- 1Laboratório de Química e Função de Proteínas e Peptídeos, Centro de Biociências e Biotecnologia, Universidade Estadual do Norte Fluminense Darcy Ribeiro, Campos dos Goytacazes, Brazil
- 2National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD, USA
- 3Laboratório de Bacteriologia, Unidade Laboratório de Análises Clínicas, Hospital de Clínicas, Universidade Federal do Paraná, Curitiba, Brazil
- 4Faculdades e Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil
Background
Enterobacter aerogenes is a motile, non-spore forming, Gram-negative bacteria from the Enterobacteriaceae family. Enterobacter spp. have emerged as multidrug-resistant (MDR) nosocomial bacteria, especially in intensive care units (Loiwal et al., 1999; Piagnerelli et al., 2002). Therefore, over the last decade Enterobacter spp. were included in the ESKAPE group, which also comprises Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa (Rice, 2008; Boucher et al., 2009). Further, bloodstream infections with MDR E. aerogenes have been associated with high mortality rates (Davin-Regli and Pagés, 2015).
Hospital outbreaks due to E. aerogenes have been reported in Europe since the mid-1990s and have been related to an epidemic extended-spectrum beta-lactamase (ESBL) clone carrying the blaTEM-24 gene (Bosi et al., 1999; Galdbart et al., 2000; Dumarche et al., 2002; Salso et al., 2003). Constitutive AmpC a (beta-lactamase) overexpression is the major cephalosporin resistance mechanism in Enterobacter spp., happening more often than the acquisition of ampC genes through the activity of mobile genetic elements (Perez-Perez and Hanson, 2002). Further, the increased expression of ESBLs led to the adoption of carbapenems to treat E. aerogenes infections (Perez-Perez and Hanson, 2002; Davin-Regli and Pagés, 2015).
Carbapenems have been considered the antibiotic of choice for treating patients infected with ESBL-producing Enterobacteriaceae (Vardakas et al., 2012). However, emergence of carbapenem-resistant E. aerogenes isolates during carbapenem therapy of hospitalized patients (Chen et al., 2008), cases of sepsis due to carbapenem-resistant E. aerogenes after liver transplantation (Chen et al., 2009) and hospital disseminations of carbapenemase-producing E. aerogenes have been recently reported in several countries (Lavigne et al., 2013; Kuai et al., 2014; Qin et al., 2014; Pulcrano et al., 2016). Acquisition and expression of carbapenemases constitute the primary mechanism underlying the development of carbapenem resistance (Rapp and Urban, 2012). Nevertheless, loss of function mutations in porin genes and increased expression of efflux pumps or their regulators have also been associated with carbapenem resistance profiles (Pradel and Pages, 2002; Yigit et al., 2002; Bornet et al., 2003).
Broad-spectrum antimicrobial-resistant E. aerogenes isolates, some resistant to carbapenems (Qin et al., 2014) and last-line therapeutic options such as colistin (Diene et al., 2013), have been responsible for outbreaks in the United States of America (Wong et al., 2010), China (Qin et al., 2014), Japan (Goshi et al., 2002), France (Diene et al., 2013), Fiji (Narayan et al., 2009) and Brazil (Tuon et al., 2015). However, few reports related to E. aerogenes epidemiology, pathogenesis, and molecular characterization have been conducted in Brazil. Recently, five panresistant E. aerogenes isolates were reported in a Brazilian teaching hospital, resulting in a high mortality rate (37.5%) among 16 infected patients (Tuon et al., 2015). We have observed high prevalence (>20%) of ESBL-producing Enterobacteriaceae spp., in particular K. pneumoniae and E. aerogenes, in our hospital since 2003 (Nogueira Kda et al., 2014, 2015). Previous molecular characterization studies conducted over 5 years in our hospital showed high prevalence of blaCTX-M2, -M15, -M59, blaSHV-2 and blaTEM genes in Enterobacter spp. isolates (Nogueira Kda et al., 2014, 2015). The presence of blaPER-2 was also detected in a few isolates (Nogueira Kda et al., 2014, 2015). Given the severity of E. aerogenes infections and the urgent need to better understand the genetic basis of multidrug resistance, here we report the whole-genome sequencing and resistance gene repertoire of four multidrug-resistant E. aerogenes isolated from hospitalized patients in Brazil.
