- Microbiology and Cell Science Department, Genetics Institute & Institute of Food and Agricultural Science, University of Florida, Gainesville, FL, USA
CLIBASIA_01510, PrbP, is a predicted RNA polymerase binding protein in Liberibacter asiaticus. PrbP was found to regulate expression of a small subset of ribosomal genes through interactions with the β-subunit of the RNA polymerase and a short, specific sequence on the promoter region. Molecular screening assays were performed to identify small molecules that interact with PrbP in vitro. Chemical hits were analyzed for therapeutic efficacy against L. asiaticus via an infected leaf assay, where the transcriptional activity of L. asiaticus was found to decrease significantly after exposure to tolfenamic acid. Similarly, tolfenamic acid was found to inhibit L. asiaticus infection in highly symptomatic citrus seedlings. Our results indicate that PrbP is an important transcriptional regulator for survival of L. asiaticus in planta, and the chemicals identified by molecular screening assays could be used as a therapeutic treatment for huanglongbing disease.
Introduction
Liberibacter asiaticus is the prevalent causative agent of huanglongbing (HLB; citrus greening), the most devastating citrus disease worldwide. Since its emergence in the early 1900's in China, L. asiaticus has been found in nearly every citrus producing region across the globe, with devastating consequences (Bove et al., 2006). In the United States, the economic impact of HLB has already been felt throughout the Florida citrus industry, where estimated losses have exceeded $9 billion since the pathogen was first detected in 2005; and the California citrus industry is preparing for similar devastation, as this pathogen continues to spread across the west coast (Gottwald, 2010).
Liberibacter asiaticus is a phloem-limited pathogen with uneven distribution throughout the plant. It is transmitted by the Asian citrus psyllid Diaphorina citri and it behaves as intracellular plant pathogen and as an insect symbiont. These characteristics has made the control and eradication of L. asiaticus a scientific challenge with no precedents. The lack of stable culturing conditions for L. asiaticus, in a laboratory setting, has severely hampered progress toward understanding the physiology and adaptive strategies of this citrus pathogen. Genome analyses have revealed a drastic reduction of the genetic information encoded by the L. asiaticus genome, suggesting this microorganism is highly adapted to life within its host (Duan et al., 2009). One of the functional protein clusters suffering the highest negative selection were transcription factors, which represent only 2% of the total proteins encoded in the genome (Duan et al., 2009; Pagliai et al., 2014). Interestingly, the small group of transcription factors preserved in L. asiaticus includes the gene systematically annotated as CLIBASIA_01510 (here named PrbP). PrbP is a member of the CarD_CdnL_TRCF superfamily (CDD cl00588). PrbP shares low sequence identity with the transcription factor CdnL in Myxococcus xanthus and with CarD in the Mycobacteriaceae family (García-Moreno et al., 2010). Members of this protein family have been linked to pathogenesis, persistence, cell viability, and resistance to both antibiotics and environmental stress (Stallings et al., 2009; Stallings and Glickman, 2011; Weiss et al., 2012).
In absence of laboratory culturing conditions for L. asiaticus, traditional methods for testing chemicals as antimicrobials in vivo cannot be applied. In this study, we explored the possibility of identifying specific ligands in vitro, that affect the activity of PrbP in vivo, as a means to disrupt gene expression, and ultimately decrease viability of L. asiaticus in the citrus host. Only three eukaryotes, Spinacia oleracea, Beta vulgaris, and Drosophila elegans, were found to carry PrbP homologs (74, 72, and 46% identity, respectively). The absence of a PrbP homolog in citrus renders PrbP a unique target for therapeutics. Furthermore, the use of ligands that inactivate specific pleiotropic transcription factors, enables the selective elimination of target species in heterogeneous populations. Taken together, PrbP represents an excellent therapeutic target for the design of antimicrobial strategies against L. asiaticus.
Materials and Methods
Bacterial Strains and Growth Conditions
Escherichia coli strains were grown at 37°C under aerobic conditions in Luria-Bertani medium (LB) (Difco) or on LB agar plates. Escherichia coli strains DH5α (Invitrogen, Carlslab, CA), TOP 10 (Invitrogen), and XL1-Blue (Stratagene, La Jolla, CA) were used to propagate the plasmids for protein purification, point mutations, and the two-hybrid system (the bacterial strains and plasmids used in this study are listed in Table 1). Escherichia coli strain BL21-Rosetta(DE3) (Novagen, Gibbstown, NJ) was used for overexpression and protein purification. When required, the medium was supplemented with ampicillin (100 μg/ml), tetracycline (10 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (25 μg/ml). All antibiotics and chemicals were purchased from Sigma (St. Louis, MO).
Liberibacter crescens BT-1 was cultured at 25°C with moderate aeration (150 RPM), in modified BM7 media (Leonard et al., 2012) containing 1% Brain Heart Infusion (Difco Laboratories, Detroit, MI), 15% Fetal Bovine Serum (Sigma, St. Louis, MO), 30% TMN-FH insect medium (Sigma), α-Ketoglutaric acid (2 mg/ml), ACES (10 mg/ml), and potassium hydroxide (3.75 mg/ml), at pH 6.9. For determinations of the antimicrobial activity of metronidazole, dimetridazole, ronidazole, and tolfenamic acid, each chemical was added to the liquid media at increasing concentrations (1–500 μM). The optical density was determined at 600 nm every 24 h during 5 days.
DNA Manipulations and Gene Cloning
Standard methods were used for chromosomal DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation (Pagliai et al., 2010). Plasmids were isolated using the QIAprep® Spin Miniprep Kit (Qiagen, Valencia, CA), and PCR products were purified using Qiaquick® Purification Kits (Qiagen). All of the primers used in this study are described in Table 2. For protein expression and purification, the prbP (CLIBASIA_01510) gene was amplified by PCR, using total DNA extractions from L. asiaticus infected tissue, and cloned into the p15TV-L plasmid as described previously (Pagliai et al., 2010).
