobia have been identified as pairs of highly related strains differing in only one plasmid, as in this A. pusense pair. No A. pusense strains have been used previously in our greenhouse. It is possible that the A. pusense strains originally harbored a symbiotic plasmid that has been lost, but is still present in close relatives such as IRBG74. As with other agrobacteria, it will be of interest to define the precise symbiotic role of our isolates (Mrabet et al., 2006).
Availability of Data
The files containing the genomic sequences and the data resulting from our analyses are available and can be accessed freely at our server by ftp (ftp://kanan.ccg.unam.mx/PGFP/Apusense). A README file explains the content and mode of use.
Author Contributions
JM conceived and coordinated the project. AA performed assembly and annotation of the genomes. AA and HP performed the homology and functional analyses. YM, RD, CV, and LG performed the isolation and identification of the strains. HP and JM drafted the manuscript. All authors have read and approved the manuscript.
Funding
This project was partially supported by grants from the Consejo Nacional de Ciencia y Tecnología–Mexico (213606 and 152776) and from DGAPA-PAPIIT-UNAM (IN208216 and IN206914). The agencies had no role in the design of the study or the analysis or interpretation of the results.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sandra Contreras, Oliver Castillo, Jadaú Sánchez, José Luis Zitlalpopoca, and Paz Salas for technical assistance; Catalina Ortiz for Eckhardt profiles; and Dr. Michael Dunn for reviewing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01720/full#supplementary-material
References
Agarwal, L., and Purohit, H. J. (2013). Genome sequence of Rhizobium lupini HPC(L) isolated from saline desert soil, Kutch (Gujarat). Genome Announc. 1, e00071–12. doi: 10.1128/genomeA.00071-12
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Aserse, A.A., Räsänen, L.A., Assefa, F., Hailemariam, A., and Lindström, K. (2012). Phylogeny and diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst. Appl. Microbiol. 35, 120–131. doi: 10.1016/j.syapm.2011.11.005
Aujoulat, F., Marchandin, H., Zorgniotti, I., Masnou, A., and Jumas-Bilak, E. (2015). Rhizobium pusense is the main human pathogen in the genus Agrobacterium/Rhizobium. Clin. Microbiol. Infect. 21, 472e471–475. doi: 10.1016/j.cmi.2014.12.005
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bourras, S., Rouxel, T., and Meyer, M. (2015). Agrobacterium tumefaciens gene transfer: how a plant pathogen hacks the nuclei of plant and nonplant organisms. Phytopathology 105, 1288–1301. doi: 10.1094/PHYTO-12-14-0380-RVW
Crook, M. B., Mitra, S., Ané, J.-M., Sadowsky, M. J., and Gyaneshwar, P. (2013). Complete genome sequence of the Sesbania symbiont and rice growth-promoting endophyte Rhizobium sp. strain IRBG74. Genome Announc. 1, e00934-13. doi: 10.1128/genomea.00934-13
Daxboeck, F., Zitta, S., Assadian, O., Krause, R., Wenisch, C., and Kovarik, J. (2002). Ochrobactrum anthropi bloodstream infection complicating hemodialysis. Am. J. Kidney Dis. 40, e17.1–e17.4. doi: 10.1053/ajkd.2002.35759
De Lajudie, P., Willems, A., Nick, G., Mohamed, S. H., Torck, U., Coopman, R., et al. (1999). Agrobacterium bv. 1 strains isolated from nodules of tropical legumes. Syst. Appl. Microbiol. 22, 119–132. doi: 10.1016/S0723-2020(99)80035-6
Encarnación, S., Dunn, M., Willms, K., and Mora, J. (1995). Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177, 3058–3066.
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Goldstein, R., Sedivy, J., and Ljungquist, E. (1982). Propagation of satellite phage P4 as a plasmid. Proc. Natl. Acad. Sci. U.S.A. 79, 515–519. doi: 10.1073/pnas.79.2.515
Han, X. Y., Pham, A. S., Tarrand, J. J., Rolston, K. V., Helsel, L. O., and Levett, P. N. (2003). Bacteriologic characterization of 36 strains of Roseomonas species and proposal of Roseomonas mucosa sp. nov., and Roseomonas gilardii subsp. rosea subsp. nov. Am. J. Clin. Pathol. 120, 256–264. doi: 10.1309/731VVGVCKK351Y4J
Hynes, M. F., and McGregor, N. F. (1990). Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4, 567–574. doi: 10.1111/j.1365-2958.1990.tb00625.x
Lassalle, F., Campillo, T., Vial, L., Baude, J., Costechareyre, D., Chapulliot, D., et al. (2011). Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 3, 762–781. doi: 10.1093/gbe/evr070
López-López, A., Rogel, M. A., Ormeño-Orrillo, E., Martínez-Romero, J., and Martínez-Romero, E. (2010). Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 33, 322–327. doi: 10.1016/j.syapm.2010.07.005
Luo, R., Liu, B., Xie, Y. X., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18
Mhamdi, R., Laguerre, G., Aouani, M. E., Mars, M., and Amarger, N. (2002). Different species and symbiotic genotypes of field rhizobia can nodulate Phaseolus vulgaris in Tunisian soils. FEMS Microbiol. Ecol. 41, 77–84. doi: 10.1111/j.1574-6941.2002.tb00968.x