Methods
Sample Collection and Identification
E. aerogenes isolates C10, D2, D3, and E9 were obtained between 2006 and 2012 from patients hospitalized in wards or intensive care units at the Hospital de Clínicas of the Universidade Federal do Paraná (Curitiba, Brazil). The main selection criterion for genome sequencing was the MDR phenotype, particularly in carbapenem resistant isolates. The negative laboratory tests for carbapenemases were also taken into account, as divergent enzymes or alternative resistance mechanisms could be relevant to the observed MDR phenotypes. C10 and D2 samples were isolated from different body sites of the same patient. Isolates were grown in selective medium with an ertapenem disk (10 ug) and stored at −80°C in trypticase soy broth containing glycerol 15%. Identification of isolates was performed using Vitek® 2 Compact (BioMérieux S.A., Marcy l'Etoile, France) and by mass spectrometry using Microflex LT instrument (Bruker Daltonics, Bremen, Germany). This study was carried out in accordance with the Brazilian legislation and was approved by the Institutional Ethics Review Board of the Hospital de Clínicas, Universidade Federal do Paraná (IRB#: 2656.263/2011-11). Our study involved only bacterial isolates and no human specimens were analyzed or stored. Further, we used no patient information other than the anatomical sites from where the isolates were collected. Therefore, the same Ethics Review Board exempted us from obtaining informed consent forms.
Resistance Profile Analysis
Antimicrobial Susceptibility Testing
Isolates were tested by agar dilution against 15 antibiotics according to the Clinical and Laboratory Standard Institute guidelines (CLSI, 2015a). Minimal inhibitory concentration (MIC) was interpreted as recommended by CLSI standards (CLSI, 2015b). Polymyxin, tigecycline and fosfomycin breakpoints were interpreted using EUCAST standards (Eucast, 2016). Modified Hodge test (MHT), double-disk synergy and hydrolysis assay were performed to determine the carbapenem resistance phenotypes, as previously described (Carvalhaes et al., 2010; Eucast, 2013).
Molecular Typing and Detection of Resistance Markers
The genetic relatedness of the E. aerogenes isolates were determined by pulsed-field gel electrophoresis (PFGE), as described elsewhere (Kaufmann, 1998). DNA fingerprints were interpreted as recommended by Tenover et al. (1995). The presence of the blaMOX, blaCMY, blaLAT, blaBIL, blaDHA, blaACC, blaMIR, blaACT, blaFOX, blaTEM, blaSHV, blaCTX-M1, -M2, -M8, -M9, -M25, blaKPC, blaGES, blaIMP, blaVIM, blaNDM, blaSPM, blaGIM, blaSIM, blaOXA-23, -48, -51, -58, and -143 was tested by PCR as previously described (Payne and Thomson, 1998; Poirel et al., 2000, 2011; Perez-Perez and Hanson, 2002; Naas et al., 2008; Higgins et al., 2009; Woodford, 2010; Nordmann et al., 2011).
Genome Sequencing, Assembly, and Annotation
Genomic DNA was extracted using DNeasy 96 Blood & Tissue Kit (QIAGEN Silicon Valley, Redwood City, USA). DNA quality was assessed using a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, USA). DNA quantification was performed using Qubit (Thermo Fisher Scientific Inc., Waltham, USA). Illumina sequencing libraries with an average fragment size of 550 bp were prepared using Illumina TruSeq DNA PCR-free LT Kit (Illumina Inc., San Diego, USA). Whole-genome sequencing of paired-end (PE) libraries was performed using a HiSeq 2500 instrument in RAPID run mode (Illumina Inc., San Diego, USA) at the Life Sciences Core Facilities of the State University of Campinas (São Paulo, Brazil). Quality-based trimming and filtering was performed using Trimmomatic version 0.32 (Bolger et al., 2014). PE reads were assembled de novo using Velvet version 1.2.10 (Zerbino and Birney, 2008) and contigs were scaffolded using SSPACE version 3.0 (Boetzer et al., 2011). Gene predictions and annotations were performed using NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP; Angiuoli et al., 2008).
Identification of Antibiotic Resistance Genes
Antibiotic resistance-related genes were predicted using the ResFinder database version 2.1 (Zankari et al., 2012) with the following parameters: “all databases” were used for antimicrobial configuration, type of reads as “assembled genomes/contigs” and thresholds of 98 identity and 80% coverage between sequences. This dataset of resistance genes was complemented with BLASTp searches against the ARDB (Antibiotic Resistance Genes Database) version 1.1 (Liu and Pop, 2009) using “resistance gene complete” database, 40% identity and e-value of 0.0001.