Two-Hybrid System
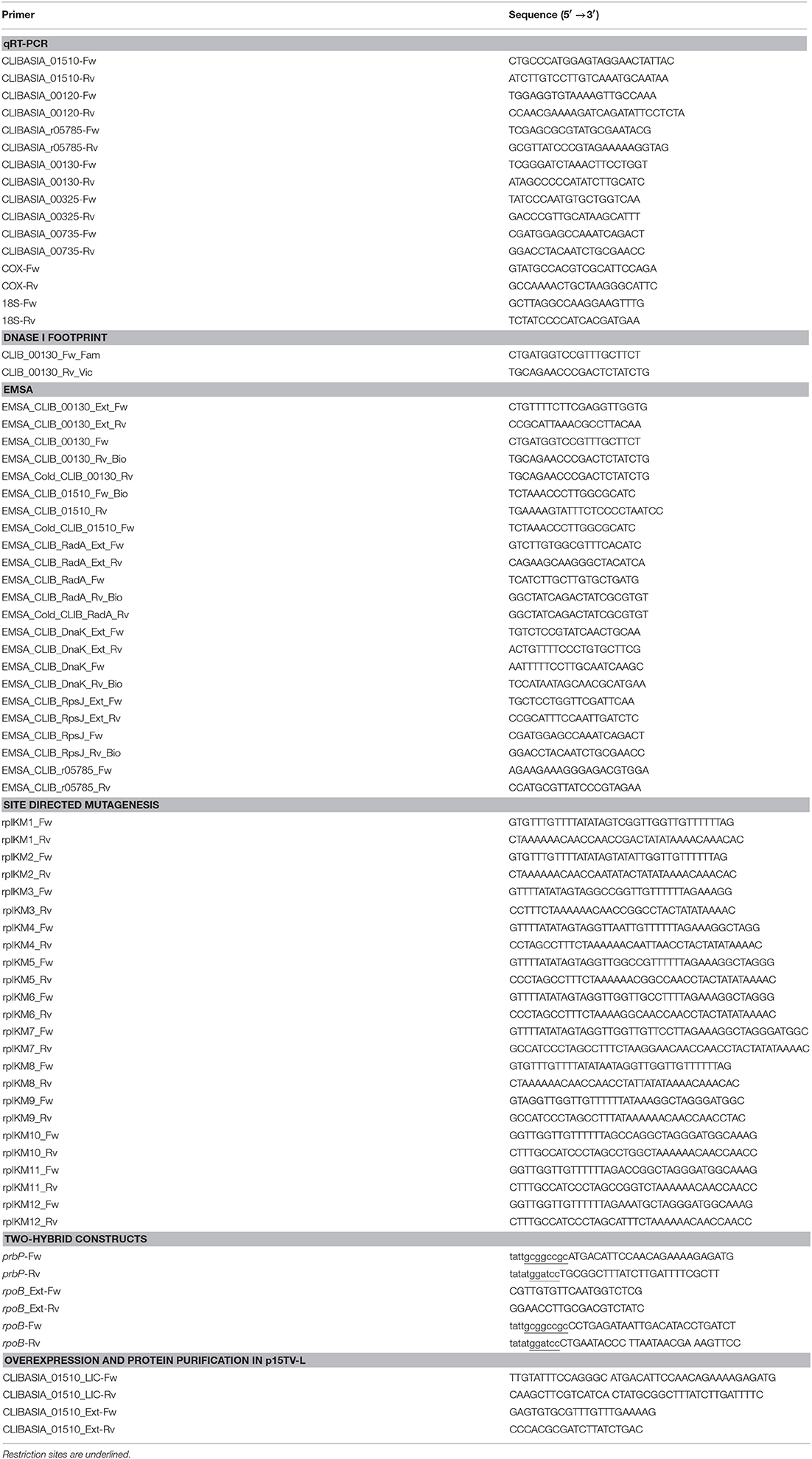
The two-hybrid system previously described by Borloo et al. (2007) was used (Borloo et al., 2007). Proteins of interest were fused to complementing β-galactosidase truncations (Δα and Δω), where the resulting level of complemented β-galactosidase activity corresponds directly to the level of interaction between the proteins. Proteins of interests were cloned into vectors pB2HΔα and pB2HΔω using the NotI and BamHI restriction sites. All subcloning steps were performed in E. coli XL-1 Blue (Stratagene, La Jolla, CA). Fusion proteins in pB2HΔα and pB2HΔω were transformed by heat shock and subsequently co-expressed in E. coli TOP10. Empty vectors were used as a control to determine baseline activity. The minimal inhibitory concentration (MIC) was determined for each chemical in E. coli (Table 3). The highest concentration that did not adversely affect the base levels in the controls was used for subsequent in vivo assays.
β-Galactosidase Assays
Escherichia coli cells were grown at 37°C in LB medium until reaching OD600 of 0.8 (late exponential phase). Cells were collected and lysed in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol; Miller, 1972). β-galactosidase activity was assayed by following the catalytic hydrolysis of chlorophenol red-β-D-galactopyranoside (Sigma-Aldrich). The absorbance at 570 nm was read continuously using a Synergy HT 96-well plate reader (BioTek, Winooski, VT). β-galactosidase activity, expressed as arbitrary units (AU), was calculated using the slope of absorbance curve normalized with the initial cell density. Preliminary β-galactosidase assays were performed with strains LB01 and LB02, in the absence and presence (0–1 mM) of each chemical to determine baseline activity. Each assay was performed in triplicates.
Protein Purification
Protein purification was performed as previously described (Lorca et al., 2007). Briefly, PrbP was cloned into vector p15TV-L. The His-tagged fusion protein was then overexpressed in E. coli BL21-Rosetta(DE3) (Novagen). Cells were grown in LB broth at 37°C, to an OD600 of 0.6. Expression was induced with 0.5 mM isopropyl -thio-β-D-galactopyranoside (IPTG). After induction, the cells were incubated at 17°C for 16 h. The cells were harvested and resuspended in binding buffer (500 mM NaCl, 5% glycerol, 50 mM HEPES, 5 mM imidazole, pH 7.5) with Roche EDTA-free protease inhibitor cocktail (Roche Applied Science, Germany). Phenylmethylsulfonyl fluoride (0.5 mM) and Tris(2-carboxyethyl)phosphine hydrochloride (0.5 mM) were added to the cells immediately before lysing. Cells were lysed using a french press. The lysates were clarified by centrifugation (30 min at 17,000 × g) and applied to a metal chelate affinity column charged with nickel. The column was washed extensively with binding buffer containing 20 mM imidazole, and the proteins were subsequently eluted from the column with elution buffer (binding buffer containing 250 mM imidazole). The purified proteins were dialyzed against 10 mM HEPES (pH 7.5), 500 mM NaCl, 2.5% glycerol, 0.5 mM TCEP, and stored at −80°C. The identity of the purified proteins was confirmed by Mass Spectrometry (as a service in the Interdisciplinary Center for Biotechnology Research, University of Florida) from protein bands isolated from SDS-PAGE gels.
Electrophoretic Mobility Shift Assays (EMSA)
Gel shift assays for PrbP were performed using aliquots of protein purified and concentrated according to the procedures described above. Fragments of the rplK, prbP, rpsJ, dnaK, and radA promoter regions were generated by PCR using pre-labeled 5′-biotin primers (Table 2). PCR products were purified using QIAquick spin columns (Qiagen). EMSA reaction mixtures (20 μL) contained 1 ng of 5′ biotin-labeled DNA probe, 12.5 ng/μL of both Poly(dI-dC) and Poly(dA-dT) nonspecific competitor DNAs, 10 mM Hepes pH 7.5, 150 mM NaCl, 5% glycerol, purified PrbP protein (0–7 μM), and ligand (0–2 mM)as indicated. Competition assays were carried out using unlabeled fragments of the promoter regions generated by PCR. After incubation at 37°C for 20 min, samples were separated on 6% acrylamide-bisacrylamide non-denaturing gels, in ice cold Tris borate-EDTA buffer, pH 8.3 (TBE). Electrophoresis was performed on ice at 100 V for 2 h. DNA was transferred to a Hybond-N+ membrane (GE Healthcare, Pittsburgh, PA) with a Semi-Dry electroblotter (Fisher Scientific, Pittsburgh, PA) at 250 mA for 45 min. Transferred DNA was cross-linked to the membrane using a Spectrolinker XL-1000 UV cross-linker equipped with 312 nm UV bulbs. Biotin labeled DNA was detected using a Phototope-Star Detection Kit (New England Biolabs, Ipswich, MA). Membranes were exposed to Kodak X-ray film.
DNase I Footprinting
Protection assays were performed on both minus and plus strands using 5′-6FAM or 5′-VIC labeled probes that were generated by PCR (primers described in Table 2). 1.5 μg of labeled PrplK probe was combined with 40 μM PrbP, 0.5 mM CaCl2, 2.5 mM MgCl2, and 0.025 U of DNase I (New England Biolabs) in a 200 μl reaction. After incubation at 37°C for 20 min, the reaction was stopped by the addition of 50 mM EDTA, pH 8.0. As a control, a digestion reaction was performed under the same conditions without PrbP. The digested DNA and the sequencing reaction products were analyzed at the Plant and Microbe Genomics facility, Ohio State University, Columbus, using a 3730 DNA analyzer. The protected regions were identified using GeneMapper software (Life Technology), as previously described (Zianni et al., 2006).
Small Molecule Screening by Differential Scanning Fluorimetry
Purified PrbP protein was screened against the Prestwick chemical library of 1152 compounds (Prestwick Chemical, France) using differential scanning fluorimetry as previously described (Vedadi et al., 2006; Niesen et al., 2007; Wrench et al., 2013). Briefly, purified PrbP protein was diluted to a final concentration of 20 μM in 100 mM HEPES, pH 7.5, 150 mM NaCl. 20 μl aliquots of a protein solution containing the chemical compounds were placed in duplicate, into 96-well plates (Bio-Rad) and heated from 25 to 80°C, at the rate of 1°C per min. A real time PCR device (iCycler IQ™, Bio-Rad) was used to monitor protein unfolding by an increase in the fluorescence of the fluorophore SYPRO Orange (Invitrogen). Fluorescence intensities were plotted against temperature for each sample well, and transition curves were fitted with the Boltzmann equation using Origin 8 software (Northampton, MA). The midpoint of each transition was calculated and compared with the midpoint calculated for the reference sample. If the difference between the midpoints was greater than 2.0°C, the corresponding compound was considered to be a “hit.”
