- 1UCIBIO/REQUIMTE, Laboratório de Microbiologia, Departamento de Ciências Biológicas, Faculdade Farmácia, Universidade do Porto, Porto, Portugal
- 2Servicio de Microbiología, Instituto Ramón y Cajal de Investigación Sanitaria, Madrid, Spain
- 3Consorcio de Investigación Biomédica en Red de Epidemiología y Salud Pública, Barcelona, Spain
- 4Faculdade de Ciências da Saúde, Universidade Fernando Pessoa, Porto, Portugal
- 5Division of Microbial Diseases, UCL Eastman Dental Institute, University College London, London, UK
- 6Unidad de Resistencia a Antibióticos y Virulencia Bacteriana (RYC-CSIC), Madrid, Spain
Ampicillin resistance has greatly contributed to the recent dramatic increase of a cluster of human adapted Enterococcus faecium lineages (ST17, ST18, and ST78) in hospital-based infections. Changes in the chromosomal pbp5 gene have been associated with different levels of ampicillin susceptibility, leading to protein variants (designated as PBP5 C-types to keep the nomenclature used in previous works) with diverse degrees of reduction in penicillin affinity. Our goal was to use a comparative genomics approach to evaluate the relationship between the diversity of PBP5 among E. faecium isolates of different phylogenomic groups as well as to assess the pbp5 transferability among isolates of disparate clonal lineages. The analyses of 78 selected E. faecium strains as well as published E. faecium genomes, suggested that the diversity of pbp5 mirrors the phylogenomic diversification of E. faecium. The presence of identical PBP5 C-types as well as similar pbp5 genetic environments in different E. faecium lineages and clones from quite different geographical and environmental origin was also documented and would indicate their horizontal gene transfer among E. faecium populations. This was supported by experimental assays showing transfer of large (≈180–280 kb) chromosomal genetic platforms containing pbp5 alleles, ponA (transglycosilase) and other metabolic and adaptive features, from E. faecium donor isolates to suitable E. faecium recipient strains. Mutation profile analysis of PBP5 from available genomes and strains from this study suggests that the spread of PBP5 C-types might have occurred even in the absence of a significant ampicillin resistance phenotype. In summary, genetic platforms containing pbp5 sequences were stably maintained in particular E. faecium lineages, but were also able to be transferred among E. faecium clones of different origins, emphasizing the growing risk of further spread of ampicillin resistance in this nosocomial pathogen.
Introduction
Infections caused by Enterococcus faecium have increasingly been reported since the early 1980s’, as have the number of antibiotic resistant isolates of this species (Top et al., 2007). Currently, most clinical strains of E. faecium are ampicillin resistant (AmpR), which often also acquire transferable mobile genetic elements, some encoding vancomycin resistance (Arias and Murray, 2012; Wagenvoort et al., 2015). Multidrug resistant E. faecium isolates are responsible for infections associated with treatment failures and high mortality rates (Arias et al., 2010; Centers for Disease Control and Prevention, 2013). However, the influence of ampicillin resistance in the population structure of E. faecium remains largely unexplored.
Enterococcus faecium is intrinsically resistant to cephalos porins and exhibit natural reduced susceptibility to penicillins. This species has six penicillin binding proteins (PBPs) belonging either to class B (monofunctional D,D-transpeptidases), or class A (bifunctional enzymes with glycosyltransferase and D,D-transpeptidase activity), some of them being associated with resistance to β-lactams (Zorzi et al., 1996; Rice et al., 2009). In E. faecium resistance to β-lactam antibiotics is conferred by the low-affinity class B PBP5, that requires the participation of class A PBPs (PonA, PbpF) to synthesize the cell wall in the presence of cephalosporins (Williamson et al., 1983; Rice et al., 2009). Resistance to high ampicillin concentrations in E. faecium was initially explained by either the enhanced production of PBP5, and/or by polymorphisms in the beta subunit of this protein (Fontana et al., 1996). Soon after, it was demonstrated that such changes were frequently strain specific and did not necessarily correlate with differences in the ampicillin MIC values (Fontana et al., 1996; Zorzi et al., 1996; Rybkine et al., 1998; Rice et al., 2001, 2004; Sifaoui et al., 2001; Belhaj et al., 2016). Further analysis of E. faecium strains with different levels of ampicillin susceptibility (0,5- > 128 mg/L) revealed that the variability of PBP5 sequences is mostly due to changes in 21 specific positions of the protein, suggesting that a sequential acquisition of mutations could have contributed to the progressive resistance to ampicillin from the early 1980s (Galloway-Peña et al., 2011; Pietta et al., 2014). Mutations in genes encoding other species-specific proteins that participate in the cell wall synthesis, such as D,D-carboxypeptidases (Ddcp and DdcY), L,D-transpeptidases (Ldtfm), glycosyltransferases (PgtA) and acetylmuramoyl-L-alanine amidase (LytG) may also slightly increase the MIC values (Rice et al., 2007; Zhang et al., 2012; Kristich et al., 2014), even in the absence of PBP5 (Sacco et al., 2014). Although the occurrence of β-lactamases has been documented in E. faecium, β-lactamases producing E. faecium remain rare (Coudron et al., 1992; Klare et al., 1992; Zhang et al., 2012; Hendrickx et al., 2013).
Enterococcus faecium population biology is dominated by two main phylogenomic groups, clade A and clade B. Most AmpR isolates belong to clade A or “hospital-associated clade” mainly comprising E. faecium from hospitalized patients (Galloway-Peña et al., 2012; Palmer et al., 2012; Lebreton et al., 2013). A subgroup within the clade A, clade A1, is enriched in mobile genetic elements and have enhanced ability to colonize and persist in human hosts due to the presence of adhesins and specific metabolic traits. In contrast, the clade B, or “community-associated E. faecium,” mostly comprises ampicillin susceptible (AmpS) isolates from healthy, non-hospitalized individuals (Willems et al., 2005; Lebreton et al., 2013; Tedim et al., 2015; Freitas et al., 2016; Guzman Prieto et al., 2016). The increasing detection of AmpR among isolates from hosts not associated with the hospital setting (Gonçalves et al., 2011; de Regt et al., 2012; Novais et al., 2013; Santos et al., 2013; Tremblay et al., 2013) is of concern, as the acquisition of transferable genes encoding AmpR might facilitate its further spread into AmpS E. faecium populations or to other less frequently recovered enterococcal species for which AmpR has been rarely documented (Raze et al., 1998). A relevant observation was the identification of a transferable chromosomal region of 60 kb comprising pbp5 and a transposon that confers resistance to glycopeptides (vanB2-CTn5386) (Carias et al., 1998; Rice et al., 2005b). Homologous recombination between similar chromosomal regions carrying pbp5 of different strains potentially associated with a conjugation mechanism has been recently suggested (García-Solache et al., 2016).
The known diversity of genotypes of AmpR E. faecium is mainly focused on the PBP5 polymorphisms analyzed in a limited number of strains (Poeta et al., 2007; Klibi et al., 2008; Galloway-Peña et al., 2011). Similarly, knowledge related to the pbp5 transferability is limited to the characterization of conjugative Tn916-like elements (CTn5382 or the interaction of CTn916 and CTn5386) in a few E. faecium strains (Carias et al., 1998; Rice et al., 2005a,b; García-Solache et al., 2016). Here, we extend previous knowledge on these issues by using a comparative genomics approach of the chromosomal regions containing pbp5, that demonstrate a relationship between the diversity of PBP5 and E. faecium phylogenomic groups. We also assess the pbp5 transferability among isolates from different clonal lineages.
Materials and Methods
Bacterial Strains
Seventy-eight isolates from a collection of 205 AmpR E. faecium (MIC ≥ 16 mg/L) recovered in Portugal from the last decades were analyzed (Novais et al., 2005a,b,c, 2006, 2013). The 78 E. faecium were selected in order to include isolates with different antibiotic resistant phenotypes, from diverse origins (39 from different patients admitted in five hospitals from different cities, 18 from swine feces and piggeries, 4 from retail poultry carcasses, 2 from healthy human feces and 15 from hospital wastewater), isolation date and, whenever possible, clonal relatedness. Susceptibility to ampicillin and 10 other antibiotics of different classes was evaluated by disk diffusion and/or agar dilution method (Clinical and Laboratory Standards Institute, 2012). β-lactamase production was tested in AmpR E. faecium isolates by the nitrocefin test (5 μl were directly placed in bacteria growing around the ampicillin disk) and PCR amplification of blaZ using primers based on the GenBank sequence no. M25257.1 (blaZF-3′-TTGCCTATGCTTCGACTTCA-5′, blaZR- 3′-AGTGAAACCGCCAAGAGTGT-5′). Clonal relatedness was established by Pulsed-Field Gel Electrophoresis (PFGE) and analysis of Multilocus Sequence Typing (MLST) data (Tenover et al., 1995; Homan et al., 2002; Freitas et al., 2013)1 using the Bayesian Analysis of Population Structure (BAPS) algorithm (Willems et al., 2012; Tedim et al., 2015).
Transferability of Ampicillin Resistance
Filter-mating assays were performed in plain Brain Heart Infusion (BHI) not supplemented with antibiotics at 37°C overnight, using a donor/recipient ratio of 1:1 (1-3 experiments per isolate) and E. faecium GE1 as the recipient strain. Further filter mating assays under the same experimental conditions but using E. faecium BM4105RF and E. faecium 64/3 strains as recipients, were performed for those field isolates able to transfer pbp5 to E. faecium GE1. The three laboratory recipient strains used differed in the susceptibility against ampicillin and the presence of pbp5: GE1 [Δpbp5; MICAmp < 0.016mg/L; tetracycline (Tet), rifampicin (Rif) and fusidic acid (Fus) resistant (R); ST515/BAPS 2.3b], BM4105RF (pbp5; RifR, FusR; MICAmp = 0.5mg/L; ST172/BAPS 1.3) and 64/3 (pbp5; RifR, FusR; MICAmp = 1mg/L; ST21/BAPS 2.3a). Recipients acquiring pbp5 are from this point named as transconjugants, based on the most likely transfer mechanism related to pbp5 (conjugation) published during the revision of this article (García-Solache et al., 2016). Transconjugants were selected on BHI agar supplemented with antibiotics (ampicillin-10 mg/L, fusidic acid-25 mg/L; rifampicin-30 mg/L) and incubated for 24 up to 96 h (37°C) to recover potential transconjugants growing at different times. Transconjugants were confirmed by comparison of their antibiotic resistance phenotype and PFGE profile with those of the donors and the recipients. Stability of acquired pbp5 chromosomal regions (from now on designated pbp5 genetic platforms) after serial daily passages (x30) on antibiotic free BHI agar was evaluated in both donor strains and transconjugants. Colonies from each passage were inoculated on plates containing the same agar medium and tested for ampicillin susceptibility by disk diffusion (Clinical and Laboratory Standards Institute, 2012).
Characterization of the Region Conferring AmpR
Total DNA from a donor strain (AmpR E. faecium clinical isolate HPH2), a recipient strain (AmpS E. faecium GE1) and the resulting GE1 transconjugant (AmpR E. faecium TCGEHPH2.1) was extracted with PureEluteTM Bacterial Genomic Kit (EdgeBio, Gaithersburg, MD, USA). DNA concentration was measured with Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA) and Qubit 2.0 fluorometer (Life Technologies). Genomic DNA (250–350 ng) was sequenced on the Illumina MiSeq system using the MiSeq reagent kit v3 and a read length configuration of 2 × 300 bp (Illumina, San Diego, CA, USA). Sequencing was carried out using a standard 2 × 71 base protocol (300–400 bp insert size) in a Genome Analyzer IIx (Illumina, San Diego, CA, USA) at the sequencing facility of the University of Newcastle (United Kingdom). The main statistics for the three sequence datasets (number of reads and coverage) analyzed are shown in Supplementary Table S1. Assembly of sequence data was done using Newbler software (454 life sciences, Roche, Branford, CT, USA). Supplementary Table S2 shows the final assembly results.