Results
Resistance Profiles
All isolates showed MDR profile and had increased MIC for at least one carbapenem. Information regarding collection date and site, clinical setting, PFGE profile and antimicrobial resistance profiles of each isolate are available in Table 1. Among the four analyzed samples, C10 and D2 were isolated from different body sites of the same patient within a short period of time (a month) and belong to the same PFGE profile. These genomes allow one to analyze the possible genome plasticity between the isolates. D3 and E9 samples were isolated from two patients with an interval of collection date greater than 5 years. D3 and E9 were also interesting because of their sensitivity to meropenem and resistance to ertapenem and imipenem. Surprisingly, E9 showed resistance to carbapenems but not to 3rd (ceftazidime and cefotaxime) and 4th generation (cefepime) cephalosporins (Table 1). All isolates possessed blaAmpC and blaTEM, as detected by PCR. The gene blaCTX-M2 was found in all isolates except E9. Phenotypic tests (i.e., Modified Hodge test and double-disk synergy) to detect carbapenemases were positive for C10, D2, and E9. However, no class A, B, and D carbapenemase encoding genes were detected by PCR. All isolates tested negative in carbapenem hydrolysis assays.
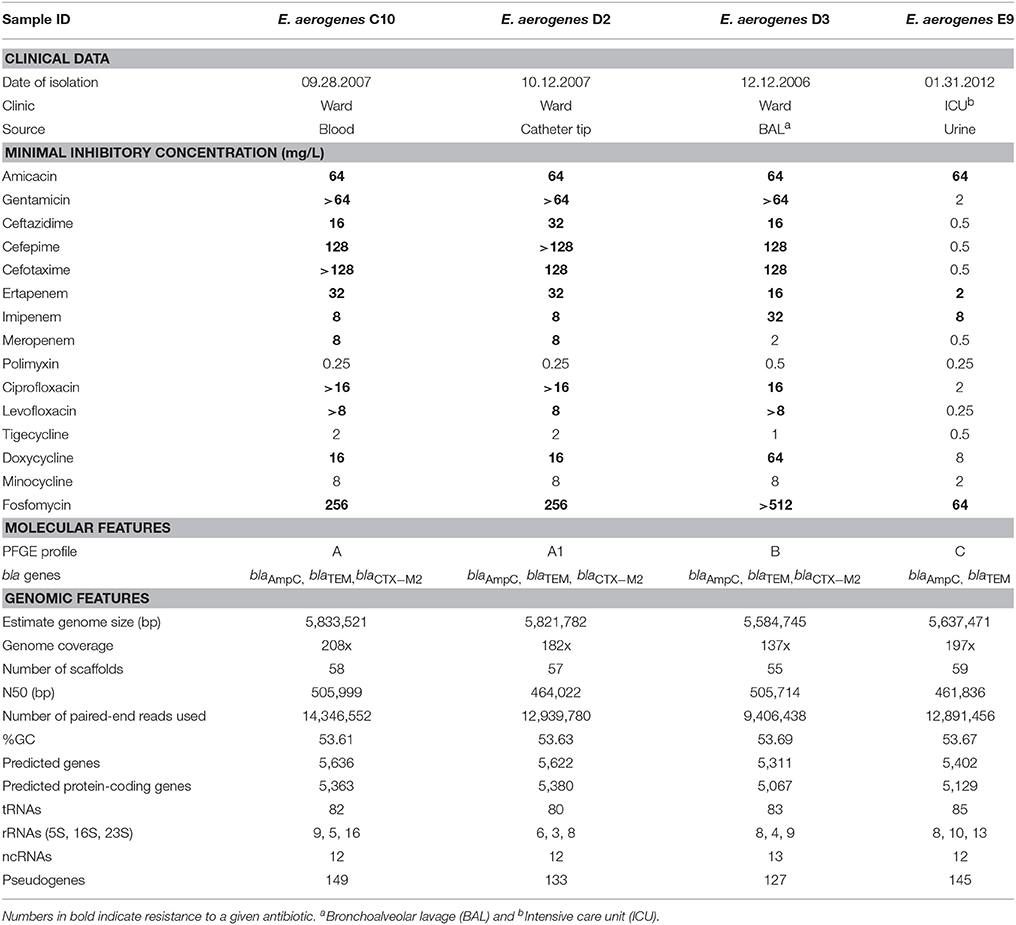
Table 1. Clinical, phenotypic, molecular data, and genomic features of the four Enterobacter aerogenes isolates reported in the present work.