Analyses of Chemicals on Infected Leaves
Leaves were collected from HLB-symptomatic Valencia Orange (C. sinensis) trees, maintained at the University of Florida main campus. All leaves used in this study were collected from new flushes on highly symptomatic branches. Prior to treatment all solutions were autoclaved or filter sterilized. 100 μM stocks of the chemical were prepared in 10 mM Tris pH 8.0. A solution of 10 mM Tris pH 8.0 was used for the controls. A scalpel was used to harvest leaves from the tree, with a horizontal cut at the base of the petiole. Each leaf was immediately suspended in 8 ml of treatment solution (with or without chemicals). Leaves were supported in a vertical position throughout the incubation period, with only the lower inch of the petiole submerged in solution (with or without chemical). Steady air flow was maintained over the leaf blades throughout the incubation period, to facilitate transpiration and the adsorption of each solution. Each treatment group consisted of 12 leaves that were processed after 24 h of incubation (with or without chemical).
For each treatment group, biological quadruplicates were prepared from the twelve leaves. The leaf midribs and petioles were collected and incubated overnight at 4°C in RNAlater solution (Life Technologies, Grand Island, NY). Following treatment with RNAlater, samples were rinsed twice with RNase free water and immediately frozen in liquid nitrogen. Samples were then freeze dried over a period of 3 days and homogenized using a GenoGrinder 2000. Homogenized samples were stored at −80°C.
RNA Purification and cDNA Synthesis
Plant and bacterial RNA was extracted from 75 mg of homogenized tissue. Extractions were carried out using the Isolate II RNA Plant Kit (Bioline, London, UK) with lysis buffer RLY. Zirconia beads (0.1 mm) were added to each sample during lysis, to aid with the disruption of bacterial cells. Purified RNA was eluted with 60 μl of RNase/DNase-free water, and subsequently treated with TURBO DNA-free DNase (Thermo Scientific, Wilmington, DE) to eliminate trace amounts of DNA. Purified RNA samples were quantified using a NanoDrop ND 1000 (Thermo Scientific, Wilmington, DE) and stored at −80°C. cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) with random hexamer primers and 0.5 μg of RNA. cDNA products were diluted 50% with DNase/RNase-free water and stored at −80°C.
Real-Time Quantitative PCR (qRT-PCR) Analysis
qRT-PCR was carried out in a iCycler IQ apparatus (Bio-Rad) using Platinum SYBR Green qPCR SuperMix for iCycler (Life Technologies) in accordance with the manufacturer's recommended protocol. Reactions were carried out using 2 μl of cDNA, in a total reaction volume of 26 μl. The genes measured for L. asiaticus included prbP (CLIBASIA_01510), L25 (CLIBASIA_01515), rplK (CLIBASIA_00130), rpsJ (CLIBASIA_00735), gyrA (CLIBASIA_00325), and 16S rRNA (CLIBASIA_r05785). The mRNA levels of rplK, CLIBASIA_r05785, rpsJ and prbP genes were normalized to the abundance of the plant genes cox2 and 18S rRNA. The expression of each plant control gene was previously examined in absence and presence of tolfenamic acid (data not shown). Quantitative reverse transcription-PCR primers are described in detail in Table 2.
Evaluation of Phytotoxicity on Sweet Orange Seedlings
Twelve-month old seedlings were randomly divided into ten groups of 8 seedlings. Tolfenamic acid was prepared in 10 mM tris buffer, and applied at concentrations of 1, 10, 100, 1000, and 10,000 μM by root soaking (with 100 ml), foliar spray (to saturation), or root soaking and foliar spray combined. A control group was also treated with 10 mM tris buffer only. Each treatment was applied twice at 2 week intervals. A non-treated control group was also maintained throughout the duration of the experiment. Seedlings were monitored for symptoms of phytotoxicity during a period of 12 months following treatment. No phytotoxic effects were observed following treatment with tolfenamic acid (up to 10 mM). Tolfenamic acid had no effect on the transcription of Cox2 or 18S rRNA (data not shown).
Evaluation of Antimicrobial Efficacy of Tolfenamic Acid against L. asiaticus
Citrus sinensis “Valencia” inoculation: Twelve-month old seedlings were graft inoculated with budwood collected from HLB-infected trees. Prior to grafting, the source of the infected tissue was analyzed by PCR to confirm the presence of viable L. asiaticus. PCR confirmation was carried out using primers for 16S rRNA (CLIBASIA_r05781) and gyrA (CLIBASIA_00325) genes (see Table 2 for primer sequences). Inoculated plants were kept in a secure greenhouse approved by the USDA Animal and Plant Health Inspection Service (APHIS). The plants were watered in accordance with the standard watering schedule for the commercial citrus industry. Citrus fertilizer (Sunniland Citrus 6-4-6, N-P-K) was applied every 2 months as instructed by the manufacturer. Two months after grafting, each plant was tested for HLB by PCR as described above.
Seedlings treatment study: After 9 months of testing positive for HLB, the highly symptomatic, infected sweet orange seedlings (described above) were randomly divided into two groups. 100 μM tolfenamic acid (TA) was applied to one group of infected seedlings as a foliar spray (to saturation), and by root soaking (with 100 ml of 100 μM TA). The buffer vehicle (10 mM tris) was applied to the control group as a foliar spray (to saturation), and by root soaking (with 100 ml of 10 mM tris). Each treatment was applied twice, at 2 week intervals. Evaluation of HLB symptoms (leaf yellowing and mottling) and L. asiaticus transcriptional activity (as a measure of viability) was performed as described above.
Statistical Analyses
qRT-PCR statistical significance was assessed using a two-tail P-value, calculated with the Mann–Whitney nonparametric test.
Results
PrbP Binds Specifically to the Promoter Region of rplK
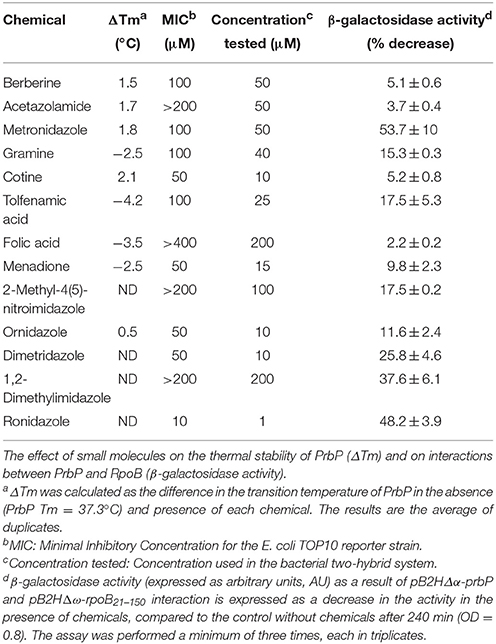
Previous studies in PrbP homologs have found that these proteins may bind non-specifically to the promoter regions of the rrnA, and rpsH genes (Gulten and Sacchettini, 2013; Srivastava et al., 2013). In this study, we examined the DNA binding properties of PrbP in L. asiaticus, using the promoter region of genes encoding the ribosomal proteins rplK (CLIBASIA_00130; PrplK) and rpsJ (CLIBASIA_00735; PrpsJ), as well as the promoter regions of 16S rRNA (CLIBASIA_r05785; P16S), prbP (CLIBASIA_01510; PprbP), dnaK (CLIBASIA_02620; PdnaK), and radA (CLIBASIA_01095; PradA). We found that PrbP was able to bind PrplK at 3.5 μM and P16S at 5 μM, while binding to PrpsJ, PprbP, PdnaK, and PradA was only observed with higher concentrations (10–15 μM) of PrbP (Figure 1). Competition assays were used to confirm the specificity of the PrbP:PrplK interaction, where a 10-fold excess of unlabeled PrplK was found to completely out-compete the labeled PrplK fragment (Figure 2). At higher concentrations (100–400-fold excess), PdnaK was found to compete with the labeled PrplK fragment, but to a lesser extent than observed with unlabeled PrplK. The addition of excess (up to 400-fold) unlabeled PradA or unlabeled PprbP had no effect (Figure 2). These results indicate that PrbP binds to a specific sequence in the promoter region of rplK, with higher affinity. DNase I footprinting was used to identify the DNA binding site for PrbP in the promoter region of rplK (Figure 3A). The 24 nucleotide protected site (GTAGGTTGGTTGTTTTTTAGAAAG), is located 31 bp from the translational start codon, on the plus strand. The critical DNA contact residues were identified by EMSA analysis, following site directed mutagenesis of the rplK promoter region (Figures 3B,C). Of the twelve mutant EMSA probes tested, only rplK-M1, rplK-M4, rplK-M5, rplK-M10, and rplK-M11 resulted in decreased binding to PrbP (Figure 3C). These results indicate that nnAnnnnGGTTnnnnnnnnnAAAn is the PrbP recognition sequence for this promoter. These results confirm that L. asiaticus PrbP interacts with a specific sequence in the promoter region.