Gene prediction was performed using GeneMark.hmm 3.05 (Besemer et al., 2001). Similarity searches for potential protein-coding regions were carried out against a UniRef100 database2 using Best Blast Hit approach (Supplementary Table S3). The sequence of the transferred region containing the pbp5 gene was predicted by a gene-by-gene comparison strategy using blastn (genes present in transconjugant strain TCGEHPH2.1 and donor strain HPH2 but not in recipient strain GE1) (Supplementary Table S4). Sequence of the putative “transferred region” was mapped against the genome of the E. faecium strain DO (RefSeq Accession: NC_017960) using Easyfig 1.2 software (Sullivan et al., 2011). In addition to E. faecium DO, the “transferred region” was also aligned against fully sequenced and closed genomes of E. faecium Aus0004, E. faecium NRRL B-2354 and E. faecium Aus0085 strains (RefSeq accessions numbers: NC_017022, NC_020207 and NC_021994, respectively) with Mauve (Darling et al., 2004). Functional annotation of the query sequences was performed using Blast2GO and Uniprot and KEGG databases3
Analysis of the pbp5 Genetic Environment
An 8–10 kb DNA fragment comprising the pbp5 gene and its boundaries was characterized by PCR assays in 15 transconjugants and 21 field isolates (Table 1). PCR conditions were adapted according to the expected amplicon size (<3 kb/>3 kb): 0.5/1 mM of each primer, 2/2.5 mM MgCl2, 1x of reaction buffer, 0.2/0.4 mM of each deoxynucleoside triphosphate, 1,25U GoTaq® Flexi DNA Polymerase [Promega Corporation, Madison, WI, USA]/2.5U Takara LA Taq polymerase (TakaraTM Bio Inc., Shiga, Japan). The amplification program was 25 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C; 1 cycle 10 min at 72°C (for fragments <3 kb) or 35 cycles of 30 s at 96°C, 1 min at 55°C, 7 min at 72°C; 1 cycle 10 min at 72°C (for fragments >3 kb). Amplicons were further discriminated by comparing the corresponding RFLP patterns obtained after digestion with DdeI or ApaI and sequences. Genomic location of pbp5 was identified by hybridization of I-CeuI and SmaI-digested genomic DNA using 23S rDNA and/or pbp5 probes (Liu et al., 1993; Freitas et al., 2013). Labeling and detection were carried out with Gene Images Alkphos direct labeling system kit following the manufacturer’s instructions (Amersham GB/GE Healthcare Life Sciences UK limited). The presence of CTn916, CTn5386 and CTn5382, previously associated with transferable AmpR, was determined by analyzing the presence of specific sequences (integrases, excisionases, non-integrase left region) (Carias et al., 1998; León-Sampedro et al., 2016). The transferred 8–10 kb genetic regions carrying pbp5 characterized in this study were compared with those of all E. faecium available genomes at the National Center for Biotechnology Information (NCBI) database (Supplementary Table S5).
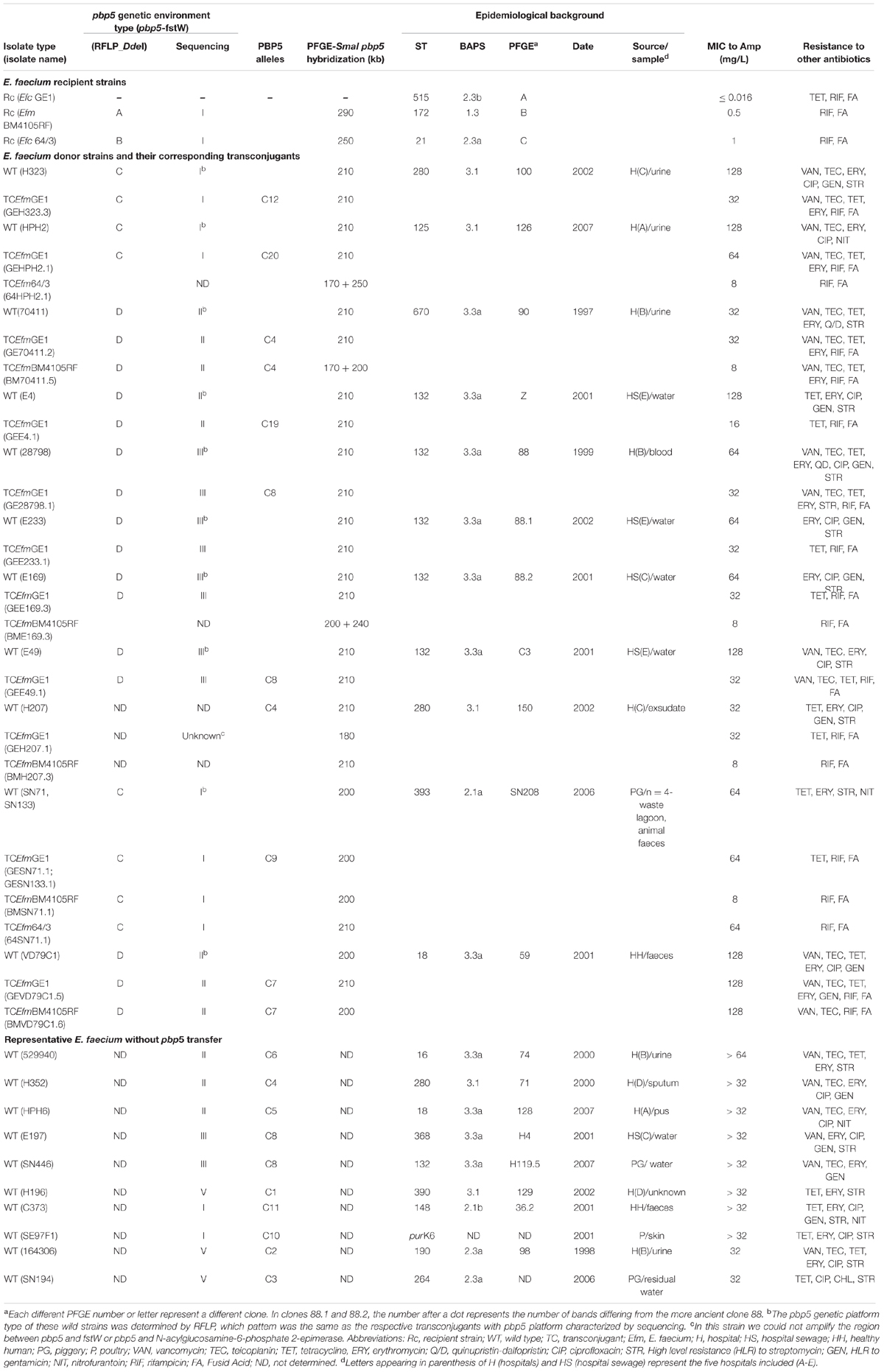
TABLE 1. Epidemiological background, pbp5 gene characterization and antibiotic resistance among E. faecium strains and transconjugants from this study.
Phylogenetic Analysis of PBP5
Comparative analysis of all PBP5 protein sequences available in the GenBank database with those identified in this study was performed using the basic local alignment sequence tool (BLAST) (Altschul et al., 1997) and the ClustalW2 program for multiple sequence alignment (Larkin et al., 2007; Goujon et al., 2010), hosted by the National Center for Biotechnology Information (NCBI) and the European Bioinformatic Centre (EBI), respectively. The PBP5 amino acid sequences were designated by a “C” followed by a number in agreement with the nomenclature used in previous works (Supplementary Table S6) (Galloway-Peña et al., 2011). Multiple sequence alignments of all PBP5 sequences were performed through MEGA 7 software4 and the topology of the phylogenetic tree was inferred by maximum-likelihood algorithm using PhyML (Guindon et al., 2010) with bootstrap analysis based in 1000 permutations and a cut-off of ≥70%.
Phylogenetic congruence of core genomes and pbp5 sequences was established by comparative analysis of their gene tree topologies. Core genomes for the 233 E. faecium strains whose draft genome sequences were available at NCBI Trace Database (last updated on February 2014), were reconstructed independently using an all-against-all reciprocal BLAST approach. Briefly, each CDS was used as a BLAST query against a local database of CDS from all genomes in the sample analyzed. Afterward, we re-annotated the chromosome sequences with GeneMarkS v2.8 (Besemer et al., 2001). The core genome was defined as the non-redundant genes present in all strains using CD-HIT (Li and Godzik, 2006) with the 80% coverage, 80% identity cut-offs, and parsed by homemade Perl script. All core genome genes were concatenated and aligned using MAFFT (Katoh and Toh, 2010). The phylogenetic tree was built by FastTree2 (Price et al., 2010). Tree comparison was carried out by cophyloplot command of APE (Analyses of Phylogenetics and Evolution) (Paradis et al., 2004) using R software.
Results
Transferability of AmpR (pbp5)
Twelve isolates (n = 12/78; 15%; Table 1) were able to transfer under our experimental conditions a chromosomal genetic platform containing pbp5 (associated with AmpR) to E. faecium strain GE1. Five of them were also able to transfer pbp5 to E. faecium BM4105RF and two, to E. faecium 64/3. All donor strains belonged to major E. faecium human lineages mostly associated with clinical isolates causing hospital infections, namely BAPS subgroups 3.3a (ST670, ST132, ST280), 3.1 (ST280, ST125), and 2.1a (ST393). They were obtained from samples of hospitalized humans but also from samples of hospital sewage, healthy humans and piggeries. β-lactamase production was not detected in any of the isolates.
All transconjugants had similar PFGE profiles to recipient strains (Figure 1), were resistant to rifampicin and fusidic acid and exhibited a variable susceptibility to ampicillin (MICs = 8–128 mg/L), which was often lower than that of their corresponding donor isolates (8 mg/L or 16 vs. 32 mg/L to >256 mg/L). Besides the AmpR phenotype, some transconjugants also exhibited resistance to vancomycin (n = 8), teicoplanin (n = 8), erythromycin (n = 6), tetracycline (n = 1; in non-GE1 transconjugants; as GE1 strain is tetracycline-resistant), streptomycin (n = 1) and/or gentamicin (n = 1). Transconjugants carrying the pbp5 genetic platform either alone or with plasmids harboring genes encoding resistance to different antibiotics were recovered from selection plates supplemented with ampicillin and not with other antibiotics (data not shown).