Genomic Features
We obtained between 16,841,714 and 25,138,390 150 bp PE reads per library. After genome assembly, 5,833,521 bp were assembled in 58 scaffolds for C10, 5,821,782 bp were assembled in 57 scaffolds for D2, 5,584,745 bp were assembled in 55 scaffolds for D3 and 5,637,471 bp were assembled in 59 scaffolds for E9. By using the NCBI Prokaryotic Annotation Pipeline, we were able to predict 5,363, 5,380, 5,067, and 5,129 protein-coding sequences in each of the genomes listed above, respectively. Genomic features of the four sequenced genomes are summarized in Table 1.
Antibiotic Resistance Genes
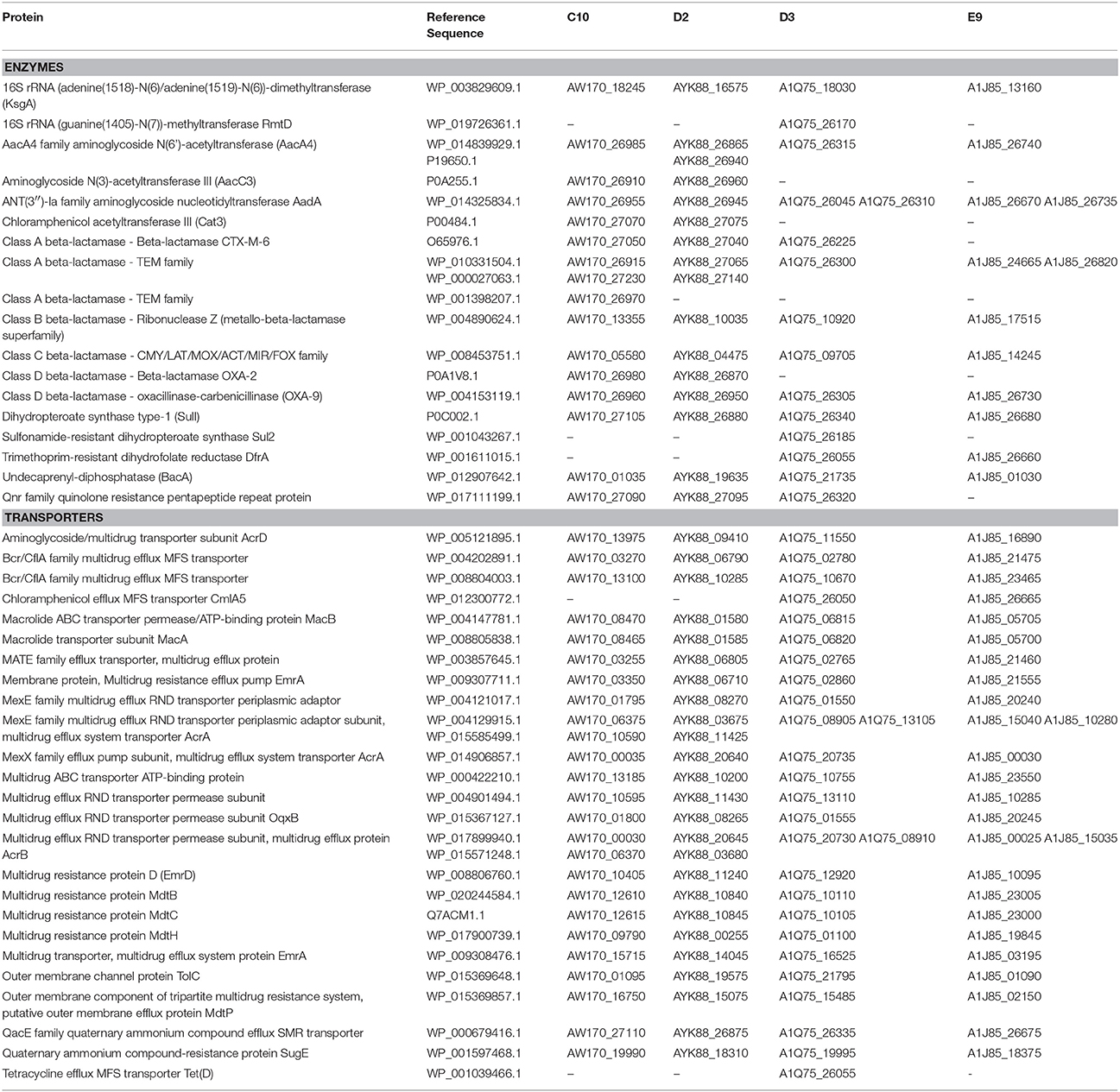
A total of 18 enzymes related to antibiotic resistance were identified using ResFinder, ARDB and PGAAP (Table 2). All isolates harbor genes related to: (i) aminoglycoside resistance (genes aacA4 and aadA); (ii) beta-lactam resistance, including genes belonging to class A beta-lactamases (TEM family), class B beta-lactamases (Ribonuclease Z), class C beta-lactamases (CMY/LAT/MOX/ACT/MIR/FOX family) and class D beta-lactamases (OXA-9); (iii) bacitracin resistance (gene bacA), and (iv) sulphonamide resistance (gene sul1; Table 2). Genes sul2 and rmtD were only identified in E. aerogenes D3. The gene sul2 has been implicated on sulphonamide resistance for inducing high expression levels of the enzyme dihydropteroate synthase (Sköld, 2001), while rmtD has been related to aminoglycoside resistance and this variant was identified for the first time in South America in a P. aeruginosa isolate in 2005 (Doi et al., 2007). Interestingly, E. aerogenes D3 was isolated in 2006, indicating that this variant has spread amongst Enterobacteriaceae in Brazil since its first report (Doi et al., 2007).