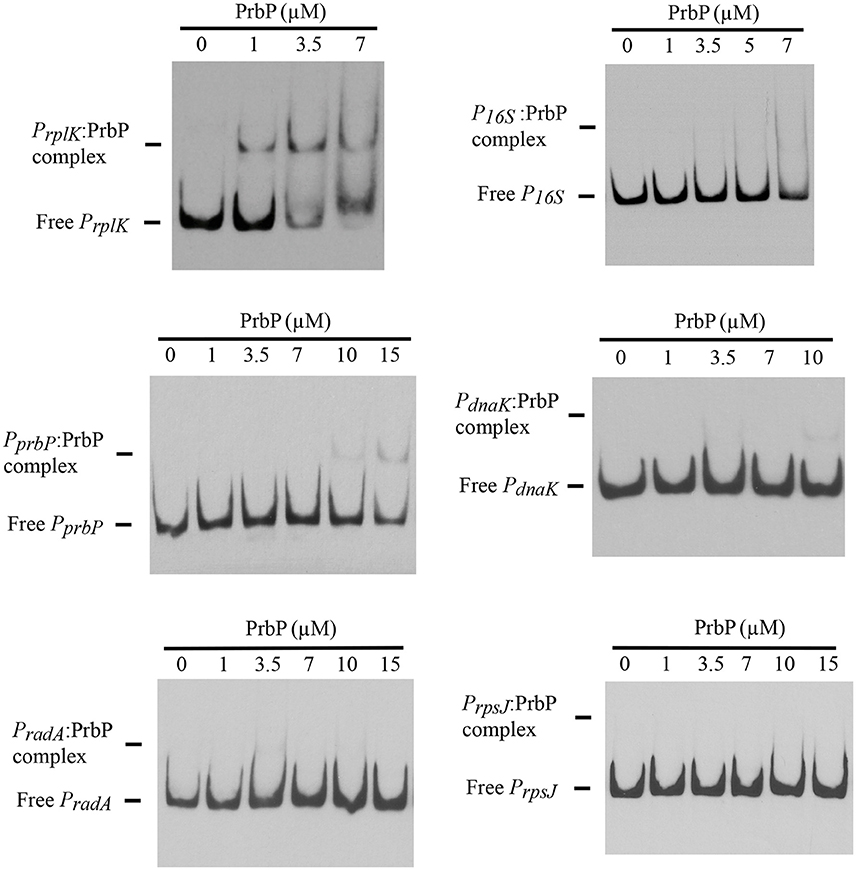
Figure 1. DNA binding assays with PrbP. EMSAs were conducted with 1 ng of biotin labeled probes PrplK, P16S, PrpsJ, PprbP, PdnaK, or PradA and increasing concentrations (0–15 μM) of PrbP, as indicated on top of each panel. No protein was added to the first lane of each gel.
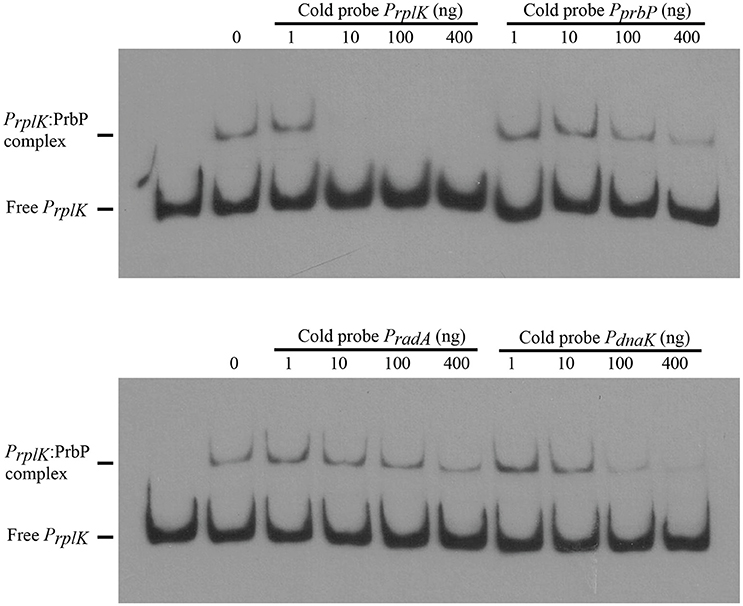
Figure 2. PrbP binds specifically to rplK promoter region. For competition experiments, a biotin labeled PrplK probe was incubated with 2.5 μM PrbP and mixed with increasing concentrations (1–400 ng) of an unlabeled, double stranded probe (PrplK, PprbP, PradA, or PdnaK), as indicated above each panel.
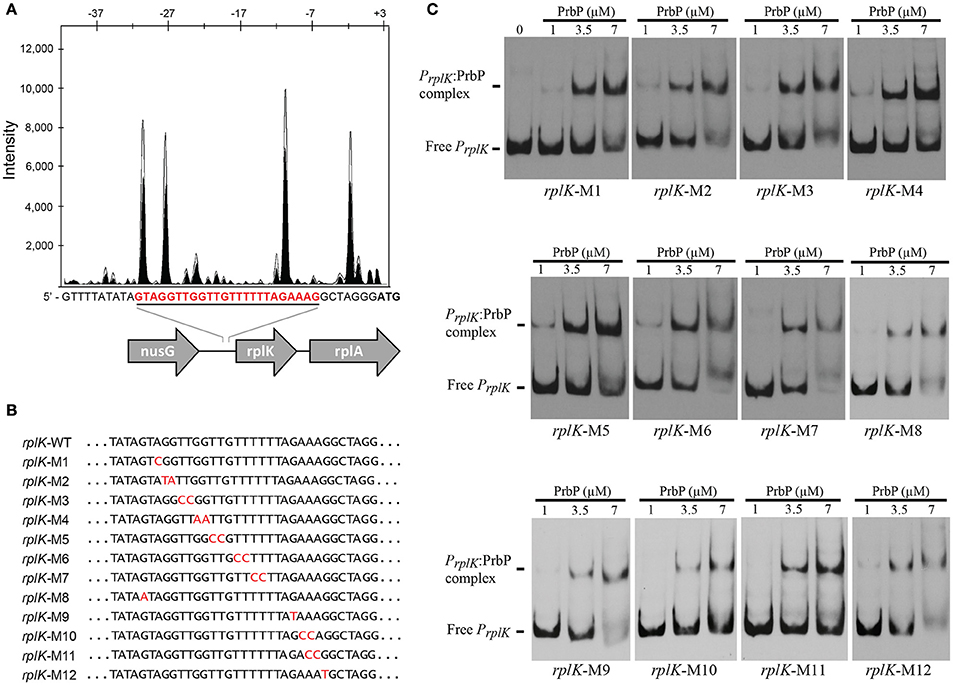
Figure 3. Identification of PrbP binding site in rplK promotor (PrplK). (A) DNAse I footprint assays identified a protected site (red) located 31 bp from the rplK translation start site. The electropherogram shows a fragment of the digested probe in absence (white) or presence (black) of PrbP, highlighting the protected region. (B) Site directed mutagenesis of the rplK promoter region. Mutated residues are indicated (in red) for each of the rplK mutants (rplK-M1–rplK-M12). (C) EMSA assays were conducted using 1 ng of each mutant PrplK probe (rplK-M1–rplK-M12) with increasing concentrations (0–7 μM) of PrbP, as indicated on top of each panel. The amount of free PrplK was used to determine the binding efficiency of PrbP with each mutant probe.