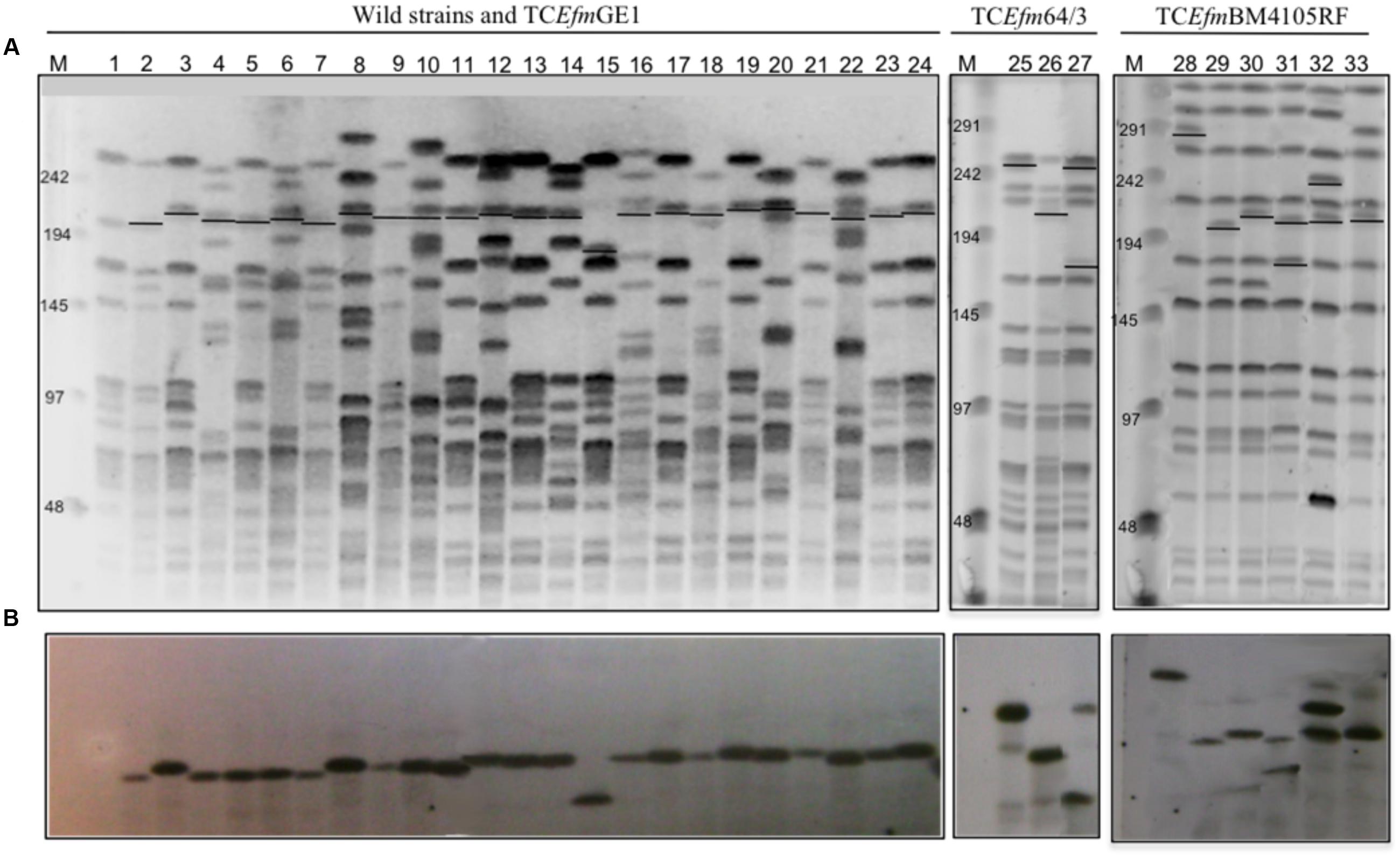
FIGURE 1. Clonality and hybridization assays with pbp5 probes of wild type, recipients and transconjugants strains. (A) PFGE SmaI digested DNA of recipient strains (1-E. faecium GE1, 25-E. faecium 64/3, 28-E. faecium BM4105RF), wild type (2-VD79C1, 4-SN71, 6-SN133, 8-HPH2, 10-H323, 12-70411, 14-H207, 16-E49, 18-E233, 20-E169, 22-E4) and transconjugants (3-GEVD79C1.5, 5-GESN71.1, 7-GESN133.1, 9-GEHPH2.1, 11-GEH323.3, 13-GE70411.2, 15-GE207.1, 17-GEE49.1, 19-GEE233.1, 21-GEE169.3, 23-GE28798.1, 24-GEE4.1, 26- 64SN71.1, 27- 64HPH2.1, 29- BMSN71.1, 30- BMH207.3, 31- BM70411.5, 32- BME169.3, 33- BMVD79C1.6). (B) Hybridization assays with a pbp5 probe (primers P1 and P2-Figure 4). M- Low Range PFGE Marker, kb (New England, BioLabs). Abbreviations: TCEfm- Transconjugant E. faecium.
The pbp5 Gene of AmpR Isolates Is Located on Large and Transferable Chromosomal Genetic Platforms Containing Metabolic Traits
Hybridization of pbp5 and 23S rDNA probes with a digested I-CeuI DNA band of high MW revealed a chromosomal location of pbp5. Further hybridization of SmaI-digested genomic DNA with the pbp5 probe showed hybridization to fragments of ∼210 kb in all but one GE1 transconjugants, for which the pbp5 probe hybridized to a band of ∼180 kb (Figure 1). Hybridization of the same probe with fragments of SmaI-digested genomic DNA of E. faecium BM4105RF and E. faecium 64/3 transconjugants of different sizes reflects independent transfer events. Similarity in the PFGE patterns of the transconjugants obtained using different donors indicates that DNA acquisition occurs within a particular region of the genome (Figure 1). The donor strains lack integrases/excisionases of Tn916-like conjugative transposons (Tn916, Tn5386, Tn5382).
For whole genome sequence a representative donor:recipient pair and its transconjugant were analyzed. The donor was the clinical HPH2 strain; the recipient, E. faecium GE1 which did not originally carry pbp5, and thus limiting the possibilities of recombination events between the acquired and natural pbp5 genetic platform in the recipient strain; and the size of the SmaI digested genomic DNA fragment, the one that most commonly hybridizing with the pbp5 probe (∼210 kb). Sequencing of the recipient (E. faecium GE1), the donor (E. faecium HPH2) and the transconjugant (E. faecium TCGEHPH2.1) allowed us to identify 7 contigs with genes present in the donor and its transconjugant but absent in the recipient strain. The fragments which comprised the pbp5 gene, represent a genetic platform of approximately 280 kb that is slightly larger than that inferred from the SmaI-PFGE gels. A more detailed analysis of the contigs revealed that two of them (contig00158 and to a lesser extent contig00068) also contained genes present in the recipient and transconjugants but not in the donor strain, which could be explained either by the partial transfer of the pbp5 genetic platform and/or post-transfer recombination events (Figure 2). The comparative analysis of the genomes sequenced in this work with the four closed E. faecium genomes available at the NCBI database at the time of writing this manuscript [DO (NC_017960.1), NRRL B-2354 (CP004063.1), Aus0004 (NC_017022.1), Aus0085 (NC_021994.1) strains], revealed a common region of 153kb for our donor/transconjugant strains and the four genomes analyzed, which was absent in the recipient E. faecium GE1 genome (Figure 3). The remaining sequence of the 280 kb “transferred chromosomal region” was variably present and located in different genomes (Figure 3).
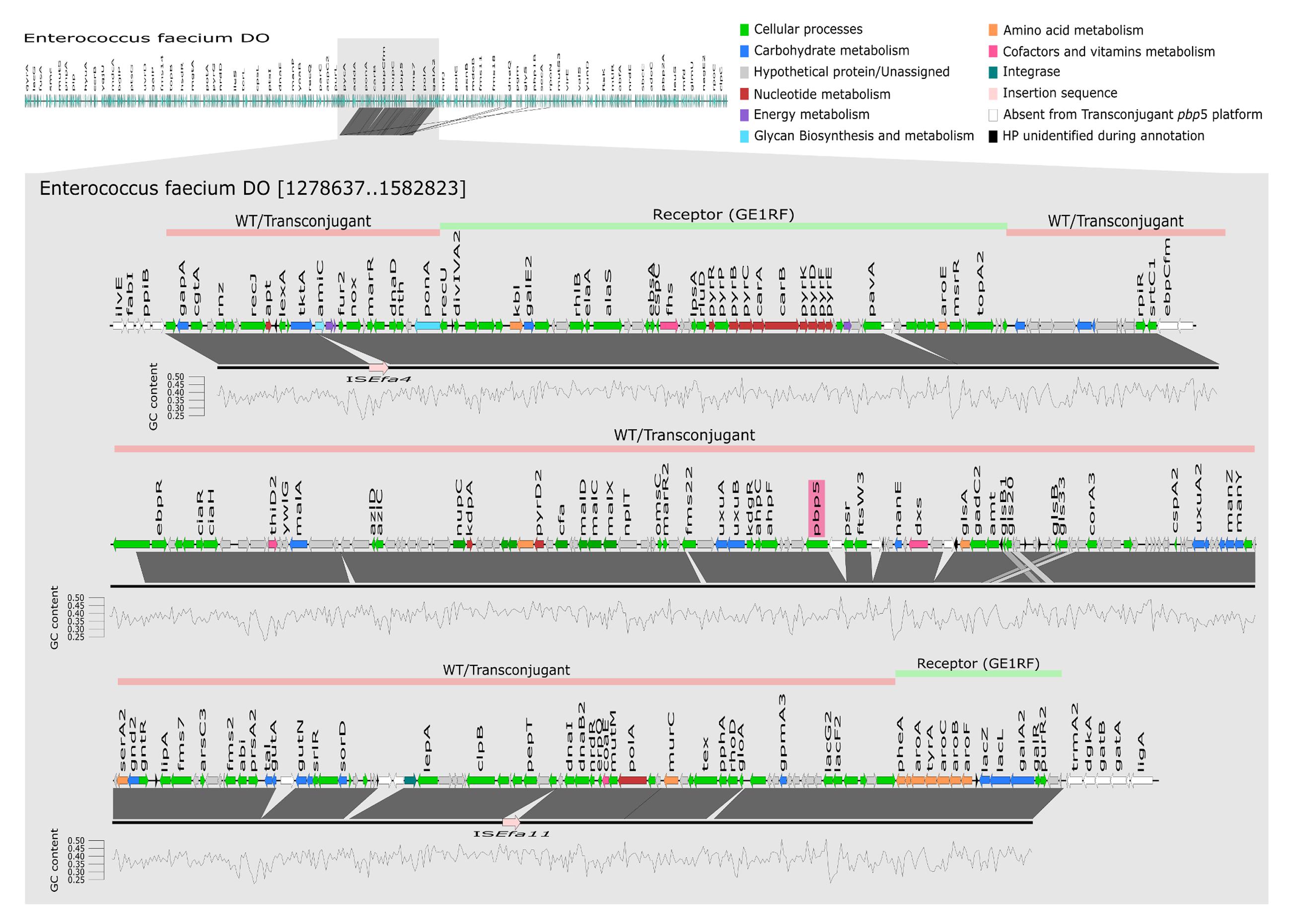
FIGURE 2. Representation of the transferable chromosomal genetic platform containing pbp5. Mapping and annotation (using KEGG database) of transferable pbp5 genetic platform of transconjugant TCGEHPH2.1 (represented by black line) using E. faecium DO as reference genome. Lines above E. faecium DO genetic structure represent the transferable pbp5 genetic platform observed in wild type and transconjugant isolates (pink) and genomic regions of E. faecium GE1 recipient and transconjugant (green). GC content was calculated using seqinr in Rstudio. The window used to calculate the GC content was 200 bp, represented in the figure by each vertices of the graph. WT, wild type.