Although the four isolates showed carbapenem-resistance, no carbapenemase gene was identified using molecular detection or in silico analysis. Hence, it is likely that these isolates employ alternative mechanisms to counter carbapenem effects. Various multidrug efflux transporters were found in the genomes described here (Table 2). They belong to four superfamilies: the major facilitator superfamily (MFS), multidrug and toxic compound extrusion (MATE), ATP-binding cassette (ABC) and resistance-nodulation-cell division (RND). RND type of transporters has been often associated with multidrug resistance of Gram-negative bacteria (Nikaido, 1998). In particular, the RND type genes forming the AcrA-AcrB-TolC efflux pump were found in multiple copies in our isolates (Table 2). Experimental evolution studies of E. aerogenes under successive imipenem exposure reported alterations in membrane permeability with complete loss of porins (e.g., Omp35 and Omp36) and overexpression of AcrAB-TolC efflux pumps (Bornet et al., 2003; Thiolas et al., 2005; Lavigne et al., 2012). As a result of efflux pump expression, the E. aerogenes isolates showed resistance to carbapenems and other antibiotics, especially fluoroquinolones (Bornet et al., 2003; Thiolas et al., 2005; Lavigne et al., 2012). Given the multiple copies of genes encoding efflux pumps in our isolates, it is possible that an increased expression of AcrAB-TolC efflux pumps could contribute to the observed carbapenem-resistant profiles.
E. aerogenes is an emergent nosocomial pathogen with a diversity of mechanisms to circumvent antimicrobial activity. Here we reported the phenotypic screens, genome sequencing, and prediction of putative resistance gene repertoires of four multidrug-resistant E. aerogenes isolated between 2006 and 2012. The data reported here may help understand the biochemistry, evolution, and epidemiology of this important pathogen. The material provided in this work may be used in future comparative genomics and molecular epidemiology studies aiming to clarify the resistance profiles and dynamics of multidrug-resistant Enterobacteriaceae species.
Data Access
The genome sequence of E. aerogenes C10, E. aerogenes D2, E. aerogenes D3 and E. aerogenes E9 have been deposited in DDBJ/EMBL/GenBank under the accession numbers LUTZ00000000, LSOH00000000, LUTT00000000, and LULD00000000, respectively. Data are available in FASTA, annotated GenBank flat file and ASN.1 formats. The respective genome versions described in this paper are LUTZ01000000, LSOH01000000, LUTT01000000, and LULD01000000. Sequencing reads (fastq format) of each isolate were deposited in Sequence Read Archive (SRA) under the accession numbers SRP083774 (E. aerogenes C10), SRP083784 (E. aerogenes D2), SRP083785 (E. aerogenes D3), and SRP083786 (E. aerogenes E9). Users can download the data for research purposes, citing the present manuscript as original reference.
Author Contributions
AG, NV, JP, LD, and TV conceived the idea and designed the study. JP performed the sample collections and wet lab experiments. AG and NV carried out the genome analysis. AG, NV, JP, LD, and TV interpreted the data and wrote the manuscript. All authors have read and approved the final version of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ; E-26/110.236/2011 and E-26/102.259/2013). This research was partially supported by the Intramural Research Program of the National Library of Medicine (NLM), National Institutes of Health (NIH). NV postdoctoral fellowship is funded by a partnership between CNPq and NIH. TV is a recipient of an established investigator fellowship award from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We thank Bruker Corporation of Brazil for performing the MALDI-TOF assay and the staff of the Life Sciences Core Facility (LaCTAD), from State University of Campinas (UNICAMP), for library preparation and genome sequencing.