Liberibacter asiaticus PrbP Interacts with the β-Subunit of the RNA Polymerase
A bacterial two-hybrid system was used to investigate potential interactions between PrbP and the β-subunit of the RNAP. Plasmids pB2HΔα and pB2HΔω were used to create fusions of the L. asiaticus genes CLIBASIA_01510 (prbP) and CLIBASIA_0011021–150 (rpoB, residues 21–150), to the β-galactosidase truncations Δα and Δω as described by Borloo et al. (2007). Protein-protein interactions were followed by β-galactosidase activity. High levels of β-galactosidase activity were observed in strains LB02 (carrying pB2HΔα-prbP and pB2HΔω-rpoB21–150) and LB03 (carrying pB2HΔα-rpoB21–150 and pB2HΔω-prbP) (5823 ± 345 AU and 3252 ± 217AU, respectively), when compared to the control strains (Figure 4). These results confirm that L. asiaticus PrbP interacts with the RNAP.
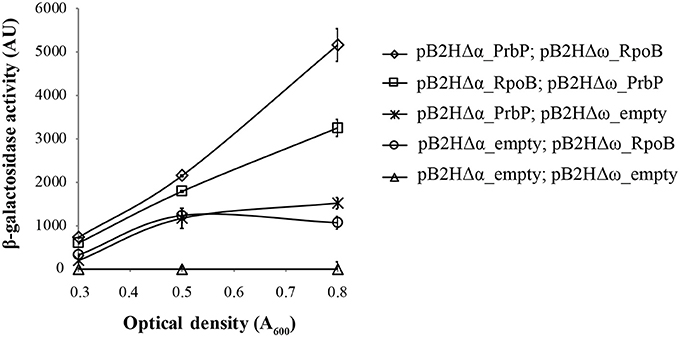
Figure 4. Analysis of PrbP interactions with RpoB. β-galactosidase activity was performed using E. coli TOP10 derivatives as reporter strains, transformed with the following plasmid combinations: LB01 (Δ) carrying the empty pB2HΔα and pB2HΔω plasmids; LB02 (♢) carrying pB2HΔα_prbP and pB2HΔω_rpoB21–150); LB03 (□) carrying pB2HΔα_rpoB21–150 and pB2HΔω_prbP; LB04 ( ) carrying pB2HΔα_prbP and the empty pB2HΔω; and LB05 (O) carrying the empty pB2HΔα and pB2HΔω_rpoB21–150. Assays of β-galactosidase activity are expressed in arbitrary units (AU), and were performed at different growth phases (as indicated by optical density, A600), in triplicates. The β-galactosidase activity for each strain was quantified after subtraction of the baseline activity.
) carrying pB2HΔα_prbP and the empty pB2HΔω; and LB05 (O) carrying the empty pB2HΔα and pB2HΔω_rpoB21–150. Assays of β-galactosidase activity are expressed in arbitrary units (AU), and were performed at different growth phases (as indicated by optical density, A600), in triplicates. The β-galactosidase activity for each strain was quantified after subtraction of the baseline activity.
Identification of Small Molecules That Bind PrbP
Our approach to study the biological role of PrbP was to identify small molecules that bind and/or interact with the protein to regulate its activity. To this end, the L. asiaticus prbP gene was cloned into vector p15TV-L, and subsequently overexpressed in E. coli BL21. The purified protein was screened against the Prestwick chemical library of small molecules, by differential scanning fluorimetry (Vedadi et al., 2006; Pagliai et al., 2010; Wrench et al., 2013). The midpoint transition temperature of PrbP was determined to be 37.3 ± 0.5°C. From the 1200 small molecules examined in the screening, 8 compounds were found to induce a shift in the midpoint transition temperature (ΔTm) of PrbP. Metronidazole, cotine, gramine, menadione, folic acid, and tolfenamic acid were found to have the strongest effect, inducing a Tm shift of 1.8°, 2.1°, −2.5°, −2.5°, −3.5°, and −4.5°C, respectively (Table 3). Berberine chloride and acetazolamide were found to interact with PrbP to a lesser degree, with a ΔTm of 1.5° and 1.7°C, respectively.
Small Molecules Modulate PrbP Interactions with the RNA Polymerase
A bacterial two-hybrid system was used to further investigate the interactions between PrbP and RNAP, in the absence and presence of each chemical. The MIC was determined for each chemical in E. coli (Table 3). Preliminary β-galactosidase assays were performed using strains LB01 and LB02, in absence and presence (1 μM−1 mM) of each chemical to determine baseline activity. The highest concentration that did not adversely affect the base levels in the controls was used for subsequent in vivo assays (Table 3). Metronidazole showed the strongest effect, decreasing PrbP/RpoB interactions by 53.7%, while berberine, acetazolamide, gramine, cotine, menadione, folic acid, and tolfenamic acid had a lesser effect, decreasing the interaction by 5.1, 3.7, 15.3, 5.2, 9.8, 2.2, and 17.5%, respectively (Table 3).
Consequently, we identified five compounds with chemical scaffolds similar to metronidazole (dimetridazole, 2-methyl-4(5)-nitroimidazole, 1,2-dimethylimidazol, ornidazole, and ronidazole) and their effect was tested on the interaction between PrbP and RpoB. The inhibition values obtained were low for ornidazole (11.6%), 2-methyl-4(5)-nitroimidazole (17.5%), and dimetridazole (25.8%). When tested at higher concentrations (200 μM), 1,2-dimethylimidazole was found to decrease PrbP/RpoB interactions by 37.6%, however, no effect was observed at lower concentrations (<50 μM). Conversely, a lower concentration (1 μM) of ronidazole was found to significantly decrease (48%) PrbP/RpoB interactions (Table 3).
Small Molecules Disrupt Interactions between PrbP and DNA
EMSA were performed with PrbP in the presence and absence of the small molecules identified by differential scanning fluorimetry. The promoter region of rplK (PrplK) was used as the target DNA in all assays. Of the compounds tested, tolfenamic acid was the only effective inhibitor of interactions between PrbP and PrplK (Figure 5A). Complete disruption of the PrbP:PrplK complex was observed with 350 μM tolfenamic acid (Figure 5B). Metronidazole, ronidazole, acetazolamide, gramine, cotine, menadione, and folic acid had no effect on the PrbP:PrplK complex when present at concentrations up to 2 mM.
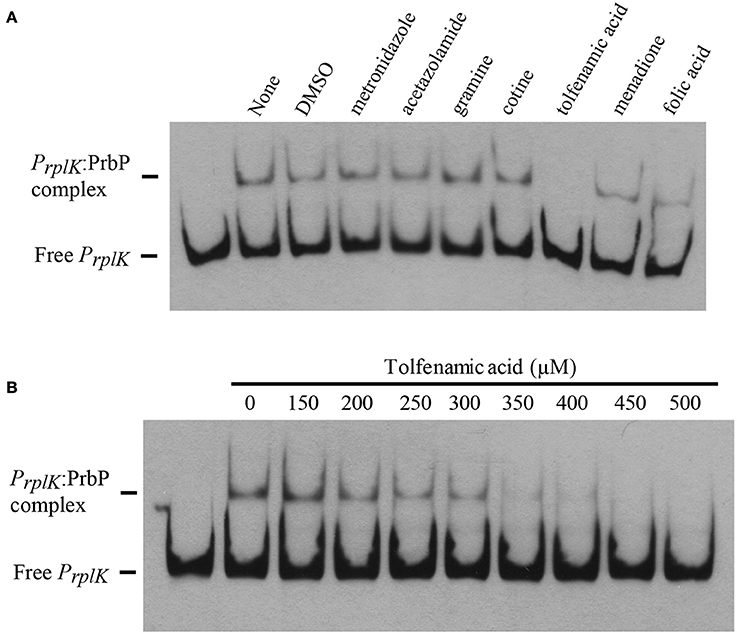
Figure 5. Small molecules decrease PrbP binding to PrplK. (A) EMSAs were conducted with the biotin labeled probe Prplk (1 ng) and PrbP (2.5 μM), in the absence (lane 2; None) and presence of 2 mM metronidazole, acetazolamide, gramine, cotine, tolfenamic acid, menadione, or folic acid. DMSO (5% final concentration) was ran as a solvent control (lane 3). No protein was added to the first lane. (B) EMSAs were conducted with probe PrplK (1 ng) and increasing concentrations (0–500 μM) of tolfenamic acid, as indicated on top of the panel. PrbP was maintained at 2.5 μM. No protein was added to the first lane.