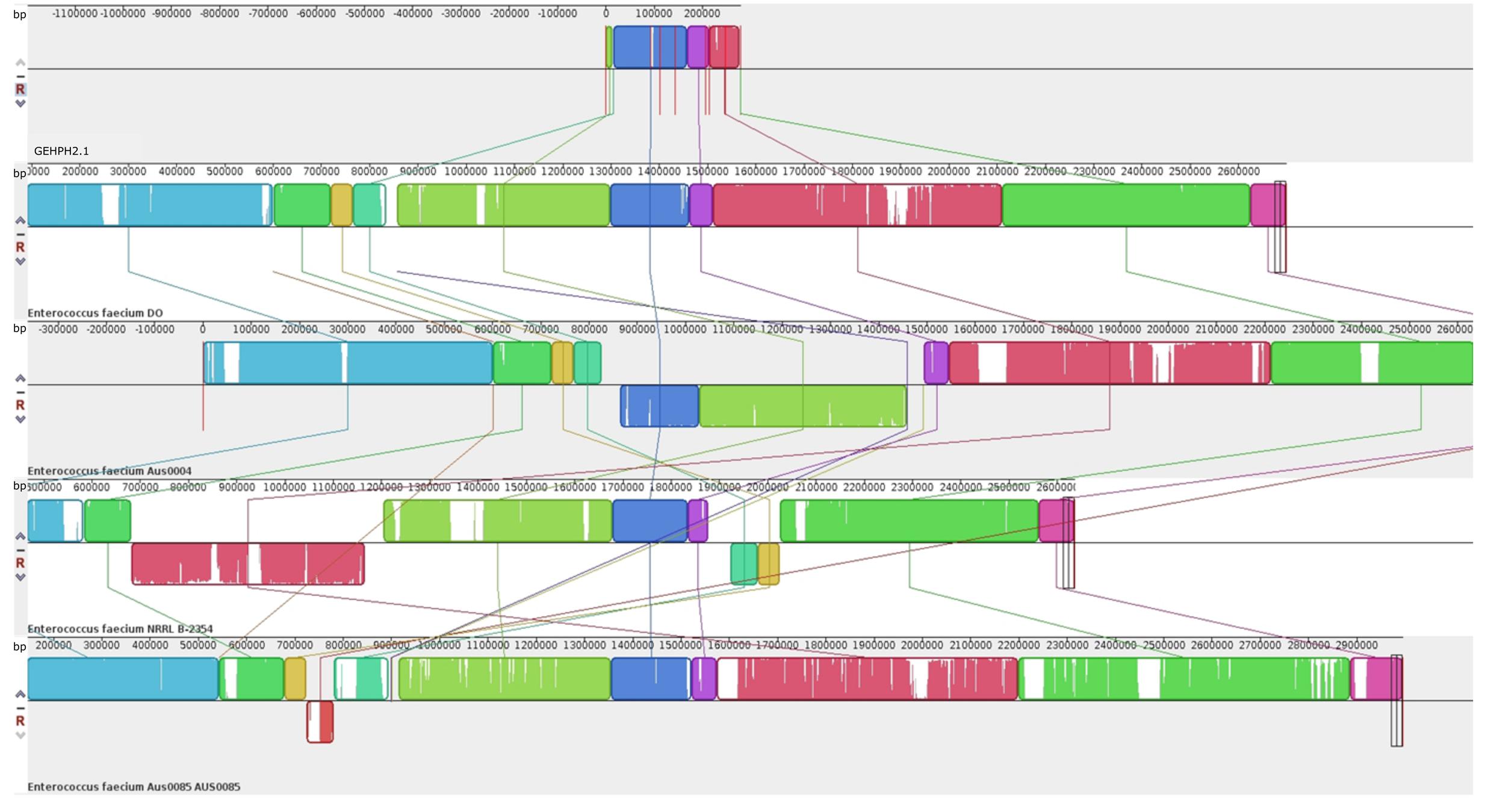
FIGURE 3. Comparison by MAUVE of the transferable genetic platform containing pbp5 (TCGEHPH2.1) with the four close E. faecium genomes (DO, Aus 0004, Aus0085 and NRRL B-2354), present in GenBank database. Each color block represents a genome region present in at least two of the sequences analyzed. These similar blocks can be identified by their color. Blank regions within the block represent point mutations or even regions that are absent in the same color blocks of the other sequences.
Figure 2 shows a detailed characterization of the 280 kb transferred genetic platform, with ORFs of a G + C content ranging from 24 to 41%. Besides the pbp5-related resistance to β-lactams, this chromosomal transferable genetic platform contains genes involved in different cellular functions including amino acids and carbohydrate transport, redox processes, survival under stressful conditions in the intestinal environment (e.g., acid and bile tolerance) and also other genes related to β-lactam resistance. Among the last category is a gene (named ponA) encoding a bifunctional class A PBP with transpeptidase and transglycosylase activity, which is involved in the synthesis of peptidoglycan, allowing the survival in the presence of cell wall inhibitors (Rice et al., 2009). A copy of the CiaRH operon, coding for a two-component signal-transduction system responsive to cell envelope lysis-stress response and restricted to Streptococcus spp. to date, was also detected. The 280 kb genetic platform containing pbp5 also had genes with presumed influence on microbe-host interactions. These include three phosphotransferase systems (PTS, namely glucitol/sorbitol, L-ascorbate and mannose/fructose/sorbose), the ABC transport and transformation system of maltodextrines (malACDX) and the N-acetylmannosamine-6-phosphate epimerase (NanE; part of a pathway that allows the usage of sialic acids, major components of glycoproteins, gangliosides, and other sialoglyco-conjugates). Other metabolic genes in the pbp5 genetic platform could be involved in tolerance to the intestinal acidic environment, including glyceraldehyde-3-phosphate dehydrogenase, ATP synthase subunit α, NADH dehydrogenase, glutaminase and genes encoding enzymes involved in the production of ammonia from glutamine and deamination or transport of branched/nitrogenated amino acids. Finally, five stress response proteins including two belonging to the Csp (cold shock proteins) system (CspA and CspC) and the small chaperone Hsp20, and Gls33 (also present in E. faecium genomes available in the GenBank databases) were also identified. They are involved in stress responses to salts, pH and ethanol exposure in Clostridium. Hsp20 is a small chaperone protein involved in survival of different abiotic stress conditions including heat (55°C) and salt (5 mM) in Bifidobacterium longum (Khaskheli et al., 2015).
To date, only two E. faecium strains lack pbp5. These are strains GE1 and D344SRF, which are AmpS isolates. The D344SRF strain is susceptible to cephalosporin and ampicillin due to the spontaneous deletion of a 170 kb genome fragment that includes pbp5 and other pbp genes. This deletion occurred by the interaction of CTn5386 (a 60 kb element that comprises pbp5 and vanB2) with Tn916 (Rice et al., 2007). The GE1 laboratory strain does only harbor the integrase of Tn5801. The causes of the loss of the pbp5 genetic platform are still unknown.
The Chromosomal Region Harboring pbp5 Exhibit Hotspots for Insertions
The occurrence of different insertion sequences (ISs), (Figure 4) in the boundaries of the pbp5 gene and the presence of a >3027 bp psr-pbp5 fragment in E. hirae (99.9% identical to that found on E. faecium) suggest the presence of hot spots that could facilitate recombination of regions containing the pbp5. To test this possibility, we further analyzed the 8–10 Kb genetic environment of pbp5 from 21 wild type and 15 transconjugant isolates included in this study as well as available enterococcal genomes. Identical RFLP patterns of these 8–10 kb fragment were observed for each pair of donor and its corresponding transconjugants, but were different from those naturally occurring in the pbp5 carrying recipient strains E. faecium BM4105RF and 64/3 (Figure 4). Sequencing of fragments representing distinct RFLP patterns and comparative analysis with similar pbp5 genetic environment in GenBank available genomes, revealed 21 variants of such 8–10 kb chromosomal fragments (designated by roman numerals), which differed in the number and type of ISs (Figure 4). Three variants (types I, II, III) corresponded to pbp5 transferable platforms described in this study (Figure 4; Table 1).
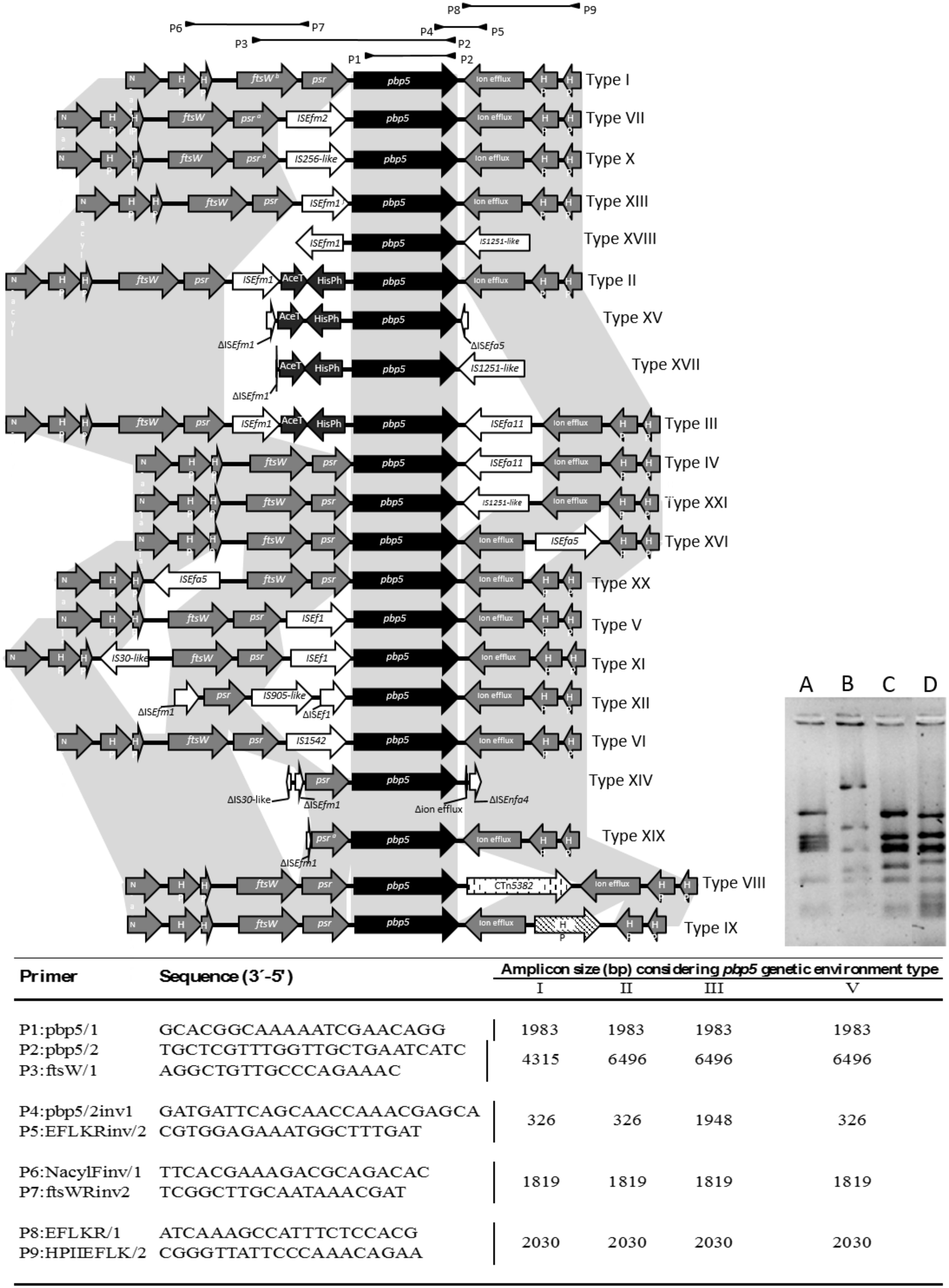
FIGURE 4. Characterization of E. faecium pbp5 genetic environment by PCR and sequencing. The Roman numbers I, II, III, and V represent the different pbp5 genetic platforms detected in E. faecium from this study. The numbers IV, VI to XXI were detected in available genomes from GenBank database. The different types were named according to diversity of insertions sequences, genomic fragments or conjugative transposons within genes or intergenic regions. Mutations or recombinations within genes or intergenic regions were not considered for type classification. The Table indicates the primers used (designed for this study; P1/P2 described by Dahl et al., 2000) and the size of PCR amplicons from genetic environment of types I–III and V. The A, B, C and D lines of the bottom right side figure represent RFLP patterns of amplicon P3-P2 of mobile platforms I (pattern C) and II/III (pattern D) of isolates included in this study, when digested with DdeI restriction enzyme. The patterns A and B correspond to the amplicons of the recipient strains E. faecium BM4105RF and 64/3, respectively. a These gene has an extra stop codon within its sequence. Abbreviations: N-acyl, (N-acyl-glucosamine-6-phosphate-2-epimerase); HP (hypothetical protein); ftsW (cell cycle protein); psr, (pbp5 synthesis repressor); pbp5 (gene encoding penicillin binding protein 5); AceT (acetiltransferase); HisPh (Histidinol Phosphate Phosphatase).