References
Angiuoli, S. V., Gussman, A., Klimke, W., Cochrane, G., Field, D., Garrity, G., et al. (2008). Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. OMICS 12, 137–141. doi: 10.1089/omi.2008.0017
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bornet, C., Chollet, R., Malléa, M., Chevalier, J., Davin-Regli, A., Pagés, J. M., et al. (2003). Imipenem and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem. Biophys. Res. Commun. 301, 985–990. doi: 10.1016/S0006-291X(03)00074-3
Bosi, C., Davin-Regli, A., Bornet, C., Mallea, M., Pages, J. M., and Bollet, C. (1999). Most Enterobacter aerogenes strains in France belong to a prevalent clone. J. Clin. Microbiol. 37, 2165–2169.
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Carvalhaes, C. G., Picão, R. C., Nicoletti, A. G., Xavier, D. E., and Gales, A. C. (2010). Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J. Antimicrob. Chemother. 65, 249–251. doi: 10.1093/jac/dkp431
Chen, H., Zhang, Y., Chen, Y. G., Yu, Y. S., Zheng, S. S., and Li, L. J. (2009). Sepsis resulting from Enterobacter aerogenes resistant to carbapenems after liver transplantation. HBPD INT 8, 320–322. Available online at: http://www.hbpdint.com/EN/Y2009/V8/I3/320#
Chen, Y. G., Zhang, Y., Yu, Y. S., Qu, T. T., Wei, Z. Q., Shen, P., et al. (2008). In vivo development of carbapenem resistance in clinical isolates of Enterobacter aerogenes producing multiple beta-lactamases. Int. J. Antimicrob. Agents 32, 302–307. doi: 10.1016/j.ijantimicag.2008.02.014
CLSI (2015a). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 10th Edn. CLSI document M7-A10. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2015b). Performance Standards for Antimicrobial Susceptibility Testing 25th Informational Supplement. CLSI document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute.
Davin-Regli, A., and Pagés, J. M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6:392. doi: 10.3389/fmicb.2015.00392
Diene, S. M., Merhej, V., Henry, M., El Filali, A., Roux, V., Robert, C., et al. (2013). The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol. Biol. Evol. 30, 369–383. doi: 10.1093/molbev/mss236
Doi, Y., de Oliveira Garcia, D., Adams, J., and Paterson, D. L. (2007). Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 51, 852–856. doi: 10.1128/AAC.01345-06
Dumarche, P., De Champs, C., Sirot, D., Chanal, C., Bonnet, R., and Sirot, J. (2002). TEM derivative-producing Enterobacter aerogenes strains: dissemination of a prevalent clone. Antimicrob. Agents Chemother. 46, 1128–1131. doi: 10.1128/AAC.46.4.1128-1131.2002
Eucast (2013). EucastGuidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Version 1.0, December, 2013. Available online at: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf
Eucast (2016). Eucast Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 6.0, May, 2016. Available online at: http://www.eucast.org
Galdbart, J. O., Lémann, F., Ainouz, D., Féron, P., Lambert-Zechovsky, N., and Branger, C. (2000). TEM-24 extended-spectrum beta-lactamase-producing Enterobacter aerogenes: long-term clonal dissemination in French hospitals. Clin. Microbiol. Infect. 6, 316–323. doi: 10.1046/j.1469-0691.2000.00092.x
Goshi, S., Taneike, I., Nakagawa, S., Kojio, S., Tamura, Y., Ohara, T., et al. (2002). DNA analysis of nosocomial infection by Enterobacter aerogenes in three cases of septicaemia in Japan. J. Hosp. Infect. 51, 221–225. doi: 10.1053/jhin.2002.1255
Higgins, P. G., Poirel, L., Lehmann, M., Nordmann, P., and Seifert, H. (2009). OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 5035–5038. doi: 10.1128/AAC.00856-09
Kaufmann, M. E. (1998). “Pulsed-field gel electrophoresis,” in Molecular Bacteriology: Protocols and Clinical Applications, eds N. Woodford and A. P. Johnson (Totowa, NJ: Humana Press Inc), 33–51.