Small Molecules as Therapeutics against L. asiaticus
As propagation of L. asiaticus still remains elusive under laboratory conditions, we used the culturable close relative L. crescens to determine the antimicrobial activity of metronidazole, dimetridazole, ronidazole, and tolfenamic acid. It was found that metronidazole, dimetridazole and ronidazole were not inhibitory at concentrations up to 500 μM. When combined with the results of the bacterial two hybrid system, these results suggest that interactions between PrbP and RNAP may not be critical for the persistence of L. crescens under the conditions tested. Conversely, growth inhibition of L. crescens was observed in presence of tolfenamic acid (70 μM), indicating interactions between PrbP and DNA are indeed essential for the survival of L. crescens (Figure 6).
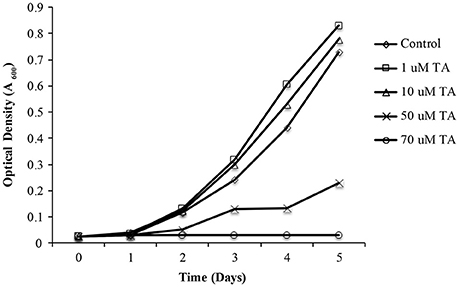
Figure 6. The effect of tolfenamic acid on Liberibacter crescens growth. L. crescens BT-1 was cultured at 25°C with moderate aeration (150 RPM), in modified BM7 media, with increasing concentrations (0–70 μM) of tolfenamic acid as indicated: (♢) Control without TA; (□) 1 μM TA; (Δ) 10 μM TA; (X) 50 μM TA; (O) 70 μM TA. Bacterial growth was determined by optical density at 600 nm (A600), over a period of 5 days.
An infected leaf assay was used to assess the efficacy of tolfenamic acid against L. asiaticus, in vitro (Pagliai et al., 2014). Leaves were collected from Valencia Orange (C. sinensis) trees infected with L. asiaticus. Upon collection, leaves were immediately immersed in 1, 10, or 100 μM tolfenamic acid. Following incubation, each sample was analyzed by qRT-PCR to determine the relative change in gene expression for 16S rRNA, DNA gyrase subunit A, L10 ribosomal, PrbP and RplK protein (encoded by CLIBASIA_r05785, gyrA, rplJ, prbP, and rplK, respectively) as viability parameters for L. asiaticus (Pagliai et al., 2014). Due to the variability in bacterial load, samples were normalized to the plant gene cox2 and 18S rRNA. Samples incubated with 10 or 100 μM tolfenamic acid showed a significant (p < 0.05) decrease in the expression of all five genes (Figure 7). These results suggest that tolfenamic acid may be inhibitory to L. asiaticus at 10 and 100 μM.
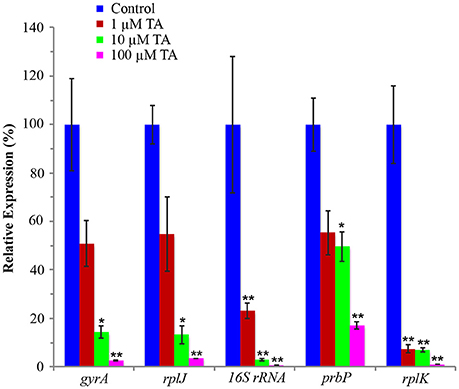
Figure 7. Tolfenamic acid modulates the activity of PrbP in vivo. The effect of tolfenamic acid on the transcriptional activity of L. asiaticus. The expression levels of gyrA, rplJ, 16S RNA, prbP and rplK were assessed after 24 h. Tolfenamic acid (TA) was tested at increasing concentrations (0–100 μM) as indicated: (red) 1 μM TA, (green) 10 μM TA, (magenta) 100 μM TA. Control samples (blue) were treated with buffer only. (*p < 0.05; **p < 0.005). The plant gene cox2 was used to normalize the expression values between samples.
The addition of 1 μM tolfenamic acid did not have a significant effect on the expression levels of gyrA, prbP, or rplJ, however, a significant (p < 0.005) decrease in expression was observed for rplK and 16S rRNA (93 and 77% reduction, respectively). These results indicate that PrbP is a transcriptional activator for rplK and the 16S ribosomal genes in L. asiaticus. Additionally, these results indicate that expression of prbP is not auto-regulated at the level of transcription, in a manner dependent on DNA binding.
Use of Tolfenamic Acid as Antimicrobial in HLB-Infected Citrus Seedlings
Prior to testing the efficacy of tolfenamic acid in L. asiaticus infected citrus trees, a phytotoxicity assessment was performed using healthy 12 month-old citrus seedlings. Following 6 months of treatment with tolfenamic acid (1, 10, and 100 μM), no phytotoxic effects were observed in any of the treatment groups. The efficacy of tolfenamic acid was subsequently determined in L. asiaticus-infected citrus seedlings (Citrus sinensis, “Valencia”). Seedlings were infected with L. asiaticus (via grafting) and maintained in a greenhouse for 12 months to allow the infection to spread throughout the entire plant. Each plant had symptoms of advanced L. asiaticus infection and the presence of L. asiaticus was confirmed by PCR prior to beginning treatments with tolfenamic acid (Figures 8, 9).

Figure 8. Tolfenamic acid is an effective antimicrobial compound against L. asiaticus in HLB-infected citrus seedlings. Recovery of root and canopy tissue in L. asiaticus-infected seedling TA-2 (upper panel) after treatment with 100 μM tolfenamic acid. The canopy and root tissue was photographed before treatment with tolfenamic acid, 2 weeks after treatment, 2 months after treatment, and 11 months after treatment. L. asiaticus-infected seedlings TA-1, TA-3, and TA-4 are shown in the lower panel. Photographs were taken before treatment began and 11 months after treatment.

Figure 9. Photographs of control seedlings (n = 4) treated with buffer vehicle (10 mM Tris, 0.1% DMSO). Seedlings were photographed before treatment began, and post treatment (for up to 11 months).
Tolfenamic acid (100 μM) was administered to infected seedlings via root soaking and as a foliar spray (to saturation). Two applications were made over a period of 2 weeks. Four infected seedlings were also maintained under the same conditions as a control. Each plant was examined and photographed every 2 weeks to monitor the canopy and root tissue for signs of recovery or disease progression. New root tissue was observed in several of the treated plants 2 weeks after treatment. Two months after treatment, healthy new flush growth began to push on several of the treated plants, and continued to grow over the next 11 months with no visible signs of infection (Figure 8). Eleven months after treatments were applied, three of the four plants treated with tolfenamic acid showed clear signs of recovery in both root and canopy tissues (Figure 8). In contrast, in the control group treated with the buffer vehicle only, three out of the four plants died (Figure 9). In the treated plants, qRT-PCR analysis of root tissue revealed the absence of L. asiaticus infection in plants TA-1, TA-2, and TA-3 while plant TA-4 showed only a 10% reduction in the expression of rplJ (Figure 10). A significant drop (80–95% reduction) in L. asiaticus titer was also observed in the canopy tissue of 75% of the treated plants (Figure 10).