The predominant 8–10 kb fragment identified in both AmpR and AmpS E. faecium analyzed in this study did not contain indels and is considered here as the pbp5 genetic environment prototype; arbitrarily named Type I (Supplementary Table S5, Figure 4 and 5). The pbp5 or other genes (e.g., psr. ftsw, ion efflux genes) were flanked by one or two insertions sequences of the IS256, ISL3 or IS982 families (ISEf1, ISEfm1, ISEfm2, IS1542, IS256-like, ISEfa11, or IS1251-like) in isolates carrying types II to XXI, with the exception of type VIII that had CTn5382 (vanB2) inserted downstream of pbp5. The identifiable boundaries of ISs detected within the 8–10 kb boundaries of pbp5, as well as the common nucleotide positions at which insertions occurred, suggested recent acquisition events at some hot-spots on an ancestral sequence (Figure 4). Despite of predominance of type I, some types seem to be more associated with specific hosts as the case of type V, predominant in strains from pigs and type II/II-like, III or type XIV-like which are frequent among clinical isolates. Epidemiological distribution of isolates appears in Supplementary Table S5.
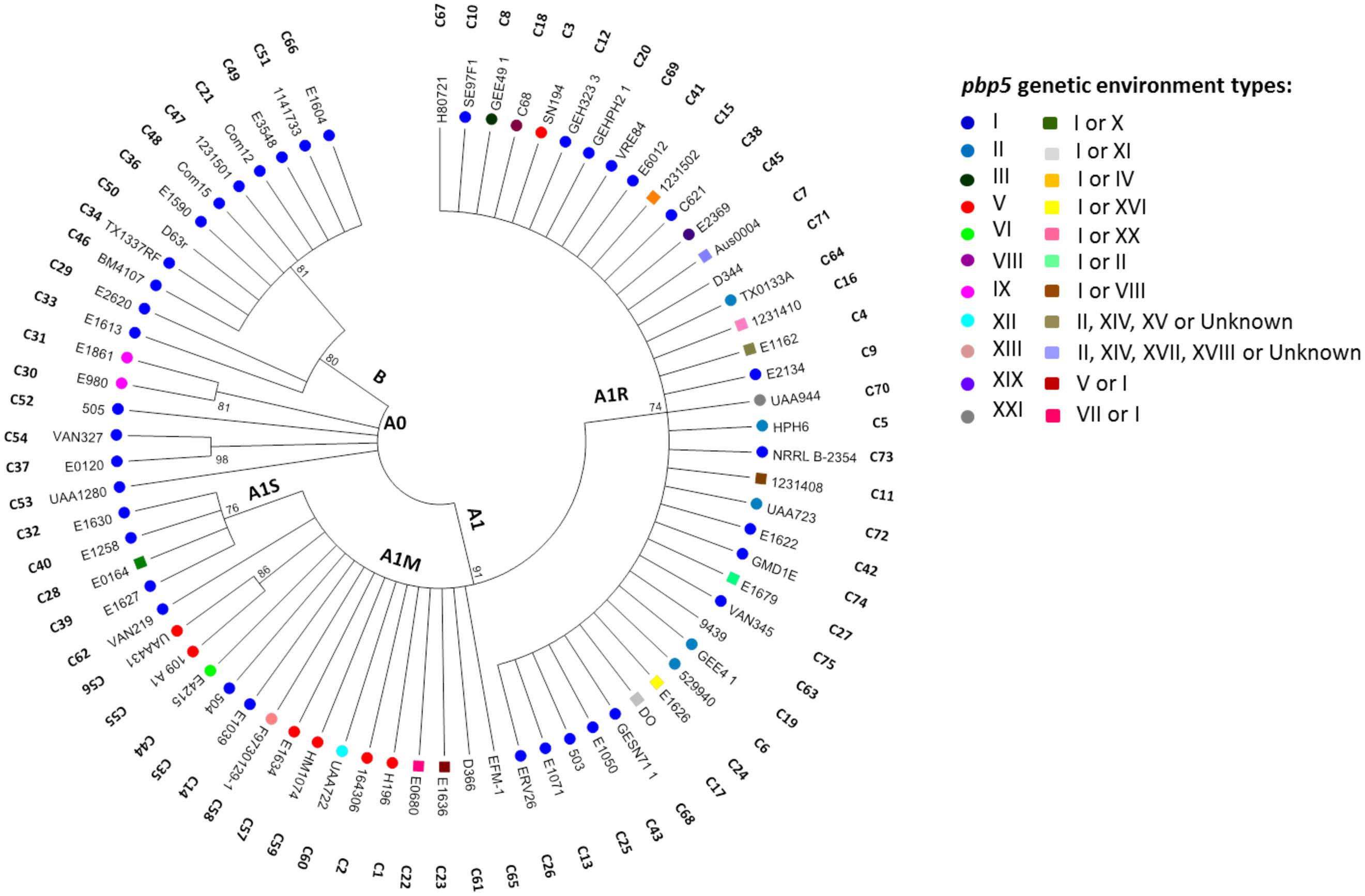
FIGURE 5. Phylogenetic analysis of PBP5 protein sequences from E. faecium isolates of this study and available in GenBank database (last update February 2014). The Maximum Likelihood tree was obtained using Mega 7 and the JTT method. The tree with the highest log likelihood (-2000,3736) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model. Then, the topology with superior log likelihood value was selected. The analysis involved 75 amino acid sequences. All positions containing gaps and missing data were eliminated of the analysis. The final dataset had a total of 445 positions. Evolutionary analyses were conducted in MEGA7. Bootstrap values are indicated and are based in 1000 permutations. The cut-off used was ≥70%. “C” followed by a number represent PBP5 amino acid combinations described in Supplementary Table S6. Only one E. faecium isolate carrying representative PBP5 sequences (each type of “C”) was included in the tree. The pbp5 genetic environments types described in Figure 4 are indicated by colored circles or by colored squares when more than one type was detected in isolates carrying the same “C.” No symbol reflects that the genetic environment was not elucidated. NA, not applicable.
Diversity of PBP5 Sequences Reflects the Phylogenomic Diversification of E. faecium
We identified 75 PBP5 protein variants (Supplementary Tables S5 and S6) corresponding both to AmpR and AmpS strains which comprise 20 of the previously described variants C4, C7, C9, C11, C15-C18, C21, C24, C46-C48, C50, C51, C61, C63, C65, C67, C71 (Rybkine et al., 1998; López et al., 2009; Galloway-Peña et al., 2011). The other 55 variants were firstly detected in this study either in strains from Portugal (n = 11) or from available genomes at NCBI database (n = 44). The C4, C7, C8, C9, C12, C19, and C20 sequences were identified in isolates able to transfer pbp5 (Table 1, Supplementary Table S6; Figure 5), which belonged mostly to ST18, its SLV ST125, ST280 and ST670 (BAPS 3.3a, 3.1 and 2.1a, respectively) (Table 1).
Figure 5 shows the phylogenetic tree constructed with all PBP5 protein sequences from this study and those available at GenBank databases. The tree is split into two major clades arbitrarily named B and A, mirroring the clades associated with populations of non-hospitalized persons and hospital isolates respectively, which were previously inferred from phylogenomic studies of E. faecium (Lebreton et al., 2013). Clade B comprises PBP5 of AmpS isolates (PBP5-S), which mainly correspond to strains of BAPS subgroups 1.2 and 1.5. Some are similar to the prototype PBP5-S C46 sequence of E. faecium BM4107 strain (Sifaoui et al., 2001) but most of them also exhibit mutations at positions T25A, S39T and D644N, which are also common to PBP5 sequences of clade A isolates (Supplementary Table S6). Two strains, isolated in 1964 and 2006, further showed changes at S27G + T324A and S27G respectively, such mutations corresponding to the PBP5-R consensus sequence (Pietta et al., 2014). The type I chromosomal region mentioned above was observed in all available isolates from this group (Figure 5; Supplementary Table S5).
Clade A includes PBP5 variants grouped in two main clusters arbitrarily designed here A0 and A1, with most isolates sharing mutations at positions V24A, S27G, E100Q, K144Q, T172A, T324A, N496K, A499T and E525D (7 of the 21 positions used to establish sequence diversity of this protein) (Pietta et al., 2014). The Clade A0, represented by C30, C31, C37, C52, C53, and C54 PBP5 sequences, corresponds to AmpS isolates of different BAPS groups (BAPS 1.2, BAPS 2.3a and BAPS 3.3b), recovered from different hosts (animals and humans), different countries and collected from 1995 to 2001 (Figure 5, Supplementary Table S6). Most isolates within clade A0 carried pbp5 within a type I fragment. Sequences of Clade A1 were classified in three subclusters that parallels E. faecium populations of BAPS groups 2 and 3 (Willems et al., 2012; Tedim et al., 2015). They were designated as A1S, comprising a subset of PBP5-AmpS variants (mainly associated with human and animal isolates of BAPS subgroup 2.1b); A1M, comprising a subset of PBP5-AmpS and PBP5-AmpR (mostly associated with animal isolates of diverse BAPS subgroups 2.1b, 2.3a, 2.3b, 3.1, 3.2, and 3.3b); and A1R, including almost all PBP5-AmpR (mostly associated with the clinical setting and BAPS subgroups 3.1, 3.3a and 2.1a, the latter two only observed in this group).
All isolates from Clades A1S, A1M and A1R share mutations R34Q, G66E, L177I and A216S but also present some differences. Mutations S39N and A401S plus A499I were specific for subclades A1S and A1M, respectively. The last two polymorphisms were previously documented in two AmpR strains (Pietta et al., 2014) but this study suggests that they were fixed in certain populations despite having been previously discarded as relevant changes. One of the A1M strains had also variations at specific positions linked to the A1R group (A68T, E85D, M485T, V586L, E629V and P667S). Of note is the PBP5 variant C23 within this cluster, which was overrepresented (n = 44).