Kuai, S., Shao, H., Huang, L., Pei, H., Lu, Z., Wang, W., et al. (2014). KPC-2 carbapenemase and DHA-1 AmpC determinants carried on the same plasmid in Enterobacter aerogenes. J. Med. Microbiol. 63, 367–370. doi: 10.1099/jmm.0.054627-0
Lavigne, J. P., Sotto, A., Nicolas-Chanoine, M. H., Bouziges, N., Bourg, G., Davin-Regli, A., et al. (2012). Membrane permeability, a pivotal function involved in antibiotic resistance and virulence in Enterobacter aerogenes clinical isolates. Clin. Microbiol. Infect. 18, 539–545. doi: 10.1111/j.1469-0691.2011.03607.x
Lavigne, J. P., Sotto, A., Nicolas-Chanoine, M. H., Bouziges, N., Pagés, J. M., and Davin-Regli, A. (2013). An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int. J. Antimicrob. Agents 41, 130–136. doi: 10.1016/j.ijantimicag.2012.10.010
Liu, B., and Pop, M. (2009). ARDB–Antibiotic Resistance Genes Database. Nucleic Acids Res. 37, D443–D447. doi: 10.1093/nar/gkn656
Loiwal, V., Kumar, A., Gupta, P., Gomber, S., and Ramachandran, V. G. (1999). Enterobacter aerogenes outbreak in a neonatal intensive care unit. Pediatr. Int. 41, 157–161. doi: 10.1046/j.1442-200X.1999.4121033.x
Naas, T., Cuzon, G., Villegas, M. V., Lartigue, M. F., Quinn, J. P., and Nordmann, P. (2008). Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52, 1257–1263. doi: 10.1128/AAC.01451-07
Narayan, S. A., Kool, J. L., Vakololoma, M., Steer, A. C., Mejia, A., Drake, A., et al. (2009). Investigation and control of an outbreak of Enterobacter aerogenes bloodstream infection in a neonatal intensive care unit in Fiji. Infect. Control Hosp. Epidemiol. 30, 797–800. doi: 10.1086/598240
Nikaido, H. (1998). Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1), S32–S41. doi: 10.1086/514920
Nogueira Kda, S., Conte, D., Maia, F. V., and Dalla-Costa, L. M. (2015). Distribution of extended-spectrum beta-lactamase types in a Brazilian tertiary hospital. Rev. Soc. Bras. Med. Trop. 48, 162–169. doi: 10.1590/0037-8682-0009-2015
Nogueira Kda, S., Paganini, M. C., Conte, A., Cogo, L. L., Taborda de Messias Reason, I., da Silva, M. J., et al. (2014). Emergence of extended-spectrum beta-lactamase producing Enterobacter spp. in patients with bacteremia in a tertiary hospital in southern Brazil. Enferm. Infecc. Microbiol. Clin. 32, 87–92. doi: 10.1016/j.eimc.2013.02.004
Nordmann, P., Poirel, L., Carrër, A., Toleman, M. A., and Walsh, T. R. (2011). How to detect NDM-1 producers. J. Clin. Microbiol. 49, 718–721. doi: 10.1128/JCM.01773-10
Payne, D. J., and Thomson, C. J. (1998). “Molecular approaches for the detection and identification of β-lactamases,” in Molecular Bacteriology: Protocols and Clinical Applications, eds N. Woodford and A. P. Johnson (Totowa, NJ: Humana Press Inc), 495–512.