Mhamdi, R., Mrabet, M., Laguerre, G., Tiwari, R., and Aouani, M. E. (2005). Colonization of Phaseolus vulgaris nodules by Agrobacterium-like strains. Can. J. Microbiol. 51, 105–111. doi: 10.1139/w04-120
Mora, Y., Díaz, R., Vargas-Lagunas, C., Peralta, H., Guerrero, G., Aguilar, A., et al. (2014). Nitrogen-fixing rhizobial strains isolated from common bean seeds: phylogeny, physiology, and genome analysis. Appl. Environ. Microbiol. 80, 5644–5654. doi: 10.1128/AEM.01491-14
Mousavi, S. A., Willems, A., Nesme, X., de Lajudie, P., and Lindström, K. (2015). Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst. Appl. Microbiol. 38, 84–90. doi: 10.1016/j.syapm.2014.12.003
Mrabet, M., Mnasri, B., Romdhane, S. B., Laguerre, G., Aouani, M.E., and Mhamdi, R. (2006). Agrobacterium strains isolated from root nodules of common bean specifically reduce nodulation by Rhizobium gallicum. FEMS Microbiol. Ecol. 56, 304–309. doi: 10.1111/j.1574-6941.2006.00069.x
Panday, D., Schumann, P., and Das, S.K. (2011). Rhizobium pusense sp. nov., isolated from the rhizosphere of chickpea (Cicer arietinum L.) Int. J. Syst. Evol. Microbiol. 61, 2632–2639. doi: 10.1099/ijs.0.028407-0
Peralta, H., Aguilar, A., Díaz, R., Mora, Y., Martínez-Batallar, G., Salazar, E., et al. (2016). Genomic studies of nitrogen-fixing rhizobial strains from Phaseolus vulgaris seeds and nodules. BMC Genomics 17:711. doi: 10.1186/s12864-016-3053-z
Ramírez-Bahena, M. H., Vial, L., Lassalle, F., Diel, B., Chapulliot, D., Daubin, V., et al. (2014). Single acquisition of protelomerase gave rise to speciation of a large and diverse clade within the Agrobacterium/Rhizobium supercluster characterized by the presence of a linear chromid. Mol. Phylogenet. Evol. 73, 202–207. doi: 10.1016/j.ympev.2014.01.005
Rozahon, M., Ismayil, N., Hamood, B., Erkin, R., Abdurahman, M., Mamtimin, H., et al. (2014). Rhizobium populi sp. nov., an endophytic bacterium isolated from Populus euphratica. Int. J. Syst. Evol. Microbiol. 64, 3215–3221. doi: 10.1099/ijs.0.061416-0
Singh, K. R., and Prasad, M. (2016). Advances in Agrobacterium tumefaciens-mediated genetic transformation of graminaceous crops. Protoplasma 253, 691–707. doi: 10.1007/s00709-015-0905-3
Sturz, A. V., Christie, B. R., Matheson, B. G., and Nowak, J. (1997). Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol. Fertil. Soils 25, 13–19. doi: 10.1007/s003740050273
Tan, Z., Hurek, T., Vinuesa, P., Mûller, P., Ladha, J. K., and Reinhold-Hurek, B. (2001). Specific detection of Bradyrhizobium and Rhizobium strains colonizing rice (Oryza sativa) roots by 16S–23S ribosomal DNA intergenic spacer-targeted PCR. Appl. Environ. Microbiol. 67, 3655–3664. doi: 10.1128/AEM.67.8.3655-3664.2001
Tanaka, K., Arafat, H. H., Urbanczyk, H., Yamamoto, S., Moriguchi, K., Sawada, H., et al. (2009). Ability of Agrobacterium tumefaciens and A. rhizogenes strains, inability of A. vitis and A. rubi strains to adapt to salt-insufficient environment, and taxonomic significance of a simple salt requirement test in the pathogenic Agrobacterium species. J. Gen. Appl. Microbiol. 55, 35–41. doi: 10.2323/jgam.55.35
Toledo, I., Lloret, L., and Martínez-Romero, E. (2003). Sinorhizobium americanus sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Syst. Appl. Microbiol. 26, 54–64. doi: 10.1078/072320203322337317
Wibberg, D., Blom, J., Jaenicke, S., Kollin, F., Rupp, O., Scharf, B., et al. (2011). Complete genome sequencing of Agrobacterium sp. H13-3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J. Biotechnol. 155, 50–62. doi: 10.1016/j.jbiotec.2011.01.010
Xi, J., Sheng, X., and He, L. (2014). Draft genome sequence of Rhizobium sp. H41, a rock-weathering bacterium from a weathered rock surface. Genome Announc. 2, e01127–14. doi: 10.1128/genomeA.01127-14
Keywords: Agrobacterium, symbiosis, Phaseolus vulgaris, nodulation
Citation: Aguilar A, Peralta H, Mora Y, Díaz R, Vargas-Lagunas C, Girard L and Mora J (2016) Genomic Comparison of Agrobacterium pusense Strains Isolated from Bean Nodules. Front. Microbiol. 7:1720. doi: 10.3389/fmicb.2016.01720
Received: 12 August 2016; Accepted: 13 October 2016;
Published: 27 October 2016.
Edited by:
Xavier Perret, University of Geneva, SwitzerlandReviewed by:
David Dowling, Institute of Technology Carlow, IrelandJulie Ardley, Murdoch University, Australia
Copyright © 2016 Aguilar, Peralta, Mora, Díaz, Vargas-Lagunas, Girard and Mora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaime Mora, am1vcmFAY2NnLnVuYW0ubXg=
 Alejandro Aguilar
Alejandro Aguilar Humberto Peralta
Humberto Peralta Yolanda Mora
Yolanda Mora Rafael Díaz
Rafael Díaz Carmen Vargas-Lagunas
Carmen Vargas-Lagunas Lourdes Girard
Lourdes Girard Jaime Mora
Jaime Mora