PBP5 variants within the A1R group exhibited six mutations (A68T, E85D, S204G, 466′S/D, M485A/T, E629V, P667S) predominant in this group, some of them located in the active site of the protein (466′S/466′D, M485A/T) and in the end of a turn between the β1 and β2 strands (E629V; P667S) (Fontana et al., 1996; Rice et al., 2004). It is of note that some AmpS isolates (BAPS subgroups 3.1, 2.3a and 3.2) with PBP5 sequences that cluster in the A1R subgroup (including the PBP5 of the recipient E. faecium 64/3) lack the mutation M485A and E629V, which confirmed that such mutations might be necessary for AmpR phenotype, as reported (Rice et al., 2004). Changes A68T, E85D and S204G were shared by AmpS and AmpR isolates of A1R group (Supplementary Table S6). Some strains exhibited the particular pattern of mutations Q408H, A558T, G582S, K632Q and, eventually, V462A, N546T and P642L. They had not previously been associated with AmpR. Distribution of different variants of the 8–10 kb boundaries of pbp5 gene is shown in Figure 5 and Supplementary Table S5. Of note, is the detection of type I in isolates of different groups of clade A1 as well as the association of types V and XI with isolates of group A1M and of the similar types II, III, XIV, XV, and XVII with isolates of group A1R.
We also analyzed the diversity of other genes previously associated with AmpR (ddcP, ddcY, ldtEfm, pgtA, lytG) in available genomes, (protein sequence per UniRef100 available at Uniprot using as reference E. faecium strain Aus0004). Single-locus phylogenetic tree of PBP5 was congruent with those of proteins codified by ldtEfm and ddcY but was non-congruent with those of ddcP, pgtA and lytG (data not shown).
Analysis of gene tree topologies (E. faecium core genomes vs. E. faecium pbp5 genes) revealed discrepancies that indicate transfer of pbp5 between E. faecium populations (Figure 6) and explains the occurrence of both the same PBP5 variants in isolates of different clonal lineages, and different PBP5 sequences in strains of the same phylogenomic groups (Figure 5, Supplementary Table S5).
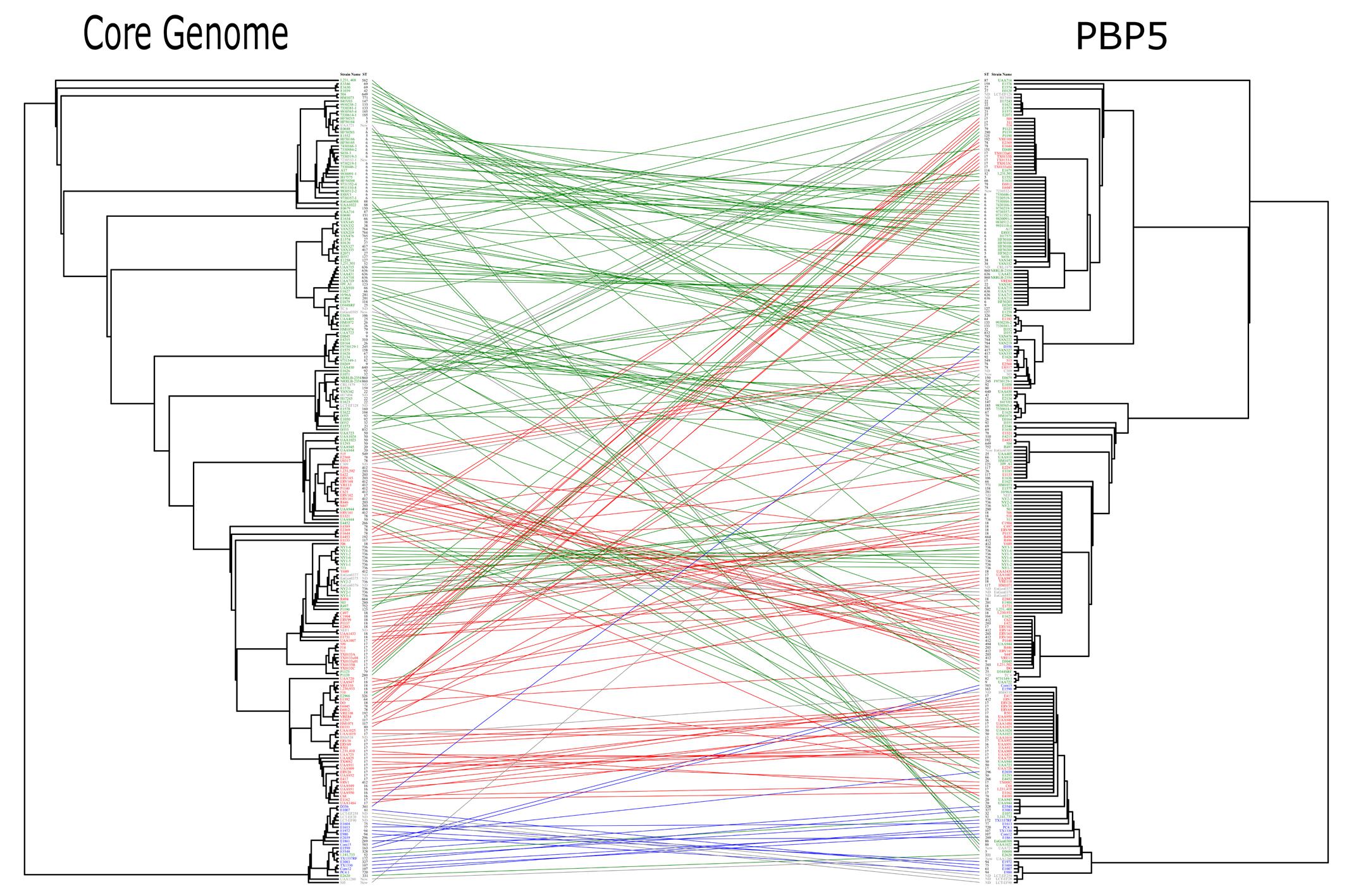
FIGURE 6. Comparison of gene tree topologies (Core Genome phylogeny vs. pbp5 phylogeny). Strains of clade B are represented in blue, those of Clade A1 in red and those Clade A2 in green. The edges join the core genome and the corresponding pbp5 of each strain. Both trees were made by ML using GTR-CAT. Tress and edges were built using APE packages. The results of Mantel test (r = -0.03 and significance of 0.761) show the rearrangement of the pbp5 in comparison with the core genome. The Mantel test (APE package) was made using the distance matrix calculated from nucleotide alignments.
Ampicillin Resistance Was Maintained Without Selective Pressure
AmpR was maintained in the wild-type strains and respective E. faecium GE1 transconjugants over 200 generations in antibiotic free BHI indicating that the acquired genetic platforms containing pbp5 may persist in absence of selective pressure in different genetic backgrounds, suggesting no-fitness cost imposed by the acquired pbp5 platforms. The morphology and growth rate of all evolved transconjugants was similar among each other and to the normal growth of the E. faecium GE1 recipient strain.
Discussion
This study documents the occurrence and diversity of a large chromosomal region containing pbp5 in almost all E. faecium genomes and a parallel diversification between the PBP5 and the E. faecium core genome. The transferability of this region observed under laboratory conditions using different clonal backgrounds would greatly enhance the possibilities of E. faecium adaptation to changing environments.
Polymorphisms in the PBP5 protein sequences allowed grouping of PBP5 variants in clusters that mimic the phylogenomic diversification of E. faecium (Galloway-Peña et al., 2012; Lebreton et al., 2013). Lebreton et al. (2013) suggested a model for evolvability of this enterococcal species consisting of a first split of “clade B” and “clade A” coincidental with human and animal co-habitation occurring 30,000 y ago, and a further split of “clade A” in subclades “A1” and “A2” after the introduction of antibiotics in the late 1940s. Two main clusters of PBP5 variants were also identified in the present study, designated as “B” (associated with the E. faecium “clade B”), and “A” that further split in subgroups A0 (including only AmpS isolates) and A1 comprising three small groups linked to different E. faecium populations differing in the susceptibility to ampicillin: A1S (AmpS from healthy humans of different BAPS groups), A1M (AmpS and AmpR from humans and animals), and A1R (AmpR from clinical isolates). Based on the apparent universal presence of pbp5 in E. faecium populations, we could speculate that pbp5 might have predated the E. faecium evolutionary split among different hosts. Although some authors have suggested a sequential acquisition of amino acid changes in PBP5 sequence (Galloway-Peña et al., 2011; Pietta et al., 2014), such diversification may indicate different evolutionary routes for AmpR in response to distinct selective pressures in different hosts, similarly to what has been observed for different β-lactamase enzymes of Gram negative organisms (Novais et al., 2010).
The full characterization of the transferable 153 kb chromosomal region that contains not only pbp5 but also genes contributing to survival in the gastrointestinal tract (e.g., resistance to acids and bile) in almost all available E. faecium genomes suggest a contribution of this genetic platform to the adaptation to the mammalian intestine and persistence in abiotic environments. Due to the scarcity of E. faecium isolates from non-mammals and non-human hosts, it would result highly speculative to hypothesize about the origin of the PBP5 and other genes located in this genetic platform. The PBP5 is a member of the Mec family, which comprises proteins with natural low affinity for β-lactams as PBP2a (mecA) of Staphylococcus aureus and PBP3r of Enterococcus hirae (99% identical to PBP5, suggesting a common origin for PBP5 and PBP3r) (Raze et al., 1998; Hiramatsu et al., 2013). For staphylococci, MecA would have been essential for the survival of ancestral members of staphylococci in the presence of β-lactam antibiotics produced by fungi and Actinobacteria and lost in the Devonian period with the emergence of mammals (more than 400 million of years ago). This would be coincidental with the separation and adaptation of staphylococcal species to different hosts (Hiramatsu et al., 2013), in any case isolating Staphylococcus populations from old β-lactam producers. Recent antibiotic pressure would have influenced the transference of PBP2a from environmental to human strains of staphylococci. It is of note that the 153 kb transferred genetic platform characterized in this study has a variable G + C content, reflecting HGT events from different sources and suggesting that a similar evolutionary route as that of PBP2a of staphylococci might have also occurred for E. faecium PBP5.
The transfer of pbp5 in E. faecium resulting of the interaction between transposons as Tn5382 or Tn916 plus Tn5386 was previously demonstrated for a few isolates (Rice et al., 2005a). During the revision process of this manuscript, a work by García-Solache et al. (2016) was published. This work suggested that transference of chromosomal regions carrying pbp5 occurs by homologous recombination between similar DNA regions of donor and recipient strains enriched in AT nucleotides with a possible involvement of plasmids (García-Solache et al., 2016). Tn916-like elements were not detected in our strains but the presence of a band of similar size in both donors and recipients and a sequence enriched in AT in the boundaries of the chromosomal transferred region (data not shown) suggested that homologous recombination might occur between E. faecium donor and recipient DNA from our study. Although the presence of plasmid genes within this acquired pbp5 genetic platform was not observed in the transconjugant we sequenced as well as in the case of some transconjugants described in the study of García-Solache et al. (2016), we cannot completely discard this possibility due to the common presence of plasmids of certain families in donors analyzed (data not shown) (Manson et al., 2010; Freitas et al., 2016).
Despite the relevance of elucidation of the mechanism of pbp5 transfer at molecular level in future studies, it is of note that disparate topologies of the phylogenetic trees of E. faecium core genomes and PBP5 sequences obtained in our study reflect how frequent the transfer of pbp5 genetic platforms under natural circumstances may occur (Tripathi and Sowdhamini, 2008). Similarly, atypical phylogenetic associations have been signaled as evidence of lateral transfer of various types of serine-proteases (PBP5 belongs to this family of enzymes) throughout the prokaryotic world (Tripathi and Sowdhamini, 2008). In fact, we present evidence of inter-chromosomal transfer for different PBP5 C-types. However, in our case, pbp5 horizontal gene transfer (HGT) events seems to preferentially happen in particular populations, which being more prone to mutation (the subclade A1 that is characterized by a higher mutation rate than clade B) would further facilitate the pbp5 fixation and evolution followed by their clonal expansion under antibiotic selective pressure, reflecting the “ex unibus plurum” evolutionary dynamics (Baquero, 2011). Heterogeneity of populations that colonize humans, designated as “clouds” elsewhere (Stanczak-Mrozek et al., 2015), leads to a possible global adaptive benefit for certain clones and finally for the overall species, that is enhanced by HGT.