Perez-Perez, F. J., and Hanson, N. D. (2002). Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40, 2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002
Piagnerelli, M., Carlier, E., Deplano, A., Lejeune, P., and Govaerts, D. (2002). Risk factors for infection and molecular typing in patients in the intensive care unit colonized with nosocomial Enterobacter aerogenes. Infect. Control Hosp. Epidemiol. 23, 452–456. doi: 10.1086/502084
Poirel, L., Le Thomas, I., Naas, T., Karim, A., and Nordmann, P. (2000). Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44, 622–632. doi: 10.1128/AAC.44.3.622-632.2000
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pradel, E., and Pagés, J. M. (2002). The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 46, 2640–2643. doi: 10.1128/AAC.46.8.2640-2643.2002
Pulcrano, G., Pignanelli, S., Vollaro, A., Esposito, M., Iula, V. D., Roscetto, E., et al. (2016). Isolation of Enterobacter aerogenes carrying bla and bla genes recovered from a hospital Intensive Care Unit. APMIS 124, 516–521. doi: 10.1111/apm.12528
Qin, X., Yang, Y., Hu, F., and Zhu, D. (2014). Hospital clonal dissemination of Enterobacter aerogenes producing carbapenemase KPC-2 in a Chinese teaching hospital. J. Med. Microbiol. 63, 222–228. doi: 10.1099/jmm.0.064865-0
Rapp, R. P., and Urban, C. (2012). Klebsiella pneumoniae carbapenemases in Enterobacteriaceae: history, evolution, and microbiology concerns. Pharmacotherapy 32, 399–407. doi: 10.1002/j.1875-9114.2012.01035.x
Rice, L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Salso, S., Culebras, E., Andrade, R., and Picazo, J. J. (2003). Outbreak of TEM-24-producing Enterobacter aerogenes in a Spanish hospital. Microb. Drug Resist. 9, 299–305. doi: 10.1089/107662903322286517
Skold, O. (2001). Resistance to trimethoprim and sulfonamides. Vet. Res. 32, 261–273. doi: 10.1051/vetres:2001123
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Thiolas, A., Bollet, C., La Scola, B., Raoult, D., and Pagés, J. M. (2005). Successive emergence of Enterobacter aerogenes strains resistant to imipenem and colistin in a patient. Antimicrob. Agents Chemother. 49, 1354–1358. doi: 10.1128/AAC.49.4.1354-1358.2005
Tuon, F. F., Scharf, C., Rocha, J. L., Cieslinsk, J., Becker, G. N., and Arend, L. N. (2015). KPC-producing Enterobacter aerogenes infection. Braz. J. Infect. Dis. 19, 324–327. doi: 10.1016/j.bjid.2015.01.003
Vardakas, K. Z., Tansarli, G. S., Rafailidis, P. I., and Falagas, M. E. (2012). Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum beta-lactamases: a systematic review and meta-analysis. J. Antimicrob. Chemother. 67, 2793–2803. doi: 10.1093/jac/dks301
Wong, M. R., Del Rosso, P., Heine, L., Volpe, V., Lee, L., Kornblum, J., et al. (2010). An outbreak of Klebsiella pneumoniae and Enterobacter aerogenes bacteremia after interventional pain management procedures, New York City, 2008. Reg. Anesth. Pain Med. 35, 496–499. doi: 10.1097/AAP.0b013e3181fa1163
Woodford, N. (2010). Rapid characterization of beta-lactamases by multiplex PCR. Methods Mol. Biol. 642, 181–192. doi: 10.1007/978-1-60327-279-7_14
Yigit, H., Anderson, G. J., Biddle, J. W., Steward, C. D., Rasheed, J. K., Valera, L. L., et al. (2002). Carbapenem resistance in a clinical isolate of Enterobacter aerogenes is associated with decreased expression of OmpF and OmpC porin analogs. Antimicrob. Agents Chemother. 46, 3817–3822. doi: 10.1128/AAC.46.12.3817-3822.2002
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Brazil, Enterobacter aerogenes, genome sequencing, multidrug resistance, carbapenem resistance
Citation: Grazziotin AL, Vidal NM, Palmeiro JK, Dalla-Costa LM and Venancio TM (2016) Genome Sequencing of Four Multidrug-Resistant Enterobacter aerogenes Isolates from Hospitalized Patients in Brazil. Front. Microbiol. 7:1649. doi: 10.3389/fmicb.2016.01649
Received: 13 June 2016; Accepted: 04 October 2016;
Published: 27 October 2016.
Edited by:
Frank T. Robb, University of Maryland, Baltimore, USAReviewed by:
Suleyman Yildirim, Istanbul Medipol University, TurkeyWeigang Qiu, Hunter College, USA
Copyright © 2016 Grazziotin, Vidal, Palmeiro, Dalla-Costa and Venancio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Laura Grazziotin, YW5hbGF1cmFncmF6emlvdGluQGdtYWlsLmNvbQ==
Libera Maria Dalla-Costa, bG1kYWxsYWNvc3RhQGdtYWlsLmNvbQ==
Thiago M. Venancio, dGhpYWdvLnZlbmFuY2lvQGdtYWlsLmNvbQ==
 Ana Laura Grazziotin
Ana Laura Grazziotin Newton M. Vidal
Newton M. Vidal Jussara K. Palmeiro
Jussara K. Palmeiro Libera Maria Dalla-Costa3,4*
Libera Maria Dalla-Costa3,4* Thiago M. Venancio
Thiago M. Venancio