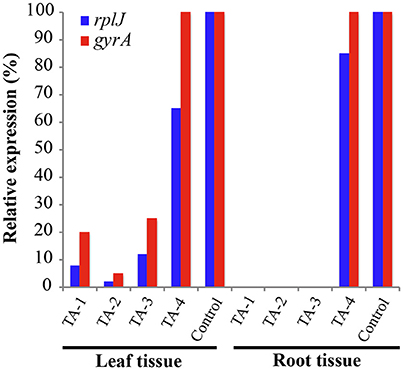
Figure 10. Gene expression in leaf and root tissue 11 months after treatment with tolfenamic acid. L. asiaticus mRNA levels for rplJ (blue) and gyrA (red) were determined in fresh tissue collected from treated seedlings (TA-1, TA-2, TA-3, and TA-4) and compared to the untreated controls. The plant gene cox2 was used to normalize the expression values between samples. The lower mRNA expression levels observed in treated plants are indicative of reduced L. asiaticus infection. No amplification was observed in root samples from plants TA-1, TA-2, and TA-3.
Discussion
Due to the extended timeframe and financial obligations associated with the development and approval of new therapeutic compounds, drug repurposing has become an appealing approach to drug discovery; the availability of pharmacokinetic and pharmacodynamic data can significantly reduce research costs, and expedite the approval process. For these reasons, we elected to use a library of FDA approved compounds as a starting point for screening protein targets. The absence of putative pathogenicity determinants in L. asiaticus, prompted the investigation of unconventional proteins as targets for the development of therapeutic strategies against this devastating pathogen. The in silico analyses revealed a small number of simple regulatory mechanisms in the L. asiaticus genome, which enable its survival during the changing growth conditions encountered in two remarkably different hosts, the psyllid and the citrus plant. As a consequence, the transcriptional regulation of several metabolic pathways must be achieved with only a few transcription factors. Based on these observations, the inactivation of a transcription factor may result in pleiotropic effects, influencing the ability for L. asiaticus to survive within the citrus host.
CarD, a low identity homolog to PrbP, is an essential RNA polymerase binding protein in Mycobacterium species. It is involved in pathogenesis, persistence, cell viability, and resistance to both antibiotics and stress (Stallings et al., 2009; Srivastava et al., 2013). CarD interacts with both the β-subunit of the RNA polymerase and, non-specifically, with DNA sequences in the promoter region of several genes (Srivastava et al., 2013). In L. asiaticus, PrbP also showed interaction with RNAP and DNA. We also found that PrbP was capable of differentially binding the rplK promoter. The putative binding site in the rplK promoter is located 31 bp upstream from the translation start site. This binding location is in agreement with the role of PrbP as a transcriptional activator that may function to stabilize the open promotor complex.
The transcription of rRNA is one of the rate-limiting steps controlling cell growth under ideal and stress conditions; during this process, formation of stable RNAP/open-promotor complexes is the most time consuming step (Zhou and Jin, 1998; Bartlett et al., 2000; Jin et al., 2012; Häkkinen et al., 2013). In Mycobacterium, CarD was recently found to increase the formation of RNAP/open promoter complexes, and stabilize RNAP open complexes by preventing transcription bubble collapse (Davis et al., 2015; Rammohan et al., 2015). Point mutations in CarD that weaken interactions with RNAP are detrimental in M. tuberculosis, resulting in reduced viability and increased sensitivity to antibiotics and oxidative stress (Weiss et al., 2012). As such, we hypothesized that the inactivation/inhibition of PrbP would cause a decrease in transcript production in L. asiaticus, potentially resulting in decreased viability and persistence within the host.
While several compounds were found to modulate interactions between PrbP and RNA polymerase, tolfenamic acid was the only small molecule that decreased interactions between PrbP and DNA (Figure 5). The effect of these chemicals was also tested in vivo, where tolfenamic acid was found to be the only small molecule that affected the overall transcriptional activity of L. crescens. The results suggest that the ligand specificity for disruption of the PrbP:PrplK complex is more stringent than the binding specificity for ligands that disrupt interactions between PrbP and RNAP.
To confirm the effect of tolfenamic acid in planta, tolfenamic acid was applied by root soaking and foliar spray to L. asiaticus infected citrus seedlings in a greenhouse setting. Each seedling was confirmed to harbor viable, L. asiaticus cells for at least 6 months prior to beginning treatment. In addition to testing positive for L. asiaticus, each plant also displayed severe signs of infection, including blotchy mottle, yellowing shoots, and severe damage to the root system. After the initial treatment, the first sign of recovery in treated seedlings was the development of new root growth. Rapid recovery of the root system was observed in 75% of seedlings that were treated with tolfenamic acid (Figures 8, 10). Although several plants showed improved root growth in as little as 14 days after treatment, the canopy tissue was slower to recover (5–11 months before substantial amounts of new growth was observed).
The extent of vascular damage caused by the presence of L. asiaticus may affect the rate that compounds (chemical treatments) are distributed throughout the canopy, and thus the rate at which each plant recovers. Previous studies have shown that HLB significantly affects the vascular flow and exchange of nutrients between root and canopy tissue (Etxeberria et al., 2009; Koh et al., 2012). Factors contributing to the reduction in vascular flow include the accumulation of starch within the xylem and phloem, the blockage of sieve elements by callose formation and bacterial cell debris, and compartmentalization as the tree attempts to isolate the pathogen (Kim et al., 2009; Koh et al., 2012; Aritua et al., 2013). During our analysis of tissue samples collected from HLB-infected seedlings, the highest expression levels of L. asiaticus genes (post-treatment) was observed in tissue collected from underdeveloped regions of the canopy, where leaf growth was stagnate. Since the static growth of these branches is indicative of severely obstructed vascular flow, it is possible that the transport of small molecules (from the injection site) to these regions of the tree was insufficient to deliver enough chemical to completely inhibit the growth of L. asiaticus. As such, the treatment of highly infected citrus trees may require repeat injections or a combined application method that will facilitate even distribution throughout the canopy tissue (such as a foliar spray with adjuvants). Combined treatments with small molecules and thermotherapy may also be a viable option, as more efficient methods of heat treatment are developed.
There are very few antibiotics approved for use in the treatment of plant diseases, and of those, only streptomycin and tetracycline have shown minimal success in the treatment of HLB, with tetracycline having phytotoxic effects at the concentrations required for treatment of L. asiaticus. The combined use of penicillin and streptomycin showed a suppressive effect on L. asiaticus (Zhang et al., 2011), however, the use of β-lactam antibiotics is highly regulated and not approved for use in agriculture. As such, the use of small molecules is a promising alternative to combat L. asiaticus.
In this study, we were able to confirm that tolfenamic acid is an effective inhibitor of L. asiaticus in planta. While additional studies are needed to fully understand the recovery process of treated trees, these encouraging results indicate the potential for tolfenamic acid to be used as a systemic antimicrobial therapy for HLB.
Author Contributions
CLG, FP, LP, LB, and MT conducted the experiments and analyzed the results. CLG, GL, and CFG conceived the idea for the project, analyzed the results, and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
A patent application has been submitted for the use of tolfenamic acid for the treatment of HLB.
Acknowledgments
We are grateful to Dr. Svetlana Folimonova for providing infected seedlings. We would also like to thank Jain Irrigation Inc. for donating the greenhouse irrigation supplies for this project. This work was supported in whole or part by the Citrus Research and Development Foundation, Inc. http://citrusrdf.org/ (award number 00080214 to CFG) and the National Institute of Food and Agriculture, U.S. Department of Agriculture http://nifa.usda.gov/ (award number 2015-70016-23029 to GL and CFG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Abbreviations
EMSA, electrophoretic mobility shift assay.