The necessary contribution of different genes for the full expression of AmpR phenotype is suggested by the lower MIC values of the transconjugants in comparison with the wild type strains in this and other studies (Rice et al., 2005b; Zhang et al., 2012). The transferable genetic platform identified here contained pbp5 and ponA but also other genes (ciaRH operon) previously associated with β-lactam resistance and virulence in S. pneumoniae, but still unexplored in enterococci (Guenzi et al., 1994; Jordan et al., 2008; Krawczyk-Balska and Markiewicz, 2016). Outside this pbp5, only ldtEfm and ddcY showed a similar non-congruent topology with that of pbp5 indicating that these genes are also under accelerated evolution, which could explain eventual AmpR phenotypes with a lack of correlation with PBP5 sequences (Zhang et al., 2012). The variable ampicillin susceptibility phenotypes observed in different transconjugants even when using the same recipient strain, suggest either a partial transfer of the pbp5 genetic platform or occurrence of recombination events leading to variation in MICs, which seem to occur frequently in commensal bacteria (Ingle et al., 2016).
Conclusion
The arsenal of adaptive traits located on the chromosomal region containing pbp5 characterized in this study suggests its involvement in the adaptation of E. faecium to the gastrointestinal tract of mammals, and its possible contribution to the spread of pbp5 by horizontal gene transfer, even in the absence of specific β-lactam exposure. However, the apparent frequent transfer events of such genetic platform among “clouds” of closely related E. faecium populations, would indicate bacterial shifts in the evolution of pathogenicity (including colonization) and antibiotic resistance that occurred in response to changes in patients‘ demographics, medical strategies and interventions, within the paradigm of the “Hamiltonian medicine” (Fraser et al., 2005). The data further highlight the increasing need for evolutionary biology to be aligned with medical challenges (Nesse et al., 2010).
Nucleotide Sequence Genbank Accession Numbers
The sequences corresponding to representative pbp5 genetic plat forms (Types I-IV) were assigned GenBank accession numbers JN208885, JN208888, JN208884 and JN208886, respectively. PBP5 amino acid were analyzed and designated by a “C” followed by a number (Table S6). New sequences correspond to GenBank accession numbers JN208883 (C6 amino acid combination), JN208889 (C19 amino acid combination); JN208887 (C8 amino acid combination); JN208886 (C3 amino acid combination), JN208882 (C2 amino acid combination), KC479673 (C1 amino acid combination), KC479675 (C5 amino acid combination), KC479676 (C10 amino acid combination) and KC479674 (C12 amino acid combination). The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MBRG00000000. The version described in this paper is version MBRG01000000.
Author Contributions
CN, LP, and TMC designed the study. CN, ARF, APR, MA-H, ES, and RE performed wet lab experiments and participated in the analysis of the data. APT and VFL performed bioinformatic analysis. FB, APR, and LP provided expertise, participated in the analysis of data, and in the revision of the manuscript. CN, APT, and TMC performed the analysis of data and wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Educacão e Ciência (MEC) through national funds and co-financed by Fundo Europeu de Desenvolvimento Regional (FEDER) under the Partnership Agreement PT2020 with the reference UID/ MULTI/04378/2013–POCI/01/0145/FERDER/007728 to CN and LP, Instituto de Salud Carlos III (Plan Estatal de I+D+i 2013-2016), Grant PI12-01581 and PI15-01307 to TMC and CIBERESP (CB06/02/0053) to FB, European Commission, Seventh Framework Program (EVOTARFP7-HEALTH-282004) to TMC and FB and the Regional Government of Madrid in Spain (PROMPT- S2010/BMD2414) to FB and TMC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editor consider relevant to the content of the manuscript have been disclosed.
Acknowledgments
APT and VFL were funded by the European Union (EvoTAR-282004). ARF and ES were supported by fellowship from FCT [grant SFRH/BPD/96148/2013 through Programa Operacional Capital Humano (POCH) and SFRH/BD/63955/2009 POPHQREN, respectively]. MA-H was supported by a Marie Curie Intra European Research Fellowship; grant number 272914, Bio-CHARGE; Investigation into the Biological Cost and Adaptation of the Host to Antibiotic Resistance on Mobile Genetic Elements in Enterococcus Species. VFL is further supported by a Research Award Grant 2016 of the European Society for Clinical Microbiology and Infectious Diseases (ESCMID).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01581
Footnotes
- ^http://pubmlst.org/
- ^http://www.uniprot.org/uniref/
- ^http://www.genome.jp/kegg/genome.html.
- ^www.megasoftware.net
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arias, C. A., Contreras, G. A., and Murray, B. E. (2010). Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 16, 555–562. doi: 10.1111/j.1198-743X.2010.03214.x
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Baquero, F. (2011). The 2010 garrod lecture: the dimensions of evolution in antibiotic resistance: ex unibus plurum et ex pluribus unum. J. Antimicrob. Chemother. 66, 1659–1672. doi: 10.1093/jac/dkr214
Belhaj, M., Boutiba-Ben Boubaker, I., and Slim, A. (2016). Penicillin-binding protein 5 sequence alteration and levels of plp5 mRNA expression in clinical isolates of Enterococcus faecium with different levels of ampicillin resistance. Microb. Drug Resist. 22, 202–210. doi: 10.1089/mdr.2015.0211
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Carias, L. L., Rudin, S. D., Donskey, C. J., and Rice, L. B. (1998). Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180, 4426–4434.
Centers for Disease Control and Prevention (2013). Antibiotic Resistance Threats in the United States. Available at: http://www.cdc.gov/drugresistance/threat-report-2013/
Clinical and Laboratory Standards Institute (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement M100-S22. Wayne, PA: CLSI.
Coudron, P. E., Markowitz, S. M., and Wong, E. S. (1992). Isolation of a beta-lactamase-producing, aminoglycoside-resistant strain of Enterococcus faecium. Antimicrob. Agents Chemother. 36, 1125–1126. doi: 10.1128/AAC.36.5.1125
Dahl, K. H., Lundblad, E. W., Rokenes, T. P., Olsvik, O., and Sundsfjord, A. (2000). Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146, 1469–1479. doi: 10.1099/00221287-146-6-1469
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
de Regt, M. J. A., van Schaik, W., van Luit-Asbroek, M., Dekker, H. A. T., van Duijkeren, E., Koning, C. J. M., et al. (2012). Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS ONE 7:e30319. doi: 10.1371/journal.pone.0030319
Fontana, R., Ligozzi, M., Pittaluga, F., and Satta, G. (1996). Intrinsic penicillin resistance in enterococci. Microb. Drug Resist. 2, 209–213. doi: 10.1089/mdr.1996.2.209
Fraser, C., Hanage, W. P., and Spratt, B. G. (2005). Neutral microepidemic evolution of bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 102, 1968–1973. doi: 10.1073/pnas.0406993102
Freitas, A. R., Novais, C., Tedim, A. P., Francia, M. V. M. V., Baquero, F., Coque, T. M., et al. (2013). Microevolutionary events involving narrow host plasmid influences local fixation of vancomycin-resistance in Enterococcus. PLoS ONE 8:e60589. doi: 10.1371/journal.pone.0060589
Freitas, A. R., Tedim, A. P., Francia, M. V., Jensen, L. B., Novais, C., Peixe, L., et al. (2016). Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012). J. Antimicrob. Chemother. doi: 10.1093/jac/dkw312 [Epub ahead of print].
Galloway-Peña, J., Roh, J. H., Latorre, M., Qin, X., and Murray, B. E. (2012). Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS ONE 7:e30187. doi: 10.1371/journal.pone.0030187
Galloway-Peña, J. R., Rice, L. B., and Murray, B. E. (2011). Analysis of PBP5 of early U.S. isolates of Enterococcus faecium: sequence variation alone does not explain increasing ampicillin resistance over time. Antimicrob. Agents Chemother. 55, 3272–3277. doi: 10.1128/AAC.00099-11
García-Solache, M., Lebreton, F., McLaughlin, R. E., Whiteaker, J. D., Gilmore, M. S., and Rice, L. B. (2016). Homologous recombination within large chromosomal regions facilitates acquisition of beta-lactam and vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00488-16 [Epub ahead of print].
Gonçalves, A., Igrejas, G., Radhouani, H., López, M., Guerra, A., Petrucci-Fonseca, F., et al. (2011). Detection of vancomycin-resistant enterococci from faecal samples of Iberian wolf and Iberian lynx, including Enterococcus faecium strains of CC17 and the new singleton ST573. Sci. Total Environ. 41, 266–268. doi: 10.1016/j.scitotenv.2011.09.074
Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699. doi: 10.1093/nar/gkq313
Guenzi, E., Gasc, A. M., Sicard, M. A., and Hakenbeck, R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12, 505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guzman Prieto, A. M., van Schaik, W., Rogers, M. R. C., Coque, T. M., Baquero, F., Corander, J., et al. (2016). Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front. Microbiol. 7:788. doi: 10.3389/fmicb.2016.00788
Hendrickx, A. P. A., van Schaik, W., and Willems, R. J. L. (2013). The cell wall architecture of Enterococcus faecium: from resistance to pathogenesis. Future Microbiol. 8, 993–1010. doi: 10.2217/fmb.13.66
Hiramatsu, K., Ito, T., Tsubakishita, S., Sasaki, T., Takeuchi, F., Morimoto, Y., et al. (2013). Genomic basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 45:117. doi: 10.3947/ic.2013.45.2.117
Homan, W. L., Tribe, D., Poznanski, S., Li, M., Hogg, G., Spalburg, E., et al. (2002). Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40, 1963–1971. doi: 10.1128/JCM.40.9.3548-3548.2002-a
Ingle, D. J., Valcanis, M., Kuzevski, A., Tauschek, M., Inouye, M., Stinear, T., et al. (2016). In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2. doi: 10.1099/mgen.0.000064
Jordan, S., Hutchings, M. I., and Mascher, T. (2008). Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32, 107–146. doi: 10.1111/j.1574-6976.2007.00091.x
Katoh, K., and Toh, H. (2010). Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26, 1899–1900. doi: 10.1093/bioinformatics/btq224
Khaskheli, G. B., Zuo, F., Yu, R., and Chen, S. (2015). Overexpression of small heat shock protein enhances heat- and salt-stress tolerance of Bifidobacterium longum NCC2705. Curr. Microbiol. 71, 8–15. doi: 10.1007/s00284-015-0811-0
Klare, I., Rodloff, A. C., Wagner, J., Witte, W., and Hakenbeck, R. (1992). Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 36, 783–787. doi: 10.1128/AAC.36.4.783
Klibi, N., Sáenz, Y., Zarazaga, M., Ben Slama, K., Masmoudi, A., Ruiz-Larrea, F., et al. (2008). Polymorphism in pbp5 gene detected in clinical Enterococcus faecium strains with different ampicillin MICs from a Tunisian hospital. J. Chemother. 20, 436–440. doi: 10.1179/joc.2008.20.4.436
Krawczyk-Balska, A., and Markiewicz, Z. (2016). The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 120, 251–265. doi: 10.1111/jam.12989
Kristich, C., Rice, L., and Arias, C. (2014). “Enterococcal infection—treatment and antibiotic resistance,” in Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, eds M. S. Gilmore, D. B. Clewell, Y. Ike, and N. Shankar (Boston, MA: Massachusetts Eye and Ear Infirmary).