References
Aritua, V., Achor, D., Gmitter, F. G., Albrigo, G., and Wang, N. (2013). Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus Liberibacter asiaticus infection. PLoS ONE 8:e73742. doi: 10.1371/journal.pone.0073742
Bartlett, M. S., Gaal, T., Ross, W., and Gourse, R. L. (2000). Regulation of rRNA transcription is remarkably robust: FIS compensates for altered nucleoside triphosphate sensing by mutant RNA polymerases at Escherichia coli rrn P1 promoters. J. Bacteriol. 182, 1969–1977. doi: 10.1128/JB.182.7.1969-1977.2000
Borloo, J., De Smet, L., Vergauwen, B., Van Beeumen, J. J., and Devreese, B. (2007). A beta-galactosidase-based bacterial two-hybrid system to assess protein-protein interactions in the correct cellular environment. J. Proteome Res. 6, 2587–2595. doi: 10.1021/pr070037j
Bove, J. M., Genomique, D. R., Pathogene, P., Recherche, C., De Bordeaux, I., and Araraquara, P. (2006). Huanglongbing : a destructive, newly-emerging, century old disease of citrus. J. Plant Pathol. 88, 7–37. doi: 10.4454/jpp.v88i1.828
Davis, E., Chen, J., Leon, K., Darst, S. A., and Campbell, E. A. (2015). Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res. 43, 433–445. doi: 10.1093/nar/gku1231
Duan, Y., Zhou, L., Hall, D. G., Li, W., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of citrus huanglongbing bacterium, “Candidatus Liberibacter asiaticus” obtained through metagenomics. Mol. Plant Microbe Interact. 22, 1011–1020. doi: 10.1094/MPMI-22-8-1011
Etxeberria, E., Gonzalez, P., Achor, D., and Albrigo, G. (2009). Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol. Mol. Plant Pathol. 74, 76–83. doi: 10.1016/j.pmpp.2009.09.004
García-Moreno, D., Abellón-Ruiz, J., García-Heras, F., Murillo, F. J., Padmanabhan, S., and Elías-Arnanz, M. (2010). CdnL, a member of the large CarD-like family of bacterial proteins, is vital for Myxococcus xanthus and differs functionally from the global transcriptional regulator CarD. Nucleic Acids Res. 38, 4586–4598. doi: 10.1093/nar/gkq214
Gottwald, T. R. (2010). Current epidemiological Understanding on citrus Huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009
Gulten, G., and Sacchettini, J. C. (2013). Structure of the Mtb CarD/RNAP β-lobes complex reveals the molecular basis of interaction and presents a distinct DNA-binding domain for Mtb CarD. Structure 21, 1859–1869. doi: 10.1016/j.str.2013.08.014
Häkkinen, A., Tran, H., Yli-Harja, O., and Ribeiro, A. S. (2013). Effects of rate-limiting steps in transcription initiation on genetic filter motifs. PLoS ONE 8:e70439. doi: 10.1371/journal.pone.0070439
Jin, D. J., Cagliero, C., and Zhou, Y. N. (2012). Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 36, 269–287. doi: 10.1111/j.1574-6976.2011.00279.x.Growth
Kim, J.-S., Sagaram, U. S., Burns, J. K., Li, J.-L., and Wang, N. (2009). Response of sweet orange (Citrus sinensis) to “Candidatus Liberibacter asiaticus” infection: microscopy and microarray analyses. Phytopathology 99, 50–57. doi: 10.1094/PHYTO-99-1-0050
Koh, E. J., Zhou, L., Williams, D. S., Park, J., Ding, N., Duan, Y. P., et al. (2012). Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacter asiaticus.” Protoplasma 249, 687–697. doi: 10.1007/s00709-011-0312-3
Leonard, M. T., Fagen, J. R., Davis-Richardson, A. G., Davis, M. J., and Triplett, E. W. (2012). Complete genome sequence of Liberibacter crescens BT-1. Stand. Genomic Sci. 7, 271–283. doi: 10.4056/sigs.3326772
Lorca, G. L., Ezersky, A., Lunin, V. V., Walker, J. R., Altamentova, S., Evdokimova, E., et al. (2007). Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J. Biol. Chem. 282, 16476–16491. doi: 10.1074/jbc.M610838200
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Springs Harbor, NY. Cold Spring Harbor Laboratory Press.
Niesen, F. H., Berglund, H., and Vedadi, M. (2007). The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protocols 2, 2212–2221. doi: 10.1038/nprot.2007.321
Pagliai, F. A., Gardner, C. L., Bojilova, L., Sarnegrim, A., Tamayo, C., Potts, A. H., et al. (2014). The transcriptional activator LdtR from “Candidatus Liberibacter asiaticus” mediates osmotic stress tolerance. PLoS Pathogens 10:e1004101. doi: 10.1371/journal.ppat.1004101
Pagliai, F. A., Gardner, C. L., Pande, S. G., and Lorca, G. L. (2010). LVIS553 transcriptional regulator specifically recognizes novobiocin as an effector molecule. J. Biol. Chem. 285, 16921–16930. doi: 10.1074/jbc.M110.111138
Rammohan, J., Ruiz Manzano, A., Garner, A. L., Stallings, C. L., and Galburt, E. A. (2015). CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 43, 3272–3285. doi: 10.1093/nar/gkv078
Srivastava, D. B., Leon, K., Osmundson, J., Garner, A. L., Weiss, L. A., Westblade, L. F., et al. (2013). Structure and function of CarD, an essential mycobacterial transcription factor. Proc. Natl. Acad. Sci. U.S.A. 110, 12619–12624. doi: 10.1073/pnas.1308270110
Stallings, C. L., and Glickman, M. S. (2011). CarD: a new RNA polymerase modulator in mycobacteria. Transcription 2, 15–18. doi: 10.4161/trns.2.1.13628
Stallings, C. L., Stephanou, N. C., Chu, L., Hochschild, A., Nickels, B. E., and Glickman, M. S. (2009). CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138, 146–159. doi: 10.1016/j.cell.2009.04.041
Vedadi, M., Niesen, F. H., Allali-Hassani, A., Fedorov, O. Y., Finerty, P. J., Wasney, G., et al. (2006). Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. U.S.A. 103, 15835–15340. doi: 10.1073/pnas.0605224103
Weiss, L., A, Harrison, P. G., Nickels, B. E., Glickman, M. S., Campbell, E. A., Darst, S. A., et al. (2012). Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J. Bacteriol. 194, 5621–5631. doi: 10.1128/JB.00879-12
Wrench, A. P., Gardner, C. L., Gonzalez, C. F., and Lorca, G. L. (2013). Identification of a small molecule that modifies MglA/SspA interaction and impairs intramacrophage survival of Francisella tularensis. PLoS ONE 8:e54498. doi: 10.1371/journal.pone.0054498
Zhang, M., Powell, C., A, Zhou, L., He, Z., Stover, E., and Duan, Y. (2011). Chemical compounds effective against the citrus Huanglongbing bacterium “Candidatus Liberibacter asiaticus” in planta. Phytopathology 101, 1097–1103. doi: 10.1094/PHYTO-09-10-0262
Zhou, Y. N., and Jin, D. J. (1998). The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 2908–2913. doi: 10.1073/pnas.95.6.2908
Keywords: transcriptional accessory protein, Liberibacter asiaticus, tolfenamic acid, antimicrobial, citrus
Citation: Gardner CL, Pagliai FA, Pan L, Bojilova L, Torino MI, Lorca GL and Gonzalez CF (2016) Drug Repurposing: Tolfenamic Acid Inactivates PrbP, a Transcriptional Accessory Protein in Liberibacter asiaticus. Front. Microbiol. 7:1630. doi: 10.3389/fmicb.2016.01630
Received: 03 August 2016; Accepted: 30 September 2016;
Published: 18 October 2016.
Edited by:
Abelardo Margolles, Spanish National Research Council, SpainReviewed by:
Vishvanath Tiwari, Central University of Rajasthan, IndiaHongxia Wang, University of Alabama at Birmingham, USA
Copyright © 2016 Gardner, Pagliai, Pan, Bojilova, Torino, Lorca and Gonzalez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Graciela L. Lorca, Z2xvcmNhQHVmbC5lZHU=
Claudio F. Gonzalez, Y2Znb256YWxlekB1ZmwuZWR1
 Christopher L. Gardner
Christopher L. Gardner Fernando A. Pagliai
Fernando A. Pagliai Lei Pan
Lei Pan Lora Bojilova
Lora Bojilova Maria I. Torino
Maria I. Torino Graciela L. Lorca
Graciela L. Lorca Claudio F. Gonzalez
Claudio F. Gonzalez