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lebreton, F., van Schaik, W., McGuire, A. M., Godfrey, P., Griggs, A., Mazumdar, V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4:e00534-13. doi: 10.1128/mBio.00534-13
León-Sampedro, R., Novais, C., Peixe, L., Baquero, F., and Coque, T. M. (2016). Diversity and evolution of the Tn5801-tet(M)-like integrative and conjugative elements among Enterococcus. Streptococcus, and Staphylococcus. Antimicrob. Agents Chemother. 60, 1736–1746. doi: 10.1128/AAC.01864-15
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Liu, S. L., Hessel, A., and Sanderson, K. E. (1993). Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U.S.A. 90, 6874–6878. doi: 10.1073/pnas.90.14.6874
López, M., Sáenz, Y., Rojo-Bezares, B., Martínez, S., del Campo, R., Ruiz-Larrea, F., et al. (2009). Detection of vanA and vanB2-containing enterococci from food samples in Spain, including Enterococcus faecium strains of CC17 and the new singleton ST425. Int. J. Food Microbiol. 133, 172–178. doi: 10.1016/j.ijfoodmicro.2009.05.020
Manson, J. M., Hancock, L. E., and Gilmore, M. S. (2010). Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U.S.A. 107, 12269–12274. doi: 10.1073/pnas.1000139107
Nesse, R. M., Bergstrom, C. T., Ellison, P. T., Flier, J. S., Gluckman, P., Govindaraju, D. R., et al. (2010). Evolution in health and medicine Sackler colloquium: Making evolutionary biology a basic science for medicine. Proc. Natl. Acad. Sci. U.S.A. 107(Suppl. 1), 1800–1807. doi: 10.1073/pnas.0906224106
Novais, A., Comas, I., Baquero, F., Cantón, R., Coque, T. M., Moya, A., et al. (2010). Evolutionary trajectories of beta-lactamase CTX-M-1 cluster enzymes: predicting antibiotic resistance. PLoS Pathog. 6:e1000735. doi: 10.1371/journal.ppat.1000735
Novais, C., Coque, T. M., Costa, M. J., Sousa, J. C., Baquero, F., and Peixe, L. V. (2005a). High occurrence and persistence of antibiotic-resistant enterococci in poultry food samples in Portugal. J. Antimicrob. Chemother. 56, 1139–1143. doi: 10.1093/jac/dki360
Novais, C., Coque, T. M., Ferreira, H., Sousa, J. C., and Peixe, L. (2005b). Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl. Environ. Microbiol. 71, 3364–3368. doi: 10.1128/AEM.71.6.3364-3368.2005
Novais, C., Coque, T. M., Sousa, J. C., and Peixe, L. V. (2006). Antimicrobial resistance among faecal enterococci from healthy individuals in Portugal. Clin. Microbiol. Infect. 12, 1131–1134. doi: 10.1111/j.1469-0691.2006.01542.x
Novais, C., Freitas, A. R., Silveira, E., Antunes, P., Silva, R., Coque, T. M., et al. (2013). Spread of multidrug-resistant Enterococcus to animals and humans: an underestimated role for the pig farm environment. J. Antimicrob. Chemother. 68, 2746–2754. doi: 10.1093/jac/dkt289
Novais, C., Sousa, J. C., Coque, T. M., and Peixe, L. V. (2005c). Molecular characterization of glycopeptide-resistant Enterococcus faecium isolates from Portuguese hospitals. Antimicrob. Agents Chemother. 49, 3073–3079. doi: 10.1128/AAC.49.7.3073-3079.2005
Palmer, K. L., Godfrey, P., Griggs, A., Kos, V. N., Zucker, J., Desjardins, C., et al. (2012). Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio 3:e00318-11. doi: 10.1128/mBio.00318-11
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Pietta, E., Montealegre, M. C., Roh, J. H., Cocconcelli, P. S., and Murray, B. E. (2014). Enterococcus faecium PBP5-S/R, the missing link between PBP5-S and PBP5-R. Antimicrob. Agents Chemother. 58, 6978–6981. doi: 10.1128/AAC.03648-14
Poeta, P., Costa, D., Igrejas, G., Saenz, Y., Zarazaga, M., Rodrigues, J., et al. (2007). Polymorphisms of the pbp5 gene and correlation with ampicillin resistance in Enterococcus faecium isolates of animal origin. J. Med. Microbiol. 56, 236–240. doi: 10.1099/jmm.0.46778-0
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Raze, D., Dardenne, O., Hallut, S., Martinez-Bueno, M., Coyette, J., and Ghuysen, J. M. (1998). The gene encoding the low-affinity penicillin-binding protein 3r in Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob. Agents Chemother. 42, 534–539.
Rice, L. B., Bellais, S., Carias, L. L., Hutton-Thomas, R., Bonomo, R. A., Caspers, P., et al. (2004). Impact of specific pbp5 mutations on expression of beta-lactam resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 48, 3028–3032. doi: 10.1128/AAC.48.8.3028-3032.2004
Rice, L. B., Carias, L. L., Hutton-Thomas, R., and Rudin, S. (2007). Interaction of related Tn916-like transposons: analysis of excision events promoted by Tn916 and Tn5386 integrases. J. Bacteriol. 189, 3909–3917. doi: 10.1128/JB.00859-06
Rice, L. B., Carias, L. L., Hutton-Thomas, R., Sifaoui, F., Gutmann, L., and Rudin, S. D. (2001). Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45, 1480–1486. doi: 10.1128/AAC.45.5.1480-1486.2001
Rice, L. B., Carias, L. L., Marshall, S., Rudin, S. D., and Hutton-Thomas, R. (2005a). Tn5386, a novel Tn916-like mobile element in Enterococcus faecium D344R that interacts with Tn916 to yield a large genomic deletion. J. Bacteriol. 187, 6668–6677. doi: 10.1128/JB.187.19.6668-6677.2005
Rice, L. B., Carias, L. L., Rudin, S., Hutton, R., Marshall, S., Hassan, M., et al. (2009). Role of class A penicillin-binding proteins in the expression of beta-lactam resistance in Enterococcus faecium. J. Bacteriol. 191, 3649–3656. doi: 10.1128/JB.01834-08
Rice, L. B., Carias, L. L., Rudin, S., Lakticová, V., Wood, A., and Hutton-Thomas, R. (2005b). Enterococcus faecium low-affinity pbp5 is a transferable determinant. Antimicrob. Agents Chemother. 49, 5007–5012. doi: 10.1128/AAC.49.12.5007-5012.2005
Rybkine, T., Mainardi, J. L., Sougakoff, W., Collatz, E., and Gutmann, L. (1998). Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J. Infect. Dis. 178, 159–163. doi: 10.1086/515605
Sacco, E., Cortes, M., Josseaume, N., Rice, L. B., Mainardi, J.-L., and Arthur, M. (2014). Serine/threonine protein phosphatase-mediated control of the peptidoglycan cross-linking L,D-transpeptidase pathway in Enterococcus faecium. MBio 5:e01446-14. doi: 10.1128/mBio.01446-14
Santos, T., Silva, N., Igrejas, G., Rodrigues, P., Micael, J., Rodrigues, T., et al. (2013). Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe 24, 25–31. doi: 10.1016/j.anaerobe.2013.09.004
Sifaoui, F., Arthur, M., Rice, L., and Gutmann, L. (2001). Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45, 2594–2597. doi: 10.1128/AAC.45.9.2594-2597.2001
Stanczak-Mrozek, K. I., Manne, A., Knight, G. M., Gould, K., Witney, A. A., and Lindsay, J. A. (2015). Within-host diversity of MRSA antimicrobial resistances. J. Antimicrob. Chemother. 70, 2191–2198. doi: 10.1093/jac/dkv119
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tedim, A. P., Ruiz-Garbajosa, P., Corander, J., Rodríguez, C. M., Cantón, R., Willems, R. J., et al. (2015). Population biology of intestinal Enterococcus isolates from hospitalized and nonhospitalized individuals in different age groups. Appl. Environ. Microbiol. 81, 1820–1831. doi: 10.1128/AEM.03661-14
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Top, J., Willems, R., Blok, H., de Regt, M., Jalink, K., Troelstra, A., et al. (2007). Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin. Microbiol. Infect. 13, 316–319. doi: 10.1111/j.1469-0691.2006.01631.x
Tremblay, C.-L., Charlebois, A., Masson, L., and Archambault, M. (2013). Characterization of hospital-associated lineages of ampicillin-resistant Enterococcus faecium from clinical cases in dogs and humans. Front. Microbiol. 4:245. doi: 10.3389/fmicb.2013.00245
Tripathi, L. P., and Sowdhamini, R. (2008). Genome-wide survey of prokaryotic serine proteases: analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genomics 9:549. doi: 10.1186/1471-2164-9-549
Wagenvoort, J. H. T., De Brauwer, E. I. G. B., Penders, R. J. R., van der Linden, C. J., Willems, R. J., Top, J., et al. (2015). Environmental survival of vancomycin-sensitive ampicillin-resistant Enterococcus faecium (AREfm). Eur. J. Clin. Microbiol. Infect. Dis. 34, 1901–1903. doi: 10.1007/s10096-015-2430-x
Willems, R. J. L., Top, J., van Santen, M., Robinson, D. A., Coque, T. M., Baquero, F., et al. (2005). Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11, 821–828. doi: 10.3201/eid1106.041204
Willems, R. J. L., Top, J., van Schaik, W., Leavis, H., Bonten, M., Sirén, J., et al. (2012). Restricted gene flow among hospital subpopulations of Enterococcus faecium. MBio 3:e00151-12. doi: 10.1128/mBio.00151-12
Williamson, R., Calderwood, S. B., Moellering, R. C., and Tomasz, A. (1983). Studies on the mechanism of intrinsic resistance to beta-lactam antibiotics in group D streptococci. J. Gen. Microbiol. 129, 813–822. doi: 10.1099/00221287-129-3-813
Zhang, X., Paganelli, F. L., Bierschenk, D., Kuipers, A., Bonten, M. J. M., Willems, R. J. L., et al. (2012). Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet. 8:e1002804. doi: 10.1371/journal.pgen.1002804
Keywords: ampicillin resistance, PBP5 mutations, PonA, chromosomal transfer, phylogenomic diversification
Citation: Novais C, Tedim AP, Lanza VF, Freitas AR, Silveira E, Escada R, Roberts AP, Al-Haroni M, Baquero F, Peixe L and Coque TM (2016) Co-diversification of Enterococcus faecium Core Genomes and PBP5: Evidences of pbp5 Horizontal Transfer. Front. Microbiol. 7:1581. doi: 10.3389/fmicb.2016.01581
Received: 18 May 2016; Accepted: 21 September 2016;
Published: 06 October 2016.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Tanel X. Tenson, University of Tartu, EstoniaLuca Guardabassi, Ross University School of Veterinary Medicine, Saint Kitts and Nevis
Copyright © 2016 Novais, Tedim, Lanza, Freitas, Silveira, Escada, Roberts, Al-Haroni, Baquero, Peixe and Coque. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa M. Coque, bWFyaWF0ZXJlc2EuY29xdWVAc2FsdWQubWFkcmlkLm9yZw==
 Carla Novais
Carla Novais Ana P. Tedim
Ana P. Tedim Val F. Lanza
Val F. Lanza Ana R. Freitas
Ana R. Freitas Eduarda Silveira
Eduarda Silveira Ricardo Escada
Ricardo Escada Adam P. Roberts
Adam P. Roberts Mohammed Al-Haroni
Mohammed Al-Haroni Fernando Baquero
Fernando Baquero Luísa Peixe
Luísa Peixe Teresa M. Coque
Teresa M. Coque